Abstract
Background
The phospholipase A2 Group 6 (PLA2G6, also known as PLA2, PARK14, and iPLA2) gene encodes a group VIA calcium-independent phospholipase A2. Genetic polymorphism of PLA2G6 has been indicated to be involved in conferring susceptibility for Parkinson’s disease (PD), whereas conclusive results have not been obtained. Thus, we intended to conduct a systematic review to determine if PLA2G6 genetic variation confers a greater susceptibility to PD.
Methods
All case-control studies that investigated the association of the PLA2G6 polymorphisms with the risk of PD published before 15 July 2018 were included. The literature was comprehensively searched and identified in five English databases (EBSCO, Pubmed, OVID, EMBASE and ISI Web of Knowledge) and four Chinese databases (Wanfang database, Chinese Biomedical Literature Database, China Academic Journals Database and VIP database). We performed analyses of study characteristics, heterogeneity, and forest plot in analyses analogous to dominant, codominant and additive models with the pooled odds ratio (OR) in fixed- or random-effects models as the measure of association.
Results
A total of 664 potentially relevant studies were retrieved with the initial search, of which eight studies fulfilled the inclusion criteria, and included 2,779 PD patients and 3,291 control participants,. Among all the reported 27 genetic variants, 15 single nucleotide polymorphisms (SNPs) were present only in patients, and only five available SNPs (rs2267369, rs140758033, c.1959T>A (Gly653Gly), rs76718524, rs199935023) were pooled in the meta-analysis. However, there was no evidence for a significant association between the five SNPs and PD risk in dominant, codominant and allele models, suggesting a lack of association between PLA2G6 genetic variation and PD susceptibility.
Conclusion
The present study assessed the association of PLA2G6 genetic polymorphism with the risk PD, and the result strongly demonstrates that PLA2G6 polymorphism is not associated with PD susceptibility.
Keywords: Parkinson’s disease, PD, PLA2G6, systematic review
Introduction
Parkinson’s disease (PD), one of the most disabling disorders of the central nervous system, frequently develops in elderly patients affecting 1% of the population above 60 years and up to 4% of the population above 85 years.1 In addition to mental symptoms, patients with PD display typical motor symptoms such as postural instability, muscle rigidity, static tremors and difficulty with walking and gait.2 PD is a complex multifactor disease characterized by the extensive presence of α-synuclein-containing Lewy bodies in the substantia nigra (SN), in the pars compacta of which loss of dopaminergic neurons leads to reduced facilitation of voluntary movements.3 Literature in the past decade has suggested that genetic factors might play an important role in the development of PD.4 Thus far, researches related to genetic mutation or variation such as candidate-gene analyses, whole-genome linkage scans as well as family-based studies have clarified 14 chromosomal loci (PARK1-PARK14) in which mutations are unequivocally linked to rare forms of PD.5
The gene of phospholipase A2 Group 6 (PLA2G6, also known as PARK14, iPLA2 and PLA2), consisting of 17 exons, locates on chromosome 22q12-13.6 The protein encoded by the PLA2G6 gene is an 85-kDa group VI calcium-independent phospholipase A2 beta (iPLA2 β, also known as PLA2G6), which selectively hydrolyses glycerophospholipids to release free fatty acids.7 The PLA2G6 protein may play an indispensable role in transmembrane ion flux, fas-mediated apoptosis and leukotriene and prostaglandin synthesis.
