Abstract
In Trichoderma reesei, carbon catabolite repression (CCR) significantly downregulates the transcription of cellulolytic enzymes, which is usually mediated by the zinc finger protein Cre1. It was found that there is a conserved region at the C-terminus of Cre1/CreA in several cellulase-producing fungi that contains up to three continuous S/T phosphorylation sites. Here, S387, S388, T389, and T390 at the C-terminus of Cre1 in T. reesei were mutated to valine for mimicking an unphosphorylated state, thereby generating the transformants Tr_Cre1S387V, Tr_Cre1S388V, Tr_Cre1T389V, and Tr_Cre1T390V, respectively. Transcription of cel7a in Tr_ Cre1S388V was markedly higher than that of the parent strain when grown in glucose-containing media. Under these conditions, both filter paperase (FPase) and p-nitrophenyl-β-D-cellobioside (pNPCase) activities, as well as soluble proteins from Tr_Cre1S388V were significantly increased by up to 2- to 3-fold compared with that of other transformants and the parent strain. The results suggested that S388 is critical site of phosphorylation for triggering CCR at the terminus of Cre1. To our knowledge, this is the first report demonstrating an improvement of cellulase production in T. reesei under CCR by mimicking dephosphorylation at the C-terminus of Cre1. Taken together, we developed a precision engineering strategy based on the modification of phosphorylation sites of Cre1 transcription factor to enhance the production of cellulase in T. reesei under CCR.
Keywords: carbon catabolite repression, transcription factor, Cre1, cellulase, phosphorylation, Trichoderma reesei
Graphical Abstract
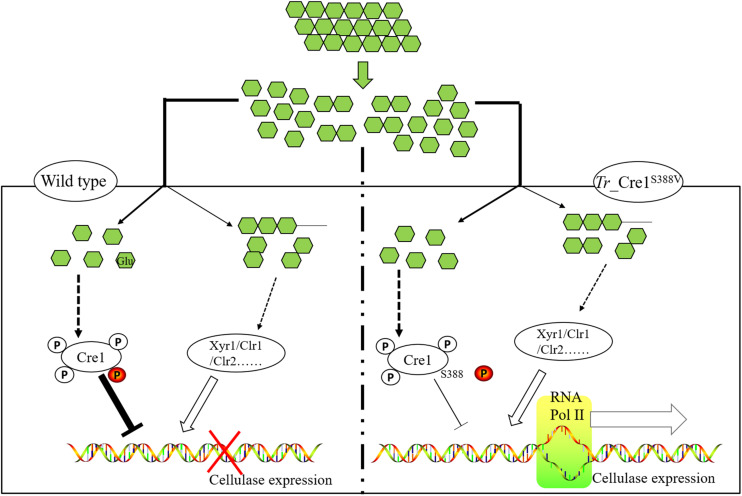
Schematic diagram on transcriptional regulation of cellulase expression in mutant and parent strains. We developed a precision engineering strategy by mimicking dephosphorylation at a key residue S388 of Cre1 to improve the production of cellulase in fungal species under CCR, highlighting that this technology can accelerate the industrial process of lignocellulosic biorefinery.
Introduction
The filamentous ascomycete Trichoderma reesei, a clonal derivative of the ascomycete Hypocrea jecorina (Kuhls et al., 1996), is widely used in industrial production of cellulases and xylanases (Derntl et al., 2019). There is a broad range of industrial applications of these cellulolytic enzymes in the food and feed, textile, and pulp and paper industries, as well as in the production of lignocellulosic bioethanol (Wilson, 2009).
Carbon catabolite repression (CCR) is a global regulatory system that is found in nearly all heterotrophic hosts (Görke and Stülke, 2008). Notably, the presence of monosaccharides, like D-glucose, during cellulolytic enzyme production triggers CCR, which is regulated mainly by the cellulase transcriptional repressor Cre1 and activators Xyr1, Clr1, Clr2, etc., and significantly downregulates the transcription of cellulolytic enzymes in T. reesei (Wang et al., 2019). Endogenous Cre1, which belongs to the group of C2H2-type zinc finger proteins, binds to the promoters of target genes, such as cel7a, and inhibits the transcription of encoded cellulase genes (Mello-de-Sousa et al., 2014).
Many reports have demonstrated that the knockout of cre1 in filamentous fungi alleviated CCR (Ilmén et al., 1996; Long et al., 2018). However, it was reported that Cre1 not only plays an essential role in correct nucleosome positioning, but also participates in several morphological functions, such as hyphal development and sporulation (Ries et al., 2014). Moreover, it was shown that the disruption of cre1 in T. reesei, or its replacement with a cre1 mutant, inhibits hypal growth (Portnoy et al., 2011), affects biomass accumulation, and leads to a decrease in cellulase production (Nakari-Setälä et al., 2009; Rassinger et al., 2018; Wang et al., 2019).
