Abstract
Snake venom metalloproteinases (SVMPs) represent a diverse group of multi-domain proteins with several biological activities such as the ability to induce hemorrhage, proteolytic degradation of fibrinogen and fibrin, induction of apoptosis and inhibition of platelet aggregation. Due to these activities, SVMPs are responsible for many of the well-known pathological phenotypes in snake envenomations caused particularly by species from the Viperidae family and the Crotalinae subfamily. These proteins have been classified based on their size and domain structure into P–I, P-II and P-III classes. Comparatively, members of the P–I SVMPs possess the simplest structures, formed by the catalytic metalloproteinase domain only; the P-II SVMPs are moderately more complex, having the canonical disintegrin domain in addition to the metalloproteinase domain; members of the P-III class are more structurally varied, comprising the metalloproteinase, disintegrin-like, and cysteine-rich domains. Proteolytic cleavage, repeated domain loss and presence of other ancillary domains are responsible for structural diversities in the P-III class. However, studies continue to unveil the relationship between the structure and function of these proteins. In this review, we recovered evidences from literature on the structural peculiarities and functional classification of Snake Venom Metalloproteinases. In addition, we reflect on diversities that exist among each class while taking into account specific and up-to-date class-based activities.
Keywords: Metalloproteinase, Snake venom, Integrin-binding motif, metalloproteinase, Disintegrin
In this review we highlight the main studies on SVMPs activities and the structure-function relationship of the protein domains.
1. Snake Venom Metalloproteinases (SVMPs)
Snake Venom Metalloproteinases (SVMPs) are zinc-dependent enzymes that have been described in the venoms of snakes, particularly from species in the Viperidae family and Crotalinae subfamily (Fox and Serrano, 2009, 2008a, 2005). Additionally, the distribution of SVMPs can be extended to the venoms of Elapidae, Atractaspididae and Colubridae snake families, although in these families, SVMPs are found in smaller concentrations (Ching et al., 2006; Georgieva et al., 2011; Kulkeaw et al., 2007; Quinton et al., 2005; Tan et al., 2019, 2015). Recent studies have showed that the presence of SVMPs in certain venoms could be related to environmental features such as the climate, which in turn influences snake foraging strategies (Vidal et al., 2019; Zancolli et al., 2019). Snakes that live in an environment with milder winters and less seasonal variation in temperature have a haemotoxic venom mostly composed by SVMPs. These data indicate that epigenetic factors influence the evolution of toxins, having an impact in the field of snake genomics and consequently affecting the relationship between SVMPs structures and its functions.
Features such as variation in domain composition, functional diversity and possibility of class recombination of SVMPs have frustrated previous attempts to resolve and harmoniously incorporate newly discovered SVMPs into the known classes. For instance, the initial classification of SVMPs divided them into four classes based on the cDNA/mRNA sequences of their precursors (N-classification or Nucleotide-based classification) or the size of the mature proteins purified from the snake venoms (P-classification or Protein-based classification) (Bjarnason and Fox, 1994; Hite et al., 1994). The N–I or Metalloproteinase class I has the simplest sequence, which is encoded by a signal sequence, a pro-domain and a metalloproteinase domain. The N–I transcripts resulted in the P–I class of proteins that had only the metalloproteinase domain. The N-II class encoded a signal, a pro-domain, a metalloproteinase and a disintegrin domains giving rise to the mature proteins of the P-II class (metalloproteinase and disintegrin domains). Differently, the N-III class encoded a disintegrin-like domain instead of the disintegrin domain and an additional cysteine-rich domain. The N-III nucleotide sequences resulted in the mature proteins of the P-III class. The last class, the N-IV, encompassed an additional lectin-like domain, giving rise to P-IV proteins (Bjarnason and Fox, 1994; Hite et al., 1994). However, N-IV transcripts have never been found in the transcriptomes of snake venom glands, so this classification is no longer used (Bjarnason and Fox, 1994; Moura-da-Silva et al., 2011).
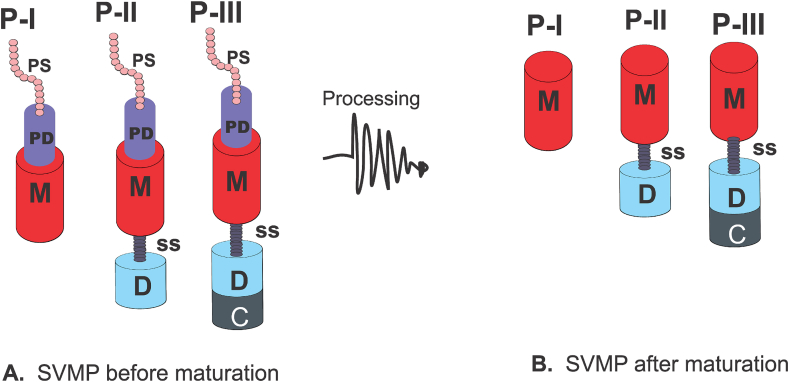
The most adopted SVMP classification dates from 2008 and it classifies the metalloproteinases into three classes: P–I (20–30 kDa) containing a metalloproteinase domain in its mature form; P-II (30–60 kDa; P-IIa, P-IIb, P-IIc, P-IId and P-IIe) containing a disintegrin domain linked to the C-terminus of the metalloproteinase domain, and P-III (60–100 kDa; P-IIIa, P-IIIb, P-IIIc and P-IIId) consisting of a metalloproteinase domain, a disintegrin-like and a cysteine rich domains (Fox and Serrano, 2008b). The lack of proteomic evidence and the non-observation of a P-IV mRNA transcript led to the merge of the P-IV class into the P-III class (P-IIId) with the suggestion that the P-IV class represents a post translational modification of the accepted P-III structure with the addition of a lectin-like domain (Fox and Serrano, 2008b, 2009; Markland and Swenson, 2013). Fig. 1 shows a general scheme of the canonical structures peculiar to each SVMP class before and after maturation.
Fig. 1.
A general scheme of the canonical structures peculiar to each SVMP class. 1A depicts immature SVMPs and it encompasses a P–I class containing peptide signal sequence (PS), pro-domain (PD) and metalloproteinase (M) domain; P-II consist of PS, PD, M, spacer sequence (SS) and disintegrin (D) domain; and P-III contains PS, PD, M, SS, disintegrin-like (D) and cysteine rich (C) domains. 1B delineates processed SVMPs (without PS and PD). Thus, canonical structure of the matured P–I consists of only the M domain; P-II contains M, SS and D domains; and P-III is comprised of M, SS, D and C domains.
1.1. SVMPs classification
1.1.1. The P–I SVMPs
The P–I class includes the smallest and simplest metalloproteinases with molecular masses ranging from 20 to 30 kDa and the presence of only a metalloproteinase (M) domain when isolated from the snake venom (Markland and Swenson, 2013; Sani et al., 2019). The M domain has a conserved zinc-binding sequence (HEXXHXXGXXH) followed by a conserved “Methionine-turn” motif (Da Silva et al., 2012; Herrera et al., 2015). P–I members have the ability to degrade basement membrane components, which corresponds to their proteolytic, hemorrhagic and edema forming activities (Preciado et al., 2019; Suvilesh et al., 2017). However, as P–I SVMPs only have the M domain, studies demonstrated that they have a diffuse localization in the extracellular matrix, and consequently a less toxic effect than P-II or P-III SVMPs (Suvilesh et al., 2017).
Many studies have characterized proteins belonging to the P–I class, including their crystal structures (Akao et al., 2010; Bernardes et al., 2013; Chou et al., 2013; Ferreira et al., 2009; Gomis-Rüth et al., 1994, 1993; Gong et al., 1998; Huang et al., 2002; Kumasaka et al., 1996; Lingott et al., 2009; Lou et al., 2005; Patiño et al., 2010; Watanabe et al., 2009; Zhang et al., 1994). Shannon et al. (1989) and Takeya et al. (1989) were the first to report the complete sequences of two representatives of the P–I class: Ht-d and H2-proteinase (Shannon et al., 1989; Takeya et al., 1989). Early characterization of Ht-d, a hemorrhagic metalloproteinase from the venom of the western diamond-black rattlesnake Crotalus atrox, showed that this P–I representative has a molecular mass of 23,234 Da (Shannon et al., 1989). In the same year, structural analysis of peptides resulting from digests with cyanogen bromide, lysyl endopeptidase, trypsin, Staphylococcus aureus V8 protease and thermolysin revealed the primary structure and the position of the disulfide linkage in the non-hemorrhagic H2-proteinase from habu snake (Trimeresurus flavoviridis) (Takeya et al., 1989). The same study also demonstrated that the P–I H2-proteinase is a non-glycosylated single chain polypeptide with an approximate size of 23 kDa (22,991 Da) and 201 amino acid residues.
In general, P–I members present high similarity in their structures, but a wide variation regarding their hemorrhagic activity (Camacho et al., 2019). Data from molecular dynamic simulations showed differences in the structures of hemorrhagic and non-hemorrhagic P–I SVMPs (Wallnoefer et al., 2010). The main difference was associated with the flexibility and rigidity of the loop near the active site. Briefly, non-hemorrhagic P–I members have great flexibility in the loop region after the Met-turn whereas hemorrhagic P–I members have higher flexibility in the loop area before the Met-turn. This differential flexibility can be responsible for the unequal hemorrhagic activity found in this class, having a role in protein-protein interaction at the basement membrane (Camacho et al., 2019; Wallnoefer et al., 2010).
An extensive review that punctuated the structure and biological activity of SVMPs mentioned the characterization of the crystal structures of 10 members of the P–I class: Atrolysin C, Acutolysin A, BapI, Fibrolase, HT-2, Atroxase, LHF-II, H2-proteinase, HR2A and Gramminelysin I; whose biological activities include hemorrhage, mionecrosis, inflammation, fibrinolysis, non-hemorrhagic proteolysis, and apoptosis (Fox and Serrano, 2005). After this review, other P–I members were characterized such as the non-hemorrhagic FII from Agkistrodon acutus (Lou et al., 2005), the non-hemorrhagic Leucurolysin-A from Bothrops leucurus (Ferreira et al., 2009), Batx-I from Bothrops atrox (Patiño et al., 2010), BpirMP from Bothrops pirajai (Bernardes et al., 2013), the hemmorrhagic okinalysin from Ovophis okinavensis (Komori et al., 2014), the fibrinolytic and hemorrhagic proteins jararafibrase III (21,400 ± 500 Da) and jararafibrase IV (21,200 ± 400 Da) from the venom of Bothrops jararaca (Maruyama et al.), bothrojararactivase (22, 829 Da), BJ-PI (~ 25 kDa) and Bj-PI2 (28 kDa) also from B. jararaca venom (Berger et al., 2008; Da Silva et al., 2012; Oliveira et al., 2009). Bothrojaractivase stands out due to its pronounced prothrombin-activating activity, while BJ-PI displays high fibrinogenolytic and caseinolytic effect. Bj-PI2 lacks myonecrotic and hemorrhagic activity although it has the ability to recruit inflammatory cells and degrade fibrin (Berger et al., 2008; Da Silva et al., 2012; Oliveira et al., 2009). In another study, two P–I SVMPs, BpMP-I and BpMP-II, were characterized from the venom of Bothrops pauloensis. The non-hemorrhagic BpMP-I is a 20 kDa P–I that exhibits fibrinogenolytic activity while BpMP-II (23 kDa) was observed to inhibit cell adhesion and angiogenesis (Naves de Souza et al., 2012). Although there was no positive correlation between the presence of BpMP-II and local tissue hemorrhage, its azocaseinolytic activity has also been reported (Achê et al., 2015). Table 1 describes a list of the most well-known P–I SVMPs.
Table 1.
P–I SVMPs representatives and their reported activities.
| SVMP (P–I) | Molecular Mass (kDa) | Snake Species | Reported Activities | References | |
|---|---|---|---|---|---|
| 1. | HR2A | 23.02 | Trimeresurus flavoviridis | Hemorrhagic | Miyata et al., 1989, Takahashi and Ohsaka, 1970 |
| 2. | HT-2 | 25 | Crotalus ruber | Hemorrhagic and fibrinogenolytic | Mori et al. (1987) |
| 3. | HT-3 | 25.50 | Crotalus ruber | Hemorrhagic and fibrinogenolytic | Mori et al. (1987) |
| 4. | Atroxase | 23.50 | Crotalus atrox | Non-hemorrhagic and fibrinolytic | Willis and Tu (1988) |
| 5. | Ht-d | 23.23 | Crotalus atrox | Hemorrhagic (degradation of fibronectin, laminin, type IV collagen, nidogen, and gelatins) | Shannon et al. (1989); Uniprot |
| 6. | H2-proteinase | 22.99 | Trimeresurus flavoviridis | Non-hemorrrhagic and proteolytic | Takeya et al. (1989) |
| 7. | LHF-II | 22.3 | Lachesis muta | Hemorrhagic, caseinolytic fibrinolytic and fibrinogenolytic | Sanchez et al. (2016) |
| 8. | Jararafibrase III | 21.4 | Bothrops jararaca | Fibrinolytic and hemorrhagic | Maruyama et al. (1993) |
| 9. | Jararafibrase IV | 21.2 | Bothrops jararaca | Fibrinolytic and hemorrhagic | Maruyama et al. (1993) |
| 10. | Adamalysin II | 24 | Crotalus adamenteus | Endopeptidase, non-hemorrhagic, and inactivates serpins by limited proteolysis of their reactive-site loops | Gomis-Rüth et al. (1993); Uniprot |
| 11. | BapI | 24 | Bothrops asper | Weakly hemorrhagic, fibrinogenolytic, caseinolytic and fibrinolytic | Gutiérrez et al. (1995) |
| 12. | Acutolysin A | 22 | Agkistrodon acutus | Hemorrhagic | Gong et al. (1998) |
| 13. | GramminelysinI | 27.02 | Trimeresurus gramineus | Fibrinogenolytic and apoptotic | Wu et al. (2001) |
| 14. | EoVMP1 | 24 | Echis ocellatus | Non-hemorrhagic and activates prothrombin (procoagulant) | Howes et al., 2003, Howes et al., 2005 |
| 15. | Agkislysin | 22 | Agkistrodon acutus | Fibrinogenolytic, fibrinolytic and prothrombin-activating | Wang et al. (2004) |
| 16. | Insularinase | 22.64 | Bothrops insularis | Fibrinogenolytic, fibrinolytic and prothrombin-activating | de Albuquerque Modesto et al. (2005) |
| 17. | FII | 26 kDa | Agkistrodon acutus | Non-hemorrhagic | Lou et al. (2005) |
| 18. | BjussuMP-II | 24 | Bothrops jararacussu | Antiplatelet, non-hemorrhagic, gelatinolytic, collagenolytic, fibrinolytic and fibrinogenolytic | Marcussi et al. (2007) |
| 19. | BnP1 | 24 | Bothrops neuwendi | Fibrinolytic and fibrinogenolytic |
Baldo et al. (2008) |
| 20. | Bothrojararactivase | 22.83 | Bothrops jararaca | Prothrombin-activating | Berger et al. (2008) |
| 21. | Bj-PI | ~ 25 | Bothrops jararaca | Fibrinogenolytic and caseinolytic | Oliveira et al. (2009) |
| 22. | Leucurolysin-A | 23 | Bothrops leucurus | Non-hemorrhagic and non-glycosylated-fibrinogenase | Ferreira et al. (2009) |
| 23. | CcH1 | 25 | Cerastes cerastes | Hemorrhagic | Boukhalfa-Abib et al. (2009) |
| 24. | Batx-1 | 23.3 | Bothrops atrox | Fibrinogenolytic and Hemorrhagic |
Patiño et al. (2010) |
| 25. | Bj-PI2 | 28.08 | Bothrops jararaca | Non-hemorrhagic and fibrinolytic. It can recruit inflammatory cells | Da Silva et al. (2012) |
| 26. | BpMP-I | 20 | Bothropoides pauloensis | Non-hemorrrhagic and fibrinogenolytic | Naves de Souza et al. (2012) |
| 27. | BpirMP | 23.1 | Bothrops pirajai | Thrombocytic, fibrinogenolytic and fibrinolytic |
Bernardes et al. (2013) |
| 28. | BmooMPα-I | 22.6 | Bothrops moojeni | Kininogenase | Okamoto et al. (2014) |
| 29. | Okinalysin | 22 | Ovophis okinavensis | Caseinolytic and hemorrhagic | Komori et al. (2014) |
| 30. | BpMP-II | 23 | Bothropoides pauloensis | Azocaseinolytic. It can inhibit cell adhesion and angiogenesis |
Achê et al. (2015) |
| 31. | Bar-I | 23.39 | Bothrops barnetti | Non-hemorrhagic, fibrinogenolytic and fibrinolytic | Sanchez et al. (2016) |
1.2. The P-II SVMPs
The P-II class is structurally more complex than the P–I class but less than those of the P-III class. Several studies from the cDNA sequences have shown that the P-II class contains multiple domains including a pro-domain (PD; non-matured proteins), a metalloproteinase (M) domain and a disintegrin (D) domain (Camacho et al., 2014; Chen et al., 2003; Fox and Serrano, 2009, 2008b; 2005; Nikai et al., 2000; Takeda et al., 2012). The derived P-II members have been only purified from the venoms of snakes belonging to the Viperidae family, which suggests that they emerged from a P-III SVMP precursor that lost its cysteine-rich domain, after the divergence of Viperidae from the Elapidae family (Carbajo et al., 2015; Sanz and Calvete, 2016). Additionally, the disintegrin domain of P-II SVMPs may have been lost on diverse moments, forming the P–I SVMPs (Casewell et al., 2011).
