Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer type characterized by dysregulated cell signalling pathways and resistance to treatment. The insulin-like growth factor (IGF) signalling pathway has been identified to have a role in tumour progression and therapy resistance. However, its regulatory roles in PDAC have remained to be fully elucidated. In the present study, dysregulated microRNAs (miRNAs) in PDAC were explored with a focus on those that may be involved in regulating the insulin/IGF signalling pathway. A total of 208 patients were recruited, comprising 112 patients with PDAC, 50 patients with chronic pancreatitis (CP) and 46 subjects as a control group (CG). miRNA-specific quantitative PCR assays were used to measure 300 candidate miRNAs. The Student's t-test was applied to compare miRNA regulation between cancer patients and controls with a false discovery rate correction using Bonferroni-type comparison procedures. The DIANA-mirPath v.3 tool and HMDD v3.0 were used to identify miRNA-mRNA interactions within specific pathways. In patients with PDAC, 42 miRNAs were significantly upregulated and 42 were downregulated compared to the CG (P<0.01). In the PDAC vs. CP analysis, 16 significantly (P<0.01) upregulated and 16 downregulated miRNAs were identified. Of note, members of the let-7 family of miRNAs were downregulated and were indicated to target several components of the insulin receptor (INSR)/IGF pathway, including receptors and binding proteins, for upregulation and thus, may enable the activation of the pathway. Downregulation of the let-7 family may help promote the INSR/IGF pathway in PDAC. It may thus be an effective target for the development of INSR/IGF pathway-specific treatment strategies.
Keywords: microRNA, let-7, insulin receptor/insulin growth factor, pancreatic ductal adenocarcinoma, dysregulated pathways
Introduction
According to 2018 GLOBOCAN statistics, pancreatic cancer had 458,918 new cases and 432,242 mortalities, which accounted for 4.5% of cancer-associated deaths worldwide (1,2). Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, has a poor prognosis with a five-year overall survival rate of 5%, which has not significantly improved over the last two decades despite improved chemotherapeutic and biological agents (3). Hence, there is a requirement to better understand the underlying molecular mechanisms of the disease as a basis for identifying novel and more effective therapeutic targets.
More than one growth factor potentiates carcinogenesis and multiple growth factor signalling pathways are consequently upregulated in cancerous cells (4–6). The epidermal growth factor family of receptors (EGFR) are overexpressed in >60% of PDAC tumours (7); however, EGFR-targeted therapies have been unsuccessful in improving overall patient survival (5,8). Hence, there must be a simultaneous upregulation of alternative pathways. One such potential pathway is the insulin/insulin-like growth factor (IGF) pathway. In PDAC, overexpression of both IGF-1 and type 1 IGF receptor (IGF-1R) has been demonstrated (9). Furthermore, EGFR has been identified to be co-expressed with IGF-1R, an interaction that confers resistance to EGFR-specific therapies (10).
The IGF signalling pathway is important for cellular growth, proliferation and differentiation, as well as for tissue integrity (11). It has been indicated to be aberrant in PDAC (12), enhancing tumourigenesis and therapeutic resistance (13); furthermore, IGF-1R is associated with increased proliferation and angiogenesis and decreased apoptosis (14–16). A recent study by our group reported on the upregulation of the IGF signalling pathway in resected pancreatic tumours obtained from South African patients (17). Furthermore, IGF-1R has been demonstrated to have predictive and prognostic value, as its overexpression has been associated with metastases and decreased overall survival in PDAC (5), highlighting the requirement to further investigate the regulation of this pathway and its mechanism of action in PDAC.
MicroRNAs (miRNAs) are non-coding RNAs ~17–24 nucleotides in length, which are involved in biological and cellular processes by regulation of mRNA translation (18,19). Circulating miRNAs have been determined to be involved in tumorigenesis serving as biomarkers and therapeutic targets (20–24). In the present study, differentially expressed miRNAs in patients with PDAC compared to patients with chronic pancreatitis (CP) and a control group (CG) were identified. Using bioinformatics analysis, the possible regulation of the insulin receptor (INSR)/IGF signalling pathway activity by miRNAs was further investigated.
Patients and methods
Patients
A total of 112 patients with cytologically or histologically confirmed treatment-naive PDAC located in the head of the pancreas were recruited at the time of diagnosis in Chris Hani Baragwanath Hospital, Johannesburg, South Africa from January 2014 to December 2016. Participants were categorized into three clinical groups according to their TNM staging (25). Group 1 consisted of participants with resectable, non-metastatic disease (TNM stage 0-IIB), Group 2 comprised patients with non-resectable disease without evidence of metastases (TNM stage III) and Group 3 included participants with metastatic disease (TNM stage IV). Participants with tumours other than PDAC were excluded. All clinical information was collected and stored using REDCap v6.7 electronic data capture tools (26).
As the CP group, patients who had completed at least one year of out-patient follow-up with the Hepatopancreatobiliary unit in Chris Hani Baragwanath Hospital, (Johannesburg, South Africa) and admitted between January 2014 to December 2016 and had an abdominal CT scan demonstrating no mass or lesion suspicious of a tumour in their pancreas within two weeks of their recruitment date were included. CP was diagnosed radiologically or endoscopically according to international guidelines (27,28). Any patient with CP and a pancreas mass or cyst was excluded.
As the CG, consecutive patients admitted to the vascular ward in Chris Hani Baragwanath Hospital (Johannesburg, South Africa) and admitted between January 2014 to December 2016 with severe peripheral arterial disease who underwent an abdominal CT scan as part of their clinical work-up were included if the scan demonstrated a healthy pancreas. Participants were excluded if they had a history of any previous malignancy or pancreatic disease, obstructive jaundice, unexplained diarrhoea or weight loss.
