Abstract
Campylobacter jejuni is a major foodborne pathogen and common cause of bacterial enteritis worldwide. A total of 622 C. jejuni isolates recovered from food animals and retail meats in the United States through the National Antimicrobial Resistance Monitoring System between 2013 and 2017 were sequenced using an Illumina MiSeq. Sequences were combined with WGS data of 222 human isolates downloaded from NCBI and analyzed by core genome multilocus sequence typing (cgMLST) and traditional MLST. cgMLST allelic difference (AD) thresholds of 0, 5, 10, 25, 50, 100 and 200 identified 828, 734, 652, 543, 422, 298 and 197 cgMLST types among the 844 isolates, respectively, and traditional MLST identified 174 ST. The cgMLST scheme allowing an AD of 200 (cgMLST200) revealed strong correlation with MLST. cgMLST200 showed 40.5% retail chicken isolates, 56.5% swine, 77.4% dairy cattle and 78.9% beef cattle isolates shared cgMLST sequence type with human isolates. All ST-8 had the same cgMLST200 type (cgMLST200-12) and 74.3% of ST-8 and 75% cgMLST200-12 were confirmed as sheep abortion virulence clones by PorA analysis. Twenty-nine acquired resistance genes, including 21 alleles of blaOXA, tetO, aph(3′)-IIIa, ant(6)-Ia, aadE, aad9, aph(2′)-Ig, aph(2′)-Ih, sat4 plus mutations in gyrA, 23SrRNA and L22 were identified. Resistance genotypes were strongly linked with cgMLST200 type for certain groups including 12/12 cgMLST200-510 with the A103V substitution in L22 and 10/11 cgMLST200-608 with the T86I GyrA substitution associated with macrolide and quinolone resistance, respectively. In summary, the cgMLST200 threshold scheme combined with resistance genotype information could provide an excellent subtyping scheme for source attribution of human C. jejuni infections.
Keywords: cgMLST, Campylobacter jejuni, source attribution, foodborne pathogens, PorA
1. Introduction
Campylobacter sp. are one of the leading causes of foodborne illness in the United States, causing an estimated 1.5 million infections each year [1,2]. Worldwide, campylobacteriosis is the most common cause of bacterial enteritis in humans with C. jejuni being the major contributor [3]. In otherwise healthy individuals, C. jejuni infections commonly result in a self-limiting diarrheal disease that is believed to commonly go unreported [4]. In immunocompromised patients, however, untreated C. jejuni infections can progress to sepsis and in rare cases meningitis [5,6]. Further, C. jejuni is associated with several sequelae including reactive arthritis, Guillain-Barre syndrome, Miller-Fisher syndrome and prolonged exposure in infants is associated with growth deficits [7,8,9]. Although considered a foodborne pathogen in humans, C. jejuni can be acquired from a wide range of sources including retail meats, raw milk, companion animals, fecal contamination, wild birds and other environmental reservoirs [10,11]. C. jejuni is also increasingly viewed as a reservoir for antibiotic resistance genes in both the environment and the food supply chain [2,12]. As a naturally competent organism, C. jejuni is capable of incorporating exogenous DNA during logarithmic growth [13]. This ability to readily acquire resistance genes from the environment enables Campylobacter strains to adapt to a range of antibiotic selective pressures found across multiple types of food animal facilities. Campylobacter species are also able to colonize food animals asymptomatically as a component of the gut microbiota and persist in the production environment with no apparent cost to the host [14,15].
Studies evaluating the prevalence of Campylobacter species in retail meats have shown higher rates in raw chicken (44%) compared to beef (0.5%) and pork (0.4%) sources [16]. While exposure to retail chicken meat is a major risk factor for campylobacteriosis, many isolates from chicken carcasses at food processing sites and retail chicken products do not show genetic relatedness to human pathogenic strains [17,18,19,20,21]. Strains recovered from beef cattle, dairy cattle, chickens, turkeys, swine, sheep and goats have all been linked to disease in humans [20,22]. This plurality of reservoirs demonstrates the need to evaluate the pathogenic contribution of each source to inform better practices and reduce the burden of campylobacteriosis on human health. The United States Department of Agriculture Food Safety and Inspection Service (USDA-FSIS) NARMS screening for foodborne pathogens in the ceca of several major food animals revealed prevalence of Campylobacter species in beef cattle (42%), dairy cattle (43%) and swine (32%) at much higher rates than found in retail meats, suggesting that infection from non-poultry animal sources may be underestimated [23]. The NARMS studies showed that high prevalence populations of C. jejuni were present in more food animal sources than previously observed. This broad range of food animal hosts combined with the potential for asymptomatic carriage and environmental contamination all confound efforts to identify the source of Campylobacter infections.
Many C. jejuni strains exhibit a narrow range of host specificity and this trait can be used to inform source attribution of strains that cause human infections [24,25]. However, strains with close genetic similarity can infect different hosts and established typing methods such as pulsed field gel electrophoresis (PFGE) or seven-gene multilocus sequence typing (MLST) lack the resolution to distinguish between them [10,18,26,27,28,29]. One proposed solution to this limitation is core genome fingerprinting, a method that expands on the seven loci of C. jejuni MLST to include 40 genomic targets conserved across C. jejuni genomes [29]. While this aids in strain differentiation, it also results in a labor-intensive set of five multiplexed PCR reactions. Analyses of the variable regions surrounding the fla gene or CRISPR loci have also been introduced, but these are often used in conjunction with MLST and incur a cost in labor [30].
