Abstract
Background
Impaired or hyperactive pancreas regeneration after injury would cause exocrine insufficiency or recurrent / chronic pancreatitis and potentially carcinogenesis. Macrophages are the most abundant immune cells in the regenerative pancreas, however their phenotype and role remain poorly defined.
Method
Using caerulein-induced acute pancreatitis (AP) model, we examined the dynamic landscape of pancreatic macrophages throughout the acute inflammation to regeneration phases by flow cytometric and RNA-seq analyses. Liposome depletion of macrophages, Il4ra−/− mice as well as inhibitors were used to elucidate the role and regulatory mechanism of macrophages during pancreatic regeneration.
Findings
We found that M1 macrophages dominated in the pro-inflammatory phase of AP, while M2-like macrophages dominated during pancreas repair/regeneration. Depletion of macrophages at early or late regenerative stage dramatically blocked the acinar-ductal metaplasia (ADM) or delayed inflammation resolution, respectively. Moreover, alternative activation of macrophages was partially dependent on IL-4RA signaling, and ECM/AKT activation in pancreatic macrophages facilitated inflammation resolution during tissue regeneration.
Interpretation
Our findings illustrate a dynamic phenotype and function of macrophages during AP repair/regeneration, helping us better understand the mechanism of pancreatic regeneration and providing clues for novel therapeutic strategy.
Keywords: Macrophage; Acute pancreatitis; Pancreatic regeneration; IL-4RA signaling, PI3K-AKT signaling
Abbreviations: AP, acute pancreatitis; ADM, acinar-ductal metaplasia; M1 macrophages, Classically activated macrophages; M2 Macrophages, Alternatively activated macrophages; GSEA, Gene set enrichment analysis
Graphical abstract
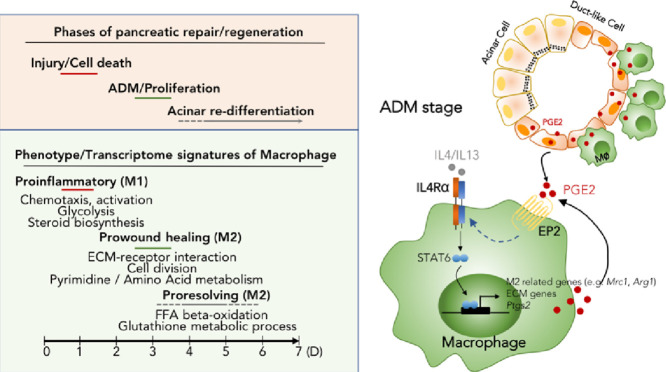
Schematic diagram of dynamic phenotype and function of macrophages following AP injury, and polarization mechanism of M2 macrophages at ADM stage.
Research in Context.
Evidence before this study
Dysregulation of regenerative program following AP injury would cause exocrine insufficiency or recurrent / chronic pancreatitis. Infiltrating leukocytes, particular macrophages, determine the extent of ADM formation.
Added value of this study
We demonstrated dynamic transcriptome profile and function of pancreatic macrophages during pancreas repair/ regeneration post AP injury. Furthermore, IL4RA signaling mediated M2 polarization during ADM stage, limiting the extent of ADM formation. Macrophage-derived PI3K-AKT signaling facilitates inflammation resolution during acinar re-differentiation.
Implications of all the available evidence
Our findings pave the way for the development of macrophage-based therapies to enhance the efficiency of regenerative process, providing a novel strategy for pancreatic disease treatment.
Alt-text: Unlabelled box
1. Introduction
Acute pancreatitis (AP) is one of the most common inflammatory gastrointestinal disorders, appearing to recover via regeneration of exocrine acinar cells both in mouse and human models. Understanding the molecular and cellular processes in restoring pancreatic acinar cell homeostasis is crucial because dysregulation of the regenerative program can lead to exocrine insufficiency, recurrent / chronic inflammation, and potentially carcinogenesis [1], [2], [3]. The regenerative process involves transient phases of inflammation, acinar-to-ductal metaplasia (ADM), and acinar re-differentiation [3], which can be driven by both intrinsic and extrinsic signals. For instance, the acinar cell NF-κB signaling is a critical determinant of pancreatic inflammation and ADM [3], whereas a number of developmental signals (e.g., Hedgehog, Notch, and Hippo) [3], [4], [5] and transcription factors (e.g., Nr5a2, Bmil1, and Ezh2) [6], [7], [8] are devoted to promoting acinar re-differentiation after injury. It has been shown that the crosstalk between acinar cells and infiltrating leukocytes, particularly macrophages, determines the initial severity of injury as well as the extent of ADM formation [9], [10], [11], [12]. A recent study of pancreatitis in Rag1 knockout mice documented a macrophage-dependent regeneration defect caused by depletion of T and B lymphocytes [13]. However, a comprehensive understanding of the dynamic phenotype and function of macrophage during pancreatic regeneration is lacking.
Macrophage is an important driver of inflammatory and tissue regenerative responses triggered by tissue damages, including infection, autoimmune disorders, mechanical or toxic injuries, and other causes [14], [15], [16]. Upon tissue injury, bone marrow (BM)-derived macrophages and/or tissue resident macrophages are activated and act in a pro-inflammatory way in response to local milieu, and then quickly transit into a wound repair phenotype [17,18]. If monocytes/macrophages recruitment or activation is blocked following tissue injury, the early inflammatory response is often diminished [19]. However, impaired or inappropriate macrophage activation may also reduce wound healing and prolonged exposure to proinflammatory stimuli, leading to incomplete tissue regeneration [20]. Hence, macrophages must be well-orchestrated following tissue injury. Macrophages could be simply divided into M1 and M2 subtypes based on stimuli and function in vitro [21]; however, in the physiological or pathological conditions, macrophages in and among tissues are highly heterogeneous with regards to their phenotype and function. Moreover, the polarization of macrophage in specific tissue could be dynamically shaped by their complicated local microenvironment.
The phenotype and role of macrophages have been well-documented in acute and chronic pancreatitis [10, 11, 22, 23], however little is known about how macrophages might regulate pancreas repair / regeneration post injury, including ADM and acinar re-differentiation. Toward this end, we used flow cytometry, immunofluorescence, and RNA sequencing to characterize the dynamic landscape of macrophages following AP injury. Throughout the acute inflammatory to recovery phases, the phenotype of pancreatic macrophage was changed from M1 to M2-like, and M2-like macrophages were the most abundant cells at the stage of acinar-to-ductal metaplasia (ADM). With macrophage depletion experiments, we demonstrated that macrophages play distinct roles in the early and late phases of regeneration respectively. Furthermore, we demonstrated that IL4Rα signaling was critical for appropriate ADM formation post AP injury, and inhibition of PI3K/AKT signaling in macrophages was essential for inflammation resolution during acinar re-differentiation. Herein, delineating the dynamic phenotype and role of macrophage during pancreatic repair /regeneration improves understanding of the pathological process of the disease and provides therapeutic strategy that can either treat and / or prevent the related disorders.
2. Materials and methods
2.1. Mice
Balb/c and C57BL/6 J mice were purchased from Shanghai Silaike Experimental Animals Co, Ltd. B6.SJL (CD45.1) and Il4ra−/− mice were purchased from The Jackson Laboratory and were bred in-house. C57BL/6 J (CD45.1/CD45.2) mice were hybridized from B6.SJL (CD45.1) and C57BL/6 J (CD45.2) mice strains. Mice were sacrificed and harvested at indicated time points post AP. All experimental mice were age- (6–8 weeks) and sex-matched, and animal experiments were approved by the guidelines of the Ren Ji Hospital Institutional Animal Care and Ethics Committee.
2.2. Mixed bone marrow chimeras
Competitive mixed BM (Bone Marrow) chimeric mice were generated by lethally irradiating CCR2WTCD45.1+CD45.2+ mice with 9.5-Gy g radiation in two doses at ~3 h apart, followed by i.v. injection of 5 × 106 BM cells comprising 1:1 mixture of cells from CCR2WTCD45.1+ and CCR2KOCD45.2+ mice. Chimeric mice were left to engraft for at least 8 weeks before further experimental manipulation.
3. Recovery acute pancreatitis (RAP) model and treatments
Acute pancreatitis was induced by caerulein injection in mice as previously described [24]. In brief, mice were given intraperitoneal injections (i.p., 10 times hourly) of saline as control or 100 μg/kg caerulein (Sigma-Aldrich), and then harvested at indicated time points during recovery.
For macrophage depletion, mice were intraperitoneally given 200μl clodronate liposome or control liposome (FormuMax Scientific) once daily and harvested at indicated time points. For macrophage PI3K-AKT inhibition experiment, mice were treated (i.p.) with 5 mg/kg TG100–115 (APExBIO) or DMSO dissolved in 200μl saline twice daily at indicated time points and harvest at D3 or D5 post AP induction.
