Abstract
Cancer upregulates glycolysis, glutaminolysis and lipogenesis, and induces a catabolic state in patients. The concurrent inhibition of both tumor anabolism and host catabolism, and the energetic consequences of such an approach, have not previously been fully investigated. In the present study, CT26.WT murine colon cancer cells were treated with the combination of anti-anabolic drugs orlistat, lonidamine and 6-diazo-5-oxo-L-norleucine (DON; OLD scheme), which are inhibitors of the de novo synthesis of fatty acids, glycolysis and glutaminolysis, respectively. In addition, the effects of OLD scheme sumplemented with the combination of anti-catabolic compounds, namely growth hormone, insulin and indomethacin (GII scheme), were also evaluated. The effects of the compounds used in combination on CT26.WT cell viability, clonogenicity and energetic metabolism were assessed in vitro. The results demonstrated that the anti-anabolic approach reduced cell viability, clonogenicity and cell cycle progression, and increased apoptosis. These effects were associated with decreased oxidative phosphorylation, glycolysis and fuel flexibility. Furthermore, the anti-catabolic scheme, alone or supplemented with anti-anabolic compounds, did not favor tumor growth. These findings indicated that the simultaneous pharmacological inhibition of tumor anabolism and host catabolism exhibits antitumor effects that should be further evaluated.
Keywords: metabolism, colon cancer, glycolysis, glutaminolysis, de novo fatty acid synthesis
Introduction
Metabolic aberrations are hallmarks of cancer. It has been reported that the majority of malignant cells reprogram their metabolism to induce anabolism via increasing glycolysis, glutaminolysis and de novo synthesis of fatty acids through hexokinase-II (HK2) (1), glutaminase (GLS) (2) and fatty acid synthase (FASN) (3) upregulation, respectively. Furthermore, cancer progression induces a catabolic state in patients, characterized by systemic inflammation, insulin resistance (4), a negative energy balance in the host (5) and proteolysis/lipolysis to support the survival of the tumor (6,7).
Colon cancer harbors oncogenic mutations, including Wnt, KRAS, MYC and TP53 (8). These tumor genes reprogram the metabolism through re-routing glucose to anabolic pathways (9), increasing the expression of FASN and promoting glutamine metabolism (8). These alterations may occur early in colon cancer development to favor the tumorigenic process (10).
Our previous studies demonstrated synergy and antitumor effects of orlistat, lonidamine and 6-diazo-5-oxo-L-norleucine (DON; termed ‘OLD’), known to inhibit FASN, HK2 and GLS, respectively (11,12) in a number of cancer cell lines but not in primary lung fibroblasts. However, no studies have been reported exploring the simultaneous effects of drug combination regimens against tumor anabolism and host catabolism. Therefore, in the present study, the OLD scheme supplemented with the anti-catabolic drugs growth hormone, insulin and indomethacin (GII scheme) were used. Furthermore, the effects of the combination of six drugs (OLD + GII schemes) in CT26.WT cells was also investigated. The results of the present study demonstrated that OLD and six-drug combination schemes resulted in reduced cell viability, clonogenic capacity and cell cycle progression, and induced apoptosis. These effects were associated with a quiescent energetic phenotype and limited substrate flexibility, while the three anti-catabolic drugs did not favor malignant growth.
Materials and methods
Cell line and culture
In the present study the CT26.WT (ATCC) cell line was employed. Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (Corning Inc.) and 1% streptomycin/amphotericin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 incubator.
Drugs
Orlistat (Psicofarma, S.A., De C.V.), lonidamine (Sigma-Aldrich; Merck KGaA), DON (Sigma-Aldrich; Merck KGaA), growth hormone (GH; Merck KGaA), insulin (Eli Lilly & Co.) and indomethacin (Sigma-Aldrich; Merck KGaA) were used. Orlistat and indomethacin were dissolved in absolute ethanol, lonidamine in DMSO (both from Sigma-Aldrich; Merck KGaA), and DON, GH and insulin in complete medium. The compounds were used in the anti-anabolic (OLD, orlistat + lonidamine + DON), anti-catabolic (GII, GH + insulin + indomethacin), or six-drugs combination (OLD + GII, named 6 drugs) schemes.
