Abstract
MicroRNAs (miRNAs) are non-coding small RNAs that can function as gene regulators and are involved in tumorigenesis. We review the commonly dysregulated miRNAs in liver tumor tissues and plasma/serum of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) patients. The frequently reported up-regulated miRNAs in liver tumor tissues include miR-18a, miR-21, miR-221, miR-222, and miR-224, whereas down-regulated miRNAs include miR-26a, miR-101, miR-122, miR-125b, miR-145, miR-199a, miR-199b, miR-200a, and miR-223. For a subset of these miRNAs (up-regulated miR-222 and miR-224, down-regulated miR-26a and miR-125b), the pattern of dysregulated circulating miRNAs in plasma/serum is mirrored in tumor tissue based on multiple independent studies. Dysregulated miRNAs target oncogenes or tumor suppressor genes involved in hepatocarcinogenesis. Normalization of dysregulated miRNAs by up- or down-regulation has been shown to inhibit HCC cell proliferation or sensitize liver cancer cells to chemotherapeutic treatment. miRNAs hold as yet unrealized potential as biomarkers for early detection of HCC and as precision therapeutic targets, but further studies in diverse populations and across all stages of HCC are needed.
Keywords: biomarkers, hepatitis B virus, hepatocellular carcinoma, microRNA, gene expression, early diagnosis, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and deadly cancers in the world (1, 2). Major risk factors for HCC are chronic infection by hepatitis B virus (HBV) or hepatitis C virus (HCV) (3). HCC is usually diagnosed at the late stages, due to the low sensitivity of the current diagnostic methods, which include imaging and quantification of alpha-fetoprotein (AFP) levels. Although recent advances in genomic technology have identified a variety of genetic alterations in HCC tissues, convenient biomarkers with sufficient sensitivity and specificity for early diagnosis of HCC are still lacking.
Detection of microRNAs (miRNAs) has recently gained increasing attention for their potential utility in the early diagnosis of HCC. miRNAs are one of the major post-transcriptional regulators of gene expression. As non-coding small endogenous RNAs with ~22 nucleotides, miRNAs silence genes by binding to the 3' untranslated region (3' UTR) of messenger RNAs (mRNAs) and triggering mRNA degradation or translational repression (4–6). To date, more than 2,600 mature human miRNAs have been listed on the miRbase database (http://www.mirbase.org). Each miRNA can target multiple mRNAs with varying effects and a single mRNA may be targeted by multiple miRNAs. miRNAs modulate various biological molecular pathways and cellular processes, including cell proliferation, differentiation, development, apoptosis, angiogenesis, metabolism, and immune responses (7–10). Dysregulated miRNAs have been implicated in the development of a variety of tumors, including HCC, and may serve as robust biomarkers for cancer diagnosis and prognosis (11–14).
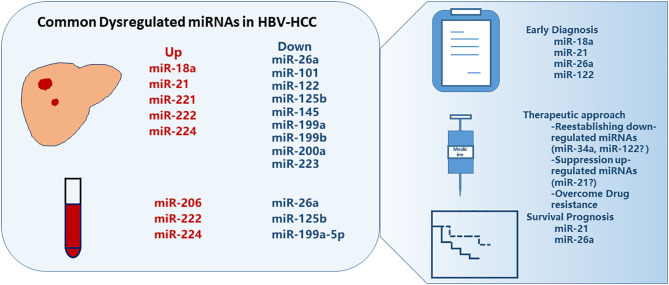
Given that miRNAs expression levels might differ among HCC patients with different etiological factors (15) and that HBV is the predominant risk factor for HCC (16), the present review focuses on miRNAs involved with HBV-related HCC (HBV-HCC). We have assessed patterns of reported dysregulated miRNAs in the HBV-HCC patients and present the mechanisms and potential applications of miRNAs in the diagnosis, prognosis, and treatment of HBV-HCC (Figure 1).
Figure 1.
Commonly dysregulated microRNAs in HBV-related HCC. Several miRNAs are up- or down-regulated in liver tumor tissues or in plasma/serum, some of which showed promise for early diagnosis and survival prognosis of HCC, and can be manipulated for treatment.
Dysregulated miRNAs IN HBV-HCC
Comparisons of HBV-HCC tumor tissue to either matched non-tumor tissue or liver tissue from healthy controls indicate that a subset of miRNAs is differentially expressed between health and tumor tissues. In Table 1, we list miRNAs that have been replicated in at least two HBV-HCC studies. Commonly reported up-regulated miRNAs include miR-18a, miR-21, miR-221, miR-222, and miR-224, whereas down-regulated miRNAs include miR-26a, miR-101, miR-122, miR-125b, miR-145, miR-199a, miR-199b, miR-200a, and miR-223 (17–36).
Table 1.
Dysregulated microRNAs in the tissue of predominantly HBV-related HCC.
| miRNAs |
Dysregulation type |
Fold change | Case vs. control | Samples details | References | |
|---|---|---|---|---|---|---|
| Size and HBV status | Underlying cirrhosis % (n) | |||||
| miR-18a | Up-regulated | 0.585a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) |
| Up-regulated | 3.223b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) | |
| miR-21 | Up-regulated | 2.29a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| Up-regulated | 3.67b | HCC vs. ANT | 115 HCC (101 HBV) | 51% (59/115) | (20) | |
| Up-regulated | NA | HCC vs. ANT | 148 HCC (82 HBV) | 41% (45/109) | (21) | |
| Up-regulated | NA | HCC vs. ANT | 31 HBV-HCC | NA | (22) | |
| Up-regulated | NA | HCC vs. ANT | 24 HBV-DNs, 29 small HBV-HCC nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) | |
| Up-regulated | 3.72b | HCC vs. ANT | 42 HBV-HCC | NA | (24) | |
| miR-221 | Up-regulated | 1.51a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| Up-regulated | NA | HCC vs. ANT | 135 HCC (96 HBV) | 95% (128/135) | (25) | |
| Up-regulated | 4.00b | HCC vs. ANT | 115 HCC (101 HBV) | 51% (59/115) | (20) | |
| Up-regulated | NA | HCC vs. ANT | 31 HBV-HCC | NA | (22) | |
| Up-regulated | NA | HCC vs. ANT | 24 HBV-DNs, 29 small HBV-HCC Nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) | |
| Up-regulated | 1.57a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) | |
| miR-222 | Up-regulated | 1.41a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) |
| Up-regulated | 4.44b | HCC vs. ANT | 115 HCC (101 HBV) | 51% (59/115) | (20) | |
| Up-regulated | 4.964b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) | |
| Up-regulated | NA | HCC vs. ANT | 42 HCC (33 HBV), 6 HCV, 3 NBNC-HCC | 85% (28/33) | (26) | |
| miR-224 | Up-regulated | NA | HCC vs. ANT | 24 HBV-DNs, 29 small HBV-HCC Nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) |
| Up-regulated | 27.231b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) | |
| Up-regulated | 0.903a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) | |
| miR-26a | Down-regulated | 0.37 b | HCC vs. ANT | 455 HCC (412 HBV) | 88% (400/455) | (27) |
| Down-regulated | −1.59a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) | |
| miR-101 | Down-regulated | NA | HCC vs. ANT | 25 HCC (20 HBV), 20 HC (HBV negative) | 72% (18/25) | (28) |
| Down-regulated | NA | HCC vs. HC HCC vs. CHB HCC vs. LC |
67 HBV-HCC, 61 HBV-LC, 79 CHB, 30 Normal control | NA | (29) | |
| Down-regulated | 0.214b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) | |
| Down-regulated | −0.958a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) | |
| miR-122 | Down-regulated | −1.67a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| Down-regulated | NA | HCC vs. ANT | 97 HCC (84 HBV) | NA | (30) | |
| Down-regulated | NA | HCC vs. ANT, HBV-HCC vs. non-HBV-HCC |
142 HCC (103 HBV) | 58% (82/142) | (31) | |
| Down-regulated | NA | HCC vs. ANT | 24 HBV-DNs, 29 small HBV-HCC Nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) | |
| Down-regulated | 0.60b | Venous metastases vs. solitary tumors | 214 HBV-HCC | 93% (199/214) | (32) | |
| miR-125b | Down-regulated | −0.893a | HCC vs. ANT | 78 HCC (62 HBV) | 51% (40/78) | (17) |
| Down-regulated | NA | HCC vs. ANT | 97 HCC (84 HBV) | NA | (30) | |
| Down-regulated | 0.58b | Venous metastases vs. solitary tumors | 214 HBV-HCC | 93% (199/214) | (32) | |
| miR-145 | Down-regulated | NA | HCC vs. ANT LGDN vs. ANT HGDN vs. ANT |
24 HBV-DNs, 29 small HBV-HCC Nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) |
| 0.28b | HCC vs. ANT | 42 HBV-HCC | NA | (24) | ||
| miR-145-5P | Down-Regulated | −2.39a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| miR-199a | Down-regulated | 0.149b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) |
| Down-regulated | NA | HCC vs. ANT | 97 HCC (84 HBV) | NA | (30) | |
| miR-199a-5P | Down-regulated | −4.51a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| miR-199a-3P | Down-regulated | −2.78a | HCC vs. ANT | 100 HCC (58 HBV, 8 HCV, 27 NBNC, 27 Unknown) | 46% (46/100) | (19) |
| miR-199b | Down-regulated | NA | HCC vs. ANT LGDN vs. ANT HGDN vs. ANT |
24 HBV-DNs, 29 small HBV-HCC Nodules, 38 HBV-ANTs | 92% (22/24) in DNs 93% (27/29) in HCC | (23) |
| miR-200a | Down-regulated | NA | HCC vs. ANT | 120 HCC (97 HBV) | 78% (93/120) | (33) |
| Down-regulated | 0.421b | HCC vs. ANT | 101 HCC (71 HBV) | NA | (34) | |
| Down-regulated | 0.522b | HCC vs. ANT | 95 HCC (78 HBV) | 47% (45/95) | (35) | |
| miR-223 | Down-regulated | −1.92a | HCC vs. ANT | 100 HCC (58 HBV) | 46% (46/100) | (19) |
| Down-regulated | 0.267b | HCC vs. ANT | 22 HCC (20 HBV) | NA | (18) | |
| Down-regulated | 0.20b | HCC vs. ANT | 42 HCC (33 HBV), 6 HCV, 3 NBNC-HCC | 85% (28/33) | (26) | |
Fold changes were based on the original report;
Log2 fold change.
