Version Changes
Revised. Amendments from Version 1
After receiving the very generous reviews, the new version of the manuscript and software reflects our attempts at improving expanding the usability of scRepertoire as a whole. As Drs. Drufuca, Bonnal, and Pagani suggested we have added scRepertoire interaction with a number of R packages and changed the definition of clonotype for B cells. Per the comments made by Dr. Stuart, we have extensively modified the code and accompanying documentation. In addition, we are in the process of submitting the package to Bioconductor. We have added an author, Gloria Kraus, who assisted in the development of the software after the paper was initially submitted. We want to thank both reviews for the suggestions, as well as a number of users, for immensely improving the scRepertoire package.
Abstract
Single-cell sequencing is an emerging technology in the field of immunology and oncology that allows researchers to couple RNA quantification and other modalities, like immune cell receptor profiling at the level of an individual cell. A number of workflows and software packages have been created to process and analyze single-cell transcriptomic data. These packages allow users to take the vast dimensionality of the data generated in single-cell-based experiments and distill the data into novel insights. Unlike the transcriptomic field, there is a lack of options for software that allow for single-cell immune receptor profiling. Enabling users to easily combine mRNA and immune profiling, scRepertoire was built to process data derived from 10x Genomics Chromium Immune Profiling for both T-cell receptor (TCR) and immunoglobulin (Ig) enrichment workflows and subsequently interacts with a number of popular R packages for single-cell expression, such as Seurat. The scRepertoire R package and processed data are open source and available on GitHub and provides in-depth tutorials on the capability of the package.
Keywords: Single-cell RNA sequencing, immune receptor profiling, R, clonotypic analysis
Introduction
The molecular resolution offered by single-cell sequencing (SCS) technologies has led to extensive investigations in the realms of developmental biology, oncology, and immunology. In terms of the latter field, SCS offers the ability to couple the exploration of transcriptomic heterogeneity in immune cells along a disease process with clonality 1. A number of methods exist for dimensional reduction of mRNA data, reviewed by Chen et al. 2 that have been implemented into R packages to assist in processing and analysis of SCS experiments. However, a gap exists in the processing of V(D)J sequencing, descriptive statistics, clonal comparisons, and repertoire diversity with the current SCS R packages.
With these limitations in mind, scRepertoire 3 was generated ( Figure 1). Built using R, scRepertoire is a toolkit to assist in the analysis of immune profiles for both B and T cells, while interacting with the popular Seurat pipeline 4– 6, as well as SingleCellExperiment and monocle3 class expression objects. scRepertoire also includes processed single-cell mRNA and V(D)J sequencing data of 12,911 tumor-infiltrating and peripheral-blood T cells derived from three renal clear cell carcinoma patient, which is characterized below to demonstrate the capabilities of the package.
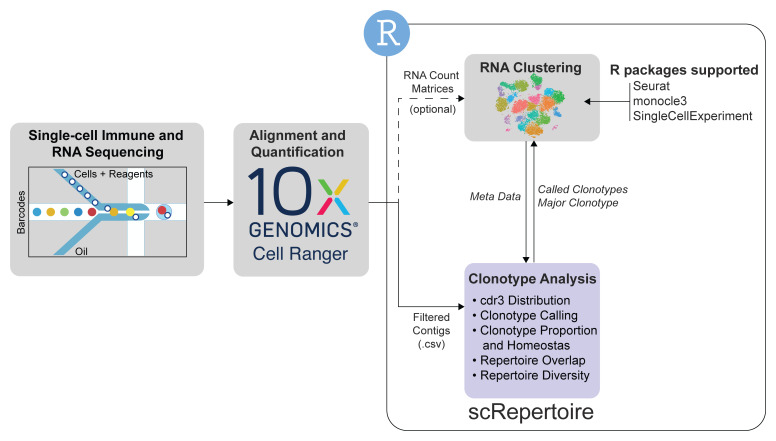
Figure 1. A general workflow for single-cell data analysis involving scRepertoire.
