Although enteroviruses are associated with a wide variety of diseases and conditions, their mode of replication is well conserved. Their genome is carried as a single, positive-sense RNA strand. At the 5′ end of the strand is an approximately 90-nucleotide self-complementary region called the 5′ cloverleaf, or the oriL. This noncoding region serves as a platform upon which host and virus proteins, including the 3B, 3C, and 3D virus proteins, assemble in order to initiate replication of a negative-sense RNA strand.
KEYWORDS: SAXS, X-ray crystallography, enterovirus, nuclear magnetic resonance, picornavirus, structure, viral replication
SUMMARY
Although enteroviruses are associated with a wide variety of diseases and conditions, their mode of replication is well conserved. Their genome is carried as a single, positive-sense RNA strand. At the 5′ end of the strand is an approximately 90-nucleotide self-complementary region called the 5′ cloverleaf, or the oriL. This noncoding region serves as a platform upon which host and virus proteins, including the 3B, 3C, and 3D virus proteins, assemble in order to initiate replication of a negative-sense RNA strand. The negative strand in turn serves as a template for synthesis of multiple positive-sense RNA strands. Building on structural studies of individual RNA stem-loops, the structure of the intact 5′ cloverleaf from rhinovirus has recently been determined via nuclear magnetic resonance/small-angle X-ray scattering (NMR/SAXS)-based methods, while structures have also been determined for enterovirus 3A, 3B, 3C, and 3D proteins. Analysis of these structures, together with structural and modeling studies of interactions between host and virus proteins and RNA, has begun to provide insight into the enterovirus replication mechanism and the potential to inhibit replication by blocking these interactions.
INTRODUCTION
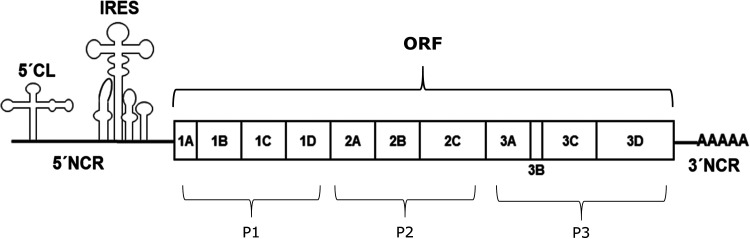
The enterovirus genome consists of a single positive-sense RNA strand of approximately 7,500 bases (Fig. 1) (1–4). This RNA strand plays three distinct types of roles. (i) Nearly 90% of this RNA strand represents a single open reading frame that codes for the virus polyprotein that is subsequently cleaved into active pieces by two virus-encoded proteases, 2Apro and 3Cpro. (ii) The strand, in its entirety, serves as a replication template for synthesis of a complementary negative-sense RNA strand, which is in turn used as a template to synthesize additional positive-sense strands. These additional positive-sense strands then act as additional mRNA, and they can be packaged into new virus particles that are released from the cell to infect other cells. (iii) Parts of the RNA positive-sense strand serve a regulatory role in the translation and replication processes, beyond simply serving as mRNA and a replication template. In particular, the 5′ end contains an extensive (∼750 bases) noncoding region (NCR) that plays a key role in control of both replication and translation (5–7). Translational control resides largely in the internal ribosomal entry site (IRES), an ∼450-nucleotide highly self-complementary region within the 5′ NCR, which recruits host cell ribosomes in order to initiate translation of the mRNA region that lies just 3′ to it. (8).
FIG 1.
Schematic representation of the enterovirus genome. The 5′ NCR includes a small (<100 nucleotide) cloverleaf that serves as an RNA replication platform and a larger IRES region that controls translation of virus proteins. The single open reading frame (ORF) codes for a single polyprotein that is later separated into 11 proteins (1A through 3D) via the action of virus-encoded proteases. This review focuses upon structural studies of the 5′-cloverleaf (5′-CL) replication platform and the four replication-linked proteins found in region P3 of the polygene (3A, 3B, 3C, and 3D).
The extreme 5′ end of the 5′ NCR contains a smaller (<100 nucleotide) self-complementary region referred to as the 5′ cloverleaf (5′-CL), or the oriL. A secondary structure prediction of the 5′-CL from rhinovirus 14 (RV14) is shown in Fig. 2 (upper left). This cloverleaf serves as a platform upon which various host and virus proteins assemble in order to initiate synthesis of the negative strand, that is, to initiate replication (9–11). Thus, the 5′-CL can be called the enterovirus replication platform, and it, along with proteins that interact with it, will be the focus of this review.
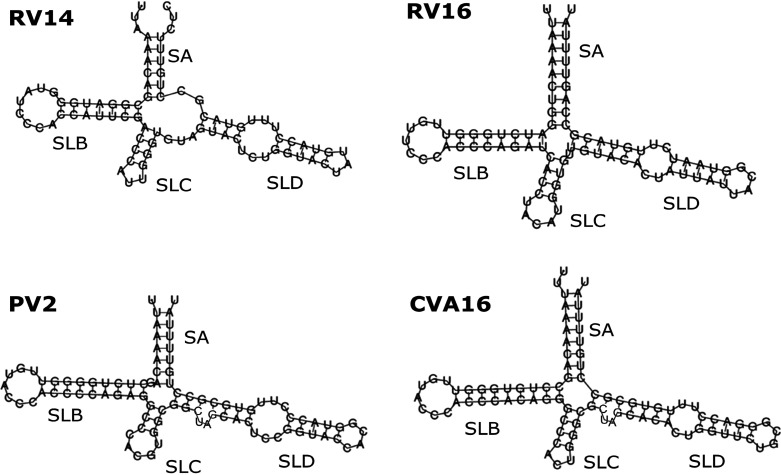
FIG 2.
Enterovirus 5′-CL predicted secondary structure. The 5′-most 83 nucleotides of the RV14 genome are predicted to form a cloverleaf with a four-way junction. The four converging structural elements are stem A (SA) and stem-loops B, C, and D (SLB, SLC, and SLD). SLB and SLD together comprise over 60% of the cloverleaf. SLD was predicted to contain a 3-by-3 U-rich bulge separating two short Watson-Crick base-paired regions. For comparison, the predicted secondary structure of three of the other enteroviruses discussed in this review are also presented. Note that in these four structures, the string of three cytosines in SLB are conserved, and two of the three pyrimidine mismatches within SLD are also conserved. The predicted lengths of the helices vary, but it appears as if the lengths of SLB and SLD may be correlated.
5′ REPLICATION ELEMENT RNA
SLD
Structural investigation of the Enterovirus 5′-CL RNA began with stem-loop D (SLD), the largest stem-loop structure in the 5′-CL, which is the oriL binding site for the virus protein 3CD. The SLD-3CD interaction is essential for effective virus replication. To date, there have been three published nuclear magnetic resonance (NMR) solution structures of SLD: one from coxsackievirus B3 (CVB3) (12), an enterovirus consensus sequence (13), and one from rhinovirus-14 (RV14) (14). Although among enteroviruses the SLD shows some sequence variability, the three-dimensional structures of these three SLD variants are relatively well conserved.
Preliminary sequence-based analysis of each of these SLD variants predicted the presence of two regions of base pairs (stem I and stem II) separated by a pyrimidine mismatch region (Fig. 3a) that was predicted to form a 3-by-3 bulge. However, upon NMR analysis, the U bases within this mismatch region showed observable imino resonances, with very strong imino-to-imino nuclear Overhauser effects (NOEs) between U bases. Together with imino proton chemical shifts well outside the normal range for a Watson-Crick base pair, the data indicated the formation of two noncanonical U-U base pairs within the predicted bulge. Evidence for a central C-U base pair was also present. Therefore, the bulge did not exist, and, instead, three pyrimidine-pyrimidine base pairs in this region connect the predicted stem I and stem II, forming a single contiguous stem.
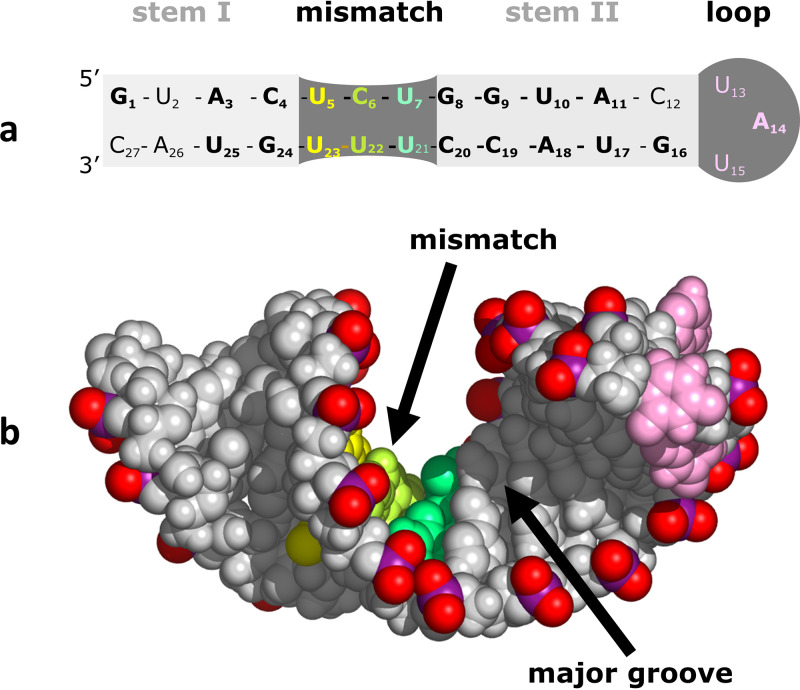
FIG 3.
RV14 SLD structure. (a) Secondary structure derived via NMR structural analysis. Nucleotides conserved between RV14, enterovirus consensus, and CVB3 SLD sequences are shown in bold. These include the mismatch region and 2 bp to the left and 4 bp to the right of the mismatch. The mismatch region is base paired, resulting in a single continuous stem spanning the regions labeled as stem I, mismatch, and stem II. (b) NMR-based structure. Due to strains apparently introduced by mismatch base pair formation, the helix departs from standard A-form geometry and presents a wide and accessible major groove.
Residual dipolar couplings (RDCs) can be used to monitor the angle between the 1H-15N or 1H-13C bond and the external magnetic field to define the relative orientations of parts of a biomolecule in solution. The consensus and RV14 SLD structures were determined with RDCs (13, 14). Without RDC constraints, the consensus SLD structure was poorly defined. In particular, the relative orientation of stem I versus that of stem II was not well defined. Inclusion of RDCs led to a better definition of global orientation as well as axial length. Incorporating RDC constraints resulted in a shortening of the overall axial length, forming a more compact structure. A similar phenomenon was present in RV14 SLD (14). With the inclusion of RDCs, the axial length decreased from 52 Å to 47 Å, and the overall root mean square deviation (RMSD) improved from 1.00 Å to 0.59 Å. The inclusion of RV14 RDCs was also critical in establishing hydrogen bond geometry for the C-U base pair in the mismatch region.
Globally, each of the SLD structures adopts a single double-helix structure (Fig. 3b) with some A-form characteristics, capped by a well-defined tetraloop (CVB3 and consensus) or a more dynamic triloop (RV14) (12–14). However, due apparently to the pyrimidine mismatch region, deviations from standard A-form RNA are present. Typical A-form RNA contains a wide (∼11 Å) but shallow minor groove and a narrow (∼3.0 Å) but deep major groove, effectively restricting access for protein interactions to the minor groove. However, all three of the SLD structures display widened major grooves (∼13 Å) atypical of A-form RNA, as well as a decrease in minor groove width by ∼2.5 to 3 Å. The atypical features occur near the pyrimidine mismatch region of the helix. These unusual features presumably arise due to the smaller pyrimidine-pyrimidine base pair sizes, creating shorter interstrand C1′-C1′ distances and yielding a narrow minor groove width and a concomitantly widened major groove. Functionally, the wide major groove is of high interest. An unusually accessible major groove could allow for the potential protein interactions that would be impossible with standard A-form RNA (15). Also, major groove surfaces, as opposed to minor groove surfaces, when accessible, present a more discriminating set of functional groups, permitting more sequence-specific recognition and interactions. This feature in SLD could be relevant to the interaction between SLD and the virus protein 3CD, as discussed below in the 3C/SLD interaction section.
SLB
The second-largest stem-loop structure in the 5′-CL RNA is stem-loop B (SLB), which interacts with the host poly(C) binding protein (PCBP2) as a step in the circularization of the virus genome. The sole Enterovirus SLB that has been structurally determined is from RV14 (16). Based on sequence analysis, SLB is predicted to form a 7-bp helix, with one wobble base pair, capped by a large 8-nucleotide loop (Fig. 2, upper left). The detection of imino resonances for G or U in five canonical Watson-Crick base pairs and the G and U for the wobble pair confirmed the presence of a stable 6-bp helical structure. The predicted closing base pair, nearest the four-way junction, is not sufficiently stable to produce an observable imino proton.