In the last several years, whole genome sequencing (WGS) technology has been implemented in many public health surveillance programs as a fast and affordable alternative to PFGE and MLST [31,32]. Using WGS data for analysis can overcome the limitations of PFGE and MLST by evaluating the entire genome containing all housekeeping, virulence and resistance genes. Specifically, cgMLST typing schemes using a core genome of 1343 loci found in >95% of C. jejuni strains have demonstrated an increased discriminatory power over traditional MLST [33]. In a comparison of strains from multiple campylobacteriosis outbreaks, this cgMLST application was able to group strains from the same outbreak event into distinct clusters. We propose to explore the cgMLST approach further by determining whether the ability of cgMLST to establish genomic relatedness could be used to inform the source attribution of human pathogenic strains of C. jejuni. In the U.S., the contributions of PulseNet and the National Antimicrobial Resistance Monitoring System (NARMS) surveillance programs to the WGS repository maintained by NCBI provide an excellent source of foodborne pathogen sequence data against which to validate our cgMLST typing scheme. The objective of this study is to evaluate the utility of a cgMLST typing scheme for the source attribution of human pathogenic strains of C. jejuni.
2. Results
2.1. MLST and cgMLST Diversity Indices
The discriminatory power of cgMLST typing was determined by the number of AD between C. jejuni strains (Table S1). We compared and evaluated traditional seven-gene MLST and cgMLST schema allowing for a range of 0 to 200 AD using Simpson’s index of diversity (Table 1). With regards to this dataset, a diversity index (D.I.) score of 1.000 indicated each sequence type contained no more than two strains while a D.I. lower than 1.000 represented a greater number of strains belonging to fewer conserved, groups. All cgMLST allelic difference thresholds evaluated resulted in a greater discriminatory power than traditional MLST. These results showed that the cgMLST scheme allowing an allelic difference of 200 (cgMLST200) had a discriminatory power most like traditional MLST with Simpson’s D.I. values of 0.964 vs. 0.963. The cgMLST200 schema classified the strains into 197 groups while traditional MLST generated 174 groups. Further, decreasing the allowable allelic differences generated a more stringent typing scheme resulting in an increasing number of highly related groups. It has been proposed that using AD less than 10 can be used for foodborne outbreak investigation [34].
Table 1.
Comparison of MLST and cgMLST Typing Methods.
| Typing Scheme | Schema Diversity | Number of Groups Generated | |||||
|---|---|---|---|---|---|---|---|
| Number of Groups Identified * | Simpson’s D.I. | Beef | Chicken | Dairy | Human | Swine | |
| MLST | 174 | 0.963 | 33 | 54 | 34 | 64 | 16 |
| cgMLST200 | 197 | 0.964 | 38 | 102 | 40 | 87 | 18 |
| cgMLST100 | 298 | 0.983 | 73 | 131 | 68 | 103 | 22 |
| cgMLST50 | 422 | 0.991 | 115 | 150 | 107 | 122 | 22 |
| cgMLST25 | 543 | 0.997 | 149 | 168 | 141 | 145 | 23 |
| cgMLST10 | 652 | 0.999 | 175 | 187 | 162 | 156 | 23 |
| cgMLST5 | 734 | 0.999 | 196 | 196 | 173 | 175 | 23 |
| cgMLST0 | 828 | 1 | 211 | 205 | 180 | 216 | 23 |
* Typing groups may be found in multiple hosts.
2.2. Typing Schema and Source Attribution
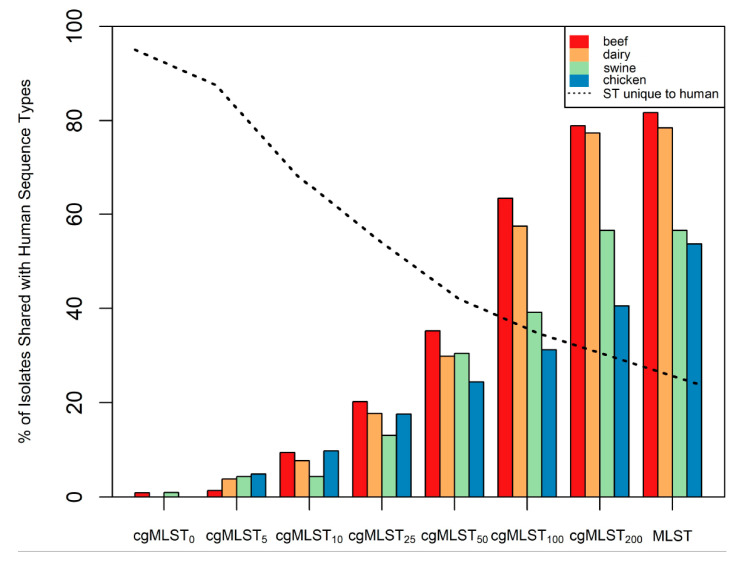
To demonstrate the utility of the different typing methods for identifying the source of infection, typing groups from cgMLST and traditional MLST were evaluated by host origin. Specifically, cgMLST groups were classified by the property of containing strains isolated from humans. The ratio of isolates recovered from humans to isolates from each of the other sources was determined using cgMLST and traditional MLST (Figure 1). Traditional MLST and cgMLST200 typing methods showed similar representation of human pathogenic strains across all four food animal sources. The cgMLST200 scheme showed that 40.5% of isolates from retail chicken, 56.5% from swine, 77.4% from dairy cattle and 78.9% from beef cattle shared the same cgMLST type with isolates recovered from humans. In comparison, traditional MLST showed 53.6% of isolates from retail chicken, 56.5% from swine, 78.4% from dairy cattle and 81.7% from beef cattle were classified in human pathogenic typing groups. The prevalence of human pathogenic strains in these food animal sources is similar to previous analyses using a PFGE typing method [18]. Further, the similar contribution of human pathogenic strains between beef cattle and dairy cattle reflect a conserved composition of C. jejuni strains, as indicated by population analysis (Figure S1).