The analysis starts with the single-cell immune and mRNA sequencing and Cell Ranger-based alignment with the 10x Genomics pipeline. With the TCR or Ig sequencing, scRepertoire can import the filtered overlapping DNA segments, or contigs. The alignments are filtered by cell type of interest and combined using the individual cell barcodes. Clonotypes can be called using the gene sequence of the immune receptor loci, CDR3 nucleotide sequence or CDR3 amino acid sequence. After clonotype assignment, more extensive clonotypic analysis can be performed at the individual sample level or across all samples. General outputs from scRepertoire can be imported into a number of single-cell expression formats to visualize clonotype data overlaid onto the cell clustering. Likewise, metadata from the expression objects can be imported into scRepertoire to analyze clonotypes by assigned clusters.
Methods
Operation
System requirements for running scRepertoire 3 include the installation of R v3.5.1 and the the Seurat R package (v3.1.2). Utilization of scRepertoire is dependent on the total number of single-cells being processed, with a base estimate of 1 Gb of random-access memory and a modern CPU.
Data
The isolation and processing of the 10x-Genomics-based single-cell mRNA and V(D)J Chromium sequencing data for immune cells has previously been described 7, 8. In addition, T cells were identified using expression values for canonical T cell markers: CD3D, CD4, CD8A, CD8B1 and previous clustering. T cells were isolated and reclustered using the integration method from the Seurat R package (v3.1.2) with 20 principal components and a resolution of 0.5 4. All code used to generate the figures appearing in the manuscript is available at https://github.com/ncborcherding/scRepertoire.
Implementation
The scRepertoire was built and tested in R v3.5.1. Analysis for scRepertoire was inspired from the bulk immune profiling tcR (v2.2.4) R package without derivations in code 9. Clonotypes can be called using the combination of immune loci genes, a more sensitive approach, or the nucleotide/amino acid sequence of the complementary-determining region 3 (CDR3). In addition to the base functions in R, data processing was performed using the dplyr (v0.8.3) and reshape2 (v1.4.3) R packages. Visualizations are generated using the ggplot2 (v3.2.1) and ggalluvial (v0.11.1) R packages with color pallets derived from the use of colorRamps (v2.3) and RColorBrewer (v1.1.2) R packages. Diversity metrics are calculated using the vegan (v2.5-6) R package. Visual outputs of functions are stored as layers of geometric or statistical ggplot layering, allowing users to easily modify presentation.
Results
Clonal analysis
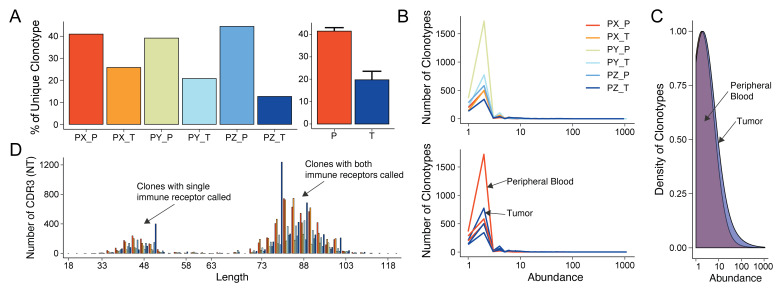
scRepertoire 3 can be used to call clonotypes using the CDR3 amino acid/nucleotide sequences, by gene usage, or by the combination of CDR3 nucleotide sequences and genes. Using the quantContig function, unique clonotypes can be visualized as raw values or scaled to the size of the library for samples or by type ( Figure 2A). The total abundance of clonotypes can also be visualized calling abundanceContig ( Figure 2B) or relative abundance of clonotypes ( Figure 2C). Additionally, the distribution of CDR3 nucleotide or amino acid sequences for clonotypes can be visualized with lengthContig ( Figure 2D). More advance distribution analysis is also available using the clonesizeDistribution function based on recent work using Jensen-Shannon divergence.
Figure 2. Basic clonotypic analysis functions in scRepertoire.
