Abstract
Cell-free systems that mimic essential cell functions, such as gene expression, have dramatically expanded in recent years, both in terms of applications and widespread adoption. Here we provide a review of cell-extract methods, with a specific focus on prokaryotic systems. Firstly, we describe the diversity of Escherichia coli genetic strains available and their corresponding utility. We then trace the history of cell-extract methodology over the past 20 years, showing key improvements that lower the entry level for new researchers. Next, we survey the rise of new prokaryotic cell-free systems, with associated methods, and the opportunities provided. Finally, we use this historical perspective to comment on the role of methodology improvements and highlight where further improvements may be possible.
Keywords: Cell-free systems, Cell-free expression, Cell-free extract, Synthetic biology, Methods
Abbreviations: CFE, Cell-Free Expression; CFPS, Cell-Free Protein Synthesis; TXTL, Transcription and Translation; GFP, Green Fluorescent Protein; OD, Optical Density; TFF, Tangential Flow Filtration
1. Introduction
Cell-free expression (CFE) systems mimic the transcription and/or translation capabilities of cells without requiring living, intact cells. These approaches have advantages over cells including mitigation of transport issues due to the disruption of membranes, funneling of cellular resources to the function of interest by removal of genomic DNA, and reduced toxicity issues by removal of cell growth constraints. CFE systems were originally used as a tool to understand basic mechanisms in biology, most notably the elucidation of the genetic code in the 1960's [1]. Over the past decade, CFE systems have found use in a range of applications, including sensors, manufacturing, and genetic prototyping; these applications have recently been reviewed extensively [[2], [3], [4]].
There are a wide variety of types of CFE systems, also referred to as cell-free protein synthesis (CFPS) and TXTL (for transcription-translation). We use CFE here in line with a recent comprehensive review of the field [4] and to be inclusive of systems that only perform transcription or translation. The creation of CFE systems is achieved either by purifying and recombining all components necessary to achieve the desired transcription/translation function (known as the PURE system and not a focus here) [[5], [6], [7]], or by using cellular extracts paired with a mixture of resources and cofactors. Many cell types have been used to make CFE extracts, including several strains of Escherichia coli, a variety of other prokaryotes, yeast, plants, and mammals [3]. For extract preparation, several cell lysis approaches have been demonstrated, including bead-beating [[8], [9], [10]], pressurized shear flow [11,12], sonication [10,13], freeze-thawing [14], and the use of lysozymes [15]. After lysis, extracts are processed through several steps, including multiple centrifugations, incubation, and dialysis. The growth conditions, extract preparation methods, and post-processing steps vary by lab, organism, and application. The resulting extracts are stored for later use where they are combined with a mixture of: (a) cofactors such as amino acids, nucleotides, salts, and an energy source (referred to here as the “reagent mix”), and (b) DNA encoding the function of interest. The content and preparation of the reagent mix has recently been reviewed by Dopp et al. [16]. The combined reaction is then incubated to perform the intended CFE function.
For decades, most method improvements for prokaryotic CFE systems have been achieved by a handful of labs. With a recent influx of new users, several publications have emerged aimed at reducing the barrier to entry to the field via cheaper or simpler methods [7,8,10,13,14,[17], [18], [19]], understanding and reducing variability [[20], [21], [22], [23], [24]], and methodology reviews or protocols explicitly aimed at enabling new users [3,20,22,25]. CFE has even been put forth as a powerful educational tool to teach the fundamentals of biology [[26], [27], [28]]. In the past two years, several reviews have summarized various aspects of CFE research [[2], [3], [4],16,[29], [30], [31], [32]]; here we examine the methods used for the production of prokaryotic extracts for CFE, which to date does not exist in the literature. We anticipate that this work will be of utility to new entrants to the field seeking to understand the differences between the myriad protocols, the relative advantages and disadvantages of each step, and the rationale behind methodological choices. We further point such readers to Gregorio et al. for an expansive introduction to CFE [3] and the aforementioned review by Dopp et al. on reagent mixes [16]; in combination with this work, these reviews provide a comprehensive primer on CFE and prokaryotic methodologies. Motivated by the publication of several new methods for E. coli in recent years [8,[12], [13], [14], [15],[17], [18], [19], [20],25,[33], [34], [35], [36], [37]], as well as the creation of extracts from at least 10 new prokaryotic organisms [[38], [39], [40], [41], [42], [43], [44], [45], [46], [47]], we trace the development of these methods over the past 20 years. For E. coli extracts, we start by surveying the strains and genetic modifications used in the CFE field, then chronicle efforts to optimize individual steps within the extract preparation process. We then detail methods for non-E. coli prokaryotic CFE extracts. Using the aggregated information, we comment on methodological steps that may be worthy of further investigation and future outlooks for the field.
2. Methods of extract preparation for E. coli
In the following sections, we review the literature on methods to produce CFE extract from E. coli. As the original and still most common organism used for CFE [3], methodological improvements stretch back approximately 60 years [48]. In this review, we focus on advances in extract production since 2000. We compile strains used and key genetic modifications, then outline extract preparation methods (Fig. 1). For each preparatory stage, we provide a timeline for advances to show the evolution of methods and highlight areas for further investigation.
Fig. 1.
Overview of workflow for CFE extract preparation methods.
2.1. Strain optimization
E. coli extracts have been produced from a number of cell strains, each engineered to achieve specific goals such as altering productivity and inclusion or removal of specific enzymes. The properties needed for new applications can be conferred from engineered strains to their extracts. E. coli strains commonly used to prepare CFE are shown in Table 1 and genes that have been manipulated to modify the properties of CFE are in Table 2. We note that most strains used only for very specific applications, such as overexpressed enzymes in a biosynthesis pathway, are not included.
Table 1.
Commonly used strains, with genotypes. Citations indicate originally developed locations and/or application.
| Strain | Genotype | Ref(s) |
|---|---|---|
| BL21-Rosetta (DE3)a | F−ompT hsdSB(rB− mB−) gal dcm (DE3)a pRARE (Novagen) | [49,50] |
| BL21-Rosetta2 (DE3)a | F−ompT hsdSB(rB− mB−) gal dcm (DE3)a pRARE2 (Novagen) | [8,33,51] |
| BL21-Star (DE3)a | F−ompT hsdSB(rB−, mB−) gal dcm rne131 (DE3)a | [25,49,50] |
| BL21-Gold-dLac (DE3)a | F−ompT hsdSB(rB− mB−) dcm gal (DE3)aendA lacZYA | [14] |
| JS006 | MG1655 araC lacI | [52,53] |
| A19 | rna gdhA2 relA1 spoT metB1 | [11,50,54,55] |
| KC1 | A19 speA tnaA tonA endA sdaA sdaB met+ | [56] |
| KC6 | KC1 gshA | [57] |
| KG6-der. | KC6 rnb ackA+ef-tu+hchA+ibpA+ibpB+if-1+if-2+if-3+ | [58] |
| KGK10 | KC6 gorB trxB-HA | [[59], [60], [61]] |
| NMR1 | A19 endA met+ | [56] |
| NMR2 | A19 speA tnaA tonA endA met+ | [56] |
| NMR4 | A19 recD endA met+ | [62] |
| NMR5 | A19 lambda phage<>recBCD met+ | [62] |
| S30BL/Dna | BL21(DE3) dnaK/J+grpE+ | [63] |
| S30BL/DsbC | BL21(DE3) dsbC+ | [63] |
| S30BL/GroE | BL21(DE3) groEL/ES+ | [63] |
| S30OB | F−ompT hsdSB(rB− mB−) gal dcm lacY1 ahpC (DE3) gor522::Tn10 trxB (Novagen) | [63] |
| S30OB/Dna | S30OB dnaK/J+grpE+ | [63] |
| S30OB/DsbC | S30OB dsbC+ | [63] |
| S30OB/GroE | S30OB groEL/ES+ | [63] |
Each of these strains is available with and without DE3 modifications, which enables induction of T7 polymerase. Some studies use DE3 strains while others do not.
Table 2.
Genes commonly modified in engineered cell-free strains. Citations indicate where more information about the gene in the context of cell-free can be found.
| Gene | Description | Ref(s) |
|---|---|---|
| ackA | acetate kinase, added to increase yield | [64] |
| araC | transcriptional activator, removed to prevent interference with AraC-expressing circuits | [52,53] |
| csdA | cold shock degradosome protein, removed to prevent mRNA decay during preparation | [65] |
| dnaJ | chaperone protein, added to assist folding with dnaK, grpE | [63] |
| dnaK | chaperone protein, added to assist folding with dnaJ, grpE | [63] |
| dsbC | disulfide isomerase, added for disulfide bond formation | [66] |
| ef-tu | translation factor, added to increase yields (most abundant protein in cell, potentially rate-limiting) | [64,67] |
| endA | endonuclease, removed for plasmid stability | [56] |
| gamS | nuclease inhibitor from lambda phage, added to protect linear DNA | [60,62,68,69] |
| gorB | glutathione reductase, removed to prevent disulfide bond persistence | [59,63] |
| groEJ | chaperone protein, added to assist folding with groEL | [63] |
| groEL | chaperone protein, added to assist folding with groEJ | [63] |
| grpE | heat shock protein, added to assist folding with dnaJ, dnaK | [63] |
| gshA | glutamate-cysteine ligase, removed to stabilize cysteine | [57] |
| hchA | chaperone protein, added to increase solubility and yield | [64] |
| ibpA | small heat shock protein (chaperone), added to increase solubility and yield | [64] |
| ibpB | small heat shock protein (chaperone), added to increase solubility and yield | [64] |
| If-1 | initiation factor 1, added to increase yield | [64] |
| If-2 | initiation factor 2, added to increase yield | [64] |
| If-3 | initiation factor 3, added to increase yield | [58,64,67,70] |
| lacI | transcriptional repressor, removed to prevent interference with LacI in circuits | [52,53] |
| lacZYA | lac operon, removed to eliminate background when using LacZ as a reporter | [14] |
| lysR | λ phage endolysin, added to disrupt the bacterial cell wall to facilitate lysis | [14] |
| mazF | MazF toxin, removed to prevent mRNA degradation at ‘ACA’ sites | [65] |
| met | P1 selection marker, engineering scar | [56] |
| pnp | PNPase, involved in mRNA degradation, removed or tagged for post-growth removal | [64,71] |
| recD | exonuclease subunit, removed to protect linear DNA (ineffective), or tagged and removed post-growth (successful) | [62,71] |
| rna | RNAse A, removed for RNA stability | [54,55] |
| rnb | RNAse II, removed for RNA stability | [65,67] |
| rpfA | release factor 1, removed to encourage noncanonical amino acid incorporation | [72] |
| sdaA | serine deaminase, removed to stabilize serine | [56] |
| sdaB | serine deaminase, removed to stabilize serine | [56] |
| speA | arginine decarboxylase, removed to stabilize arginine | [56] |
| tnaA | tryptophanase, removed to stabilize tryptophan | [56] |
| tonA | outer membrane protein, engineering scar | [56] |
| trxB | thioredoxin reductase, removed post-growth with HA tag to prevent disulfide bond persistence | [59,63] |
Common strains used throughout biotechnology are natural starting points for CFE extracts as their properties already reflect the needs of researchers. Lab strains such as K19, first introduced in 1966 [54], have been frequently used [55,73], as well as MRE-600 [74]; BL21-derivatives such as CP strains [9], Rosetta and Rosetta 2 strains [8,33,[49], [50], [51],68], DE3 strains [13,19,25,50,75], and Star strains [25,49]; Origami strains [63], and K12 MG1655 strains [13,35,37]. These strains are chosen for generally favorable properties such as rapid growth, rare tRNAs (Rosetta), T7 RNA polymerase (DE3 lysogens), reduced mRNA degradation (“Star”), disulfide bond formation (txrB/gor deletions in Origami), or general optimization for protein production (BL21). The properties of these base strains are not mutually exclusive; in particular, DE3 lysogens are present in many strains. Specific examples follow on the strain-level optimization of protein production, and biomedical and sensing application cases.