Cell viability and colony formation ability assays
CT26.WT cells were seeded into 6-well plates (Costar; Corning Inc.) at a density of 3×104 cells/well in 2 ml complete medium. Following 24 h pre-incubation, cells were treated for an additional 72 h with OLD, GII or the six-drug combination schemes. Optimal doses of the OLD and GII schemes used in the present study were chosen according to our previous study (11) and pharmacokinetic data of human studies (13–15), respectively. The OLD and GII scheme doses are listed in Table SI. Control cells treated with the same volume of the corresponding drug vehicles were used to normalize each drug condition. Fresh complete medium supplemented with drugs/vehicles was replaced every 24 h. Following 72 h, cells were detached using a 0.25% trypsin-EDTA solution (Gibco; Thermo Fisher Scientific, Inc.) and cell viability was evaluated via trypan blue (Life Technologies; Thermo Fisher Scientific, Inc.) and the TC10™ Unity Automated Cell Counter (Bio-Rad Laboratories, Inc.). The cytotoxic effect of each treatment was expressed as the percentage of cell viability relative to control cells. Subsequently, 1,000 cells/condition were recovered and plated into new 6-well plates. Fresh complete medium was replaced every 48 h for 14 days to allow colony formation. Finally, colonies were fixed with a methanol/acetic acid solution (3:1 v/v), dyed with a violet crystal solution (Sigma-Aldrich; Merck KGaA) and counted with the ImageJ v2.0 software (National Institutes of Health) (16).
Flow cytometry
A total of 3×104 cells/well were seeded into 6-well plates and treated as mentioned above. Subsequently, cells were recovered and dyed with propidium iodide (Sigma-Aldrich; Merck KGaA) for 1 h. Then, 20,000 cells/sample were analyzed using the BD FACSCanto™ II flow cytometer (BD Biosciences). Cell cycle analysis was performed with the ModFit LT v2.0 software (Verity Software House, Inc.). In independent assays, following treatment for 72 h, cells were recovered and dyed with the Annexin V-FLUOS Staining Kit (Roche). Apoptosis and necrosis rates were simultaneously analyzed using flow cytometry (10,000 events/sample) with the BD FACSDiva™ v6.1.3 software (BD Biosciences). Each condition was compared to its control.
Total protein extraction, western blot analysis and densitometry
After the cells were treated with OLD, GII and six-drug combination and controls for 72 h, cells were washed once with 1X PBS and then harvested with a 0.05% trypsin-0.025% EDTA solution. Detached cells were washed once again with 1X PBS, and proteins were extracted with radioimmunoprecipitation buffer (150 mM NaCl; 1.0% IGEPAL CA-630; 0.5% sodium deoxycholate; 0.1% SDS; 50 mM Tris, pH 8.0) in the presence of proteinase inhibitors (catalog no. p8340; Sigma-Aldrich, Merck KGaA). Protein concentration was determined using a bicinchoninic acid assay and the integrity was assessed by Coomassie staining. A total of 30 µg protein was separated by 15% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (cat no. 162-0177; Bio-Rad Laboratories, Inc.). The membrane was blocked with 5% skim milk in 1X PBS for 1 h at room temperature, and subsequently incubated with the antibody against Cleaved Caspase-3 (catalog no. 9664; 1:1,000 from Cell Signaling Technology, Inc.), and anti-actin peroxidase (A3854; 1:10,000; Sigma-Aldrich; Merck KGaA) in blocking solution (5% skim milk in TBS + 0.1% Tween-20), overnight at 4°C. The secondary antibody was bovine anti-rabbit, (sc-2370, Santa Cruz Biotechnology, Inc.), which was diluted 1:1,000 and the incubation was performed for 1 h at room temperature. Protein bands were visualized using the chromogenic substrate Clarity Western Enhanced Chemiluminescence Substrate (catalog no. 1705060; Bio-Rad Laboratories, Inc.). Bands were densitometrically quantified using the ImageJ software, version 1.50f (National Institutes of Health).
Oxygen consumption and extracellular acidification rates
Oxidative phosphorylation, glycolysis and fuel flexibility were assessed by quantifying oxygen consumption (OCR) and extracellular acidification rates (ECAR) via the Seahorse Bioscience Extracellular Flux Analyzer XF96e (Seahorse Bioscience). Briefly, 3×103 cells/well were seeded into XF96 culture microplates (Seahorse Bioscience) with 100 µl complete medium using a Viaflo Assist robot (Integra Biosciences). Following 24 h pre-incubation, cells were treated for 14 h with the pharmacological drug combinations or their controls. Subsequently, 1 h prior to each Seahorse assay, cells were equilibrated with bicarbonate-free low buffered medium (Seahorse Bioscience), pH 7.4, without any supplements, at 37°C in a non-CO2 incubator. All required reagents for each experiment were prepared in Seahorse assay medium and loaded into the cartridges with Viaflo Assist for 30 min into Seahorse plates as the experiment progressed. All results were normalized to the respective controls according to cellular confluence immediately after each assay. The cellular confluence was measured by scanning the plate with IncuCyte® ZOOM equipment (Essen Bioscience), and the results were analyzed using the Wave software (Seahorse Bioscience).
Oxidative phosphorylation and glycolysis assays
Oxidative phosphorylation and glycolysis rates were evaluated using the XF Cell Mito Stress Test and XF Glycolysis Stress Test, respectively. The experimental design for both assays was performed as described by Zaytseva et al (17). Both OCR (pmoles/min) and ECAR (mpH/min) were measured to indicate oxidative phosphorylation, while only ECAR was considered for glycolysis. A total of 12 measurements/assay were conducted and each drug condition was compared to its control.
