Abstract
Bovine leukemia virus (BLV) belongs to the genus, Deltaretrovirus of the family, Retroviridae and it is the causative agent of enzootic bovine leukosis. The prevalence of BLV in three provinces in the Red River Delta Region in the North of Vietnam, Hanoi, Vinhphuc and Bacninh was studied from April 2017 to June 2018. A total of 275 blood samples collected from cattle were used for serum isolation and DNA extraction. Of these samples, 266 sera were subjected to ELISA test for detecting antibody against BLV gp51 protein and 152 DNA samples were used to detect the 444 bp fragment corresponding to a part of the gp51 region of the env by nested PCR. The results showed that 16.5% (n=44) and 21.1% (n=32) of samples were positive for BLV gp51 antibody and BLV proviral DNA, respectively. Phylogenetic analysis of the partial (423 bp) and complete (913 bp) BLV env-gp51 gene indicated that Vietnamese strains were clustered into genotypes 1, 6 and 10 (G1, G6 and G10). Of those genotypes, G1 genotype was dominant; G6 strains were designated as G6e and G6f subgenotypes; the existence of genotype 10 was confirmed for the first time in Vietnam. The present study provides important information regarding the prevalence of BLV infection and genetic characteristics of BLV strains identified in Vietnam, contributing to promote the establishment of disease control and eradication strategies in Vietnam.
Keywords: bovine leukemia virus (BLV), cattle, detection, genotype, Vietnam
Bovine leukemia virus (BLV) belongs to the genus, Deltaretrovirus of the family, Retroviridae that is the causative agent of enzootic bovine leukosis. Most BLV-infected animals are asymptomatic virus carriers. Only 30–70% of the infected cattle develop persistent lymphocytosis and 0.1–10% of them develop tumors [7, 30].
BLV infection has been reported to occur worldwide in cattle populations and the prevalence of infection varies among and within countries [33, 38]. Moreover, BLV infection causes serious economic damage to the livestock industry due to the decrease in milk production, reproduction rates, shortening cow longevity and increase heifer replacement costs [3, 35, 36]. Therefore, the majority of the western European countries, Australia and New Zealand established eradication programs and control measurement resulting in BLV infection rates negligible [1, 15, 22, 27, 33]. However, most countries are still confronting the burden of BLV infection. Across Asian countries, BLV infection rate remains a wide range from 3.9% to 70% among Japan [29], Korea [19], China [42, 43], Taiwan [41], Thailand [20], Philippines [32], Cambodia [24], Mongolia [28], Myanmar [31], and Vietnam [9].
The BLV genome consists of gag, pro, pol, and env gene, which encode structural proteins and enzymes, the regulatory genes tax and rex, and the accessory genes R3 and G4, and two identical long terminal repeats [40]. The env gene encodes the envelope protein complex composing gp51 surface glycoprotein (SU) and gp30 transmembrane (TM) protein. As the env-gp51 plays an essential and indispensable role for viral life cycle and viral infectivity, such as cell entry and production of neutralizing antibodies [17, 21], the env-gp51 gene had been widely become a target gene for diagnosis, molecular characterization and genotyping of BLV [12, 19, 20, 32, 34, 43]. Based on the analysis of gp51 gene sequences, Rodriguez et al. demonstrated that BLV strains can be classified into 7 distinct genotypes [39]. Subsequently, on the basis of phylogenetic analysis of gp51 gene sequences, a study reported the existence of genotype 8 in BLV samples from Croatia [2]. The novel genotypes, genotypes 9 and genotype 10 were confirmed in Bolivia [34], Thailand [20] and Myanmar [31]. Finally, in 2019, the newest BLV genotype, genotype 11 was discovered in China [43].
The studies related to BLV have been increasing worldwide, however, few have attempted to conduct serological and genotyping studies of BLV infection in Vietnam. Therefore, this study aims to determine the prevalence of BLV in Vietnam by enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR). Furthermore, we have aimed to reveal the sequence variability of Vietnamese BLV strains by performing DNA sequencing and phylogenetic analysis of both partial and complete env-gp51 gene sequences.
MATERIALS AND METHODS
Ethical statement
The blood samples were collected by the Vietnam National University of Agriculture in strict accordance following the guidelines of National technical regulation on Animal diseases−General requirements for sample collection, storage and shipment (QCVN01-83:2011/BNNPTNT). Consent was obtained from the farm owners before animal sampling.
Study population and animals
The study population was comprised of cattle kept in three provinces, namely, Hanoi, Vinhphuc, and Bacninh, in the Red River Delta region in the Northern part of Vietnam (Fig. 1). The population of cattle in the Red River Delta region in 2016 was 493,100 [14]. The present investigation included 275 blood samples collected from either dairy or beef cattle kept on 80 farms, including 43 farms (n=168) in Hanoi, 29 farms (n=77) in Vinhphuc and 8 farms (n=30) in Bacninh between April 2017 to June 2018. The farms were selected for convenience sampling, e.g. geographical location and cooperative farms [10]. The farm holding less than 10 cattle was considered as small sized farm. The farm having 10 to 45 cattle was considered as medium sized farm. This present research included 53 dairy cattle farms keeping Holstein Friesian (HF), Jersey, Lai HF (Holstein Friesian and Blanc Bleu Belge cross) breed, 24 beef cattle holding Lai Sind (Vietnamese native cattle and Red Sindhi cross), Red Sindhi, Lai Brahma (Vietnamese native cattle and Brahma cross), Blanc Bleu Belge (BBB), Vietnamese native cattle and the 3 remained mixed breed farms. The animals included 241 female cattle (203 HF, 1 Yersey, 22 Lai Sind, 10 Vietnamese native, 5 Lai Brahma) and 34 male cattle (1 HF, 2 Lai HF, 23 Lai Sind, 4 BBB, 1 Vietnamese native, 3 Red Sindhi). The animals were selected based on their owner’s willingness to provide the samples. At least one blood sample was collected from each herd. Age of these cattle ranged from 4 months to 9 years old.