Figure 1.
Proportion of C. jejuni from each source sharing a typing group with strains known to cause disease in humans. Column height indicates the percentage of strains from each source (beef, dairy, chicken or swine) that were classified in a group that contained human pathogenic strains of C. jejuni. The dashed line indicates the percentage of strains found in typing groups unique to humans.
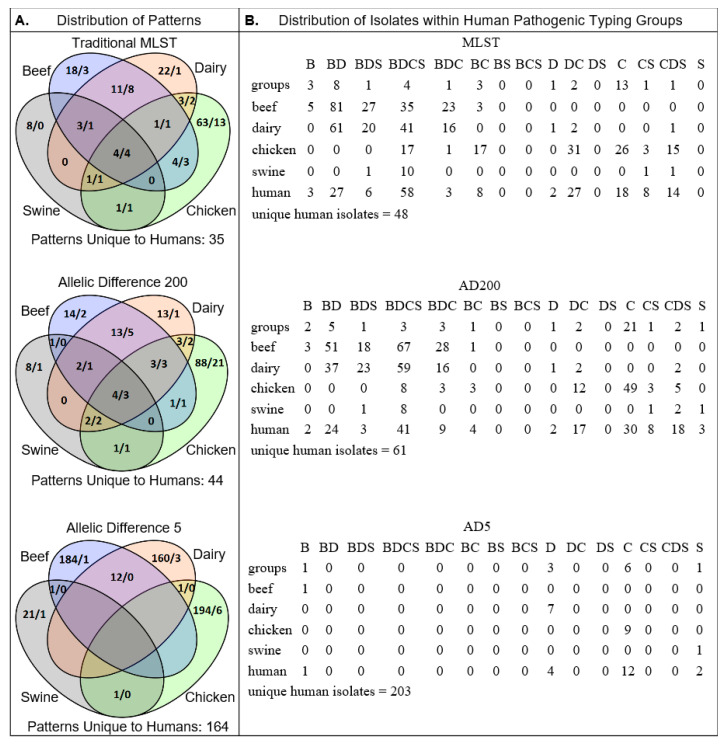
The similarities between traditional MLST and cgMLST were consistent with the diversity indices calculated earlier (Table 1). The stringency of grouping criteria (i.e., the threshold of differences between groups) reflected the number of isolates from non-human sources categorized into human pathogenic groups (Figure S3). Specific to groups containing isolates from humans, the cgMLST200 scheme identified 87 groups encompassing 404 strains, 222 of which were isolated from a human source (55%) while the more stringent cgMLST5 scheme identified 175 groups with 243 strains, 222 from a human source (91.4%). This trend of increased stringency favoring a more homologous source composition was conserved for all C. jejuni sources evaluated (Figure 2).
Figure 2.
(A) Venn diagrams showing the number of cgMLST patterns associated with each source for traditional MLST (upper), cgMLST200 (middle) and cgMLST5 (lower). Patterns are numbered by total (left) followed by those shared with human pathogenic strains (right). (B) Distribution of individual strains belonging to the human pathogenic patterns in A. Isolates recovered from humans found in typing groups unique to humans are listed below for each typing scheme. Group totals correspond with A. and source identification is as follows: Beef (B), Dairy (D), Swine (S) and Chicken (C).
2.3. Validation of Typing Schema
One test of the utility of a typing scheme is its ability to categorize strains with phenotypically distinct traits into discrete groups. Traditional MLST has been shown to successfully categorize the sheep abortion (SA) strains of C. jejuni into ST-8 [35]. To validate the typing utility of the cgMLST scheme, we evaluated the distribution of strains with the SA-associated PorA sequence across both cgMLST200 and seven-gene MLST sequence types. All 74 strains identified as ST-8, as sequence type linked to the SA-associated PorA sequence, were classified into a single cgMLST200 group, cgMLST200-12. Within the entire population, 87/844 (10.3%) of the strains encoded for the SA-associated PorA sequence. The MLST-8 group contained 74 strains, 55 (74.3%) of which encoded the SA-associated PorA sequence (Table 2). The remaining 13 strains encoding for the SA-associated PorA sequence were found in ST 577, 61, 1244, 459 and 922 (Table S2). The cgMLST200-12 group contained 76 strains, 57 of which (75%) had the SA-associated PorA, showing the cgMLST200-12 to be slightly better for classifying SA clones in this set of 844 Campylobacter genomes. Overlay of PorA annotation data onto a minimum spanning tree of cgMLST type revealed two clusters harboring the majority of strains encoding for the SA-associated PorA sequence: cgMLST200-12 and cgMLST200-50 (Figure S2). When the allelic difference threshold was decreased from 200 to 50, stronger associations between typing groups were observed. For example, 51/55 (92.7%) of cgMLST50-136, 9/10 (90%) of cgMLST50-15 and 6/7 (85.7%) cgMLST50-51 strains encoded for the SA-associated PorA sequence. The cgMLST50-136 group contained one of the isolates with the SA-associated PorA sequence not accounted for in the traditional ST-8 group. Together, these results indicate that cgMLST200-12 and ST-8 predominate but not exclusively harbor the SA-associated PorA variant.
Table 2.