Using NMR-based analysis, including residual dipolar couplings, RV14 SLB was shown to be a 6-bp A-form helix (Fig. 4), with a highly dynamic eight-base loop. The helical region is well defined, with an RMSD value of 0.415 Å, and contains a minor groove (∼10.5 Å wide) typical of A-form RNA. However, its major groove (∼8.7 Å wide) shares some characteristics with the major groove of B-form DNA, as will be discussed further below. The overall SLB RMSD of 2.249 Å reflects the high degree of conformational variability in the regions outside the helix. Weak H1′-H2′ couplings were observed for nucleotides in the helix, suggesting C3′-endo sugar conformation, a characteristic consistent with A-form geometry. In addition, strong H1′-H2′couplings observed for each of the loop and terminal nucleotides are consistent with a C2′-endo sugar conformation. The SLB loop nucleotides display narrow intense resonances with chemical shift values similar to those of nucleoside monophosphates (NMPs), along with relatively small RDC values. These data strongly suggest a highly flexible and disordered loop region. Based on these criteria, the most flexible region within the loop appears to be the string of four consecutive pyrimidine bases (nucleotides 12 to 15). These nucleotides are solvent exposed in most of the final calculated structures, suggesting that all or some of these nucleotides would be immediately accessible for protein interactions.
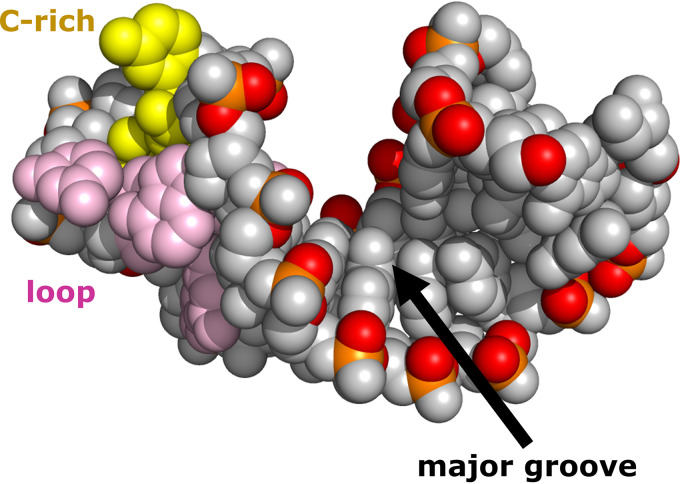
FIG 4.
RV14 SLB structure. A 6-bp A-form helix is capped by a highly dynamic 8-bp loop (pink and yellow bases), with a C-rich patch (yellow bases) in the loop near the stem. This C-rich region would be accessible to the host poly(C) binding protein (PCBP2). Due to the limited length of the helix, the major groove is accessible.
Typically, for an A-form RNA helix, the shortest phosphorus distance measurements that define major groove width are Pi to Pi−6. This measurement requires a minimum of 7 bp. The RV14 SLB helix contains only 6 bp, indicating that SLB is too short to close off access to the major groove. The shortest phosphorus distances measured were Pi to Pi−5, yielding an average major groove width of 8.7 Å, atypically large for A-form RNA, even though other helical parameters are consistent with A-form geometry. Thus, the SLB major groove is more accessible than the major groove in a longer A-form stem. This accessibility could play a role in interactions with PCBP2 or other unidentified proteins and molecules. It is interesting that both SLB and SLD contain accessible major grooves. However, major groove accessibility in SLD is apparently due to a pyrimidine-pyrimidine base pairing and not due to the length of the helix, as is the case for SLB. Yet both of the widened major grooves of the stem-loops have the potential to enhance interactions with host or virus proteins.
Intact 5′ Cloverleaf
Currently, two solution structures of the full RV14 5′-CL are available (17). Determining the high-resolution structure of large RNA by NMR alone is difficult because of spectral overlap combined with broadening due to rapid relaxation. Therefore, a combined small-angle X-ray scattering (SAXS)/NMR analysis was used. The SAXS/NMR approach can provide high-resolution structural information, particularly when separate high-resolution structures are available for parts of a complex to be studied (18–20). In this case the 5′-CL can be thought of as a complex between stem A and three stem-loops, the largest two of which (SLB and SLD) have high-resolution structures available, as discussed above. Together, SLB and SLD comprise over 60% of the total cloverleaf. Chemical shift analysis, together with RDC and SAXS fitting, was used to confirm that the SLB and SLD structures were largely conserved upon incorporation into the cloverleaf.
The most notable change upon incorporation of SLB and SLD into the 5′-CL involves the stem terminus of SLB. In the absence of magnesium, the C10-G31 base pair becomes stabilized in the 5′-CL. This extends the helix of SLB in the 5′-CL by 1 bp, matching the predicted length of SLB. In addition, the imino resonance of U29 appears to split into two or more resonances, suggesting the presence of two or more slowly interconverting states. However, in the presence of magnesium, the imino resonance of G31 disappears, suggesting a destabilization of the C10-G31 base pair, returning to the state seen in the isolated SLB structure. The two or more resonances observed for U29 also collapse to form a single sharp imino resonance. This evidence clearly indicates that, in the presence of magnesium, SLB in the RV14 5′-CL adopts a single stable conformation that is similar to that of the isolated SLB structure.
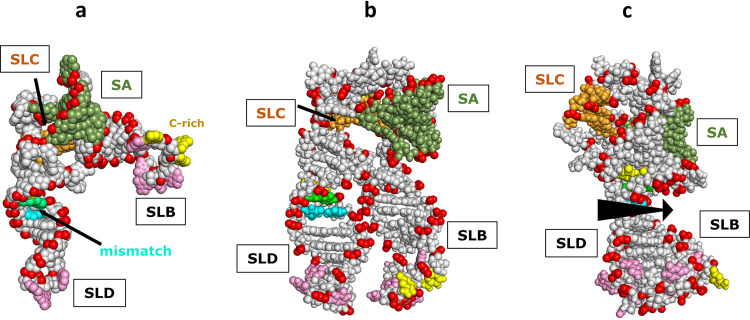
The global structure of the 5′-CL is also very different in the absence and the presence of magnesium (17). The positive charge associated with metal cations can shield the electrostatic repulsion between backbone phosphate groups, allowing RNA to fold into a more stable and compact tertiary conformation (21–23). The most abundant divalent cation in cells is magnesium, and it is considered the most important divalent cation for RNA stabilization. In the absence of magnesium, the RV14 5′-CL adopts an open conformation, with SLB and SLD approximately perpendicular to one another (Fig. 5a). In contrast, in the presence of magnesium, SLB and SLD swing toward each other to form a more compact and more ordered structure (Fig. 5b). Since the loops of SLB and SLD are known to bind PCBP2 and 3CD, respectively, the possible interactions between these two proteins would stand to be affected by the presence of magnesium. In addition, it is possible that since magnesium can induce this structural change, protein binding may also affect a similar change so that SLB and SLD may approach each other even in the absence of magnesium in certain circumstances that are yet to be determined.
FIG 5.
RV14 5′-CL NMR/SAXS-based structure. (a) Open conformation observed in the absence of magnesium. SLB and SLD are approximately at a right angle to each other. SLC is obscured by stem A (SA) in this view. The positions of the three SLD mismatch base pairs and the SLB C-rich loop region are indicated. (b) Closed conformation observed in the presence of magnesium. SLB and SLD are parallel and in close contact, facilitated by a magnesium counter ion(s); SLC is again obscured. (c) Closed conformation from panel b, rotated 90° to show that the accessible major grooves of SLB and SLD align to create an extensive accessible major groove surface (arrow). SLB is largely obscured by SLD in this view.
Interestingly, in the compact magnesium-induced structure, when SLB and SLD come together, they do so in an orientation that aligns the accessible major grooves that were discussed in the previous section (Fig. 5c). This suggests that if proteins do indeed bind into these grooves as part of the replication process, there may be some interaction of a protein across both grooves or interaction between the two proteins that bind separately into each groove. Additional studies will be necessary to elucidate the potential participation of this major groove surface in the replication process.
In the context of the above structure, it is instructive to compare the predicted 5′-CL secondary structure from RV14 with that of three of the other representative enteroviruses to be discussed in this review. As can be seen in Fig. 2, the overall predictions are similar, with three stem-loops and a stem converging at a four-way junction. The string of three consecutive cytosines in the SLB loop, which interact with PCBP2, are conserved. Within SLD, the pyrimidine-rich mismatch region is somewhat conserved, with either two or three mismatches present. This suggests that the unique characteristics of SLD giving rise to an accessible major groove may be conserved. However, SLB is predicted to be longer in the other enteroviruses than in RV14, and this could result in closing off access to the SLB major groove. Interestingly, the length of SLB appears to be correlated with the length of SLD, with both being extended near the four-way junction, not near the loop. So, while the overall sizes of the 5′-CL structures may vary, it would appear that the relative positioning of the SLB and SLD loops may be conserved, which would conserve the relative positioning of PCBP2 and 3CD bound to each loop.
VIRUS-ENCODED 3A, 3B, 3C, AND 3D PROTEINS
A schematic view of the enterovirus genome was given in Fig. 1. The single open reading frame codes for a single polyprotein that contains three regions, denoted P1, P2, and P3. The P1 proteins encode structural proteins, while regions P2 and P3 encode replication-related proteins. This review will focus on the structure of proteins from the P3 region which perform the bulk of the replication functions.
The enterovirus precursor protein P3 (3ABCD) can be cleaved into fragments of various sizes (8, 24–26). The major pathway is cleavage into 3AB and 3CD. In the minor pathway, P3 is cleaved into 3A and 3BCD, the latter of which is subsequently cleaved into 3BC and 3D, and finally 3BC can be cleaved into 3B and 3C. Thus, molecular species found in the cell can include 3A, 3B, 3C, 3D, 3AB, 3BC, 3CD, 3BCD, and 3ABCD. High-resolution structural results are available for each of the isolated proteins 3A (soluble fragment only), 3B, 3C, and 3D, as well as for the 3CD protein. These five proteins will be discussed in turn below.
3A
Enterovirus replication takes place on remodeled host cell membrane surfaces (27, 28). It is thought that membrane materials are recruited from host cell secretory or autophagy organelles. The membrane-associated 3A protein (29) plays a central role in membrane remodeling, in part via direct interaction with GBF1 and indirect interaction with Arf1 (30–34). This remodeling not only creates the appropriate surface for replication but also disturbs the secretory pathway, which in turn suppresses immune function by impairing both cytokine secretion and expression of major histocompatibility complex (MHC) class I on the host cell surface (35–37). In addition, 3A recruits PI4KIIIβ to the replication membrane surface via mutual interactions with the GPC60 protein (38, 39). This recruitment increases levels of the lipid phosphatidylinositol 4-phosphate (PI4P), which in turn may help to recruit the virus polymerase and/or PH-domain proteins with additional membrane-altering properties.
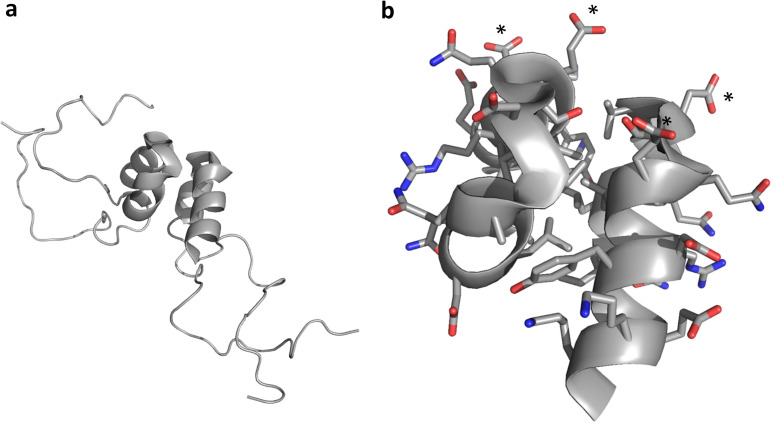
Structural biology of membrane-associated proteins entails significant challenges. The poliovirus (PV) 3A protein consists of 87 residues: an N-terminal 58-residue soluble region (3A-N) followed by a 22-residue hydrophobic region (membrane-associated) and seven additional C-terminal residues. The solution structure of 3A-N was published in 2003 (40). 3A-N forms a symmetric dimer (Fig. 6a). Each monomer unit contains a two-helix hairpin spanning residues 23 to 41, consisting of a two-turn helix connected by a short, nearly 180°, turn to a three-turn helix. Mostly disordered N- and C-terminal regions flank residues 23 to 41. Mutational analysis has been performed to attempt to relate 3A structure and function (11, 30, 40, 41). These studies suggest that while the C-terminal disordered region of 3A-N may simply physically tether the 3A-N fragment to the C-terminal membrane-associated part of 3A, the N-terminal region of 3A-N appears to play a more critical and specific role in both 3A and 3B activity.