Furthermore, it was reported that CCR is achieved with the phosphotransferase system (Wang et al., 2017; Liu et al., 2020). Post-translational modifications, especially phosphorylation, of the proteins involved, including Cre1, play an essential role in signal transduction to achieve CCR (Jiang et al., 2018; Horta et al., 2019). Horta et al. (2019) revealed that there is large-scale phosphorylation and dephosphorylation of substrate-specific proteins, including Cre1, in Neurospora crassa, according to MS/MS-based peptide analysis. Nguyen et al. (2016) suggested that phosphorylation of Cre1 plays a role in the onset of CCR induction and sensing of the carbon source. Cziferszky et al. (2002) reported that phosphorylation of Cre1 at S241 in T. reesei has a significant effect on the repression of cellulolytic enzyme genes at the transcriptional level under CCR. Thus, the modification of specific phosphorylation sites in Cre1/CreA may be a rational strategy for releasing or attenuating CCR to achieve improvement of cellulase production.
In this study, the modification of phosphorylation sites at the C-terminus of Cre1 from T. reesei was performed for mimicking a dephosphorylated state, generating four transformants. Growth behavior, transcriptional profiles, and enzymatic activities of these transformants were investigated. Obtained data proved that the mutation of S388 to V388 at the C-terminus of Cre1 results in great improvement of cellulase production in the presence of glucose. Furthermore, it suggested that loss of phosphorylation at S388 at the continuous phosphorylation motif (SSTT) plays an important role in alleviating CCR.
Materials and Methods
Strains and Reagents
The T. reesei strain (CCTCC M2015804) that was deposited in the China Center for Type Culture Collection (CCTCC) was used as the parent strain (Zhang et al., 2019) in this study, and pNPC was purchased from Aladdin (Shandong, China). All polymerase chain reaction amplifications were performed using DNA polymerase (Vazyme Co., Ltd., Nanking, China). RNAisoTM reagent and PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time) were purchased from Takara Bio Inc. (Shiga, Japan). FastStart Essential DNA Green Master was purchased from Roche (Basel, Switzerland). All other chemicals were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). ABclonal MultiF Seamless Assembly Mix was purchased from ABclonal (Wuhan, China). Primers were synthesized by Personalbio Biotech Co., Ltd. (Shanghai, China).
Construction of cre1 Mutation Replacement Cassettes and Propagation of Transformant Strains
All plasmids were constructed based on homologous recombination, which was mediated by the ABclonal MultiF Seamless Assembly Mix. The primers used for the construction of cre1 mutation replacement cassettes are listed in Supplementary Table S1. All the plasmids and linearized expression cassettes used or constructed in this study were listed in Supplementary Table S2. The transformation of replacement cassettes into T. reesei was performed using previously described methods (Wang et al., 2019). The transformants were selected on plates containing minimal medium (MM) supplemented with 2% glucose and 200 μg/mL hygromycin. Propagation of transformant strains was performed based on a protocol published previously (Wang et al., 2019).
Analysis of Growth Phenotype and Transcriptional Profile
The method used for culturing fungi for growth phenotype, transcript analysis, and RT-qPCR assays was already described previously (Wang et al., 2019). The primers used for RT-qPCR are listed in Supplementary Table S1.
Microscopic observation of hyphal morphology was performed based on a previously described method (Wang et al., 2019). Approximately, 1 × 103 spores were inoculated on slides with solidified medium containing PDA, wheat bran, MM with 2% glucose, or MM with 2% glycerol at 30°C for 48 h. Microscopic images of hyphae with 400 × magnification (Nikon Eclipse E100, Tokyo, Japan) were then captured using a Nikon D5000 camera.
Cellulase Production and Determination of Enzymatic Activities, Soluble Protein, and Biomass
The parent strain or mutants (approximately 1 × 107 spores) was inoculated in 300 ml flask including MM solution containing 1% glucose, 1% trytone, 1% wheat bran, and 0.5% CaCO3 at 30°C and 200 rpm. After 48 h of cultivation, the 10 ml media as the seed was added into 500 ml flask including 100 ml liquid medium. In this study, batch cultures and pulse fed-batch cultures were cultivated at 30°C and 200 rpm for 7 days. The initial pH value was adjusted at pH 5.0 with 3 M NaOH. Batch culture was performed in the medium with MM solution containing 2% glucose based on a protocol published previously (Wang et al., 2019). Pulse fed-batch culture: For MM solution containing 2% avicel, 0.6% glucose was pulse-fed on the second and third days. Cultures were timely collected to measure pNPCase activity, as well as extracellular and intracellular protein concentrations. Protein and biomass concentrations were estimated according to a previously described method (Wang et al., 2019).
