Abstract
Background
Differences in carcinogenesis and therapeutic efficacy according to ethnicity have been reported for lung cancer, and understanding differences in genetic mutation profiles among ethnicities is important for interpreting the results of clinical trials, preventing carcinogenesis, and individualizing treatment. However, no studies have focused on differences in mutation profiles among different ethnicities using large-scale genomic analysis data with detailed information on smoking history, the main cause of lung cancer.
Methods
To clarify the differences in genetic mutation profiles between Caucasian and Japanese subjects, we compared data from The Cancer Genome Atlas, which mainly included Caucasians, with results from the Japan Molecular Epidemiology for lung cancer study, which is an epidemiological study only involving Japanese subjects. We divided the participants into four groups according to smoking status and performed comparative analysis by tissue type (lung adenocarcinoma and squamous cell lung cancer).
Results
In patients with lung adenocarcinoma, the frequency of EGFR mutations was lower in Caucasian subjects than in Japanese subjects (14.6% vs. 51.1%), whereas the frequencies of mutations in other genes, namely KRAS (32.9% vs. 9.3%), TP53 (45.2% vs. 20.7%), BRAF (9.6% vs. 1.3%), PIK3CA (5.9% vs. 2.6%), KEAP1 (17.8% vs. 0.5%), NF1 (10.9% vs. 0.5%), STK11 (17.8% vs. 0.7%), RBM10 (8.7% vs. 0.1%), and MET (7.8% vs. 0.1%), were higher in Caucasian subjects. Among patients with squamous cell carcinoma, TP53 (81.2% vs. 49.1%), PIK3CA (14.5% vs. 6.8%), KEAP1 (12.7% vs. 0.9%), and NFE2L2 mutations (15.8% vs. 13.6%) were more common in Caucasian subjects.
Conclusions
Ethnicity is an important and complex characteristic that must be recognized and considered, even in the era of precision medicine. We should collaborate to share data for different ethnicities and incorporate them into clinical practice and the design of global clinical studies. Carefully designed molecular epidemiological studies focusing on ethnic differences are warranted.
Keywords: Lung cancer, ethnicity, Asian, Caucasian, mutation
Introduction
Lung cancer is the most common cancer in the world, both in terms of the numbers of new cases (1.8 million cases, 12.9% of all cancer cases) and deaths (1.6 million deaths, 19.4%) (1). The incidence of lung cancer differs by sex and ethnicity. Women comprise approximately 45% of patients with lung cancer in the US, versus only 25–30% in Eastern Asia (2,3). It has been recognized that ethnic differences contribute to disparities in carcinogenesis and treatment outcomes in lung cancer (4). The disparities can be attributed to the variety of mutations triggered by evolutionary forces over time and across populations. Although several single-nucleotide polymorphisms as significant genome-wide signals can be associated with these mutations, environmental factors including smoking, dust exposure, obesity, and potential viral infections (human papillomavirus) are also essential to the genomic diversity.
Many biomarkers, including epidermal growth factor receptor (EGFR), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), and v-Ki-ras2 Kirsten rat sarcoma (KRAS) mutations and anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1) fusions, have been examined in clinical trials and clinical practice. EGFR mutations are more frequently found in female patients who are never-smokers and who have adenocarcinoma histology, whereas KRAS mutations are more common in smokers (5-7). With the progress of molecular targeted therapy, the importance of the relationship between various gene mutations involved in carcinogenesis and specific factors in the development of therapeutics targeting those genes has attracted attention.
In recent years, the development of next-generation sequencing (NGS) technology has made it possible to detect various mutations; thus, some large-scale epidemiological investigations of genetic mutations associated with lung cancers have been reported (5-7). However, comparisons of genetic mutation frequency by ethnicity have not been performed, and the exact mechanisms behind these differences are not clear. These ethnic differences should be considered when assessing global clinical trials that include different ethnic populations, particularly Asian and non-Asian patients, for whom stratified analysis by ethnicity and ethnicity will be needed. We focused on the Japan Molecular Epidemiology for lung cancer (JME) study and The Cancer Genome Atlas (TCGA) to clarify differences in mutation profiles for lung cancer by ethnicity because those investigations had large sample sizes and detailed information on smoking status (5-7). The JME study only targeted Japanese patients, whereas TCGA predominantly targeted Caucasian patients, thereby permitting comparisons of mutation frequency by ethnicity.