PLA2G6 genetic mutation has previously been considered to cause autosomal recessive inheritance of infantile neuroaxonal dystrophy (INAD) in 2006,8 and was also characterized as a causative gene of neurodegeneration with brain iron accumulation (NBIA).6 Lately, the PLA2G6 gene mutation has been identified as a locus for parkinsonism.9–11 The neuropathological finding revealed widespread α-synuclein-containing Lewy body pathology and loss of dopaminergic neurons in the SN, supporting an association between PLA2G6 gene mutation and PD.9 The finding reported by Kauther et al12 displayed that the nonsynonymous single nucleotide polymorphisms (SNPs) rs139093920 (Ala781Thr) and 2339A>G (Asn780Ser) was only present in patients of L-dopa-responsive sporadic early-onset PD (EOPD), while not in any of the control participants yet,12 implying a possible association between PLA2G6 and the EOPD pathogenesis. More recently, Tan et al13 in Singapore distinguish a Pro806Arg substitution (rs140758033, c.2417C>G) in exon 17 in one of 96 PD EOPD patients, while this mutation was not found in any other 100 healthy controls.13 This specific finding emphasized the potential role of PLA2G6 gene mutation acting as a possible pathogenic risk factor for PD, and this is confirmed by other studies from Taiwan,14 Germany,12 India,15 Japan,11,16 Iran,10 and China,17–20 implying that PLA2G6 mutations may be widely distributed in parkinsonian patients of various populations. In addition, Gui et al19 identified four rare PLA2G6 mutations in 4 of 250 PD patients and in none of 550 controls, and they further revealed in subsequent experiments that the catalytic activity of phospholipids-hydrolyzing function was impaired by the four mutations they reported (c.1791delC (P.His597fx69), c.1959T>A (Gly653Gly), c.2077C>G (Leu693Val), c.1966C>G (Leu656Val)), supporting a possible role of PLA2G6 gene mutations played in the pathogenesis of PD-causing in Chinese Han populations.19
However, controversial results have been obtained and whether PLA2G6 gene mutation is a risk factor for PD remains conflicting. The reported mutations such as rs140758033 which was reported to be associated with PD susceptibility13 have also been found in control participants (10/310) as well as in PD patients (12/379),16 leading to the speculation that PLA2G6 gene possibly does not play an important role in PD.16,18 Whether PLA2G6 genetic mutation could contribute to PD, and whether single heterozygous mutations in PLA2G6 gene is also a risk factor for PD remains intriguing.13 Thus, this systematic review was conducted to address the possible association between PLA2G6 genetic variation and PD susceptibility.
Methods
Literature Search and Collection
The systematic review and meta-analysis was conducted according to PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) statement21 including literature search strategy, study selection criteria, raw data extraction, and data analysis. The literature were comprehensively searched and identified in five English databases (OVID, Pubmed, EBSCO, EMBASE, and Web of Knowledge,) and four Chinese databases (Wanfang database, Chinese Biomedical Literature Database, China Academic Journals Database, and VIP database). All case-control studies that investigated the association between PLA2G6 polymorphisms and the risk of PD published before July 15, 2018 were included. The search term we used in this study was (“Parkinsonism” or “Parkinson’s disease” or PD or “Parkinsons disease”) and (“phospholipase A2 group VI” or GVI or PLA2 or INAD1 or NBIA2 or iPLA2 or NBIA2A or NBIA2B or PARK14 or PNPLA9 or CaI-PLA2 or IPLA2-VIA or iPLA2beta or PLA2G6) and (mutation or variation or variant or polymorphisms or polymorphism). An available literature search, reference lists as well as supplemental materials were performed independently, or subsequently identified manually to recognize potentially relevant studies by two investigators. In addition, the website PDGene (http://www.pdgene.org) was also consulted in this study.
Inclusion and Exclusion Criteria
All the available studies had to meet the following inclusion criteria: 1) a case-control design that was conducted to evaluate the association between PLA2G6 gene mutation and PD risk; 2) sufficient original data such as genotype frequency or number could be obtained directly in the published paper, be calculated from the text, or provided by the authors after request to calculate the odds ratio (OR) with its 95% confidence interval (CI) and P-value; 3) raw data not being the same as other studies; 4) be published in an English or Chinese peer-reviewed journal. Studies were excluded if they met the following exclusion criteria: 1) animal studies; 2) without case-control design; 3) contained overlapping data with another study; 4) cases only or without controls; 5) editorials, comments, reviews and abstracts; and 6) sufficient data could not be obtained in the published version nor provided, even after requesting from authors.
The included studies were determined by two reviewers working independently to identify by title, key words, abstract, and full-text. Relevant to recommendations for the genetic association reports, the scientific quality of the included studies was evaluated according to the HuGENET guidelines22 and study quality was assessed using the Reporting of Observational Studies in Epidemiology (STROBE) tool.23 The STROBE statement, a 22-item tool specifically designed to evaluate observational studies quality, does not provide ways to clearly define a score allowing one to rate the quality of the study. As a general rule, the higher the score, the higher the quality of the study. According to the reports of Masini et al24 and Biagi et al,25 the cut-offs for three levels of score was adopted: 0–14 as poor quality, 15–25 as intermediate quality and 26–33 as good quality of the study in this report.
