Abstract
Liquid biopsy refers to the sampling and molecular analysis of the biofluids of circulating tumor cells, extracellular vesicles, nucleic acids, and so forth. Exosomes are small extracellular vesicles with sizes between 30–150 nm. They are secreted by multivesicular bodies through exocytosis in live cells and can participate in intercellular communication due to their contents, including nucleic acids, proteins, and lipids. Herein, we investigate publication frequencies on exosomes over the past 10 years, and review recent clinical studies on liquid biopsy of exosomes in the fields of oncology, pregnancy disorders, cardiovascular diseases, and organ transplantation. We also describe the advantages of exosomes as an effective liquid biopsy tool and the progression of exosome extraction methods. Finally, we depict the commercial development of exosome research and discuss the future role of exosomes in liquid biopsy.
Subject terms: Molecular biology, Tumour biomarkers
Introduction
Exosomes, which are small extracellular vesicles with sizes between 30–150 nm, are a subtype of extracellular vesicles (EVs), which are secreted by all cell types and are responsible for intercell communication.1 The International Society for Extracellular Vesicles (ISEV) recommends using “EVs” instead of exosomes or microvesicles and gives terms for EV subtypes based on the physical properties.2 We quoted the original statement out of respect for their research. However, most articles cited in this review did not clarify the definition clearly and were only capable of enriching rather than purifying exosomes with current methods. Exosomes were originally described to be released from sheep reticulocytes.3 With the large number of studies that followed, exosomes were found to exist in almost all body fluids, primarily blood,4 urine,5 cerebrospinal fluid,6 saliva,7 pleural effusion,8 ascites fluid,9 amniotic fluid,10 breast milk,11 and bronchoalveolar lavage fluid (BALF).12 Exosomes, originating from the endosomal pathway via the formation of late endosomes or multivesicular bodies,1 enclose a variable spectrum of molecules characterized by parent cells, including nucleic acids (DNA, mRNA, microRNA (miRNA), lncRNA, circRNA, etc.), proteins, and lipids, which can be transported over distances within the protection of a lipid bilayer-enclosed structure.1 Recently, a contradictory point was raised in a study published in Cell that based on proteomic profiles, double-stranded DNA and DNA-binding histones were absent in exosomes or any other type of small EVs.13 An autophagy- and multivesicular endosome-related pathway was the driver of extracellular DNA secretion instead of exosome-dependent pathway.13 However, Yokoi et al.14 subsequently confirmed the presence of double-stranded DNA in exosomes by imaging flow cytometry and described how nuclear content loaded into exosomes. Since the DNA detection methods applied in these studies are different, it is unclear whether genomic DNA exists in exosomes, although we still recognize the value of the studies on exosomal DNA (exoDNA).
Researchers around the world express great enthusiasm for exosomes as biomarkers in liquid biopsy. Based on a PubMed search in January 2020, we statistically analyzed the number of publications regarding the diagnostic efficacy of exosomes in the past 5 years with the MeSh Terms “exosomes” OR “small extracellular vesicles” (microparticles and microvesicles are not MeSh Terms in PubMed), “diagnosis” OR “biomarker”, “mutation” OR “copy number” OR “DNA”, “RNA” (“mRNA”, “microRNA”, “lncRNA”, “circRNA”), “protein”, and “liquid biopsy”. There were 88 relevant publications on DNA, 695 on RNA (including 74 about mRNA, 534 about microRNA, 52 about lncRNA, 14 about circRNA), and 824 on protein. Protein is the most-studied content of exosomes, followed by miRNA. It attracted the greatest interest in exosome research as a biomarker carrier for diagnosis over the past 10 years, especially in 2018 (Fig. 1).
Fig. 1.
Publication frequencies of studies investigating different contents of exosomes as liquid biopsy for disease diagnosis over the past 10 years based on the PubMed search, January 2020
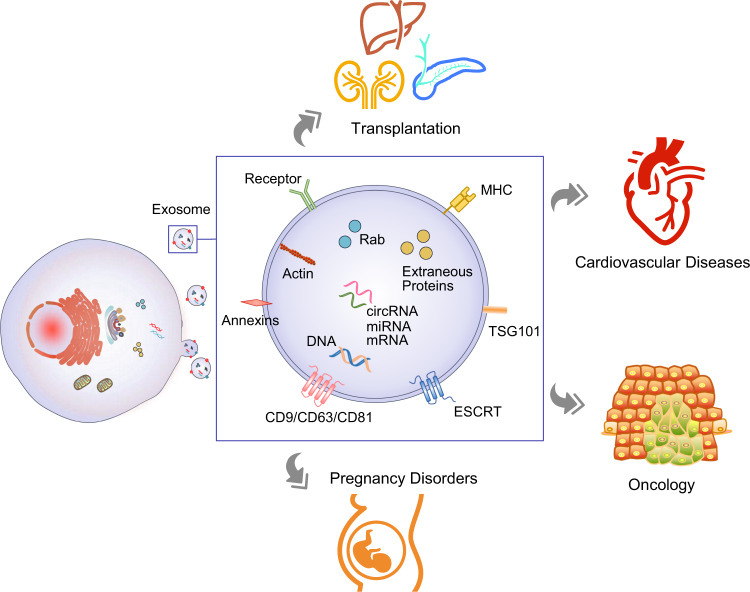
Simultaneously, exosomes play critical roles in various physiological and pathological processes, including cancer, pregnancy disorders, cardiovascular diseases, and immune responses.15 By virtue of the exponential evolution of liquid biopsy in recent decades, traditional solid biopsy shows considerably more limitations. It is imperative to introduce liquid biopsy to clinical practice to reduce invasive operations and promote more precise medical intervention.16 Herein, we mainly introduce the advantages of exosomes as liquid biopsy and their application as a potential complement to personalized medicine in some common malignant tumors, pregnancy disorders, cardiovascular diseases, and organ transplantation (Fig. 2). Owing to the great prospects of exosomes in clinical applications, a commercial chain of exosome research-related technologies has been formed and is still under development.
Fig. 2.
Biogenesis, secretion, composition, and application of exosomes as liquid biopsy. Exosomes, originating from the endosomal pathway via the formation of late endosomes or multivesicular bodies, enclose a variable spectrum of molecules characterized by parent cells, including nucleic acids (DNA, mRNA, miRNA, lncRNA, circRNA, etc.), proteins, and lipids, which shows great promise in clinical applications in cancer, pregnancy disorders, cardiovascular diseases and organ transplantation
Advantages of exosomes in liquid biopsy
Exosomes show significant superiority over other sources of liquid biopsy. First, exosomes exist in almost all body fluids and possess high stability encapsulated by lipid bilayers. Similar quality of exosomal markers exists in samples stored at 4 °C for 24 h and then at −80 °C, samples immediately stored at −80 °C and fresh urine samples.17 And it is better to keep exosomes at 4 °C within 24 h but at −80 °C for long-term storage. Exosomes prefer to be isolated in pH 7 solution rather than in acidic environments.18 The high biological stability can reduce the cost of sample short-term storage and the difficulty of transportation, which greatly enhances the clinical applicability of exosomes. Second, exosomes are secreted by living cells and contain biological information from the parental cells and be more representative than cell-free DNA (cfDNA), which is secreted during necrosis or apoptosis.19 Third, exosome identification is clear and simple. Exosomes express specific proteins such as CD63, ALIX, TS101, and HSP70,20 which can be used as markers to effectively distinguish exosomes from other vesicles, and their specific cup-shaped characteristic makes them identifiable by electron microscopy.21 Fourth, exosomes can present specific surface proteins from parental cells22 and even target cells, which can realize the isolation of origin-specific exosomes and predict organ-specific metastasis.23 Fifth, exosomes show superiority to conventional serum-based biomarkers, such as carcinoembryonic antigen, in diagnostic accuracy.24 Sixth, compared with circulating tumor cells (CTCs), exosomes are relatively convenient to obtain from almost any body fluids. Additionally, there are many recognized classic extraction methods, such as ultracentrifugation,25 and a considerable number of novel methods under development, for instance, commercial isolation kits, indicating more feasibility for clinical application than CTCs, while CTC collection is still tough and complicated.26 In addition, with regard to cfDNA, most human plasma cfDNA is located in exosomes,27 and the copy number of mitochondrial DNA is higher in exosomes than in plasma or peripheral blood from patients with advanced serous epithelial ovarian cancer.28 Meanwhile, the detection sensitivity and specificity of exoDNA mutation frequency are higher than those of cfDNA29–31 and exoDNA has shown greater prognostic value as well.29,32 Generally, despite the slightly more complicated DNA extraction procedures, exosomal DNA possesses more abundant biological information and higher accuracy for prognosis prediction than cfDNA.
