Abstract
Cells of the human breast gland express an array of keratins, of which some are used for characterizing both normal and neoplastic breast tissue. However, the expression pattern of certain keratins has yet to be detailed. Here, the expression of a differentiation marker of epidermal epithelium, keratin 10 (K10), was investigated in the human breast gland. While in normal breast tissue generally less than 1% of luminal epithelial cells expressed K10, in women >30 years of age glandular structures with K10-positive (K10pos) cells were found at higher frequency than in younger women. K10pos cells belong to a mature luminal compartment as they were negative for cKIT, positive for Ks20.8, and mostly non-cycling. In breast cancer, around 16% of primary breast carcinomas tested were positive for K10 by immunohistochemistry. Interestingly, K10pos tumor cells generally exhibit features of differentiation similar to their normal counterparts. Although this suggests that K10 is a marker of tumor differentiation, data based on gene expression analysis imply that high levels of K10 dictate a worse outcome for breast cancer patients. These findings can form the basis of future studies that should unravel which role K10 may play as a marker of breast cancer:
Keywords: estrogen receptor, immunohistochemistry, KRT10, Ks20.8, mammary gland
Introduction
The female human breast gland consists of an epithelium organized in a differentiation hierarchy that has yet to be fully understood. Unraveling the organization of this hierarchy is complicated by the fact that several events can lead to changes in the composition of the gland with increased age, including repeated cycles of menstruation, pregnancy and lactation, as well as postmenopausal regression. Acquiring a detailed insight into the various cellular subsets that populate the epithelial compartment in the normal gland may be key to understand the mechanisms that underlie the development of different breast cancer subtypes.
The distinct expression patterns of keratin intermediary filaments have widely been used to characterize the epithelia of various organs. In a number of epithelia, a subcellular change of keratin expression marks a transit between differentiation states. For example, in the epidermis there is a switch from expression of K5 and K14 in cells of the basal layer that provide the stem cell compartment to K1 and K10 in the differentiating suprabasal keratinocytes.1 Thus, a distinct distribution of keratins within an epithelium may be involved in compartmentalizing various differentiation states.
The luminal epithelial cells that line the ducts and lobular ductules in the human breast gland express a selection of keratins that characterize simple epithelia, including K7,2 K8,3 K18,3 and K19.4 However, the expression pattern is more complex as subsets of cells also express several keratins characteristic of stratified epithelia, including K5,5 K6a,6 K14,7 K15,6 and K17.7 Documentation of the presence of other stratified epithelial keratin markers in the breast gland is limited.8 For K10, its expression has not been described in the normal gland, and in breast cancer it has been reported to be present in rare cases.9–11
This study reveals that in fact rare K10pos cells are found in the normal human breast gland and that structures that contain these cells are more frequently present in women >30 years of age. Marker expression analysis demonstrated that K10pos cells are part of a differentiated compartment in both normal breast and breast cancer tissue, and that a significant subset of tumors are in fact K10pos.
Materials and Methods
Human Breast Tissue Samples
Normal human breast tissue was donated by consenting women undergoing reduction mammoplasty for cosmetic reasons performed at Capio CFR Hospitaler at Lyngby or Hellerup, Denmark. All personal information on donors was confidential except for the date and year of birth. In some cases, only the year of birth was available. Material from some of the biopsies has been used in previous studies.12–15 Samples of breast cancer were obtained completely anonymously from patients at the State University Hospital, Copenhagen. The storage and use of human material have been reviewed and approved by the Regional Scientific Ethical Committees (H-2-2011-052 and H-3-2010-095) and the Danish Data Protection Agency (2011-41-6722) and have been handled according to established guidelines in subsequent experiments.
Immunostaining
Cryopreserved tissue specimens were cut at 6 μm on a cryostat (Microm HM560; Axlab, Vedbæk, Denmark) and were subsequently fixed either in ice cold methanol for 5 min and air-dried or in 3.7% formaldehyde (Merck; Darmstadt, Germany), followed by permeabilization with Triton X-100 detergent (Sigma-Aldrich, Denmark). For peroxidase staining, specimens were incubated with primary antibody for 1 hr, and Ultravision ONE Detection System was used according to the manufacturer’s instructions (Labvision; Thermo Fisher Scientific, Denmark). Nuclei were stained with hematoxylin. For immunofluorescence, the specimens were incubated for 2 hr with primary antibody followed by incubation with secondary isotype-specific Alexa Fluor antibodies for 30 min. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Cultured cells were fixed and stained as described above. For staining of smears, cells were first dissociated to single cells from breast epithelial organoids isolated as described previously.12,16 Apart from classifying them by the presence of estrogen receptor α (ERα), breast carcinomas were also subdivided into molecular phenotypes by staining for progesterone receptor and HER2: Luminal A (ERαpos and/or PRpos, no HER2 overexpression), Luminal B (ERαpos and/or PRpos, HER2 overexpression), HER2-enriched (ERαneg, PRneg, HER2 overexpression), and Triple-Negative (ERαneg, PRneg, no HER2 overexpression). Antibodies and dilutions are listed in Table 1.
Table 1.