FIG 6.
3A-N structure. (a) A soluble N-terminal 59-residue fragment of the PV1 3A protein forms a symmetric homodimer in solution. Each monomer contains a helical hairpin, flanked by largely disordered N and C termini. (b) Zoomed view of the 3A-N dimer interface, showing a hydrophobic dimerization surface that includes residues I-22, L-25, L-26, V-29, V-34, Y-37, C-38, and W-43. The nondimer surfaces are highly charged, with a negative charge cluster (D-29 and E-32 from each monomer) indicated by asterisks. Side chain oxygen atoms are shown in red. Side chain nitrogen atoms are shown in blue.
The highly ordered 3A helical hairpin is amphipathic, providing a hydrophobic face for dimerization and polar faces that interact with solvent (Fig. 6b). While the polar faces contain both positive and negative charges, a cluster of four negative residues (D-29 and E-32 from each monomer) appears near the 180° turn between helices. This negative patch may play a role in interacting with a positive patch in 3B, helping to position 3B correctly for interaction with 3D. The role of dimerization is not yet clear though it does result in a doubling of the 22-residue membrane-associated region and, hence, likely helps to stabilize interactions with membranes (40).
3B
The enterovirus 3B peptide becomes uridylated (42) on its third amino acid (tyrosine). This uridylated peptide then serves as a primer for RNA synthesis during virus replication. As a result, 3B is found covalently linked to the 5′ end of the viral genome, for which reason it is also called VPg (virion protein genome linked).
The PV1 3B peptide consists of 22 amino acids. NMR analysis of PV1 3B in aqueous solution shows it to be disordered and flexible in low-salt buffer at neutral pH (43). Interestingly, uridylation does appear to decrease disorder in VPg to some extent, relative to the structure of the unuridylated VPg, as evidenced by the appearance of medium- and long-range NOE cross-peaks, but no regular secondary structural elements are introduced (44). In contrast, the addition of trimethylamine N-oxide (TMAO) induces two turns of an alpha helix at the VPg C terminus though the functional consequences of this observation are unclear (43). 3B structure is again discussed below in the context of replication complexes involving 3Dpol.
3Cpro
The first enterovirus protein to be studied structurally was the 3C protease (3Cpro). This viral protease performs the bulk of virus polyprotein processing by cleaving the peptide bond at Gln-Gly junctions to create individual virus proteins (4, 8). 3Cpro similarly cleaves a large number of host proteins, effecting sweeping cellular changes such as disruption of transcription (45–49), translation (50–56), nucleo-cytoplasmic transport (57–61), and Golgi compartments (62). 3Cpro-induced cleavage also triggers apoptosis (49, 63–66), which is required for release of nascent virus particles. A second distinct function for 3Cpro is binding to RNA. Specifically, 3Cpro or its precursors bind to oriL, oriR, and oriI, which are located, respectively, within the 5′ NCR, the 3′ NCR, and the coding region of the 2C protein (9, 67–72). The oriL interaction is the best understood of these: in this interaction, 3Cpro binds stem-loop D (SLD) of the 5′ cloverleaf (5′-CL) and, in so doing, brings along the still covalently attached 3D polymerase, which is the topic of the next section (73). In addition, 3Cpro binds phosphoinositides (74) and so plays a role in membrane association. The 3Cpro protein also appears to play a role in VPg uridylation (69, 75).
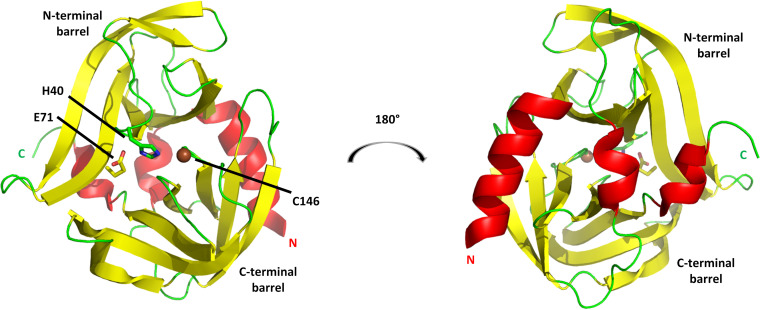
The first 3Cpro structure determination was that of rhinovirus (RV14) (76). Crystallography revealed that this 182-amino-acid protein consists primarily of two topologically equivalent six-strand beta barrels that form a proteolytic pocket at their interface (Fig. 7, left). The architecture is very similar to that of trypsin-like serine proteases (77). However, the active-site catalytic triad consists of His-40, Glu-71, and Cys-146, in place of His, Asp, and Ser in trypsin. Therefore, although structurally similar to serine proteases, 3Cpro is a cysteine protease.
FIG 7.
3Cpro from RV14. The cysteine protease structure consists principally of two six-stranded beta barrels connected by a long linker. (Left) View of the proteolytic active-site face, with the position of the three catalytic triad residues indicated. (Right) View of the opposite face, a surface that consists largely of three helices: a long alpha helix at the N terminus (marked by N), a short 310 helix near the C terminus (marked by C), and a short alpha helix at the center of the linker connecting the N-terminal and C-terminal barrels. This opposite face interacts with RNA and phosphoinositides. Models are colored according to secondary structure: helices are red, beta strands are yellow, and other elements are green. The catalytic C-146 sulfur atom is shown as a brown sphere.
Both His-40 and Glu-71 are positioned in the top (N-terminal) beta barrel, while Cys-146 is from the bottom (C-terminal) barrel. More specifically, both His-40 and Cys-146 lie within the central loop (connecting strands 3 and 4) of their respective barrels, while Glu-71 is positioned within the sixth strand of the N-terminal barrel. Although differences in backbone positions of the catalytic triad residues relative to those of serine proteases are seen, due for instance to the increased length of Glu relative to that of Asp, the functional parts of the side chains are in equivalent relative positions. Therefore, it is straightforward to infer that the substantial knowledge of the serine protease mechanism can be applied to 3Cpro. It is therefore safe to assume that His-40 and Glu-71 help to position and deprotonate the Cys-146 side chain, causing it to become a powerful nucleophile that targets the substrate Gln carbonyl carbon.
There are, however, significant structural differences between 3Cpro and trypsin. The most obvious difference is the positioning of alpha helices. Trypsin and 3Cpro each contain three conserved helices. However, the largest 3Cpro helix (3.5 turns) occurs at its extreme N terminus and may play a role in binding phosphatidyl inositol phosphates (PIPs) in intracellular membranes (74). There is no analogous helix in trypsin. The largest trypsin helix (4 turns) occurs at its C terminus, while 3Cpro contains only a short (5-residue) 310 helix near its C terminus. In 3Cpro, the linker connecting the two barrels contains a third helix, a 1.5-turn alpha helix, near its center. This helix is absent in trypsin. Some 3Cpro forms (e.g., PV [78]) also contain a short ∼1-turn alpha helix in the loop that contains the catalytic His-40 residue. An analogous helix is present in trypsin at this position. Trypsin also contains an additional two-turn alpha helix between strands 2 and 3 of the C-terminal beta barrel, which has no analog in 3Cpro.
Together, the three conserved 3Cpro helices, one from each terminus and one from the linker, form a distinct helical surface on the face opposite from the active site (Fig. 7, right). This face, which contains significant basic character, is the site for RNA interaction (9, 79, 80). In fact, the RNA and PIP-binding surfaces overlap, resulting in competitive binding to 3Cpro (74). It is also interesting that the 3Cpro proteolytic active site and the RNA binding surface each contain residues from both of the two 3Cpro beta barrels. Thus, it would not be surprising that binding to either one of these faces could allosterically affect activity at the opposite face, perhaps by affecting the spatial relationship between the two barrels. Indeed, functional evidence (81) along with chemical shift mapping (82) has confirmed allosteric control of each of these faces by binding to the opposite face.
A more subtle difference between trypsin and the 3Cpro structure is that many of the loops connecting beta strands are of different lengths in the two proteins. In general, the loops in 3Cpro are shorter. For instance, trypsin has a lengthy (∼11-residue) loop between strands 4 and 5 of its N-terminal barrel, which is used for calcium binding. In contrast, 3Cpro has only a short turn between these two strands. Similarly, an extended autolysis loop connecting strands 1 and 2 from the trypsin C-terminal barrel is also absent in 3Cpro. The two-turn helix between strands 2 and 3 of the C-terminal barrel, found only in trypsin, was mentioned in the previous paragraph. Due mostly to these differences in the number of residues connecting beta strands, the length of the bovine pancreatic trypsin protease exceeds that of 3Cpro by 63 residues although the two proteins share a core structure and topology. The shorter loops in 3Cpro would be expected to reduce the exposure to proteolytic degradation.
An additional conclusion of 3Cpro structural analysis is that one 3Cpro molecule cannot cut itself free of its flanking 3B and 3D proteins: the 3B-3C and 3C-3D cut sites (at the N and C termini, respectively, of 3Cpro) would be too distant from the active site. Thus, 3Cpro must be cut in trans; that is, a second 3Cpro molecule (or 3C-containing larger molecule) must be used.
Since this first description of 3Cpro structure, several other enterovirus 3C protease structures have been determined, including those of two additional rhinoviruses (RV2 [83] and RV16 [84]), poliovirus (78), two forms of foot-and-mouth disease virus (FMDV) (85, 86), two forms of hand-foot-and-mouth disease (HFMD)-linked EV71 (reference 87 and PDB entry 5HXF), two coxsackieviruses (the cardiovirus CVB3 [88] and CVA16 [89], which also causes HFMD), and two additional enteroviruses linked to non-polio paralysis (EVB93 [90] and EV68 [91]). These structures show very high similarity to the 3Cpro structure of RV14. (The FMDV picornavirus belongs to the subclassification aphthovirus, not enterovirus, but FMDV has been included in this review due to the insights that FMDV structural biology has provided that apply also to the closely related enteroviruses.)
Efforts to inhibit the proteolytic activity of 3Cpro are beyond the scope of this review, but, briefly, structures of 3Cpro complexed with either covalently or noncovalently bound inhibitors or target peptides have been published for each of the 3Cpro forms listed above (84, 91, 92). Complexes with peptides mimicking target sites were mostly accomplished via mutation of the active-site Cys residue to alanine to prevent cleavage of the target. Inhibitors are in general designed to fit into the proteolytic pocket but to resist cleavage. Inhibitors have not yet been developed to target the opposite face of 3Cpro.
3Dpol
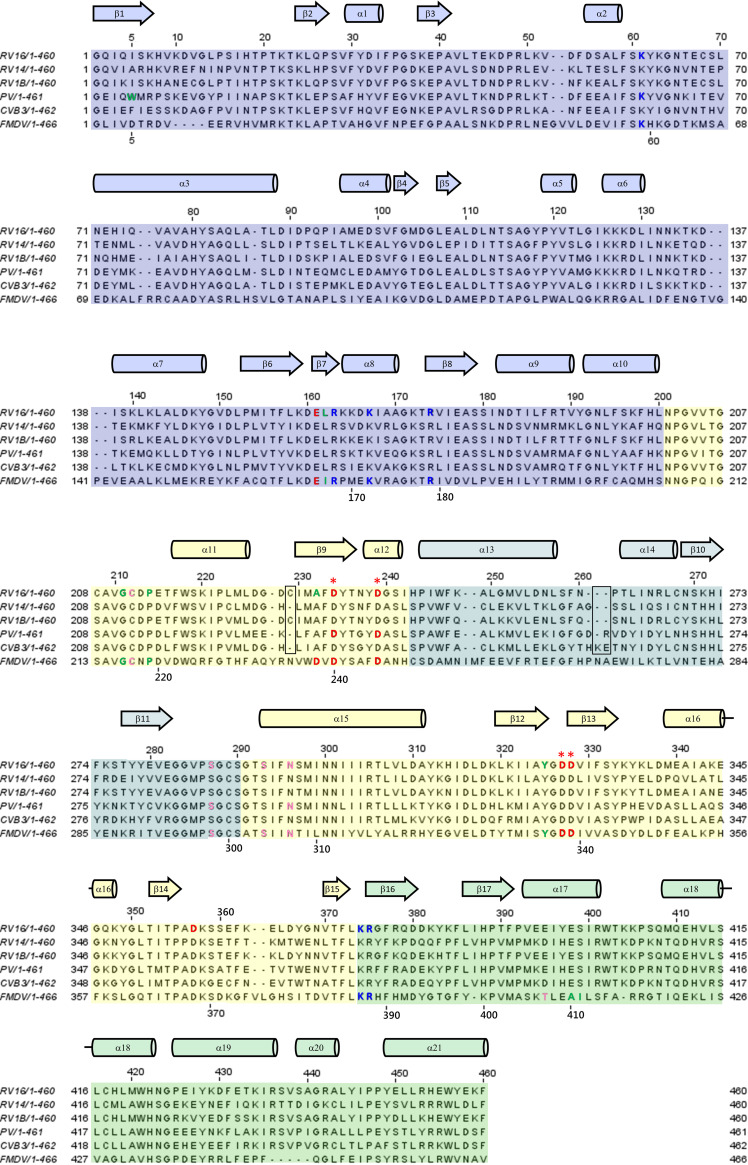
A sequence alignment of all 3Dpol variants discussed in this review is presented in Fig. 8. As can be seen, the numbering for each of these variants differs by at most two, apart from the nonenterovirus FMDV 3Dpol variant, which has multiple inserts and deletions relative to the enterovirus sequences. In this section, for consistency, the amino numbering for 3Dpol from RV16 is used throughout. The corresponding residue numbers for each 3Dpol variant discussed in this review can be ascertained from Fig. 8.