In silico Prediction of Protein Domains and Homologous Sequence Alignment
Partial in silico domain prediction of Cre1 (Trichoderma reesei NCBI Accession ID: AAB01677.1) was performed as previously described (Rassinger et al., 2018). Cre1 (Trichoderma reesei, NCBI Accession ID: AAB01677.1) and its homologs in Penicillium oxalicum (NCBI Accession ID: EPS28222.1), Aspergillus oryzae (NCBI Accession ID: AAK11189.1), Aspergillus nidulans (NCBI Accession ID: XP_663799.1), Aspergillus niger (NCBI Accession ID: AAA32690.1), Aspergillus fumigatus (NCBI Accession ID: XP_755510.1), Thermoascus aurantiacus (NCBI Accession ID: AAT34979.1), Talaromyces emersonii (NCBI Accession ID: AAL33631.4), Thermothelomyces thermophilus (NCBI Accession ID: XP_003665891.1), and Neurospora crassa (NCBI Accession ID: EAA32758.1) were analyzed using ClustalX2.
Results
Phosphorylation of Cre1
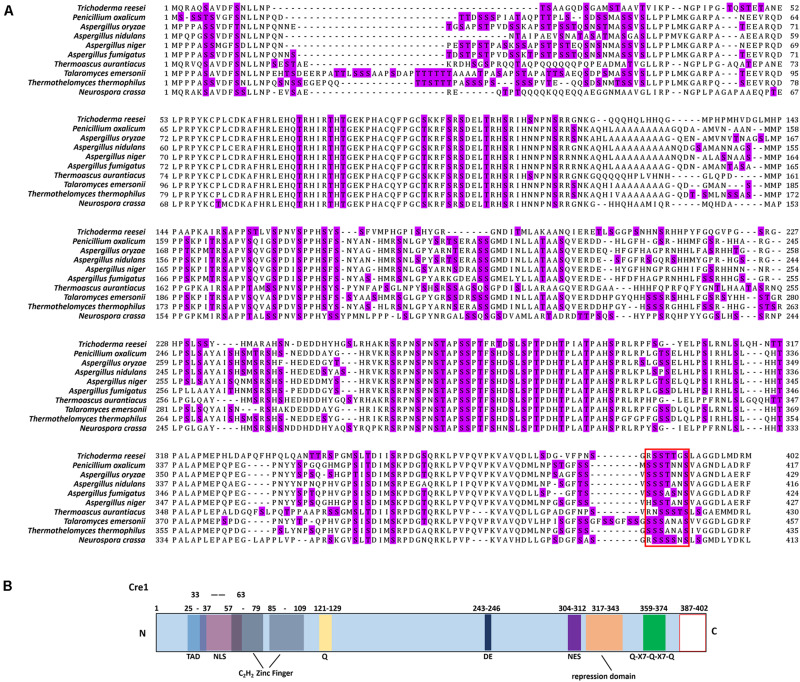
As shown in Figure 1A, we found that there is a conserved region (red box) at the C-terminus of Cre1/CreA/CreT in Trichoderma reesei, Penicillium oxalicum, Aspergillus oryzae, Aspergillus nidulans, Aspergillus fumigatus, Aspergillus niger, Thermoascus aurantiacus, Talaromyces emersonii, Thermothelomyces thermophilus, and Neurospora crassa using homology alignment. Most of these conserved regions contain up to three continuous S/T phosphorylation sites. Moreover, phosphorylation of the peptide sequence extending from amino acids 387 to 402 (SSTTGSLAGGDLMDRM) at the C-terminus of Cre1 from T. reesei (Tr_Cre1) was identified with LC-MS/MS by Nguyen et al. (2016). To our knowledge, however, there is so far no report on the function of the C-terminus of Cre1 from T. reesei (Figure 1B).
FIGURE 1.
Amino acid sequence alignments of Cre1/CreA and in silico domain prediction of Cre1. (A) Amino acid sequence alignments of Cre1/CreA from Trichoderma reesei, Penicillium oxalicum, Aspergillus oryzae, Aspergillus nidulans, Aspergillus fumigatus, Aspergillus niger, Thermoascus aurantiacus, Talaromyces emersonii, Thermothelomyces thermophilus, and Neurospora crassa; S/T phosphorylation sites are highlighted; (B) in silico domain prediction of Cre1. Numbers indicate the amino acid (aa) positions and colored boxes indicate identified C2H2 zink finger domains (gray); pink, nuclear localization signal (NLS); light blue, transactivation domain (TAD); yellow, glutamine (Q); dark blue, aspartic (D) and glutamic (E) acids; green, Q-X7- Q-X7-Q; violet, nuclear export signal (NES); orange, repression domain; blank, C-terminus of Cre1/CreA. The putative domains of Cre1were predicted by a number of in silico prediction tools and alignment algorithms alignment algorithms as described in the section “Materials and Methods.”