Exosomes in tumor diagnosis, prognosis prediction, and treatment response assessment
At present, solid biopsy is still the gold standard for pathological diagnosis and is mostly the basis for treatment of cancer. However, solid biopsy is invasive, sometimes unable to perform, and tumor heterogeneity is inevitable. Noninvasive liquid biopsy shows great advantages for individualized and precise diagnosis and treatment.16 Tumor-derived exosomes (TDEs) are critically related to tumor progression, metastatic niche formation, and immune evasion,33 which indicates that TDEs may hold great promise for cancer diagnosis, prognosis and treatment response assessment.
Exosomes in early tumor diagnosis
Early screening and accurate diagnosis are undoubtedly the primary issues for patients with tumors or precancerous lesions in reducing mortality and increasing the recovery rate. In pancreatic cancer, a high probability of KRAS mutation in circulating exoDNA was found in the early-stage.29 Given that the elevated level of GPC1+-circulating exosomes obviously occurred in patients with pancreatic ductal carcinoma (PDAC)34 and colorectal cancer (CRC)35 compared to healthy controls, this could serve as an early detection tool for tumors in the digestive system. A clinical trial (NCT03032913) conducted by Etienne BUSCAIL completed the recruitment of 20 PDAC patients and 20 noncancer patients, whose blood samples were collected to detect CTCs and GPC1+ exosomes for diagnosis accuracy assessment and comparison. Diverse forms of exosomal RNAs also promisingly take part in the early diagnosis of cancers. A panel consisting of two mRNAs (KRTAP5-4 and MAGEA3) and one lncRNA (BCAR4) was a promising candidate for the CRC diagnosis.36 A database of exosome-containing RNA (including 18,333 mRNAs, 15,501 lncRNAs, and 58,330 circRNAs) in human blood provided a platform for further discovery and clinical application of circulating exosomal biomarkers.37 In lung cancer, detection of exosome-based EGFR T790M has shown great potential for clinical diagnosis to avoid unnecessary tumor biopsies in non-small cell lung cancer (NSCLC).38 Multiple proteins in exosomes exert powerful efficacy in distinguishing cancerous and noncancerous patients. By using an EV array containing 49 antibodies that could capture and detect exosomes in plasma, Sandfeld-Paulsen et al.39 revealed that CD151, CD171, and tetraspanin 8 were the most significant molecules to separate patients with all histological lung cancer from cancer-free individuals. Of note, based on the development of mass spectrometry (MS) technology and proteome profiles, thousands of proteins can be captured from one sample of micro quantity. Chen et al.40 used MS to compare breast cancer (BC) patient-derived exosomes with those of noncancer patients and identified 144 distinctly elevated exosome phosphorylated proteins. Subsequently, they utilized MS and parallel reaction monitoring techniques to validate four of them: PKG1, RALGAPA2, NFX1, and TJP2. In further study and clinical application of liquid biopsy, the combination of various contents of exosomes as biomarkers can provide a more effective guarantee for the accuracy of cancer diagnosis and prognosis.41,42 The studies described above primarily focused on peripheral blood-derived exosomes, while as common malignant tumors, urinary system- and genitourinary tract-related cancers are preferentially detected through urine-based testing.43 In renal cell carcinoma, after MS profiling and a literature search, Raimondo et al.44 selected ten proteins as a panel to distinguish cancer and healthy control. Furthermore, multiple miRNAs carried by exosomes, such as miR-21-5p,45,46 miR-4454 and miR-720/3007a, were elevated in bladder cancer patient urine,46 which could potentially serve as early diagnosis biomarkers for bladder cancer.
Exosomes in tumor prognosis prediction
With longitudinal monitoring in the treatment course of patients with metastatic PDAC by plasma-based exoDNA detection, Bernard et al.32 indicated that KRAS mutation detected at baseline by digital droplet PCR (ddPCR) and a mutation frequency above 5% indicated poor clinical outcome. In another study, exosomal KRAS mutations were proven to be better than CA 19-9 levels for the prognostic surveillance of patients with PDAC.29 Keklikoglou et al.47 revealed why cytotoxic chemotherapy promotes metastasis in BC. They found that annexin A6-enriched exosomes were largely secreted after cytotoxic chemotherapy and transferred to endothelial cells (ECs) in the lung, thus inducing a premetastatic niche to lay the foundation of lung metastasis. Exosome-associated Annexin II48 and L-plastin49 also play a vital role in metastasis and can be potential candidates for the prognosis of advanced BC. In lung cancer, FLI1 exonic circular RNAs was identified as a novel carcinogenic driver contributing to the metastasis of small cell lung cancer (SCLC), which can be a potential biomarker for the prognosis and surveillance of SCLC.50 In prostate cancer, based on the MS proteomic profile, Bijnsdorp et al.51 revealed that urine-derived exosomal ITGA3 and ITGB1 were upregulated in metastatic patients compared with those with benign tumors and early-stage cancer. Recently, it was revealed that circulating exosomal PD-L1, but not soluble PD-L1, was associated with tumor progression in head and neck cancer52 and NSCLC.53 Simultaneously, serum-based exosomal PD-L1 possessed the ability to predict unfavorable prognosis in PDAC.54
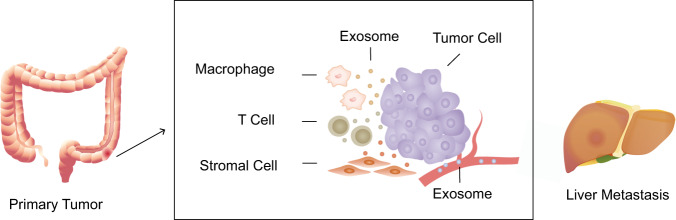
Liver metastasis of colorectal cancer is still a major problem that needs to be overcome. Multiple works have been conducted to determine the mechanism of liver metastasis, including the exosome-mediated hypothesis. Teng et al.55 demonstrated that miR-193a, a tumor suppressor miRNA, was selectively sorted into TDEs by major vault protein, which led to higher expression of oncogenic miRNAs in the tumor cells and promoted colon cancer progression. Overexpression of miR-193a in circulating exosomes can be a promising biomarker for the liver metastasis in colon cancer. Zeng et al.56 illustrated that TDE miR-25-3p regulated the liver metastasis of CRC by inducing vascular permeability and angiogenesis and thus creating a premetastatic niche. A prospective clinical trial on rectal cancer was carried out and found that plasma-based exosomal miR-141-3p and miR-375 were significantly increased in patients with liver metastasis compared to those without.57 Recently, Hu et al.58 revealed another mechanism of liver metastasis of CRC via cancer-associated fibroblast-derived exosomes, which elevated the expression of miR-92a-3p and enhanced epithelial-mesenchymal transition and cell stemness in CRC cells. Complementarily, miR-21,59 miR-18a, miR-17-5p,60 and miR‐548c‐5p61 can be used as early screening markers for liver metastasis of CRC (Fig. 3). In summary, miRNAs are critical elements in regulating liver metastasis of colorectal cancer, whereas the specific targets are still under exploration, and an accurate exosomal miRNA panel for prognosis prediction makes sense in clinical applications.
Fig. 3.
Exosomes in liver metastasis of colorectal cancer. Stromal cell-derived exosomes can carry bioactive molecules (e.g., miR-92a-3p) to CRC cells and enhance epithelial-mesenchymal transition and cell stemness to promote liver metastasis. Many other miRNAs (e.g., miR-193a, miR-21, miR-25-3p, miR-18a, miR-17-5p, miR-141-3p, miR-548c-5p, miR-375, and miR-6803-5p) encapsulated in exosomes from primary CRC cells, which flow to the liver via blood circulation and lead to liver metastasis, can be candidates for prognosis
Exosomes in tumor treatment response assessment
Exosomes also play a potentially useful role in treatment response assessment, especially drug resistance, which is a major obstacle for advanced malignant tumors. TDEs can carry various drug resistance-associated molecules to target cells62 and thus induce EMT, promote antiapoptotic pathways, alter signal transduction alterations, and regulate specific targets in cancer cells63 to facilitate treatment failure. Wei et al.64 demonstrated that exosomal miR-222-3p could promote the expression of gemcitabine resistance and a malignant phenotype by targeting the promoter of SOCS3, which could be used as a predictor of treatment response for NSCLC patients. Qu et al.65 found a novel lncRNA-lncARSR (lncRNA activated in renal cell carcinoma with sunitinib resistance), as a biomarker for treatment, as it is transferred through tumor-derived exosomes to promote sunitinib resistance by upregulating AXL and c-MET expression. Nevertheless, there are also some favorable contents that enhance sensitivity to chemotherapy. Liu et al.66 revealed that miR-128-3p-containing exosomes derived from normal intestinal cells would be transported to oxaliplatin-resistant CRC cells and thus increase the treatment response. Upregulated exosomal miR-567 could reverse trastuzumab resistance in BC as well.67 Another mechanism of the drug resistance is the drug‐efflux ability of cancer cells via exosomes to decrease intracellular drug accumulation, including direct drug export68 and transfer of drug efflux pumps (e.g., P-glycoproteins, ATP-transporter A2/A3, multidrug-resistant protein-1).69 Importantly, tumor-associated macrophage-derived exosomes also contribute to drug resistance by targeting cancer cells to inactivate sensitivity to therapy.70,71
On the other hand, as a hotspot in cancer therapy, exosomal PD-L1 has attracted considerable attention from researchers for its potential in anti-PD-1/PD-L1 therapy response prediction. In melanoma, upregulated by interferon-γ, an increasing number of PD-L1-containing exosomes were secreted by malignant cells, indicating resistance to anti-PD-1 therapy in patients without previous immunotherapy but denoting a favorable response in patients on pembrolizumab treatment since nonresponsive patients hardly experienced changes in exosomal PD-L1 levels during immunotherapy.72 To date, most studies on cancer therapy response assessment remain at the level of cancer cells in vitro, and more clinical trials are needed to validate the clinical capacity of these biomarkers (Table 1).
