Abstract
Cancer causes most of the mortality and morbidity worldwide, with a significant increase in incidence during recent years. MicroRNAs (miRNAs/miRs) are non-coding small RNAs capable of regulating gene expression. They regulate crucial cellular processes, including proliferation, differentiation, metastasis and apoptosis. Therefore, abnormal miRNA expression is associated with multiple diseases, including cancer. There are two types of cancer-associated miRNAs, oncogenic and tumor suppressor miRNAs, depending on their roles and expression patterns in cancer. Accordingly, miRNAs are considered to be targets for cancer prevention and treatment. miR-33a controls cellular cholesterol uptake and synthesis, which are both closely associated with carcinogenesis. The present review thoroughly describes the roles of miR-33a in more than a dozen types of cancer and the underlying mechanisms. Accordingly, the present review may serve as a guide for researchers studying the involvement of miR-33a in diverse cancer settings.
Keywords: microRNAs, biomarkers, oncogenes, tumor suppressors, carcinogenesis
1. Introduction
MicroRNAs (miRNAs/miRs) are endogenous non-coding small RNAs that are 19–24 nucleotides in length and are widely found in eukaryotes (1). In general, they bind to complementary sites on the 3′-untranslated regions of mRNAs, which are consequently destabilized or translationally suppressed (1). This post-transcriptional regulation serves essential roles physiologically and pathologically. miRNAs regulate important biological processes, including proliferation and apoptosis (2,3). Numerous miRNAs are involved in the tumorigenic transformation or migration of cells (4). Recent advancements in molecular biology technologies have introduced novel approaches to study the role of miRNAs in cancer and have thereby led to novel findings regarding cancer pathogenesis, prognosis and treatment (5,6).
Cancer is one of the leading causes of mortality and morbidity globally. According to the latest data published in the GLOBOCAN database in 2018, 18.1 million cancer cases and 9.6 million cancer-associated deaths were reported worldwide (7). Asia has the highest number of patients with cancer in the world, which accounts for 48.4% of the global cancer population (7). The five most frequent types of cancer worldwide in descending order are lung cancer, breast carcinoma, colorectal cancer, prostate carcinoma and gastric cancer (7). The five types of cancer with the highest mortality rates worldwide are lung cancer, colorectal carcinoma, gastric cancer, liver carcinoma and breast cancer (7). The types of cancer with the highest cancer-associated morbidity and mortality in men and women worldwide are lung and breast cancer, respectively (7). It has been estimated that there will be 22 million new cancer cases and 13 million cancer deaths per year by 2030 (8). Therefore, the identification of effective approaches for the prevention, diagnosis and treatment of cancer is required, in addition to the traditional approaches of surgery, chemotherapy and radiotherapy.
Abnormal miRNA expression is associated with tumorigenesis, and more than half of all miRNAs in the human genome are located in cancer-associated regions or fragile sites (9). In 2006, Calin et al (10) reported that the miRNA cluster miR-15/miR-16-1 was eliminated or repressed in 69% of patients with chronic lymphocytic leukemia. The aforementioned study was the first to demonstrated that miRNAs are closely associated with cancer (10). Abnormally expressed miRNAs in cancer can be functionally classified as oncogenic, such as miR-17-92 (11), or tumor suppressors, such as let-7 (12), which are upregulated and downregulated in tumor cells, respectively. Accordingly, the upregulation of the oncogenic miRNAs and the downregulation of the tumor suppressor miRNAs may promote tumor development. However, a miRNA may serve different roles in different types of cancer. For example, miR-221 acts as an oncogene in hepatocellular carcinoma (13), but it is a tumor suppressor in erythroblastic leukemia (14). In addition to serving as biomarkers for tumorigenesis, miRNAs are associated with tumor chemoresistance. Upregulation of some miRNAs, such as miR-19 (15) and miR-21 (16), can enhance tumor chemoresistance, whereas other miRNAs, such as miR-130a (17) and miR-298 (18), are downregulated in chemoresistant tumor cells. Therefore, it may be possible to use miRNA expression profiles of tumors for diagnostic, prognostic and therapeutic purposes. However, this requires an in-depth analysis of miRNA-associated mechanisms in various types of tumor.
miR-33a is located in the 16th intron of the human sterol regulatory element binding transcription factor 2 (SREBF-2) gene on chromosome 22 (19). This highly conserved intronic miRNA regulates the genes that control cholesterol uptake or synthesis (20). SREBFs activate the expression of >30 genes involved in the synthesis or cellular uptake of cholesterol, triglycerides, phospholipids and fatty acids, along with the NADPH cofactors required to synthesize these biomolecules (21). In addition, the SREBF signaling pathway regulates various cellular processes, including phagocytosis and cell cycle progression (22). In healthy tissues, transcriptional activation of SREBF-2 upregulates miR-33a, consequently enhancing cellular lipid metabolism, including that of cholesterol (22). Studies have demonstrated that cholesterol is involved in apoptosis, whereas the SREBP/miR-33 axis participates in cell growth and cell cycle progression (23,24). Therefore, dysregulation of miR-33a levels may contribute to tumorigenesis by affecting cholesterol levels. In this case, tumors may be treated by correcting the miR-33a levels or associated mechanisms.
The present review describes the available literature regarding miR-33a and cancer and provides an overview of miR-33a-associated mechanisms in hepatocellular, gastric, colorectal, pancreatic and gallbladder carcinomas, as well as osteosarcoma, melanoma, glioma, esophageal and tongue squamous cell cancer, and lung, laryngeal, renal, thyroid, prostate and breast cancer. Additionally, 38 target genes of miR-33a and the possibility of their involvement in cancer are examined, providing a basis for future research investigating the role of miR-33a in cancer.
2. miR-33a and its role in cancer
Digestive system cancers
Hepatocellular carcinoma
Hepatocellular carcinoma is the third most common type of cancer globally (25–27). Hepatitis B and C virus infections are considered to be the main causes of hepatocellular carcinoma, which can also be caused by non-alcoholic liver disease or alcohol abuse (28).
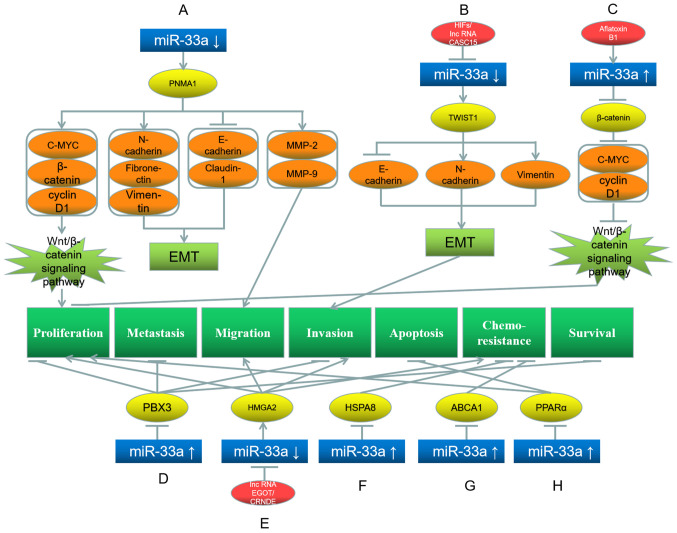
In this type of cancer, miR-33a can act as a tumor suppressor gene. Liu et al (29) have demonstrated that miR-33a-5p is downregulated, while its target gene paraneoplastic antigen Ma1 (PNMA1) is upregulated in hepatoma cells. Consequently, the downstream proteins of PNMA1, such as C-MYC, β-catenin and cyclin D1, are upregulated; these proteins activate the Wnt signaling pathway, which facilitates the proliferation of hepatoma cells (29). Additionally, PNMA1 upregulates N-cadherin, fibronectin and vimentin expression, but downregulates E-cadherin and claudin-1 expression, thereby promoting the epithelial-mesenchymal transition (EMT) of the cells, while also promoting their migration through the upregulation of matrix metalloproteinases-2/9 (Fig. 1A) (29). Similarly, Guo et al (30) and Li et al (31) have demonstrated that hypoxia-inducible factors (HIFs) and long non-coding RNA (lncRNA) cancer susceptibility 15 (CASC15) decrease the miR-33a level in hepatoma cells. Additionally, the miR-33a target gene Twist basic helix-loop-helix transcription factor 1 (TWIST1) is consequently upregulated, which subsequently downregulates E-cadherin and upregulates N-cadherin and vimentin expression, thereby further promoting EMT and invasion of hepatoma cells (Fig. 1B) (30,31).
Figure 1.
Roles of miR-33a and its target genes in hepatocellular carcinoma, and the underlying mechanisms. (A) miR-33a downregulation upregulates its target gene PNMA1, which further upregulates C-MYC, β-catenin, N-cadherin, fibronectin, vimentin, MMP-2 and MMP-9, while downregulates E-cadherin and claudin-1, promoting EMT, proliferation and migration. (B) HIFs and lncRNA CASC15 downregulate miR-33a and further upregulate the target gene TWIST1, which upregulates N-cadherin and vimentin, while downregulates E-cadherin, promoting EMT and invasion. (C) Aflatoxin B1 upregulates miR-33a, which inhibits its target gene β-catenin and further downstream proteins C-MYC and cyclin D1, leading to proliferation inhibition. (D) miR-33a upregulation downregulates its target gene PBX3, leading to the inhibition of proliferation, metastasis, invasion and survival. (E) LncRNA EGOT and lncRNA CRNDE downregulate miR-33a, which further upregulates its target gene HMGA2, promoting proliferation, migration, invasion and chemoresistance. (F) miR-33a upregulation downregulates its target gene HSPA8, inhibiting chemoresistance. (G) miR-33a upregulation downregulates its target gene ABCA1, inhibiting chemoresistance. (H) miR-33a upregulation downregulates its target gene PPARα, promoting proliferation and inhibiting apoptosis. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; PNMA1, paraneoplastic antigen Ma1; MMP, matrix metalloproteinase; TWIST, Twist basic helix-loop-helix transcription factor 1; EMT, epithelial-mesenchymal transition; PBX3, pre-B-cell leukemia transcription factor 3; HSPA8, heat shock protein family A (Hsp70) member 8; ABCA1, ATP-binding cassette transporter A1; PPARα, peroxisome proliferator activated receptor α.
By contrast, Fang et al (32) have revealed that aflatoxin B1 upregulates miR-33a in the hepatoma Chang (IC50, 40 µg/ml) and HepG2 (IC50, 77 µg/ml) cell lines. Consequently, β-catenin and its downstream proteins C-MYC and cyclin D1 are downregulated, whereby the Wnt signaling pathway is suppressed, and so is the proliferation of hepatoma cells (Fig. 1C) (32). Similarly, Han et al (33) have observed that miR-33a-3p is downregulated in hepatoma cells, and its forced expression suppresses the proliferation, metastasis and invasion of these cells by downregulating the target gene pre-B-cell leukemia transcription factor 3 (Fig. 1D). Furthermore, low miR-33a expression is associated with a low survival rate in patients with hepatocellular carcinoma (33). Similarly, Wu et al (34) and Han et al (35) have illustrated that lncRNA eosinophil granule ontogeny transcript and lncRNA colorectal neoplasia differentially expressed in hepatoma cells inhibit miR-33a expression, which upregulates the target gene high mobility group AT-hook 2 (HMGA2), thereby promoting the proliferation, migration, invasion and chemoresistance of hepatoma cells (Fig. 1E).
Furthermore, miR-33a can affect the chemoresistance of hepatoma cells. Meng et al (36) have demonstrated that hepatoma cells with high miR-33a-5p levels are more sensitive to cisplatin than those with low expression levels, and this cisplatin resistance can be decreased by downregulating heat shock protein family A (Hsp70) member 8 through miR-33a-5p expression (Fig. 1F). Similarly, Hou et al (37) have reported that miR-33a overexpression in Lgr5+ hepatoma-cancer stem cells decreases ATP-binding cassette transporter A1 (ABCA1) expression, thereby reducing the resistance of these cells to doxorubicin (Fig. 1G). Accordingly, the aforementioned study indicated that increasing the levels of miR-33a in hepatoma cells may reduce their resistance to doxorubicin (37).
Although most studies have illustrated that miR-33a serves as a tumor suppressor in hepatoma cells, Chang et al (38) have observed the opposite effect, reporting that miR-33a upregulation and the consequent downregulation of its target peroxisome proliferator activated receptor α promote proliferation, while inhibiting the apoptosis of hepatoma cells (Fig. 1H). The reason underlying these opposing observations is unclear.
Gastric cancer
Gastric cancer is one of the most common digestive system cancers and causes major public health problems worldwide (39). Dietary factors are the main cause of this type of cancer, although genetic or environmental factors, such as stress, are also closely associated with it (40).