Data Extraction
For all the included studies we pooled to in the systematic review, the following information was collected from each of all eligible studies, including the first author, year of publication, ethnicity, number of PD cases and controls, number of the EOPD and LOPD patients, gender and sex ratio, age at onset and examination, and disease duration, gene scanning area, genotyping methods, genotype counts among cases and controls, and Hardy-Weinberg equilibrium (HWE) in controls (P-value). If overlapped samples were used in more than one paper, the one that contained the largest sample size and the most detailed were kept. If key data was missing from the published paper or only expressed graphically, we tried to contact the first author or the corresponding author to inquire for further information or to calculate it by ourselves if feasible. Two authors worked independently to evaluate all the trials and evaluate all the data. If the two participants could not reach a consensus, another specialized investigator was consulted to resolve the disagreement.
Statistical Analysis
An estimate of the combined effect sizes utilizing OR with a random effects model or a fixed effects model was used to calculate the frequency of genotype and alleles of PLA2G6 and estimate the risks of the genotypes on PD. Codominant model of heterozygote comparison (Aa vs AA), dominant model (aa+Aa vs AA), and additive model (allele a vs allele A) was adopted respectively to estimate the risks of the genotypes on PD, in which “A” and “a” represented the major and the minor allele, respectively. Heterogeneity across individual studies was assessed with the value of I2 or a chi-square-based Q-test.26 A random-effects model was used when P-value of the Q-test <0.1 or I2 > 50%, otherwise, a fixed-effects model was applied.27 Sensitivity analysis was performed by deleting one individual study in turn from the total and reanalyzing the remainder to evaluate the stability of the results. Egger’s regression test28 was applied to investigate the potential publication bias. The Review Manger 5 Version 5.0 and Comprehensive Meta-Analysis Version 2.2 were used to perform all the statistical analyses. Pooled OR with 95% CI was adopted to measure the strength of the association between PLA2G6 genetic variation and PD risk. Probability of P-value less than 0.05 was considered statistically significant.
Results
Characteristics of the Study
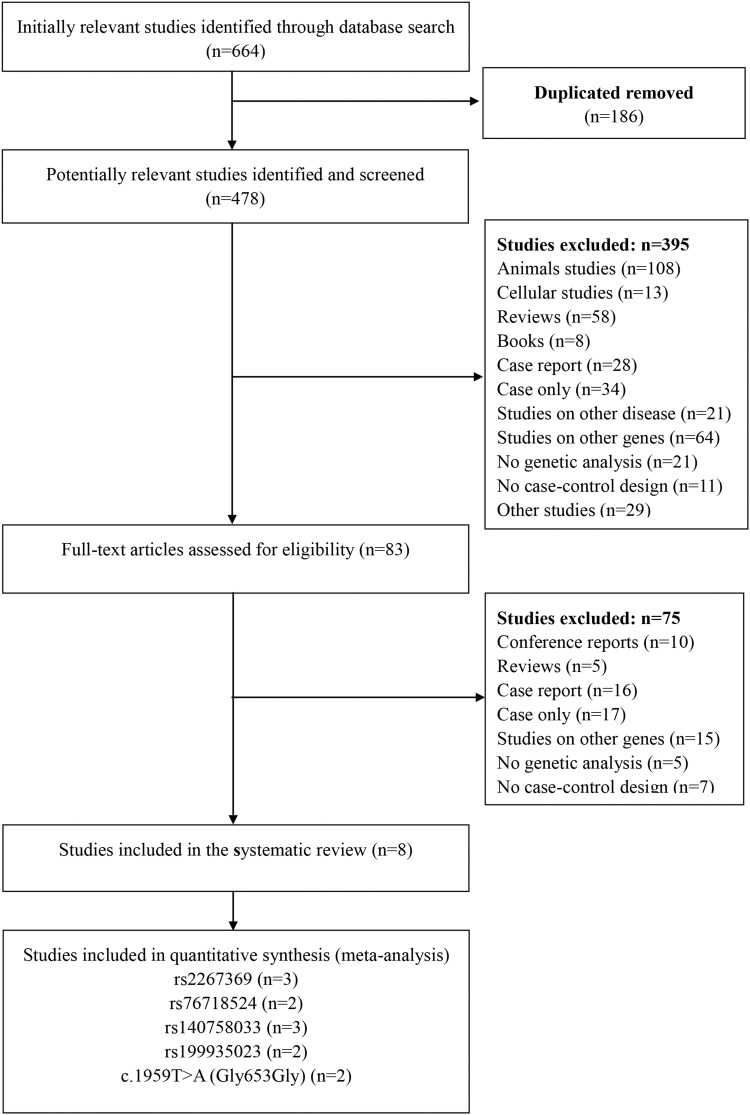
The flow chart of literature collection and selection is shown in Figure 1. A total of 664 potentially relevant articles were identified from nine databases according to the search strategy. After removing 186 duplicated articles, 478 potentially relevant studies were further identified after going through the titles and abstracts. After reading through the full text articles, 83 studies were preliminarily selected for further assessment for eligibility, and 5 reviews, 16 case reports and 10 academic proceedings were excluded. Also, another 44 studies were abandoned because they were without case-control design (n=7), not related to PLA2G6 gene (n=15), without proper control subjects (n=17) and insufficient published genotype data (n=5). Thus, eight eligible studies12–14,16–20 fulfilling the inclusion criteria included 2,779 PD patients and 3,291 controls.
