Abstract
The development of Pseudomonas strains for industrial production of fuels and chemicals will require the integration of heterologous genes and pathways into the chromosome. Finding the most appropriate integration site to maximize strain performance is an essential part of the strain design process. We characterized seven chromosomal loci in Pseudomonas putida KT2440 for integration of a fluorescent protein expression construct. Insertion in five of the loci did not affect growth rate, but fluorescence varied by up to 27-fold. Three sites displaying a diversity of phenotypes with the fluorescent reporter were also chosen for the integration of a gene encoding a muconate importer. Depending on the integration locus, expression of the importer varied by approximately 3-fold and produced significant phenotypic differences. This work demonstrates the impact of the integration location on host viability, gene expression, and overall strain performance.
Highlights
-
•
Pseudomonas putida KT2440 chromosomal loci were characterized as potential insertion targets for heterologous genes.
-
•
Integration location had a significant effect on heterologous protein expression and host phenotype.
-
•
The identification of an appropriate chromosomal insertion location is essential to optimize genetic engineering design.
1. Introduction
Pseudomonas putida has become a desirable target organism for the production of industrial products due to several attributes of its native physiology (Ankenbauer et al., 2020). P. putida has a naturally diverse catabolism, with the capacity to metabolize aliphatic, aromatic and heterocyclic compounds in addition to glucose (Schmid et al., 2001), lending to the potential to metabolize carbon from complex feedstocks for commercial chemical production. An unusual cyclic pathway organization for catabolism of hexose sugars provides P. putida with a highly flexible metabolism that can adapt to the demands of diverse metabolic pathways (Nikel et al., 2015). Multiple degradation pathways have been identified and characterized in P. putida including the homo-proto-catechuate (Ornston and Stanier, 1966), phenylacetyl-CoA (Olivera et al., 1998), gentisate (Bayly and Barbour, 1984), homogentisate (Arias-Barrau et al., 2004), and beta-ketoadipate pathways (Nichols and Harwood, 1995; Poblete-Castro et al., 2012). Additionally, select strains of pseudomonads have been found to house plasmids carrying genes enabling the degradation of chemicals that can be toxic to many other organisms such as naphthalene (Dunn and Gunsalus, 1973), 4-chloronitrobenzene (Zhen et al., 2006), phenol (Kivisaar et al., 1990), toluene (Zylstra and Gibson, 1989), and 2,4-xylenol (Chen et al., 2014). Tolerance to inhibitory chemicals further extends the capacity of the organism to survive in industrial conditions.
Genetic modification of pseudomonads for industrial bioprocessing requires the ability to stably express heterologous genes and pathways. Furthermore, it is important that heterologous constructs be integrated into the chromosome (Tyo et al., 2009), as plasmid maintenance requires the maintenance of a plasmid-specific selection pressure such as the use of antibiotics. Generating the correct expression stoichiometry of the heterologous genes is equally important and can be challenging. Design parameters used to control protein expression are typically gene copy number (Morlino et al., 1999), promoter (Samuelson, 2011), and terminator type (Curran et al., 2015). However, the chromosomal integration location can affect heterologous protein expression independently of these parameters. The three dimensional structure of the chromosome (Krogh et al., 2018), the order, and the orientation of the genes play a role in determining the expression of each gene (Slager and Veening, 2016). The physical bending and condensing of the chromosome can bridge delocalized genes together (Le et al., 2013), possibly contributing to the rate and order of transcription, and thus, the expression of each gene. DNA supercoiling in response to environmental factors has been shown to affect protein expression (Hulton et al., 1990), and to play a major role in the evolutionary conservation of gene order in bacteria (Sobetzko, 2016). Integrating a heterologous segment of DNA can potentially disrupt this sequence, order and topology, disabling the ability of the host to grow by changing essential gene expression patterns. Transcription into and out of the heterologous segment may also occur, altering native protein expression. Additionally, appropriate expression of the heterologous DNA could be altered due to interference by regional protein and DNA interactions. Introduction of heterologous DNA into the chromosome would benefit from the characterization of insertion loci in which the expression of the heterologous DNA can be optimized without disrupting the natural sequence and topology of the chromosome that is required for normal cell functioning.
The characterization of chromosomal integration loci for this purpose has been explored to various degrees in several organisms such as Escherichia coli (Sousa et al. 1997; Bassalo et al., 2016), Bacillus subtilis (Sauer et al., 2016), Saccharomyces cerevisiae (Flagfeldt et al., 2009; Yamane et al., 1998), Lactococcus lactis (Thompson and Gasson, 2001), P. putida KT2440 (Domröse et al., 2019), and Drosophila melanogaster (Markstein et al., 2008). In this study, we have rationally selected seven insertion loci on the P. putida KT2440 chromosome to test for the expression of a fluorescent reporter gene and the effect of the insertion of the reporter on the growth of the host. A transmembrane transporter protein for the polymer precursor cis,cis-muconate was integrated into these three sites to further verify the effect of the insertion site on protein expression and organismal phenotype.