Comparison of Phenotypic Characterization and Transcriptional Levels
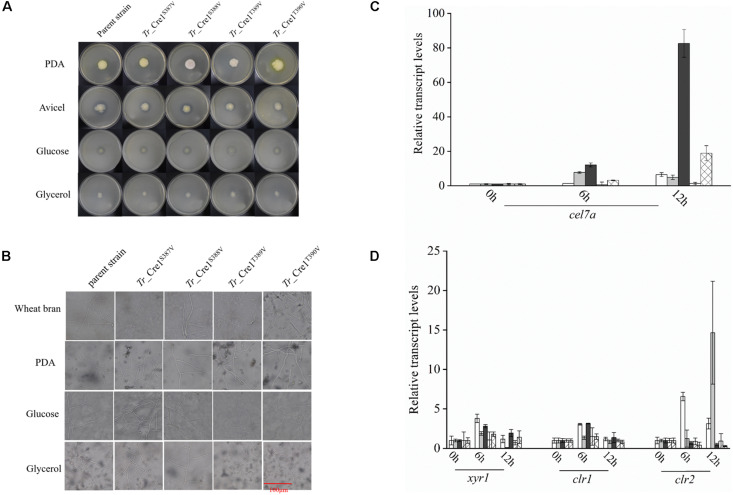
Amino acid residues S387, S388, T389, and T390 of Tr_Cre1 of the parent strain were mutated to valine to mimic their dephosphorylation, generating the transformants named Tr_Cre1S387V, Tr_ Cre1S388V, Tr_ Cre1T389V, and Tr_ Cre1T390V, respectively. The mutation of transformants was verified using PCR and DNA sequencing (Supplementary Figure S1). Phenotypic analysis of each strain was performed after incubating the plates for 6 days at 30°C. Figures 2A,B shows that no morphological changes on the colonies and hyphae were observed among the transformants and parent strain. It was proved that the substitution of S387, S388, T389, or T390 in Tr_Cre1 for valine had no significant effect on the growth behavior of transformants.
FIGURE 2.
Analysis of phenotypic characterization and transcriptional levels. (A) Morphologies of the parent strain and transformants on plates containing different carbon sources. For growth assays, transformants and parent strain were grown on plates with 2% (w/v) potato dextrose agar (PDA), avicel, glucose, or glycerol as a sole carbon source. Plates were incubated at 30°C and pictures were taken after 144 h. (B) Microscopic observation of hyphae of parent strain and transformants. Approximately, 1 × 103 spores were inoculated on slides with solidified medium containing PDA, wheat bran, MM with 2% glucose, or MM with 2% glycerol at 30°C for 48 h. Scar bar: 100 μm. (C) Transcriptional levels of cel7a in the parent strain and transformants. Transcriptional levels of cel7a in Tr_Cre1S387V (gray), Tr_ Cre1S388V (dark), Tr_ Cre1S389V (oblique line), Tr_ Cre1S390V (gridlines) transformants and parent strain (blank) grown in presence of a mixture of glucose and avicel as carbon source. Gene expression levels were normalized (2–ΔΔCT analysis) to that of actin. Mean values are shown; error bars indicate the standard deviation of three independently grown cultures. (D) Transcriptional levels of xyr1, clr1, and clr2 in the parent strain and transformants. Transcriptional levels of xyr1, clr1, and clr2 in Tr_Cre1S387V (gray), Tr_ Cre1S388V (dark), Tr_ Cre1S389V (oblique line), Tr_ Cre1S390V (gridlines) transformants and parent strain (blank) grown in presence of a mixture of glucose and avicel as carbon source. Gene expression levels were normalized (2–ΔΔCT analysis) to that of actin. Mean values are shown; error bars indicate the standard deviation of three independently grown cultures.
Subsequently, the expression levels of cel7a, xyr1, clr1, and clr2 in the parent and transformant strains were investigated using a mixture of glucose and avicel as carbon source (Figures 2C,D). No significant changes in xyr1, clr1, and clr2 expression were observed at the transcriptional level among the transformants and the parent strain, each grown in a mixture of glucose and avicel. However, transcription of cel7a in Tr_ Cre1S388V was markedly higher than in the parent strain when using a mixture of glucose and avicel as carbon source.