List of Antibodies Used for Immunostaining.
| Antibody Target | Clone | Company/Cat. No. | Peroxidase | Fluorescence |
|---|---|---|---|---|
| cKIT | K45 | NeoMarkers/MS-289 | 1:100 | 1:25 |
| Estrogen receptor α | 1D5 | Dako/M7047 | 1:100 | — |
| HER2 | TAB250 | Invitrogen/28-0003Z | 1:200 | — |
| Keratin 10 | DE-K10 | Invitrogen/MA5-13705 | 1:50 | 1:25 |
| Keratin 10 | RKSE60 | Millipore/MAB3230 | 1:10 | — |
| Keratin Ks20.8 | Ks20.8 | Dako/M7019 | — | 1:10 |
| Keratin | CAM5.2 | BD Biosciences/345779 | — | 1:25 |
| Ki67 | SP6 | Thermo Scientific/RM-9106 | — | 1:25 |
| Progesterone receptor | Pgr636 | Dako/M3569 | 1:100 | — |
| Alexa Fluor 488 goat anti-mouse IgG1 | — | Invitrogen/A21151 | — | 1:500 |
| Alexa Fluor 568 goat anti-mouse IgG2a | — | Invitrogen/A21134 | — | 1:500 |
| Alexa Fluor 568 goat anti-rabbit IgG | — | Invitrogen/A11036 | — | 1:500 |
Microscopy and Image Acquisition
A lobular or ductal segment with the presence of one or more K10-expressing cells after performing immunohistochemistry (IHC) was assigned as K10pos. For breast carcinomas, a tumor was considered positive for K10, cKIT, Ks20.8, ERα, or PR when >1% of the neoplastic cells expressed the given marker. Quantification of smear stainings and sections stained for Ki67 was performed on images that were acquired and digitally stored. Images were acquired on a Leica DM550B microscope equipped with a DFC550 digital camera or a Zeiss LSM 700 confocal microscope located at the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen. In smears, 1000 CAM5.2-positive (CAM5.2pos) luminal cells were quantified for each biopsy. For quantification of Ki67-positive cells in cultures, 200 cells were counted (either within the K10pos population or on the overall population) in each of three different culture flasks. On tissue sections, 200 K10pos cells or CAM5.2pos cells were evaluated for Ki67 expression for each of three tumors.
Cell Culture
MCF10A is a spontaneously immortalized breast epithelial–derived non-tumorigenic cell line.17 MCF10A cells were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient mixture F-12 (DMEM/F12, 1:1; Life Technologies, Denmark) supplemented with 2 mmol/l glutamine, 20 ng/ml epidermal growth factor (PeproTech, Germany), 10 µg/ml insulin, 0.5 µg/ml hydrocortisone, 10 ng/ml cholera toxin (Sigma-Aldrich), and 5% fetal bovine serum (FBS). MCF7 breast cancer cells were cultured in DMEM 1965 supplemented with 2 mmol/l glutamine, 7 non-essential amino acids (NEA), 6 ng/ml insulin (Sigma-Aldrich), and 5% FBS. All cells were cultured at 37C in a humidified incubator with an atmosphere of 5% CO2.
RNA Isolation and Real-time Quantitative PCR
For each biopsy, 10 tissue slices with a thickness of 6 to 8 µm were sectioned on a cryostat and dissolved in Trizol (Life technologies). Total RNA was isolated by a spin column method (Zymo Research, CA). The quality and integrity of the extracted RNA was evaluated by a spectrophotometer (Nanodrop 1000). RNAs were reverse transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Denmark). Quantified real-time PCR was performed using Taqman gene expression assays (Applied Biosystems) on Bio-Rad CFX manager 3.0 and thermocycler (Bio-Rad, Denmark). Expression of KRT10 (Hs00166289_m1) was normalized to GAPDH (Hs02758991_g1) and EPCAM (Hs00901885_m1).
Kaplan–Meier Plots
A web tool, KM-plotter (kmplot.com), was used to generate Kaplan–Meier plots based on data sets from more than 6000 breast cancer patients.18 Analysis of expression of KRT10 in the data sets was performed using the JetSet best probe set (Affymetrix ID: 213287_s_at). Expression data were trichotomized to compare the patients with the highest levels of KRT10 tumor expression (upper tertile) vs the lowest (lower tertile) for relapse-free survival (RFS) and overall survival (OS). Patients with ERα-positive and ERα-negative tumors were analyzed separately, as well as in total. Furthermore, more detailed subcluster analysis was performed by including parameters such as molecular subtype, tumor grade, lymph node status, and treatment modality. The number of patients analyzed can be found in Supplemental Table S4.
Results
Rare K10pos Luminal Cells Are Most Frequently Present in Breast Gland Structures of Women >30 Years of Age
Here, we analyzed the expression of K10 in human normal breast glands by IHC on cryostat sections using a well-characterized antibody, clone DE-K10.19,20 A minor subset of glandular structures demonstrated the presence of K10pos cells. Typically, when present, K10pos cells were located as single cells or in small foci as part of the luminal compartment (Fig. 1A). The staining pattern was validated by comparison with another often-used monoclonal antibody against K10, RKSE60,21–23 in serial sections (Supplemental Fig. S1). The actual proportion of luminal cells expressing K10 was quantified on trypsinized cellular smears derived from breast gland tissue from five women (age range: 19–46 years). In all cases, less than 1% of luminal cells were positive (Supplemental Fig. S2).
Figure 1.