FIG 8.
Sequence alignment of the 3Dpol variants discussed in this review. The f1 and f2 regions of the fingers domain are shaded blue and teal, respectively. The palm and thumb domains are shaded yellow and green, respectively. Residues discussed in the text are indicated by text coloring according to the polarity of their side chains: red for acidic, blue for basic, pink for polar, and green for hydrophobic. The four conserved Asp residues from the palm domain that are most prominently discussed in the text are marked by red asterisks. Secondary structure is depicted as helices (cylinders) and beta strands (block arrows). Residue numbering above each row of the alignment corresponds to that of RV16. FMDV numbering is provided along the bottom of each row in areas near residues that are discussed in the text. The two boxed regions indicate the regions where the enterovirus sequences contain differing numbers of residues. The positions of these minor gaps can vary slightly in different alignments, but these two small regions account for the minor difference in numbering between the depicted enteroviruses. FMDV is not an enterovirus and has multiple insertions and deletions relative to sequences of the enteroviruses, resulting in larger differences in numbering.
Enteroviruses replicate their RNA genome directly, with no DNA intermediate. Thus, they must code for an RNA-dependent RNA polymerase (RdRp). For brevity, this 52-kDa polymerase is called 3Dpol (70). Similar to other nucleotide polymerases (93, 94), the catalytic mechanism is effectively a phosphoryl transfer reaction: the 3′ OH group of the nascent lengthening strand (primer strand) is deprotonated. The resultant strongly nucleophilic 3′ O group attacks the α phosphate group of the incoming nucleoside triphosphate (NTP). The α phosphate group and the attached base are then transferred onto the primer chain, and the β-γ diphosphate group is released from the ribonucleotide triphosphate (rNTP). Two divalent metal ions (typically Mg+2) are involved. These ions help to orient the incoming rNTP in an active conformation, stabilize the deprotonated 3′ O group of the primer strand, and stabilize a transition state in which the rNTP is transiently bound to the primer strand, prior to the release of the β-γ diphosphate group (95). Various segments of the polymerase help to position the metal ions and help to position and move the template strand, primer strand, and the incoming NTP. The discussion of 3Dpol structure below should be viewed in light of this conserved mechanism.
The 3Dpol enzyme displays self-associative properties: 3Dpol-3Dpol interactions are detected in yeast two-hybrid and cross-linking studies (96), and 3Dpol activity displays a concentration dependence that also suggests intermolecular cooperativity (97). Thus, 3Dpol-3Dpol interactions can affect crystallization, leading to a loss of density along and near interfacial regions (98).
The first enterovirus 3Dpol structure determination effort, published in 1997 (98), targeted poliovirus (PV). Due to intermolecular contacts in the crystal, the structure of only approximately 70% of the protein could be determined. However, the results were sufficient to show that enterovirus 3Dpol shares a common fold with the three other categories of nucleic acid polymerases: DNA-dependent DNA polymerases (DdDp; replication), DNA-dependent RNA polymerases (DdRp; transcriptase) and RNA-dependent DNA polymerases (RdDp; reverse transcriptase). The fold common to all four of these polymerase categories is referred to as a “right hand,” first used to describe the large (Klenow) fragment of Escherichia coli DNA polymerase I (99).
The 3Dpol hand consists of three domains, a palm, fingers, and thumb domain, as shown in Fig. 9. For clarity, the full structure of the RV16 3Dpol structure, published in 2004 along with RV14 and RV1B 3Dpol structures (100), is used in Fig. 9, and as mentioned, RV16 numbering will be used in this section. In another report published in 2004, mutation of one of the interfacial regions of the PV 3Dpol, very near the C terminus was used to successfully disrupt intermolecular interactions and allow full structure determination of the PV 3Dpol (101). The FMDV 3Dpol structure was also determined in 2004 (102). Other 3Dpol structures have followed more recently, with similar results, as part of complex studies with RNA, which will be discussed further in the following section.
FIG 9.
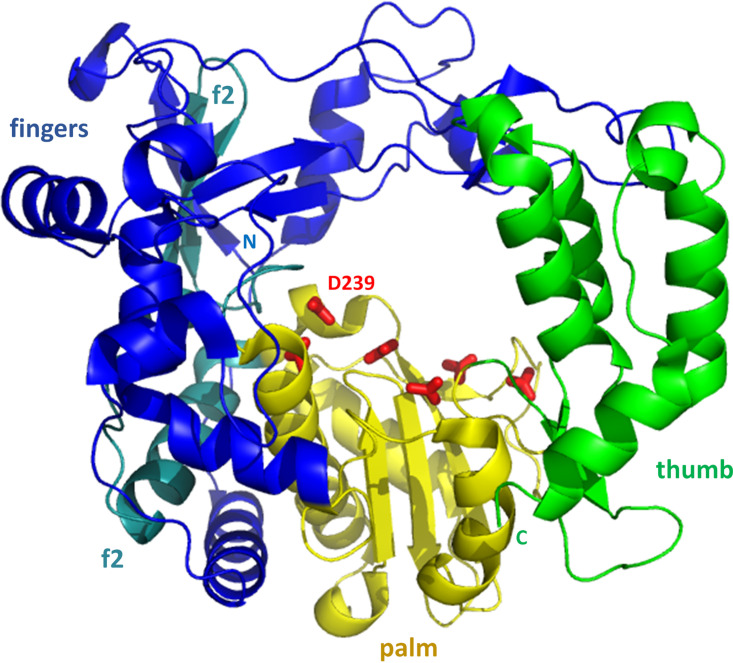
3Dpol from RV16. The structure resembles a right hand (facing the reader), with a central palm domain (yellow) connected to an N-terminal fingers domain (blue and teal) and a C-terminal thumb domain (green). The fingers and thumb are in spatial contact, closing the circle and forming a channel through the polymerase. Side chains of Asp and Asn residues that coordinate catalytically required magnesium ions are shown as red sticks along the top of the palm domain. D-239 also hydrogen bonds to the 2′ OH group of the template, helping to select for an RNA template. The two f2 labels identify residues 243 to 290 (teal) of the fingers domain, which are sequentially inserted between two regions of the palm domain. The five-strand beta sheet from the fingers domain lies below the top f2 label. The positions of the N and C termini are indicated.
The PV, FMDV, and RV 3Dpol structures are all very similar. The palm domain is central to the right-hand fold, sequentially, structurally, and functionally. Using RV16 numbering, the 14-kDa palm consists of residues 201 to 373, minus an intervening sequence from the fingers domain at residues 243 to 290. Its core structure, present in all four categories of polymerase (RdRp, DdDp, RdDp, and DdRp), is two alpha helices packed behind a three-stranded beta sheet (Fig. 9). The most extensive additional secondary structural element is a three-turn helix that packs above the beta sheet and connects the palm to the thumb region to the right. This additional helix was previously observed only in RdDps (and now in RdRps). The extensive fingers domain is to the left. A channel through 3Dpol is evident, and passage of RNA through this channel will be discussed in subsequent sections. Residues known to be important for binding the two catalytically required magnesium ions are observed on the top of the palm, facing the channel. These include Asp-234, -239, -327, -328, and -357, along with Asn-296 (shown as red sticks in Fig. 9). Asp-239 is also involved in selecting for an RNA template, by hydrogen bonding to the 2′-OH group.
The fingers domain is the largest of the three domains. It consists of residues 1 to 200, along with the aforementioned stretch of residues 243 to 290 that sequentially partitions the palm domain. Altogether, this domain is approximately 28 kDa, representing more than one half of the total polymerase. For discussion, the fingers domain can be divided into three regions: (i) the inner finger region, consisting of five helices (also known as five fingers) near the palm; (ii) the outer finger region, further from the palm, consisting of a five-strand beta sheet capped by several surface-exposed helices, along with an additional helix that supports the position of the N terminus; (iii) the N terminus (residues 1 to 54), which has limited regular secondary structure and reaches across to contact the thumb, thereby completing the circle surrounding the channel through the polymerase.
The smallest domain, the 10-kDa thumb domain, is comprised of the C-terminal residues 374 to 460. It primarily consists of a five-helix bundle that contains four long and one short helix. The bundle is sequentially connected to the palm domain by a small two-strand beta sheet and connected spatially to the N terminus region (of the finger domain) via the top of the helix bundle.
Three additional PV 3Dpol crystal structures were also published in 2004 (101): (i) an inactive G1A mutation that helped to establish the importance of positioning of the N terminus; (ii) a 68-residue N-terminal truncation mutant; (iii) a cocrystal in which ribo-GTP (rGTP) was diffused into a preexisting 3Dpol crystal. The last helped to confirm the role of Asp-239, which, as mentioned, hydrogen bonds to the 2′ OH group, selecting for an RNA template. Via comparison of the structures with and without rGTP and comparison with structures of the other classes of polymerases, it appears that relatively modest conformational rearrangement of 3Dpol is necessary in order to accommodate the substrate and product RNA molecules.
The conservation of 3Dpol structure among enteroviruses was further confirmed via structural analysis of 3Dpol from CVB3 (103). An additional fact that became apparent upon solving the CVB3 3Dpol structure is that a distortion in the N-terminal beta strand, part of the five-strand beta sheet in the outer region of the fingers domain, is conserved in PV, RV, and CVB3. This distortion, which results in solvent exposure of the hydrophobic side chain of residue Phe-5, was originally thought to be a crystal-packing artifact when it was seen in PV. However, functional and mutational analysis that was undertaken based on structural conservation indicates that this distortion is necessary for polymerase activity. It was proposed that exposure of a hydrophobic residue at position 5 is necessary for subsequent burial of the N terminus as a required step in replication. Results from mutational analysis of the FMDV 3Dpol N terminus (104), along with crystallography and RNA binding, appear to confirm both that the 3Dpol N terminus is conformationally flexible and that this flexibility is important in function.
As mentioned above, experiments suggest that intermolecular 3Dpol-3Dpol interactions not only affect crystallization but may also assist in replication. Electron microscopy (EM) has been used to visualize two-dimensional PV 3Dpol arrays that may form on virus-induced host cell membranes (105–107). These arrays utilize 3Dpol-3Dpol contacts that were first identified as poliovirus 3Dpol crystallographic contacts, along with additional contacts. EM studies have identified a lysine-rich patch that forms at the interface between adjacent 3Dpol filaments within the two-dimensional array and may help attract the RNA template to the filament (108). The concept that replication may take place on a surface array of 3Dpol molecules (surface catalysis) would help to explain the cooperative nature of RNA replication by 3Dpol and may be related to the variable VPg binding sites discussed in the initiation complex section (see below).
3CD
The 3CD protein, consisting of the covalently linked 3Cpro and 3Dpol molecules, is a functional precursor to the protease and polymerase. The 3CD protein displays protease activity but with somewhat altered specificity relative to that of 3Cpro (109, 110). However, 3CD does not function as a polymerase (111). In addition, 3CD has recently been shown to induce biogenesis of membrane surfaces required for replication (112).
The first structural studies of 3CD protein were performed with PV. In order to produce viable crystals of PV 3CD (113), it was necessary once again to mutate several residues from the self-associating surface near the 3D C terminus, as was done to assist PV 3D crystallography. In addition, four mutations were made in the 3C region: three acidic residues were mutated to prevent self-association of 3C (E55A, D58A, and E63A), and the active-site cysteine was mutated (C147A) to prevent cleavage of 3CD into 3C and 3D.