Regular fold change.
ANT, adjacent non-cancerous tissue; CHB, chronic hepatitis B; DN, dysplastic nodules; HBV, hepatitis B virus; HC, healthy control; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HGDN, high-grade dysplastic nodule; LC, liver cirrhosis; LGDN, low-grade dysplastic nodule; NA, not available; NBNC, non-HBV non-HCV.
Due to limited liver tissue accessibility and the invasive nature of biopsy, studies assessing circulating miRNAs in plasma or serum from patients with HBV-HCC have increased dramatically in recent years. Cellular miRNAs from tumors leak into the circulation system following cell injury, apoptosis, and necrosis or by secretion through cell-derived exosomes and shedding vesicles (37). Circulating miRNAs in serum or plasma are stable (38), suggesting that circulating miRNAs may be accessible and quantifiable cancer diagnostic or prognostic biomarkers. Commonly reported dysregulated circulating miRNAs from patients with HBV-HCC include miR-21, miR-26, miR-122, miR-125b, miR-192, miR-206, miR-222, miR-223, and miR-224 (28, 29, 39–46) (Table 2).
Table 2.
Dysregulated microRNAs in the plasma/serum of patients with HBV-related HCC.
| miRNAs | Dysregulation type | Fold change | Case vs. control | Samples details | References |
|---|---|---|---|---|---|
| miR-18a | Up-regulated | NA | HCC vs. HC, HCC vs. (CHB + LC) | 101 HBV-HCC, 30 CHB or HBV-LC, 60 HC | (44) |
| miR-192 | Up-regulated | 1.4b | HCC vs. (LC+CHB+HC) | 457 HBV-HCC, 141 HBV-LC, 169 CHB, 167 HC | (45) |
| miR-206 | Up-regulated | 9.94b | HCC vs. HC | 261 HBV-HCC, 173 HC | (46) |
| Up-regulated | 3.51b | HCC vs. LC | 261 HBV-HCC, 233 HBV-LC | (46) | |
| Up-regulated | 2.98 ± 3.94b | HCC vs. matched control | 55 HBV-HCC, 50 age and gender-matched control | (39) | |
| miR-221 | Up-regulated | 4.83b | HCC vs. HC | 46 HCC (30 HBV), 20 HC | (41) |
| miR-222 | Up-regulated | NA | HCC vs. HC | 70 HBV-HCC, 48 CHB, 34 HC | (43) |
| Up-regulated | 3.01b | HCC vs. HC | 46 HCC (30 HBV), 20 HC | (41) | |
| miR-224 | Up-regulated | 1.88b | HCC vs. HC | 46 HCC (30 HBV), 20 HC | (41) |
| miR-21 | Up-regulated | 1.9b | HCC vs. (LC+CHB+HC) | 457 HBV-HCC, 141 HBV-LC, 169 CHB, 167 HC | (45) |
| Up-regulated | NA | HCC vs. HC | 97 HCC (60 HBV), 30 HC | (47) | |
| Up-regulated | 2.85b | HCC vs. HC | 46 HCC (30 HBV), 20 HC | (41) | |
| Up-regulated | NA | HCC vs. HC | 101 HCC (76 HBV), 48 CHB, 89 HC | (42) | |
| Down-regulated | NA | HCC vs. CHB | 101 HCC (76 HBV), 48 CHB, 89 HC | (42) | |
| miR-122 | Up-regulated | 4.09 ± 5.38 b | HCC vs. HBV (ASC +CHB) | 65 HBV-HCC, 160 controls | (39) |
| Up-regulated | NA | HCC vs. HC | 70 HBV-HCC, 48 CHB, 34 HC | (43) | |
| Up-regulated | NA | HCC vs. HC | 101 HCC (76 HBV), 48 CHB, 89 HC | (42) | |
| Down-regulated | 0.7b | HCC vs. (LC+CHB+HC) | 457 HBV-HCC, 141 HBV-LC, 169 CHB, 167 HC | (45) | |
| Down-regulated | NA | HCC vs. CHB | 101 HCC (76 HBV), 48 CHB, 89 HC | (42) | |
| miR-192-5p | Up-regulated | 1.71b | HCC vs. HC | 212 HBV-HCC, 110 HC | (48) |
| Up-regulated | 1.97b | HCC vs. LC | 212 HBV-HCC, 106 HBV-LC | (48) | |
| Down-regulated | 0.85b | HCC vs. LC | 261 HBV-HCC, 233 HBV-LC | (46) | |
| Down-regulated | 0.77b | HCC vs. HC | 261 HBV-HCC, 173 HC | (46) | |
| miR-223 | Up-regulated | NA | HCC vs. HC | 101 HCC (76 HBV), 89 HC | (42) |
| Up-regulated | 2.97 ± 1.67 b | HCC vs. HC | 65 HBV-HCC, 160 controls | (39) | |
| Up-regulated | NA | HCC vs. HC | 70 HBV-HCC, 34 HC | (43) | |
| No difference | NA | HCC vs. CHB | 101 HCC (76 HBV), 48 CHB | (42) | |
| Down-regulated | 0.3b | HCC vs. (LC+CHB+HC) | 457 HBV-HCC, 141 HBV-LC, 169 CHB, 167 HC | (45) | |
| Down-regulated | NA | HCC vs. HBV (ASC +CHB) | 65 HBV-HCC, 135 HBV (55 ASC+ 80 CHB) | (39) | |
| miR-26a | Down-regulated | 0.2b | HCC vs. (LC+CHB+HC) | 457 HBV-HCC, 141 HBV-LC, 169 CHB, 167 HC | (45) |
| miR-26a-5p | Down-regulated | 0.65b | HCC vs. HC | 261 HBV-HCC, 173 HC | (46) |
| Down-regulated | 0.54b | HCC vs. LC | 261 HBV-HCC, 233 HBV-LC | (46) | |
| miR-122-5p | Down-regulated | 0.27b | HCC vs. HC | 261 HBV-HCC, 173 HC | (46) |
| Down-regulated | 0.54b | HCC vs. LC | 261 HBV-HCC, 233 HBV-LC | (46) | |
| miR-125b | Down-regulated | 0.26 ± 0.46b | HCC vs. HBV (ASC +CHB) | 65 HBV-HCC, 135 HBV (55 ASC+ 80 CHB) | (39) |
| Down-regulated | NA | HCC vs. LC | 30 HCC (28 HBV), 30 LC (27 HBV) | (40) | |
| Down-regulated | NA | HCC vs. CHB | 30 HCC (28 HBV), 30 CHB | (40) | |
| miR-199a-5p | Down-regulated | 0.58b | HCC vs. HC | 261 HBV-HCC, 173 HC | (46) |
| Down-regulated | 0.87b | HCC vs. LC | 261 HBV-HCC, 233 HBV-LC | (46) |
Fold changes were based on the original report;
Log2 fold change.
Regular fold change.
ASC, asymptomatic carrier; CHB, chronic hepatitis B; HBV, Hepatitis B virus; HBV-HCC, HBV-related HCC; HBV-LC, HBV-related LC; HC, healthy control; HCC, hepatocellular carcinoma; LC, liver cirrhosis; NA, not available.
For a subset of the miRNAs [e.g., up-regulated miR-18a, miR-221, miR-222, miR-224, and down-regulated miR-26a and miR-125b (Table 3)], dysregulated patterns were consistent among multiple independent studies and between tumor tissue and serum/plasma. These microRNAs may be of more translational value in the diagnosis, differential diagnosis or even therapy for HBV-HCC. However, other miRNAs showed inconsistent or contrasting profiles of dysregulation among studies or between tumor tissue and serum/plasma (Tables 1, 2). For example, downregulation of miR-122 was common in HCC tissue (19, 23, 32), but circulating miRNA levels were upregulated in some studies (39, 42, 43) and downregulated in others (45). Based on the observation that increased serum miR-122 is presented in both HCC patients and chronic hepatitis patients, some researchers speculate that higher levels of miR-122 in serum may result from liver injury rather than HCC itself (42, 43). It is also likely that factors governing the expression of miRNAs in the tissues and sera of HCC patients might differ. Additional factors that may contribute to discordant findings among these results include differences in patient selection, tumor stage, biological sample handling, and storage, miRNA probes employed, sample size, or genetic background of study populations (49).
Table 3.
Common consistently dysregulated microRNAs between tumor tissue and serum/plasma in HBV- HCC.
Mechanism of miRNA Dysregulation IN HCC
It's not fully understood if miRNA dysregulation in HCC is the cause, consequence of HCC development or both. Accumulating evidence indicates that some dysregulated miRNAs are active players in tumor initiation and progression. The direct targets of miRNAs may be protein-coding genes involved in any or all pathophysiological mechanisms of cancer development, including cell growth, apoptosis, invasion, and metastasis. miRNAs may function as either tumor promoters or tumor suppressors depending on their target genes (50). miRNAs in HCC that target and suppress oncogenes may be down-regulated, while miRNAs that target suppressor genes may be up-regulated during tumor development (Figure 2). The miR-122 expression is largely liver-specific and under transcriptional control by the liver-enriched transcription factors HNF1A, HNF3A, and HNF3B (51). miR-122 can function as a tumor suppressor by suppressing HCC growth, invasion, migration, angiogenesis and by increasing HCC apoptosis and cell cycle arrest (52). miRNA-122 targets multiple genes, including BCL9, Bcl-w, NDRG3, cyclin G1, ADAM17, ADAM10, G6PD, and pituitary tumor-transforming gene 1 (PTTG1) binding factor (PBF), all of which have been implicated in tumor development (53–60). Other miRNAs such as miRNA-21 function as oncogenes by stimulating HCC growth, invasion, and migration (23, 61, 62). The inhibition of miR-21 suppresses HCC tumor growth (63).