Fig. 1.
A map of the Red River Delta region showing the number of cattle (heads) sampled from three provinces involved in the present study. n, indicates the total number of cattle in each province.
Sample collection and serum preparation
Peripheral blood was obtained from the jugular vein with or without anticoagulant. Clotted blood samples were centrifuged for 5 min at 3,000 rpm and each serum sample was collected in 1.5 ml centrifuge tubes and stored at −30°C for further analysis.
Enzyme-linked immunosorbent assay (ELISA) test
The antibody against BLV was detected by ELISA kit (JNC Inc., Tokyo, Japan) according to the manufacturer’s instructions. In brief, serum samples were diluted 1:50 in diluent buffer that provided in the kit and used for ELISA assay. Subsequently, optical density (OD) values were measured by Chromate Microplate Reader (Awareness Technology, Inc., Palm City, FL, USA). The sample-to-positive (S/P) ratio was used to determine the antibody-positive sample. The sample was interpreted as positive if the value of S/P ratio was equal to or greater than 0.3 and the value of the positive control was greater than 0.6.
DNA extraction
DNA was extracted from 200 µl ethylenediaminetetraacetic acid (EDTA)-treated whole blood sample using QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s recommended protocol. Afterwards, the extracted DNA concentration was measured by NanoDropTM Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −30°C until required for PCR.
Detection of BLV provirus by Nested polymerase chain reaction (nested PCR)
The extracted DNA samples were used as a template to detect the partial gp51 (env) gene of BLV using two sets of primers previously reported by Fechner et al. [11] (External primers env5037F/env5613R and internal primers env5104F/env5526R; Table 1). External primers and internal primers resulted in the amplification of 598 bp and 444 bp DNA fragments in the gp51 region of env gene, respectively. For both the first and the second round PCR, the same component of the mixture was used. In brief, each 20 µl reaction buffer contained 10 µl Buffer KOD FX Neo 2x; 4 µl dNTPs 2 mM; 3.8 µl distilled water; 0.8 µl of external primers mixture for the first round PCR or 0.8 µl of internal primers mixture for the second round PCR and 0.4 µl of KOD FX Neo Polymerase (Toyobo, Osaka, Japan); One µl of DNA (20 ng/µl) extracted from blood was added to the first round PCR and 1 µl of the first round PCR product was added to the nested PCR and. The conditions for the first round PCR amplification were as follows: an initial denaturation at 98°C for 1 min; 40 cycles of denaturing at 98°C for 10 sec, annealing at 58°C for 30 sec and extension at 72°C for 1 min. The reaction parameters for the second round PCR were 1 min at 98°C for initial denaturation, followed by 35 cycles of 10 sec at 98°C for denaturing, 30 sec at 70°C for annealing and 1 min at 72°C for extension. The amplification reactions were performed in a S1000TM Thermal Cycler (Bio Rad Laboratories, Inc.; Hercules, CA, USA). Both rounds of the PCR products were analyzed by electrophoresis on 2% agarose gel containing Gel Red (Biotium, Inc., Fremont, CA, USA) in Tris-acetate-EDTA buffer.
Table 1. Primers used in this study.
| Primer name | Sequence (5′–3′) | Target band | Reference |
|---|---|---|---|
| 1. Nested PCR bovine leukemia virus env-gp51 | |||
| Env5037F | TCTGTGCCAAGTCTCCCAGATA | 598 bp | Fechner et al. [11] |
| Env5613R | AACAACAACCTCTGGGAAGGGT | ||
| Env5104F | CCCACAAGGGCGGCGCCGGTTT | 444 bp | |
| Env5526R | GCGAGGCCGGGTCCAGAGCTGG | ||
| 2. Sequence of nested PCR product env-gp51 | |||
| Pol4605F | TCAGAGGGCGGAGAAACAC | 1,558 bp | Dao et al. [9] |
| Env6190R | GGTCAAGCATTTTATCAGG | ||
| Pol4794F | TGGGTTCCCTGGCGTTT | 1,375 bp | |
| Env6148R | AAAAAGGGCTAATAGGAACAGG | ||
Nucleotide sequence and phylogenetic analysis
The samples, which were satisfied by both the nested PCR and ELISA tests, were further used for complete gp51 sequencing. Two sets of primers for the nested PCR previously described [9] were used to amplify 1,558 bp and 1,375 bp sequences of the BLV env-gp51 gene (Table 1). The conditions for the first round PCR amplification were as follows: an initial denaturation at 94°C for 2 min; 40 cycles of denaturing at 98°C for 10 sec, annealing at 57°C for 30 sec and extension at 72°C for 1 min; additional cycle was run at 72°C for 10 min. The reaction parameters for the second round PCR were 2 min at 94°C for initial denaturation, followed by 35 cycles of 10 sec at 98°C for denaturing, 30 sec at 56°C for annealing and 1 min at 72°C for extension; the last cycle was done at 72°C for 10 min. The final PCR products were separated on 1% agarose gels and purified using NucleSpin® Gel and PCR Clean up kit (Macherey-Nagel GmbH & Co., KG, Germany) following the manufacturer’s recommendations. The purified PCR products were then sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and 3130xl Genetic Analyzer (Thermo Fisher Scientific), according to the manufacturer’s instructions. Obtained sequences were analyzed using MEGA 7 software [18].