Figure 1.
Flow diagram of the study selection process.
In Table 1, we provide the main characteristics, and summarize the quality of all the included studies in Supplementary Table 1. For detailed literature evaluation and scoring methods, please refer to the Supplementary Table 2 excerpted from relevant literature.23 In this report, we included five intermediate quality studies14,17-20 and three poor quality studies.12,13,16 The year of publication was distributed from 2010 to 2013. In terms of the sample sizes of all the studies, it ranged from 72 to 981 cases and 100 to 802 controls. The participants in those included studies were from different countries or areas, specifically four studies from China,17–20 one from Taiwan,14 and the other three from Japan,16 Germany12 and Singapore,13 respectively. PD patients including sPD patients as well as fPD patients were mainly diagnosed based on the United Kingdom Parkinson’s Disease Brain Bank criteria. Among all those included studies, one study17 included fPD patients, five studies13,16,18-20 included only sPD patients, and the other two12,14 included both sPD patients and fPD patients. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP),20 PCR and DNA direct sequencing,12,13,16-19 and DNA sequencing combined with TaqMan together with multiple-ligation probe amplification (MLPA) analysis14 were the main genotyping methods adopted in these included studies. The scanning areas were determined mainly by SNP specific,20 the exons16,19 or the exons and exon-intron boundaries.12–14,17,18 The age distribution showed no significant difference between PD patients and controls in all the included eight studies (Table 1). Finally, all the control subjects showed no deviation from HWE with all P-values > 0.05 (Supplementary Table 3).
Table 1.
Characteristics of the Eligible Studies Included in the Systematic Review
| Study | Genotyping Method | Scanning Area | Ethnicity | Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Male (%) | fPD (%) | Age at Onset (Range) | Age at Examination (Range) | Disease Duration | n | Male (%) | Age at Examination (Range) | ||||
| Gui, Y. X. et al 201319 | PCR and DNA sequencing | All exons | Chinese | 250 | 145 (58.0) | NA | 55.4±10.2 | 60.6±10.5 | 60.6±10.5 | 550 | 324 (58.9) | 61.4±11.3 |
| Zhou, Y. et al 201220 | PCR–RFLP | SNP specific | Chinese | 202 | 106 (52.5) | NA | 57.3±11.1 (27–83) | 62.7±10.7 (24–84) | 3.9±3.3 | 212 | 125(59.0) | 62.7±10.4 (34–86) |
| Tian, J. Y. et al 201217 | PCR and DNA sequencing | All the exons and exon-intron boundaries | Chinese | 72 | 42 (58.3) | 72(100) | 32.7±6.4 (13–40) | NA | 4.2±3.8 (1–16) | 500 | NA | NA |
| Lv, Z.Y. et al 201218 | PCR and DNA sequencing | SNP specific | Chinese | 531 | 302 (56.9) | 0(0) | 55.0±11.7 (19–81) | NA | NA | 561 | 280 (50.0) | 53.3±16.4 |
| Lu, C. S. et al 201214 | DNA sequencing, TaqMan and MLPA analysis | All the exons and exon-intron boundaries | Taiwanese | 981 | 582 (59.3) | 25 (0.25) | 56.8±11.6 (31–91) | NA | NA | 802 | 569 (70.9) | 59.0±16.2 (22–91) |
| Tomiyama, H. et al 201116 | PCR and DNA sequencing | A the exons | Japanese | 379 | 181 (47.8) | 0(0) | 52.7±14.3 (7–88) | 60.2±14.0 (12–92) | 7.4±5.6 (0–40) | 310 | 122 (39.3) | 58.5±13.2 (23–98) |
| Kauther, K. M. et al 201112 | PCR and DNA sequencing | All the exons and exon-intron boundaries | German | 268 | 110 (41.0) | 41(15.3) | 42.4 (18–50) | NA | NA | 256 | 119 (46.4) | 47.6 (19–83) |
| Tan, E. K. et al 201013 | PCR and DNA sequencing | All the exons and exon-intron boundaries | Singaporean | 96 | NA | NA | NA | NA | NA | 100 | NA | NA |
Abbreviations: NA, not available; fPD, familial Parkinson’s disease; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; MLPA, multiple-ligation probe amplification; SNP, single nucleotide polymorphism.
