Abstract
Objective
To analyze the active compounds, potential targets, and diseases of JianPi Fu Recipe (JPFR) based on network pharmacology and bioinformatics and verify the potential biological function and mechanism of JPFR in vitro and in vivo.
Methods
Network pharmacology databases including TCMSP, TCM-PTD, TCMID, and DrugBank were used to screen the active compounds and potential drug targets of JPFR. Cytoscape 3.7 software was applied to construct the interaction network between active compounds and potential targets. The DAVID online database analysis was performed to investigate the potential effective diseases and involved signaling pathways according to the results of the GO function and KEGG pathways enrichment analysis. To ensure standardization and maintain interbatch reliability of JPFR, High Performance Liquid Chromatography (HPLC) was used to establish a “chemical fingerprint.” For biological function validation, the effect of JPFR on the proliferation and migration of CRC cells in vitro was investigated by CCK-8 and transwell and wound healing assay, and the effect of JPFR on the growth and metastasis of CRC cells in vivo was detected by building a lung metastasis model in nude mice and in vivo imaging. For the potential mechanism validation, the expressions of MALAT1, PTBP-2, and β-catenin in CRC cells and transplanted CRC tumors were detected by real-time PCR, western blot, and immunohistochemical staining analysis.
Results
According to the rules of oral bioavailability (OB) > 30% and drug-likeness (DL) > 0.18, 244 effective compounds in JPFR were screened out, as well as the corresponding 132 potential drug targets. By the analysis of DAVID database, all these key targets were associated closely with the cancer diseases such as prostate cancer, colorectal cancer, bladder cancer, small cell lung cancer, pancreatic cancer, and hepatocellular carcinoma. In addition, multiple signaling pathways were closely related to JPFR, including p53, Wnt, PI3K-Akt, IL-17, HIF-1, p38-MAPK, NF-κB, PD-L1 expression and PD-1 checkpoint pathway, VEGF, JAK-STAT, and Hippo. The systematical analysis showed that various active compounds of JPFR were closely connected with Wnt/β-catenin, EGFR, HIF-1, TGFβ/Smads, and IL6-STAT3 signaling pathway, including kaempferol, isorhamnetin, calycosin, quercetin, medicarpin, phaseol, spinasterol, hederagenin, beta-sitosterol, wighteone, luteolin, and isotrifoliol. For in vitro experiments, the migration and growth of human CRC cells were inhibited by the JPFR extract in a dose-dependent way, and the expression of MALAT1, PTBP-2, β-catenin, MMP7, c-Myc, and Cyclin D1 in CRC cells were downregulated by the JPFR extract in a dose-dependent way. For in vivo metastasis experiments, the numbers of lung metastasis were found to be decreased by the JPFR extract in a dose-dependent manner, and the expressions of metastasis-associated genes including MALAT1, PTBP-2, β-catenin, and MMP7 in the lung metastases were downregulated dose dependently by the JPFR extract. For the orthotopic transplanted tumor experiments, the JPFR extract could inhibit the growth of orthotopic transplanted tumors and downregulate the expression of c-Myc and Cyclin D1 in a dose-dependent manner. Moreover, the JPFR extract could prolong the survival time of tumor-bearing mice in a dose-dependent manner.
Conclusions
Through effective network pharmacology analysis, we found that JPFR contains many effective compounds which may directly target cancer-associated signaling pathways. The in vitro and in vivo experiments further confirmed that JPFR could inhibit the growth and metastasis of CRC cells by regulating β-catenin signaling-associated genes or proteins.
1. Introduction
Traditional Chinese medicine (TCM) has become an important method for comprehensive treatment of different cancer patients. Clinical studies and meta-analyses indicated that longer duration of TCM herbal use is associated with improved survival outcomes, lower incidence of neurotoxicity, and higher quality of life in later period colorectal cancer patients in China [1–3]. JianPi Fu Recipe (JPFR) is a TCM compound from long-term clinical experience, which is composed of Astragalus propinquus Schischkin (Leguminosae), Sophora flavescens Aiton (Leguminosae), Codonopsis pilosula (Franch.) Nannf (Campanulaceae), Atractylodes japonica Koidz. ex Kitam. (Compositae), Poria cocos (Schw) Wolf (Polyporaceae), Epimedium brevicornu Maxim. (Berberidaceae), Pinellia ternata (Thunb.) Makino (Araceae), Citrus reticulata Blanco (Rutaceae), Glycyrrhiza uralensis Fisch (Leguminosae), and Curcuma phaeocaulis Valeton (Zingiberaceae).
Chinese herbal formulae are complicated in compounds, targets, and action mechanisms. The emergence of databases, such as TCMSP [4], TCMID [5], TCM-PTD [6], and DrugBank [7] and the development of network pharmacology [8, 9] and bioinformatics [10] provide new directions to screen the active components and potential targets and predict the function and mechanism of JPFR. Wu et al. applied the methods of molecular docking and computer network pharmacology to screen the active ingredients of Chinese herbs for coronary heart disease treatment and constructed a drug-target-disease network to investigate the regularity of TCM on the complex network in the body [11]. Li et al. applied the network pharmacology method to explain the material bases of the cold syndrome-heat syndrome [12–15]. This study firstly aims to explore the effective active compounds, effective targets, and involved pathways of JPFR basing on multiple online databases and network pharmacology analysis and verify the potential biological function and mechanism of JPFR in vitro and in vivo, providing scientific evidence for the clinical application of JPFR.
2. Materials and Methods
2.1. Network Pharmacology Analysis of JPFR
The active compounds of JPFR were mainly mined from the online databases TCMSP (http://lsp.nwsuaf.edu.cn/tcmsp.php), TCMID (http://www.megabionet.org/tcmid), and TCM-PTD (http://tcm.zju.edu.cn/ptd). All the targets of the screened active compounds were obtained from Drugbank 6.8 database (http://tcm.zju.edu.cn/ptd). Cytoscape software 3.7 was applied to construct the active compound-target network for JPFR. The KEGG enrichment analysis was performed using the DAVID database, including biological functions and signaling pathways. Top 12 cancers and 12 KEGG enrichment pathways were listed in graphs (P < 0.05). Based on the active compound-target network of JPFR constructed above and the searching of literatures, the systematical analysis of the potential regulatory signaling pathways of the most active compounds was provided.
