Abstract
Exosomal microRNA (miR) can affect signaling pathways in various physiological and pathological conditions, including ovarian cancer (OC). miR-34b, the first microRNA targeted in a human clinical trial for cancer treatment, exhibited decreased expression in several cancer types. However, the biological function of exosomal miR-34b in OC has not been elucidated. In the present study, using reverse transcription-quantitative PCR, it was reported that exosomal miR-34b is downregulated in OC cells. Exosomal miR-34b reduced cell proliferation and epithelial-mesenchymal transition (EMT) in the OC cell line SKOV3. In addition, it was confirmed that Notch2, which is upregulated in SKOV3 cells, is a target of miR-34b. Moreover, exosomal miR-34b and Notch2 levels were found to be negatively correlated. The present data highlights the importance of exosomal miR-34b-mediated inhibition of cell proliferation and EMT, suggesting that exosomal miR-34b has value as a diagnostic biomarker and a potential molecular target for the treatment of OC.
Keywords: exosomal miR-34b, ovarian cancer, proliferation, epithelial-mesenchymal transition, Notch2
Introduction
Ovarian cancer (OC) is one of the most fatal malignancies in gynecologic cancer worldwide owing to its frequent detection at an advanced stage (1,2). Based on the statistics from The American Cancer Society in 2013, OC accounts for 4% of all cancer types diagnosed in women, with an estimated 22,000 new cases and 14,000 deaths per annum in the United States alone (3,4). OC has a low 5-year survival rate, and up to 70% of patients exhibit invasion and metastasis (5). The epithelial-mesenchymal transition (EMT) and collective cell migration to neighboring tissues are key steps in OC progression and metastasis, although the precise underlying mechanisms are unclear (6).
MicroRNAs (miRNAs/miRs) are small, single-stranded non-coding RNAs that regulate gene expression by binding to specific target mRNA sequences, thereby degrading mRNAs or inhibiting their translation (7). A previous study reported an association between epithelial ovarian cancer (EOC) and aberrant miRNA regulation (8). Various miRNAs are known to be dysregulated in the advanced stages of OC, suggesting that they are involved in malignancy and metastasis (9).
Additional studies have shown that miRNAs can be secreted into the extracellular space, mostly in the form of exosomes, and function in intercellular communication (10–12). Exosomal miRNAs are also important biomarkers of diagnosis and progression of OC (13). For instance, the expression of members of the miR-373 and miR-200 families were significantly upregulated in patients with OC compared with healthy women (14). A previous study reported that exosomal miR-30a-5p may be a potential biomarker for OC (15). miR-34b belongs to the miR-34 family and is directly transactivated by the p53 tumor suppressor (16). In general, miR-34b can promote apoptosis and cell cycle arrest resulting in p53 activation, thereby acting as a mediator of tumor suppression by p53 (17). Several studies have shown that miR-34b is downregulated in OC, but its mechanism of action and precise role in intercellular communication are poorly understood (18,19).
The present study examined the expression and biological functions of exosomal miR-34b and investigated the mechanisms underlying its role in OC.
Materials and methods
Cell culture
The human ovarian surface epithelium cell line, IOSE-80 and the OC cell lines SKOV3, A2780 and OVCAR3 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, and cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS, 100 units/ml penicillin and 100 µg/ml streptomycin in 5% CO2 at 37°C.
Exosome purification
Exosomes were isolated from the culture medium by differential centrifugation. Briefly, the conditioned medium was centrifuged at 1,500 × g for 20 min at 4°C to separate the cells, followed by centrifugation at 12,000 × g for 35 min at room temperature and filtration through a 0.22-µm filter to remove cell debris. Exosomes were pelleted by ultracentrifugation at 120,000 × g for 90 min at room temperature, and washed with PBS. Subsequently, the exosomes were pelleted at 120,000 × g for 70 min at room temperature and resuspended in 100 µl of PBS.
Nanoparticle tracking analysis (NTA)
The Nanosight NTA NS300 (Malvern Instruments, Inc.) was used to identify the concentration and size of isolated exosomes by tracking the Brownian motion of particles. The samples were captured for 60 sec at room temperature with manual shutter.
Electron microscopy
For electron microscopy analysis, exosomes isolated from the cell line, IOSE-80 dissolved in PBS were loaded to carbon-coated nickel grids and negatively stained with 2% methylamine tungstate (Sigma-Aldrich, Inc; Merck KGaA) at 37°C for 10 min. After drying at room temperature for 20 min, the samples were viewed using a JEM-1230 transmission electron microscope (Nikon Corporation) at 80 kV.
