Abstract
Background
Many case series on Corona Virus Disease (COVID-19) have reported gastrointestinal (GI) and hepatic manifestations in a proportion of cases; however, the data is conflicting. The relationship of GI and hepatic involvement with severe clinical course of COVID-19 has also not been explored.
Objectives
The main objectives were to determine the frequency of GI and hepatic manifestations of COVID-19 and to explore their relationship with severe clinical course.
Methods
We searched PubMed for studies published between January 1, 2020, and March 25, 2020, with data on GI and hepatic manifestations in adult patients with COVID-19. These data were compared between patients with severe and good clinical course using the random-effects model and odds ratio (OR) as the effect size. If the heterogeneity among studies was high, sensitivity analysis was performed for each outcome.
Results
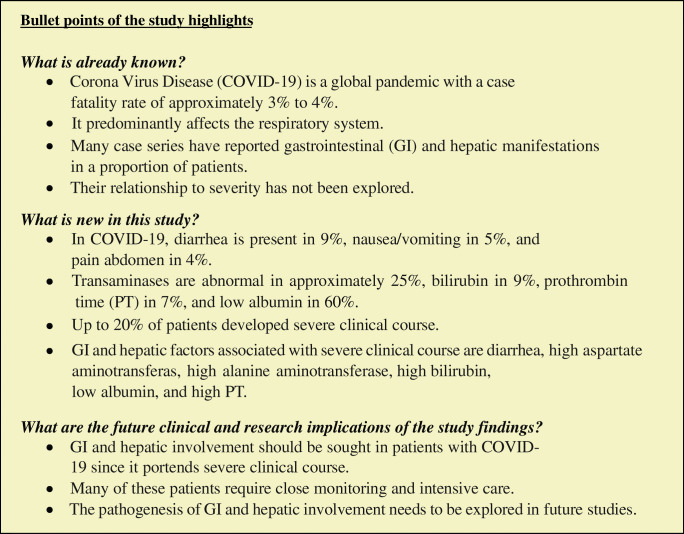
We included 62 studies (8301 patients) in the systematic review and 26 studies (4676 patients) in the meta-analysis. Diarrhea was the most common GI symptom (9%), followed by nausea/vomiting (5%) and abdominal pain (4%). Transaminases were abnormal in approximately 25%, bilirubin in 9%, prothrombin time (PT) in 7%, and low albumin in 60%. Up to 20% patients developed severe clinical course, and GI and hepatic factors associated with severe clinical course were as follows: diarrhea (OR 2), high aspartate aminotransferase (OR 1.4), high alanine aminotransferase (OR 1.6), high bilirubin (OR 2.4), low albumin (OR 3.4), and high PT (OR 3).
Conclusions
GI and hepatic involvement should be sought in patients with COVID-19 since it portends severe clinical course. The pathogenesis of GI and hepatic involvement needs to be explored in future studies.
Electronic supplementary material
The online version of this article (10.1007/s12664-020-01058-3) contains supplementary material, which is available to authorized users.
Keywords: 2019-nCoV, Coronavirus, COVID-19, nCoV-2019, Novel coronavirus, SARS-CoV-2
Introduction
Corona Virus Disease (COVID-19) is a new disease, which within 3 months of its origin, has now spread to more than two hundred countries and territories around the world, affecting more than 4,342,000 people and caused more than 292,000 deaths, as of 13 May 2020 [1]. On 11 March 2020, the World Health Organization (WHO) had declared COVID-19 a pandemic because of alarming levels of its spread and severity [2]. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is sufficiently genetically divergent from SARS-CoV to be considered a new human-infecting betacoronavirus [3]. It mainly affects the respiratory tract, and the illness ranges in severity from asymptomatic or mildly symptomatic to severe or critical disease. Although the current estimate of the case fatality rate of COVID-19 is < 5%, up to 15% to 18% of patients may become severe or critically ill, some of them requiring intensive care unit (ICU) care and mechanical ventilation [4].
Since COVID-19 is a new disease, knowledge about this disease is still incomplete and evolving. Apart from the respiratory system involvement, many case series have also reported variable involvement of gastrointestinal (GI) and hepatic systems. However, most of these case series have small sample size, and the data in these are heterogenous and conflicting. In addition, the relationship of GI and hepatic involvement with the severity and outcome of COVID-19 is also not clear.
Hence, this systematic review and meta-analysis was conducted with the following objectives: (1) to study the frequency of GI and hepatic manifestations in COVID-19, and (2) to determine whether GI or hepatic manifestations are associated with severe clinical course of COVID-19.
Methods
Since this is a systematic review and meta-analysis, an institutional review board or an ethics committee approval was not required. The PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) and MOOSE (Meta-analysis of Observational Studies in Epidemiology) guidelines were consulted during the stages of design, analysis, and reporting of this meta-analysis [5, 6]. The protocol of this meta-analysis is registered with the International Prospective Register of Systematic Reviews (PROSPERO) vide registration number CRD42020179482 and is available in full on the NIHR (National Institute for Health Research) website (https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020179482).
Study selection
Relevant papers in English language were searched in PubMed database using the following keywords: “2019-nCoV,” “nCoV-2019,” “novel Coronavirus 2019,” “SARS-CoV-2,” “COVID-19,” “coronavirus,” “coronavirus covid-19,” and “corona virus.” Since the first report of COVID-19 disease was published on 31 December 2019 [7], we limited our search to articles published since 1 January 2020. The last search was performed on March 25, 2020. Since there is a high likelihood of duplicate publications on COVID-19 [8], especially, the same set of patients being reported in English as well as Chinese or other languages, we restricted our search to papers published in English language only. For the same reason, we restricted our search to PubMed database only and did not search other databases.
The eligibility criteria for the inclusion of studies were as follows:
The studies should be in English language.
The studies should be published in full; studies published only in abstract form (such as conference abstracts) were excluded.
The study design should be retrospective or prospective, observational, or case-control study with data on GI and/or hepatic manifestations of COVID-19 disease. Interventional studies such as controlled or uncontrolled drug trials were excluded. Case reports, meta-analyses, and systematic reviews were also excluded.
The participants should be adult patients with COVID-19 disease. Studies describing exclusively pediatric population were excluded; however, studies which had both adult and pediatric patients were included. Studies describing exclusively pregnant women were also excluded.
Data extraction
The following data were extracted from each study: date of online publication, number of patients, study setting, demographic data, comorbidities, presenting symptoms (including GI symptoms), laboratory features (including liver function tests), number of patients having severe and good clinical course. The following criteria were used to identify patients having “severe clinical course”:
Patients requiring ICU care
Patients developing acute respiratory distress syndrome (ARDS), shock, respiratory failure, or those requiring mechanical ventilation as defined according to the guidance of the WHO for COVID-19 [9]
Patients categorized as severe or critical groups according to the diagnostic and treatment guideline for SARS-CoV-2 issued by the National Health Commission of the People’s Republic of China (version 3–5) [10, 11]
Patients not surviving
Patients not having any of the features of “severe clinical course” were categorized into “good clinical course.” The baseline clinical and laboratory features of patients with severe clinical course and good clinical course were tabulated separately and compared using meta-analytical tools. For studies with missing data, the corresponding authors of those studies were contacted with a request to provide the missing data.
Assessment of quality of studies
For the assessment of quality of studies including risk of bias, the National Institute of Health (NIH) tools were used, which were developed jointly by the National Heart, Lung and Blood Institute (NHLBI) and the Research Triangle Institute International [12]. For the first objective of this systematic review, i.e. to study the GI and hepatic manifestations of COVID-19, the NIH tool for case series was used. It uses nine domains; and based on these, each study was given an overall rating of good, fair, or poor, depending on “yes” response in ≥ 7 domains, 4–6 domains, and < 4 domains, respectively. For the second objective of this meta-analysis, i.e. to determine the GI and hepatic factors associated with severe clinical course of COVID-19, the NIH tool for case-control studies was used. It uses twelve domains, and based on these, each study was given an overall rating of good, fair, or poor, depending on “yes” response in ≥ 8 domains, 6–7 domains, and < 6 domains, respectively.