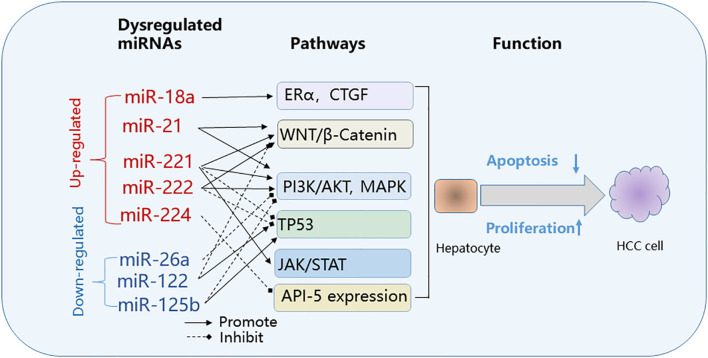
Figure 2.
Common pathways targeted by dysregulated microRNAs in HBV-related HCC. Effect on cancer pathways by dysregulated miRNAs: upregulated (red); down-regulated (blue). API-5, apoptosis inhibitor-5; CTGF, connective tissue growth factor; ERα, estrogen receptorα; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; JAK/STAT, Janus kinase/signal transducer; PI3K/MAPK, phosphoinositide 3-kinase/mitogen-activated protein kinase; WNT/β-Catenin, wingless-related integration site/beta-catenin.
Dysregulated miRNAs affect key cellular pathways that play a role in the pathogenesis of HBV-HCC (Figure 2). The commonly targeted pathways by dysregulated miRNAs in HBV-HCC include the Janus kinase/signal transducer (JAK/STAT), phosphoinositide 3-kinase/mitogen-activated protein kinase (PI3K)/AKT and MAPK, Wingless-related integration site/beta-catenin (WNT/β-Catenin) and TP-53 pathways (40, 64–73).
Interaction of miRNAs and HBV in HBV-HCC
HBV can directly regulate cellular miRNAs levels. miR-122 is targeted and inhibited by HBV mRNA, which harbors a miR-122 complementary site, leading to the upregulation of the PTTG1-binding protein and promotion of HCC tumor growth and cell invasion (57). Down-regulation of miR-122 occurs mainly in HBV-HCCs but not in HCV-infected HCCs (74). HBV downregulates miR-101 expression by directly inhibiting its promoter activity (75). Hepatitis B X antigen (HBx) increases the expression of miR-21 and subsequently promotes the progression of HCC by targeting PTEN and the tumor suppressor PDCD4 (61). HBx suppresses p53-mediated activation of miR-148a thereby promoting tumor growth and metastasis; expression of miR-148a reduced tumor growth and invasion. In patients with HBV-HCC, miR-148a was down-regulated. These results suggest that activation of miRNA-148a or down-regulation of its targeted pathways may have a role in HCC treatment (76).
In contrast, cellular miRNAs, including miR-122, and miR-125 and miR-199 family members, affect HBV replication (77). miR-125a-5p, markedly downregulated in HCC, inhibits HBsAg expression and secretion (78).
Using RNA deep sequencing and northern blotting, HBV-encoded miRNAs were recently identified. HBV-miR-3 was shown to restrict HBV replication, by targeting the region of HBV 3.5-kb mRNA encoding HBV core antigen (HBc) (79). Another HBV-encoded miRNA, HBV-miR-2, can promote the oncogenic activity of liver cancer cells (80). HBV-encoded miRNAs likely contribute to HBV-specific HCC development.
The complex interactions and molecular interactions among cellular miRNAs and HBV have been reviewed in (73, 81–83).
Dysregulated miRNAs in Liver Cancer Stem Cells (LCSCs)
Cancer stem cells are small subpopulations of tumor-initiating cells within tumors that capable of self-renewal, differentiation, and proliferation. LCSCs can be identified by several highly expressed stem cell surface markers including epithelial cell adhesion molecule (EpCAM), CD90, CD44, CD133, and CD13 (84, 85). The other reported LCSC surface markers include OV6, DLK1, ABCG2, ALDH, and CD24 (84–87). LCSCs are responsible for tumor initiation, metastasis, relapse, and chemo- and radiation-therapy resistance in liver cancer (87). The specific influence of HBV on LCSCs remains largely unknown. Liver inflammatory damage induced by chronic HBV and HCV infection and liver toxins can induce somatic mutations, genomic instability, and epigenetic perturbations, resulting in the deregulation of self-renewal and differentiation signaling pathways of activated liver progenitor cells, which promotes the transformation of liver progenitor cells into LCSCs (84). It has been reported that HBx promotes the stem-like properties of OV6+ CSCs in HBV-related HCC via MDM2 independent of p53 (88). Concomitant elevated expression of HBx and OV6 predicts a poor prognosis for patients with HBV- HCC (88).
Multiple miRNAs have been reported to regulate a variety of biological behaviors of LCSCs, including let-7, miR-200, miR-122, miR-181, miR-1246, miR-152, miR-145, miR-217, miR-500a-3p, and miR-148 (87). miRNAs affect the CSC phenotype by regulating the expression of oncogenes and stem cell-related genes (85). These miRNAs target key molecules in the following pathways involved in carcinogenesis: Wnt/beta-catenin signaling, TGF-beta signaling, JAK/STAT signaling, epithelial-mesenchymal transition (EMT) in LCSCs (87). miRNA profiling comparisons between CSC+ and CSC− HCCs, as separated by hepatic CSC biomarkers (EpCAM, CD133, CD90, CD44, and CD24), identified aberrant downregulation of liver-specific miR-192-5p in HCC cells, which correlated with increased CSC populations with stemness features and shorter survival in HCC patients (89). Over-expression of miR-192-5p inhibited the stemness features of human liver cancer cell lines, with decreased spheroid formation, decreased CSC number and diversity and decreased expression of CSC biomarkers and increased expression of genes related to hepatocyte metabolism (89). Hepatitis B virus X protein (HBx) induces expression of EpCAM by upregulating miR-181 to promote stemness in hepatocarcinogenesis (90, 91). The knockdown of miR-181 significantly reduces the EpCAM+ LCSCs and tumor-initiating ability (92).
Targeting the regulation of these miRNAs or their pathways may serve as a potential therapeutic strategy to inhibit or eradicate LCSCs (87). Restoring of miR-122 has been demonstrated to suppresses stem-like HCC cells (93). It would be interesting to explore the clinical utility of restoring the miR-192-5p for riding of LCSCs (89).
Epigenetic Alterations and miRNAs in HCC
Epigenetic alterations such as DNA methylation and histone modification are essential for chromatin remodeling and regulation of both coding genes and miRNAs. Abnormal DNA methylation patterns of a number of miRNAs in HCC have been reported for hypermethylation of miR-1, miR-9, miR-10a, miR-10b, miR-124, miR-125b, miR-132, miR-148a, miR-195, miR-196b, miR-203, miR-320, miR-375, miR-378, miR-497, miR-596, miR-663, and miR-1247, and for hypomethylation of miR-23a, miR-25, miR-27a, miR-93, and miR-106b (94). Among these miRNAs, only miR-125b presents consistent dysregulation pattern of expression, and was down-regulated both in tissue (17, 30, 32) and serum (39, 40) of patients with HCC. The expression of miR-125b was significantly increased by the methylation inhibitor 5-aza-2'-deoxycytidine in HCC cells, suggesting the epigenetically modulation of the expression of miR-125b (95).
Histone modifications, including acetylation, methylation, and phosphorylation of lysine residues, play an important role in expression regulation of genes including miRNAs in HCC tumor tissue. For example, levels of hsa-miR-449a in HCC cell lines was enhanced significantly by inhibiting histone deacetylases (HDACs), which were up-regulated in HCC tissue (96). Reduced expression of miR-199a/b-3p, one of the consistently and markedly decreased miRNA in HCC, is mediated by histone methylation and independent of DNA methylation (97). On the other hand, some miRNAs have been reported to be involved in hepatocarcinogenesis by regulating histone deacetylases (HDACs), including miR-1, miR-22, and miR-200a targeting HDAC4, miR-31 and miR-145 targeting HDAC2, miR-221 targeting HDAC6, miR-29c targeting SIRT1, miR-125a-5p, and miR-125b targeting SIRT7, suggesting the potential use of miRNA-based therapies in HCC (98).
Chromatin modifiers or remodelers regulate accessibility to chromatin and positioning of nucleosome in the DNA. Upregulated enhancer of zeste homolog 2 (EZH2) in HCC, a well-studied chromatin modifier which mediates gene silencing in HCC, represses miR-622 by enhancing H3K27 trimethylation, and is correlated with unfavorable HCC prognosis (32). CCCTC-binding factor (CTCF) is a highly conserved insulator-binding protein with an enhancer-blocking function and contributes to the epigenetic regulation of some miRNAs (99). In breast cancer cells, disruption of CTCF binding at miR-125b1 CpG island (CGI) is associated with CGI methylation and the gain of the repressive histone marks including H3K9me3 and H3K27me3, and induces silencing of miR-125b1 expression (100). Considering the miR-125b is consistently down-regulated in HCC tissue (17, 30, 32) and serum (39, 40), disruption of CTCF binding might modulate HCC development.
Circular RNAs (circRNAs) are a class of highly conserved, stable and abundant non-coding RNAs (ncRNAs) that can regulate gene expression at transcriptional or post-transcriptional levels. The majority of circRNAs function as sponges of miRNA (101) and deregulation of a number of circRNAs have been reported in HCC. For example, circHIPK3 can sponge 9 miRNAs with 18 potential binding sites, including directly binding to the well-known tumor suppressor miR-124, reducing its activity (102). circTRIM33–12 acts as the sponge of miR-191 to suppress HCC (103). Artificial circRNAs which bind and sponge specific miRNAs can be constructed to achieve better inhibitory effects on oncogenic or pathogenic miRNAs, indicating a promising strategy to treat HCC.
Regulating miRNA as a Therapeutic Approach for HCC
Normalization of dysregulated miRNAs in patients with HBV-HCC, by either up- or down-regulation of dysregulated miRNAs, is a plausible therapeutic approach in treating HCC.