2.1.1. Strains to improve CFE production
Increasing protein production has been one of the main focuses of cell-free optimization. A common strain-level strategy to facilitate this is the induction of T7 RNA polymerase from DE3 strains (avoiding the need for exogenous addition) [19,25,50,75]. More CFE-specific strain modification findings in this area were pioneered by Swartz and colleagues from 2004 onwards. Earlier efforts on engineering cell-free systems focused on protocol improvements [55,74] and energy regeneration [73,76]. However, in 2004 an influential paper from Michel-Reydellet et al. described deletions in genes encoding amino acid degradation enzymes, thereby stabilizing amino acid supply and protein production [56]. The paper identified four limiting amino acids: arginine, serine, tryptophan, and cysteine. Arginine was stabilized by removing speA, a gene encoding an arginine decarboxylase, thereby inhibiting the conversion of arginine to putrescine. Serine was stabilized by removing serine deaminases sdaA and sdaB, inhibiting the conversion of serine to pyruvate. Tryptophan was stabilized by removing tnaA, which encodes a tryptophanase. Attempts to stabilize cysteine via deletions of tnaA and yfhQ did not succeed. A follow-up paper identified gshA, a glutamate-cysteine ligase, as the cysteine degradation culprit [57]. The resulting gshA deletion strain was named KC6 [56,57].
High-throughput approaches for determining positive and negative factors for cell-free expression have also been employed. Expression of 55 E. coli genes from linear DNA templates in NMR5 extract [70], led to a study analyzing the impact of 49 genes affecting transcription, folding, energy, and cell-division on cell-free yields [67]. Later, Airen (in unpublished but peer-reviewed thesis work) expressed 3789 E. coli open reading frames, identifying 79 positive and 60 negative effectors of CFE yield [64]. Using this information on negative effectors, four mutant strains were made that, when combined with (a) supplementation with positive effectors, (b) stabilization of pH, (c) substrate replenishment, and (d) mRNA stabilization, were able to increase expression 4-fold. While strains with four negative effectors (pnp, rnb, raiA, and mazG) removed did not result in significantly increased expression, supplementation with ibpA, ibpB, if-1, if-2, if-3, and ef-tu did demonstrate increased yields [64,67].
To stabilize linear DNA templates in CFE reactions, the lambda-phage cluster has also been inserted into strains made into cell-free extracts [62], creating the NMR5 strain. Earlier efforts had revealed Gam to be the main RecBCD inhibitor and showed stabilization of linear DNA when Gam was added in purified form [68]. Later, Seki et al. after first observing that decreased temperatures increased yields via reduced degradation of linear DNA template [77], demonstrated improved yields by creating a strain where pnp, a gene involved in mRNA degradation, and recD, involved in degradation of linear DNA, were both tagged with a streptavidin binding peptide such that these enzymes could be removed after lysis [71].
Other efforts focused on the overexpression of molecular chaperones capable of reducing aggregation and improving solubility of eukaryotic proteins such as human erythropoietin [63]. Plasmids were used to overexpress chaperone and heat-shock genes groEL/ES, dnaK/J and grpE, or dsbC. The Kim group also explored the creation of extracts from the Origami strain (Novagen), intended to promote disulfide bond formation. The roles of proteins TrxB, Gor, and DsbC would later be formally explored in the context of disulfide bond formation in work by Knapp et al. [59].
A final example notable for its novelty is that of Didovyk et al. where they engineered a strain to lyse by freeze-thawing [14]. In their system, lambda phage endolysin gene R, which degrades the cell wall, is expressed prior to harvest. Cells expressing R grow normally until the inner membrane is disrupted, which can be achieved by freeze-thaw or chemical means. The authors capitalize on this effect to achieve efficient CFE extracts from a highly simplified protocol. This is an example of using strain-level modifications to streamline the protocol by which a CFE system is produced, and may be an area of opportunity for further exploration.
2.1.3. Application-specific strains
In many cases, strains are engineered to achieve features related to specific uses for CFE. In this section, we highlight strain modifications for key CFE applications but do not offer an exhaustive review. The first example, echoing the original use of CFE for fundamental research, is the development of strains that facilitate the incorporation of noncanonical monomers into polypeptides. The use of MAGE-recoded [78] strains lacking release factor 1 [79] combined with removal of negative effectors rna, rnb, csdA, mazF, and endA have led to improved incorporation of noncanonical amino acids into proteins made by CFE [65,80]. Other work in this space has focused on aspects other than strain engineering to enhance incorporation [[81], [82], [83]].
One of the more mature areas of strain engineering for CFE is in facilitating the expression of proteins with disulfide bonds, a subset of proteins (most notably antibodies) with biotechnological utility. Disulfide bonds are a common feature of mammalian proteins but are difficult to implement in cell-free systems due to rapid reduction in vitro [84]. While iodoacetamide treatment can inactivate thiols responsible for reducing disulfide bonds [66], the treatment globally targets –SH groups and can inactivate critical enzymes (such as DsbC and G-3PDH) [59]. A more effective strategy was the creation of a hemagglutinin tagged trxB (thioredoxin reductase) and gor (glutathione reductase) deletion strain, supplemented with dsbC. TrxB is tagged to allow for it to be present during cell growth but removed after cell-free processing, however significant glutathione reductase activity in CFE was still observed subsequent to its removal [59]. It is noted that this genotype closely represents the Origami strain (Novagen) that contains knockouts of trxB and gor with suppressor mutations in ahpC, and was demonstrated successfully for cell-free production two years prior [63]. The resulting strain (KGK10) and findings that accompanied its production form the basis for current production efforts of disulfide bond proteins. CFE derived from variants of this strain that also overexpress chaperone proteins were found to effectively express immunoglobulin proteins despite coming from a prokaryotic background [85]. Commercially, Sutro Biopharma utilizes variants of the strain for producing cytokine rhGM-CSF at 200 L scale [61] and producing antibody fragment light and heavy chains [60]. A recent addition is the use of the commercial SHuffle T7 Express lysY strain of E. coli that expresses T7 RNAP and DsbC isomerase enzymes to rapidly prototype proteins with disulfide bonds [86].
In a similar application space, engineered strains have been utilized to glycosylate proteins expressed in CFE. Addition of lipid-linked oligosaccharides and purified PglB from Campylobacter jejuni was shown to produce N-linked glycosylations in multiple proteins using both commercial S30 (Promega) and PURE (NEB) CFE systems [87]. This general approach was later implemented using the glycosylation-optimized E. coli strain CLM24 [88] as a host strain in which a glycan biosynthesis pathway and oligosaccharyltransferase had been expressed prior to harvest [89]. A further-engineered strain (CLM24 ΔlpxM) producing lower endotoxin levels was demonstrated in a modular scheme to produce multiple vaccine-type antigens in pursuit of point-of-need drug manufacturing [90]. Recently, CFE from endotoxin-free ClearColi cells was recently used to produce a therapeutic protein [91], which increases the potential of CFE in medical applications. Combined with recent strategies using CFE to probe glycosylation site specificity [92] and exploration of glycan diversity [93], CFE systems may soon offer the capability to synthesize specific glycopeptides with potential therapeutic applications.
Strains engineered for genetic circuit prototyping have also been used to generate extracts. For example, cells with lacI and araC knockouts and lacking tetR, such as strain JS006 [53] have been made into extracts to build oscillators that require exogenous LacI [52]. Conversely, ExpressIQ (lacIqQ) has been used to shut down operons that are LacI sensitive [94]. Commercially, cells optimized for 1,4-butanediol production were used by Genomatica as the starting strain for lysis to test expression efficiency hypotheses [95,96]. In another example, Marshall et al. expressed Cas enzymes dSpyCas9 or dFnCpf1 in extract strains to enable screening of CRISPR-repression designs [97]. .
Related to circuit prototyping, strain engineering can also benefit sensing applications. One clear example is knocking out endogenous expression of LacZ [14] to reduce background signal where LacZ is used as an eye-readable colorimetric reporter [98,99]. Another example is enrichment of enzymes used to process an analyte of interest into another product for which a known sensor exists, as has been recently demonstrated for phloroglucinol [100] and atrazine [101].
2.2. Pre-lysis processing
The first major stage in production of extracts for CFE is the growth of cell mass, which we call “pre-lysis processing” here. Within this stage, there are a series of optimization points: starter culturing, growth scale, culture conditions, induction/harvest timing, and pelleting (Fig. 2). In this section we lay out the progression of advances in pre-processing steps for CFE.
Fig. 2.
Historical evolution of pre-lysis processing methods. Note that the y-axis does not indicate a linear timescale.
A driving consideration across pre-lysis processing is maximization of active translational machinery. Several studies, reviewed below, have aimed to maximize ribosome concentration by maximizing growth rate, which has been shown to correlate directly with growth rate [102,103]. However, detailed examinations have shown that translation rates in CFE are more complicated than simple ribosome counts [35,104,105]. Quantitative analyses have shed some light on ribosome counts and rates in CFE [104,106], including through dynamic modeling [38]. While we do not review these mechanistic studies here, it is important to acknowledge them as context for pre-lysis processing methodology optimization efforts.
2.2.1. Starter culturing
Until recently, starter culturing was one of the few aspects of CFE systems that had not been clearly explored in the literature for optimization. All approaches follow traditional microbiological techniques where plates are streaked from a glycerol stock, single colonies are used to inoculate a starter culture, and the starter culture is used to inoculate either a second starter culture or the primary culture (next section). With one recent exception, no studies present data comparing the outcomes of different starter culturing approaches, likely because these are considered standard techniques with minor variations in volume and timing. Presumably, the different protocols are designed to facilitate consistent workflows and are not expected to impact extract activity. Recent results showing high productivity from cells grown to high densities enables more flexible experimental timing [35]. Building on this result, Levine et al. demonstrated that primary cultures could be started directly from picked colonies without loss of activity [19], which allows a 24 h workflow from a colony on a plate to CFE reaction.
2.2.2. Growth scale and vessels
Historically, fermenters were used to produce cell biomass. The original protocols utilized fermenters of up to 10 L to grow cells [107]. Building off of this, Zawada et al. demonstrated at 10 L scale (20 g/L wet pellet cell mass) an alternative fermentation strategy to accelerate fermentation times and produce extracts from cells with higher ribosome concentrations [108]. The same protocol is cited by Sutro Biopharma for use in a 200 L bioreactor that is custom-retrofitted with baffles [61]. Other groups continue to use fermenters at the 10–30 L scale [13,36].
Biomass can also be produced at shake-flask scale (1 L of cell culture in a 2.8–4 L Erlenmeyer flask). The use of shake flasks was first described in 2004 by Kigawa et al. [9]. Subsequently, two studies have demonstrated equivalent yields for shake flasks and fermenters [75,108], and most studies in recent years have used shake flasks, presumably due to the reduced labor, expertise, and cost of equipment compared to fermenters. Flasks may also be more amenable to exploration of protocol optimizations. Typical yields for cell cultures of 1 L are about 1–2 mL of crude extract [8]. Because most protocols are focused on maintaining fast growth and aeration before capture at culture mid-log phase, growth-maximizing baffled flasks are frequently used (e.g. TunAir or Ultra Yield flasks). Unless otherwise stated, for the studies referenced in this review, shaking was used in all cases and ranged from 160 to 280 rpm.
For even smaller volumes, Kwon and Jewett demonstrated the first rapid production of extract using 10 mL culture tubes, allowing for the exploration of ~100 strains per day using basic, readily available equipment (a sonicator, small shaker, and tabletop centrifuge) [13]. They documented equivalent expression from extracts generated using different culture volumes and vessels (test tubes at 10 mL; shake flasks at 50, 100, 500, and 1000 mL; and fermenter at 10 L) and demonstrated the utility of CFE for exploring multiple rapidly-engineered strains or conditions.
2.2.3. Growth conditions
While the original CFE protocols utilized 28 °C for growth [107], current protocols incubate at 37 °C to optimize protein production in the extract through increased translation machinery at maximum growth rate [103]. There is evidence that temperature, affecting growth rate, has a direct correlation with extract productivity. In particular, Seki et al. found a positive linear correlation between productivity and culture temperature, with yields increasing 66% from 20 °C to 37 °C when using plasmid DNA (trends were different for linear DNA due to temperature-dependent DNA degradation) [77]. Similarly, Yamane et al. showed that higher growth rates at 42 °C using a supplemented media yielded 40% increased activity compared to 37 °C growth [109]. While most studies use 37 °C, a 2017 study did show increased yield at 25 °C compared to 37 °C; however, many other methodological details varied between the tests [36].
