Abstract
The present study was undertaken to find novel genes associated with colistin resistance in Klebsiella pneumoniae. Five colistin-resistant mutants were derived from four colistin-susceptible parental K. pneumoniae strains belonging to different clones. Whole-genome sequencing was performed for the nine K. pneumoniae strains to screen altered candidate genes. Expression levels of genes with amino acid alterations in derivative strains were determined using quantitative real-time Polymerase chain reaction (PCR). Colistin susceptibility was examined in a parental strain complemented with altered candidate genes. Overall, 13 genetic alterations were identified in five pairs of isogenic K. pneumoniae strains. Genetic alterations related to KP1_3468, including the insertion of an IS5-like element in an intergenic or coding region and amino acid substitutions, were identified in three separate derivative strains. Amino acid substitutions and deletion of PhoQ were determined in one derivative strain. With inactivation of CrrA and substituted CrrB, amino acid substitutions and deletion were identified in a repressor of galETK operon (KP1_0061) and hypothetical protein (KP1_3620), respectively. Decreased colistin susceptibility was observed in a parental strain complemented with KP1-0061, but not a KP1-3620 gene. This study demonstrated diverse genetic paths to colistin resistance in K. pneumoniae. Our results suggest that a repressor of galETK operon may play an important role in colistin resistance in K. pneumoniae.
Keywords: Klebsiella pneumoniae, colistin resistance, CrrAB
1. Introduction
Klebsiella pneumoniae is an opportunistic pathogen causing urinary tract infections (UTIs), pneumonia, and bacteremia, particularly in intensive care units. Recently, infections by multidrug-resistant (MDR) or carbapenem-resistant K. pneumoniae isolates have become a major concern worldwide [1]. The increase in MDR or carbapenem-resistant K. pneumoniae infections has reduced the efficacy of antimicrobial treatments and has raised mortality and morbidity. The limited availability of antimicrobial agents to treat infections caused by Gram-negative pathogens, including K. pneumoniae, prompted the resurgence of old antimicrobial agents such as polymyxins (polymyxn B and colistin) [2]. However, the use of colistin induced the emergence of colistin resistance, and even pandrug-resistant isolates, which are resistant to all available antimicrobial agents [3].
Polymyxins are polycationic antimicrobial peptides that interact with the hydrophobic lipid A component of lipopolysaccharides (LPS) in Gram-negative bacteria, which cause bacterial death by disrupting the outer cell membrane [4]. It is known that the modification of LPS following the addition of 4-amino-4-deoxy-l-arabinose to lipid A is related to colistin resistance in K. pneumoniae [5]. The modification of LPS is associated with the pbgPE operon, which is regulated by two-component regulatory systems, such as PmrAB and PhoPQ [6]. In addition, the eight types of plasmid-mediated mcr genes are associated with the addition of phosphoethanolamine to lipid A, which results in colistin resistance [7]. Loss of LPS has been reported in colistin-resistant Acinetobacter baumannii isolates [8], but not in K. pneumoniae. Recently, insertional inactivation of MgrB, a negative regulator of PhoPQ, was detected in colistin-resistant K. pneumoniae isolates [9]. However, mutations in PmrAB, PhoPQ, and MgrB were not found in all colistin-resistant K. pneumoniae isolates, and the colistin resistance mechanism has not been fully elucidated.
Recently, genomic analysis of antimicrobial-susceptible and -resistant bacterial isolates has been used to reveal the mechanisms of antimicrobial resistance [10]. Although several genomic analyses have highlighted colistin resistance mechanisms in A. baumannii and Pseudomonas aeruginosa [11,12,13], there is a paucity of data regarding the acquired colistin resistance in K. pneumoniae [14]. This study focused on identifying genetic alterations associated with colistin resistance among five pairs of K. pneumoniae isogenic strains using whole-genome sequencing.
2. Results
2.1. Antimicrobial Susceptibility
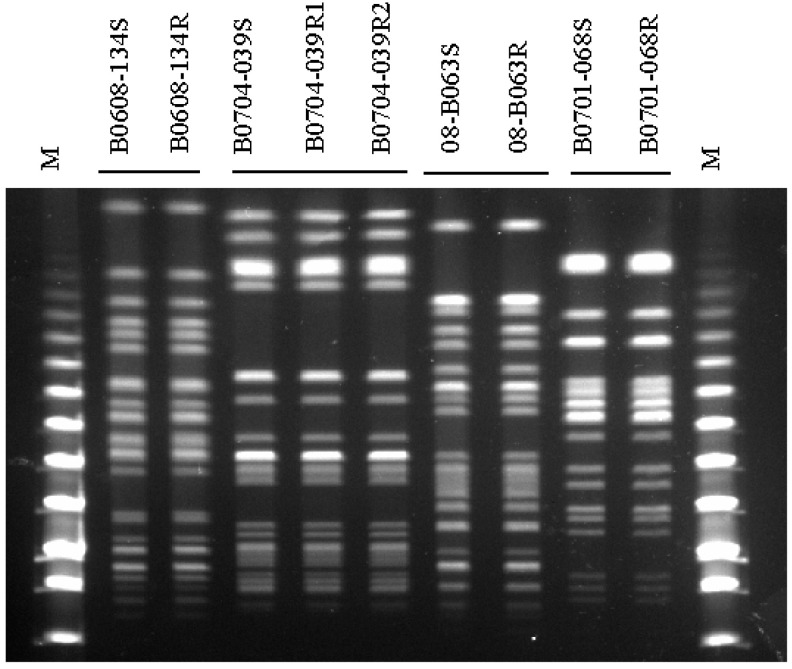
Parental and isogenic mutant strains showed the same sequence type (ST) and the same patterns in pulsed-field gel electrophoresis (PFGE) analysis with XbaI (Figure 1). The antimicrobial susceptibilities of the nine K. pneumoniae strains are presented in Table 1. While four colistin-susceptible parental strains showed colistin and polymyxin B minimum inhibitory concentrations (MICs) of from 0.5 to 2 mg/L, the colistin and polymyxin B MICs for their isogenic colistin-resistant strains were from >64 to >8196 mg/L. Although the MICs of ciprofloxacin and trimethoprim/sulfamethoxazole were increased in 08-B063R, no significant increase or decrease in the MICs of the other antimicrobial agents was observed in the induced colistin-resistant strains.
