Abstract
Cobalamin C (cblC) deficiency, the most common inborn error of intracellular cobalamin metabolism, is caused by mutations in MMACHC, a gene responsible for the processing and intracellular trafficking of vitamin B12. This recessive disorder is characterized by a failure to metabolize cobalamin into adenosyl- and methylcobalamin, which results in the biochemical perturbations of methylmalonic acidemia, hyperhomocysteinemia and hypomethioninemia caused by the impaired activity of the downstream enzymes, methylmalonyl-CoA mutase and methionine synthase. Cobalamin C deficiency can be accompanied by a wide spectrum of clinical manifestations, including progressive blindness, and, in mice, manifests with very early embryonic lethality. Because zebrafish harbor a full complement of cobalamin metabolic enzymes, we used genome editing to study the loss of mmachc function and to develop the first viable animal model of cblC deficiency. mmachc mutants survived the embryonic period but perished in early juvenile life. The mutants displayed the metabolic and clinical features of cblC deficiency including methylmalonic acidemia, severe growth retardation and lethality. Morphologic and metabolic parameters improved when the mutants were raised in water supplemented with small molecules used to treat patients, including hydroxocobalamin, methylcobalamin, methionine and betaine. Furthermore, mmachc mutants bred to express rod and/or cone fluorescent reporters, manifested a retinopathy and thin optic nerves (ON). Expression analysis using whole eye mRNA revealed the dysregulation of genes involved in phototransduction and cholesterol metabolism. Zebrafish with mmachc deficiency recapitulate the several of the phenotypic and biochemical features of the human disorder, including ocular pathology, and show a response to established treatments.
Introduction
Cobalamin (vitamin B12) is an essential micronutrient for vertebrates, and a complex set of pre- and post-absorptive pathways has evolved to capture the vitamin from the environment and transport into the cell, where it is metabolized into either 5′-deoxyadenosylcobalamin (AdoCbl) or methylcobalamin (MeCbl), the active cofactors for the methylmalonyl-CoA mutase (MMUT, EC 5.4.99.2) or methionine synthase (MTR, EC 1.16.1.8) reactions (1). Genetic and cellular studies of patients with disorders of cobalamin metabolism have afforded the identification of many proteins, both enzymes and regulators, that are involved in the transport, trafficking, processing and synthesis of intracellular AdoCbl and MeCbl (2,3). Combined methylmalonic acidemia and homocystinuria, cblC-type (cblC) (OMIM #277400), the most common disorder of intracellular cobalamin metabolism (4), is caused by pathogenic variants in MMACHC, an enzyme involved in the decyanation, dealkyation and intracellular trafficking of vitamin B12 (5–7).
The phenotypic manifestations of cblC deficiency are remarkable in terms of clinical and pathological heterogeneity (4,8). Patients with severe forms of cblC typically manifest non-head sparing intrauterine growth restriction and subsequent failure to thrive during early life (9). Congenital heart disease, including non-compaction cardiomyopathy, anemia and seizures are variably present (10–12). Some patients can even experience metabolic acidosis and hyperammonemia (13). If the disorder is not promptly recognized and treated with high doses of injectable hydroxocobalamin (OHCbl), it can be fatal (9,14). While medical management with OHCbl, betaine, folinic acid and optimal protein intake can improve clinical symptoms and laboratory parameters (15–17), many patients still develop an ocular syndrome characterized by a ‘bulls-eye’ maculopathy with a progressive retinal degeneration and blindness (18,19). In the USA and many other countries, cblC deficiency is detected through expanded newborn screening (12,20,21), which adds urgency to efforts aimed at understanding disease mechanisms and the development of optimal treatment regimens.
Despite the recognition of cblC deficiency as a clinical entity for over four decades, there are limited pathological data (22–25), and descriptive cellular studies have proposed that impaired methylation, homocysteine toxicity, glutathione deficiency, oxidative stress, endoplasmic reticulum stress and mislocalization of RNA binding proteins may contribute to the disease manifestations (26–33). A previous effort to create mice with cblC deficiency using an Mmachc gene trap allele revealed that after day E3.5, the cblC embryos failed to develop (34). Similarly, mice that harbor the loss of function mutations in other genes involved in intracellular cobalamin metabolism, such as Mtrr (cblE), Mtr (cblG) and Lmbrd1 (cblF), have also displayed embryonic lethality, highlighting the critical role of vitamin B12 metabolism in embryonic development (35–37).
Given that fish are known to require vitamin B12 for growth (38), we identified the zebrafish orthologue of MMACHC and then used zinc finger nucleases (ZFNs) to create germline mutant mmachc alleles that were bred to model cblC deficiency. Indeed, mmachc homozygous fish display phenotypic and biochemical features similar to those seen in cblC patients such as growth impairment, developmental delay and organic acid accretion; respond to established small molecule therapies; and manifest a retinopathy that we further characterize using transgenesis and transcriptomics. mmachc zebrafish present the first tractable model of cblC deficiency and demonstrate a critical role of a vitamin B12 metabolic enzyme for survival and retinal morphology.
Results
Genes involved in cobalamin metabolism are present in zebrafish
We identified putative zebrafish orthologues of genes involved in human propionyl-CoA and cobalamin metabolism using an in silico approach (Table 1). Many were highly conserved, with each sharing between 32 and 84% amino acid identity with the corresponding human protein (Table 1). The zebrafish orthologue of MMACHC, mmachc, shares 51% sequence identity with the human protein but diverges at the C-terminus, resulting in a shorter protein (250 versus 282 amino acids in human).
Table 1.
Danio rerio orthologs of genes in the propionyl-CoA and cobalamin metabolism pathway
| Human gene | D. rerio ortholog | D. rerio ref gene # | D. rerio ref protein # | Protein sequence identity with human (%) | Other names; function | Reference |
|---|---|---|---|---|---|---|
| LRP2 | lrp2 | NM_001194987.1 | NP_001181916.1 | 66 | Megalin; Uptake of cobalamin into the kidney epithelium | (49) |
| CUBN | cubn | XM_002666678.4 | XP_002666724.2 | 53 | Cubulin; Uptake of cobalamin into the intestinal epithelium | (49) |
| AMN | amn | XM_002661272.3 | XP_002661318.2 | 38 | Amnionless; Uptake of cobalamin into the intestinal epithelium | (49) |
| GIF | tcnba | NM_001128735.3 | NP_001122207.1 | NA | Intrinsic factor; Binding of cobalamin | (47,49) |
| TCN1 | tcnbb | NM_001252649.1 | NP_001239578.1 | NA | Haptocorrin, R binders; Binding of cobalamin | (47) |
| TCN2 | tcn2 | NM_001123231.2 | NP_001116703.1 | 32 | Transcobalamin; Binding of cobalamin | (47,48) |
| CD320 | — | — | — | — | Transcobalamin receptor; Cellular uptake of cobalamin | |
| LMBRD1 | lmbrd1 | NM_001008612.1 | NP_001008612.1 | 74 | Lysosomal export of cobalamin | |
| ABCD4 | abcd4 | NM_001076717.2 | NP_001070185.2 | 68 | Lysosomal export of cobalamin | |
| HCFC1 | hcfc1a hcfc1b | NM_001045064.1 NM_001128537.1 | NP_001038529.1 NP_001122009.1 | 71 75 | Transcriptional co-regulator of MMACHC | (73) |
| THAP11 | thap11 | NM_213102.1 | NP_998267.1 | 76 | Transcriptional co-regulator of MMACHC | |
| ZNF143 | znf143a znf143b | NM_001007441.1 NM_200979.1 | NP_001007442.1 NP_957273.1 | 61 74 | Transcriptional co-regulator of MMACHC | |
| PRDX1 | prdx1 | NM_001013471.2 | NP_001013489.2 | 82 | Peroxiredoxin 1, antioxidant enzyme, mutation in humans causes epi-cblC | (74) |
| MMACHC | mmachc | NM_001114889.1 | NP_001108361.1 | 51 | Decyanation and dealkylation of cobalamin | |
| MMADHC | mmadhc | NM_205594.1 | NP_991157.1 | 55 | Binding partner of MMACHC | |
| MTRR | mtrr | XM_684065.4 | XP_689157.2 | 56 | Conversion of reduced cobalamin to methylcobalamin | |
| MTR | mtr | NM_198072.1 | NP_932338.1 | 78 | Formation of methionine from homocysteine | (75) |
| CBS | cbsa cbsb | NM_001111232.1 NM_001014345.2 | NP_001104702.1 NP_001014367.2 | 78 78 |
Formation of cystathionine from homocysteine and serine | (46) |
| PCCA | pcca | NM_001002746.2 | NP_001002746.1 | 77 | Conversion of priopionyl-CoA to D-methylmalonyl-CoA | |
| PCCB | pccb | NM_212925.1 | NP_998090.1 | 83 | Conversion of priopionyl-CoA to D-methylmalonyl-CoA | |
| MCEE | mcee | NM_001020766.1 | NP_001018602.1 | 76 | Isomerization of D-methylmalonyl-CoA to L-methylmalonyl-CoA | |
| MMUT | mut | NM_001099226.1 | NP_001092696.1 | 84 | Isomerization of L-methylmalonyl-CoA to succinyl-CoA | |
| MMAA | mmaa | NM_001105112.1 | NP_001098582.1 | 67 | Protection and reactivation MUT | |
| MMAB | mmab | NM_001082809.1 | NP_001076278.1 | 66 | ATR, Conversion reduced cobalamin to adenosylcobalamin |
Phenotype of mmachc morphants and creation of ZFN mutant lines
Because cblC deficiency can present with congenital malformations such as intrauterine growth retardation, microcephaly and cardiac defects, we first used translation-blocking or splice-blocking morpholino antisense oligonucleotides to knockdown mmachc and observed a very severe embryonic phenotype that included growth impairment, developmental delay (delayed hatching from the chorion and a persistent yolk sac), abnormalities of axis development as evidenced by a curved body, impaired swimming, severe pericardial effusion and brain edema (Supplementary Material, Figure S1). This complex developmental phenotype was more severe in the initiation site versus splice junction morphants, dose-dependent (Supplementary Material, Figure S1) and reproducible when studied on a p53 knock-out background (not presented).