2.2. Cell Line and Cell Culture
The colorectal cancer cell line used in this study was LoVo (human colon, Dukes' type C, grade IV, colorectal adenocarcinoma) from ATCC. The culture medium contained F-12K (SIGMA, UK) supplemented with 10% FBS and 100 U/ml penicillin and 100 g/ml streptomycin. LoVo cells were incubated in a couveuse at 37°C with 5% CO2, with high humidity.
2.3. Preparation of the JianPi Fu Recipe Extract
JianPi Fu Recipe (JPFR) is composed of dried medicinal herbs including Astragalus propinquus Schischkin (Leguminosae), Sophora flavescens Aiton (Leguminosae), Codonopsis pilosula (Franch.) Nannf (Campanulaceae), Atractylodes japonica Koidz. ex Kitam. (Compositae), Poria cocos (Schw) Wolf (Polyporaceae), Epimedium brevicornu Maxim. (Berberidaceae), Pinellia ternata (Thunb.) Makino (Araceae), Citrus reticulata Blanco (Rutaceae), Glycyrrhiza uralensis Fisch (Leguminosae), and Curcuma phaeocaulis Valeton (Zingiberaceae). The preparation of the alcohol extract of JPFR is divided into two steps: firstly, soaking dried traditional Chinese herbal medicine with 10 times volume 95% ethanol for 1 hour and, then, heating reflux extraction for 1 hour, filtrating them and, secondly, adding 8 times volume 95% ethanol reflux extraction for 1 hour, repeating the aforementioned procedure. Finally, these two filtered solutions are combined, ethanol is retrieved, and we get the JPFR alcohol extract. To prepare 20 mg/ml JPFR solution, a 0.24 g JPFR alcohol extract was weighed using electronic scales and dissolved in a 12 ml F-12K medium supplemented with 10% fetal bovine serum (FBS), following by ultrasonic mixing solution overnight and filtering bacteria with a 0.22 μm filter. In the following experiments, we diluted the 20 mg/ml JPFR solution to the required concentration using the F-12K medium supplemented with 10% FBS.
2.4. High Performance Liquid Chromatography (HPLC) Analysis
HPLC analysis was conducted by using the Agilent 1260 infinity (ZK-001).The separation was carried out on an Alltech Uitrasphere C18 column. In order to establish the High Performance Liquid Chromatography (HPLC) method for the determination of astragaloside IV, matrine, and oxymatrine in JPFR, an Alltech Uitrasphere C18 (Alltech Ultrasphere C18) was used with acetonitrile-water (32 : 68) as the mobile phase. The flow rate was 1.0 ml/min, and the detection wavelength was at 220 mm. The column temperature was 30°C.
JPFR was handled by low temperature drying and, then, made into lyophilized powder. The precisely weighed 5 g lyophilized powder was added into 20 ml purified water. It was carefully weighed again after adding water, followed by ultrasonic processing for 30 minutes. Then, it is weighed again, and purified water is used to make up to the lost weight. We waited until the powder was fully dissolved. The solution was filtered, and the 25 ml overfiltrate was precisely measured. The saturated n-butanol was used to extract the overfiltrate 4 times, 25 ml each time. The n-butanol solution was combined and washed 3 times with ammonia solution, 30 ml each time, and the washing solution was discarded. The n-butanol solution was evaporated, the residue was dissolved with methanol, transferred to a 5 ml volumetric flask, methanol was added to scale, and shook well. All the tested solutions were filtered through Millex 0.22 μm nylon membrane syringe filters (Millipore, USA). The contents of astragaloside IV, matrine, and oxymatrine in each sample were calculated using standard curves of astragaloside IV, matrine, and oxymatrine.
2.5. CCK-8 Analysis
The LoVo cells in the logarithmic growth period were inoculated in 96-well plates at the concentration of 2000–3000 cells per well. After the cells completely adhered, the serum-free medium was changed to starve overnight. Adding the alcohol extract of JPFR, the concentration of JPFR was prepared with 12.5 μg/mL, 25 μg/mL, 50 μg/mL, 100 μg/mL, 200 μg/mL, 300 μg/mL, 400 μg/mL, and 500 μg/mL. A control group was established. After 48 hours, the cellcountingkit-8 solution was added to each pore at the ratio of 10%. After the solution mixed completely, cells were cultured at 37°C for 4 hours. The absorption of the enzyme was measured at 490 nm/630 nm (double wavelength).
2.6. Colony Formation Analysis
The cells which were starved for 2 days were incubated with JPFR in a conditioned medium for 6 days. After washing in the conditioned medium 2 times, the cells were suspended in the conditioned medium. The cell suspension concentration was adjusted to 1 × 103/ml. The cell suspension of 0.4 ml per well was cultured in a 6-well culture plate for 2 weeks, and the colony formation was observed. When the cells formed visible clones (about 50–150 cells per clone), cells were terminated cultured. The cells were incubated with PBS 2 times, and then, carbinol 1 ml/well was added. After fixing for 15 min, 1ml giemsa was added to each hole for 30 mins. The culture plate was placed in the gel imaging system. With the visible light condition, the number of clones was counted by software carried by the machine. Images were scanned and saved. Clone formation rate = clone number/number of inoculated cells × 100%. Adjusted clone formation rate = Clone formation rate (%)/Clone formation rate in the control group (%).
2.7. Transwell Assay
The serum-free medium was diluted with 1 : 4, and 40 μL was added to the room of the 24-orifice plate transwell chamber, and the room temperature was maintained overnight until the gel solidified. The surface of the transwell chamber was incubated on the surface of the membrane with fibronectin as a cytochemotactic factor. 100 μL, 1 × 105 cell suspension that resuspended in a serum-free medium was added to the transwell chamber, and the next chamber was added the medium of 10% fetal bovine serum. The cells in each group had 3 complex holes, and the methods of observation and analysis were the same as those of the migration experiment.