Methods
Study design
We investigated the differences of mutational profiles between Caucasian and Japanese populations by comparing TCGA and JME study data.
TCGA is a series of cross-sectional, comprehensive genomic studies using NGS and whole-exome sequencing in the US. Approximately 70% of the patients were Caucasian, whereas 2% were Asian. Most patients had early-stage non-small cell lung carcinoma (NSCLC) (6,7). They targeted 384 patients for whom smoking status information was available, including 219 patients with lung adenocarcinoma and 165 patients with lung squamous cell carcinoma.
The JME study is a prospective and multicenter molecular epidemiology study designed to elucidate the mechanism of carcinogenesis in smokers and never-smokers with early-stage NSCLC. In total, 876 surgical samples from 441 ever-smokers and 435 never-smokers (651 patients with lung adenocarcinoma and 118 patients with lung squamous cell carcinoma) with early-stage NSCLC that were subjected to molecular analyses for 72 cancer-associated genes using NGS were examined in the current study.
Smoking status was classified into four groups according to the time of smoking cessation to compare data from TCGA and JME studies (Table 1: A, never-smokers; B, former smoker for >15 years; C, former smoker for ≤15 years; and D, current smokers). We compared mutation data from the two studies for each smoking status separately for lung adenocarcinoma and lung squamous cell carcinoma. In both studies, there were only a few nonsmokers with squamous cell carcinoma, and thus, comparisons of genetic profiles among nonsmokers were not performed.
Table 1. The frequencies of gene mutations in TCGA and JME studies.
| Gene | A (%) | B (%) | C (%) | D (%) | Total (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA | JME | TCGA | JME | TCGA | JME | TCGA | JME | TCGA | JME | P | |||||
| AD | N=32 | N=417 | N=69 | N=68 | N=73 | N=92 | N=45 | N=74 | N=219 | N=651 | |||||
| EGFR | 31.3 | 61.6 | 20.3 | 44.1 | 8.2 | 29.3 | 4.4 | 25.7 | 14.6 | 51.1 | <0.001 | ||||
| KRAS | 15.6 | 4.3 | 33.3 | 13.2 | 37.0 | 21.7 | 37.8 | 18.9 | 32.9 | 9.3 | <0.001 | ||||
| TP53 | 25.0 | 14.1 | 42.0 | 23.5 | 45.2 | 35.9 | 64.4 | 36.5 | 45.2 | 20.7 | <0.001 | ||||
| BRAF | 0.0 | 1.0 | 13.0 | 1.5 | 8.2 | 1.1 | 13.3 | 2.7 | 9.6 | 1.3 | <0.001 | ||||
| PIK3CA | 3.1 | 2.4 | 5.8 | 4.4 | 5.5 | 3.3 | 8.9 | 1.4 | 5.9 | 2.6 | 0.034 | ||||
| HER2 | 9.4 | 0.2 | 1.4 | 0.0 | 2.7 | 0.0 | 0.0 | 0.0 | 2.7 | 0.1 | 0.015 | ||||
| KEAP1 | 6.3 | 0.0 | 20.3 | 2.9 | 20.5 | 1.1 | 17.8 | 0.0 | 17.8 | 0.5 | <0.001 | ||||
| NF1 | 0.0 | 0.5 | 7.2 | 0.0 | 17.8 | 0.0 | 13.3 | 1.4 | 10.9 | 0.5 | <0.001 | ||||
| STK11 | 6.3 | 0.2 | 14.5 | 0.0 | 27.4 | 1.1 | 15.6 | 4.1 | 17.8 | 0.7 | <0.001 | ||||
| RBM10 | 6.3 | 0.2 | 7.2 | 0.0 | 6.8 | 0.0 | 15.6 | 0.0 | 8.7 | 0.1 | <0.001 | ||||