Due to concerns regarding off-target effects in morpholino studies (39), ZFNs were used to target exon 2 for mutagenesis because this location is close to the homologous position of a severe and common human pathogenic variant, c.271dupA p.Arg91Lysfs*14 (5). Two mmachc alleles were recovered for phenotyping: hg12 (4 bp insertion, c.127_130dup p.Lys44Profs*21) and hg13 (a complex indel resulting in a net 10 bp deletion; c.95_132delins28 p.Gly32Valfs*48) (Fig. 1). Both alleles are frameshift mutations that are predicted to cause nonsense mediated decay of the corresponding mRNA. Quantitative PCR (qPCR) showed that mmachc RNA levels were significantly reduced in mmachchg12/hg12 fish, undetectable in mmachchg13/hg13 fish (P < 0.0001) (Fig. 1B and C) and reduced by approximately 50% in the mmachchg12 and mmachchg13 heterozygotes compared with wild-type fish (P < 0.001).
Figure 1.

mmachc alleles, gene expression and survival. (A) Exon 2 of mmachc was targeted by Zn finger mutagenesis; Two alleles were identified for further study: hg12 a 4 bp insertion c.127_130dup p.Lys44Profs*21 and hg13 a complex indel with a net 10 bp deletion c.95_132delins28 p.Gly32Valfs*48. (B and C) qPCR showed that mmachc RNA levels were reduced in mmachchg12/hg12 fish (7.4 ± 1.6% of WT) and undetectable in mmachchg13/hg13 mutants (P < 0.0001) at 7 dpf. The level of mmachc transcript in the heterozygotes was reduced by approximately 50% relative to wild-type fish (P < 0.0001) (+/hg12: 52.4 ± 5.5%; +/hg13:48.6 ± 2.9%). (D and E) mmachchg12/hg12 fish and mmachchg13/hg13 fish were present in expected ratios during the larval period 7, 14, 21 and 28 dpf (P > 0.4). By the time they reached the juvenile period at 35 dpf, >90% of mutants perished and no mutants were detected past 42 dpf (***P < 0.001) N = 104–256, each genotype shown.
Survival, growth and morphology
In stark contrast to the mmachc morphants, mmachchg12/hg12 and mmachchg13/hg13 fish survived the embryonic period (0–6 day post-fertilization; dpf). During the larval period (7–28 dpf), wild-type, heterozygotes and homozygous mutants were present at the expected Mendelian ratios (chi square P > 0.4; Supplementary Material, Tables S1 and S2). At 35 dpf, more than 90% of the homozygous mutants perished, resulting in a skewed genotype distribution; no homozygous mutants survived past 42 dpf (Fig. 1D and E, Supplementary Material, Figure S1 and Supplementary Material, Table S2; chi square P < 0.001).
To characterize the morphological phenotype of the cblC mutants, we observed the fish throughout the embryonic and larval periods and found that the mutant fish were approximately half the length of an unaffected clutchmate, at 28 dpf before they perished (Fig. 2A). Fish were photographed at weekly intervals (7, 14, 21 and 28 dpf), genotyped and measured to quantify growth (40). In the homozygous mutant fish, the standard length (SL) and height at the anterior of the anal fin (HAA) were indistinguishable from heterozygote and wild-type fish at 7 dpf (Supplementary Material, Figure S2). Beginning at 14 dpf, a modest but statistically significant reduction in SL and HAA of the mutants relative to controls was noted (P < 0.01) (Supplementary Material, Figure S2). The growth of the cblC mutants arrested between 14 and 28 dpf, and by 28 dpf, all mmachc homozygous mutants demonstrated severe growth impairment compared with unaffected clutchmates (P < 0.0001) (Fig. 2B and C). In addition to growth impairment, the mutant fish were delayed in developmental morphology in all parameters assessed (Supplementary Material, Figures S3 and S4), consistent with a failure to thrive phenotype (41).
Figure 2.

Growth and biochemical measurements. (A) Photograph of mmachc+/+ fish (top) and mmachchg13/hg13 fish (bottom) showing severe growth impairment at 28 dpf. Scale bar = 2 mm. (B) To quantify growth, SL and height at the anterior of the anal fin (HAA) were measured at 28 dpf in wild-type, heterozygous and mutant fish (N = 11–27). Mutant SL was 57–58% of controls and (C) HAA was 23–34% of controls (****P < 0.0001). There was no statistically significant difference in SL and HAA measurements between heterozygous and wild-type fish. (D and E) Selected metabolites were measured by GC-MS in hg12 fish tissue extract (N = 3 pools) and metabolite levels were normalized to protein content. (D) MMA was extremely elevated at all time points compared with controls and at 28 dpf, the [MMA] were 340-fold elevated. (E) Methylcitrate levels were also significantly elevated at all time points. Data shown are mean + SD (****P < 0.0001).
mmachc expression during development and in adult tissues
The striking discrepancy between the morphant and germline mutants prompted us to explore the expression of mmachc during development in wild-type fish. qPCR analyses of mmachc expression in adult tissues revealed highest expression in liver, kidney, muscle and spinal cord (Supplementary Material, Figure S5) with lower levels detected in the brain, eye, gill, heart and intestine. The wild-type mmachc transcript abundance decreased rapidly relative to β-actin in early embryogenesis (0–72 hpf), falling to 5.1% of the early level by 72 hpf (Supplementary Material, Figure S5) but then increased during the juvenile period reaching 60% of the initial level at 56 dpf.
In an attempt to exacerbate the embryonic phenotype, we raised cblC zebrafish embryos in water or water containing 1 mM propionate, a precursor of the metabolic pathway used to chemically stress the mutants. However, mutants and controls had normal morphology and survival at 5 dpf (Supplementary Material, Figure S5), suggesting maternal mmachc may have a protective effect.
Primary and secondary biochemical perturbations
We measured the characteristic metabolites of cblC in whole fish extracts using gas chromatography/mass spectrometry (GC-MS) (42). Although homocysteine was too low to measure accurately in mmachc fish extracts, methylmalonic acid (MMA) was easily detected and quantitated. Despite displaying normal growth and gross morphology at 7 dpf, mmachchg12/hg12 and mmachchg13/hg13 mutants displayed the typical organic acid accumulation that is seen in cblC patients, specifically elevated MMA as well as increased total methylcitrate, a shunt metabolite derived from propionate metabolism (P < 0.0001) (hg12Fig. 2D and E; hg13Supplementary Material, Figure S6). At 14 and 21 dpf, MMA concentrations continued to increase, and by 28 dpf, the metabolite levels in the mutant extracts were over 300-fold higher than clutchmates (P < 0.0001). Methionine, serine, cysteine and glycine were decreased at 21 and 28 dpf (Supplementary Material, Figure S6).
To evaluate potential disease mechanisms, we measured global DNA methylation and observed no significant differences between mutant and control fish at 28 dpf (Supplementary Material, Figure S7). We also measured glutathione and found that the total glutathione concentrations, normalized to protein content, were reduced by 51–66% in 28 dpf mutants (hg12 P < 0.01, hg13P < 0.001) and the GSSG/GSH (oxidized/free) ratio was abnormal (hg12P < 0.05, hg13P < 0.001) (Supplementary Material, Figure S7).