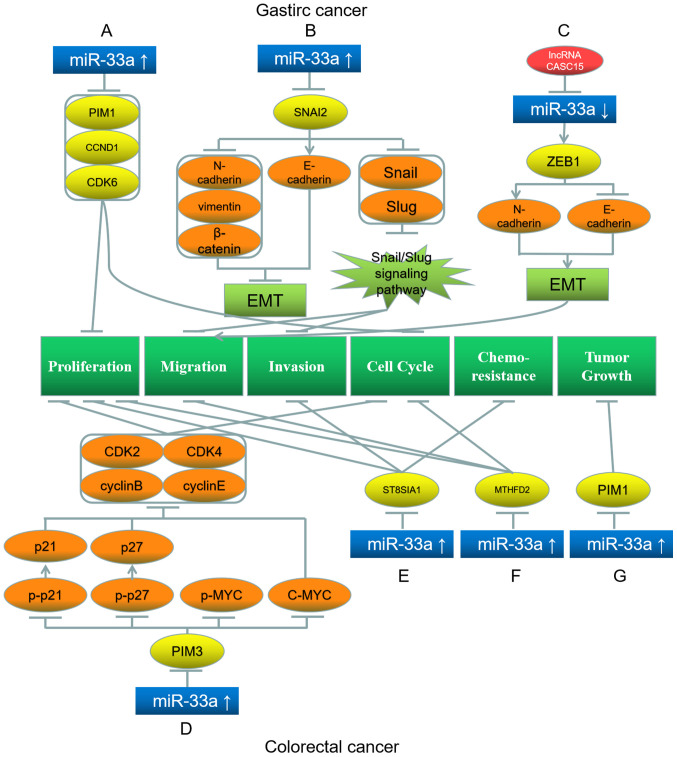
miR-33a serves a role in suppressing oncogenes in gastric cancer. Wang et al (41), have illustrated that miR-33a expression is decreased in gastric tumors. Increased miR-33a downregulates Pim-1 proto-oncogene (PIM1), cyclin D1 and cyclin-dependent kinase (CDK6) to impede the proliferation of gastric cancer cells, causing them to be arrested at the G1 stage (Fig. 2A) (41). Among these three target genes, the interaction of miR-33a with PIM1 has the greatest impact on gastric cancer (41). Additionally, Chen et al (42) have demonstrated that miR-33a is downregulated in gastric tumor cells. miR-33a overexpression inhibits Snail family transcriptional repressor 2 (SNAI2), thereby upregulating the downstream protein E-cadherin and downregulating N-cadherin, vimentin and β-catenin, which suppresses EMT in gastric tumor cells (42). In addition, miR-33a downregulates Snail and Slug levels, and inhibition of the Snail/Slug signaling pathway hinders migration and invasion of these cells (Fig. 2B) (42). Wu et al (43) have revealed that lncRNA CASC15 is a miRNA sponge in gastric cancer cells, as in hepatoma cells. CASC15 reduces miR-33a-5p expression, thereby upregulating the miR-33a target gene zinc finger E-Box binding homeobox 1 (ZEB1), which subsequently upregulates N-cadherin and downregulates E-cadherin, promoting the EMT and migration of gastric cancer cells (Fig. 2C) (43).
Figure 2.
Roles of miR-33a and its target genes in gastric (top) and colorectal (bottom) cancer, and the underlying mechanisms. (A) miR-33a upregulation downregulates its target genes PIM1, CCND1 and CDK6, which inhibit proliferation and cell cycle. (B) miR-33a upregulation inhibits its target gene SNAI2, which further downregulates N-cadherin, vimentin, β-catenin, Snail and Slug, while upregulates E-cadherin, leading to the inhibition of EMT, migration and invasion. (C) LncRNA CASC15 inhibits miR-33a and further upregulates its target gene ZEB1, which upregulates N-cadherin and downregulates E-cadherin, promoting EMT and proliferation. (D) miR-33a upregulation inhibits its target gene PIM3, which further downregulates the downstream proteins p-p21 (upregulating p21), p-p27 (upregulating p27), p-MYC, C-MYC, CDK2, CDK4, cyclin B and cyclin E, leading to proliferation and cell cycle inhibition. (E) miR-33a upregulation downregulates its target gene ST8SIA1, inhibiting proliferation, invasion and chemoresistance. (F) miR-33a upregulation downregulates its target gene MTHFD2, inhibiting proliferation, migration and cell cycle. (G) miR-33a upregulation downregulates its target gene PIM1, inhibiting tumor growth. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; EMT, epithelial-mesenchymal transition; p, phosphorylated; PIM1/3, Pim-1/3 proto-oncogene; CCND1, cyclin D1; CDK, cyclin-dependent kinase; SNAI2, Snail family transcriptional repressor 2; ZEB1, Zinc finger E-Box binding homeobox 1; ST8SIA1, ST8α-N-acetyl-neuraminide-α-2,8-sialyltransferase 1; MTHFD2, methylenetetrahydrofolate dehydrogenase 2.
Colorectal cancer
Colorectal cancer is the third most common malignancy and the fourth most common principal cause of tumor mortality in the world (44). According to a cancer research carried out in 2019, a considerable proportion (21%) of colorectal tumor cases is detected at the advanced stage (45). The occurrence of colorectal cancer is strongly associated with a high fat-diet, especially with a cholesterol-rich diet (46–49).
miR-33a activates the transcription of genes associated with cholesterol metabolism. Accordingly, it serves as a tumor suppressor gene in colorectal carcinoma. Wang et al (50) have reported that miR-33a can neutralize the effect of excessive cholesterol intake on the proliferation of colorectal cancer cells. While cholesterol downregulates miR-33a and consequently upregulates the miR-33a target PIM3, downregulation of PIM3 by miR-33a inhibits the phosphorylation of p21, p27 and p-MYC, and C-MYC expression, which deactivate the cell cycle-associated proteins CDK2, CDK4, cyclin B and cyclin E (50). Consequently, the proliferation and cell cycle progression of colorectal cancer cells are inhibited (Fig. 2D) (50). Shan et al (51) have demonstrated that miR-33a and let-7e are downregulated in colorectal cancer cells. These two miRNAs can bind to ST8α-N-acetyl-neuraminide-α-2,8- sialyltransferase 1 (ST8SIA1) and consequently further inhibit the proliferation, invasion and chemoresistance of colorectal cancer cells (Fig. 2E) (51). ST8SIA1 overexpression suppresses these inhibitory effects of miR-33a and let-7e (51). Therefore, these two miRNAs and ST8SIA1 affect colorectal cancer cells through mutual restrictions (51). Similarly, Yan et al (52) have illustrated that miR-33a-5p is downregulated, while its target gene methylenetetrahydrofolate dehydrogenase 2 is upregulated in colorectal carcinoma (52). Additionally, the proliferation, migration and cell cycle progression of colorectal carcinoma cells can be repressed by overexpressed miR-33a-5p (Fig. 2F) (52).
Notably, Ibrahim et al (53) have demonstrated that miRNAs and polyethyleneimine (PEI) can form biocompatible complexes that can be used in miRNA replacement therapies. Delivery of the miR-33a/PEI complex into colorectal cancer cells suppresses tumor growth via the downregulation of PIM1 (Fig. 2G) (53). This observation indicates the efficacy of such a biocomplex, which may be used as a novel treatment in colorectal cancer (53).
Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC), also known as pancreatic cancer, is one of the most lethal malignancies in the world (54). In 2018, PDAC exhibited the lowest 5-year relative survival rate compared with all other solid tumors, and caused 432,242 deaths worldwide (55). Early-stage PDAC is asymptomatic; therefore, the manifestation of the illness is delayed (54). Consequently, ~50% patients are already at the metastatic stage at initial diagnosis (54).
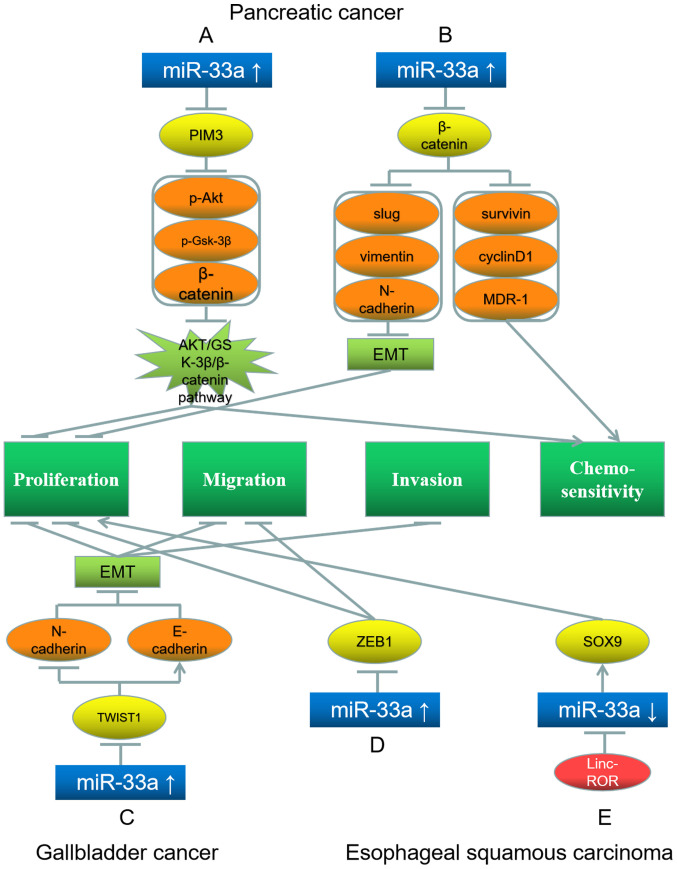
miR-33a is a tumor suppressor in PDAC. Liang et al (56) have revealed that increasing the expression of downregulated miR-33a in pancreatic cancer inhibits PIM3 expression and downregulates the downstream proteins phosphorylated (p)-AKT, p-glycogen synthase kinase (GSK)-3β and β-catenin. Consequently, the AKT/GSK-3β/β-catenin signaling pathway is repressed, and the proliferation and gemcitabine-resistance of pancreatic cancer cells are inhibited (Fig. 3A) (56). In another study, Liang et al (57) demonstrated that high levels of miR-33a suppress the nuclear translocation of β-catenin, thereby repressing the expression levels of downstream proteins, such as slug, vimentin, N-cadherin, survivin, cyclin D1 and multidrug resistance 1. Consequently, the gemcitabine-sensitivity of pancreatic cancer cells is increased, and the proliferation and EMT of pancreatic tumor cells are suppressed (Fig. 3B) (57).
Figure 3.
Roles of miR-33a and its target genes in pancreatic cancer (top), and gallbladder (bottom left) and esophageal squamous (bottom middle and right) carcinoma, and the underlying mechanisms. (A) miR-33a upregulation inhibits its target gene PIM3; this further downregulates p-AKT, p-GSK-3β and β-catenin, leading to proliferation inhibition and chemosensitivity promotion. (B) miR-33a upregulation inhibits its target gene β-catenin, further downregulating slug, vimentin, N-cadherin, survivin, cyclin D1 and MDR-1, leading to EMT inhibition and chemosensitivity promotion. (C) miR-33a upregulation inhibits its target gene TWIST1, further downregulating N-cadherin and upregulating E-cadherin, leading to inhibition of EMT, proliferation, migration and invasion. (D) miR-33a upregulation downregulates its target gene ZEB1, inhibiting proliferation and migration. (E) Linc-ROR downregulates miR-33a, further upregulating its target gene SOX9, which promotes proliferation. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; EMT, epithelial-mesenchymal transition; p-, phosphorylated; PIM3, Pim-3 proto-oncogene; GSK, glycogen synthase kinase; MDR-1, multidrug resistance 1; SOX9, SRY-box 9; TWIST1, Twist basic helix-loop-helix transcription factor 1; ZEB1, Zinc finger E-Box binding homeobox 1.
Gallbladder cancer
Gallbladder cancer is a rare malignancy. The incidence of gallbladder cancer is higher in developing countries compared with developed countries (58).
miR-33a represses carcinogenesis in gallbladder cancer. Zhang et al (59) have demonstrated that upregulating miR-33a in IL-6-induced gallbladder carcinoma cells downregulates the downstream protein N-cadherin and upregulates E-cadherin via the downregulation of the miR-33a target TWIST1. Consequently, the EMT, proliferation, migration and invasion of gallbladder tumor cells are inhibited (Fig. 3C) (59).
Esophageal squamous cell carcinoma (ESCC)
ESCC accounts for ~90% of esophageal cancer cases (60). The causes of ESCC are associated with economic status and unhealthy habits, such as smoking and alcohol abuse (61).
miR-33a acts as a tumor suppressor in ESCC. It targets and downregulates ZEB1 and thereby suppresses ESCC proliferation and migration (Fig. 3D) (62). Notably, lncRNA differentiation antagonizing non-protein coding RNA (DANCR) downregulates miR-33a-5p levels through miRNA sponging in ESCC (62). Wang et al (63) have detected the regulatory network linc-ROR-miRNA-SRY-box 9 (SOX9) in ESCC. Linc-ROR inhibits the expression of multiple miRNAs, including miR-33a, and consequently upregulates their target gene SOX9, which confers stem cell-like properties and promotes cell proliferation (Fig. 3E) (63).