2.8. Wound Healing Assay
Cells (4 × 105) in the 10% FBS-containing medium were seeded on a six-well plate to make a confluent monolayer. The monolayer cells were scratched by a plastic tip and washed with PBS to remove cell debris. An F-12K medium containing 0.5% FBS was, then, added to each well, and the scratched monolayer was incubated in a 37°C incubator with 5% CO2 for 24 hours. Wound closure was measured in five random fields at ×200 magnification using a DMI3000B inverted microscope (Leica, USA). The wound healing percentage was calculated as follows: migrated cell surface area/total surface area × 100%, in which migrated cell surface area = length of cell migration (mm) × 2 × length of defined areas, total surface area = beginning width × length of defined areas.
2.9. In Vivo Experiment
For the lung metastasis model, the single-cell suspension was prepared by the logarithmic long-term fluorescent LoVo cells, and the cell concentration was diluted to 2.5 × 107/ml by PBS (phosphate buffer saline). After the activity of cells was evaluated more than 95% by trypan blue dyeing, 200 μL cell suspension was injected to the tail vein of BALB/c nude mice. The growth and metastasis of tumor in the body were observed by using the American fine real living imaging system (IVIS 200). Each nude mouse was injected with 150 μL 30 mg/ml fluorescein substrate 10–25 min before observation.
For the orthotopic transplantation tumor model, the single-cell suspension was prepared by the LoVo cells from the logarithmic growth period, and the cell concentration was adjusted to 1 × 107/ml by PBS. After the activity of cells was evaluated more than 95% by trypan blue dyeing, the orthotopic transplantation tumor model began to establish. 10 BALB/c nude mice (4-6-week old, 20 g ± 2 g body weight), half male and half female, were prepared. The subcutaneous of the armpit of the right forelimb of nude mice was routinely sterilized. After modeling, the size of tumor in the tumor-bearing mice was observed, and the tumor grew to about 1–1.2 cm in diameter. After being executed, the tumor was removed and cut into 1 mm3 and, then, inoculated in the cecum of nude mice. After the operation, the abdominal junction was sutured with an operative thread. Under the condition of SPF (Specific Pathogen Free) in an isolated squirrel cage, it was kept at constant temperature and humidity.
Forty mice were prepared for the lung metastasis model and orthotopic transplantation tumor model, respectively. For each model, the mice were randomly divided into 5 groups, 8 mice in each group: (1) a blank control group (model group): intragastric administration of normal saline (once a day, for 7 days); (2) a low-dose JPFR group: JPFR decoction gavage (once a day, a total of 28 days); (3) a medium dose JPFR group: JPFR decoction gavage (once a day, a total of 28 days); (4) a high-dose JPFR group: JPFR decoction gavage (once a day, a total of 28 days); and (5) a positive control group (L-OHP): intraperitoneal injection of oxaliplatin 0.6 mg/kg mL (once every other day, for 28 days). After 28 days, the nude mice were sacrificed. The primary tumors and the lung tissues of the mice were surgically removed and investigated. All experimental protocols were reviewed and approved by the animal ethics committee of Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine.
2.10. Real-Time PCR
The total RNA of cells was extracted by the one-step method by Trizol, and then, 2 μg was used for reverse transcription. Quantitative PCR was carried out using the 20 μl reaction system. The sample tube and the internal tube were equipped with a double tube. The gene expression levels were analyzed by quantitative real-time PCR using the ABI7500 instrument (ABI, USA). The reaction conditions were 94°C for 2 min, 94°C for 30 s, 60°C for 30 s, and a total of 40 cycles. After the end of the reaction, ABI 7500 SDS software automatically analyzed the fluorescence signal and converted it to the Ct value. The sequences of the primers used are listed in Table 1.
Table 1.
Primer sequences for real-time PCR.
| Gene | Primer sequences | Amplified fragments (nt) |
|---|---|---|
| MALAT1 | F:5-AGGCGTTGTGCGTAGAGGA-3 R:5-GGATTTTTACCAACCACTCGC-3 |
80 |
| GAPDH | F:5-AGAAGGCTGGGGCTCATTTG-3 R:5-AGGGGCCATCCACAGTCTTC-3 |
258 |
2.11. Western Blot
Cells were washed by PBS three times, and then, the cell lysate was added. The extracted proteins were quantified and loaded for SDS-PAGE gel electrophoresis, transferred to PVDF membranes, and blocked with 10% milk. Subsequently, the membranes were incubated with the primary antibodies and the following HRP-conjugated secondary antibodies. All the resulting immunocomplexes were visualized by enhanced chemiluminescence, followed by directly photographing and quantitative analyzing.
2.12. Immunohistochemical Staining Analysis
Paraffin-embedded tissues were sectioned for IHC (immunohistochemistry) staining. In detail, the experiments were performed using the first antibody, HRP-conjugated secondary antibody, and DAB (diaminobenzidine) detection reagents. The DMI3000B microscope was used for photographing. All the data were evaluated and classified blindly by two investigators from the pathology department of Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine.
2.13. Statistical Analysis
The data were represented by means ± SD or median with 95% CI (confidence interval). Statistic analysis was performed using Student's t-test, one-way AVOVA analysis, the Mann–Whitney test, or the Kruskal-Wallis test, as appropriate, with the significance level at P < 0.05. The results were analyzed by SPSS 22.0 software (IBM, USA).