Determining Cellulase Activity in the Presence of Glucose
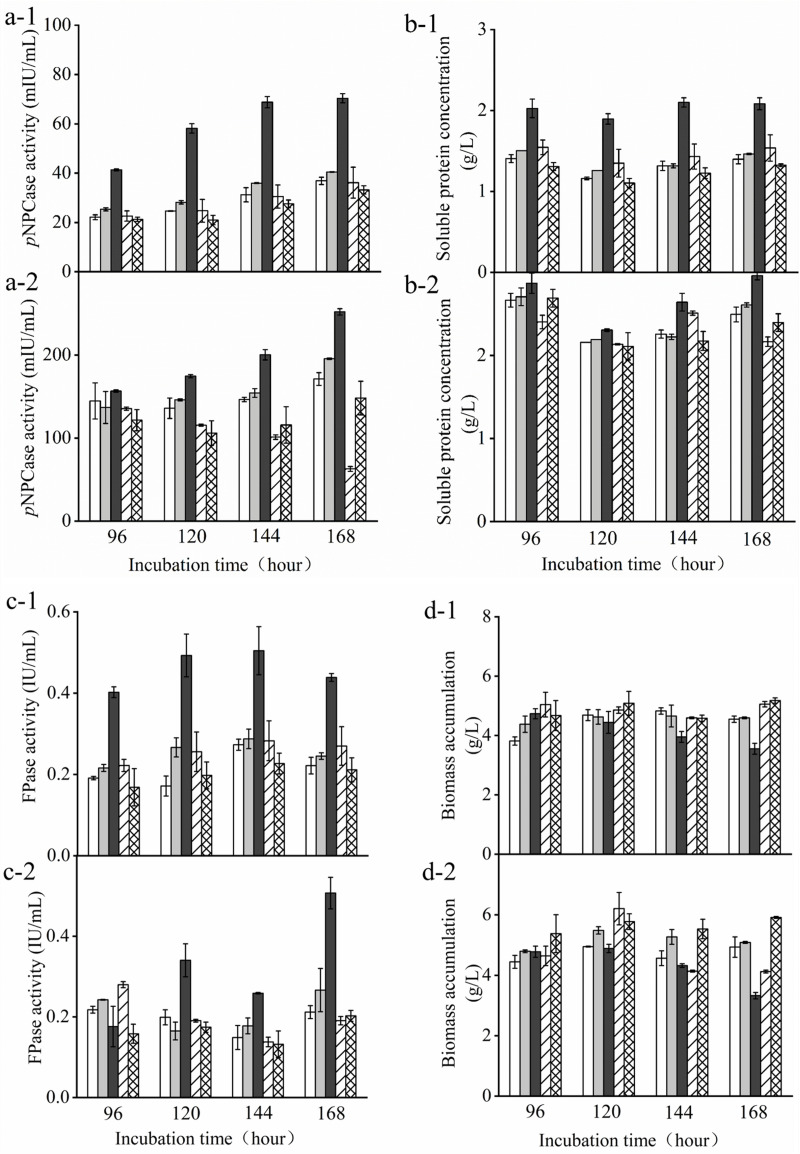
Furthermore, we compared the transformants and the parent strain cultured in either glucose-containing initial media in the batch culture or the supplemental media in the pulse fed-batch culture containing a mixture of glucose and avicel as carbon source. FPase and pNPCase (CBHI) activities, biomass, and soluble protein present in the supernatant of transformant and parent strain cultures were determined over 7 days of cultivation under either condition (Figure 3). There was no obvious difference in biomass among the transformants and the parent strain grown with glucose or the mixture of glucose and avicel before day 4. However, the biomass accumulation of Tr_ Cre1S388V was somewhat decreased after 6 days culture. After 7 days, FPase and pNPCase activities, as well as soluble protein from Tr_ Cre1S388V were significantly increased by 2. 25-, 2. 45-, and 1.93-fold, respectively, compared with those of the other transformants or the parent strain grown in glucose as a sole carbon source (Figure 3). Accordingly, FPase and pNPCase activities, and also soluble protein from Tr_ Cre1S388V, were significantly increased by 3. 50-, 2. 19-, and 1.78-fold upon 7 days of culture with the mixture of glucose and avicel, respectively, compared with those of the other transformants and the parent strain (Figure 3). Our findings proved that the mutation of S388 at the C-terminus of Cre1 results in an improvement of cellulase production in the presence of glucose. In line with this, amino acid residues 387–390 at the C-terminus of Cre1 were predicted by the NetPhos 3.1 Server1 and KinasePhos2 software as potential targets for phosphorylation. Additionally, it was found that S388 maybe be phosphorylated by cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA), a conserved key factor of a nutrient-sensing pathway that acts in parallel to the MAP kinase pathway (Ortiz-Urquiza and Keyhani, 2015), whereas S387, T389, and T390 are not. Furthermore, we suggest that dephosphorylation of S388 at the continuous phosphorylation motif (SSTT) plays an important role in alleviating CCR in the presence of glucose.
FIGURE 3.