RNA preparation and reverse transcription quantitative (RT-q)PCR
Total RNA was extracted from cells or using TRIzol® reagent (Takara Biotechnology Co., Ltd). Briefly, 1 µg of total RNA was reverse transcribed into cDNA using the Mir-X™ miRNA FirstStrand Synthesis kit (Takara Biotechnology Co., Ltd.) at 37°C for 60 min and then 85°C for 5 sec. qPCR analyses were conducted using the SYBR Prime Script miRNA RT-PCR kit (Takara Biotechnology Co., Ltd), following the manufacturers procedure. All reagents used for RT-qPCR were obtained from Takara Biotechnology Co., Ltd. The universal miRNA Reverse and U6 primers were also obtained from Takara Biotechnology Co., Ltd. The miR-34b, the universal miRNA Reverse, U6 and mRNA primer sequences are shown in Table I.
Table I.
Primer sequences used in reverse transcription-quantitative PCR.
| Gene | Primer sequence (5′ → 3′) |
|---|---|
| miR-34b | |
| Forward | CAATCACTAACTCCACTGCCAT |
| U6 | |
| Forward | CGCAAGGATGACACGCAAATTCG |
| Universal Reverse | CCAGTGCAGGGTCCGAGGT |
| Notch2 | |
| Forward | GGGACCCTGTCATACCCTCT |
| Reverse | GAGCCATGCTTACGCTTTCG |
| E-cadherin | |
| Forward | AGGCTAGAGGGTCACCGCGTC |
| Reverse | GCTTTGCAGTTCCGACGCCAC |
| N-cadherin | |
| Forward | AGTCAACTGCAACCGTGTGT |
| Reverse | AGCGTTCCTGTTCCACTCAT |
| Snail | |
| Forward | CCTCCC TGTCAGATGAGGAC |
| Reverse | CCAGG CTGAGGTATTCCTTG |
| β-actin | |
| Forward | AGCCATGTACGTAGCCATCC |
| Reverse | CTCTCAGCTGTGGTGGTGAA |
miR, microRNA.
Western blot analysis
Proteins were extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) and the total protein content was measured using the bicinchoninic acid assay (Beyotime Biotechnology, Inc) by Fluoroskan (Thermo Fisher Scientific Inc.). A total of 20 µg protein per lane was loaded onto a 12% SDS-PAGE gel. Proteins were then transferred to polyvinylidene difluoride membranes, and blocked using 5% BSA at room temperature for 1 h. Then, immunoblotting was performed with primary antibodies against TSG101 (1:2,000; cat. no. ab125011; Abcam), CD63 (1;2000, cat. no. sc-5275; Santa Cruz Biotechnology Inc.), Notch2 (cat. no. 5732S), E-cadherin (cat. no. 14472), N-cadherin (cat. no. 13116), Snail (cat. no. 3879S) and β-actin (cat. no. 3700S) (1:2,000; all Cell Signaling Technology, Inc.) at 4°C overnight. After washing, the membranes were incubated with goat anti-rabbit or anti-mouse antibody conjugated to horseradish peroxidase (1:2,000; Cell Signaling Technology) for 1 h at room temperature. An enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific Inc.) was used to detect the bands. Densitometry was performed using ImageJ v1.8.0 software (National Institutes of Health).
Proliferation assay
Cell Counting Kit (CCK)-8 assays (Dojindo Molecular Technologies, Inc.) were used to evaluate cell viability. Briefly, exponentially growing cells were counted and seeded in 96-well plates at 1×104 cells/ well. After 72 h, CCK-8 was added to each well according to the manufacturers instructions and the plates were incubated for at 37°C for 2 h. Then, the absorbance was measured with Fluoroskan (Thermo Fisher Scientific Inc.) at 490 nm.
Wound healing assay
For the wound healing assay, SKOV3 cells were seeded in 6-cm culture dishes. Following serum starvation for 24 h, a straight scratch was created using a 200-µl pipette tip and the wound was visualized under a light microscope (magnification, ×100). Images of cells were captured under the microscope and the percentage of the scratch healing was calculated.
Transfection
SKOV3 cells were seeded on 6-well plates, and transfected using TurboFect Transfection Reagent (Thermo Fisher Scientific, Inc.) according to the manufacturers instructions. anti-miR-34b (100 nM), miR-34b mimics (50 nM) and small interfering (si)-Notch2 (100 nM) were used to regulate the expression of miR-34b and Notch2. Scrambled miR-control and silencer negative control 1 siRNA were used as negative controls. Cells were collected 48 h post-transfection for further experimentation. The sequences used were as follows: miR-34b mimics sense, 5-AGGCAGUGUAAUUAGCUGAUUGU-3 and antisense, 5-AAUCAGCUAAUUACACUGCCUUU-3; anti-miR-34b 5-ACAAUCAGCUAAUUACACUGCCU-3; and si-Notch2 sense, 5-GGAGGUCUCAGUGGAUAUATT-3 and antisense, 5-UAUAUCCACUGAGACCUCCTT-3.