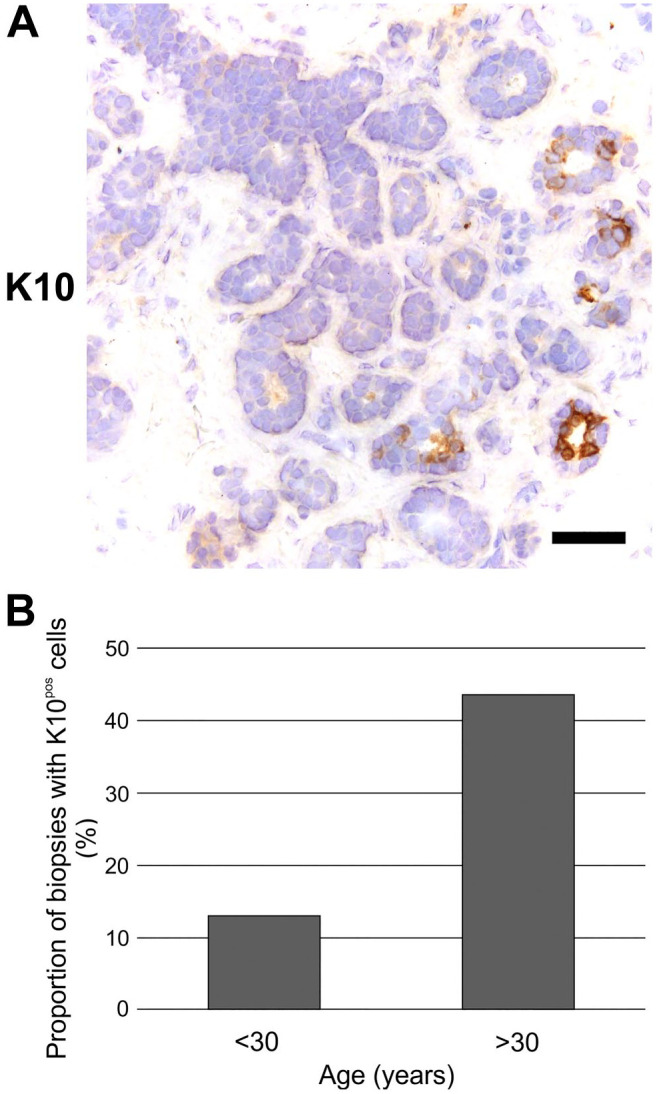
Rare K10pos cells are most frequently present in breast glands of women >30 years of age. (A) Immunoperoxidase staining demonstrating expression of K10 in a small subset of luminal cells. Scale bar, 50 µm. (B) Proportion of biopsies with K10pos cells in breast gland structures from women divided into two age groups. Note that the number of K10pos biopsies are significantly higher in women aged >30 years (10 of 23) compared with those aged <30 years (3 of 23) (Pearson’s Chi-square test, p<0.05). Abbreviation: K10pos, K10-positive.
To obtain a more comprehensive expression pattern of K10 in the normal breast gland, tissue sections from 45 women aged 13 to 74 years were immunostained. A total of 956 glandular structures were evaluated, of which 54 (5.6%) contained K10pos cells (Supplemental Table S1). Overall, K10pos structures showed a fairly even distribution between lobules (38 of 604 lobules, 6.3%) and ducts (16 of 352 ducts, 4.5%) (Table 2; Supplemental Table S1).
Table 2.
Expression of K10 in the Normal Breast Gland by Immunohistochemistry.
| Fraction of Structures With K10pos Cells | ||
|---|---|---|
| n | % | |
| Lobules | 38/604 | 6.3 |
| Ducts | 16/352 | 4.5 |
| Total | 54/956 | 5.6 |
Abbreviation: K10pos, K10-positive.
Dividing the biopsies into two age groups revealed that for women <30 years, 3 of 23 biopsies (13.0%) had K10pos structures, whereas they were present in 10 of 23 biopsies (43.5%) from women >30 years (Fig. 1B, p<0.05 by Pearson’s Chi-square test).
Thus, rare K10pos luminal cells are more frequently found in normal breast gland structures in women >30 years of age.
K10pos Cells Belong to a Mature Luminal Compartment
We have previously used markers that define various differentiation states within the luminal lineage by IHC.6,13,24 Whereas cKIT defines luminal progenitor cells,14,24–26 Ks20.8 is a marker that encompasses a mature luminal compartment expressing ERα, GATA3, and CD166.13
In tissue sections double-stained for K10 and cKIT by immunofluorescence, the two markers generally segregated into different cells (Fig. 2A). Moreover, when staining for K10 and Ks20.8, the two markers co-stained, albeit with Ks20.8 more widely present in luminal cells (Fig. 2B). Furthermore, no K10pos cells were found to express the cell cycle marker Ki67 (Fig. 2C). Due to the low frequency of K10pos cells in the tissue sections, it was not possible to estimate the actual frequency of cells in cycle in comparison with luminal cells in general. Instead, a normal-derived breast epithelial cell line MCF10A containing a minor population of K10pos cells was tested. Although around 88% of MCF10A cells in general were cycling by evaluating Ki67, only around 9% of K10pos cells expressed Ki67 (Supplemental Fig. S3).
Figure 2.
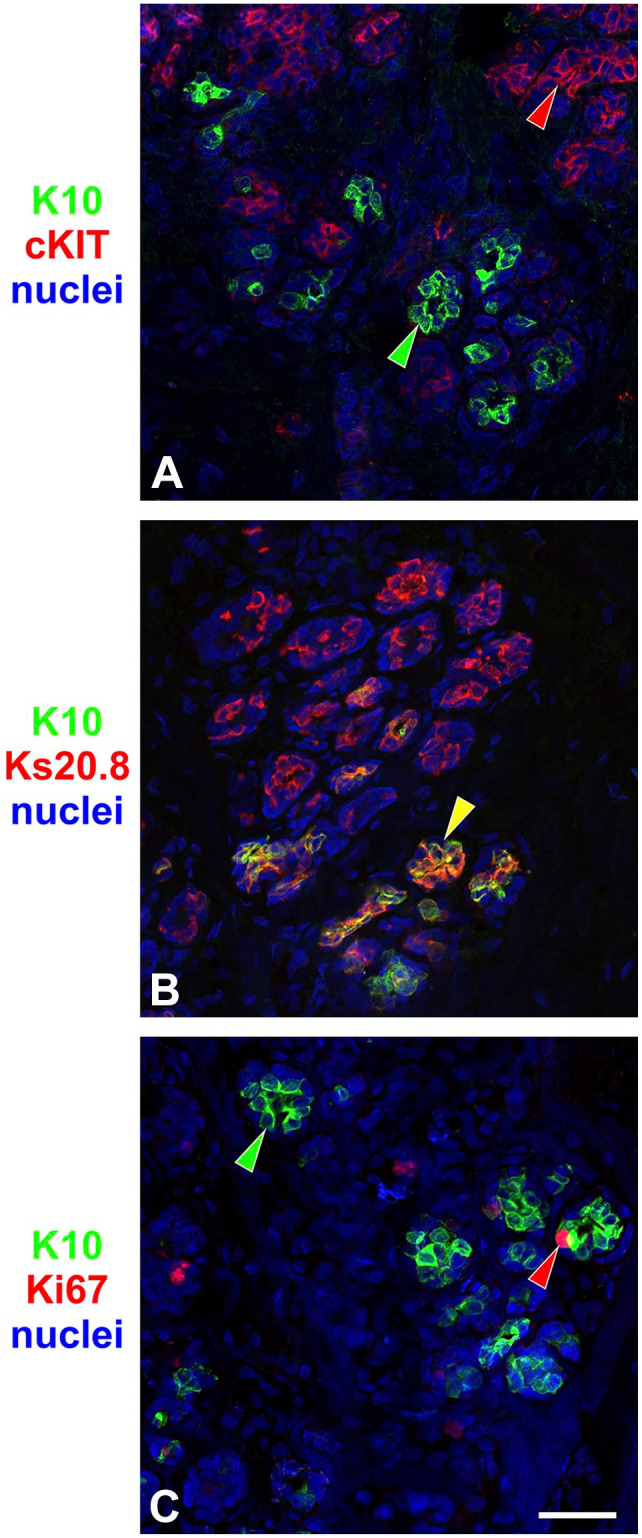
K10-positive (K10pos) cells belong to a mature luminal compartment. Immunofluorescence on normal breast tissue sections stained for K10 (green) and (A) cKIT, (B) Ks20.8, and (C) Ki67 (red). Although staining of K10 and cKIT or Ki67 segregated into different cells, K10pos cells were confined to a subset of Ks20.8pos cells. Arrowheads indicate examples of cells that are single-positive (green or red) or double-positive (yellow). Scale bar, 50 µm.