Growth media can vary between extract preparations, though typically media are undefined, (e.g. LB, 2xYT) [110]. Media may be supplemented; for example, asparagine, glutamine, and tryptophan have been added to a complex medium to encourage faster growth [109]. For fermenter growth, glucose and amino acid concentration can be selectively monitored and fed to prevent acetate accumulation [108]. In 2000, Kim and Choi identified the addition of phosphate and glucose to a 2xYT medium (named 2xYT-PG) to be suppressive of phosphatase activity in the resulting extracts [111]. Phosphatase activity was found to consume phosphoenol pyruvate (PEP) and the amino acid cysteine, reducing yields. This medium has formed the basis for most cell-free preparations for years, with the notable split that some groups omit glucose (”2xYT-P”) while others do not [13,14,18,25,35,37,61]. Silverman et al. recently directly tested the performance impact of including glucose and found a significant productivity drop from a native bacterial promoter [20]; it is unclear in recent protocols if glucose is detrimental when T7 transcription is used instead. One additional study explored the impact of different media on the glycolysis pathway in CFE, but did not examine protein production [112].
A noteworthy modification to the standard 2xYT-P or 2xYT-PG media is a formulation that merges 2xYT-PG medium with autoinduction (AI) medium [113] to form a new cell-free autoinduction (CFAI) medium, which enables harvesting cells at high culture densities without loss of CFE activity [19]. The authors first found that AI medium, which is similar to 2xYT-PG but uses lactose and glycerol as the primary carbon sources instead of glucose, showed slightly higher activity when harvested at standard densities, but both media had lower productivity when harvested at higher densities (see section 2.2.4 for detailed discussion of harvest densities). By increasing the buffering capacity and lactose in the medium, the authors then showed high activity from cells harvested at high culture densities. Moreover, they found that additional glycerol, tryptone, and yeast extract did not change performance. This ability to harvest at high densities without productivity loss is useful for obtaining more extract per volume of culture.
2.2.4. Harvest and induction timing
A major point of optimization in the pre-processing phase has been the harvest point. In traditional protocols, slow growth rates were used to limit acetate accumulation while maximizing total cell mass through high culture density; however, in 2005 Zawada et al. presented a modified strategy to reach high densities with accelerated growth rates, resulting in slightly more active extracts in far less time [108]. Most current protocols define a harvest point via an optical density measurement at 600 nm (hereafter referred to as OD) within the mid-log phase where translation machinery is most abundant [103]. Sun et al. state that harvesting in mid-log phase is critical for extract quality but do not provide any data [8]. In 2015, Kwon and Jewett showed that the optimal harvest point varies by strain, noting insensitivity to harvest OD between 2.5 and 5.5 for BL21-Star (DE3) cells but a significant drop in activity for C495 cells harvested at an OD above 3.5 [13]. Other recent studies have suggested extract activity has slight dependence on culture density within exponential phase when tested between OD of 2.7 and 4 [19] or 5, 7, and 8 h of growth [37]. Dopp et al. observe that OD measurements can vary significantly between instruments and labs [114], and therefore recommend that each lab optimize the harvest OD for their own process [25].
Some methods additionally include an induction step to express T7 polymerase to avoid adding purified polymerase later. Kim et al. first demonstrated the approach in 2006 by adding 1 mM IPTG to growing BL21-Star (DE3) cells at 0.6 OD to induce expression of T7 polymerase; the study showed active CFE extract, but made no comparison to non-induced extracts [50]. Later, others confirmed that the method yielded fully active extract compared to adding purified T7 polymerase to uninduced cells [13,75]. More recently, Dopp et al. explored the impact of varying both harvest time (3–5 h) and IPTG induction (35–85 min before harvest) [25]. They found that, for their conditions, IPTG induction at 1 h before harvest was optimal, though a stronger dependence on growth time was observed. Most recently, Levine et al. induced T7 polymerase expression using AI medium (see section 2.2.3), saving effort by obviating the need for a manual induction step [19].
Three recent studies offer a major departure from the focus on mid-exponential phase harvest points by growing cells to higher densities. Harvesting at high densities potentially offers more cell mass per volume of medium, less need to continually monitor OD, more flexible experimental designs due to less sensitive harvest timing, and expression of alternative sigma factors. First, Katsura et al. demonstrated productive extracts from cells grown to stationary phase in 30 L fermenters [36]. Their motivation was to improve consistency of harvest conditions, though they did not present any data for comparison; moreover, they introduced a number of innovations making it difficult to disentangle the relative contributions of each change to productivity. Next, Failmezger et al. demonstrated that, contrary to earlier studies, yields were consistent for cultures grown in shake flasks to exponential or stationary phase [35]. This result calls into question the assumption that CFE is most active when cells are harvested during exponential phase due to higher ribosome concentrations [115], and indeed the authors found a reduction in the concentration of ribosomes for the extracts harvested during stationary phase. While this result is not fully explained, the authors suggest a higher fraction of active ribosomes for cells harvested in stationary phase as a likely explanation. In contrast to these results showing equivalent yields, Kim et al. report a statistically significant 32.8% drop in activity for cells grown to stationary phase compared to mid-exponential (14 and 7 h growth, respectively) using the same 2xYT-PG medium [37]. More recently, Levine et al. expanded on the results of Failmezger et al. by modifying the media (see section 2.2.3). Initially, they found ~50% reduction in activity when cells were harvested at a high density (OD of 10) compared to a more standard density (OD of 2.5), whether using 2xYT-PG or AI media. Noting a pH drop for cells harvested at stationary phase (also noted by Failmezger et al.), the authors showed that media with increased buffering capacity (and additional lactose for induction of T7 polymerase) produced fully productive extract compared to cultures harvested during exponential phase [19]. It is possible that in addition to pH during growth, differences in extract processing steps between the three studies, which include homogenization [35] vs. sonication [19,37] and differing exposures to buffers at multiple stages, could impact the protein synthesis activity.
2.2.5. Pelleting, washing, and storage
At harvest, all extract preparation protocols involve centrifugation followed by washing. Centrifugation protocols vary across studies with a shift around 2013. Prior to 2013, a 30 min centrifugation was typical, though speeds decreased over time (16,000×g in 2004 [9], 9000×g in 2005 [11], and 8000×g [10] and 6000×g [75] in 2012). Then beginning in 2013, methods began to show reduced centrifugation times as well as speeds (Sun et al.: 5000×g for 12 min in 2013 [8]; Kwon et al.: 5000×g for 15 min in 2015 [13]; Krinsky et al.: 7000×g for 10 min in 2016 [34]; Levine et al.: 5000×g for 10 min in 2019 [18]). The decreasing centrifugation speeds and times are not discussed in any of these studies, nor any data provided comparing centrifugation times or speeds. One explanation may be that users consider pelleting of cells to be standard technique and simply use settings that adequately perform the step with their centrifuge model. Decreased centrifugation speeds and times could also decrease forces experienced by cells and make cell resuspension steps easier. Nearly all studies perform centrifugation at 4 °C, with one recent exception at 10 °C [18].
The initial centrifugation is followed by washing in buffer and re-centrifugation, again maintaining cold temperatures. The wash buffer used (called variously “Buffer A″, “S30 Buffer”, or “S30A Buffer”) is largely consistent throughout extract preparation protocols, containing 14 mM magnesium and 60 mM potassium salts, 5–50 mM Tris, and 0–10 mM DTT at a final of pH of 7.7–8.2. Typical protocols perform three wash steps; however, in 2005 Liu et al. first showed that a single wash step resulted in the same productivity as two or three washes [11]. Nevertheless, most protocols continued to follow protocols with three washes, including recent publications [18,37], though some have used one [10] or two [61] washes. Two new studies have confirmed the result of Liu et al. that a single wash step has the same yield [19,25]. Each wash step is performed by resuspending the pellet in the wash buffer and then centrifuging. Within each study, the specific protocols for centrifugation after each wash are typically the same as the initial centrifugation, though not always. In total, optimization of the pelleting steps can drop centrifugation time from 2 h 45 min [11] to 20 min [18,34].
For large-scale production of CFE extracts, Katsura et al. applied tangential flow filtration (TFF) instead of centrifugation to concentrate and wash cells. With TFF, the culture is pumped along a filtration membrane, allowing media to flow out as waste [36]. While they did not present data comparing this approach to traditional centrifugation and washes, their process did result in extract activities on par with other methods.
The final aspect of the pelleting process is storage. Early studies flash-froze cells in liquid nitrogen and stored at ≤ −80 °C before lysis [11,107]. Subsequent protocols have flash frozen pellets [13], while others have moved immediately into cell lysis [8]. Until recently, little data on the effects of freezing has been available apart from Kigawa et al. stating in 2005 that in their experience frozen cells lose activity after three days. Since then, Dopp et al. state that they found no difference between flash-freezing in liquid nitrogen, freezing the cells at −80 °C without flash-freezing in liquid nitrogen, or directly processing the cells [25]. In addition, Silverman et al. presented data in 2019 that there was no difference between flash-freezing in liquid nitrogen and using the cells directly [20]. No studies to date have clearly explored the impact of storing washed cells over time.
2.3. Lysis
Considerations of note when selecting a lysis method are efficiency, scale, ease of use, and preservation of cell components. In addition, the ratio of buffer volume to cell pellet mass prior to lysis has been shown to impact the productivity of the CFE system [13]. Accordingly, published protocols have cell disruption methods tailored to their use case. As lysis involves resuspension of cells, the ratio of buffer to cell mass is a factor in most protocols. Commonly, ≈1 mL of buffer (typically S30) is used to suspend 1 g of wet cell mass after the final wash step, presumably because this ratio is the minimum which yields a viscosity low enough for processing. This ratio has been explored for sonication and it was confirmed that ≈1 mL of buffer per gram of cell mass was optimal for sfGFP expression [13]. However, some studies use a fixed volume for resuspension [34] or another ratio, e.g. 2 mL/g [61].
Another cross-cutting concern is localized sample heating that may denature native proteins. While at least one study did not find a correlation between temperature and activity during lysis by several methods [10], sample heating may still be an issue. Several approaches perform lysis in bursts with cool down stages on ice to limit these effects.
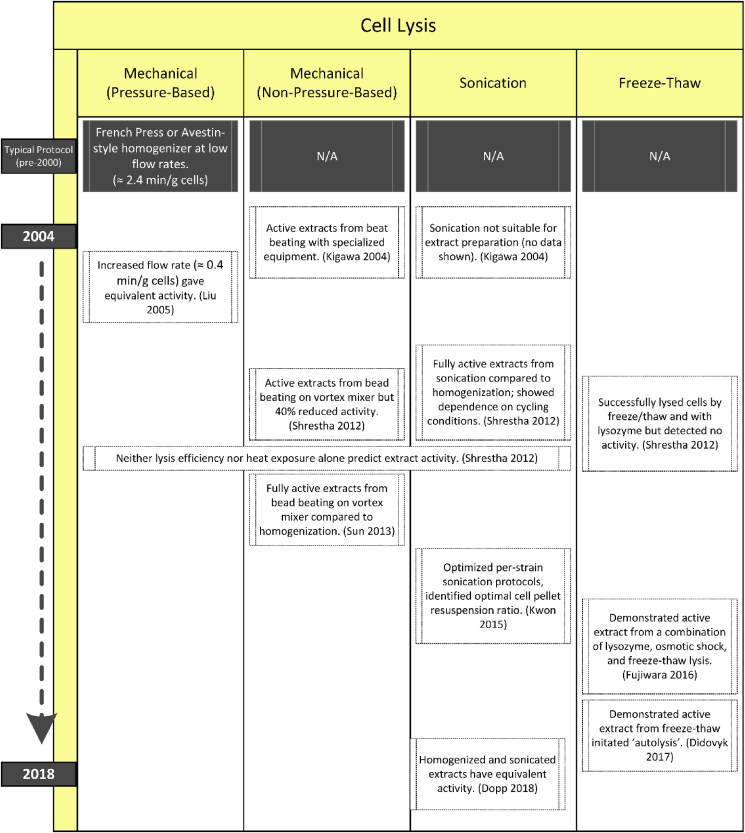
We divide our discussion of lysis methods into five categories: mechanical (pressure-based), mechanical (non-pressure-based), sonication, temperature, and chemical (Fig. 3).