Statistical analysis
The demographic, clinical, and laboratory data was displayed as n (%) or mean (standard deviation, [SD]). For data with median, interquartile range (IQR), or range, the method described by Wan et al. was used to calculate mean and SD [13]. The meta-analysis was performed using odds ratios (ORs) with 95% confidence intervals (CIs) as the effect sizes of dichotomous data and a p-value of < 0.05 to show a meaningful difference in the outcomes. To obtain ORs from continuous data (i.e. from standardized mean difference), Hasselblad and Hedges’ method was used [14]. To assess the heterogeneity among studies, I2 statistic was calculated. An I2 value > 50% indicated substantial heterogeneity. To take care of heterogeneity among the studies, and to calculate a more conservative result, the ORs were pooled using only the random-effects model. In addition, if the heterogeneity among the studies was ≥ 50%, a sensitivity analysis was also performed after removing the outlier studies. Review Manager version 5.3.5 software (The Nordic Cochrane Centre, Copenhagen, Denmark) and Microsoft Excel (version 16.35) were employed for the meta-analysis and statistical analyses.
Results
Study selection and data collection
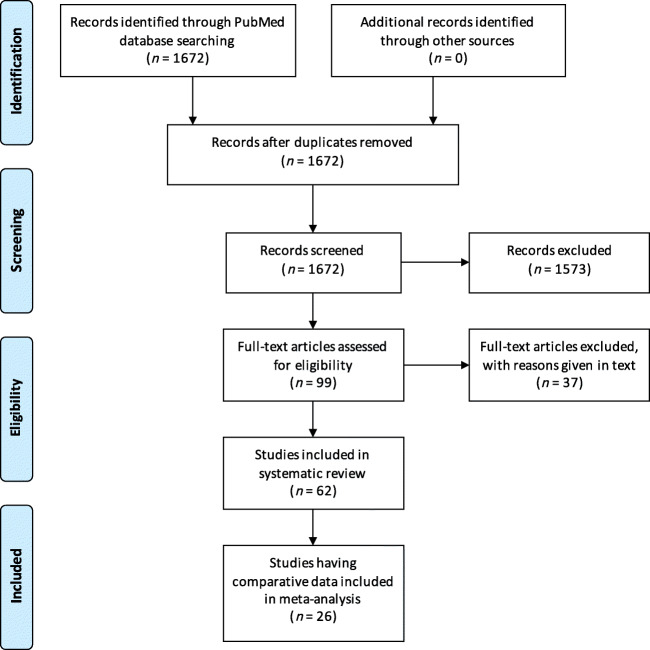
Using the keywords “2019-nCoV,” “nCoV-2019,” “novel Coronavirus 2019,” “SARS-CoV-2,” “COVID-19,” “coronavirus,” “coronavirus covid-19,” and “corona virus” and limiting the Entrez date from 1 January 2020, through 25 March 2020, initially 1672 publications in English language were retrieved from the PubMed database, which were screened for relevance (Fig. 1). After carefully going through the abstracts and full texts (if needed), of these publications, only 99 potentially relevant studies were evaluated in detail for potential inclusion. Of these, 37 studies were excluded because of the following reasons: (1) 15 studies did not have any data on GI or hepatic manifestations of COVID-19; (2) 9 studies had overlapping patients with other included studies; (3) 4 studies had only radiological or epidemiological data without any clinical data; (4) 2 studies included only pregnant women; (5) 1 study included only those patients who had GI symptoms and excluded patients without GI symptoms; (6) 1 study included only COVID-19-influenza co-infected patients; (7) 1 study included only cancer patients with COVID-19; and (8) 4 studies had data not relevant to present study. Hence, the remaining 62 studies were included in the systematic review. Of these 62 studies, only 26 had comparative data between severe and good clinical course; hence, these 26 studies were included in the meta-analysis (Fig. 1).
Fig. 1.
PRISMA flow diagram depicting the flow of information through different phases of the systematic review. PRISMA Preferred Reporting Items for Systematic Reviews and Meta-Analyses
Characteristics and quality of the included studies
The study characteristics of the included 62 studies are given in Table 1. The online publication date of the studies in the PubMed database was from January 24, 2020, through March 25, 2020. Thirty-eight out of 62 (61%) studies were from single centers, while the remaining 24 (39%) were multi-center studies. Most studies (56/62, 90%) were from mainland China, and of the remaining 6 studies, two were from Singapore (3%), and one each (1.6%) from Hong Kong, USA, Australia, and Europe. The total included patients were 8301, and of them, 8076 (97%) were reported from mainland China. The median number of patients included in the studies was 74 (IQR: 34–137). The largest study was by Guan et al. [15], which recruited 1099 patients from 552 hospitals in 30 provinces, autonomous regions, and municipalities of mainland China.
Table 1.
Characteristics and quality of studies included in systematic review and meta-analysis
| First author | PMID | Date of publication | Setting | N | Clinical course | Quality of study | ||
|---|---|---|---|---|---|---|---|---|
| Severe | Good | Included in systematic review | Included in meta-analysis | |||||
| Chan et al. [29] | 31986261 | 24-Jan-20 | Single center in Shenzhen, Guangdong Province, China | 6 | 8 | |||
| Huang et al. [30] | 31986264 | 24-Jan-20 | Single center in Wuhan, Hubei Province, China | 41 | 13 | 28 | 8 | 9 |
| Chen et al. [31] | 32007143 | 30-Jan-20 | Single center in Wuhan, Hubei Province, China | 99 | 8 | |||
| Song et al. [32] | 32027573 | 06-Feb-20 | Single center in Shanghai, China | 51 | 8 | |||
| Chang et al. [33] | 32031568 | 07-Feb-20 | Three hospitals in Beijing, China | 13 | 8 | |||
| Wang et al. [34] | 32031570 | 07-Feb-20 | Single center in Wuhan, Hubei Province, China | 138 | 36 | 102 | 8 | 9 |
| Liu et al. [35] | 32044814 | 07-Feb-20 | Nine tertiary hospitals in Hubei Province, China | 137 | 8 | |||
| Liu et al. [36] | 32048163 | 09-Feb-20 | Single center in Shenzhen, Guangdong Province, China | 12 | 8 | |||
| Ren et al. [37] | 32004165 | 11-Feb-20 | Single center in Wuhan, Hubei Province, China | 5 | 8 | |||
| Xu et al. [38] | 32075786 | 19-Feb-20 | Seven hospitals in Zhejiang Province, China | 62 | 8 | |||
| Zhang et al. [39] | 32077115 | 19-Feb-20 | Single center in Wuhan, Hubei Province, China | 140 | 58 | 82 | 8 | 9 |
| Wu et al. [40] | 32091414 | 21-Feb-20 | Two hospitals in Chongqing, China | 80 | 8 | |||
| Yang et al. [41] | 32105632 | 24-Feb-20 | Single center in Wuhan, Hubei Province, China | 52 | 32 | 20 | 8 | 9 |
| Shi et al. [42] | 32105637 | 24-Feb-20 | Two centers in Wuhan, Hubei Province, China | 81 | 8 | |||
| Xu et al. [43] | 32109443 | 25-Feb-20 | Single center in Beijing, China | 50 | 13 | 37 | 8 | 9 |
| Yang et al. [44] | 32112884 | 26-Feb-20 | Three tertiary hospitals in Wenzhou City, Zhejiang Province, China | 149 | 8 | |||
| Huang et al. [45] | 32114074 | 27-Feb-20 | Single center in Wuhan, Hubei Province, China | 34 | 8 | |||
| Xu et al. [46] | 32107577 | 28-Feb-20 | Single center in Guangdong Province, China | 90 | 8 | |||