| SMARCA4 | 0.0 | 0.2 | 4.3 | 0.0 | 6.8 | 0.0 | 8.9 | 0.0 | 5.5 | 0.1 | <0.001 | ||||
| ARID1A | 6.3 | 0.0 | 11.6 | 0.0 | 4.1 | 0.0 | 6.7 | 0.0 | 7.3 | 0.0 | <0.001 | ||||
| U2AF1 | 0.0 | 0.0 | 2.9 | 0.0 | 4.1 | 1.1 | 6.7 | 1.4 | 3.7 | 0.3 | <0.001 | ||||
| RB1 | 6.3 | 0.2 | 5.8 | 0.0 | 4.1 | 0.0 | 2.2 | 1.4 | 4.6 | 0.3 | <0.001 | ||||
| MET | 15.6 | 0.2 | 5.8 | 0.0 | 9.6 | 0.0 | 2.2 | 0.0 | 7.8 | 0.1 | <0.001 | ||||
| HRAS | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.5 | 0.0 | 0.252 | ||||
| NRAS | 0.0 | 0.2 | 0.0 | 0.0 | 1.4 | 1.1 | 0.0 | 0.0 | 0.5 | 0.3 | 1 | ||||
| AKT1 | 0.0 | 0.0 | 1.4 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 | 0.4 | 0.2 | 0.440 | ||||
| SQ | N=50 | N=19 | N=87 | N=63 | N=28 | N=36 | N=165 | N=118 | |||||||
| EGFR | 2.0 | 5.3 | 3.4 | 1.6 | 3.6 | 0.0 | 3.0 | 1.7 | 0.703 | ||||||
| KRAS | 0.0 | 0.0 | 2.3 | 3.2 | 0.0 | 2.8 | 1.2 | 2.6 | 0.653 | ||||||
| TP53 | 72.0 | 68.4 | 85.1 | 47.6 | 85.7 | 41.7 | 81.2 | 49.1 | <0.001 | ||||||
| BRAF | 2.0 | 0.0 | 4.6 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.078 | ||||||
| PIK3CA | 22.0 | 10.5 | 12.6 | 6.3 | 7.1 | 5.6 | 14.5 | 6.8 | 0.065 | ||||||
| HER2 | 0.0 | 0.0 | 3.4 | 0.0 | 3.6 | 0.0 | 2.4 | 0.0 | 0.143 | ||||||
| KEAP1 | 12.0 | 0.0 | 12.6 | 1.6 | 14.3 | 0.0 | 12.7 | 0.9 | 0.001 | ||||||
| NF1 | 10.0 | 0.0 | 13.8 | 1.6 | 12.3 | 0.0 | 12.4 | 0.9 | <0.001 | ||||||
| PTEN | 6.0 | 0.0 | 11.5 | 1.6 | 3.6 | 0.0 | 8.5 | 0.9 | 0.005 | ||||||
| STK11 | 2.0 | 5.3 | 2.3 | 0.0 | 0.0 | 0.0 | 1.8 | 0.9 | 0.643 | ||||||
| SMARCA4 | 4.0 | 0.0 | 3.4 | 0.0 | 7.1 | 0.0 | 4.2 | 0.0 | 0.044 | ||||||
| RB1 | 4.0 | 0.0 | 9.2 | 0.0 | 0.0 | 0.0 | 6.1 | 0.0 | 0.006 | ||||||
| HRAS | 0.0 | 0.0 | 3.4 | 1.6 | 7.1 | 0.0 | 3.0 | 0.9 | 0.406 | ||||||
| NRAS | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 1 | ||||||
| AKT1 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.6 | 0.6 | 1.7 | 0.573 | ||||||
| NFE2L2 | 14.0 | 0.0 | 13.8 | 12.7 | 25.0 | 22.2 | 15.8 | 13.6 | 0.731 | ||||||
| FBXW7 | 4.0 | 0.0 | 5.7 | 0.0 | 10.7 | 5.6 | 6.0 | 1.7 | 0.081 | ||||||
| SMAD4 | 4.0 | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.078 | ||||||
| APC | 6.0 | 0.0 | 4.6 | 0.0 | 3.6 | 0.0 | 4.9 | 0.0 | 0.023 | ||||||
| FGFR2 | 2.0 | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 0.143 | ||||||
| CUL3 | 8.0 | 0.0 | 5.7 | 0.0 | 0.0 | 0.0 | 5.4 | 0.0 | 0.012 | ||||||
| RASA1 | 4.0 | 0.0 | 9.2 | 0.0 | 0.0 | 0.0 | 6.1 | 0.0 | 0.006 | ||||||
| TSC2 | 4.0 | 0.0 | 4.6 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | 0.043 | ||||||
A, never smokers; B, current reformed smokers for >15 years; C, current reformed smokers for ≤15 years; D, current smokers; TCGA, The Cancer Genome Atlas; JME, Japan Molecular Epidemiology for lung cancer; AD, adenocarcinoma; SQ, squamous cell carcinoma.