Figure 1.
Pulsed-field gel electrophoresis (PFGE) profiles obtained with restriction enzyme XbaI.
Table 1.
Antimicrobial susceptibility profiles in four pairs of isogenic Klebsiella pneumoniae strains.
| Isolate | ST a | Minimum Inhibitory Concentration(mg/L) b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COL | PB | AMP | AZ | CTX | CAZ | CIP | GEN | IMI | P/T | SXT | ||
| B0608-134 | 730 | 1 | 2 | 64 | ≤0.06 | ≤0.06 | 1 | ≤0.06 | 1 | 0.125 | 16/4 | 0.06/1.18 |
| B0608-134R | 256 | >64 | 64 | ≤0.06 | ≤0.06 | 1 | ≤0.06 | 2 | 0.25 | 16/4 | 0.06/1.18 | |
| B0704-039 | 11 | 0.5 | 2 | >64 | >64 | >64 | >64 | >64 | >64 | 1 | >64/4 | 4/76 |
| B0704-039R1 | >8192 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 1 | >64/4 | 4/76 | |
| B0704-039R2 | >128 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 0.5 | >64/4 | 4/76 | |
| 08-B063 | 23 | 0.5 | 2 | 64 | ≤0.06 | ≤0.06 | 0.5 | ≤0.06 | 0.5 | 0.125 | 8/4 | 0.12/2.37 |
| 08-B063R | 512 | >64 | >64 | ≤0.06 | ≤0.06 | 0.5 | 0.125 | 0.5 | 0.125 | 16/4 | 0.5/9.5 | |
| B0701-068 | 152 | 0.5 | 2 | 64 | ≤0.06 | ≤0.06 | 1 | ≤0.06 | 1 | 0.25 | 16/4 | 0.06/1.18 |
| B0701-068R | 1024 | >64 | >64 | ≤0.06 | ≤0.06 | 1 | ≤0.06 | 1 | 0.25 | 16/4 | 0.125/2.37 | |
a ST, Sequence type in multi-locus sequence typing. b COL, colistin; PB, polymyxin B; AMP, ampicillin; AZ, aztreonam; CTX, cefotaxime; CAZ, ceftazidime; CIP, ciprofloxacin; GEN, gentamicin; IMI, imipenem; P/T, piperacillin/tazobactam; SXT, trimethoprim/sulfamethoxazole.
2.2. Mutations in Colistin-Resistant Isolates
Next-generation sequencing produced 13–17 million nucleotides for each of the nine strains. They covered 90.37–98.37-fold. We were able to map reads from three isolates to 68.61–88.82% and 72.44–81.42% of the K. pneumoniae NTUH-K2044 and K. pneumoniae MGH 78578 (NC_009648.1) reference genomes, respectively. Overall, the genomic analyses of five pairs of isogenic K. pneumoniae strains showed 13 nucleotide alterations, including six single nucleotide polymorphisms (SNPs) and seven short insertions and deletions (INDELs). Among the nucleotide alterations, eleven were located in coding sequences, and two were identified in intergenic sequences (IGSs) (Table 2).
Table 2.
Nucleotide and amino acid alterations and mRNA expression changes among five pairs of isogenic Klebsiella pneumoniae strains.