Luciferase reporter assay
SKOV3 cells were evaluated using the recombinant pMIR-reporter luciferase vector (Guangzhou RiboBio Co., Ltd.) with a luciferase reporter assay (Promega Corporation). The same transfection methodology was used as described in the previous paragraph. Briefly, cells were transfected with the Notch2 3 untranslated region (UTR) wildtype (WT) and Notch2 3UTR mutant (Mut) plasmids, together with the negative control RNA and miR-34b mimics using TurboFect Transfection reagent (Thermo Fisher Scientific, Inc.) in 24-well plates. pRL-CMV luciferase plasmids served as the internal control. At 48 h after transfection, cells were harvested and lysed. Luminescence was determined using the dual-luciferase reporter assay system (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
Prediction of targets of miR-34b
Three online prediction algorithms PicTar (https://pictar.mdc-berlin.de/), TargetScan (http://www.targetscan.org/vert_71/) and miRDB (http://mirdb.org/) were used to identified targets gene of miR-34b and then the common genes selected from the 3 algorithms were identified as the candidates.
Statistical analysis
All data are presented as mean ± standard error of the mean from three individual experiments. GraphPad Prism version 7 (GraphPad Software) was used for data analyses. Paired Students t-test and a one-way analysis of variance (ANOVA) with Tukeys post hoc test were used to assess the differences. Pearsons correlation analysis was used to analyze the correlation between the expression of exosomal miR-34b and Notch2 mRNA. P<0.05 was considered to indicate a statistically significant difference.
Results
Exosomal miR-34b expression is downregulated in OC cells
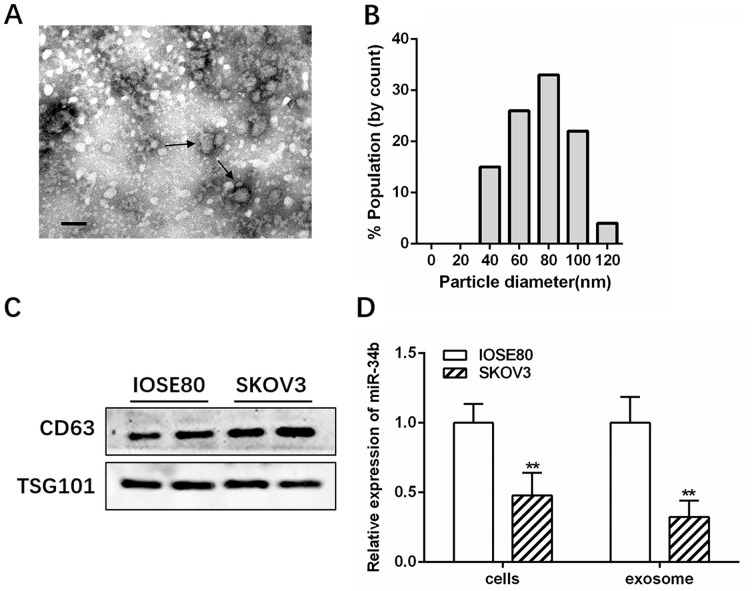
Transmission electron microscopy and NanoSight analysis showed that IOSE-80 exosomes had a double-layer membrane and were primarily 40–100 nm in diameter (Fig. 1A and B). Western blotting further confirmed the presence of exosomes by identifying the well-established markers, CD63 and TSG101 (Fig. 1C). RT-qPCR revealed that miR-34b was highly expressed in the IOSE-80 cell line, and had decreased levels in SKOV3 cells compared with several other OC cell lines (A2780 and OVCAR3) (Fig. S1; *P<0.05; **P<0.01). Therefore, IOSE-80 and SKOV3 cells were selected for further study. The RT-qPCR results showed that miR-34b levels were significantly lower in SKOV3-derived exosomes (SK-Exos) compared with those in the IOSE-80-derived exosomes (IO-Exos), consistent with the cellular expression (Fig. 1D; **P<0.01).
Figure 1.
Exosomal miR-34b is downregulated in ovarian cancer. (A) Electron microscopy analysis of exosomes isolated from the SKOV3 conditioned medium. Arrows indicate the exosomes. Scale bar, 100 nm. (B) Size distribution graph of exosomes isolated from the SKOV3 conditioned medium and quantified using nanoparticle tracking analysis. (C) Western blotting of the exosomal markers, CD63 and TSG101, in exosomes isolated from IOSE-80 and SKOV3 conditional medium. (D) Relative expression levels of miR-34b in IOSE-80 and SKOV3 cells and exosomes. **P<0.01 vs. IOSE-80. miR, microRNA.