Respiratory system cancers
As one of the most common types of cancer in the world, lung cancer causes the majority of tumor-associated deaths; in 2018, 2.1 million patients with lung cancer were diagnosed, and lung cancer caused 1.8 million deaths worldwide (7). In China, the main causes of lung carcinoma are air pollution and smoking (64). There are two types of lung cancer, small cell lung carcinoma and non-small cell lung carcinoma (NSCLC), and the latter can be further divided into three subtypes: Squamous cell cancer, adenocarcinoma and large-cell carcinoma. Most cases of lung cancer (80-85%) are NSCLC (64).
NSCLC
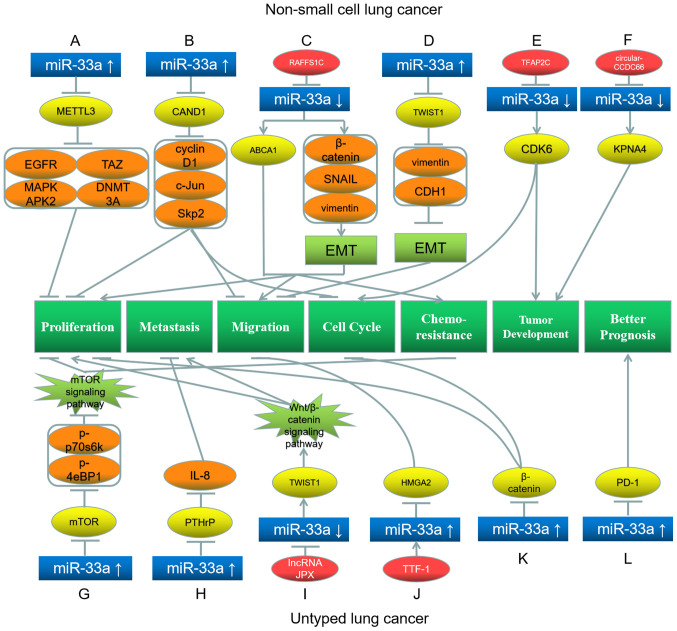
Du et al (65) have indicated that miR-33a is downregulated in NSCLC cells, and that the proliferation of NSCLC cells is subdued by upregulating miR-33a, which downregulates its target gene methyltransferase-like 3 and the subsequent downstream proteins epidermal growth factor receptor, tafazzin, mitogen-activated protein kinase-activated protein kinase 2 and DNA methyltransferase 3A (Fig. 4A). Additionally, Kang et al (66) have observed that miR-33a is downregulated in NSCLC cells, and its upregulation represses the proliferation, migration and cell cycle progression of these cells by downregulating its target gene Cullin associated and neddylation dissociated 1 and subsequent downstream proteins cyclin D1, c-Jun and S-phase kinase associated protein 2 (Fig. 4B). Amaar and Reeves (67) have demonstrated that the oncogene Ras association domain family member 1 (RAFFS1C) downregulates miR-33a-5p, whereby the miR-33a-5p target ABCA1 is upregulated, enhancing the chemoresistance of NSCLC cells. Additionally, the aforementioned study has indicated that RAFFS1C promotes the EMT, proliferation and migration of NSCLC cells by downregulating miR-33a-5p and upregulating β-catenin, SNAIL and vimentin (Fig. 4C) (67). Furthermore, Yang et al (68) have observed that miR-33a is downregulated in NSCLC. miR-33a targets TWIST1, whereby the downstream proteins vimentin and cadherin-1 are downregulated, and the EMT and migration of NSCLC cells are suppressed (Fig. 4D) (68). Kang et al (69) have demonstrated that the oncogene transcription factor AP-2 γ is overexpressed in NSCLC cells and downregulates miR-33a. Consequently, the miR-33a target CDK6 is upregulated, whereby the cell cycle progression and tumor development of NSCLC cells are enhanced (Fig. 4E) (69). Wang et al (70) have revealed that overexpression of circular-CCDC66 downregulates miR-33a-5p in NSCLC cells, consequently upregulating the miR-33a-5p target karyopherin subunit α4 and promoting tumorigenesis and development of NSCLC (Fig. 4F).
Figure 4.
Roles of miR-33a and its target genes in non-small cell lung cancer (top) and untyped lung cancer (bottom), and the underlying mechanisms. (A) miR-33a upregulation inhibits its target gene METTL3, further downregulating EGFR, TAZ, MAPKAPK2 and DNMT3A, leading to proliferation inhibition. (B) miR-33a upregulation inhibits its target gene CAND1, further downregulating cyclin D1, c-Jun and Skp2, leading to inhibition of proliferation, migration and cell cycle. (C) RAFFS1C inhibits miR-33a and further upregulates its target gene ABCA1, as well as β-catenin, SNAIL and vimentin, leading to promotion of EMT, proliferation, migration and chemoresistance. (D) miR-33a upregulation inhibits its target gene TWIST1, further downregulating vimentin and CDH1, leading to inhibition of EMT and migration. (E) TFAP2C inhibits miR-33a and further upregulates its target gene CDK6, which promotes cell cycle progression and tumor development. (F) circular-CCDC66 inhibits miR-33a and further upregulates its target gene KPNA4, which promotes tumor development. (G) miR-33a upregulation inhibits its target gene mTOR, downregulating p-p70S6K and p-4EBP1, leading to inhibition of proliferation and chemoresistance. (H) miR-33a upregulation inhibits its target gene PTHrP, downregulating IL-8 and leading to metastasis inhibition. (I) lncRNA JPX inhibits miR-33a and further upregulates its target gene TWIST1, leading to promotion of proliferation and metastasis. (J) TTF-1 upregulates miR-33a and further downregulates its target gene HMGA2, which inhibits migration. (K) miR-33a upregulation downregulates its target gene β-catenin, which inhibits proliferation and cell cycle. (L) miR-33a upregulation downregulates its target gene PD-1, resulting in a better prognosis. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; EMT, epithelial-mesenchymal transition; p-, phosphorylated; DNMT3A, DNA methyltransferase 3A; MAPKAPK2, mitogen-activated protein kinase-activated protein kinase 2; PTHrP, parathyroid hormone-related protein; TAZ, tafazzin; Skp2, S-phase kinase associated protein 2; METTL3, methyltransferase-like 3; EGFR, epidermal growth factor receptor; CAND1, Cullin associated and neddylation dissociated 1; ABCA1, ATP-binding cassette transporter A1; TWIST1, Twist basic helix-loop-helix transcription factor 1; CDH1, cadherin 1; CDK6, cyclin-dependent kinase 6; KPNA4, karyopherin subunit α4; IL-8, interleukin-8; HMGA2, high mobility group AT-hook 2; PD-1, programmed cell death protein 1.
Untyped lung cancer
Li et al (71) have confirmed that miR-33a-5p is downregulated in lung tumor cells. By acting on its target gene mTOR and downstream proteins p-p70S6K and p-eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), miR-33a-5p inhibits the mTOR signaling pathway, inhibits proliferation and reduces celastrol resistance in lung cancer cells (Fig. 4G) (71). Additionally, Kuo et al (72) have demonstrated that miR-33a is downregulated in lung cancer and that its overexpression represses bone metastasis by downregulating parathyroid hormone-related protein and its downstream protein interleukin (IL)-8 (Fig. 4H). Pan et al (73) have illustrated that lncRNA JPX transcript, XIST activator downregulates miR-33a-5p in lung cancer cells through miRNA sponging. This downregulation upregulates the target gene TWIST1 and activates the Wnt/β-catenin signaling pathway, subsequently promoting lung cancer cell proliferation and metastasis (Fig. 4I) (73). Rice et al (74) have observed that thyroid transcription factor-1 binds to the promoter of SREBF2, which is the host gene of miR-33a, and activates miR-33a transcription. miR-33a subsequently downregulates its target, HMGA2, whereby the migration of lung cancer cells is suppressed (Fig. 4J) (74). Additionally, Zhu et al (75) have demonstrated that miR-33a expression is decreased in lung cancer cells, while increasing miR-33a expression downregulates β-catenin and causes G1/S phase arrest, thereby suppressing cancer cell proliferation (Fig. 4K). Boldrini et al (76) have indicated that patients with lung cancer with high miR-33a expression and low programmed cell death protein 1 expression in the tumor tissues have an improved prognosis and a higher survival rate (Fig. 4L). Therefore, high levels of miR-33a may be a favorable prognostic indicator for lung cancer (76).
Head and neck cancer
Laryngeal carcinoma
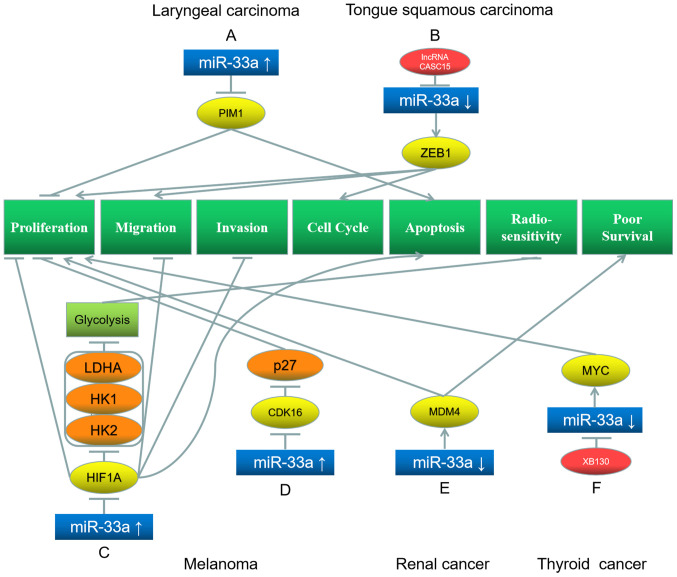
Laryngeal cancer is one of the most frequent head and neck malignancies (77). Most patients with laryngeal cancer (95%) have the habit of smoking or consuming alcohol (78). Karatas (79) demonstrated that high levels of miR-33a inhibit the proliferation of laryngeal carcinoma cells and promote apoptosis via downregulation of PIM1 (Fig. 5A).
Figure 5.
Roles of miR-33a and its target genes in laryngeal (top left) and tongue squamous (top right) carcinoma, melanoma (bottom left and middle), renal cancer (bottom middle) and thyroid cancer (bottom right), and the underlying mechanisms. (A) miR-33a upregulation downregulates its target gene PIM1, inhibiting proliferation and promotes apoptosis. (B) LncRNA CASC15 downregulates miR-33a and further upregulates its target gene ZEB1, which promotes proliferation, migration and cell cycle. (C) miR-33a upregulation inhibits its target gene HIF1A, downregulating LDHA, HK1 and HK2, leading to inhibition of glycolysis, proliferation, migration, invasion and promotion of apoptosis. (D) miR-33a upregulation inhibits its target gene CDK16, downregulating p27, leading to inhibition of proliferation. (E) miR-33a downregulation upregulates its target gene MDM4, which promotes proliferation and results in poor survival. (F) XB130 downregulates miR-33a and further upregulates its target gene MYC, which promotes proliferation. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; PIM1, Pim-1 proto-oncogene; ZEB1, Zinc finger E-Box binding homeobox 1; HIF1A, hypoxia inducible factor 1 subunit α; LDHA, lactate dehydrogenase A; HK, hexokinase; CDK, cyclin-dependent kinase; MDM4, mouse double minute 4.
Tongue squamous carcinoma (TSCC)
TSCC is one of the most aggressive oral cancers, accounting for ~41% of oral cancer cases (80). miR-33a acts as a tumor suppressor in TSCC. Zuo et al (81) have reported that lncRNA CASC15 downregulates miR-33a-5p by miRNA sponging, thereby upregulating the miR-33a-5p target ZEB1 and enhancing the proliferation, migration and cell cycle of TSCC cells (Fig. 5B).
Skin cancer
Melanoma
Melanoma is the fifth and seventh most common malignant tumor in males and females, respectively (82). Melanoma-associated mortality accounts for 80% of skin cancer-associated deaths, and the occurrence has notably increased in previous years (83). The cause of this disease is associated with both environmental and genetic factors, such as sex, family history, geographical location and sun exposure (84).
miR-33a suppresses melanoma. Zhou et al (83) have illustrated that miR-33a overexpression inhibits the proliferation, invasion, migration and promotes apoptosis of malignant melanoma cells by downregulating its target gene HIF1A (Fig. 5C). Cao et al (85) have indicated that miR-33a-5p expression is decreased in melanoma cells. High miR-33a-5p expression downregulates HIF1A and the downstream proteins lactate dehydrogenase A, hexokinase 1 (HK1) and HK2, thereby repressing the glycolysis process and increasing the radiation sensitivity of melanoma cells (Fig. 5C) (85). Tian et al (86) have revealed that miR-33a targets and downregulates CDK16 in melanoma, consequently subduing the phosphorylation of p27. As a result, the degradation of p27 is reduced, and the proliferation of melanoma cells is inhibited (Fig. 5D) (86). Lv et al (87) have demonstrated that lncRNA colon cancer associated transcript 1 downregulates miR-33a through miRNA sponging to promote the proliferation, migration and invasion of melanoma cells. Additionally, Fu et al (88) have illustrated that lncRNA paternally expressed 10 (PEG10) inhibits melanoma cell proliferation, migration and invasion by downregulating miR-33a and activating the PI3K/AKT and mTOR signaling pathways.