2. Methods and materials
2.1. Integration site selection
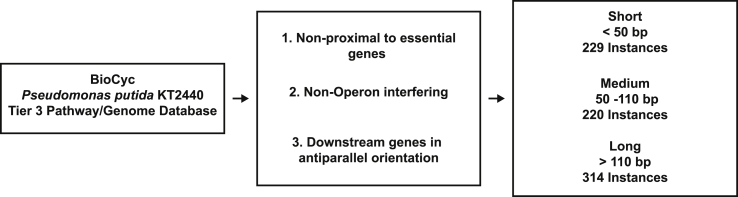
Insertion regions for heterologous gene insertion were manually curated from the P. putida KT2440 genome that were not adjacent to essential genes, were not between genes within the same operon, and were in anti-parallel orientation from the downstream gene. These sites were identified using the BioCyc P. putida KT2440 Tier 3 Pathway/Genome Database (PGDB) (Karp et al., 2019). The selected insertion regions were then categorized based on length, short (≤50 bp), medium (>50 bp and <110 bp), or long (≥110 bp), and then finally down-selected to a list of 7 insertion regions to encompass a range of all three lengths, representing subsections of the full genome. Integration location sites were named based on the upstream gene identifier.
2.2. Plasmid design and assembly
Plasmids were designed for integration into Pseudomonas putida KT2440 by homologous recombination, inserting the heterologous DNA without removing any endogenous genome sequence. The mKate fluorescent reporter gene and the flanking 400 nucleotide base pair sequences upstream and downstream of each integration site was synthesized by Integrated DNA Technologies (Integrated DNA Technologies (IDT), Coralville, IA) and inserted into the pK18mob-sacB suicide vector carrying a kanamycin resistant cassette and sacB gene which confers sensitivity to sucrose (Elmore et al., 2017; Johnson and Beckham, 2015; Marx, 2008) using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs (NEB), Ipswitch, MA). Plasmid constructs were transformed into NEB 5-alpha F’Iq (NEB) Eschericia coli competent cells by heat shock, plated onto LB agar plates supplemented with 50 μg/mL kanamycin sulfate, and incubated at 37 °C for 16–24 h, or until distinct single colony formation was visible. PCR was conducted on single colonies to identify the presence of successfully assembled plasmid transformants using primers synthesized by IDT and the Quick-Load Taq 2X Master Mix (NEB) system. Positive colonies were transferred into LB liquid medium supplemented with 50 μg/mL kanamycin sulfate and incubated at 37 °C, shaking at 200 rpm, for 16 h. Plasmids were purified from the cell culture using the Qiagen miniprep kit, quantified, and Sanger sequenced by Eurofins Genomics. Plasmid sequences are provided in Supplementary Materials.
The allelic exchange vector pK18sB Genbank accession number MH166772 (Jayakody et al., 2018) was used for integration of mucK at the 5042 site. Briefly, recombination sites upstream and downstream of the 5042 insertion site were amplified by PCR from P. putida KT2440 genomic DNA, and a Pc promoter-mNeonGreen-T4Terminator fragment was amplified from a derivative of the plasmid pSW002-Pc-mNeonGreen (Wilton et al., 2017). The three PCR fragments were cloned into EcoRI and HindIII linearized pK18sB using the GeneArt Seamless Cloning and Assembly Enzyme Mix (Thermo Fisher Scientific, Waltham, MA), creating the vector pK18sB-5042::Pc-mNeonGreen-T4T. The fluorescent protein mNeonGreen was then replaced by MucKADP1 using restriction-ligation cloning. Briefly, MucKADP1 was codon and expression optimized (Blue Heron Biotechnology Codon Optimizer tool) and synthesized as a gBlock (IDT). The gBlock was amplified by PCR, digested with XbaI and XhoI, and cloned into pK18sB-5042::Pc-mNeonGreen-T4T digested with the same enzymes. This created the vector pK18sB-5042::Pc-MucKADP1-T4T. Plasmids for integration of mucK at the 1642 and 2224 sites were constructed as follows: the Pc-MucKADP1 segment of vector pK18sB-5042::Pc-MucKADP1-T4T was amplified by PCR and cloned into PCR-linearized vectors pK18mobsacB_1642 and pK18mobsacB_2224 using GeneArt Seamless Cloning.
2.3. Pseudomonas putida transformation
P. putida KT2440 was used as the wild-type strain. Wild type liquid cultures were grown from plated single colonies in 25 mL of LB medium overnight at 30 °C, shaking at 200 rpm, to saturation. The following day, the cultures were harvested by centrifugation at 10,000 g, washed three times in 15 mL of 10% glycerol, and resuspended in a 50-fold smaller volume of 10% glycerol to generate electrocompetent cells for subsequent transformations.
Transformation was implemented by electroporation in the presence of 500–1000 ng plasmid DNA (0.1 cm cuvette, 1.6 kV, 25 μF, 200 Ω) as previously described (Elmore et al., 2017). Cells were then incubated in 950 μL SOC medium at 30 °C for 2 h, followed by plating on LB agar supplemented with 50 μg/mL kanamycin sulfate at 30 °C for 16 h. Resultant single colonies were re-streaked to single colonies on fresh LB agar +50 μg/mL kanamycin sulfate plates at 30 °C for 16 h to remove any enduring wild-type cells. Resultant colonies were streaked to single colony on YT-25% sucrose agar (10 g/L yeast extract, 20 g l/L tryptone, 250 g/L sucrose, 18 g/L agar) for counter-selection against the plasmid backbone, incubating at 30 °C for ~30–48 h. Surviving colonies were then re-streaked onto fresh YT-25% sucrose plates and incubated at 30 °C for 16 h to remove any enduring sacB-containing cells. All final colonies were screened for complete mKate or MucK integration and plasmid backbone excision by colony PCR, and whole genome resequencing. Three lines of each resistant transformant strain were subsequently re-streaked onto LB Plates, followed by long term archiving in 20% glycerol/LB at −80 °C.