Determination of cellulase activity in the presence of glucose. T. reesei transformants and the parent strain were cultivated in liquid medium supplemented with 2% (w/v) (x-1) glucose or (x-2) a mixture of glucose and avicel as carbon source. Activity of p-nitrophenyl-β−D-cellobioside (pNPCase) (a), soluble protein (b), filter paperase (FPase) activity (c), and biomass accumulation (d) of each culture were measured in biological and technical duplicates. Enzymatic activities are given as mean values, with error bars indicating the standard deviation. Symbols: blank, parent strain; light gray, Tr_Cre1S387V; dark gray, Tr_ Cre1S388V; oblique line, Tr_ Cre1T389V; gridline, Tr_ Cre1T390V.
Discussion
Carbon catabolite repression is a global regulatory system found in nearly all heterotrophic organisms (Liu et al., 2020). However, the substrate pairs inducing its expression as well as its underlying regulatory mechanisms need to be analyzed on a case-by-case basis, making it difficult to develop universal strategies to overcome its undesirable effects. A better fundamental understanding is still required to enable smarter designs for facilitating multiple substrate utilization.
Thus, several attempts were made to alleviate CCR by introducing modifications in Cre1 for improving cellulase production. It was proved that replacement of the endogenous transcription factor Cre1/CreA by an artificial minimal transcriptional activator, such as Cre1-96 or other Cre1/CreA mutants, leads to the release or attenuation of CCR, which in turn supports cellulase production in T. reesei and A. nidulans (Shroff et al., 1997; Mello-de-Sousa et al., 2014; Rassinger et al., 2018; Zhang et al., 2018). However, Liu et al. (2019) demonstrated that full-length Cre1 might be necessary for the rapid growth of T. reesei. Our results suggested that dephosphorylation at the C-terminus of Cre1 has no effect on growth behavior because of the retaining of full-length Cre1 (Figures 2A,B). Meanwhile, it was assumed that phosphorylation of Cre1 in T. reesei affects its repressing activity on cellulase genes under CCR (Cziferszky et al., 2002; Nguyen et al., 2016).
Among the potential phosphorylation target sites in the region covering amino acids 387–390 at the C-terminus of Cre1, S388 maybe be phosphorylated by PKA, in contrast to S387, T389, and T390. As a conserved nutrient-sensing pathway, the cAMP-PKA pathway plays an important role in the response to environmental carbon availability to activate or repress the relevant downstream genetic pathways (Ortiz-Urquiza and Keyhani, 2015; Hinterdobler et al., 2019). In Aspergillus fumigatus and Neurospora crassa, PKA is activated in the presence of glucose, thereby inhibiting alternative carbon source usage due to CCR (Brown et al., 2014; Huberman et al., 2016). In accordance with a previous study (Ribeiro et al., 2019), the pkaA deletion strain showed a significant improvement in cellulase activity in Aspergillus nidulans.
The results shown in Figures 2, 3 proved that the phosphorylation status at S388 of Cre1 in T. reesei plays an essential role in the regulation of transcription and synthesis of cellulolytic enzymes. Thus, we speculated that S388 of Cre1 in T. reesei was phosphorylated by one of the kinases acting in the PKA pathway in the presence of glucose, leading to CCR and, finally, to the CCR-mediated inhibition of the expression of cellulolytic enzymes. Our hypothesis is consistent with the report by Ribeiro et al. (2019), except for the phosphorylation site.
Here, we focused on the continuous phosphorylation of a particular motif (SSTT) at the C-terminus of Cre1 (Figure 1A) and analyzed mutants mimicking dephosphorylation of these sites. According to the results presented in Figure 3, an improvement of FPase and pNPCase activities contributed to a partial release from CCR. Cziferszky et al. (2002) reported that phosphorylation of another motif (HSNDEDD) was abolished when E244 was mutated to valine, following which CCR in T. reesei was depressed. Moreover, there is little change at the transcriptional levels of xyr1, clr1, and clr2 expression in Tr_ Cre1S388V, though FPase and pNPCase activities were greatly enhanced. Thus, we suggest that dephosphorylation of the motif (SSTT) has a great influence on the binding of target genes rather than the co-regulated gene, that is, xyr1, clr1, or clr2.
Therefore, we suggest that dephosphorylation modification of specific sites of Cre1 is a good strategy to reduce the CCR effect, and to enhance enzyme activity without affecting the growth of microorganisms. However, in order for this strategy to be effective it is crucial to identify the important phosphorylation sites, which requires a clear understanding of the regulatory pathway. Thus, this strategy will be hampered by the fact that the regulatory system is complex and currently not well understood (Ortiz-Urquiza and Keyhani, 2015; Adnan et al., 2017; Kunitake et al., 2019). Therefore, it is necessary to start from the downstream target, in this case Cre1, to correctly identify the pivotal sites, providing a simple way to enhance the production of cellulase in fungal species exhibiting CCR.