Table 1.
Exosomes in common malignant tumors
| Applications | No. of patients | Source | Volume of body fluid | Targets | Exosome extraction | Extraction method | Detection method | References |
|---|---|---|---|---|---|---|---|---|
| Early diagnosis | ||||||||
| Pancreatic cancer | 263 | Plasma | 0.9–1.5 ml | KRAS | Ultracentrifugation | MagAttract High Molecular Weight DNA kit | ddPCR | 29 |
| 221 | Serum | 250 μl | GPC1 | Ultracentrifugation + sucrose gradient centrifugation | Affinity capture | Flow cytometry analysis | 34 | |
| 85 | Serum | – | CKAP4 | PS Capture Exosome ELISA Kit | PS Capture Exosome ELISA Kit | PS Capture Exosome ELISA Kit | 170 | |
| Colorectal cancer | 140 | Serum | 300 μl | lncRNA | Ultracentrifugation | TRIzol | qPCR | 36 |
| 40 | Serum | 250 μl | miRNA | ExoQuick-TC™ Exosome Precipitation Solution kit | miRNeasy Serum/Plasma kit | qRT–PCR | 160 | |
| 102 | Tissue homogenate | – | GPC1 | ExoCapTM Exosome Isolation, Enrichment kit | Affinity capture | Flow cytometry analysis | 35 | |
| Plasma | miR‐96‐5p, miR‐149 | TRIzol | qPCR | |||||
| 116 | Serum | 250 μl | CEA | ExoQuickTM Exosome Precipitation Solution | ELISA | ELISA | 171 | |
| 124 | Plasma | 500 μl | Copine III | Ultracentrifugation | PIPA buffer | ELISA | 24 | |
| Lung cancer | 210 | Plasma | 1–2 ml | EGFR T790M | Ultracentrifugation | ExoLution Plus platform | Allele-specific qPCR assay | 38 |
| 105 | Plasma | – | miRNAs | Ultracentrifugation | mirVanaTM miRNA Isolation Kit | qPCR | 172 | |
| BALF | ||||||||
| 581 | Plasma | – | Proteins | Extracellular vesicle array | Extracellular vesicle array | Extracellular vesicle array | 39 | |
| 171 | Serum | – | Proteins | Ultracentrifugation | Lysis buffer | Immunoblotting | 173 | |
| Breast cancer | 32 | Plasma | 250 μl | miR-21, miR-1246 | Exoquick-TCTM reagent | TRIzol | qRT–PCR | 174 |
| 38 | Serum | – | miR-105 | Ultracentrifugation | TRIzol | qRT–PCR | 175 | |
| 240 | Serum or Plasma | 500 μl | CD82 | ExoQuickTM exosome precipitation solution | Strong RIPA lysate | ELISA/western blot | 176 | |
| 44 | Plasma | 5.5 ml | Phosphoproteins | Ultracentrifugation | – | LC-MS/MS | 40 | |
| Renal cell carcinoma | 52 | Urine | – | Proteins | Differential centrifugation + density gradient ultracentrifugation/ultrafiltration | – | LC-MS/MS | 44 |
| Bladder cancer | 69 | Urine | 38.5 ml | miRNAs | Differential centrifugation | miRNeasy Mini Kit/RNA MS2/Clean-up Kit | miRNA microarray | 45 |
| Cholangio-carcinoma | 134 | Serum | 1 ml | Proteins | Ultracentrifugation | – | MS | 177 |
| Prognosis | ||||||||
| Pancreatic cancer | 194 | Plasma | 15 ml | KRAS | Ultracentrifugation | QIAmp Circulating Nucleic Acid Kit | ddPCR | 32 |
| 91 | Serum | 250 μl | PD-L1, c-MET | Invitrogen Total Exosome Isolation Reagent | – | Flow cytometry | 54 | |
| Colorectal cancer | 255 | Serum | 1 ml | miR-19a | Ultracentrifugation/Total Exosome Isolation Kit | miRNeasy mini kit | qRT–PCR | 178 |
| 28 | Serum | 1 ml | miR-17-5p, miR-92a-3p | qEV Size Exclusion Columns | HiPure Liquid RNA, miRNA Kit | qRT–PCR | 60 | |
| 108 | Serum | 100 μl–1 ml | miR‐548c‐5p | Invitrogen™ Total Exosome Isolation Kit | miRNeasy mini kit | qPCR | 61 | |
| 93 | Plasma | 500 μl /1 ml | miR-141-3p miR-375 | ExiqonTM miRCURY Exosome Isolation Kit | ExiqonTM miRCURY RNA Isolation Kit | qPCR | 57 | |
| 87 | Serum | – | miR-25-3p | Ultracentrifugation | TRIzol | qPCR | 56 | |
| Lung cancer | 106 | Serum | up to 4 ml | FLI1 exonic circular RNAs | ExoEasy Maxi Kit | TRIzol | qRT–PCR | 50 |
| 276 | Plasma | 10 μl | NY‐ESO‐1 | Extracellular Vesicle Array | Extracellular Vesicle Array | Extracellular Vesicle Array | 179 | |
| 85 | Serum | 4 ml | PD-L1 | Invitrogen™ Total Exosome Isolation Kit | RIPA | ELISA | 53 | |
| 20 | Plasma | 5 ml | Amphiregulin | Ultracentrifugation | – | ELISA | 180 | |
| Head and neck cancer | 40 | Plasma | 1 ml | PD-L1 | Mini size exclusion chromatography | – | Flow cytometry | 52 |
| Breast cancer | 53 | Serum | 5 ml | miR-222 | Density gradient centrifugation | Maxwell® 16 miRNA Tissue kit | qPCR | 181 |
| Melanoma | 56 | Serum | 200 μl | miRNA-125b | ExoQuickTM precipitation solution | TRIzol | qPCR | 155 |
| 96 | Serum | 250 μl | S100B, MIA | ExoQuickTM precipitation solution | – | Immuno assays | 182 | |
| Esophageal carcinoma | 602 | Saliva | 3–5 ml | Transcriptionally induced chimeric RNAs | ExoQuick™ exosomes precipitation solution | TRIzol | qRT–PCR | 183 |
| Prostate cancer | 13 | Urine | 5 ml | ITGA3, ITGB1 | Ultracentrifugation | Lysis buffer | Western blot | 51 |
| Treatment response | ||||||||
| Lung cancer | 84 | Plasma | 3 ml | EGFR (RNA) | ExoLution™ Plus | ExoLution™ Plus | NGS | 41 |
| 50 | Serum | – | miR-222-3p | Ultracentrifugation | RNeasy Kit | PCR | 64 | |
| Breast cancer | 53 | Serum | 5 ml | miR-21 | Density gradient centrifugation | Maxwell® 16 miRNA Tissue kit | qPCR | 181 |
| Melanoma | – | Plasma | 1 ml | PD-L1 | Ultracentrifugation + exosome isolation kit | Reverse phase protein array | Reverse phase protein array | 72 |
Recent clinical studies on exosomes as biomarkers for early diagnosis, prognosis and treatment response monitoring of cancers, including cohort scale, source of body fluid, functional targets of exosomes, exosome extraction methods, target isolation and detection methods
Exosomes in pregnancy disorders
Exosomes show great capacity for the diagnosis of pregnancy disorders, including hypertension and hyperglycemia during gravidity, and prenatal screening. Although the utilization of exosomes in maternal peripheral blood, urine and amniotic fluid during pregnancy is still under exploration, the established facts that placental cells can release exosomes to communicate with the maternal body and the biogenesis and secretion of placenta-derived exosomes (PDEs) are regulated by the microenvironment, such as glucose concentration and oxygen tension,73 make exosomes a noninvasive and promising tool for the early diagnosis and prognosis of pregnancy disorders.