Statistical analysis
The correlation between the proportion of patients carrying each driver mutation and ethnicity was analyzed using χ2 tests, except when a small sample size (<5) required the use of Fisher’s exact test. The association of the frequency of each mutation with smoking status was evaluated through multivariate logistic regression testing. Statistical significance was assumed for a two-tailed P value of <0.05.
Results
Comparison of the mutation profiles between TCGA and JME studies
The frequencies of gene mutations in TCGA and JME studies are shown in Table 1. Differences in gene frequencies between the studies were observed for not only EGFR, KRAS, and TP53, which have relatively high mutation frequencies, but also other genes including PIK3CA, KEAP1, NF1, STK11, ARID1A, MET, and NFE2L2.
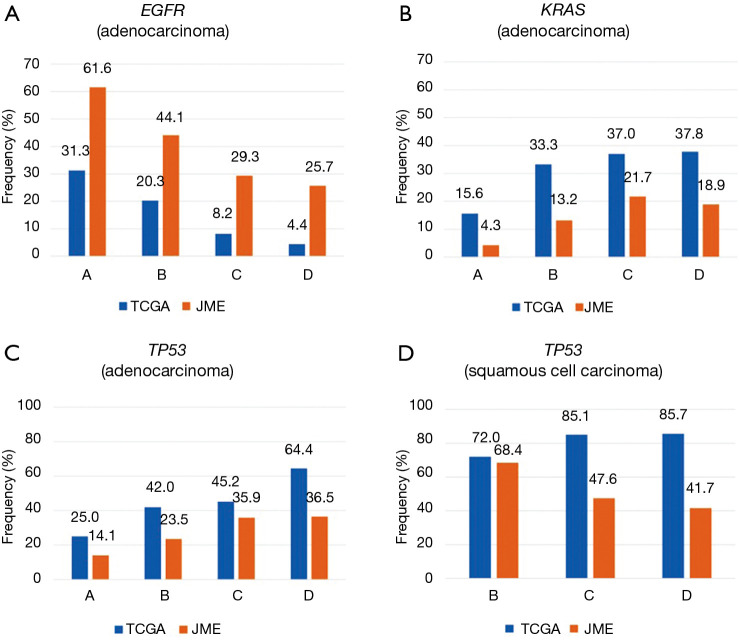
Among Japanese patients in the JME study, the overall prevalence of EGFR mutations was 43.6%, including rates of 51.1% among patients with adenocarcinoma and 61.6% among never-smokers. Among the mostly Caucasian patients in the TCGA study, the overall EGFR mutation prevalence was 9.6%, including rates of 14.6% among patients with adenocarcinoma and 31.3% among never-smokers (Table 1). EGFR mutations were more frequently found in patients with adenocarcinoma and never-smokers regardless of ethnicity. Meanwhile, among patients with adenocarcinoma, a shorter period since smoking cessation was linked to a lower frequency of EGFR mutations in both Japanese [odds ratio (OR) =0.562; 95% confidence interval (CI), 0.479–0.659; P<0.001] and Caucasian subjects (OR =0.451; 95% CI, 0.292–0.695; P<0.001) (Figure 1A). EGFR mutation rates were always higher in Japanese subjects than in Caucasian subjects regardless of smoking status; thus, a significant ethnic difference in mutation prevalence was confirmed (P<0.001). EGFR mutations were less frequent in patients with squamous cell carcinoma, and it was not possible to assess ethnic differences.
Figure 1.
Frequency of somatic mutations by smoking status. The frequencies of EGFR (A), KRAS (B), and TP53 (C) mutations significantly increased as the duration of the nonsmoking period decreased in patients with adenocarcinoma. There was no significant interaction between the TP53 mutation rate and the nonsmoking period in squamous carcinoma (D).
Another major ethnic difference among patients with adenocarcinoma was that the KRAS mutation rate was 9.3% in the JME study, compared with 32.9% in the TCGA study. KRAS mutations were significantly less common in Japanese subjects than in Caucasian subjects regardless of the smoking status (P<0.001). When examining the association between the frequency of KRAS mutations and smoking status, a short nonsmoking period was a risk factor for KRAS mutations in Japanese subjects (OR =1.83; 95% CI, 1.47–2.28; P<0.001), whereas the association was not significant in Caucasian subjects (OR =1.33; 95% CI, 0.991–1.80; P=0.057) (Figure 1B). Among patients with squamous cell carcinoma, KRAS mutations were less common among ever-smokers, although we could not assess ethnic differences.