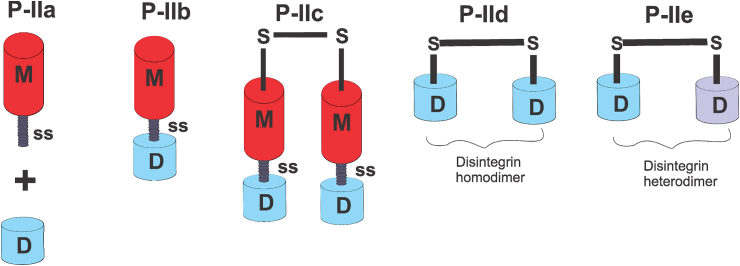
P-II class members have molecular masses ranging from 30 to 60 kDa and a number of proteins belonging to this class have already been described (Casewell et al., 2011). Although the arrangement of the domains is the same in all the P-II members, there is a variation in the length of the spacer sequence between the metalloproteinase and disintegrin domains and in the pattern of cysteines. P-II SVMPs possess 12 cysteine residues that are uniquely patterned, but some members such as agkistin (Wang et al., 2003) and albolamin (Jangprasert and Rojnuckarin, 2014) have two additional cysteine residues. A report in the literature revealed that the disintegrin domain, which abutted the C-terminal segment of the metalloproteinase domain could be vulnerable to proteolytic release (Hite et al., 1992). However, this proteolytic processing could be prevented by the presence of 2-additional cysteine residues capable of forming disulfide bonds, which gives stability to the structure, thereby hindering auto-proteolysis of the C-terminal segment of the metalloproteinase domain (Fox and Serrano, 2005). Additionally, the pattern of cysteine in different domains of P-II SVMPs can be responsible for their diversities as such pattern, in addition to stability, may contribute to unique folding and dimerization. As previously noted, the current classification scheme (Fig. 2) recognizes 5 sub-classes of P-II SVMPs including P-IIa, P-IIb, P-IIc, P-IId and P-IIe (Fox and Serrano, 2008b).
Fig. 2.
Sub-classification of matured P-II SVMPs. The P-IIa consists of a processed fragment of metalloproteinase (M) and another fragment of disintegrin (D). The P-IIb is canonical and it is comprised of M, spacer sequence (SS) and D domain. Members of the P-IIc sub-class are dimers; each monomer is a canonical form. Dimerization of two identical disintegrins give rises to the P-IId sub-class; whereas in the P-IIe, non-identical disintegrins form dimers.
One unique feature of the members of the P-II class is the presence of an integrin-binding motif usually called RGD-motif (RGD stands for arginine-glycine-aspartate). The RGD-motif is not totally conserved in all P-II members. There are copious reports of RGD variants such as KGD (lysine-glycine-aspartate), VGD (valine-glycine-aspartate), WGD (tryptophan-glycine-aspartate), MLD (methionine-leucine-aspartate), RTS (arginine-threonine-serine), KTS (lysine-threonine-serine) and so on (Calvete et al., 2005; Cesar et al., 2019; Rivas-Mercado and Garza-Ocañas, 2017). The RGD-motif is embedded in a flexible loop constituted by the oligopeptide chain of 13 aminoacyl residues, which interact significantly with the integrin αIIBβ3/GpIIb/IIIa inhibiting platelet aggregation (Chung et al., 1999; Masuda et al., 2000).
In some P-II structures, additional intra-chain interaction culminate to structural reinforcement and stability, thus, preventing the proteolytic release of their disintegrins. Sequence alignment and phylogenetic analysis revealed this feature in jerdonitin, a 36 kDa protein purified from the venom of Trimeresurus jerdonii (Chen et al., 2003). Additionally, the presence of the RGD-motif in jerdonitin enhanced its ability to inhibit ADP-induced human platelet aggregation in a dose-dependent fashion (Chen et al., 2003). In another report, two P-II proteins, TJM-1 and TJM-2, were isolated from the venom of Trimeresurus jerdonii and comparative analysis has revealed the presence of additional Cys-407 and Cys-426 in TJM-2. More recently, Jangprasert and Rojnuckarin (2014) reported the cloning of albolamin from a cDNA library obtained from the green pit viper Cryptelytrops albolaris venom gland (Jangprasert and Rojnuckarin, 2014). Western blot probe using anti-polyhistidine antibody revealed that the recombinant protein has a molecular mass of approximately 35 kDa and it belongs to the P-II class SVMP. In addition to the conserved RGD-integrin binding motif present in the disintegrin domain, albolamin also contains two additional cysteine residues in its structure, which may prevent auto-proteolysis of the protein (Jangprasert and Rojnuckarin, 2014). Activity report showed that albolamin could degrade human collagen type IV and inhibit collagen-induced platelet aggregation in a dose dependent manner (Jangprasert and Rojnuckarin, 2014).
Continuous advancements in recombinant DNA technology have provided scientists with tools for the characterization of more protein structures. For instance, there was the successful cloning of the ahpfibrase from the venom of Gloydius halys (Zhang et al., 2010). This P-II protein has 32 kDa and contains the canonical RGD-motif, which is responsible for platelet aggregation inhibition, besides its fibrinolytic activity (Zhang et al., 2010). In addition, Wang et al. (2003) cloned the P-II agkistin from the venom of Agkistrodon halys and demonstrated that this protein of 57 kDa (as shown by Western blot profiling) inhibited ADP-induced platelet aggregation and induced apoptosis in Human Uterine Microvascular Endothelial Cells (HUMEC) (Wang et al., 2003). Sequence determination and structural analysis revealed the presence of the RGD motif in the disintegrin domain of agkistin and the presence of two cysteine residues (Cys 407 and Cys 426) in addition to the 12 uniquely patterned cysteine residues of P-II members (Wang et al., 2003).
As mentioned previously, there are several reports showing that some members of the P-II SVMP class do not contain the canonical RGD motif (Calvete et al., 2005; Cesar et al., 2019; Rivas-Mercado and Garza-Ocañas, 2017). While some motifs share similarities, others are too varied and since the RGD motif is responsible for inhibition of platelet aggregation, members lacking this motif may have different activities and functions. For example, the primary structure of bilitoxin-1, a P-II SVMP of 32 kDa from Agkistrodon bilineatus, revealed that this hemorrhagic protein lacks platelet aggregation inhibitory activity and this may be due to the presence of a MGD binding sequence contrary to the canonical RGD motif (Nikai et al., 2000). Notwithstanding, the MGD motif has been shown to impair the function of α5β1 integrin (Calvete et al., 2005). On the other hand, peptide mass fingerprinting and protein sequence revealed that the presence of the KGD motif in the disintegrin domain of stejnitin did not affect its ability to inhibit ADP-induced platelet aggregation (Han et al., 2007). This 35 kDa protein was purified from the venom of Trimeresurus stejnegeri. DNA fragmentation alongside flow cytometry analysis demonstrated that it could trigger apoptosis in ECV304 cells (Han et al., 2007). Sighamatr and Rojnuckarin (2007) reported the isolation of the recombinant P-II albolatin from the venom of Trimeresurus albolaris. Expression analysis of the D domain showed the presence of a KGD motif that does not interfere with the inhibition of collagen-induced platelet aggregation (Singhamatr and Rojnuckarin, 2007). In a similar case, the dimeric BlatH1 (84 kDa), isolated from Bothriechis lateralis, hydrolyzed fibrinogen, gelatin and azocasein and it inhibited both ADP- and collagen-induced platelet aggregation (Camacho et al., 2014). In fact, this protein lacks the conventional RGD-integrin-binding motif, which was replaced by the TDN (threonine-aspartate-asparagine) motif (Camacho et al., 2014). CcMP-II, a 35 kDa hemorrhagic P-II protein purified from Cerastes cerastes snake venom, has the ability to inhibit platelet aggregation and it has been shown to hydrolyze ECM components such as type IV collagen and laminin, besides possessing α-fibrinogenase activity (Boukhalfa-Abib and Laraba-Djebari, 2015).
Several studies have cloned and expressed the disintegrin domain of P-II SVMPs. For instance, DisBa-01 (Ramos et al., 2008), bothrostatin (Fernandez et al., 2005), BnMPIIx (neuwiedin) (Lima-dos-Santos et al., 2015) and CamVMPII (r-cam-dis) (Suntravat et al., 2013) were identified from cDNA libraries constructed from the venom transcriptome of four different snake species: Bothrops alternatus, Bothrops jararaca, Bothrops neuwiedi and Crotalus adamanteus, respectively. SDS-PAGE analyses showed that bothrostatin, CamVMPII and DisBa-01 are proteins of approximately 8 kDa (Ramos et al., 2008). Although MALDI-TOF analysis showed that BnMPIIx has a molecular mass of 8 kDa, data from SDS-PAGE fingerprint correlated this protein to a molecular mass of 14 kDa (Lima-dos-Santos et al., 2015). It was demonstrated that BnMPIIx inhibited ADP-induced platelet aggregation whereas bothrostatin inhibited collagen-induced platelet aggregation. CamVMPII inhibited both processes (Suntravat et al., 2013). The inability of BnMPIIx to inhibit collagen-induced platelet aggregation may be due to a unique cysteine sequence and the substitution of some residues at the C-terminal of its RGD binding loop. DisBa-01, a recombinant RGD-disintegrin, demonstrated a αvβ3 integrin blocking effect (Ramos et al., 2008). In fact, recent data from Danilucci et al. (2019) demonstrated that DisBa-01 interfered in the αvβ3/VEGFR2 crosstalk, culminating into inhibition of HUVEC adhesion, migration and VEGF-mediated angiogenesis (Danilucci et al., 2019). Other examples of cloned disintegrins containing a RGD-motif are: trigamin (Trimeresurus gramineus), a platelet aggregation inhibitor (Neeper and Jacobson, 1990), jerdonin (Trimeresurus jerdonii), an antagonist of the platelet GPIIb-IIIa receptor (Zhou et al., 2004), bitistatin (Bothrops arietans), a platelet aggregation inhibitor that binds to the αIIbβ3 integrin ((Knight and Romano, 2005), insularin (Bothrops insularis), an inhibitor of endothelial cell adhesion (Della-Casa et al., 2011) and salmosin 1 (A. h. brevicandus), that inhibited the binding of αIIbβ3 integrin to fibrinogen (Kang et al., 1998; Park et al., 1998). A summary of P-II SVMPs members is described in Table 2.
Table 2.
P-II SVMP members and their reported activities.