| Guan et al. [15] | 32109013 | 28-Feb-20 | 552 hospitals in 30 provinces, autonomous regions, municipalities, China | 1099 | 173 | 926 | 8 | 9 |
| Liu et al. [47] | 32118640 | 28-Feb-20 | Three tertiary hospitals in Wuhan, Hubei Province, China | 78 | 11 | 67 | 8 | 9 |
| Wu et al. [48] | 32109279 | 29-Feb-20 | Multi-center in Jiangsu Province, China | 80 | 8 | |||
| Li et al. [49] | 32118615 | 29-Feb-20 | Single center in Chongqing, China | 83 | 25 | 58 | 8 | 9 |
| Cao et al. [50] | 32123993 | 02-Mar-20 | Single center in Wuhan, Hubei Province, China | 102 | 18 | 84 | 8 | 9 |
| Zhang et al. [51] | 32124995 | 03-Mar-20 | Single center in Jinhua, Zhejiang Province, China | 14 | 5 | |||
| Young et al. [52] | 32125362 | 03-Mar-20 | Four hospitals in Singapore | 18 | 6 | 12 | 8 | 9 |
| Ruan et al. [53] | 32125452 | 03-Mar-20 | Two centers in Wuhan, Hubei Province, China | 150 | 68 | 82 | 8 | 9 |
| Zhao et al. [54] | 32125873 | 03-Mar-20 | Four institutions in Hunan Province, China | 101 | 14 | 87 | 8 | 9 |
| Xiong et al. [55] | 32134800 | 03-Mar-20 | Single center in Wuhan, Hubei Province, China | 42 | 8 | |||
| Xiao et al. [56] | 32142773 | 03-Mar-20 | Single center in Guangdong Province, China | 73 | 5 | |||
| Hu et al. [57] | 32146694 | 04-Mar-20 | Single center in Nanjing, Jiangsu Province, China | 24 | 8 | |||
| Zhou et al. [58] | 32134681 | 05-Mar-20 | Single center in Wuhan, Hubei Province, China | 62 | 8 | |||
| Wang et al. [59] | 32139464 | 05-Mar-20 | Single center in Zhengzhou, Henan Province, China | 18 | 6 | |||
| Spiteri et al. [60] | 32156327 | 05-Mar-20 | WHO European region except UK | 38 | 4 | |||
| CNIRST [61] | 32156224 | 11-Mar-20 | Multi-center in Australia | 71 | 4 | |||
| Zhou et al. [62] | 32171076 | 11-Mar-20 | Two hospitals in Wuhan, Hubei Province, China | 191 | 54 | 137 | 8 | 9 |
| Liu et al. [63] | 32171866 | 11-Mar-20 | Single center in Hainan Province, China | 56 | 8 | |||
| Chen et al. [64] | 32171869 | 11-Mar-20 | Single center in Shanghai, China | 249 | 22 | 227 | 8 | 9 |
| Zhao et al. [65] | 32161968 | 12-Mar-20 | Two hospitals in Anhui Province, China | 19 | 8 | |||
| Chen et al. [66] | 32162699 | 12-Mar-20 | Single center in Anhui Province, China | 9 | 8 | |||
| Zhu et al. [67] | 32167181 | 13-Mar-20 | Two emergency departments in Anhui Province, China | 32 | 8 | |||
| Wu et al. [68] | 32167524 | 13-Mar-20 | Single center in Wuhan, Hubei Province, China | 201 | 84 | 117 | 8 | 9 |
| Cheng et al. [69] | 32174128 | 14-Mar-20 | Single center in Shanghai, China | 11 | 8 | |||
| Mo et al. [70] | 32173725 | 16-Mar-20 | Single center in Wuhan, Hubei Province, China | 155 | 85 | 70 | 8 | 9 |
| Wang et al. [71] | 32176772 | 16-Mar-20 | Single center in Wuhan, Hubei Province, China | 69 | 14 | 55 | 8 | 9 |
| Wang et al. [72] | 32179910 | 17-Mar-20 | Single center in Shenzhen, Guangdong Province, China | 55 | 8 | |||
| Han et al. [73] | 32181672 | 17-Mar-20 | Single center in Wuhan, Hubei Province, China | 108 | 7 | |||
| Qian et al. [74] | 32181807 | 17-Mar-20 | Five hospitals in Zhejiang Province, China | 91 | 9 | 82 | 8 | 9 |
| Gao et al. [75] | 32181911 | 17-Mar-20 | Single center in Fuyang, Anhui Province, China | 43 | 15 | 28 | 8 | 9 |
| Shi et al. [76] | 32188484 | 18-Mar-20 | Multi-center in Zhejiang Province, China | 487 | 49 | 438 | 7 | 8 |
| Arentz et al. [77] | 32191259 | 19-Mar-20 | Single center in Washington State, USA | 21 | 21 | 0 | 7 | |
| Wang et al. [78] | 32191587 | 19-Mar-20 | Single center in Wuhan, Hubei Province, China | 90 | 7 | |||
| Wu et al. [79] | 32199469 | 19-Mar-20 | Single center in Zhuhai, Guangdong Province, China | 74 | 18 | 56 | 7 | |
| Liu et al. [80] | 32199493 | 19-Mar-20 | Single center in Nanchang, Jiangxi Province, China | 76 | 30 | 46 | 6 | 9 |
| Zhang et al. [81] | 32205284 | 20-Mar-20 | Multi-center in Zhejiang Province, China | 645 | 64 | 581 | 8 | 9 |
| Deng et al. [82] | 32209890 | 20-Mar-20 | Two tertiary hospitals in Wuhan, Hubei Province, China | 225 | 109 | 116 | 8 | 8 |
| Wan et al. [83] | 32198776 | 21-Mar-20 | Multi-center in Chongqing, China | 135 | 40 | 95 | 8 | 9 |
| Zhou et al. [84] | 32209382 | 21-Mar-20 | Single center in Wuhan, Hubei Province, China | 34 | 8 | 26 | 7 | 9 |
| To et al. [85] | 32213337 | 23-Mar-20 | Two hospitals in Hong Kong | 23 | 10 | 13 | 7 | 9 |
| Jin et al. [86] | 32213556 | 24-Mar-20 | Multi-center in Zhejiang Province, China | 651 | 64 | 587 | 8 | |
| Sun et al. [87] | 32211755 | 25-Mar-20 | Single center in Singapore | 54 | 7 | |||
| Shi et al. [88] | 32211816 | 25-Mar-20 | Single center in Wuhan, Hubei Province, China | 416 | 8 | |||
| Lian et al. [89] | 32211844 | 25-Mar-20 | Multi-center in Zhejiang Province, China | 788 | 78 | 710 | 8 | |
| 8301 | 1241/6210 (20%) | 4969/6210 (80%) | ||||||
PMID PubMed identification number, N number
The quality of study was assessed using the NIH tool for case series [12], and the results are shown in Table 1. The scores were as follows: 8/9 (48 studies [77%]); 7/9 (8 studies [13%]); 6/9 (2 studies [3%]); 5/9 (2 studies [3%]); and 4/9 (2 studies [3%]). Thus, 56 studies (90%) were of good quality (scores of ≥ 7) and the remaining 6 studies (10%) were of fair quality (scores 4–6). None of the included study was judged poor.
Twenty-six studies (4676 patients), which were included in the meta-analysis, were evaluated for quality by NIH tool for case-control studies [12] (Table 1). The scores were as follows: 9/12 (24 studies [92%]) and 8/12 (2 studies [8%]). Thus, all the studies were of good quality with scores ≥ 8. Out of the twelve domains assessed by this tool, the three domains in which all the studies were given “0” score were sample size justification, blinding of assessors, and adjusting for confounding variables.
Demographic data and clinical presentation of patients
Table 2 shows the clinical presentation of the included patients. The total number of patients was 8301, and of these, 4443 (54%) were males. Thus, the male to female ratio was approximately 1.2:1. The pooled mean age of patients was 48.7 ± 16.5 years.
Table 2.