Exosomes in hypertensive disorders of pregnancy
Hypertensive disorders of pregnancy, especially preeclampsia (PE) and eclampsia, are the major risks for the health of women and their infants.74 PE affects 3–5% of pregnancies, leading to severe maternal-fetal mortality.75 Placental hypoxia, which can be caused by PE, increases the secretion of exosomes from placental cells and varies the components.76 In addition to conventional markers, placental alkaline phosphatase (PLAP) is a placenta-specific marker for the extraction and quantification of PDEs.77,78 Pillay et al.79 demonstrated that the ratio of PDEs to the total number of exosomes (PLAP+ exosome ratio) was strikingly reduced in early-onset PE and late-onset PE, whereas the relative concentration of PDEs was significantly increased compared to that in normotensive patients. Biro et al.80 collected plasma samples from pregnant women diagnosed with PE, gestational hypertension or chronic hypertension, and healthy controls. Plasma-based exosomal miRNA analysis through reverse transcription polymerase chain reaction (RT-PCR) revealed that the levels of total miRNA and hypoxia-sensitive miR-210 in circulating exosomes were markedly higher in the PE patients than other groups, especially in severe PE. A study on placental exosome changes in PE women across gestation conducted by Carlos Salomon identified exosomal miRNAs by next generation sequencing and finally found that miR-486-1-5p, miR-486-2-5p, and exosome concentration were strikingly higher in PE than in healthy controls and that the two miRNAs selected could serve as potential candidates to predict the occurrence of PE,81 which could greatly improve the management of pregnancy hypertension.
Exosomes in gestational diabetes mellitus
Hyperglycemia is another major factor affecting the bioactivity of placental exosomes. Insulin resistance and hyperinsulinemia can be induced by pregnancy-related hormones (e.g., diabetogenic autacoids) in normal pregnancies.82,83 Gestational diabetes mellitus (GDM) is characterized as glucose intolerance for the first diagnosis during pregnancy.84 GDM affects up to 25% of pregnancies worldwide,85 and the incidence is rapidly increasing due to the global increase in type 2 diabetes and obesity.86 The release and bioactivity of PDEs are regulated by glucose in the first trimester of gestation.87 Salomon et al.88 carried out a retrospectively stratified study on the changes in PDEs in pregnant women with GDM during gestation. They established that the level of PDEs was elevated in both normal and GDM pregnancies, while higher in GDM, especially in early gestation (11–14 weeks). Early diagnosis of GDM and timely pharmacological interventions could reduce the long-term damage on mothers and fetuses.89
Exosomes in prenatal screening
As well, the detection of exosomes can play a role in prenatal screening. PLAP+ exosome ratio from maternal plasma was a potential marker of fetal growth and placental function, as it was obviously lower in patients with fetal growth restriction than in healthy controls.90 Combined with type-B ultrasonic examination and physical examination, plasma exosome detection could allow much more precise diagnosis and monitoring of fetal growth restriction before parturition. Additionally, downregulated miR-300 and miR-299-5p in amniotic fluid-based exosomes could serve as biomarkers for the diagnosis of congenital obstructive nephropathy. Regrettably, invasive amniocentesis remained.91
In summary, maternal circulating exosomal miRNAs, particularly PDEs, are the most popular biomarkers studied in pregnancy disorders, prenatal screening and preterm birth monitoring.92 Urinary exosomes were also affected by maternal changes in gestation, which possessed potential for the diagnosis of intrahepatic cholestasis93 and hypertension.94 Blood and urine testing are already regular during pregnancy, and exosomal biomarkers could be a supplement to detect and predict disorders in pregnant women and fetuses.
Exosomes in cardiovascular diseases
Cardiovascular diseases (CVDs) are one of the major concerns in human health, especially coronary artery disease (CAD), which remains the leading cause of global mortality.95 Currently, circulating biomarkers of CVDs, such as total cholesterol levels and low-density lipoproteins (LDL), and myocardial infarction (MI) prognostic biomarkers, including high-sensitivity C-reactive protein, high-sensitivity cardiac troponin and creatine kinase MB, can only roughly evaluate the risk of the occurrence and progression of the disease but cannot precisely predict whether the process starts or develops.96,97 In this sense, novel blood-based exosome biopsy can offer a promising platform to assist clinical diagnosis and prediction more accurately. Mounting works have proven that exosomal miRNAs possess promising protective functions in CVDs.98–100 Simultaneously, circulating exosomes have shown great potential for diagnosis and risk assessment in CVDs.95
Exosomes in coronary artery disease
In CAD, exosomal miR-133a was elevated in injured myocardium and dead cardiomyocytes in particular.101 Furthermore, miR-146a-abounded exosomes from cardiosphere-derived cells were revealed to promote angiogenesis and inhibit apoptosis, which indicates the therapeutic efficacy of exosomes.102 Exosomes containing miR-210, miR-132, miR-181,103 and miR-378b, miR-623, miR-941 (associated with ejection fraction improvement), miR-1256, miR-384 (associated with fibrosis reduction), miR-525-3p, miR-5155p, miR-1224 (associated with angiogenesis induction)104 and GATA4-responsive-miR-451105 derived from cardiac progenitor cells also possess the same cardioprotective function. Recently, Liu et al.106 made substantial progress in identifying the therapeutic role of circulating endothelial cell-derived microvesicle miRNAs, particularly miR-92a-3p, in regulating the phenotypes of ECs and vascular smooth muscle cells under atherosclerotic conditions, which could be a candidate for the prognosis of CAD. In contrast, miR-939-5p was downregulated in serum-based exosomes from patients with MI and inhibited angiogenesis via the nitric oxide signaling pathway.107 Apart from miRNAs, exosomal proteins also play a significant role in CVD. P-selectin-expressing microparticles,108 CD3+/CD45+, SMA-α+-circulating exosome levels,109 and exosomal Cystatin C, Serpin F2, CD14 levels110 were correlated with a high risk for incident CVD and mortality. Based on liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), Cheow et al.111 identified 252 upregulated EV proteins after MI and created a potential panel for the early diagnosis of MI, including apolipoprotein C-III, apolipoprotein D, platelet glycoprotein Ib alpha chain, complement C1q subcomponent subunit A, and complement C5.
Exosomes in heart failure
In heart failure (HF), miR-22, miR-320a, miR-423-5p, and miR-92b were overexpressed in both serum and serum exosomes and can be uesd as specific biomarkers for the diagnosis and prognosis of systolic HF.112 Several serum-based exosomes containing p53-responsive miRNAs, such as miR-34a, miR-192, and miR-194, were upregulated in HF patients within 1 year of acute MI onset.113 Moreover, it was demonstrated that an increased ratio of endothelial apoptotic microparticles (CD31+/Annexin V+) to mononuclear progenitor cells was related to adverse clinical outcome in patients with acutely decompensated chronic HF.114
In addition, exosomes are also related to multiple other CVDs, such as stroke,115 cardiomyopathy,116,117 cardiac arrhythmia,118 and valvular heart disease.119 Generally, miRNAs are the most prevalent molecules in CVD-associated exosomes as well, revealing superiority as a diagnostic biomarker and a promising therapeutic tool.
Exosomes in organ transplantation
Transplantation of organs from living donors is a feasible way to cure patients with advanced organ failure. However, the recognition of the allograft by the recipients’ immune system, and thus the subsequent rejection, is a major obstacle in organ transplantation therapy. Allograft rejection is mediated by T lymphocytes in the recipient’s secondary lymphoid organs that recognize donor major histocompatibility (MHC) antigens through direct (donor MHC and peptides) or indirect (recipient MHC and donor-derived peptides) pathways.120 The activation of T lymphocytes is ascribed to the migration of the donor antigen from professional antigen-presenting cells to recipient lymphoid tissues and directly communication with T lymphocytes.121 In recent years, it has been discovered that allograft rejection depends on the transmission of EVs from the donor graft organ, which carries donor MHC to recipient antigen-presenting cells and activates the immune response to allografts.122 The T-cell activation pathway via exosomes is termed the semidirect pathway.123,124 Accurate and early diagnosis and longitudinal monitoring of immunologic rejection are essential in the prevention and treatment of clinical transplantation. Current detection methods can sense immune rejection, but by then, the transplanted organ has often undergone irreversible damage. Consequently, there is an urgent need for convenient and noninvasive tools to detect chronic allograft rejection, which affects the survival of most transplant recipients. Thus, the existence of donor-specific exosomes and exosomal changes caused by immunologic rejection over time125 can serve as a surrogate biomarker in acute or chronic rejection of solid organ allografts.
Exosomes in lung transplantation
In lung transplantation, serum/BALF-based exosomes presenting donor HLA, SAgs and immunoregulatory miRNAs from recipients might contribute to acute rejection (AR) and predict early diagnosis of allograft rejection.126 Besides, analysis of BALF-based exosomal mRNAs was significantly different between lung transplantation recipients with or without AR, and the upregulated molecules in AR samples showed a substantial trend toward an inflammatory environment related to both innate and adaptive immune responses.127
Exosomes in heart transplantation
In heart transplantation, Kennel et al.128 demonstrated that circulating exosomal protein content varied in heart transplantation recipients with allograft rejection and that fifteen proteins were strikingly different and primarily related to the immune response. Consequently, exosomal protein analysis could be a powerful tool for post-transplantation monitoring. Besides, Sukma et al.129 demonstrated a new view on the mechanism of acute cellular rejection in cardiac allografts in which miR-142-3p-incorporated exosomes from heart transplant patients were transferred to ECs and undermined endothelial barrier function via downregulation of RAB11FIP2. Recently, Saha et al.104 proved the protective role of cardiac progenitor cell-derived exosomes in MI. They demonstrated that quantitative and cargo profiles of exosomes from circulating cardiac progenitor cells or cardiosphere-derived cells could be a potential tool for noninvasive surveillance after heart transplantation.