| SVMP (P-II) |
M Mass (kDa) | Domain Composition (As Studied) | Snake Species | Integrin-binding Motif | Reported Activities | References | |
|---|---|---|---|---|---|---|---|
| 1. | Trigamin | 9 | Disintegrin | Trimeresurus gramineus | RGD | Inhibition of platelet aggregation | Huang et al., 2002, Neeper and Jacobson, 1990 |
| 2. | Salmosin 1 | 7.5 | Disintegrin | A. h. brevicandus) | RGD | Inhibition of αIIbβ3 integrin binding to fibrinogen | Kang et al., 1998, Park et al., 1998 |
| 3. | Atrolysin E/D | 7.4 | Disintegrin | Crotalus atrox | MVD | Inhibition of both ADP- and collagen-induced platelet aggregations | Shimokawa et al. (1997) |
| 4. | Bilitoxin-1 | 32.28 | Dimeric; each monomer contains metalloproteinsase, and disintegrin | Agkistrodon bilineatus | MGD | Hemorrhagic | Nikai et al. (2000) |
| 5. | Jerdonitin | 36 | Metalloproteinase and disintegrin | Trimeresurus jerdonii | RGD | Inhibition of ADP-induced human platelet aggregation | Chen et al. (2003) |
| 6. | Agkistin | ~57 | Metalloproteinase and disintegrin. | Agkistrodon halys | RGD | Inhibition of ADP-induced platelet aggregation and induction of apoptosis in HUMEC | Wang et al. (2003) |
| 7. | Jerdonin | 7.5 | Disintegrin | Trimeresurus jerdonii | RGD | Antagonist of the platelet's GPIIb-IIIa receptor | Zhou et al. (2004) |
| 8. | Bitistatin | 9 | Disintegrin | Bothrops arietans | RGD | Inhibition of platelet aggregation via binding to the αIIbβ3 integrin | Knight and Romano (2005) |
| 9. | Bothrostatin | 8 | Disintegrin | Bothrops jararaca | RGD | Inhibition of collagen-induced platelet aggregation | Fernandez et al. (2005) |
| 10. | Stejnitin | 35 | Metalloproteinase and disintegrin | Trimeresurus stejnegeri | KGD | Inhibition of ADP-induced platelet aggregation and induction of apoptosis in ECV304 cells | Han et al. (2007) |
| 11. | Albolatin (only the disintegrin domain) |
11 | Homodimeric disintegrins | Trimeresurus albolaris | KGD | Inhibition of collagen-induced platelet aggregation | Singhamatr and Rojnuckarin (2007) |
| 12. | DisBa-01 | 8 | Disintegrin | Bothrops alternatus | RGD | αvβ3 – blocking effect and inhibition of angiogenesis in HUVEC | Danilucci et al., 2019, Ramos et al., 2008 |
| 13. | Ahpfibrase | 32 | Metalloproteinase and disintegrin | Gloydius halys | RGD | Fibrinolytic activity and inhibition of platelet aggregation | Zhang et al. (2010) |
| 14. | Insularin | 14 | Disintegrin | Bothrops insularis | RGD | Inhibition of endothelial cell adhesion | Della-Casa et al. (2011) |
| 15. | CamVMPII (r-cam-dis) | 8 | Disintegrin | Crotalus adamanteus | RGD | Inhibition of both ADP- and collagen-induced platelet aggregation | Suntravat et al. (2013) |
| 16. | Albolamin | 35 | Metalloproteinase and disintegrin | Cryptelytrops albolaris | RGD | Degradation of human collagen type IV and inhibition of collagen-induced platelet aggregation | Jangprasert and Rojnuckarin (2014) |
| 17. | BlatH1 | 84 | Dimeric; each monomer contains metalloproteinsase, and disintegrin | Bothriechis lateralis | RGD | Inhibition of both ADP- and collagen-induced platelet aggregation; hemorrhagic, fibrinogenolytic, gelatinolytic and azocaseinolytic |
Camacho et al. (2014) |
| 18. | BnMPIIx (neuwiedin) | 8.18 | Disintegrin | Bothrops neuwiedi | RGD | Inhibition of ADP-induced platelet aggregation | Lima-dos-Santos et al. (2015) |
| 190. | CcMP-II Boukhalfa- |
35 | Metalloproteinsase, and disintegrin | Cerastes | Hemorrhagic and α-fibrinogenase activity |
Boukhalfa-Abib and Laraba-Djebari (2015) |
1.3. The P-III SVMPs
The P-III class of SVMP encompasses a group of diverse proteins with higher molecular masses, usually ranging from 60 to 100 kDa and having a more complex structure than P–I and P-II SVMPs (Markland and Swenson, 2013). Members of the P-III class are widely distributed in the venoms of the Viperidae, Elapidae, Atractaspididae and Colubridae families (Casewell et al., 2011), supporting the studies that indicate that P-III SVMPs are derived from an early recruitment, duplication and neofunctionalization of an ancestral ADAM gene, before the radiation of the advanced snakes (~ 60 million years ago) (Carbajo et al., 2015; Casewell, 2012; Fry, 2005). P-III class comprise a pro-domain (PD), metalloproteinase (M), disintegrin-like (D) and cysteine-rich (C) domains. The presence of the non-catalytic disintegrin-like domain and the cysteine rich domain in the P-III class accounts for their potent hemorrhagic activities (Escalante et al., 2011). For instance, VaH4 is a hemorrhagin purified from the venom of Vipera ammodytes ammodytes. This protein is extremely hemorrhagic – hydrolyzing extracellular matrix components such as fibronectin, nidogen and the α-chain of fibrinogen – and possesses a potent cytotoxic activity against HeLa cells (Leonardi et al., 2014).
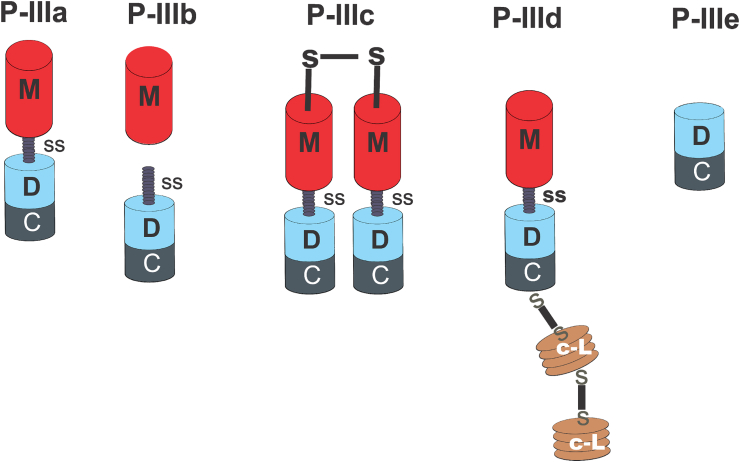
Due to its structural complexity, the P-III class was further classified into four subclasses: P-IIIa, P-IIIb, P-IIIc and P-IIId (Fox and Serrano, 2008b). The P-IIIa is considered the canonical subclass and it encompasses members containing a metalloproteinase, disintegrin-like and cysteine rich domains after post-translational exclusion of its pro-domain (Fox and Serrano, 2008b). A representative member of this subclass is the protein ohagin, a 50 kDa fibrinogenolytic protease isolated from the venom of Ophiophagus Hannah (Guo et al., 2007). The P-IIIb subclass comprises proteins that had undergone post-translational cleavage and/or proteolytic processing, which releases a mature form containing a disintegrin-like domain (with a D/ECD motif) and a cysteine-rich domain Fox and Serrano (2008b); Markland and Swenson (2013). A representative member of this subclass is the protein Alternagin isolated from the venom of Bothrops alternatus (Souza et al., 2000). Alternagin inhibits α2β1 integrin, thereby decreasing cell adhesion to type I collagen (Selistre-de-Araujo et al., 2005) and interferes with the α2β1/VEGFR2 crosstalk, thus inhibiting angiogenesis in HUVECs (dos Santos et al., 2020). Other example of the P-IIIb subclass is the protein TSV-DM purified from Trimeresurus stejnegeri venom wich potentially induces apoptosis on vascular endothelial cells (Wan et al., 2006a). In the P-IIIc subclass of SVMPs, some members are able to form dimers. VaH3 is a P-IIIc hemorrhagin purified from the venom of Vipera ammodytes ammodytes (Sajevic et al., 2013). Data from MALDI/TOF analysis suggest that it has a molecular mass of 104 kDa in its non-reduced form, while it has a molecular mass of 53.7 kDa following chemical reduction by S-carbamoylmethylation (Sajevic et al., 2013). VaH3 cleaves basement membrane (BM) and extracellular matrix (ECM) proteins including type IV collagen, fibronectin and nidogen (Sajevic et al., 2013). This data with the report of Leonardi et al. (2014) suggest that members of the P-IIIc class may be proteolytically potent against basal lamina and ECM proteins, although VaH3 can also cleave prothrombin and factor X without activating them (Leonardi et al., 2014; Sajevic et al., 2013). In addition, the dimers formed by the P-IIIc subclass can be constituted by two homologous monomers as in the case of VaH3 (Sajevic et al., 2013) or by a heterodimer (Wan et al., 2006b). Complexation of some P-III class members with C-type lectin-like protein (Snaclecs) has been identified in the P-IIId subclass. In this subclass, a C-type lectin like domain is connected to the canonical P-III structure by a disulfide bridge (Bjarnason and Fox, 1995). RVV-X is a representative of the P-IIId subclass and it can be found in the Russel's viper venom (Daboia russelii). RVV-X sequence was completely reported by Takeya et al. (1992) (Takeya et al., 1992). The unique 3D-structure of RVV-X has a hook-spanner-wrench configuration capable of binding to membrane receptors and coagulation factors instead of carbohydrates, demonstrating how important the relationship between structure and function is (Takeda et al., 2007). Moreover, RVV-X potently inhibited both ADP- and collagen-induced platelet aggregations, thereby causing intravascular coagulation in preys (Takeya et al., 1992). The complex 3D-structures of PIIIc and P-IIId members are related to enhanced pharmacological activity and toxicity (Doley and Kini, 2009).
The four subclasses (P-IIIa, b, c, and d) are well-known and documented in the literature. However, studies suggested the existence of a P-IIIe subclass comprising proteins that lack the catalytic metalloproteinase domain (Balija et al., 2020; Leonardi et al., 2019). Although Casewell et al. (2011) had previously correlated the evolutionary history of serpents to a repeated domain loss (Casewell et al., 2011), recent data from proteomic and transcriptomic analysis of Vipera ammodytes ammodytes venom identified Vaa-MPIII-3 as a member of the new P-IIIe subclass (Leonardi et al., 2019). This protein is an apparent 21 kDa glycoprotein homologous to P-III SVMPs, lacking a metalloproteinase domain, thus consisting of only a partial disintegrin-like domain and an adjacent cysteine-rich domain (Leonardi et al., 2019). Fig. 3 represents the sub classification of the P-III class of SVMPs.
Fig. 3.
Classification of the P-III SVMPs. P-IIIa has a metalloproteinase (M), a disintegrin-like (D) and a cysteine-rich (C) domain, displaying the canonical structure of P-III SVMPs. The D adjacent to the M domain of P-IIIb subclass is vulnerable to proteolytic cleavage. Members of the P-IIIc subclass form homo- or heterodimers connected by a disulfide bridge formed between the M domains of the two monomers. The P-IIId subclass represents a complexed P-III SVMP in which the C-terminal of the cysteine-rich domain is covalently linked to a tandem arrangement of two covalently bridged Snake C-type lectin-like regions-Snaclecs (c-L). The M domain is absent in the novel P-IIIe subclass and the processed form is a didomain containing D and C domains.
Previously in our laboratory, we reported the three dimensional structure of the protein bothropasin a P-III isolated from Bothrops jararaca venom (Muniz et al., 2008). Sequence alignment and comparative analysis with 41 other P-III SVMPs from NCBI revealed a significant pattern of sequence arrangement in the hyper-variable region (HVR) of members of this class (Muniz et al., 2008). A sub-group in the aligned sequences was highly homologous to bothropasin and showed a conserved sequence in the region of the so-called HVR, therefore referred to as highly conserved region – HCR. This conserved sequence was very distinct from the HVR of the sub-group that encompasses the protein VAP1, suggesting that this HVR vary from one group to the other, but it is conserved within those of bothropasin. Considering these data, we proposed a new classification for the P-III class into two sub-groups, the P-III-HCR and the P-III-HVR (Muniz et al., 2008). Additionally, we have reported the transcriptome analysis of bioactive agents from the venom gland of Bothrops alternatus (De Paula et al., 2014). Although consistent with a previous report from literature (Vonk et al., 2013), we identified copies of venom prolyl endopeptidases in the venom gland. It is noteworthy to mention the presence of P73 and P123 prolyl residues preceded by conserved K72 and H122 residues in members of the P-III class (De Paula et al., 2014). This data suggest the presence of two prolyl endopeptidase cleavage sites only in P-III members, thereby further distinguishing the P-III class from the P–I and P-II classes. Additionally, we hypothesized that the P-III SVMPs may be cognate substrates of the venom prolyl endopeptidases (De Paula et al., 2014).
The ECD (glutamic, cystein, aspatic) motif is a putative collagen interacting motif present in the disintegrin-like domain of most of the P-III class members (Fox and Serrano, 2008b; Markland and Swenson, 2013). Stejnihagin A and stejnihagin B are P-III representatives that were cloned from the venom of Trimeresurus stejnegeri (Wan et al., 2006b). Stejnihagin A has a SECD collagen interacting motif whereas stejnihagin B has a TECD collagen interacting motif. On the contrary, a few members of the P-III class contain the canonical RGD motif typical of the P-II class (Mazzi et al., 2007). Even though they contain a motif peculiar to the disintegrins, till date there is no available data that have tentatively excluded them from P-III SVMPs. An example is BjussuMP-I, a P-III hemorrhagic metalloproteinase from Bothrops jararacussu venom (Mazzi et al., 2007). The primary structure of this protein showed a conserved RGD-motif in the disintegrin-like domain. Activity reports showed that BjussuMP-I can induce lyses in fibrin clots and inhibit ADP- and collagen-induced platelet aggregations (Mazzi et al., 2007).
Most members of the P-III class have a conserved stretch of six cysteinyl residues in their metalloproteinase domain as observed in stejnihagin A and B – Cys123, Cys163, Cys165, Cys170, Cys187 and Cys203 (Wan et al., 2006b). However, studies have identified the seventh cysteinyl residue in some SVMPs, which may be responsible for additional activity of the proteins (Fox and Serrano, 2005). Some studies have correlated the presence of the seventh cysteinyl residue to vascular apoptosis-inducing activity (Trummal et al., 2005; You et al., 2003). Additionally, this seventh cysteinyl residue may be conserved in the P-IIIb subclass since representatives of this subclass (including VAP1) have been demonstrated to potently induce apoptosis of vascular endothelial cells. Fig. 4 shows the amino acid sequence of bothropasin, a PIII isolated from B.jararaca venom, with emphasis on the disulfide bridges (Muniz et al., 2008). Table 3 shows the sub-classification of the P-III SVMPs and some examples of these proteins.
Fig. 4.
Amino acid sequence of bothropasin, a PIII isolated from B. jararaca venom (Muniz et al., 2008). Cys residues that are in the form of disulfide bonds are shaded gray, numerated and connected by continuous lines. The free cysteine 189 is shaded in black. Calcium-binding sites as well as zinc-binding site are indicated and connected by dashed lines. Metalloproteinase (M), disintegrin (D) and cysteine-rich (C) domains.
Table 3.