Fig. 3.
Historical evolution of lysis methods for CFE. Note that the y-axis does not indicate a linear timescale.
2.3.1. Mechanical (pressure-based)
High pressure disruption mechanisms such as impingement homogenizers are among the earliest and most widely utilized methods for lysis for extract preparation [116]. These instruments work by forcing cell suspensions through a narrow aperture under high pressure. The high-velocity flow of cells either impinges on an opposite high-pressure stream of cells or a rigid valve/nozzle surface. The resulting shear forces and rapid decompression are thought to be critical in the formation of inverted membrane vesicles in the resultant extracts [117]. Because the enzymes essential to the oxidative phosphorylation pathway must be membrane-associated to function, creating these vesicles is a theoretical advantage of this method via increases in the metabolic efficiency of extracts.
For E. coli extracts, different types of impingement homogenizers are currently in use, ranging from French press-style homogenization [12,50,55] to Avestin™-type homogenization [11,36,68,75,117]. Both types of homogenizers allow for scaling of batches; French-press homogenizers scale up to the size of the press (typically 30 mL), while Avestin™-type homogenizers allow for continuous flow. Specifics of impingement homogenization have not been thoroughly explored, notably the pressure of lysis. One observation by Liu et al. was that the rate of flow, previously kept low over concerns that sheared genomic DNA would not sediment properly during subsequent centrifugations, did not affect the activity of the extract [11]. Similar findings by Dopp et al. showed equivalent activity when using a homogenizer or sonicator [25].
2.3.2. Mechanical (non-pressure-based)
Non-pressure-based mechanical methods utilize a grinding mechanism, typically by agitation of a mixture of suspended cells and ceramic/glass beads, the motion of which break apart the cells and efficiently shear DNA [118]. Industrial scale bead-mills have been employed for cellular lysis [116], as have the use of “bead-beater” type desktop devices [119]. For cell-free protocols, bead-beaters and even standard vortex mixers have been utilized [[8], [9], [10]]. Beads are easily separated from the lysate by centrifugation or filtration and no expensive equipment is required, reducing the financial barrier of entry. These protocols also have utility in lysing non-E. coli cells such as cyanobacteria [120] and environmental samples from soil [121]. To maintain high protein concentrations necessary for cell-free expression, beads can also be filtered out of solutions after processing [8]. Protocols alternate between agitation and incubation on ice to avoid excessive sample heating [8,10]. Katsura et al. recently presented work on scale up in which they compared the use of a low throughput bead-beating device, an industrial-scale bead mill device, and a high-pressure homogenizer [36]. Across several conditions they found that the homogenizer gave higher yields and offered easier scalability. Bead-beating remains useful for low-cost entry to the field and for the ability to work in small volumes, enabling processing of small batches to rapidly explore different cell lines or preparation methods.
2.3.3. Sonication
Sonication, or acoustic lysis, relies on ultrasound energy (15–20 kHz) to disrupt cells in solution. The mechanism of lysis is cavitation, a phenomena where microbubbles form at nucleation sites, absorb energy, and burst, releasing mechanical shock waves that disrupt the cell wall and can shear DNA [116]. Until 2012, the only documented use of sonication for CFE extracts in the literature was a comment by Kigawa et al. that in their tests the method “is not suitable for extract preparation, due to sample heating and difficulty of management”, though no data was provided [9]. Nevertheless, Shrestha et al. showed in 2012 that by optimizing burst and cooling times they could achieve protein yields comparable to high-pressure homogenization [10]. Significantly, they also showed that sample temperature during lysis did not predict activity across lysis methods, including noting that pressure-based homogenization heated the samples more than sonication. This work was followed by a study from Kwon and Jewett that further optimized the method across volumes, cycling conditions, and total energy for two different strains, identifying that optimal protocols can vary between strains [13]. This study and another by Dopp et al. [25] independently confirmed the result from Shrestha et al. [10] that CFE yields were equivalent for extracts lysed by sonication or high-pressure homogenization, establishing sonication as a low-cost and high-throughput method compatible with volumes ranging from 100 μL to 30 mL. At the time of this writing, Kwon and Jewett's study has been cited 158 times, reflecting the wide adoption of sonication as a method for producing cell-free extract both for E. coli and non-E. coli prokaryotes (see Section 3 below).
2.3.4. Freeze-thaw
Disruption of cellular membranes by freeze-thaw cycles is one of the easiest methods of cellular disruption for producing purified proteins [122,123]. This lysis can take place with or without chemical or enzymatic assistance such as lysozymes. The first published attempt to use this approach for CFE was in 2012 by Shrestha et al. where 99.6%–99.9% lysis efficiency was achieved but no protein production was observed [10]. More recently, Didovyk et al. demonstrated that expression of a phage endolysin protein during growth makes cells susceptible to lysis by freeze-thawing or by rehydration after lyophilization [14]. This novel approach offers significant advantages over traditional lysis methods as it lacks a requirement for specialized equipment and decreases the need for technical labor. Lysis via rehydration after lyophilization could be particularly advantageous in certain applications, such as the preparation of paper-based sensors [14,[98], [99], [100]].
2.3.5. Chemical lysis
Chemical lysis refers to the use of enzymes or detergents to disrupt cell walls, typically used in the context of protein purification. In such applications, enzymes such as lysozyme (degrades the peptidoglycan layer in E. coli) or benzonase (nuclease to remove DNA and RNA) are commonly utilized with detergents such as Tween-20, Triton-X, or RIPA buffer or commercial mixtures such as BugBuster (Novagen) or CellLytic X (Sigma). Initial attempts to use lysozymes for lysis successfully lysed the cells, but did not result in active extracts [10]. Extracts prepared via treatment with lysozyme followed by osmotic shock and a freeze-thaw cycle did, however, lead to active CFE reactions with yields similar to other methods [15], although this approach has not been widely adopted to date.
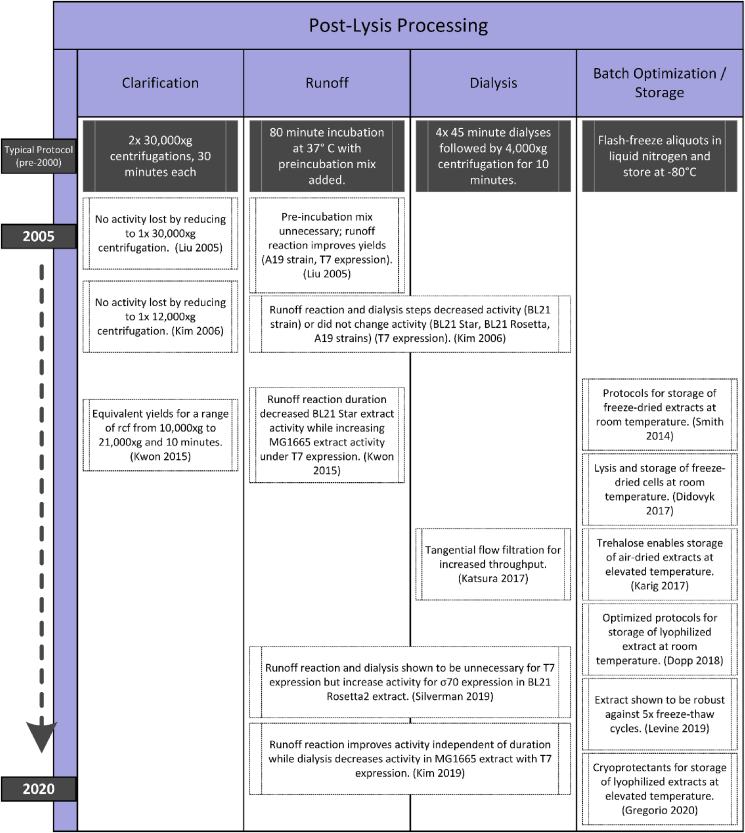
2.4. Post-lysis
The final phase of extract preparation is a series of post-lysis processing steps to prepare cell lysates for use in CFE reactions, including clarification, run-off, dialysis, and batch optimization and storage (Fig. 4).
Fig. 4.
Historical evolution of post-lysis processing methods. Note that the y-axis does not indicate a linear timescale.
2.4.1. Clarification
Lysates are typically viscous and difficult to manipulate. To remove insoluble material, the lysis step is always followed by a clarification step in which the lysate is centrifuged to separate debris from the enzymes, small molecules, and co-factors necessary to drive coupled transcription-translation. Crude extract can be used with no clarification step, though issues arise with handling the highly viscous solution and one study showed 20% lower activity [50]. Early clarifications consisted of two successive 30 min centrifugations at 30,000×g [9,107,124]. Then in 2005, Liu et al. found that a single centrifugation step gave equivalent performance [11], and in 2006 Kim et al. demonstrated that one 10 min 12,000×g centrifugation gave increased expression [50], leading them to name their method S12 for the decreased centrifugation speed compared to traditional S30 extract nomenclature (we note that the method also incorporates alterations to the runoff reaction). The S12 method was reproduced independently, demonstrating a 30% increased yield using S12 over S30 [125]. Kwon et al. further explored this variable, demonstrating the same activity for a range of speeds between 10,000 and 21,000×g [13]. Subsequently, 12,000×g spins have become widely adopted for preparing cell-free systems from compatible strains [8,10,13,20,25,37], with some exceptions [18,34,35]. To limit confusion, we note that while S12 is a useful naming convention here, it is not widely used in the later literature; S30 is still frequently referred to regardless of centrifugation method, especially to describe the buffers used.
2.4.2. Runoff reaction
A runoff reaction is often conducted after clarification of the extract with the intention of releasing ribosomes from bound mRNA and degrading sheared host mRNA and DNA [48,126]. Runoff reactions typically lead to visible precipitation [8] and are usually followed with an additional centrifugation using the same or similar protocol as the previous clarification step. Early runoff reactions included the addition of a pre-incubation mix containing Tris, Mg, ATP, DTT, amino acids, PEP, and pyruvate kinase to the clarified extract prior incubation at 37 °C for 80 min. However, Liu et al. first reported in 2005 that the pre-incubation mix did not increase expression using an A19 strain but that reduced incubation times did reduce activity [11]. In addition, ribosome release as the reason for the runoff is called into question, with a new hypothesis that the runoff reaction activates activators or degrades inhibitors. In 2006, Kim et al. showed that the pre-incubation step was unnecessary for a BL21 strain, yet in contrast to Liu et al. they showed only marginal yield dependence on runoff reaction duration, noting a peak at 30 min [50]. They then compared the activity across three additional strains: Rosetta (DE3), BL21-Star (DE3), and A19, finding that in all cases the traditional longer incubation did not improve yields (with the possible exception of A19). Zawada et al. then showed a strong activity dependence on both duration and temperature of the runoff reaction, this time for a derivative of the A19 strain [61]. At the same time, in non-peer-reviewed work, Roy et al. conducted a proteomics analysis on cell-free preparations with different runoff times and added proteins, and concluded that the runoff reaction primarily serves to remove cold-shock proteins accumulated during cell-free preparations [127]. Follow-on work concerning this hypothesis was not found outside this patent, making this conclusion a potential area of further inquiry. Later, Kwon and Jewett identified a strain-specific runoff property, with activity of extracts from BL21-Star (DE3) showing decreased activity with increasing runoff incubation times, while a MG1655 strain showed low activity without runoff and an optimal runoff incubation time of 60 min [13]. Similarly, Kim et al. showed strain-dependent performance for different runoff incubation times using another MG1655 strain [37].
A recent study by Silverman et al. shed some additional light on the topic by demonstrating that a runoff reaction significantly improved performance for expression from a native promoter [20]. All studies in the previous paragraph relied on T7 expression. Other studies using native promoters [8,12,33,51,106] all include a runoff reaction. Recent protocols focusing on expression from T7 promoters remain split between including [18,35] and omitting the runoff reaction [19,25,34]. In all, the results for studies exploring the impact of a runoff reaction are perhaps the most conflicting of the variables reviewed here. While various studies speculate on the possible reasons for observed differences, the underlying mechanisms driving the differing effects observed across strains and polymerases remain largely unexplained.