Alignment of PorA SA Determinant Region between Sequence Types.
| Sequence a,b,c | cgMLST200-12 vs. MLST-8 d |
Description | |
|---|---|---|---|
| 170- | * * * * * * * * * * * | −209 | Conserved residues |
| NCTC11168 | FMAAEQGADLLEHSNISTTS NQAPFKVDSVGNLY | Reference sequence | |
| N48272F | FMAEEQGADLLGKST ISTTQKAAPFQADSLGNLY | 55:53 | SA clonal sequence |
| N43804F | FMAEEQGADLLGKST ISIT QKAAPFQADSLGNLY | 1:1 | Substitution not in SA loci |
| N45191FR | FMAEEQGT DLLGKST ISTTQKAAPFQADSLGNLY | 1:1 | Substitution not in SA loci |
| N46355F | FMAEEQGADLLGKSTISTT QKAAPFQTNSLGNLY | 1:1 | Substitution in SA loci |
| N49694F | FMEKEQ I S DLVG SNSSTFNVDSI GNLY | 16:16 | Deletion and substitutions in SA loci |
| N44409F | FMAAEQSS DLVG ANGSAFKVDSI ENLY | 1:1 | Deletion and substitutions in SA loci |
| N45200F | FMAKEQGSDLVG ANGSAFNVDSIGNLY | 1:1 | Deletion and substitutions in SA loci |
a * indicates conserved residue at sequence position, b Bold formatting indicates amino acid substitution associated with the SA phenotype, c Underline formatting indicates amino acid substitution not associated with the SA phenotype, d cgMLST200-12 (left) and MLST-8 (right) isolates identified with the corresponding PorA aa sequence.
2.4. Antimicrobial Resistance Genotypes and cgMLST
A total of 29 unique antimicrobial resistance genes were identified in the 844 Campylobacter genomes, including 21 alleles of blaOXA, tetO, aph(3′)-IIIa, aadE, aad9, aph(2′)-Ih, aph(2′)-h and sat4. Antimicrobial resistance mutations and substitutions were observed among 271 strains, including GyrA at aa 68 (n = 168), 23SrRNA at bp 2074 (n = 3), 23SrRNA at bp 2075 (n = 8), 50SL22 at aa 103 (n = 112), RpsL at aa 43 (n = 2) and RpsL at aa 88 (n = 2). Certain cgMLST200 groups had conserved antimicrobial resistance genotypes. In total, 12/12 cgMLST200-4 and 6/6 cgMLST200-584 strains carried both blaOXA-193 and tetO, 12/12 cgMLST200-510 had the A103V substitution in L22, and 10/11 cgMLST200-608 the T86I substitution in GyrA. The tetO gene was also present in 74/76 cgMLST200-12 isolates. Within the population of 844 Campylobacter isolates, 636 encoded for a blaOXA gene. The most prevalent allele was blaOXA-193, present in 398 strains in all five host types evaluated. The remaining blaOXA alleles demonstrated specificity towards certain human sequence types. Of the Campylobacter strains isolated from humans, 10/10 cgMLST200-86 encoded blaOXA-61, 8/8 cgMLST200-510 encoded blaOXA-603, 5/5 cgMLST200-76 encoded blaOXA-449 and 4/4 cgMLST200-435 encoded blaOXA-447 (Table S1).
The above antibiotic resistance patterns were then evaluated to determine if antibiotic resistance determinants could be used in conjunction with sequence type as an indicator of isolate source. The cgMLST200-4 group isolates were obtained from all host types evaluated, including beef, chicken, dairy, swine and human while the cgMLST200-584 group were obtained from either chicken or human. cgMLST200-510 isolates were obtained from chicken, swine and human sources and the cgMLST200-608 type was only found in chicken. Evaluating the blaOXA family of genes, only blaOXA-61 was able to help inform host source as no dairy cattle from that group encoded blaOXA-61. These data indicate that while antibiotic resistance determinants were restricted to sequence types, the distribution of alleles within a sequence type was not limited to a specific host.
3. Discussion
The cgMLST scheme described here provides a convenient strategy for strain typing by evaluating the 1343 loci present in the core genome of C. jejuni. This increased resolution over traditional seven-gene MLST allows cgMLST typing schemes to more accurately inform the source of human pathogenic C. jejuni strains. One factor that complicates the use of traditional MLST for source attribution is the ability of C. jejuni to infect multiple host types [36]. Using the cgMLST approach, we have demonstrated that allowing fewer allelic differences between isolates segregates strains from different sources into discrete groups (Figure 2). Further, source composition analysis of the typing groups reveals which sources harbor the greatest number of human pathogenic strains. In addition to reducing the number of potential sources, the allelic threshold informs the degree of genetic relatedness between human pathogenic strains and human non-pathogenic strains from known sources (Table 3).
Table 3.
Source Attribution of C. jejuni Isolates across Multiple Typing Schema.
| Strain ID | MLST | cgMLST200 | cgMLST5 | cgMLST0 | ||||
|---|---|---|---|---|---|---|---|---|
| Group ID | Source * | Group ID | Source | Group ID | Source | Group ID | Source | |
| N46804F | 982 | B,D,H | 28 | B,C,D,H,S | 57 | D,H | 250 | D |
| SRR5217159 | 982 | B,D,H | 28 | B,C,D,H,S | 57 | D,H | 754 | H |
| N45809F | 982 | B,D,H | 28 | B,C,D,H,S | 209 | D,H | 209 | D |
| SRR5878335 | 982 | B,D,H | 28 | B,C,D,H,S | 209 | D,H | 826 | H |
| N48272F | 8 | B,C,D,H,S | 12 | B,C,D,H,S | 223 | D,H | 291 | D |
| SRR2970498 | 8 | B,C,D,H,S | 12 | B,C,D,H,S | 223 | D,H | 528 | H |
| N50152F | 806 | B,D,H,S | 18 | B,D,H,S | 362 | B,H | 362 | B |
| SRR5604241 | 806 | B,D,H,S | 18 | B,D,H,S | 362 | B,H | 806 | H |
| SRR5152202 | 50 | C,D,H | 100 | C,D,H | 400 | C,H | 719 | C |
| SRR1794080 | 50 | C,D,H | 100 | C,D,H | 400 | C,H | 400 | H |
| SRR2075414 | 45 | B,C,D,H,S | 407 | C,H | 407 | C,H | 407 | C |
| SRR3029056 | 45 | B,C,D,H,S | 407 | C,H | 407 | C,H | 407 | H |
| SRR2094236 | 353 | C,D,H | 412 | C,H | 412 | C,H | 412 | C |
| SRR2182579 | 353 | C,D,H | 412 | C,H | 412 | C,H | 444 | H |
| SRR3092138 | 52 | C,H,S | 510 | C,H,S | 512 | S,H | 822 | S |
| SRR5754156 | 52 | C,H,S | 510 | C,H,S | 512 | S,H | 551 | H |
| SRR5152177 | 2083 | C,H | 584 | C,H | 593 | C,H | 714 | C |
| SRR5932391 | 2083 | C,H | 584 | C,H | 593 | C,H | 833 | H |
* Beef (B), Dairy (D), Human (H), Chicken (C) and Swine (S).