Sub-classification of P-III SVMPs and their activities.
| SVMP | Sub-class | Domains in processed forms | Snake Species | Activities | References | |
|---|---|---|---|---|---|---|
| 1. | Ohagin | P-IIIa | Metalloproteinase, disintegrin-like and cysteine rich domains | Ophiophagus hannah | Fibrinogenolytic activity and inhibition of ADP-induced platelet aggregation | Guo et al. (2007) |
| 2. | VaF1 | P-IIIa | Metalloproteinase, disintegrin-like and cysteine rich domains | Vipera ammodytes ammodytes | α-fibrinogenase activity | Leonardi et al. (2015) |
| 3. | HR-Ele1 | P-IIIa | Metalloproteinase, disintegrin-like and cysteine rich domains | Protobothrops elegans | Hemorrhagic, αβ-fibrinogenase, type I collagenase | Oyama and Takahashi (2015) |
| 4. | Atrolysin A | P-IIIa | Metalloproteinase, disintegrin-like and cysteine rich domains | Crotalus atrox | Hemorrhagic, proteolytic, degrades plasma fibronectin, type-I collagenase, type-IV collagenase, fibrinogenase, gelatinase, sheddase of glycoprotein VI, | Fox and Bjarnason, 1995, Pinto et al., 2007 |
| 5. | Albocollagenase | P-IIIa | Metalloproteinase, disintegrin-like and cysteine rich domains | Cryptelytrops albolaris | Inhibition of collagen-induced platelet aggregation, type IV collagenase | Pinyachat et al. (2011) |
| 6. | Alternagin-C | P-IIIb | Disintegrin-like domain linked to a cysteine-rich domain | Bothrops alternatus | Inhibition of integrin-collagen interaction, inhibition of angiogenesis in HUVECs | Cominetti et al., 2004, dos Santos et al., 2020, Selistre-de-Araujo et al., 2005, Souza et al., 2000 |
| 7. | Catrocollastatin | P-IIIb | Disintegrin-like domain linked to a cysteine-rich domain | Crotalus atrox | Inhibition of collagen-induced platelet aggregation | Fox and Serrano, 2008b, Zhou et al., 1995 |
| 8. | Baltergin-C | P-IIIb | Disintegrin-like domain linked to a cysteine-rich domain | Bothrops alternatus | Hemorrhagic, type IV collagenase | Gay et al. (2009) |
| 9. | SV-PAD-2 | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Protobothrops elegans | αβγ-fibrinogenase inhibition of ADP- and collagen-induced inhibition of platelet aggregation |
Oyama and Takahashi (2015) |
| 10. | AHPM | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Agkistrodon halys pallas | α-fibrinogenase, inhibition of ADP-induced platelet aggregation | Song et al. (2013) |
| 11. | VaH3 | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Vipera ammodytes ammodytes | Proteolytic cleavage of prothrombin, X factor as well as BM and ECM protein components | Sajevic et al. (2013) |
| 12. | VaH4 | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Vipera ammodytes ammodytes | Hemorrhagic (hydrolysis of ECM proteins) and cytotoxic activities against Hela cells | Leonardi et al. (2014) |
| 13. | HV1 | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Trimeresurus flavoviridis | Apoptosis of vascular endothelial cell in vitro | Masuda et al. (2001) |
| 14. | VAP1 | P-IIIc | Dimeric; each monomer contains: a metalloproteinase, a disintegrin-like and a cysteine-rich domain | Crotalus atrox | Fibrinogenolytic, Apoptosis of vascular endothelial cell in vitro | Masuda et al. (2000) |
| 15. | VLFXAs | P-IIId | Metalloproteinase, disintegrin-like, cysteine-rich and two C-type lectin-like domains | Vipera ammodytes ammodytes | Activation of coagulation factor X to Xa, inhibition of collagen-induced platelet aggregation | Leonardi et al. (2008) |
| 16. | RVV-X | P-IIId | Metalloproteinase, disintegrin-like, cysteine-rich and two C-type lectin-like domains | Daboia russelli | Inhibition of both ADP- and collagen-induced platelet aggregations | Takeya et al. (1992) |
| 17. | Vaa-MPIII-3 | P-IIIe | Disintegrin-like and cysteine-rich domains | Vipera ammodytes ammodytes | Hemorrhagic | Leonardi et al. (2019) |
2. Domains functions
The resolution of the three-dimensional structures of SVMPs domains were important to the characterization and classification of these metalloproteinases as well as to elucidate the relationship between the domains and their specific functions during snake envenomation. Table 4 shows the SVMPs available in the Protein Data Bank (PDB), which 3D-structures were determined.
Table 4.
Three-dimensional structures of SVMPs available in the Protein Data Bank (PDB).
| Protein (PDB) | Snake Species | Class SVMP | Method | References |
|---|---|---|---|---|
| Acutolysin A (1BUD) | Deinagkistrodon acutus | P-I | X-ray | Gong et al. (1998) |
| Acutolysin-C (1QUA) | Deinagkistrodon acutus | P-I | X-ray | Zhu et al. (1999) |
| FII (1YP1) | Deinagkistrodon acutus | P-I | X-ray | Lou et al. (2005) |
| AaHIV (3HDB) | Deinagkistrodon acutus | P-III | X-ray | Zhu et al. (2009) |
| Disintegrin (2M75) | Calloselasma rhodostoma | P-IIa (D-domain) | NMR | Chuang et al. (2013) |
| Rhodostomin (Rho) (2M7F, 2M7H, 4R5R, 3UCI, 4RQG, 4M4C, 4R5U, 14Y, 1Q7I, 1Q7J, 2LJV, 2M75, 2PJF, 2PJI) | Calloselasma rhodostoma | P-II (recombinant D-domain with and without mutations) | NMR and X-ray | Chen et al., 2009, Huang et al., 2015, Shiu et al., 2015 |
| Atragin (3K7L) | Naja atra | P-III | X-ray | Guan et al. (2010) |
| K-like protein (3K7N) | Naja atra | P-III | X-ray | Guan et al. (2010) |
| Leucurolysin-A (4Q1L) | Bothrops leucurus | P-I | X-ray | Ferreira et al. (2015) |
| TM-1 (4J4M) | Protobothrops mucrosquamatus | P-I | X-ray | Chou et al. (2013) |
| BmooMPalpha-I (3GBO) | Bothrops moojeni | P-I | X-ray | Akao et al. (2010) |
| TM-3 complexed with pEKW (1KUK) | Protobothrops mucrosquamatus | P-I | X-ray | Huang et al. (2002) |
| TM-3 (1KUF) | Protobothrops mucrosquamatus | P-I | X-ray | Huang et al. (2002) |
| Bothropasin (3DSL) | Bothrops jararaca | P-III | X-ray | Muniz et al. (2008) |
| VAP1 (2ERQ, 2ERP, 2ERO)) | Crotalus atrox | P-III | X-ray | Takeda et al. (2006) |
| VAP2 (2DW0, 2DW1, 2DW2) | Crotalus atrox | P-III | X-ray | Igarashi et al. (2007) |
| Disintegrin (1TEJ) | Echis carinatus | P-IId | X-ray | Bilgrami et al. (2005) |
| Acostatin (3C05) | Agkistrodon contortrix contortrix | P-IIe | X-ray | Moiseeva et al. (2008) |
| Schistatin (1RMR) | Echis carinatus | P-IId | X-ray | Bilgrami et al. (2004) |
| Trimestatin (1JL2) | Protobothrops flavoviridis | P-IIa (D-domain) | X-ray | Fujii et al. (2003) |
| Bitistatin A (2MOP) | Bitis arietans | P-IIa (D-domain) | NMR | Carbajo et al. (2015) |
| Bitistatin B (2MP5) | Bitis arietans | P-IIa (D-domain) | NMR | Carbajo et al. (2015) |
| Salmosin (1L3X) | Gloydius brevicaudus | P-IIa (D-domain) | NMR | Shin et al. (2003) |
| Echistatin (1RO3) | Echis carinatus | P-IIa (D-domain) | NMR | Monleón et al. (2005) |
| Obtustatin (1MPZ) | Macrovipera lebetina obtusa | P-IIa (D-domain) | NMR | Moreno-Murciano et al. (2003) |
| Kistrin (1N4Y) | Calloselasma rhodostoma | P-IIa (D-domain) | NMR | Adler et al. (1991) |
| Jerdostatin (2W9V, 2W9O, 2W9U, 2W9W) | Protobothrops jerdonii | P-IIa (recombinant D-domain) | NMR | Carbajo et al. (2011) |
| RVV-X (2E3X) | Daboia siamensis | P-III | X-ray | Takeda et al. (2007) |
| Atrolysin-C (1HTD/1ATL) | Crotalus atrox | P-I | X-ray | Zhang et al. (1994) |
| BaP1 (1ND1) | Bothrops asper | P-I | X-ray | Watanabe et al. (2009) |
| BaP1 complexed with a peptidomimetic (2W14) | Bothrops asper | P-I | X-ray | Lingott et al. (2009) |
| Adamalysin II (1IAG) | Crotalus adamanteus | P-I | X-ray | Gomis-Rüth et al. (1993) |
| Flavoridin (1FVL) | Protobothrops flavoviridis | P-IIa (D-domain) | NMR | Senn and Klaus (1993) |
| H2-proteinase (1WN1) | Protobothrops flavoviridis | P-I | X-ray | Kumasaka et al. (1996) |
| Disintegrin (1Z1X) | Echis carinatus | P-IIa (D-domain) | X-ray | Hassan et al. (2005) |
As previously described, studies have shown that some activities of SVMPs attributed to one of the domains are enhanced by the presence of another domain. In addition, we will provide a general description of the activities assigned to each domain in the following subsections.
2.1. Signal peptide and pro-domain
Unprocessed SVMPs contain a signal sequence that serves as an export signal and a molecular address label (Jia et al., 1996). Meanwhile the pro-domain contains a conserved stretch of PKMCGVT that controls maturation of SVMP by a cysteine switch mechanism (Ompraba et al., 2010; Takeda et al., 2012).
2.2. Metalloproteinase domain
The metalloproteinase domain of the SVMP classes represents the catalytically active domain of all the identified members. Three-dimensional structures of SVMPs revealed that this domain has an oblate ellipsoidal format with a small lower region and an upper main region containing the active site. Interesting, the C-terminal of the lower region is formed by a helix preceded by an irregular folded domain that is considered essential for substrate recognition (Takeda et al., 2012).
In the early part of this review, we have coalesced several reports that identified the presence of a zinc-binding scaffold in the M domain (Bernardes et al., 2013; Chou et al., 2013; Da Silva et al., 2012; Herrera et al., 2015; Komori et al., 2014; Lingott et al., 2009; Patiño et al., 2010). This identification validates that the biological activities of such domain solely depend on a zinc-aided catalysis. To further support this claim, researchers highlighted the presence of a unique HEXXHXXGXXH consensus sequence, which is responsible for coordinating the putative binding of zinc metal. The report that punctuated the loss of a metalloproteinase domain in the new P-IIIe subclass revealed its lack of metalloproteinase specific function/activities (Leonardi et al., 2019). Put together, the function of this domain depends on the pentahedrally coordinated zinc reinforced by the unique sequence of methionine called the “methionine turn” (Met-turn) (Markland and Swenson, 2013).
Recently, an immunological probe assay unveiled the presence of a conserved sequence of 21 amino acids (KARMYELANIVNEILRYLYMH) constituting a B-cell epitope in the M domain (Molina et al., 2018). This study hypothesized that the function of the M domain may be linked to immunological reaction involving migration of inflammatory cytokines and activation of various pathways underlining the pathological phenotype of snake envenomation (Molina et al., 2018). However, more data are necessary to validate this assumption.
One of the most studied functions mediated by the metalloproteinase domain during snake envenomation is its hemorrhagic activity, which is marked by hydrolysis of basement membrane proteins, such as laminin and type IV collagen that surround endothelial cells in the capillaries leading to local hemorrhage and in some cases causing a systemic effect (bleeding) (De Souza et al., 2016). Interestingly, Herrera et al. (2015) showed through an ex vivo model of mouse cremaster muscle tissue that P-II and P-III SVMPs co-localize with type IV collagen in capillaries and arterioles (microvessels) whereas P–I SVMPs has a widespread localization throughout the tissue. This difference in tissue localization is probably responsible for the high hemorrhagic activity of P-II and P-III members compared to the P–I class (Herrera et al., 2015). Moreover, the metalloproteinase domain is also important in non-hemorrhagic proteins, being associated with fibrino(geno)lytic and apoptosis inducing activities. In in vitro experiments, endothelial cell apoptosis is more pronounced after envenomation by non-hemorrhagic SVMPs (Baldo et al., 2008), and it occurs when the catalytic domain disrupts the molecular assembly formed between focal adhesions of endothelial cells and the extracellular matrix (Díaz et al., 2005). Such apoptotic mechanism may be underlined by alterations in the endothelial cell-integrin-extracellular matrix assembly resulted from metalloproteinase proteolytic activity. When such assembly is intact, the activities of certain proteins capable of triggering apoptosis remain suppressed.
Studies have also revealed the fibrino(geno)lytic activity of the metalloproteinase domain. There have been several reports of alteration in ligand-receptor interaction; such alteration corresponds to the ectodomain sheddase activities of this domain (Ho et al., 2002; Moura-da-Silva et al., 1996; Moura-da-Silva and Baldo, 2012). For instance, Oliveira et al. (2009) showed how atroxlysin-III from the Peruvian pit viper snake (Bothrops oligolepis) induced glycoprotein VI shedding, culminating into platelet dysfunction (Oliveira et al., 2019). Previously, the sheddase activity of atrolysin a, a P-III SVMP from Crotalus atrox venom, was demonstrated against collagen IV and annexin in cultured human fibroblasts (Pinto et al., 2007). Similar effects were suggested for jararhagin, a P-III SVMP from Bothrops jararaca upon TNF-α precursor (Moura-da-Silva et al., 1996).
2.3. Disintegrin and disintegrin-like domains
The disintegrin and the disintegrin-like domains have been briefly described at the early part of this review. We have also provided evidences from literature about the presence of an integrin-binding motif in these domains, its peculiarity and its variants. It is well-know that almost all the P-III SVMPs have a disintegrin-like domain that contains a D/ECD-conserved motif instead of the canonical RGD-motif of the disintegrins (P-II class).
The function of both disintegrin and disintegrin-like domains is marked by the presence of the conserved motif. For example, the disintegrin domain of P-II SVMPs is responsible for platelet aggregation inhibition due to its binding to αIIbβ3 integrin (Masuda et al., 2000; Ramos et al., 2008). Since integrins represent adhesive frameworks (although not exclusive); connecting the cellular cytoskeleton to extracellular matrix components, the interaction between disintegrins and integrins may also limit cell to cell and cell-ECM interactions. Interestingly, most of the activities of the disintegrins are marked by their interaction with integrins. Structurally, disintegrins can vary depending on the type of integrin-binding motif present in them and such variation may account for differences in target selection and substrate diversity (Muniz et al., 2008). Disintegrin-integrin interaction also depends on amino acid sequences present in the disintegrin loop (a structural feature that encloses the integrin binding motif), and in the C-terminus of the disintegrin. The presence of these two sequences culminated to variation in the integrin binding (Chang et al., 2017).
In general, disintegrins can inhibit adhesion and migration of several cell lines. Recently, Montealegre-Sanchez and co-workers reported that Lansbermin-l, a low molecular weight RGD-disintegrin from Porthidium lansbergii lansbergii, inhibited the adhesion and migration of MCF7 and MDA-DB 231 breast cancer cells by interfering with α2 or β1-containing integrins (Montealegre-Sánchez et al., 2019). Furthermore, Chalier et al. (2020) characterized the RGD-disintegrin Dabmaurin-1 isolated from Daboia mauritanica snake. Dabmaurin-1 inhibited HMEC proliferation, adhesion and migration to several ECM components, as well as tube formation after interaction with β1-containing integrins (Chalier et al., 2020). Additionally, in our laboratory, we demonstrated that DisBa-01, a RGD-disintegrin, inhibited cell migration and adhesion in murine breast tumor cell line 4T1BM2. Our data confirmed that DisBa-01 hampered cell cycle progression at S phase and induced autophagy (Lino et al., 2019). In addition, we demonstrated that the blockade of αvβ3 integrin in oral squamous carcinoma cells by DisBa-01 inhibits cell directionality of migration (Montenegro et al., 2017).
The presence of the non-catalytic disintegrin-like and cysteine-rich domains is strongly responsible for the hemorrhagic activity of the P-III class members (Escalante et al., 2011). Escalante and co-workers revealed that the activities of P-III SVMPs are not limited to their catalytic domain and that most of the activities may be in tandem or cumulatively mediated by more than one domain. This means that P-III SVMPs containing an intact metalloproteinase domain and a disintegrin-like domain may be more potent in their activities than those lacking the non-catalytic domain (Escalante et al., 2011). The ECD-containing disintegrin-like proteins of the P-III class have also been shown to inhibit the adhesion of cells to collagen (De Luca et al., 1995; Jia et al., 2000; Souza et al., 2000; Suntravat et al., 2016; Tanjoni et al., 2010; Zhou et al., 1996).