2.4. Genome re-sequencing
Genomic DNA was purified from saturated liquid cultures using Qiagen’s DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Nextera XT libraries (Illumina, San Diego, CA) were produced according to the manufacturer’s protocol (15031942 v03). Final library validation was conducted on an Agilent Bioanalyzer (Agilent, Santa Clara, CA) using a DNA7500 chip and the concentration was determined using an Invitrogen Qubit (Waltham, MA) with the broad range double stranded DNA assay. Barcoded libraries were pooled and prepared for sequencing following the manufacturer’s recommended protocol (15039740v09, Standard Normalization). One paired end sequencing run (2 × 301) was completed on an Illumina MiSeq instrument (Illumina, San Diego, CA) using v3 chemistry.
2.5. Growth and fluorescence assays
Three lines of each transformant strain were inoculated from single colonies into 5 mL of LB and grown overnight at 30 °C, shaking at 200 rpm, for 16 h. Liquid cultures were then centrifuged and washed in the target assay growth medium (M9 + 30 mM glucose). Each well in a black walled, μClear flat-bottom, 96-well plate (Greiner Bio-One, Kremsmunster, Austria) was inoculated at 1%, into a final volume of 100 μL of the assay growth medium. Three replicates of each line, for each strain were measured simultaneously. Each plate was incubated at 30 °C, with continuous shaking, in a Synergy Mx plate reader (Bioteck, Winooski, VT), measuring OD700 nm and F588, 635 nm every 10 min for 48 h.
Growth rates were calculated using the LINEST function (fit to 10 measurements) in Microsoft Excel (2016) to determine the slope of the exponential growth phase, at mid-log. Fluorescence readings were normalized to the optical density and reported at mid-log of growth.
2.6. Live cell fluorescence microscopy
The P. putida KT2440 wild type, and P. putida KT2440 strains with mKate inserted at PP_1642, PP_2224, and PP_5322 were selected from single colonies and grown to saturation in LB overnight at 30 °C, shaking at 200 rpm. Saturated cultures were diluted 1/100 into fresh M9 minimal medium supplemented with 30 mM glucose and allowed to grow at 30 °C with continuous shaking until early log phase was reached (OD 0.1–0.2). Cells were imaged using a Zeiss Axio Imager microscope (Carl Zeiss, Gottingen, Germany) with a Canon EOS RebelT1i (Canon, Tokyo, Japan) attached. Images were captured at 240X magnification under bright field light conditions, and with a fluorescent filter. Samples were exposed for approximately 2.5 s.
2.7. Analysis of strain growth on cis,cis-muconate and MucK expression levels
Cis,cis-muconic acid was purchased from ACROS Organics (AC297760100). Stock solutions with a maximum concentration of about 800 mM were prepared by dissolving the powdered reagent in water containing 2 molar equivalents of NaOH. The pH was adjusted to 7.0 with NaOH, as needed, to maintain solubility and prevent isomerization. After filter sterilization, final concentrations were determined by absorbance at 260 nm using an extinction coefficient of 16,900 M−1cm−1 (Sistrom and Stanier, 1954).
Overnight cultures of wild type P. putida KT2440 and strains carrying the mucK insertion were inoculated into 10 mL LB medium to give a starting culture density of 0.2 OD600 nm and were grown with shaking at 30 °C until the culture density reached 1.0 OD600 nm. Cells were harvested by centrifugation and were washed twice in M9 salts before resuspension in the same buffer. For growth curve measurements, the washed cells were resuspended in M9 medium at a final OD600 nm of 0.1. 575 μL aliquots were dispensed into a 48-well, lidded microplate and 25 μL of glucose or cis,cis-muconate stock solution was added to give a final concentration of 30 mM carbon source. The plate was incubated at 30 °C in an Epoch 2 microplate reader (BioTek) with continuous double orbital shaking at 548 cpm. OD600 nm measurements were taken every 15 min. Each growth curve represents the average of four replicates. For proteomics experiments, the washed cells were used to inoculate 50 mL M9 medium containing 30 mM glucose (for wild-type KT2440) or 30 mM cis,cis-muconate (for strains containing mucK insertions) to a starting culture density of 0.1 OD600 nm and grown until OD600 nm reached 0.7. Cells were harvested by centrifugation and were washed one time with ice cold PBS. The washed cell pellets were weighed and then flash frozen in liquid nitrogen. All proteomics samples were prepared in triplicate.
2.8. Targeted proteomics
Sample protein extraction was based on a previously established protocol (Nakayasu et al., 2016). Each cell pellet was suspended to 200 μL using H2O and transferred to a solvent resistant tube (Sorenson). To the cell suspension, 800 μL of cold (−20 °C) chloroform:methanol mix (prepared 2:1 (v/v)) was added and vigorously vortexed for 30 s. The sample was then placed on ice for 5 min and then vortexed again for 30 s followed by centrifugation at 15,000 × g for 5 min at 4 °C. The upper, water-soluble metabolite phase and the lower, lipid soluble phase were removed. The remaining protein interlayer had 1 mL of cold 100% methanol added, vortexed and centrifuged again at 15,000×g for 5 min at 4 °C to pellet the protein. The methanol was then decanted off and the samples were placed open in a fume hood to dry for ~10 min.
