Abstract
In advanced stage non-small cell lung cancer (NSCLC) patients, Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) testing may soon acquire a predictive significance to select patients for AMG510 treatment. Since tissue samples are not always available, liquid biopsy may represent a viable option for KRAS testing. Here, we review the last three years clinical practice performed on 194 plasma based liquid biopsies by next generation sequencing (NGS) SiRe® panel. In particular, 36 (18.6%) KRAS mutated cases were identified, with an overall median allelic frequency of 5.0% (ranging between 0.2% and 46.8%). No concomitant mutations were observed in the other NSCLC clinical relevant genes included in the SiRe® panel, such as epidermal growth factor receptor (EGFR) and v-Raf murine sarcoma viral oncogene homolog B (BRAF). Exon 2 p.G12C was the most common detected mutation (13/36, 36.1%). In conclusion, our data update and confirm that SiRe® NGS panel represents a robust analytical tool to assess KRAS mutational status on circulating tumor DNA. Further investigation is required to design more cost-effective diagnostic algorithms to harmonize clinical relevant biomarker testing on tissue and blood in advanced stage NSCLC clinical practice.
Keywords: Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS), next generation sequencing (NGS), lung cancer, AMG510, G12C, basal setting, liquid biopsy
Introduction
Lung cancer is the most frequent cause of cancer death, worldwide (1). In the last years, several clinical trials have defined the pivotal role of the molecular assessment of different biomarkers, such as epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 receptor tyrosine kinase (ROS1), v-Raf murine sarcoma viral oncogene homolog B (BRAF), and programmed death-ligand 1 (PD-L1), in order to administrate either tyrosine kinase inhibitors (TKIs) or immune-checkpoint inhibitors to improve survival and quality of life of advanced stage non-small cell lung cancer (NSCLC) patients (2-14). For this reason the international guideline from the College of American Pathologists (CAP), the International Association for the Study of Lung Cancer (IASLC), and the Association for Molecular Pathology (AMP), the National Comprehensive Cancer Network (NCCN) and the American Society of Clinical Oncology (ASCO) guidelines established a minimum panel of “must test genes”, represented by mutations in EGFR, and BRAF, and gene fusions affecting ALK and ROS1, in addition to evaluation of the expression of PD-L1 (15-17).
Beyond these biomarkers, other genes are currently being evaluated as predictive indicators (18-21). In particular, Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) mutations, occurring in 25–30% of NSCLC patients, have recently acquired a relevant interest, not only as a negative prognostic biomarker, but also as actionable targets (22). As a matter of the fact, a great deal of interest was raised by KRAS exon 2 p.G12C (occurring in about 13% of NSCLC patients). In fact, the preliminary results, presented at the 2019 ASCO annual meeting, on the performance of the small molecule AMG510 (Amgen, Thousand Oaks, CA) in a phase 1 clinical trial were promising both in terms of efficacy and safety (23,24). Currently, also other drugs targeting different KRAS mutations are under investigation (25).
In order to simultaneously detect all these relevant biomarkers, high throughput devices should be implemented in molecular predictive pathology laboratories. Next generation sequencing (NGS) is a fascinating tool that enables the analysis of different biomarkers for different patients at the same time (26). In our Institution we routinely adopt NGS to assess NSCLC biomarkers; to this end, we designed a customized NGS panel, that makes cost-effective the batching of different tumor samples. In fact, this panel, termed SiRe®, is meant to provide a unique workflow of samples for different specimen types (tissue and liquid biopsies) (27-30). Thus, whenever tissue is not adequate or available for NGS testing, which is a common occurrence for advanced stage NSCLC patients, SiRe® NGS plasma based analysis is a viable option to assess EGFR mutational status (31). In a previous study, we showed that SiRe® panel analytical parameters, such as number of total reads, mean read length, number of mapped reads, percentage of reads on target, average reads per amplicon and uniformity of amplicon coverage, are more than satisfactory to reliable assess EGFR mutational status (29). Beyond EGFR testing, the SiRe® panel also generate data on KRAS mutational status. Thus, in this study, we reviewed our NGS records, generated in the last three years from plasma specimens. While for EGFR testing a well defined diagnostic algorithm has been established, no specific data are available to evaluate quality benchmark for plasma based KRAS testing. This study was undertaken firstly to assess whether, in patients without available or adequate tissue, liquid biopsy is a feasible and reliable tool; secondly, to evaluate the overall KRAS mutational rate and the occurrence of single hotspot alterations in exon 2 and 3. The final goal is to build data derived from clinical practice to be useful to designed specific diagnostic algorithm that could harmonize KRAS tissue and blood based assays.