3. Results
3.1. Potential Effect Mechanism Mining of JPFR Basing on Network Pharmacology Analysis
Active compounds of JPFR were screened using the network pharmacology databases, including TCMSP, TCMID, and TCM-PTD. The results showed that a total of 245 active ingredients were screened out in JPFR according to the cutoff value of OB > 30% and DL > 0.18, including 20 compounds in Astragalus propinquus Schischkin (Leguminosae), 7 compounds in Atractylodes japonica Koidz. ex Kitam. (Compositae), 5 compounds in Citrus reticulata Blanco (Rutaceae), 21 compounds in Codonopsis pilosula (Franch.) Nannf. (Campanulaceae), 92 compounds in Glycyrrhiza uralensis Fisch (Leguminosae), 13 compounds in Pinellia ternata (Thunb.) Makino (Araceae), 15 compounds in Poria cocos (Schw.) Wolf (Polyporaceae), 45 compounds in Sophora flavescens Aiton (Leguminosae), 23 compounds in Epimedium brevicornu Maxim. (Berberidaceae), and 3 compounds in Curcuma phaeocaulis Valeton (Zingiberaceae) (Table S1). TCMSP and Drugbank database were used to screen the targets for each active compound in JPFR. We have found 953 targets in Astragalus propinquus Schischkin (Leguminosae), 774 targets in Atractylodes japonica Koidz. ex Kitam. (Compositae), 479 targets in Citrus reticulata Blanco (Rutaceae), 911 targets in Codonopsis pilosula (Franch.) Nannf. (Campanulaceae), 2506 targets in Glycyrrhiza uralensis Fisch (Leguminosae), 1302 targets in Pinellia ternata (Thunb.) Makino (Araceae), 121 targets in Poria cocos (Schw.) Wolf (Polyporaceae), 963 targets in Sophora flavescens Aiton (Leguminosae), 24 targets in Curcuma phaeocaulis Valeton (Zingiberaceae), and 510 targets in Epimedium brevicornu Maxim. (Berberidaceae). By removing duplicate targets, 132 targets were screened for the following analysis (Table S1). To determine the relationship between the compounds in 10 Chinese herbal medicines of JPFR and the corresponding targets, a compound-target network was constructed by Cytoscape 3.7 software. Figure 1(a) shows that the compound-target network includes 10 herbs, 244 compounds, and 132 targets in total. In the network, some targets interact with more compounds than others. This suggests that many targets can be regulated by a variety of compounds rather than just one compound. For example, prostaglandin G/H synthase 1/2 (PTGS1/2), peroxisome proliferator-activated receptor gamma (PPARG), serine/threonine-protein kinase Chk1 (CHEK1), and glycogen synthase kinase-3β (GSK-3β) are composed of various active substances.
Figure 1.

Potential effect mechanism mining of JPFR basing on network pharmacology analysis. (a) Active compounds of JPFR were screened using the network pharmacology databases, including TCMSP, TCMID, and TCM-PTD. TCMSP and Drugbank database were used to screen the targets for each active compound in JPFR. The compound-target network was constructed by Cytoscape 3.7 software. The red octagon represents the traditional Chinese medicine compound, the purple circle represents the compound, the green diamond represents the target, and the black line represents the relationship between the compound active ingredients and the corresponding targets. (b) PPI (Protein-Protein Interaction) Network Diagram according to screening results from (a). The circles in the PPI network represent the interaction of different targets for JPFR, and the lines represent interactions with each other. (c-d) DAVID database was used to mine the effective cancer diseases and potential signaling pathways of JPFR. Fifty-five target genes were enriched in the GO function and KEGG signaling pathway analysis. (e) Systematical analysis of the potential regulatory mechanism of active ingredients basing on the active compound-target network of JPFR constructed before and the searched literatures.
As shown in Figure 1(b), the circles in the PPI network represent the interaction of different targets for JPFR, and the lines represent interactions with each other. The network consists of 54 nodes and 532 edges, indicating that there are 532 relationships among the 54 target genes. The more the number of connected lines (i.e., the degree), the more important the targets in the network, such as the targets CASP3 (degree = 41), VEGFA (degree = 41), EGFR (degree = 40), IL6 (degree = 40), and MAPK8 (degree = ) with a large median value (not less than 30). 40) and CCND1 (degree = 39), MYC (degree = 39), ERBB2 (degree = 33), and CYCS (degree = 39).
DAVID database was used to mine the potentially effective diseases of JPFR. Fifty-four target genes were enriched in the GO function and KEGG pathway enrichment analysis. According to the statistical principle P < 0.05, the KEGG enrichment analysis results show that the top 12 cancer diseases that JPFR might have potential effective efficacy are presented in Figure 1(c), such as prostate cancer, colorectal cancer, bladder cancer, small cell lung cancer, pancreatic cancer, and hepatocellular carcinoma. The DAVID Functional Annotation database was applied to further explore the molecular mechanism of the active compounds of JPFR in different cancer diseases. The data showed that multiple signaling pathways including p53, Wnt, PI3K-Akt, IL-17, HIF-1, p38-MAPK, NF-κB, PD-L1 expression and PD-1 checkpoint pathway, VEGF, JAK-STAT, and Hippo were closely related to JPFR (Figure 1(d)). Based on the active compound-target network of JPFR constructed before, the systematical analysis showed that various active compounds of JPFR, such as kaempferol, isorhamnetin, calycosin, quercetin, medicarpin, phaseol, spinasterol, hederagenin, beta-sitosterol, wighteone, luteolin, and isotrifoliol, were closely connected with Wnt/β-catenin, EGFR, HIF-1, TGFβ/Smads, and IL6-STAT3 signaling pathway (Figure 1(e)).
3.2. Effects of JPFR on the Proliferation and Migration of CRC Cells In Vitro
Our clinical practice has demonstrated that JPFR can reduce distant metastasis of CRC and improve the life quality of CRC patients. Therefore, herein, we focus our research on the underlying function and mechanism of JPFR in CRC cells. As shown in Figure 2, HPLC data indicated that JPFR has a stable ingredient of astragaloside IV, matrine, and oxymatrine, indicating the consistent quality of JPFR for the following in vitro and in vivo experiments. Firstly, the CCK-8 assay was performed to observe the effect of JPFR on the proliferation of CRC cells. The results showed that JPFR has an obvious inhibitory effect on the proliferation of LoVo cells in a dose-dependent manner (Figure 3(a)). The subsequent colony formation experiments further confirmed that JPFR can inhibit the colony-forming ability of CRC cells in a dose-dependent manner (Figures 3(b) and 3(c)).
Figure 2.