Sample processing
From each participant, venous blood samples were collected in 4.5-ml tubes containing EDTA and stored at room temperature. Samples were centrifuged for 15 min at 1,500 × g at room temperature within 4 h of collection. The plasma was aliquoted into 1-ml Eppendorf tubes and stored at −80°C. Analysis was performed within 12 months of sample collection and storage.
Reverse transcription-quantitative PCR (RT-qPCR)
Plasma miRNA was extracted using the miRNeasy serum/plasma miRNA isolation kit (Qiagen GmbH). The ID3EAL miRNA knowledge panel 384 Target kit (MiRXES) was used and experiments were performed according to the manufacturer's instructions. The kit contained the ID3EAL Panel RT Primer Pool for the miRNAs with a set of three proprietary spike-in RNAs [ID3EAL Panel RNA Spike-In (MiRXES)] to normalize variations in RNA isolation efficiency. Isolated serum RNA was subjected to RT using the ID3EAL Reverse Transcriptase kit (MiRXES). A 6-log serial dilution of synthetic templates for each miRNA was concurrently reversed-transcribed. Using the miRNA-specific qPCR assays-ID3EAL miRNA qPCR Master Mix (MiRXES), >300 candidate miRNAs were measured in each complementary DNA sample. The following cycling conditions were used: 95°C for 10 min, 40°C for 5 min, and then 45 cycles of 95°C for 10 sec and 60°C for 30 sec. Absolute copy numbers of each miRNA were determined by interpolation of the Cq values to that of the synthetic miRNA standard curves and adjusted for RT-qPCR efficiency variation. The 2−ΔΔCq method was used for quantification (29).
Data normalization and statistical analysis
A set of three endogenous reference miRNAs were identified using geNorm and NormFinder (30,31). The geometric mean of the three normalizers was used to normalize the miRNA expression data across subjects. Student's t-test was applied to compare miRNA expression between cancer and control groups, with false discovery rate correction using Bonferroni-type comparison procedures (32). The Sequential Forward Floating Search algorithm (33) was used to optimize the biomarker selection during the internal cross-validation process for the discovery study and AUC values were set as the optimization target. The linear support vector machine was used to construct multi-variant biomarker panels with optimal performance in classifying control and cancer groups. Demographic data and clinical characteristics of the three patient groups were compared using one-way analysis of variance. Bonferroni's test was used for post hoc analysis. P<0.05 was considered to indicate statistical significance. The Stata v14 statistical programme (StataCorp) was used for analyses and values expressed as n (%) or median (range). For values represented as n (%), a student's t-test was performed for the normally distributed continuous variables, and P<0.05 was considered significant.
Bioinformatics analysis
Dysregulated miRNAs were considered if they had a significance of P<0.01 and absolute fold change of >1.5. The DIANA-mirPath v.3 tool was used for interaction and pathway analyses (34). The tool uses Tarbase v.8 to identify miRNA-mRNA interactions and probes the Kyoto Encyclopedia of Genes and Genomes for pathway analysis (35,36). GeneMania and HMDD v3.0 were used to visualize interactions of target genes and observe miRNA-interacting partners, respectively (37,38). The highlighting of key genes of interest on pathways and network was drawn using Microsoft PowerPoint v2013 (Microsoft).
Results
Clinicopathological characteristics of study participants
The study included 208 subjects, comprising 112 patients with PDAC, 50 patients with CP and 46 patients with critical limb ischaemia as a CG. The demographic features and traditional risk factors of the subjects are provided in Table I. In the PDAC group, there were 15 patients in Group 1, 57 patients in Group 2 and 40 in Group 3. The 6-month overall survival rate for Groups 1, 2 and 3 was 80.0, 91.2, and 32.5%, respectively (Table SI). There were 35 patients with survival for <6 months, 27 of which were from Group 3. The body mass index of all of the patients was determined and was not significantly different between the three study groups.
Table I.
Demographic and clinical characteristics of patients recruited for the study.
| Variable | PDAC (n=112) | CP (n=50) | Control (n=46) | P-value (PDAC vs. CP) | P-value (PDAC vs. control) |
|---|---|---|---|---|---|
| Male/female ratio | 1.65:1 | 4.2:1 | 1.4:1 | 0.179 | 0.901 |
| Agea, years | 59 (33–85) | 52 (38–69) | 61 (35–89) | P<0.001 | 0.704 |
| Chronicb illness | 69 (62) | 16 (32) | 31 (67.3) | 0.095 | 0.692 |
| Smoking statusb | |||||
| Ever smoked | 72 (64) | 44 (88) | 26 (57.1) | P<0.003 | 0.120 |
| Current smoking | 29 (26) | 34 (68) | 7 (15) | P<0.001 | P<0.02 |
| Alcohol useb | 78 (70) | 45 (90) | 23 (50) | P<0.004 | 0.088 |
| Body mass indexa, kg/m2 | 21.4 (15.6–29.2) | 19.0 (14.8–24.5) | 22.1 (16.1–26.3) | 0.366 | 0.423 |
Values are expressed as
median (range)
n (%). PDAC, pancreatic ductal adenocarcinoma; CP, chronic pancreatitis.
Differential expression of miRNA
miRNA expression was compared between the PDAC group and the CG and 42 significantly upregulated and 42 downregulated miRNAs were identified (P<0.01; Table SII). When comparing the PDAC group to both the control and chronic pancreatitis groups combined together (PDAC vs. CG+CP), 16 significantly upregulated and 16 downregulated miRNAs in PDAC were identified (P<0.01; Fig. 1; Table SIII). Of note, the downregulated miRNAs included members of the let-7 family, including Homo sapiens (hsa)-let-7f-5p and hsa-let-7a-5p. Comparison of the PDAC group with solely the CP group provided only three significantly upregulated and four downregulated miRNAs (P<0.01; Table SIV).