Exosomes in kidney transplantation
In kidney transplantation (KTx), Park et al.130 developed a detective and analytic method–integrated kidney exosome analysis (iKEA). iKEA showed high detection accuracy in clinical urine samples from patients with kidney transplant rejection, and its portability simplifies transplant recipient monitoring. Recently, Lim et al.131 conducted a proteomic analysis of urinary exosomes derived from KTx patients. Based on MS, 169 urinary exosome proteins were identified, among which 46 were upregulated in stable recipients and 17 were overexpressed in AR patients. Finally, they selected tetraspanin-1 and hemopexin, which were remarkably elevated in patients with AR, as potential markers for the diagnosis of AR in KTx recipients. Plasma exosomes also play a role in KTx. Zhang et al.132 demonstrated that plasma exosome mRNA-based analysis can be a potential tool for the early diagnosis of allograft rejection in KTx patients.
Exosomes in islet transplantation
In islet transplantation, Vallabhajosyula et al.125 demonstrated that proteomic and RNA signatures, quantity and other signal changes in donor HLA exosomes could indicate early injury/loss of islet mass; therefore, transplant islet exosomes could be a reliable biomarker for monitoring patients undergoing islet transplantation over long-term follow-up. Recently, Korutla et al.133 reported a relevant case confirming that circulating transplant islet-specific exosomes could be a potential biomarker for distinguishing between pancreatic β cell damage secondary to autoimmune relapse or immune rejection in islet-transplanted patients diagnosed with autoimmune type 1 diabetes.
In conclusion, immune response-related proteins and immunoregulatory miRNAs varied significantly in exosomes derived from patients after organ transplantation. Peripheral blood and urine-based exosomes may serve as a practical tool for the early detection of AR and long-term monitoring to prevent chronic allograft rejection (Table 2).
Table 2.
Exosomes in pregnancy disorders, cardiovascular diseases, and organ transplantation
| Fields | No. of patients | Source | Volume of body fluid | Targets | Exosome extraction | Extraction method | Detection method | Findings | References |
|---|---|---|---|---|---|---|---|---|---|
| Pregnancy disorders | |||||||||
| Pregnancy hypertension | 98 | Plasma | 500 μl | miR-210 | Exosome precipitation solution | NucleoSpin miRNA Plasma Kit | qPCR | Diagnosis | 80 |
| Preeclampsia | 47 | Plasma | 1 ml | miR-486-1-5p, miR-486-2-5p | Ultracentrifugation + ultrafiltration + OptiPrep gradient centrifugation | miRNeasy Mini Kit, TRIzol | NGS | Diagnosis | 81 |
| 45 | Plasma | 1 ml | PLAP | Ultracentrifugation | ELISA | ELISA | Diagnosis | 79 | |
| Preterm birth | 20 | Plasma | 1 ml | miRNAs | Ultracentrifugation | RNeasy Mini Kit | NGS | Prognosis | 92 |
| Gestational diabetes mellitus | 20 | Plasma | 1 ml | CD63, PLAP | Ultracentrifugation + ultrafiltration + OptiPrep gradient centrifugation | ELISA | ELISA | Prognosis | 88 |
| Congenital obstructive nephropathy | 8 | Amniotic fluid | 10 ml | miR-300, miR-299-5p | Ultracentrifugation + sucrose gradient centrifugation | TRIzol | miRNA microarray | Diagnosis | 91 |
| Intrauterine growth restriction | 30 | Plasma | 1 ml | PLAP | Ultracentrifugation + iodixanol gradient centrifugation | Quantum dots | ELISA | Diagnosis | 90 |
| Cardiovascular diseases | |||||||||
| Myocardial infarction | 20 | Serum | 1 ml | miR-939 | Ultracentrifugation | TRIzol | qPCR | Prognosis | 107 |
| 35 | Plasma | 5 ml | proteins | Ultracentrifugation | Trypsin | LC-MS/MS | Diagnosis | 111 | |
| Coronary artery disease | 180 | Plasma | 250 μl | miR-92a-3p | Ultracentrifugation | TRIzol | qPCR | Prognosis | 106 |
| Vascular disease | 1060 | Plasma | 150 μl | Cystatin C, Serpin F2, CD14 | ExoQuick™ | Roche Complete Lysis-M | Immunoassay | Prognosis | 110 |
| Heart failure | 20 | Serum | – | miR-423-5p, miR-22, miR-320a, miR-92b | ExoQuick™ Exosome Precipitation Solution | Ethanol precipitation | qRT–PCR | Prognosis | 112 |
| 100 | Serum | 250 μl | miRNA | ExoQuick™ Exosome Precipitation Solution | ISOGEN II | qPCR | Prognosis | 113 | |
| Stroke | 131 | Serum | – | miR-9, miR-124 | ExoQuickTM Solution | Exosome RNA Purification Kit | qPCR | Diagnosis | 115 |
| Cardiac arrhythmia | 37 | Serum | 30 μl | IL-1 β, P-selectin | Affinity capture | ELISA | ELISA | Prognosis | 118 |
| Organ transplantation | |||||||||
| Lung | 30 | BALF | – | SAgs, Collagen-V | Ultracentrifugation | – | Western blot | Diagnosis | 126 |
| Serum | 1 ml | Total Exosome Isolation Reagent kit | |||||||
| 12 | BALF | 20-60 ml | mRNAs | Ultracentrifugation | miRNeasy, miRCURY | RNA-Seq | Diagnosis | 127 | |
| Heart | 10 | Serum | – | miR-142-3p | Ultracentrifugation | microRNeasy mini kit | qRT–PCR | Diagnosis | 129 |
| 48 | Serum | 200 μl | Proteins | Total Exosome Isolation | Trypsin | LC-MS/MS | Prognosis | 128 | |
| Kidney | 44 | Urine | 15 ml | CD3 | Ultracentrifugation | iKEA | iKEA | Prognosis | 130 |
| 47 | Urine | – | Tetraspanin-1, Hemopexin | Ultracentrifugation | RIPA buffer | LC-MS/MS | Diagnosis | 131 | |
| 64 | Plasma | – | mRNAs | exoRNeasy Serum/Plasma Midi Kit | exoRNeasy Serum/Plasma Midi Kit | qPCR | Diagnosis | 132 | |
Cohort scale, source of body fluid, functional targets of exosomes, exosome extraction methods, target isolation, and detection methods of recent clinical studies on exosomes as biomarkers for diagnosis or prognosis of these diseases
Progression in exosome extraction methods
Thery et al.25 firstly proposed the classic method of ultracentrifugation for isolating exosomes. After that, differential ultracentrifugation became the most common method for exosome isolation. According to a global survey in October 2015, >80% of researchers used this method for EV separation.134 Exosome extraction methods are mainly based on their physical characteristics and components.16 Importantly, exosome quality will be greatly affected by different exosome extraction procedure.135 Currently, these methods for exosome extraction can be commonly divided into differential ultracentrifugation, density gradient/cushion centrifugation, size exclusion chromatography (SEC), precipitation, (immuno-) affinity capture, microfluidic approaches, etc.136 While differential ultracentrifugation is the most commonly applied method for exosome isolation with relatively satisfactory purity, it is time-consuming and requires costly instruments, which are nevertheless disadvantages that cannot be ignored, making it untoward for clinical application.137,138 Density gradient centrifugation can further increase the exosome purity and has been recommended as a standard to validate an EV experiment,139 whereas it is more time consuming and of low yield.2 SEC excludes nonexosomal particles by vesicle size.140 However, some samples may contain a large proportion of lipoprotein particles of similar size to exosomes, and thus, they cannot be distinguished.141 Kaloyan et al.142 compared the difference between murine plasma exosomes extracted by ultracentrifugation and SEC. They found that the latter extracted a large number of exosomes that were enriched with protein but also with many chylomicron-positive lipid particles and nonvesicle-associated proteins.142 With a high yield of exosomes, the precipitation method provides the least pure exosomes compared to size exclusion isolation and density gradient purification.143 It often precipitates viruses, proteins, and other substances together with exosomes, which may influence subsequent experiments. In contrast, affinity capture methods often provide high purity, although the recovery seems unsatisfactory.2
As mentioned above, purity improvement is an issue that most methods need to consider, especially regarding lipoprotein contamination (including high-density lipoproteins (HDL), LDL, very low-density lipoproteins (VLDL), and chylomicrons) in blood samples.140 The similar density of EVs and HDL, as well as the tremendous gap in abundance between EVs and LDL, makes it difficult to separate them by density gradient ultracentrifugation. SEC can purify EVs from HDL/LDL by size differences, but not chylomicrons or VLDL, which possess similar sizes to EVs.140,144 The combination of ultracentrifugation and SEC140 or the combination of density gradient fractionation and immunoaffinity capture13 could greatly reduce contamination. Essentially, the most common way applied to decrease contamination is dilution, which is a necessary step before ultracentrifugation.145 What also caught our attention is that during the preparation of proteins/peptides for MS technology, a solid-phase extraction method will be utilized for lipoprotein removal,146 which may also be suitable for exosome purification. Minimal Information for Studies of Extracellular Vesicles guidelines (2018) put forward the classification of existing extraction methods based on recovery and specificity of exosomes,2 which provided a way for researchers to select appropriate and stable methods in subsequent studies. However, we are still going to explore a better methodology for substantial yields and reliable quality of exosomes.