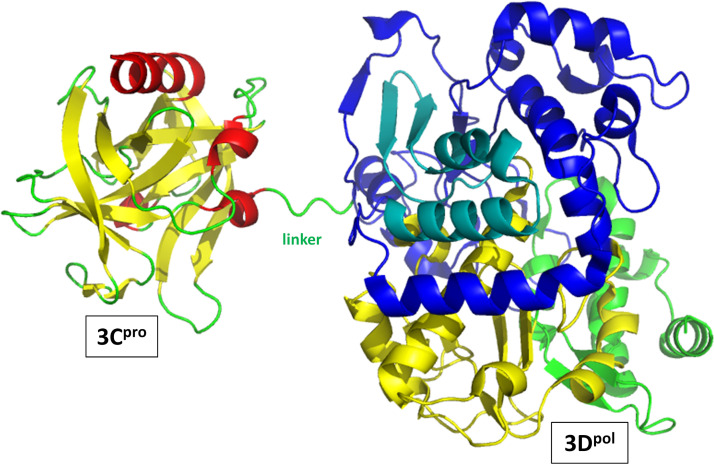
With these alterations, a 3CD crystal form was determined that showed no intramolecular contact between the 3C and 3D regions of a single molecule (Fig. 10). These two regions were joined by a poorly ordered and thus highly accessible seven-residue linker (residues 180 to 186) that contains the target site for separation of 3C and 3D, the Q-183/G-184 peptide bond. Thus, the linker contains four residues from 3C and three residues from 3D. The structure confirms that in the context of 3CD, the 3C proteolytic active site is too distant from the Q-183/G-184 site to be capable of cutting itself free from 3CD: cleavage must be performed by a distinct 3C (or 3C-containing) molecule.
FIG 10.
3CD from poliovirus. Orientation showing the linker between the 3C and 3D domains, emphasizing that the 3C (left) and 3D (right) domains within a 3CD molecule do not contact each other in the 3CD crystal. The 3C domain makes the closest approach to the N-terminal fingers region of 3D, to which it is covalently attached via the linker. As discussed in the text, in solution, the relative positioning and interactions of the 3C and 3D moieties are dynamic. Coloring is as described in the legends of Fig. 7 and 9, except that no side chains are shown and the linker between 3C and 3D is shown in green.
Globally, PV 3CD has very high structural similarity to the two separate molecules: main chain RMSD is 0.6 Å between 3C and the 3C domain of 3CD and 0.49 Å and 0.67 Å between 3D and the two symmetry-related 3D domains in the 3CD crystal. However, there are significant localized changes in both 3C and 3D upon incorporation into 3CD, with most of these occurring near the N terminus of each domain. It is not surprising that there would be changes near the Q-183/G-184 cut site: after cleavage, the last three residues of 3Cpro become more disordered. In contrast, the first three residues of 3Dpol change position and conformation but become well ordered in free 3Dpol due to a new set of hydrogen bonds. These bonds result in a lengthening of the N-terminal beta strand of the five-strand sheet in the fingers domain. As mentioned in the section discussing 3Dpol, hydrophobic residue 5 (a tryptophan in PV 3Dpol) is thought to be critical for activity. While Trp-5 is in a similar position in 3Dpol and the 3D domain of 3CD, the lengthening of the beta strand after cleavage occurs just N-terminal to Trp-5 (residues 2 to 4). Thus, the environment of Trp-5 changes significantly. Interestingly, the N-terminal helix of 3Cpro, which is far from the Q-183/G-184 site, also changes position slightly, though the biological relevance of this change is unclear.
Other significant localized changes are sprinkled throughout the 3Dpol N-terminal fingers domain, and these could help to explain why 3CD cannot function as a polymerase; release of the N terminus after cleavage at Q-183/G-184 appears to trigger these additional changes even though the extreme N terminus structure does not change to a great extent. Perhaps flexibility of the N terminus, which is not possible when 3D is covalently bound to 3C, is required to trigger these additional changes and activate the polymerase.
Even with the introduced mutations that assist crystal formation by reducing 3D-3D and 3C-3C contacts, intermolecular contacts are abundant in the PV 3CD crystal. Each 3D domain contacts other 3D domains and a 3C domain from another molecule. These extensive interactions further support the concept that larger complexes may be important in replication. Furthermore, results from a molecular dynamics and small-angle X-ray scattering analysis of PV 3CD suggest that the intramolecular relationship (orientation and distance) between the 3C and 3D regions in 3CD may be dynamic in solution, allowing for direct contact between 3C and 3D within one 3CD molecule (114). This dynamic relationship may be important for function.
Finally, it should be noted that in addition to 3C, 3D, and 3CD, the larger protein precursors 3BCD and 3ABCD are also found in infected host cells. As discussed above, the activity of 3CD differs from that of the separate 3C and 3D molecules. Less is known regarding 3BCD and 3ABCD function, and structural information is not yet available for either. However, evidence suggests that 3ABCD may be the preferred form of these proteins in replication and that separate 3ABCD molecules may provide the activities of 3B (VPg), 3C, and 3D to a particular replication nucleus (115). This concept is consistent with the surface array concept mentioned in the 3Dpol section: it is conceivable that arrays of 3ABCD may form on intracellular membranes, priming and cleaving each other to initiate replication.
PROTEIN/RNA COMPLEXES
Interactions between virus RNA and virus proteins are central to the replication complex. As such, a considerable number of structures are available detailing interactions between the 3Dpol replicase and RNA strands representing parts of the virus genome. In addition, a single study has been performed to determine the structure of the complex between the 3Cpro protease and part of the enterovirus 5′-CL RNA.
3Cpro/SLD Complex
A combined SAXS/NMR approach was used to investigate the interaction between 3Cpro and SLD from the 5′-cloverleaf RNA, using RV14 as a model system (82). The RV14 3Cpro structure was not at the time available in the Protein Data Bank (PDB). Instead, an RV14 3Cpro PDB file was generated via an approach using NMR-based molecular replacement. In this approach, an RV14 3Cpro homology model was first generated based upon the RV2 3Cpro structure (83). The model was then refined to fit RV14 3Cpro backbone 15N,1H-residual dipolar couplings. This structure was confirmed via visual inspection of figures in a previous publication (76). Interactions with RV14 SLD were then mapped via chemical shift perturbations in 3Cpro upon addition of SLD.
As expected, the perturbations mapped strongly to the surface opposite from the proteolytic active site, particularly the N-terminal half of the linker joining the two beta barrels and nearby residues from the N-terminal beta barrel. In addition, HN resonances from most of the 3.5-turn N-terminal alpha helix were broadened beyond the detection limit, indicating instability on a millisecond-microsecond time scale in the conformation or the environment of this helix upon exposure to SLD. The chemical shift mapping information was combined with additional information regarding the RNA-binding surface of 3Cpro obtained via sequence conservation and mutagenesis analysis (67). These combined interaction data were then used as restraints while generating atomic models that fit the SAXS data of a 1:1 complex of SLD-3Cpro.
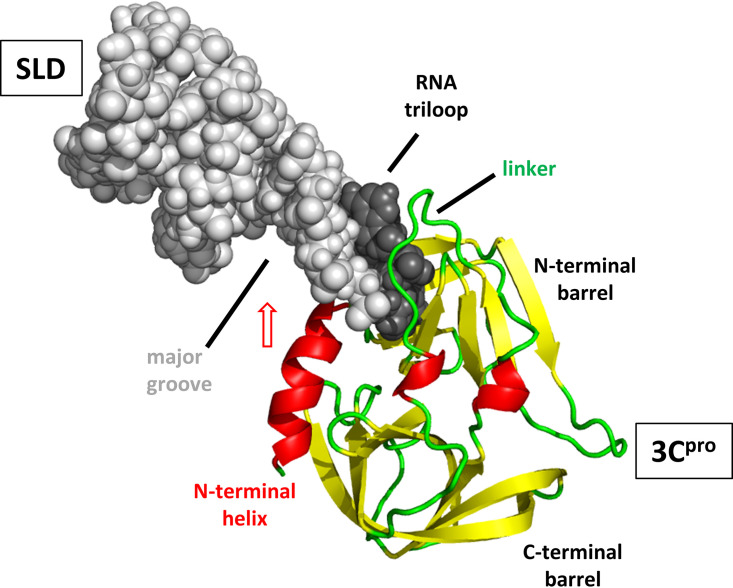
The resulting structure is shown in Fig. 11. In this structure, the loop region of SLD inserts between the N-terminal half of the linker and the C-terminal half of the broadened N-terminal alpha helix. One interesting possibility that arises from inspection of this complex structure is that the 3Cpro N-terminal alpha helix appears to be in position to swing up and into the unusually accessible major groove from SLD. If such a motion occurs on a millisecond-microsecond time scale, this could explain the observed resonance broadening described for the N-terminal helix. It could also help to explain the roles of both the wide major groove of SLD (14) and the N-terminal alpha helix of 3Cpro. In addition, small perturbations were observed near the proteolytic active site. It is not difficult to imagine that disturbances of the N-terminal barrel by RNA binding could affect the proteolytic active site, perhaps by altering either the relationship between the catalytic triad residues or the geometry of other parts of the binding surface for the target peptide. A recent analysis of chemical shift and 15N relaxation experiments also showed changes on each of the two PV 3Cpro surfaces when target molecules bind to the opposite face (81). These alterations may be an important element of regulation of the viral replication cycle.
FIG 11.
RV14 3Cpro/SLD complex. 3Cpro is shown as a cartoon and oriented and colored as described in the legend of Fig. 7 (right panel). SLD is shown as spheres. The SLD triloop is shaded darker than the stem regions. The structure indicates that the SLD triloop inserts into 3Cpro between the linker joining the two beta barrels and the N-terminal alpha helix. Broadening of NMR resonances from the N-terminal helix suggests that it may swing into the accessible major groove of SLD (red arrow).
Replication Complexes
Replication of the enterovirus RNA can be broadly divided into two major steps: initiation and elongation. The dividing line between these two processes is subject to definition. For the purposes of this review, initiation will be considered processes involving VPg, which serves as an RNA primer through the addition of two uridine bases to its Tyr-3 side chain. Elongation complexes involve only RNA and 3Dpol. Elongation complexes will be discussed first because they began to be studied earlier.
3Dpol/RNA: elongation complexes.
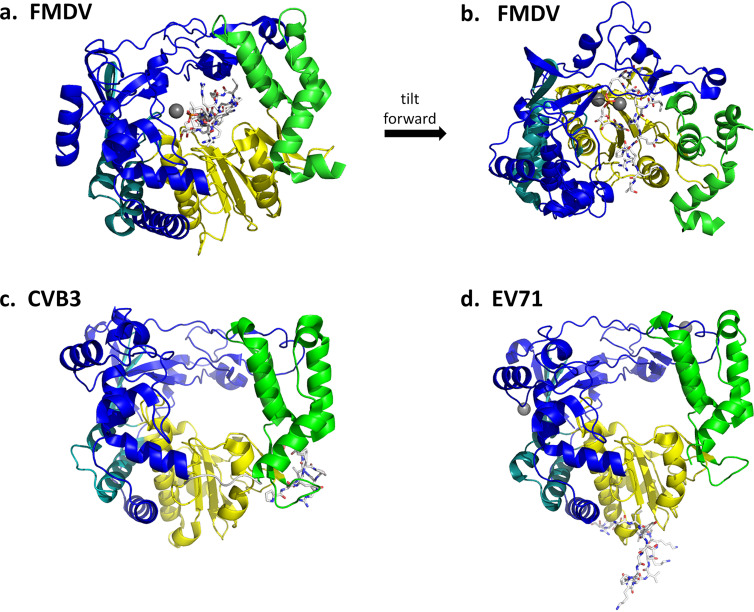
Substantial efforts have been made to use structural biology of enterovirus 3Dpol/RNA complexes to further understand the process of RNA replication. In 2004, the structure of FMDV 3Dpol complexed with a dimerized self-complementary RNA 10-mer was published (102). In its dimerized form, the RNA 10-mer was designed to serve as both a template and a primer strand and so help to elucidate the positioning of both strands during replication, as well as identifying/confirming the important 3Dpol residues for catalysis and positioning.
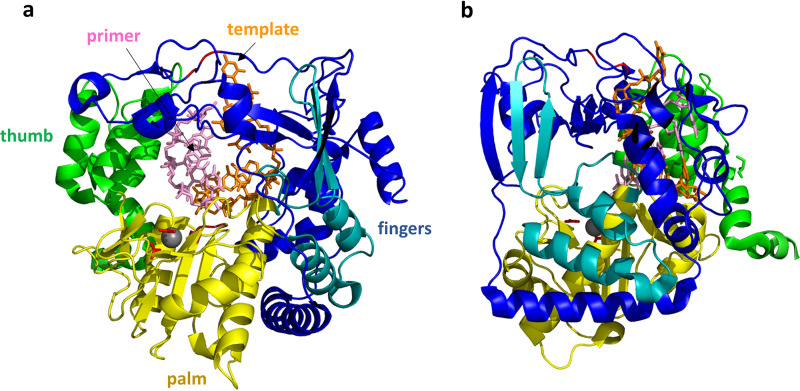
Globally, the structure of FMDV 3Dpol changed very little upon interaction with the RNA (Fig. 12a). The RNA double helix, as expected, inserts into the cavity, with the template strand interacting mostly with the fingers domain while the primer strand interacts extensively with both the palm and the thumb domains. Among the residues implicated/confirmed to be important for catalysis in FMDV 3Dpol are Asp-238 and Asp-240 which, via their Oδ atoms, coordinate magnesium in the crystal structure while Asp-338 and Asp-339 are positioned to be the catalytic carboxylate residues (note that FMDV residue numbering is used here; the equivalent residues in RV16 are Ala-232 which obviously has no Oδ atom, Asp-234, Asp-327, and Asp-328). Each of these four aspartic acid residues is from the palm domain, which is accordingly viewed as the main catalytic domain. The RNA double helix resides near the surface of the polymerase, as can be seen in Fig. 12b, with the 3′ end of the primer strand (the strand to be catalytically extended) to the left. Thus, to proceed with lengthening the primer strand, the polymerase must move to the left or, alternatively, move the RNA to the right. Residues were identified that stabilize the position of the 3′ end by interacting with other nucleotides in the primer strand.