Growth and metabolic parameters of mmachc mutant fish improve with cobalamin and amino acid treatments
To assess the effectiveness of selected therapies employed to treat patients with cblC, we raised mmachchg13 fish in water supplemented with one of three different cobalamin derivatives [OHCbl, cyanocobalamin (CNCbl) and methylcobalamin (MeCbl)], methionine or betaine, and measured their growth and metabolite profiles at 28 dpf. There was a statistically significant improvement in both growth parameters, SL and HAA, in mmachchg13/hg13 fish treated with OHCbl, MeCbl, methionine and betaine compared with the untreated mutants (P < 0.0001) (Fig. 3). Cyanocobalamin treatment marginally improved SL (P < 0.01) but did not improve HAA. MeCbl and methionine, demonstrated the most improvement in HAA compared with the standard treatment, OHCbl (Fig. 3). None of the cofactors or amino acids altered the growth of wild-type or heterozygous clutchmates.
Figure 3.

Treatment studies. Fish from mmachchg13/hg13 incrosses were grown in independent tanks with system water or water supplemented with either hydroxocobalamin (OHCbl 0.5 mg/ml), methylcobalamin (MeCbl 0.5 mg/ml), cyanocobalamin (CNCbl 0.5 mg/ml), methionine (Met 2 mg/ml) or betaine (10 mg/ml) and were sacrificed at 28 dpf for growth measurements. (A) SL of the mutant fish improved with all treatments; however, the growth improvement was modest (1.9–11.1% increase). Data shown are % of clutchmate controls (+/+, +/hg13). (B) Height at the anterior of the anal fin (HAA) improved with all treatments (5.5–13.6% increase) except for cyanocobalamin. N = 20–22 mutants; N = 51–55 controls. Data shown are % of clutchmate controls (+/+, +/hg13). (C) Treatment with hydroxocobalamin and cyanocobalamin decreased MMA levels (77.4 and 18.7% respectively), and methionine increased [MMA] by 36.5%. (D) Treatment with hydroxocobalamin decreased methylcitrate production by 39.6%, whereas methionine increased methylcitrate levels 81%. One way ANOVA **P < 0.01 ***P < 0.001; ****P < 0.0001.
Index metabolites were also measured in fish extracts after treatments. MMA concentrations, which were massively elevated at baseline, decreased 77% in mutants treated with OHCbl (P < 0.0001) and 19% with CNCbl (P < 0.01) (Fig. 3C), while methylcitrate decreased 29% only after OHCbl exposure (P < 0.05) (Fig. 3D). As expected, methionine supplementation increased propionate-derived metabolites, MMA and total methylcitrate (Fig. 3D). Despite the fact that MeCbl did not improve MMA and methylcitrate levels, it was the most effective treatment for improving the growth of the cblC mutants, and superior to OHCbl with the HAA measurement, a proxy of growth (% increase: MeCbl 13.6% and OHCbl 5.5%).
Histological findings in cblC fish
We performed a general histologic survey of both mmachc fish lines at 7 and 28 dpf. The morphology of the brain, kidney and heart was within normal limits at 7 and 28 dpf (data not presented). However, a complete absence of gill secondary lamellae, the primary site of gas and ion exchange was noted in both mutant lines at 28 dpf (43) (Supplementary Material, Figure S8). Gill morphology was normal at 7 dpf.
Given the retinal degeneration observed in cblC patients, we also examined the cellular appearance of the retina and observed thin ON (Fig. 4B), decreased thickness of the outer nuclear layer (ONL) and shortened outersegments of the photoreceptors (PRs) (Fig. 4D) at 28 dpf, but not at earlier timepoints. Heterozygote mmachc animals were then crossed with transgenic lines expressing fluorescent markers in the rods TG(rho:EGFP) or cones TG(3.2TαCP-tdTomato) to create double transgenic mmachc fish [(mmachc hg13/hg13; TG(rho:EGFP); TG(3.2TαCP-tdTomato)] to visualize the ONL/PRs in more detail (44,45). Confocal microscopy in the double transgenic 28 dpf mmachc mutants revealed a striking decrease in the tdTomato (cone) fluorescence in the outersegments of the PRs (Fig. 4F). A single layer of GFP positive rods was observed in the ONL, which appeared decreased in numbers compared with controls (Fig. 4E and F). Flow cytometry indeed confirmed that there were decreased GFP positive cells (rods) in mutants compared with heterozygote eyes at 28 dpf (P < 0.0001) yet did not detect a difference in the number of tdTomato positive cells (cones) (Supplementary Material, Figure S9). In aggregate, the histology and flow sorting results confirm that there is PR cell loss in the cblC fish, implicating the PRs, and possibly the retinal pigmented epithelium (RPE), in the pathogenesis of cblC retinopathy.
Figure 4.

Retinal histology. hg12 fish were sacrificed at 28 dpf for retinal H&E; control heterozygote (A and C) and mutant (B and D). The mutants had small eyes, thin ON and thinning of the ONL and PR layer which contains the inner and outersegments of the rods and cones; 20× magnification. (E and F) The mutant lines (hg13 shown) were crossed with transgenic fish that express fluorescent markers in rod PRs (rhodopsin: TG(rho:EGFP)) and cone PR-specific protein [transducin-alpha: TG(3.2TαCP-tdTomato)] and were sectioned and examined with confocal microscopy. mmachchg13/hg13 (F) showed a decreased number of rod PRs in the ONL (green) and less cone transducin-alpha fluorescence in the PR layer (arrow) compared with heterozygous controls (E). Scale bar = 50 microns.
Transcriptional analysis of the mmachc eye
To gain insight into the molecular pathophysiology of cblC retinopathy, microarray analysis of whole-eye RNA from untreated mmachc+/hg13 and mmachchg13/hg13 fish was performed at two time points: 7 dpf when there were no apparent histological retinal changes and at 28 dpf when there was significant retinal pathology. Principal component analysis (PCA) and hierarchical clustering showed that each genotype and timepoint was separate and distinct from one another (Fig. 5). About 98 transcripts were differentially expressed between the mmachc fish and controls at 7 dpf. A subset of these (N = 54) were unique and had annotation available (Supplementary Material, Table S3). About 320 transcripts were differentially expressed at 28 dpf, of which 131 were unique and annotated (Supplementary Material, Table S4). There was no overlap between the two gene lists.
Figure 5.
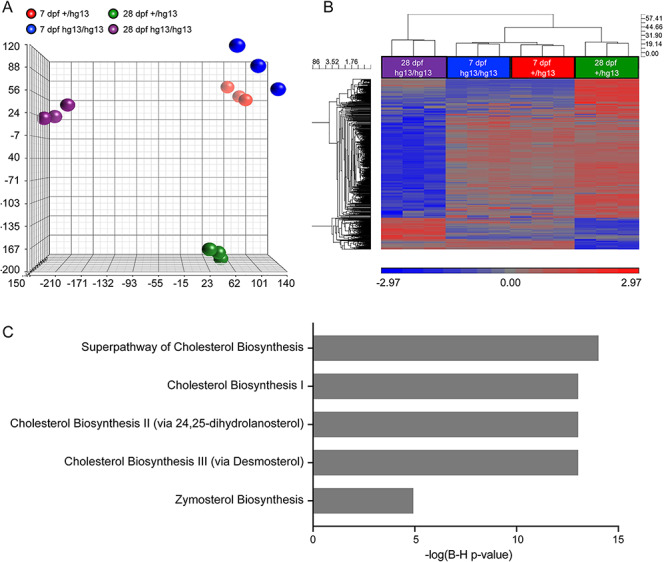
Microarray study. Microarrays were performed using RNA from mmachc+/hg13 and mmachchg13/hg13 eyes. (A) PCA shows the clustering of the samples into distinct groups. (B) Heatmap of microarray dataset using cutoff of 2-fold and P value < 0.05 indicated clustering of the 7 dpf samples with the 28 dpf mutants as a distinct group being far apart, which is consistent with the abnormal retinal histology, (C) Ingenuity pathway analysis of 28 dpf samples showed the dysregulation of several cholesterol biosynthesis pathways.
We validated several of the dysregulated genes by qPCR: arntl2, prkg2, guca1c and per3 at 7 dpf (Supplementary Material, Figure S10) and mvd, sqle, lss, igf2bp1 and pdk2 at 28 dpf (Supplementary Material, Figure S11). Ingenuity pathway analysis of the 28 dpf dataset revealed that the pathways of cholesterol biosynthesis were dysregulated (Fig. 5C); however, in the 7 dpf dataset, there were no pathways identified perhaps due to a smaller number of dysregulated genes.