Fuel flexibility assay
Fuel flexibility assay was performed using the XF Mito Fuel Flex Test. This test reveals the dependence of fuel mitochondrial respiration on glucose, glutamine and fatty acids and its ability to employ a substrate when the other two are inhibited. The measurements were performed according to the manufacturer's protocol and only the OCR values were considered. The injection order of each inhibitor/pair of inhibitors required to evaluate the capacity, dependency and energetic flexibility are presented in Table SII. A total of 15 measurements/assay were performed and each treatment was compared to its control.
Statistical analysis
All experiments were independently performed in triplicate, with three internal replicates. Significant differences were determined using multiple t-tests with Holm-Sidak correction. The results were analyzed using the GraphPad Prism v6 software (GraphPad Software, Inc.). Data are expressed as mean ± standard error of the mean (SEM), and P<0.05 was considered to indicate a statistically significant difference.
Results
The anti-anabolic scheme reduces CT26.WT cells viability and clonogenic capacity
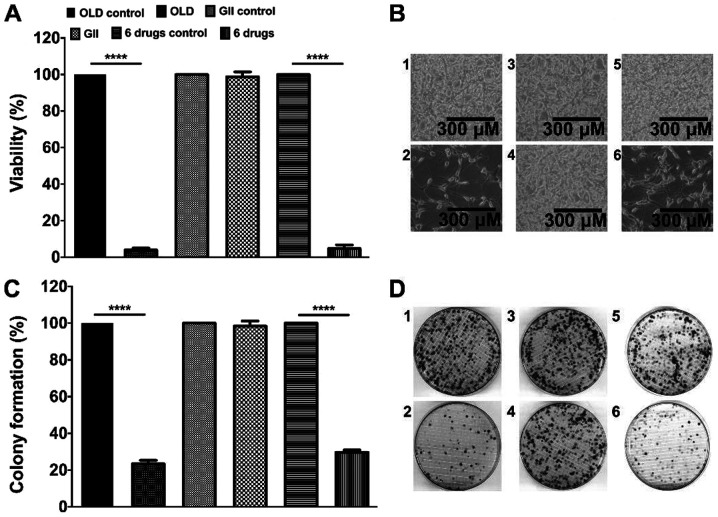
The present study investigated whether the treatment of cells for 72 h with the anti-anabolic (OLD), anti-catabolic (GII) or six-drug (OLD + GII) schemes affected cell viability. The results demonstrated that both OLD and six-drug schemes reduced ~95% of the cell viability (Fig. 1A). Subsequently, clonogenic assays demonstrated that following culture for 14 days the colony numbers for both OLD and six-drug schemes were decreased to ~25% (Fig. 1C). Representative images for each evaluated condition of cells following treatment for 72 h and colonies from the clonogenic assays are presented in Fig. 1B and D. No statistically significant effects were detected in cells treated with the GII scheme compared to the control cells (Fig. 1A-D).
Figure 1.
The inhibition of the de novo synthesis of fatty acids, glycolysis and glutaminolysis diminishes cell viability and clonogenicity. (A) Percentage of cell viability after 72 h of treatment with each scheme. (B) Treated cells with either the OLD control (1), OLD (2), GII control (3), GII (4), 6 drugs control (5), or 6 drugs (6) conditions, after 72 h. (C) Percentage of colony formation 14 days after the 72 h treatment with each scheme. (D) CT26.WT plates from either the OLD control (1), OLD (2), GII control (3), GII (4), 6 drugs control (5), or 6 drugs (6) conditions, after 14 days. Data are expressed as mean ± SEM. Scale bars, 300 µm. ****P<0.0001. OLD, orlistat + lonidamine + DON; GII, growth hormone + insulin + indomethacin; 6 drugs, OLD + GII.