Materials and methods
Patient characteristics
To establish the rate of KRAS mutated cases in NSCLC as detected by the SiRe® panel on blood, a computerized search was carried out. Patients who performed SiRe® NGS plasma based analysis, from January 2016 to December 2018, were selected by using the following inclusion criteria: (I) cyto-histological evidence of advanced stage lung adenocarcinoma, (II) unavailability of adequate tissue samples for molecular tests, and (III) naïve to any treatment. This search yielded a total of 194 patients, with a preponderance of male (123/194, 63.4%); the overall KRAS mutation rate and the distribution of the single mutation occurring in exon 2 and 3 was assessed in this population.
All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Helsinki Declaration (http://www.wma.net/en/30publications/10policies/b3/). According to the aforementioned National Guidelines, the study did not require an Ethical Committee approval since it did not modify the patient clinical management.
NGS analysis
As a general rule, in our clinical practice, liquid biopsy is performed upon oncologist request. Patients are received directly in our Institution where a nurse, who is a permanent member of our molecular laboratory staff, collect in EDTA Vacutainer tubes (BD, Plymount, UK) ten mL of peripheral blood, as previously described (27,29,30). By this in-house approach, samples are immediately processed in order to avoid the risk of circulating tumor DNA (ctDNA) degradation. Two centrifugation steps (2,300 rpm for 10 min) are performed to obtain at least 1.2 mL of plasma for each sample. Subsequently, ctDNA is extracted by adopting the QIAsymphony DSPVirus/Pathogen Midi Kit on the QIAsymphony robot (Qiagen, Venlo Limburg), following the manufacturer instructions. After the extraction, ctDNA is analyzed by NGS. A customized NGS panel, called SiRe®, that covers 568 clinical relevant mutations in six genes [EGFR, KRAS, Neuroblastoma RAS Viral Oncogene Homolog (NRAS), BRAF, KIT Proto-Oncogene, Receptor Tyrosine Kinase (KIT) and Platelet Derived Growth Factor Receptor Alpha (PDGFRA)] is adopted to perform molecular analysis, as previously reported (27-30). In addition to automatic variant calling analysis, carried out by using SiRe® panel specific optimized variant caller plug-in (v.5.0.2.1) parameters, BAM files are visually inspected with the Golden Helix Genome Browser v.2.0.7 (Bozeman, MT, USA).
In this current study, to assess the robustness and the reliability of SiRe® panel on blood, cases showing KRAS mutations were evaluated from an analytical point of view, taking into account several run metric parameters; in particular, number of total reads, mean read length, number of mapped reads, percentage of reads on target, average reads per amplicon and uniformity of amplicon coverage were evaluated for each KRAS mutated case.
Results
SiRe® NGS analysis
On the overall, 36 (18.6%) out of the 194 liquid biopsies harbored a KRAS point mutation either in exon 2 or in exon 3 (Table 1). No concomitant mutations were observed in the other NSCLC clinical relevant genes included in the SiRe® panel, such as EGFR and BRAF.
Table 1. Detection of KRAS mutational status by next generation sequencing approach on liquid biopsy in basal setting.
| KRAS mutated cases/total cases (%) | p.G12A (%) | p.G12C (%) | p.G12D (%) | p.G12S (%) | p.G12V (%) | p.G13D (%) | p.G13S (%) | p.A59V (%) | p.Q61H (%) |
|---|---|---|---|---|---|---|---|---|---|
| 36/194 (18.6) | 1/36 (2.8) | 13/36 (36.1) | 5/36 (13.9) | 3/36 (8.3) | 3/36 (8.3) | 4/36 (11.1) | 4/36 (11.1) | 1/36 (2.8) | 2/36 (5.6) |
KRAS, Kirsten Rat Sarcoma Viral Oncogene Homolog.
Clinical data of KRAS mutant cases are reported in Table 2. Briefly, patients’ age widely ranged (36 to 89 years), with a median value of 66.9 years (Table 2). Moreover, KRAS mutations were more frequent in male than female patients (24 vs. 12; 66.7% vs. 33.3%). The overall median allelic frequency (AF) was 5.0% (ranging between 0.2% and 46.8%) (Table 3).