Preliminary studies suggest that reestablishing the expression of down-regulated miRNAs might restore the tumor-suppressing function of miRNAs. In a first-in-human Phase 1 trial of a miRNA therapy using a liposomal miR-34a mimic in patients with advanced solid tumors including HBV-HCC, the miR-34a mimic showed antitumor activity (104). In another study upregulation of miR-122, which is frequently down-regulated in HCC patients, suppressed the proliferation and invasion capability of HCC-derived cells and increased sensitivity to chemotherapy (31, 105–107). Restoring miR-122 in stem-like HCC cells was shown to decrease cell proliferation and reduce tumor size in a mouse model (93). Besides miR-122, other miRNAs may have value in treating HCC. A recent study showed that injection of exosomal miR-335-5p, a tumor suppressor, can inhibit HCC cell proliferation and invasion as well as result in slower cancer growth (108). On the other hand, suppression of miR-21, which is frequently up-regulated in patients with HCC, leads to increased sensitivity to chemotherapeutic drugs (21).
In addition to direct targeting of miRNA, modulating the upstream genes that control miRNA expression is another therapeutic strategy. Upregulation of miR-122 by activating the farnesoid X receptor transcription factor (FXR), suppressed the proliferation of HCC cells in vitro and reduced the growth of HCC xenografts in vivo (109).
The crosstalk between epigenetics and miRNA related to HCC provides new opportunities for the development of more effective therapy for HCC by targeting epigenetic modulation of miRNAs as discussed above. Restoring the expression of tumor suppressor miRNA by inhibitors of DNA methylation and histone deacetylase, and inhibiting the expression of oncogenes by artificial circRNAs sponging specific miRNAs may be promising therapeutic strategies for HCC.
Regulating miRNA-mediated immune response in HCC may prove to be a promising therapeutic strategy. Most recently, Tian's group demonstrated that HBV mediates PD-L1-induced T cell immune exhaustion through the interaction of the oncofetal gene SALL4 and miR-200c (110). They showed that miR-200c controls PD-L1 expression by directly targeting the 3′-UTR of PD-L1 and that overexpression of miR-200c antagonizes HBV-mediated PD-L1 expression and reverses antiviral CD8+ T cell exhaustion.
A group of miRNAs are involved either directly or indirectly in drug resistance and either suppressing or activating miRNAs may reduce drug resistance. For example, a recent study reported that some miRNAs contribute to drug resistance to sorafenib. Targeting these miRNAs by the artificial long non-coding RNA improved treatment response in patients with HCC (111). Other studies found that restoration of miR-122 can sensitize HCC cancer cells to adriamycin and vincristine (112) as well as reverse doxorubicin-resistance in HCC cells (113). MiR-101 was shown to sensitize liver cancer cells to chemotherapeutic treatment (114).
The risk of undesirable effects of miRNA targeting, due in large part to off-target binding, is challenging. Adverse events were common in the miR-34a mimic trial, the first clinical trial for the treatment of HBV-HCC (104) and the trial was recently terminated due to immune-related serious adverse events (115). Of the clinical trials using miRNAs that are dysregulated in HBV-HCC, one phase II trial of miR-122 as a treatment modality for HCV has been completed and a miR-21 phase II trial for Alports syndrome was suspended (115). The application of miRNA-targeting therapy has strong potential in personalized medicine, although off-target effects remains a significant hurdle.
miRNAs as Biomarkers in HBV-HCC
Early diagnosis of HCC, crucial for treatment outcome, remains challenging. The limitations of imaging technology and AFP detection to diagnose small and atypical HCC calls for more sensitive and specific biomarkers. Based on reports that many miRNAs are expressed differentially in HBV-HCC patients (Tables 1, 2) and miRNAs dysregulation is an early event in hepatocarcinogenesis occurring in pre-malignant dysplastic nodules (23, 47), the detection of miRNAs, especially circulating miRNAs levels, is gaining increasing recognition and attention for their potential clinical utility as biomarkers in screening and early diagnosis of HBV-HCC and predicting HCC prognosis as well.
miRNAs as Diagnostic Biomarkers in HBV-HCC
Potential single miRNAs and miRNA panels that have been proposed as early diagnostic biomarkers for HBV-HCC are summarized in Table 4. Circulating miRNAs, including miR-18a, miR-21, miR-101, miR-122, miR-139, miR-223, and some miRNA panels may have diagnostic utility in distinguishing HBV-HCC patients from patients with chronic HBV infection (CHB) or liver cirrhosis (LC). Complicating the consensus and interpretation of the results of the studies (Table 4) are the differences in control groups employed [i.e., HBV-negative or HBV infected persons (CHB or LC) (22, 29, 42–46, 48, 116–118)].
Table 4.
Diagnostic value of miRNAs in HBV-related HCC.
| miRNA | Sample details | Diagnostic value | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample type | Size of case | Underlying cirrhosis, % | Control | Specificity (%) | Sensitivity (%) | AUC | CI of AUC | ||
| miR-18a | S | 101 HBV-HCC | NA | 60 HC | 75.0 | 86.1 | 0.881 | 0.829–0.933 | (44) |
| 101 HBV-HCC | NA | 30 CHB or LC | 70.0 | 77.2 | 0.775 | 0.681–0.869 | (44) | ||
| miR-21 | S | 101 HCC (76 HBV) | NA | 89 HC | 73.5 | 84 | 0.87 | 0.81–0.93 | (42) |
| 57 HBV-HCC | NA | 30 HC + 29 HBV | 71.2 | 89.5 | 0.865 | NA | (116) | ||
| miR-101 | S | 67 HBV-HCC | NA | 30 HC | 70.0 | 76.1 | 0.788 | 0.693–0.865 | (29) |
| 67 HBV-HCC | NA | 79 CHB | 62.0 | 88.1 | 0.777 | 0.701–0.842 | (29) | ||
| 67 HBV-HCC | NA | 61 HBV-LC | 90.2 | 95.5 | 0.976 | 0.931–0.995 | (29) | ||
| miR-122 | S | 70 HBV-HCC | 75% (51/68) | 34 HC | 83.3 | 81.6 | 0.869 | 0.786–0.952 | (43) |
| 70 HBV-HCC | 75% (51/68) | 48 CHB | 57.8 | 77.6 | 0.63 | 0.516–0.743 | (43) | ||
| 101 HCC (76 HBV) | NA | 89 HC | 69.1 | 70.7 | 0.79 | 0.71–0.86 | (42) | ||
| miR-139 | P | 31 HBV-HCC | NA | 31 CHB | 58.1 | 80.6 | 0.761 | 0.643–0.878 | (22) |
| miR-223 | S | 101 HCC (76 HBV) | NA | 89 HC | 80 | 76.5 | 0.86 | 0.80–0.92 | (42) |
| miR-15b and miR-130b | S | 57 HBV-HCC | NA | 30 HC + 29 HBV | 91.5 | 98.3 | 0.981 | NA | (116) |
| miR-27b-3p, miR-192-5p | S | 212 HBV-HCC | NA | 110 HC + 106 HBV- LC | 91.2 | 68.6 | 0.836 | 0.783–0.880 | (48) |
| 110 HC | 95.2 | 68.5 | 0.823 | 0.748–0.866 | |||||
| 106 HBV-LC | 79.3 | 78.5 | 0.859 | 0.804–0.906 | |||||
| miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, miR-505 | S | 153 HBV-HCC | NA | 60 HC + 68 CHB + 71 HBV-LC | 88.9 | 74.5 | 0.817 | 0.769–0.865 | (117) |
| 153 HBV-HCC | NA | 68 CHB + 71 HBV-LC | 89.9 | 74.5 | 0.822 | 0.772–0.873 | |||
| 49 HBV-HCC | NA | 48 HC + 42 inactive HBsAg carrier + | 91.1 | 85.7 | 0.884 | 0.818–0.951 | |||
| 49 HBV-HCC | NA | 42 inactive HBsAg carrier | 83.3 | 85.7 | 0.845 | 0.758–0.932 | |||
| 27 HBV-HCC | NA | 135 matched CHB | 80.0 | 70.4 | 0.752 | 0.644–0.860 | |||
| miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, miR-801 | P | 457 HBV-HCC | NA | 141 HBV-LC + 169 CHB + 167 HC | 83.5 | 81.8 | 0.888 | 0.852–0.917 | (45) |
| 167 HC | 93.9 | 83.2 | 0.941 | 0/905–0.966 | |||||
| 169 CHB | 76.4 | 79.1 | 0.842 | 0.792–0.883 | |||||
| 141 HBV-LC | 91.1 | 75 | 0.884 | 0.838–0.921 | |||||
| miR122, miR1228, miR141, miR192, miR199a, miR206, miR26a, miR433 | S | 261 HBV-HCC | NA | 173 HC + 233 HBV-LC | 76.2 | 90.3 | 0.879 | 0.842–0.941 | (46) |
| 173 HC | 83.3 | 82.8 | 0.893 | 0.849–0.94 | |||||
| 233 HBV-LC | 84.6 | 81.6 | 0.892 | 0.844–0.939 | |||||
| miR-20a-5p, miR-25-3p, miR-30a-5p, miR-92a-3p, miR-132-3p, miR-185-5p, miR-320a, miR-324-3p | P | 67 HBV-HCC | NA | 82 HBV | 64.6 | 86.6 | 0.802 | NA | (118) |
When data from training set and validation set are available, only the latter is presented. AUC, area under the curve; CHB, chronic hepatitis B; CI, confidence interval; HBV, Hepatitis B virus; HC, healthy control; HCC, hepatocellular carcinoma; LC, liver cirrhosis; NA, not available; P, plasma; S, serum.
A recent study revealed that a seven-miRNA classifier (miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505) had significantly higher sensitivity than AFP to discriminate between HCC and healthy controls, inactive HBsAg carriers, CHB patients, and HBV-cirrhosis patients. Critically this miRNA classifier was the first biomarker to diagnosis preclinical HCC, which was detected in eight of 27 HBV infected individuals 12 months before clinical diagnosis of HCC. This miRNA classifier holds promise for improving clinical outcomes by early HCC detection and curative treatment (117).