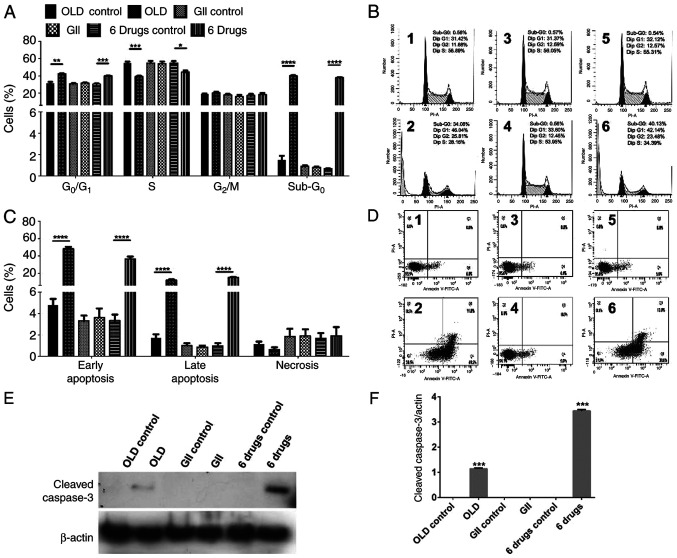
The anti-anabolic drug combination inhibits cell cycle progression and induces apoptosis-related cell death in CT26.WT-treated cells
Subsequently, the present study aimed to identify possible treatment-induced changes on cell cycle. Both OLD and six-drug schemes induced G0/G1 cell cycle arrest and decreased the proportion of S phase cells. In addition, both schemes induced sub-G0 phase in ~40% of cells, indicating cell death (Fig. 2A-B). Furthermore, apoptosis and necrosis rates were evaluated. OLD and six-drug treatments promoted cell apoptosis, mainly in its early form, without significantly increasing necrosis (Fig. 2C-D). Representative figures of cell cycle assays (Fig. 2B) and apoptosis/necrosis rates (Fig. 2D) are shown for each evaluated condition. In line with cell viability and clonogenic assays, no changes were observed in cells treated with the GII scheme. To further corroborate that cells underwent apoptotic death, the evaluation by western blot showed the presence of caspase-3 following OLD and six-drug treatments but not with GII (Fig. 2E and F).
Figure 2.
The anti-anabolic drug combinations block cell cycle progression and stimulates apoptosis. (A) Percentage of cells in each phase of cell cycle after 72 h of treatment with each scheme. (B) ModFit diagrams showing cell cycle distribution from either the OLD control (1), OLD (2), GII control (3), GII (4), 6 drugs control (5), or 6 drugs (6) conditions. (C) Percentage of cells either alive, on early apoptosis, on late apoptosis, or on necrosis, after 72 h of treatment with each scheme. (D) Diva diagrams showing alive (Q1), necrotic (Q2), late apoptotic (Q3), or early apoptotic (Q4) cells, from either the OLD control (1), OLD (2), GII control (3), GII (4), 6 drugs control (5), or 6 drugs (6) conditions. (E and F) Western blot evaluation (E) and densitometric analysis (F) of cleaved caspase-3 among all the schemes. Data are expressed as mean ± SEM. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. OLD, orlistat + lonidamine + DON; GII, growth hormone + insulin + indomethacin; 6 drugs, OLD + GII.
The anti-anabolic drug combination affects the energetic metabolism of CT26.WT cells
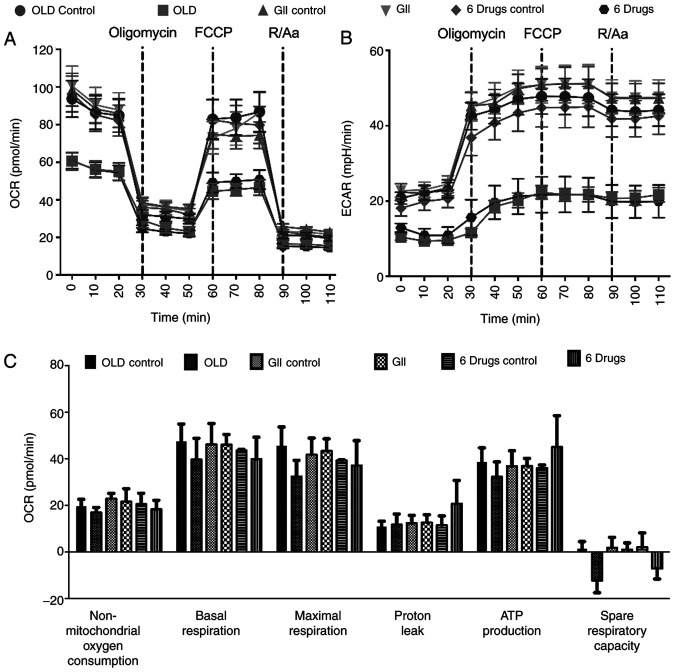
As the employed compounds modulated metabolism, the cells energy production was subsequently investigated. Therefore, both OCR and ECAR rates were evaluated. First, the treatment timepoint that caused prolonged metabolic alterations was determined. The results indicated that cells treated with the compounds for 14 h exhibited the same effects on oxidative phosphorylation and glycolysis compared with those noted following treatment for 72 h (data not shown). Therefore, 14 h treatment was selected for metabolic analysis.
The evaluation of oxidative phosphorylation demonstrated that both OLD and six-drug schemes showed reduced OCR and ECAR rates. These effects were preserved following sequential injections of oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) and rotenone/antimycin A (Fig. 3A and B). Although no significant changes were observed in any of the individual oxidative phosphorylation parameters, treated cells failed to increase OCR beyond basal levels after FCCP injection, resulting in decreased OCR values in the spare respiratory capacity of OLD- and six-drug-treated cells (Fig. 3C).
Figure 3.