Exosomal miR-34b attenuates proliferation and EMT in ovarian cancer
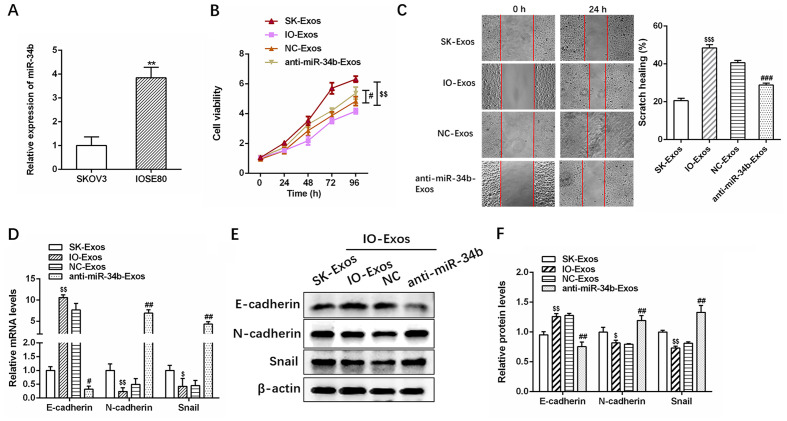
RT-qPCR revealed that miR-34b expression was significantly upregulated in SKOV3 co-cultured with IOSE-80 cells compared with that of SKOV3 cells alone (Fig. 2A; **P<0.01), suggesting that miR-34b containing exosomes were delivered from IOSE-80 cells in the upper Transwell chamber to the recipient SKOV3 cells in the lower chamber. RT-qPCR also revealed anti-miR-34b transfection significantly decreased miR-34b expression in exosomes compared with IOSE-80 exosomes (NC-Exos) cells (Fig. S2). The CCK 8 and wound healing assays indicated that IO-Exos markedly attenuated the proliferation and invasion of SKOV3 cells, while exosomes derived from miR-34b-antagonized IOSE-80 cells decreased IOSE-80 (IO)-Exos-induced target cell proliferation and invasion (Fig. 2B and C; #P<0.05; ##P<0.01; $$P<0.01 and $$$P<0.001).
Figure 2.
Exosomal miR-34b suppresses cell proliferation and EMT in ovarian cancer cells. (A) Relative expression levels of miR-34b in SKOV3 cells co-cultured with SKOV3 or IOSE-80. (B) Cell viability assays and (C) cell invasion assays in SKOV3 cells cultured with exosomes isolated from SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (D) Relative expression levels of EMT markers (E-cadherin, N-cadherin and Snail) in SKOV3 cells co-cultured with SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (E) and (F) Western blotting analysis of EMT markers (E-cadherin, N-cadherin, and Snail) (20,21) in SKOV3 cells co-cultured with SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. **P<0.01 vs. SKOV3. $P<0.05, $$P<0.01 and $$$P<0.001 vs. SK-Exos, #P<0.05, ##P<0.01 vs. NC-Exos. EMT, epithelial-mesenchymal transition; NC, negative control; miR, microRNA; SK, SKOV3; IO, IOSE-80; Exos, exosomes.
Furthermore, as demonstrated by RT-qPCR and western blotting, IO-Exos was associated with elevated expression of E-cadherin (epithelial marker) (20) and reduced expression of N-cadherin and Snail (mesenchymal markers) (21) at both the mRNA and protein levels in SKOV3 cells (Fig. 2D-F). In contrast, anti-miR-34b-Exos was associated with reduced E-cadherin and increased N-cadherin and Snail expression (Fig. 2D-F; #P<0.05; ##P<0.01; $P<0.05 and $$P<0.01). In summary, the aforementioned results demonstrated that exosomal miR-34b attenuates the proliferation and EMT in OC cells.
Notch2 is a direct target of miR-34b
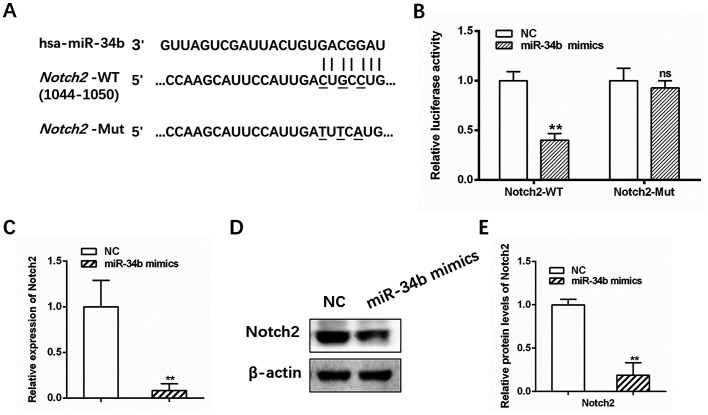
miRNAs are typically involved in the regulation of target genes. Three online prediction algorithms (PicTar, TargetScan and miRDB) identified Notch2 as a potential target gene of miR-34b, with a putative binding site at base pairs 1044–1050 (Fig. 3A). Elevated Notch signaling activity has been reported in previous studies investigating OC (22,23). The dual-luciferase reporter assay showed that luciferase activity for the WT Notch2 3-UTR was significantly suppressed in cells transfected with miR-34b mimics compared with the control (Fig. 3B; **P<0.01), while no significant change in activity was observed for Mut Notch2 (Fig. 3B; **P<0.01). Furthermore, Notch2 mRNA and protein levels were decreased following treatment with miR-34b mimics (Fig. 3C-E; **P<0.01). Collectively, these results demonstrated that Notch2 is a direct target of miR-34b.