The BLV genome sequences from Vietnam were aligned in parallel with BLV env sequences from each reference sequence of the eleven known BLV genotypes available in GenBank of National Center for Biotechnology Information (NCBI). Phylogenetic analysis of the partial (423 bp) and complete (913 bp) sequence of the env-gp51 gene was performed using MEGA 7 software. Kimura 2 parameter model with gamma distribution (K2 + G) was found as the model with the best appropriate to analyze the BLV env-gp51 gene sequences by using the “Find Best DNA/Protein Models” tool of MEGA 7. Phylogenetic trees were constructed using the maximum likelihood (ML) algorithm with the Kimura-2 parameter model of nucleotide of substitution. The reliability of phylogenetic relationships was evaluated using bootstrap analysis with 1,000 replicates.
Statistical analysis
BLV prevalence determined by ELISA was analyzed in relation to individual level (location, breed, gender, and age) and farm level (farm scale and farm type). Chi-square (χ2) test of independence was used to determine the presence of significant relationship between two variables (gender and breed). All statistical analysis were performed with Excel software.
RESULTS
Detection of BLV infection by ELISA and nested PCR
A total of 275 blood samples collected from three provinces, including Hanoi, Vinhphuc and Bacninh, as shown in Fig. 1, were screened for BLV infection. Of these samples, 266 were subjected to ELISA test for detecting the antibody against BLV gp51 protein and 152 extracted DNA samples were used in nested PCR as template to amplify the 444 bp fragment corresponding to a part of the env-gp51 by nested PCR.
The results of ELISA test for detecting the BLV antibody are summarized in Table 2. The prevalence in individual level of BLV in the research area was 16.5% (44/266) by ELISA test. Of the 159 samples collected from different farms in Hanoi, 20 (12.6%) were found positive. Of the 77 screened samples collected in Vinhphuc, 15 were antibody positive (19.5% prevalence). When compared between three provinces as mentioned above, samples collected from Bacninh indicated the highest level of BLV infection rate of 30% (9/30). There was a significant difference between dairy and beef cattle groups (P<0.01). No positive sample was observed among the beef cattle group, whereas all positive cattle detected in this study belonged to the dairy cattle group. The percentage of BLV infection in the dairy cattle group was 22.1% (44/199). The prevalence between male and female group was significantly different (P<0.01). Among 232 female cattle involved in this research, 44 (19.0%) were found BLV positive by ELISA. In contrast, no BLV positive cattle was found in the male group. Five of the 49 cattle (10.2%) at 1 year old or younger were positive, whereas the prevalence of BLV in older cattle was found to be higher. However, the difference was not significant (P>0.05). The BLV positive proportion in farm level was determined as 28.8% (23/80). Among the two sized farm groups, 15 out of 61 farms (24.6%) in the small sized farm group had at least one positive cattle, whereas a higher positive proportion (42.1%) was observed in the medium sized group. However, the prevalence between small and medium farms was not significantly different (P>0.05). BLV herd level seroprevalence was 43.4% in the dairy cattle farm. Antibody against BLV was not detected in beef and mixed breed cattle farms included in this research.
Table 2. Bovine leukemia virus infection in cattle in individual and farm levels as detected by ELISA.
| Categories | No. tested | No. positive | Prevalence (%) | ||
|---|---|---|---|---|---|
| Individual level | Location | Hanoi a) | 159 | 20 | 12.6 |
| Bacninh a) | 30 | 9 | 30.0 | ||
| Vinh Phuc | 77 | 15 | 19.5 | ||
| Breed | Dairy cattle b) | 199 | 44 | 22.1 | |
| Beef cattle b) | 67 | 0 | 0.0 | ||
| Gender | Male c) | 34 | 0 | 0.0 | |
| Female c) | 232 | 44 | 19.0 | ||
| Age | ≤1 year old | 49 | 5 | 10.2 | |
| >1; ≤2 years old | 75 | 13 | 17.3 | ||
| >2; ≤3 years old | 74 | 13 | 17.6 | ||
| >3 years old | 67 | 13 | 19.4 | ||
| Undetermined | 1 | 0 | 0.0 | ||
| Total | 266 | 44 | 16.5 | ||
| Farm level | Scale | Small sized farm | 61 | 15 | 24.6 |
| Medium sized farm | 19 | 8 | 42.1 | ||
| Type | Dairy farm d) | 53 | 23 | 43.4 | |
| Beef farm d) | 24 | 0 | 0.0 | ||
| Mixed farm | 3 | 0 | 0.0 | ||
| Total | 80 | 23 | 28.8 | ||
a–d) Prevalence was significantly different (a) P=0.0152, b) P<0.001, c) P=0.0054, d) P<0.001). b–d) There was dependency between gender distribution and breed distribution (χ2=90.08, P<0.05); between gender distribution and farm type distribution (χ2=87.05, P<0.05), indicating that these differences might have been caused by breed effect.
Out of 152 DNA samples tested by nested PCR, 444 bp env-gp51 fragment was yielded in 32 samples (21.1%).
Phylogenetic analysis of partial and complete sequences of BLV env-gp51 of Vietnamese strains
The BLV env-gp51 gene (913 bp) sequence of the representative BLV Vietnamese strains identified in the present study were submitted to DDBJ/EMBL/Genbank databases under accession numbers LC512445 to LC512452.
The partial env-gp51 gene sequence (423 bp) of Vietnamese isolates were aligned in parallel with 53 corresponding sequences from all of the 11 BLV genotypes available in GenBank. Thereafter, a ML phylogenetic tree based on Kimura-2 model of nucleotide substitution with gamma distribution (K2 + G) was constructed (Fig. 2).
Fig. 2.