Figure 1.
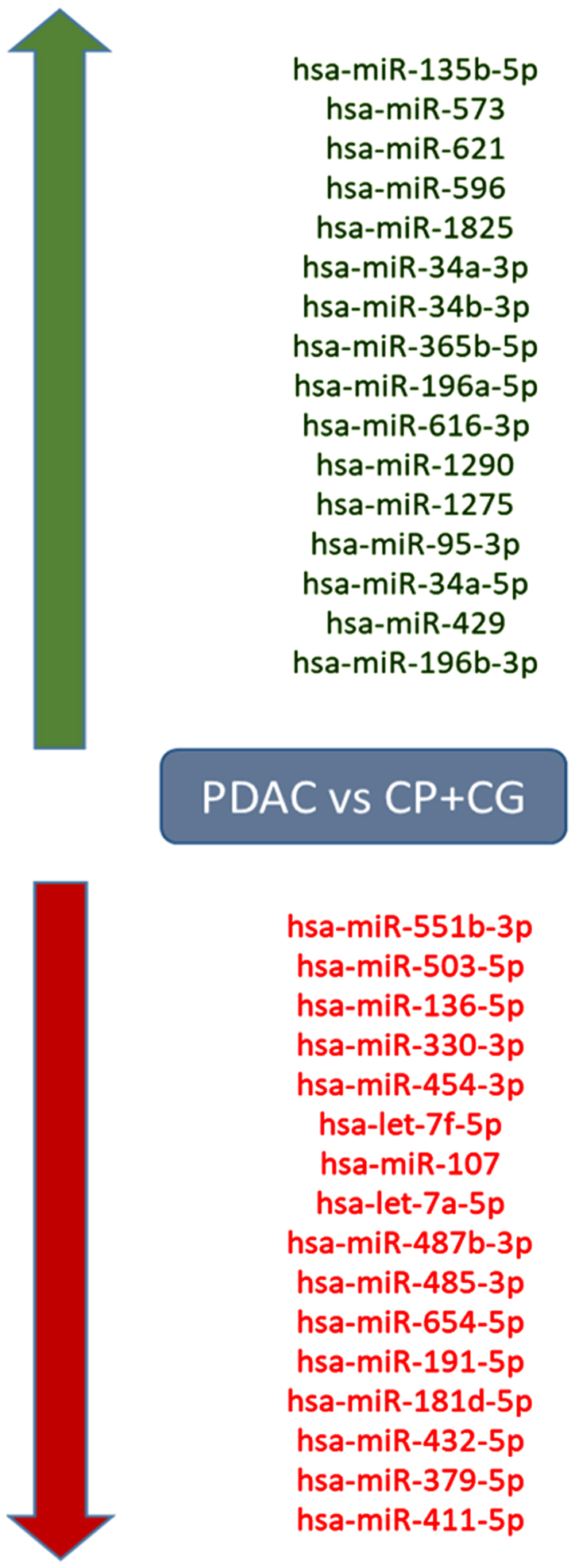
Dysregulated miRNAs in patients with PDAC. Compared to patients with CP and the control group, patients with PDAC had 16 significantly upregulated and 16 downregulated miRNAs (P<0.01). The red colour indicates downregulation and green denotes upregulation. CP, chronic pancreatitis; CG, control group; PDAC, pancreatic ductal adenocarcinoma; hsa, Homo sapiens; miRNA/miR, microRNA.
Let-7 miRNAs target the INSR and IGF signalling pathways
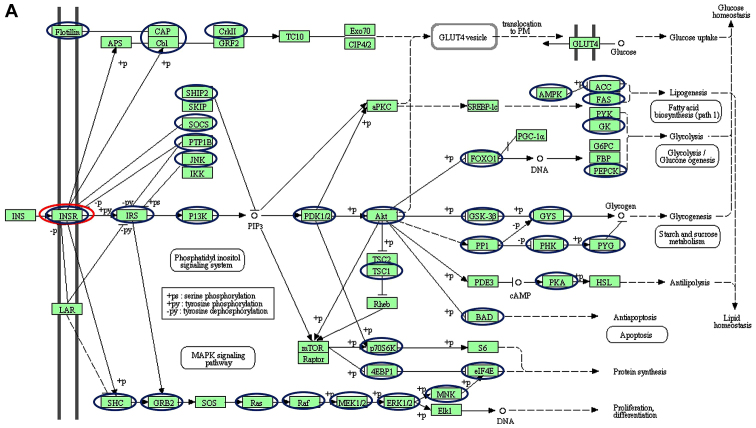
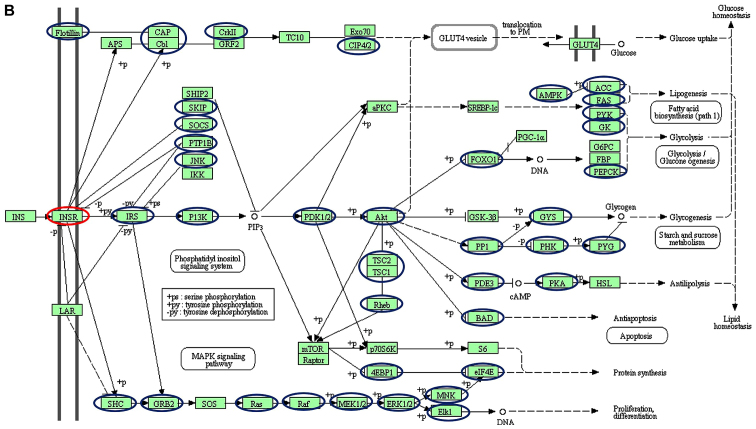
Using a targeted bioinformatics analysis, it was demonstrated that the downregulated and upregulated miRNAs targeted several components of the insulin pathway. However, only the downregulated miRNAs were observed to target INSR (Figs. 2 and 3). Amongst the downregulated miRNAs, members of the let-7 family, hsa-let-7f-5p and hsa-let-7a-5p, were implicated in targeting INSR.