2.4.3. Dialysis
After the runoff reaction, extracts are sometimes dialyzed against a final S30 run buffer at 4 °C [8]. Dialysis steps vary from one cycle of either 3 h [8] or 18 h [107] to four cycles of 45 min [9]. However, Liu et al. found no significant difference between 0 and 4 dialysis cycles [11]. This result was confirmed in 2006 by Kim et al. who found dialysis unnecessary across four strains [50]. Another more recent study found that the dialysis step decreased activity for an MG1655 strain [37]. As with the runoff reaction, Silverman et al. found that dialysis is important for expression from native promoters [20], and other previous studies using native promoters all include dialysis steps [8,12,33,51,106]. Recent methods vary both in the details of dialysis protocols and inclusion [8,20,35,75] or omission of the step [10,13,25,61]. The dialysis itself is typically followed by an additional centrifugation that either matches the protocol used for clarification [8,20] or uses a shorter protocol of 4000×g for 20 min [9,11,35,124]. To increase throughput, Katsura et al. recently introduced the use of a TFF device for this step and demonstrated active extract [36]. As with the runoff reaction, detailed understanding of the effect of dialysis on extract composition and performance remains unclear.
2.4.4. Batch optimization and storage
Some protocols include additional steps aimed at adjustments to account for batch-to-batch variability between extract preparations. A common first measurement is a Bradford assay to determine the total the protein concentration in the extract [8,18,35,37]. This test serves as both a quality control check and allows dilution to a consistent concentration between batches. Other methods determine the optimal amount of certain reagent mix components for that batch of extract, including magnesium [8,12,18], potassium [8], and DTT [8].
The majority of methods complete the extract preparation by aliquoting, flash-freezing in liquid nitrogen, and storage at −80 °C. However, recent studies have explored lyophilization as a storage method, including optimizing lyophilization protocols [25,128], separate processing of extract and reagent mix [[129], [130], [131]], use of cryoprotectants [129,131,132], and even as a combination storage and lysis step [14]. Robust performance after storage at elevated temperatures of 37 °C for several months has been shown [129]. Lyophilized CFE has been leveraged for applications in sensing [14,[98], [99], [100], [101],[133], [134], [135], [136], [137], [138]], point-of-need manufacturing [99,134,138,139], and education [[26], [27], [28]].
3. Non-E. coli prokaryotes
While E. coli is the current standard for CFE, similar systems derived from other non-model bacterial species have been created due to their advantages in particular applications. For instance, the rapid growth rate and associated high protein production [140] of the halophilic marine-dwelling Vibrio natriegens has led to the creation of V. natriegens CFE extracts by several laboratories [40,43,44,141] in order to develop a platform for maximal protein output. A number of CFE systems have been generated for prototyping the function of genetic components in those organisms [[38], [39], [40]]. Streptomyces species have been explored for their applications in biofuel production and natural product synthesis, and it has been demonstrated that Streptomyces CFE has advantages over E. coli CFE for the expression and solubility of certain native Streptomyces proteins [41,46,142], possibly because Streptomyces genes have a high GC content that benefit from Streptomyces expression machinery. A recent review extensively covers the applications of alternative-chassis CFE [3]; we provide here a brief summary of non-E. coli prokaryotic CFE extracts and their applications in Table 3, noting that an emphasis is placed on the most recent methods available for each organism.
Table 3.
Summary of CFE systems from non-E. coli prokaryotes and their applications.
| Organism | Type | CFE Application | Proteins Made | Ref(s) |
|---|---|---|---|---|
| Bacillus megaterium | Gram positive | Prototyping RBSs, promoters | GFP, mCherry | [38] |
| Bacillus subtilis | Gram positive | Prototyping promoters | GFPmut3b, renilla luciferase | [39,40] |
| Clostridium autoethanogenum | Gram positive | Prototyping genetic parts | Luciferase, metabolic enzymes | [47] |
| Corynebacterium glutamicum | Gram positive | Prototyping promoters | eGFP | [40] |
| Escherichia fergusonii | Gram negative | Prototyping promoters | eGFP | [40] |
| Klebsiella oxytoca | Gram negative | Prototyping promoters | eGFP | [40] |
| Lactococcus lactis | Gram positive | Prototyping promoters | None (only transcription active) | [40] |
| Pantoea agglomerans | Gram negative | Prototyping promoters | eGFP | [40] |
| Pseudomonas fluorescens | Gram negative | Protein synthesis at low temperatures | GFP, apolipoprotein, pancreatic RNase, p37a, glucokinase, peptidases | [143] |
| Pseudomonas putida | Gram negative | Prototyping genetic parts for bioremediation | sfGFP | [40,45] |
| Salmonella enterica | Gram negative | Prototyping promoters | eGFP | [40] |
| Streptomyces species | Gram positive | Natural product biosynthesis | eGFP, sfGFP, metabolic proteins | [41,46,142,144,145] |
| Sulfolobus solfataricus | Archaeal thermophile | Elucidation of archaeal translation mechanisms | ORF 104, ORF 143 | [146,147] |
| Sulfolobus tokodaii | Archaeal thermophile | Translation of single stranded DNA | Polyphenylalanine | [148] |
| Thermococcus kodakaraensis | Archaeal thermophile | Production of thermostable proteins | Chitinase | [149] |
| Thermus thermophilus | Gram negative | Studying mechanisms of thermophilic protein synthesis | Polyphenylalanine | [148,150] |
| Vibrio natriegens | Gram negative | High protein yields | sfGFP, eGFP | [40,[42], [43], [44]] |
Despite the diversity of non-E. coli species that have been used to generate CFE systems, the general methodology used in recent years for preparing these cell extracts (Table 4) and supplementing the reactions have primarily followed gold standard methods from E. coli [8,13]. In general, the choice of culture medium and temperature during cell growth matched standard culturing procedures for that species. The culture medium chosen [43,44,141] and OD at cell harvest [44,45] sometimes had a substantial impact on the final yield of the CFE reaction, and optimization of these parameters contributed to more productive CFE systems in certain cases. Sonication was used most often as the method for cell lysis during the extract preparation process (Table 4), likely due in part to the ability to scale down the volume of cells required and to increase throughput when testing parameters in order to optimize a novel CFE platform. The duration, energy input, and other sonication parameters were optimized for each bacterial species examined.
Table 4.
Recent publications of extract preparation methods for non-E. coli prokaryotes and their reported GFP variantc yields from CFE reactions.
| Organism | Cell Growth Conditions (medium, temp., OD) | Lysis Method | Runoff/Dialysis | GFP (ng/μL) | Ref (s) |
|---|---|---|---|---|---|
| Bacillus megaterium | 2xYT, 37 °C, OD 2.0 | Sonication | Y/N | 134 | [38] |
| Bacillus subtilus | 2xYT-P, 30 °C, OD ~3 | Sonication | Y/Y | 21.6 | [39] |
| Bacillus subtilis | 2xYT-P, 30 °C, OD 1.2–1.6 | Sonication | Y/Y | 0.308 | [40] |
| Clostridium autoethanogenum | Gas with CO, 37 °C, ODb | Sonication | Y/Y | 236 | [47] |
| Corynebacterium glutamicum | BHI, 30 °C, OD 1.2–1.6 | Sonication | Y/Y | 3.32 | [40] |
| Escherichia fergusonii | 2xYT-P, 37 °C, OD 1.2–1.6 | Sonication | Y/Y | 51.7 | [40] |
| Klebsiella oxytoca | 2xYT-P, 30 °C, OD 1.2–1.6 | Sonication | Y/Y | 4.21 | [40] |
| Lactococcus lactis | MRS, 37 °C, OD 1.2–1.6 | Sonication | Y/Y | 0.149 | [40] |
| Pantoea agglomerans | 2xYT-P, 30 °C, OD 1.2–1.6 | Sonication | Y/Y | 172 | [40] |
| Pseudomonas putida | LB, 26 °C, OD 2.5 | Sonication | N/N | 198 | [45] |
| Pseudomonas putida | 2xYT-P, 30 °C, OD 1.2–1.6 | Sonication | Y/Y | 10.4 | [40] |
| Salmonella enterica | 2xYT-P, 37 °C, OD 1.2–1.6 | Sonication | Y/Y | 0.263 | [40] |
| Streptomyces coelicolorISP-5233 | YEME, 30 °C, mid-log | Sonication | N/N | 30a | [142] |
| Streptomyces coelicolorISP-5233 | YEME, 30 °C, mid-log | Pressure | N/N | 30a | [46] |
| Streptomyces coelicolorM1152 | YEME, 30 °C, mid-log | Pressure | N/N | 20a | [46] |
| Streptomyces lividans66 | YEME, 30 °C, mid-log | Pressure | N/N | 45a | [46] |
| Streptomyces lividans 66 | YEME, 30 °C, mid-log | Sonication | N/N | 84.7 | [142] |
| Streptomyces lividans B12275 | YEME, 30 °C, mid-log | Pressure | N/N | 55a | [46] |
| Streptomyces lividans B12275 | YEME, 30 °C, mid-log | Sonication | N/N | 117 | [142] |
| Streptomyces rimosusB2659 | YEME, 30 °C, mid-log | Sonication | N/N | 100a | [142] |
| Streptomyces roseosporus | YEME, 30 °C, mid-log | Sonication | N/N | 60a | [142] |
| Streptomyces species F4474 | YEME, 30 °C, mid-log | Sonication | N/N | 40a | [142] |
| Streptomyces venezuelaeDSM40230 | GYM, 28 °C, OD 4.0 | Sonication | Y/N | 35.1 | [41] |
| Streptomyces venezuelae ATCC15439 | YEME, 30 °C, mid-log | Sonication | N/N | 45a | [142] |
| Vibrio natriegens | BHIN, 37 °C, log | Pressure | Y/Y | 400 | [43] |
| Vibrio natriegens | BHI, 37 °C, OD 6.5–7.5 | Sonication | Y/N | 1600 | [44] |
| Vibrio natriegens | LB + v2, 30 °C, OD 1.0 | Sonication | N/N | 260 | [42] |
| Vibrio natriegens | BHI + v2, 37 °C, OD 1.2–1.6 | Sonication | Y/Y | 78.5 | [40] |
Value was approximated from a figure in the corresponding reference.
OD value at harvest not reported.
Reported variants included eGFP, sfGFP, GFPmut3b, and GFP.
For some species, a runoff reaction significantly increased the productivity of the CFE system [38,39,41]; in other cases, it had no impact [46,142] or a deleterious effect [44,141]. Runoff had a negative effect on the V. natriegens CFE system as incubation time increased, as reported by two laboratories that independently prepared and optimized the system. As such, much like the strain-dependence observed for E. coli, it seems likely that the impact of including runoff reactions and dialysis steps during extract preparation is dependent on the species in question and may need to be optimized on a case-by-case basis.
4. Discussion
We have provided here a review of methodological developments for prokaryotic CFE extracts over the past 20 years, broken down into E. coli strains used, pre-lysis processing, lysis, post-lysis processing, and non-E. coli strains used. In addition to offering consolidated information to new entrants to the CFE field, the review serves to highlight portions of the methodology that remain ripe for investigation.
Workhorse E. coli strains are unsurprisingly popular for CFE work, particularly as recent work has elucidated ‘best practices’ for their use in a variety of applications. This does not diminish contributions made in the optimization of strains specifically for CFE, and points to an area of opportunity to more systematically compare strains and genetic modifications that have been used, perhaps even consolidating advances into a single ‘super’ strain. Beyond efficient general expression, considerable options are available at the strain level for the production of proteins with disulfide bonds [59,63], proteins with noncanonical amino acids incorporated [65,72,80], and glycoproteins [89,90], which together enable more facile production of important biotechnology products like antibodies and vaccine antigens. Sensing and prototyping applications of CFE have also seen benefits from purpose-built strains.
Within pre-lysis processing, recent studies showing productive extracts from cells in stationary phase offer a major simplification of growth protocols [19], yet highlight gaps in the understanding of what makes a productive extract [35]. While multiple studies showed that a single wash step gave the same yield as the traditional three [11,19], curiously we did not find any evidence that even a single wash step increases yield over moving directly from the initial centrifugation to lysis. As a minor point, while centrifugation speeds and durations for cell pelleting have steadily dropped from 16,000×g for 30 min [9] to 5000×g for 10 min [18], no studies have looked at this variable directly, nor has a lower limit been identified.