Demographic data and clinical presentation of patients
| First author | N | Age | Gender | Comorbidities | General symptoms | Gastro-intestinal symptoms | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Male | Male% | Any | CLD | Fever | Cough | Sputum | Dyspnea | Diarrhea | Nausea Vomiting | Pain abdomen | ||
| Chan et al. [29] | 6 | 46.2 | 22.5 | 3 | 50 | 4 | – | 5 | 4 | 1 | 0 | 2 | – | – |
| Huang et al. [30] | 41 | 49.3 | 13.1 | 30 | 73 | 13 | 1 | 40 | 31 | (11/39) | (22/40) | (1/38) | – | – |
| Chen et al. [31] | 99 | 55.5 | 13.1 | 67 | 68 | 50 | – | 82 | 81 | – | 31 | 2 | 1 | – |
| Song et al. [32] | 51 | 49.0 | 16.0 | 25 | 49 | 11 | 1 | 49 | 24 | 10 | 7 | 5 | 3 | – |
| Chang et al.[33] | 13 | 38.7 | 11.6 | 10 | 77 | – | – | 12 | 6 | 2 | – | 1 | – | – |
| Wang et al. [34] | 138 | 55.3 | 19.5 | 75 | 54 | 64 | 4 | 136 | 82 | 37 | 43 | 14 | 14 | 3 |
| Liu et al. [35] | 137 | 54.2 | 12.1 | 61 | 45 | 27 | – | 112 | 66 | 6 | 26 | 11 | – | – |
| Liu et al. [36] | 12 | 53.7 | 18.0 | 8 | 67 | 7 | 0 | 10 | 11 | – | – | 2 | 2 | – |
| Ren et al. [37] | 5 | 53.6 | 9.5 | 3 | 60 | 2 | 1 | 5 | 5 | 1 | 4 | 0 | – | – |
| Xu et al. [38] | 62 | 41.7 | 15.2 | 35 | 56 | 20 | 7 | 48 | 50 | 35 | 2 | 3 | – | – |
| Zhang et al. [39] | 140 | 56.5 | 11.8 | 71 | 51 | 90 | 8 | (110/120) | (90/120) | – | (44/120) | (18/139) | (24/139) | (8/139) |
| Wu et al. [40] | 80 | 44.0 | 11.0 | 42 | 53 | 15 | – | 61 | 58 | 11 | 7 | 7 | – | 7 |
| Yang et al. [41] | 52 | 59.7 | 13.3 | 35 | 67 | 21 | – | 51 | 40 | – | 33 | – | 2 | – |
| Shi et al. [42] | 81 | 49.5 | 11.0 | 42 | 52 | 21 | 7 | 59 | 48 | 15 | 34 | 3 | 4 | – |
| Xu et al. [43] | 50 | 43.9 | 16.8 | 29 | 58 | – | – | 43 | 20 | 7 | 4 | 1 | – | – |
| Yang et al. [44] | 149 | 45.1 | 13.4 | 81 | 54 | – | 8 | 114 | 87 | 48 | 2 | 11 | 2 | – |
| Huang et al. [45] | 34 | 56.2 | 17.1 | 14 | 41 | 16 | 1 | 32 | 17 | 8 | 5 | 5 | – | – |
| Xu et al. [46] | 90 | 51.0 | 13.8 | 39 | 43 | 45 | – | 70 | 57 | 11 | – | 5 | 5 | – |
| Guan et al. [15] | 1099 | 46.7 | 17.1 | 640 | 58 | 261 | 23 | 975 | 745 | 370 | 205 | 42 | 55 | – |
| Liu et al. [47] | 78 | 42.7 | 18.1 | 39 | 50 | – | – | 57 | 34 | – | – | – | – | – |
| Wu et al. [48] | 80 | 46.1 | 15.4 | 39 | 49 | 38 | 1 | 63 | 51 | – | 30 | 1 | 1 | – |
| Li et al. [49] | 83 | 45.5 | 12.3 | 44 | 53 | 15 | – | 72 | 65 | 15 | 9 | 7 | – | 7 |
| Cao et al. [50] | 102 | 52.7 | 22.6 | 53 | 52 | 47 | 2 | 83 | 50 | – | – | 11 | – | – |
| Zhang et al. [51] | 14 | 46.8 | 20.2 | 7 | 50 | – | – | 13 | 10 | – | – | 0 | 0 | – |
| Young et al. [52] | 18 | 49.5 | 11.5 | 9 | 50 | 5 | – | 13 | 15 | – | 2 | 3 | – | – |
| Ruan et al. [53] | 150 | 57.7 | 12.5 | 102 | 68 | 77 | 4 | 127 | 110 | 54 | 110 | – | – | – |
| Zhao et al. [54] | 101 | 44.4 | 11.6 | 56 | 55 | 30 | – | 79 | 63 | – | 1 | 3 | 2 | – |
| Xiong et al. [55] | 42 | 49.5 | 14.1 | 25 | 60 | 13 | – | 36 | 27 | – | 8 | 10 | – | – |
| Xiao et al. [56] | 73 | 41.2 | 16.2 | 41 | 56 | – | – | – | 53 | – | – | 26 | – | – |
| Hu et al. [57] | 24 | 36.2 | 29.9 | 8 | 33 | – | 0 | 5 | 2 | – | – | – | – | – |
| Zhou et al. [58] | 62 | 52.8 | 12.2 | 39 | 63 | – | – | 54 | 28 | 28 | 15 | 9 | – | – |
| Wang et al. [59] | 18 | 41.0 | 20.9 | 10 | 56 | 6 | – | 17 | 10 | – | 4 | 3 | 1 | – |
| Spiteri et al. [60] | 38 | 41.8 | 18.5 | 25 | 66 | (2/7) | (0/7) | (20/31) | (14/31) | – | (2/31) | (1/31) | (1/31) | – |
| CNIRST [61] | 71 | 46.0 | 19.8 | 36 | 51 | – | – | (22/34) | (24/34) | – | (3/34) | (9/34) | (2/34) | – |
| Zhou et al. [62] | 191 | 56.3 | 15.7 | 119 | 62 | 91 | – | 180 | 151 | 44 | 56 | 9 | 7 | – |
| Liu et al. [63] | 56 | 52.1 | 14.7 | 31 | 55 | – | 1 | 44 | 21 | 21 | 4 | – | 10 | – |
| Chen et al. [64] | 249 | 50.3 | 20.9 | 126 | 51 | 90 | 2 | 217 | 91 | – | 19 | 8 | – | – |
| Zhao et al. [65] | 19 | 43.7 | 23.2 | 11 | 58 | 3 | 1 | 15 | 9 | – | – | 1 | – | – |
| Chen et al. [66] | 9 | 42.1 | 15.3 | 5 | 56 | 0 | – | 8 | 7 | – | 0 | 2 | – | 0 |
| Zhu et al. [67] | 32 | 44.3 | 13.2 | 15 | 47 | – | 2 | 27 | 21 | 5 | – | 1 | – | – |
| Wu et al. [68] | 201 | 51.3 | 12.7 | 128 | 64 | – | 7 | 188 | 163 | 83 | 80 | – | – | – |
| Cheng et al. [69] | 11 | 50.4 | 15.5 | 8 | 73 | – | – | 8 | 7 | 3 | 1 | 1 | – | – |
| Mo et al. [70] | 155 | 54.0 | 18.0 | 86 | 55 | 71 | 7 | 126 | 97 | – | 50 | 7 | 3 | 3 |
| Wang et al. [71] | 69 | 46.3 | 20.4 | 32 | 46 | 25 | 1 | 60 | 38 | 20 | 20 | 10 | 3 | – |
| Wang et al. [72] | 55 | 42.2 | 14.7 | 22 | 40 | – | 1 | 7 | 7 | – | – | – | – | – |
| Han et al. [73] | 108 | 45.0 | 13.6 | 38 | 35 | – | – | 94 | 65 | – | – | 15 | – | – |
| Qian et al. [74] | 91 | 47.8 | 15.4 | 37 | 41 | – | – | 65 | 55 | 30 | 10 | 21 | 11 | – |
| Gao et al. [75] | 43 | 43.7 | 12.1 | 26 | 60 | – | – | – | – | – | – | – | – | – |
| Shi et al. [76] | 487 | 46.0 | 19.0 | 259 | 53 | – | 22 | – | – | – | – | – | – | – |
| Arentz et al. [77] | 21 | 70.0 | 13.0 | 11 | 52 | 18 | 1 | 11 | 11 | – | 17 | – | – | – |
| Wang et al. [78] | 90 | 45.0 | 14.0 | 33 | 37 | – | – | 55 | 20 | – | – | 6 | – | 2 |
| Wu et al. [79] | 74 | 43.5 | 3.8 | 39 | 53 | – | – | 45 | 37 | – | 9 | 26 | – | – |
| Liu et al. [80] | 76 | 48.3 | 15.7 | 48 | 63 | – | – | 63 | 35 | – | 9 | 2 | 2 | – |
| Zhang et al. [81] | 645 | 45.3 | 14.3 | 328 | 51 | (177/468) | (25/620) | 540 | 425 | 225 | 26 | 53 | 22 | – |
| Deng et al. [82] | 225 | 55.4 | 19.0 | 124 | 55 | 127 | – | 189 | 85 | 49 | 99 | 33 | – | – |
| Wan et al. [83] | 135 | 46.0 | 14.2 | 72 | 53 | 43 | 2 | 120 | 102 | 12 | 18 | 18 | 4 | – |
| Zhou et al. [84] | 34 | 64.7 | 8.8 | 17 | 50 | – | – | – | – | – | – | – | – | – |
| To et al. [85] | 23 | 58.4 | 11.6 | 13 | 57 | 11 | – | 22 | 5 | – | 4 | 2 | 1 | – |
| Jin et al. [86] | 651 | 45.2 | 14.4 | 331 | 51 | 178 | 25 | 545 | 435 | 227 | 27 | – | – | – |
| Sun et al. [87] | 54 | 44.8 | 13.9 | 29 | 54 | 5 | 0 | – | 36 | 13 | 7 | 20 | – | – |
| Shi et al. [88] | 416 | 61.0 | 12.5 | 205 | 49 | – | 4 | 334 | 144 | 23 | 117 | 16 | – | – |
| Lian et al. [89] | 788 | 45.8 | 14.9 | 407 | 52 | 218 | 31 | 636 | 506 | 265 | 37 | 88 | – | – |
| 8301 | 48.7 | 16.5 | 4443 | 54% |
1957 /5785 (34%) |
200 /6183 (3%) |
6274 /7546 (83%) |
4651 /7673 (61%) |
1690 /5894 (29%) |
1263 /6962 (18%) |
555 /6401 (9%) |
182 /3629 (5%) |
30 /694 (4%) |
|
N number, SD standard deviation, CLD chronic liver disease
Table 2 also shows the clinical presentation and underlying comorbidities of the included patients. Thirty-four percent (1957/5785) of patients had one or more of some chronic underlying comorbidity such as hypertension, diabetes, cardiovascular disease, and chronic kidney disease (individual data not shown). Notably, chronic liver disease (including chronic hepatitis B or C, non-alcoholic steatohepatitis [NASH], or cirrhosis) was present in just 3% (200/6183) of patients.