Apart from these classic extraction methods, multiple new extraction and detection technologies have been reported in recent years, such as the microfluidic chip147,148 and a method of integrated extraction and quantitative analysis of exosomal nucleic acids and proteins,149 with high specificity and intact yield of exosomes, small required sample volume and simple, time-saving operation. Moreover, a novel urine-based EV extraction and enrichment method was established by Woo et al.150. This new technology takes only 30 min to enrich EVs from 4 ml urine. Following mRNA extraction, AR-V7 and androgen receptor full-length mRNA detection via ddPCR, they proposed exosomal AR-V7 transcript as a promising biomarker for the clinical application of urinary biopsy in prostate cancer.150 Additionally, a rapid and simple Vn96-peptide-based EV isolation method was established, which could capture heat shock proteins abounded on the surface of EVs.151 Bijnsdorp et al.152 optimized this time- (~1.5 h) and cost-efficient method and indicated similar efficacy compared to ultracentrifugation when extracting urinary EVs. At the same time, a size-based exosome total isolation chip, which was easy to operate, with higher yield and similar purity compared to ultracentrifugation and required small sample volumes (10–100 μl), was designed to simplify EV extraction from clinical biofluids, such as plasma, urine, and lavage.153 Complementarily, Chen et al.154 proposed an anion-exchange-based method for separating exosomes directly from plasma or cell culture media by anion magnetic beads within 30 min.
More importantly, the exosome isolation kit, the most representative commercial product, is becoming more prevalent recently and includes the ExoQuick precipitation solution,155,156 Total Exosome Isolation Reagent kit126 and ME™ kit157 based on precipitation; Exo-Spin™158 based on precipitation and size exclusion; and exoEasy Maxi Kit,55 ExoCapTM Exosome Isolation and Enrichment kit, and Exo-FlowTM159 based on immunocapture. It has been reported that kit-based exosome isolation methods are more convenient and effective160 and obtain similar exosome recovery and purity comparable to the ultracentrifugation method.128,161 However, Tian et al.138 recently demonstrated that a large proportion of contaminants existed in products extracted by commercial isolation kits and that the products exhibited much lower purity than those extracted with ultracentrifugation. The use of commercial kits is still controversial, and more technical refinement is urged for clinical application. Finally, we summarized some cost-convenient exosome extraction methods, which possessed equivalent or higher exosome yield and purity compared with ultracentrifugation, potentially feasible for clinical use in Table 3.
Table 3.
Exosome extraction methods potentially feasible for clinical application. Some novel and cost-convenient exosome extraction methods, which possessed equivalent or higher exosome yield and purity compared with ultracentrifugation, were summarized in terms of time, source, minimum volume of biofluid, exosome quality identification, technical principles and clinical field applied
| Extraction method | Time | Source | Sample volume | Quality identification | Principle | Clinical field |
|---|---|---|---|---|---|---|
| Vn96-peptide-based | 1.5 h | Plasma, culture media/urine | 1 ml | Proteomics | Vn96-peptide binding to heat shock proteins- based affinity capture |
Breast cancer151 Prostate cancer152 Nephronophthisis-related ciliopathies184 |
| Exosome total isolation chip | 5 ml/h (culture media), 1 h (plasma) | Culture media, plasma, urine, BALF | 10–100 μl | miRNA sequence, proteomics | Nanoporous membrane-based filtration | Lung cancer153 |
| Anion-exchange -based | 30 min | Culture media, plasma | 500 μl | Proteomics | Anion-exchange-based magnetic beads | Prostate cancer154 |
| Microfluidic chip | Overnight for device functionalization, <20 min for capture and release | Culture media, serum | <100 μl | Proteomics | CD63/CD9 and EpCAM based immunoaffinity capture | Ovarian cancer148,185 |
Commercial development of exosomes
A multitude of companies have been established to exploit biotechnology development in exosomes. Founded in 2015, Codiak BioSciences is a pioneer biotechnology company developing exosome treatments for various diseases and is headquartered in Cambridge, Massachusetts. Codiak BioSciences has developed the engEx™ Platform to engineer exosomes to express and deliver therapeutic drug candidates. ExoSTING is a major and promising immune therapeutic candidate targeting cancer. Compared to free STING agonists, exoSTING is highly potent with minimal toxic potential. It is rarely affected by serum systemic cytokines and preserves the vitality of effector T cells and antigen-presenting cells in tumors to maintain sustained immune protection. Recently, exoSTING is being developed for solid tumor therapy that activates the “STING” receptor in immune cells. Relevant clinical trials will be carried out in the first half of 2020.162 Exosome Diagnostics is a revolutionary developer of molecular diagnostics based on biological fluids, which was acquired by Bio-Techne last year. They are aimed at developing novel and precise exosome technology mainly in liquid biopsy of multiple cancers, including lung38,41 and prostate cancer.163,164 The ExoDx Prostate® (IntelliScore) (EPI) test is the star production of exosome diagnostics. This is a urine-based and completely noninvasive test designed to assist physicians in assessing whether an individual patient over 50 years old tested with 2–10 ng/ml prostate-specific antigen, which presenting for a needle biopsy, is at greater risk for high-grade prostate cancer; therefore, the patient can avoid unnecessary biopsy and, instead, continue to follow up.163 Moreover, the commercial exosome isolation kit ExoLution Plus Isolation Kit owned by Exosome Diagnostics is widely used in exosome research. Recently, Avalon GloboCare and its subsidiary Genexosome Technologies clarified the establishment and development of the first salivary-based exosome miRNA biomarker-miR-185 as a dual target for the diagnosis and treatment of oral cancer.165 Additionally, Avalon GloboCare is working on the identification of human angiogenic exosomes.166 PureTech Health167 collaborated with Roche to impel the advancement of technology for oral administration of antisense oligonucleotides with PureTech’s milk exosome-based technology168 to transform conventional intravenous injection therapy for improved efficacy and reduced toxicity.
Conclusions
With the progression of precise and individual medicine, conventional solid biopsy has gradually shown considerable limitations, whereas the occurrence of liquid biopsy greatly makes up for it and provides a promising platform for noninvasive diagnosis and prognosis. Undoubtedly, exosomes play an important role in various physiological and pathological processes, and compelling evidence has proven exosomes to be a potential tool for clinical application, including liquid biopsy and therapy, with the powerful advantages of existence in all body fluids, stable biological activity, higher sensitivity and specificity in diagnosis and prognosis, and organotropic characteristics.23 Meanwhile, various sources of biofluid are applied in different diseases according to human anatomy and pathophysiology. For example, nervous system diseases will prefer cerebrospinal fluid,169 prostate cancer, and urinary system diseases will benefit more from urine and most solid tumors, pregnancy disorders and CVDs are prone to blood testing.
However, there are still some barriers between basic research and real clinical practice. First, a standardization of the classification and extraction method of exosomes for different body liquids is urgently needed. More efficient methods with a low biofluid volume requirement and high purity and yield are the foundation of subsequent applications. Second, the identification of specific subtypes of EVs is urgently needed, as different vesicles may exert various biological effects. Current methods to extract exosomes (as shown in Tables 1 and 2) are too diverse to confirm the purity of the product. Therefore, it is necessary to standardize the protocols and identification methods when attempting to use exosomes widely in clinical testing. Additionally, more reliable biomarkers should be confirmed. Although many molecules carried by exosomes have been documented to serve as potential biomarkers, little of them are qualified for application. It may be a better direction to validate documented biomarkers on a larger scale to create new panels for multiple fields. Last but not the least, as potential therapeutic cargo, the biological safety, targeted efficacy, and adverse effects of exosomes must be confirmed before clinical use.
Acknowledgements
This work was supported by National Key R&D Program of China (No. 2016YFC1302803), National Natural Science Foundation of China (No.81500115, No. 81672342), and Zhejiang Provincial Natural Science Foundation of China (No. LY19H030012).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Biting Zhou, Kailun Xu, Xi Zheng
Contributor Information
Yingkuan Shao, Email: ykshao@zju.edu.cn.
Shu Zheng, Email: zhengshu@zju.edu.cn.