The overall prevalence of TP53 mutations in patients with NSCLC was 25.1% (20.7% in patients with adenocarcinoma, 49.1% in patients with squamous cell carcinoma) in the JME study, versus 60.7% in the TCGA study (45.2% in patients with adenocarcinoma, 81.2% in patients with squamous cell carcinoma), illustrating that TP53 mutations were significantly more common in Caucasian patients than in Japanese patients regardless of the histology (P<0.001). In addition, the TP53 mutation rate increased as the duration of the nonsmoking period decreased among patients with adenocarcinoma (TCGA: OR =1.62; 95% CI, 1.21–2.16; P=0.0011; JME: OR =1.60; 95% CI, 1.36–1.88; P<0.001) (Figure 1C); by contrast, there was no significant interaction between the mutation rate and nonsmoking period in subjects with squamous carcinoma (TCGA: OR =1.70; 95% CI, 0.927–3.13; P=0.086; JME: OR =0.611; 95% CI, 0.350–1.07; P=0.083) (Figure 1D).
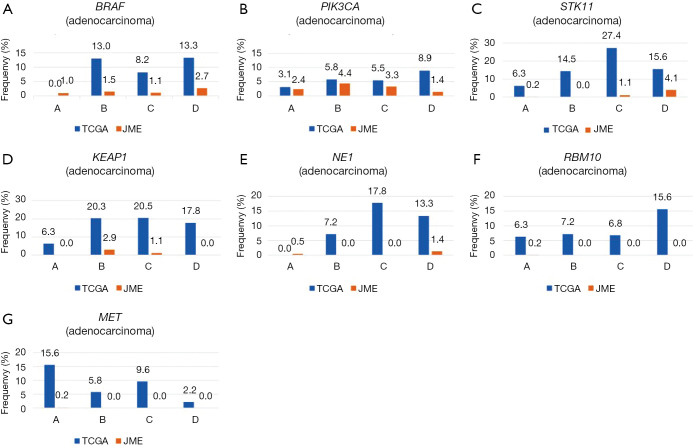
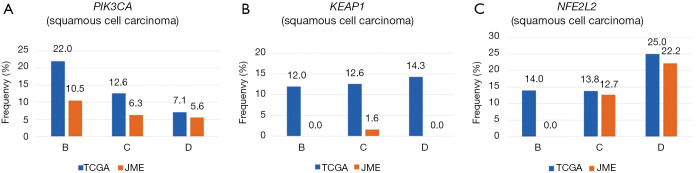
Other mutations were uncommon in both ethnicities, but because gene mutations are generally rare in Japanese patients, the mutation rates appeared to be higher in Caucasian patients for both adenocarcinoma (BRAF, 9.6% vs. 1.3%; PIK3CA, 5.9% vs. 2.6%; KEAP1, 17.8% vs. 0.5%; NF1, 10.9% vs. 0.5%; STK11, 17.8% vs. 0.7%; RBM10, 8.7% vs. 0.1%; MET, 7.8% vs. 0.1%) and squamous cell carcinoma (PIK3CA, 14.5% vs. 6.8%; KEAP1, 12.7% vs. 0.9%; NFE2L2, 15.8% vs. 13.6%) (Table 1, Figures S1,S2).
Figure S1.
Frequency of somatic mutations (BRAF, PIK3CA, STK11, KEAP1, NF1, BRM10, and MET) based on smoking status in adenocarcinoma. All mutation rates were always higher in Caucasian subjects than in Japanese subjects, regardless of the smoking status.
Figure S2.
Frequency of somatic mutations (PIK3CA, KEAP1, and NFE2L2) based on smoking status in squamous cell carcinoma. All mutation rates were always higher in Caucasian subjects than in Japanese subjects, regardless of the smoking status.
Discussion
NSCLC has become a prominent example of a solid tumor malignancy for which precision medicine has been employed. In addition, treatments for NSCLC targeting various genetic mutations have been developed, and many clinical trials have been designed to test these. Although, there are several reports that examined the influence of ethnic differences on the frequencies of EGFR, KRAS, and TP53 mutations (8-10), it is often unclear whether the other relatively rare mutations or their frequencies depend only on ethnicity. We, thus, investigated the differences in the mutation profiles of Caucasian and Japanese patients based on two large-scale epidemiological studies (TCGA and JME study).