Table 2. KRAS mutated patients characteristics evaluated in all instances.
| Characteristics | Value |
|---|---|
| Patients, n | 36 (100.0%) |
| Male | 24 (66.7%) |
| Female | 12 (33.3%) |
| Age range | 36–89 |
| Median age | 66.9 |
KRAS, Kirsten Rat Sarcoma Viral Oncogene Homolog.
Table 3. Correlation among patients’ age, sex and KRAS mutational status.
| Samples | Age | Sex | KRAS mutation (AF) |
|---|---|---|---|
| 1 | 36 | F | p.G12D (1.5%) |
| 2 | 64 | M | p.G12C (1.3%) |
| 3 | 48 | M | p.G12C (0.6%) |
| 4 | 59 | F | p.G12D (1.3%) |
| 5 | 89 | M | p.G12D (7.2%) |
| 6 | 62 | M | p.G12C (5.6%) |
| 7 | 64 | M | p.G13S (0.2%) |
| 8 | 76 | F | p.G13D (0.3%) |
| 9 | 69 | M | p.G12S (6.4%) |
| 10 | 65 | M | p.G12C (3.3%) |
| 11 | 84 | M | p.G13S (0.2%) |
| 12 | 74 | F | p.G12C (24.0%) |
| 13 | 73 | F | p.G12C (0.9%) |
| 14 | 49 | M | p.G12S (1.4%) |
| 15 | 61 | F | p.G12C (4.6%) |
| 16 | 69 | M | p.G12C (46.8%) |
| 17 | 70 | M | p.G12S (7.1%) |
| 18 | 65 | F | p.G12V (24.0%) |
| 19 | 68 | M | p.G12D (0.9%) |
| 20 | 58 | M | p.G12C (3.9%) |
| 21 | 65 | F | p.G12C (2.5%) |
| 22 | 69 | M | p.G13D (0.4%) |
| 23 | 73 | M | p.G12C (2.9%) |
| 24 | 84 | M | p.G13S (0.6%) |
| 25 | 70 | M | p.G13S (0.7%) |
| 26 | 65 | M | p.G13D (4.2%) |
| 27 | 63 | M | p.G12C (4.7%) |
| 28 | 78 | M | p.G12D (2.0%) |
| 29 | 77 | M | p.G12A (0.7%) |
| 30 | 81 | F | p.G12V (1.0%) |
| 31 | 51 | F | p.G12C (3.1%) |
| 32 | 68 | M | p.G13D (0.7%) |
| 33 | 52 | F | p.G12V (4.6%) |
| 34 | 70 | M | p.Q61H (0.2%) |
| 35 | 77 | M | p.Q61H (4.7%) |
| 36 | 64 | F | p.A59V (0.2%) |
KRAS, Kirsten Rat Sarcoma Viral Oncogene Homolog; AF, allelic frequency; F, female; M, male.
Most KRAS mutations occurred in exon 2 (33/36, 91.7%) whereas exon 3-point mutations were less frequently (3/36, 8.3%) (Tables 1 and 3). KRAS exon 2 p.G12C was the most common mutation (13/36, 36.1%). The other mutations are reported in Tables 1 and 3. KRAS exon 2 p.G12C was identified mostly in male patients (8/13, 61.5%), whereas female were a lower number of mutated cases was evaluated (5/13, 38.5%). The median age of patients harboring KRAS exon 2 p.G12C was 63.5 (ranging between 48 and 74 years). The median AF of KRAS exon 2 p.G12C was 8.0% (ranging between 0.6% and 46.8%) (Table 3).
NGS analytical performance
NGS run parameters in KRAS mutated cases were evaluated in order to assess the analytical performance of the SiRe® panel. Taking into account all 36 mutant cases, an average of 152371.03 (ranging from 21306.00 to 477906.00) reads per sample was obtained. The median read length was of 133.92 bp (ranging from 95.00 to 209.00). Concerning the number of mapped reads, an average of 151697.61 (ranging from 21126.00 to 476909.00) was obtained, with a 95.64% reads on target (ranging from 86.60% to 99.08%). Considering amplicon parameters, an average reads per amplicon of 3416.26 (ranging from 488.50 to 10048.00) was evaluated with a 98.28% uniformity of amplicon coverage (ranging from 95.24% to 100.00%) (Table 4).