3.1. Histology and immunofluorescence
Mice were killed by CO2 inhalation, and then their pancreata were rapidly removed. Pancreas pieces were immediately fixed in 10% formalin or frozen in Tissue-Tek OCT compound. Fixed tissues were sectioned and used for hematoxylin and eosin (H&E). Histology score was evaluated as previous reported [13]. In brief, 3–5 random images from every mouse pancreas slide were evaluated. The degree of tissue integrity, ADM and immune cell infiltration were individually scored from 0 to 5, and then were combined as total histology score.
Frozen tissues were sectioned for immunofluorescence staining with F4/80 (Bio-Rad, MCA497GA), AF488-CD206 (BioLegend, clone C068C2), Ki67 (Abcam, ab15580), PGE2 (Abcam, ab2318) and CK19 (DSHB, TROMA-III), and analyzed with confocal microscopy (LSM700; Zeiss).
4. Antibodies and flow cytometry
All antibodies used for flow cytometry were purchased from BioLegend unless indicated. For surface staining, murine cells were stained with the following Abs: APC—CD45.2 (clone 104), AF700-CD45.2 (clone 104), APC/Cy7-CD45.1 (clone A20), PE/Cy7-CD4 (clone RM4–5), Percp/Cy5.5-CD11b (clone M1/70), BV421-F4/80 (clone BM8), APC/Cy7-CD11c (clone N418), AF488-Ly-6C (clone HK1.4), PE-Ly6G (clone 1A8), PE-IL4RA (clone 1015F8), AF488-CD206 (clone C068C2), AF700-MHCII (clone M5/114.15.2), and APC—CCR2 (clone 475,301,R&D Systems). For intracellular staining, cells were stained with surface markers first and then fixed and permeabilized using a Foxp3 Transcription Factor Staining Buffer Set (eBioscience). PE-Ki67 (clone 16A8) and COX-2 (AF4198, R&D Systems) together with donkey anti-goat AF488 (Invitrogen) were used for intracellular staining. For EdU staining, BMDMs (Bone Marrow-Derived Macrophages) were conducted as the manufacturer's instruction of Click-iT™ EdU AF488 Flow Cytometry Assay Kit (Life Technology, C10632). Cells were acquired on a BD LSRFortessa (BD Biosciences) and analyzed with FlowJo software (TreeStar).
4.1. Cell preparation and in vitro cultures
Pancreatic leukocytes were isolated using a collagenase digestion method described previously for flow cytometry analysis [24]. In brief, pancreas was cut into pieces and digested in FACS Buffer (HBSS + 10% FCS) containing 2 mg/ml collagenase type IV. The tissues were incubated in a shaker at 37 °C for 15 min and vortexed at a low speed for 20 s before passing through a 70 μm filter. Murine pancreatic macrophages were enriched by CD45+ magnetic beads (Miltenyi Biotec) and then sorted based on CD11b and F4/80 expression.
For BM cell preparation [24], both ends of the femur and tibia were cut and flushed with a syringe filled with FACS Buffer (HBSS + 10% FCS) to extrude BM cells into a sterile petri dish. Then cells were cultured in cRPMI medium with 50 ng/ml M-CSF for 6 days for BMDMs preparation. For M2 polarization, BMDMs were treated with 10–20 ng/ml IL4/IL13.
The solution of PGE2 (Cayman) was replaced from methyl acetate to DMSO before usage. PGE2 (1 µM, 10 µm), TG100–115(0 µM, 10 µM, 50 µM) and EPi (10 µM, Cayman) were used to treat BMDMs as indicate.
4.2. Quantitative rt-pcr
Pancreas tissues or cells were lysed with Trizol reagent (Invitrogen) for total RNA extraction. Total RNA (5 µg) was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and mRNA expression of the indicated genes was assessed by real-time PCR using FastStart Universal SYBR Green Master (Rox) (Roche) using an ABI StepOnePlus System. Data were analyzed using the 2−ΔCT method, and all gene expression levels were normalized to Gapdh. The following primers were used for quantitative PCR:
mIl4, forward, 5′-GGTCTCAACCCCCAGCTAGT-3′,
mIl4, reverse, 5′-GCCGATGATCTCTCTCAAGTGAT-3′;
mIl13, forward, 5′-CCTGGCTCTTGCTTGCCTT-3′,
mIl13, reverse, 5′-GGTCTTGTGTGATGTTGCTCA-3′;
mIl4ra, forward, 5′-TGGATCTGGGAGCATCAAGGT-3′,
mIl4ra, reverse, 5′-TGGAAGTGCGGATGTAGTCAG-3′;
mMrc1, forward, 5′-CTCTGTTCAGCTATTGGACGC-3′,
mMrc1, reverse, 5′-CGGAATTTCTGGGATTCAGCTTC-3′;
mRelma, forward, 5′-AGGAACTTCTTGCCAATCCA-3′,
mRelma, reverse, 5′-ACAAGCACACCCAGTAGCAG-3′;
mChil3, forward, 5′-CAGGTCTGGCAATTCTTCTGAA-3′,
mChil3, reverse, 5′-GTCTTGCTCATGTGTGTAAGTGA-3′;
mArg1, forward, 5′-CTCCAAGCCAAAGTCCTTAGAG-3′,
mArg1, reverse, 5′-AGGAGCTGTCATTAGGGACATC-3′;
mHbegf, forward, 5′-CGGGGAGTGCAGATACCTG-3′,
mHbegf, reverse, 5′-TTCTCCACTGGTAGAGTCAGC-3′;
mPtgs1, forward, 5′-ATGAGTCGAAGGAGTCTCTCG-3′,
mPtgs1, reverse, 5′-GCACGGATAGTAACAACAGGGA-3′;
mPtgs2, forward, 5′-TTCAACACACTCTATCACTGGC-3′,
mPtgs2, reverse, 5′-AGAAGCGTTTGCGGTACTCAT-3′;
mGapdh, forward, 5′-AGGTCGGTGTGAACGGATTTG-3′,
mGapdh, reverse, 5′-TGTAGACCATGTAGTTGAGGTCA-3′.
5. RNA-seq and analyses
Pancreatic leukocytes were enriched by CD45+ magnetic beads (Miltenyi Biotec) first, then pancreatic macrophages (CD11b+F4/80+ cells) were sorted via flow cytometry based on surface markers. RNA from sorted cells was isolated by TRIzol Reagent (Invitrogen). RNA purity was checked using the Nano Photometer® spectrophotometer (IMPLEN, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for Transcriptome sequencing: A total amount of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations and index codes were added to attribute sequences to each sample. After quality control, we selected Hisat2 as the mapping tool for that Hisat2 can generate a database of splice junctions based on the gene model annotation file and thus a better mapping result than other non-splice mapping tools. Feature Counts v1.5.0-p3 was used to count the reads numbers mapped to each gene. And then, FPKM (fragment per kilo base per million) of each gene was calculated based on the length of the gene and reads count mapped to this gene. An FPKM filtering cutoff of 1.0 in at least one of the six samples was used to determine expressed transcripts. Prior to differential gene expression analysis, for each sequenced library, the read counts were adjusted by edgeR program package through one scaling normalized factor.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes database (KEGG) pathway analysis: The overall DEGs (q-value ≤0.05 and FC ≥ 1.5) were analyzed for enrichment of Gene Ontology (GO) terms (biological process (BP) and molecular function (MF) and Kyoto Encyclopedia of Genes and Genomes database (KEGG) pathways using DAVID Bioinformatics Resources. For GO analysis, values of P ≤ 0.05 identified differentially enriched terms, and genes with a q-value < 0.05 among the DEGs were considered significantly enriched. Significant differentially enriched KEGG pathways were considered as those with a value of P ≤ 0.05.
Short time-series expression miner (STEM) analysis:
The detection of gene clustering profiles were conducted by using the STEM [25, 26] clustering algorithm to identify temporal gene expression profiles during the RAP, with the maximum number of model profiles set to 50 and maximum unit change in model profiles between time points set to 2. The values of the gene expression were normalized following FPKM calculation method. After each gene was assigned to the filtering criteria of model profiles, the correlation coefficient was determined. We performed standard hypothesis testing using the true ordering of time points and determined the p-value using the number of genes assigned to the model profile and the expected number of assigned genes (adjusted p-value, 0.05 by Bonferroni correction). We analyzed all the colored boxes which were statistically significant. To further understand the biological functions of DEGs clustered in STEM, GO and KEGG analyses were also performed using DAVID Bioinformatics Resources.