Of the presenting symptoms, fever was the most common, present in 83% (6274/7546) of patients, followed by cough in 61% (4651/7673), expectoration (sputum) in 29% (1690/5894), and dyspnea in 18% (1263/6962) patients. Among the GI symptoms, diarrhea was the most common, present in 9% (555/6401), followed by nausea/vomiting in 5% (182/3629) and abdominal pain in just 4% (30/694) patients (Table 2).
Liver function tests of patients
Only 23 studies had data on liver function tests (Table 3). The transaminases, aspartate aminotransferase (AST) and alanine aminotransferase (ALT), were elevated in 25% (559/2226) and 23% (477/2107) patients, respectively. Serum bilirubin was elevated in 9% (130/1471), and prothrombin time (PT) was elevated in just 7% (55/750) patients. Very few studies had data on serum albumin; however, it was below the normal range in 60% (491/817) patients.
Table 3.
Liver function tests of patients
| First author | N | High AST | High ALT | High bilirubin | Low albumin | High PT |
|---|---|---|---|---|---|---|
| Chan et al. [29] | 6 | 0 | 0 | 0 | 0 | 0 |
| Huang et al. [30] | 41 | 15 | – | – | – | – |
| Chen et al. [31] | 99 | 35 | 28 | 18 | 97 | 5 |
| Liu et al. [36] | 12 | 2 | 2 | 0 | 6 | – |
| Ren et al. [37] | 5 | – | 1 | – | – | 1 |
| Xu et al. [38] | 62 | 10 | – | – | – | – |
| Yang et al. [41] | 52 | 15 | 15 | – | – | – |
| Shi et al. [42] | 81 | 43 | – | – | – | – |
| Yang et al. [44] | 149 | 27 | 18 | 4 | 9 | 17 |
| Huang et al. [45] | 34 | 7 | 8 | 3 | 25 | 17 |
| Guan et al. [15] | 1099 | (168/757) | (158/741) | (76/722) | – | – |
| Wu et al. [48] | 80 | 3 | 3 | 1 | 2 | 0 |
| Ruan et al. [53] | 150 | (54/147) | (43/146) | (18/147) | (114/148) | – |
| Hu et al. [57] | 24 | 0 | 2 | 0 | – | – |
| Wang et al. [59] | 18 | (4/16) | (4/16) | – | – | – |
| Zhou et al. [62] | 191 | – | (59/189) | – | – | (11/182) |
| Zhao et al. [65] | 19 | (5/18) | (5/18) | – | – | – |
| Wu et al. [68] | 201 | (59/198) | (43/198) | (10/198) | (195/198) | (4/195) |
| Mo et al. [70] | 155 | 54 | 54 | – | – | – |
| Wang et al. [71] | 69 | 19 | 23 | – | – | – |
| Qian et al. [74] | 91 | 9 | 7 | – | 43 | – |
| Wan et al. [83] | 135 | 30 | – | – | – | – |
| To et al. [85] | 23 | – | 4 | – | – | – |
| 559/2226 (25%) | 477/2107 (23%) | 130/1471 (9%) | 491/817 (60%) | 55/750 (7%) |
AST aspartate aminotransferase, ALT alanine aminotransferase, PT prothrombin time, N number
Clinical outcome
Since, at the time of publication of most of these studies, many of the included patients were still admitted in the hospital, with many in the ICUs, it was not possible by many studies to provide the mortality data. However, many studies did classify the patients into severe and non-severe groups based on pre-defined criteria [9–11]. Overall, 30 studies reported that 80% (4969/6210) of their patients had good clinical course, while the remaining 20% (1241/6210) patients had severe clinical course based on pre-defined severity criteria [9–11], including admission to ICU, development of ARDS, ventilatory requirement, or mortality (Table 1).
Factors associated with severe clinical course: age and comorbidity
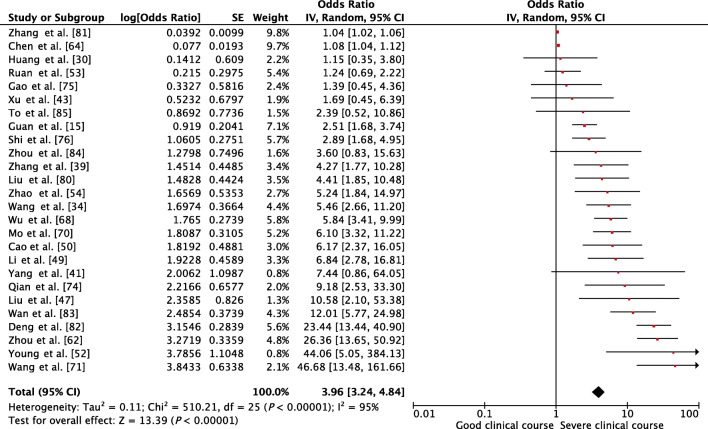
Age was compared between patients with severe clinical course and good clinical course in 26 studies. The mean age in patients with severe clinical course was found to be significantly higher than in patients with good clinical course (60 ± 16 vs. 46 ± 16 years; p < 0.01). The pooled OR of patients of advanced age (≥ 60 years) having severe clinical course was 3.96 (95% CI, 3.24–4.84; p < 0.01) (Fig. 2). The heterogeneity among the studies was substantial with an I2 of 95%. A sensitivity analysis after removing 7 outlier studies revealed a higher pooled odds ratio of 4.19 (95% CI, 3.18–5.53; p < 0.01) and an acceptable I2 of 42%.
Fig. 2.
Forest plot showing pooled odds ratio for patients with higher age (≥ 60 years) developing severe clinical course
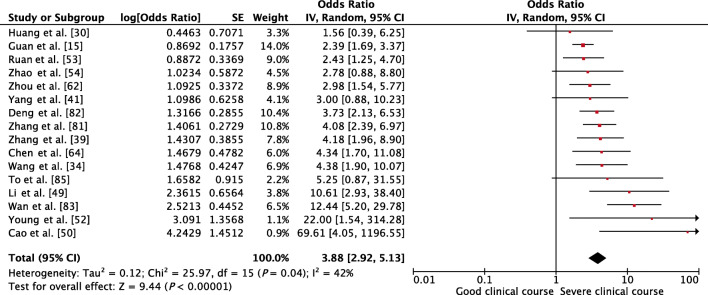
Presence of any comorbidity was assessed as a factor associated with severe clinical course in 16 studies. The pooled OR of 3.88 (95% CI, 2.92–5.13; p < 0.01) suggested that the presence of any comorbidity was significantly associated with severe clinical course (Fig. 3). The heterogeneity among studies was acceptable with an I2 of 42%.
Fig. 3.
Forest plot showing pooled odds ratio for patients having any comorbidity developing severe clinical course
GI and hepatic factors associated with severe clinical course
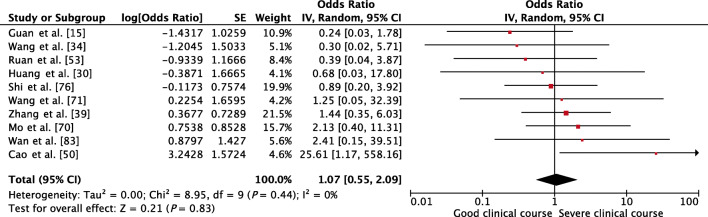
Presence of underlying chronic liver disease, as a factor associated with severe clinical course, was assessed in 10 studies, and it was not found to be significantly associated (pooled OR 1.07 [95% CI, 0.55–2.09; p = 0.83]) (Fig. 4). There was no heterogeneity among the studies (I2 = 0%).
Fig. 4.