( A) Scaled unique clonotypes by total number of TCRs sequenced by patient and type of sample (peripheral, P; tumor, T), using the quantContig function. ( B) Total abundance of clonotypes by sample and type using the abundanceContig function. ( C) Relative abundance of clonotypes using density comparing peripheral blood to tumor samples. ( D) CDR3 nucleotide length analysis by sample using the lengthContig function. The bimodal nature of the curve is a function of calling clonotypes for cells with both one and two immune receptors sequenced.
Proportional analysis and diversity measures
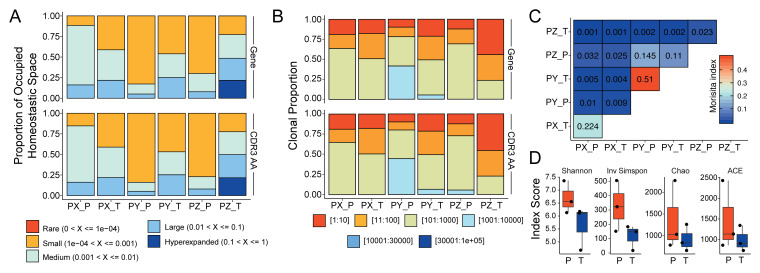
More in depth analysis of clonal architecture is available. Within the framework of scRepertoire, analysis of clonal homeostasis, or the clonal space occupied by clonotypes of specific proportions, can be visualized by clonalHomeostasis function ( Figure 3A). Similarly, clonalProportion can be called to look at the proportion of clonal space occupied by specific clonotypes ( Figure 3B). Overlap between the samples can be calculated and visualized with clonalOverlap, using either the overlap coefficient or Morisita index methods ( Figure 3C). Measured of diversity across samples or groups can be quantified with the clonalDiversity function, demonstrating an overall reduction in clonal diversity in tumor samples ( Figure 3D).
Figure 3. Advanced clonal measures between samples.
( A) Clonal homeostatic space representations across all six samples using the gene and CDR3 AA sequence for clonotype calling. ( B) Relative proportional space occupied by specific clonotypes across all six samples using the gene and CDR3 AA sequence for clonotype calling. ( C) Morisita overlap quantifications for clonotypes across all six samples. ( D) Diversity measures based on clonotypes by sample type using Shannon, Inverse Simpson, Chao, and abundance-based coverage estimator (ACE) indices.
Expression interaction
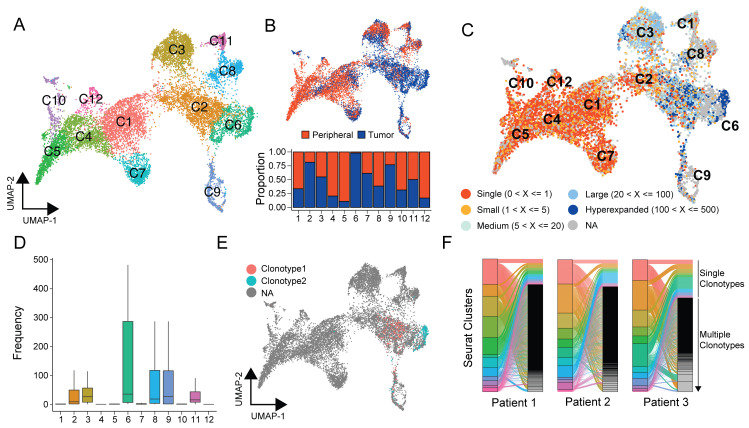
After the processing and analysis of the TCR repertoire with the base features, the next step is using scRepertoire to interact with the single-cell mRNA data. The expression data for the 12,911 cells built into the package have already been clusters ( Figure 4A), with a clear distribution of the clusters into peripheral-blood- versus tumor-predominant ( Figure 4B). Using the combineExpression function in scRepertoire, we can look at the clonotypic frequencies of cells that comprise the UMAP-based clusters ( Figure 4C). This function also works with the SingleCellExperiment and monocle3 class of expression objects. ( Figure 4D). In addition to clonal distribution, we can also use highlightClonotypes to set specific sequences of clonotypes to be visualized ( Figure 4D), with clonotype 1 referring to the amino acid sequence “CAVNGGSQGNLIF_CSAEREDTDTQYF” and clonotype 2 for the amino acid sequence "NA_CATSATLRVVAEKLFF". Interesting clonotype 2 is restricted to a subcluster of the C6 cluster ( Figure 4D). After combining both the clonotype and expression data, interaction between categories, such as cluster label and clonotype frequency can be visualized with the alluvialClonotypes function ( Figure 4E). This function can also be used to examine the dynamics of single or multiple expanded clonotypes across the categorical variables ( Figure 4E). Further, after the attachment of the expression information to a single-cell expression object, the function, expression2List() allows users generate analyses based on any categorical variable in the meta data..