RNA-seq raw sequence tags and processed bed files have been submitted to the National Center for Biotechnology Gene Expression Omnibus (GEO) database under accession GSE138134.
5.1. Statistical analysis
The unpaired Student t-test or ANOVA was used to determine statistical significance, and a P value <0.05 was considered significant. Values are expressed as mean ± SEM (Prism 6; GraphPad Software). Unless indicated otherwise, the results are from two or three independent experiments.
6. Results
6.1. Phenotypic change of pancreatic macrophage following AP injury
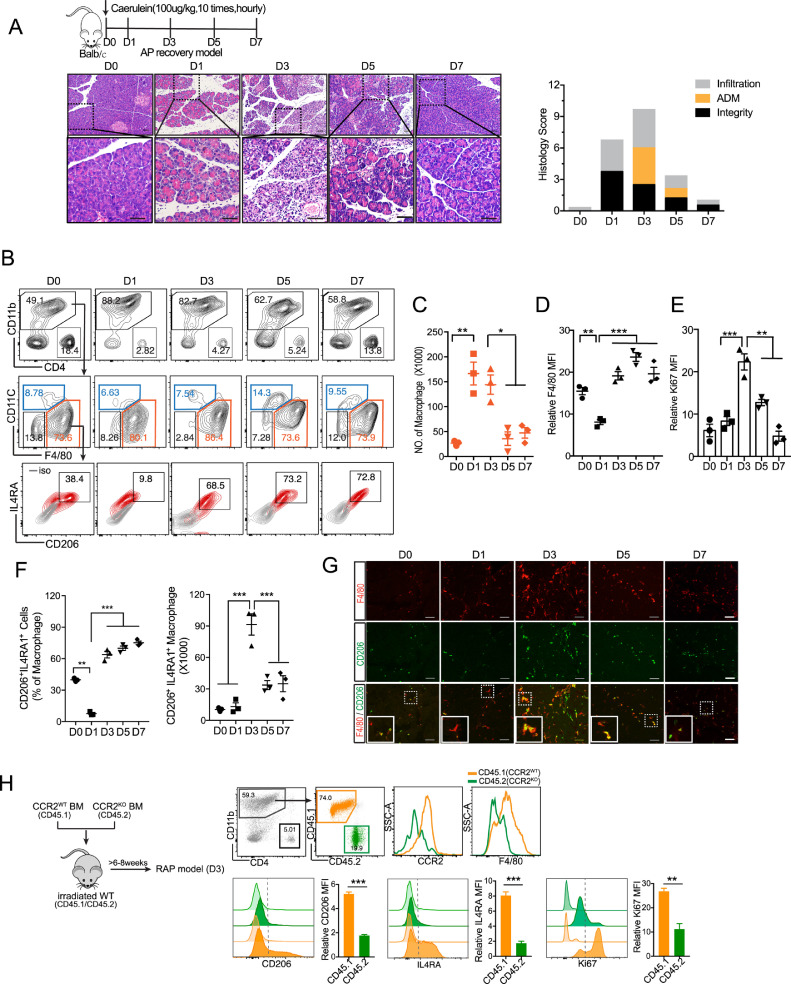
Studies on pathogenesis of inflammation and repair / regeneration in human acute pancreatitis (AP) are limited due to the unavailability of pancreas tissues. Therefore, animal models, especially caerulein-induced AP model, have been well used to investigate the initiation and progression of AP. To investigate the repair / regeneration process following AP, we induced AP with higher dose of caerulein (100 μg / kg, 10 times hourly) and collected the pancreas at the different time points within one week (Fig. 1A). Consistent with previous report [27], mice undergoing caerulein treatment revealed a histological sign of acinar necrosis and leukocyte infiltration as well as elevated serum amylase at 24 h after AP induction (Fig. 1A, and Supplementary Fig. 1A). Typical acinar-to-ductal metaplasia (ADM) structure appeared on day 3, which quickly recover to normal acini around day 7. In addition to ADM, acinar cell proliferation also contributed to pancreas repair / regeneration [28, 29], in which Ki67+ staining peaked on day 3 and gradually dropped as recovery (Supplementary Fig. 1B).
Fig. 1.
Phenotypic change of pancreatic macrophage following AP injury.
(A) Balb/c mice were intraperitoneally injected with 100 μg/kg caerulein (AP) or saline as control (D0) hourly for 10 times, and then sacrificed on D1, D3, D5, D7 post AP induction. Representative H&E staining of the pancreas at indicated time point is shown. Scale bar, 50 µm. (B) Phenotypic change of pancreatic macrophages (CD11b+F4/80+CD11c−) post AP injury by flow cytometric analysis. (C-E) The total number of macrophages, the level of F4/80 and Ki67 in macrophages are shown. (F) Percentage and total number of M2-like macrophages (CD206+IL4RA+) are shown. (G) Representative immunofluorescence images of the pancreas from mice post AP injury stained with F4/80 (macrophage), CD206 (M2 marker) and DAPI (nuclear). Scale bars, 50 µm. (H) CCR2WTCD45.1+CD45.2 + C57BL/6 mice were lethally irradiated and reconstituted with a 1:1 mixture of BM cells from CCR2WTCD45.1+ and CCR2KOCD45.2+mice over 8 weeks. Mice were injected with caerulein and harvested 3 days post AP induction. CD11b+CD4− cells were analyzed using flow cytometry following pancreatic leukocyte isolation. Representative flow cytometry plots and bar graphs depicting the proportion of macrophages and the expression level of F4/80, CD206, IL4RA and Ki67 originating from CCR2WTCD45.1+ versus from CCR2KOCD45.2+ are shown. Data are means ± SEM (n = 3 for each group) *P<0.05, **P<0.01, ***P<0.001(unpaired two-tailed Student's t-test).
Accumulating evidence has shown that the number and activation of macrophages determine the severity of pancreatitis [24, 30, 31]. During the early stage of AP, inflammatory monocytes (CD11b+Ly6ChiCCR2+) are mobilized from bone marrow to inflamed pancreas [24], and then differentiate into M1 polarized macrophages in response to damage-associated molecular patterns (DAMPs) stimulation in a Toll-like receptors (TLRs)-dependent manner [30]. However, little is known about the fate of these macrophages and whether macrophages also contribute to the regeneration/repair process after AP injury. To this end, we isolated and analyzed the pancreatic leukocytes following AP induction (Fig. 1B). Obviously, the most abundant immune cells within inflamed pancreas were macrophages (CD11b+F4/80+CD11c−). The number of pancreatic macrophages analyzed with flow cytometry was markedly increased on day 1 and day 3 (Fig. 1C).
We noticed that pancreatic macrophages were gradually maturated from day 1 to day 3 based on F4/80 level (Fig. 1D). Macrophage accumulation in tissue occurs in two ways: recruitment of monocytes and proliferation of resident macrophages. As previously reported, the majority of macrophages at acute inflammatory phase of AP were derived from the infiltration of monocytes. According to our findings, pancreatic macrophages on day 3 had higher proliferative capacity compared to those on day 1 (Fig. 1E), which might partially explain the increase in macrophages on day 3. Further, flow cytometric data revealed that the percentage of M2-like macrophages (CD206+IL4RA+) was dropped on day 1 (acute inflammatory phase) and increased since day 3 (ADM phase), while the number of these M2-like macrophages peaked on day 3 and quickly decreased afterward (Fig. 1B & F). Moreover, flow cytometric data were consistent with the immunofluorescence analysis, in which F4/80+CD206+ cells were the most abundant on day 3 (Fig. 1G)
Next, we wondered whether these CCR2+ monocytes/macrophages could be reprogrammed to M2-like macrophages following pancreas injury. To this end, we set up a competitive BM chimera by lethally irradiating recipient CCR2WTCD45.1+CD45.2 + C57BL/6 mice and reconstituting them with a 1:1 mixture of BM-derived from CCR2 wildtype (CCR2WTCD45.1+) and CCR2 knockout (CCR2KOCD45.2+) mice, so that macrophages derived from CCR2KO and CCR2WT mice could be distinguished from each other as well as from those of the recipient mice based on the allotypic CD45 markers. Following 8 weeks of engraftment and induction of AP, we found that the proliferative macrophages (Ki67+) on day 3 mainly come from CCR2WT mice. Macrophages from CCR2WT mice exhibited a higher expression level of CD206 and IL4RA when compared to macrophages from CCR2KO mice (Fig. 1H). Taken together, the results indicate that M2-like macrophages at ADM stage are mainly derived from CCR2+ monocytes/macrophages, which are recruited at early stage of AP.