Discussion
Survival and growth
In the present work, we have exploited the genetic and genomic resources available to the zebrafish research community to create the first viable animal model of an important, but historically difficult to study, inborn error of vitamin B12 metabolism, which manifests early embryonic lethality in mice (34). Using a ZFN-based genome editing approach, we created mutant alleles of mmachc, which are adjacent to two variants seen in cblC patients, p.Pro42Thrfs*19 and p.Leu43Trpfs*28 (5). While clear discrepancies between morphant and ZFN mmachc loss-of-function mutant phenotypes were observed consistent with previous reports (39), we believe that the relatively high levels of maternal mmachc RNA in the unfertilized eggs may have rescued the mutant fish from the very early lethality displayed by the mmachc morphants (Supplementary Material, Figure S5). The mmachc morphants had axial developmental defects similar to a zebrafish model of a related disorder with elevated homocysteine, cystathionine β-synthase, suggesting that the transsulfuration pathway may play a role (46). Our findings further highlight the critical role of vitamin B12 in zebrafish for development and metabolism (38) and validate the previous efforts to characterize proteins involved in cobalamin absorption and transport (47–49).
Both lines of mmachc mutant fish perished during the early juvenile period similar to the reduced survival observed in untreated patients with cblC deficiency (9,14). While we were not able to determine the exact cause of mortality, we note that the secondary lamellae in the fish gills that normally develop between 7 and 12 dpf failed to develop. It is unknown how mmachc or B12 contributes to zebrafish gill development, but the secondary lamellae contain mitochondria-rich cells (ionocytes) and are involved in acid–base balance in early development similar to the kidney in humans (43). In addition, the lethality coincided with the early juvenile period where there was physiological increase in mmachc expression in wild-type fish and may reflect increased requirements for cobalamin. In addition to poor survival, cblC fish showed arrested development. Of note, patients with cblC deficiency can have growth impairment in utero, and feeding difficulties and failure to thrive are common, even while on treatment (14).
Metabolic perturbations: primary and secondary
Metabolite elevations in the cblC zebrafish model recapitulated some of those seen in affected individuals with cblC deficiency (4,50). Although measurements of homocysteine were too low to interpret by our methods, organic acids, including MMA and 2-methylcitric acid, were up to 300-fold elevated. The MMA concentrations are similar to those observed in tissues from mice with a lethal form of mut-type methylmalonic acidemia (51), suggesting MMA may contribute to the observed mortality in the juvenile period.
Increased oxidative stress, mediated through the formation of reactive oxygen species (ROS), has been hypothesized to contribute to the pathophysiology of cblC deficiency (10) as biomarkers of oxidative stress were increased in urine and patient-derived fibroblasts (26,27). Glutathione, a key scavenger of ROS, and cysteine, an amino acid essential for glutathione synthesis, have been reported to be decreased in cblC patients’ serum (30). We therefore measured the total glutathione pool and, note a reduction in total glutathione and increased GSSG/GSH ratio in the mmachc mutants. Whether this is due to low cysteine as has been observed in mice with a related disorder (52), or another mechanism remains uncertain but illustrates a potential avenue for future study and perhaps therapeutic intervention using cblC zebrafish.
We also explored the potential effects on global DNA methylation. In cblC deficiency, the failure to synthesize MeCbl results in impaired remethylation of homocysteine to methionine and, possibly, a subsequent decrease in S-adenosylmethionine (SAM) (8). SAM is an essential methyl donor involved in the synthesis of small molecules (e.g. creatine and phosphatidylcholine), lipids (sphingomyelin) as well as protein and DNA methylation (8). The small size of our mutant fish limited our ability to measure SAM and other downstream metabolites, and global protein methylation. Although we did not observe significant changes in global DNA methylation in whole fish extracts, it remains possible that the analysis of individual organs, and even cells, may reveal tissue-specific hypomethylation as it has been observed in related mouse models of Cd320 and Mthfr deficiencies (53,54).
Response to established and potential small molecule therapies
The established treatment guidelines for cblC deficiency recommend parenteral hydroxocobalamin (OHCbl), betaine and other supplements aimed at improving biochemical parameters (55), and OHCbl treatment has been documented to improve biochemical parameters, survival and growth (9,14). However, studies regarding dosing, frequency and clinical outcomes are limited and are primarily based on case reports and physician preferences (10,17,55). We therefore utilized the zebrafish model to explore, in an unbiased fashion, some of the current treatments, including cobalamin derivatives and the amino acids, methionine and betaine.
Patients with cblC do not respond to CNCbl (56), likely because MMACHC is involved in its decyanation (57). We confirmed that CNCbl had limited effect on our cblC mutants’ growth parameters and [MMA] did not decrease. In contrast, OHCbl, the standard therapy used in patients, decreased [MMA] but surprisingly did not improve HAA as well as other treatments aimed at the reduction of homocysteine and the improvement of methionine deficiency (MeCbl, betaine and methionine). MeCbl emerged as the most effective treatment to improve growth parameters in the cblC zebrafish despite having no effect on the metabolites measured. MeCbl deficiency is a characteristic biochemical property of cblC fibroblasts, yet MeCbl treatment has only been documented in a few case reports (58–60). This suggests that the further exploration of MeCbl treatment in patients could be considered but combination with OHCbl would likely be necessary as MeCbl alone showed only a small decrease in [MMA] in the cblC zebrafish.
It must be noted that there are several limitations of our treatment studies, including that B12 metabolism and transport in zebrafish may be significantly different than in humans, the route of administration was likely oral and through the epidermis compared to intramuscular injections in humans, and that neither B12 nor amino acid levels were measured in fish following treatment to confirm absorption and we were limited in our ability to measure homocysteine. In addition, it is possible that the cofactors could have been degraded during the treatment period despite frequent renewal. However, because the concentrations of our treatments were in the superphysiologic range, e.g. OHCbl 0.5 mg/ml versus cobalamin in human serum < 1000 pg/ml and methionine 13 mM versus human serum < 50 μM, we believe that extra-corporeal metabolism of cofactors likely did not contribute to the changes observed. Nevertheless, we were able to confirm metabolic and phenotypic effects from cofactors and supplements known to be clinically effective (OHCbl) and, likewise, failed to observe benefits from those that are not therapeutic (CNCbl).
Exploring the pathophysiology and transcriptome of cblC associated eye disease
The maculopathy and retinal degeneration experienced by many cblC patients remains one of the most mysterious and challenging clinical findings in this disorder (61). The progressive retinal degeneration is typically preceded by severe degeneration of the macula in early infancy and can even manifest a macular ‘coloboma’ which may be due to abnormal ocular development or early rapid degeneration (18). Using the zebrafish model, we sought to explore cblC-associated ocular pathology and determined that mmachc mutant retinas were histologically normal during the embryonic period, but developed pathological changes at the end of the larval stage, consistent with a degenerative process. The findings in the mmachchg13/hg13 zebrafish are very similar to what has been reported in a single patient autopsy where thinning of the outernuclear layer, loss of PRs with swollen mitochondria on EM and hypo/hyperpigmentation of the RPE were documented (25). While the RPE appeared grossly intact, the dramatic reduction of cone derived fluorescent material present in the mmachc hg13/hg13; TG(rho:EGFP); TG(3.2TαCP-tdTomato) mutants suggests that the PRs have shortened outersegments and/or the RPE may be unable to phagocytose discarded opsins and other material from the PRs following light exposure.
In an attempt to characterize the molecular basis of the retinal disease, we explored the transcriptome of whole-eye RNA. At 7 dpf, when the retina was histologically normal, we found that the guanylyl cyclase-activating protein (GCAP) genes, guca1a, guca1c and guca1d, were dysregulated. These genes encode GCAPs, calcium sensors involved in regulating guanylate cyclases during phototransduction. Although the mechanism connecting mmachc and potential GCAP dysregulation is uncertain, in humans, mutations in GUCA1A result in autosomal dominant cone, cone-rod and macular dystrophies (62,63). Most pathogenic variants are missense and have been shown to alter Ca2+ sensitivity, resulting in the persistent stimulation of GC1 in the dark, and subsequent PR death. In addition, slc16a8, a transporter for monocarboxylates (lactate, pyruvate, branched-chain amino acids and ketone bodies) was downregulated. This gene was identified in a genome wide association study for age-related macular degeneration (64) and may represent another target locus for cblC-related eye disease, which initially involves the macula. However, at 28 dpf when significant retinal pathology was histologically apparent and disease-related changes may be obscured, numerous changes where noted (Fig. 5C, Supplementary Material, Table S5). Cholesterol metabolism and one carbon metabolism pathway genes, including gnmt and mthfd1l, emerged as differentially expressed. In addition, the transcriptional changes we observed in the eyes of the mutant cblC zebrafish may also be a result of global abnormalities in RNA trafficking as it have been described in cblC patient fibroblasts (32), cell lines and animal models of related cobalamin disorders (31–33,65). In the future, this zebrafish model could be used for further exploration of this potential disease mechanism.