It is important to highlight that 3D-structures of P-III SVMPs showed that the metalloproteinase domain alongside with the disintegrin-like and the cysteine-rich domains (MDC) form a C-shaped structure that places the HVR region of the cysteine-rich domain near to the catalytic site of the metalloproteinase domain. This is important to substrate recognition, because this C-shaped configuration allows a flexibility between the catalytic site and the exosite of P-III throughout the catalytic cycle (Takeda, 2016).
2.4. Cysteine-rich domain
Structural studies have shown that the cysteine-rich domain (C-domain) of bothropasin is formed by two α-helices, 4 β-strands and loops (Muniz et al., 2008; Takeda et al., 2006). The loop is stabilized by the presence of a disulfide bond found in the C-domain, which contain the hyper-variable region (HVR) or the highly-conserved region (HCR), depending on the specific subclass of P-III SVMPs (Muniz et al., 2008). Structural studies indicated that the C-domain could bind to a free- or membrane-bound target protein positioning the M domain to its substrate for proteolytic attack (Takeda et al., 2006). In addition, the HVR can have an essential role in protein-protein interactions, suggesting a structural correlation for the variety of biological activities found in the P-III class (Takeda, 2016). As mentioned before, the HVR localizes opposite to the catalytic site in the C-shaped MDC structure, thus it has been addressed with the function of substrate/peptide recognition (Takeda, 2016; Takeda et al., 2012). For instance, Menezes et al. (2008) showed that the HVR of the C-domain formed a protein-to-protein adhesive framework capable of inducing leucocyte rolling in microcirculation (Menezes et al., 2008). In another study, they demonstrated through site directed mutagenesis that the HVR of this domain inhibited platelet aggregation (Menezes et al., 2011). Additionally, Serrano et al. (2007) showed that the von Willebrand factor-mediated platelet aggregation was inhibited by the C-domain (Serrano et al., 2007). Solid-phase binding assay using type I collagen and vWF revealed that this domain contains exosites that can act as a cell-surface-receptor-binding site (Serrano et al., 2005), thereby revealing the role of the C-domain in the promotion of hemorrhage through substrate recognition, target selection and positioning of the metalloproteinase domain for its proteolytic activity (Serrano et al., 2006).
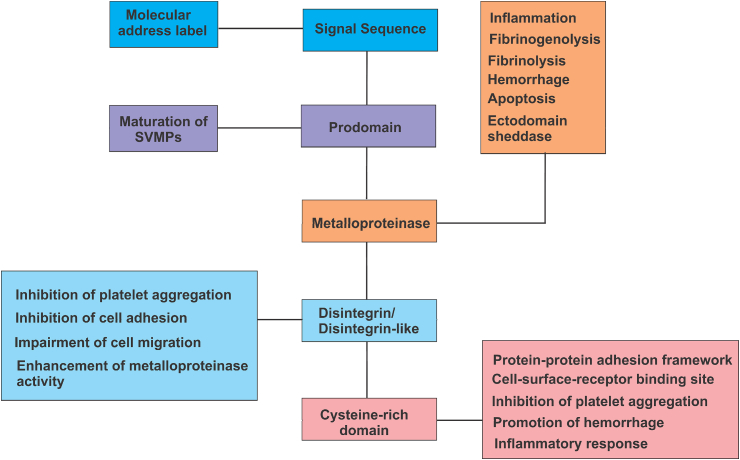
The C-domain also plays a critical role in inflammation. In functional association with the disintegrin-like domain, it has been shown to sufficiently cause leucocyte rolling and release of pro-inflammatory cytokines in the early stage of acute inflammatory response (Clissa et al., 2006). However, a recent report showed that the catalytic metalloproteinase domain of SVMPs is the principal effector of leucocyte recruitment, suggesting that the C-domain could also present an inflammatory response only in the absence of the M domain (Zychar et al., 2020). Fig. 5 shows the summary of the SVMP domains and its functions.
Fig. 5.
Summary of SVMP domains and their functions.
3. Concluding remark
Snake Venom Metalloproteinases are diverse in both structure and activities. The most recent classification recognizes three classes: P–I, P-II and P-III, including sub-classification of P-II and P-III classes. Furthermore, advancement in protein characterization and sophistication in molecular techniques continue to avail scientists the opportunity to identify novel SVMPs and correlate activities to specific domains. However, the exact sub-domain structures responsible for these activities have only been speculated in some cases. In addition, despite the identification of novel SVMPs, not all have been properly classified, especially into the P-II and P-III sub-classes. Therefore, as the field of molecular toxinology advances, scientists should maximize the use of available tools to characterize and identify the exact sub-domain structures that are responsible for their specific activities; as this will not only permit proper classification, but also expand the current understanding on the contribution of SVMPs to snake envenomation. Snake genomics, transcriptomics and proteomics contribute qualitatively and quantitatively to studies of structure-function relationship of venom components, adding new information that ultimately help to characterize toxins, notably SVMPs. As the Venomics develops, our knowledge evolves, opening doors to new discoveries in the field.
Author contributions
Original Draft Preparation, O.T.O. and D.H.F.S.; Writing - review & editing, O.T.O., D.H.F.S., P.K.S., H.S.S; Funding acquisition D.H.F.S. and H.S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grant Nº 306,225/2017–4 from CNPq and grants No. 2018/06297–9 and 2019/11,437–7 from the São Paulo Research Foundation (FAPESP). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) Finance Code 001.
Declaration of competing interest
The authors declare no conflict of interest.
References
- Achê D.C., Gomes M.S.R., de Souza D.L.N., Silva M.A., Brandeburgo M.I.H., Yoneyama K.A.G., Rodrigues R.S., Borges M.H., Lopes D.S., Rodrigues V. de M. Biochemical properties of a new PI SVMP from Bothrops pauloensis: inhibition of cell adhesion and angiogenesis. Int. J. Biol. Macromol. 2015;72:445–453. doi: 10.1016/j.ijbiomac.2014.08.050. [DOI] [PubMed] [Google Scholar]
- Adler M., Lazarus R.A., Dennis M.S., Wagner G. Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist. Science. 1991;253:445–448. doi: 10.1126/science.1862345. 80. [DOI] [PubMed] [Google Scholar]
- Akao P.K., Tonoli C.C.C., Navarro M.S., Cintra A.C.O., Neto J.R., Arni R.K., Murakami M.T. Structural studies of BmooMPα-I, a non-hemorrhagic metalloproteinase from Bothrops moojeni venom. Toxicon. 2010;55:361–368. doi: 10.1016/j.toxicon.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Baldo C., Tanjoni I., León I.R., Batista I.F.C., Della-Casa M.S., Clissa P.B., Weinlich R., Lopes-Ferreira M., Lebrun I., Amarante-Mendes G.P., Rodrigues V.M., Perales J., Valente R.H., Moura-da-Silva A.M. BnP1, a novel P-I metalloproteinase from Bothrops neuwiedi venom: biological effects benchmarking relatively to jararhagin, a P-III SVMP. Toxicon. 2008;51:54–65. doi: 10.1016/j.toxicon.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Balija M.L., Leonardi A., Brgles M., Sviben D., Kurtovic T., Halassy B., Krizaj I. Biological activities and proteomic profile of the venom of Vipera ursinii ssp., a very rare karst viper from Croatia. Toxins. 2020;12:1–16. doi: 10.3390/toxins12030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M., Pinto A.F.M., Guimarães J.A. Purification and functional characterization of bothrojaractivase, a prothrombin-activating metalloproteinase isolated from Bothrops jararaca snake venom. Toxicon. 2008;51:488–501. doi: 10.1016/j.toxicon.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bernardes C.P., Menaldo D.L., Camacho E., Rosa J.C., Escalante T., Rucavado A., Lomonte B., Gutiérrez J.M., Sampaio S.V. Proteomic analysis of Bothrops pirajai snake venom and characterization of BpirMP, a new P-I metalloproteinase. J. Proteomics. 2013;80:250–267. doi: 10.1016/j.jprot.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Bilgrami S., Tomar S., Yadav S., Kaur P., Kumar J., Jabeen T., Sharma S., Singh T.P. Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 Å resolution. J. Mol. Biol. 2004;341:829–837. doi: 10.1016/j.jmb.2004.06.048. [DOI] [PubMed] [Google Scholar]
- Bilgrami S., Yadav S., Kaur P., Sharma S., Perbandt M., Betzel C., Singh T.P. Crystal structure of the disintegrin heterodimer from saw-scaled viper (Echis carinatus) at 1.9 Å resolution. Biochemistry. 2005;44:11058–11066. doi: 10.1021/bi050849y. [DOI] [PubMed] [Google Scholar]
- Bjarnason J.B., Fox J.W. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Bjarnason J.B., Fox J.W. Snake venom metalloendopeptidases: reprolysins. Methods Enzymol. 1995;248:345–368. doi: 10.1016/0076-6879(95)48023-4. [DOI] [PubMed] [Google Scholar]
- Boukhalfa-Abib H., Meksem A., Laraba-Djebari F. Purification and biochemical characterization of a novel hemorrhagic metalloproteinase from horned viper (Cerastes cerastes) venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009;150:285–290. doi: 10.1016/j.cbpc.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Boukhalfa-Abib H., Laraba-Djebari F. CcMP-II, a new hemorrhagic metalloproteinase from Cerastes cerastes snake venom: purification, biochemical characterization and amino acid sequence analysis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015;167:65–73. doi: 10.1016/j.cbpc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Marcinkiewicz C., Monleón D., Esteve V., Celda B., Juárez P., Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Camacho E., Villalobos E., Sanz L., Pérez A., Escalante T., Lomonte B., Calvete J.J., Gutiérrez J.M., Rucavado A. Understanding structural and functional aspects of PII snake venom metalloproteinases: characterization of BlatH1, a hemorrhagic dimeric enzyme from the venom of Bothriechis lateralis. Biochimie. 2014;101:145–155. doi: 10.1016/j.biochi.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Camacho E., Escalante T., Remans K., Gutiérrez J.M., Rucavado A. Site mutation of residues in a loop surrounding the active site of a P[sbnd]I snake venom metalloproteinase abrogates its hemorrhagic activity. Biochem. Biophys. Res. Commun. 2019;512:859–863. doi: 10.1016/j.bbrc.2019.03.152. [DOI] [PubMed] [Google Scholar]
- Carbajo R.J., Sanz L., Mosulén S., Pérez A., Marcinkiewicz C., Pineda-Lucena A., Calvete J.J. NMR structure and dynamics of recombinant wild type and mutated jerdostatin, a selective inhibitor of integrin α1β1. Proteins Struct. Funct. Bioinforma. 2011;79:2530–2542. doi: 10.1002/prot.23076. [DOI] [PubMed] [Google Scholar]
- Carbajo R.J., Sanz L., Perez A., Calvete J.J. NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins. FEBS J. 2015;282:341–360. doi: 10.1111/febs.13138. [DOI] [PubMed] [Google Scholar]
- Casewell N.R., Wagstaff S.C., Harrison R.A., Renjifo C., Wüster W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 2011;28:2637–2649. doi: 10.1093/molbev/msr091. [DOI] [PubMed] [Google Scholar]
- Casewell N.R. On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon. 2012;60:449–454. doi: 10.1016/j.toxicon.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Cesar P.H.S., Braga M.A., Trento M.V.C., Menaldo D.L., Marcussi S. Snake venom disintegrins: an overview of their interaction with integrins. Curr. Drug Targets. 2019 doi: 10.2174/1389450119666181022154737. [DOI] [PubMed] [Google Scholar]
- Chalier F., Mugnier L., Tarbe M., Aboudou S., Villard C., Kovacic H., Gigmes D., Mansuelle P., Pomyers H. De, Luis J., Mabrouk K. Isolation of an Anti-tumour Disintegrin: Dabmaurin-1, a peptide lebein-1-like, from Daboia mauritanica venom. Toxins. 2020;12:1–20. doi: 10.3390/toxins12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.T., Shiu J.H., Huang C.H., Chen Y.C., Chen C.Y., Chang Y.S., Chuang W.J. Effects of the RGD loop and C-terminus of rhodostomin on regulating integrin αIIbβ3 recognition. PloS One. 2017;12:1–19. doi: 10.1371/journal.pone.0175321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Shiu J.H., Hsieh Y.H., Liu Y.C., Chen Yen Chin, Chen Yi Chun, Jeng W.Y., Tang M.J., Lo S.J., Chuang W.J. Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: importance of the interactions between the D residue and integrin. Proteins Struct. Funct. Bioinforma. 2009;76:808–821. doi: 10.1002/prot.22387. [DOI] [PubMed] [Google Scholar]
- Chen R.Q., Jin Y., Wu J.B., Zhou X.D., Lu Q.M., Wang W.Y., Xiong Y.L. A new protein structure of P-II class snake venom metalloproteinases: it comprises metalloproteinase and disintegrin domains. Biochem. Biophys. Res. Commun. 2003;310:182–187. doi: 10.1016/j.bbrc.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Ching A.T.C., Rocha M.M.T., Paes Leme A.F., Pimenta D.C., de Fátima D., Furtado M., Serrano S.M.T., Ho P.L., Junqueira-de-Azevedo I.L.M. Some aspects of the venom proteome of the Colubridae snake Philodryas olfersii revealed from a Duvernoy's (venom) gland transcriptome. FEBS Lett. 2006;580:4417–4422. doi: 10.1016/j.febslet.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Chou T.L., Wu C.H., Huang K.F., Wang A.H.J. Crystal structure of a Trimeresurus mucrosquamatus venom metalloproteinase providing new insights into the inhibition by endogenous tripeptide inhibitors. Toxicon. 2013;71:140–146. doi: 10.1016/j.toxicon.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Chuang W., Chang Y., Shiu J., Chen C., Chen Y. The C-terminal region of disintegrin modulate its 3D conformation and cooperate with RGD loop in regulating recognitions of integrins. PDB. 2013 https://www.rcsb.org/structure/2M75 [WWW Document] [Google Scholar]
- Chung C.H., Au L.C., Huang T.F. Molecular cloning and sequence analysis of aggretin, a collagen-like platelet aggregation inducer. Biochem. Biophys. Res. Commun. 1999;263:723–727. doi: 10.1006/bbrc.1999.1457. [DOI] [PubMed] [Google Scholar]
- Clissa P.B., Lopes-Ferreira M., Della-Casa M.S., Farsky S.H., Moura-da-Silva A.M. Importance of jararhagin disintegrin-like and cysteine-rich domains in the early events of local inflammatory response. Toxicon. 2006;47:591–596. doi: 10.1016/j.toxicon.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Cominetti M.R., Terruggi C.H.B., Ramos O.H.P., Fox J.W., Mariano-Oliveira A., De Freitas M.S., Figueiredo C.C., Morandi V., Selistre-de-Araujo H.S. Alternagin-C, a disintegrin-like protein, induces vascular endothelial cell growth factor (VEGF) expression and endothelial cell proliferation in vitro. J. Biol. Chem. 2004;279:18247–18255. doi: 10.1074/jbc.M311771200. [DOI] [PubMed] [Google Scholar]