Urinary system cancer
Renal cancer
Renal cancer incidence in males is double as that in females (89). The morbidity of renal cancer has been increasing in recent years (90). Jiang et al (91) have demonstrated that miR-33a serves as a tumor suppressor in renal cancer. Low miR-33a levels in renal cancer cells lead to the upregulation of its target gene mouse double minute 4 (Fig. 5E) (91). Consequently, the proliferation of the cancer cells increases, causing poor overall survival in patients with renal cancer (91).
Endocrine system cancer
Thyroid cancer
Thyroid cancer incidence has notably increased in the past few years, and according to a study published in 2020, it has been predicted that thyroid cancer will be the fourth leading type of cancer in the world (92). Takeshita et al (93) have illustrated that actin filament associated protein 1 like 2, a novel protein, suppresses miR-33a expression in thyroid cancer cells, thereby upregulating the miR-33a target MYC, which facilitates the proliferation of thyroid cancer cells (Fig. 5F).
Nervous system cancer
Glioma
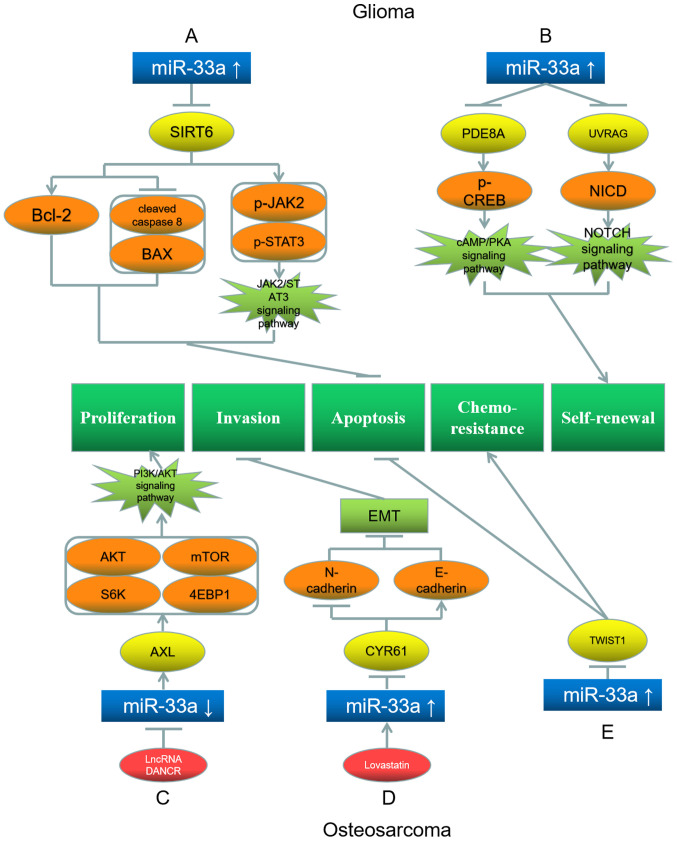
Glioma is the most common and deadly malignancy of the central nervous system (94). The main causes of glioma are congenital factors and carcinogenic environmental exposure (94). Studies have indicated that miR-33a acts as an oncogene in glioma. Chang et al (95) have revealed that miR-33a is upregulated in glioma cells. Consequently, the miR-33a target sirtuin 6 (SIRT6) is downregulated, reducing the levels of cleaved caspase 8 and BAX, and promoting Bcl-2 expression (95). Downregulation of SIRT6 increases the phosphorylation level of Janus kinase 2 (JAK2) and STAT3, enhancing the JAK2/STAT3 pathway, which attenuates H2O2-induced oxidative stress and suppresses apoptosis in glioma cells, and thus miR-33a overexpression inhibits apoptosis in glioma (Fig. 6A) (95). Wang et al (96) have demonstrated that miR-33a overexpression downregulates phosphodiesterase 8A (PDE8A) and UV radiation resistance associated (UVRAG), and upregulates their downstream proteins p-CREB and Notch intracellular domain, respectively, in glioma cells. Consequently, the cAMP/protein kinase A (PKA) and NOTCH signaling pathways are promoted, which enhance the self-renewal of glioma-initiating cells (Fig. 6B) (96).
Figure 6.
Roles of miR-33a and its target genes in glioma (top) and osteosarcoma (bottom), and the underlying mechanisms. (A) miR-33a upregulation inhibits its target gene SIRT6, upregulating Bcl-2, p-JAK2 and p-STAT3, while downregulating cleaved caspase 8 and BAX, leading to apoptosis inhibition. (B) miR-33a upregulation downregulates its target genes PDE8A and UVRAG, upregulating p-CREB and NICD, leading to self-renewal promotion. (C) LncRNA DANCR downregulates miR-33a and further upregulates its target gene AXL, which upregulates AKT, mTOR, S6K and 4EBP1, leading to proliferation promotion. (D) Lovastatin upregulates miR-33a and inhibits its target gene CYR61, downregulating N-cadherin and upregulating E-cadherin, leading to inhibition of EMT and invasion. (E) miR-33a upregulation downregulates TWIST1, promoting chemoresistance and inhibiting apoptosis. Red represents the factor causing miR-33a dysregulation; blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; EMT, epithelial-mesenchymal transition; SIRT6, sirtuin 6; p-, phosphorylated; JAK2, Janus kinase 2; PDE8A, phosphodiesterase 8A; PKA, protein kinase A; TWIST1, Twist basic helix-loop-helix transcription factor 1; UVRAG, UV radiation resistance associated; NICD, Notch intracellular domain; AXL, AXL receptor tyrosine kinase; 4EBP1, eukaryotic translation initiation factor 4E binding protein 1; S6K, ribosomal protein S6K; CYR61, cysteine-rich angiogenic inducer 61.
Musculoskeletal system cancer
Osteosarcoma
Osteosarcoma is a primary bone tumor, which accounts for 60% of all patients with sarcoma (97), and is more common in children and young adults than in adults (98). The role of miR-33a in osteosarcoma is controversial. Jiang et al (99) have observed that lncRNA DANCR is a miRNA sponge in osteosarcoma cells. DANCR targets and downregulates miR-33a-5p, resulting in the upregulation of AXL receptor tyrosine kinase (AXL), a target gene of miR-33a-5p (99). Consequently, the PI3K/AKT signaling pathway is activated, with the upregulation of the downstream genes AKT, mTOR, ribosomal protein S6K (S6K) and 4EBP1 (99). These molecular changes eventually promote cell proliferation and osteosarcoma development (Fig. 6C) (99). Huang et al (100) have illustrated that lovastatin enhances miR-33a expression in osteosarcoma. Therefore, the miR-33a target cysteine-rich angiogenic inducer 61 and downstream protein N-cadherin are downregulated, while E-cadherin is upregulated, whereby the EMT and invasion of tumor cells are suppressed (Fig. 6D) (100). Zhang et al (101) have illustrated that miR-33a-5p expression is downregulated in osteosarcoma cells compared with in normal tissues, and overexpressing miR-33a-5p suppresses cell proliferation in osteosarcoma.
In contrast to the aforementioned studies, Zhou et al (102) have observed miR-33a upregulation in osteosarcoma. Consequently, the miR-33a target TWIST1 is downregulated, whereby the cisplatin resistance of osteosarcoma cells is enhanced, while their apoptosis is suppressed (Fig. 6E) (102).
Male and female cancer
Prostate cancer (PC)
PC is the second most common cause of male-cancer-associated deaths worldwide (103). Androgen is generally considered as the main inducer of PC. High androgen levels are accordingly associated with a high risk of PC. In addition, PC is associated with carcinogenic environmental exposure, an unhealthy diet and genetic factors (104,105).
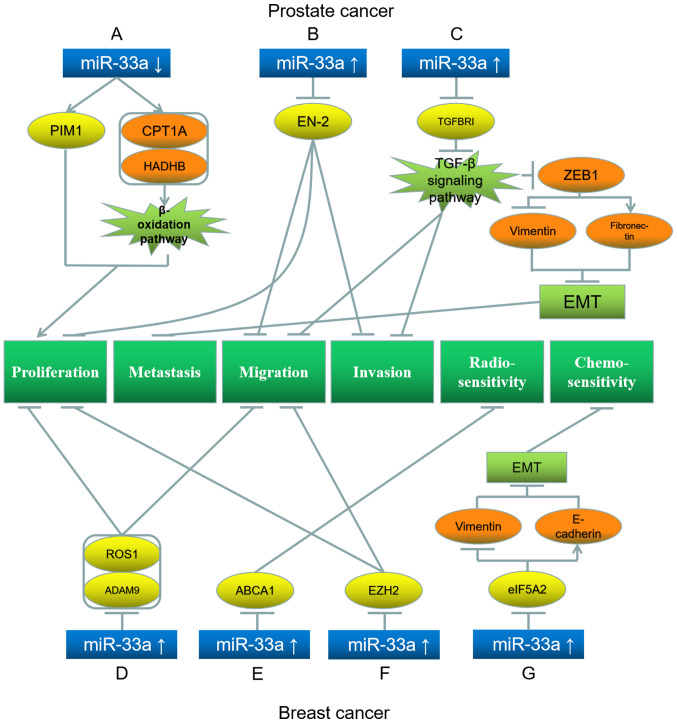
Abnormal downregulation of miR-33a is observed in PC. Karatas et al (106) have revealed that miR-33a downregulation increases the expression levels of PIM1 and other lipid β-oxidation pathway proteins, such as carnitine palmitoyltransferase 1A and hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit β, thereby enhancing the proliferation of PC cells (Fig. 7A). Li et al (107) have demonstrated that miR-33a is a tumor suppressor in PC, and high levels of miR-33a downregulate its target gene engrailed homeobox 2, thereby suppressing the proliferation, migration and invasion of PC cells (Fig. 7B). Dai et al (108) have demonstrated that miR-33a-5p is downregulated in PC cells. miR-33a-5p overexpression in PC cells downregulates its target gene transforming growth factor β receptor 1 (TGFBRI), leading to the repression of the TGF-β signaling pathway and suppressing PC invasion and migration (108). Additionally, ZEB1, a downstream protein of the TGF-β signaling pathway, is consequently downregulated, suppressing bone metastasis (108). Furthermore, ZEB1 downregulation inhibits EMT by downregulating vimentin and upregulating fibronectin in PC cells (Fig. 7C) (108).
Figure 7.
Roles of miR-33a and its target genes in prostate (top) and breast cancer (bottom), and the underlying mechanisms. (A) miR-33a downregulation upregulates its target gene PIM1, as well as CPT1A and HADHB, which promote β-oxidation pathway and proliferation. (B) miR-33a upregulation downregulates EN-2, inhibiting proliferation, migration and invasion. (C) miR-33a upregulation inhibits its target gene TGFBRI, which further downregulates ZEB1, downregulating vimentin and upregulating fibronectin, leading to inhibition of EMT, metastasis, invasion and migration. (D) miR-33a upregulation downregulates its target genes ROS1 and ADAM9, inhibiting proliferation and migration. (E) miR-33a upregulation downregulates its target gene ABCA1, inhibiting radiosensitivity. (F) miR-33a upregulation downregulates its target gene EZH2, inhibiting proliferation and migration. (G) miR-33a upregulation inhibits its target gene eIF5A2, downregulating vimentin and upregulating E-cadherin, leading to inhibition of EMT and chemoresistance. Blue represents miR-33a; yellow represents the target genes of miR-33a; orange represents the downstream proteins; light green represents the signaling pathways and EMT processes; dark green represents the biological processes of cancer cells. miR, microRNA; EMT, epithelial-mesenchymal transition; PIM1, Pim-1 proto-oncogene; EN-2, engrailed homeobox 2; TFGBRI, transforming growth factor β receptor 1; ZEB1, Zinc finger E-Box binding homeobox 1; ADAM9, ADAM metallopeptidase domain 9; ABCA1, ATP-binding cassette transporter A1; EZH2, enhancer of zeste homolog 2; eIF5A2, eukaryotic translation initiation factor 5A2; CPT1A, carnitine palmitoyltransferase 1A; HADHB, hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit β; ROS1, ROS proto-oncogene 1, receptor tyrosine kinase.