In summary, we report the first viable model of cblC deficiency that replicates several known clinical and pathological manifestations seen in affected patients, including retinal pathology. Transcriptional profiling identified several possible candidate genes for further exploration including guca1a and slc16a8, and new pathways, such as formate metabolism, to consider for small molecule interventions. Finally, mmachc mutant zebrafish were responsive to known therapeutics and, therefore, will be useful to test the efficacy of current and proposed therapies for cblC deficiency.
Materials and Methods
Zebrafish maintenance
Zebrafish (Danio rerio) were maintained under approved National Institutes of Health animal use protocols (# G05-5 and G16-3) in accordance with the Zebrafish Book (66). Embryos were maintained at 28.5°C in E3 embryo medium supplemented with 1% methylene blue. All experiments were performed on zebrafish from the Ekkwill (EK) genetic background. After 5 dpf, fish were raised on a circulating water system at a stocking density of 30 fish per liter during the first month and then at 10–15 fish per liter and 14 hour light cycle. Fish were fed once per day with rotifers enriched with RotiGrow Plus (Reed Mariculture, Campbell, CA) from 5–9 dpf, twice per day with San Francisco strain brine shrimp eggs (Brine Shrimp Direct, Ogden, UT) from 9 to 14 dpf, once per day with Larval AP100 150–250 microns larval food supplement (Zieglar Bros., Gardners, PA) and once per day with Hatchfry Encapsulon 250–450 microns larval food supplement (Argent Laboratories, Redmond, WA) from 15 to 30 dpf, twice per day with Aquarium Flake: Chrill (3:1) Food liquid mix (Ocean Start International, Burlingame, CA) from 30 to 90 dpf and once per day with Ziegler crumble (Ziegler Bros., Gardners, PA) after 90 dpf.
Morpholino Injections
Fluorescein-tagged morpholinos (MOs) were purchased in lyophilized form (Gene Tools. Philomath, OR), re-suspended in distilled water and quantified spectrophotometrically (NanoDrop Technologies, Wilminton, DE). A standard negative control MO purchased from Gene Tools and gene-specific translation blocking (mmachc-atg MO: 5′-GCGTTCACTGGAGATCGCCATTTTT-3′) and splice blocking morpholinos designed at the exon 3-intron 3 splice junction (mmachc-spMO: 5′-GGCATATAAAGACTGACCTTCTCTC-3′) were microinjected into the yolk sac of single-cell stage embryos using a pneumatic pico pump (World Precision Instruments, Sarasota, FL). Phenotypes were captured with a Leica DC500 (Leica Microsystems, Buffalo Grove, IL) camera attached to a Leica MZ16F microscope (Leica Microsystems, Buffalo Grove, IL). RTPCR primers used to amplify the abnormally spliced isoforms: mmachc forward 5′-CGCACCATGGCGATCTCCAGTGAACGCGTGG-3′ and reverse 5′-TCAGCTCTGCGTGTCGGTCAGGTATCCCCACTG-3′ and gapdh forward 5′-ACTCCACTCATGGCCGTTAC-3′ and reverse 5′-TCTTCTGTGTGGCGGTGTAG-3′.
Targeted mutagenesis using ZFNs
CompZr ZFNs to target mmachc were designed and characterized by Sigma-Aldrich (St. Louis, MO) and two mutant alleles, mmachchg12 and mmachchg13, were generated as previously described (67). Fish were genotyped by fluorescent PCR and capillary electrophoresis on genomic DNA extracted from embryos or fin biopsies as described in detail previously (67,68). The following primers were used for fluorescent PCR genotyping: M13 labeled forward primer 5′-TGTAAAACGACGGCCAGTATACTGGTCTCCGAGGGACTG-3′ and PIG tailed reverse primer 5′-GTGTCTTAGAGATGCAGGCAGACAGTG-3′ with a product size of 268 bp. PCR products were analyzed by fragment separation using an ABI sequencer (Supplementary Material, Figure S12) (67,68).
Determination of growth and developmental parameters
Images were captured using an Axio Zoom V16 microscope with AxioCam HRc camera or digital camera and processed using AxioVision 4.8 software (Carl Zeiss, Jena, Germany). Measurements, SL and height at anterior of anal fin (HAA), were collected for each fish and analyzed using ImageJ (69). Developmental staging of postembryonic zebrafish has been previously described (40). For pelvic fin scoring, we used the following scale: 0 no pelvic bud present, 1 presence of bud and 2 presence of pelvic fin. For caudal fin scoring, we used the following scale: 3 = dispersed melanophores, no interior black stripe condensation; 4 = some striping but black and yellow stripes do not span entire length of tail with sparse melanophores in yellow stripe; 5 = black and yellow stripe span the entire length of tail with X1v yellow stripe appearing; and 6 = adult caudal fin pigmentation (40). Melanophore pattern score was used as described (40).
Propionate sensitivity
One zebrafish embryo (24 hpf) was placed in each well of a 96-well plate with 200 μL of E3 embryo water (control) or E3 embryo water supplemented with 1 mM sodium propionate. Plates were placed in an incubator at 28.5°C and monitored daily. Viability was scored visually based on the presence of a heartbeat until 5 dpf.
Histology
Tissues were fixed in 4% paraformaldehyde overnight at 4°C, dehydrated in 70% ethanol, embedded into paraffin blocks, sectioned at 20, 50 or 100 μm and stained with H&E (HistoServ, Germantown, MD). Images were captured using an Axio Imger.D2 microscope and Axion Cam HRc camera and analyzed with Zen 2011 software (Carl Zeiss, Jena, Germany). Fish expressing fluorescent markers were prepared as previously described (70) and cryosectioned at 20 μm (HistoServ, Germantown, MD). Slides were rehydrated with PBS for 10 min at room temperature and mounted with ProLong Gold Antifade Reagent with DAPI (Invitrogen, Carlsbad, CA) to visualize the nuclei. Confocal images were captured with a Leica SP5 confocal microscope and analyzed using LASAF software (Leica, Solms, Germany).
Treatment studies
Fish were grown in Mini Bow 2.5 Aquaria tanks with a circulating waterfall system (Aqueon, Franklin, WI) without a charcoal filter due to the possible charcoal absorption of B12 (71). Fish were stocked at a density of 30 fish per liter and grown for 28 days. Water was supplemented with: 0.5 mg/ml hydroxocobalamin (College Pharmacy, Colorado Springs, CO), 0.5 mg/ml cyanocobalamin (Sigma-Aldrich, St. Louis, MO), 0.5 mg/ml methylcobalamin (College Pharmacy, Colorado Springs, CO), 2 mg/ml methionine (Sigma-Aldrich, St. Louis, MO) or 10 mg/ml betaine (Sigma-Aldrich, St. Louis, MO). Fish grown only in system water served as the control. Ten percent of the total water volume was replenished every 3 days and included the supplements. Water quality was monitored using a Fresh Water Aquaculture Model AQ-2 Test Kit (LaMotte, Chestertown, MD).
RNA extraction, RT-PCR and qPCR
Total RNA was extracted from three independent pools of embryos (n = 3 controls and n = 10–15 mutants), whole fish and adult tissues (controls n = 1) with Trizol Reagent (Invitrogen, Carlsbad, CA). About 1.5 μg of RNA was reverse transcribed with random hexamer primers using High Capacity cDNA Reverse Transcription Kit (Invitrogen, Carlsbad, CA). For standard PCR, cDNA was amplified with AmpliTaq Gold DNA Polymerase (Invitrogen, Carlsbad, CA) using primers designed against full-length mmachc: 5′-CGCACCATGGCGATCTCCAGTGAACGCGTGG-3′ and 5′-TCAGCTCTGCGTGTCGGTCAGGTATCCCCACTG-3′ and gapdh: 5′-TCTTCTGTGTGGCGGTGTAG-3′ and 5′-ACTCCACTCATGGCCGTTAC-3′ and visualized by standard gel-electrophoresis. For real-time qPCR, cDNA was quantified using primers listed in Supplementary Material, Table S6 and FastStart Universal SYBR Green master mix (Roche, Indianapolis, IN) on a 7500 Fast Real-Time system in accordance with the manufacturer’s protocol (Applied Biosystems, Foster City, CA). Relative expression was determined using the comparative Ct method (ΔΔCt) and normalized to β-actin. All samples were analyzed in duplicate.