The Summary of PLA2G6 Gene SNPs and Quantitative Synthesis
The study comprehensively summarized the genotype and allele frequencies of a total of 27 SNPs in PLA2G6 gene that have been reported by case-control studies meeting the inclusion criteria to date, among which, seven SNPs (rs4375, rs132985, rs2284063,18 rs11570597 (−130C>T), rs11570680 (1027G>A, Ala343Thr), −20C>A and rs138683183 (2340C>T, Asn780Asn)12) exist in PD patients as well as in control participants, the other 15 SNPs were only found in PD patients (Supplementary Table 3), and only five SNPs (rs2267369, rs76718524, rs140758033, c.1959T>A (Gly653Gly) and rs199935023) reported by two or more studies were available for meta-analysis.
Related to the five SNPs available for quantitative synthesis, three papers with 660 PD patients and 758 control participants concerning rs2267369,12–14,16,18 three studies with 675 patients of PD and 610 control participants concerning rs140758033,13,14,16 two studies with 470 patients of PD and 468 control participants concerning rs76718524,12,20 two studies with 346 patients and 650 controls concerning c.1959T>A,13,19 and two studies with 1361 PD patients and 1112 control participants concerning rs199935023,14,16 respectively, were pooled in the meta-analysis (Table 2 and Supplementary Table 3).
Table 2.
Summary of the Meta-Analysis Results Related to the Pooled PLA2G6 Variants and Parkinson’s Disease
| Variant | No. of Studies | Total Participants (Cases/Controls) | Genetic Model | Pooled OR (95% CI) | P-value | I2 |
|---|---|---|---|---|---|---|
| rs2267369 | 3 | 1418 (660/758) | A | 1.21 (0.93–1.56) | 0.15 | 0% |
| D | 1.28 (0.97–1.69) | 0.08 | 0% | |||
| C | 1.32 (0.99–1.75) | 0.06 | 0% | |||
| rs76718524 | 2 | 938 (470/468) | A | 0.96 (0.06–15.31) | 0.97 | NA |
| D | 0.96 (0.06–15.31) | 0.97 | NA | |||
| C | 0.96 (0.06–15.31) | 0.97 | NA | |||
| rs140758033 | 3 | 1285 (675/610) | A | 0.99 (0.46–2.12) | 0.97 | 0% |
| D | 0.99 (0.46–2.13) | 0.97 | 0% | |||
| C | 0.99 (0.46–2.13) | 0.97 | 0% | |||
| rs199935023 | 2 | 2473 (1361/1112) | A | 1.73 (0.56–5.31) | 0.34 | NA |
| D | 1.55 (0.49–4.86) | 0.46 | NA | |||
| C | 1.36 (0.42–4.40) | 0.60 | NA | |||
| c.1959T>A (Gly653Gly) | 2 | 996 (346/650) | A | 4.50 0.46–43.57) | 0.19 | NA |
| D | 4.51 (0.46–43.85) | 0.19 | NA | |||
| C | 4.51 (0.46–43.85) | 0.19 | NA |
Note: I2 represents the variation in OR attributable to heterogeneity.
Abbreviations: CI, confidence interval; OR, odds ratio; A, allele model; D, dominant model; C, codominant model of heterozygote comparison; NA: not available.
The genotype frequency is presented in Supplementary Table 3. The meta-analyses between rs2267369, rs140758033, rs76718524, c.1959T>A, rs199935023 and PD have been conducted, and the main results of meta-analysis are shown in Table 2. Generally, a fixed-effects model was used under the allele model, dominant model and codominant model of heterozygote comparison due to the absence of heterogeneity among the individual studies included in this meta-analysis. The pooled meta-analysis results showed that no significant associations existed between all the five single SNPs and PD susceptibility under all the three genetic models (Table 2 and Supplementary Figure 1).