The lack of cKIT, the presence of Ks20.8, and the low number of cycling K10pos cells suggest that these cells belong to a subset of mature luminal cells in the normal breast gland.
K10pos Breast Carcinomas Mimic a Normal Luminal Differentiation Pattern
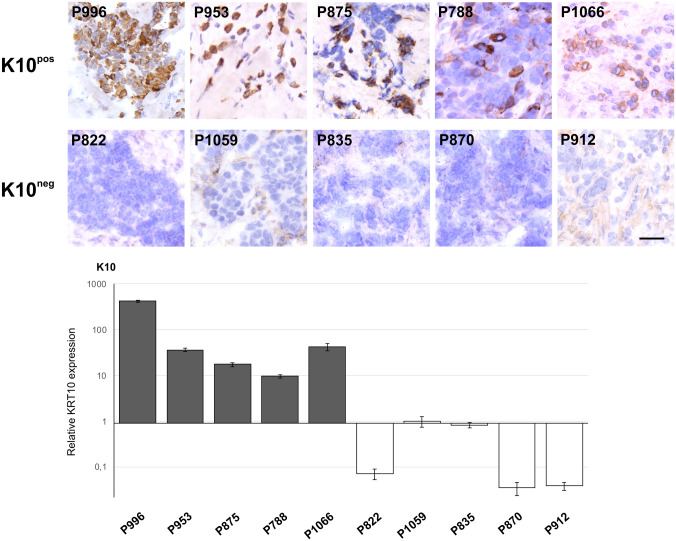
As carcinomas often reflect the epithelium of origin with regard to keratin expression,1 we stained a sample of breast carcinomas for K10 (Fig. 3B). In total, 22 of 135 tumors (16.3%) were positive (Fig. 3B; Supplemental Table S2). When subdividing the stained carcinomas into ERα-positive and ERα-negative, respectively, 17 of 84 (20.2%) and 5 of 51 (9.8%) were K10pos (Fig. 3B; Supplemental Table S2). Further classification of tumors into molecular phenotypes revealed that 13 of 74 (17.6%) Luminal A, 4 of 10 (40%) Luminal B, 1 of 16 (6.3%) HER2-enriched, and 4 of 35 (11.4%) Triple-Negative were K10pos. Detection of K10 by IHC was further corroborated by its gene expression in breast tumors verified by real-time quantitative reverse transcriptase PCR (Fig. 4).
Figure 3.
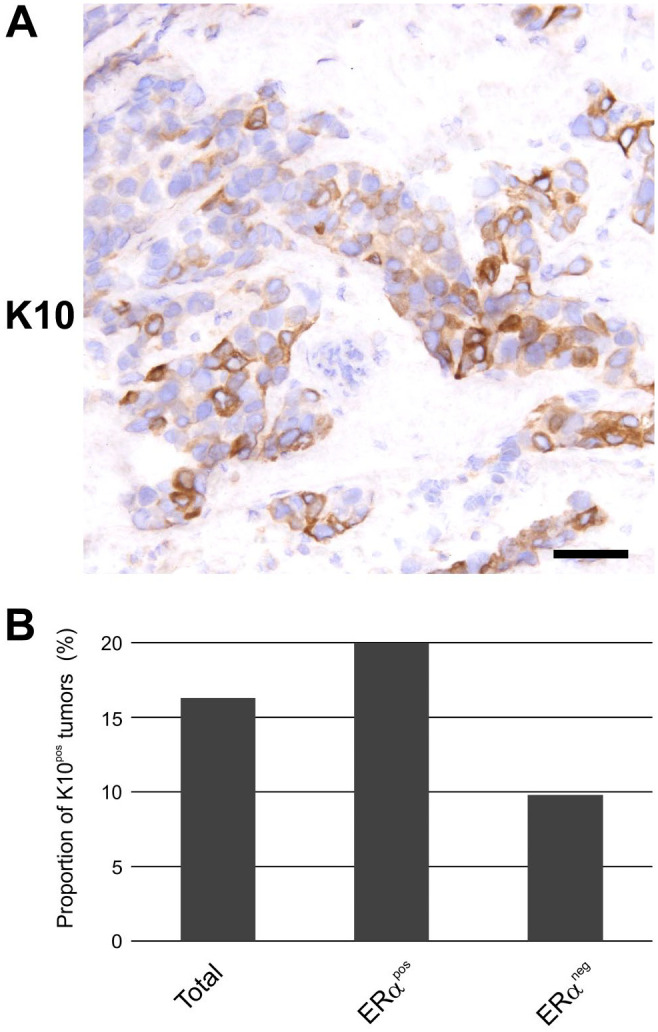
K10 is expressed in a subset of breast carciniomas. (A) Immunoperoxidase staining demonstrating heterogeneous expression of K10 in a breast carcinoma. Scale bar, 50 µm. (B) Proportion of K10pos breast carcinomas in total (n=135), as well as those divided into ERαpos (n=84) and ERαneg (n=51). Abbreviations: ERαpos, estrogen receptor α–positive; ERαneg, estrogen receptor α–negative; K10pos, K10-positive.