6.2. Distinct role of macrophages at different stages of AP recovery
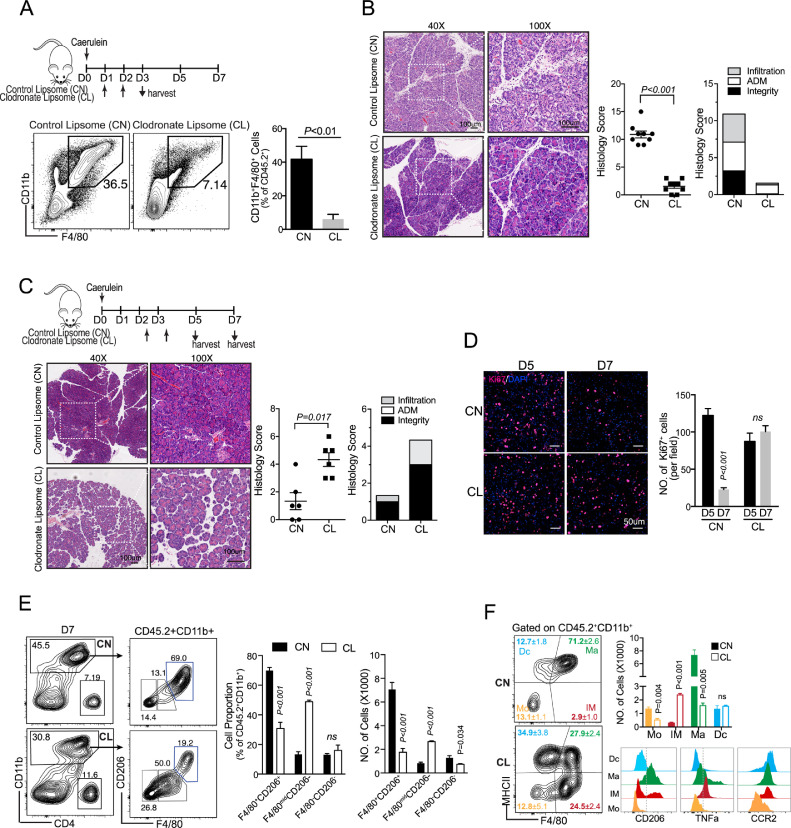
In view of the phenotypic change of macrophage during AP recovery, we next sought to examine the role of macrophages at different stages of repair/ regeneration by clearance of macrophages with liposome. First, we depleted pancreatic macrophages right after acute inflammatory response (from day 1 to day 2) and examined pancreas on day 3. As shown, depletion of macrophages before day 3 could largely reduce duct-like structures, suggesting a key role of macrophages in the formation of ADM (Fig. 2A & B).
Fig. 2.
Distinct roles of macrophage in different stages of AP recovery
(A) Pancreatic macrophages were depleted by intraperitoneal injection of clodronate liposome (CL) or control liposome (CN) prior to ADM formation (on D1 & D2). Representative flow cytometry plots and bar graphs depicting the proportion of macrophages from two groups are shown. (B) Representative of pancreas H&E on D3 from indicated group. Scale bar, 100 µm. (C) Pancreatic macrophages were depleted during ADM (on D2.5&3.5), and mice were harvested on D5 or D7. Representative of pancreas H&E on D7 from indicated group. (D) Representative Ki67 staining of the pancreas from indicated group. The statistics of Ki67+ cells are shown as bar graph. (E) Representative flow cytometry plots and bar graphs depicting the proportion and number of M2-like macrophages (CD11b+F4/80+CD206+), immature macrophages (CD11b+F4/80midCD206−) and monocytes (CD11b+F4/80−CD206−) from CN- and CL- treated groups on D7. (F) Representative flow cytometry plots and bar graphs depicting the proportion and number of macrophages (Ma, F4/80+MHCII+), immature macrophages (IM, F4/80midMHCII−), monocytes (Mo, F4/90-MHCII-) and dendritic cells (Dc, F4/80−MHCII+) among total myeloid cells. The expression level of CD206, TNFα and CCR2 from Dc, Ma, IM and Mo are shown. Data presented as means ± SEM (n = 3–5, unpaired two-tailed Student's t-test).
Next, we found that the depletion of macrophages on day 3 after AP injury significantly delayed pancreatic repair according to histology (Fig. 2C). As previously reported, Ki67+ cell peaked on day 3, and gradually dropped as pancreas recovery (Fig. S1B). In consistent with histological results, we found Ki67+ cells greatly dropped from day 5 to day 7 in CN-treated group, but not in CL-treated group, indicating that the repair process was impaired in CL-treated mice (Fig. 2D). Furthermore, flow cytometric analysis confirmed that CD11b+F4/80+CD206+ macrophages in pancreas were greatly decreased on day 7, which was consistent with Immunofluorescence data (Fig. 2E and Sup Fig. 2). Of total CD11b+ monocytes/macrophages, the mature macrophages (F4/80+MHCIIhi) were reduced, while immature macrophages (F4/80midMHCII−) were increased on day 7 when transient depletion of pancreatic macrophages at ADM stage (day 3) (Fig. 2F). The F4/80midMHCII− macrophages expressed a higher level of CCR2 and TNFα, indicating that they were differentiated from inflammatory monocytes (CD11b+Ly6ChiCCR2+ monocytes) and could cause tissue damage (Fig. 2F). The above results indicated that the depletion of pancreatic macrophages at ADM stage (dominant as M2-like phenotype), will trigger the influx of inflammatory monocytes into pancreas again, thereby causing tissue damage on day 7, as shown in Fig. 2C.
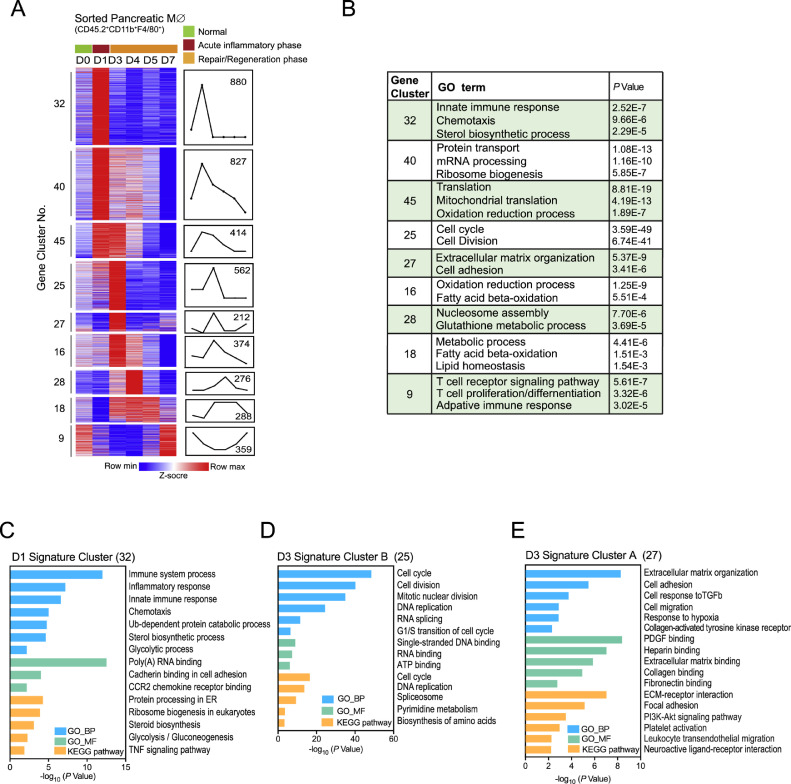
6.3. Dynamic transcriptome profiles of pancreatic macrophage in regenerative pancreas
To further understand the phenotype and function of macrophages during AP recovery, pancreatic macrophages were isolated for RNA-seq analysis. To capture the dynamic of gene expression, we clustered genes according to their expression levels quantified as reads per kilobase per million mapped reads (RPKM) in RNA-seq profiles. Clusters with significance were shown as heatmap (Fig. 3A). To assess the function of these gene clusters, we performed functional annotation analyses using the web tools DAVID Gene Ontology (GO). Enriched signature GO terms of gene clusters are summarized in the table (Fig. 3B). Of note, genes that are highly expressed in acute inflammatory phase (cluster 32, D1 signature cluster) are specifically enriched for inflammatory response and CCR2 dependent chemotaxis (Fig. 3C), which is consistent with our previous finding [24]. Cluster 25 and 27, which contain genes with high expression at ADM stage (day 3), have distinct expression patterns and functions, indicating macrophage heterogeneity in this phase. For example, genes related to ECM-receptor interaction and PI3K-AKT signaling pathway are enriched in cluster 27, while genes related to cell cycle are enriched in cluster 25 (Fig 3. D&E).
Fig. 3.