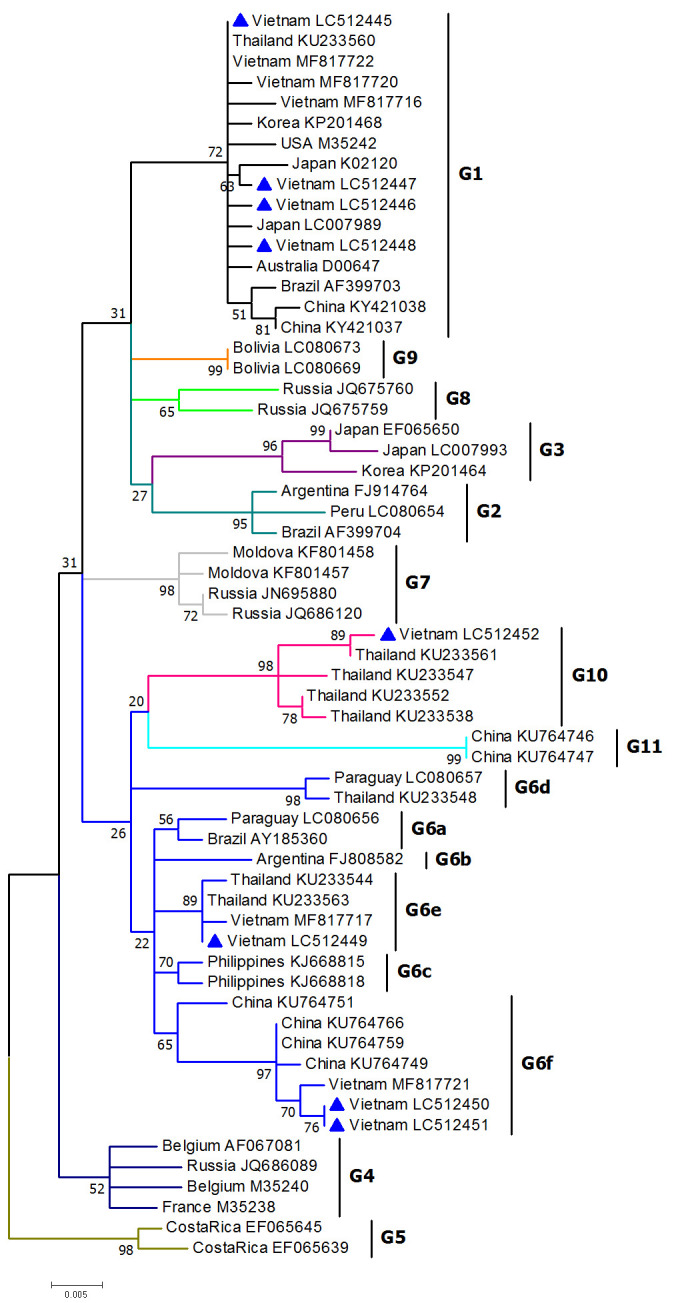
A maximum likelihood (ML) phylogenetic tree built based on the nucleotide sequences of the partial BLV env-gp51 gene (423 bp) from Vietnam and strains representing all known BLV genotypes. BLV strains identified in this research are indicated by filled triangles and the remaining strains are shown by country name and GenBank accession number. All genotypes are indicated by vertical lines with the symbol “G”. The numbers at the branches show ML bootstrap support values. The bar at the bottom of the figure denotes the distance. The tree was rooted on G5.
The phylogenetic tree of the env-gp51 gene revealed that Vietnamese isolates belonged to genotypes 1, 6 and 10, supported by high bootstrap value ≥96 for each group (Fig. 2 and Supplementary Fig. 1). Genotype 1 was dominant among the samples analyzed in the present study. Two isolates from Bacninh province (LC512450, LC512451) and one isolate from Hanoi (LC512449) were assigned to genotype 6 with the supporting value of 96 in ML analysis (Supplementary Fig. 1). The Vietnamese G6 strain (LC512449) belonged to the subgenotype G6e. Remarkably, two G6 genotype isolates from Bacninh (LC512450, LC512451) were grouped into an independent branch which also included G6f strains. This subgrouping is supported by bootstrap value ≥65 (Fig. 2). Noticeably, the phylogenetic analysis revealed the existence of genotype 10 in Vietnamese cattle and Vietnamese strains were found genetically more closely related to the strains circulating in Thailand.
In order to confirm the genotyping results found in 423 bp env-gp51 gene sequences analysis, we performed another phylogenetic analysis based on complete gp51 gene sequence (913 bp) of Vietnamese isolates and corresponding sequences from ten BLV genotypes (genotype 11 strains were not included) (Supplementary Fig. 1). The ML phylogenetic tree based on complete gp51 gene sequence was consistent with that of the partial env-gp51 sequences, i.e., the Vietnamese BLV strains were clustered into genotypes 1, 6, and 10.
Nucleotide and amino acid substitutions of BLV env-gp51 in Vietnamese strains
Partial env-gp51 nucleotide sequences (423 bp) from the seven typical Vietnamese strains were aligned with that of the reference G1, G6 and G10 strains (Supplementary Fig. 2A). Twenty-nine substitutions were identified in our obtained sequences. Of these substitutions, 23 were silent and the others were nonsynonymous substitutions (indicated by numbers and black filled triangles in Supplementary Fig. 2A). The six nonsynonymous substitution sites consisted of residues 106 (nt 317), 133 (nt 397), 143 (nt 428), 144 (nt 431), 177 (nt 529), and 189 (nt 566). Our BLV strain grouped in subgenotype G6f shared a unique nonsynonymous substitution, which was distinguished from the other G6 subgenotypes, in the third base of residue 189 (nt 566) with two subgenotype G6f strains from Vietnam and China. The alignment of nucleotide sequences revealed the existence of genotype-10-specific amino acid substitutions at residues 106 (nt 317), 133 (nt 397), 143 (nt 428), 144 (nt 431), and 177 (nt 529) in Vietnamese strains.