Among these miRNAs and miRNA panels, miR-122 is the most replicated miRNA biomarker in HCC, which has a sensitivity ranging from 71 to 81%, specificity from 59 to 83%, and an AUC from 0.63 to 0.87 to distinguish HBV-HCC from controls (42, 43). miR-122 is also included in two miRNA panels for HBV-HCC (45, 46). However, the diagnostic utility of miR-122 in HBV-HCC also extends to other HCCs (119).
Multiple approaches may be taken to improve the diagnostic performance of miRNA biomarkers in HBV-HCC. The type of biological sample is one of the key factors influencing sensitivity and specificity.
Exosomes are secreted by most cell types including cancer cells. Serum exosomes are highly enriched in miRNAs and exosomes can transfer miRNAs between cells, thus affecting HCC cancer proliferation, migration, metastasis, drug resistance (120). A meta-analysis published in 2019 suggested that exosomal miRNAs have superior diagnostic value in prostate cancer patients (121). With regard to diagnosis of HBV-HCC, recent studies indicate that exosomal miRNAs might also be a better choice than miRNAs from whole serum or plasma for early diagnosis. Wang et al. found that the detection of exosomal miR-21, which is enriched in exomes, had improved sensitivity over the whole serum (122). Similarly, miR-125b levels in exosomes were significantly lower than in serum from patients with HBV-HCC when compared to patients with CHB or LC, which explains, at least in part, why miR-125b levels in exosomes, but not in serum, independently predict HCC progression (40). Another study comparing HBV-HCC to CHB or LC, found a greater difference in miRNA levels in exosomes compared to whole serum (123). Combinations of miRNAs with other classic serum markers, i.e., AFP, is another approach to increase sensitivity and specificity of blood-based early detection of HBV-HCC (117, 118), especially for atypical HCC cases with lower serum AFP levels. The better performance of this add-on strategy was demonstrated in HCC cases caused by non-HBV factors as well (124).
miRNAs as Biomarkers for HBV-HCC Prognosis
Expression levels of several miRNAs in liver tissue or circulation were correlated with disease severity and survival of HBV-HCC patients. Commonly reported single miRNAs and miRNA biomarker panels in predicting the survival of HBV-HCC are summarized in Table 5. Single miRNAs and miRNA panels associated with shorter survival include miR-21, miR-221, and two 20-mer miRNA signature profiles (20, 21, 25, 32, 47, 128, 129); miR-26a, miR-26b, miR-122, miR-125b, and miR-203 were associated with longer survival (27, 31, 40, 130). Among these miRNAs, miR-21 was the most replicated with a hazard ratio (HR) ranging from 1.4 to 2.2 in predicting the long-term progression of HBV-HCC (Table 5); miR-21 was also associated with HCCs (131). Given the enrichment of miRNAs in serum exosomes, detection of serum exosomal miRNAs can be used to predict prognosis of HCC patients (40).
Table 5.
Prognostic value of miRNAs in HBV-related HCC.
| Sample | miRNA panels |
Risk/protective factor |
HR/RR | CI | P-value | Outcome | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Type | Size of case | Underlying cirrhosis, % | |||||||
| P/S | 97 HCC (60 HBV) | 32.0% (31/97) | miR-21 | Risk | 2.229 | 1.328–3.743 | 0.002 | OS | (47) |
| P/S | 136 HCC (129 HBV) | NA | miR-200a | Risk | 1.75 | 1.45–2.11 | <0.001 | OS | (125) |
| P/S | 62 HCC (40 HBV) | NA | miR-1246 | Risk | 10.24 | 1.39–75.67 | 0.023 | OS | (126) |
| P/S | 62 HCC (40 HBV) | NA | miR-1246 | Risk | 10.12 | 1.45–70.47 | 0.020 | DFS | (126) |
| P/S | 120 HBV-HCC | 85.8% (103/120) | miR-26a | Protective | 0.29 | 0.11–0.76 | 0.011 | LT-free survival | (127) |
| P/S | 120 HBV-HCC | 85.8% (103/120) | miR-29a | Protective | 0.36 | 0.15–0.91 | 0.030 | LT-free survival | (127) |
| Exosome | 128 HCC (121 HBV) | 76% (97/128) | miR-125b | Protective | 0.36 | 0.18–0.74 | 0.005 | OS | (40) |
| T | 148 HCC (82 HBV) | 41% (45/109) | miR-21 | Risk | NA | 1.19–1.47 | 0.004 | DFS | (21) |
| T | 140 HBV-HCC | NA | miR-21 | Risk | 1.509 | 1.079–2.112 | 0.016 | 3-years OS | (128) |
| T | 108 HBV-HCC | NA | miR-21 | Risk | 1.416 | 1.057–1.897 | 0.020 | 5-years OS | (128) |
| T | 115 HCC (101 HBV) | 51% (59/115) | miR-221 | Risk | 2.09 | 1.09–4.04 | 0.027 | MFS | (20) |
| T | 135 HCC (96 HBV) | 95% (128/135) | miR-221 | Risk | 2.846 | 1.564–5.181 | 0.001 | DFS | (25) |
| T | 135 HCC (96 HBV) | 95% (128/135) | miR-221 | Risk | 2.969 | 1.629–5.408 | <0.001 | OS | (25) |
| T | 166 HCC (146 HBV) | 84% (139/166) | 20-miRNA prognostic signature* | Risk | 2.75 | 1.58–4.79 | <0.001 | OS | (129) |
| T | 214 HBV-HCC | 93% (199/214) | 20-miRNA metastasis signature# | Risk | 2.1 | 1.2–3.6 | 0.01 | OS | (32) |
| T | 455 HCC (412 HBV) | 88% (400/455) | miR-26a | Protective | 0.48 | 0.21–1.0 | 0.05 | OS | (27) |
| T | 455 HCC (412 HBV) | 88% (400/455) | miR-26b | Protective | 0.48 | 0.20–0.91 | 0.04 | OS | (27) |
| T | 142 HCC (103 HBV) | 58% (82/142) | miR-122 | Protective | NA | NA | 0.001 | OS | (31) |
| T | 120 HCC (97 HBV) | 78% (93/120) | miR-200a | Protective | 0.382 | 0.215–0.896 | 0.004 | OS | (33) |
| T | 101 HCC (71 HBV) | NA | miR-200a | Protective | 0.403 | 0.242–0.670 | <0.001 | OS | (34) |
| T | 66 HCC (64 HBV) | NA | miR-203 | Protective | 0.202 | 0.064–0.638 | 0.006 | RFS | (130) |
| T | 66 HCC (64 HBV) | NA | miR-203 | Protective | 0.332 | 0.139–0.793 | 0.013 | OS | (130) |
miR-708, miR-34c-3p, miR-584, miR-4310, miR-744, miR-141, let-7d, miR-15a, miR-142-3p, miR-10b, let-7e, miR-28-3p, miR-193b, miR-101, miR-451, miR-142-5p, miR-26b, miR-497, miR-29c, miR-140-3p.
miR-338, miR-219-1, miR-207, miR-185, miR-30c-1, miR-1-2, miR-34a, miR-19a, miR-148a, miR-124a-2, miR-9-2, miR-148b, miR-122a, miR-125b-2, miR-194, miR-30a, miR-126, let-7g, miR-15a, miR-30e.
CI, confidence interval; DFS, disease-free survival; HBV, Hepatitis B virus; HR, hazard ratio; HCC, hepatocellular carcinoma; LT, liver transplantation; MFS, metastasis-free survival; OS, overall survival; P/S, plasma or serum; Ref, reference; RFS, recurrence-free survival; RR, relative risk; T, tissue.
It should also be noted that other studies found no significant associations with survival between HBV-HCC patients with high or low levels of miRNAs, (i.e., miR-21, miR-122, and miR-125b) (126, 132, 133). These disparate results may be due to differences in study design, analysis, and participant characteristics. For example, the cut-off value used to divide high and low miRNA-expressed population varies among studies and can be quite arbitrary [e.g., using a fixed value or average value or optimal cut-off value from Youden index analysis, or a ratio comparison to adjacent non-tumor tissue] (20, 21, 40, 126). The outcome events also varied, including overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS), and liver transplantation (LT)-free survival. These differences among studies make comparison challenging. These limitations will need to be addressed to establish reliable diagnostic and prognostic miRNA biomarker panels for HBV-HCC.
Dysregulated miRNAs in HCV-HCC
Since effective HCV-curative, direct-acting antiviral agents (DAA) are widely used worldwide in recent years (134, 135), fewer cases of HCC will be caused by HCV infection in the future. Subsequently, HBV infection will likely be the predominant cause of HCC worldwide. The pattern of dysregulated miRNAs in HCV-HCC, nevertheless, may still shed insights on the HBV-HCC pathogenesis as the comparison may reveal pathogen-specific and pathogen-independent tumorigenic pathways.
Several miRNAs showed similar dysregulation patterns in HCV- HCC and HBV-HCC (Table 6), including up-regulation of miR-18 (136), miR-221 (137) and miR-224 (15, 138, 139), and down-regulation of miR-199a-5p (136). These miRNAs may be involved in key cancer pathways that are shared by HBV- and HCV-HCC, including the WNT/β-Catenin and TP53 pathways. These miRNA and pathways may, therefore, be putative common targets for diagnostic, prognostic, and therapeutic interventions. Direct comparisons of miRNAs in HBV- and HCV-HCCs are lacking. In a small study comparing HBV-HCC and HCV-HCC tumor samples, the abundance of miR-122 was significantly reduced in HBV-HCC but not HCV-HCC, providing evidence of pathogen-specific dysregulation of miRNAs (74).
Table 6.
Common microRNAs dysregulated consistently in HBV-HCC and HCV-HCC.
Challenges and Future Directions
Accumulating evidence indicates that miRNAs, which function as gene regulators at the post-transcriptional level, are involved in the development of HBV-HCC. The expression levels of some single miRNAs or miRNA panels have the specificity and sensitivity to diagnose HCC and to predict survival; therefore, miRNA profiling panels are promising biomarkers for early diagnosis and survival prediction of HCC (Figure 1). Clinical trials to establish the utility of these panels in clinical practice are warranted.