Dynamic transcriptome profiles of pancreatic macrophage in regenerating pancreas
Pancreatic macrophages (CD11b+F4/80+) from D0 to D7 post AP induction were isolated and sorted for RNA-seq analysis. (A) Heat map of gene expression patterns of differentially transcribed genes from pancreatic macrophages on D0, D1, D3, D4, D5 and D7 post AP induction by STEM clustering analysis. (B) Top enriched GO terms of genes from each gene cluster are summarized in the table. (C-E) GO and KEGG pathway analyses for indicated gene clusters.
Studying the metabolism of macrophages in recent years has emphasized the close link existing between metabolic state and the phenotype/function of those cells. Here in our study, we observed metabolic reprogramming in pancreatic macrophage from acute inflammatory phase to ADM and acinar re-differentiation phase. Consistent with previous reports, activated glycolysis is coming along with M1 polarization of macrophages in acute inflammatory phase. While at ADM stage, activated amino acid metabolism and pyrimidine metabolism in macrophages indicate their M2 polarization and highly proliferative capacity, respectively, which is consistent with our flow cytometric data. During the acinar re-differentiation phase, macrophages exhibit an inflammation resolving phenotype accompanied by activated glutathione metabolism and fatty acid beta-oxidation. Thus, dynamic metabolic profiles of macrophages following AP injury provide a potential strategy for targeting pancreatic macrophages from different stages.
6.4. PI3K-AKT activation in macrophage promotes inflammation resolution after ADM phase
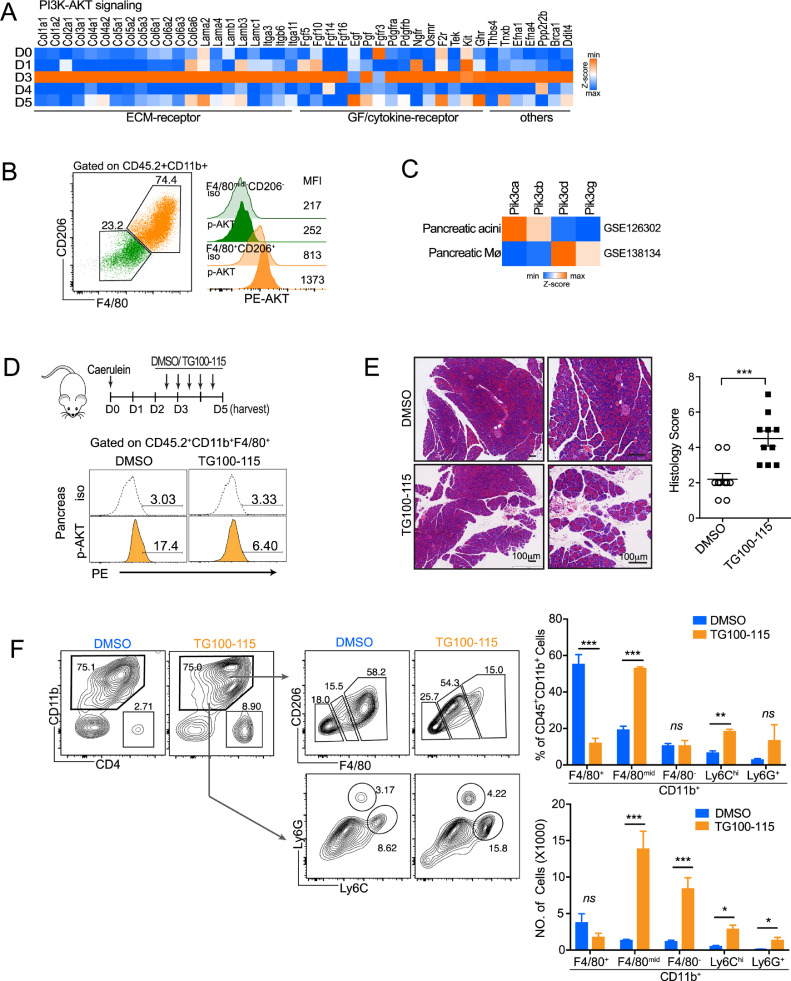
Based on RNA-seq data, PI3K-AKT signaling was significantly enriched in macrophages during ADM. When we took a close look at these elevated genes in PI3K-AKT pathway, we found that more than 80% of genes were related to extracellular matrix (ECM)-receptor interaction, growth factor / cytokine-receptor interaction, indicating their involvement in the crosstalk between macrophages with neighboring cells (Fig. 4A). Meanwhile, the higher AKT activation was observed in CD11b+F4/80+CD206+ pancreatic macrophages on day 3 (Fig. 4B). As extracellular matrix components were involved in wound closure [32], [33], [34], [35], we next sought to investigate whether targeting PI3K-AKT signaling in macrophages would affect AP recovery. Given that PI3K δ/γ (Pik3cd and Pik3cg) were dominantly expressed on pancreatic macrophages (Fig. 4C), a specific inhibitor of PI3K δ/γ (TG100–115) was used to targeting macrophages in the course of ADM. The PI3K-AKT signaling axis has been shown to play roles in macrophage activation and polarization [36, 37]. When inhibiting PI3K-AKT prior to the ADM stage, neither ADM formation nor M2 activation was changed on day 3 (Supplemental Fig. 3). However, when we inhibited PI3K-AKT activation of macrophages at ADM stage, pancreatic repair/regeneration was obviously hindered (Fig. 4D&E). By flow cytometric analysis, we found that the number of M2-like macrophages (CD11b+F4/80+CD206+) in pancreas had less changed, while the number of immature macrophages (CD11b+F4/80mid), inflammatory monocytes (CD11b+Ly6Chi) and neutrophils (CD11b+Ly6G+) were elevated significantly (Fig. 4F). Taken together, blocking PI3K-AKT signaling in pancreatic macrophages will trigger pro-inflammatory response and tissue injury, indicating an anti-inflammatory or pro-resolving phenotype of these macrophages at ADM stage (on day 3).
Fig. 4.
Blocking macrophage PI3K-AKT activation during ADM triggers reflux of inflammatory cells and tissue damage.
(A) Heat map lists normalized gene expression related to PI3K-AKT signaling, enriched in gene cluster 27. (B) The level of p-AKT in pancreatic macrophages on day 3 was analyzed by flow cytometry. (C) Relative expression of Pik3ca, Pik3cb, Pik3cd and Pik3cg in pancreatic acini (GSE126302) and pancreatic macrophage (GSE138134). (D) Mice were administrated with DMSO/TG100–115 (5 mg/kg, i.p., twice daily) since D2.5 post AP induction, and then harvested on D5. The level of p-AKT in pancreatic macrophage on day 3 were shown. (E) Representative H&E staining of pancreas (day 5) from DMSO- or TG100–115- treated mice. Scale bar,100 µm. (F) The proportion and number of CD11b+F4/80mid, CD11b+F4/80−, CD11b+Ly6Chi and CD11b+Ly6G+ cells from different treatment groups are shown. *P<0.05, **P<0.01, ***P<0.001, ns, not significant. Data presented as means ± SEM (unpaired two-tailed Student's t-test).