FIG 12.
FMDV 3Dpol/RNA elongation complex. (a) The view of 3Dpol is rotated 180° from the view shown in Fig. 9. The RNA template strand (orange) and primer strand (pink) are shown as sticks, while the protein is shown as a cartoon colored as described in the legend of Fig. 9. The 3Dpol structure largely resembles that of RV14 shown in Fig. 9. The FMDV 3Dpol structure was determined in the presence and absence of RNA, and little structural rearrangement was detected. The template RNA strand interacts mainly with the fingers region, while the primer strand interacts with the thumb and palm. A magnesium ion that binds near the catalytic site is shown as a gray sphere, with side chains of the coordinating D-238 and D-240 residues shown as red sticks. The positions of the two catalytic Asp residues, D-338 and D-339, are also indicated by the red loop to the right of the magnesium ion. (b) This view of the structure is rotated 90°. From this side view, it can be seen that the RNA double helix is positioned close to the right-hand surface, which is the top surface of the structure shown in panel a. During replication, new bases would be added to the 3′ end of the primer strand, extending that strand to the left. Thus, the double helix would need to proceed to the right (or, relatively speaking, the polymerase proceeds to the left) in order to processively position the new RNA 3′ end in the polymerase active site.
Subsequent crystal structures of RNA elongation complexes helped to confirm the above results and add new detail. In these studies, an active polymerase/RNA template/RNA primer complex was first crystallized. Then various rNTPs or mutagenic analogs were soaked into the crystal. The addition of these rNTPs/analogs results in lengthening of the primer chain as directed by the template sequence but only to the point where the next required rNTP is missing. In this way, various stop points could be studied, giving insight into the elongation process.
Crystallographic elongation studies of FMDV 3Dpol (116) and PV 3Dpol (76) helped to confirm elongation mechanism information that had been previously established by other means (117, 118). The FMDV 3Dpol crystal structure showed a hydrogen bond between the 3′ OH of the primer strand and the catalytic Asp-338 from the palm domain (Asp-327 in RV16). Again, only one metal ion (magnesium) was seen, bound to Asp-338 and coordinating the triphosphate group of the incoming rNTP. The magnesium that should coordinate to the 3′ OH, to help it ionize, was absent. It is believed that this second magnesium will bind at this site but only transiently during certain steps in the polymerization process. Three additional conserved interactions were observed between different primer molecules and the palm domain; these involved residues Tyr-336, Lys-387, and Arg-388 (Tyr-325, Lys-374, and Arg-375 in RV16), the last two of which hydrogen bond to the primer sugar-phosphodiester backbone. Recognition and proper positioning of the incoming rNTP/analog involve the palm domain residues Asp-245 and Asn-307 (Asp-239 and Asn-296 in RV16). In addition, the stretch of residues Ser-298 to Ser-304 (Ser-288 to Ser-293 in RV16), from the loop connecting the palm to the finger region, is involved in interactions with both the template and the incoming rNTP.
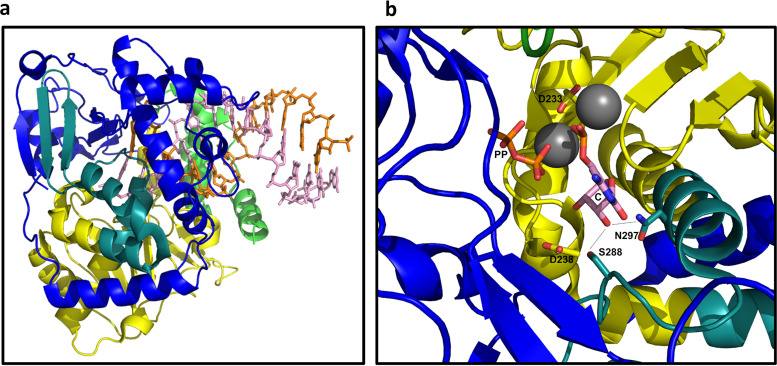
A similar study of PV elongation complexes (Fig. 13a) was used to propose a six-state model for the elongation cycle (119). These states are the following: (1) an open active site with no incoming rNTP bound, (2) an open active site with rNTP now bound, (3) an active site closed with rNTP bound, and then, after catalysis, (4) a pretranslocation complex with the RNA about to move but with the active site still closed, (5) a pretranslocation complex but with the active site now open, and (6) a translocation intermediate with an unknown structure. Then the cycle begins again with state 1 for the next nucleotide. Here, closure of the active site refers to a concerted conformational change that can be thought of as a chain reaction. First, the +1 template base (the one about to be paired) and its incoming rNTP both pivot down toward the active site. This pushes the side chain of Asp-238 (Asp-239 in RV16) and causes it to break its contacts with Asn-297 (Asn-296 in RV16), replacing these with new interactions to Lys-61 and Ser-288 (Lys-61 and Ser-287 in RV16). As a result, Ser-288 and Asn-297 are both now in position to hydrogen bond to the rNTP 2′ OH group (Fig. 13b), which positions the rNTP properly for catalysis. This movement also stabilizes a 3-strand beta sheet in the palm domain that causes Asp-233 (Asp-234 in RV16) to swing toward the RNA where it helps to coordinate both catalytic magnesium ions (Fig. 13b).
FIG 13.
Poliovirus 3Dpol/RNA elongation complex. (a) The lengthening RNA double helix protrudes from the right side of the polymerase. Orientation and coloring are as shown Fig. 12b. (b) Zoomed image of the active site in its closed conformation. The incoming rNTP (C) and its paired template base (not shown) swing down, displacing D-238, which frees up N-297 and positions S-288 so that both residues hydrogen bond (dashed lines) to the 2′ OH group of the incoming rNTP. In addition, a three-strand beta sheet (upper right) is stabilized, positioning D-233 to coordinate both catalytic magnesium ions (spheres). One of the magnesium ions stabilizes the leaving pyrophosphate group (PP).
A subsequent study of PV, CVB3, and RV16 elongation complexes (120) helped to establish conserved features of enterovirus replication complexes, including that 8 bp of the elongating RNA are held tightly in position within the cavity region, that there is a conserved binding pocket for the template +2 nucleotide, and that there is a conserved nonstandard backbone conformation of the template strand within the active site. A more recent study of EV71 (hand-foot-and-mouth disease) established additional details regarding the replication process, including rNTP recognition and active-site closure (121). But perhaps the most interesting implication of the EV71 study is that the two strands, the template and primer, may translocate asymmetrically. Intermediates were identified in which the primer and most of the template had translocated, but various stretches of the template strand had not. Based on this observation, it was proposed that translocation of the primer strand may, at least in part, precede translocation of the template strand. However, the caveat must be considered that crystal packing may influence the movement of the RNA strands as the reaction proceeds in the crystal.
3Dpol/RNA/VPg: initiation complexes.
(i) FMDV.
The first structure determination of a picornavirus initiation complex was from FMDV. The FMDV genome codes for three VPgs (VPg1, VPg2, and VPg3) in series, and each can act as a primer for RNA replication (122, 123). Vpg1 was chosen for this study. Two initiation complex forms were crystallized (124), each starting from a mixture of FMDV 3Dpol, VPg1, UTP, magnesium, and manganese. For the second crystal, an RNA oligonucleotide of 10 adenine bases (A10) was also added. Interestingly, for the second crystal, VPg1 and UTP were found combined into VPg1-UMP. However, the A10 oligonucleotide itself was disordered and is not present in the PDB file. In the first crystal, the UTP molecule was not visible. A single magnesium ion and one manganese ion were visible near the 3Dpol active site only in the second crystal.
The conformations of 3Dpol and VPg1 were similar in each structure, and so only the second crystal structure (in the presence of A10) will be discussed (Fig. 14a). The N-terminal 15 residues of VPg1 are found within the 3Dpol cavity, in an ordered and mostly extended conformation but without regular secondary structure. These 15 residues extend through the full length of the 3Dpol cavity (Fig. 14b), in a similar position as the template RNA strand from elongation complex studies. The disordered C-terminal end of VPg1 apparently extends beyond the range of the 3Dpol cavity. The VPg1 N terminus is near the NTP entry site (Fig. 14a, back side), and the lengthy uridylated Tyr-3 side chain extends toward the C terminus of the peptide (Fig. 14a, forward), which places it close to the polymerase active site. Vpg1 makes contact with all three domains of 3Dpol.
FIG 14.
3D/VPg initiation complexes. VPg can bind at three different positions. (a) FMDV 3Dpol (cartoon), with VPg1 (sticks) bound in the central cavity. (b) Same structure as shown in panel a but rotated to show that VPg1 extended completely through the cavity. (c) CVB3 3Dpol with VPg bound to the outside of the thumb/palm junction region. (d) EV71 3Dpol with VPg bound to the outside of the palm.
Changes in the conformation of FMDV 3Dpol due to VPg1 are relatively modest. A few side chains involved in uridylation, such as those of Arg-179 (Arg-174 in RV16) of the fingers domain and Asp-338 (Asp-327 in RV16) of the palm domain, are conformationally rearranged. Arg-179 maintains double salt bridges with Glu-166 (Glu-161 in RV16) in both crystals and also interacts with the hydroxyl group of Tyr-3 in nonuridylated VPg1 and with one of the oxygens from the UMP phosphate group when it is complexed with uridylated VPg1.
Other FMDV 3D polymerase residues involved in interactions with the VPg1 N terminus include Glu-166, Ile-167, Arg-168, Lys-172, and Arg-179 (residues 161 to 163, 167, and 174 in RV16; Ile-167 is replaced by Leu-162) in the fingers domain and Thr-407, Ala-410, and Ile-411 (not conserved in RV16) of the thumb domain, which together stabilize the Tyr-3 conformation at the active site. Two main chain-side chain hydrogen bonds were observed from the backbone oxygen atoms of Pro-2 and Tyr-3 of VPg1 to the side chains of Lys-172 and Arg-168. The main chain oxygen atoms of Pro-6 and Leu-7 of VPg1 are hydrogen bonded to side chain of Lys-387 (Lys-374 in RV16) from the palm domain. The Leu-7 hydrophobic side chain also contacts a hydrophobic region of the 3Dpol cavity formed by the side chains of Glu-166, Ile-167 of the fingers domain and Thr-407, Ala-410, and Ile-411 of the thumb. Arg-388 (Arg-375 in RV16) of the palm interacts with Glu-8, Arg-9, and Gln-10 of VPg1; Tyr-336 (Tyr-325 in RV16) of the palm and Gly-216, Cys-217, and Pro-219 (residues 211, 212 and 214 in RV16) from the fingers domain contact Arg-11 of VPg1 near the exit of the polymerase cavity.
(ii) CVB3 and EV71.
The structure of a 3Dpol-VPg complex from CVB3 was published in 2008 (125). Interestingly, VPg was found not in the polymerase cavity but on the outside surface of 3Dpol, binding across the palm/thumb junction region. VPg residues 7 to 15 have the highest electron density and are assumed to form the main binding surface for 3Dpol. The meaning of this second binding site for VPg is uncertain, but its presence is supported by binding and mutational studies (126–128), which point to the possibility that VPg may be uridylated while in this position. However, in this position, VPg is too far from the polymerase active site to serve as a primer or to be uridylated by the same 3Dpol molecules. Two hypotheses have arisen. The first (transuridylation model) supposes that a second 3Dpol molecule uridylates this VPg molecule. The second hypothesis (array model) also involves additional 3Dpol molecules but in an array and supposes that a set of replication-inactive 3Dpol molecules with VPg in this position help to stabilize the position or conformation of a second set of 3Dpol molecules, in which replication takes place.