| Set | Gene Name | Function | Nucleotide Alteration | Amino Acid Alteration | Fold Change of mRNA Expression |
|---|---|---|---|---|---|
| B0608-134 versus B0608-134R | Intergenic region between KPN_02065 and KPN_02066 | KPN_02065, putative transport protein KPN_02066, putative response regulator consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain | Insertion of Insertion sequence (IS)5-like element | - | 208.99 ± 48.27-fold increase for KPN_02065 |
| 4.48 ± 0.1-fold increase for KPN_02066 | |||||
| B0704-039 versus B0704-039R1 | Intergenic region between KP1_3468 and KP1_3469 |
KP1_3468; conserved hypothetical protein KP1_3469; putative outer membrane |
Insertion of IS5-like element | - | 2.51 ± 0.49-fold increase for KP1_3468 |
| 1.03 ± 0.11 fold increase for KP1_3469 | |||||
| B0704-039 versus B0704-039R2 | KP1_3468 | Hypothetical protein | T26A, C108G, A128T | L9Q, S36R, K43I | 7.91 ± 0.31-fold increase |
| mgrB | PhoPQ regulatory protein | Insertion of IS5-like element | Frameshift | - | |
| B08-063 versus B08-063R | phoQ | Sensor protein PhoQ | A803C | Y268S | 7.49 ± 0.52-fold increase |
| 12-bp deletion at 341st nucleotide | Deletion of 5 amino acids at position 114 | ||||
| KP1_3620 | Hypothetical protein | 1 bp deletion at 51st nucleotide | Premature stop at position 14 | - | |
| KP1_0061 | Repressor of galETK operon | T104G | V35G | 2.11 ± 0.35-fold increase | |
| B0701-068 versus B0701-068R | KP1_3468 | Hypothetical protein | Insertion of IS5-like element | Frameshift | 0.9 ± 0.14-fold increase |
| KPN_02067 | Sensor histidine protein kinase (RstA regulator) | G593T | G198V | 1.36 ± 0.05-fold increase | |
| mgrB | PhoPQ regulatory protein | Insertion of IS5-like element | Frameshift | - |
In B0608-134R and B0704-039R1, only IGS alterations were found. In B0608-134R, an IS5-like element was inserted between KPN_02065 and KPN_02066. In B0704-039R1, an IS5-like element was inserted between KP1_3468 and KP1_3469.
Alterations in KP1_3468 were also seen in B0704-039R2, a colistin-resistant strain derived from the same parent strain as B0704-039R1. However, these changes were in the coding region. Specifically, three nucleotide substitutions (T26A, C108G, and A128T) led to the amino acid substitutions L9Q, S36R, and K43I, respectively. This strain also contained an IS5-like element insertion in mgrB, which encodes a PhoPQ regulatory protein.
In B08-063R, four nucleotide alterations were found in three genes, phoQ, KP1_3620, and KP1_0061. In phoQ, two alterations were found: A803C, leading to a Y268S amino acid substitution, and a 12-bp deletion at the 341st nucleotide position. In KP1_3620, which encodes a hypothetical protein, a 1-bp deletion at the 51st nucleotide position caused a premature stop codon at amino acid position 14. In KP1_0061, which encodes a repressor of the galETK operon, a single nucleotide substitution, T104G, caused the amino acid change V35G.
As in B0704-039R2, B0701-068R showed disruptions in MgrB and KP1_3468 caused by IS5-like element insertions. In addition, one nucleotide alteration (G593T), leading to the amino acid substitution G198V, was found in KPN_02067, which encodes a RstA regulator (sensor histidine protein kinase).
2.3. Orthologs Involved in Colistin Resistance
The KPN_02065, KPN_02066, and KPN_02067 genes were aligned with the crrAB region (Locus ID H239_3059-H239_3062) of the UHKPC45 strain. The KPN_02066 genes were uniquely matched against H239_3061 (crrA). In addition, KPN_02065 and KPN_02067 genes had a 1-bp mismatch to H239_3062 and 6-bp mismatch to the H239_3060 (crrB), respectively. The KP1_3468 gene was matched against the mgrB gene (Gene ID 17167876) of the K. pneumoniae CG43 stain. The sequence of the KP1_3468 gene is identical to the mgrB gene.
2.4. Change of mRNA Levels in Candidate Genes Associated with Colistin Resistance
For candidate genes associated with colistin resistance based on genome analysis, mRNA expression levels in colistin-resistant derivatives were compared with those from the susceptible parental strains (Table 2). Genes disrupted by insertion of IS5-like elements, such as mgrB in B0704-039R2 and B0701-068R, and KP1_3620 in B08-063R, were not included in this analysis.
For mutations in IGS regions, mRNA expression of the upstream and downstream genes was analyzed to reveal whether mutations in the IGS region could influence nearby gene expression. In B0608-134R, with its IS5-like element insertion, KPN_02065 and KPN_02066 expression levels were increased by 208.99- and 4.48-fold respectively, compared with B0608-134. In B0704-039R1, expression of KP1_3468 mRNA was increased 2.51-fold compared with its susceptible parental strain, but KP1_3469 exhibited no significant expression change.
Expression of KP1_3468 mRNA was increased by 7.91-fold in B0704-039R2 compared with B0704-039. In B08-063R, increased expression was seen in phoQ (7.49-fold) and the galETK operon repressor, KP1_0061 (2.11-fold), compared to its parental strain. In isolate B0701-068R, there was increased expression of the RstA regulator, KPN_02067 (1.36-fold), but no expression change in KP1_3468 was observed compared with its susceptible parental strain.