Forest plot showing pooled odds ratio for patients with chronic liver disease developing severe clinical course
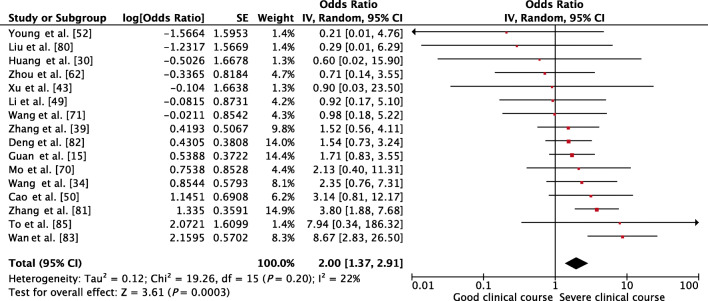
Among the GI symptoms, diarrhea was found in 9% of cases. Whether it was associated with severe clinical course was assessed in 16 studies. A pooled OR of 2.00 (95% CI, 1.37–2.91; p < 0.01) indicated that it was significantly associated with severe clinical course (Fig. 5). There was non-significant heterogeneity among studies with an I2 of 22%.
Fig. 5.
Forest plot showing pooled odds ratio of diarrhea being associated with severe clinical course
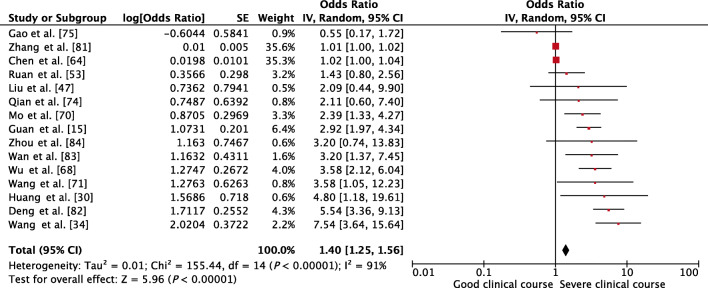
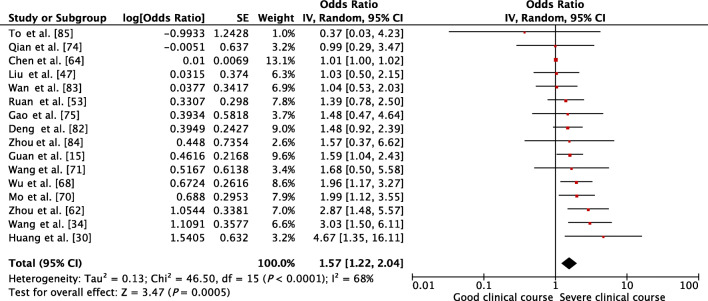
Transaminases, AST and ALT values, were reported to be associated with severe clinical course by many individual studies. AST was evaluated in 15 studies, and higher than normal values of AST was found to be significantly associated with severe clinical course (pooled OR 1.40 [95% CI, 1.25–1.56]; p < 0.01) (Fig. 6). The heterogeneity among studies was substantial (I2 of 91%) so a sensitivity analysis was done after removing four outlier studies, which again revealed a significant pooled OR of 2.45 (95% CI, 1.83–3.28; p < 0.01) and an acceptable I2 of 31%. ALT was evaluated in 16 studies, and higher than normal values of ALT was found to be significantly associated with severe clinical course (pooled OR 1.57 [95% CI, 1.22–2.04; p < 0.01]) (Fig. 7). Since the heterogeneity among the studies was high (I2 68%), a sensitivity analysis was done after removing one outlier study, which still revealed a significant pooled OR of 1.68 (95% CI, 1.39–2.04; p < 0.01) and an I2 of just 8%.
Fig. 6.
Forest plot showing pooled odds ratio of higher than normal values of aspartate aminotransferase being associated with severe clinical course
Fig. 7.
Forest plot showing pooled odds ratio of higher than normal values of alanine transaminase being associated with severe clinical course
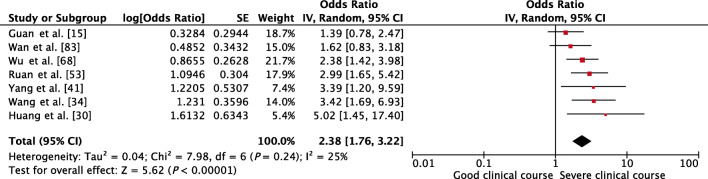
Serum bilirubin was assessed in 7 studies, and higher than normal values of serum bilirubin was found to be significantly associated with severe clinical course with a pooled OR of 2.38 (95% CI, 1.76–3.22; p < 0.01) (Fig. 8). The heterogeneity among the studies was non-significant with an I2 of 25%.
Fig. 8.
Forest plot showing pooled odds ratio of higher than normal values of serum bilirubin being associated with severe clinical course
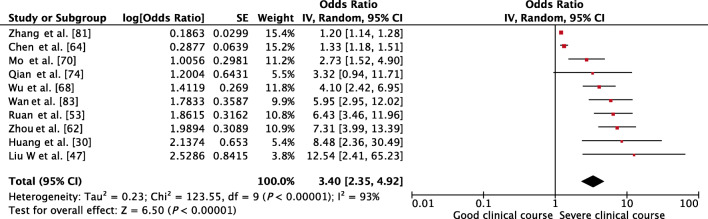
Serum albumin was assessed in 10 studies, and lower than normal values of serum albumin was found to be significantly associated with severe clinical course with a pooled OR of 3.40 (95% CI, 2.35–4.92; p < 0.01) (Fig. 9). Since the heterogeneity among studies was substantial with an I2 of 93%, a sensitivity analysis was done after removing two outlier studies, which again revealed a significant pooled OR of 5.09 (95% CI, 3.75–6.91; p < 0.01) and an acceptable I2 of 25%.
Fig. 9.
Forest plot showing pooled odds ratio of lower than normal values of serum albumin being associated with severe clinical course
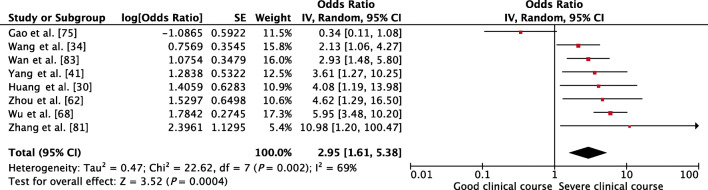
PT as a factor associated with severe clinical course was evaluated in 8 studies. Higher than control values of PT was found to be significantly associated with severe clinical course with a pooled OR of 2.95 (95% CI, 1.61–5.38; p < 0.01) (Fig. 10). Since the heterogeneity among studies was significant with an I2 of 69%, a sensitivity analysis was done after removing one outlier study, which again showed a significant pooled OR of 3.81 (95% CI, 2.68–5.41; p < 0.01) and non-significant I2 of 13%.
Fig. 10.
Forest plot showing pooled odds ratio of higher than normal values of prothrombin time being associated with severe clinical course
Discussion
To summarize the results of this systematic review and meta-analysis, the mean age of patients with COVID-19 is 48.7 ± 16.5 years with a male to female ratio of 1.2:1. Common symptoms and their frequency are fever (83%), cough (61%), expectoration (29%), and dyspnea (18%). Among the GI symptoms, diarrhea was the most common, present in 9%, followed by nausea/vomiting in 5%, and pain abdomen in just 4% of patients. Transaminases were abnormal in approx. 25%, serum albumin was low in 60%, bilirubin was high in 9%, and prothrombin time was abnormal in 7%. Twenty percent of patients developed severe clinical course, and two of the most common factors associated with severe clinical course are age ≥ 60 years (OR approximately [approx.] 4) and underlying chronic comorbidity (OR approx. 4). However, underlying chronic liver disease was not associated with severe clinical course. Presence of diarrhea was associated with severe clinical course (OR 2). Liver function abnormalities were associated with severe clinical course: high AST (OR approx. 2), high ALT (OR approx. 1.7), high bilirubin (OR approx. 2), low albumin (OR approx. 5), and high PT (OR approx. 4).
The strength of this systematic review and meta-analysis is that it included a large number of studies: 62 studies involving 8301 patients for the data on demographic and clinical features (especially GI and hepatic) and 26 studies involving 4676 patients for meta-analysis of GI and hepatic factors associated with severe clinical course. This is one of the first meta-analyses on factors associated with severe clinical course. The results of our study show that, even though COVID-19 is a disease of predominantly respiratory system, however, the involvement of the digestive system is also common, and it portends severe clinical course, including ICU requirement, or even mortality.