Quality control of JPFR extracts basing on the HPLC analysis of astragaloside IV, matrine, and oxymatrine. HPLC (High Performance Liquid Chromatography) analysis was performed to characterize the ingredients of astragaloside IV in Astragalus propinquus Schischkin (Leguminosae)(one component of JPFR) and matrine and oxymatrine in Sophora flavescens Aiton (Leguminosae)(one component of JPFR).
Figure 3.

Effects of JPFR on the proliferation and migration of CRC cells in vitro. (a) CCK-8 assay was performed to observe the effect of JPFR on the proliferation of CRC cells. (b, c) Colony formation and quantitative analysis were performed to observe the effect of JPFR on the colony-forming ability of CRC cells. (d, e) Transwell and quantitative assay were performed to evaluate the migration ability of LoVo cells. (f, g) Wound healing and quantitative assay were performed to evaluate the migration ability of LoVo cells. ∗P < 0.05, ∗∗P < 0.01 versus the control group.
Subsequently, the transwell and wound healing experiments were performed to see the effect of JPFR on the migration of CRC cells. The transwell assay demonstrated that JPFR inhibited the migration of LoVo cells in a concentration-dependent manner (Figures 3(d) and 3(e)). In addition, the wound healing assay further demonstrated that JPFR could inhibit the migration of LoVo cells in a concentration-dependent manner (Figures 3(f) and 3(g)). All the abovementioned data demonstrated that JPFR has obvious inhibitory effects on the proliferation and migration of CRC cells in vitro.
3.3. Effects of JPFR on CRC Metastasis- and Growth-Related Genes and Proteins In Vitro
Since JPFR has inhibitory effects on the proliferation and migration of CRC cells in vitro, we further investigated whether JPFR could affect the expression of some metastasis- and growth-related genes and proteins. In Table 2, by network pharmacology analysis, we have showed the intersection genes between JPFR and intestinal cancer targets, such as GSK3B, CCND1, HIF-1A, MYC, CCNB1, and CD44, implying the vital regulatory effect of JPFR on Wnt/β-catenin signaling. MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is a lncRNA that plays an important role in the growth and metastasis of CRC and associated closely with Wnt/β-catenin signaling [16]. Quantitative real-time PCR was used to observe the effects of JPFR on the expression levels of MALAT1 in CRC cells. The results showed that JPFR could decrease the mRNA levels of MALAT1 in a dose-dependent manner (Figure 4(a)). PTBP-2, β-catenin, and MMP7 have been reported as the downstream targeted proteins of MALAT1 [16, 17]. Our western blot assay further demonstrated that JPFR could downregulate the expression of PTBP-2, β-catenin, and MMP7 in a dose-dependent manner (Figures 4(b) and 4(c)).
Table 2.
Intersection genes between JPFR and intestinal cancer targets.
| Symbol | entrezID |
|---|---|
| PGR | 5241 |
| PTGS1 | 5742 |
| AR | 367 |
| CHEK1 | 1111 |
| CHRM3 | 1131 |
| ADH1B | 125 |
| PPARG | 5468 |
| GSK3B | 2932 |
| IKBKB | 3551 |
| BCL2 | 596 |
| CASP3 | 836 |
| MAPK8 | 5599 |
| CYP3A4 | 1576 |
| CYP1A1 | 1543 |
| ICAM1 | 3383 |
| SELE | 6401 |
| VCAM1 | 7412 |
| ALOX5 | 240 |
| GSTP1 | 2950 |
| AHR | 196 |
| GSTM1 | 2944 |
| EGFR | 1956 |
| VEGFA | 7422 |
| CCND1 | 595 |
| FOS | 2353 |
| CASP9 | 842 |
| PLAU | 5328 |
| RB1 | 5925 |
| IL6 | 3569 |
| TP63 | 8626 |
| NFKBIA | 4792 |
| CASP8 | 841 |
| HIF1A | 3091 |
| ERBB2 | 2064 |
| CAV1 | 857 |
| MYC | 4609 |
| BIRC5 | 332 |
| DUOX2 | 50506 |
| NOS3 | 4846 |
| HSPB1 | 3315 |
| CCNB1 | 891 |
| NFE2L2 | 4780 |
| NQO1 | 1728 |
| PARP1 | 142 |
| CRP | 1401 |
| RASSF1 | 11186 |
| IGF2 | 3481 |
| ERBB3 | 2065 |
| TIMP1 | 7076 |
| MDM2 | 4193 |
| PCNA | 5111 |
| MCL1 | 4170 |
| CYCS | 54205 |
| CD44 | 960 |
Figure 4.
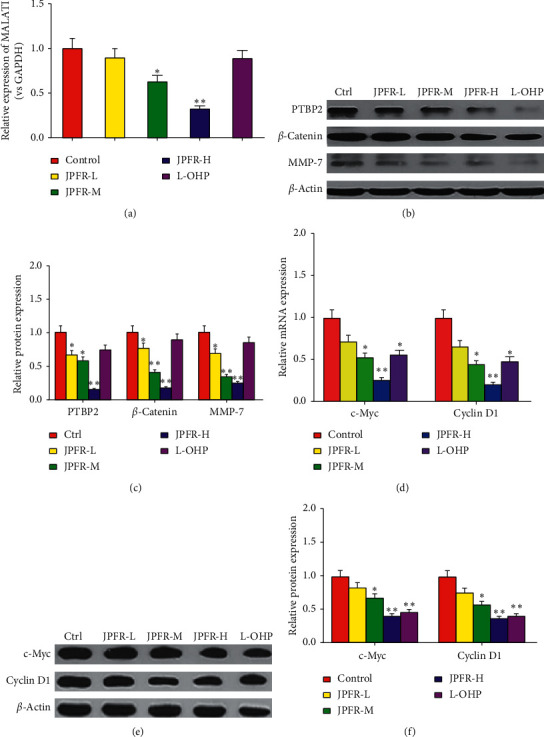
Effects of JPFR on CRC metastasis- and growth-related genes and proteins in vitro. (a) Real-time PCR was performed to observe the effect of JPFR on the relative MALAT1 expression. (b-c) Western blot was performed to study the effect of JPFR on the protein expression of PTBP-2, β-catenin, and MMP7. (d) Real-time PCR was performed to observe the effect of JPFR on the relative mRNA expression of c-Myc and Cyclin D1. (e-f) Western blot was performed to study the effect of JPFR on the protein expression of c-Myc and Cyclin D1. ∗P < 0.05, ∗∗P < 0.01 versus the control group.