2.5. Confirmation of the Role of KP1_0061 and KP1_3620 Gene in Colistin Resistance
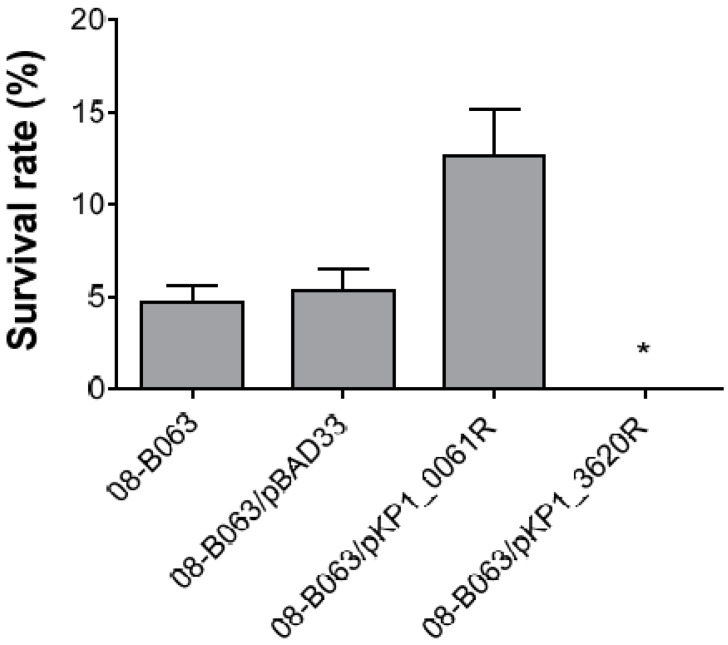
To examine whether altered novel candidate genes, KP1_0061 and KP1_3620, from 08-B063R strain were associated with colistin resistance, susceptibility testing to colistin was conducted in the 08-B063 strain harboring a pBAD33 vector containing altered KP1_0061 or KP1_3620 gene. All of the 08-B063 strains harboring a pBAD33 vector induced by 1% L-arabinose showed similar colistin MICs compared to their wild strain, but not 08-B063/pKP1_3620R strain (Table 3). Colistin MIC of 08-B063/pKP1_3620R strain was decreased by 4-fold compared to its parental strain. After the 1 h of incubation with colistin (1 mg/L), 4.7% and 5.3% of bacteria survived in 08-B063 and 08-B063/pBAD33 strains, respectively (Figure 2). The viability of the 08-B063/pKP1_0061R strain against colistin was 2.7-fold higher than its wild-type strain. The viability of the 08-B063/pKP1_3620R strain against colistin could not be determined due to values less than a detection limit of 50 colony-forming unit (CFU)/mL.
Table 3.
Colistin MICs in 08-B063 strain harboring a pBAD33 vector containing altered genes.
| Isolates | Colistin MICs(mg/L) |
|---|---|
| 08-B063 | 0.5 |
| 08-B063/pBAD33 | 0.5 |
| 08-B063/pKP1_0061R | 0.5 |
| 08-B063/pKP1_3620R | 0.125 |
Figure 2.
Colistin susceptibility testing in the 08-B063 strain carrying the pKP1_0061R or pKP1_3620R vector. The strains were challenged with 1 mg/L of colistin for 1 h. * After 1 h incubation with colistin, survival of 08-B063/pKP1_3620R could not be detected due to values less than a detection limit of 50 CFU/mL.
3. Discussion
In this study, we used comparative genome analysis to screen for candidate genes associated with colistin resistance in pairs of isogenic strains belonging to four different K. pneumoniae clones. ST11 is a dominant clone in Extended-Spectrum β-Lactamase (ESBL)-producing K. pneumoniae isolates and is a known cause of UTIs and bacteremia in Korea [15]. ST23 is reported to be a hyper-virulent clone that causes pyogenic liver abscesses with a hyper-mucoviscous phenotype [16]. Although ST152 and ST730 are not seen as frequently as ST11 and ST23, they were used in this study to compare the genetic paths to colistin resistance among K. pneumoniae clones. In addition, two colistin-resistant derivatives from one susceptible parental strain were included to determine whether the genetic path to colistin resistance is unique to each strain.
We identified a total of 13 genetic alterations in five colistin-resistant K. pneumoniae mutants, with one to four alterations per strain. Of these, amino acid substitutions in PhoQ and inactivation of MgrB by IS5-like elements are well-known mechanisms associated with colistin resistance in K. pneumoniae [5]. In this study, PhoQ variations and MgrB inactivation were identified in only one and two colistin-resistant derivatives, respectively. No PhoQ variation or MgrB inactivation was identified in two of the mutants (B0608-134R and B0704-039R1). Instead, these mutants had genetic alterations in intergenic regions.
In B0608-134R, an IS5-like element was inserted into the intergenic region between KPN_02065 and KPN_02066. These two genes, which encode a putative transport protein and response regulator, were highly expressed in a colistin-resistant mutant. This suggests that the insertion of an IS5-like element in the IGS may affect the expression and/or function of these two proteins. KPN_02066 encodes a response regulator harboring a CheY-like receiver domain. Response regulators with a CheY-like receiver domain are known to be associated with signal transduction and transcription in Deinococcus radiodurans [17]. Although the precise mechanism is unknown, the modification of signal transduction and transcription may be associated with colistin resistance in this strain.
One of the interesting findings in this study is that genetic alterations related to KP1_3468 occurred repeatedly in either the nearby IGS region (B0704-039R1) or as amino acid substitutions in the coding sequence (B0704-039R2). Although B0704-039R1 contained only a single IGS insertion between KP1_3468 and KP1_3469, KP1_3468 alterations in B0704-039R2 and B0701-068R were accompanied by disruptions such as MgrB inactivation. Thus, the association of KP1_3468 with colistin resistance is not clear, and further investigation is required. It is also notable that two colistin-resistant derivatives from one susceptible parental strain showed different genetic variations in MgrB. Colistin-resistant mutants derived from the same susceptible strain showed different amino acid substitutions reported in A. baumannii [18].