Figure 2.
Schematic of the insulin signalling pathways, including the different genes targeted by (A) downregulated and (B) upregulated miRNAs. Blue and red nodes indicate genes in the pathways determined by the KEGG software to be targeted by dysregulated miRNAs. Of interest, INSR is only targeted by the downregulated let-7 family (red nodes). The figure was generated from the Kyoto Encyclopedia of Genes and Genomes pathways. The nodes (circles) were drawn onto the KEGG-generated figure using Microsoft Powerpoint v2013 to highlight targeted genes. INSR, insulin receptor; miRNA, microRNA; CAP, catabolite gene activator protein; Cb1; cannabinoid receptor interacting protein 1; CRKII, CRK proto-oncogene II; GRF2, general regulatory factor 2; TC10, TC10 protein; EXO70; exocyst complex protein; CIP, Cdc42-interacting protein; GLUT4, glucose transporter type 4; Lar, low antibody response; SHIP2 (INPPL1), inositol polyphosphate phosphatase like 1; SKIP, skeletal muscle and kidney-enriched inositol phosphatase; SOCS (CISH), cytokine inducible SH2 containing protein; PTPIB, peptidylprolyl isomerase B; JNK, c-Jun NH2-terminal kinase; IKK, I-κ B kinase; P13K, phosphoinositide 3-kinase; αPKC, protein kinase Cα; PDK, pyruvate dehydrogenase kinase; SREBP-1C, SREBP-sterol regulatory element binding protein 1C; PRKAA1 (AMPK), protein kinase AMP-activated catalytic subunit α1; ACC, acetyl-CoA carboxylase; FAS, Fas cell surface death receptor; PyK, pyruvate kinase; GK, glycerol kinase; G6PC, glucose-6-phosphatase catalytic subunit; FBP, fructose-1,6-bisphosphatase 1; PEPCK, phosphoenolpyruvate carboxykinase; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1-α; FOXO1, Forkhead box protein O1; AKT (PKB), protein kinase B; GSK3B, glycogen synthase kinase 3β; GYS, glycogen synthase; PP1, serine/threonine protein phosphatase; PHKA2 (PHK), phosphorylase kinase regulatory subunit α 2; PYG, glycogen phosphorylase; TSC1, TSC complex subunit 1; PDE3, phosphodiesterase 3; PKA, protein kinase; HSL, hormone-sensitive lipase; BAD, BCL2-associated agonist of cell death; RPS6KB1 (P70S6K), ribosomal protein S6 kinase B1; MTOR, mechanistic target of rapamycin kinase; EIF4EBP1 (4EBP1), eukaryotic translation initiation factor 4E binding protein 1; S6, ribosomal protein S6; EIF4E, eukaryotic translation Initiation factor 4E; MNK, mitogen-activated protein kinase (MAPK) interacting protein kinase; ELK1, ETS transcription factor; ERK, extracellular signal-regulated kinase; MEK (MAP2K), mitogen-activated protein kinase kinase; RAF, RAF proto-oncogene serine/threonine-protein kinase; RAS proto-oncogene; GRB2, growth factor receptor bound protein 2; SHC, SHC adaptor protein 1; SOS, SOS Ras/Rac guanine nucleotide exchange factor.
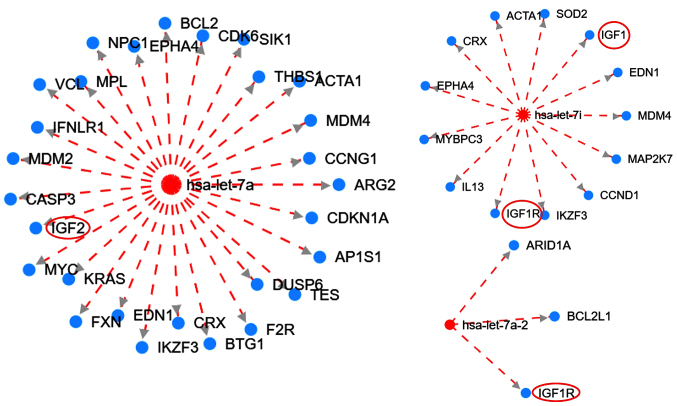
Figure 3.
Interaction network of the let-7 family targeting the IGF family members generated from HMDD version 3. Red circles (added with Microsoft PowerPoint v2013) indicate members of the IGF signaling pathway. The broken red lines indicate downregulation. Red and blue nodes are microRNAs and genes, respectively. IGF1R, type 1 insulin-like growth factor receptor; IGF, insulin-like growth factor; ARID1A, AT-rich interaction domain 1A; BCL2L1, BCL2 like 1; EDN1, endothelin 1; MDM4, MDM4 regulator of p53; MAP2K7, mitogen-activated protein kinase kinase 7; CCND1, cyclin D1; IKZF3, IKAROS family zinc finger 3; IL13, interleukin 13; MYBPC3, myosin binding protein C3; EPHA4, EPH receptor A4; CRX, cone-rod homeobox; ACTA1, actin α1 skeletal muscle; SOD2, superoxide dismutase 2; BCL2, BCL2 apoptosis regulator; CDK6, cyclin dependent kinase 6; SIK1, salt inducible kinase 1; THBS1, thrombospondin 1; CCNG1, cyclin G1; ARG2, arginase 2; CDKN1A, cyclin dependent kinase inhibitor 1A; AP1S1, adaptor related protein complex 1 subunit σ1; DUSP6, dual specificity phosphatase 6; TES, testin LIM domain protein; F2R, coagulation factor II thrombin receptor; BTG1, BTG anti-proliferation factor 1; FXN, frataxin; KRAS, KRAS proto-oncogene; MYC, MYC proto-oncogene; CASP3, caspase 3; MDM2, MDM2 proto-oncogene; IFNLR1, interferon λ receptor 1; VCL, vinculin; MPL, MPL proto-oncogene; EPHA4, EPH receptor A4; hsa, Homo sapiens.