Adding to the intrigue, the crystal structure of an EV71 3Dpol-VPg complex was reported in 2013. In this crystal, VPg was found at a third binding site, on the bottom of the palm domain. VPg is in a V-shaped confirmation, with the top two edges of the V, comprised of residues 1 to 3 and 9 to 20, bound to the palm domain. Interestingly, these VPg regions are complementary to the region binding to CVB3 3Dpol. Again, mutational studies suggest that this binding site is important and may be involved in VPg uridylation (10). The suggestion was made that picornaviruses may be divided into two groups: those that use a trans mechanism to uridylate VPg bound to the outside of a second 3Dpol molecule (e.g., CVB3 and EV71) and those that use a cis mechanism in which VPg enters the cavity where it is uridylated and elongated by the same 3Dpol molecule. These theories remain controversial. Clearly, while great strides have been made, additional studies will be required to more fully elucidate the process of replication in enteroviruses.
PERSPECTIVES
Great strides have been made using structural biology to help understand picornavirus replication. Structural information regarding the isolated 3A, 3B, 3C, and 3D proteins is now available, along with a structure of the 5′-CL RNA. These studies provide three-dimensional structures of the building blocks. In addition, a large number of crystal structures are available for 3Dpol complexed with primer and template RNA molecules of various lengths and/or with VPg. A complex between 3C and SLD from the 5′-CL RNA has been studied. The 3CD active precursor protein has also been studied. The combined results from biochemical and genetic studies, analyses of similarities between 3Cpro and serine proteases and between 3Dpol and other polynucleotide polymerases, and the discussed structural works have provided an understanding of protein cleavage by 3Cpro and RNA polymerization by 3Dpol, including aspects of the role played by 3B (VPg). However, much is still unknown. Below is a brief discussion of a few of the more immediate outstanding questions that structural biology might help to answer.
First, the 5′-CL structure has been determined only for RV14. While sequence analysis suggests a high degree of structural conservation, it is not yet known whether specific features, such as the alignment of the accessible major grooves of SLB and SLD in the closed form (with Mg+2), are conserved across the enteroviruses. Nor is it known whether this major groove surface functions in accommodating protein groups, such as perhaps the N-terminal helix of 3Cpro (or 3CD or other active precursors that include 3Cpro). Is the open or closed state of the 5′-CL conformation the active state? Or are both states needed? Is the ability of the 5′-CL RNA to make this conformational switch conserved and required for replication?
Although we know that the 3C/3D relationship in 3CD can be dynamic in solution, we do not yet know how this relationship may change during replication nor how it may affect or be affected by the replication process. While the 3Cpro/SLD complex structure begins to build a picture of replication assembly on the 5′-CL, no structures of the full 5′-CL complexed with any viral or host protein, including 3CD and PCBP2, are yet available. Is it possible that binding of these proteins helps to trigger a conformational change similar to that triggered by magnesium in vitro? Even with these and other unanswered questions, the interaction of 3Cpro with the 5′-CL (oriL) is at this point better characterized than its interactions with oriI and oriR. Furthermore, structures of 3AB, 3BC, 3BCD, and 3ABCD are not yet available, let alone complex structures involving these precursors and RNA.
In addition to the need to investigate larger RNA/protein pieces of the replication complex, a major looming challenge is presented by the fact that replication takes place on remodeled host cell membrane surfaces. To this point, for instance, structural information for 3A is available only for its soluble domain. The membrane-associated domain remains to be structurally determined, along with structural information regarding how it helps the system to interact with membranes. These interactions include host proteins such as GBF1 and GPC60. Solid-state NMR may play a role in elucidating these interactions. However, this is just the tip of the membrane iceberg. In addition to 3A, many of the other discussed enterovirus proteins also interact with remodeled host cell membranes or membrane-associated molecules, and thus studies in the absence of membranes are destined to provide only a part of the story. As the studied complexes become larger, NMR and crystallographic analyses become more challenging while SAXS and various microscopies become more useful. Due to the limited resolution of the last two techniques, it will remain necessary to fill in the fine detail via crystallographic and NMR studies of isolated components and smaller complexes.
These are some of the questions that remain regarding the early steps of replication involving synthesis of a negative strand from the positive-sense RNA genome. The nascent negative strands are then used as templates in a second round of polymerization to create a large number of new positive-sense strands. Even less is known about this second half of the replication process, which is arguably the central replication step since it results in new genetic material and in much larger numbers than the intermediary negative strand. While some of the building blocks will be the same as in negative-strand synthesis, there are bound to be differences in positive-strand synthesis that provide additional opportunities for intervention in the highly conserved enterovirus replication process.
REFERENCES
- 1.Kitamura N, Adler C, Wimmer E. 1980. Structure and expression of the picornavirus genome. Ann N Y Acad Sci 354:183–201. doi: 10.1111/j.1749-6632.1980.tb27967.x. [DOI] [PubMed] [Google Scholar]
- 2.Agol VI. 1980. Structure, translation, and replication of picornaviral genomes. Prog Med Virol 26:119–157. [PubMed] [Google Scholar]
- 3.Wimmer E, Kuhn RJ, Pincus S, Yang CF, Toyoda H, Nicklin MJ, Takeda N. 1987. Molecular events leading to picornavirus genome replication. J Cell Sci Suppl 7:251–276. doi: 10.1242/jcs.1987.supplement_7.18. [DOI] [PubMed] [Google Scholar]
- 4.Skern T, Duechler M, Sommergruber W, Blaas D, Kuechler E. 1987. The molecular biology of human rhinoviruses. Biochem Soc Symp 53:63–73. [PubMed] [Google Scholar]
- 5.Trono D, Andino R, Baltimore D. 1988. An RNA sequence of hundreds of nucleotides at the 5’ end of poliovirus RNA is involved in allowing viral protein synthesis. J Virol 62:2291–2299. doi: 10.1128/JVI.62.7.2291-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agol VI. 1991. The 5′-untranslated region of picornaviral genomes. Adv Virus Res 40:103–180. doi: 10.1016/s0065-3527(08)60278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohll JB, Percy N, Ley R, Evans DJ, Almond JW, Barclay WS. 1994. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol 68:4384–4391. doi: 10.1128/JVI.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krausslich HG, Nicklin MJ, Lee CK, Wimmer E. 1988. Polyprotein processing in picornavirus replication. Biochimie 70:119–130. doi: 10.1016/0300-9084(88)90166-6. [DOI] [PubMed] [Google Scholar]
- 9.Leong LE, Walker PA, Porter AG. 1993. Human rhinovirus-14 protease 3C (3Cpro) binds specifically to the 5′-noncoding region of the viral RNA. Evidence that 3Cpro has different domains for the RNA binding and proteolytic activities. J Biol Chem 268:25735–25739. [PubMed] [Google Scholar]
- 10.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang W, Cuconati A, Paul AV, Cao X, Wimmer E. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 12.Ohlenschlager O, Wohnert J, Bucci E, Seitz S, Hafner S, Ramachandran R, Zell R, Gorlach M. 2004. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Du Z, Yu J, Ulyanov NB, Andino R, James TL. 2004. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry 43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- 14.Headey SJ, Huang H, Claridge JK, Soares GA, Dutta K, Schwalbe M, Yang D, Pascal SM. 2007. NMR structure of stem-loop D from human rhinovirus-14. RNA 13:351–360. doi: 10.1261/rna.313707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka Y, Fujii S, Hiroaki H, Sakata T, Tanaka T, Uesugi S, Tomita K-i, Kyogoku Y. 1999. A′-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC). Nucleic Acids Res 27:949–955. doi: 10.1093/nar/27.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warden MS, Tonelli M, Cornilescu G, Liu D, Hopersberger LJ, Ponniah K, Pascal SM. 2017. Structure of RNA stem loop B from the picornavirus replication platform. Biochemistry 56:2549–2557. doi: 10.1021/acs.biochem.7b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warden MS, Cai K, Cornilescu G, Burke JE, Ponniah K, Butcher SE, Pascal SM. 2019. Conformational flexibility in the enterovirus RNA replication platform. RNA 25:376–387. doi: 10.1261/rna.069476.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornilescu G, Didychuk AL, Rodgers ML, Michael LA, Burke JE, Montemayor EJ, Hoskins AA, Butcher SE. 2016. Structural analysis of multi-helical RNAs by NMR-SAXS/WAXS: application to the U4/U6 di-snRNA. J Mol Biol 428:777–789. doi: 10.1016/j.jmb.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke JE, Sashital DG, Zuo X, Wang Y-X, Butcher SE. 2012. Structure of the yeast U2/U6 snRNA complex. RNA 18:673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grishaev A, Ying J, Canny MD, Pardi A, Bax A. 2008. Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J Biomol NMR 42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodson SA. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol 9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Tinoco I, Bustamante C. 1999. How RNA folds. J Mol Biol 293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez RL, Tinoco I. 2001. Identification and characterization of metal ion binding sites in RNA. Methods Enzymol 338:421–443. doi: 10.1016/s0076-6879(02)38231-4. [DOI] [PubMed] [Google Scholar]
- 24.Palmenberg AC. 1987. Picornaviral processing: some new ideas. J Cell Biochem 33:191–198. doi: 10.1002/jcb.240330306. [DOI] [PubMed] [Google Scholar]
- 25.Hellen CU, Krausslich HG, Wimmer E. 1989. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry 28:9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- 26.Lawson MA, Semler BL. 1990. Picornavirus protein processing–enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol 161:49–87. [PubMed] [Google Scholar]
- 27.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belov GA, van Kuppeveld FJ. 2012. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr Opin Virol 2:740–747. doi: 10.1016/j.coviro.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lama J, Carrasco L. 1995. Mutations in the hydrophobic domain of poliovirus protein 3AB abrogate its permeabilizing activity. FEBS Lett 367:5–11. doi: 10.1016/0014-5793(95)00523-c. [DOI] [PubMed] [Google Scholar]
- 30.Doedens JR, Kirkegaard K. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J 14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doedens JR, Giddings TH Jr, Kirkegaard K. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol 71:9054–9064. doi: 10.1128/JVI.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KH, Klumperman J, Henke A, Jackson CL, Melchers WJ, van Kuppeveld FJ. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev Cell 11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Wessels E, Duijsings D, Lanke KH, van Dooren SH, Jackson CL, Melchers WJ, van Kuppeveld FJ. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J Virol 80:11852–11860. doi: 10.1128/JVI.01225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondratova AA, Neznanov N, Kondratov RV, Gudkov AV. 2005. Poliovirus protein 3A binds and inactivates LIS1, causing block of membrane protein trafficking and deregulation of cell division. Cell Cycle 4:1403–1410. doi: 10.4161/cc.4.10.2041. [DOI] [PubMed] [Google Scholar]
- 36.Dodd DA, Giddings TH Jr, Kirkegaard K. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J Virol 75:8158–8165. doi: 10.1128/jvi.75.17.8158-8165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci U S A 97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. 2012. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIβ. J Virol 86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss DM, Glustrom LW, Wuttke DS. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J Mol Biol 330:225–234. doi: 10.1016/S0022-2836(03)00577-1. [DOI] [PubMed] [Google Scholar]
- 41.Berstein HD, Baltimore D. 1988. Poliovirus mutant that contains a cold-sensitive defect in viral RNA synthesis. J Virol 62:2922–2928. doi: 10.1128/JVI.62.8.2922-2928.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambros V, Baltimore D. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem 253:5263–5266. [PubMed] [Google Scholar]
- 43.Schein CH, Oezguen N, Volk DE, Garimella R, Paul A, Braun W. 2006. NMR structure of the viral peptide linked to the genome (VPg) of poliovirus. Peptides 27:1676–1684. doi: 10.1016/j.peptides.2006.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schein CH, Oezguen N, van der Heden van Noort GJ, Filippov DV, Paul A, Kumar E, Braun W. 2010. NMR solution structure of poliovirus uridylyated peptide linked to the genome (VPgpU). Peptides 31:1441–1448. doi: 10.1016/j.peptides.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee R, Weidman MK, Navarro S, Comai L, Dasgupta A. 2005. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J Gen Virol 86:2315–2322. doi: 10.1099/vir.0.80817-0. [DOI] [PubMed] [Google Scholar]
- 46.Kundu P, Raychaudhuri S, Tsai W, Dasgupta A. 2005. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J Virol 79:9702–9713. doi: 10.1128/JVI.79.15.9702-9713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yalamanchili P, Datta U, Dasgupta A. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J Virol 71:1220–1226. doi: 10.1128/JVI.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yalamanchili P, Weidman K, Dasgupta A. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]
- 49.Weidman MK, Yalamanchili P, Ng B, Tsai W, Dasgupta A. 2001. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology 291:260–271. doi: 10.1006/viro.2001.1215. [DOI] [PubMed] [Google Scholar]