c-Myc and Cyclin D1 are both important regulators for tumor growth [18, 19]. Our quantitative real-time PCR data demonstrated that JPFR could decrease the mRNA expression of c-Myc and Cyclin D1 in a dose-dependent manner (Figure 4(d)). Western blot assay further demonstrated that JPFR could downregulate the protein expression of c-Myc and Cyclin D1 in a dose-dependent manner (Figures 4(e) and 4(f)). All the abovementioned results demonstrated that JPFR inhibited the proliferation and migration of CRC cells partly through regulating the expression of MALAT1 and downstream targets PTBP-2, β-catenin, and MMP7, as well as c-Myc and Cyclin D1.
3.4. Effects of JPFR on CRC Lung Metastasis in Nude Mice
In order to investigate the inhibitory effects of JPFR on lung metastases in vivo, the experimental lung metastasis model was built in nude mice by tail vein injection of Luc-labeled CRC LoVo cells. In Figures 5(a) and 5(b), the luciferase intensity of Luc-labeled LoVo cells in lung metastatic lesions in the positive control L-OHP group was significantly decreased relative to the blank control group. The treatment of low-dose JPFR can decrease the luciferase intensity of Luc-labeled LoVo cells in lung metastatic lesions. Moreover, the luciferase intensity of Luc-labeled LoVo cells in lung metastatic lesions in middle-dose and high-dose JPFR groups were found to be dramatically decreased, compared to the control group and low-dose JPFR group. After 28 days, the nude mice were sacrificed, and the lung metastatic lesions of the mice were surgically removed and investigated. The results in Figure 5(c) demonstrated that JPFR reduced the numbers of lung metastatic lesions in a dose-dependent manner relative to the control group. In addition, the positive control L-OHP group also significantly reduced the numbers of lung metastatic lesions relative to the blank control group (Figure 5(c)).
Figure 5.

Effects of JPFR on CRC lung metastasis and orthotopic transplanted tumors growth in nude mice. (a-c) Effect of JPFR on the lung metastasis of human colon cancer cells in nude mice. (d-e) Effect of JPFR on the orthotopic transplanted CRC tumors growth in nude mice. ∗P < 0.05, ∗∗P < 0.01 versus the control group. (f) Effect of JPFR on the survival time of nude mice with orthotopic transplanted CRC tumors.
3.5. Effects of JPFR on the Orthotopic Transplanted Tumors Growth of CRC in Nude Mice
To observe the effects of JPFR on the tumors growth of CRC in vivo, we built the orthotopic transplanted tumors model by implanting the subcutaneous tumor into the cecum of the nude mice. After administration of JPFR for 7 weeks, parts of the nude mice were killed and the tumors were weighed. In Figures 5(d) and 5(e), the results showed that JPFR could significantly reduce the sizes and weights of orthotopic transplanted tumors of CRC in a dose-dependent manner. In addition, our results showed that JPFR could significantly increase the survival time of nude mice with orthotopic transplanted tumors of CRC in a dose-dependent manner (Figure 5(f)). All the abovementioned data implied that JPFR can inhibit the growth of orthotopic transplanted tumors of CRC and prolong the survival time of tumor-bearing mice.
3.6. Regulatory Effects of JPFR on Metastasis- and Growth-Related Genes and Proteins In Vivo
Quantitative real-time PCR was used to detect the lncRNA expression of the metastasis-related gene MALAT1 in the lung metastases. The results showed that JPFR could decrease the lncRNA expression of MALAT1 in a dose-dependent manner (Figure 6(a)). Western blot assay further demonstrated that JPFR could downregulate the expression of MALAT1 downstream targeted proteins PTBP-2, β-catenin, and MMP7 in a dose-dependent manner (Figures 6(b) and 6(c)). This result was further confirmed by the immunohistochemical staining detection of PTBP-2, β-catenin, and MMP7 in the lung metastases (Figures 6(d) and 6(e)).
Figure 6.

Regulatory effects of JPFR on metastasis-related genes and proteins in vivo. (a) Real-time PCR assay of the expression level of MALAT1. (b-c) Western blot and quantitative analysis of the protein expressions of PTBP2, β-catenin, and MMP7 in the lung metastases. (d-e) Immunohistochemistry and quantitative analysis of the expressions of PTBP-2, β-catenin, and MMP7 in the lung metastases. ∗P < 0.05, ∗∗P < 0.01 versus the control group.
In addition, the quantitative real-time PCR results showed that JPFR could dose-dependently decrease the mRNA expression of c-Myc and CyclinD1 in the orthotopic transplanted tumors (Figure 7(a)). Western blot assay further demonstrated that JPFR could downregulate the expression of c-Myc and CyclinD1 in a dose-dependent manner (Figures 7(b) and 7(c)), which was further confirmed by immunohistochemical staining detection of c-Myc and CyclinD1 in the orthotopic transplanted tumors (Figures 7(d) and 7(e)).
Figure 7.

Regulatory effects of JPFR on tumor growth-related genes and proteins in vivo. (a) Real-time PCR assay of the expression level of c-Myc and Cyclin D1. (b-c) Western blot and quantitative analysis of the protein expressions of c-Myc and Cyclin D1 in the orthotopic transplanted tumors. (d-e) Immunohistochemistry and quantitative analysis of the protein expressions of c-Myc and Cyclin D1 in the orthotopic transplanted tumors. ∗P < 0.05, ∗∗P < 0.01 versus the control group.