For the lysis stage, the proliferation of the use of sonication is notable for its use of more affordable equipment and its ability to explore more preparation conditions by lysing smaller batches of cells [10,13]. The freeze-thaw method described by Didovyk et al. requires even less specialized equipment and is even simpler than sonication [14], yet to date has not been used in other publications. The adoption and optimization of this approach has significant potential to further streamline preparation of CFE extracts going forward. Clear paths forward include testing the approach in other strains and species or using alternatives to freeze-thawing to puncture the inner membrane for cell lysis.
The post-lysis processing stages remain a major area for further investigation to better understand when each step is helpful. In addition to improved methods, elucidating why the steps impact performance could make a significant contribution to fundamental understanding of CFE systems. As a minor point, while reduction of centrifugation speeds and durations for the clarification step have been shown not to impact performance, no lower limit has been observed. Finally, continued development of extract storage via lyophilization will drive the expansion of applications for CFE.
CFE systems derived from prokaryotes other than E. coli have been created to enable new capabilities, such as the ability to express proteins at lower temperatures or to prototype the function of genetic parts for in vivo use in organisms other than E. coli. The preparation of extracts for such systems generally follows procedures that have been widely adopted in recent years for E. coli extract preparation, though with the exception of the highly productive V. natriegens, yields do not match those attained with E. coli. Enhancements to these systems may include deviating more substantially from the standard E. coli extract preparation protocols at certain steps (such as harvest OD, lysis method, inclusion of runoff or other post-processing steps, etc.) or producing genetic modifications to the organism. These optimizations are likely to be species-dependent with careful considerations to the biological requirements of each organism.
A common attribute of the majority of the studies referenced here is the use of the production of GFP as the metric for the activity of an extract. While extremely useful as a standard across labs due to ease of measurement, the approach hides potentially large and complex underlying differences between methods [35], ignores the fraction of non-functional protein produced [151], and could poorly predict performance for some applications. The recent application of proteomics [112,[152], [153], [154]] and metabolomics [155] tools are important advances to better understand the underlying differences between extracts and CFE systems more generally.
It is striking that across the methodological improvements reviewed here, nearly all studies report equivalent yields using a simplified protocol rather than improved yields. This observation suggests that the productivity of E. coli CFE systems are limited by factors other than the extract preparation method. Likely candidates are the reagent mix used [16], or factors related to getting the most activity from the components present, such as the percent of active ribosomes [35]. As these other factors are engineered to increase yields, the variables discussed here may need to be revisited as activity becomes sensitive to choices made during extract preparation.
It is worth highlighting the progression of primary labs contributing improvements to CFE extract methods for E. coli and other prokaryotes. For about 15 years from the mid-1990's until 2010, advances were mostly made by groups led by Yokoyama, Kim, Swartz, or Kigawa. Then, between 2010 and 2015, new contributions were offered by the labs of Noireaux, Bundy, Jewett, and Sutro Biopharma, Inc. Since 2015, however, we identified contributions from 13 additional labs to the development of prokaryotic CFE methods. This rapid expansion mirrors the expansion of applications of CFE [[2], [3], [4]], and also underscores the impact of the methodological improvements made by the pioneer labs. Major advances such as equivalent yields for growth in flasks compared to fermenters [9,75,108], the use of affordable sonication for lysis [10,13], and reduction of processing steps [11,35,50], have all led to recent work describing a 24 h workflow from colony to CFE reaction that requires minimal capital investment [19]. This dramatic leap forward from traditional methods acclaims decades of work by the community. Given that several recent publications continue to explicitly cite lowering the barrier of entry for new labs as a primary motivation for their work [3,14,[18], [19], [20],22,25], we anticipate this trend of expansion for the field will continue. Ideally, all of these contributions will eventually yield to extremely-cheap, commercially-available CFE systems, making the tools available to an even wider range of labs and applications than is possible today.
CRediT authorship contribution statement
Stephanie D. Cole: Data curation, Writing - original draft. Aleksandr E. Miklos: Data curation, Writing - original draft. Abel C. Chiao: Data curation, Writing - original draft. Zachary Z. Sun: Data curation, Writing - original draft. Matthew W. Lux: Data curation, Writing - original draft.
Declaration of competing interest
ZZS and ACC have ownership in Synvitrobio, Inc. dba Tierra Biosciences, a company commercializing the applications of cell-free systems.
Acknowledgements
We thank Richard M. Murray for review of earlier versions of the manuscript and Casey B. Bernhards, Marilyn S. Lee, Caitlin E. Sharpes, Patricia E. Buckley, and Nathan D. Mcdonald for helpful edits and feedback. We acknowledge our funding sources: the US Office of the Secretary of Defense Applied Research for the Advancement of S&T Priorities program (SDC, AEM, MWL), DARPA SBIR to Synvitrobio, Inc. dba Tierra Biosciences (ACC, ZSS), contract No: W911NF-16-P-0003, and a Caltech Grubstake Grant (ACC, ZSS).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Nirenberg M.W., Matthaei J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci Unit States Am. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tinafar A., Jaenes K., Pardee K. Synthetic biology goes cell-free. BMC Biol. 2019;17:1–14. doi: 10.1186/s12915-019-0685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregorio N.E., Levine M.Z., Oza J.P. A user's guide to cell-free protein synthesis. Methods and Protocols. 2019;2:24. doi: 10.3390/mps2010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman A.D., Karim A.S., Jewett M.C. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21:151–170. doi: 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 6.Li J., Zhang C., Huang P., Kuru E., Forster-Benson E.T.C., Li T. Dissecting limiting factors of the protein synthesis using recombinant elements (PURE) system. Translation. 2017;5 doi: 10.1080/21690731.2017.1327006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavickova B., Maerkl S.J. A simple, robust, and low-cost method to produce the PURE cell-free system. ACS Synth Biol. 2019;8:455–462. doi: 10.1021/acssynbio.8b00427. [DOI] [PubMed] [Google Scholar]
- 8.Sun Z.Z., Hayes C.A., Shin J., Caschera F., Murray R.M., Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. JoVE. 2013:1–14. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kigawa T., Yabuki T., Matsuda N., Matsuda T., Nakajima R., Tanaka A. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genom. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 10.Shrestha P., Holland T.M., Bundy B.C. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques. 2012;53:163–174. doi: 10.2144/0000113924. [DOI] [PubMed] [Google Scholar]
- 11.Liu D.V., Zawada J.F., Swartz J.R. Streamlining Escherichia Coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol Prog. 2005;21:460–465. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 12.Caschera F., Noireaux V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription–translation system. Biochimie. 2014;99:162–168. doi: 10.1016/j.biochi.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 13.Kwon Y.C., Jewett M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci Rep. 2015;5:8663. doi: 10.1038/srep08663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didovyk A., Tonooka T., Tsimring L., Hasty J. Rapid and scalable preparation of bacterial lysates for cell-free gene expression. ACS Synth Biol. 2017;6:2198–2208. doi: 10.1021/acssynbio.7b00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara K., Doi N. Biochemical preparation of cell extract for cell-free protein synthesis without physical disruption. PloS One. 2016;11 doi: 10.1371/journal.pone.0154614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dopp B.J.L., Tamiev D.D., Reuel N.F. Cell-free supplement mixtures: elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol Adv. 2019;37:246–258. doi: 10.1016/j.biotechadv.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Dopp J.L., Rothstein S.M., Mansell T.J., Reuel N.F. Rapid prototyping of proteins: mail order gene fragments to assayable proteins within 24 hours. Biotechnol Bioeng. 2019;116:667–676. doi: 10.1002/bit.26912. [DOI] [PubMed] [Google Scholar]
- 18.Levine M.Z., Gregorio N.E., Jewett M.C., Watts K.R., Oza J.P. Escherichia coli-Based Cell-Free Protein Synthesis: protocols for a robust, flexible, and accessible platform technology. JoVE : JoVE. 2019:1–11. doi: 10.3791/58882. [DOI] [PubMed] [Google Scholar]
- 19.Levine M.Z., So B., Mullin A.C., Watts K.R., Oza J.P. Redesigned upstream processing enables a 24-hour workflow from E. coli cells to cell-free protein synthesis. 2019. [DOI]
- 20.Silverman A.D., Kelley-Loughnane N., Lucks J.B., Jewett M.C. Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry. ACS Synth Biol. 2019;8:403–414. doi: 10.1021/acssynbio.8b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole S.D., Beabout K., Turner K.B., Smith Z.K., Funk V.L., Harbaugh S.V. Quantification of interlaboratory cell-free protein synthesis variability. ACS Synth Biol. 2019;8:2080–2091. doi: 10.1021/acssynbio.9b00178. [DOI] [PubMed] [Google Scholar]
- 22.Dopp J.L., Jo Y.R., Reuel N.F. Methods to reduce variability in E. Coli-based cell-free protein expression experiments. Synthetic and Systems Biotechnology. 2019;4:204–211. doi: 10.1016/j.synbio.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi M.K., Hayes C.A., Chappell J., Sun Z.Z., Murray R.M., Noireaux V. Characterizing and prototyping genetic networks with cell-free transcription-translation reactions. Methods. 2015;86:60–72. doi: 10.1016/j.ymeth.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 24.Chizzolini F., Forlin M., Yeh Martin N., Berloffa G., Cecchi D., Mansy S.S. Cell-free translation is more variable than transcription. ACS Synth Biol. 2017;6:638–647. doi: 10.1021/acssynbio.6b00250. [DOI] [PubMed] [Google Scholar]
- 25.Dopp J.L., Reuel N.F. Process optimization for scalable E. coli extract preparation for cell-free protein synthesis. Biochem Eng J. 2018;138:21–28. doi: 10.1016/j.bej.2018.06.021. [DOI] [Google Scholar]
- 26.Huang A., Nguyen P.Q., Stark J.C., Takahashi M.K., Donghia N., Ferrante T. Biobits™ explorer: a modular synthetic biology education kit. Science Advances. 2018;4:1–11. doi: 10.1126/sciadv.aat5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark J.C., Huang A., Nguyen P.Q., Dubner R.S., Hsu K.J., Ferrante T.C. BioBits™ Bright: a fluorescent synthetic biology education kit. Science Advances. 2018;4:1–11. doi: 10.1126/sciadv.aat5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark J.C., Huang A., Hsu K.J., Dubner R.S., Forbrook J., Marshalla S. BioBits health: classroom Activities exploring engineering, biology, and human health with fluorescent readouts. ACS Synth Biol. 2019;8:1001–1009. doi: 10.1021/acssynbio.8b00381. [DOI] [PubMed] [Google Scholar]
- 29.Noireaux V., Liu A.P. The new age of cell-free biology. Annu Rev Biomed Eng. 2020;22:51–77. doi: 10.1146/annurev-bioeng-092019-111110. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.H., Kim D.M. Recent advances in development of cell-free protein synthesis systems for fast and efficient production of recombinant proteins. FEMS Microbiol Lett. 2018;365:1–7. doi: 10.1093/femsle/fny174. [DOI] [PubMed] [Google Scholar]