References
- 1.Yáñez-Mó M, et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750–1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 4.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 5.Pisitkun, T., Shen, R. F. & Knepper, M. A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl Acad. Sci. USA101, 13368–13373 (2004). [DOI] [PMC free article] [PubMed]
- 6.Street JM, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palanisamy V, et al. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bard MP, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 9.Runz S, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 10.Keller S, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 11.Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 12.Kulshreshtha A, Ahmad T, Agrawal A, Ghosh B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 2013;131:1194–1203. doi: 10.1016/j.jaci.2012.12.1565. [DOI] [PubMed] [Google Scholar]
- 13.Jeppesen DK, et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e418. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoi A, et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019;5:eaax8849. doi: 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 16.Macias M, et al. Liquid biopsy: from basic research to clinical practice. Adv. Clin. Chem. 2018;83:73–119. doi: 10.1016/bs.acc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Cheruvanky A, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 2007;292:F1657–1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Zeng Q, Han Q, Xia W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell. 2019;10:295–299. doi: 10.1007/s13238-018-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Janku F, Zhan Q, Fan JB. Accessing genetic information with liquid biopsies. Trends Genet. 2015;31:564–575. doi: 10.1016/j.tig.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Xu R, Greening DW, Zhu H-J, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J. Clin. Investig. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 22.Tannetta D, Masliukaite I, Vatish M, Redman C, Sargent I. Update of syncytiotrophoblast derived extracellular vesicles in normal pregnancy and preeclampsia. J. Reprod. Immunol. 2017;119:98–106. doi: 10.1016/j.jri.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun B, et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell. Physiol. 2019;234:1416–1425. doi: 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- 25.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 26.Tayoun T, et al. CTC-derived models: a window into the seeding capacity of circulating tumor cells (CTCs) Cells. 2019;8:1145. doi: 10.3390/cells8101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE. 2017;12:e0183915. doi: 10.1371/journal.pone.0183915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keseru JS, et al. Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian cancer. J. Biotechnol. 2019;298:76–81. doi: 10.1016/j.jbiotec.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Allenson K, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017;28:741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohrmann L, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin. Cancer Res. 2018;24:181–188. doi: 10.1158/1078-0432.CCR-17-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y, et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018;29:2379–2383. doi: 10.1093/annonc/mdy458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard, V. et al. Circulating nucleic acids associate with outcomes of patients with pancreatic cancer. Gastroenterology.156, 108–118 (2019). [DOI] [PMC free article] [PubMed]
- 33.Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J. Hematol. Oncol. 2019;12:84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell. Mol. Med. 2017;21:838–847. doi: 10.1111/jcmm.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong L, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2016;25:1158–1166. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 37.Li S, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–d112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellanos-Rizaldos E, et al. Exosome-based detection of EGFR T790M in plasma from non–small cell lung cancer patients. Clin. Cancer Res. 2018;24:2944–2950. doi: 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 39.Sandfeld-Paulsen B, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016;11:1701–1710. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 40.Chen IH, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl Acad. Sci. USA. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krug AK, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018;29:2143. doi: 10.1093/annonc/mdy261. [DOI] [PubMed] [Google Scholar]
- 42.Madhavan B, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 43.Street JM, Koritzinsky EH, Glispie DM, Star RA, Yuen PS. Urine exosomes: an emerging trove of biomarkers. Adv. Clin. Chem. 2017;78:103–122. doi: 10.1016/bs.acc.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Raimondo F, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013;9:1220–1233. doi: 10.1039/c3mb25582d. [DOI] [PubMed] [Google Scholar]
- 45.Matsuzaki K, et al. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget. 2017;8:24668–24678. doi: 10.18632/oncotarget.14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keklikoglou I, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 2019;21:190–202. doi: 10.1038/s41556-018-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maji S, et al. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol. Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiedemann K, et al. Exosomal release of L-plastin by breast cancer cells facilitates metastatic bone osteolysis. Transl. Oncol. 2019;12:462–474. doi: 10.1016/j.tranon.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 2019;25:1302–1317. doi: 10.1158/1078-0432.CCR-18-1447. [DOI] [PubMed] [Google Scholar]
- 51.Bijnsdorp, I. V. et al. Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J. Extracell. Vesicles.2, 1–10 (2013). [DOI] [PMC free article] [PubMed]
- 52.Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical significance of PD-L1(+) exosomes in plasma of head and neck cancer patients. Clin. Cancer Res. 2018;24:896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li C, et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J. Transl. Med. 2019;17:355. doi: 10.1186/s12967-019-2101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lux, A., Kahlert, C., Grützmann, R. & Pilarsky, C. c-Met and PD-L1 on circulating exosomes as diagnostic and prognostic markers for pancreatic cancer. Int. J. Mol. Sci.20, 3305 (2019). [DOI] [PMC free article] [PubMed]
- 55.Teng Y, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng Z, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meltzer S, et al. Circulating exosomal miR-141-3p and miR-375 in metastatic progression of rectal cancer. Transl. Oncol. 2019;12:1038–1044. doi: 10.1016/j.tranon.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu JL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao Y, et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39:1368–1379. doi: 10.1093/carcin/bgy115. [DOI] [PubMed] [Google Scholar]
- 60.Fu F, Jiang W, Zhou L, Chen Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol. 2018;11:221–232. doi: 10.1016/j.tranon.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng, Z. Y., Gu, R. H. & Yan, B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J. Cell. Biochem.120, 1457–1463 (2018). [DOI] [PubMed]
- 62.Wei Y, et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014;147:423–431. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 63.Mashouri L, et al. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei F, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol. Cancer. 2017;16:132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qu L, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 66.Liu T, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer. 2019;18:43–43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Han M, et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020;11:43. doi: 10.1038/s41419-020-2250-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Expulsion of small molecules in vesicles shed by cancer cells: association with gene expression and chemosensitivity profiles. Cancer Res. 2003;63:4331–4337. [PubMed] [Google Scholar]
- 69.Sharma A. Chemoresistance in cancer cells: exosomes as potential regulators of therapeutic tumor heterogeneity. Nanomedicine (Lond., Engl.) 2017;12:2137–2148. doi: 10.2217/nnm-2017-0184. [DOI] [PubMed] [Google Scholar]
- 70.Zheng P, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017;36:53. doi: 10.1186/s13046-017-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Binenbaum Y, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78:5287–5299. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 72.Chen G, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salomon C, Scholz K, Kobayashi M. Hypoxia regulates the response of trophoblast-derived exosomes to hyperglycemia and displays a difference placental exosome profile in plasma from patients with gestational diabetes mellitus. Reprod. Sci. 2015;22:257–258. [Google Scholar]
- 74.American College of Obstetricians and Gynecologists. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet. Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 75.Mol BWJ, et al. Pre-eclampsia. Lancet (Lond., Engl.) 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 76.Salomon C, et al. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS ONE. 2013;8:e79636. doi: 10.1371/journal.pone.0079636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dragovic RA, et al. Isolation of syncytiotrophoblast microvesicles and exosomes and their characterisation by multicolour flow cytometry and fluorescence nanoparticle tracking analysis. Methods (San. Diego, Calif.) 2015;87:64–74. doi: 10.1016/j.ymeth.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salomon C, et al. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS ONE. 2014;9:e98667. doi: 10.1371/journal.pone.0098667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pillay P, Maharaj N, Moodley J, Mackraj I. Placental exosomes and pre-eclampsia: maternal circulating levels in normal pregnancies and, early and late onset pre-eclamptic pregnancies. Placenta. 2016;46:18–25. doi: 10.1016/j.placenta.2016.08.078. [DOI] [PubMed] [Google Scholar]
- 80.Biro O, et al. Various levels of circulating exosomal total-miRNA and miR-210 hypoxamiR in different forms of pregnancy hypertension. Pregnancy Hypertens. 2017;10:207–212. doi: 10.1016/j.preghy.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Salomon C, et al. Placental exosomes as early biomarker of preeclampsia: potential role of exosomal microRNAs across gestation. J. Clin. Endocrinol. Metab. 2017;102:3182–3194. doi: 10.1210/jc.2017-00672. [DOI] [PubMed] [Google Scholar]
- 82.Barbour LA, et al. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care. 2007;30:S112–119. doi: 10.2337/dc07-s202. [DOI] [PubMed] [Google Scholar]
- 83.Lain KY, Catalano PM. Metabolic changes in pregnancy. Clin. Obstet. Gynecol. 2007;50:938–948. doi: 10.1097/GRF.0b013e31815a5494. [DOI] [PubMed] [Google Scholar]
- 84.American Diabetes Association. Standards of medical care in diabetes–2014. Diabetes Care37, Suppl 1, S14–80, 10.2337/dc14-S014 (2014). [DOI] [PubMed]
- 85.Simmons D. in A Practical Manual of Diabetes in Pregnancy (eds McCance D., Maresh, M., Sacks D. A.) (London: Blackwell Publishing, London, 2010).