However, there are several limitations and knowledge gaps in the current literature. In HBV-HCC, most HCC arise from cirrhotic tissues, thus miRNA changes may originate from either or both HCC and cirrhotic tissues. Underlying cirrhosis was present in 45–95% of HCC cases among studies that reported this information (Table 1), other studies did not report cirrhosis status. How miRNA profiles differ between cirrhotic and non-cirrhotic HBV-HCC remains largely unexplored (140).
The heterogeneity of methodologies in control selection, miRNA detecting technologies, case and control characteristics, and biostatistical analyses in studies also contribute to different results among studies. Failure to replicate findings may be due to small sample size affecting power leading to type 1 and type 2 errors. A major confounder among the studies is the selection of control tissue or sample. For example, comparisons may be made between tumor and non-tumor tissues from the same patients or different individuals. qRT-PCR quantification methods and platforms for miRNAs vary in their sensitivity and breadth. Technical replication to control for between and within-sample variation was lacking in some studies (42). Although most studies use internal controls to normalize miRNA expression levels of target genes (e.g., U6 SnRNA, GAPDH, miR-16, RNU43, cel-miRNA-39, or synthetic cel-miR-67), no universal internal references are used making comparisons among studies challenging (32, 36, 42, 47, 117, 122, 141, 142). Reviewers and journals are aware that a lack of replication in clinical research is a growing area of concern. A common set of internal controls would facilitate the replication and validation of informative miRNAs. Another source of failure to replicate is that the coverage of the miRNA arrays varies by more than 2-fold (308 to 829 miRNAs) (17–19, 32, 45). Definitions of differential expression vary from >2-fold change to <1.5 change in others. Over conservative cut-offs tend to lead to type 2 errors while less conservative cut-offs tend to increase type 1 errors. Next-generation sequencing is particularly prone to mis-annotations of microRNAs, which may lead to false-positive (143) or false-negative findings (144).
Before miRNAs can be used in a clinical setting, standardized methods for sample collection and handling should be implemented. Clinical trials will need to be conducted to assess the performance of miRNA biomarkers in addition to or in place of current diagnostic methods before their acceptance into surveillance or screening programs or for clinical management of HCC. We consider design issues and knowledge gaps that warrant attention in future investigations.
Sample size: is a major factor affecting power and validity. Since most miRNA have a moderate (<3-fold) difference between cases and controls and both large intra-individual and inter-individual variation, large sample sizes are required for sufficient power to minimize type 1 and II errors. Replication using public datasets [e.g., the Cancer Genome Atlas (TCGA) database] may provide additional supporting evidence (145, 146).
Validation for circulating miRNAs: To develop liquid biopsies for detection, diagnosis, and prognosis, miRNAs identified from serum/plasma should be validated to miRNAs obtained from tumor tissue before clinical evaluation as biomarkers. Non-specific circulating miRNAs may originate from other high blood-flow organs and tissue (147).
Clinical trials: Promising miRNAs markers must be tested for efficacy vs. standard of care (imaging and AFP levels) in randomized clinical trials before entering clinical practice.
HCC early detection: Since HCC is usually diagnosed mid to late-stage HCC, early HCC is rarely studied for miRNAs. Data comparing miRNAs expression levels in LC and early HCC groups is scarce and is urgently needed, as most HBV-HCCs develop from cirrhotic liver tissue. Clinical trials for miRNA early-diagnosis should focus on patients with HBV, HCV, or liver cirrhosis at high risk for HCC.
miRNA profiling for HBV-HCC: Evaluation of differences and commonalities of miRNA profiles in HCCs arising from HBV and other underlying liver diseases
Personalized medicine: Basic and clinical investigations for the clinical utility of precision miRNA-targeting therapies.
Diversity of miRNA investigations: Most HBV-HCC studies have enrolled Asian patients because of their high carrier rate for HBV. However, it is unknown if miRNA results are similar across diverse populations, particularly in Africa where HBV prevalence is also high (73). The generalizability of findings in Asians needs to be tested in other global populations.
Taken together, the recent studies in miRNAs provide encouraging evidence that miRNAs detection may aid in the diagnosis, survival prediction, and treatment of HBV-HCC. More well-designed and well-powered case-control or longitudinal studies in diverse populations are critically needed to validate the utility of miRNAs in HCC and translate miRNA into clinical use.
Author Contributions
JX and PA conceived idea, prepared the tables, and wrote the manuscript. YY and CW revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Brean Derrett, an NIH CRTA Postbaccalaureate Fellow, for technical assistance.
Glossary
Abbreviations
- 3′ UTR
3′ untranslated region
- AFP
alpha-fetoprotein
- ANT
adjacent non-cancerous tissue
- ASC
asymptomatic carrier
- API-5
apoptosis inhibitor-5
- AUC
area under curve
- CCNG1
cyclin G1
- CGI
CpG island
- CHB
chronic hepatitis B
- CI
confidence interval
- CTCF
CCCTC-binding factor
- CTGF
connective tissue growth factor
- DFS
disease-free survival
- DN
dysplastic nodules
- EMT
epithelial-mesenchymal transition
- ERα
estrogen receptorα
- EZH2
Enhancer of zeste homolog 2
- FXR
farnesoid X receptor
- HBV
hepatitis B virus
- HBV-HCC
HBV-related HCC
- HBV-LC
HBV-related LC
- HBx
HBV X protein
- HC
healthy control
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HDAC
histone deacetylases
- HGDN
high-grade dysplastic nodule
- HR
hazard ratio
- JAK/STAT
Janus kinase/signal transducer
- LC
liver cirrhosis
- LGDN
low-grade dysplastic nodule
- LT
liver transplantation
- miRNAs
microRNAs
- mRNAs
messenger RNAs
- NA
not available
- NBNC
non-HBV non-HCV
- onco-miRNA
oncogenic miRNA
- OR
odds ratio
- OS
overall survival
- PBF
pituitary tumor-transforming gene 1 binding factor
- PTTG1
pituitary tumor-transforming gene 1
- P
plasma
- P/S
plasma or serum
- PI3K/MAPK
phosphoinositide 3-kinase/mitogen-activated protein kinase
- RFS
recurrence-free survival
- ROC
receiver-operator characteristic curve
- RFS
recurrence-free survival
- RR
relative risk
- S
serum
- T
tissue
- WNT/β-Catenin
wingless-related integration site/beta-catenin.
Footnotes
Funding. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported in part by the China 13th 5-years science and technology major project on the prevention and treatment of major infectious diseases (2017ZX10202202).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. (2011) 365:1118–27. 10.1056/NEJMra1001683 [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular Carcinoma. Gastroenterology. (2016) 150:835–53. 10.1053/j.gastro.2015.12.041 [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 6.Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. (2015) 25:651–65. 10.1016/j.tcb.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Jin Y. MicroRNA in cell differentiation and development. Sci China C Life Sci. (2009) 52:205–11. 10.1007/s11427-009-0040-5 [DOI] [PubMed] [Google Scholar]
- 8.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. (2016) 16:279–94. 10.1038/nri.2016.40 [DOI] [PubMed] [Google Scholar]
- 9.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. (2015) 6:8474–90. 10.18632/oncotarget.3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landskroner-Eiger S, Moneke I, Sessa WC. miRNAs as modulators of angiogenesis. Cold Spring Harb Perspect Med. (2013) 3:a006643. 10.1101/cshperspect.a006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Claret FX, Wu W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma. Front Oncol. (2019) 9:756. 10.3389/fonc.2019.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandini E, Fanini F. MicroRNAs and androgen receptor: emerging players in breast cancer. Front Genet. (2019) 10:203. 10.3389/fgene.2019.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers. (2019) 11:888. 10.3390/cancers11060888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki R, Osaki M, Okada F. MicroRNA-based diagnosis and treatment of metastatic human osteosarcoma. Cancers. (2019) 11:553. 10.3390/cancers11040553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. (2008) 47:1955–63. 10.1002/hep.22256 [DOI] [PubMed] [Google Scholar]
- 16.WHO Global Hepatitis Report 2017. Geneva: World Health Organization; (2017). Available online at: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (accessed July 22, 2017). [Google Scholar]
- 17.Li W, Xie L, He X, Li J, Tu K, Wei L, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. (2008) 123:1616–22. 10.1002/ijc.23693 [DOI] [PubMed] [Google Scholar]
- 18.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. (2009) 69:1135–42. 10.1158/0008-5472.CAN-08-2886 [DOI] [PubMed] [Google Scholar]
- 19.Thurnherr T, Mah WC, Lei Z, Jin Y, Rozen SG, Lee CG. Differentially expressed miRNAs in hepatocellular carcinoma target genes in the genetic information processing and metabolism pathways. Sci Rep. (2016) 6:20065. 10.1038/srep20065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon SO, Chun SM, Han EH, Choi J, Jang SJ, Koh SA, et al. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum Pathol. (2011) 42:1391–400. 10.1016/j.humpath.2010.12.010 [DOI] [PubMed] [Google Scholar]
- 21.He X, Li J, Guo W, Liu W, Yu J, Song W, et al. Targeting the microRNA-21/AP1 axis by 5-fluorouracil and pirarubicin in human hepatocellular carcinoma. Oncotarget. (2015) 6:2302–14. 10.18632/oncotarget.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Yin J, Yuan L, Wang S, Yang L, Du X, et al. Downregulation of microRNA-139 is associated with hepatocellular carcinoma risk and short-term survival. Oncol Rep. (2014) 31:1699–706. 10.3892/or.2014.3032 [DOI] [PubMed] [Google Scholar]
- 23.Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO. Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol. (2011) 54:1177–84. 10.1016/j.jhep.2010.09.023 [DOI] [PubMed] [Google Scholar]
- 24.Dundar HZ, Aksoy F, Aksoy SA, Tasar P, Ugras N, Tunca B, et al. Overexpression of miR-21 is associated with recurrence in patients with Hepatitis B virus-mediated hepatocellular carcinoma undergoing liver transplantation. Transplant Proc. (2019) 51:1157–61. 10.1016/j.transproceed.2019.01.089 [DOI] [PubMed] [Google Scholar]
- 25.Chen F, Li XF, Fu DS, Huang JG, Yang SE. Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomark. (2017) 18:209–14. 10.3233/CBM-161671 [DOI] [PubMed] [Google Scholar]