6.5. Decreased M2-like macrophages and dysregulated regeneration in Il4ra-deficient mice
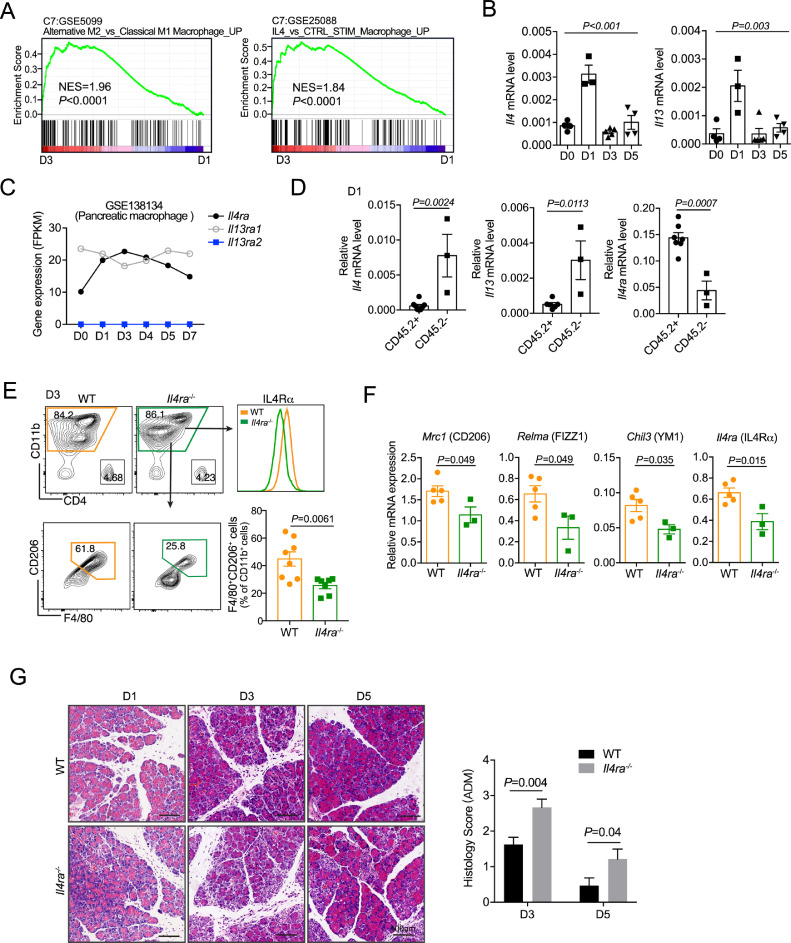
To explore the regulatory mechanisms of pancreatic macrophages during regeneration, we performed gene set enrichment analysis (GSEA) to compare transcriptome profiles of macrophages from day 3 and day 1. In line with our results and previous reports, M2 activation of macrophages and IL4 stimulation signatures were enriched in macrophages from day 3 (Fig. 5A). IL4 and IL13 induce M2 activation via IL4 receptor (a heterodimeric receptor composed of IL4RA and IL13RA1) in a STAT6 dependent manner [38, 39]. Here we found that the expression of Il4 and Il13 were increased on day 1, and the expression of Il4ra was elevated since day 1 and peaked on day 3 (Fig. 5B & C). To further define the cell source of the receptor and ligands, we isolated and compared CD45.2+ and CD45.2− cells from pancreas one day post AP injury. Interestingly, Il4ra1 was dominantly expressed on immune cells (CD45.2+), while Il4 and Il13 were mainly expressed on parenchymal cells (CD45.2−) (Fig. 5D, Supplemental Fig. 4). Furthermore, by using Il4ra-deficient mice, we demonstrated that IL4Rα signaling mediated the polarization of M2 macrophage during pancreatic repair/regeneration after AP injury (Fig. 5F). Notably, the pancreas from Il4ra−/- mice had more ADM structure on day 3 and day 5 as compared with their counterparts (Fig. 5G). Collectively, these findings indicate that IL4/IL4Rα axis is required for M2 polarization of pancreatic macrophages during AP recovery, exerting anti-inflammatory function to control the extent of ADM formation.
Fig. 5.
Decreased M2-like macrophages and dysregulated regeneration in Il4ra-deficient mice
(A) Gene set enrichment analysis (GSEA) of gene expression between day 3(D3) and day 1(D1). (B) Relative mRNA level of Il4 and Il13 in pancreas during AP recovery were detected with qPCR assay. (C) Il4ra, Il13ra1 and Il13ra2 mRNA level in GSE138134 are shown. (D) CD45.2+ and CD45.2− cells from D1 pancreas were sorted respectively for Il4, Il13 and Il4ra expression determination. (E) Wildtype (WT) and IL4Rα deficiency (Il4ra−/−) mice were subjected to AP. Pancreatic leukocytes were isolated 3 days post AP induction for analysis. Representative flow cytometry plots and bar graphs depicting the proportion of F4/80+CD206+ cells in total myeloid cells. (F) Total myeloid cells (CD11b+ cells) were sorted from E for M2-related genes detection. (G) WT and Il4ra−/− mice were subjected to AP, and pancreas was harvested on day1(D1), day 3(D3) and day 5(D5) post AP injury. Representative H&E staining of the pancreas from indicated time points. Scale bar, 50 µm. Data presented as means ± SEM, n = 3–8 mice per group (One-way ANOVA for B; unpaired two-tailed Student's t-test for d-F).
6.6. PGE2 augments IL4Rα signaling to enhance M2 activation of macrophage
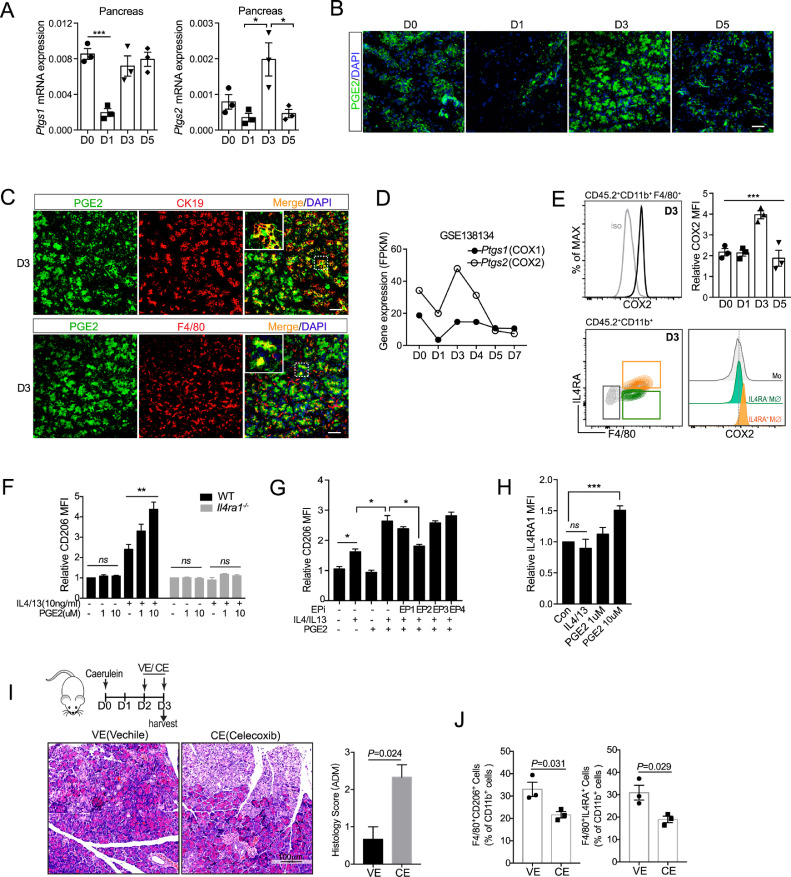
Macrophages are able to switch from one functional phenotype to another in response to variable local microenvironmental signals. Besides canonical signaling cascades (e.g., IL4/IL4Rα), macrophages could integrate metabolic cues triggered by extracellular stimuli for their M2 activation. For example, it has been shown that prostaglandin E2 (PGE2) can act in concert with IL-4 to promote M2 activation through cyclic AMP (cAMP) pathway in adipose tissue [40]. Several studies have documented the role of PGE2 in tissue regeneration [41], [42], [43], [44], [45]. Here we wondered whether PGE2 also participated in M2 polarization of pancreatic macrophages following AP injury. To this end, we first examined the dynamic expression of PGE2 following AP injury. COX1 and COX2 are two key enzymes for producing prostaglandins. Here we found the expression of Ptgs2 (COX2) peaked on day 3, while the expression of Ptgs1 (COX1) dropped on day 1 and back to basal level on day 3 (Fig. 6A). Consistently, immunofluorescence analysis showed that PGE2 was elevated on day 3, and rapidly returned to basal level on day 5 (Fig. 6B). Of note, PGE2 was dominantly co-expressed with duct-like cells (CK19+cells) on day 3, as well as partly co-expressed with macrophages (F4/80+ cells) (Fig. 6C). Furthermore, we found that COX2 expression in macrophages also peaked on day 3 according to our RNA-seq data (Fig. 6D) and flow cytometric analysis (Fig. 6E). More interestingly, IL4Rα+ macrophages expressed a higher level of COX2, as compared to IL4Rα− macrophages and monocytes (Fig. 6E).
Fig. 6.
PGE2 augments IL4Rα signaling to enhance M2 activation of pancreatic macrophages
(A) Ptgs1 and Ptgs2 mRNA expression in pancreas following AP injury (n = 3 for each group). (B) Representative immunofluorescence images of the pancreas from mice post AP injury stained with PGE2 and DAPI. Scale bars, 50 µm. (C) Representative immunofluorescence images of day 3(D3) pancreas co-stained with PGE2, together with CK19 (duct-like cell) or F4/80 (macrophage). (D) Ptgs1 and Ptgs2 mRNA level in pancreatic macrophages during AP recovery are shown (GSE138134). (E) COX2 expression in pancreatic macrophages was analyzed by flow cytometry (n = 3). (F) BMDMs from WT or Il4ra−/− mice were treated with different doses of PGE2 with or without IL4/IL13 (10 ng/ml) for 2 days. Relative CD206 expression was determined by flow cytometry. (G) BMDMs were treated with IL4/IL13, PGE2, as well as EP inhibitor (EPi) for 2 days. Relative CD206 expression was determined by flow cytometry. (H) Relative IL4Rα expression of BMDMs upon indicated treatment for 24 h was examined with flow cytometric assay. (I) Vehicle (VE) or COX2 inhibitor Celecoxib (CE, 20 mg/kg) was administrated via gavage daily from day 2(D2) and harvested on day 3(D3). Representative H&E staining of pancreas from VE- or CE- treated mice. Scale bar,100 µm. (J) Pancreatic leukocytes from 2 indicated groups were isolated and analyzed. Bar graphs depict the proportion of indicated cells. *P<0.05, **P<0.01, ***P<0.001. Data are shown as means ± SEM (One-way ANOVA for E; unpaired two-tailed Student's t-test for A, F-J).