- 50.Belsham GJ, McInerney GM, Ross-Smith N. 2000. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol 74:272–280. doi: 10.1128/jvi.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Breyne S, Bonderoff JM, Chumakov KM, Lloyd RE, Hellen CU. 2008. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 378:118–122. doi: 10.1016/j.virol.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly (rC)-binding protein. J Virol 81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B, Seitz S, Kusov Y, Zell R, Gauss-Müller V. 2007. RNA interaction and cleavage of poly (C)-binding protein 2 by hepatitis A virus protease. Biochem Biophys Res Commun 364:725–730. doi: 10.1016/j.bbrc.2007.09.133. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M, Arias C, Garabedian A, Palmenberg AC, Mohr I. 2012. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol 86:10686–10694. doi: 10.1128/JVI.00896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P, Han L, Chu F, Jagdeo J, Jan E, Fujita T, Luo H. 2013. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 8:e79546. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence P, Schafer EA, Rieder E. 2012. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 425:40–52. doi: 10.1016/j.virol.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 57.Flather D, Semler BL. 2015. Picornaviruses and nuclear functions: targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front Microbiol 6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caly L, Ghildyal R, Jans DA. 2015. Respiratory virus modulation of host nucleocytoplasmic transport; target for therapeutic intervention? Front Microbiol 6:848. doi: 10.3389/fmicb.2015.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker EJ, Younessi P, Fulcher AJ, McCuaig R, Thomas BJ, Bardin PG, Jans DA, Ghildyal R. 2013. Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS One 8:e71316. doi: 10.1371/journal.pone.0071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghildyal R, Jordan B, Li D, Dagher H, Bardin PG, Gern JE, Jans DA. 2009. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J Virol 83:7349–7352. doi: 10.1128/JVI.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park N, Katikaneni P, Skern T, Gustin KE. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Z, Mogensen MM, Powell PP, Curry S, Wileman T. 2013. Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol 87:11721–11729. doi: 10.1128/JVI.01355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shubin AV, Demidyuk IV, Lunina NA, Komissarov AA, Roschina MP, Leonova OG, Kostrov SV. 2015. Protease 3C of hepatitis A virus induces vacuolization of lysosomal/endosomal organelles and caspase-independent cell death. BMC Cell Biol 16:4. doi: 10.1186/s12860-015-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chau DH, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. 2007. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12:513–524. doi: 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 65.Li ML, Hsu TA, Chen TC, Chang SC, Lee JC, Chen CC, Stollar V, Shih SR. 2002. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 293:386–395. doi: 10.1006/viro.2001.1310. [DOI] [PubMed] [Google Scholar]
- 66.Barco A, Feduchi E, Carrasco L. 2000. Poliovirus protease 3Cpro kills cells by apoptosis. Virology 266:352–360. doi: 10.1006/viro.1999.0043. [DOI] [PubMed] [Google Scholar]
- 67.Andino R, Rieckhof GE, Achacoso PL, Baltimore D. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J 12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem 269:27004–27014. [PubMed] [Google Scholar]
- 69.Pathak HB, Arnold JJ, Wiegand PN, Hargittai MRS, Cameron CE. 2007. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J Biol Chem 282:16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pathak HB, Ghosh SKB, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol): a surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem 277:31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 71.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin J, Paul AV, Wimmer E, Rieder E. 2003. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J Virol 77:5152–5166. doi: 10.1128/jvi.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiang W, Harris KS, Alexander L, Wimmer E. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol 69:3658–3667. doi: 10.1128/JVI.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shengjuler D, Chan YM, Sun S, Moustafa IM, Li ZL, Gohara DW, Buck M, Cremer PS, Boehr DD, Cameron CE. 2017. The RNA-binding site of poliovirus 3C protein doubles as a phosphoinositide-binding domain. Structure 25:1875–1886.e7. doi: 10.1016/j.str.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen M, Wang Q, Yang Y, Pathak HB, Arnold JJ, Castro C, Lemon SM, Cameron CE. 2007. Human rhinovirus type 14 gain-of-function mutants for oriI utilization define residues of 3C(D) and 3Dpol that contribute to assembly and stability of the picornavirus VPg uridylylation complex. J Virol 81:12485–12495. doi: 10.1128/JVI.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 77.Wlodawer A, Walter J, Huber R, Sjolin L. 1984. Structure of bovine pancreatic trypsin inhibitor. Results of joint neutron and x-rays refinement crystal form II. J Mol Biol 180:301–329. doi: 10.1016/S0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]
- 78.Mosimann SC, Cherney MM, Sia S, Plotch S, James M. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol 273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 79.Blair WS, Nguyen JH, Parsley TB, Semler BL. 1996. Mutations in the poliovirus 3CD proteinase S1-specificity pocket affect substrate recognition and RNA binding. Virology 218:1–13. doi: 10.1006/viro.1996.0160. [DOI] [PubMed] [Google Scholar]
- 80.Shih SR, Chiang C, Chen TC, Wu CN, Hsu JT, Lee JC, Hwang MJ, Li ML, Chen GW, Ho MS. 2004. Mutations at KFRDI and VGK domains of enterovirus 71 3C protease affect its RNA binding and proteolytic activities. J Biomed Sci 11:239–248. doi: 10.1007/bf02256567. [DOI] [PubMed] [Google Scholar]
- 81.Chan YM, Moustafa IM, Arnold JJ, Cameron CE, Boehr DD. 2016. Long-range communication between different functional sites in the picornaviral 3C protein. Structure 24:509–517. doi: 10.1016/j.str.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Claridge JK, Headey SJ, Chow JY, Schwalbe M, Edwards PJ, Jeffries CM, Venugopal H, Trewhella J, Pascal SM. 2009. A picornaviral loop-to-loop replication complex. J Struct Biol 166:251–262. doi: 10.1016/j.jsb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matthews DA, Dragovich PS, Webber SE, Fuhrman SA, Patick AK, Zalman LS, Hendrickson TF, Love RA, Prins TJ, Marakovits JT, Zhou R, Tikhe J, Ford CE, Meador JW, Ferre RA, Brown EL, Binford SL, Brothers MA, DeLisle DM, Worland ST. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc Natl Acad Sci U S A 96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Namoto K, Sirockin F, Sellner H, Wiesmann C, Villard F, Moreau RJ, Valeur E, Paulding SC, Schleeger S, Schipp K, Loup J, Andrews L, Swale R, Robinson M, Farady CJ. 2018. Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors. Bioorg Med Chem Lett 28:906–909. doi: 10.1016/j.bmcl.2018.01.064. [DOI] [PubMed] [Google Scholar]
- 85.Birtley JR, Knox SR, Jaulent AM, Brick P, Leatherbarrow RJ, Curry S. 2005. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem 280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 86.Yang J, Leen EN, Maree FF, Curry S. 2016. Crystal structure of the 3C protease from Southern African Territories type 2 foot-and-mouth disease virus. PeerJ 4:e1964. doi: 10.7717/peerj.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cui S, Wang J, Fan T, Qin B, Guo L, Lei X, Wang J, Wang M, Jin Q. 2011. Crystal structure of human enterovirus 71 3C protease. J Mol Biol 408:449–461. doi: 10.1016/j.jmb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, Shih SR, Liang PH, Wang AH. 2009. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem 284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu G, Qi J, Chen Z, Xu X, Gao F, Lin D, Qian W, Liu H, Jiang H, Yan J, Gao GF. 2011. Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J Virol 85:10319–10331. doi: 10.1128/JVI.00787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Costenaro L, Kaczmarska Z, Arnan C, Janowski R, Coutard B, Sola M, Gorbalenya AE, Norder H, Canard B, Coll M. 2011. Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. J Virol 85:10764–10773. doi: 10.1128/JVI.05062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan J, George S, Kusov Y, Perbandt M, Anemuller S, Mesters JR, Norder H, Coutard B, Lacroix C, Leyssen P, Neyts J, Hilgenfeld R. 2013. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J Virol 87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van der Linden L, Wolthers KC, van Kuppeveld FJ. 2015. Replication and Inhibitors of Enteroviruses and Parechoviruses. Viruses 7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O'Reilly EK, Kao CC. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 94.Kirkegaard K. 1992. Genetic analysis of picornaviruses. Curr Opin Genet Dev 2:64–70. doi: 10.1016/s0959-437x(05)80324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Steitz TA. 1998. A mechanism for all polymerases. Nature 391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 96.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol 72:6732–6741. doi: 10.1128/JVI.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pata JD, Schultz SC, Kirkegaard K. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen JL, Long AM, Schultz SC. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 99.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. 1985. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 100.Love RA, Maegley KA, Yu X, Ferre RA, Lingardo LK, Diehl W, Parge HE, Dragovich PS, Fuhrman SA. 2004. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 12:1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 101.Thompson AA, Peersen OB. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J 23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmís C, Domingo E, Verdaguer N. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem 279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 103.Campagnola G, Weygandt M, Scoggin K, Peersen O. 2008. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J Virol 82:9458–9464. doi: 10.1128/JVI.00647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ferrer-Orta C, de la Higuera I, Caridi F, Sanchez-Aparicio MT, Moreno E, Perales C, Singh K, Sarafianos SG, Sobrino F, Domingo E, Verdaguer N. 2015. Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition. J Virol 89:6848–6859. doi: 10.1128/JVI.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lyle JM, Bullitt E, Bienz K, Kirkegaard K. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218–2222. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- 106.Tellez AB, Wang J, Tanner EJ, Spagnolo JF, Kirkegaard K, Bullitt E. 2011. Interstitial contacts in an RNA-dependent RNA polymerase lattice. J Mol Biol 412:737–750. doi: 10.1016/j.jmb.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Spagnolo JF, Rossignol E, Bullitt E, Kirkegaard K. 2010. Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA 16:382–393. doi: 10.1261/rna.1955410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Lyle JM, Bullitt E. 2013. Surface for catalysis by poliovirus RNA-dependent RNA polymerase. J Mol Biol 425:2529–2540. doi: 10.1016/j.jmb.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davis GJ, Wang QM, Cox GA, Johnson RB, Wakulchik M, Dotson CA, Villarreal EC. 1997. Expression and purification of recombinant rhinovirus 14 3CD proteinase and its comparison to the 3C proteinase. Arch Biochem Biophys 346:125–130. doi: 10.1006/abbi.1997.0291. [DOI] [PubMed] [Google Scholar]
- 110.Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 111.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banerjee S, Aponte-Diaz D, Yeager C, Sharma SD, Ning G, Oh HS, Han Q, Umeda M, Hara Y, Wang RYL, Cameron CE. 2018. Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein. PLoS Pathog 14:e1007086. doi: 10.1371/journal.ppat.1007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. 2007. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol 81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moustafa IM, Gohara DW, Uchida A, Yennawar N, Cameron CE. 2015. Conformational ensemble of the poliovirus 3CD precursor observed by MD simulations and confirmed by SAXS: a strategy to expand the viral proteome? Viruses 7:5962–5986. doi: 10.3390/v7112919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spear A, Ogram SA, Morasco BJ, Smerage LE, Flanegan JB. 2015. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology 485:492–501. doi: 10.1016/j.virol.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ferrer-Orta C, Arias A, Pérez-Luque R, Escarmís C, Domingo E, Verdaguer N. 2007. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci U S A 104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gohara DW, Arnold JJ, Cameron CE. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43:5149–5158. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arnold JJ, Gohara DW, Cameron CE. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gong P, Peersen OB. 2010. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gong P, Kortus MG, Nix JC, Davis RE, Peersen OB. 2013. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS One 8:e60272. doi: 10.1371/journal.pone.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shu B, Gong P. 2016. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc Natl Acad Sci U S A 113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Forss S, Schaller H. 1982. A tandem repeat gene in a picornavirus. Nucleic Acids Res 10:6441–6450. doi: 10.1093/nar/10.20.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Falk MM, Sobrino F, Beck E. 1992. VPg gene amplification correlates with infective particle formation in foot-and-mouth disease virus. J Virol 66:2251–2260. doi: 10.1128/JVI.66.4.2251-2260.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferrer-Orta C, Arias A, Agudo R, Pérez-Luque R, Escarmís C, Domingo E, Verdaguer N. 2006. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J 25:880–888. doi: 10.1038/sj.emboj.7600971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gruez A, Selisko B, Roberts M, Bricogne G, Bussetta C, Jabafi I, Coutard B, De Palma AM, Neyts J, Canard B. 2008. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J Virol 82:9577–9590. doi: 10.1128/JVI.00631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hope DA, Diamond SE, Kirkegaard K. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol 71:9490–9498. doi: 10.1128/JVI.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lyle JM, Clewell A, Richmond K, Richards OC, Hope DA, Schultz SC, Kirkegaard K. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem 277:16324–16331. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- 128.Tellez AB, Crowder S, Spagnolo JF, Thompson AA, Peersen OB, Brutlag DL, Kirkegaard K. 2006. Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer. J Mol Biol 357:665–675. doi: 10.1016/j.jmb.2005.12.044. [DOI] [PubMed] [Google Scholar]