- 26.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. (2008) 135:257–69. 10.1053/j.gastro.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 27.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. (2009) 361:1437–47. 10.1056/NEJMoa0901282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Wei X, Tang C, Li J, Liu R, Shen A, et al. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol Lett. (2013) 6:1811–15. 10.3892/ol.2013.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Y, Yao Q, Butt AM, Guo J, Tian Z, Bao X, et al. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, hepatocellular carcinoma. Cancer Biol Ther. (2014) 15:1248–55. 10.4161/cbt.29688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. (2010) 6:402. 10.1038/msb.2010.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, et al. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. (2015) 290:1170–85. 10.1074/jbc.M114.601203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. (2008) 47:897–907. 10.1002/hep.22160 [DOI] [PubMed] [Google Scholar]
- 33.Xiao F, Zhang W, Zhou L, Xie H, Xing C, Ding S, et al. microRNA-200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6. Oncol Rep. (2013) 30:2203–10. 10.3892/or.2013.2715 [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao Y, et al. MicroRNA-200a suppresses metastatic potential of side population cells in human hepatocellular carcinoma by decreasing ZEB2. Oncotarget. (2015) 6:7918–29. 10.18632/oncotarget.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. (2015) 15:62. 10.1186/s12935-015-0214-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. (2008) 173:856–64. 10.2353/ajpath.2008.080096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. (2008) 18:997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 39.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. (2010) 70:9798–807. 10.1158/0008-5472.CAN-10-1001 [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. (2017) 10:3843–51. 10.2147/OTT.S140062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. (2011) 406:70–3. 10.1016/j.bbrc.2011.01.111 [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. (2011) 50:136–42. 10.1002/mc.20712 [DOI] [PubMed] [Google Scholar]
- 43.Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS ONE. (2011) 6:e28486. 10.1371/journal.pone.0028486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. (2012) 57:2910–6. 10.1007/s10620-012-2317-y [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. (2011) 29:4781–8. 10.1200/JCO.2011.38.2697 [DOI] [PubMed] [Google Scholar]
- 46.Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS ONE. (2014) 9:e107986. 10.1371/journal.pone.0107986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Zhang J, Zhou L, Lu P, Zheng ZG, Sun W, et al. Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): clinical analyses of patients and an HCC rat model. Int J Clin Exp Pathol. (2015) 8:1466–78. Available online at: http://www.ijcep.com/files/ijcep0004608.pdf [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu HT, Liu RB, Liang YY, Hasan AME, Wang HY, Shao Q, et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. (2017) 37:888–96. 10.1111/liv.13356 [DOI] [PubMed] [Google Scholar]
- 49.Fornari F, Ferracin M, Trere D, Milazzo M, Marinelli S, Galassi M, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS ONE. (2015) 10:e0141448. 10.1371/journal.pone.0141448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. (2009) 28:3526–36. 10.1038/onc.2009.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122–a key factor and therapeutic target in liver disease. J Hepatol. (2015) 62:448–57. 10.1016/j.jhep.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 53.Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, et al. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. (2011) 26:1281–6. 10.3892/or.2011.1375 [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. (2012) 55:730–41. 10.1002/hep.24809 [DOI] [PubMed] [Google Scholar]
- 55.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. (2009) 49:1571–82. 10.1002/hep.22806 [DOI] [PubMed] [Google Scholar]
- 56.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. (2009) 284:32015–27. 10.1074/jbc.M109.016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. (2013) 87:2193–205. 10.1128/JVI.02831-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. (2008) 375:315–20. 10.1016/j.bbrc.2008.07.154 [DOI] [PubMed] [Google Scholar]
- 59.Luna JM, Barajas JM, Teng KY, Sun HL, Moore MJ, Rice CM, et al. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Mol Cell. (2017) 67:400–10.e7. 10.1016/j.molcel.2017.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barajas JM, Reyes R, Guerrero MJ, Jacob ST, Motiwala T, Ghoshal K. The role of miR-122 in the dysregulation of glucose-6-phosphate dehydrogenase (G6PD) expression in hepatocellular cancer. Sci Rep. (2018) 8:9105. 10.1038/s41598-018-27358-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. (2007) 133:647–58. 10.1053/j.gastro.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. (2008) 27:2128–36. 10.1038/sj.onc.1210856 [DOI] [PubMed] [Google Scholar]
- 63.Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, et al. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res. (2015) 13:1009–21. 10.1158/1541-7786.MCR-14-0703 [DOI] [PubMed] [Google Scholar]
- 64.Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. (2009) 136:683–93. 10.1053/j.gastro.2008.10.029 [DOI] [PubMed] [Google Scholar]
- 65.Liu X, Zhang Y, Wang P, Wang H, Su H, Zhou X, et al. HBX protein-induced downregulation of microRNA-18a is responsible for upregulation of connective tissue growth factor in hbv infection-associated hepatocarcinoma. Med Sci Monit. (2016) 22:2492–500. 10.12659/MSM.895943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen JJ, Tang YS, Huang SF, Ai JG, Wang HX, Zhang LP. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBVrelated hepatocellular carcinoma by targeting estrogen receptor-alpha. Oncol Rep. (2015) 33:792–8. 10.3892/or.2014.3647 [DOI] [PubMed] [Google Scholar]
- 67.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. (2013) 13:21. 10.1186/1471-2407-13-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang S, Zhou D, Li YX, Ming ZY, Li KZ, Wu GB, et al. In vivo and in vitro effects of microRNA-221 on hepatocellular carcinoma development and progression through the JAK-STAT3 signaling pathway by targeting SOCS3. J Cell Physiol. (2019) 234:3500–14. 10.1002/jcp.26863 [DOI] [PubMed] [Google Scholar]
- 69.Bandopadhyay M, Banerjee A, Sarkar N, Panigrahi R, Datta S, Pal A, et al. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. (2014) 14:721. 10.1186/1471-2407-14-721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY, Cao Y, et al. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int J Clin Exp Med. (2014) 7:893–902. Available online at: http://www.ijcem.com/files/ijcem0000090.pdf [PMC free article] [PubMed] [Google Scholar]
- 71.Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. (2010) 16:867–75. 10.1158/1078-0432.CCR-09-1840 [DOI] [PubMed] [Google Scholar]
- 72.Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. (2008) 100:1134–43. 10.1093/jnci/djn243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sartorius K, Sartorius B, Kramvis A, Singh E, Turchinovich A, Burwinkel B, et al. Circulating microRNA's as a diagnostic tool for hepatocellular carcinoma in a hyper endemic HIV setting, KwaZulu-natal, South Africa: a case control study protocol focusing on viral etiology. BMC Cancer. (2017) 17:894 10.1186/s12885-017-3915-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spaniel C, Honda M, Selitsky SR, Yamane D, Shimakami T, Kaneko S, et al. microRNA-122 abundance in hepatocellular carcinoma and non-tumor liver tissue from Japanese patients with persistent HCV versus HBV infection. PLoS ONE. (2013) 8:e76867. 10.1371/journal.pone.0076867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, et al. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. (2014) 159:2397–410. 10.1007/s00705-014-2084-5 [DOI] [PubMed] [Google Scholar]
- 76.Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. (2013) 123:630–45. 10.1172/JCI64265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamontagne J. Hepatitis B virus and microRNAs: complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol. (2015) 21:7375. 10.3748/wjg.v21.i24.7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. (2011) 39:5157–63. 10.1093/nar/gkr067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, et al. Hepatitis B virus-encoded microRNA controls viral replication. J Virol. (2017) 91:e01919–16. 10.1128/JVI.01919-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao L, Zhou Y, Sui Z, Zhang Y, Liu Y, Xie H, et al. HBV-encoded miR-2 functions as an oncogene by downregulating TRIM35 but upregulating RAN in liver cancer cells. EBioMedicine. (2019) 48:117–29. 10.2139/ssrn.3365052 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Zhang X, Hou J, Lu M. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet. (2013) 4:202. 10.3389/fgene.2013.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu WH, Yeh SH, Chen PJ. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim Biophys Acta. (2011) 1809:678–85. 10.1016/j.bbagrm.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 83.Sagnelli E, Potenza N, Onorato L, Sagnelli C, Coppola N, Russo A. Micro-RNAs in hepatitis B virus-related chronic liver diseases and hepatocellular carcinoma. World J Hepatol. (2018) 10:558–70. 10.4254/wjh.v10.i9.558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou G, Wilson G, George J, Qiao L. Targeting cancer stem cells as a therapeutic approach in liver cancer. Curr Gene Ther. (2015) 15:161–70. 10.2174/1566523214666141224095938 [DOI] [PubMed] [Google Scholar]