Figure 3.
Notch2 is a direct target gene of miR-34b. (A) Predicted miR-34b binding sites in the 3′ UTR of Notch2 mRNA. (B) Luciferase activity in cells 48 h after transfection. (C) Relative expression levels of Notch2 in SKOV3 cells transfected with NC or miR-34b mimics. (D) and (E) Western blotting of Notch2 in SKOV3 cells transfected with NC or miR-34b mimics. **P<0.01 vs. NC. miR, microRNA; UTR, untranslated region; NC, negative control; ns, not significant.
Notch2 is negatively correlated with exosomal miR-34b
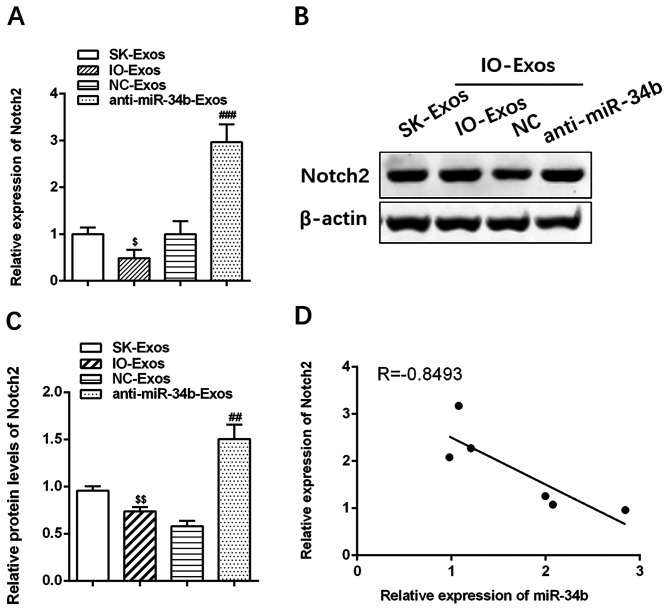
The mRNA and protein levels of Notch2 were lower in cells co-cultured with IOSE-80 compared with those in SKOV3 cells alone (Fig. 4A-C; ##P<0.01; ###P<0.001; $P<0.05 and $$P<0.01). In contrast, Notch2 mRNA and protein levels were increased in cells treated with anti-miR-34b-Exos (Fig. 4A-C). These results confirmed that Notch2 is negatively correlated with exosomal miR-34b (r=−0.8493, Fig. 4D).
Figure 4.
Notch2 is negatively associated with exosomal miR-34b. (A) Relative expression levels of Notch2 in SKOV3 cells cultured with exosomes isolated from SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (B) and (C) Western blotting of Notch2 in SKOV3 cells co-cultured with SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (D) Notch2 is negatively correlated with exosomal miR-34b. $P<0.05, $$P<0.01 vs. SK-Exos. ##P<0.01, ###P<0.001 vs. NC-Exos. miR, microRNA; SK, SKOV3; IO, IOSE-80; Exos, exosomes.
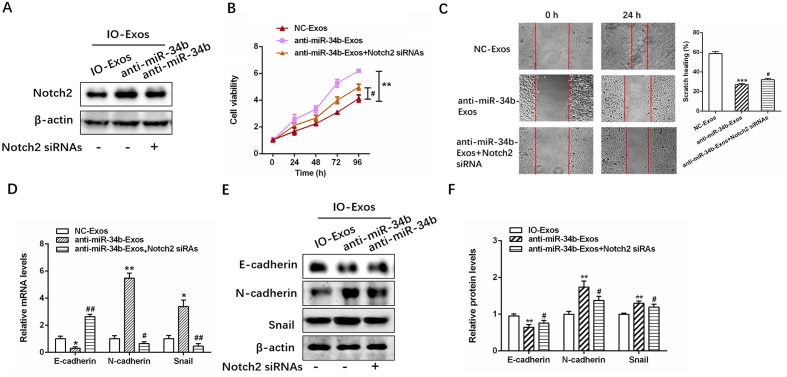
Notch2 knockdown decreases the effect of exosomal anti-miR-34b on cell proliferation and EMT in OC
The efficiency of Notch2 siRNA transfection was examined in SKOV3 cells (Fig. S3), and then successfully transfected SKOV3 cells were treated with IO-Exos-NC or anti-miR-34b IO-Exos (Fig. 5A). As demonstrated by the CCK-8 and wound healing assays, Notch2 knockdown significantly inhibited the proliferation and invasion induced by exosomal anti-miR-34b (Fig. 5B and C; **P<0.01; ***P<0.001 #P<0.05 and ##P<0.01). In addition, exosomal anti-miR-34b-induced EMT inhibition was partly decreased by Notch2-knockdown in SKOV3 cells (Fig. 5D-F; *P<0.05; **P<0.01; #P<0.05 and ##P<0.01). Together, these results suggested that exosomal miR-34b mediates cell proliferation and EMT via Notch2.