Furthermore, the HDMM tool was used to illustrate the interaction between the let-7 family of miRNAs with several genes, including members of the IGF signalling pathway (Fig. 3). The analysis suggested that let-7 targets the IGF signalling pathway for downregulation. Let-7 has an inhibitory effect on the IGF pathway and is downregulated, suggesting the subsequent upregulation of this pathway.
Expression of the let-7 family in different stages of PDAC
The levels of let-7 miRNAs were differentially expressed in different groups of PDAC (Table SVa). Further analysis across the PDAC groups indicated that 5 miRNAs were significantly upregulated in Group 3 compared to Groups 1 and 2 combined (P<0.05; Table SVb). Although the differences in expression levels were not significant between ‘Group 3 vs. 1+2’, certain members of the let-7 miRNAs were differentially expressed across the various groups. When comparing Group 3 to a combination of Groups 1 and 2, hsa-let-7b-5p, hsa-let-7a-3p and hsa-let-7d-3p were upregulated and hsa-let-7a-5p and hsa-let-7f-5p were downregulated. Furthermore, hsa-let-7b-3p, hsa-let-7d-3p, hsa-let-7a-5p and hsa-let-7e-3p were upregulated in Group 3 vs. Group 2 but downregulated in Group 2 vs. 1. On the other hand, hsa-let-7a-5p and hsa-let-7f-5p were downregulated in Group 3 vs. 2 but upregulated in Group 2 vs. 1. The potential prognostic value of the let-7 miRNAs was also assessed by comparing levels observed in patients with a ‘poor prognosis’ and those with a ‘good prognosis’ (Table SI). A poor or good prognosis was defined by the survival of patients in months; and since patients were followed up for only one year, those with a survival time of <12 months were categorized as having a ‘poor prognosis’. Comparing patients in the poor vs. good prognosis group, a total of 20 miRNAs were associated with poor prognosis, as they were significantly upregulated in the poor prognosis group (P<0.05). However, none of the let-7 family members was significantly dysregulated (Table SVI).
Discussion
In the present study, the number of dysregulated miRNAs in patients with PDAC compared with the CG was higher than that in the PDAC compared with the CP group. The decreased number of significantly dysregulated miRNAs between the CP and the PDAC group may be due to shared pathological molecular mechanisms, as CP is a risk factor for the development of PDAC (39). The present study indicated that members of the let-7 miRNA family are dysregulated in PDAC. Of note, the let-7 family members were significantly downregulated in patients with PDAC. Let-7a-5p and let-7f-5p were also downregulated in the late stages of PDAC. This finding corroborates the observation that downregulation of let-7 in gemcitabine-resistant pancreatic cancer cells was associated with activation of the epithelial-to-mesenchymal pathway characteristic of metastasis (40).
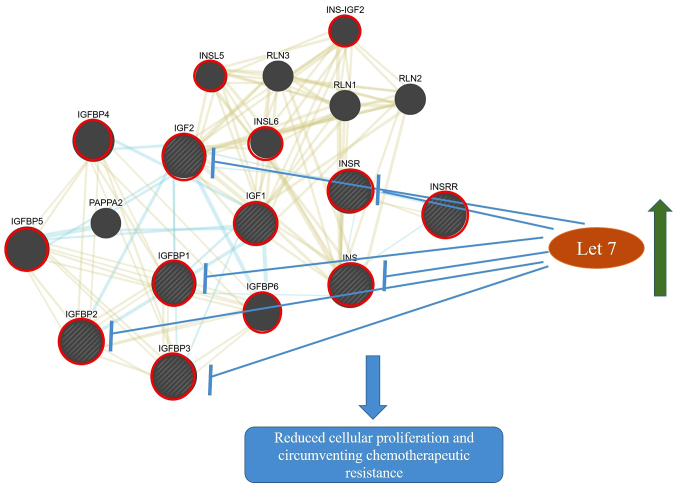
The let-7 family inhibits the expression of components of the INSR/IGF signalling pathway (Figs. 2 and 3) and their downregulation may result in increased activity of this pathway. This suggests that hypothetically, an increase in let-7 may inhibit the INSR/IGF signalling pathway and subsequently inhibit cellular proliferation and promote chemotherapeutic sensitivity (Fig. 4). The let-7 family was initially identified in the nematode Caenorhabditis elegans. In humans, the let-7 family has 13 members, which have been demonstrated to have a significant role in the process of carcinogenesis (41). In 2016, Encarnación et al (42) indicated that overexpression of let-7b in patients with breast cancer resulted in increased DNA repair capacity. Aberrant DNA repair capacity is characteristic of PDAC (43,44). Previous studies have suggested that expression of the let-7 family was significantly reduced in PDAC tumor cells as compared with normal acinar cells and that induction of let-7 expression inhibited cellular proliferation (45). One study indicated that treatment with diflourinated curcumin, a curcumin analogue with anti-oxidant properties, inhibited tumor growth and this was associated with increased levels of let-7 (46).
Figure 4.