The repeated finding of IS5-like element insertions in colistin-resistant derivatives is also noteworthy. As an IS5-like element is already present in the genome of susceptible parental strains (data not shown), it is likely to originate endogenously [5]. It is thought that IS elements play beneficial roles in adaptive evolution in bacteria by generating genetic diversity [19]. Our results suggest that mobile IS5-like elements can facilitate the development of colistin resistance after colistin exposure in K. pneumoniae.
In B08-063R, amino acid substitution and upregulation of KP1_0061, which encodes a galETK operon repressor, were identified in addition to PhoQ alterations. Although there were no changes in MICs between a parental strain with or without complementation with altered KP1_0061, decreased susceptibility in the presence of 1 mg/L colistin was observed in complementation strains. The galETK operon is associated with galactose metabolism, and the product Uridine diphosphate (UDP)-galactose contributes to the early stages of LPS biosynthesis in K. Pneumoniae [20]. Upregulation of KP1_0061 may repress the galETK operon, leading to obstruction of LPS synthesis. It is speculated that obstruction of LPS synthesis due to repression of the galETK operon might decrease the interaction between colistin and the outer membrane. Unfortunately, one of the candidate genes, KP1_3620, found through next-generation sequencing failed to reduce susceptibility to colistin. Further studies are needed to determine whether these genes were associated with decreased susceptibility to trimethoprim-sulfamethoxazole in 08-B063R in this study.
The discovery of an amino acid substitution in a RstA regulator in B0701-068R was also interesting because the RstA operon is activated by the PhoPQ system in low Mg2+. It is also associated with virulence factors such as biofilm formation or iron transport in Negative bacteria [21,22]. It is hypothesized that overexpression of the PhoPQ system due to the inactivation of MgrB may result in colistin resistance, while downstream, the RstAB system may lead to increased fitness of a colistin-resistant strain.
In this study, genetic alterations in colistin-resistant derivatives of K. pneumoniae were investigated by whole-genome sequencing. Five pairs of isogenic colistin-susceptible and -resistant strains did not show identical genetic alterations, implying that there are diverse genetic paths to colistin resistance. In addition to well-known phoQ, mgrB, and crrAB associated with colistin resistance, other candidate genes such as KP1_3468, and KP1_0061 (encoding a galETK operon repressor) were found to be associated with colistin resistance in K. pneumoniae. We also showed that an IS5-like element might play a role in the evolution of antimicrobial resistance. Further study is required to investigate the roles of these genes and IS in the emergence of colistin resistance.
4. Materials and Methods
4.1. Bacterial Strains
Four colistin-susceptible K. pneumoniae strains (B0608-134, B0704-039, 08-B063, and B0701-068) isolated from patients during 2006–2007 in Korea were used in this study. To determine colistin resistance mechanisms in diverse lineages of K. pneumoniae, four strains belonging to different clones (ST730, ST11, ST23, and ST152) according to multi-locus sequence typing (MLST) analysis were used (Table 1) [23]. To induce resistance to colistin, the strains were repeatedly cultured with increasing concentrations of colistin in lysogeny broth (LB). The colistin-resistant mutants (B0608-134R, B0704-039R1, B0704-039R2, 08-B063R, and B0701-068R) were selected in LB containing colistin ranging from 2 to 64 mg/L. B0704-039R1 and B0704-039R2 were derived from the same susceptible strain in different flasks.
4.2. Pulsed-Field Gel Electrophoresis (PFGE)
To investigate the clonal relationship between parental and mutant strains, PFGE was carried out as previously described [24]. Briefly, Genomic DNA was digested with XbalI (New England Biolabs, Beverly, MA, USA) and the fragments were separated in a 1% agarose gel using CHEF DR II-apparatus (Biorad, Hercules, CA, USA). The electrophoresis conditions were a constant voltage of 6 V/cm with pulse times from 5 to 90 s for 18 h. Lambda ladders (New England Biolabs, Beverly, MA, USA) were applied as molecular size markers. PFGE gels were stained with Ethidium bromide, visualized by ultraviolet light (BioRad, Berkeley, CA, USA) and converted to TIFF files.
4.3. In Vitro Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) of antimicrobial agents were determined by Clinical and Laboratory Standards Institute (CLSI) broth microdilution methods [25]. Eleven antimicrobial agents were tested: colistin, polymyxin B, ampicillin, aztreonam, cefotaxime, ceftazidime, ciprofloxacin, gentamicin, imipenem, piperacillin/tazobactam, and trimethoprim/sulfamethoxazole. The results were interpreted based on susceptibility interpretive criteria established in the CLSI standard M100-S28 [25]. Quality control was performed using E. coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853 in three independent experiments. K. pneumoniae isolates would be considered susceptible to colistin and polymyxin B at breakpoints of ≤2 mg/L [26,27].
4.4. Genome Sequencing and Confirmation of Genetic Variants
Genomic DNA was extracted from overnight cultures of nine K. pneumoniae strains by using a G-spinTM Genomic DNA extraction Kit (iNtRON Biotechnology, Inc., South Korea). Purified DNA with a 260/280 purity ratio of 1.75–2.0 was sequenced using Illumina Hiseq2000 Preliminary Performance Parameters, with 100X coverage (Macrogen; Seoul, South Korea). After sequencing, raw sequence data were filtered based on quality score and then assembled onto the K. pneumoniae NTUH-K2044 (NC_012731.1) and K. pneumoniae MGH 78578 (NC_009648.1) reference genomic sequences using Burrows-Wheeler Aligner v0.5.9-r16. Sequence features, including SNPs and INDELs, in colistin-resistant strains were identified through next-generation sequencing and confirmed through Sanger sequencing, using the primers listed in Table 4.