Earlier approaches to increase the resolution of sequence typing methods have incorporated genomic determinants of antibiotic resistance [24,25]. In this study, 102 unique combinations of antibiotic resistance determinants were present in our population of C. jejuni isolates, with tet(O) and blaoxa alleles present in isolates obtained from each source evaluated (Table S3). Our data revealed an interesting trend in antibiotic resistance between the food animal groups. Specifically, none of the determinants of macrolide resistance in the AMRFinder database were found in either beef or dairy cattle, food animal sources where macrolides are commonly administered by injection Although macrolides (e.g., tylosin) can apply selective pressure on C. jejuni during food animal production, the associated resistance determinants were absent in isolates recovered from the beef and dairy cattle populations in our study [37,38]. Interestingly, while two-thirds of the strains with macrolide resistance determinants were isolated from chicken sources, these same substitutions have been shown to incur a fitness cost and growth rate decrease in chickens in the absence of antibiotic selection pressure [39]. Strains lacking these substitutions have been shown to outcompete macrolide-resistant mutant strains [40,41]. In light of this, the absence of macrolide resistance determinants in isolates from our cattle populations compared to a 52.7% prevalence in our population of chicken isolates suggests that the distribution of macrolide resistance between these animal sources is not random.
One limitation of this study is that we do not have direct epidemiological evidence linking a pathogen from a specific source and sequence type to human disease. cgMLST shows strong genetic relatedness between human and non-human sourced isolates of C. jejuni; however, we currently lack the epidemiological metadata necessary to determine causality. We may conclude that there are strains of C. jejuni isolated from humans very closely related to strains isolated from food animal sources, differing only by the sequence of five alleles. A further limitation of the study is the underrepresentation of C. jejuni isolates from swine. While the sample size is too small to draw conclusions from our swine strains, we have demonstrated that a subset of isolates capable of infecting swine share a close genetic relatedness (i.e., AD < 5) with isolates that also infect humans, beef cattle, dairy cattle and chickens. The genomic similarity of these strains from different hosts suggests that these sequence types identify strains with a less restricted host range.
Source attribution of C. jejuni infections allows food industry professionals to tailor their practices to prevent the spread of foodborne pathogens. It also enables healthcare professionals to more efficiently use resources to contain the source of the disease. Similar to previous work, we used cgMLST to identify groups of genetically related strains of human pathogenic C. jejuni [33]. Expanding on this, we were also able to evaluate the cooccurrence of cgMLST groups, host source and antibiotic resistance determinants to identify which food sources harbor strains most closely related to pathogens that cause disease in humans. Our data demonstrate that evaluating Campylobacter strains across the 1343 loci of the cgMLST typing scheme provides a higher resolution method to determine genomic relatedness compared to traditional seven-gene MLST. Additionally, cgMLST typing has the potential to inform clinical decisions as the associations between cgMLST type and antibiotic resistance are established. Finally, as WGS data become more widely available, cgMLST typing serves as a cost-effective method for comparing multiple stains of C. jejuni. In conclusion, the cgMLST200 threshold combined with resistance genotype could provide an excellent subtyping scheme for source attribution of human C. jejuni infections.
4. Materials and Methods
4.1. Bacterial Strains and Sequencing
A total of 622 C. jejuni isolates recovered from food animals and retail meats were collected through NARMS starting in 2013 [42]. C. jejuni strains were obtained from the following sources: beef cattle cecal isolates (n = 213), dairy cattle cecal isolates (n = 181), chicken cecal isolates (n = 11), chicken retail meat isolates (n = 194) and swine cecal isolates (n = 23). Genomic DNA was extracted using the Qiagen DNeasy Blood and Tissue kit (Qiagen, Gaithersburg, MD) and genomes were sequenced on an Illumina MiSeq using v3 chemistry (Illumina, San Diego, CA). C. jejuni genomes were assembled using the de Brujin graph assembler from the CLC Genomics Workbench version 8.0 (CLC bio Aarhus, Denmark). These sequence data with a mean coverage of 51.37 were added to a collection of 222 sequences of human pathogenic C. jejuni that were submitted to the National Center for Biotechnology Information (NCBI) by the Centers for Disease Control and Prevention (CDC) to generate the pool of 844 genomes used in this project.
4.2. Traditional Multilocus Sequence Typing
The analysis of traditional seven-gene MLST was performed using whole-genome sequencing (WGS) data. Campylobacter multilocus sequence typing (MLST) allelic profiles and sequences were downloaded from the PubMLST database [43]. A total of 9098 profiles for seven different loci were used for the MLST analysis. The SRST2 pipeline was used to determine the MLST type for our Campylobacter isolates [44].