Crosstalk between the insulin/IGF signalling pathways and the potential targeting of let-7. The upregulation of let-7 microRNAs may block the overexpression of components of the INSR/IGF pathways resulting in reduced cell proliferation and increase sensitivity to therapy. Blue and yellow networks indicate shared pathways (INSR/IGF pathway) and protein domains (N- and C-domains), respectively. The node sizes are inversely proportional to the gene score rank determined by GeneMANIA. Shaded nodes indicate genes inputted into the software. Red circles are used to highlight key genes involved in the INSR/IGF pathway. The interacting network was generated from GeneMania. Microsoft PowerPoint v2013 was also used to make additional drawings including let-7 node, arrows indicating upregulation, inhibition and the subsequent possible effect of this inhibition INSR, insulin receptor; IGF1, insulin-like growth factor 1; IGF2, insulin-like growth factor 2; IGFBP, Insulin-like growth factor-binding protein; INS-IGF2, Insulin, isoform 2; INSL, Insulin-like; INSRR, insulin receptor-related receptor; INS, insulin; PAPPA2, pappalysin-2; RLN, relaxin.
Early-onset diabetes and obesity are risk factors for PDAC (47). It is known that obesity-linked upregulation of the insulin/IGF signalling pathway occurs in carcinogenesis via an increase in the NF-κβ pathway, which induces inflammation, cellular proliferation, migration and metastasis (48). A study on transgenic let-7 mice indicated decreased body weight and increased glucose levels linked to inhibition of components of INSR/IGF and the PI3K/mTOR pathway (49).
Several receptors of the insulin and IGF family such as INSR and IGF-1R were also indicated to be targeted for downregulation by let-7 members (Figs. 2 and 3). The receptors of insulin and IGF are related, belonging to a family of receptor tyrosine kinases. Each has two receptors, INSR-A and INSR-B for insulin and IGF-1R and IGF-2R for IGF (50,51). These receptors are overexpressed in PDAC and exhibit complex cross-talk with each other (52). The INSR potentiates tumor progression, as it was indicated to be significantly overexpressed along with progression and increased tumor stage (53). The IGF-1R is primarily responsible for mediating cellular responses to the IGFs (54). Both INSR and IGF-1R are highly homologous and are able to hybridize, enhancing tumor growth (53). In a recent study, overexpression of IGF-1R was observed in urothelial carcinoma, indicating its utility as a therapeutic target (55). Hence, IGF-1R overexpression correlates with the tumor stage and its co-expression with EGFR has been associated with reduced overall survival (5). Similarly, IGF-2R is overexpressed in PDAC compared to normal ductal cells (56). Knockdown of the IGFR pathway in a mouse model inhibited cellular proliferation and promoted apoptosis of PDAC cells (57). Regrettably, several drugs developed to target both INSRs and IGF have been unsuccessful in inhibiting PDAC development, possibly due to heterodimerization of the IR and its interaction with IGF-1R, resulting in resistance to these drugs (13).
The present study further suggested that let-7 targets IGF-binding proteins (IGFBPs) which tightly regulate the expression of IGFs. The IGFBPs were determined to be elevated in patients with PDAC compared to those with chronic pancreatitis (58) and may contribute to exacerbating pancreatic tumourigenesis. IGF2BP1 was indicated to promote cellular proliferation in pancreatic cancer cells via the AKT signalling pathway (59). A recent study performed by our group confirmed the upregulation of the Akt pathway in resected pancreatic tumours (17). Similarly, IGF2BP2 expression was determined to promote tumour progression and to be correlated with poor survival of patients with pancreatic cancer (60). It may therefore be concluded that IGFBPs may serve as a biomarker for the disease. Since let-7 targets IGFBPs, it may be hypothesized that strategies targeting let-7 may provide an avenue for evading therapeutic resistance observed in pancreatic cancer treatment.
It has also been suggested that IGF signalling is important in the formation of the dense avascular stroma characteristic of PDAC (12), another crucial factor involved in chemotherapy resistance (61). Therefore, upregulation of let-7 may inhibit stromal formation, increasing tumor chemosensitivity.
In conclusion, the present study demonstrated downregulation of certain members of the let-7 family in plasma samples of patients with PDAC compared to patients with CP, a pre-malignant condition, as well as to a CG with significant risk factors common to patients with PDAC. Since the in silico analysis indicated that let-7 targets the INSR/IGF pathway, its downregulation may increase the expression of the INSR/IGF pathway and may therefore be an effective target for the development of INSR/IGF pathway-specific treatment strategies. Future studies investigating the effects of let-7 in modulating the IGF pathway in pancreatic cancer by using in vitro and in vivo systems should be performed to validate this hypothesis. Additionally, it would be pertinent to conduct a longer follow-up Kaplan Meier analyses for overall survival between patients that have high or low let-7 expression.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Lihan Zhou and Dr Cheng He of MiRXES PTY Ltd, National University Singapore, for assistance with the miRNA analyses.
Funding
This study was funded by the South African National Research Foundation (grant no. 91508, ref. no. CSUR13091741850). EEN was funded by a South African Medical Research Council Grant awarded to the Wits Common Epithelial Cancer Research Centre.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MB was responsible for the conceptualization of the study, drafted and revised the manuscript. EEN performed data analyses, drafted and revised the manuscript. Both authors read and approved the final manuscript.