Conclusion
This study precisely identifies the dephosphorylation/phosphorylation sites of Cre1 and the key kinase participating in CCR in T. reesei. Our work provides a potential workflow for creating T. reesei strains resistant to CCR when full-length Cre1 was retained. Above all, we developed a precision engineering strategy to improve the production of cellulases in fungal species under CCR, highlighting that the application of this technology could accelerate the industrial adoption of lignocellulosic biorefinery methods.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in NCBI database (https://www.ncbi.nlm.nih.gov/) under the accession numbers: AAB01677.1, EPS28222.1, AAK11189.1, XP_663799.1, AAA32690.1, XP_755510.1, AAT34979.1, AAL33631.4, and EAA32758.1.
Author Contributions
LH, YT, WM, and KN performed the experiments. LH and XF wrote the manuscript and conceived the study. LH, YT, WM, SH, YL, and XF were involved in analysis and interpretation of experimental data. XF coordinated the project. All authors contributed to the article and approved the submitted version.
Conflict of Interest
SH was employed by the company Shandong Henglu Biological Technology Co., Ltd., Part of data in this manuscript have been issued in China patent application (CN 201911135400.2). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Prof. Luying Xun’s help in revising this manuscript. The part contents in this article have been released as a pre-print at BioRxiv (Han et al., 2020).
Abbreviations
- cAMP
cyclic adenosine monophosphate
- CCR
carbon catabolite repression
- FP
filter paper
- PKA
protein kinase A
- RT-qPCR
reverse transcription quantitative polymerase chain reaction.
Funding. This work was supported by the National Key R&D Program of China (No. 2018YFB1501701), the National Natural Science Foundation of China (Nos. 31570040 and 31870785), the Key Technologies R&D Program of Shandong Province (No. 2018GSF121021), and the State Key Laboratory of Microbial Technology Open Projects Fund.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00852/full#supplementary-material
References
- Adnan M., Zheng W., Islam W., Arif M., Abubakar Y. S., Wang Z., et al. (2017). Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 19:48. 10.3390/ijms19010048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. A., Ries L., Goldman G. H. (2014). How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. FG B. 72 48–63. 10.1016/j.fgb.2014.06.012 [DOI] [PubMed] [Google Scholar]
- Cziferszky A., Mach R. L., Kubicek C. P. (2002). Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277 14688–14694. 10.1074/jbc.M200744200 [DOI] [PubMed] [Google Scholar]
- Derntl C., Mach R. L., Mach-Aigner A. R. (2019). Fusion transcription factors for strong, constitutive expression of cellulases and xylanases in Trichoderma reesei. Biotechnol. Biofuels 12:231. 10.1186/s13068-019-1575-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B., Stülke J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- Han L., Tan Y., Ma W., Niu K., Hou S., Guo W., et al. (2020). Precision engineering of the transcription factor Cre1 in Hypocrea jecorina (Trichoderma reesei) for efficient cellulase production in the presence of glucose. bioRxiv [Preprint]. 10.1101/2020.03.08.982249v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterdobler W., Schuster A., Tisch D., Özkan E., Bazafkan H., Schinnerl J., et al. (2019). The role of PKAc1 in gene regulation and trichodimerol production in Trichoderma reesei. Fungal. Biol. Biotechnol. 6:12. 10.1186/s40694-019-0075-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horta M., Thieme N., Gao Y., Burnum-Johnson K. E., Nicora C. D., Gritsenko M. A., et al. (2019). Broad substrate-specific phosphorylation events are associated with the initial stage of plant cell wall recognition in Neurospora crassa. Front. Microbiol. 10:2317. 10.3389/fmicb.2019.02317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman L. B., Liu J., Qin L., Glass N. L. (2016). Regulation of the lignocellulolytic response in filamentous fungi. Fungal. Biol. Rev. 30 101–111. 10.1016/j.fbr.2016.06.001 [DOI] [Google Scholar]
- Ilmén M., Thrane C., Penttilä M. (1996). The glucose repressor genecre1 of Trichoderma: Isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251 451–460. 10.1007/bf02172374 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liu K., Guo W., Zhang R., Liu F., Zhang N., et al. (2018). “Lignocellulase formation, regulation, and secretion mechanisms in Hypocrea jecorina (Trichoderma reesei) and other filamentous fungi,” in Fungal Cellulolytic Enzymes, eds Fang X., Qu Y. (Singapore: Springer; ). [Google Scholar]