Effects of the anti-anabolic drug schemes on the oxidative phosphorylation. (A and B) Mitochondrial respiration measured with the XF Cell Mito Stress Test. (C) Individual parameters for mitochondrial respiration. All the measurements were done 14 h after treatment. Data are expressed as mean ± SEM. OLD, Orlistat + lonidamine + DON; GII, growth hormone + insulin + indomethacin; 6 drugs, OLD + GII; OCR, oxygen consumption rate; ECAR, extracellular acidification rate.
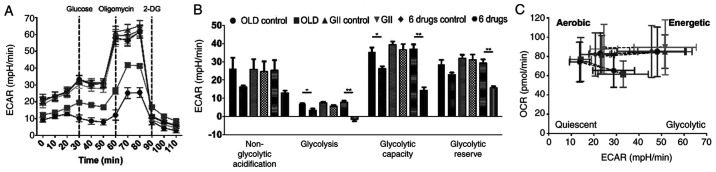
However, glycolysis analysis showed reduced ECAR following treatment with OLD or six-drug schemes. This effect was more pronounced in cells treated with six-drug combination (Fig. 4A) and this scheme exhibited significantly higher effects on individual parameters of glycolysis. Therefore, glycolysis, glycolytic capacity and glycolytic reserve were reduced (Fig. 4B). Besides, energetic phenotype charts were constructed using data emerged from the oxidative phosphorylation assays, including basal and FCCP-induced maximal mitochondrial stress measurements. The results demonstrated that cells treated with either OLD or six-drug schemes exhibited decreased basal OCR and ECAR rates and a quiescent metabolic phenotype following treatment with FCCP (Fig. 4C). However, the GII treatment did not increase OCR and ECAR values as compared with those noted in the control groups.
Figure 4.
The anti-anabolic combinations limit the energetic production from glycolysis and modify the energetic phenotype. (A) Energetic production through glycolysis, measured with the XF Glycolysis Stress Test. (B) Individual parameters for glycolysis. (C) Energetic phenotype chart involving OCR and ECAR, under basal measurements and after the maximal stress induced by FCCP. All the measurements were done 14 h after treatment. Data are expressed as mean ± SEM. *P<0.05, and **P<0.01. OLD, orlistat + lonidamine + DON; GII, growth hormone + insulin + indomethacin; 6 drugs, OLD + GII; OCR, oxygen consumption rate; ECAR, extracellular acidification rate.
OLD limits the fuel flexibility in CT26.WT-treated cells
Since the OLD scheme inhibited glycolysis, glutaminolysis and de novo synthesis of fatty acids, the dependence and capacity of treated cells to alter their preferred energetic substrate when the three metabolic pathways were blocked, were further investigated (Fig. 5A and B). The results obtained revealed that control cells exhibited increased glucose and decreased glutamine dependency and flexibility (Fig. 5C). Therefore, the dependency rate on basal conditions was 36.72, 28.5 and 10.83% for glucose, fatty acids and glutamine, respectively. In addition, the flexibility rates were 38.21, 11.02 and 10.04%, respectively. However, following treatment with OLD scheme for 14 h, cells dramatically changed their metabolic flexibility from glucose and glutamine and increased their dependency on glucose (Fig. 5D). The metabolic dependency from both glutamine and fatty acids increased to comparable levels, at 41.37 and 36.9%, respectively. Finally, the flexibility from glucose, glutamine and fatty acids was reduced to approximately negative values.
Figure 5.
The OLD scheme induces an increase in dependency towards glucose. (A and B) Cellular respiration corresponding to substrate dependency (A) and capacity (B) obtained with the XF Mito Fuel Flex Test. (C and D) Fuel oxidation diagrams representing flexibility and dependency towards the three energetic substrates with OLD control (C) or OLD treatment (D). The sum of flexibility and dependency indicates the capacity. Data are expressed as mean ± SEM. All the measurements were done 14 h after treatment. OLD, Orlistat + lonidamine + DON; GlucD, glucose dependency; LD, long-chain fatty acid dependency; GlutD, glutamine dependency; GlucC, glucose capacity; LC, long-chain fatty acid capacity; GlutC, glutamine capacity; OCR, oxygen consumption rate.
Discussion
The present study demonstrated that the simultaneous use of OLD inhibitors and the six-drug scheme exhibited antitumor effects in vitro, associated with reduced cellular energy production.
Currently, targeting the abnormal metabolism in cancer as a novel form of therapy is gaining momentum (18,19). Although cancer metabolism inhibitors have not yet been applied in routine clinical practice, pre-clinical and early-phase clinical studies with lonidamine and DON demonstrate they were well-tolerated in cancer patients (20). Intravenous orlistat has not being tested in humans, but it was well-tolerated in mice at therapeutical plasma concentration (21,22).