Figure 4.
The presence of K10 in breast carcinomas by immunohistochemistry correlates to the expression of KRT10 transcript. A selection of five K10pos and five K10neg tumors (top panel) were analyzed for relative KRT10 expression by real-time quantitative reverse transcriptase PCR. Scale bar, 50 µm. Abbreviations: K10pos, K10-positive; K10neg, K10-negative.
To analyze whether K10pos cells in breast tumors retain a phenotype mimicking a mature luminal differentiation program found in normal tissue, we verified the expression of cKIT and Ks20.8 in all K10pos tumors. Nineteen of 22 tumors were negative for luminal progenitor marker cKIT (Supplemental Table S3). None of the three cKIT-positive tumors coexpressed K10 and cKIT when tested by immunofluorescence (Fig. 5A).
Figure 5.
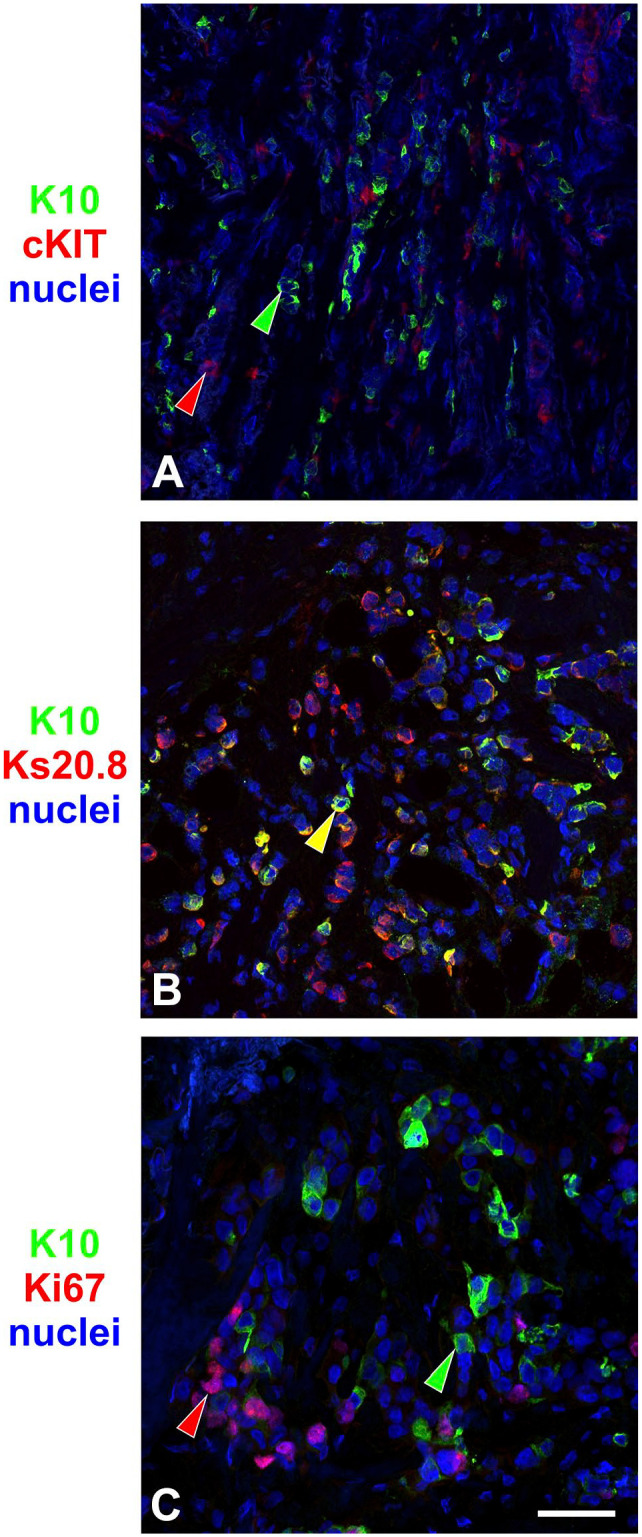
K10-positive (K10pos) breast carcinomas mimic a normal luminal differentiation pattern. Immunofluorescence on breast cancer tissue sections stained for K10 (green) and (A) cKIT, (B) Ks20.8, and (C) Ki67 (red). Like in normal tissue, staining of K10 and cKIT segregated into different cells, whereas K10 stained a subset of Ks20.8pos cells. Ki67 rarely overlapped with K10pos cells. Arrowheads mark examples of cells that are single-positive (green or red) or double-positive (yellow). Scale bar, 50µm.
Of the 22 K10pos tumors, 21 (95%) also expressed the mature luminal marker Ks20.8 (Supplemental Table S3). Testing a sample of four Ks20.8-positive tumors revealed that K10pos cells generally also expressed Ks20.8 (Fig. 5B).
Furthermore, the presence of cell cycle marker Ki67 was less frequent in K10pos cells compared with the overall fraction of cycling neoplastic cells when performing quantification on three tumor biopsies (Supplemental Fig. S4A–C). Immunostaining of breast cancer cell line MCF7 revealed a similar pattern (Supplemental Fig. S4D).