4.3. Core Genome MLST
The cgMLST scheme of 1343 loci, defined from 2742 Campylobacter isolates from Oxfordshire, UK, between 2011 and 2014, was downloaded from the PubMLST online database and queried against our collection of 844 C. jejuni using a BLAST search [33,43]. As the core genome represents genes present in >95% C. jejuni strains, up to 100 missing alleles were allowed for each genome [33]. The core-genome sequence type (cgST) for each isolate was determined based on the cgMLST scheme and a new cgST was assigned to a genome if the combination of alleles for the genome was not found in the cgMLST scheme. The cgST and allele composition for all genomes were assigned to the allelic profiles where missing alleles were represented with an “N”. Graphical representations of the population as minimum spanning trees from cgMLST data were generated using GrapeTree [45].
4.4. Hierarchical Clustering of cgMLST Data
Distance between genomes was calculated based on the pairwise distance between allelic profiles and was measured as allelic difference (AD). All genomes were clustered on seven different levels of AD: 0, 5, 10, 25, 50, 100 and 200 using the single-linage clustering method. With this method, a genome was linked to a group if the genome had less than or equal to the threshold number of AD with at least one other genome of that group. Genomes clustered together at each AD threshold were assigned the same group ID.
4.5. Analysis of Population Composition
Underlying population composition was determined using STRUCTURE software v2.3.4 to analyze allele profiles across the 1343 loci of the C. jejuni core genome for the 844 strains present in this study. Software parameters were informed by prior source attribution studies and are described as follows [46]. The program was run using 50,000 burn-in iterations followed by 50,000 iterations applying a no-admixture model and assuming allele frequencies were independent. Source population was used to inform the analysis, however strains originating from humans were not used in model generation.
4.6. Identification of Antimicrobial Resistance Genotypes
Antimicrobial resistance genes were identified using the AMRFinder tool [47] that uses a curated AMR gene reference database of 4579 antimicrobial resistance proteins, more than 560 hidden Markov models (HMMs) and a curated protein family hierarchy. AMRFinder was also used to identify antibiotic resistance determinants in Campylobacter, including mutations and substitutions in the gyrA gene, 23S ribosomal RNA, 50S ribosomal protein L22 and 30S ribosomal protein S12 RpsL.
4.7. PorA Analysis
The sequence variation of the major outer membrane protein (PorA) is implicated in pathogenesis of C. jejuni [48]. The PorA sequence of the reference strain NCTC11168 was downloaded from NCBI GenBank as a wild-type comparator. This sequence was compared to our collection of 844 C. jejuni using an in-house perl script to determine the population-wide distribution of allelic variations of the loop 4 region in PorA, a region associated with a C. jejuni phenotype that can result in abortions in sheep. Isolates encoding a PorA sequence with less than 70% identity to the comparator strain NCTC11168 were excluded, leaving 739 Campylobacter isolates used in the PorA analysis. The loop 4 region of the sheep abortion (SA) associated PorA sequence was queried against the remaining C. jejuni sequences using blastx, and the results were aligned with ClustalW. The sequences were then visualized using MAFFT through EMBL to identify isolates with the SA-associated PorA sequence [49]. Sequence typing results from MLST and cgMLST were compared against the presence of the SA-associated PorA sequence to evaluate the ability of each typing scheme to identify SA strains of C. jejuni.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/7/532/s1, Figure S1: Cluster Distribution by Population. Distribution of conserved population clusters as determined by analysis of cgMLST allele profiles. Cluster frequencies are shown both by (A) aggregate population and (B) source animal contribution. (C) Proportional contribution of clusters illustrates differences in population composition between animal sources, Figure S2: Minimum spanning trees of C. jejuni isolates as constructed from cgMLST profiles represented by (A) isolation source, (B) most populous cgMLST200 group and (C) presence of SA-associated PorA sequence, Figure S3: Distribution of sequence type groups (large lettering, above) and isolate composition of human pathogenic groups (small lettering, below) across multiple food animal sources for different cgMLST thresholds. The total number of groups for each food source is shown to the left of the “/” and the number of groups that contain human pathogenic isolates are on the right. Isolate-level composition of human pathogenic groups below are displayed in the order of “a/b/c/d/e” corresponding to the number of isolates from (a) beef cattle, (b) dairy cattle, (c) chicken, (d) swine and (e) human, Table S1: Sequence Type and Metadata for C. jejuni Isolates, Table S2: MLST Allelic Profiles of Sequence Types Containing Strains Encoding for the SA-associated PorA Sequence, Table S3: Distribution of C. jejuni AMR Determinant Profiles across Source Animal.
Author Contributions
Conceptualization, C.-H.H. and S.Z.; Data curation, C.-H.H., L.H. and S.M.; Formal analysis, C.-H.H., L.H., S.M. and S.Z.; Funding acquisition, E.S., P.M. and S.Z.; Investigation, C.-H.H., L.H., S.M. and S.Z.; Methodology, C.-H.H., L.H. and S.Z.; Project administration, E.S., P.M. and S.Z.; Resources, E.S., P.M. and S.Z.; Software, C.-H.H.; Supervision, E.S., P.M. and S.Z.; Validation, C.-H.H., L.H. and S.M.; Visualization, C.-H.H. and L.H.; Writing—original draft, L.H. and S.Z.; Writing—review & editing, C.-H.H., L.H., S.M., E.S., P.M., Q.Z. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding and the APC was funded by the FDA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tack D.M., Marder E.P., Griffin P.M., Cieslak P.R., Dunn J., Hurd S., Scallan E., Lathrop S., Muse A., Ryan P. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2015–2018. Morb. Mortal. Wkly. Rep. 2019;68:369. doi: 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC . In: Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services, editor. CDC; Atlanta, GA, USA: 2019. [Google Scholar]
- 3.WHO Campylobacter. [(accessed on 3 January 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/campylobacter.