Table 4. NGS parameters.
| Samples | Reads | Mean read length (bp) | Number of mapped reads | Percent reads on target (%) | Average reads per amplicon | Uniformity of amplicon coverage (%) |
|---|---|---|---|---|---|---|
| 1 | 163,218.00 | 128.00 | 162,752.00 | 98.06 | 3,800.00 | 97.62 |
| 2 | 151,445.00 | 127.00 | 150,370.00 | 97.03 | 3,474.00 | 97.62 |
| 3 | 103,023.00 | 128.00 | 102,573.00 | 97.79 | 2,388.00 | 97.62 |
| 4 | 71,556.00 | 129.00 | 71,360.00 | 97.54 | 1,657.00 | 97.62 |
| 5 | 21,306.00 | 127.00 | 21,126.00 | 97.12 | 488.50 | 97.62 |
| 6 | 113,202.00 | 130.00 | 112,930.00 | 98.01 | 2,635.00 | 97.62 |
| 7 | 348,181.00 | 135.00 | 347,148.00 | 97.91 | 8,093.00 | 97.62 |
| 8 | 72,635.00 | 128.00 | 72,364.00 | 97.77 | 1,685.00 | 97.62 |
| 9 | 133,275.00 | 132.00 | 132,908.00 | 97.57 | 3,087.00 | 97.62 |
| 10 | 92,859.00 | 126.00 | 91,962.00 | 96.90 | 2,122.00 | 97.62 |
| 11 | 121,037.00 | 129.00 | 120,379.00 | 97.44 | 2,793.00 | 97.62 |
| 12 | 372,007.00 | 128.00 | 371,444.00 | 99.08 | 8,762.00 | 100.00 |
| 13 | 261,348.00 | 128.00 | 260,930.00 | 97.99 | 5,485.00 | 100.00 |
| 14 | 77,389.00 | 125.00 | 77,178.00 | 98.13 | 1,735.00 | 100.00 |
| 15 | 477,906.00 | 128.00 | 476,909.00 | 98.56 | 10,048.00 | 100.00 |
| 16 | 246,262.00 | 146.00 | 245,509.00 | 97.77 | 5,715.00 | 100.00 |
| 17 | 82,079.00 | 124.00 | 81,537.00 | 94.06 | 1,826.00 | 100.00 |
| 18 | 96,798.00 | 128.00 | 94,712.00 | 94.23 | 2,125.00 | 100.00 |
| 19 | 77,020.00 | 123.00 | 76,284.00 | 93.81 | 1,704.00 | 100.00 |
| 20 | 88,066.00 | 123.00 | 87,374.00 | 95.04 | 1,977.00 | 100.00 |
| 21 | 140,493.00 | 209.00 | 139,914.00 | 90.27 | 3,007.00 | 95.24 |
| 22 | 167,791.00 | 204.00 | 166,706.00 | 87.79 | 3,485.00 | 95.24 |
| 23 | 90,000.00 | 202.00 | 89,342.00 | 86.60 | 1,842.00 | 95.24 |
| 24 | 72,172.00 | 127.00 | 71,371.00 | 96.28 | 1,636.00 | 100.00 |
| 25 | 37,987.00 | 95.00 | 37,234.00 | 96.33 | 854.00 | 100.00 |
| 26 | 123,333.00 | 129.00 | 122,705.00 | 93.95 | 2,745.00 | 97.62 |
| 27 | 51,115.00 | 126.00 | 50,886.00 | 95.43 | 1,156.00 | 95.24 |
| 28 | 318,977.00 | 129.00 | 317,125.00 | 96.63 | 7,296.00 | 95.24 |
| 29 | 175,337.00 | 128.00 | 174,508.00 | 94.37 | 3,921.00 | 97.61 |
| 30 | 250,685.00 | 126.00 | 249,571.00 | 92.27 | 5,483.00 | 100.00 |
| 31 | 167,650.00 | 128.00 | 167,391.00 | 94.62 | 3,771.00 | 100.00 |
| 32 | 97,953.00 | 126.00 | 97,331.00 | 91.84 | 2,128.00 | 100.00 |
| 33 | 281,066.00 | 133.00 | 280,645.00 | 92.92 | 6,209.00 | 100.00 |
| 34 | 173,021.00 | 127.00 | 172,314.00 | 97.55 | 4,002.00 | 97.62 |
| 35 | 47,845.00 | 130.00 | 47,653.00 | 97.32 | 1,104.00 | 97.62 |
| 36 | 119,320.00 | 130.00 | 118,669.00 | 97.21 | 2,747.00 | 97.62 |
NGS, next generation sequencing; bp, base pair.