4. Discussion
In the past few years, traditional Chinese medicine (TCM) has become an important method for comprehensive treatment of cancer patients. JianPi Fu Recipe (JPFR) is a TCM compound from long-term clinical experience, which is composed of Astragalus propinquus Schischkin (Leguminosae), Sophora flavescens Aiton (Leguminosae), Codonopsis pilosula (Franch.) Nannf (Campanulaceae), Atractylodes japonica Koidz. ex Kitam. (Compositae), Poria cocos (Schw) Wolf (Polyporaceae), Epimedium brevicornu Maxim. (Berberidaceae), Pinellia ternata (Thunb.) Makino (Araceae), Citrus reticulata Blanco (Rutaceae), Glycyrrhiza uralensis Fisch (Leguminosae), and Curcuma phaeocaulis Valeton (Zingiberaceae). The clinical application of TCM in cancer diseases needs the material basis support, which promotes better understanding of the underlying effect mechanism of JPFR in CRC treatment.
Generally, TCM is complicated in compounds, targets, and molecular mechanism. The emergences of databases including TCMSP [4], TCMID [5], TCM-PTD [6], and DrugBank [7], as well as the development of network pharmacology [8, 9] and bioinformatics [10], provide new strategies to screen the active components and potential targets and predict the potential mechanism of JPFR. In our study, we firstly explore the effective active compounds, effective targets, and involved pathways of JPFR based on multiple online databases and network pharmacology analysis. According to the rules of oral bioavailability (OB) > 30% and drug-likeness (DL) > 0.18, 244 effective compounds in JPFR were screened out, as well as the corresponding 132 potential drug targets. By the analysis of DAVID database [20], all these key targets were associated closely with the cancers such as prostate cancer, colorectal cancer, bladder cancer, small cell lung cancer, pancreatic cancer, and hepatocellular carcinoma. In addition, multiple signaling pathways were closely related to JPFR, including p53, Wnt, PI3K-Akt, IL-17, HIF-1, p38-MAPK, NF-κB, PD-L1 expression and PD-1 checkpoint pathway, VEGF, JAK-STAT, and Hippo. The systematical analysis showed that various active compounds of JPFR were closely connected with Wnt/β-catenin, EGFR, HIF-1, TGFβ/Smads, and the IL6-STAT3 signaling pathway, such as kaempferol, isorhamnetin, calycosin, quercetin, medicarpin, phaseol, spinasterol, hederagenin, beta-sitosterol, wighteone, luteolin, and isotrifoliol.
CRC is a heterogeneous and molecularly complex disease [21]. One of the leading challenges for CRC is that the clinical diagnosis of CRC may be late [22]. Approximately 20% of CRC patients have distant metastases at the time of diagnosis, and an additional 20% develop metastases during the follow-up [23]. The 5-year survival rate of distant metastases in CRC was only 12.5% [24]. Researchers strived to investigate the mechanisms to reduce the mortality of CRC in the past decades. Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs composed of >200 nucleotides [25]. As an important lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been demonstrated to be crucial in the processing of pre-mRNAs and was revealed to be upregulated in CRC [26]. It was found that MALAT1 could promote CRC growth and metastasis through regulating the MALAT1/PTBP-2/β-catenin signaling pathway. PTBP2 has been found highly expressed in cancer cells and can promote cancer cell proliferation [27, 28]. Galbán et al. found that PTBP2 and HUR jointly promoted the translation of hypoxia-inducible factor 1 α (HIF-1α) and regulated several downstream cancer-related genes, such as vascular endothelial growth factor (VEGF) and β-catenin [29]. Many studies have demonstrated that Wnt/β-catenin signaling connected closely with the progression and development of different cancers. Upon stimulation of upstream activators and the helping of some transcription factors such as BCL9, β-catenin protein translocates to the nucleus from the cytoplasm, binds to the promoter of TCF/LEF, and activates the transcription of its target genes such as c-Myc, cyclinD1, and MMP7, leading to tumor initiation and development [18, 19, 30, 31].
Clinical studies and meta-analyses have indicated that longer duration of TCM herbal use is associated with improved survival outcomes, lower incidence of neurotoxicity and higher quality of life in later period CRC patients in China [1–3]. Our clinical practice has demonstrated that JPFR can reduce distant metastasis of CRC and improve the life quality of CRC patients. For in vitro experiments, the migration and growth of human CRC cells were inhibited by JPFR extract in a dose-dependent way, and the expression of MALAT1, PTBP-2, β-catenin, MMP7, c-Myc, and Cyclin D1 in CRC cells was downregulated by JPFR extract in a dose-dependent way. For in vivo metastasis experiments, the numbers of lung metastasis were found to be decreased by JPFR extract in a dose-dependent manner, and the expressions of metastasis-associated genes including MALAT1, PTBP-2, β-catenin, and MMP7 in the lung metastasis were downregulated dose dependently by JPFR extract. For the orthotopic transplanted tumors experiments, JPFR extract could inhibit the growth of orthotopic transplanted tumors in a dose-dependent manner, and JPFR extract could downregulate the expression of c-Myc and CyclinD1 in a dose-dependent manner. Moreover, JPFR extract could prolong the survival time of tumor-bearing mice in a dose-dependent manner.
5. Conclusions
In summary, we demonstrated that JPFR contains many effective compounds which may directly target to the cancer-associated signaling pathways. The in vitro and in vivo experiments further confirmed that JPFR could inhibit the growth and metastasis of colorectal cancer LoVo cells by regulating β-catenin signaling-associated genes or proteins.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (81473628, 81873279), Special Clinical Research Project of Shanghai Health and Planning Commission (201840325), Shanghai Science and Technology Commission Hospital guidance Project (19401935200), and Shanghai University of Traditional Chinese Medicine Budgeted Project (18LK057).
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
J. L. Ren conceived and designed the experiments; X. Y. Lu, X. L. Wu, L. Jing, and L. J. Tao performed the experiments; L. Jing, and L. J. Tao analyzed the data; Y. X. Zhang, R. K. Huang, and G. Zhang contributed reagents/materials/analysis tools; X. Y. Lu, X. L. Wu, and J. L. Ren wrote the paper. All authors read and approved the final manuscript.
Supplementary Materials
Table S1 is “potential effect mechanism mining of JPFR basing on network pharmacology analysis.” Other explanatory text of figures and tables has already been listed in the separated Word.