Ethics approval and informed consent to participate
A prospective study including black South African participants was undertaken. Ethics approval was obtained from the University of the Witwatersrand Human Research Ethics Committee (Medical), certificate no. M130551. All patients recruited for the study provided written informed consent prior to sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J Oncol. 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ferrone CR, Brennan MF, Gonen M, Coit DG, Fong Y, Chung S, Tang L, Klimstra D, Allen PJ. Pancreatic adenocarcinoma: The actual 5-year survivors. J Gastrointest Surg. 2008;12:701–706. doi: 10.1007/s11605-007-0384-8. [DOI] [PubMed] [Google Scholar]
- 4.Trajkovic-Arsic M, Kalideris E, Siveke JT. The role of insulin and IGF system in pancreatic cancer. J Mol Endocrinol. 2013;50:R67–R74. doi: 10.1530/JME-12-0259. [DOI] [PubMed] [Google Scholar]
- 5.Valsecchi ME, McDonald M, Brody JR, Hyslop T, Freydin B, Yeo CJ, Solomides C, Peiper SC, Witkiewicz AK. Epidermal growth factor receptor and insulinlike growth factor 1 receptor expression predict poor survival in pancreatic ductal adenocarcinoma. Cancer. 2012;118:3484–3493. doi: 10.1002/cncr.26661. [DOI] [PubMed] [Google Scholar]
- 6.Nandy D, Mukhopadhyay D. Growth factor mediated signaling in pancreatic pathogenesis. Cancers (Basel) 2011;3:841–871. doi: 10.3390/cancers3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faller BA, Burtness B. Treatment of pancreatic cancer with epidermal growth factor receptor-targeted therapy. Biologics. 2009;3:419–428. [PMC free article] [PubMed] [Google Scholar]
- 8.Zielinski R, Przytycki PF, Zheng J, Zhang D, Przytycka TM, Capala J. The crosstalk between EGF, IGF, and Insulin cell signaling pathways-computational and experimental analysis. BMC Syst Biol. 2009;3:88. doi: 10.1186/1752-0509-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann U. Insulin-like growth factor I overexpression in human pancreatic cancer: Evidence for autocrine and paracrine roles. Cancer Res. 1995;55:2007–2011. [PubMed] [Google Scholar]
- 10.Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62:200–207. [PubMed] [Google Scholar]
- 11.LeRoith D, Roberts Jr CT. The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/S0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 12.Mutgan AC, Besikcioglu HE, Wang S, Friess H, Ceyhan GO, Demir IE. Insulin/IGF-driven cancer cell-stroma crosstalk as a novel therapeutic target in pancreatic cancer. Mol Cancer. 2018;17:66. doi: 10.1186/s12943-018-0806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh P, Alex JM, Bast F. Insulin receptor (IR) and insulin-like growth factor receptor 1 (IGF-1R) signaling systems: Novel treatment strategies for cancer. Med Oncol. 2014;31:805. doi: 10.1007/s12032-013-0805-3. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Bloom DA, Cance WG, Kurenova EV, Golubovskaya VM, Hochwald SN. FAK and IGF-IR interact to provide survival signals in human pancreatic adenocarcinoma cells. Carcinogenesis. 2008;29:1096–1107. doi: 10.1093/carcin/bgn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Sawai H, Matsuo Y, Ochi N, Yasuda A, Takahashi H, Wakasugi T, Funahashi H, Sato M, Takeyama H. IGF-1 mediates PTEN suppression and enhances cell invasion and proliferation via activation of the IGF-1/PI3K/Akt signaling pathway in pancreatic cancer cells. J Surg Res. 2010;160:90–101. doi: 10.1016/j.jss.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Neid M, Datta K, Stephan S, Khanna I, Pal S, Shaw L, White M, Mukhopadhyay D. Role of insulin receptor substrates and protein kinase C-zeta in vascular permeability factor/vascular endothelial growth factor expression in pancreatic cancer cells. J Biol Chem. 2004;279:3941–3948. doi: 10.1074/jbc.M303975200. [DOI] [PubMed] [Google Scholar]
- 17.Nweke E, Ntwasa M, Brand M, Devar J, Smith M, Candy G. Increased expression of plakoglobin is associated with upregulated MAPK and PI3K/AKT signalling pathways in early resectable pancreatic ductal adenocarcinoma. Oncol Lett. 2020;19:4133–4141. doi: 10.3892/ol.2020.11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan MA, Zubair H, Srivastava SK, Singh S, Singh AP. Insights into the role of microRNAs in pancreatic cancer pathogenesis: Potential for diagnosis, prognosis, and therapy. Adv Exp Med Biol. 2015;889:71–87. doi: 10.1007/978-3-319-23730-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 20.Gong L, Wang C, Gao Y, Wang J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed Pharmacother. 2016;83:58–63. doi: 10.1016/j.biopha.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 21.Ono S, Lam S, Nagahara M, Hoon DS. Circulating microRNA biomarkers as liquid biopsy for cancer patients: Pros and cons of current assays. J Clin Med. 2015;4:1890–1907. doi: 10.3390/jcm4101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vafaee F, Diakos C, Kirschner MB, Reid G, Michael MZ, Horvath LG, Alinejad-Rokny H, Cheng ZJ, Kuncic Z, Clarke S. A data-driven, knowledge-based approach to biomarker discovery: Application to circulating microRNA markers of colorectal cancer prognosis. NPJ Syst Biol Appl. 2018;4:20. doi: 10.1038/s41540-018-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bratulic S, Gatto F, Nielsen J. The translational status of cancer liquid biopsies. Regen Eng Transl Med. 2019 Nov 25; doi: 10.1007/s40883-019-00141-2. [DOI] [Google Scholar]
- 24.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 25.Chun YS, Pawlik TM, Vauthey JN. 8th Edition of the AJCC Cancer Staging Manual: Pancreas and Hepatobiliary cancers. Ann Surg Oncol. 2018;25:845–847. doi: 10.1245/s10434-017-6025-x. [DOI] [PubMed] [Google Scholar]