Figure 5.
Notch2 knockdown abates the effect of exosomal anti-miR-34b on cell proliferation and the EMT in ovarian cancer cells. (A) Relative expression levels of Notch2 in SKOV3 cells co-cultured with IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (B) Cell viability and (C) cell invasion assays in SKOV3 cells cultured with exosomes isolated from SKOV-3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (D) Relative expression levels of EMT markers (E-cadherin, N-cadherin and Snail) in SKOV3 cells cultured with exosomes isolated from SKOV3, IOSE-80, IOSE-80+NC or IOSE-80+anti-miR-34b. (E) and (F) western blotting of E-cadherin, N-cadherin and Snail in SKOV3 cells co-cultured with IOSE-80, IOSE-80+NC, or IOSE-80+anti-miR-34b. **P<0.01, ***P<0.001 vs. NC-Exos. #P<0.05, ##P<0.01 vs. anti-miR-34b-Exos. EMT, epithelial-mesenchymal transition; NC, negative control; Exos, exosomes; IO, IOSE-80; SK, SKOV3; si, small interfering.
Discussion
Despite improvements in surgical approaches, OC still has a high mortality rate, which can be primarily attributed to late-stage diagnosis and a high rate of metastasis (24). Previous studies have demonstrated the critical roles of miRNAs in tumorigenesis, progression and metastasis in OC (25,26). Approximately 400 dysregulated miRNAs have been identified in OC, some of which are secreted in exosomes, and are involved in diverse biological functions, including cell proliferation, migration and resistance to paclitaxel and Cisplatin® (27,28). In addition, exosomal miRNAs, including miR-145, miR-940 and Let-7, have been identified as potential diagnostic biomarkers for OC (29–31). Therefore, understanding the roles of dysregulated exosomal miRNAs in OC may help improve diagnosis and treatment. The present study demonstrated that exosomal miR-34b expression is decreased in OC and has an inhibitory effect on cell proliferation and EMT. These results may improve our existing understanding of the fundamental mechanisms underlying the pathogenesis of OC.
miR-34b can act as a tumor suppressor via several mechanisms, including a positive feedback loop between miR-34b and p53 (32). Decreased expression of miR-34b has been detected in various types of cancer, such as colorectal, pancreatic, gastric and ovarian cancer, due to the inactivation of its promoter by CpG methylation (19,33–35). Consequently, miR-34b has become the first miR target to reach the phase 1 clinical trials for solid tumors, such as lung, colon, prostate as well as other disorders, such as cardiac fibrosis, cardiometabolic disease and chronic heart failure (36,37). Moreover, miR-34b has been investigated in body fluids, such as plasma and urine and tissues and has been identified as a biomarker for clear cell renal cell carcinoma (38,39). Several studies have shown that miR-34b expression is lower in OC tissues compared with that in normal ovaries (16,30). However, the functions and underlying mechanisms of action of exosomal miR-34b in OC have not been explored. The present study demonstrated that exosomal miR-34b is highly dysregulated in OC cells. Exosomal miR-34b can suppress cell proliferation and expression of the epithelial adhesion molecule, E-cadherin, and increase the expression of the mesenchymal markers, N-cadherin and Snail, suggesting that miR-34b inhibits EMT in OC (20,40).
Notch2, a member of the Notch family of proteins, is a conserved cell surface receptor that mediates cellular interactions, and is associated with the initiation and development of various types of tumors, such as liver, brain and gastric tumors (41,42). Indeed, Notch signaling has a wide range of functions, and, depending on the cancer type, can function as either a tumor promoter or suppressor, highlighting its complex role in cancer (43). In ovaries, Notch signaling regulates granulosa cell proliferation and coordinates follicular growth; therefore, it is involved in ovarian follicle development (44). In addition, increased Notch2 expression is significantly correlated with poor progression-free survival rate in OC, suggesting that it has prognostic value (45). The present study is the first to report that Notch2 is a direct target gene of miR-34b in OC, to the best of our knowledge. Furthermore, Notch2 was significantly negatively associated with exosomal miR-34b levels, and knockdown of Notch2 abated the decreased cell proliferation and EMT mediated by exosomal anti-miR-34b in OC.