Breast cancer
Breast carcinoma is the leading cause of female-cancer-associated deaths worldwide (109). It is mainly associated with age, long-term excessive alcohol consumption, genetic factors and long-term use of exogenous estrogen (110,111).
miR-33a acts as a tumor suppressor in breast carcinoma. Zhang et al (112) have demonstrated that miR-33a is downregulated in breast cancer cells. Increased miR-33a levels downregulate its target genes ROS1 and ADAM metallopeptidase domain 9, and thus inhibits tumor cell proliferation and migration (Fig. 7D) (112). Additionally, Wolfe et al (113) have demonstrated that miR-33a is downregulated in inflammatory breast tumor cells. Notably, miR-33a can decrease high-density-lipoprotein (HDL)-induced radio-sensitivity by downregulating ABCA1 in breast cancer cells (Fig. 7E) (113). Zeng et al (114) have demonstrated that miR-33a expression is downregulated in triple-negative breast cancer (TNBC) cells, whereas the miR-33a target enhancer of zeste homolog 2 is upregulated. The high levels of miR-33a in TNBC cells can inhibit the proliferation and migration of these cells (Fig. 7F) (114). Guan et al (115) have illustrated that compared with non-TNBC cells, miR-33a-5p expression is significantly lower in TNBC cells, and miR-33a-5p overexpression downregulates its target gene eukaryotic translation initiation factor 5A2 (eIF5A2) and increases the sensitivity of TNBC cells to doxorubicin. Additionally, eIF5A2 downregulation decreases vimentin expression, while increasing that of E-cadherin, thereby inhibiting the EMT of TNBC cells (Fig. 7G) (115).
3. Discussion
The involvement of miRNAs in cancer is an exciting research field, but the association and mechanism of interaction between miRNAs and cancer remain at the preliminary stages of research. Studies have indicated that miR-33a affects cell proliferation, migration, metastasis, invasion and other biological processes by acting on 38 different target genes, thereby affecting the cell cycle, apoptosis, self-renewal and resistance to traditional chemotherapy and radiation therapy in cancer cells. Therefore, the prognosis and survival rate of patients with cancer may be predicted based on the levels of miR-33a.
Diverse roles of miR-33a in cancer
To date, studies have revealed that miR-33a is a tumor suppressor gene and is downregulated in numerous types of cancer, such as liver carcinoma, lung cancer, colorectal carcinoma, PC and melanoma, whereas it is upregulated in glioma cells and acts as an oncogene. In addition, it is important to note that miR-33a serves different roles in osteosarcoma. Three studies (99–101) have illustrated that miR-33a is downregulated in osteosarcoma, and that its overexpression suppresses the proliferation, invasion and migration of osteosarcoma cells. By contrast, Zhou et al (102) reported that miR-33a is upregulated in osteosarcoma and inhibits the apoptosis of osteosarcoma cells with cisplatin-resistance. However, these opposing observations in osteosarcoma cells are not specific to miR-33a, and there are studies presenting contrasting expression patterns in the same types of cancer for other miRNAs. For example, Jang et al (116) have revealed that miR-708 is an oncogene in NSCLC, and its increased expression in NSCLC is associated with a low survival rate in non-smoker patients with lung cancer. By contrast, Wu et al (117) have demonstrated that miR-708-5p is a tumor suppressor gene in NSCLC, and that miR-708-5p overexpression inhibits the invasion and metastasis of NSCLC cells. Abnormal miRNA expression in cancer may occur via various mechanisms, but the precise mechanistic details of aberrant miR-33a expression in tumors have not been clearly determined, and further research is required. Furthermore, it remains unclear why there are opposing observations regarding the expression patterns of miR-33a in osteosarcoma cells.
Target genes of miR-33a in malignancies
Among the 38 target genes mentioned in the literature (Table I), 8 have been verified in different types of cancer: TWIST1 in hepatocellular carcinoma, gallbladder cancer, lung cancer and osteosarcoma; β-catenin in hepatocellular carcinoma, pancreatic cancer and lung cancer; HMGA2 in hepatocellular carcinoma and lung cancer; ABCA1 in hepatocellular carcinoma, NSCLC and breast carcinoma; CDK6 in gastric and lung cancer; PIM1 in gastric, colorectal, laryngeal and prostate cancer; PIM3 in pancreatic and colorectal cancer; and ZEB1 in gastric cancer, esophageal squamous carcinoma and tongue squamous cell carcinoma. These target genes may be reliable therapeutic targets as the target genes of miR-33a. In addition, future studies should continue to investigate this miRNA and its eight target genes in other types of cancer that have not been analyzed yet, to explore whether these genes have similar interactions and mechanisms affecting these other types of cancer. The remaining 30 target genes are currently only validated in one type of cancer each, and thus, these target genes have great research potential. The expression levels, mechanism and reliability of miR-33a and its association with these targets in other types of cancer should be further explored. The reliability of their interactions and underlying mechanistic details in cancer should also be assessed.
Table I.
miR-33a expression and roles in different types of cancer.
| A, Digestive system: Hepatocellular carcinoma | |||||
|---|---|---|---|---|---|
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Liu et al, 2018 | Down | PNMA1 | Up | EMT, proliferation, migration | (29) |
| Guo et al, 2016; Li et al, 2019 | Down | TWIST1 | Up | EMT, invasion, proliferation | (30,31) |
| Fang et al, 2013 | Down | β-catenin | Up | Proliferation | (32) |
| Han et al, 2016 | Down | PBX3 | Up | Proliferation, metastasis, invasion, low survival | (33) |
| Wu et al, 2019 | Down | HMGA2 | Up | Proliferation, migration, invasion, drug resistance | (34,35) |
| Han et al, 2019 | |||||
| Meng et al, 2017 | Down | HSPA8 | Up | Cisplatin resistance | (36) |
| Hou et al, 2017 | Down | ABCA1 | Up | Doxorubicin resistance | (37) |
| Chang et al, 2017 | Up | PPARα | Down | Proliferation, apoptosis | (38) |
| B, Digestive system: Gastric cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Wang et al, 2015 | Down | CDK6 | Up | Proliferation, cell cycle | (41) |
| CCND1 | |||||
| PIM1 | |||||
| Chen et al, 2019 | Down | SNAI2 | Up | EMT, invasion, migration | (42) |
| Wu et al, 2018 | Down | ZEB1 | Up | EMT, migration | (43) |
| C, Digestive system: Colorectal cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Wang et al, 2019 | Down | PIM3 | Up | Proliferation, cell cycle | (50) |
| Shan et al, 2017 | Down | ST8SIA1 | Up | Proliferation, invasion, drug resistance | (51) |
| Yan et al, 2019 | Down | MTHFD2 | Up | Proliferation, migration, cell cycle | (52) |
| Ibrahim et al, 2011 | Down | PIM1 | Up | Tumor growth | (53) |
| D, Digestive system: Pancreatic cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Liang et al, 2015 | Down | PIM3 | Up | Proliferation, gemcitabine sensitivity | (56) |
| Liang et al, 2015 | Down | β-catenin | Up | EMT, proliferation, gemcitabine sensitivity | (57) |
| E, Digestive system: Gallbladder cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Zhang et al, 2016 | Down | TWIST1 | Up | EMT, proliferation, migration, invasion | (59) |
| F, Digestive system: Esophageal squamous carcinoma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Zhang et al, 2019 | Down | ZEB1 | Up | Proliferation, migration | (62) |
| Wang et al, 2017 | Down | SOX9 | Up | Proliferation | (63) |
| G, Respiratory system: Non-small cell lung cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Du et al, 2017 | Down | METTL3 | Up | Proliferation | (65) |
| Kang et al, 2018 | Down | CAND1 | Up | Proliferation, migration Cell cycle | (66) |
| Amaar and Reeves et al, 2019 | Down | ABCA1 | Up | EMT, proliferation, migration, chemoresistance | (67) |
| Yang et al, 2015 | Down | TWIST1 | Up | Migration, EMT | (68) |
| Kang et al, 2017 | Down | CDK6 | Up | Cell cycle, tumor development | (69) |
| Wang et al, 2020 | Down | KPNA4 | Up | Tumorigenesis, tumor development | (70) |
| H, Respiratory system: Untyped lung cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Li et al, 2018 | Down | mTOR | Up | Proliferation, celastrol resistance | (71) |
| Kuo et al, 2013 | Down | CDK6 | Up | Bone metastasis | (72) |
| Pan et al, 2020 | Down | TWIST1 | Up | Proliferation, metastasis | (73) |
| Rice et al, 2013 | Down | HMGA2 | Up | Migration | (74) |
| Zhu et al, 2015 | Down | β-catenin | Up | Proliferation, cell cycle | (75) |
| Boldrini et al, 2017 | Down | PD-1 | Up | Prognosis | (76) |
| I, Head and neck: Laryngeal carcinoma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Karatas et al, 2018 | Down | PIM1 | Up | Proliferation, apoptosis | (79) |
| J, Head and neck: Tongue squamous carcinoma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Zuo et al, 2018 | Down | ZEB1 | Up | Proliferation, migration, cell cycle | (81) |
| K, Skin: Melanoma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Zhou et al, 2015 Cao et al, 2017 | Down | HIF1A | Up | Proliferation, migration, invasion, apoptosis, radiosensitivity | (83,85) |
| Tian et al, 2016 | Down | CDK16 | Up | Proliferation | (86) |
| Lv et al, 2018 | Down | N/A | N/A | Proliferation, migration, invasion | (87,88) |
| Fu et al, 2019 | |||||
| L, Urinary system: Renal cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Jiang et al, 2019 | Down | MDM4 | Up | Proliferation, survival | (91) |
| M, Endocrine system: Thyroid cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Takeshita et al, 2013 | Down | MYC | Up | Proliferation | (93) |
| N, Nervous system: Glioma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Chang et al, 2017 | Up | SIRT6 | Down | Apoptosis | (95) |
| Wang et al, 2014 | Up | PDE8A, UVRAG | Down | Self-renewal | (96) |
| O, Musculoskeletal system: Osteosarcoma | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Jiang et al, 2017 | Down | AXL | Up | Proliferation | (99) |
| Huang et al, 2018 | Down | CYR61 | Up | EMT, invasion | (100) |
| Zhang et al, 2015 | Down | N/A | N/A | Proliferation | (101) |
| Zhou et al, 2014 | Up | TWIST1 | Down | Cisplatin resistance, apoptosis | (102) |
| P, Reproductive system: Prostate cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Karatas et al, 2017 | Down | PIM1 | Up | Proliferation, invasion, migration | (106) |
| Li et al, 2017 | Down | EN-2 | Up | Proliferation, migration, invasion | (107) |
| Dai et al, 2019 | Down | TGFBRI | Up | EMT, invasion, migration, metastasis | (108) |
| Q, Reproductive system: Breast cancer | |||||
| First author, year | miR-33a expression | Target gene | Target gene expression | Function | (Refs.) |
| Zhang et al, 2015 | Down | ADAM9 | Up | Proliferation, migration | (112) |
| ROS1 | |||||
| Wolfe et al, 2016 | Down | ABCA1 | Up | Radiosensitivity | (113) |
| Weihua et al, 2020 | Down | EZH2 | Up | Proliferation, migration | (114) |
| Guan et al, 2019 | Down | eFI5A2 | Up | EMT, doxorubicin sensitivity | (115) |
miR, microRNA; EMT, epithelial-mesenchymal transition; ABCA1, ATP-binding cassette transporter A1; AXL, AXL receptor tyrosine kinase; CAND1, Cullin associated and neddylation dissociated 1; CCND1, cyclin D1; CDK, cyclin-dependent kinase; CYR61, cysteine-rich angiogenic inducer 61; EN-1, engrailed homeobox 2; EZH2, enhancer of zeste homolog 2; HIF1A, hypoxia inducible factor 1 subunit α; HMGA2, high mobility group AT-hook 2; HSPA8, heat shock protein family A (Hsp70) member 8; MDM4, mouse double minute 4; METTL3, methyltransferase-like 3; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; PBX3, pre-B-cell leukemia transcription factor 3; PD-1, programmed cell death protein 1; PDE8A, phosphodiesterase 8A; PIM1/3, Pim-1/3 proto-oncogene; PNMA1, paraneoplastic antigen Ma1; PPARα, peroxisome proliferator activated receptor α; SIRT6, sirtuin 6; SNAI2, Snail family transcriptional repressor 2; SOX9, SRY-box 9; ST8SIA1, ST8α-N-acetyl-neuraminide-α-2,8-sialyltransferase 1; TFGBRI, transforming growth factor β receptor 1; TWIST1, Twist basic helix-loop-helix transcription factor 1; UVRAG, UV radiation resistance associated; ZEB1, zinc finger E-Box binding homeobox 1.
Signaling pathways affected by miR-33a in carcinogenesis
The mechanisms underlying miR-33a-mediated carcinogenesis are extremely complex and involve multiple signaling pathways. There are nine signaling pathways mentioned in the literature, namely the Wnt/β-catenin, Snail/Slug, AKT/GSK-3β/β-catenin, mTOR, TGF-β, JAK2/STAT3, PKA, NOTCH and PI3K/AKT signaling pathways.