Our study showed that although GI symptoms are not very common, they can still occur in up to 10% of patients. However, it is still unclear how the SARS-CoV-2 virus induces GI symptoms and whether SARS-CoV-2 can be transmitted through the GI tract [16]. The SARS-CoV-2 virus has been documented in fecal samples as well as in intestinal mucosa of infected patients, and this may indicate that GI symptoms are a result of invasion of GI tract by SARS-CoV-2 as an alternative route of infection. SARS-CoV-2 uses angiotensin-converting enzyme-2 (ACE2) as a viral receptor to enter host cells [17], and these receptors are not only highly expressed in the small intestine, especially in the proximal and distal enterocytes [18], but also an important regulator of intestinal inflammation [19]. The SARS-CoV-2 infection of the ACE2-expressing enterocytes leads to mucosal inflammation, malabsorption, unbalanced intestinal secretion, and activation of the enteric nervous system, resulting in diarrhea. Other indirect mechanisms of GI involvement in COVID-19 could be damage of the GI mucosa through a chain of inflammatory responses; drugs-induced (especially antibiotics) diarrhea; and changes in the composition and function of intestinal microbiota that mutually affect the respiratory tract through immune regulation, the so-called “gut-lung axis” [20, 21]. Whatever be the mechanism of GI involvement, the implication of diarrhea and prolonged fecal shedding of SARS-CoV-2 viral particles (even after clinical recovery), is the potential for fecal-oral transmission of COVID-19, especially in countries with poor sanitation [22, 23]. Hence, a negative stool sample should be a mandatory criterion before discharging a patient from the hospital.
The pathogenesis of hepatic involvement may also be multi-factorial. It has been shown previously that cholangiocytes may too express ACE2 receptors [24]; and so it has been speculated that SARS-CoV-2 infection might cause bile duct involvement rather than hepatocellular involvement. However, significant increases in circulating levels of serum gamma-glutamyl transferase, alkaline phosphatase, or bilirubin have been rarely reported in COVID-19 patients. In addition, liver histopathologic features from COVID-19 patients also did not show any significant damage in hepatocytes or bile duct cells. For this reason, it is hypothesized that COVID-19-related liver dysfunction is more likely due to secondary liver damage rather than primary cholangiocyte or hepatocyte damage by SARS-CoV-2. The secondary mechanisms of COVID-19-related liver dysfunction could be systemic inflammatory response syndrome (SIRS); sepsis; hypoxia-reperfusion dysfunction; and drug-induced liver injury (DILI) as a result of hepatotoxic drugs used to treat COVID-19 [25]. SIRS is a result of sudden initiation of an inflammatory cascade due to the activation of both natural and cellular immunity triggered by COVID-19 infection. Hyperactivated immune responses and cytokine storm–related systemic inflammation in SARS-CoV-2 infection can affect and damage many organs, including the gut and liver [26]. In addition to SIRS, sepsis is also not uncommon in severe and critical COVID-19 cases. The pathophysiology of sepsis-related liver injury includes cholestasis due to altered bile metabolism, hepatocellular injury due to drug toxicity, or overwhelming inflammatory hypoxic liver injury due to ischemia and shock [27]. Furthermore, severe hypoxia and hypovolemia could be the major causes of ischemic liver injury in COVID-19 cases with hypotension and shock. Ischemic hepatic injury is associated with metabolic acidosis, calcium overloading, and changes of mitochondrial membrane permeability, and has thus far usually manifested as high aminotransferase concentrations in serum [26]. In addition, many patients with COVID-19 have a history of simultaneous use of multiple antiviral, antimalarial, and antibiotic drugs, which are potentially hepato-toxic, such as oseltamivir, abidol, lopinavir/ritonavir, hydroxychloroquine, and azithromycin. Thus, DILI may be an additional factor in liver function derangement [28].
Our meta-analysis had a few limitations. The first limitation was that several of the identified factors may be confounding variables, and which of these factors are independently associated with severe clinical course could not be determined. Since our meta-analysis included only retrospective case series with a lot of heterogeneity, identification of independent factors was not possible. Only a well-designed prospective study can identify independent factors associated with severe clinical course or mortality. The second limitation was the non-uniformity of the clinical endpoint. It may be argued that clubbing studies that compared patients with and without ICU admission with studies that used mortality as endpoint may not be statistically justified. However, in the present circumstances, when many of the included patients are still admitted and sufficient follow-up time has still not elapsed to determine their final outcome, using a composite endpoint of “severe clinical course” seems to be the most acceptable endpoint in our meta-analysis. The third limitation is that most studies in this meta-analysis are from China. COVID-19 is now a global disease; however, at the time of data extraction and analysis very few non-China studies were published. This meta-analysis will need updating when more studies from other countries become available.
In conclusion, our systematic review and meta-analysis has shown that, even though GI and hepatic manifestation are not very common in COVID-19, their presence portends a severe clinical course. GI symptoms, especially diarrhea, should be enquired from patients because it not only indicates severe disease, these patients are more likely to have fecal shedding of virus with potential infectivity to others. In addition, liver function tests in COVID-19 patients need to be monitored, which are likely to be abnormal in up to one-fourth of patients, especially those with severe clinical course.
Electronic supplementary material
(DOC 64 kb)
Compliance with ethical standards
Conflict of interest
AK, AA, PS, SAA, NB, VS, SK, and AS declare that they have no conflict of interest.
Ethics statement
The study was performed conforming to the Helsinki declaration of 1975, as revised in 2000 and 2008 concerning human and animal rights, and the authors followed the policy concerning informed consent as shown on Springer.com.
Disclaimer
The authors are solely responsible for the data and the contents of the paper. In no way, the Honorary Editor-in-Chief, Editorial Board Members, or the printer/publishers are responsible for the results/findings and content of this article.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coronavirus [Internet]. [cited 2020 Apr 11]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.Mahase E. Covid-19: WHO declares pandemic because of “alarming levels” of spread, severity, and inaction. BMJ. 2020;368:m1036. doi: 10.1136/bmj.m1036. [DOI] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–74. [DOI] [PMC free article] [PubMed]
- 4.Sun P, Qie S, Liu Z, Ren J, Li K, Xi J. Clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis. J Med Virol. 2020 ;92:612–7.. [DOI] [PMC free article] [PubMed]
- 5.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. [DOI] [PMC free article] [PubMed]
- 7.武汉市卫生健康委员会 [Wuhan Municipal Health Committee] [Internet]. [cited 2020 Mar 23]. Available from: http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989
- 8.Bauchner H, Golub RM, Zylke J. Editorial concern-possible reporting of the same patients with COVID-19 in different reports. JAMA. 2020. 10.1001/jama.2020.3980. [DOI] [PubMed]
- 9.Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected [Internet]. [cited 2020 Mar 28]. Available from: https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected
- 10.Diagnosis and treatment of pneumonitis with a new coronavirus infection (Trial version 3) [Internet]. [cited 2020 Mar 28]. Available from: http://www.nhc.gov.cn/xcs/zhengcwj/202001/f492c9153ea9437bb587ce2ffcbee1fa/files/39e7578d85964dbe81117736dd789d8f.pdf
- 11.关于印发新型冠状病毒感染的肺炎诊疗方案(试行第五版)的通知 国家中医药管理局政府网站 [Notice on Issuing a New Coronavirus Infected Pneumonia Diagnosis and Treatment Plan (Trial Version 5)] [Internet]. [cited 2020 Mar 28]. Available from: http://bgs.satcm.gov.cn/zhengcewenjian/2020-02-06/12847.html
- 12.Study Quality Assessment Tools | National Heart, Lung, and Blood Institute (NHLBI) [Internet]. [cited 2020 Mar 28]. Available from: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools
- 13.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasselblad V, Hedges LV. Meta-analysis of screening and diagnostic tests. Psychol Bull. 1995;117:167–178. doi: 10.1037/0033-2909.117.1.167. [DOI] [PubMed] [Google Scholar]
- 15.Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. [DOI] [PMC free article] [PubMed]
- 16.Ng SC, Tilg H. COVID-19 and the gastrointestinal tract: more than meets the eye. Gut. 2020;69:973–974. doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. [DOI] [PMC free article] [PubMed]
- 18.Liang W, Feng Z, Rao S, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–3. [DOI] [PubMed]
- 19.Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–81. [DOI] [PMC free article] [PubMed]
- 20.Musa S. Hepatic and gastrointestinal involvement in coronavirus disease 2019 (COVID-19): what do we know till now? Arab J Gastroenterol. 2020;21:3–8. [DOI] [PMC free article] [PubMed]
- 21.Budden KF, Gellatly SL, Wood DLA, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. [DOI] [PubMed]
- 22.Chen Y, Chen L, Deng Q, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833-40.. [DOI] [PubMed]
- 23.Ding S, Liang TJ. Review: Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission: a COVID-19 virological and clinical review. Gastroenterology. 2020;S0016-5085(20)30571-0. [DOI] [PMC free article] [PubMed]
- 24.Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv. Cold Spring Harbor Laboratory. 2020;2020:03.931766.