Flow cytometry
Fish eyes were microdissected, pooled (N = 6 eyes control and N = 20–30 mutant) and dissociated in 0.25% Trypsin (Invitrogen, Carlsbad, CA) at room temperature and passed through 70 and 40 μm cell strainers (BioExpress, Kaysville, UT). Fetal bovine serum (Invitrogen, Carlsbad, CA) was added at a concentration of 5% to terminate the reaction. To visualize the nuclei, cells were diluted to 3 × 105 cells/ml and incubated in 1 μL Vybrant DyeCycle Ruby stain (Invitrogen, Carlsbad, CA) for 15 min at 37°C. Cells were immediately placed on ice and analyzed using the BD FACSCalibur (BD Biosciences, San Jose, CA) with 488 and 633 nm lasers. Data were analyzed using FlowJo software (TreeStar, Ashland, OR).
Metabolite analysis
Fish were pooled (3 fish per sample at 7 and 14 dpf; 3 controls and 10–15 mutants per sample at 21 and 28 dpf due to their very small size), homogenized in water and centrifuged at 10 000× g for 10 min at 4°C. Supernatant extracts were analyzed by gas chromatography–mass spectrometry (Metabolite Laboratories Inc., Denver, CO) as previously described (72). Total protein concentration was determined using the BCA Protein Assay (Thermo Fischer Scientific, Rockford, IL). Metabolite values were normalized to protein concentration to account for the size difference and the number of mutant and control fish.
Microarray and data analysis
Total RNA were harvested from three independent pools of mmachc+/hg13 and mmachchg13/hg13 eyes with QIAzol Lysis Reagent and the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacter’s protocol. Agilent 2100 Bioanalyzer (Santa Clara, CA) evaluated RNA concentration and quality. Three-hundred nanograms were amplified, reverse transcribed and biotinylated using the GeneChip WT PLUS Reagent Kit (Affymetrix, Santa Clara, CA). cDNA was hybridized to Zebrafish Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA) according to manufacturer’s protocol.
The microarray data were analyzed with the gene expression workflow found in Partek Genomics Suite (St. Louis, MO). PCA was performed in Partek and showed that mmachchg13/hg13 and mmachc+/hg13 are separate and distinct from each other. To find the optimal fold change cutoff, the fold-change between all pair-wise comparisons of the biological replicates in the control group over all probesets was calculated. Since the fold-change distribution is normal across all probesets, we calculate the upper threshold by the mean plus 2 SD. Based on this analysis, genes were called differentially expressed if they had a fold change greater than 2.0 for 7 dpf and 2.5 for 28 dpf and only genes that had annotation were reported. The Ensembl Biomart was used to annotate genes that were not identified in Partek. Ingenuity Pathway Analysis (Qiagen) calculated enriched pathways with Benjamini-Hochberg correction for the differentially expressed gene list. The data were deposited in the Gene Expression Omnibus database: GSE130433.
Quantification of total 5-mC content
Genomic DNA was isolated from three pools of fish (3 fish per sample at 7 and 14 dpf; 3 controls and 10–15 mutants per sample at 21 and 28 dpf due to their very small size) using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). DNA concentration and purity was determined spectrophotometrically by comparing the absorbance values at 260, 280 and 230 nm. A total of 100 ng of DNA was analyzed in duplicate using the Methyl Flash Methylated DNA Quantification Kit according to the manufacturer’s protocol (Epigentek Group Inc., New York, NY).
Quantification of GSH/GSSG
Fish were pooled in triplicate and homogenized in 5% sulfosalicylic acid (SSA) (Sigma-Aldrich, St. Louis, MO) and centrifuged at 10 000× g for 10 min at 4°C. The supernatant was collected, diluted in H2O to reduce the SSA concentration to 0.5% and assayed in duplicate using the GSSG/GSH Quantification Kit according to the manufacturer’s protocol (Dojindo Laboratories, Rockville, MD). Pellets were resuspended in T-PER containing a protease-inhibitor cocktail (Thermo Fischer Scientific) and clarified by centrifugation at 10 000× g for 10 min at 4°C. Protein content was determined using the BCA Protein Assay (Thermo Fischer Scientific, Rockford, IL). Total glutathione levels were normalized to protein content.
Statistical analysis
Statistical analysis was performed with Prism 4.0 (GraphPad, La Jolla, CA). Data are presented as the means ± SD. Statistical comparisons were performed using chi square, two-tailed unpaired Student’s t-test, one-way ANOVA or two-way ANOVA, followed by Bonferroni post hoc test for multiple comparisons. In all instances, P values were considered statistically significant if the value was <0.05.
Supplementary Material
Acknowledgements
We are thankful to Blake Carrington for technical assistance in maintaining and genotyping mutant fish and the zebrafish husbandry staff for excellent care of the animals. We thank Julia Fekecs for her assistance with the figures.
Funding
This work was supported by the Michael Clapcich Fund for Retinal Research and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Conflict of Interest statement. The authors have no conflict of interests to declare.
References
- 1. Banerjee R., Gherasim C. and Padovani D. (2009) The tinker, tailor, soldier in intracellular B12 trafficking. Curr. Opin. Chem. Biol., 13, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cooper B.A. and Rosenblatt D.S. (1987) Inherited defects of vitamin B12 metabolism. Annu. Rev. Nutr., 7, 291–320. [DOI] [PubMed] [Google Scholar]
- 3. Quadros E.V. (2010) Advances in the understanding of cobalamin assimilation and metabolism. Br. J. Haematol., 148, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sloan J.L., Carrillo N., Adams D. and Venditti C.P. (2008) Disorders of intracellular cobalamin metabolism. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K. and Amemiya A. (eds), GeneReviews®, Seattle (WA): University of Washington, Seattle; 1993–2020. [PubMed] [Google Scholar]
- 5. Lerner-Ellis J.P., Anastasio N., Liu J., Coelho D., Suormala T., Stucki M., Loewy A.D., Gurd S., Grundberg E., Morel C.F. et al. (2009) Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype-phenotype correlations. Hum. Mutat., 30, 1072–1081. [DOI] [PubMed] [Google Scholar]
- 6. Lerner-Ellis J.P., Tirone J.C., Pawelek P.D., Dore C., Atkinson J.L., Watkins D., Morel C.F., Fujiwara T.M., Moras E., Hosack A.R. et al. (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet., 38, 93–100. [DOI] [PubMed] [Google Scholar]
- 7. Gherasim C., Ruetz M., Li Z., Hudolin S. and Banerjee R. (2015) Pathogenic mutations differentially affect the catalytic activities of the human B12-processing chaperone CblC and increase futile redox cycling. J. Biol. Chem., 290, 11393–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrillo-Carrasco N. and Venditti C.P. (2012) Combined methylmalonic acidemia and homocystinuria, cblC type. II. Complications, pathophysiology, and outcomes. J. Inherit. Metab. Dis., 35, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenblatt D.S., Aspler A.L., Shevell M.I., Pletcher B.A., Fenton W.A. and Seashore M.R. (1997) Clinical heterogeneity and prognosis in combined methylmalonic aciduria and homocystinuria (cblC). J. Inherit. Metab. Dis., 20, 528–538. [DOI] [PubMed] [Google Scholar]
- 10. Martinelli D., Deodato F. and Dionisi-Vici C. (2011) Cobalamin C defect: natural history, pathophysiology, and treatment. J. Inherit. Metab. Dis., 34, 127–135. [DOI] [PubMed] [Google Scholar]
- 11. Profitlich L.E., Kirmse B., Wasserstein M.P., Diaz G.A. and Srivastava S. (2009) High prevalence of structural heart disease in children with cblC-type methylmalonic aciduria and homocystinuria. Mol. Genet. Metab., 98, 344–348. [DOI] [PubMed] [Google Scholar]
- 12. Weisfeld-Adams J.D., Bender H.A., Miley-Akerstedt A., Frempong T., Schrager N.L., Patel K., Naidich T.P., Stein V., Spat J., Towns S. et al. (2013) Neurologic and neurodevelopmental phenotypes in young children with early-treated combined methylmalonic acidemia and homocystinuria, cobalamin C type. Mol. Genet. Metab., 110, 241–247. [DOI] [PubMed] [Google Scholar]