To provide greater insights into the effects of nucleotide substitutions found in Vietnamese BLV strains, alignment of deduced amino acid sequences of the seven partial env-gp51 sequences of the representative genotype 1, 6, and 10 strain sequences were performed. As shown in Supplementary Fig. 2B, we found one substitution in the neutralizing domain 1 (ND1) (97–106), one in the CD4+ T cell Epitope (84–113), three in the neutralizing domain 2 (ND2) (131–150), two in the zinc binding zone (ZB) (137–156), one in the CD8+ T cell Epitope (154–183) and two in the E Epitope region (175–194). However, G Epitope, neutralizing domain 3 (ND3) and B Epitope in all the Vietnamese strains were found fully conserved.
DISCUSSION
Prevalence of BLV infection differs among countries. Epidemiological and molecular studies on BLV have been increasing worldwide, however, very few attempts have been conducted to investigate BLV infection in Vietnam. In the present study, 275 cattle sampled from 3 provinces in the Red River Delta Region of Vietnam were screened for BLV prevalence and 31 Vietnamese isolates were characterized.
Our results demonstrated that the percentage of BLV infection in Vietnam was 16.5% as determined by ELISA tested and 21.1% by nested PCR. Different prevalence rates were observed according to the cattle breeds and gender. BLV seroprevalences of male and female cattle were significantly different (P<0.01). Dairy cattle had a greater prevalence of BLV (22.1%) compared with beef cattle in Vietnam (P<0.01). Similarly, the number of BLV-infected dairy farms compared with BLV-infected beef breeding farms was significantly higher (P<0.001). However, our statistic analysis suggested that there was dependency between gender distribution and breed distribution (χ2=90.08, P<0.05); between gender distribution and farm type distribution (χ2=87.05, P<0.05), indicating that these differences might have been caused by breed effect. Interestingly, no serological positive sample was found in beef cattle (predominant breed, Lai Sind) suggesting that dairy cattle in Vietnam were more severely affected by BLV infection than beef cattle. Similarly, in the previous studies, higher BLV prevalence within dairy cattle was observed when compared with beef cattle [4, 5, 16, 23, 25, 26, 42]. This phenomenon may be explained by the association between breed and genomic factor playing a role in the susceptibility of cattle to BLV infection [42]. Besides, the difference of the BLV infection prevalence in dairy and beef cattle farm type may have been caused by distinct in husbandry practices [25, 42]. Considering the feeding calves by pooled colostrum and milk, sharing milking machine among cows, and shorter keeping time of fattening beef animals may be the factors that caused difference of BLV infection between dairy and beef cattle in Vietnam.
BLV induces a persistent infection in cattle and higher prevalence within older cattle has been observed [41]. It is possible that the older cattle have potentially been infected with BLV for a long period. Although we found that older cattle had a higher prevalence of BLV infection compared with younger animals (≤ one year old), the difference was not significant.
Our data showed that BLV infection was widespread in cattle population in whole research area, particularly in Bacninh province (30.0%, 9/30) that had significantly higher infection rate than Hanoi (12.6%, 20/159). However, the difference may be caused by breed effect whereas all cattle in Bacninh province were dairy cattle (data not shown).
The BLV infection percentage in Vietnam was higher than that of the other neighboring countries, such as Philippines (9.7%) [32], Myanmar (9.1%) [31] and Cambodia (5.3%) [24]. Whereas, the BLV infection percentage in Vietnam was lower than the results observed in other Asian countries such as China (49.1%) [42], Korea (54.2%) [19], Thailand (58.7%) [20] and Japan (73.3%) [29]. Varieties in the percentage of BLV infection are likely to occur among countries and areas within the same country. A most recent research related to BLV in Vietnam published in 2019 indicated that BLV prevalence circulating among small and medium holding farms in the North of Vietnam was 35.48% as determined by both ELISA test (targeted gp51) and real time PCR (targeted the BLV pol and tax gene) [9]. The prevalence of BLV infection observed in this study may not represent the actual prevalence of BLV in the studied locations because the samples were not collected randomly. However, the present research provides evidence that BLV infection in Vietnamese cattle is endemic.
The samples satisfied by both ELISA and nested PCR test were further used for sequencing to determine their genotypes. Three genotypes 1, 6, and 10 were identified in this study. Recent phylogenetic studies using BLV env gene sequences from the isolated strains have demonstrated the existence of 11 genotypes distributing worldwide. Genotype 1 is the most dominant genotype distributing worldwide including the United States, South American countries, Asian countries, and Australia. In the present study, genotype 1 was confirmed in total of 28 BLV isolates (90%, 28/31). Thus, genotype 1 is suggested to be the predominant genotype in Vietnam. The same result was also obtained in the previous research [9].
Phylogenetic analysis of the partial or complete sequence of the env-gp51 gene indicated the existence of six subgenotypes or subgroups within BLV genotype 6: G6a, G6b, G6c [32], G6d, G6e [13, 20], and a recently designated subgenotype G6f [9, 43]. Two Vietnamese BLV isolates (LC512450, LC512451) from the present research were clustered into subgroup G6f with the reported subgenotype G6f strains (GenBank accession numbers: KU764766, KU764759, KU764749 and MF817721) identified in the recent studies of Vietnam and China [9, 43]. Three strains (KU764766, KU764759, KU764749) were assigned as subgenotype G6e in the previous research [43]. However, the results of our phylogenetic analysis based on 423 bp of env-gp51 suggested that these strains should be clustered into subgenotype G6f rather than G6e, supported by bootstrap value of 65 in ML analysis (Fig. 2).