The results of our research were consistent with those of a previous report, which states that EGFR mutations occur at a significantly higher frequency in lung adenocarcinoma among women, never-smokers, and East Asian subjects (9). Further, we demonstrated that ethnic differences had a considerable effect on EGFR mutation prevalence, although this prevalence changed based on the duration of the nonsmoking period. However, previous studies indicated that many Asian patients have types of polymorphisms that lead to a decreased intrinsic production of the EGFR protein (8); thus, the reasons for the high frequency of mutations among Asian patients remain unclear. If a certain critical level of EGFR is required to drive cells toward a malignant phenotype, another mechanism, involving the occurrence of activating mutations of EGFR and/or the autonomous activation of its downstream signaling may be required for the development of lung cancer among Asians. Further, it has been reported that the development of EGFR mutations was proportional to the dose of environmental tobacco smoke (ETS) exposure in never-smokers (11,12). Japanese women are more likely to be exposed to ETS, which may explain the high EGFR mutation frequency in Japanese patients. However, there are conflicting data published regarding this relationship (13,14); EGFR mutations can also be found in other histological subtypes of NSCLC as well as in former and current smokers, thus requiring further research on the mutation.
In our research, KRAS mutations were found to be significantly more common in Caucasian subjects than in Japanese subjects, and we demonstrated that a short nonsmoking period was a risk factor for KRAS mutations, especially in Japanese patients. In contrast to EGFR mutations, KRAS mutations were initially identified in patients with lung adenocarcinoma who had a history of heavy cigarette smoking; they were considered uncommon in patients without any history of cigarette smoking (15). Nonetheless, in the TCGA study, the KRAS mutation rate was 15.6% among patients with lung adenocarcinoma, even among Caucasian never-smokers. It was reported that, compared with former or current smokers, never-smokers were significantly more likely to have transition mutations (G to A) rather than transversion mutations (G to T or G to C), the latter of which are known to be smoking-related (16). Thus, we may need to approach the study of KRAS mutations differently for ever-smokers and never-smokers. On the contrary, among patients with lung adenocarcinoma, this research revealed KRAS mutations in 4.3% of Japanese patients without a history of smoking; the mutations were significantly less frequent in Japanese patients than in Caucasian patients, even among ever-smokers. Meanwhile, KRAS mutations were extremely uncommon in both Caucasian and Japanese patients with squamous cell carcinomas, regardless of their smoking history. A mutually exclusive relationship exists between KRAS and EGFR mutations; the population with KRAS mutations differs from that with EGFR mutations because, unlike the latter, KRAS mutations are closely related with smoking. However, there are differences in the KRAS mutation frequency that cannot be explained by smoking alone, and these mutations are also considered to be independently associated with ethnic differences.
We also revealed here, the frequency of the TP53 mutation for each ethnicity, and this frequency differed significantly. In NSCLC, the TP53 mutation frequency varies considerably among different populations and studies, ranging from 18% to 60% (17); therefore, our findings help clarify the frequency of the TP53. Additionally, we clarified the association between the TP53 mutation rate and smoking status. Although TP53 point mutations were scattered along the coding sequence, they tended to cluster at certain locations in exons 5–8 as hot spot codons, which in NSCLC, were associated with smoking (18). This study also found that TP53 mutations occurred at a significantly higher frequency in ever-smokers than in never-smokers. It was reported that pesticide exposure, cement housing, and the use of an indoor kitchen were risk factors for TP53 mutations; pesticides, which are widely used in the US, are probable human lung carcinogens (19). There is evidence that distinct mutation patterns of TP53 are linked with environmental carcinogens (18); however, there is little information on the genetic factors found in this region that affect lung cancer. We could not confirm the impact of environmental factors, excluding smoking, on NSCLC and, therefore, this is a subject of future investigation.