- Da Silva I.R.F., Lorenzetti R., Rennó A.L., Baldissera L., Zelanis A., Serrano S.M.D.T., Hyslop S. BJ-PI2, A non-hemorrhagic metalloproteinase from Bothrops jararaca snake venom. Biochim. Biophys. Acta Gen. Subj. 2012;1820:1809–1821. doi: 10.1016/j.bbagen.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Danilucci T.M., Santos P.K., Pachane B.C., Pisani G.F.D., Lino R.L.B., Casali B.C., S S de Araujo H. Recombinant RGD-disintegrin DisBa-01 blocks integrin αvβ3 and impairs VEGF signaling in endothelial cells. Cell Commun. Signal. 2019;1:1–15. doi: 10.1186/s12964-019-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque Modesto J.C., Junqueira-de-Azevedo I.L.M., Neves-Ferreira A.G.C., Fritzen M., Oliva M.L.V., Ho P.L., Perales J., Chudzinski-Tavassi A.M. Insularinase A, a prothrombin activator from Bothrops insularis venom, is a metalloprotease derived from a gene encoding protease and disintegrin domains. Biol. Chem. 2005;386:589–600. doi: 10.1515/BC.2005.069. [DOI] [PubMed] [Google Scholar]
- De Luca M., Ward C.M., Ohmori K., Andrews R.K., Berndt M.C. Jararhagin and jaracetin: novel snake venom inhibitors of the integrin collagen receptor, alpha 2 beta 1. Biochem. Biophys. Res. Commun. 1995;206:570–576. doi: 10.1006/bbrc.1995.1081. [DOI] [PubMed] [Google Scholar]
- De Paula F.F.P., Ribeiro J.U., Santos L.M., De Souza D.H.F., Leonardecz E., Henrique-Silva F., Selistre-De-Araújo H.S. Molecular characterization of metalloproteases from Bothrops alternatus snake venom. Comp. Biochem. Physiol. Genom. Proteonomics. 2014;12:74–83. doi: 10.1016/j.cbd.2014.09.001. [DOI] [PubMed] [Google Scholar]
- De Souza R.A., Díaz N., Nagem R.A.P., Ferreira R.S., Suárez D. Unraveling the distinctive features of hemorrhagic and non-hemorrhagic snake venom metalloproteinases using molecular simulations. J. Comput. Aided Mol. Des. 2016;30:69–83. doi: 10.1007/s10822-015-9889-5. [DOI] [PubMed] [Google Scholar]
- Della-Casa M.S., Junqueira-de-Azevedo I., Butera D., Clissa P.B., Lopes D.S., Serrano S.M.T., Pimenta D.C., Magalhães G.S., Lee Ho P., Moura-da-Silva A.M. Insularin, a disintegrin from Bothrops insularis venom: inhibition of platelet aggregation and endothelial cell adhesion by the native and recombinant GST-insularin proteins. Toxicon. 2011;57:125–133. doi: 10.1016/j.toxicon.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Díaz C., Valverde L., Brenes O., Rucavado A., Gutiérrez J.M. Characterization of events associated with apoptosis/anoikis induced by snake venom metalloproteinase BaP1 on human endothelial cells. J. Cell. Biochem. 2005;94:520–528. doi: 10.1002/jcb.20322. [DOI] [PubMed] [Google Scholar]
- Doley R., Kini R.M. Protein complexes in snake venom. Cell. Mol. Life Sci. 2009;66:2851–2871. doi: 10.1007/s00018-009-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos P.K., Altei W.F., Danilucci T.M., Lino R.L.B., Pachane B.C., Nunes A.C.C., Selistre-de-Araujo H.S. Alternagin-C (ALT-C), a disintegrin-like protein, attenuates alpha2beta1 integrin and VEGF receptor 2 signaling resulting in angiogenesis inhibition. Biochimie. 2020;174:144–158. doi: 10.1016/j.biochi.2020.04.023. [DOI] [PubMed] [Google Scholar]
- Escalante T., Rucavado A., Fox J.W., Gutiérrez J.M. Key events in microvascular damage induced by snake venom hemorrhagic metalloproteinases. J. Proteomics. 2011;74:1781–1794. doi: 10.1016/j.jprot.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Fernandez J.H., Silva C.A., Assakura M.T., Camargo A.C.M., Serrano S.M.T. Molecular cloning, functional expression, and molecular modeling of bothrostatin, a new highly active disintegrin from Bothrops jararaca venom. Biochem. Biophys. Res. Commun. 2005;329:457–464. doi: 10.1016/j.bbrc.2005.01.148. [DOI] [PubMed] [Google Scholar]
- Ferreira R.N., Rates B., Richardson M., Guimarães B.G., Sanchez E.O.F., Pimenta A.M.D.C., Nagem R.A.P. Complete amino-acid sequence, crystallization and preliminary X-ray diffraction studies of leucurolysin-a, a nonhaemorrhagic metalloproteinase from Bothrops leucurus snake venom. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009;65:798–801. doi: 10.1107/S1744309109025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R.N., Rates B., Richardson M., Guimaraes B.G., Sanchez E.O.F., Pimenta A.M.C., Nagem R.A.P. Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW) PDB. 2015 https://www.rcsb.org/structure/4q1l [WWW Document] [Google Scholar]
- Fox J.W., Bjarnason J.B. Atrolysins: metalloproteinases from Crotalus atrox venom. Methods Enzymol. 1995;248:368–387. doi: 10.1016/0076-6879(95)48024-2. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Serrano S.M.T. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Serrano S.M.T. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8:909–920. doi: 10.1002/pmic.200700777. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Serrano S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Serrano S.M.T. Timeline of key events in snake venom metalloproteinase research. J. Proteomics. 2009;72:200–209. doi: 10.1016/j.jprot.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Fry B.G. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Okuda D., Fujimoto Z., Horii K., Morita T., Mizuno H. Crystal structure of trimestatin, a disintegrin containing a cell adhesion recognition motif RGD. J. Mol. Biol. 2003;332:1115–1122. doi: 10.1016/s0022-2836(03)00991-4. [DOI] [PubMed] [Google Scholar]
- Gay C.C., Maruñak S.L., Teibler P., Ruiz R., Acosta de Pérez O.C., Leiva L.C. Systemic alterations induced by a Bothrops alternatus hemorrhagic metalloproteinase (baltergin) in mice. Toxicon. 2009;53:53–59. doi: 10.1016/j.toxicon.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Georgieva D., Seifert J., Öhler M., Von Bergen M., Spencer P., Arni R.K., Genov N., Betzel C. Pseudechis australis venomics: adaptation for a defense against microbial pathogens and recruitment of body transferrin. J. Proteome Res. 2011;10:2440–2464. doi: 10.1021/pr101248e. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth F.X., Kress L.F., Bode W. First structure of a snake venom metalloproteinase: a prototype for matrix metalloproteinases/collagenases. EMBO J. 1993;12:4151–4157. doi: 10.1002/j.1460-2075.1993.tb06099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth F.X., Kress L.F., Kellermann J., Mayr I., Lee X., Huber R., Bode W. Refined 2.0 Å X-ray crystal structure of the snake venom zinc-endopeptidase adamalysin II: primary and tertiary structure determination, refinement, molecular structure and comparison with astacin, collagenase and thermolysin. J. Mol. Biol. 1994 doi: 10.1006/jmbi.1994.1392. [DOI] [PubMed] [Google Scholar]
- Gong W., Zhu X., Liu S., Teng M., Niu L. Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus. J. Mol. Biol. 1998;283:657–668. doi: 10.1006/jmbi.1998.2110. [DOI] [PubMed] [Google Scholar]
- Guan H.-H., Goh K.-S., Davamani F., Wu P.-L., Huang Y.-W., Jeyakanthan J., Wu W., Chen C.-J. Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J. Struct. Biol. 2010;169:294–303. doi: 10.1016/j.jsb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.X., Zeng L., Lee W.H., Zhang Y., Jin Y. Isolation and cloning of a metalloproteinase from king cobra snake venom. Toxicon. 2007;49:954–965. doi: 10.1016/j.toxicon.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J., Romero M., Díaz C., Borkow G., Ovadia M. Isolation and characterization of a metalloproteinase with weak hemorrhagic activity from the venom of the snake Bothrops asper (terciopelo) Toxicon. 1995;33:19–29. doi: 10.1016/0041-0101(94)00138-X. [DOI] [PubMed] [Google Scholar]
- Han Y.P., Lu X.Y., Wang X.F., Xu J. Isolation and characterization of a novel P-II class snake venom metalloproteinase from Trimeresurus stejnegeri. Toxicon. 2007;49:889–898. doi: 10.1016/j.toxicon.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Hassan M.I., Ethayathulla A.S., Bilgrami S., Singh B., Yadav S., Singh T.P. Crystal Structure of a novel disintegrin from Saw-scaled viper at 3.2A resolution. PDB. 2005 https://www.rcsb.org/structure/1Z1X [WWW Document] [Google Scholar]
- Herrera C., Escalante T., Voisin M.B., Rucavado A., Morazán D., Macêdo J.K.A., Calvete J.J., Sanz L., Nourshargh S., Gutiérrez J.M., Fox J.W. Tissue localization and extracellular matrix degradation by PI, PII and PIII snake venom metalloproteinases: clues on the mechanisms of venom-induced hemorrhage. PLoS Neglected Trop. Dis. 2015;9:1–20. doi: 10.1371/journal.pntd.0003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite L.A., Shannon J.D., Bjarnason J.B., Fox J.W. Sequence of a cDNA clone encoding the zinc metalloproteinase hemorrhagic toxin e from Crotalus atrox: evidence for signal, zymogen, and disintegrin-like structures. Biochemistry. 1992;31:6203–6211. doi: 10.1021/bi00142a005. [DOI] [PubMed] [Google Scholar]
- Hite L.A., Jia L.-G., Bjarnason J.B., Fox J.W. cDNA sequences for four snake venom metalloproteinase.pdf. Arch. Biochem. Biophys. 1994 doi: 10.1006/abbi.1994.1026. [DOI] [PubMed] [Google Scholar]
- Ho P.L., De Toledo Serrano S.M., Chudzinski-Tavassi A.M., Da Silva A.M.M., Mentele R., Caldas C., Oliva M.L.V., De Fátima Correia Batista I., De Oliveira M.L.S. Angiostatin-like molecules are generated by snake venom metalloproteinases. Biochem. Biophys. Res. Commun. 2002;294:879–885. doi: 10.1016/S0006-291X(02)00567-3. [DOI] [PubMed] [Google Scholar]
- Howes J.M., Wilkinson M.C., Theakston R.D.G., Laing G.D. The purification and partial characterisation of two novel metalloproteinases from the venom of the West African carpet viper, Echis ocellatus. Toxicon. 2003;42:21–27. doi: 10.1016/S0041-0101(03)00096-5. [DOI] [PubMed] [Google Scholar]
- Howes J.M., Kamiguti A.S., Theakston R.D.G., Wilkinson M.C., Laing G.D. Effects of three novel metalloproteinases from the venom of the West African saw-scaled viper, Echis ocellatus on blood coagulation and platelets. Biochim. Biophys. Acta Gen. Subj. 2005;1724:194–202. doi: 10.1016/j.bbagen.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Huang C.H., Shiu J.H., Chang Y.T., Jeng W.Y., Chuang W.J. Effects of the regions adjacent to the RGD motif in disintegrins on their inhibitory activities and structures. PDB. 2015 http://www.rcsb.org/structure/4R5R [WWW Document] [Google Scholar]
- Huang K.-F., Chiou S.-H., Ko T.-P., Yuann J.-M., Wang A.H.-J. The 1.35 A structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: elucidation of a TNF alpha-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1118–1128. doi: 10.1107/s090744490200656x. [DOI] [PubMed] [Google Scholar]
- Igarashi T., Araki S., Mori H., Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581:2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Jangprasert P., Rojnuckarin P. Molecular cloning, expression and characterization of albolamin: a type P-IIa snake venom metalloproteinase from green pit viper (Cryptelytrops albolabris) Toxicon. 2014;79:19–27. doi: 10.1016/j.toxicon.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Jia L.G., Shimokawa K., Bjarnason J.B., Fox J.W. Snake venom metalloproteinases: structure, function and relationship to the ADAMs family of proteins. Toxicon. 1996;34:1269–1276. doi: 10.1016/s0041-0101(96)00108-0. [DOI] [PubMed] [Google Scholar]
- Jia L.G., Wang X.M., Shannon J.D., Bjarnason J.B., Fox J.W. Inhibition of platelet aggregation by the recombinant cysteine-rich domain of the hemorrhagic snake venom metalloproteinase, atrolysin A. Arch. Biochem. Biophys. 2000;373:281–286. doi: 10.1006/abbi.1999.1517. [DOI] [PubMed] [Google Scholar]
- Kang I.C., Chung K.H., Lee S.J., Yun Y., Moon H.M., Kim D.S. Purification and molecular cloning of a platelet aggregation inhibitor from the snake (Agkistrodon halys brevicaudus) venom. Thromb. Res. 1998;91:65–73. doi: 10.1016/S0049-3848(98)00053-X. [DOI] [PubMed] [Google Scholar]
- Knight L.C., Romano J.E. Functional expression of bitistatin, a disintegrin with potential use in molecular imaging of thromboembolic disease. Protein Expr. Purif. 2005;39:307–319. doi: 10.1016/j.pep.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Komori Y., Murakami E., Uchiya K., Nonogaki T., Nikai T. Okinalysin, a novel P-I metalloproteinase from Ovophis okinavensis: biological properties and effect on vascular endothelial cells. Toxins. 2014;6:2594–2604. doi: 10.3390/toxins6092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkeaw K., Chaicumpa W., Sakolvaree Y., Tongtawe P., Tapchaisri P. Proteome and immunome of the venom of the Thai cobra, Naja kaouthia. Toxicon. 2007;49:1026–1041. doi: 10.1016/j.toxicon.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Kumasaka T., Yamamoto M., Moriyama H., Tanaka N., Sato M., Katsube Y., Yamakawa Y., Omori-Satoh T., Iwanaga S., Ueki T. Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis. J. Biochem. 1996;119:49–57. doi: 10.1093/oxfordjournals.jbchem.a021215. [DOI] [PubMed] [Google Scholar]
- Leonardi A., Fox J.W., Trampuš-Bakija A., Križaj I. Two coagulation factor X activators from Vipera a. ammodytes venom with potential to treat patients with dysfunctional factors IXa or VIIa. Toxicon. 2008;52:628–637. doi: 10.1016/j.toxicon.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Leonardi A., Sajevic T., Kovačič L., Pungerčar J., Lang Balija M., Halassy B., Trampuš Bakija A., Križaj I. Hemorrhagin VaH4, a covalent heterodimeric P-III metalloproteinase from Vipera ammodytes ammodytes with a potential antitumour activity. Toxicon. 2014;77:141–155. doi: 10.1016/j.toxicon.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Leonardi A., Sajevic T., Latinović Z., Pungerčar J., Lang Balija M., Trampuš Bakija A., Vidmar R., Halassy B., Križaj I. Structural and biochemical characterisation of VaF1, a P-IIIa fibrinogenolytic metalloproteinase from Vipera ammodytes ammodytes venom. Biochimie. 2015;109:78–87. doi: 10.1016/j.biochi.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Leonardi A., Sajevic T., Pungerčar J., Križaj I. Comprehensive study of the proteome and transcriptome of the venom of the most venomous European viper: discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019;18:2287–2309. doi: 10.1021/acs.jproteome.9b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-dos-Santos I., Della-Casa M.S., Portes-Junior J.A., Calabria P.A.L., Magalhães G.S., Moura-da-Silva A.M. Characterization of Neuwiedin, a new disintegrin from Bothrops neuwiedi venom gland with distinct cysteine pattern. Toxicon. 2015;104:57–64. doi: 10.1016/j.toxicon.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Lingott T., Schleberger C., Gutiérrez J.M., Merfort I. High-resolution crystal structure of the snake venom metalloproteinase BaP1 complexed with a peptidomimetic: insight into inhibitor binding. Biochemistry. 2009;48:6166–6174. doi: 10.1021/bi9002315. [DOI] [PubMed] [Google Scholar]