The present study aimed to investigate the antitumor effects of the combination of lonidamine, an inhibitor of glycolysis (23); DON, an inhibitor of glutaminolysis (24); and orlistat, which inhibits the de novo synthesis of fatty acids (25). It has been reported that these anabolic pathways are hyper-functioning in malignancies, thus these drug schemes were used to inhibit cancer anabolism (18,19). In addition, tumor progression induces a pro-inflammatory and catabolic state in the host, resulting in cancer-associated cachexia (26,27). Therefore, the OLD scheme in combination with GH, insulin and indomethacin (GII scheme) was employed to reduce systemic inflammation and lipolysis, and to promote protein biosynthesis and glucose internalization (28–35).
OLD and six-drug schemes decreased cancer growth by inhibiting cell cycle progression at G0/G1 and S phases, and promoting cell apoptosis. To the best of our knowledge, the application of a triple drug scheme that inhibits tumor anabolism has not yet been reported. A study demonstrated that the concomitant treatment with lonidamine and DON showed higher anti-leukemia effects compared to those noted when each inhibitor was administered separately (36). Additionally, in non-small cell lung cancer combined lonidamine and glutaminase inhibitor-968 treatment induced potent cytotoxicity and growth inhibition (37). Regarding the GII combination treatment, although the GH-cancer relationship has been a subject of discussion (38), previous in vitro and in vivo studies have demonstrated that it does not exhibit pro-tumor effects (39–41). Furthermore, although insulin promotes malignant cell proliferation and invasiveness in vitro (42,43), however, contradictory effects have been reported regarding cancer survival and risk in patients (44–46). According to these studies, in the present model, GII treatment did not show pro-tumor effects, although GII-treated cells expressed GH and insulin receptors, as well as cyclooxygenase (data not shown). All these molecules are targeted by the GII treatment scheme. Therefore, the absence of tumor-promoting effects cannot be attributed to the lack of GH and insulin receptors, and cyclooxygenase expression.
Interestingly, the OLD and six-drug schemes significantly reduced glycolysis, as demonstrated by diminished ECAR values. These findings suggested that OLD and six-drug, but not GII treatment, shifted the energy response to a more quiescent metabolic state. The extent of glycolysis inhibition was equivalent to that noted in a previous study with lonidamine treatment in non-small cell lung cancer, employing the Seahorse methodology as well (35). On the other hand, OLD treatment increased cellular dependence on the oxidation of glucose and glutamine, and reduced fuel flexibility on both substrates. These findings are well-suited to a more quiescent energetic phenotype.
In summary, the present study demonstrated that the concomitant blockade with OLD scheme exhibited antitumor effects and dramatically affected the energetic machinery of CT26.WT colon cancer cells. Currently, several novel inhibitors of glycolysis, glutaminolysis and de novo synthesis of fatty acids, as well as anti-cachectic agents, are under pre-clinical and clinical investigation. This study revealed the antitumor effects of OLD treatment, in order to inhibit tumor anabolism, and the lack of pro-tumor effects of the GII scheme.
Supplementary Material
Acknowledgements
Dr Alejandro Schcolnik-Cabrera is a student belonging to the Plan de Estudios Combinados en Medicina (PECEM), UNAM. The authors would like to thank Ms. Rocío Morales-Bárcenas from the National Cancer Institute (Mexico City, Mexico) for her technical support with the flow cytometer.
Funding
The current work was funded by CONACyT (grant no. 140654) and supported by the CONACyT scholarship (no. 439704), provided to Alejandro Schcolnik-Cabrera.
Availability of data and materials
All data generated or analyzed during this study are included in the present article.
Author's contributions
ASC performed all the experiments, analyzed the data, and wrote the manuscript. MJ helped to perform clonogenic assays. ASC, ACB, GDG, DL, SH, ART, JDC and ADG designed the experiments. ADG conceived the project, wrote the manuscript and together with DL, SH and ART, provided infrastructure and research facilities to execute the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This work was approved by the Ethic and Scientific committees of the National Cancer Institute of Mexico (protocol nos. 017/009/IBI and CEI/1055/17), in Mexico City, Mexico.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: Cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erickson JW, Cerione RA. Glutaminase: A hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 4.Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: Possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 5.Tayek JA, Brasel JA. Failure of anabolism in malnourished cancer patients receiving growth hormone: A clinical research center study. J Clin Endocrinol Metab. 1995;80:2082–2087. doi: 10.1210/jcem.80.7.7608259. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Cheng T. Q's next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tijerina AJ. The biochemical basis of metabolism in cancer cachexia. Dimens Crit Care Nurs. 2004;23:237–243. doi: 10.1097/00003465-200411000-00001. [DOI] [PubMed] [Google Scholar]