- 85.Qiu L, Li H, Fu S, Chen X, Lu L. Surface markers of liver cancer stem cells and innovative targeted-therapy strategies for HCC. Oncol Lett. (2018) 15:2039–48. 10.3892/ol.2017.7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. (2007) 132:2542–56. 10.1053/j.gastro.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 87.Lou W, Liu J, Gao Y, Zhong G, Ding B, Xu L, et al. MicroRNA regulation of liver cancer stem cells. Am J Cancer Res. (2018) 8:1126–41. Available online at: http://www.ajcr.us/files/ajcr0079739.pdf [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C, Wang MD, Cheng P, Huang H, Dong W, Zhang WW, et al. Hepatitis B virus X protein promotes the stem-like properties of OV6(+) cancer cells in hepatocellular carcinoma. Cell Death Dis. (2017) 8:e2560. 10.1038/cddis.2016.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gu Y, Wei X, Sun Y, Gao H, Zheng X, Wong LL, et al. miR-192-5p silencing by genetic aberrations is a key event in hepatocellular carcinomas with cancer stem cell features. Cancer Res. (2019) 79:941–53. 10.1158/0008-5472.CAN-18-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arzumanyan A, Friedman T, Ng IO, Clayton MM, Lian Z, Feitelson MA. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. (2011) 71:3701–8. 10.1158/0008-5472.CAN-10-3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji J, Zheng X, Forgues M, Yamashita T, Wauthier EL, Reid LM, et al. Identification of microRNAs specific for epithelial cell adhesion molecule-positive tumor cells in hepatocellular carcinoma. Hepatology. (2015) 62:829–40. 10.1002/hep.27886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. (2009) 50:472–80. 10.1002/hep.22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boix L, Lopez-Oliva JM, Rhodes AC, Bruix J. Restoring miR122 in human stem-like hepatocarcinoma cells, prompts tumor dormancy through Smad-independent TGF-beta pathway. Oncotarget. (2016) 7:71309–29. 10.18632/oncotarget.11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nasr MA, Salah RA, Abd Elkodous M, Elshenawy SE, El-Badri N. Dysregulated microRNA fingerprints and methylation patterns in hepatocellular carcinoma, cancer stem cells, and mesenchymal stem cells. Front Cell Dev Biol. (2019) 7:229. 10.3389/fcell.2019.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alpini G, Glaser SS, Zhang JP, Francis H, Han Y, Gong J, et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J Hepatol. (2011) 55:1339–45. 10.1016/j.jhep.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. (2012) 143:811–20.e15. 10.1053/j.gastro.2012.05.033 [DOI] [PubMed] [Google Scholar]
- 97.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. (2011) 19:232–43. 10.1016/j.ccr.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 98.Kim HS, Shen Q, Nam SW. Histone deacetylases and their regulatory microRNAs in hepatocarcinogenesis. J Korean Med Sci. (2015) 30:1375–80. 10.3346/jkms.2015.30.10.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saito Y, Saito H. Role of CTCF in the regulation of microRNA expression. Front Genet. (2012) 3:186. 10.3389/fgene.2012.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soto-Reyes E, Gonzalez-Barrios R, Cisneros-Soberanis F, Herrera-Goepfert R, Perez V, Cantu D, et al. Disruption of CTCF at the miR-125b1 locus in gynecological cancers. BMC Cancer. (2012) 12:40. 10.1186/1471-2407-12-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiu L, Xu H, Ji M, Shang D, Lu Z, Wu Y, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. (2019) 231:116660. 10.1016/j.lfs.2019.116660 [DOI] [PubMed] [Google Scholar]
- 102.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA circTRIM33-12 acts as the sponge of microRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. (2019) 18:105. 10.1186/s12943-019-1031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. (2017) 35:180–8. 10.1007/s10637-016-0407-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu Q, Zhang M, Tu J, Pang L, Cai W, Liu X. MicroRNA-122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncol Rep. (2015) 34:2054–64. 10.3892/or.2015.4175 [DOI] [PubMed] [Google Scholar]
- 106.Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. (2017) 360:210–7. 10.1016/j.yexcr.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 107.Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. (2009) 69:5761–7. 10.1158/0008-5472.CAN-08-4797 [DOI] [PubMed] [Google Scholar]
- 108.Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. (2018) 67:940–54. 10.1002/hep.29586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He J, Zhao K, Zheng L, Xu Z, Gong W, Chen S, et al. Upregulation of microRNA-122 by farnesoid X receptor suppresses the growth of hepatocellular carcinoma cells. Mol Cancer. (2015) 14:163. 10.1186/s12943-015-0427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun C, Lan P, Han Q, Huang M, Zhang Z, Xu G, et al. Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun. (2018) 9:1241. 10.1038/s41467-018-03584-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tang S, Tan G, Jiang X, Han P, Zhai B, Dong X, et al. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget. (2016) 7:73257–69. 10.18632/oncotarget.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. (2011) 310:160–9. 10.1016/j.canlet.2011.06.027 [DOI] [PubMed] [Google Scholar]
- 113.Pan C, Wang X, Shi K, Zheng Y, Li J, Chen Y, et al. MiR-122 reverses the doxorubicin-resistance in hepatocellular carcinoma cells through regulating the tumor metabolism. PLoS ONE. (2016) 11:e0152090. 10.1371/journal.pone.0152090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. (2014) 60:590–8. 10.1016/j.jhep.2013.10.028 [DOI] [PubMed] [Google Scholar]
- 115.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. (2019) 10:478. 10.3389/fgene.2019.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. (2012) 2:e000825. 10.1136/bmjopen-2012-000825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. (2015) 16:804–15. 10.1016/S1470-2045(15)00048-0 [DOI] [PubMed] [Google Scholar]
- 118.Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. (2015) 137:1679–90. 10.1002/ijc.29544 [DOI] [PubMed] [Google Scholar]
- 119.Huang JT, Liu SM, Ma H, Yang Y, Zhang X, Sun H, et al. Systematic review and meta-analysis: circulating miRNAs for diagnosis of hepatocellular Carcinoma. J Cell Physiol. (2016) 231:328–35. 10.1002/jcp.25135 [DOI] [PubMed] [Google Scholar]
- 120.Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. The role of microRNAs in hepatocellular Carcinoma. J Cancer. (2018) 9:3557–69. 10.7150/jca.26350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang B, Xiong WY, Hou HJ, Xu Q, Cai XL, Zeng TX, et al. Exosomal miRNAs as biomarkers of cancer: a meta-analysis. Clin Lab. (2019) 65:181011. 10.7754/Clin.Lab.2018.181011 [DOI] [PubMed] [Google Scholar]
- 122.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. (2014) 2014:864894. 10.1155/2014/864894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. (2015) 47:e184. 10.1038/emm.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. (2012) 56:167–75. 10.1016/j.jhep.2011.04.026 [DOI] [PubMed] [Google Scholar]
- 125.Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H, et al. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS ONE. (2014) 9:e109347. 10.1371/journal.pone.0109347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ng KT, Lo CM, Wong N, Li CX, Qi X, Liu XB, et al. Early-phase circulating miRNAs predict tumor recurrence and survival of hepatocellular carcinoma patients after liver transplantation. Oncotarget. (2016) 7:19824–39. 10.18632/oncotarget.7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cho HJ, Kim SS, Nam JS, Kim JK, Lee JH, Kim B, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol. (2017) 41:181–9. 10.1016/j.clinre.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 128.Shi KQ, Lin Z, Chen XJ, Song M, Wang YQ, Cai YJ, et al. Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget. (2015) 6:25093–108. 10.18632/oncotarget.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. (2013) 19:4780–91. 10.1158/1078-0432.CCR-12-2728 [DOI] [PubMed] [Google Scholar]
- 130.Chen HY, Han ZB, Fan JW, Xia J, Wu JY, Qiu GQ, et al. miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med Oncol. (2012) 29:1859–65. 10.1007/s12032-011-0031-9 [DOI] [PubMed] [Google Scholar]
- 131.Wang WY, Zhang HF, Wang L, Ma YP, Gao F, Zhang SJ, et al. miR-21 expression predicts prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. (2014) 38:715–9. 10.1016/j.clinre.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 132.Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH, Kim BW, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem. (2015) 48:1073–8. 10.1016/j.clinbiochem.2015.06.019 [DOI] [PubMed] [Google Scholar]
- 133.Kim SS, Nam JS, Cho HJ, Won JH, Kim JW, Ji JH, et al. Plasma micoRNA-122 as a predictive marker for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. (2017) 32:199–207. 10.1111/jgh.13448 [DOI] [PubMed] [Google Scholar]
- 134.American Association for the Study of Liver Diseases and Infectious Diseases Society of America HCV Guidance: Recommendations for Testing, Managing, and Treating Hepatitis C. Alexandria, VA: AASLD-IDSA (2020). Available online at: http://www.hcvguidelines.org (accessed January 2, 2020).
- 135.Chinese Society of Hepatology and Chinese Society of Infectious Diseases and Chinese Medical Association . Guidelines for the prevention and treatment of hepatitis C (2019 version). Zhonghua Gan Zang Bing Za Zhi. (2019) 27:962–79. 10.3760/cma.j.issn.1007-3418.2019.12.008 [DOI] [PubMed] [Google Scholar]
- 136.Morita K, Shirabe K, Taketomi A, Soejima Y, Yoshizumi T, Uchiyama H, et al. Relevance of microRNA-18a and microRNA-199a-5p to hepatocellular carcinoma recurrence after living donor liver transplantation. Liver Transpl. (2016) 22:665–76. 10.1002/lt.24400 [DOI] [PubMed] [Google Scholar]
- 137.El-Garem H, Ammer A, Shehab H, Shaker O, Anwer M, El-Akel W, et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol. (2014) 6:818–24. 10.4254/wjh.v6.i11.818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. (2013) 133:816–24. 10.1002/ijc.28075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. (2006) 25:2537–45. 10.1038/sj.onc.1209283 [DOI] [PubMed] [Google Scholar]
- 140.Fittipaldi S, Vasuri F, Bonora S, Degiovanni A, Santandrea G, Cucchetti A, et al. miRNA signature of hepatocellular carcinoma vascularization: how the controls can influence the signature. Dig Dis Sci. (2017) 62:2397–407. 10.1007/s10620-017-4654-3 [DOI] [PubMed] [Google Scholar]
- 141.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. (2013) 52:297–303. 10.1002/mc.21864 [DOI] [PubMed] [Google Scholar]
- 142.Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer. (2017) 17:805. 10.1186/s12885-017-3816-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schopman NC, Heynen S, Haasnoot J, Berkhout B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. (2010) 7:573–6. 10.4161/rna.7.5.13141 [DOI] [PubMed] [Google Scholar]
- 144.Hansen TB, Bramsen JB, Kjems J. Re-inspection of small RNA sequence datasets reveals several novel human miRNA genes. PLoS ONE. (2010) 5:e10961. 10.1371/journal.pone.0010961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhen Y, Xinghui Z, Chao W, Yi Z, Jinwen C, Ruifang G, et al. Several microRNAs could predict survival in patients with hepatitis B-related liver cancer. Sci Rep. (2017) 7:45195. 10.1038/srep45195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mei Y, You Y, Xia J, Gong JP, Wang YB. Identifying differentially expressed micrornas between cirrhotic and non-cirrhotic hepatocellular carcinoma and exploring their functions using bioinformatic analysis. Cell Physiol Biochem. (2018) 48:1443–56. 10.1159/000492254 [DOI] [PubMed] [Google Scholar]
- 147.Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. (2015) 21:9853–62. 10.3748/wjg.v21.i34.9853 [DOI] [PMC free article] [PubMed] [Google Scholar]