- 26.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conwell DL, Lee LS, Yadav D, Longnecker DS, Miller FH, Mortele KJ, Levy MJ, Kwon R, Lieb JG, Stevens T, et al. American pancreatic association practice guidelines in chronic pancreatitis: Evidence-based report on diagnostic guidelines. Pancreas. 2014;43:1143–1162. doi: 10.1097/MPA.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Löhr JM, Dominguez-Munoz E, Rosendahl J, Besselink M, Mayerle J, Lerch MM, Haas S, Akisik F, Kartalis N, Iglesias-Garcia J, et al. United European Gastroenterology evidence-based guidelines for the diagnosis and therapy of chronic pancreatitis (HaPanEU) United European Gastroenterol J. 2017;5:153–199. doi: 10.1177/2050640616684695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Lanoix D, Lacasse AA, St-Pierre J, Taylor SC, Ethier-Chiasson M, Lafond J, Vaillancourt C. Quantitative PCR Pitfalls: The case of the human placenta. Mol Biotechnol. 2012;52:234–243. doi: 10.1007/s12033-012-9539-2. [DOI] [PubMed] [Google Scholar]
- 31.St-Pierre J, Grégoire JC, Vaillancourt C. A simple method to assess group difference in RT-qPCR reference gene selection using GeNorm: The case of the placental sex. Sci Rep. 2017;7:16923. doi: 10.1038/s41598-017-16916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen SY, Feng Z, Yi X. A general introduction to adjustment for multiple comparisons. J Thorac Dis. 2017;9:1725–1729. doi: 10.21037/jtd.2017.05.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reneker J, Shyu CR. Applying sequential forward floating selection to protein structure prediction with a study of HIV-1 RP. AMIA Annu Symp Proc. 2006;2006:1072. [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, Zhou Y, Cui Q. HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019;47:D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirkegård J, Mortensen FV, Cronin-Fenton D. Chronic pancreatitis and pancreatic cancer Risk: A systematic review and meta-analysis. Am J Gastroenterol. 2017;112:1366–1372. doi: 10.1038/ajg.2017.218. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 42.Encarnación J, Ortiz C, Vergne R, Vargas W, Coppola D, Matta JL. High DRC levels are associated with Let-7b overexpression in women with breast cancer. Int J Mol Sci. 2016;17:865. doi: 10.3390/ijms17060865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perkhofer L, Illing A, Gout J, Frappart PO, Kleger A. Precision medicine meets the DNA damage response in pancreatic cancer. Oncoscience. 2018;5:6–8. doi: 10.18632/oncoscience.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, Boardman LA, Petersen GM. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928–4935. doi: 10.1158/0008-5472.CAN-07-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, Buscail L, Cordelier P. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 46.Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, Kong D, Ahmad A, Li Y, Padhye S, Sarkar FH. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bracci PM. Obesity and pancreatic cancer: Overview of epidemiologic evidence and biologic mechanisms. Mol Carcinog. 2012;51:53–63. doi: 10.1002/mc.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowers LW, Rossi EL, O'Flanagan CH, deGraffenried LA, Hursting SD. The role of the Insulin/IGF system in cancer: Lessons learned from clinical trials and the energy balance-cancer link. Front Endocrinol (Lausanne) 2015;6:77. doi: 10.3389/fendo.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailyes EM, Navé BT, Soos MA, Orr SR, Hayward AC, Siddle K. Insulin receptor/IGF-I receptor hybrids are widely distributed in mammalian tissues: Quantification of individual receptor species by selective immunoprecipitation and immunoblotting. Biochem J. 1997;327:209–215. doi: 10.1042/bj3270209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 52.Ohmura E, Okada M, Onoda N, Kamiya Y, Murakami H, Tsushima T, Shizume K. Insulin-like growth factor I and transforming growth factor alpha as autocrine growth factors in human pancreatic cancer cell growth. Cancer Res. 1990;50:103–107. [PubMed] [Google Scholar]
- 53.Ulanet DB, Ludwig DL, Kahn CR, Hanahan D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proc Natl Acad Sci USA. 2010;107:10791–10798. doi: 10.1073/pnas.0914076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brahmkhatri VP, Prasanna C, Atreya HS. Insulin-Like growth factor system in cancer: Novel targeted therapies. Biomed Res Int. 2015;2015:538019. doi: 10.1155/2015/538019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eich ML, Tregnago AC, Faraj SF, Palsgrove DN, Fujita K, Bezerra SM, Munari E, Sharma R, Chaux A, Netto GJ. Insulin-like growth factor-1 receptor expression in upper tract urothelial carcinoma. Virchows Arch. 2019;474:21–27. doi: 10.1007/s00428-018-2468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishiwata T, Bergmann U, Kornmann M, Lopez M, Beger HG, Korc M. Altered expression of insulin-like growth factor II receptor in human pancreatic cancer. Pancreas. 1997;15:367–373. doi: 10.1097/00006676-199711000-00006. [DOI] [PubMed] [Google Scholar]
- 57.Tian X, Hao K, Qin C, Xie K, Xie X, Yang Y. Insulin-like growth factor 1 receptor promotes the growth and chemoresistance of pancreatic cancer. Dig Dis Sci. 2013;58:2705–2712. doi: 10.1007/s10620-013-2673-2. [DOI] [PubMed] [Google Scholar]
- 58.Karna E, Surazynski A, Orłowski K, Łaszkiewicz J, Puchalski Z, Nawrat P, Pałka J. Serum and tissue level of insulin-like growth factor-I (IGF-I) and IGF-I binding proteins as an index of pancreatitis and pancreatic cancer. Int J Exp Pathol. 2002;83:239–245. doi: 10.1046/j.1365-2613.2002.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan BS, Cheng M, Zhang L. Insulin-like growth factor 2 mRNA-binding protein 1 promotes cell proliferation via activation of AKT and is directly targeted by microRNA-494 in pancreatic cancer. World J Gastroenterol. 2019;25:6063–6076. doi: 10.3748/wjg.v25.i40.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dahlem C, Barghash A, Puchas P, Haybaeck J, Kessler SM. The insulin-like growth factor 2 mRNA binding protein IMP2/IGF2BP2 is overexpressed and correlates with poor survival in pancreatic cancer. Int J Mol Sci. 2019;20:3204. doi: 10.3390/ijms20133204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.