- 25.Feng G, Zheng KI, Yan Q-Q, et al. COVID-19 and liver dysfunction: current insights and emergent therapeutic strategies. J Clin Transl Hepatol. 2020;8:18–24. [DOI] [PMC free article] [PubMed]
- 26.Li J, Fan J-G. Characteristics and mechanism of liver injury in 2019 coronavirus disease. J Clin Transl Hepatol. 2020;8:13–17. doi: 10.14218/JCTH.2020.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strnad P, Tacke F, Koch A, Trautwein C. Liver - guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 28.Boeckmans J, Rodrigues RM, Demuyser T, Piérard D, Vanhaecke T, Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol. 2020;94:1367–9. [DOI] [PMC free article] [PubMed]
- 29.Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–23. [DOI] [PMC free article] [PubMed]
- 30.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed]
- 31.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. [DOI] [PMC free article] [PubMed]
- 32.Song F, Shi N, Shan F, et al. Emerging coronavirus 2019-nCoV pneumonia. Radiology. 2020;200274.
- 33.Chang D, Lin M, Wei L, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA. 2020;323:1092–3. [DOI] [PMC free article] [PubMed]
- 34.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. [DOI] [PMC free article] [PubMed]
- 35.Liu K, Fang Y-Y, Deng Y, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl). 2020;133:1025–31. [DOI] [PMC free article] [PubMed]
- 36.Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–74. [DOI] [PMC free article] [PubMed]
- 37.Ren L-L, Wang Y-M, Wu Z-Q, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133:1015–24. [DOI] [PMC free article] [PubMed]
- 38.Xu X-W, Wu X-X, Jiang X-G, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. [DOI] [PMC free article] [PubMed]
- 39.Zhang J-J, Dong X, Cao Y-Y, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–41. [DOI] [PubMed]
- 40.Wu J, Wu X, Zeng W, et al. Chest CT findings in patients with corona virus disease 2019 and its relationship with clinical features. Investig Radiol. 2020;55:257–61. [DOI] [PMC free article] [PubMed]
- 41.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–81. [DOI] [PMC free article] [PubMed]
- 42.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–34. [DOI] [PMC free article] [PubMed]
- 43.Xu Y-H, Dong J-H, An W-M, et al. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J Infect. 2020;80:394–400. [DOI] [PMC free article] [PubMed]
- 44.Yang W, Cao Q, Qin L, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):a multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80:388–93. [DOI] [PMC free article] [PubMed]
- 45.Huang Y, Tu M, Wang S, et al. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: a retrospective single center analysis. Travel Med Infect Dis. 2020;101606. [DOI] [PMC free article] [PubMed]
- 46.Xu X, Yu C, Qu J, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47:1275–80. [DOI] [PMC free article] [PubMed]
- 47.Liu W, Tao Z-W, Lei W, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl). 2020;133:1032–8. [DOI] [PMC free article] [PubMed]
- 48.Wu J, Liu J, Zhao X, et al. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020;ciaa199. [DOI] [PMC free article] [PubMed]
- 49.Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Investig Radiol. 2020;55:327–31. [DOI] [PMC free article] [PubMed]
- 50.Cao J, Hu X, Cheng W, et al. Clinical features and short-term outcomes of 18 patients with corona virus disease 2019 in intensive care unit. Intensive Care Med. 2020;46:851–3. [DOI] [PMC free article] [PubMed]
- 51.Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488. [DOI] [PMC free article] [PubMed]
- 53.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–8. [DOI] [PMC free article] [PubMed]
- 54.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am J Roentgenol. 2020;214:1072–7. [DOI] [PubMed]
- 55.Xiong Y, Sun D, Liu Y, et al. Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Investig Radiol. 2020;55:332–9. [DOI] [PMC free article] [PubMed]
- 56.Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–3.e3. [DOI] [PMC free article] [PubMed]
- 57.Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–11. [DOI] [PMC free article] [PubMed]
- 58.Zhou S, Wang Y, Zhu T, Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol.2020;214:1287–94. [DOI] [PubMed]
- 59.Wang L, Gao Y-H, Lou L-L, Zhang G-J. The clinical dynamics of 18 cases of COVID-19 outside of Wuhan, China. Eur Respir J. 2020;55:2000398. doi: 10.1183/13993003.00398-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spiteri G, Fielding J, Diercke M, et al. First cases of coronavirus disease 2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Euro Surveill. 2020;25:2000178. [DOI] [PMC free article] [PubMed]
- 61.COVID-19 National Incident Room Surveillance Team. COVID-19, Australia: Epidemiology Report 6 (Reporting week ending 19:00 AEDT 7 March 2020). Commun Dis Intell. 2018;2020:44. [DOI] [PubMed]
- 62.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62. [DOI] [PMC free article] [PubMed]
- 63.Liu K, Chen Y, Lin R, Han K. Clinical feature of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. 2020;80:e14–8. [DOI] [PMC free article] [PubMed]
- 64.Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–6. [DOI] [PMC free article] [PubMed]
- 65.Zhao D, Yao F, Wang L, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. 2020:ciaa247. [DOI] [PMC free article] [PubMed]
- 66.Chen Q, Quan B, Li X, et al. A report of clinical diagnosis and treatment of nine cases of coronavirus disease 2019. J Med Virol. 2020;92:683–7.. [DOI] [PMC free article] [PubMed]
- 67.Zhu W, Xie K, Lu H, Xu L, Zhou S, Fang S. Initial clinical features of suspected coronavirus disease 2019 in two emergency departments outside of Hubei, China. J Med Virol. 2020. 10.1002/jmv.25763. [DOI] [PMC free article] [PubMed]
- 68.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020:e200994. [DOI] [PMC free article] [PubMed]
- 69.Cheng Z, Lu Y, Cao Q, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. AJR Am J Roentgenol. 2020;215:121-6. [DOI] [PubMed]
- 70.Mo P, Xing Y, Xiao Y, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020:ciaa270.
- 71.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020:ciaa272. [DOI] [PMC free article] [PubMed]
- 72.Wang Y, Liu Y, Liu L, et al. Clinical outcome of 55 asymptomatic cases at the time of hospital admission infected with SARS-Coronavirus-2 in Shenzhen. China J Infect Dis. 2020;221:1770–4. [DOI] [PMC free article] [PubMed]
- 73.Han R, Huang L, Jiang H, Dong J, Peng H, Zhang D. Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia. AJR Am J Roentgenol. 2020:1–6. 10.2214/AJR.20.22961. [DOI] [PubMed]
- 74.Qian G-Q, Yang N-B, Ding F, et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID-19 in Zhejiang, China: A retrospective, multi-centre case series. QJM. 2020;hcaa089. [DOI] [PMC free article] [PubMed]
- 75.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. [DOI] [PMC free article] [PubMed]
- 76.Shi Y, Yu X, Zhao H, et al. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24:108. [DOI] [PMC free article] [PubMed]
- 77.Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. 2020;323:1612. [DOI] [PMC free article] [PubMed]
- 78.Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020;200843. [DOI] [PMC free article] [PubMed]
- 79.Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–5. [DOI] [PMC free article] [PubMed]
- 80.Liu Y, Yan L-M, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–7. [DOI] [PMC free article] [PubMed]
- 81.Zhang X, Cai H, Hu J, et al. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int J Infect Dis. 2020;94:81–7. [DOI] [PMC free article] [PubMed]
- 82.Deng Y, Liu W, Liu K, et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 (COVID-19) in Wuhan, China: a retrospective study. Chin Med J (Engl). 2020;133:1261–7. [DOI] [PMC free article] [PubMed]
- 83.Wan S, Xiang Y, Fang W, et al. Clinical features and treatment of COVID-19 patients in Northeast Chongqing. J Med Virol. 2020l;92:797–806. [DOI] [PMC free article] [PubMed]
- 84.Zhou B, She J, Wang Y, Ma X. The clinical characteristics of myocardial injury 1 in severe and very severe patients with 2019 novel coronavirus disease. J Inf Secur. 2020. [DOI] [PMC free article] [PubMed]
- 85.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–74. [DOI] [PMC free article] [PubMed]
- 86.Jin X, Lian J-S, Hu J-H, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–9. [DOI] [PMC free article] [PubMed]
- 87.Sun Y, Koh V, Marimuthu K, et al. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020:ciaa322. [DOI] [PMC free article] [PubMed]
- 88.Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020:e200950. [DOI] [PMC free article] [PubMed]
- 89.Lian J, Jin X, Hao S, et al. Analysis of epidemiological and clinical features in older patients with Corona virus disease 2019 (COVID-19) out of Wuhan. Clin Infect Dis. 2020;ciaa242. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 64 kb)