- 4.Fischer G.H., Paterek E. StatPearls [Internet] StatPearls Publishing; Treasure Island, FL, USA: 2019. Campylobacter. [Google Scholar]
- 5.Gallo M.T., Di Domenico E.G., Toma L., Marchesi F., Pelagalli L., Manghisi N., Ascenzioni F., Prignano G., Mengarelli A., Ensoli F. Campylobacter jejuni fatal sepsis in a patient with Non-Hodgkin’s lymphoma: Case report and literature review of a difficult diagnosis. Int. J. Mol. Sci. 2016;17:544. doi: 10.3390/ijms17040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusulja M., Santini M., Margetić K., Guzvinec M., Šoprek S., Butić I., Tambić Andrašević A. Meningitis caused by Campylobacter jejuni: A case presentation and literature review. Acta Clin. Belg. 2020:1–6. doi: 10.1080/17843286.2020.1721133. [DOI] [PubMed] [Google Scholar]
- 7.Ueno T., Kon T., Kurihara A.-I., Tomiyama M. Unilateral oculomotor nerve palsy following Campylobacter infection: A mild form of Miller Fisher syndrome without ataxia. Intern. Med. 2017;56:2929–2932. doi: 10.2169/internalmedicine.8824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St Charles J., Bell J., Gadsden B., Malik A., Cooke H., Van de Grift L., Kim H., Smith E., Mansfield L. Guillain Barré Syndrome is induced in Non-Obese Diabetic (NOD) mice following Campylobacter jejuni infection and is exacerbated by antibiotics. J. Autoimmun. 2017;77:11–38. doi: 10.1016/j.jaut.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Rouhani S., Griffin N.W., Yori P.P., Olortegui M.P., Siguas Salas M., Rengifo Trigoso D., Moulton L.H., Houpt E.R., Barratt M.J., Kosek M.N., et al. Gut microbiota features associated with Campylobacter burden and postnatal linear growth deficits in a Peruvian birth cohort. Eur. PMC. 2019 doi: 10.1093/cid/ciz906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thépault A., Méric G., Rivoal K., Pascoe B., Mageiros L., Touzain F., Rose V., Béven V., Chemaly M., Sheppard S.K. Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl. Environ. Microbiol. 2017;83:e03085–e030816. doi: 10.1128/AEM.03085-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thépault A., Rose V., Queguiner M., Chemaly M., Rivoal K. Dogs and Cats: Reservoirs for Highly Diverse Campylobacter jejuni and a Potential Source of Human Exposure. Animals. 2020;10:838. doi: 10.3390/ani10050838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mourkas E., Florez-Cuadrado D., Pascoe B., Calland J.K., Bayliss S.C., Mageiros L., Méric G., Hitchings M.D., Quesada A., Porrero C. Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ. Microbiol. 2019;21:4597–4613. doi: 10.1111/1462-2920.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beauchamp J.M., Leveque R.M., Dawid S., DiRita V.J. Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni. Proc. Natl. Acad. Sci. USA. 2017;114:E8053–E8061. doi: 10.1073/pnas.1703331114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Sánchez L., Melero B., Jaime I., Hänninen M.-L., Rossi M., Rovira J. Campylobacter jejuni survival in a poultry processing plant environment. Food Microbiol. 2017;65:185–192. doi: 10.1016/j.fm.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Mao S., Zhang M., Liu J., Zhu W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015;5:16116. doi: 10.1038/srep16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FDA . National Antimicrobial Resistance Monitoring System-Enteric Bacteria (NARMS): 2007 Executive Report. Department of Health and Human Services, US FDA; Rockville, MD, USA: 2010. [Google Scholar]
- 17.Hänninen M.-L., Pajarre S., Klossner M.-L., Rautelin H. Typing of Human Campylobacter jejuniIsolates in Finland by Pulsed-Field Gel Electrophoresis. J. Clin. Microbiol. 1998;36:1787–1789. doi: 10.1128/JCM.36.6.1787-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyson G.H., Tate H.P., Abbott J., Tran T.-T., Kabera C., Crarey E., Young S., McDERMOTT P.F., Sprague G., Campbell M. Molecular subtyping and source attribution of Campylobacter isolated from food animals. J. Food Prot. 2016;79:1891–1897. doi: 10.4315/0362-028X.JFP-16-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyhs U., Katzav M., Isohanni P., Heiska H., Maijala R. The temporal, PFGE and resistance pattern associations suggest that poultry products are only a minor source of human infections in western Finland. Food Microbiol. 2010;27:311–315. doi: 10.1016/j.fm.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Mughini Gras L., Smid J.H., Wagenaar J.A., de Boer A.G., Havelaar A.H., Friesema I.H., French N.P., Busani L., van Pelt W. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: A combined case-control and source attribution analysis. PLoS ONE. 2012;7:e42599. doi: 10.1371/journal.pone.0042599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Giannatale E., Garofolo G., Alessiani A., Di Donato G., Candeloro L., Vencia W., Decastelli L., Marotta F. Tracing back clinical Campylobacter jejuni in the Northwest of Italy and assessing their potential source. Front. Microbiol. 2016;7:887. doi: 10.3389/fmicb.2016.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris N.V., Kimball T.J., Bennett P., Johnson Y., Wakely D., Nolan C.M. Campylobacter jejuni enteritis associated with raw goat’s milk. Am. J. Epidemiol. 1987;126:179–186. doi: 10.1093/aje/126.2.179. [DOI] [PubMed] [Google Scholar]
- 23.The National Antimicrobial Resistance Monitoring System: Enteric Bacteria. FDA; Silver Spring, MD, USA: 2013. [Google Scholar]