- Kuhls K., Lieckfeldt E., Samuels G. J., Kovacs W., Meyer W., Petrini O., et al. (1996). Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. U.S.A. 93 7755–7760. 10.1073/pnas.93.15.7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitake E., Li Y., Uchida R., Nohara T., Asano K., Hattori A., et al. (2019). CreA-independent carbon catabolite repression of cellulase genes by trimeric G-protein and protein kinase A in Aspergillus nidulans. Curr. Genet. 65 941–952. 10.1007/s00294-019-00944-4 [DOI] [PubMed] [Google Scholar]
- Liu N., Santala S., Stephanopoulos G. (2020). Mixed carbon substrates: a necessary nuisance or a missed opportunity? Curr. Opin. Biotechnol. 62 15–21. 10.1016/j.copbio.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Liu P., Lin A., Zhang G., Zhang J., Chen Y., Shen T., et al. (2019). Enhancement of cellulase production in Trichoderma reesei RUT-C30 by comparative genomic screening. Microb. Cell. Fact. 18:81. 10.1186/s12934-019-1131-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Cheng Y., Cui J., Liu J., Gan L., Zeng B., et al. (2018). Enhancing cellulase and hemicellulase production in Trichoderma orientalis EU7-22 via knockout of the creA. Mol. Biotechnol. 60 55–61. 10.1007/s12033-017-0046-3 [DOI] [PubMed] [Google Scholar]
- Mello-de-Sousa T. M., Gorsche R., Rassinger A., Poças-Fonseca M. J., Mach R. L., Mach-Aigner A. R. (2014). A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei. Biotechnol. Biofuels. 7:129. 10.1186/s13068-014-0129-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakari-Setälä T., Paloheimo M., Kallio J., Vehmaanperä J., Penttilä M., Saloheimo M. (2009). Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl. Environ. Microb. 75 4853–4860. 10.1128/AEM.00282-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen E. V., Imanishi S. Y., Haapaniemi P., Yadav A., Saloheimo M., Corthals G. L., et al. (2016). Quantitative site-specific phosphoproteomics of Trichoderma reesei signaling pathways upon induction of hydrolytic enzyme production. J. Proteome. Res. 15 457–467. 10.1021/acs.jproteome.5b00796 [DOI] [PubMed] [Google Scholar]
- Ortiz-Urquiza A., Keyhani N. O. (2015). Stress response signaling and virulence: insights from entomopathogenic fungi. Curr. Genet. 61 239–249. 10.1007/s00294-014-0439-9 [DOI] [PubMed] [Google Scholar]
- Portnoy T., Margeot A., Linke R., Atanasova L., Fekete E., Sándor E., et al. (2011). The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genom. 12:269. 10.1186/1471-2164-12-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassinger A., Gacek-Matthews A., Strauss J., Mach R. L., Mach-Aigner A. R. (2018). Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 5 1–18. 10.1186/s40694-018-0059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro L. F., Chelius C., Boppidi K. R., Naik N. S., Hossain S., Ramsey J. J., et al. (2019). Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. mBio 10:e02825-18. 10.1128/mBio.02825-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries L., Belshaw N. J., Ilmén M., Penttilä M. E., Alapuranen M., Archer D. B. (2014). The role of CRE1 in nucleosome positioning within the cbh1 promoter and coding regions of Trichoderma reesei. Appl. Microbiol. Biot. 98 749–762. 10.1007/s00253-013-5354-3 [DOI] [PubMed] [Google Scholar]
- Shroff R. A., O’Connor S. M., Hynes M. J., Lockington R. A., Kelly J. M. (1997). Null alleles of creA, the regulator of carbon catabolite repression in Aspergillus nidulans. Fungal. Genet. Biol. 22 28–38. 10.1006/fgbi.1997.0989 [DOI] [PubMed] [Google Scholar]
- Wang F., Zhang R., Han L., Guo W., Du Z., Niu K., et al. (2019). Use of fusion transcription factors to reprogram cellulase transcription and enable efficient cellulase production in Trichoderma reesei. Biotechnol. Biofuels 12 1–12. 10.1186/s13068-019-1589-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang M., Li L., Dong Y., Jiang Y., Liu K., et al. (2017). Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation. Biotechnol. Biofuels 10:99. 10.1186/s13068-017-0789-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. B. (2009). Cellulases and biofuels. Curr. Opin. Biotech. 20 295–299. 10.1016/j.copbio.2009.05.007 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang G., Wang W., Wei D. (2018). Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microbial. Cell. Fact. 17:75. 10.1186/s12934-018-0926-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu Y., Zhang Y., Feng D., Hou S., Guo W., et al. (2019). Identification of a thermostable fungal lytic polysaccharide monooxygenase and evaluation of its effect on lignocellulosic degradation. Appl. Microbiol. Biot. 103:5739. 10.1007/s00253-019-09928-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in NCBI database (https://www.ncbi.nlm.nih.gov/) under the accession numbers: AAB01677.1, EPS28222.1, AAK11189.1, XP_663799.1, AAA32690.1, XP_755510.1, AAT34979.1, AAL33631.4, and EAA32758.1.