A solution of 8 M urea in 100 mM NH4HCO3 was added to the protein pellets and vortexed into solution. A bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL) was performed to determine protein concentration. Following the assay, dithiothreitol (DTT) was added to the samples to the final concentration of 5 mM and incubated at 60 °C for 30 min with constant shaking at 850 rpm. The samples were alkylated by the addition of iodoacetamide (IAA) to a final concentration of 40 mM and incubated at 37 °C for 1 h with 850 rpm shaking in the dark. Samples were then diluted 10-fold for digestion preparation with 100 mM NH4HCO3 and 1 mM CaCl2 (final concentration). USB trypsin was added to all protein samples at a 1:50 (w/w) trypsin-to-protein ratio and incubated for 3 h at 37 °C. Digested samples were desalted using 1 mL Discovery C18 SPE columns (Supelco, Bellefonte, PA) using the following protocol: 3 mL of methanol was added for conditioning the column followed by 2 mL of 0.1% trifluoroacetic acid (TFA) in H2O. The samples were then loaded onto each column followed by 4 mL of 95:5 water:acetonitrile, 0.1% TFA. Samples were eluted with 1 mL 80:20 acetonitrile:water, 0.1% TFA. The samples were concentrated down to ~100 μL using a Speed Vac, and a final BCA was performed to determine the peptide concentration. Samples were diluted to 0.20 μg/μL with nanopure water for targeted proteomics analysis.
Targeted Proteomics was performed via a Liquid Chromatography (LC) - Selected Reaction Monitoring (SRM) approach. Three fully-tryptic peptides (GTAIGGAYNVGR, LIFQDK, ILAGFMADK) of MucK protein were initially selected based on their SRM suitability scores predicated by CONSeQuence (Eyers et al., 2011) and Prego (Searle et al., 2015) software tools. All the peptides were further compared on BLAST to ensure their uniqueness to target proteins in the organism. Crude (minimum purity) synthetic heavy isotope-labeled (e.g., 13C/15N on C-terminal lysine and arginine) peptides were purchased from New England Peptide (Gardner, MA). Upon receiving, the crude synthetic heavy peptides were mixed together and diluted with 0.1% formic acid in 15% acetonitrile in water to obtain a nominal concentration of 2 pmol/μL for each individual peptide. The heavy peptide mixture stock solution was aliquoted and stored at −80 °C until further use.
Analysis of the targeted proteomics assay was conducted via LC-SRM. Each SRM precursor-fragment ion pair (i.e., transitions) was first analyzed with LC-SRM by spiking heavy peptides into test samples. Even though all the three endogenous peptides can be detected in strains carrying the mucK insertion, two peptides (GTAIGGAYNVGR and LIFQDK) had the best inter-peptide correlation and were selected for final interpretation of protein abundance. Three transitions per peptide were selected in a final assay based on their MS response and transition interferences. Collision energies of transitions were obtained using empirical equations provided in the Skyline software package (MacLean et al., 2010).
To facilitate protein quantification, crude heavy peptide mixture stock solution was spiked in the 0.20 μg/μL peptide samples at a nominal concentration of 50 fmol/μL for each peptide. LC-SRM analysis utilized a nanoACQUITY UPLC® system (Waters Corporation, Milford, MA) coupled online to a TSQ Altis™ triple quadrupole mass spectrometer (Thermo Fisher Scientific). The UPLC® system was equipped with an ACQUITY UPLC BEH 1.7 μm C18 column (100 μm i.d. × 10 cm) and the mobile phases were (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. 2 μL of sample (i.e., 0.4 μg of peptides) were loaded onto the column and separated using a 110-min gradient profile as follows (min:flow-rate-μL/min:%B): 0:0.4:1, 6:0.6:1, 7:0.4:1, 9:0.4:6, 40:0.4:13, 70:0.4:22, 80:0.4:40, 85:0.4:95, 91:0.5:95, 92:0.5:95, 93:0.5:50, 94:0.5:95, 95:0.6:1, 98:0.4:1. The LC column was operated at a temperature of 42 °C. The TSQ Altis™ triple quadrupole mass spectrometer was operated with ion spray voltages of 2100 ± 100 V and a capillary inlet temperature of 350 °C. Tube lens voltages were obtained from automatic tuning and calibration without further optimization. Both Q1 and Q3 were set at unit resolution of 0.7 FWHM and Q2 gas pressure was optimized at 1.5 mTorr. The transitions were scanned with a dwell time of 0.625 msec
LC-SRM data were imported into Skyline and the peak boundaries were manually inspected to ensure correct peak assignment and peak boundaries. Peak detection and integration were determined based on two criteria: 1) the same LC retention time and 2) approximately the same relative peak intensity ratios across multiple transitions between the light peptides and heavy peptide standards. The total peak area ratios of endogenous light peptides and their corresponding heavy isotope-labeled internal standards were then exported from Skyline as Ratio-to-Standard. For each peptide, the total peak area ratios of individual samples were normalized to the average total peak area ratio of all the samples. For each sample, protein abundance was calculated as an average of the normalized total peak area ratios of all three peptides of a protein.
3. Results and discussion
3.1. Integration site selection and construction
The P. putida KT2440 chromosome was computationally screened for potential heterologous DNA integration loci that were not adjacent to essential genes, did not integrate within an operon, and were in an antiparallel orientation relative to the downstream gene [Fig. 1].
Fig. 1.
Process for computationally selecting integration loci.