The existence of genotype 1 and 6 was reported in the recent study about BLV circulating among small and medium holding cattle farms in the North of Vietnam [9]. Interestingly, in the present research, we identified an additional genotype 10, bringing together, the total number of confirmed BLV genotypes in Vietnam into three. BLV infection may be diverse in Vietnam, not only restricted in genotype 1 and 6 as found in the recent research [9]. However, an only limited number of sample was identified as genotype 10 (one strain isolated in Vinhphuc province).
Interestingly, G6 and G10 are suggested to be separated in phylogenetic tree, constructed based on the complete BLV env-gp51 (913 bp) than that of the shorter sequence (423 bp) (Fig. 2 and Supplementary Fig. 1).
Remarkably, our phylogenetic tree also revealed that Vietnamese BLV isolates of genotype 6 and 10 was genetically more closely related with the BLV 6 and 10 strains circulating in Thailand. Genotype 10 was first reported in Thailand [20]. Moreover, Vietnam imported live cattle from Thailand in order to supply the growing demand for meat consumption and breeding [6]. The importation of BLV-infected animals has been considered as the main source of BLV introduction [8]. Therefore, we hypothesize that Vietnamese cattle may have been infected by BLV which came from outside Vietnam and then subsequently transmitted throughout the domestic cattle population.
The identification of 29 nucleotide substitutions of BLV Vietnamese strains were confirmed by analyzing of partial BLV env-gp51 sequences, 23 of which were silent substitutions, and 6 were amino acid substitutions. Nucleotide substitutions were observed at residues 106 (overlapping ND1 and CD4+ T cell Epitope), 133, 143, 144 (between the second ND and overlapping of ND2 and ZB), 177 (overlapping CD8+ T cell Epitope and E Epitope) and 189 (between E Epitope). Thus, all of the amino acid changes occurred within epitope regions. This result was consistent with previous findings that most amino acid substitutions in env-gp51 were found within epitope regions rather than at the random locations [37, 44]. Alignment of partial BLV env-gp51 showed a unique nucleotide and amino acid substitution (189-nt 566) to subgenotype G6f (Supplementary Fig. 2B). Nevertheless, the biological functions of that substitution in the E Epitope was unraveled.
Amino acid substitutions were found in overlapping CD4+ T cell Epitope and ND1 (106), ND2, overlapping ND2-ZB (133, 143, 144) and overlapping CD8+ Epitope and E Epitope (177) in the Vietnamese genotype 10 strains. These substitutions have also been previously observed in other BLV genotype 10 strains showing the specific characteristics of this genotype [20, 31].
Although a limited number of samples from three provinces was analyzed in the present study, both serological (ELISA) and molecular (nested PCR) analysis demonstrated the existence of BLV infection in cattle kept in Vietnam. Based on genetic analysis of the partial (423 bp) and complete (913 bp) BLV env-gp51 gene of the Vietnamese isolates, the present study revealed the existence of three genotypes 1, 6, and 10 in Vietnamese cattle population. Additionally, phylogenetic analysis indicated a close relationship between Vietnamese strains and BLV genotypes 1, 6, and 10 strains circulating in Thailand. The present research provides important information about BLV infection levels and genetic characteristics of Vietnamese strains. A comprehensive screening in the larger location should be conducted. More attention and providing supplementary information of BLV is also required to establish and implement an effective preventive method to eradicate BLV infection in Vietnam.
Supplementary
Acknowledgments
This work was partly supported by JSPS KAKENHI Grant Number 20H03142.
REFERENCES
- 1.Acaite J., Tamosiunas V., Lukauskas K., Milius J., Pieskus J.2007. The eradication experience of enzootic bovine leukosis from Lithuania. Prev. Vet. Med. 82: 83–89. doi: 10.1016/j.prevetmed.2007.05.010 [DOI] [PubMed] [Google Scholar]
- 2.Balić D., Lojkić I., Periškić M., Bedeković T., Jungić A., Lemo N., Roić B., Cač Z., Barbić L., Madić J.2012. Identification of a new genotype of bovine leukemia virus. Arch. Virol. 157: 1281–1290. doi: 10.1007/s00705-012-1300-4 [DOI] [PubMed] [Google Scholar]
- 3.Bartlett P. C., Norby B., Byrem T. M., Parmelee A., Ledergerber J. T., Erskine R. J.2013. Bovine leukemia virus and cow longevity in Michigan dairy herds. J. Dairy Sci. 96: 1591–1597. doi: 10.3168/jds.2012-5930 [DOI] [PubMed] [Google Scholar]
- 4.Bauermann F. V., Ridpath J. F., Dargatz D. A.2017. Bovine leukemia virus seroprevalence among cattle presented for slaughter in the United States. J. Vet. Diagn. Invest. 29: 704–706. doi: 10.1177/1040638717702183 [DOI] [PubMed] [Google Scholar]
- 5.Baumgartener L. E., Olson C., Miller J. M., Van Der Maaten M. J.1975. Survey for antibodies to leukemia (C-type) virus in cattle. J. Am. Vet. Med. Assoc. 166: 249–251. [PubMed] [Google Scholar]
- 6.Bunmee T., Chaiwang N., Kaewkot C., Jaturasitha S.2018. Current situation and future prospects for beef production in Thailand - A review. Asian-Australas. J. Anim. Sci. 31: 968–975. doi: 10.5713/ajas.18.0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burny A., Cleuter Y., Kettmann R., Mammerickx M., Marbaix G., Portetelle D., van den Broeke A., Willems L., Thomas R.1988. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Vet. Microbiol. 17: 197–218. doi: 10.1016/0378-1135(88)90066-1 [DOI] [PubMed] [Google Scholar]