- 31.Lim H.J., Kim D.M. Cell-free metabolic engineering: recent developments and future prospects. Methods Protoc. 2019;2:33. doi: 10.3390/mps2020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bundy B.C., Hunt J.P., Jewett M.C., Swartz J.R., Wood D.W., Frey D.D. Cell-free biomanufacturing. Current Opinion in Chemical Engineering. 2018;22:177–183. doi: 10.1016/j.coche.2018.10.003. [DOI] [Google Scholar]
- 33.Shin J., Noireaux V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 34.Krinsky N., Kaduri M., Shainsky-Roitman J., Goldfeder M., Ivanir E., Benhar I. A simple and rapid method for preparing a cell-free bacterial lysate for protein synthesis. PloS One. 2016;11 doi: 10.1371/journal.pone.0165137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Failmezger J., Rauter M., Nitschel R., Kraml M., Siemann-Herzberg M. Cell-free protein synthesis from non-growing, stressed Escherichia coli. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-16767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsura K., Matsuda T., Tomabechi Y., Yonemochi M., Hanada K., Ohsawa N. A reproducible and scalable procedure for preparing bacterial extracts for cell-free protein synthesis. J Biochem. 2017;162:357–369. doi: 10.1093/jb/mvx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J., Copeland C.E., Padumane S.R., Kwon Y.C. A crude extract preparation and optimization from a genomically engineered Escherichia coli for the cell-free protein synthesis system: practical laboratory guideline. Methods Protoc. 2019;2:68. doi: 10.3390/mps2030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore S.J., MacDonald J.T., Wienecke S., Ishwarbhai A., Tsipa A., Aw R. Rapid acquisition and model-based analysis of cell-free transcription–translation reactions from nonmodel bacteria. Proc Natl Acad Sci Unit States Am. 2018;115:4340–4349. doi: 10.1073/pnas.1715806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelwick R., Webb A.J., MacDonald J.T., Freemont P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab Eng. 2016;38:370–381. doi: 10.1016/j.ymben.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Yim S.S., Johns N.I., Park J., Gomes A.L., McBee R.M., Richardson M. Multiplex transcriptional characterizations across diverse bacterial species using cell‐free systems. Mol Syst Biol. 2019;15:1–15. doi: 10.15252/msb.20198875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore S.J., Lai H.E., Needham H., Polizzi K.M., Freemont P.S. Streptomyces venezuelae TX-TL – a next generation cell-free synthetic biology tool. Biotechnol J. 2017;12:1–7. doi: 10.1002/biot.201600678. [DOI] [PubMed] [Google Scholar]
- 42.Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth Biol. 2018;7:2245–2255. doi: 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- 43.Failmezger J., Scholz S., Blombach B., Siemann-Herzberg M. Cell-free protein synthesis from fast-growing Vibrio natriegens. Front Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiegand D.J., Lee H.H., Ostrov N., Church G.M. Establishing a cell-free Vibrio natriegens expression system. ACS Synth Biol. 2018;7:2475–2479. doi: 10.1021/acssynbio.8b00222. [DOI] [PubMed] [Google Scholar]
- 45.Wang H., Li J., Jewett M.C. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synthetic Biology. 2018;3:1–7. doi: 10.1093/synbio/ysy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Wang H., Kwon Y.C., Jewett M.C. Establishing a high yielding streptomyces-based cell-free protein synthesis system. Biotechnol Bioeng. 2017;114:1343–1353. doi: 10.1002/bit.26253. [DOI] [PubMed] [Google Scholar]
- 47.Kruger A., Mueller A.P., Rybnicky G.A., Engle N.L., Yang Z.K., Tschaplinski T.J. Development of a clostridia-based cell-free system for prototyping genetic parts and metabolic pathways. Metab Eng. 2020 doi: 10.1016/j.ymben.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Nirenberg M.W. Methods enzymol. 1963. Cell-free protein synthesis directed by messenger RNA; pp. 17–23. [Google Scholar]
- 49.Ahn J.H., Chu H.S., Kim T.W., Oh I.S., Choi C.Y., Hahn G.H. Cell-free synthesis of recombinant proteins from PCR-amplified genes at a comparable productivity to that of plasmid-based reactions. Biochem Biophys Res Commun. 2005;338:1346–1352. doi: 10.1016/j.bbrc.2005.10.094. [DOI] [PubMed] [Google Scholar]
- 50.Kim T.W., Keum J.W., Oh I.S., Choi C.Y., Park C.G., Kim D.M. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol. 2006;126:554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 51.Shin J., Noireaux V. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J Biol Eng. 2010;4:9. doi: 10.1186/1754-1611-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niederholtmeyer H., Sun Z.Z., Hori Y., Yeung E., Verpoorte A., Murray R.M. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife. 2015;4 doi: 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gesteland R.F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966;16:67–84. doi: 10.1016/S0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- 55.Kim D.M., Kigawa T., Choi C.Y., Yokoyama S. A highly efficient cell-free, protein synthesis system from Escherichia coli. Eur J Biochem. 1996;239:881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 56.Michel-Reydellet N., Calhoun K., Swartz J. Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome. Metab Eng. 2004;6:197–203. doi: 10.1016/j.ymben.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Calhoun K.A., Swartz J.R. Total amino acid stabilization during cell-free protein synthesis reactions. J Biotechnol. 2006;123:193–203. doi: 10.1016/j.jbiotec.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 58.Airen I., Swartz J.R. United States Patent.US20130316397A1; 2013. Cell-free polypeptide synthesis. [Google Scholar]
- 59.Knapp K.G., Swartz J.R. Evidence for an additional disulfide reduction pathway in Escherichia coli. J Biosci Bioeng. 2007;103:373–376. doi: 10.1263/jbb.103.373. [DOI] [PubMed] [Google Scholar]
- 60.Yin G., Garces E.D., Yang J., Zhang J., Tran C., Steiner A.R. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs. 2012;4:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zawada J.F., Yin G., Steiner A.R., Yang J., Naresh A., Roy S.M. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michel-Reydellet N., Woodrow K., Swartz J. Increasing PCR fragment stability and protein yields in a cell-free system with genetically modified Escherichia coli extracts. J Mol Microbiol Biotechnol. 2005;9:26–34. doi: 10.1159/000088143. [DOI] [PubMed] [Google Scholar]
- 63.Kang S.H., Kim D.M., Kim H.J., Jun S.Y., Lee K.Y., Kim H.J. Cell-free production of aggregation-prone proteins in soluble and active forms. Biotechnol Prog. 2005;21:1412–1419. doi: 10.1021/bp050087y. [DOI] [PubMed] [Google Scholar]
- 64.Airen I.O. Stanford University; 2011. Genome-wide functional genomic analysis for physiological investigation and improvement of cell-free protein synthesis [dissertation] [Google Scholar]
- 65.Hong S.H., Kwon Y.-C., Martin R.W., Des Soye B.J., de Paz A.M., Swonger K.N. Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. Chembiochem. 2015;16:844–853. doi: 10.1002/cbic.201402708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin G., Swartz J.R. Enhancing multiple disulfide bonded protein folding in a cell-free system. Biotechnol Bioeng. 2004;86:188–195. doi: 10.1002/bit.10827. [DOI] [PubMed] [Google Scholar]
- 67.Woodrow K.A., Swartz J.R. A sequential expression system for high-throughput functional genomic analysis. Proteomics. 2007;7:3870–3879. doi: 10.1002/pmic.200700471. [DOI] [PubMed] [Google Scholar]
- 68.Sitaraman K., Esposito D., Klarmann G., Le Grice S.F., Hartley J.L., Chatterjee D.K. A novel cell-free protein synthesis system. J Biotechnol. 2004;110:257–263. doi: 10.1016/j.jbiotec.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 69.Sun Z.Z., Yeung E., Hayes C.A., Noireaux V., Murray R.M. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth Biol. 2014;3:387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 70.Woodrow K.A., Airen I.O., Swartz J.R. Rapid expression of functional genomic libraries. J Proteome Res. 2006;5:3288–3300. doi: 10.1021/pr050459y. [DOI] [PubMed] [Google Scholar]
- 71.Seki E., Matsuda N., Kigawa T. Multiple inhibitory factor removal from an Escherichia coli cell extract improves cell-free protein synthesis. J Biosci Bioeng. 2009;108:30–35. doi: 10.1016/j.jbiosc.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 72.Hong S.H., Kwon Y.C., Jewett M.C. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis. Frontiers in Chemistry. 2014;2:1–7. doi: 10.3389/fchem.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kigawa T., Yabuki T., Yoshida Y., Tsutsui M., Ito Y., Shibata T. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/S0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 74.Spirin A., Baranov V., Ryabova L., Ovodov S., Alakhov Y. A continuous cell-free translation system capable of producing polypeptides in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 75.Yang W.C., Patel K.G., Wong H.E., Swartz J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol Prog. 2012;28:413–420. doi: 10.1002/btpr.1509. [DOI] [PubMed] [Google Scholar]
- 76.Ryabova L.A., Desplancq D., Spirin A.S., Plückthun A. Functional antibody production using cell-free translation: effects of protein disulfide isomerase and chaperones. Nat Biotechnol. 1997;15:79–84. doi: 10.1038/nbt0197-79. [DOI] [PubMed] [Google Scholar]
- 77.Seki E., Matsuda N., Yokoyama S., Kigawa T. Cell-free protein synthesis system from Escherichia coli cells cultured at decreased temperatures improves productivity by decreasing DNA template degradation. Anal Biochem. 2008;377:156–161. doi: 10.1016/j.ab.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lajoie M.J., Rovner A.J., Goodman D.B., Aerni H.R., Haimovich A.D., Kuznetsov G. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin R.W., Des Soye B.J., Kwon Y.C., Kay J., Davis R.G., Thomas P.M. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh-Blom A., Hughes R.A., Ellington A.D. An amino acid depleted cell-free protein synthesis system for the incorporation of non-canonical amino acid analogs into proteins. J Biotechnol. 2014;178:12–22. doi: 10.1016/j.jbiotec.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 82.Lee K.-H., Catherine C., Kim D.-M. Enhanced production of unnatural amino acid-containing proteins in a cell-free protein synthesis system. J Ind Eng Chem. 2016;37:90–94. doi: 10.1016/j.jiec.2016.03.008. [DOI] [Google Scholar]
- 83.Lee J., Schwieter K.E., Watkins A.M., Kim D.S., Yu H., Schwarz K.J. Expanding the limits of the second genetic code with ribozymes. Nat Commun. 2019;10:5097. doi: 10.1038/s41467-019-12916-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swartz J. Developing cell-free biology for industrial applications. J Ind Microbiol Biotechnol. 2006;33:476–485. doi: 10.1007/s10295-006-0127-y. [DOI] [PubMed] [Google Scholar]
- 85.Groff D., Armstrong S., Rivers P.J., Zhang J., Yang J., Green E. Engineering toward a bacterial "endoplasmic reticulum" for the rapid expression of immunoglobulin proteins. mAbs. 2014;6:671–678. doi: 10.4161/mabs.28172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dopp J.L., Reuel N.F. Simple, functional, inexpensive cell extract for in vitro prototyping of proteins with disulfide bonds. BioRxiv. 2020 doi: 10.1101/2019.12.19.883413. [DOI] [Google Scholar]
- 87.Guarino C., DeLisa M.P. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology. 2012;22:596–601. doi: 10.1093/glycob/cwr151. [DOI] [PubMed] [Google Scholar]
- 88.Feldman M.F., Wacker M., Hernandez M., Hitchen P.G., Marolda C.L., Kowarik M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jaroentomeechai T., Stark J.C., Natarajan A., Glasscock C.J., Yates L.E., Hsu K.J. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat Commun. 2018;9:2686. doi: 10.1038/s41467-018-05110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stark J.C., Jaroentomeechai T., Moeller T.D., Dubner R.S., Hsu K.J., Stevenson T.C. On-demand, cell-free biomanufacturing of conjugate vaccines at the point-of-care. bioRxiv. 2019 doi: 10.1101/681841. [DOI] [Google Scholar]
- 91.Wilding K.M., Hunt J.P., Wilkerson J.W., Funk P.J., Swensen R.L., Carver W.C. Endotoxin-free E. Coli-based cell-free protein synthesis: pre-expression endotoxin removal approaches for on-demand cancer therapeutic production. Biotechnol J. 2019;14 doi: 10.1002/biot.201800271. [DOI] [PubMed] [Google Scholar]
- 92.Kightlinger W., Lin L., Rosztoczy M., Li W., DeLisa M.P., Mrksich M. Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases. Nat Chem Biol. 2018;14:627–635. doi: 10.1038/s41589-018-0051-2. [DOI] [PubMed] [Google Scholar]