In our study, we observed that the frequencies of other gene mutations, including those of BRAF, PIK3CA, HER2, KEAP1, NF1, STK11, RBM10, SMARCA4, ARID1A, U2AF1, RB1, and MET in adenocarcinoma, and those of KEAP1, NF1, PTEN, SMARCA4, RB1, APC, CUL3, RASA1, and TSC2 in squamous cell carcinoma were extremely low in Japanese patients; therefore, the frequencies were significantly higher in the Caucasian patients regardless of their smoking status (Table 1). These gene mutations were also being studied for their relation to the carcinogenesis, treatment, and prognosis of lung cancer. For example, the Keap1-nuclear factor erythroid 2-related factor-2 (Nrf2) pathway is a key determinant for cells to cope with oxidative stress (20). Elevated Nrf2 levels and Keap1 dysfunction have been frequently identified in lung cancer, and it is possible that these are associated with tumor progression, cytoprotection, resistance to chemotherapeutic drugs, and poor prognosis (21). In addition, PIK3CA mutations represent a distinctive genetic feature of NSCLC with chronic obstructive pulmonary disease (22). However, there have been few reports that have assessed the frequency of these mutations, and there is variation in the observed frequency depending on the report. Further, no reports have assessed the influence of ethnic differences on those infrequent mutations that cause lung cancer. The results of our study suggested that Caucasian patients have higher mutation risks, although we did not assess how many mutation coincidences occurred in each case. One of the reasons is that some mutations may often commonly occur with KRAS mutations like KEAP1, and ethnic differences may affect the frequency of KRAS mutations. In addition, it is possible that the EGFR mutation, which is more common in Asians, may be mutually exclusive with other gene mutations. Furthermore, other causes may include the standard of living in each region.
It was reported that when the tumor mutation burden (TMB) was dichotomized as high vs. low to intermediate, Caucasian ethnicity was associated with a high TMB, while Hispanic ethnicity was associated with a low to intermediate TMB (23). The TMB may be associated with ethnicity, but there are few studies for other ethnicities, including Asians. The association between TMB and immunotherapy response was reported previously (23,24) and, thus, the assessment of infrequent mutations would also be important.
In our study, although the frequencies of various other gene mutations were found to differ by ethnicity, the exact mechanism by which ethnic differences cause this diversity was unclear. Further, we did not investigate the effect of environmental factors other than smoking on the variation in mutation frequency. In the future, clarifying the differences in gene mutation profiles among all ethnicities and environmental factors will lead to better oncology prevention approaches and individualized treatments.
There are some limitations to our research; the main limitation of our research is that TCGA includes population of various ethnicities, although most patients were Caucasian. Thus, this research is not a pure comparison of Caucasian and Japanese populations. Another potential limitation is the differences in the methods used to detect gene mutations among studies.
However, the comparison of such large-scale trials is important for clarifying differences in gene profiles among ethnicities. To the best of our knowledge, this is the largest integrative genomic analysis study focusing on the relationship between ethnicity and oncogenic driver mutations. Our research will help clarify the relationship between biological differences in ethnicity and mutation profiles to interpret clinical trial results, inhibit carcinogenesis, and provide personalized treatments.
Conclusions
Ethnicity is an important and complex characteristic that needs to be recognized in the design and conduct of global clinical studies, as safety, tolerability, and response may vary between Asian and non-Asian patients. We clarified the differences in the frequencies of genetic mutations by ethnicity (Japanese vs. Caucasian) based on two large-scale epidemiological studies. These results should be of great help in terms of cancer prevention and individualized treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and and informed consent was taken from all individual participants.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Tomoya Kawaguchi and Phillip C. Mack) for the Series “Ethnic Difference in Lung Cancer” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2019.08.61). The series “Ethnic Difference in Lung Cancer” was commissioned by the editorial office without any funding or sponsorship. TK served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Thoracic Disease from Dec 2018 to Nov 2020. The authors have no other conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Ahn MJ, Lee J, Park YH, et al. Korean ethnicity as compared with white ethnicity is an independent favorable prognostic factor for overall survival in non-small cell lung cancer before and after the oral epidermal growth factor receptor tyrosine kinase inhibitor era. J Thorac Oncol 2010;5:1185-96. 