- 8.Camargos M. F., Pereda A., Stancek D., Rocha M. A., dos Reis J. K., Greiser-Wilke I., Leite R. C.2007. Molecular characterization of the env gene from Brazilian field isolates of Bovine leukemia virus. Virus Genes 34: 343–350. doi: 10.1007/s11262-006-0011-x [DOI] [PubMed] [Google Scholar]
- 9.Dao T. D., Nguyen H. T., Than S. T., Bui V. N., Ogawa H., Imai K.2019. Bovine leukemia virus genotype 1 and 6 are circulating among dairy and beef cattle of small and medium holding farms in northern Vietnam. Jpn. J. Vet. Res. 67: 83. [Google Scholar]
- 10.Dohoo I. R., Martin W., Stryhn H. E.2010. Veterinary Epidemiologic Research, 2nd ed., VER Inc, Charlottetown, Prince Edward Island, Canada. [Google Scholar]
- 11.Fechner H., Blankenstein P., Looman A. C., Elwert J., Geue L., Albrecht C., Kurg A., Beier D., Marquardt O., Ebner D.1997. Provirus variants of the bovine leukemia virus and their relation to the serological status of naturally infected cattle. Virology 237: 261–269. doi: 10.1006/viro.1997.8784 [DOI] [PubMed] [Google Scholar]
- 12.Felmer R., Muñoz G., Zúñiga J., Recabal M.2005. Molecular analysis of a 444 bp fragment of the bovine leukaemia virus gp51 env gene reveals a high frequency of non-silent point mutations and suggests the presence of two subgroups of BLV in Chile. Vet. Microbiol. 108: 39–47. doi: 10.1016/j.vetmic.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 13.Gautam S., Mishra N., Kalaiyarasu S., Jhade S. K., Sood R.2018. Molecular Characterization of Bovine Leukaemia Virus (BLV) Strains Reveals Existence of Genotype 6 in Cattle in India with evidence of a new subgenotype. Transbound. Emerg. Dis. 65: 1968–1978. doi: 10.1111/tbed.12979 [DOI] [PubMed] [Google Scholar]
- 14.General Statistics Office of Vietnam. 2016. Number of cattle as of annual 1st October by province. http://www.gso.gov.vn/default_en.aspx?tabid=778 [accessed on January 9, 2020].
- 15.Hinrich V.2012. Reports from industry surveillance and disease control programmes−New Zealand enzootic bovine leukosis (EBL) control scheme. Surveillance 37: 33–34. [Google Scholar]
- 16.Hopkins S. G., DiGiacomo R. F.1997. Natural transmission of bovine leukemia virus in dairy and beef cattle. Vet. Clin. North Am. Food Anim. Pract. 13: 107–128. doi: 10.1016/S0749-0720(15)30367-4 [DOI] [PubMed] [Google Scholar]
- 17.Johnston E. R., Radke K.2000. The SU and TM envelope protein subunits of bovine leukemia virus are linked by disulfide bonds, both in cells and in virions. J. Virol. 74: 2930–2935. doi: 10.1128/JVI.74.6.2930-2935.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S., Stecher G., Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee E., Kim E. J., Joung H. K., Kim B. H., Song J. Y., Cho I. S., Lee K. K., Shin Y. K.2015. Sequencing and phylogenetic analysis of the gp51 gene from Korean bovine leukemia virus isolates. Virol. J. 12: 64. doi: 10.1186/s12985-015-0286-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E., Kim E. J., Ratthanophart J., Vitoonpong R., Kim B. H., Cho I. S., Song J. Y., Lee K. K., Shin Y. K.2016. Molecular epidemiological and serological studies of bovine leukemia virus (BLV) infection in Thailand cattle. Infect. Genet. Evol. 41: 245–254. doi: 10.1016/j.meegid.2016.04.010 [DOI] [PubMed] [Google Scholar]
- 21.Mamoun R. Z., Morisson M., Rebeyrotte N., Busetta B., Couez D., Kettmann R., Hospital M., Guillemain B.1990. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 64: 4180–4188. doi: 10.1128/JVI.64.9.4180-4188.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maresca C., Costarelli S., Dettori A., Felici A., Iscaro C., Feliziani F.2015. Enzootic bovine leukosis: report of eradication and surveillance measures in Italy over an 8-year period (2005–2012). Prev. Vet. Med. 119: 222–226. doi: 10.1016/j.prevetmed.2015.02.024 [DOI] [PubMed] [Google Scholar]
- 23.Marín C., de López N. M., Alvarez L., Lozano O., España W., Castaños H., León A.1978. Epidemiology of bovine leukemia in Venezuela. Ann. Rech. Vet. 9: 743–746. [PubMed] [Google Scholar]
- 24.Meas S., Ohashi K., Tum S., Chhin M., Te K., Miura K., Sugimoto C., Onuma M.2000. Seroprevalence of bovine immunodeficiency virus and bovine leukemia virus in draught animals in Cambodia. J. Vet. Med. Sci. 62: 779–781. doi: 10.1292/jvms.62.779 [DOI] [PubMed] [Google Scholar]
- 25.Murakami K., Kobayashi S., Konishi M., Kameyama K., Tsutsui T.2013. Nationwide survey of bovine leukemia virus infection among dairy and beef breeding cattle in Japan from 2009–2011. J. Vet. Med. Sci. 75: 1123–1126. doi: 10.1292/jvms.12-0374 [DOI] [PubMed] [Google Scholar]
- 26.Murakami K., Kobayashi S., Konishi M., Kameyama K., Yamamoto T., Tsutsui T.2011. The recent prevalence of bovine leukemia virus (BLV) infection among Japanese cattle. Vet. Microbiol. 148: 84–88. doi: 10.1016/j.vetmic.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 27.Nuotio L., Rusanen H., Sihvonen L., Neuvonen E.2003. Eradication of enzootic bovine leukosis from Finland. Prev. Vet. Med. 59: 43–49. doi: 10.1016/S0167-5877(03)00057-6 [DOI] [PubMed] [Google Scholar]
- 28.Ochirkhuu N., Konnai S., Odbileg R., Nishimori A., Okagawa T., Murata S., Ohashi K.2016. Detection of bovine leukemia virus and identification of its genotype in Mongolian cattle. Arch. Virol. 161: 985–991. doi: 10.1007/s00705-015-2676-8 [DOI] [PubMed] [Google Scholar]
- 29.Ohno A., Takeshima S. N., Matsumoto Y., Aida Y.2015. Risk factors associated with increased bovine leukemia virus proviral load in infected cattle in Japan from 2012 to 2014. Virus Res. 210: 283–290. doi: 10.1016/j.virusres.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 30.OIE World Organization for Animal Health. 2018. Manual of diagnostic tests and vaccines for terrestrial animals. https://www.oie.int/en/standard-setting/terrestrial-manual/ [accessed on January 29, 2020].