Liu et al (29) have revealed that PNMA1, a target gene of miR-33a, is upregulated in hepatocellular carcinoma. PNMA1 upregulation enables β-catenin to translocate to the nucleus of hepatoma cells, stimulating Wnt/β-catenin signal transduction, and consequently promoting the proliferation and EMT of hepatoma cells. Fang et al (32) have illustrated that miR-33a also directly targets β-catenin and thus inhibits the proliferation of hepatoma cells. In general, GSK-3β regulates the phosphorylation and degradation of β-catenin (118), but a study by Fang et al (32) has indicated that GSK-3β is not associated with the miR-33a-mediated downregulation of β-catenin in hepatoma cells. Liang et al (56) have revealed that PIM3, a target of miR-33a, may regulate the AKT and Wnt/β-catenin signaling pathways. PIM3 upregulation in pancreatic cancer increases the phosphorylation of GSK-3β and AKT, and the expression levels of β-catenin (56). Therefore, miR-33a and PIM3 may act on the AKT/Wnt/β-catenin signaling pathway to inhibit the proliferation and gemcitabine-resistance of pancreatic tumors (56). In lung cancer, Pan et al (73) have demonstrated that miR-33a downregulation upregulates its target gene TWIST1 and activates the Wnt/β-catenin signaling pathway, subsequently promoting lung cancer cell proliferation and metastasis. TWIST1 upregulation, GSK-3β downregulation and activation of the Wnt/β-catenin signaling pathway have also been observed in NSCLC by Zheng et al (119). According to the aforementioned studies, the association between miR-33a and the Wnt/β-catenin signaling pathway in hepatocellular carcinoma, pancreatic cancer and lung cancer remains unclear and requires further clarification in the future.
Chen et al (42) have revealed that miR-33a inhibits the EMT, invasion and migration of gastric tumor cells by targeting SNAI2 to deactivate the Snail/Slug signaling pathway, which also serves a similar role in tongue squamous cell carcinoma (120) and thyroid cancer (121). Cao et al (122) have reported that, in invasive ductal carcinoma, SNAI2 promotes the EMT process by inhibiting E-cadherin expression. Slug is encoded by the SNAI2 gene, and the two constitute the Snail/Slug signaling pathway. Nevertheless, the specific mechanism of miR-33a in this signaling pathway has not been thoroughly clarified yet.
Liang et al (56) have demonstrated that miR-33a upregulation in pancreatic cancer downregulates PIM3 and inhibits the AKT/GSK-3β/β-catenin signaling pathway, thus suppressing the proliferation and gemcitabine-resistance in pancreatic cells. A study by Carreras-Torres et al (123) has demonstrated that type 2 diabetes is one of the risk factors of pancreatic cancer. Furthermore, the inhibition of the AKT/GSK-3β/β-catenin signaling pathway can prevent the development of type 2 diabetes (124). In addition, Wijesekara et al (125) have reported that miR-33a overexpression in pancreatic tissues decreases insulin secretion, and increases blood cholesterol levels. Although miR-33a is known to suppress the AKT/GSK-3β/β-catenin signaling pathway in pancreatic cancer cells, the mechanism requires further detailed study.
mTOR can activate p70S6K and phosphorylation of 4EBP1, which are strongly associated with cancer cell proliferation (126). Li et al (71) have illustrated that miR-33a targets mTOR and downregulates p-p70S6K and p-4EBP1 in lung cancer, indicating that miR-33a suppresses the proliferation of lung cancer cells via the mTOR signaling pathway. In addition, Xia and Xu (127) have illustrated that the PI3K/AKT/mTOR signaling pathway can act altogether, and inhibiting this axis can impede the self-renewal of tumor cells, as well as the clone formation and differentiation of tumor stem cells. mTOR is a downstream protein of AKT, and phosphorylation of AKT increases mTOR expression (128,129). Consequently, the expression levels of the downstream protein p70S6K and phosphorylation of 4EBP1 are upregulated, promoting the proliferation of cancer cells (126). Fu et al (88) have demonstrated that lncRNA PEG10 inhibits miR-33a and promotes melanoma cell proliferation, migration and invasion by activating the PI3K/AKT and mTOR signaling pathways. In osteosarcoma, Jiang et al (99) have demonstrated that lncRNA DANCR upregulates the target protein AXL in osteosarcoma cells by downregulating miR-33a-5p, thereby increasing the expression levels of the downstream proteins PI3K and AKT. Therefore, this novel regulatory network of DANCR-miR-33a-AXL-PI3K/AKT/mTOR is expected to provide a novel therapeutic target for osteosarcoma (99). All aforementioned studies demonstrate that miR-33a is closely associated with the proteins in the PI3K/AKT/mTOR signaling pathway, which serves a vital role in tumor development.
Dai et al (108) have demonstrated that increasing miR-33a expression in PC cells downregulates the target gene TGFBRI, leading to repression of the TGF-β signaling pathway and suppression of bone metastasis. Additionally, Fournier et al (130) have observed that blocking the TGF-β signaling pathway suppresses bone metastasis in PC. However, the inhibitory effect of miR-33a on bone metastasis in PC via suppression of the TGF-β signaling pathway remains unclear, and further research is required.
Chang et al (95) and Feng et al (131) have demonstrated that SIRT6, a target protein of miR-33a, inhibits JAK2/STAT3 signaling pathway activation in glioma cells by decreasing the phosphorylation of JAK2 and STAT3. This signaling pathway is highly activated in numerous malignant cells, including glioma cells, and its repression can inhibit tumor growth, invasion and EMT (132,133). Tang et al (134) have indicated that the glioma tumor suppressor miR-876-3p downregulates its target gene kinesin family member 20A, whereby the JAK2/STAT3 signaling pathway is repressed; consequently, the proliferation, invasion and EMT of glioma cells are suppressed. High SIRT6 expression can block the growth of these cancer cells and facilitate their apoptosis. In addition, in glioma cancer cells, miR-33a targets PDE8A and UVRAG, whose overexpression activates the cAMP/PKA and NOTCH signaling pathways, respectively (96). Notably, the two signaling pathways can promote the self-renewal of glioma initiation cells only when simultaneously activated (91). In glioma, Tanabe et al (135) have revealed that the adenylyl cyclase/cAMP/PKA signaling pathway increases IL-1β-induced IL-6 synthesis by enhancing the JAK2/STAT3 signaling pathway. Liu et al (136) have illustrated that the oncogene TRPM7 promotes the proliferation, migration and invasion of glioma cells by activating the JAK2/STAT3 and Notch signaling pathways. Overall, the JAK2/STAT3, cAMP/PKA and NOTCH signaling pathways are critical in glioma. The mechanisms underlying the regulation of these signaling pathways by miR-33a require further study.
miR-33a in tumor prognosis and treatment
Numerous studies have reported the crucial role of miR-33a in carcinogenesis. Accordingly, miR-33a may be used for the prognosis or treatment of tumors. Xie et al (137) have illustrated that low miR-33a expression is associated with tumorigenesis and poor prognosis in hepatoma. Hou et al (138) have demonstrated that adjuvant chemotherapy improves the overall and disease-free survival rates of patients with NSCLC with high miR-33a expression. Therefore, miR-33a may be used as a biomarker for NSCLC prognosis. In addition to the experimental evidence, bioinformatics studies have predicted that miR-33a expression patterns may be used as a prognostic marker in cancer. For example, Pan et al (139) have demonstrated that a two-miRNA signature consisting of miR-33a-5p and miR-128 may serve as a novel biomarker for early NSCLC detection. Kandimalla et al (140) have identified an eight-miRNA signature, which includes miR-33a, that achieved excellent prediction for colorectal tumor recurrence in stages two and three.
As aforementioned, miRNAs may be used as novel therapeutic targets for cancer (9). The involvement of miR-33a in carcinogenesis enables cancer treatment by modulating the expression levels of miR-33a. Previous studies have indicated that high miR-33a levels affect the chemosensitivity of hepatocellular carcinoma (26,36,37), glioma (141) and osteosarcoma (102), as well as colorectal (51), pancreatic (56,57), lung (67,71) and breast cancer (115). The radiosensitivity of cancer cells can also be affected by miR-33a expression; high levels of miR-33a decrease HDL-induced radiosensitivity of breast cancer (113), whereas in melanoma, miR-33a increases the radiosensitivity by repressing the glycolysis process (85).
Overall, miR-33a is a potential cancer-associated miRNA that deserves further investigation. At present, only a small part of the literature mentions the signaling pathways affected by miR-33a and its target genes in tumors. Most of the miR-33a-modulated signaling pathways associated with cancer remain unclear. Once the action mechanisms of miR-33a in cancer are clarified, it may be possible to determine high-risk cancer cases and predict prognosis according to the levels of miR-33a and its targets in the cancer cells of the patients, and effectively design targeted therapies. Finally, to the best of our knowledge, the present paper is the first review of the involvement of miR-33a and its target genes in diverse types of cancer, thereby providing comprehensive information for future research on the involvement of miR-33a in cancer.
4. Conclusions
Abnormal expression levels of miRNAs, such as miR-33a, are associated with various diseases, including cancer. In the present review, the roles of miR-33a and its 38 target genes were summarized in more than a dozen types of cancer. Among the 38 target genes of this miRNA, 8 have been confirmed in various types of cancer, while the rest have been validated in only one type of cancer each.
miR-33a and its target genes affect different biological behaviors of tumor cells, such as proliferation, metastasis, migration, invasion, cell cycle, apoptosis, self-renewal, chemoresistance and radioresistance, as well as patient outcomes, such as prognosis and survival. miR-33a serves diverse regulatory roles in cancer. It is a tumor suppressor in most types of cancer, whereas it acts as an oncogene in glioma, and it possibly serves controversial roles in osteosarcoma. These different roles of miR-33a imply that it may affect different types of cancer via different molecular mechanisms.
miR-33a participates in nine signaling pathways that are associated with cancer. These pathways are the Wnt/β-catenin, Snail/Slug, AKT/GSK-3β/β-catenin, mTOR, TGF-β, JAK2/STAT3, PKA, NOTCH and PI3K/AKT pathways, although the mechanistic details of their involvement in cancer remain elusive. Therefore, further studies are required to explore the precise mechanistic roles of miR-33a in these types of cancer, so that novel targeted treatments can be designed and developed to improve patient outcomes.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- AXL
AXL receptor tyrosine kinase
- CDK
cyclin-dependent kinase
- eIF5A2
eukaryotic translation initiation factor 5A2
- HIF1A
hypoxia inducible factor 1 subunit α
- HK1/2
hexokinase 1/2
- HMGA2
high mobility group AT-hook 2
- IL
interleukin
- JAK2
Janus kinase 2
- miRNA/miR
microRNA
- PDE8A
phosphodiesterase 8A
- PIM1/3
Pim-1/3 proto oncogene
- PNMA1
paraneoplastic antigen Ma1
- RAFFS1C
Ras association domain family member 1
- SIRT6
sirtuin 6
- SNAI1/2
Snail family transcriptional repressor 1/2
- SOX9
SRY-box 9
- ST8SIA1
ST8α-N-acetyl-neuraminide-α-2,8-sialyltransferase 1
- S6K
ribosomal protein S6K
- TGFBRI
transforming growth factor β receptor 1
- TWIST1
Twist basic helix-loop-helix transcription factor 1
- UVRAG
UV radiation resistance associated
- ZEB1
zinc finger E-Box binding homeobox 1
- 4EBP1
eukaryotic translation initiation factor 4E binding protein 1
Funding
The present review was supported by the National Natural Science Foundation of China (grant no. 31770774) and the Provincial Major Project of Basic or Applied Research in Natural Science, Guangdong Provincial Education Department (grant no. 2016KZDXM038).
Availability of data and materials
Not applicable.