Table 4.
Primers used in this study.
| Primer | Sequence (5′→ 3′) | Amplicon Size (bp) |
Reference |
|---|---|---|---|
| Primers for sequencing | |||
| 134-target1-F | TGAATGCACAAGGTAAAGCCAGG | 500 | This study |
| 134-target1-R | CCGTCTTCGGCTCTTAAGGTTTT | ||
| 039-target1-F | GACACAGCCAGCGATGCCAG | 416 | This study |
| 039-target1-R | TTGTATGATCCATGGCGTGA | ||
| 063-target1,2-F | ATGCAGGTAATAACCAATCT | 751 | This study |
| 063-target1,2-R | ATCCCGGAAAACCTGAATAT | ||
| 063-target3-F | CCATAATAAAAAAAATAATC | 362 | This study |
| 063-target3-R | GCCCCTCCCCACACATCTTT | ||
| 063-target4-F | ACGGCCGAGCGCCAGAAACG | 720 | This study |
| 063-target4-R | TCGCATCCGCATCCGCCAGG | ||
| 068-target1-F | TATGATGCACACCTGTCGGG | 780 | This study |
| 068-target1-R | CACTGTGGAATAACACCCCA | ||
| 068-target2-F | GGCACTGAATAATGCGTAAG | 480 | This study |
| 068-target2-R | GCCTGAAACAAATCTCTCAG | ||
| mgrB-ext-F | CAGCCAGCGATGCCAGATTT | 380 | [5] |
| mgrB-ext-R | CCTGGCGTGATTTTGACACGA | ||
| Primers for qRT-PCR | |||
| RT-KPN_02065-F | CCCCGGGGTGCTATCTCAC | 135 | This study |
| RT-KPN_02065-R | CCGCCCAGGATGGCATTTAT | ||
| RT-KPN_02066-F | TCGCTGCCATACTGACAGGCT | 148 | This study |
| RT-KPN_02066-R | TTCCAGCCATCGTACACGGG | ||
| RT-KP1_3468-F | AAAAATTACGGTGGGTTTTACTGA | 109 | This study |
| RT-KP1_3468-R | ATGCCGCTGAAAAACTGAAC | ||
| RT-KP1_3469-F | TGGATTGGCGTATTATTGAGC | 122 | This study |
| RT-KP1_3469-R | TGAACATAAAGTGCGGTGCT | ||
| RT-phoQ-F | TGCCAGGGAAGCGGACTAC | 100 | This study |
| RT-phoQ-R | GCGGCGGATCAGTGATAAAC | ||
| RT-KP1_0061-F | TTATCACCGACTGCAACACC | 138 | This study |
| RT-KP1_0061-R | CGACATCGGTCTCAATGATG | ||
| RT-KPN-02067-F | GGGAAACCTTAGTGCCAGAGC | 111 | This study |
| RT-KPN-02067-R | TGCGCATCCAGCGTCTGTAA | ||
| RT-rpoB-F | CGCGTATGTCCGATCGAAA | 100 | This study |
| RT-rpoB-F | GCGTCTCAAGGAAGCCATATTC | ||
4.5. Identification of Orthologous Genes
To exclude genes known to be the cause of colistin resistance in K. pneumoniae from complementation analysis, KPN_02065, KPN_02066, KPN-02067, and KP1_3468 genes were matched against the crrAB region and mgrB gene [5,28]. The crrAB region and mgrB sequence database was provided by the National Center for Biotechnology Information (NCBI). A comparison of the orthologous sequence was analyzed by DNAstar EditSeq and MegAline (DNASTAR, Inc., Madison, WI, USA).
4.6. mRNA Expression Analysis
Based on the whole-genome sequencing data, the expression levels of candidate genes associated with colistin resistance were investigated using quantitative real-time PCR in five pairs of isogenic K. pneumoniae strains, as previously described [29]. Briefly, total RNA was used as the template for reverse transcription according to the manufacturer’s protocol of the Omniscript reverse transcriptase kit (Qiagen, Hilden, Germany). SYBR green PCR master mix (Applied Biosystems, CA, USA) was used for the amplifications in a Thermal Cycler Dice Real-Time System TP800 (Takara Bio Inc., Shiga, Japan). The thermal cycling conditions included an initial denaturation step at 95 °C for 10 min, followed by 30 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The fold changes in expression in mutant strains compared to parental strains were determined using the primers listed in Table 4. Gene expression was analyzed as a relative mRNA level normalized to a housekeeping gene, rpoB. Three independent experiments were performed for analysis.