Sensitivity Analysis and Potential Bias
Sensitive analyses on rs2267369 and rs140758033, the only two SNPs which were included in three or more studies available for meta-analysis, was conducted by sequentially excluding each case-control study, and the results of all the three genetic models were consistent, suggesting the credibility and stability of this meta-analysis of the overall population and subgroup. Besides, publication bias was evaluated by the Egger’s test, and no significant results were observed (data not shown).
Discussions
Main Findings
The existing study lacks pooled assessments on the association between PLA2G6 genic variation and PD susceptibility, even though numerous studies on the impact of PLA2G6 gene mutation on PD risk have been reported.19,29-31 This specific study consequently has been designed to uncover whether this relationship is warranted. In this study, a comprehensive study search was conducted and eight articles with a total of 6,070 PD patients and control participants were included to investigate the relationship between PLA2G6 genetic variants and the risk of PD. The genotype distribution of a total of 27 PLA2G6 genetic variants were summarized, and five available variants (rs2267369, rs140758033, rs76718524, rs199935023, and c.1959T>A) were pooled in the meta-analysis. Overall, our quantitative analysis failed to reveal significant association between PLA2G6 genetic variation and susceptibility of PD depending on the available academic evidence so far.
According to the available literature, apart from INAD8 and NBIA,6 PLA2G6 genetic variation is indicated to be related with Type 2 diabetes risk (rs132984 and rs2284060)32 and it has also been reported to be associated with some other brain diseases such as bipolar disorder (rs3788533)33 and Alzheimer’s disease.34 PLA2G6 gene variation was considered to be relevant with the underlying risk for PD,19 which disrupted cell membrane structure, promoted apoptosis,6,18 destroyed in lipid and glucose metabolism35 and thus probably led to PD. Besides, patients with PLA2G6-related dystonia-parkinsonism typically show prominent parkinsonism with other signs such as dystonia, severe cognitive decline, pyramidal signs, and psychiatric features.9 Thus, to some extent, PLA2G6 has been termed as one of the risk factors for both fPD patients,36 which consist of about 10% of PD patients, as well as idiopathic PD.14,31
On the contrary, our meta-analysis results obtained from this study did not reach any clear consensus, and also did not reveal any significant association between all the five available SNPs of PLA2G6 and PD risk (Table 2, Supplementary Figure 1), even though 15 SNPs were present only in PD patients (Supplementary Table 2). Besides, the literature on the relationship between all the total 27 SNPs reported by case-control studies up to now and PD is replete with small studies that report controversial findings. Furthermore, it is worth noting that seven SNPs were present both in PD patients and controls (Supplementary Table 2), suggesting them to be physiological genetic variants rather than causal mutations. To sum up, our present study failed to reveal significant association between PLA2G6 polymorphism and susceptibility of PD. Through this present study, to elucidate the etiology of PD, instead of this specific kind of research related to a single genetic mutation which may be not prospective in terms of elucidating the pathological mechanism of PD directly, more studies with larger sample size are warranted to be conducted in future.
Limitations
Some limitations to this study should be discussed. Firstly, the genetic background of ethnic groups was not categorized due to the limited number of the included feasible studies, and a gene-environment or gene-gene interaction which may potentially exert an effect on the statistical results was not investigated. Secondly, the published literature was not sufficiently large for a meta-analysis on most of the reported variants, especially for the five SNPs pooled to the meta-analysis. Finally, the limitations of the observational nature of the original data and the lack of uniformity among studies could not be overlooked, for example, the negative results were less often published compared with positive results, and some studies lacked some of the detailed clinical presentation information such as treatment medication, duration of disease, co-morbid conditions, which may cause serious confounding bias.
Conclusions
The present systematic review and meta-analysis assessed the association of PLA2G6 genetic polymorphism with the risk PD. We found 15 variants were present only in PD patients, but failed to detect a significant association between the available five SNPs of PLA2G6 and the risk of PD. The results strongly demonstrate that PLA2G6 polymorphism is not associated with susceptibility to PD.
Acknowledgment
We gratefully acknowledge Prof. Nobutaka Hattori for replying to our request for original data.