- 8.La Vecchia S, Sebastián C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev Biol. 2020;98:63–70. doi: 10.1016/j.semcdb.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Lewis NE, Abdel-Haleem AM. The evolution of genome-scale models of cancer metabolism. Front Physiol. 2013;4:237. doi: 10.3389/fphys.2013.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci USA. 2017;114:E7697–E7706. doi: 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schcolnik-Cabrera A, Dominguez-Gómez G, Chávez-Blanco A, Ramírez-Yautentzi M, Morales-Bárcenas R, Chávez-Díaz J, Taja-Chayeb L, Dueáas-González A. A combination of inhibitors of glycolysis, glutaminolysis and de novo fatty acid synthesis decrease the expression of chemokines in human colon cancer cells. Oncol Lett. 2019;18:6909–6916. doi: 10.3892/ol.2019.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cervantes-Madrid D, Dueñas-González A. Antitumor effects of a drug combination targeting glycolysis, glutaminolysis and de novo synthesis of fatty acids. Oncol Rep. 2015;34:1533–1542. doi: 10.3892/or.2015.4077. [DOI] [PubMed] [Google Scholar]
- 13.Maimaiti M, Tanahashi Y, Mohri Z, Fujieda K. Development of a bioassay system for human growth hormone determination with close correlation to immunoassay. J Clin Lab Anal. 2012;26:328–335. doi: 10.1002/jcla.21527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Peña A, Riddle M, Morrow LA, Jiang HH, Linnebjerg H, Scott A, Win KM, Hompesch M, Mace KF, Jacobson JG, et al. Pharmacokinetics and pharmacodynamics of high-dose human regular U-500 insulin versus human regular U-100 insulin in healthy obese subjects. Diabetes Care. 2011;34:2496–2501. doi: 10.2337/dc11-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manvelian G, Daniels S, Altman R. A phase I study evaluating the pharmacokinetic profile of a novel, proprietary, nano-formulated, lower-dose oral indomethacin. Postgrad Med. 2012;124:197–205. doi: 10.3810/pgm.2012.01.2524. [DOI] [PubMed] [Google Scholar]
- 16.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaytseva YY, Harris JW, Mitov MI, Kim JT, Butterfield DA, Lee EY, Weiss HL, Gao T, Evers BM. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891–18904. doi: 10.18632/oncotarget.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clem BF, O'Neal J, Klarer AC, Telang S, Chesney J. Clinical development of cancer therapeutics that target metabolism. QJM. 2016;109:367–372. doi: 10.1093/qjmed/hcv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes-Madrid D, Romero Y, Dueñas-González A. Reviving lonidamine and 6-diazo-5-oxo-l-norleucine to be used in combination for metabolic cancer therapy. BioMed Res Int. 2015;2015:690492. doi: 10.1155/2015/690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kridel SJ, Axelrod F, Rozenkrantz N, Smith JW. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64:2070–2075. doi: 10.1158/0008-5472.CAN-03-3645. [DOI] [PubMed] [Google Scholar]
- 22.Schcolnik-Cabrera A, Chávez-Blanco A, Domínguez-Gómez G, Taja-Chayeb L, Morales-Barcenas R, Trejo-Becerril C, Perez-Cardenas E, Gonzalez-Fierro A, Dueñas-González A. Orlistat as a FASN inhibitor and multitargeted agent for cancer therapy. Expert Opin Investig Drugs. 2018;27:475–489. doi: 10.1080/13543784.2018.1471132. [DOI] [PubMed] [Google Scholar]
- 23.Bhutia YD, Babu E, Ganapathy V. Re-programming tumour cell metabolism to treat cancer: No lone target for lonidamine. Biochem J. 2016;473:1503–1506. doi: 10.1042/BCJ20160068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thangavelu K, Chong QY, Low BC, Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA) Sci Rep. 2014;4:3827. doi: 10.1038/srep03827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pemble CW IV, Johnson LC, Kridel SJ, Lowther WT. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat Struct Mol Biol. 2007;14:704–709. doi: 10.1038/nsmb1265. [DOI] [PubMed] [Google Scholar]
- 26.Schcolnik-Cabrera A, Chávez-Blanco A, Domínguez-Gómez G, Dueñas-González A. Understanding tumor anabolism and patient catabolism in cancer-associated cachexia. Am J Cancer Res. 2017;7:1107–1135. [PMC free article] [PubMed] [Google Scholar]
- 27.Mondello P, Mian M, Aloisi C, Famà F, Mondello S, Pitini V. Cancer cachexia syndrome: Pathogenesis, diagnosis, and new therapeutic options. Nutr Cancer. 2015;67:12–26. doi: 10.1080/01635581.2015.976318. [DOI] [PubMed] [Google Scholar]