Discussion
Our data obtained in real life, generated from a review of routine clinical reports, underlined the technical feasibility of KRAS mutation assessment in plasma samples. In fact, in advanced stage NSCLC treatment-naïve patients, the customized NGS SiRe® panel offers a valuable option to select, for targeted treatments, patients that despite the morphological evidence of NSCLC do not have tissue availability for molecular diagnosis (Figure 1). Diagnostic algorithms are well designed for EGFR testing, as liquid biopsy is recommended as the initial procedure whenever tissue is not available (32). Conversely, clinical procedures are less well established to detect KRAS exon 2 p.G12C as an actionable biomarker. To date, KRAS testing in advanced stage NSCLC patients has been performed either to provide prognostic information (33), or to rule out less common driver alterations, to make lung biomarker testing more effective. More recently, KRAS exon 2 mutations also acquired a novel predictive significance. In particular, promising results have been shown by a novel drug (AMG510) administered to advanced stage NSCLC patients harboring a KRAS exon 2 p.G12C point mutation (24). Were these very promising data confirmed in a phase III clinical trial, it would be conceivable that KRAS mutational testing will soon be mandatory in clinical practice. Currently, in the experience of our laboratory, a referral center for lung cancer biomarker testing, tissue specimens are often (17%) rejected for insufficient cellularity (34). Thus, it is conceivable that KRAS testing will be often performed on ctDNA. In this setting, multiplexing is crucial to ensure a complete genomic profile. In addition, when evaluating cases wild type for KRAS mutations, the possibility to detect EGFR and BRAF mutations would be very useful to distinguish between real KRAS wild type samples from those tumors that do not shed ctDNA into the bloodstream. In other words, testing plasma for KRAS mutations has a double role: first, to select patients for AMG510; second, to make the molecular status assessment of EGFR and BRAF more cost-effective. In fact, considering our data, a not negligible subset of patients (6.7%; 13/194) of cases showing a KRAS exon 2 p.G12C point mutation may undergo a KRAS TKI treatment, while an additional 11.9% (23/194) of cases, showing other common KRAS alterations, would not need an EGFR and BRAF molecular test.
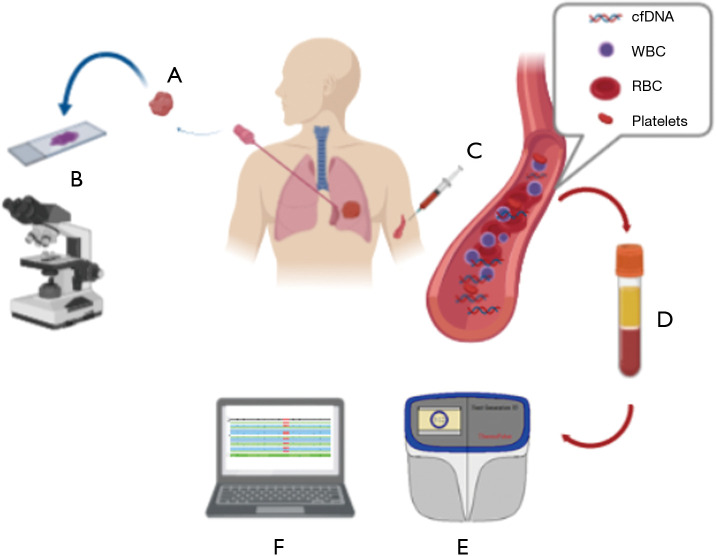
Figure 1.
If tissue samples (histological or cytological) were inadequate or insufficient at morphological evaluation for molecular analysis (A, B), liquid biopsy may represent a useful tool (C). In our study, after blood withdrawn (C), plasma was obtained after centrifugation (D) and extracted ctDNA was further analyzed by using our next generation sequencing panel (SiRe®) on S5 platform (Thermo Fisher Scientific, Waltham, MA) (E). Visual inspection of the sequencing reads was performed by using the Golden Helix Genome Browser v.2.0.7 (Bozeman, MT, USA) (F). (Credit: Created with Biorender).