References
- 1.Xu Y., Mao J. J., Sun L., et al. Association between use of traditional Chinese medicine herbal therapy and survival outcomes in patients with stage II and III colorectal cancer: a multicenter prospective cohort study. JNCI Monographs. 2017;2017(52):p. lgx015. doi: 10.1093/jncimonographs/lgx015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCulloch M., Ly H., Broffman M., See C., Clemons J., Chang R. Chinese herbal medicine and fluorouracil-based chemotherapy for colorectal cancer: a quality-adjusted meta-analysis of randomized controlled trials. Integrative Cancer Therapies. 2016;15(3):285–307. doi: 10.1177/1534735416638738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen M., May B. H., Zhou I. W., Xue C. C. L., Zhang A. L. FOLFOX 4 combined with herbal medicine for advanced colorectal cancer: a systematic review. Phytotherapy Research. 2014;28(7):976–991. doi: 10.1002/ptr.5092. [DOI] [PubMed] [Google Scholar]
- 4.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue R., Fang Z., Zhang M., et al. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Research. 2012;41(D1):D1089–D1095. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao J. L., Ren J. L., Ji Q., et al. Study on network pharmacology of Jianpi Bushen Formula for treating colon cancer. China Journal of Traditional Chinese Medicine and Pharmacy. 2017;32:1772–1774. [Google Scholar]
- 7.Law V., Knox C., Djoumbou Y., et al. DrugBank 4.0: sheddingnew light on drug metabolism. Nucleic Acids Research. 2014;42:1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 9.Zhang G. B., Li Q. Y., Chen Q.-L, Su S. B. Network pharmacology: a new approach for Chinese herbal medicine research. Evidence-Based Complementary and Alternative Medicine. 2013;2013:9. doi: 10.1155/2013/621423.621423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Kampen A. H. C., Moerland P. D. Taking bioinformatics to systems medicine. Methods in Molecular Biology. 2016;1386:17–41. doi: 10.1007/978-1-4939-3283-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu D. H., Qiu X. H., Zhu W., et al. Study on mechanism of clearing-heat herbs on coronary disease in molecular level by network pharmacology. China Journal of Traditional Chinese Medicine and Pharmacy. 2011;26:1004–1008. [Google Scholar]
- 12.Li S., Wu L., Zhang Z. Constructing biological networks through combined literature mining and microarray analysis: a LMMA approach. Bioinformatics. 2006;22(17):2143–2150. doi: 10.1093/bioinformatics/btl363. [DOI] [PubMed] [Google Scholar]
- 13.Li S., Zhang Z. Q., Wu L. J., Zhang X. G., Wang Y. Y., Li Y. D. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Systems Biology. 2007;1(1):51–60. doi: 10.1049/iet-syb:20060032. [DOI] [PubMed] [Google Scholar]
- 14.Zhao S. W., Li S. Network-based relating pharmacological and Genomic spaces for drug target identification. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011764.el1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R., Ma T., Liang X., Li S. Imbalanced network biomarkers for traditional Chinese medicine Syndrome in gastritis patients. Scientific Reports. 2013;3(1):p. 1543. doi: 10.1038/srep01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Q., Liu X., Fu X., et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0078700.e78700 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Ji Q., Zhang L., Liu X., et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. British Journal of Cancer. 2014;111(4):736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z.-R., Wu Y.-F., Ma C.-Y., Nie S.-D., Mao X.-H., Shi Y.-Z. Down-regulation of c-Myc expression inhibits the invasion of bile duct carcinoma cells. Cell Biology International. 2011;35(8):799–802. doi: 10.1042/cbi20110099. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Gill A. J. M., Issacs J. D., et al. The Wnt/β-catenin pathway drives increased cyclin D1 levels in lymph node metastasis in papillary thyroid cancer. Human Pathology. 2012;43(7):1044–1050. doi: 10.1016/j.humpath.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G., Jr., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(9):p. P3. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 21.Punt C. J. A., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nature Reviews Clinical Oncology. 2017;14(4):235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 22.Beck D. E., Roberts P. L., Saclarides T. J., et al. The ASCRS Textbook of Colon and Rectal Surgery. NewYork, NY, USA: Springer; 2011. [Google Scholar]
- 23.Da Silva F. C., Wernhoff P., Dominguez-Barrera C., Dominguez-valentin M. Update on hereditary colorectal cancer. Anticancer Research. 2016;36(9):4399–4406. doi: 10.21873/anticanres.10983. [DOI] [PubMed] [Google Scholar]
- 24.Tuohy T. M. F., Rowe K. G., Mineau G. P., Pimentel R., Burt R. W., Samadder N. J. Risk of colorectal cancer and adenomas in the families of patients with adenomas: a population-based study in Utah. Cancer. 2014;120(1):35–42. doi: 10.1002/cncr.28227. [DOI] [PubMed] [Google Scholar]
- 25.Bhan A., Soleimani M., Mandal S. S. Long noncoding RNA and cancer: a new paradigm. Cancer Research. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.can-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L., Wu Y., Tan D., et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. Journal of Experimental & Clinical Cancer Research. 2015;34(1):p. 7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton J. G., Mayer S. A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes & Development. 1991;5(7):1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 28.He X., Pool M., Darcy K. M., et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26(34):4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galbán S., Kuwano Y., Pullmann R., Jr., et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1 α. Molecular and Cellular Biology. 2008;28(1):93–107. doi: 10.1128/mcb.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villar J., Cabrera N. E., Valladares F., et al. Activation of the Wnt/β-Catenin signaling pathway by mechanical ventilation is associated with ventilator-induced pulmonary fibrosis in healthy lungs. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023914.e23914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeilstra J., Joosten S. P. J., Dokter M., Verwiel E., Spaargaren M., Pals S. T. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Research. 2008;68(10):3655–3661. doi: 10.1158/0008-5472.can-07-2940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 is “potential effect mechanism mining of JPFR basing on network pharmacology analysis.” Other explanatory text of figures and tables has already been listed in the separated Word.
Data Availability Statement
The data used to support the findings of this study are included within the article.