4.7. Complementation Assay
To determine whether the mutation in novel candidate genes was necessary to confer colistin resistance, the pBAD33-derived plasmids were purified from Escherichia coli DH5α (American Type Culture Collection, VA, USA). The plasmid vectors carrying the altered KP1_0061 or KP1_3620 gene were introduced into the 08-B063 strain. To construct the pKP1_0061R vector, PCR-generated fragments harboring the KP1_0061 coding region of the 08-B063R strain were cloned into the SacI and HindIII (New England Biolabs, MA, USA) sites of multiple cloning site (MCS) in the pBAD33 vector. To construct the pKP1_3620R vector, PCR-generated fragments harboring the KP1_3620 coding region of the 08-B063R strain were cloned into the SmaI and PstI site of multiple cloning sites (MCS) in pBAD33 vectors. These vectors were used to transform E. coli DH5α cells. The vectors were purified from the transformants and confirmed using restriction analysis with enzyme treatments and Sanger sequencing. Electroporation of vector into the 08-B063 strain was conducted as described [30]. The transformed cells were selected on an agar plate containing 100 mg/L chloramphenicol. The presence of these plasmids in the colonies was confirmed using restriction analysis and PCR, with the primers pBAD-MCS-F and pBAD-MCS-R. The strains each carrying pBAD33, pKP1_0061R, or pKP1_3620R were grown in the presence of both chloramphenicol 25 mg/L and 1% L-arabinose.
4.8. Colistin Susceptibility Testing
Colistin susceptibility testing was performed as described with some modifications [6]. In brief, the overnight-grown 08-B063 strain carrying the pBAD33, pKP1_0061R, or pKP1_3620R vector was obtained from Mueller Hinton Broth (MHB) in the presence of both chloramphenicol 25 mg/L and 1% L-arabinose. The bacteria were then washed twice and a suspension containing ca. 1.5 × 106 CFU/mL in MHB was prepared. Then, 100 μL of the suspension was placed in each well of a 96-well micro-titer plate and 100 μL MHB or MHB-diluted colistin was added to each well to final concentrations of 1 mg/L of colistin. The plate was incubated at 37 °C for 1 h. Subsequently, 100 μL of the suspension was directly plated on Mueller Hinton agar plates and incubated at 37 °C overnight to determine the number of viable bacteria. The survival rates were expressed as colony counts divided by the number of the same culture treated with MHB and multiplied by 100. The results were shown as the average ± standard deviation. Changes in the colistin susceptibility of mutants complemented with pKP1_0061R or pKP1_3620R vectors were examined using the quantitative bactericidal assay.
5. Conclusions
In all mutants derived from different clones, genetic alterations were observed in well-known phoQ, mgrB, and crrAB genes associated with acquired colistin resistance. In addition, an intrinsic IS5-like element was found in intergenic or intragenic regions of those genes. However, two novel candidate genes such as KP1_0061 and KP1_3620 were discovered in one colistin-resistant mutant. Together with the divergent alterations between two mutants originated from the same parental strain, these results indicate diverse genetic paths to colistin resistance in K. pneumoniae. This study will provide a better understanding of bacterial evolution against colistin resistance in K. pneumoniae.
Acknowledgments
Klebsiella pneumoniae isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID) (Seoul, South Korea).
Author Contributions
Conceptualization, M.C. and K.S.K.; methodology, M.C. and K.S.K.; software, M.C. and K.S.K.; validation, M.C. and K.S.K.; formal analysis, M.C. and K.S.K.; investigation, M.C. and K.S.K.; data curation, M.C. and K.S.K.; writing—original draft preparation, M.C. and K.S.K.; writing—review and editing, M.C. and K.S.K.; visualization, M.C. and K.S.K.; resources, K.S.K.; supervision, K.S.K.; project administration, K.S.K.; funding acquisition, K.S.K. All authors have contributed equally to the writing and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (grant NRF-2019R1A2C2004879).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tzouvelekis L.S., Markogiannakis A., Psichogiou M., Tassios P.T., Daikos G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012;25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nation R.L., Li J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2010;22:535–543. doi: 10.1097/QCO.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park Y.K., Peck K.R., Cheong H.S., Chung D.R., Song J.H., Ko K.S. Extreme drug resistance in Acinetobacter baumannii infections in intensive care units, South Korea. Emerg. Infect. Dis. 2009;15:1325–1327. doi: 10.3201/eid1508.080772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanazawa K., Sato Y., Ohki K., Okimura K., Uchida Y., Shindo M., Sakura N. Contribution of each amino acid residue in polymyxinB(3) to antimicrobial and lipopolysaccharide binding activity. Chem. Pharm. Bull. (Tokyo) 2009;57:240–244. doi: 10.1248/cpb.57.240. [DOI] [PubMed] [Google Scholar]
- 5.Cannatelli A., D’Andrea M.M., Giani T., Di Pilato V., Arena F., Ambretti S., Gaibani P., Rossolini G.M. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 2013;57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H.Y., Chen Y.F., Peng H.L. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 2010;17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalmolin T.V., Lima-Morales D., Barth A.L. Plasmid-mediated colistin resistance: What do we know? J. Infect. 2018;1:16–22. doi: 10.29245/2689-9981/2018/2.1109. [DOI] [Google Scholar]