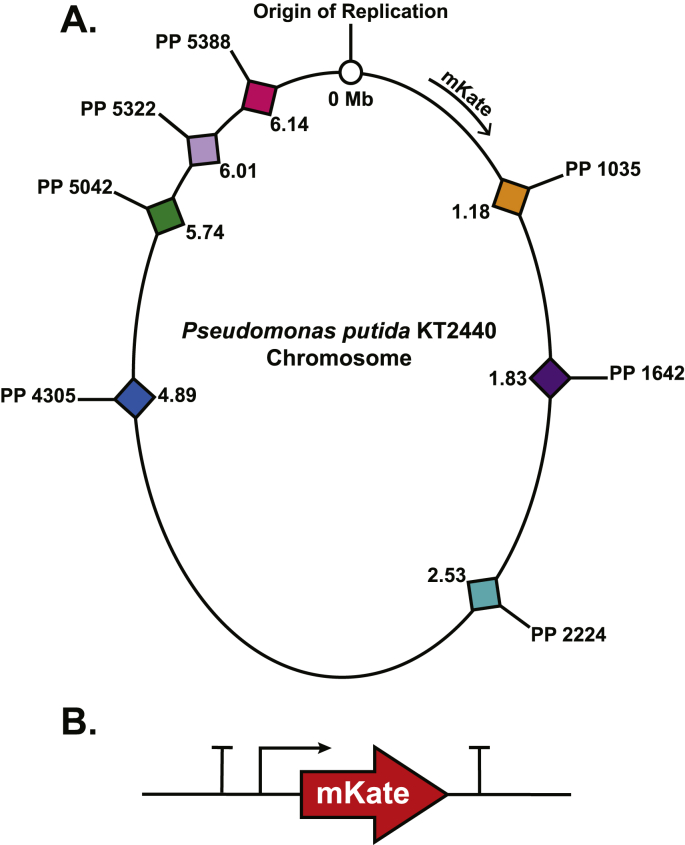
From the generated list of possibilities, we down-selected to seven sites for the practical and manageable generation of single reporter strains by homologous recombination [Fig. 2A]. These sites represented different regions of the chromosome and varied in total intergenic length (12–211 bp). The annotated functions of the genes upstream and downstream of each loci are provided in Supplemental Table 1. We designed and synthesized an mKate reporter construct driven by the E. coli Ptac promoter, which was flanked by terminators on both the 5′ and 3′ ends to insulate the construct from outside transcription readthrough, and to prevent readthrough from the mKate gene into the downstream gene [Fig. 2B]. The mKate fluorescent reporter construct was introduced into the seven different insertion locations on the P. putida KT2440 chromosome by homologous recombination, always in the same orientation (clockwise, as drawn in Fig. 2A).
Fig. 2.
[A] Map of P. putida KT2440 chromosome indicating the Origin of Replication, and the seven genes directly upstream of the integration loci selected for reporter gene insertion. The mKate construct was integrated in the same clockwise orientation at each locus. [B] Map of the mKate reporter construct under the control of the E. coli Ptac promoter and flanked outside the promoter and the 3′ end by terminators.
Whole genome resequencing of the engineered strains showed that each mKate construct was integrated into the targeted genome region. Several minor differences were noted relative to the original integration design. For one locus, PP_5388, the mKate construct integrated into the gene directly downstream of the target site, PP_5734, which encodes a hypothetical protein of unknown function. For two loci, PP_1035 and PP_1642, there were short base pair deletions from the intergenic region, 40 and 28 bp respectively. Finally, a point mutation occurred in the 5’ terminator of the PP_1035 strain.
3.2. Effects of integration location on growth and expression
Growth and fluorescence of the seven mKate expression strains were measured in M9 minimal medium supplemented with 30 mM glucose. The different integration loci had significant effects on both growth rate and mKate expression. At sites PP_1035, PP_4305, PP_5042, PP_5322, and PP_5388, insertion of mKate did not have a negative effect on growth, and all strains grew similarly to the wild type. The strains with mKate integration at PP_1642 or PP_2224 grew significantly slower, at 30% of the rate of the wild type [Fig. 3]. The specific mKate expression, as measured by Fluorescence/OD, varied up to 27-fold between integration sites, even for locations that did not affect growth. Microscopy of wild-type and engineered cells identified no morphological differences due to fluorescent protein expression, as well as expected levels of cell-to-cell variability in fluorescence (Supplemental Fig. 1).
Fig. 3.
P. putida KT2440 strains each with the same mKate construct integrated at different insertion loci (named by the upstream gene) around the chromosome were grown in M9 minimal medium supplemented with 30 mM glucose for 48 h. The growth rate of each strain measured by optical density at 700 nm is plotted relative to the respective fluorescence emission/OD700 nm at mid-log of growth. Each circle represents one biological replicate.
Potential integration loci such as PP_1642 and PP_2224, which impose significant fitness costs on the host, have obvious tradeoffs. Even among the neutral sites, though, a 27-fold variation in protein expression could have significant effects on the function of a heterologous pathway. Understanding and accommodating these differences will greatly improve the reliability of pathway design. There are several possibilities that may help explain the variances in host growth or protein expression. Copy number of the genes may be temporarily duplicated during DNA replication (Slager and Veening, 2016), especially close to the origin of replication. This variation may partially explain the expression levels at some sites such as PP_5322 having the greatest mKate expression (close to the oriC), and PP_2224 being one of the lowest mKate expression sites (close to the terminus). Although the integration of PP_5388 interrupted the coding sequence of a hypothetical protein, this did not alter normal growth rates.
A previous study (Domröse et al., 2019) used Tn5 transposition to randomly integrate a 21 kb prodigiosin gene cluster across the P. putida KT2440 chromosome. In that example, it was found that the mutants with the highest expression were biased to one specific location, one of seven rrn operons. The authors propose that the optimized insertion at this locus is due to both the redundancy of the rrn operons (preventing interference of normal cell functioning) and the essentiality (high transcription levels) of the operon. However, high expression is not always desirable, and there are many situations where the ability to fine-tune expression through the choice of integration locus would be valuable. In E. coli (Bryant et al., 2014) significant differences were found in the expression of a gfp reporter under the control of a lac promoter integrated at fourteen rationally decided locations. The authors attribute the approximate 300-fold variation in expression across the sites to gene silencing at transcriptionally silent Extended Protein Occupancy Domains (tsEPODs), and transcriptional rate variations due to DNA supercoiling. This work further evidences the importance of the integration location as a major contributor to both maintaining normal cell functioning and generating the quantity of protein expression.