- Lino R.L.B., dos Santos P.K., Pisani G.F.D., Altei W.F., Cominetti M.R., Selistre-de-Araújo H.S. Alphavbeta3 integrin blocking inhibits apoptosis and induces autophagy in murine breast tumor cells. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1–13. doi: 10.1016/j.bbamcr.2019.118536. [DOI] [PubMed] [Google Scholar]
- Lou Z., Hou J., Liang X., Chen J., Qiu P., Liu Y., Rao Z., Yan G. Crystal structure of a non-hemorrhagic fibrin(ogen)olytic metalloproteinase complexed with a novel natural tri-peptide inhibitor from venom of Agkistrodon acutus. J. Struct. Biol. 2005;152:195–203. doi: 10.1016/j.jsb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Marcussi S., Bernardes C.P., Santos-Filho N.A., Mazzi M.V., Oliveira C.Z., Izidoro L.F.M., Fuly A.L., Magro A.J., Braz A.S.K., Fontes M.R.M., Giglio J.R., Soares A.M. Molecular and functional characterization of a new non-hemorrhagic metalloprotease from Bothrops jararacussu snake venom with antiplatelet activity. Peptides. 2007;28:2328–2339. doi: 10.1016/j.peptides.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Markland F.S., Swenson S. Snake venom metalloproteinases. Toxicon. 2013;62:3–18. doi: 10.1016/j.toxicon.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Maruyama M., Tanigawa M., Sugiki M., Yoshida E., Mihara H. Purification and characterization of low molecular weight fibrinolytic/hemorrhagic enzymes from snake (Bothrops jararaca) venom. Enzyme Protein. 1993;47:124–135. doi: 10.1159/000468668. [DOI] [PubMed] [Google Scholar]
- Masuda S., Ohta T., Kaji K., Fox J.W., Hayashi H., Araki S. cDNA cloning and characterization of vascular apoptosis-inducing protein 1. Biochem. Biophys. Res. Commun. 2000;278:197–204. doi: 10.1006/bbrc.2000.3770. [DOI] [PubMed] [Google Scholar]
- Masuda S., Hayashi H., Atoda H., Morita T., Araki S. Purification, cDNA cloning and characterization of the vascular apoptosis-inducing protein, HV1, from Trimeresurus flavoviridis. Eur. J. Biochem. 2001;268:3339–3345. doi: 10.1046/j.1432-1327.2001.02246.x. [DOI] [PubMed] [Google Scholar]
- Mazzi M.V., Magro A.J., Amui S.F., Oliveira C.Z., Ticli F.K., Stábeli R.G., Fuly A.L., Rosa J.C., Braz A.S.K., Fontes M.R.M., Sampaio S.V., Soares A.M. Molecular characterization and phylogenetic analysis of BjussuMP-I: a RGD-P-III class hemorrhagic metalloprotease from Bothrops jararacussu snake venom. J. Mol. Graph. Model. 2007;26:69–85. doi: 10.1016/j.jmgm.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Menezes M.C., Paes Leme A.F., Melo R.L., Silva C.A., Della Casa M., Bruni F.M., Lima C., Lopes-Ferreira M., Camargo A.C.M., Fox J.W., Serrano S.M.T. Activation of leukocyte rolling by the cysteine-rich domain and the hyper-variable region of HF3, a snake venom hemorrhagic metalloproteinase. FEBS Lett. 2008;582:3915–3921. doi: 10.1016/j.febslet.2008.10.034. [DOI] [PubMed] [Google Scholar]
- Menezes M.C., de Oliveira A.K., Melo R.L., Lopes-Ferreira M., Rioli V., Balan A., Paes Leme A.F., Serrano S.M.T. Disintegrin-like/cysteine-rich domains of the reprolysin HF3: site-directed mutagenesis reveals essential role of specific residues. Biochimie. 2011;93:345–351. doi: 10.1016/j.biochi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Miyata T., Takeya H., Ozeki Y., Arakawa M., Tokunaga F., Iwanaga S., Omori-satoh T. Primary structure of hemorrhagic protein, HR2a, isolated from the venom of Trimeresurus flavoviridis. J. Biochem. 1989;105:847–853. doi: 10.1093/oxfordjournals.jbchem.a122756. [DOI] [PubMed] [Google Scholar]
- Moiseeva N., Bau R., Swenson S.D., Markland F.S., Choe J.Y., Liu Z.J., Allaire M. Structure of acostatin, a dimeric disintegrin from Southern copperhead (Agkistrodon contortrix contortrix), at 1.7 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008;64:466–470. doi: 10.1107/S0907444908002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina Molina D.A., Guerra-Duarte C., Naves de Souza D.L., Costal-Oliveira F., Ávila G.R. de, Soccol V.T., Machado-de-Ávila R.A., Chávez-Olórtegui C. Identification of a linear B-cell epitope in the catalytic domain of bothropasin, a metalloproteinase from Bothrops jararaca snake venom. Mol. Immunol. 2018;104:20–26. doi: 10.1016/j.molimm.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Monleón D., Esteve V., Kovacs H., Calvete J.J., Celda B. Conformation and concerted dynamics of the integrin-binding site and the C-terminal region of echistatin revealed by homonuclear NMR. Biochem. J. 2005;387:57–66. doi: 10.1042/BJ20041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre-Sánchez L., Gimenes S.N.C., Lopes D.S., Teixeira S.C., Solano-Redondo L., Jiménez-Charris V. de M.R. Antitumoral potential of Lansbermin-I, a novel disintegrin from Porthidium lansbergii lansbergii venom on breast cancer cells. Curr. Top. Med. Chem. 2019 doi: 10.2174/1568026619666190806151401. E. [DOI] [PubMed] [Google Scholar]
- Montenegro C.F., Casali B.C., Lino R.L.B., Pachane B.C., Santos P.K., Horwitz A.R., Selistre-De-Araujo H.S., Lamers M.L. Inhibition of αvβ3 integrin induces loss of cell directionality of oral squamous carcinoma cells (OSCC) PloS One. 2017;12:1–10. doi: 10.1371/journal.pone.0176226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Murciano M.P., Monleón D., Marcinkiewicz C., Calvete J.J., Celda B. NMR solution structure of the non-RGD disintegrin obtustatin. J. Mol. Biol. 2003;329:135–145. doi: 10.1016/S0022-2836(03)00371-1. [DOI] [PubMed] [Google Scholar]
- Mori N., Nikai T., Sugihara H., Tu A.T. Biochemical characterization of hemorrhagic toxins with fibrinogenase activity isolated from Crotalus ruber ruber venom. Arch. Biochem. Biophys. 1987;253:108–121. doi: 10.1016/0003-9861(87)90643-6. [DOI] [PubMed] [Google Scholar]
- Moura-da-Silva A.M., Laing G.D., Paine M.J.I., Dennison J.M.T.J., Politi V., Crampton J.M., Theakston R.D.G. Processing of pro-tumor necrosis factor-α by venom metalloproteinases: a hypothesis explaining local tissue damage following snake bite. Eur. J. Immunol. 1996;26 doi: 10.1002/eji.1830260905. 2000–2005. [DOI] [PubMed] [Google Scholar]
- Moura-da-Silva A.M., Furlan M.S., Caporrino M.C., Grego K.F., Portes-Junior J.A., Clissa P.B., Valente R.H., Magalhães G.S. Diversity of metalloproteinases in Bothrops neuwiedi snake venom transcripts: evidences for recombination between different classes of SVMPs. BMC Genet. 2011;12:94. doi: 10.1186/1471-2156-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-da-Silva A.M., Baldo C. Jararhagin, a hemorrhagic snake venom metalloproteinase from Bothrops jararaca. Toxicon. 2012;60:280–289. doi: 10.1016/j.toxicon.2012.03.026. [DOI] [PubMed] [Google Scholar]
- Muniz J.R.C., Ambrosio A.L.B., Selistre-de-Araujo H.S., Cominetti M.R., Moura-da-Silva A.M., Oliva G., Garratt R.C., Souza D.H.F. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: insights for a new classification of snake venom metalloprotease subgroups. Toxicon. 2008;52:807–816. doi: 10.1016/j.toxicon.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Naves de Souza D.L., Gomes M.S.R., Ferreira F.B., Rodrigues R.S., Achê D.C., Richardson M., Borges M.H., Rodrigues V.M. Biochemical and enzymatic characterization of BpMP-I, a fibrinogenolytic metalloproteinase isolated from Bothropoides pauloensis snake venom. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012;161:102–109. doi: 10.1016/j.cbpb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Neeper M.P., Jacobson M.A. Sequence of a cDNA encoding the platelet aggregation inhibitor trigramin. Nucleid Acids Res. 1990;18:4255. doi: 10.1093/nar/18.14.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikai T., Taniguchi K., Komori Y., Masuda K., Fox J.W., Sugihara H. Primary structure and functional characterization of bilitoxin-1, a novel dimeric P-II snake venom metalloproteinase from Agkistrodon bilineatus venom. Arch. Biochem. Biophys. 2000;378:6–15. doi: 10.1006/abbi.2000.1795. [DOI] [PubMed] [Google Scholar]
- Okamoto D.N., Kondo M.Y., Oliveira L.C.G., Honorato R.V., Zanphorlin L.M., Coronado M.A., Araújo M.S., Da Motta G., Veronez C.L., Andrade S.S., Oliveira P.S.L., Arni R.K., Cintra A.C.O., Sampaio S.V., Juliano M.A., Juliano L., Murakami M.T., Gouvea I.E. P-I class metalloproteinase from Bothrops moojeni venom is a post-proline cleaving peptidase with kininogenase activity: insights into substrate selectivity and kinetic behavior. Biochim. Biophys. Acta Protein Proteonomics. 2014;1844:545–552. doi: 10.1016/j.bbapap.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Oliveira A.K., Paes Leme A.F., Assakura M.T., Menezes M.C., Zelanis A., Tashima A.K., Lopes-Ferreira M., Lima C., Camargo A.C.M., Fox J.W., Serrano S.M.T. Simplified procedures for the isolation of HF3, bothropasin, disintegrin-like/cysteine-rich protein and a novel P-I metalloproteinase from Bothrops jararaca venom. Toxicon. 2009;53:797–801. doi: 10.1016/j.toxicon.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Oliveira L.S., Estevão-Costa M.I., Alvarenga V.G., Vivas-Ruiz D.E., Yarleque A., Lima A.M., Cavaco A., Eble J.A., Sanchez E.F. Atroxlysin-III , A metalloproteinase from the venom of the Peruvian pit viper snake Bothrops atrox (jergón) induces glycoprotein VI shedding and impairs platelet function. Molecules. 2019;3489:1–21. doi: 10.3390/molecules24193489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ompraba G., Chapeaurouge A., Doley R., Devi K.R., Padmanaban P., Venkatraman C., Velmurugan D., Lin Q., Kini R.M. Identification of a novel family of snake venom proteins veficolins from cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 2010;9:1882–1893. doi: 10.1021/pr901044x. [DOI] [PubMed] [Google Scholar]
- Oyama E., Takahashi H. Purification and characterization of two high molecular mass snake venom metalloproteinases (P-III SVMPs), named SV-PAD-2 and HR-Ele-1, from the venom of Protobothrops elegans (Sakishima-habu) Toxicon. 2015;103:30–38. doi: 10.1016/j.toxicon.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Park D., Kang I., Kim H., Chung K., Kim D.S., Yun Y. Cloning and characterization of novel disintegrins from Agkistrodon halys venom. Mol. Cell. 1998;8:578—584. [PubMed] [Google Scholar]
- Patiño A.C., Pereañez J.A., Núñez V., Benjumea D.M., Fernandez M., Rucavado A., Sanz L., Calvete J.J. Isolation and biological characterization of Batx-I, a weak hemorrhagic and fibrinogenolytic PI metalloproteinase from Colombian Bothrops atrox venom. Toxicon. 2010;56:936–943. doi: 10.1016/j.toxicon.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Pinto A.F.M., Ma L., Dragulev B., Guimaraes J.A., Fox J.W. Use of SILAC for exploring sheddase and matrix degradation of fibroblasts in culture by the PIII SVMP atrolysin A: identification of two novel substrates with functional relevance. Arch. Biochem. Biophys. 2007;465:11–15. doi: 10.1016/j.abb.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Pinyachat A., Rojnuckarin P., Muanpasitporn C., Singhamatr P., Nuchprayoon S. Albocollagenase, a novel recombinant P-III snake venom metalloproteinase from green pit viper (Cryptelytrops albolabris), digests collagen and inhibits platelet aggregation. Toxicon. 2011;57:772–780. doi: 10.1016/j.toxicon.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Preciado L.M., Pereañez J.A., Comer J. Potential of matrix metalloproteinase inhibitors for the treatment of local tissue damage induced by a type P-I snake venom metalloproteinase. Toxins. 2019;12 doi: 10.3390/toxins12010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton L., Le Caer J.P., Phan G., Ligny-Lemaire C., Bourdais-Jomaron J., Ducancel F., Chamot-Rooke J. Characterization of toxins within crude venoms by combined use of Fourier transform mass spectrometry and cloning. Anal. Chem. 2005;77:6630–6639. doi: 10.1021/ac050575k. [DOI] [PubMed] [Google Scholar]
- Ramos O.H.P., Kauskot A., Cominetti M.R., Bechyne I., Salla Pontes C.L., Chareyre F., Manent J., Vassy R., Giovannini M., Legrand C., Selistre-De-Araujo H.S., Crépin M., Bonnefoy A. A novel αvβ3-blocking disintegrin containing the RGD motive, DisBa-01, inhibits bFGF-induced angiogenesis and melanoma metastasis. Clin. Exp. Metastasis. 2008;25:53–64. doi: 10.1007/s10585-007-9101-y. [DOI] [PubMed] [Google Scholar]
- Rivas-Mercado E.A., Garza-Ocañas L. Disintegrins obtained from snake venom and their pharmacological potential. Med. Univ. 2017;19:32–37. doi: 10.1016/j.rmu.2017.02.004. [DOI] [Google Scholar]
- Sajevic T., Leonardi A., Kovačič L., Lang-Balija M., Kurtović T., Pungerčar J., Halassy B., Trampuš-Bakija A., Križaj I. VaH3, one of the principal hemorrhagins in Vipera ammodytes ammodytes venom, is a homodimeric P-IIIc metalloproteinase. Biochimie. 2013;95:1158–1170. doi: 10.1016/j.biochi.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Sanchez E.F., Richardson M., Gremski L.H., Veiga S.S., Yarleque A., Niland S., Lima A.M., Estevao-Costa M.I., Eble J.A. A novel fibrinolytic metalloproteinase, barnettlysin-I from Bothrops barnetti (barnett's pitviper) snake venom with anti-platelet properties. Biochim. Biophys. Acta Gen. Subj. 2016;1860:542–556. doi: 10.1016/j.bbagen.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Sani I., Umar R.A., Hassan S.W., Faruq U.Z., Abdulhamid A., Bello F., Fakai I.M. Major enzymes from snake venoms: mechanisms of action and pharmacological applications. Asian J. Bio. Sci. 2019;12:396–403. doi: 10.3923/ajbs.2019.396.403. [DOI] [Google Scholar]