- 8.Moffatt J.H., Harper M., Harrison P., Hale J.D., Vinogradov E., Seemann T., Henry R., Crane B., St Michael F., Cox A.D., et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannatelli A., Giani T., D’Andrea M.M., Pilato V.D., Arena F., Conte V., Tryfinopoulou K., Vatopoulos A., Rossolini G.M., COLGRIT Study Group MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 2014;58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köser C.U., Ellington M.J., Peacock S.J. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández L., Alvarez-Ortega C., Wiegand I., Olivares J., Kocíncová D., Lam J.S., Martínez J.L., Hancock R.E.W. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013;57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snitkin E.S., Zelany A.M., Gupta J., NISC Comparative Sequencing Program. Palmore T.N., Murray P.R., Segre J.A. Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res. 2013;23:1155–1162. doi: 10.1101/gr.154328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.Y., Na I.Y., Park Y.K., Ko K.S. Genomic variations between colistin-susceptible and -resistant Pseudomonas aeruginosa clinical isolates and their effects on colistin resistance. J. Antimicrob. Chemother. 2014;69:1248–1256. doi: 10.1093/jac/dkt531. [DOI] [PubMed] [Google Scholar]
- 14.López-Camacho E., Gómez-Gil R., Tobes R., Tobes R., Manrique M., Lorenzo M., Galván B., Salvarelli E., Moatassim Y., Salanueva I.J., et al. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J. Antimicrob. Chemother. 2014;69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 15.Ko K.S., Lee J.Y., Baek J.Y., Suh J.Y., Lee M.Y., Choi J.Y., Yeom J.S., Kim Y.S., Jung S.I., Shin S.Y., et al. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 2010;59:822–828. doi: 10.1099/jmm.0.018119-0. [DOI] [PubMed] [Google Scholar]
- 16.Chung D.R., Lee H., Park M.H., Jung S.I., Chang H.H., Kim Y.S., Son J.S., Moon C., Kwon K.T., Ryu S.Y., et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:481–486. doi: 10.1007/s10096-011-1334-7. [DOI] [PubMed] [Google Scholar]
- 17.Hua X., Wang H., Wang C., Shen H., Song Y., Wang X., Shao S., Hua X., Cui L. Global effect of an RNA polymerase β-subunit mutation on gene expression in the radiation-resistant bacterium Deinococcus radiodurans. Sci. China Life Sci. 2011;54:854–862. doi: 10.1007/s11427-011-4209-3. [DOI] [PubMed] [Google Scholar]
- 18.Park Y.K., Choi J.Y., Shin D., Ko K.S. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2011;37:525–530. doi: 10.1016/j.ijantimicag.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Lev A.I., Astashkin E.I., Shaikhutdinova R.Z., Platonov M.E., Kartsev N.N., Volozhantsev N.V., Ershova O.N., Svetoch E.A., Fursova N.K. Identification of IS1R and IS10R elements inserted into ompk36 porin gene of two multidrug-resistant Klebsiella Pneumoniae hospital strains. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx072. [DOI] [PubMed] [Google Scholar]
- 20.Peng H.L., Fu T.F., Liu S.F., Chang H.Y. Cloning and expression of the Klebsiella pneumoniae galactose operon. J. Biochem. 1992;112:604–608. doi: 10.1093/oxfordjournals.jbchem.a123947. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., Li S., Li W., Wang P., Ding P., Li L., Wang J., Yang P., Wang Q., Xu T., et al. RstA, a two-component response regulator, plays important roles in multiple virulence-associated processes in enterohemorrhagic Escherichia coli O157:H7. Gut Pathog. 2019;11:53. doi: 10.1186/s13099-019-0335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y.C., Chang C.K., Chang C.F., Cheng Y.H., Fang P.J., Yu T., Chen S.C., Li Y.C., Hsiao C.D., Huang T. Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element. Nucleic Acids Res. 2014;42:8777–8778. doi: 10.1093/nar/gku572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diancourt L., Passet V., Verhoet J., Grimont P.A.D., Brisse S. Multilocus Sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee J.Y., Song J.H., Ko K.S. Current situation of antimicrobial resistance and genetic differences in Stenotrophomonas maltophilia complex isolates by multilocus variable number of tandem repeat analysis. Infect. Chemother. 2016;48:285–293. doi: 10.3947/ic.2016.48.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute . Performance Standards for Antimicobial Susceptibility Testing: 23rd Informational Supplement M100-S28. CLSI; Wayne, PA, USA: 2018. [Google Scholar]
- 26.Rojas L.J., Salim M., Cober E., Richter S.S., Perez F., Salata R.A., Kalayjian R.C., Watkins R.R., Marshall S., Rudin S.D., et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: Laboratory detection and impact on mortality. Clin. Infect. Dis. 2017;64:711–718. doi: 10.1093/cid/ciw805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behera B., Mathur P., Das A., Kapil A., Gupta B., Bhoi S., Farooque K., Sharma V., Misra M.C. Evaluation of susceptibility testing methods for polymyxin. Int. J. Infect. Dis. 2010;14:e596–e601. doi: 10.1016/j.ijid.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Wright M.S., Suzuki Y., Jones M.B., Marshall S.H., Rudin S.D., van Duin D., Kay K., Jacobs M.R., Bonomo R.A., Adams M.D. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 2015;59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S.J., Ko K.S. Diverse genetic alterations responsible for post-exposure colistin resistance in populations of the same strain of Klebsiella pneumoniae. Int. J. Antimicrob Agents. 2018;52:425–429. doi: 10.1016/j.ijantimicag.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Pontel L.B., Pezza A., Soncini F.C. Copper stress targets the rcs system to induce multiaggregative behavior in a copper-sensitive Salmonella strain. J. Bacteriol. 2010;192:6287–6290. doi: 10.1128/JB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]