3.3. Effects of integration locus on function of a heterologous transporter
In order to further test the effect of the integration locus on function of a heterologous gene, we integrated a functional protein, a muconate transporter, into three of the seven sites and measured the resulting variation in growth and protein expression. The integration sites selected for further characterization each displayed different phenotypes with mKate, such as slow growth and low fluorescence (PP 2224), slow growth and high fluorescence (PP 1642), or wild type growth and medium fluorescence (PP 5042).
Cis,cis-muconate is a potential value-added bioproduct that can be chemically converted into adipic acid for use in nylon synthesis (Johnson et al., 2016). Biologically, cis,cis-muconate could be further metabolized to products such as β-keto adipic acid, but these pathways will require further optimization. P. putida KT2440 can metabolize cis,cis-muconate for growth, as it is a key intermediate in the β-ketoadipate pathway for microbial dissimilation of aromatic compounds (Harwood and Parales, 1996; Ornston and Stanier, 1966). However, KT2440 is not capable of growing on exogenously supplied cis,cis-muconate, as it lacks import mechanisms for this metabolite. Assimilation of exogenous cis,cis-muconate would simplify downstream pathway optimization, but will require expression and tuning of a muconate transporter.
Here we incorporated a codon-optimized copy of mucK, the gene coding for the muconate importer from Acinetobacter baylyi ADP1 (Williams and Shaw, 1997), into three different insertion site loci. The same plasmids used for mKate insertion were used for mucK, including the homology arms and the insulating terminators. Expression of mucK was driven by the strong, constitutive Pc promoter from the class III integron of Delftia acidovorans C17 (Xu et al., 2013) and the gene 10 ribosome binding site from bacteriophage T7 (Olins et al., 1988). The three strains were then challenged to grow with muconate for functional assessment of the insertion sites.
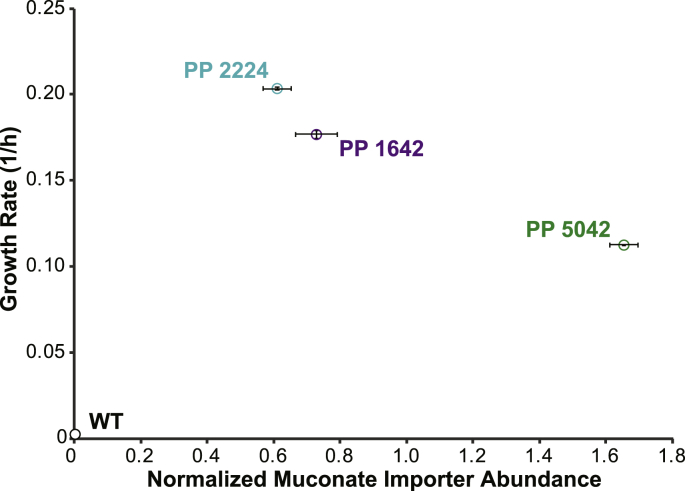
Expression of MucK conferred growth with muconate as the sole carbon source, and different growth rates were observed based on the locus of insertion [Fig. 4]. We used targeted proteomics (Supplemental Fig. 2) to compare MucK expression levels among strains with mucK inserted at different loci [Fig. 4]. Unexpectedly, the insertion site that was correlated with the lowest growth and mKate expression (PP_2224), had the highest growth rate with muconate and the lowest MucK expression. The other two sites had the inverse effect in MucK with respect to mKate. The PP_1642 site had low protein expression and higher growth rates with muconate, while the PP_5042 site had the highest protein expression and the lowest growth rate. Although high levels of heterologous membrane protein expression can be toxic, growth with glucose was not inhibited by MucK expression (Supplemental Fig. 3). It has been shown that the accumulation of high levels of intracellular muconate can be toxic to bacterial cells (Gaines et al., 1996). It may be that rapid muconate import in the presence of a highly expressed transporter could inhibit cell growth. Indeed, the strain with the slowest growth on muconate displayed the highest levels (approximately 2-fold higher) of MucK expression.
Fig. 4.
P. putida KT2440 strains with a muconate transporter gene (mucK) inserted at three different loci, were grown in M9 minimal medium supplemented with 30 mM cis, cis-muconic acid. Targeted proteomics quantified the amount of MucK in each strain (Y-axis error bars represent standard deviation of three replicate samples) and is plotted with respect to the growth rate as measured by optical density at 600 nm (X-axis error bars represent standard error of four replicate samples).
These experiments demonstrate that variation due to integration locus is not unique to fluorescent protein expression, but also affects functional proteins such as a transporter. In contrast to the previous integration locus analysis (Domröse et al., 2019) where high expression was desirable and integration loci were chosen accordingly, utility of mucK was highest when expression was moderate. Notably, growth on glucose with the insertion of mKate at PP_2224 and PP_1642 was substantially decreased, where growth on glucose in all three mucK strains was sustained at wild type levels. The expression of mKate at PP_5322 was equivalent to the expression at PP_1642 however the PP_5322 strain was able to maintain wild-type growth rates. The expression of heterologous proteins needs to be tuned for particular purposes, and the choice of integration locus will substantially affect both the final protein expression and, in some cases, cell growth.