Overall, K10pos cells in breast carcinomas retain features that resemble mature luminal cells of the normal breast gland.
Expression of KRT10 Correlates to a Worse Outcome for Breast Cancer Patients
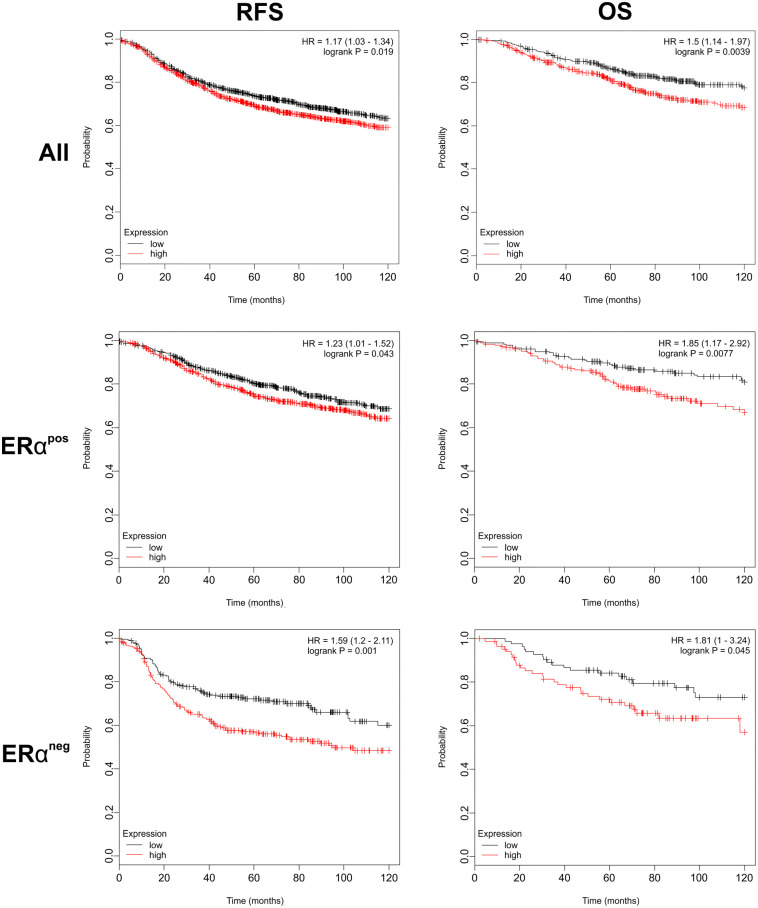
Using a web-based tool and publicly available data,27 we compared breast cancer patients with the highest and lowest levels of expression of the K10 gene, KRT10. Surprisingly, the data revealed that when compared with low levels of KRT10 in breast tumors, high levels of KRT10 correlated to a worse RFS as well as OS for breast cancer patients (Fig. 6; Supplemental Table S4A and B). This correlation was evident for both ERα-positive and ERα-negative tumors. Further subsegmentation of the available data revealed similar correlation within the Luminal A subtype, for grade 3 tumors, and for lymph node–positive tumors that were ER-negative (Supplemental Fig. S5 and Table S4C).
Figure 6.
Higher levels of KRT10 in breast cancer correlates to a worse outcome. Kaplan–Meier plots of RFS and OS of breast cancer patients stratified by RNA expression of KRT10 using a web-based survival analysis tool (kmplot.com). Patients were trichotomized into the highest tertile (red) and lowest tertile (black) for KRT10 expression and are shown for all tumors (All), ERαpos, and ERαneg. Abbreviations: ERαpos, estrogen receptor α–positive; ERαneg, estrogen receptor α–negative; OS, overall survival; RFS, relapse-free survival; HR, hazard ratio.
Thus, transcriptome analysis suggests that K10 is a marker for poor prognosis in breast cancer.
Discussion
In epidermis, K10 is a marker of cells that exit cell cycle to enter terminal differentiation.28,29 Thus, it is tempting to speculate that the presence of K10 in cells of the breast gland reflects a corresponding placement in a differentiation hierarchy. The features of K10pos luminal cells of the breast do support a placing in a similar differentiated compartment, as they are negative for cKIT, positive for Ks20.8, and generally non-cycling. However, the low frequency of K10pos cells can also lead to speculations that formation of these cells is an aberrant reminiscence of epidermal differentiation rather than a functional part of the normal differentiation program of the breast gland. In culture-based assays, myoepithelial cells derived from normal breast glands can undergo squamous cell differentiation, expressing K10.14,30 Although rare, squamous cell metaplasia can also be found in the breast gland, often with prominent histological features.31,32 However, when identified in the sample set of normal-derived biopsies that was analyzed in this study, K10pos cells did not accumulate in characteristic metaplastic structures. Rather, K10pos cells manifested themselves as infrequent single cells or distinct foci embedded as part of the normal luminal layer of glandular structures. Further investigations are required to determine whether the presence of K10 correlates to underlying metaplastic changes.
As age is the only disclosed information on the donors of the breast tissue in this study, other possible confounding events such as parity cannot fully be taken into account. In Denmark, the average age of primiparous women is 29 years,12 so the presence of an increased frequency of K10pos breast gland structures in women >30 years of age may be a reflection of age-related accumulation, of parity, or both. Although the majority of women with K10pos breast gland structures were in the age range of 43 to 53 years, the present material does not allow us to conclude whether the frequency drops in postmenopausal women.