- 13. Martinelli D., Dotta A., Massella L., Picca S., Di Pede A., Boenzi S., Aiello C. and Dionisi-Vici C. (2011) Cobalamin C defect presenting as severe neonatal hyperammonemia. Eur. J. Pediatr., 170, 887–890. [DOI] [PubMed] [Google Scholar]
- 14. Fischer S., Huemer M., Baumgartner M., Deodato F., Ballhausen D., Boneh A., Burlina A.B., Cerone R., Garcia P., Gokcay G. et al. (2014) Clinical presentation and outcome in a series of 88 patients with the cblC defect. J. Inherit. Metab. Dis., 37, 831–840. [DOI] [PubMed] [Google Scholar]
- 15. Carmel R., Bedros A.A., Mace J.W. and Goodman S.I. (1980) Congenital methylmalonic aciduria—homocystinuria with megaloblastic anemia: observations on response to hydroxocobalamin and on the effect of homocysteine and methionine on the deoxyuridine suppression test. Blood, 55, 570–579. [PubMed] [Google Scholar]
- 16. Mamlok R.J., Isenberg J.N., Rassin D.K., Norcross K. and Tallan H.H. (1986) A cobalamin metabolic defect with homocystinuria, methylmalonic aciduria and macrocytic anemia. Neuropediatrics, 17, 94–99. [DOI] [PubMed] [Google Scholar]
- 17. Bartholomew D.W., Batshaw M.L., Allen R.H., Roe C.R., Rosenblatt D., Valle D.L. and Francomano C.A. (1988) Therapeutic approaches to cobalamin-C methylmalonic acidemia and homocystinuria. J. Pediatr., 112, 32–39. [DOI] [PubMed] [Google Scholar]
- 18. Brooks B.P., Thompson A.H., Sloan J.L., Manoli I., Carrillo-Carrasco N., Zein W.M. and Venditti C.P. (2016) Ophthalmic manifestations and long-term visual outcomes in patients with cobalamin C deficiency. Ophthalmology, 123:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weisfeld-Adams J.D., McCourt E.A., Diaz G.A. and Oliver S.C. (2015) Ocular disease in the cobalamin C defect: a review of the literature and a suggested framework for clinical surveillance. Mol. Genet. Metab., 114, 537–546. [DOI] [PubMed] [Google Scholar]
- 20. Huemer M., Kozich V., Rinaldo P., Baumgartner M.R., Merinero B., Pasquini E., Ribes A. and Blom H.J. (2015) Newborn screening for homocystinurias and methylation disorders: systematic review and proposed guidelines. J. Inherit. Metab. Dis., 38, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weisfeld-Adams J.D., Morrissey M.A., Kirmse B.M., Salveson B.R., Wasserstein M.P., McGuire P.J., Sunny S., Cohen-Pfeffer J.L., Yu C., Caggana M. et al. (2010) Newborn screening and early biochemical follow-up in combined methylmalonic aciduria and homocystinuria, cblC type, and utility of methionine as a secondary screening analyte. Mol. Genet. Metab., 99, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dayan A.D. and Ramsey R.B. (1974) An inborn error of vitamin B12 metabolism associated with cellular deficiency of coenzyme forms of the vitamin. Pathological and neurochemical findings in one case. J. Neurol. Sci., 23, 117–128. [DOI] [PubMed] [Google Scholar]
- 23. Dillon M.J., England J.M., Gompertz D., Goodey P.A., Grant D.B., Hussein H.A., Linnell J.C., Matthews D.M., Mudd S.H., Newns G.H. et al. (1974) Mental retardation, megaloblastic anaemia, methylmalonic aciduria and abnormal homocysteine metabolism due to an error in vitamin B12 metabolism. Clin. Sci. Mol. Med., 47, 43–61. [DOI] [PubMed] [Google Scholar]
- 24. McCully K.S. (1969) Vascular pathology of homocysteinemia: implications for the pathogenesis of arteriosclerosis. Am. J. Pathol., 56, 111–128. [PMC free article] [PubMed] [Google Scholar]
- 25. Traboulsi E.I., Silva J.C., Geraghty M.T., Maumenee I.H., Valle D. and Green W.R. (1992) Ocular histopathologic characteristics of cobalamin C type vitamin B12 defect with methylmalonic aciduria and homocystinuria. Am. J. Ophthalmol., 113, 269–280. [DOI] [PubMed] [Google Scholar]
- 26. Mc Guire P.J., Parikh A. and Diaz G.A. (2009) Profiling of oxidative stress in patients with inborn errors of metabolism. Mol. Genet. Metab., 98, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richard E., Jorge-Finnigan A., Garcia-Villoria J., Merinero B., Desviat L.R., Gort L., Briones P., Leal F., Perez-Cerda C., Ribes A. et al. (2009) Genetic and cellular studies of oxidative stress in methylmalonic aciduria (MMA) cobalamin deficiency type C (cblC) with homocystinuria (MMACHC). Hum. Mutat., 30, 1558–1566. [DOI] [PubMed] [Google Scholar]
- 28. Caterino M., Pastore A., Strozziero M.G., Di Giovamberardino G., Imperlini E., Scolamiero E., Ingenito L., Boenzi S., Ceravolo F., Martinelli D. et al. (2015) The proteome of cblC defect: in vivo elucidation of altered cellular pathways in humans. J. Inherit. Metab. Dis., 38, 969–979. [DOI] [PubMed] [Google Scholar]
- 29. Hannibal L., DiBello P.M., Yu M., Miller A., Wang S., Willard B., Rosenblatt D.S. and Jacobsen D.W. (2011) The MMACHC proteome: hallmarks of functional cobalamin deficiency in humans. Mol. Genet. Metab., 103, 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pastore A., Martinelli D., Piemonte F., Tozzi G., Boenzi S., Di Giovamberardino G., Petrillo S., Bertini E. and Dionisi-Vici C. (2014) Glutathione metabolism in cobalamin deficiency type C (cblC). J. Inherit. Metab. Dis., 37, 125–129. [DOI] [PubMed] [Google Scholar]
- 31. Battaglia-Hsu S.F., Ghemrawi R., Coelho D., Dreumont N., Mosca P., Hergalant S., Gauchotte G., Sequeira J.M., Ndiongue M., Houlgatte R. et al. (2018) Inherited disorders of cobalamin metabolism disrupt nucleocytoplasmic transport of mRNA through impaired methylation/phosphorylation of ELAVL1/HuR. Nucleic Acids Res., 46, 7844–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghemrawi R., Arnold C., Battaglia-Hsu S.F., Pourie G., Trinh I., Bassila C., Rashka C., Wiedemann A., Flayac J., Robert A. et al. (2019) SIRT1 activation rescues the mislocalization of RNA-binding proteins and cognitive defects induced by inherited cobalamin disorders. Metabolism, 101, 153992. [DOI] [PubMed] [Google Scholar]
- 33. Ghemrawi R., Battaglia-Hsu S.F. and Arnold C. (2018) Endoplasmic reticulum stress in metabolic disorders. Cell, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreno-Garcia M.A., Pupavac M., Rosenblatt D.S., Tremblay M.L. and Jerome-Majewska L.A. (2014) The Mmachc gene is required for pre-implantation embryogenesis in the mouse. Mol. Genet. Metab., 112, 198–204. [DOI] [PubMed] [Google Scholar]
- 35. Buers I., Pennekamp P., Nitschke Y., Lowe C., Skryabin B.V. and Rutsch F. (2016) Lmbrd1 expression is essential for the initiation of gastrulation. J. Cell. Mol. Med., 20, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swanson D.A., Liu M.L., Baker P.J., Garrett L., Stitzel M., Wu J., Harris M., Banerjee R., Shane B. and Brody L.C. (2001) Targeted disruption of the methionine synthase gene in mice. Mol. Cell. Biol., 21, 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elmore C.L., Wu X., Leclerc D., Watson E.D., Bottiglieri T., Krupenko N.I., Krupenko S.A., Cross J.C., Rozen R., Gravel R.A. et al. (2007) Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol. Genet. Metab., 91, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansen A.C., Waagbo R. and Hemre G.I. (2015) New B vitamin recommendations in fish when fed plant-based diets. Aquac. Nutr., 21, 507–527. [Google Scholar]
- 39. Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S., Impel A., Kirchmaier B.C., Peterson-Maduro J., Kourkoulis G., Male I. et al. (2015) Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell, 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parichy D.M., Elizondo M.R., Mills M.G., Gordon T.N. and Engeszer R.E. (2009) Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn., 238, 2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McMenamin S.K., Chandless M.N. and Parichy D.M. (2016) Working with zebrafish at postembryonic stages. Methods Cell Biol., 134, 587–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen R.H., Stabler S.P., Savage D.G. and Lindenbaum J. (1993) Elevation of 2-methylcitric acid I and II levels in serum, urine, and cerebrospinal fluid of patients with cobalamin deficiency. Metabolism, 42, 978–988. [DOI] [PubMed] [Google Scholar]
- 43. Rombough P. (2007) The functional ontogeny of the teleost gill: which comes first, gas or ion exchange? Comp. Biochem. Physiol. A Mol. Integr. Physiol., 148, 732–742. [DOI] [PubMed] [Google Scholar]