- 28.Manson JM, Wilmore DW. Positive nitrogen balance with human growth hormone and hypocaloric intravenous feeding. Surgery. 1986;100:188–197. [PubMed] [Google Scholar]
- 29.Ward HC, Halliday D, Sim AJ. Protein and energy metabolism with biosynthetic human growth hormone after gastrointestinal surgery. Ann Surg. 1987;206:56–61. doi: 10.1097/00000658-198707000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cersosimo E, Pisters PW, Pesola G, Rogatko A, Vydelingum NA, Bajorunas D, Brennan MF. The effect of graded doses of insulin on peripheral glucose uptake and lactate release in cancer cachexia. Surgery. 1991;109:459–467. [PubMed] [Google Scholar]
- 31.Pearlstone DB, Wolf RF, Berman RS, Burt M, Brennan MF. Effect of systemic insulin on protein kinetics in postoperative cancer patients. Ann Surg Oncol. 1994;1:321–332. doi: 10.1007/BF02303571. [DOI] [PubMed] [Google Scholar]
- 32.Moley JF, Morrison SD, Norton JA. Insulin reversal of cancer cachexia in rats. Cancer Res. 1985;45:4925–4931. [PubMed] [Google Scholar]
- 33.Noguchi Y, Nomura K, Yoshikawa T, Fukuzawa K, Makino T, Tsuburaya A, Matsumoto A. Role of insulin resistance in decreasing lipoprotein lipase activity in tumor-bearing rats. Surg Today. 1996;26:271–275. doi: 10.1007/BF00311587. [DOI] [PubMed] [Google Scholar]
- 34.Hitt A, Graves E, McCarthy DO. Indomethacin preserves muscle mass and reduces levels of E3 ligases and TNF receptor type 1 in the gastrocnemius muscle of tumor-bearing mice. Res Nurs Health. 2005;28:56–66. doi: 10.1002/nur.20057. [DOI] [PubMed] [Google Scholar]
- 35.Lundholm K, Daneryd P, Körner U, Hyltander A, Bosaeus I. Evidence that long-term COX-treatment improves energy homeostasis and body composition in cancer patients with progressive cachexia. Int J Oncol. 2004;24:505–512. [PubMed] [Google Scholar]
- 36.Griffiths M, Keast D, Patrick G, Crawford M, Palmer TN. The role of glutamine and glucose analogues in metabolic inhibition of human myeloid leukaemia in vitro. Int J Biochem. 1993;25:1749–1755. doi: 10.1016/0020-711X(88)90303-5. [DOI] [PubMed] [Google Scholar]
- 37.Meijer TWH, Peeters WJM, Dubois LJ, van Gisbergen MW, Biemans R, Venhuizen JH, Span PN, Bussink J. Targeting glucose and glutamine metabolism combined with radiation therapy in non-small cell lung cancer. Lung Cancer. 2018;126:32–40. doi: 10.1016/j.lungcan.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Cohen P, Clemmons DR, Rosenfeld RG. Does the GH-IGF axis play a role in cancer pathogenesis? Growth Horm IGF Res. 2000;10:297–305. doi: 10.1054/ghir.2000.0171. [DOI] [PubMed] [Google Scholar]
- 39.Chen JY, Liang DM, Gan P, Zhang Y, Lin J. In vitro effects of recombinant human growth hormone on growth of human gastric cancer cell line BGC823 cells. World J Gastroenterol. 2004;10:1132–1136. doi: 10.3748/wjg.v10.i8.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang DM, Chen JY, Zhang Y, Gan P, Lin J, Chen AB. Effects of recombinant human growth hormone on growth of human gastric carcinoma xenograft model in nude mice. World J Gastroenterol. 2006;12:3810–3813. doi: 10.3748/wjg.v12.i24.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison LE, Blumberg D, Berman R, Ng B, Hochwald S, Brennan MF, Burt M. Effect of human growth hormone on human pancreatic carcinoma growth, protein, and cell cycle kinetics. J Surg Res. 1996;61:317–322. doi: 10.1006/jsre.1996.0123. [DOI] [PubMed] [Google Scholar]
- 42.Lu CC, Chu PY, Hsia SM, Wu CH, Tung YT, Yen GC. Insulin induction instigates cell proliferation and metastasis in human colorectal cancer cells. Int J Oncol. 2017;50:736–744. doi: 10.3892/ijo.2017.3844. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Liang H, Song Q, Xu X, Cao D. Insulin promotes progression of colon cancer by upregulation of ACAT1. Lipids Health Dis. 2018;17:122. doi: 10.1186/s12944-018-0773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulskas A, Patasius A, Linkeviciute-Ulinskiene D, Zabuliene L, Smailyte G. A cohort study of antihyperglycemic medication exposure and survival in patients with gastric cancer. Aging (Albany NY) 2019;11:7197–7205. doi: 10.18632/aging.102245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baglia ML, Cui Y, Zheng T, Yang G, Li H, You M, Xu L, Murff H, Gao YT, Zheng W, et al. Diabetes medication use in association with survival among patients of breast, colorectal, lung, or gastric cancer. Cancer Res Treat. 2019;51:538–546. doi: 10.4143/crt.2017.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlstad O, Starup-Linde J, Vestergaard P, Hjellvik V, Bazelier MT, Schmidt MK, Andersen M, Auvinen A, Haukka J, Furu K, et al. Use of insulin and insulin analogs and risk of cancer - systematic review and meta-analysis of observational studies. Curr Drug Saf. 2013;8:333–348. doi: 10.2174/15680266113136660067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the present article.