10.1097/JTO.0b013e3181e2f624 [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi T, Matsumura A, Fukai S, et al. Japanese ethnicity compared with Caucasian ethnicity and never-smoking status are independent favorable prognostic factors for overall survival in non-small cell lung cancer: a collaborative epidemiologic study of the National Hospital Organization Study Group for Lung Cancer (NHSGLC) in Japan and a Southern California Regional Cancer Registry databases. J Thorac Oncol 2010;5:1001-10. 10.1097/JTO.0b013e3181e2f607 [DOI] [PubMed] [Google Scholar]
- 4.Soo RA, Kawaguchi T, Loh M, et al. Differences in outcome and toxicity between Asian and caucasian patients with lung cancer treated with systemic therapy. Future Oncol 2012;8:451-62. 10.2217/fon.12.25 [DOI] [PubMed] [Google Scholar]
- 5.Kawaguchi T, Koh Y, Ando M, et al. Prospective Analysis of Oncogenic Driver Mutations and Environmental Factors: Japan Molecular Epidemiology for Lung Cancer Study. J Clin Oncol 2016;34:2247-57. 10.1200/JCO.2015.64.2322 [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine I, Yamamoto N, Nishio K, et al. Emerging ethnic differences in lung cancer therapy. Br J Cancer 2008;99:1757-62. 10.1038/sj.bjc.6604721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou W, Christiani DC. East meets West: ethnic differences in epidemiology and clinical behaviors of lung cancer between East Asians and Caucasians. Chin J Cancer 2011;30:287-92. 10.5732/cjc.011.10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schabath MB, Cress D, Munoz-Antonia T. Racial and Ethnic Differences in the Epidemiology and Genomics of Lung Cancer. Cancer Control 2016;23:338-46. 10.1177/107327481602300405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaguchi T, Ando M, Kubo A, et al. Long exposure of environmental tobacco smoke associated with activating EGFR mutations in never-smokers with non-small cell lung cancer. Clin Cancer Res 2011;17:39-45. 10.1158/1078-0432.CCR-10-1773 [DOI] [PubMed] [Google Scholar]
- 12.Soo RA, Kubo A, Ando M, et al. Association Between Environmental Tobacco Smoke Exposure and the Occurrence of EGFR Mutations and ALK Rearrangements in Never-smokers With Non-Small-cell Lung Cancer: Analyses From a Prospective Multinational ETS Registry. Clin Lung Cancer 2017;18:535-42. 10.1016/j.cllc.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 13.Taga M, Mechanic LE, Hagiwara N, et al. EGFR somatic mutations in lung tumors: radon exposure and passive smoking in former- and never-smoking U.S. women. Cancer Epidemiol Biomarkers Prev 2012;21:988-92. 10.1158/1055-9965.EPI-12-0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YJ, Cho BC, Jee SH, et al. Impact of environmental tobacco smoke on the incidence of mutations in epidermal growth factor receptor gene in never-smoker patients with non-small-cell lung cancer. J Clin Oncol 2010;28:487-92. 10.1200/JCO.2009.24.5480 [DOI] [PubMed] [Google Scholar]
- 15.Rodenhuis S, van de Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med 1987;317:929-35. 10.1056/NEJM198710083171504 [DOI] [PubMed] [Google Scholar]
- 16.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731-4. 10.1158/1078-0432.CCR-08-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jassem E, Niklinski J, Rosell R, et al. Types and localisation of p53 gene mutations: a report on 332 non-small cell lung cancer patients. Lung Cancer 2001;34 Suppl 2:S47-51. 10.1016/S0169-5002(01)00344-0 [DOI] [PubMed] [Google Scholar]
- 18.Hussain SP, Hofseth LJ, Harris CC. Tumor suppressor genes: at the crossroads of molecular carcinogenesis, molecular epidemiology and human risk assessment. Lung Cancer 2001;34 Suppl 2:S7-15. 10.1016/S0169-5002(01)00339-7 [DOI] [PubMed] [Google Scholar]
- 19.Alavanja MC, Dosemeci M, Samanic C, et al. Pesticides and lung cancer risk in the agricultural health study cohort. Am J Epidemiol 2004;160:876-85. 10.1093/aje/kwh290 [DOI] [PubMed] [Google Scholar]
- 20.Barrera-Rodríguez R. Importance of the Keap1-Nrf2 pathway in NSCLC: Is it a possible biomarker? Biomed Rep 2018;9:375-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollrath V, Wielandt AM, Iruretagoyena M, et al. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J 2006;395:599-609. 10.1042/BJ20051518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawa K, Koh Y, Kawaguchi T, et al. PIK3CA mutation as a distinctive genetic feature of non-small cell lung cancer with chronic obstructive pulmonary disease: A comprehensive mutational analysis from a multi-institutional cohort. Lung Cancer 2017;112:96-101. 10.1016/j.lungcan.2017.07.039 [DOI] [PubMed] [Google Scholar]
- 23.Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as