Funding Statement
This study was supported by a Grant from the Natural Science Foundation of China (81801338, 81771465), the Western medicine guidance project of Shanghai Science and Technology Commission (17411970200), Youth project of Shanghai Municipal Health and Planning Commission (20154Y0045, 20154Y0194), Shanghai Mental Health Center Medical Youth Talents “Flying Plan” (2018-FX-01, 2018-FX-03), Shanghai Key Project of Science and Technology (2018SHZDZX05), the Key Project of Clinical Research Center of Shanghai Mental Health Center (CRC2018ZD02) and Shanghai Key Laboratory of Psychotic Disorders (13dz2260500).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Keener AM, Paul KC, Folle A, Bronstein JM, Ritz B. Cognitive impairment and mortality in a population-based Parkinson’s disease cohort. J Parkinsons Dis. 2018;8(2):353–362. doi: 10.3233/JPD-171257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opara J, Malecki A, Malecka E, Socha T. Motor assessment in Parkinson`s disease. Ann Agric Environ Med. 2017;24(3):411–415. doi: 10.5604/12321966.1232774 [DOI] [PubMed] [Google Scholar]
- 3.Tysnes OB, Storstein A. Epidemiology of Parkinson’s disease. J Neural Transm. 2017;124(8):901–905. doi: 10.1007/s00702-017-1686-y [DOI] [PubMed] [Google Scholar]
- 4.Lotharius J, Brundin P. Pathogenesis of Parkinson’s disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3(12):932–942. doi: 10.1038/nrn983 [DOI] [PubMed] [Google Scholar]
- 5.Bras J, Simon-Sanchez J, Federoff M, et al. Lack of replication of association between GIGYF2 variants and Parkinson disease. Hum Mol Genet. 2009;18(2):341–346. doi: 10.1093/hmg/ddn340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan NV, Westaway SK, Morton JE, et al. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat Genet. 2006;38(7):752–754. doi: 10.1038/ng1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma ZM, Turk J. The molecular biology of the group VIA Ca2+-independent phospholipase A(2). Prog Nucleic Acid Res Mol Biol. 2001;67:1–33. [DOI] [PubMed] [Google Scholar]
- 8.Khateeb S, Flusser H, Ofir R, et al. PLA2G6 mutation underlies infantile neuroaxonal dystrophy. Am J Hum Genet. 2006;79(5):942–948. doi: 10.1086/508572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paisan-Ruiz C, Bhatia KP, Li A, et al. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65(1):19–23. doi: 10.1002/ana.21415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sina F, Shojaee S, Elahi E, Paisan-Ruiz CR. 632W mutation in PLA2G6 segregates with dystonia-parkinsonism in a consanguineous Iranian family. Eur J Neurol. 2009;16(1):101–104. doi: 10.1111/j.1468-1331.2008.02356.x [DOI] [PubMed] [Google Scholar]
- 11.Yoshino H, Tomiyama H, Tachibana N, et al. Phenotypic spectrum of patients with PLA2G6 mutation and PARK14-linked parkinsonism. Neurology. 2010;75(15):1356–1361. doi: 10.1212/WNL.0b013e3181f73649 [DOI] [PubMed] [Google Scholar]
- 12.Kauther KM, Hoft C, Rissling I, Oertel WH, Moller JC. The PLA2G6 gene in early-onset Parkinson’s disease. Mov Disord. 2011;26(13):2415–2417. doi: 10.1002/mds.23851 [DOI] [PubMed] [Google Scholar]
- 13.Tan EK, Ho P, Tan L, Prakash KM, Zhao Y. PLA2G6 mutations and Parkinson’s disease. Ann Neurol. 2010;67(1):148. doi: 10.1002/ana.21663 [DOI] [PubMed] [Google Scholar]
- 14.Lu C-S, Lai S-C, Wu R-M, et al. PLA2G6 mutations in PARK14-linked young-onset parkinsonism and sporadic Parkinson’s disease. Am J Med Genet Part B Neuropsychiatr Genet. 2012;159B(2):183–191. doi: 10.1002/ajmg.b.32012 [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal A, Schneider SA, Houlden H, et al. Indian-subcontinent NBIA: unusual phenotypes, novel PANK2 mutations, and undetermined genetic forms. Mov Disord. 2010;25(10):1424–1431. doi: 10.1002/mds.23095 [DOI] [PubMed] [Google Scholar]
- 16.Tomiyama H, Yoshino H, Ogaki K, et al. PLA2G6 variant in Parkinson’s disease. J Hum Genet. 2011;56(5):401–403. doi: 10.1038/jhg.2011.22 [DOI] [PubMed] [Google Scholar]
- 17.Tian JY, Tang BS, Shi CH, et al. Analysis of PLA2G6 gene mutation in sporadic early-onset parkinsonism patients from Chinese population. Neurosci Lett. 2012;514(2):156–158. doi: 10.1016/j.neulet.2012.02.078 [DOI] [PubMed] [Google Scholar]