Authors' contributions
ZH and CG conceptualized the present review. CG, JW and TT contributed to the formal analysis and investigation. CG wrote the original draft. ZH revised and edited the original draft, and provided funding acquisition. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: Regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kappel A, Keller A. miRNA assays in the clinical laboratory: Workflow, detection technologies and automation aspects. Clin Chem Lab Med. 2017;55:636–647. doi: 10.1515/cclm-2016-0467. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Lancet, corp-author. GLOBOCAN 2018: Counting the toll of cancer. Lancet. 2018;392:985. doi: 10.1016/S0140-6736(18)32252-9. [DOI] [PubMed] [Google Scholar]
- 8.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Z, Li Y, Huang K, Wagar N, Shim H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res. 2011;28:3091–3100. doi: 10.1007/s11095-011-0570-y. [DOI] [PubMed] [Google Scholar]
- 16.Shi GH, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, Shen YJ, Zhu Y, Zhu YP, Xiao WJ, Ma CG. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sin. 2010;31:867–873. doi: 10.1038/aps.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol. 2012;180:2490–2503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 22.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: Cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Boren J, Brindle KM. Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ. 2012;19:1561–1570. doi: 10.1038/cdd.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gharipour M, Sadeghi M. Pivotal role of microRNA-33 in metabolic syndrome: A systematic review. ARYA Atheroscler. 2013;9:372–376. [PMC free article] [PubMed] [Google Scholar]
- 25.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: A systematic review. Curr Genomics. 2011;12:130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karkampouna S, van der Helm D, Gray PC, Chen L, Klima I, Grosjean J, Burgmans MC, Farina-Sarasqueta A, Snaar-Jagalska EB, Stroka DM, et al. CRIPTO promotes an aggressive tumour phenotype and resistance to treatment in hepatocellular carcinoma. J Pathol. 2018;245:297–310. doi: 10.1002/path.5083. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Liu H, Liu ZG, Wang HS, Zhang F, Wang H, Zhang J, Chen JJ, Huang HJ, Tan Y, et al. Histone deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation and stabilization of Snail to promote metastasis of hepatoma cells. Cancer Lett. 2018;420:1–13. doi: 10.1016/j.canlet.2018.01.068. [DOI] [PubMed] [Google Scholar]
- 28.Gingold JA, Zhu D, Lee DF, Kaseb A, Chen J. Genomic profiling and metabolic homeostasis in primary liver cancers. Trends Mol Med. 2018;24:395–411. doi: 10.1016/j.molmed.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Chen B, Gu Y, Liu Q. PNMA1, regulated by miR-33a-5p, promotes proliferation and EMT in hepatocellular carcinoma by activating the Wnt/β-catenin pathway. Biomed Pharmacother. 2018;108:492–499. doi: 10.1016/j.biopha.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 30.Guo XF, Wang AY, Liu J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:3011–3016. [PubMed] [Google Scholar]
- 31.Li Y, Chen G, Yan Y, Fan Q. CASC15 promotes epithelial to mesenchymal transition and facilitates malignancy of hepatocellular carcinoma cells by increasing TWIST1 gene expression via miR-33a-5p sponging. Eur J Pharmacol. 2019;860:172589. doi: 10.1016/j.ejphar.2019.172589. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Feng Y, Wu T, Srinivas S, Yang W, Fan J, Yang C, Wang S. Aflatoxin B1 negatively regulates Wnt/β-catenin signaling pathway through activating miR-33a. PLoS One. 2013;8:e73004. doi: 10.1371/journal.pone.0073004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han SY, Han HB, Tian XY, Sun H, Xue D, Zhao C, Jiang ST, He XR, Zheng WX, Wang J, et al. MicroRNA-33a-3p suppresses cell migration and invasion by directly targeting PBX3 in human hepatocellular carcinoma. Oncotarget. 2016;7:42461–42473. doi: 10.18632/oncotarget.9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, Ai H, Zhang K, Yun H, Xie F. Long non-coding RNA EGOT promotes the malignant phenotypes of hepatocellular carcinoma cells and increases the expression of HMGA2 via down-regulating miR-33a-5p. Onco Targets Ther. 2019;12:11623–11635. doi: 10.2147/OTT.S218308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Han S, Han B, Li Z, Sun D. Downregulation of long noncoding RNA CRNDE suppresses drug resistance of liver cancer cells by increasing microRNA-33a expression and decreasing HMGA2 expression. Cell Cycle. 2019;18:2524–2537. doi: 10.1080/15384101.2019.1652035. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Meng W, Tai Y, Zhao H, Fu B, Zhang T, Liu W, Li H, Yang Y, Zhang Q, Feng Y, Chen G. Downregulation of miR-33a-5p in hepatocellular carcinoma: A possible mechanism for chemotherapy resistance. Med Sci Monit. 2017;23:1295–1304. doi: 10.12659/MSM.902692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou H, Kang Y, Li Y, Zeng Y, Ding G, Shang J. miR-33a expression sensitizes Lgr5+ HCC-CSCs to doxorubicin via ABCA1. Neoplasma. 2017;64:81–91. doi: 10.4149/neo_2017_110. [DOI] [PubMed] [Google Scholar]
- 38.Chang W, Zhang L, Xian Y, Yu Z. MicroRNA-33a promotes cell proliferation and inhibits apoptosis by targeting PPARα in human hepatocellular carcinoma. Exp Ther Med. 2017;13:2507–2514. doi: 10.3892/etm.2017.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol. 2014;20:13842–13862. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Zhou X, Shan B, Han J, Wang F, Fan X, Lv Y, Chang L, Liu W. Downregulation of microRNA-33a promotes cyclin- dependent kinase 6, cyclin D1 and PIM1 expression and gastric cancer cell proliferation. Mol Med Rep. 2015;12:6491–6500. doi: 10.3892/mmr.2015.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen DD, Cheng JT, Chandoo A, Sun XW, Zhang L, Lu MD, Sun WJ, Huang YP. microRNA-33a prevents epithelial-mesenchymal transition, invasion, and metastasis of gastric cancer cells through the Snail/Slug pathway. Am J Physiol Gastrointest Liver Physiol. 2019;317:G147–G160. doi: 10.1152/ajpgi.00284.2018. [DOI] [PubMed] [Google Scholar]
- 43.Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng W, Shi M, Wang Y. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12:799–813. doi: 10.1002/1878-0261.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 45.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 46.Herbey II, Ivankova NV, Katkoori VR, Mamaeva OA. Colorectal cancer and hypercholesterolemia: Review of current research. Exp Oncol. 2005;27:166–178. [PubMed] [Google Scholar]
- 47.Gray RT, Loughrey MB, Bankhead P, Cardwell CR, McQuaid S, O'Neill RF, Arthur K, Bingham V, McGready C, Gavin AT, et al. Statin use, candidate mevalonate pathway biomarkers, and colon cancer survival in a population-based cohort study. Br J Cancer. 2017;116:1652–1659. doi: 10.1038/bjc.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murai T. Cholesterol lowering: Role in cancer prevention and treatment. Biol Chem. 2015;396:1–11. doi: 10.1515/hsz-2014-0194. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs RJ, Voorneveld PW, Kodach LL, Hardwick JC. Cholesterol metabolism and colorectal cancers. Curr Opin Pharmacol. 2012;12:690–695. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Liu C, Hu L. Cholesterol regulates cell proliferation and apoptosis of colorectal cancer by modulating miR-33a-PIM3 pathway. Biochem Biophys Res Commun. 2019;511:685–692. doi: 10.1016/j.bbrc.2019.02.123. [DOI] [PubMed] [Google Scholar]
- 51.Shan Y, Liu Y, Zhao L, Liu B, Li Y, Jia L. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int J Biochem Cell Biol. 2017;90:48–58. doi: 10.1016/j.biocel.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 52.Yan Y, Zhang D, Lei T, Zhao C, Han J, Cui J, Wang Y. MicroRNA-33a-5p suppresses colorectal cancer cell growth by inhibiting MTHFD2. Clin Exp Pharmacol Physiol. 2019;46:928–936. doi: 10.1111/1440-1681.13125. [DOI] [PubMed] [Google Scholar]
- 53.Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 54.Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: A report of treatment and survival trends for 100,313 patients diagnosed from 1985–1995, using the national cancer database. J Am Coll Surg. 1999;189:1–7. doi: 10.1016/S1072-7515(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 55.Moffat GT, Epstein AS, O'Reilly EM. Pancreatic cancer-A disease in need: Optimizing and integrating supportive care. Cancer. 2019;125:3927–3935. doi: 10.1002/cncr.32423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang C, Yu XJ, Guo XZ, Sun MH, Wang Z, Song Y, Ni QX, Li HY, Mukaida N, Li YY. MicroRNA-33a-mediated downregulation of Pim-3 kinase expression renders human pancreatic cancer cells sensitivity to gemcitabine. Oncotarget. 2015;6:14440–14455. doi: 10.18632/oncotarget.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang C, Wang Z, Li YY, Yu BH, Zhang F, Li HY. miR-33a suppresses the nuclear translocation of β-catenin to enhance gemcitabine sensitivity in human pancreatic cancer cells. Tumour Biol. 2015;36:9395–9403. doi: 10.1007/s13277-015-3679-5. [DOI] [PubMed] [Google Scholar]
- 58.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23:3978–3998. doi: 10.3748/wjg.v23.i22.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang M, Gong W, Zuo B, Chu B, Tang Z, Zhang Y, Yang Y, Zhou D, Weng M, Qin Y, et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget. 2016;7:78640–78652. doi: 10.18632/oncotarget.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 61.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C, Wang L, Yang J, Fu Y, Li H, Xie L, Cui Y. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur J Pharmacol. 2019;861:172590. doi: 10.1016/j.ejphar.2019.172590. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Yu X, Zhang Z, Pang L, Xu J, Jiang J, Liang W, Chai Y, Hou J, Li F. Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J Exp Clin Cancer Res. 2017;36:182. doi: 10.1186/s13046-017-0658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: #pidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, Wang D, Huang J, Gao S, Gao Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–589. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 66.Kang M, Li Y, Zhao Y, He S, Shi J. miR-33a inhibits cell proliferation and invasion by targeting CAND1 in lung cancer. Clin Transl Oncol. 2018;20:457–466. doi: 10.1007/s12094-017-1730-2. [DOI] [PubMed] [Google Scholar]
- 67.Amaar YG, Reeves ME. RASSF1C regulates miR-33a and EMT marker gene expression in lung cancer cells. Oncotarget. 2019;10:123–132. doi: 10.18632/oncotarget.26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang L, Yang J, Li J, Shen X, Le Y, Zhou C, Wang S, Zhang S, Xu D, Gong Z. MircoRNA-33a inhibits epithelial-to- mesenchymal transition and metastasis and could be a prognostic marker in non-small cell lung cancer. Sci Rep. 2015;5:13677. doi: 10.1038/srep13677. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Kang J, Kim W, Lee S, Kwon D, Chun J, Son B, Kim E, Lee JM, Youn H, Youn B. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183- and miR-33a-mediated cell cycle regulation. Oncogene. 2017;36:1585–1596. doi: 10.1038/onc.2016.328. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. doi: 10.1016/j.biopha.2020.110019. [DOI] [PubMed] [Google Scholar]
- 71.Li YJ, Sun YX, Hao RM, Wu P, Zhang LJ, Ma X, Ma Y, Wang PY, Xie N, Xie SY, Chen W. miR-33a-5p enhances the sensitivity of lung adenocarcinoma cells to celastrol by regulating mTOR signaling. Int J Oncol. 2018;52:1328–1338. doi: 10.3892/ijo.2018.4276. [DOI] [PubMed] [Google Scholar]
- 72.Kuo PL, Liao SH, Hung JY, Huang MS, Hsu YL. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochim Biophys Acta. 2013;1830:3756–3766. doi: 10.1016/j.bbagen.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 73.Pan J, Fang S, Tian H, Zhou C, Zhao X, Tian H, He J, Shen W, Meng X, Jin X, Gong Z. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer. 2020;19:9. doi: 10.1186/s12943-020-1133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice SJ, Lai SC, Wood LW, Helsley KR, Runkle EA, Winslow MM, Mu D. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1) J Biol Chem. 2013;288:16348–16360. doi: 10.1074/jbc.M113.474643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu C, Zhao Y, Zhang Z, Ni Y, Li X, Yong H. MicroRNA-33a inhibits lung cancer cell proliferation and invasion by regulating the expression of β-catenin. Mol Med Rep. 2015;11:3647–3651. doi: 10.3892/mmr.2014.3134. [DOI] [PubMed] [Google Scholar]
- 76.Boldrini L, Giordano M, Niccoli C, Melfi F, Lucchi M, Mussi A, Fontanini G. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int. 2017;17:105. doi: 10.1186/s12935-017-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M, Zhao X, Liu Y, An J, Xiao H, Wang C. Aberrant expression of CDK8 regulates the malignant phenotype and associated with poor prognosis in human laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2017;274:2205–2213. doi: 10.1007/s00405-017-4484-0. [DOI] [PubMed] [Google Scholar]
- 78.Marioni G, Marchese-Ragona R, Cartei G, Marchese F, Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat Rev. 2006;32:504–515. doi: 10.1016/j.ctrv.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 79.Karatas OF. Antiproliferative potential of miR-33a in laryngeal cancer Hep-2 cells via targeting PIM1. Head Neck. 2018;40:2455–2461. doi: 10.1002/hed.25361. [DOI] [PubMed] [Google Scholar]