- Sanz L., Calvete J.J. Insights into the evolution of a snake venom multi-gene family from the genomic organization of Echis ocellatus SVMP genes. Toxins. 2016;8 doi: 10.3390/toxins8070216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selistre-de-Araujo H.S., Cominetti M.R., Terruggi C.H.B., Mariano-Oliveira A., De Freitas M.S., Crepin M., Figueiredo C.C., Morandi V. Alternagin-C, a disintegrin-like protein from the venom of Bothrops alternatus, modulates a2b1 integrin-mediated cell adhesion, migration and proliferation. Braz. J. Med. Biol. Res. 2005;38:1505–1511. doi: 10.1590/S0100-879X2005001000007. [DOI] [PubMed] [Google Scholar]
- Senn H., Klaus W. The nuclear magnetic resonance solution structure of flavoridin, an antagonist of the platelet GP IIb-IIIa receptor. J. Mol. Biol. 1993;232:907–925. doi: 10.1006/jmbi.1993.1439. [DOI] [PubMed] [Google Scholar]
- Serrano S.M.T., Jia L.G., Wang D., Shannon J.D., Fox J.W. Function of the cysteine-rich domain of the haemorrhagic metalloproteinase atrolysin A: targeting adhesion proteins collagen I and von Willebrand factor. Biochem. J. 2005;391:69–76. doi: 10.1042/BJ20050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano S.M.T., Kim J., Wang D., Dragulev B., Shannon J.D., Mann H.H., Veit G., Wagener R., Koch M., Fox J.W. The cysteine-rich domain of snake venom metalloproteinases is a ligand for von Willebrand factor A domains: role in substrate targeting. J. Biol. Chem. 2006;281:39746–39756. doi: 10.1074/jbc.M604855200. [DOI] [PubMed] [Google Scholar]
- Serrano S.M.T., Wang D., Shannon J.D., Pinto A.F.M., Polanowska-Grabowska R.K., Fox J.W. Interaction of the cysteine-rich domain of snake venom metalloproteinases with the A1 domain of von Willebrand factor promotes site-specific proteolysis of von Willebrand factor and inhibition of von Willebrand factor-mediated platelet aggregation. FEBS J. 2007;274:3611–3621. doi: 10.1111/j.1742-4658.2007.05895.x. [DOI] [PubMed] [Google Scholar]
- Shannon J.D., Baramova E.N., Bjarnason J.B., Fox J.W. Amino acid sequence of a Crotalus atrox venom metalloproteinase which cleaves type IV collagen and gelatin. J. Biol. Chem. 1989;264:11575–11583. [PubMed] [Google Scholar]
- Shimokawa K.I., Shannon J.D., Jia L.G., Fox J.W. Sequence and biological activity of catrocollastatin-C: a disintegrin- like/cysteine-rich two-domain protein from Crotalus atrox venom. Arch. Biochem. Biophys. 1997;343:35–43. doi: 10.1006/abbi.1997.0133. [DOI] [PubMed] [Google Scholar]
- Shin J., Hong S.Y., Chung K., Kang I., Jang Y., Kim D.S., Lee W. Solution structure of a novel disintegrin, salmosin, from agkistrondon halys venom. Biochemistry. 2003;42:14408–14415. doi: 10.1021/bi0300276. [DOI] [PubMed] [Google Scholar]
- Shiu J.H., Huang C.H., Chang Y.T., Jeng W.Y., Chuang W.J. Effects of the regions adjacent to the RGD motif in disintegrins on their inhibitory activities and structures. PDB. 2015 http://www.rcsb.org/structure/4RQG [WWW Document] [Google Scholar]
- Singhamatr P., Rojnuckarin P. Molecular cloning of albolatin, a novel snake venom metalloprotease from green pit viper (Trimeresurus albolabris), and expression of its disintegrin domain. Toxicon. 2007;50:1192–1200. doi: 10.1016/j.toxicon.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Song J., Xu X., Zhang Y., Guo M., Yan X., Wang S., Gao S. Purification and characterization of AHPM, a novel non-hemorrhagic P-IIIc metalloproteinase with α-fibrinogenolytic and platelet aggregation-inhibition activities, from Agkistrodon halys pallas venom. Biochimie. 2013;95:709–718. doi: 10.1016/j.biochi.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Souza D.H.F., Iemma M.R.C., Ferreira L.L., Faria J.P., Oliva M.L.V., Zingali R.B., Niewiarowski S., Selistre-de-Araujo H.S. The disintegrin-like domain of the snake venom metalloprotease alternagin inhibits α2β1 integrin-mediated cell adhesion. Arch. Biochem. Biophys. 2000;384:341–350. doi: 10.1006/abbi.2000.2120. [DOI] [PubMed] [Google Scholar]
- Suntravat M., Jia Y., Lucena S.E., Sánchez E.E., Pérez J.C. cDNA cloning of a snake venom metalloproteinase from the eastern diamondback rattlesnake (Crotalus adamanteus), and the expression of its disintegrin domain with anti-platelet effects. Toxicon. 2013;64:43–54. doi: 10.1016/j.toxicon.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntravat M., Helmke T.J., Atphaisit C., Cuevas E., Lucena S.E., Uzcátegui N.L., Sánchez E.E., Rodriguez-Acosta A. Expression, purification, and analysis of three recombinant ECD disintegrins (r-colombistatins) from P-III class snake venom metalloproteinases affecting platelet aggregation and SK-MEL-28 cell adhesion. Toxicon. 2016;122:43–49. doi: 10.1016/j.toxicon.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvilesh K.N., Urs A.N.N., Savitha M.N., Gowda M.D.M., Vishwanath B.S. Snake venom proteinases as toxins and tools. In: Chakraborti S., Dhalla N.S., editors. Proteases in Physiology and Pathology. Springer Nature Singapore; 2017. pp. 1–619. [DOI] [Google Scholar]
- Takahashi T., Ohsaka A. Purification and some properties of two hemorrhagic principles (HR2a and HR2b) in the venom of Trimeresurus flavoviridis; complete separation of the principles from proteolytic activity. BBA - Protein Struct. 1970;207:65–75. doi: 10.1016/0005-2795(70)90137-6. [DOI] [PubMed] [Google Scholar]
- Takeda S., Igarashi T., Mori H., Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Igarashi T., Mori H. Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 2007;581:5859–5864. doi: 10.1016/j.febslet.2007.11.062. [DOI] [PubMed] [Google Scholar]
- Takeda S., Takeya H., Iwanaga S. Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta Protein Proteonomics. 2012;1824:164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Takeda S. ADAM and ADAMTS family proteins and snake venom metalloproteinases: a structural overview. Toxins. 2016;8:8–11. doi: 10.3390/toxins8050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya H., Arakawa M., Miyata T., Iwanaga S., Omori-Satoh T. Primary structure of H2-proteinase, a non-hemorrhagic metalloproteinase, isolated from the venom of the habu snake, Trimeresurus flavovidis. J. Biochem. 1989;106:151–157. doi: 10.1093/oxfordjournals.jbchem.a122805. [DOI] [PubMed] [Google Scholar]
- Takeya H., Nishida S., Miyata T., Kawada S., Yukari S., Morita T., Iwanaga S. Coagulation factor X activating enzyme from Russell's viper venom (RVV-X) J. Biol. Chem. 1992;267:14109–14117. [PubMed] [Google Scholar]
- Tan C.H., Tan K.Y., Lim S.E., Tan N.H. Venomics of the beaked sea snake, Hydrophis schistosus: a minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J. Proteomics. 2015;126:121–130. doi: 10.1016/j.jprot.2015.05.035. [DOI] [PubMed] [Google Scholar]
- Tan C.H., Tan K.Y., Ng T.S., Sim S.M., Tan N.H. Venom proteome of spine-bellied sea snake (hydrophis curtus) from penang, malasya: toxicity correlation, immunoprofiling and cross-neutralization by sea snake antivenom. Toxins. 2019;11:1–19. doi: 10.3390/toxins11010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanjoni I., Evangelista K., Della-Casa M.S., Butera D., Magalhães G.S., Baldo C., Clissa P.B., Fernandes I., Eble J., Moura-da-Silva A.M. Different regions of the class P-III snake venom metalloproteinase jararhagin are involved in binding to alpha2beta1 integrin and collagen. Toxicon. 2010;55:1093–1099. doi: 10.1016/j.toxicon.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Trummal K., Tõnismägi K., Siigur E., Aaspõllu A., Lopp A., Sillat T., Saat R., Kasak L., Tammiste I., Kogerman P., Kalkkinen N., Siigur J. A novel metalloprotease from Vipera lebetina venom induces human endothelial cell apoptosis. Toxicon. 2005;46:46–61. doi: 10.1016/j.toxicon.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Vidal N., Jackson T., Jouanne H. Snake venom in context: neglected clades and concepts. Front. Ecol. Evol. 2019;7:1–18. doi: 10.3389/fevo.2019.00332. [DOI] [Google Scholar]
- Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J.R., Kerkkamp H.M.E., Vos R.A., Guerreiro I., Calvete J.J., Wüster W., Woods A.E., Logan J.M., Harrison R.A., Castoe T.A., De Koning J., Pollock D.D., Yandell M., Calderon D., Renjifo C., Currier R.B., Salgado D., Pla D., Sanz L., Hyder A.S., Ribeiro J.M.C., Arntzen J.W., Van Den Thillart G.E.E.J.M., Boetzer M., Pirovano W., Dirks R.P., Spaink H.P., Duboule D., McGlinn E., Kini R.M., Richardson M.K. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallnoefer H.G., Lingott T., Gutiérrez J.M., Merfort I., Liedl K.R. Backbone flexibility controls the activity and specificity of a protein-protein interface: specificity in snake venom metalloproteases. J. Am. Chem. Soc. 2010;132:10330–10337. doi: 10.1021/ja909908y. [DOI] [PubMed] [Google Scholar]
- Wan S.G., Jin Y., Lee W.H., Zhang Y. Cloning of two novel P-III class metalloproteinases from Trimeresurus stejnegeri venom gland. Toxicon. 2006;47:465–472. doi: 10.1016/j.toxicon.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wan S.G., Jin Y., Lee W.H., Zhang Y. A snake venom metalloproteinase that inhibited cell proliferation and induced morphological changes of ECV304 cells. Toxicon. 2006;47:480–489. doi: 10.1016/j.toxicon.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Wang S.H., Shen X.C., Yang G.Z., Wu X.F. cDNA cloning and characterization of Agkistin, a new metalloproteinase from Agkistrodon halys. Biochem. Biophys. Res. Commun. 2003;301:298–303. doi: 10.1016/S0006-291X(02)03001-2. [DOI] [PubMed] [Google Scholar]
- Wang W.J., Shih C.H., Huang T.F. A novel P-I class metalloproteinase with broad substrate-cleaving activity, agkislysin, from Agkistrodon acutus venom. Biochem. Biophys. Res. Commun. 2004;324:224–230. doi: 10.1016/j.bbrc.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Watanabe L., Shannon J.D., Valente R.H., Rucavado A., Alape-Girón A., Kamiguti A.S., Theakston R.D.G., Fox J.W., Gutiérrez J.M., Arni R.K. Amino acid sequence and crystal structure of BaP1, a metalloproteinase from Bothrops asper snake venom that exerts multiple tissue-damaging activities. Protein Sci. 2009;12:2273–2281. doi: 10.1110/ps.03102403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis T.W., Tu A.T. Purification and biochemical characterization of atroxase, a nonhemorrhagic fibrinolytic protease from western diamondback rattlesnake venom. Biochemistry. 1988;27:4769–4777. doi: 10.1021/bi00413a028. [DOI] [PubMed] [Google Scholar]
- Wu W.B., Chang S.C., Liau M.Y., Huang T.F. Purification, molecular cloning and mechanism of action of graminelysin I, a snake-venom-derived metalloproteinase that induces apoptosis of human endothelial cells. Biochem. J. 2001;357:719–728. doi: 10.1042/0264-6021:3570719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You W.K., Seo H.J., Chung K.H., Kim D.S. A novel metalloprotease from Gloydius halys venom induces endothelial cell apoptosis through its protease and disintegrin-like domains. J. Biochem. 2003;134:739–749. doi: 10.1093/jb/mvg202. [DOI] [PubMed] [Google Scholar]
- Zancolli G., Calvete J.J., Cardwell M.D., Greene H.W., Hayes W.K., Hegarty M.J., Herrmann H.W., Holycross A.T., Lannutti D.I., Mulley J.F., Sanz L., Travis Z.D., Whorley J.R., Wüster C.E., Wüster W. When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. R. Soc. B Biol. Sci. 2019;286 doi: 10.1098/rspb.2018.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Botos I., Gomis-Rüth F.X., Doll R., Blood C., Njoroge F.G., Fox J.W., Bode W., Meyer E.F. Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d) Proc. Natl. Acad. Sci. U.S.A. 1994;91:8447–8451. doi: 10.1073/pnas.91.18.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.T., Lu P., Qin Y.F., Chen S.J., Guo A.G. Cloning and identification of a novel P-II class snake venom metalloproteinase from Gloydius halys. Appl. Biochem. Biotechnol. 2010;162:1391–1402. doi: 10.1007/s12010-010-8911-6. [DOI] [PubMed] [Google Scholar]
- Zhou O., Smith B.J., Grossman M.H. Molecular cloning and expression of catrocollastatin, a snake-venom protein from Crotalus atrox (western diamondback rattlesnake) which inhibits platelet adhesion to collagen. Biochem. J. 1995;307:411–417. doi: 10.1042/bj3070411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Dangelmaier C., Smith J.B. The hemorrhagin catrocollastatin inhibits collagen-induced platelet aggregation by binding to collagen via its disintegrin-like domain. Biochem. Biophys. Res. Commun. 1996;219:720–726. doi: 10.1006/bbrc.1996.0301. [DOI] [PubMed] [Google Scholar]
- Zhou X., Jin Y., Chen R., Lu Q., Wu J., Wang W., Xiong Y. Purification, cloning and biological characterization of a novel disintegrin from Trimeresurus jerdonii venom. Toxicon. 2004;43:69–75. doi: 10.1016/j.toxicon.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Zhu X., Teng M., Niu L. Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases. Acta Crystallogr. D. 1999;55:1834–1841. doi: 10.1107/S0907444999010306. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Gao Y., Zhu Z., Yu Y., Zhang X., Zang J., Teng M., Niu L. Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins. Biochem. Biophys. Res. Commun. 2009;386:159–164. doi: 10.1016/j.bbrc.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Zychar B.C., Clissa P.B., Carvalho E., Baldo C., Gonçalves L.R.C. Leukocyte recruitment induced by snake venom metalloproteinases: role of the catalytic domain. Biochem. Biophys. Res. Commun. 2020;521:402–407. doi: 10.1016/j.bbrc.2019.10.144. [DOI] [PubMed] [Google Scholar]