- 18.Lv Z, Guo J, Sun Q, et al. Association between PLA2G6 gene polymorphisms and Parkinson’s disease in the Chinese Han population. Parkinsonism Relat Disord. 2012;18(5):641–644. doi: 10.1016/j.parkreldis.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 19.Gui YX, Xu ZP, Wen L, Liu HM, Zhao JJ, Hu XY. Four novel rare mutations of PLA2G6 in Chinese population with Parkinson’s disease. Parkinsonism Relat Disord. 2013;19(1):21–26. doi: 10.1016/j.parkreldis.2012.07.016 [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Li F, Tian X, et al. Association of Parkinson’s disease with six snps located on four parks genes in northern han Chinese population. Parkinsonism Relat Disord. 2012;18:S173–S174. doi: 10.1016/S1353-8020(11)70755-6 [DOI] [Google Scholar]
- 21.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little J, Higgins JP, Ioannidis JP, et al. STrengthening the REporting of Genetic Association studies (STREGA): an extension of the STROBE Statement. Ann Intern Med. 2009;150(3):206–215. doi: 10.7326/0003-4819-150-3-200902030-00011 [DOI] [PubMed] [Google Scholar]
- 23.von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The strengthening the reporting of observational studies in epidemiology (STROBE) Statement: guidelines for reporting observational studies. Int J Surg. 2014;12(12):1495–1499. doi: 10.1016/j.ijsu.2014.07.013 [DOI] [PubMed] [Google Scholar]
- 24.Masini A, Marini S, Gori D, Leoni E, Rochira A, Dallolio L. Evaluation of school-based interventions of active breaks in primary schools: A systematic review and meta-analysis. J Sci Med Sport. 2019;23:377–384. [DOI] [PubMed] [Google Scholar]
- 25.Biagi C, Nunzio MD, Bordoni A, Gori D, Lanari M. Effect of adherence to mediterranean diet during pregnancy on children’s health: a systematic review. Nutrients. 2019;11(5):5. doi: 10.3390/nu11050997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau J, Ioannidis JPA, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008 [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie F, Cen Z, Ouyang Z, Wu S, Xiao J, Luo W. Homozygous p.D331Y mutation in PLA2G6 in two patients with pure autosomal-recessive early-onset parkinsonism: further evidence of a fourth phenotype of PLA2G6-associated neurodegeneration. Parkinsonism Relat Disord. 2015;21(4):420–422. doi: 10.1016/j.parkreldis.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 30.Lai SC, Yeh TH, Chiu CC, et al. The pathogenic mechanism of PLA2G6 mutations in Parkinson’s disease. Mov Disord. 2014;29:S19–S20. [Google Scholar]
- 31.Malaguti MC, Melzi V, Di Giacopo R, et al. A novel homozygous PLA2G6 mutation causes dystonia-parkinsonism. Parkinsonism Relat Disord. 2015;21(3):337–339. doi: 10.1016/j.parkreldis.2015.01.001 [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Hu C, Jiang F, et al. Genetic variants of PLA2G6 are associated with Type 2 diabetes mellitus and triglyceride levels in a Chinese population. Diabet Med. 2015;32(2):280–286. doi: 10.1111/dme.12587 [DOI] [PubMed] [Google Scholar]
- 33.Xu C, Warsh JJ, Wang KS, Mao CX, Kennedy JL. Association of the iPLA2 beta gene with bipolar disorder and assessment of its interaction with TRPM2 gene polymorphisms. Psychiatr Genet. 2013;23(2):86–89. doi: 10.1097/YPG.0b013e32835d700d [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer EL, Gattaz WF. Cholinergic and glutamatergic alterations beginning at the early stages of Alzheimer disease: participation of the phospholipase A(2) enzyme. Psychopharmacology. 2008;198(1):1–27. [DOI] [PubMed] [Google Scholar]
- 35.Bao S, Jacobson DA, Wohltmann M, et al. Glucose homeostasis, insulin secretion, and islet phospholipids in mice that overexpress iPLA(2)beta in pancreatic beta-cells and in iPLA(2)beta-null mice. Am J Phys Endocrinol Metab. 2008;294(2):E217–E229. doi: 10.1152/ajpendo.00474.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi C, Tang B, Wang L, et al. PLA2G6 gene mutation in autosomal recessive early-onset parkinsonism in a Chinese cohort. Neurology. 2011;77(1):75–81. doi: 10.1212/WNL.0b013e318221acd3 [DOI] [PubMed] [Google Scholar]