- 24.Korczak B.M., Zurfluh M., Emler S., Kuhn-Oertli J., Kuhnert P. Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J. Clin. Microbiol. 2009;47:1996–2007. doi: 10.1128/JCM.00237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragimbeau C., Colin S., Devaux A., Decruyenaere F., Cauchie H.-M., Losch S., Penny C., Mossong J. Investigating the host specificity of Campylobacter jejuni and Campylobacter coli by sequencing gyrase subunit A. BMC Microbiol. 2014;14:205. doi: 10.1186/s12866-014-0205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovanen S., Rossi M., Pohja-Mykrä M., Nieminen T., Raunio-Saarnisto M., Sauvala M., Fredriksson-Ahomaa M., Hänninen M.-L., Kivistö R. Population genetics and characterization of Campylobacter jejuni isolates from western jackdaws and game birds in Finland. Appl. Environ. Microbiol. 2019;85:e02365–e023618. doi: 10.1128/AEM.02365-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosner B.M., Schielke A., Didelot X., Kops F., Breidenbach J., Willrich N., Gölz G., Alter T., Stingl K., Josenhans C. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci. Rep. 2017;7:5139. doi: 10.1038/s41598-017-05227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gripp E., Hlahla D., Didelot X., Kops F., Maurischat S., Tedin K., Alter T., Ellerbroek L., Schreiber K., Schomburg D. Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genom. 2011;12:584. doi: 10.1186/1471-2164-12-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravel A., Hurst M., Petrica N., David J., Mutschall S.K., Pintar K., Taboada E.N., Pollari F. Source attribution of human campylobacteriosis at the point of exposure by combining comparative exposure assessment and subtype comparison based on comparative genomic fingerprinting. PLoS ONE. 2017;12:e0183790. doi: 10.1371/journal.pone.0183790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frazão M.R., de Souza R.A., Medeiros M.I.C., da Silva Duque S., Cao G., Allard M.W., Falcão J.P. Molecular typing of Campylobacter jejuni strains: Comparison among four different techniques. Braz. J. Microbiol. 2020;51:519–525. doi: 10.1007/s42770-019-00218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietzka A. Expert Opinion on the Introduction of Next-Generation Typing Methods for Food-and Waterborne Diseases in the EU and EEA. ECDC; Solna, Sweden: 2015. [Google Scholar]
- 32.Nadon C., Van Walle I., Gerner-Smidt P., Campos J., Chinen I., Concepcion-Acevedo J., Gilpin B., Smith A.M., Kam K.M., Perez E. PulseNet International: Vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Eurosurveillance. 2017;22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cody A.J., Bray J.E., Jolley K.A., McCarthy N.D., Maiden M.C. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J. Clin. Microbiol. 2017;55:2086–2097. doi: 10.1128/JCM.00080-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llarena A.-K., Taboada E., Rossi M. Whole-genome sequencing in epidemiology of Campylobacter jejuni infections. J. Clin. Microbiol. 2017;55:1269–1275. doi: 10.1128/JCM.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahin O., Fitzgerald C., Stroika S., Zhao S., Sippy R.J., Kwan P., Plummer P.J., Han J., Yaeger M.J., Zhang Q. Molecular evidence for zoonotic transmission of an emergent, highly pathogenic Campylobacter jejuni clone in the United States. J. Clin. Microbiol. 2012;50:680–687. doi: 10.1128/JCM.06167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dearlove B.L., Cody A.J., Pascoe B., Méric G., Wilson D.J., Sheppard S.K. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J. 2016;10:721–729. doi: 10.1038/ismej.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wegener H.C. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2003;6:439–445. doi: 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Lin J., Yan M., Sahin O., Pereira S., Chang Y.-J., Zhang Q. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob. Agents Chemother. 2007;51:1678–1686. doi: 10.1128/AAC.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao H., Dai M., Wang Y., Peng D., Liu Z., Yuan Z. 23S rRNA mutation A2074C conferring high-level macrolide resistance and fitness cost in Campylobacter jejuni. Microb. Drug Resist. 2009;15:239–244. doi: 10.1089/mdr.2009.0008. [DOI] [PubMed] [Google Scholar]
- 40.Han F., Pu S., Wang F., Meng J., Ge B. Fitness cost of macrolide resistance in Campylobacter jejuni. Int. J. Antimicrob. Agents. 2009;34:462–466. doi: 10.1016/j.ijantimicag.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 41.Luangtongkum T., Shen Z., Seng V.W., Sahin O., Jeon B., Liu P., Zhang Q. Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agents Chemother. 2012;56:1300–1308. doi: 10.1128/AAC.05516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.USADA . FSIS Cecal Sampling under the National Antimicrobial Resistance Monitoring System (NARMS) Surveillance Program. USADA; Washington, DC, USA: 2019. p. 8. [Google Scholar]
- 43.Jolley K.A., Bray J.E., Maiden M.C. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org. website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inouye M., Dashnow H., Raven L.-A., Schultz M.B., Pope B.J., Tomita T., Zobel J., Holt K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Z., Alikhan N.-F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kittl S., Heckel G., Korczak B.M., Kuhnert P. Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS ONE. 2013;8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.-H., McDermott P.F. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019;63:e00483–e004819. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Z., Periaswamy B., Sahin O., Yaeger M., Plummer P., Zhai W., Shen Z., Dai L., Chen S.L., Zhang Q. Point mutations in the major outer membrane protein drive hypervirulence of a rapidly expanding clone of Campylobacter jejuni. Proc. Natl. Acad. Sci. USA. 2016;113:10690–10695. doi: 10.1073/pnas.1605869113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R., Potter S.C., Finn R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.