EMT is generally associated with the invasion and migration ability of cells (46). Wound healing and Transwell assays are methods to analyze cell migration. The present study, revealed that exosomal miR-34b attenuates EMT in OC by downregulation of E-cadherin and upregulation of N-cadherin and Snail. However, the Transwell assay was not performed using Matrigel, therefore invasion could not be analyzed, which is a limitation of the present study and worthy of further investigation. In addition, the critical role of miR-34b was only explored in OC cell lines, relevant OC animal models should be studied to further confirm the regulatory function of miR-34b in OC.
Overall, the regulatory network of exosomal miR34b and the mechanisms underlying its suppressive effects on cell proliferation and EMT in OC were identified. It was demonstrated that miR-34b acts by directly binding to and regulating Notch2 expression. These findings highlight the important role of exosomal miR-34b in OC, suggesting that it could act as a promising diagnostic and therapeutic target in OC in the future.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was funded by the Changhai Hospital Youth Startup Fund (grant no. 2018QNB004)
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors contributions
SL and WL designed the study and performed the experiments. HS participated in the experimental design and analysis of data, and drafted and critically revised the manuscript. HZ participated in the experiment design and the subject establishment of this article, in addition he wrote and critically revised the manuscript and gave final approval for the manuscript to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Karimi-Zarchi M, Mortazavizadeh SMR, Bashardust N, Zakerian N, Zaidabadi M, Yazdian-Anari P, Teimoori S. The clinicopathologic characteristics and 5-year survival rate of epithelial ovarian cancer in Yazd, Iran. Electron Physician. 2015;7:1399–1406. doi: 10.14661/1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer Statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Hannibal CG, Cortes R, Engholm G, Kjaer SK. Survival of ovarian cancer patients in Denmark: Excess mortality risk analysis of five-year relative survival in the period 1978–2002. Acta Obstet Gynecol Scand. 2008;87:1353–1360. doi: 10.1080/00016340802483000. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 5.Ozols RF, Bookman MA, Connolly DC, Daly MB, Godwin AK, Schilder RJ, Xu X, Hamilton TC. Focus on epithelial ovarian cancer. Cancer Cell. 2004;5:19–24. doi: 10.1016/S1535-6108(04)00002-9. [DOI] [PubMed] [Google Scholar]
- 6.Chebouti I, Kasimir-Bauer S, Buderath P, Wimberger P, Hauch S, Kimmig R, Kuhlmann JD. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget. 2017;8:48820–48831. doi: 10.18632/oncotarget.16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragomir MP, Knutsen E, Calin GA. SnapShot: unconventional miRNA functions. Cell. 2018;174:1038–1038.e1. doi: 10.1016/j.cell.2018.07.040. [DOI] [PubMed] [Google Scholar]
- 8.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Cecco L, Bagnoli M, Canevari S, Califano D, Perrone F, Pignata S, Mezzanzanica D. miRNA-based signature for predicting epithelial ovarian cancer recurrence. Transl Cancer Res 6 (S1) 2017:S232–S234. doi: 10.21037/tcr.2017.02.44. [DOI] [Google Scholar]
- 10.Lutgendorf SK, Thaker PH, Arevalo JM, Goodheart MJ, Slavich GM, Sood AK, Cole SW. Biobehavioral modulation of the exosome transcriptome in ovarian carcinoma. Cancer. 2018;124:580–586. doi: 10.1002/cncr.31078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, Li H, Zhu X, Yao L, Zhang J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147:423–431. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 13.Pan C, Stevic I, Müller V, Ni Q, Oliveira-Ferrer L, Pantel K, Schwarzenbach H. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. 2018;12:1935–1948. doi: 10.1002/1878-0261.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7:16923–16935. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, Wang J, Liu Y, Chen P, Wu X, et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33:2915–2923. doi: 10.3892/or.2015.3937. [DOI] [PubMed] [Google Scholar]