To investigate whether PGE2 contributed to M2 activation, BM-derived macrophages (BMDMs) from wildtype (WT) and Il4ra−/− mice were subjected to IL4/IL13 together with PGE2 in vitro. PGE2 alone did not affect M2 activation, but greatly augmented IL4/IL13-triggered M2 activation (Fig. 6F and Supplementary Fig. 5). More interestingly, IL4/IL13 promoted ptgs2 expression in macrophages, which could be further elevated in the presence of PGE2 (Supplementary Fig. 4). PGE2 functions by binding to 4 different G-coupled receptors named E- prostanoid (EP) 1–4 receptor. The subtype of different receptors determines the specific physiological response in different cell types. By using specific inhibitors for EPs, we determined that PGE2 promoted M2 polarization was mainly mediated by the EP2 receptor (Fig. 6G). Moreover, PGE2 could promote IL4Rα expression, thereby enhancing the IL4/IL4Rα signaling (Fig. 6H). Furthermore, inhibition of PGE2 synthesis right before ADM stage (day 2) largely decreased M2 activation and increased ADM structure in the pancreas on day 3 (Fig. 6I&J). Together, these results suggest that PGE2 released by duct-like cells and pancreatic macrophages on day 3, strengthens M2 activation, as well as limits the ADM formation in the regenerative pancreas.
7. Discussion
Given the central role of macrophages in orchestrating tissue repair in health and disease, therapies aimed at modulating macrophage activation may prove useful in promoting tissue repair, limiting fibrosis and even carcinogenesis. Proof-of-concept studies have shown that manipulating the phenotype of endogenous macrophages may be a promising therapeutic option for improving tissue repair. In addition, cell therapy with exogenous macrophages has proved beneficial for the healing of a variety of tissues. However, there are several limitations for the development of macrophages-based intervention: 1) macrophages exhibit dynamic transitions in phenotype and function as tissue repair progress, and the precise factors regulating these transitions in vivo still remain elusive; 2) the diversity of macrophages varies greatly between different tissues or under different physiological and pathological conditions of the same tissue. Macrophages are most abundant infiltrated immune cells in regenerative pancreas, however their phenotype and role in pancreatic regeneration remains poorly defined. In particular, the pancreas has a unique repair/regeneration process as compared to other tissues, hence may have a more unique mechanism that deserves our in-depth investigation. Here we took flow cytometric analysis and RNA sequencing approach to determine the dynamic phenotype of pancreatic macrophages following AP injury. Transcriptome profiles and functional assays reveal the dynamic phenotype and function of macrophages post AP injury, from pro-inflammatory, pro-wound healing to pro-resolving phases. More important, we explored the signals that had potential to regulate these transitions in vivo. We also revealed that the metabolic state of macrophages are also altered in different phases of AP, this finding provides a new gist for defining macrophages in RAP. Our findings pave the way for development of macrophage-based intervention to enhance the efficiency of regenerative process.
For pancreas, the repair progresses go through overlapping phases of inflammation, proliferation/ADM, and remodeling. Macrophages appear to be important orchestrators and effectors of this progression, altering their function and phenotype to meet the needs of the regenerative process. Macrophages present during a given stage of regeneration orchestrate a transition to the next phase. For instance, macrophages of the inflammatory phase (D1) initiate subsequent cell proliferation/ADM, whereas macrophages of the ADM phase trigger acinar re-differentiation and tissue remodeling. In all, through the current phenotype of macrophage, we could predict the stage and efficiency of tissue repair.
The unique contribution of resident tissue macrophages verse recruited monocytes/macrophages has become an important area of research, as some studies suggest nonredundant role of macrophages from different monocytes/macrophages population. For example, Kupffer cells, resident macrophages of liver, orchestrates liver damages. However, our bone marrow chimera experiment suggested that recruited CCR2+Ly6Chi monocytes/macrophages, instead of resident tissue macrophages, mainly constituted M2-like macrophages with proliferative capacity existing in the ADM phase. After ADM stage, whether there are Ly6C− monocytes or resident macrophages involved in the later stage of pancreatic regeneration still needs further investigation.
Macrophages are highly heterogeneous cells that can rapidly change their activation status and function in response to local microenvironment signals [46]. Our previous studies revealed the distinct role and regulatory mechanism of macrophages in acute and chronic pancreatitis respectively [22, 24, 30]. Upon AP injury, monocytes derived from BM are recruited into inflamed pancreas and then activated into M1 macrophages in response to DAMPs released by necrotic acinar cells. While, in the context of chronic pancreatitis (CP), the crosstalk between macrophages and pancreatic stellate cells (PSCs) promotes their M2 activation via IL4 / IL4Rα axis [22]. In this study, we sought to reveal the mechanism of polarization state transition of macrophage during ADM. ADM is characterized by the formation of duct-like cells in response to pancreatic inflammation. We wondered whether the crosstalk between macrophages and duct-like cells contributed to their polarization. As expected, we found duct-like cells expressed a higher level of PGE2, promoting macrophage M2 activation and extracellular matrix synthesis via augmenting of IL4Rα signals on macrophages. In all, we have defined the distinct mechanism of macrophage polarization in the different pathological stage of pancreas.
The essential role of macrophages in ADM triggered by oncogenic Kras has been well illustrated [9, 47]. However, little is known about the role of macrophages at inflammation-induced ADM and especially the acinar re-differentiation stage. Here we found that elevated PI3K/AKT signals at ADM stage in pancreatic macrophages didn't contribute to their M2 polarization but rather promoted inflammation resolution in the subsequent acinar re-differentiation. Besides, genes involved in extracellular matrix (ECM)-receptor interactions account for the majority genes in elevated PI3K/AKT pathway. Macrophage-derived ECM is for inflammation resolution in tissue regeneration, while during pathologically hyperactivated regenerative processes such as chronic pancreatitis, sustained ECM formation by macrophages would contribute to fibrogenesis. Thus, unraveling the molecular/ cellular bases of exocrine regeneration may better understand the pathological process of chronic pancreatitis.
In summary, our findings highlight the dynamic phenotype and role of macrophages in the process of pancreatic regeneration post AP injury. We elucidated the regulatory mechanism of polarized macrophages during ADM, as well as revealed that macrophage-derived PI3K-AKT signaling was essential for inflammation resolution during acinar re-differentiation. Our data further support the development of macrophage-based therapies to enhance the efficiency of regenerative process as a novel strategy for pancreas disease treatment.
Funding
This work was supported by Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support No.20161312 (J. Xue); The Shanghai Youth Talent Support Program (J. Xue); Shanghai Rising-Star Program No.19QA1408300 (N. Ning); Natural Science Foundation of China No.81702938, No.81770628 and No. 81,970,553 (J. Xue), No.81802307(P. Lu). The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Declaration of interests
All authors have declared that no conflict of interest exists.
Author contributors
J.X., L.P. and NN. N. designed experiment; J.X. and JH. W. interpreted data; JH.W. and L.Z performed most of the experiments; JJ. S., RZ.H., YW. J. assisted in some experiments and data analysis; A.H. assisted in the discussion; J.X. and JH. W. wrote the manuscript; J.X. provided the overall guidance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102920.
Contributor Information
Ningning Niu, Email: niunn2014@outlook.com.
Ping Lu, Email: luping314@126.com.
Jing Xue, Email: jingxue@sjtu.edu.cn, xuejing0904@126.com.