- 80.Karatas OF, Oner M, Abay A, Diyapoglu A. MicroRNAs in human tongue squamous cell carcinoma: From pathogenesis to therapeutic implications. Oral Oncol. 2017;67:124–130. doi: 10.1016/j.oraloncology.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 81.Zuo Z, Ma L, Gong Z, Xue L, Wang Q. Long non-coding RNA CASC15 promotes tongue squamous carcinoma progression through targeting miR-33a-5p. Environ Sci Pollut Res Int. 2018;25:22205–22212. doi: 10.1007/s11356-018-2300-z. [DOI] [PubMed] [Google Scholar]
- 82.Trotter SC, Sroa N, Winkelmann RR, Olencki T, Bechtel M. A global review of melanoma follow-up guidelines. J Clin Aesthet Dermatol. 2013;6:18–26. [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, Wang S, Chen X, Su J, Zhou X, et al. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1α. Cancer Biol Ther. 2015;16:846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dimitriou F, Krattinger R, Ramelyte E, Barysch MJ, Micaletto S, Dummer R, Goldinger SM. The world of melanoma: Epidemiologic, genetic, and anatomic differences of melanoma across the globe. Curr Oncol Rep. 2018;20:87. doi: 10.1007/s11912-018-0732-8. [DOI] [PubMed] [Google Scholar]
- 85.Cao K, Li J, Chen J, Qian L, Wang A, Chen X, Xiong W, Tang J, Tang S, Chen Y, et al. microRNA-33a-5p increases radiosensitivity by inhibiting glycolysis in melanoma. Oncotarget. 2017;8:83660–83672. doi: 10.18632/oncotarget.19014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian F, Wei H, Tian H, Qiu Y, Xu J. miR-33a is downregulated in melanoma cells and modulates cell proliferation by targeting PCTAIRE1. Oncol Lett. 2016;11:2741–2746. doi: 10.3892/ol.2016.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lv L, Jia JQ, Chen J. The lncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26:201–208. doi: 10.3727/096504017X14920318811749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu Y, Bi Y, Wang F, Chen X, Liu H. Declination of long noncoding RNA paternally expressed gene 10 inhibits A375 cells proliferation, migration, and invasion via mediating microRNA-33a. J Cell Biochem. 2019;120:19868–19877. doi: 10.1002/jcb.29292. [DOI] [PubMed] [Google Scholar]
- 89.Oltean MA, Matuz R, Sitar-Taut A, Mihailov A, Rednic N, Tantau A, Toganel R, Minciuna IA, Orasan O, Muresan F, Cozma A. Renal cell carcinoma with extensive tumor thrombus into the inferior vena cava and right atrium in a 70-year-old man. Am J Mens Health. 2019;13:1557988319846404. doi: 10.1177/1557988319846404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, Gore JL, Sun M, Wood C, Russo P. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang K, Sun F, Zhu J, Luo G, Ban Y, Zhang P. miR-33a inhibits cell growth in renal cancer by downregulation of MDM4 expression. Mol Genet Genomic Med. 2019;7:e833. doi: 10.1002/mgg3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat Rev Endocrinol. 2020;16:17–29. doi: 10.1038/s41574-019-0263-x. [DOI] [PubMed] [Google Scholar]
- 93.Takeshita H, Shiozaki A, Bai XH, Iitaka D, Kim H, Yang BB, Keshavjee S, Liu M. XB130, a new adaptor protein, regulates expression of tumor suppressive microRNAs in cancer cells. PLoS One. 2013;8:e59057. doi: 10.1371/journal.pone.0059057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Awad AJ, Burns TC, Zhang Y, Abounader R. Targeting MET for glioma therapy. Neurosurg Focus. 2014;37:E10. doi: 10.3171/2014.9.FOCUS14520. [DOI] [PubMed] [Google Scholar]
- 95.Chang M, Qiao L, Li B, Wang J, Zhang G, Shi W, Liu Z, Gu N, Di Z, Wang X, Tian Y. Suppression of SIRT6 by miR-33a facilitates tumor growth of glioma through apoptosis and oxidative stress resistance. Oncol Rep. 2017;38:1251–1258. doi: 10.3892/or.2017.5780. [DOI] [PubMed] [Google Scholar]
- 96.Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S, Chen R, McLendon RE, Friedman AH, Keir ST, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 98.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 100.Huang Y, Zhang J, Shao H, Liu J, Jin M, Chen J, Zhao H. miR-33a mediates the anti-tumor effect of lovastatin in osteosarcoma by targeting CYR61. Cell Physiol Biochem. 2018;51:938–948. doi: 10.1159/000495396. [DOI] [PubMed] [Google Scholar]
- 101.Zhang J, Wang D, Xiong J, Chen L, Huang J. MicroRNA-33a-5p suppresses growth of osteosarcoma cells and is downregulated in human osteosarcoma. Oncol Lett. 2015;10:2135–2141. doi: 10.3892/ol.2015.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaudreau PO, Stagg J, Soulières D, Saad F. The present and future of biomarkers in prostate cancer: Proteomics, genomics, and immunology advancements. Biomark Cancer. 2016;8(Suppl 2):S15–S33. doi: 10.4137/BIC.S31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nelson WG, DeWeese TL, DeMarzo AM. The diet, prostate inflammation, and the development of prostate cancer. Cancer Metastasis Rev. 2002;21:3–16. doi: 10.1023/A:1020110718701. [DOI] [PubMed] [Google Scholar]
- 105.Marshall JR. Diet and prostate cancer prevention. World J Urol. 2012;30:157–165. doi: 10.1007/s00345-011-0810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karatas OF, Wang J, Shao L, Ozen M, Zhang Y, Creighton CJ, Ittmann M. miR-33a is a tumor suppressor microRNA that is decreased in prostate cancer. Oncotarget. 2017;8:60243–60256. doi: 10.18632/oncotarget.19521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Q, Lu S, Li X, Hou G, Yan L, Zhang W, Qiao B. Biological function and mechanism of miR-33a in prostate cancer survival and metastasis: Via downregulating Engrailed-2. Clin Transl Oncol. 2017;19:562–570. doi: 10.1007/s12094-016-1564-3. [DOI] [PubMed] [Google Scholar]
- 108.Dai Y, Wu Z, Lang C, Zhang X, He S, Yang Q, Guo W, Lai Y, Du H, Peng X, Ren D. Copy number gain of ZEB1 mediates a double-negative feedback loop with miR-33a-5p that regulates EMT and bone metastasis of prostate cancer dependent on TGF-β signaling. Theranostics. 2019;9:6063–6079. doi: 10.7150/thno.36735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett. 2014;351:1–5. doi: 10.1016/j.canlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 110.Shield KD, Soerjomataram I, Rehm J. Alcohol use and breast cancer: A critical review. Alcohol Clin Exp Res. 2016;40:1166–1181. doi: 10.1111/acer.13071. [DOI] [PubMed] [Google Scholar]
- 111.Liang J, Shang Y. Estrogen and cancer. Annu Rev Physiol. 2013;75:225–240. doi: 10.1146/annurev-physiol-030212-183708. [DOI] [PubMed] [Google Scholar]
- 112.Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6:881–889. doi: 10.1007/s13238-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolfe AR, Bambhroliya A, Reddy JP, Debeb BG, Huo L, Larson R, Li L, Ueno NT, Woodward WA. MiR-33a decreases high-density lipoprotein-induced radiation sensitivity in breast cancer. Int J Radiat Oncol Biol Phys. 2016;95:791–799. doi: 10.1016/j.ijrobp.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng W, Zuo G, Cao X, Li W. MiR-33a functions as a tumor suppressor in triple-negative breast cancer by targeting EZH2. Cancer Cell Int. 2020;20:85. doi: 10.1186/s12935-020-1160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guan X, Gu S, Yuan M, Zheng X, Wu J. MicroRNA-33a-5p overexpression sensitizes triple-negative breast cancer to doxorubicin by inhibiting eIF5A2 and epithelial-mesenchymal transition. Oncol Lett. 2019;18:5986–5994. doi: 10.3892/ol.2019.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin Cancer Res. 2012;18:3658–3667. doi: 10.1158/1078-0432.CCR-11-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu X, Liu T, Fang O, Dong W, Zhang F, Leach L, Hu X, Luo Z. MicroRNA-708-5p acts as a therapeutic agent against metastatic lung cancer. Oncotarget. 2016;7:2417–2432. doi: 10.18632/oncotarget.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 119.Zheng S, Zhang X, Wang X, Li J. MIR31HG promotes cell proliferation and invasion by activating the Wnt/β-catenin signaling pathway in non-small cell lung cancer. Oncol Lett. 2019;17:221–229. doi: 10.3892/ol.2018.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng M, Jiang YP, Chen W, Li KD, Liu X, Gao SY, Feng H, Wang SS, Jiang J, Ma XR, et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015;6:6797–6810. doi: 10.18632/oncotarget.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Buehler D, Hardin H, Shan W, Montemayor-Garcia C, Rush PS, Asioli S, Chen H, Lloyd RV. Expression of epithelial-mesenchymal transition regulators SNAI2 and TWIST1 in thyroid carcinomas. Mod Pathol. 2013;26:54–61. doi: 10.1038/modpathol.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cao YW, Wan GX, Sun JP, Cui XB, Hu JM, Liang WH, Zheng YQ, Li WQ, Li F. Implications of the Notch1-Snail/Slug-epithelial to mesenchymal transition axis for lymph node metastasis in infiltrating ductal carcinoma. Kaohsiung J Med Sci. 2015;31:70–76. doi: 10.1016/j.kjms.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carreras-Torres R, Johansson M, Gaborieau V, Haycock PC, Wade KH, Relton CL, Martin RM, Davey Smith G, Brennan P. The role of obesity, type 2 diabetes, and metabolic factors in pancreatic cancer: A mendelian randomization study. J Natl Cancer Inst. 2017;109:djx012. doi: 10.1093/jnci/djx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ercin M, Sancar-Bas S, Bolkent S, Gezginci-Oktayoglu S. Tub and β-catenin play a key role in insulin and leptin resistance-induced pancreatic beta-cell differentiation. Biochim Biophys Acta Mol Cell Res. 2018;1865:1934–1944. doi: 10.1016/j.bbamcr.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 125.Wijesekara N, Zhang LH, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, Verchere CB, Hayden MR. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lekmine F, Uddin S, Sassano A, Parmar S, Brachmann SM, Majchrzak B, Sonenberg N, Hay N, Fish EN, Platanias LC. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J Biol Chem. 2003;278:27772–27780. doi: 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- 127.Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. Am J Cancer Res. 2015;5:1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 128.Yu JS, Cui W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 129.Yip PY. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:165–176. doi: 10.3978/j.issn.2218-6751.2015.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fournier PG, Juárez P, Jiang G, Clines GA, Niewolna M, Kim HS, Walton HW, Peng XH, Liu Y, Mohammad KS, et al. The TGF-β signaling regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell. 2015;27:809–821. doi: 10.1016/j.ccell.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Feng J, Yan PF, Zhao HY, Zhang FC, Zhao WH, Feng M. SIRT6 suppresses glioma cell growth via induction of apoptosis, inhibition of oxidative stress and suppression of JAK2/STAT3 signaling pathway activation. Oncol Rep. 2016;35:1395–1402. doi: 10.3892/or.2015.4477. [DOI] [PubMed] [Google Scholar]
- 132.Liu SC, Huang CM, Bamodu OA, Lin CS, Liu BL, Tzeng YM, Tsai JT, Lee WH, Chen TM. Ovatodiolide suppresses nasopharyngeal cancer by targeting stem cell-like population, inducing apoptosis, inhibiting EMT and dysregulating JAK/STAT signaling pathway. Phytomedicine. 2019;56:269–278. doi: 10.1016/j.phymed.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 133.Liu K, Tian T, Zheng Y, Zhou L, Dai C, Wang M, Lin S, Deng Y, Hao Q, Zhai Z, Dai Z. Scutellarin inhibits proliferation and invasion of hepatocellular carcinoma cells via down-regulation of JAK2/STAT3 pathway. J Cell Mol Med. 2019;23:3040–3044. doi: 10.1111/jcmm.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tang J, Xu J, Zhi Z, Wang X, Wang Y, Zhou Y, Chen R. MiR-876-3p targets KIF20A to block JAK2/STAT3 pathway in glioma. Am J Transl Res. 2019;11:4957–4966. [PMC free article] [PubMed] [Google Scholar]
- 135.Tanabe K, Kozawa O, Iida H. cAMP/PKA enhances interleukin-1β-induced interleukin-6 synthesis through STAT3 in glial cells. Cell Signal. 2016;28:19–24. doi: 10.1016/j.cellsig.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 136.Liu M, Inoue K, Leng T, Guo S, Xiong ZG. TRPM7 channels regulate glioma stem cell through STAT3 and Notch signaling pathways. Cell Signal. 2014;26:2773–2781. doi: 10.1016/j.cellsig.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xie RT, Cong XL, Zhong XM, Luo P, Yang HQ, Lu GX, Luo P, Chang ZY, Sun R, Wu TM, et al. MicroRNA-33a downregulation is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2018;15:4571–4577. doi: 10.3892/ol.2018.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hou LK, Ma YS, Han Y, Lu GX, Luo P, Chang ZY, Xie RT, Yang HQ, Chai L, Cai MX, et al. Association of microRNA-33a molecular signature with non-small cell lung cancer diagnosis and prognosis after chemotherapy. PLoS One. 2017;12:e0170431. doi: 10.1371/journal.pone.0170431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pan J, Zhou C, Zhao X, He J, Tian H, Shen W, Han Y, Chen J, Fang S, Meng X, et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep. 2018;8:16699. doi: 10.1038/s41598-018-35139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kandimalla R, Gao F, Matsuyama T, Ishikawa T, Uetake H, Takahashi N, Yamada Y, Becerra C, Kopetz S, Wang X, Goel A. Genome-wide discovery and identification of a novel miRNA signature for recurrence prediction in stage II and III colorectal cancer. Clin Cancer Res. 2018;24:3867–3877. doi: 10.1158/1078-0432.CCR-17-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochem Int. 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.