Although it has been suggested that K10 can be expressed in breast cancer,10 previously published studies indicate that it is a rare event at most.9,33,34 A more recent reporting on K10 expression in breast tumors was only concerning rare types of carcinomas with clear squamous differentiation.11 The fact that overall around 16% of breast carcinomas in our sample set were K10pos clearly demonstrates that the expression of K10 is not confined to metaplastic tumors. The relatively frequent observation of K10pos carcinomas by immunohistochemical stainings may relate to our use of cryosections rather than paraffin-embedded tissue, which could reduce antigen masking. Interestingly, K10pos tumors were found in both ERα-positive and ERα-negative subgroups. However, as all but one of the 22 K10pos carcinomas were also positive for luminal differentiation marker Ks20.8, it could be indicative of a certain stage of differentiation. Although based on a very small sample set of only 10 tumors, it does stand out that 40% of Luminal B carcinomas were K10pos.
As the expression of K10 correlates to a more differentiated phenotype in normal tissue, it is somewhat surprising that apparently breast cancer with high KRT10 expression generally leads to a worse outcome than tumors with low levels of KRT10. We have previously reported on the presence of increased tumor-initiating capacity in a more differentiated subset of cells within a cancer stem cell hierarchy.35,36 Also, the fact that most of K10pos tumors demonstrate considerable intratumoral heterogeneity with respect to this marker may reflect the phenotypic plasticity of the tumor cells rather than the differentiation level of the tumor per se. It should be noted that in hepatocellular carcinoma it has been shown that K10 actually is a marker for poor prognosis.37 Based on this, we suggest that further investigations on the role of K10 in breast cancer are initiated.
Supplemental Material
Supplemental material, 2019-00222R1_Production_Supplemental_Material_online_supp for The Expression Pattern of Epidermal Differentiation Marker Keratin 10 in the Normal Human Breast and Breast Cancer Cells by Jiyoung Kim and René Villadsen in Journal of Histochemistry & Cytochemistry
Acknowledgments
We thank Lena Kristensen and Anita Sharma Friismose for expert technical assistance. Vera Timmermans, Pathology Department, Rigshospitalet, is acknowledged for providing tumor biopsies, and Benedikte Thuesen, Capio CFR Hospitaler, is acknowledged for providing normal breast biopsy material. The Core Facility for Integrated Microscopy (University of Copenhagen) is acknowledged for confocal microscope accessibility.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: RV conceived the project, designed the study, performed the experiments, analyzed the data, and wrote the manuscript. JK designed the study, performed the experiments, analyzed the data, and wrote the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Anita og Tage Therkelsens Fond, Læge Sophus Carl Emil Friis og hustru Olga Friis’ Legat, Oda og Hans Svenningens Fond, Tora og Viggo Groves Mindelegat, and Kong Christian den Tiendes Fond (to R.V.) and by Familien Erichsens Mindefond and Vera og Carl Johan Michaelsens Legat (to J.K.).
Contributor Information
Jiyoung Kim, Department of Cellular and Molecular Medicine, University of Copenhagen, Copenhagen, Denmark; Novo Nordisk Foundation Center for Stem Cell Biology (DanStem), Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
René Villadsen, Department of Cellular and Molecular Medicine, University of Copenhagen, Copenhagen, Denmark.
Literature Cited
- 1. Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramaekers F, Huysmans A, Schaart G, Moesker O, Vooijs P. Tissue distribution of keratin 7 as monitored by a monoclonal antibody. Exp Cell Res. 1987;170:235–49. [DOI] [PubMed] [Google Scholar]
- 3. Lane EB. Monoclonal antibodies provide specific intramolecular markers for the study of epithelial tonofilament organization. J Cell Biol. 1982;92:665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartek J, Taylor-Papadimitriou J, Miller NMR. Patterns of expression of keratin 19 as detected with monoclonal antibodies in human breast tissues and tumours. Int J Cancer. 1985;36:299–306. [PubMed] [Google Scholar]
- 5. Boecker W, Buerger H. Evidence of progenitor cells of glandular and myoepithelial cell lineages in the human adult female breast epithelium: a new progenitor (adult stem) cell concept. Cell Prolif. 2003;36(Suppl 1):73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Villadsen R, Fridriksdottir AJ, Rønnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wetzels RH, Kuijpers HJ, Lane EB, Leigh IM, Troyanovsky SM, Holland R, van Haelst UJ, Ramaekers FC. Basal cell-specific and hyperproliferation-related keratins in human breast cancer. Am J Pathol. 1991;138:751–63. [PMC free article] [PubMed] [Google Scholar]
- 8. Moll R. Cytokeratins as markers of differentiation in the diagnosis of epithelial tumors. Subcell Biochem. 1998;31:205–62. [PubMed] [Google Scholar]
- 9. Heatley M, Maxwell P, Whiteside C, Toner P. Cytokeratin intermediate filament expression in benign and malignant breast disease. J Clin Pathol. 1995;48:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naz S, Yang HJ, Lu Q, Zhou H, Li H, Xue H. Breast cancer: search of hidden culprits. Breast J. 2010;16:215–7. [DOI] [PubMed] [Google Scholar]
- 11. Boecker W, Stenman G, Loening T, Andersson MK, Berg T, Lange A, Bankfalvi A, Samoilova V, Tiemann K, Buchwalow I. Squamous/epidermoid differentiation in normal breast and salivary gland tissues and their corresponding tumors originate from p63/K5/14-positive progenitor cells. Virchows Arch. 2015;466:21–36. [DOI] [PubMed] [Google Scholar]