Appendix. Supplementary materials
Reference
- 1.Kolodecik T., Shugrue C., Ashat M., Thrower E.C. Risk factors for pancreatic cancer: underlying mechanisms and potential targets. Front Physiol. 2013;4:415. doi: 10.3389/fphys.2013.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Momi N., Kaur S., Krishn S.R., Batra S.K. Discovering the route from inflammation to pancreatic cancer. Minerva Gastroenterol Dietol. 2012;58(4):283–297. [PMC free article] [PubMed] [Google Scholar]
- 3.Murtaugh L.C., Keefe M.D. Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 2015;77:229–249. doi: 10.1146/annurev-physiol-021014-071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siveke J.T., Lubeseder-Martellato C., Lee M., Mazur P.K., Nakhai H., Radtke F. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134(2):544–555. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Fendrich V., Esni F., Garay M.V., Feldmann G., Habbe N., Jensen J.N. Hedgehog signaling is required for effective regeneration of exocrine pancreas. Gastroenterology. 2008;135(2):621–631. doi: 10.1053/j.gastro.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda A., Morris JPt, Hebrok M. Bmi1 is required for regeneration of the exocrine pancreas in mice. Gastroenterology. 2012;143(3) doi: 10.1053/j.gastro.2012.05.009. 821-31 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallen-St Clair J., Soydaner-Azeloglu R., Lee K.E., Taylor L., Livanos A., Pylayeva-Gupta Y. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012;26(5):439–444. doi: 10.1101/gad.181800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen N.M., Neesse A., Dyck M.L., Steuber B., Koenig A.O., Lubeseder-Martellato C. Context-Dependent Epigenetic Regulation of Nuclear Factor of Activated T Cells 1 in Pancreatic Plasticity. Gastroenterology. 2017;152(6) doi: 10.1053/j.gastro.2017.01.043. 1507-20 e15. [DOI] [PubMed] [Google Scholar]
- 9.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nature reviews Gastroenterology & hepatology. 2017;14(5):296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava P., Bhatia M. Essential role of monocytes and macrophages in the progression of acute pancreatitis. World J. Gastroenterol. 2010;16(32):3995–4002. doi: 10.3748/wjg.v16.i32.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gea-Sorli S., Closa D. Role of macrophages in the progression of acute pancreatitis. World J Gastrointest Pharmacol Ther. 2010;1(5):107–111. doi: 10.4292/wjgpt.v1.i5.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liou G.Y., Doppler H., Necela B., Krishna M., Crawford H.C., Raimondo M. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 2013;202(3):563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folias A.E., Penaranda C., Su A.L., Bluestone J.A., Hebrok M. Aberrant innate immune activation following tissue injury impairs pancreatic regeneration. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannella K.M., Wynn T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017;79:593–617. doi: 10.1146/annurev-physiol-022516-034356. [DOI] [PubMed] [Google Scholar]
- 15.Mescher A.L. Macrophages and fibroblasts during inflammation and tissue repair in models of organ regeneration. Regeneration. 2017;4(2):39–53. doi: 10.1002/reg2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wynn T.A., Vannella K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A., Sinha M., Datta S., Abas M., Chaffee S., Sen C.K. Monocyte and macrophage plasticity in tissue repair and regeneration. Am. J. Pathol. 2015;185(10):2596–2606. doi: 10.1016/j.ajpath.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X., Leak R.K., Shi Y., Suenaga J., Gao Y., Zheng P. Microglial and macrophage polarization-new prospects for brain repair. Nature reviews Neurology. 2015;11(1):56–64. doi: 10.1038/nrneurol.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffield J.S., Forbes S.J., Constandinou C.M., Clay S., Partolina M., Vuthoori S. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115(1):56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna S., Biswas S., Shang Y., Collard E., Azad A., Kauh C. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLoS ONE. 2010;5(3):e9539. doi: 10.1371/journal.pone.0009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue J., Sharma V., Hsieh M.H., Chawla A., Murali R., Pandol S.J. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat Commun. 2015;6:7158. doi: 10.1038/ncomms8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Xue J., Jaffee E.M., Habtezion A. Role of immune cells and immune-based therapies in pancreatitis and pancreatic ductal adenocarcinoma. Gastroenterology. 2013;144(6):1230–1240. doi: 10.1053/j.gastro.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J., Zhang R., Hu G., Zhu H.H., Gao W.Q., Xue J. Carbon Monoxide Impairs CD11b(+)Ly-6C(hi) Monocyte Migration from the Blood to Inflamed Pancreas via Inhibition of the CCL2/CCR2 Axis. J Immunol. 2018;200(6):2104–2114. doi: 10.4049/jimmunol.1701169. [DOI] [PubMed] [Google Scholar]
- 25.Ernst J., Nau G.J., Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. 2005;21(Suppl 1):i159–i168. doi: 10.1093/bioinformatics/bti1022. [DOI] [PubMed] [Google Scholar]
- 26.Ernst J., Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerch M.M., Gorelick F.S. Models of acute and chronic pancreatitis. Gastroenterology. 2013;144(6):1180–1193. doi: 10.1053/j.gastro.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Q., Melton D.A. Pancreas regeneration. Nature. 2018;557(7705):351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai B.M., Oliver-Krasinski J., De Leon D.D., Farzad C., Hong N., Leach S.D. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet beta cell, regeneration. J Clin Invest. 2007;117(4):971–977. doi: 10.1172/JCI29988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue J., Habtezion A. Carbon monoxide-based therapy ameliorates acute pancreatitis via TLR4 inhibition. J Clin Invest. 2014;124(1):437–447. doi: 10.1172/JCI71362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sendler M., Weiss F.U., Golchert J., Homuth G., van den Brandt C., Mahajan U.M. Cathepsin B-Mediated Activation of Trypsinogen in Endocytosing Macrophages Increases Severity of Pancreatitis in Mice. Gastroenterology. 2018;154(3) doi: 10.1053/j.gastro.2017.10.018. 704-18 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahdavian Delavary B., van der Veer W.M., van Egmond M., Niessen F.B., Beelen R.H. Macrophages in skin injury and repair. Immunobiology. 2011;216(7):753–762. doi: 10.1016/j.imbio.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Xue M., Jackson C.J. Extracellular Matrix Reorganization During Wound Healing and Its Impact on Abnormal Scarring. Adv Wound Care (New Rochelle) 2015;4(3):119–136. doi: 10.1089/wound.2013.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maquart F.X., Monboisse J.C. Extracellular matrix and wound healing. Pathol. Biol. 2014;62(2):91–95. doi: 10.1016/j.patbio.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. Wound repair and regeneration. Nature. 2008;453(7193):314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 36.Vergadi E., Ieronymaki E., Lyroni K., Vaporidi K., Tsatsanis C. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol. 2017;198(3):1006–1014. doi: 10.4049/jimmunol.1601515. [DOI] [PubMed] [Google Scholar]
- 37.Luyendyk J.P., Schabbauer G.A., Tencati M., Holscher T., Pawlinski R., Mackman N. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol. 2008;180(6):4218–4226. doi: 10.4049/jimmunol.180.6.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Dyken S.J., Locksley R.M. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji Y., Sun S., Xu A., Bhargava P., Yang L., Lam K.S. Activation of natural killer T cells promotes M2 Macrophage polarization in adipose tissue and improves systemic glucose tolerance via interleukin-4 (IL-4)/STAT6 protein signaling axis in obesity. J Biol Chem. 2012;287(17):13561–13571. doi: 10.1074/jbc.M112.350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luan B., Yoon Y.S., Le Lay J., Kaestner K.H., Hedrick S., Montminy M. CREB pathway links PGE2 signaling with macrophage polarization. Proc Natl Acad Sci U S A. 2015;112(51):15642–15647. doi: 10.1073/pnas.1519644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuka S., Aoyama T., Furu M., Ito K., Jin Y., Nasu A. PGE2 signal via EP2 receptors evoked by a selective agonist enhances regeneration of injured articular cartilage. Osteoarthritis and cartilage. 2009;17(4):529–538. doi: 10.1016/j.joca.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Desai A., Yang S.Y., Bae K.B., Antczak M.I., Fink S.P. TISSUE REGENERATION. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science. 2015;348(6240):aaa2340. doi: 10.1126/science.aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter R.L., Georger M.A., Bromberg O., McGrath K.E., Frisch B.J., Becker M.W. Prostaglandin E2 increases hematopoietic stem cell survival and accelerates hematopoietic recovery after radiation injury. Stem Cells. 2013;31(2):372–383. doi: 10.1002/stem.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelus L.M., Hoggatt J., Singh P. Pulse exposure of haematopoietic grafts to prostaglandin E2 in vitro facilitates engraftment and recovery. Cell Prolif. 2011;44(Suppl 1):22–29. doi: 10.1111/j.1365-2184.2010.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Hu B., Lv X., Zhu S., Zhen G., Wan M. Prostaglandin E2 mediates sensory nerve regulation of bone homeostasis. Nat Commun. 2019;10(1):181. doi: 10.1038/s41467-018-08097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liou G.Y., Doppler H., Necela B., Edenfield B., Zhang L., Dawson D.W. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5(1):52–63. doi: 10.1158/2159-8290.CD-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.