- 16.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosseinpour Z, Salehi Z, Talesh Sasani S, Aminian K. p53 and miR-34b/c genetic variation and their impact on ulcerative colitis susceptibility. Br J Biomed Sci. 2018;75:46–49. doi: 10.1080/09674845.2017.1362729. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Lu Z, Unruh AK, Ivan C, Baggerly KA, Calin GA, Li Z, Bast RC, Jr, Le XF. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res. 2015;13:393–401. doi: 10.1158/1541-7786.MCR-14-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogt M, Munding J, Grüner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 20.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaw SY, Abdul Majeed A, Dalley AJ, Chan A, Stein S, Farah CS. Epithelial to mesenchymal transition (EMT) biomarkers--E-cadherin, beta-catenin, APC and Vimentin--in oral squamous cell carcinogenesis and transformation. Oral Oncol. 2012;48:997–1006. doi: 10.1016/j.oraloncology.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Groeneweg JW, Foster R, Growdon WB, Verheijen RH, Rueda BR. Notch signaling in serous ovarian cancer. J Ovarian Res. 2014;7:95. doi: 10.1186/s13048-014-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang CP, Yang JL, Zhang J, Li L, Huang L, Ji SY, Hu ZY, Gao F, Liu YX. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152:2437–2447. doi: 10.1210/en.2010-1182. [DOI] [PubMed] [Google Scholar]
- 24.Yeung TL, Leung CS, Yip KP, Au Yeung CL, Wong ST, Mok SC. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and molecular processes in cancer metastasis. Am J Physiol Cell Physiol. 2015;309:C444–C456. doi: 10.1152/ajpcell.00188.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava AK, Banerjee A, Cui T, Han C, Cai S, Liu L, Wu D, Cui R, Li Z, Zhang X, et al. Inhibition of miR-328-3p impairs cancer stem cell function and prevents metastasis in ovarian cancer. Cancer Res. 2019;79:2314–2326. doi: 10.1158/0008-5472.CAN-18-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao W, Han T, Li B, Ma Q, Yang P, Li H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J Ovarian Res. 2019;12:121. doi: 10.1186/s13048-019-0589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: An overview. J Cell Physiol. 2018;233:3846–3854. doi: 10.1002/jcp.26095. [DOI] [PubMed] [Google Scholar]
- 28.Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christenson LK. MicroRNA control of ovarian function. Anim Reprod. 2010;7:129–133. [PMC free article] [PubMed] [Google Scholar]
- 30.Xie YL, Yang YJ, Tang C, Sheng HJ, Jiang Y, Han K, Ding LJ. Estrogen combined with progesterone decreases cell proliferation and inhibits the expression of Bcl-2 via microRNA let-7a and miR-34b in ovarian cancer cells. Clin Transl Oncol. 2014;16:898–905. doi: 10.1007/s12094-014-1166-x. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Ying X, Wang X, Wu X, Zhu Q, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol Rep. 2017;38:522–528. doi: 10.3892/or.2017.5697. [DOI] [PubMed] [Google Scholar]
- 32.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: The feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 33.Hiyoshi Y, Schetter AJ, Okayama H, Inamura K, Anami K, Nguyen GH, Horikawa I, Hawkes JE, Bowman ED, Leung SY, et al. Increased microRNA-34b and −34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancer. PLoS One. 2015;10:e0124899. doi: 10.1371/journal.pone.0124899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129:2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Cheng H, Shi S, Cui X, Yang J, Chen L, Cen P, Cai X, Lu Y, Wu C, et al. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr Mol Med. 2013;13:467–478. doi: 10.2174/1566524011313040001. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung WJ, Koh Y, Kim S, Park H, Yoon SS. Activity of microRNA replacement reagent, MRX34, in multiple myeloma in vivo model. Cancer Res. 2016;76:1081–1081. [Google Scholar]
- 38.Butz H, Nofech-Mozes R, Ding Q, Khella HWZ, Szabó PM, Jewett M, Finelli A, Lee J, Ordon M, Stewart R, et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell-Cell Communication in Renal Cell Carcinoma. Eur Urol Focus. 2016;2:210–218. doi: 10.1016/j.euf.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Ramezani A, Devaney JM, Cohen S, Wing MR, Scott R, Knoblach S, Singhal R, Howard L, Kopp JB, Raj DS. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: A pilot study. Eur J Clin Invest. 2015;45:394–404. doi: 10.1111/eci.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 42.Xiu MX, Liu YM. The role of oncogenic Notch2 signaling in cancer: A novel therapeutic target. Am J Cancer Res. 2019;9:837–854. [PMC free article] [PubMed] [Google Scholar]
- 43.Koch U, Radtke F. Dual function of notch signaling in cancer: oncogene and tumor suppressor. In: Miele L, Artavanis-Tsakonas S, editors. Targeting Notch in Cancer. Springer; New York, NY: 2018. pp. 55–86. [DOI] [Google Scholar]
- 44.Vanorny DA, Prasasya RD, Chalpe AJ, Kilen SM, Mayo KE. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol Endocrinol. 2014;28:499–511. doi: 10.1210/me.2013-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Wang X, Huang S, Wang L, Han L, Yu S. Prognostic roles of Notch receptor mRNA expression in human ovarian cancer. Oncotarget. 2017;8:32731–32740. doi: 10.18632/oncotarget.16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.