- 93.Kightlinger W., Duncker K.E., Ramesh A., Thames A.H., Natarajan A., Stark J.C. A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat Commun. 2019;10:5404. doi: 10.1038/s41467-019-12024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun Z., Kim J., Singhal V., Murray R.M. Protein degradation in a TX-TL cell-free expression system using ClpXP protease. bioRxiv. 2015 doi: 10.1101/019695. [DOI] [Google Scholar]
- 95.Fischer S. Cell break: how cell-free biology is finally putting the engineering back in bioengineering. IEEE Pulse. 2016;7:13–16. doi: 10.1109/MPUL.2016.2514881. [DOI] [PubMed] [Google Scholar]
- 96.Schilling C. BASF Science Symposium; 2015. Accelerated development of biobased processes new developments in platform technology. [Google Scholar]
- 97.Marshall R., Maxwell C.S., Collins S.P., Jacobsen T., Luo M.L., Begemann M.B. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol Cell. 2018;69:146–157. doi: 10.1016/j.molcel.2017.12.007. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pardee K., Green A.A., Ferrante T., Cameron D.E., Daleykeyser A., Yin P. Paper-based synthetic gene networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 100.Meyer A., Saaem I., Silverman A., Varaljay V.A., Mickol R., Blum S. Organism engineering for the bioproduction of the triaminotrinitrobenzene (TATB) precursor phloroglucinol (PG) ACS Synth Biol. 2019;8:2746–2755. doi: 10.1021/acssynbio.9b00393. [DOI] [PubMed] [Google Scholar]
- 101.Silverman A.D., Akova U., Alam K.K., Jewett M.C., Lucks J.B. Design and optimization of a cell-free atrazine biosensor. ACS Synth Biol. 2020;9:671–677. doi: 10.1021/acssynbio.9b00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 103.Bosdriesz E., Molenaar D., Teusink B., Bruggeman F.J. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 2015;282:2029–2044. doi: 10.1111/febs.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Underwood K.A., Swartz J.R., Puglisi J.D. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 105.Niess A., Failmezger J., Kuschel M., Siemann-Herzberg M., Takors R. Experimentally validated model enables debottlenecking of in vitro protein synthesis and identifies a control shift under in vivo conditions. ACS Synth Biol. 2017;6:1913–1921. doi: 10.1021/acssynbio.7b00117. [DOI] [PubMed] [Google Scholar]
- 106.Garamella J., Marshall R., Rustad M., Noireaux V. The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth Biol. 2016;5:344–355. doi: 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- 107.Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- 108.Zawada J., Swartz J. Maintaining rapid growth in moderate-density Escherichia coli fermentations. Biotechnol Bioeng. 2005;89:407–415. doi: 10.1002/bit.20369. [DOI] [PubMed] [Google Scholar]
- 109.Yamane T., Ikeda Y., Nagasaka T., Nakano H. Enhanced cell-free protein synthesis using a S30 extract from Escherichia coli grown rapidly at 42°C in an amino acid enriched medium. Biotechnol Prog. 2005;21:608–613. doi: 10.1021/bp0400238. [DOI] [PubMed] [Google Scholar]
- 110.Spirin A.S., Swartz J.R. Wiley-VCH; Weinheim, Germany: 2008. Cell-free protein synthesis: methods and protocol. [Google Scholar]
- 111.Kim R.G., Choi C.Y. Expression-independent consumption of substrates in cell-free expression system from Escherichia coli. J Biotechnol. 2000;84:27–32. doi: 10.1016/S0168-1656(00)00326-6. [DOI] [PubMed] [Google Scholar]
- 112.Garcia D.C., Mohr B.P., Dovgan J.T., Hurst G.B., Standaert R.F., Doktycz M.J. Elucidating the potential of crude cell extracts for producing pyruvate from glucose. Synthetic Biology. 2018;3:1–9. doi: 10.1093/synbio/ysy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 114.Myers J.A., Curtis B.S., Curtis W.R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC Biophys. 2013;6:4. doi: 10.1186/2046-1682-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Piir K., Paier A., Liiv A., Tenson T., Ü Maiväli. Ribosome degradation in growing bacteria. EMBO Rep. 2011;12:458–462. doi: 10.1038/embor.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chisti Y., Moo-Young M. Disruption of microbial cells for intracellular products. Enzym Microb Technol. 1986;8:194–204. doi: 10.1016/0141-0229(86)90087-6. [DOI] [Google Scholar]
- 117.Jewett M.C., Calhoun K.A., Voloshin A., Wuu J.J., Swartz J.R. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller D.N., Bryant J.E., Madsen E.L., Ghiorse W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thompson J., Chassy B.M. Uptake and metabolism of sucrose by Streptococcus lactis. J Bacteriol. 1981;147:543–551. doi: 10.1128/jb.147.2.543-551.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mehta K.K., Evitt N.H., Swartz J.R. Chemical lysis of cyanobacteria. J Biol Eng. 2015;9:10. doi: 10.1186/s13036-015-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yeates C., Gillings M.R., Davison A.D., Altavilla N., Veal D.A. Methods for microbial DNA extraction from soil for PCR amplification. Biol Proced Online. 1998;1:40–47. doi: 10.1251/bpo6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ron E.Z., Kohler R.E., Davis B.D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 123.Johnson B.H., Hecht M.H. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Biotechnology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- 124.Pratt J.M. Transcription and translation: a practical approach. IRL Press; New York: 1984. Coupled transcription-translation in prokarytoic cell-free systems; pp. 179–209. [Google Scholar]
- 125.Pedersen A., Hellberg K., Enberg J., Karlsson B.G. Rational improvement of cell-free protein synthesis. N Biotech. 2011;28:218–224. doi: 10.1016/j.nbt.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 126.Jermutus L., Ryabova L.A., Plückthun A. Recent advances in producing and selecting functional proteins by using cell-free translation. Curr Opin Biotechnol. 1998;9:534–548. doi: 10.1016/s0958-1669(98)80042-6. [DOI] [PubMed] [Google Scholar]
- 127.Roy S.M., Bajad S., Evan G., Heinsohn H. United States Patent. US9040253B2; 2015. Method for enhancing recombinant protein production by cell-free protein expression. [Google Scholar]
- 128.Smith M.T., Berkheimer S.D., Werner C.J., Bundy B.C. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques. 2014;56:186–193. doi: 10.2144/000114158. [DOI] [PubMed] [Google Scholar]
- 129.Karig D.K. Cell-free synthetic biology for environmental sensing and remediation. Curr Opin Biotechnol. 2017;45:69–75. doi: 10.1016/j.copbio.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 130.Lee M.S., Raig R.M., Gupta M.K., Lux M.W. Lyophilized cell-free systems display tolerance to organic solvent exposure. ACS Synth Biol. 2020 doi: 10.1021/acssynbio.0c00267. [DOI] [PubMed] [Google Scholar]
- 131.Lee M.S., Hung C.S., Phillips D.A., Buck C.C., Gupta M.K., Lux M.W. Silk fibroin as an additive for cell-free protein synthesis. Synth Syst Biotechnol. 2020;5:145–154. doi: 10.1016/j.synbio.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gregorio N.E., Kao W.Y., Williams L.C., Hight C.M., Patel P., Watts K.R. Unlocking applications of cell-free biotechnology through enhanced shelf life and productivity of E. coli extracts. ACS Synth Biol. 2020 doi: 10.1021/acssynbio.9b00433. acssynbio.9b00433. [DOI] [PubMed] [Google Scholar]
- 133.Duyen T.T., Matsuura H., Ujiie K., Muraoka M., Harada K., Hirata K. Paper-based colorimetric biosensor for antibiotics inhibiting bacterial protein synthesis. J Biosci Bioeng. 2017;123:96–100. doi: 10.1016/j.jbiosc.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 134.Pardee K. Perspective: solidifying the impact of cell-free synthetic biology through lyophilization. Biochem Eng J. 2018;138:91–97. doi: 10.1016/j.bej.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salehi A.S.M., Tang Mj Shakalli, Smith M.T., Hunt J.M., Law R.A., Wood D.W. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal Chem. 2017;89:3395–3401. doi: 10.1021/acs.analchem.6b04034. [DOI] [PubMed] [Google Scholar]
- 136.McNerney M.P., Zhang Y., Steppe P., Silverman A.D., Jewett M.C., Styczynski M.P. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci Adv. 2019;5 doi: 10.1126/sciadv.aax4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grawe A., Dreyer A., Vornholt T., Barteczko U., Buchholz L., Drews G. A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PloS One. 2019;14 doi: 10.1371/journal.pone.0210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hunt J.P., Yang S.O., Wilding K.M., Bundy B.C. The growing impact of lyophilized cell-free protein expression systems. Bioengineered. 2017;8:325–330. doi: 10.1080/21655979.2016.1241925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Salehi A.S., Smith M.T., Bennett A.M., Williams J.B., Pitt W.G., Bundy B.C. Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system. Biotechnol J. 2016;11:274–281. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- 140.Aiyar S.E., Gaal T., Gourse R.L. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J Bacteriol. 2002;184:1349–1358. doi: 10.1128/jb.184.5.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth Biol. 2018;7:2245–2255. doi: 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- 142.Li J., Wang H., Jewett M.C. Expanding the palette of Streptomyces -based cell-free protein synthesis systems with enhanced yields. Biochem Eng J. 2018;130:29–33. doi: 10.1016/j.bej.2017.11.013. [DOI] [Google Scholar]
- 143.Nakashima N., Tamura T. Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter. Biochem Biophys Res Commun. 2004;319:671–676. doi: 10.1016/j.bbrc.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 144.Jones G.H. Macromolecular synthesis in Streptomyces antibioticus: in vitro systems for aminoacylation and translation from young and old cells. J Bacteriol. 1975;124:364–372. doi: 10.1128/jb.124.1.364-372.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Thompson J., Rae S., Cundliffe E. Coupled transcription — translation in extracts of Streptomyces lividans. Mol Gen Genet MGG. 1984;195:39–43. doi: 10.1007/BF00332721. [DOI] [PubMed] [Google Scholar]
- 146.Ruggero D., Creti R., Londei P. In vitro translation of archaeal natural mRNAs at high temperature. FEMS Microbiol Lett. 1993;107:89–94. doi: 10.1111/j.1574-6968.1993.tb06009.x. [DOI] [Google Scholar]
- 147.Condo I., Ciammaruconi A., Benelli D., Ruggero D., Londei P. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 1999;34:377–384. doi: 10.1046/j.1365-2958.1999.01615.x. [DOI] [PubMed] [Google Scholar]
- 148.Uzawa T., Yamagishi A., Oshima T. Polypeptide synthesis directed by DNA as a messenger in cell-free polypeptide synthesis by extreme thermophiles, thermus thermophilus HB27 and sulfolobus tokodaii strain 7. J Biochem. 2002;131:849–853. doi: 10.1093/oxfordjournals.jbchem.a003174. [DOI] [PubMed] [Google Scholar]
- 149.Endoh T., Kanai T., Sato Y.T., Liu D.V., Yoshikawa K., Atomi H. Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile. J Biotechnol. 2006;126:186–195. doi: 10.1016/j.jbiotec.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 150.Uzawa T., Hamasaki N., Oshima T. Effects of novel polyamines on cell-free polypeptide synthesis catalyzed by Thermus thermophilus HB8 extract. J Biochem. 1993;114:478–486. doi: 10.1093/oxfordjournals.jbchem.a124203. [DOI] [PubMed] [Google Scholar]
- 151.Iskakova M.B., Szaflarski W., Dreyfus M., Remme J., Nierhaus K.H. Troubleshooting coupled in vitro transcription-translation system derived from Escherichia coli cells: synthesis of high-yield fully active proteins. Nucleic Acids Res. 2006;34:e135. doi: 10.1093/nar/gkl462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hurst G.B., Asano K.G., Doktycz C.J., Consoli E.J., Doktycz W.L., Foster C.M. Proteomics-based tools for evaluation of cell-free protein synthesis. Anal Chem. 2017;89:11443–11451. doi: 10.1021/acs.analchem.7b02555. [DOI] [PubMed] [Google Scholar]
- 153.Foshag D., Henrich E., Hiller E., Schäfer M., Kerger C., Burger-Kentischer A. The E. coli S30 lysate proteome: a prototype for cell-free protein production. N Biotech. 2018;40:245–260. doi: 10.1016/j.nbt.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 154.Garenne D., Beisel C.L., Noireaux V. Characterization of the all-E. coli transcription-translation system myTXTL by mass spectrometry. Rapid Commun Mass Spectrom. 2019;33:1036–1048. doi: 10.1002/rcm.8438. [DOI] [PubMed] [Google Scholar]
- 155.Miguez A.M., McNerney M.P., Styczynski M.P. Metabolic profiling of Escherichia coli-based cell-free expression systems for process optimization. Ind Eng Chem Res. 2019;58:22472–22482. doi: 10.1021/acs.iecr.9b03565. [DOI] [PMC free article] [PubMed] [Google Scholar]