Figure 4. Interaction of scRepertoire with the single-cell expression R packages.
( A) UMAP projection from Seurat of the ccRCC T cells (n=12,911) into 12 distinct clusters. ( B) UMAP projection with peripheral blood (red) and tumor (blued) populations highlighted and an accompanying relative proportion composition of each cluster, scaled by the total number of peripheral blood and tumor cells, respectively. ( C) Using the combineExpression function places individual cells into groups by the number of clonotypes, which then can be displayed overlaid with the UMAP projection. ( D) After combining the clonotype information with the Seurat object, highlightClonotypes can be used to specifically highlight the individual clonotypes of interest using the sequence information. ( E) Interaction of clonotypes between multiple categories can be examined using the alluvialClonotypes function.
Conclusions
scRepertoire 3 is a R-based toolkit for the analysis of single-cell immune receptor profiling. The package is able to take the annotated filtered outputs from the 10x Genomics Cell Ranger platform and provide analysis a number of modalities, including calling clonotypes, clonal space/homeostasis, clonal diversity, and repertoire overlap between samples. Outputs from scRepertoire can combined with dimensional reduction strategies for single-cell RNA quantifications, allowing users to analyze mRNA and immune profiles together. Visualization functions in scRepertoire have a parameter, exportTable, allowing users to examine the quantifications underlying the generation of the graphs. Under the creative commons v4.0 license, the scRepertoire package is freely available from the GitHub repository and is extensively annotated to assist in implementation and modification.
Data availability
Source data
Zenodo: scRepertoire. https://doi.org/10.5281/zenodo.3856827 3.
Folder ‘Data’ contains all data required to run the vignettes described in the Results. This is also available on GitHub.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Software availability
Source code is available from GitHub: https://github.com/ncborcherding/scRepertoire.
Archived source code at the time of publication: https://doi.org/10.5281/zenodo.3856827 3.
Acknowledgements
We would like to thank Davide Angeletti of the University of Gothenburg and Jae Seung Moon of Stanford University for extensive beta testing and suggestions for improvement.
Funding Statement
Funding for this project was provided from National Institute of Health F30 fellowship CA206255.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- 1. Papalexi E, Satija R: Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- 2. Chen G, Ning B, Shi T: Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front Genet. 2019;10:317. 10.3389/fgene.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borcherding N, Bormann NL: scRepertoire (Version 1.2.0). Zenodo. 2020. 10.5281/zenodo.3856827 [DOI] [PMC free article] [PubMed]
- 4. Stuart T, Butler A, Hoffman P, et al. : Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macosko EZ, Basu A, Satija R, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler A, Hoffman P, Smibert P, et al. : Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borcherding N, Ahmed KK, Voigt AP, et al. : Transcriptional heterogeneity in cancer-associated regulatory T cells is predictive of survival. bioRxiv. 2018; 478628. 10.1101/478628 [DOI] [Google Scholar]
- 8. Vishwakarma A, Bocherding N, Chimenti MS, et al. : Mapping the Immune Landscape of Clear Cell Renal Cell Carcinoma by Single-Cell RNA-seq. bioRxiv. 2019; 824482. 10.1101/824482 [DOI] [Google Scholar]
- 9. Nazarov VI, Pogorelyy MV, Komech EA, et al. : tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics. 2015;16:175. 10.1186/s12859-015-0613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]