- 31.Polat M., Moe H. H., Shimogiri T., Moe K. K., Takeshima S. N., Aida Y.2017. The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch. Virol. 162: 425–437. doi: 10.1007/s00705-016-3118-y [DOI] [PubMed] [Google Scholar]
- 32.Polat M., Ohno A., Takeshima S. N., Kim J., Kikuya M., Matsumoto Y., Mingala C. N., Onuma M., Aida Y.2015. Detection and molecular characterization of bovine leukemia virus in Philippine cattle. Arch. Virol. 160: 285–296. doi: 10.1007/s00705-014-2280-3 [DOI] [PubMed] [Google Scholar]
- 33.Polat M., Takeshima S. N., Aida Y.2017. Epidemiology and genetic diversity of bovine leukemia virus. Virol. J. 14: 209. doi: 10.1186/s12985-017-0876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polat M., Takeshima S. N., Hosomichi K., Kim J., Miyasaka T., Yamada K., Arainga M., Murakami T., Matsumoto Y., de la Barra Diaz V., Panei C. J., González E. T., Kanemaki M., Onuma M., Giovambattista G., Aida Y.2016. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 13: 4. doi: 10.1186/s12977-016-0239-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollari F. L., DiGiacomo R. F., Evermann J. F.1993. Use of survival analysis to compare cull rates between bovine leukemia virus seropositive and seronegative dairy cows. Am. J. Vet. Res. 54: 1400–1403. [PubMed] [Google Scholar]
- 36.Pollari F. L., Wangsuphachart V. L., DiGiacomo R. F., Evermann J. F.1992. Effects of bovine leukemia virus infection on production and reproduction in dairy cattle. Can. J. Vet. Res. 56: 289–295. [PMC free article] [PubMed] [Google Scholar]
- 37.Portetelle D., Couez D., Bruck C., Kettmann R., Mammerickx M., Van der Maaten M., Brasseur R., Burny A.1989. Antigenic variants of bovine leukemia virus (BLV) are defined by amino acid substitutions in the NH2 part of the envelope glycoprotein gp51. Virology 169: 27–33. doi: 10.1016/0042-6822(89)90037-8 [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez S. M., Florins A., Gillet N., de Brogniez A., Sánchez-Alcaraz M. T., Boxus M., Boulanger F., Gutiérrez G., Trono K., Alvarez I., Vagnoni L., Willems L.2011. Preventive and therapeutic strategies for bovine leukemia virus: lessons for HTLV. Viruses 3: 1210–1248. doi: 10.3390/v3071210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez S. M., Golemba M. D., Campos R. H., Trono K., Jones L. R.2009. Bovine leukemia virus can be classified into seven genotypes: evidence for the existence of two novel clades. J. Gen. Virol. 90: 2788–2797. doi: 10.1099/vir.0.011791-0 [DOI] [PubMed] [Google Scholar]
- 40.Sagata N., Yasunaga T., Ohishi K., Tsuzuku-Kawamura J., Onuma M., Ikawa Y.1984. Comparison of the entire genomes of bovine leukemia virus and human T-cell leukemia virus and characterization of their unidentified open reading frames. EMBO J. 3: 3231–3237. doi: 10.1002/j.1460-2075.1984.tb02283.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C. T.1991. Bovine leukemia virus infection in Taiwan: epidemiological study. J. Vet. Med. Sci. 53: 395–398. doi: 10.1292/jvms.53.395 [DOI] [PubMed] [Google Scholar]
- 42.Yang Y., Fan W., Mao Y., Yang Z., Lu G., Zhang R., Zhang H., Szeto C., Wang C.2016. Bovine leukemia virus infection in cattle of China: Association with reduced milk production and increased somatic cell score. J. Dairy Sci. 99: 3688–3697. doi: 10.3168/jds.2015-10580 [DOI] [PubMed] [Google Scholar]
- 43.Yu C., Wang X., Zhou Y., Wang Y., Zhang X., Zheng Y.2019. Genotyping bovine leukemia virus in dairy cattle of Heilongjiang, northeastern China. BMC Vet. Res. 15: 179. doi: 10.1186/s12917-019-1863-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X., Buehring G. C.2007. Natural genetic variations in bovine leukemia virus envelope gene: possible effects of selection and escape. Virology 366: 150–165. doi: 10.1016/j.virol.2007.03.058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