4. Conclusion
Chromosomal integration of heterologous genes and pathways is the ideal method for metabolic engineering of industrial strains. In this study, we have selected integration sites for heterologous DNA insertion that are non-proximal to essential genes, non-operon interfering, and in antiparallel orientation to the downstream genes. We measured and characterized a small subset of possible insertion loci for the integration of heterologous DNA, showing that different integration loci can have significant effects on protein expression and the resulting organismal phenotype. This work is an important first step in optimizing the placement of target genes and pathways. Future work is needed to characterize many more sites throughout the genome, test different construct orientations, and examine operons or multigene constructs to determine the biological mechanisms responsible for expression variation. Reliable, predictable expression tuning by choice of integration site will provide additional control for metabolic engineering in this important chassis organism.
CRediT authorship contribution statement
Julie E. Chaves: Investigation, Formal analysis, Validation, Methodology, Visualization, Writing - original draft. Rosemarie Wilton: Investigation, Formal analysis, Validation, Methodology, Visualization, Writing - original draft. Yuqian Gao: Methodology, Investigation, Validation, Formal analysis, Visualization, Writing - original draft. Nathalie Munoz Munoz: Methodology, Investigation, Writing - review & editing. Meagan C. Burnet: Investigation, Writing - original draft. Zachary Schmitz: Investigation, Writing - review & editing. John Rowan: Investigation, Methodology, Writing - review & editing. Leah H. Burdick: Investigation, Writing - review & editing. Joshua Elmore: Conceptualization, Investigation. Adam Guss: Conceptualization, Methodology, Supervision, Project administration, Funding acquisition, Writing - review & editing. Dan Close: Conceptualization, Methodology, Supervision, Project administration. Jon K. Magnuson: Funding acquisition, Writing - review & editing. Kristin E. Burnum-Johnson: Methodology, Supervision, Writing - review & editing. Joshua K. Michener: Conceptualization, Methodology, Investigation, Supervision, Project administration, Funding acquisition, Visualization, Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Research was sponsored by the US Department of Energy BioEnergy Technologies Office through the Agile Biofoundry. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under contract DE-AC05-00OR22725. A portion of this research was performed at Pacific Northwest National Laboratory (PNNL) using EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. PNNL is a multiprogram national laboratory operated by Battelle for the Department of Energy (DOE) under Contract DE-AC05-76RLO 1830. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00139.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ankenbauer A., Schäfer R.A., Viegas S.C., Pobre V., Voß B., Arraiano C.M., Takors R. Pseudomonas putida KT2440 is naturally endowed to withstand industrial-scale stress conditions. Microb. Biotechnol. 2020 doi: 10.1111/1751-7915.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Barrau E., Olivera E.R., Luengo J.M., Fernández C., Galán B., García J.L., Díaz E., Minambres B. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 2004;186:5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassalo M.C., Garst A.D., Halweg-Edwards A.L., Grau W.C., Domaille D.W., Mutalik V.K., Arkin A.P., Gill R.T. Rapid and Efficient One-Step Metabolic Pathway Integration in E. coli. ACS Synth. Biol. 2016;5(7):561–568. doi: 10.1021/acssynbio.5b00187. [DOI] [PubMed] [Google Scholar]
- Bayly R.C., Barbour M.G. The degradation of aromatic compounds by the meta and gentisate pathways: biochemistry and regulation. Microbial degradation of organic compounds. Microbiol. 1984;3:253–294. [Google Scholar]
- Bryant J.A., Sellars L.E., Busby S.J.W., Lee D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;42:11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-F., Chao H., Zhou N.-Y. The catabolism of 2,4-xylenol and p-cresol share the enzymes for the oxidation of para-methyl group in Pseudomonas putida NCIMB 9866. Appl. Microbiol. Biotechnol. 2014;98:1349–1356. doi: 10.1007/s00253-013-5001-z. [DOI] [PubMed] [Google Scholar]
- Curran K.A., Morse N.J., Markham K.A., Wagman A.M., Gupta A., Alper H.S. Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synth. Biol. 2015;4:824–832. doi: 10.1021/sb5003357. [DOI] [PubMed] [Google Scholar]
- Domröse A., Hage-Hülsmann J., Thies S., Weihmann R., Kruse L., Otto M., Wierckx N., Jaeger K.-E., Drepper T., Loeschcke A. Pseudomonas putida rDNA is a favored site for the expression of biosynthetic genes. Sci. Rep. 2019;9:7028. doi: 10.1038/s41598-019-43405-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N.W., Gunsalus I.C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J.R., Furches A., Wolff G.N., Gorday K., Guss A.M. Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metab. Eng. Commun. 2017;5:1–8. doi: 10.1016/j.meteno.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers C.E., Lawless C., Wedge D.C., Lau K.W., Gaskell S.J., Hubbard S.J. CONSeQuence: prediction of reference peptides for absolute quantitative proteomics using consensus machine learning approaches. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagfeldt D.B., Siewers V., Huang L., Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- Gaines G.L., 3rd, Smith L., Neidle E.L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J. Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C.S., Parales R.E. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- Hulton C.S., Seirafi A., Hinton J.C., Sidebotham J.M., Waddell L., Pavitt G.D., Owen-Hughes T., Spassky A., Buc H., Higgins C.F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- Jayakody L.N., Johnson C.W., Whitham J.M., Giannone R.J., Black B.A., Cleveland N.S., Klingeman D.M., Michener W.E., Olstad J.L., Vardon D.R., Brown R.C., Brown S.D., Hettich R.L., Guss A.M., Beckham G.T. Thermochemical wastewater valorization via enhanced microbial toxicity tolerance. Energy Environ. Sci. 2018;11:1625–1638. [Google Scholar]