- 12. Ísberg ÓG, Kim J, Fridriksdottir AJ, Morsing M, Timmermans-Wielenga V, Rønnov-Jessen L, Petersen OW, Villadsen R. A CD146 FACS protocol enriches for luminal keratin 14/19 double positive human breast progenitors. Sci Rep. 2019;9:14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fridriksdottir AJ, Klitgaard MC, Kim J, Hopkinson BM, Petersen OW, Rønnov-Jessen L. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nat Commun. 2015;6:8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fridriksdottir AJ, Villadsen R, Morsing M, Klitgaard MC, Kim J, Petersen OW, Rønnov-Jessen L. Proof of region-specific multipotent progenitors in human breast epithelia. Proc Natl Acad Sci U S A. 2017;114:E10102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balk-Møller E, Kim J, Hopkinson B, Timmermans-Wielenga V, Petersen OW, Villadsen R. A marker of endocrine receptor-positive cells, CEACAM6, is shared by two major classes of breast cancer: luminal and HER2-enriched. Am J Pathol. 2014;184:1198–208. [DOI] [PubMed] [Google Scholar]
- 16. Rønnov-Jessen L, Petersen OW. Induction of alpha-smooth muscle actin by transforming growth factor-beta 1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab Invest. 1993;68:696–707. [PubMed] [Google Scholar]
- 17. Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 18. Nagy Á, Lánczky A, Menyhárt O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivanyi D, Ansink A, Groeneveld E, Hageman PC, Mooi WJ, Heintz APM. New monoclonal antibodies recognizing epidermal differentiation-associated keratins in formalin-fixed, paraffin-embedded tissue. Keratin 10 expression in carcinoma of the vulva. J Pathol. 1989;159:7–12. [DOI] [PubMed] [Google Scholar]
- 20. Borowiec AS, Delcourt P, Dewailly E, Bidaux G. Optimal differentiation of in vitro keratinocytes requires multifactorial external control. PLoS ONE. 2013;8:e77507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramaekers FC, Puts JJ, Moesker O, Kant A, Huysmans A, Haag D, Jap PH, Herman CJ, Vooijs GP. Antibodies to intermediate filament proteins in the immunohistochemical identification of human tumours: an overview. Histochem J. 1983;15:691–713. [DOI] [PubMed] [Google Scholar]
- 22. Berge U, Kristensen P, Rattan SIS. Kinetin-induced differentiation of normal human keratinocytes undergoing aging in vitro. Ann N Y Acad Sci. 2006;1067:332–6. [DOI] [PubMed] [Google Scholar]
- 23. Shin JW, Choi HR, Nam KM, Lee HS, Kim SA, Joe HJ, Kazumi T, Park KC. The co-expression pattern of p63 and HDAC1: a potential way to disclose stem cells in interfollicular epidermis. Int J Mol Sci. 2017;18:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J, Villadsen R. Expression of luminal progenitor marker CD117 in the human breast gland. J Histochem Cytochem. 2018;66:879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garbe JC, Pepin F, Pelissier FA, Sputova K, Fridriksdottir AJ, Guo DE, Villadsen R, Park M, Petersen OW, Borowsky AD, Stampfer MR, LaBarge MA. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 2012;72:3687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim E, Wu D, Pal B, Bouras T, Asselin-Labat M-L, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31. [DOI] [PubMed] [Google Scholar]
- 28. Paramio JM, Casanova ML, Segrelles C, Mittnacht S, Lane EB, Jorcano JL. Modulation of cell proliferation by cytokeratins K10 and K16. Mol Cell Biol. 1999;19:3086–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Santos M, Paramio JSM, Bravo A, Ramirez A, Jorcano JL. The expression of keratin k10 in the basal layer of the epidermis inhibits cell proliferation and prevents skin tumorigenesis. J Biol Chem. 2002;277:19122–30. [DOI] [PubMed] [Google Scholar]
- 30. Petersen OW, Van Deurs B. Growth factor control of myoepithelial-cell differentiation in cultures of human mammary gland. Differentiation. 1988;39:197–215. [DOI] [PubMed] [Google Scholar]
- 31. Habif DV, Perzin KH, Lipton R, Lattes R. Subareolar abscess associated with squamous metaplasia of lactiferous ducts. Am J Surg. 1970;119:523–26. [DOI] [PubMed] [Google Scholar]
- 32. Hurt MA, Díaz-Arias AA, Rosenholtz MJ, Havey AD, Stephenson HE. Posttraumatic lobular squamous metaplasia of breast. An unusual pseudocarcinomatous metaplasia resembling squamous (necrotizing) sialometaplasia of the salivary gland. Mod Pathol. 1988;1:385–90. [PubMed] [Google Scholar]
- 33. Santini D, Ceccarelli C, Taffurelli M, Pileri S, Marrano D. Differentiation pathways in primary invasive breast carcinoma as suggested by intermediate filament and biopathological marker expression. J Pathol. 1996;179:386–91. [DOI] [PubMed] [Google Scholar]
- 34. Bratthauer GL, Miettinen M, Tavassoli FA. Cytokeratin immunoreactivity in lobular intraepithelial neoplasia. J Histochem Cytochem. 2003;51:1527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim J, Villadsen R, Sorlie T, Fogh L, Gronlund SZ, Fridriksdottir AJ, Kuhn I, Rank F, Wielenga VT, Solvang H, Edwards PAW, Borresen-Dale A-L, Ronnov-Jessen L, Bissell MJ, Petersen OW. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci U S A. 2012;109:6124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bagger SO, Hopkinson BM, Pandey DP, Bak M, Brydholm AV, Villadsen R, Helin K, Rønnov-Jessen L, Petersen OW, Kim J. Aggressiveness of non-EMT breast cancer cells relies on FBXO11 activity. Mol Cancer. 2018;17:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xin-RongYang Xu Y, Shi GM, Fan J, Zhou J, Ji Y, Sun HC, Qiu SJ, Yu B, Gao Q, He YZ, Qin WZ, Chen RX, Yang GH, Wu B, Lu Q, Wu ZQ, Tang ZY. Cytokeratin 10 and cytokeratin 19: predictive markers for poor prognosis in hepatocellular carcinoma patients after curative resection. Clin Cancer Res. 2008;14:3850–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, 2019-00222R1_Production_Supplemental_Material_online_supp for The Expression Pattern of Epidermal Differentiation Marker Keratin 10 in the Normal Human Breast and Breast Cancer Cells by Jiyoung Kim and René Villadsen in Journal of Histochemistry & Cytochemistry