Despite our efforts to provide an accurate and comprehensive evaluation of the role of KRAS testing on liquid biopsy, the main limitation of this study, relies on its retrospective design. In addition, it was not possible to correlate the mutational status assessed on plasma with information relative to tissue, since matched cytohistological samples were not available. However, the aim of this study was to provide a snapshot of KRAS mutational evaluation on liquid biopsy in advanced stage NSCLC patients in routine setting. In fact, data relative to the previous validation study performed on matched tissue and blood samples strongly suggested that SiRe® NGS panel can be a reliable tool in clinical practice even when tissue samples are not available.
In conclusion, our data confirm that SiRe® NGS panel represents a robust analytical tool to assess KRAS mutational status on ctDNA. Further investigation is required to design more cost-effective diagnostic algorithms to harmonize clinical relevant biomarker testing on tissue and blood in advanced stage NSCLC clinical practice.
Supplementary
The article’s supplementary files as
Acknowledgments
This study was funded by the Department of Public Health of the University of Naples Federico II.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Declaration of Helsinki (as revised in 2013). According to the aforementioned National Guidelines, the study did not require an Ethical Committee approval since it did not modify the patient clinical management.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Alfredo Tartarone) for the Series “Improving Outcomes in Lung Cancer Through Early Diagnosis and Smoking Cessation” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.01.19). The series “Improving Outcomes in Lung Cancer Through Early Diagnosis and Smoking Cessation” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29. 10.3322/caac.20138 [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 5.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 8.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 9.Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. 10.1056/NEJMoa1704795 [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. 10.1056/NEJMoa1406766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planchard D, Besse B, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. 10.1016/S1470-2045(16)30146-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307-16. 10.1016/S1470-2045(17)30679-4 [DOI] [PubMed] [Google Scholar]
- 13.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 14.Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 15.Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. 10.5858/arpa.2017-0388-CP [DOI] [PubMed] [Google Scholar]
- 16.Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 2018;36:911-9. 10.1200/JCO.2017.76.7293 [DOI] [PubMed] [Google Scholar]
- 17.Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-SmallCell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. 10.6004/jnccn.2018.0062 [DOI] [PubMed] [Google Scholar]
- 18.Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. 10.1016/S1470-2045(16)30562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. 10.1056/NEJMoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. 10.1093/annonc/mdx523 [DOI] [PubMed] [Google Scholar]
- 22.El Osta B, Behera M, Kim S, et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2019;14:876-89. 10.1016/j.jtho.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AMG 510 First to Inhibit "Undruggable" KRAS. Cancer Discov 2019;9:988-9. [DOI] [PubMed] [Google Scholar]
- 24.Fakih M, O'Neil B, Price TJ, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol 2019;37;abstr 3003.
- 25.Hallin J, Engstrom LD, Hargis L, et al. The KRASG12C Inhibitor, MRTX849, Provides Insight Toward Therapeutic Susceptibility of KRAS Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54-71. 10.1158/2159-8290.CD-19-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagahashi M, Shimada Y, Ichikawa H, et al. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci 2019;110:6-15. 10.1111/cas.13837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. 10.1038/bjc.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pepe F, De Luca C, Smeraglio R, et al. Performance analysis of SiRe next-generation sequencing panel in diagnostic setting: focus on NSCLC routine samples. J Clin Pathol 2019;72:38-45. 10.1136/jclinpath-2018-205386 [DOI] [PubMed] [Google Scholar]
- 29.Pisapia P, Pepe F, Smeraglio R, et al. Cell free DNA analysis by SiRe(®) next generation sequencing panel in non small cell lung cancer patients: focus on basal setting. J Thorac Dis 2017;9:S1383-90. 10.21037/jtd.2017.06.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisapia P, Rocco D, Pepe F, et al. EGFR exon 19 deletion switch and development of p.L792Q mutation as a new resistance mechanism to osimertinib: a case report and literature review. Transl Cancer Res 2019;8:S64-9. 10.21037/tcr.2018.09.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. 10.1016/j.jtho.2018.05.030 [DOI] [PubMed] [Google Scholar]
- 32.Pisapia P, Malapelle U, Troncone G. Liquid Biopsy and Lung Cancer. Acta Cytol 2019;63:489-96. 10.1159/000492710 [DOI] [PubMed] [Google Scholar]
- 33.Pan W, Yang Y, Zhu H, et al. KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes in NSCLC: A meta-analysis of 41 studies. Oncotarget 2016;7:8373-88. 10.18632/oncotarget.7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. 10.1002/cncy.21322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as