- Johnson C.W., Beckham G.T. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab. Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Salvachúa D., Khanna P., Smith H., Peterson D.J., Beckham G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016;3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q., Ong W.K., Paley S.M., Subhraveti P. The BioCyc collection of microbial genomes and metabolic pathways. Briefings Bioinf. 2019;20:1085–1093. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisaar M., Hõrak R., Kasak L., Heinaru A., Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- Krogh T.J., Møller-Jensen J., Kaleta C. Impact of Chromosomal Architecture on the Function and Evolution of Bacterial Genomes. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.B.K., Imakaev M.V., Mirny L.A., Laub M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C.J. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlino G.B., Tizzani L., Fleer R., Frontali L., Bianchi M.M. Inducible amplification of gene copy number and heterologous protein production in the yeast Kluyveromyces lactis. Appl. Environ. Microbiol. 1999;65:4808–4813. doi: 10.1128/aem.65.11.4808-4813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu E.S., Nicora C.D., Sims A.C., Burnum-Johnson K.E., Kim Y.-M., Kyle J.E., Matzke M.M., Shukla A.K., Chu R.K., Schepmoes A.A., Jacobs J.M., Baric R.S., Webb-Robertson B.-J., Smith R.D., Metz T.O. 2016. MPLEx: a robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols N.N., Harwood C.S. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J. Bacteriol. 1995;177:7033–7040. doi: 10.1128/jb.177.24.7033-7040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel P.I., Chavarría M., Fuhrer T., Sauer U., de Lorenzo V. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 2015;290:25920–25932. doi: 10.1074/jbc.M115.687749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins P.O., Devine C.S., Rangwala S.H., Kavka K.S. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988;73:227–235. doi: 10.1016/0378-1119(88)90329-0. [DOI] [PubMed] [Google Scholar]
- Olivera E.R., Miñambres B., García B., Muñiz C., Moreno M.A., Ferrández A., Díaz E., García J.L., Luengo J.M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L.N., Stanier R.Y. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida : I. Biochemistry. J. Biol. Chem. 1966;241:3776–3786. [PubMed] [Google Scholar]
- Poblete-Castro I., Becker J., Dohnt K., dos Santos V.M., Wittmann C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012;93:2279–2290. doi: 10.1007/s00253-012-3928-0. [DOI] [PubMed] [Google Scholar]
- Samuelson J.C. Recent developments in difficult protein expression: a guide to E. coli strains, promoters, and relevant host mutations. Methods Mol. Biol. 2011;705:195–209. doi: 10.1007/978-1-61737-967-3_11. [DOI] [PubMed] [Google Scholar]
- Sauer C., Syvertsson S., Bohorquez L.C., Cruz R., Harwood C.R., van Rij T., Hamoen L.W. Effect of Genome Position on Heterologous Gene Expression in Bacillus subtilis: An Unbiased Analysis. ACS Synth. Biol. 2016;5(9):942–947. doi: 10.1021/acssynbio.6b00065. [DOI] [PubMed] [Google Scholar]
- Schmid A., Dordick J.S., Hauer B., Kiener A., Wubbolts M., Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Searle B.C., Egertson J.D., Bollinger J.G., Stergachis A.B., MacCoss M.J. Using data independent acquisition (DIA) to model high-responding peptides for targeted proteomics experiments. Mol. Cell. Proteomics. 2015;14:2331–2340. doi: 10.1074/mcp.M115.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistrom W.R., Stanier R.Y. The mechanism of formation of β-ketoadipic acid by bacteria. J. Biol. Chem. 1954;210:821–836. [PubMed] [Google Scholar]
- Slager J., Veening J.-W. Hard-wired control of bacterial processes by chromosomal gene location. Trends Microbiol. 2016;24:788–800. doi: 10.1016/j.tim.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobetzko P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016;44:1514–1524. doi: 10.1093/nar/gkw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C., de Lorenzo V., Cebolla A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology. 1997;143:2071–2078. doi: 10.1099/00221287-143-6-2071. [DOI] [PubMed] [Google Scholar]
- Thompson A., Gasson M.J. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2001;67:3434–3439. doi: 10.1128/AEM.67.8.3434-3439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo K.E.J., Ajikumar P.K., Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 2009;27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- Williams P.A., Shaw L.E. mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J. Bacteriol. 1997;179:5935–5942. doi: 10.1128/jb.179.18.5935-5942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton R., Ahrendt A.J., Shinde S., Sholto-Douglas D.J., Johnson J.L., Brennan M.B., Kemner K.M. A new suite of plasmid vectors for fluorescence-based imaging of root colonizing pseudomonads. Front. Plant Sci. 2017;8:2242. doi: 10.3389/fpls.2017.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Tao F., Ma C., Xu P. New constitutive vectors: useful genetic engineering tools for biocatalysis. Appl. Environ. Microbiol. 2013;79:2836–2840. doi: 10.1128/AEM.03746-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane S., Yamaoka M., Yamamoto M., Maruki T., Matsuzaki H., Hatano T., Fukui S. Region specificity of chromosome III on gene expression in the yeast Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 1998;44:275–281. doi: 10.2323/jgam.44.275. [DOI] [PubMed] [Google Scholar]
- Zhen D., Liu H., Wang S.-J., Zhang J.-J., Zhao F., Zhou N.-Y. Plasmid-mediated degradation of 4-chloronitrobenzene by newly isolated Pseudomonas putida strain ZWL73. Appl. Microbiol. Biotechnol. 2006;72:797–803. doi: 10.1007/s00253-006-0345-2. [DOI] [PubMed] [Google Scholar]
- Zylstra G.J., Gibson D.T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J. Biol. Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.