- 44. Hamaoka T., Takechi M., Chinen A., Nishiwaki Y. and Kawamura S. (2002) Visualization of rod photoreceptor development using GFP-transgenic zebrafish. Genesis, 34, 215–220. [DOI] [PubMed] [Google Scholar]
- 45. Kennedy B.N., Alvarez Y., Brockerhoff S.E., Stearns G.W., Sapetto-Rebow B., Taylor M.R. and Hurley J.B. (2007) Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Invest. Ophthalmol. Vis. Sci., 48, 522–529. [DOI] [PubMed] [Google Scholar]
- 46. Prabhudesai S., Koceja C., Dey A., Eisa-Beygi S., Leigh N.R., Bhattacharya R., Mukherjee P. and Ramchandran R. (2018) Cystathionine beta-synthase is necessary for Axis development in vivo. Front. Cell Dev. Biol., 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Benoit C.R., Stanton A.E., Tartanian A.C., Motzer A.R., McGaughey D.M., Bond S.R. and Brody L.C. (2018) Functional and phylogenetic characterization of non-canonical vitamin B12-binding proteins in zebrafish suggests involvement in cobalamin transport. J. Biol. Chem., 293:17606–17621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Greibe E., Fedosov S. and Nexo E. (2012) The cobalamin-binding protein in zebrafish is an intermediate between the three cobalamin-binding proteins in human. PLoS One, 7, e35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansen A.C., Olsvik P.A. and Hemre G.I. (2013) Effect of different dietary vitamin B12 levels on their retention in the body of zebrafish Danio rerio and on the gene expression of vitamin B12 binding proteins. Aquac. Nutr., 19, 413–420. [Google Scholar]
- 50. Carrillo-Carrasco N., Chandler R.J. and Venditti C.P. (2012) Combined methylmalonic acidemia and homocystinuria, cblC type. I. Clinical presentations, diagnosis and management. J. Inherit. Metab. Dis., 35, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chandler R.J., Sloan J., Fu H., Tsai M., Stabler S., Allen R., Kaestner K.H., Kazazian H.H. and Venditti C.P. (2007) Metabolic phenotype of methylmalonic acidemia in mice and humans: the role of skeletal muscle. BMC Med. Genet., 8, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maclean K.N., Jiang H., Aivazidis S., Kim E., Shearn C.T., Harris P.S., Petersen D.R., Allen R.H., Stabler S.P. and Roede J.R. (2018) Taurine treatment prevents derangement of the hepatic gamma-glutamyl cycle and methylglyoxal metabolism in a mouse model of classical homocystinuria: regulatory crosstalk between thiol and sulfinic acid metabolism. FASEB J., 32, 1265–1280. [DOI] [PubMed] [Google Scholar]
- 53. Fernandez-Roig S., Lai S.C., Murphy M.M., Fernandez-Ballart J. and Quadros E.V. (2012) Vitamin B12 deficiency in the brain leads to DNA hypomethylation in the TCblR/CD320 knockout mouse. Nutr. Metab. (Lond.), 9, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Z., Karaplis A.C., Ackerman S.L., Pogribny I.P., Melnyk S., Lussier-Cacan S., Chen M.F., Pai A., John S.W., Smith R.S. et al. (2001) Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum. Mol. Genet., 10, 433–443. [DOI] [PubMed] [Google Scholar]
- 55. Huemer M., Diodato D., Schwahn B., Schiff M., Bandeira A., Benoist J.F., Burlina A., Cerone R., Couce M.L., Garcia-Cazorla A. et al. (2017) Guidelines for diagnosis and management of the cobalamin-related remethylation disorders cblC, cblD, cblE, cblF, cblG, cblJ and MTHFR deficiency. J. Inherit. Metab. Dis., 40, 21–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andersson H.C. and Shapira E. (1998) Biochemical and clinical response to hydroxocobalamin versus cyanocobalamin treatment in patients with methylmalonic acidemia and homocystinuria (cblC). J. Pediatr., 132, 121–124. [DOI] [PubMed] [Google Scholar]
- 57. Li Z., Gherasim C., Lesniak N.A. and Banerjee R. (2014) Glutathione-dependent one-electron transfer reactions catalyzed by a B(1)(2) trafficking protein. J. Biol. Chem., 289, 16487–16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ribes A., Briones P., Vilaseca M.A., Lluch M., Rodes M., Maya A., Campistol J., Pascual P., Suormala T. and Baumgartner R. (1990) Methylmalonic aciduria with homocystinuria: biochemical studies, treatment, and clinical course of a Cbl-C patient. Eur. J. Pediatr., 149, 412–415. [DOI] [PubMed] [Google Scholar]
- 59. Linnell J.C., Miranda B., Bhatt H.R., Dowton S.B. and Levy H.L. (1983) Abnormal Cobalamin metabolism in a Megaloblastic child with homocystinuria, cystathioninuria and methylmalonic aciduria. J. Inherit. Metab. Dis., 6, 137–139.6422152 [Google Scholar]
- 60. Smith S.E., Kinney H.C., Swoboda K.J. and Levy H.L. (2006) Subacute combined degeneration of the spinal cord in cblC disorder despite treatment with B12. Mol. Genet. Metab., 88, 138–145. [DOI] [PubMed] [Google Scholar]
- 61. Weisfeld-Adams J.D., McCourt E.A., Diaz G.A. and Oliver S.C. (2015) Ocular disease in the cobalamin C defect: a review of the literature and a suggested framework for clinical surveillance. Mol. Genet. Metab., 114:537–546. [DOI] [PubMed] [Google Scholar]
- 62. Chen X., Sheng X., Zhuang W., Sun X., Liu G., Shi X., Huang G., Mei Y., Li Y., Pan X. et al. (2017) GUCA1A mutation causes maculopathy in a five-generation family with a wide spectrum of severity. Genet. Med., 19, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang L. and Baehr W. (2010) GCAP1 mutations associated with autosomal dominant cone dystrophy. Adv. Exp. Med. Biol., 664, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cascella R., Strafella C., Caputo V., Errichiello V., Zampatti S., Milano F., Potenza S., Mauriello S., Novelli G., Ricci F. et al. (2018) Towards the application of precision medicine in age-related macular degeneration. Prog. Retin. Eye Res., 63, 132–146. [DOI] [PubMed] [Google Scholar]
- 65. Ghemrawi R., Pooya S., Lorentz S., Gauchotte G., Arnold C., Gueant J.L. and Battaglia-Hsu S.F. (2013) Decreased vitamin B12 availability induces ER stress through impaired SIRT1-deacetylation of HSF1. Cell Death Dis., 4, e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Westerfield M. (2000) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). University of Oregon Press, Eugene. [Google Scholar]
- 67. Sood R., Carrington B., Bishop K., Jones M., Rissone A., Candotti F., Chandrasekharappa S.C. and Liu P. (2013) Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS One, 8, e57239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Varshney G.K., Carrington B., Pei W., Bishop K., Chen Z., Fan C., Xu L., Jones M., LaFave M.C., Ledin J. et al. (2016) A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc., 11, 2357–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schneider C.A., Rasband W.S. and Eliceiri K.W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barthel L.K. and Raymond P.A. (1990) Improved method for obtaining 3-microns cryosections for immunocytochemistry. J. Histochem. Cytochem., 38, 1383–1388. [DOI] [PubMed] [Google Scholar]
- 71. Lindemans J., Kapel J. and Abels J. (1979) Preparation of vitamin B-12-free serum for the use in vitamin B-12 radioassays. Clin. Chim. Acta, 95, 29–33. [DOI] [PubMed] [Google Scholar]
- 72. Marcell P.D., Stabler S.P., Podell E.R. and Allen R.H. (1985) Quantitation of methylmalonic acid and other dicarboxylic acids in normal serum and urine using capillary gas chromatography-mass spectrometry. Anal. Biochem., 150, 58–66. [DOI] [PubMed] [Google Scholar]
- 73. Quintana A.M., Geiger E.A., Achilly N., Rosenblatt D.S., Maclean K.N., Stabler S.P., Artinger K.B., Appel B. and Shaikh T.H. (2014) Hcfc1b, a zebrafish ortholog of HCFC1, regulates craniofacial development by modulating mmachc expression. Dev. Biol., 396, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gueant J.L., Chery C., Oussalah A., Nadaf J., Coelho D., Josse T., Flayac J., Robert A., Koscinski I., Gastin I. et al. (2018) A PRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat. Commun., 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee M.S., Bonner J.R., Bernard D.J., Sanchez E.L., Sause E.T., Prentice R.R., Burgess S.M. and Brody L.C. (2012) Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol., 12, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