- 86.Chu SY, et al. Maternal obesity and risk of gestational diabetes mellitus. Diabetes Care. 2007;30:2070–2076. doi: 10.2337/dc06-2559a. [DOI] [PubMed] [Google Scholar]
- 87.Mitchell MD, et al. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015;213:S173–181. doi: 10.1016/j.ajog.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 88.Salomon C, et al. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65:598–609. doi: 10.2337/db15-0966. [DOI] [PubMed] [Google Scholar]
- 89.Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet (Lond., Engl.) 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 90.Miranda J, et al. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction -Liquid biopsies to monitoring fetal growth. Placenta. 2018;64:34–43. doi: 10.1016/j.placenta.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Xie J, et al. The relationship between amniotic fluid miRNAs and congenital obstructive nephropathy. Am. J. Transl. Res. 2017;9:1754–1763. [PMC free article] [PubMed] [Google Scholar]
- 92.Menon, R. et al. Circulating exosomal miRNA profile during term and preterm birth pregnancies-a longitudinal study. Endocrinology, 160, 249–275 (2018). [DOI] [PMC free article] [PubMed]
- 93.Jiang PY, et al. MicroRNAs derived from urinary exosomes act as novel biomarkers in the diagnosis of intrahepatic cholestasis of pregnancy. Am. J. Transl. Res. 2019;11:6249–6261. [PMC free article] [PubMed] [Google Scholar]
- 94.Nielsen MR, et al. Urine exosomes from healthy and hypertensive pregnancies display elevated level of α-subunit and cleaved α- and γ-subunits of the epithelial sodium channel-ENaC. Pflug. Arch. 2017;469:1107–1119. doi: 10.1007/s00424-017-1977-z. [DOI] [PubMed] [Google Scholar]
- 95.Ghafarian F, et al. The clinical impact of exosomes in cardiovascular disorders: from basic science to clinical application. J. Cell. Physiol. 2019;234:12226–12236. doi: 10.1002/jcp.27964. [DOI] [PubMed] [Google Scholar]
- 96.Howard BV, et al. LDL cholesterol as a strong predictor of coronary heart disease in diabetic individuals with insulin resistance and low LDL: The Strong Heart Study. Arterioscler. Thromb. Vasc. Biol. 2000;20:830–835. doi: 10.1161/01.atv.20.3.830. [DOI] [PubMed] [Google Scholar]
- 97.Ndrepepa G, et al. High-sensitivity troponin T and mortality after elective percutaneous coronary intervention. J. Am. Coll. Cardiol. 2016;68:2259–2268. doi: 10.1016/j.jacc.2016.08.059. [DOI] [PubMed] [Google Scholar]
- 98.Wang K, et al. Enhanced cardioprotection by human endometrium mesenchymal stem cells driven by exosomal MicroRNA-21. Stem Cells Transl. Med. 2017;6:209–222. doi: 10.5966/sctm.2015-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ong SG, et al. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 2014;130:S60–69. doi: 10.1161/CIRCULATIONAHA.113.007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Emanueli C, Shearn AI, Angelini GD, Sahoo S. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vasc. Pharmacol. 2015;71:24–30. doi: 10.1016/j.vph.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuwabara Y, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 2011;4:446–454. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 102.Ibrahim AG, Cheng K, Marban E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014;2:606–619. doi: 10.1016/j.stemcr.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barile L, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 2014;103:530–541. doi: 10.1093/cvr/cvu167. [DOI] [PubMed] [Google Scholar]
- 104.Saha, P. et al. Circulating exosomes derived from transplanted progenitor cells aid the functional recovery of ischemic myocardium. Sci. Transl. Med.11, eaau1168 (2019). [DOI] [PMC free article] [PubMed]
- 105.Chen L, et al. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2013;431:566–571. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, et al. Atherosclerotic conditions promote the packaging of functional MicroRNA-92a-3p into endothelial microvesicles. Circulation Res. 2019;124:575–587. doi: 10.1161/CIRCRESAHA.118.314010. [DOI] [PubMed] [Google Scholar]
- 107.Li H, et al. Coronary serum exosomes derived from patients with myocardial ischemia regulate angiogenesis through the miR-939-mediated nitric oxide signaling pathway. Theranostics. 2018;8:2079–2093. doi: 10.7150/thno.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Christersson C, Thulin A, Siegbahn A. Microparticles during long-term follow-up after acute myocardial infarction. Association to atherosclerotic burden and risk of cardiovascular events. Thromb. Haemost. 2017;117:1571–1581. doi: 10.1160/TH16-11-0837. [DOI] [PubMed] [Google Scholar]
- 109.Chiva-Blanch G, et al. CD3(+)/CD45(+) and SMA-alpha(+) circulating microparticles are increased in individuals at high cardiovascular risk who will develop a major cardiovascular event. Int. J. Cardiol. 2016;208:147–149. doi: 10.1016/j.ijcard.2016.01.211. [DOI] [PubMed] [Google Scholar]
- 110.Kanhai DA, et al. Microvesicle protein levels are associated with increased risk for future vascular events and mortality in patients with clinically manifest vascular disease. Int. J. Cardiol. 2013;168:2358–2363. doi: 10.1016/j.ijcard.2013.01.231. [DOI] [PubMed] [Google Scholar]
- 111.Cheow ES, et al. Plasma-derived extracellular vesicles contain predictive biomarkers and potential therapeutic targets for myocardial ischemic (MI) injury. Mol. Cell. Proteom. 2016;15:2628–2640. doi: 10.1074/mcp.M115.055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goren Y, et al. Serum levels of microRNAs in patients with heart failure. Eur. J. Heart Fail. 2012;14:147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 113.Matsumoto S, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circulation Res. 2013;113:322–326. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 114.Berezin AE, et al. Predictive value of apoptotic microparticles to mononuclear progenitor cells ratio in advanced chronic heart failure patients. J. Cardiol. 2015;65:403–411. doi: 10.1016/j.jjcc.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 115.Ji Q, et al. Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS ONE. 2016;11:e0163645. doi: 10.1371/journal.pone.0163645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Halkein J, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J. Clin. Investig. 2013;123:2143–2154. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vandergriff AC, et al. Intravenous cardiac stem cell-derived exosomes ameliorate cardiac dysfunction in doxorubicin induced dilated cardiomyopathy. Stem Cells Int. 2015;2015:960926. doi: 10.1155/2015/960926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang H, et al. Increased serum levels of microvesicles in nonvalvular atrial fibrillation determinated by ELISA using a specific monoclonal antibody AD-1. Clin. Chim. Acta. 2010;411:1700–1704. doi: 10.1016/j.cca.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 119.Kasner M, et al. Circulating exosomal microRNAs predict functional recovery after MitraClip repair of severe mitral regurgitation. Int. J. Cardiol. 2016;215:402–405. doi: 10.1016/j.ijcard.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 120.Benichou G, Valujskikh A, Heeger PS. Contributions of direct and indirect T cell alloreactivity during allograft rejection in mice. J. Immunol. 1999;162:352–358. [PubMed] [Google Scholar]
- 121.Lakkis FG, Arakelov A, Konieczny BT, Inoue Y. Immunologic ‘ignorance’ of vascularized organ transplants in the absence of secondary lymphoid tissue. Nat. Med. 2000;6:686–688. doi: 10.1038/76267. [DOI] [PubMed] [Google Scholar]
- 122.Liu Q, et al. Donor dendritic cell-derived exosomes promote allograft-targeting immune response. J. Clin. Investig. 2016;126:2805–2820. doi: 10.1172/JCI84577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morelli AE, Bracamonte-Baran W, Burlingham WJ. Donor-derived exosomes: the trick behind the semidirect pathway of allorecognition. Curr. Opin. Organ Transplant. 2017;22:46–54. doi: 10.1097/MOT.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gonzalez-Nolasco B, Wang M, Prunevieille A, Benichou G. Emerging role of exosomes in allorecognition and allograft rejection. Curr. Opin. Organ Transplant. 2018;23:22–27. doi: 10.1097/MOT.0000000000000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vallabhajosyula P, et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Investig. 2017;127:1375–1391. doi: 10.1172/JCI87993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gunasekaran M, et al. Donor-derived exosomes with lung self-antigens in human lung allograft rejection. Am. J. Transplant. 2017;17:474–484. doi: 10.1111/ajt.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gregson AL, et al. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am. J. Respir. Crit. Care Med. 2015;192:1490–1503. doi: 10.1164/rccm.201503-0558OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kennel PJ, et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J. Heart Lung Transplant. 2018;37:409–417. doi: 10.1016/j.healun.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 129.Sukma Dewi I, et al. Exosomal miR-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc. Res. 2017;113:440–452. doi: 10.1093/cvr/cvw244. [DOI] [PubMed] [Google Scholar]
- 130.Park J, et al. Integrated kidney exosome analysis for the detection of kidney transplant rejection. ACS Nano. 2017;11:11041–11046. doi: 10.1021/acsnano.7b05083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lim JH, et al. Novel urinary exosomal biomarkers of acute T cell-mediated rejection in kidney transplant recipients: a cross-sectional study. PLoS ONE. 2018;13:e0204204. doi: 10.1371/journal.pone.0204204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang H, et al. Plasma exosomes from HLA-sensitized kidney transplant recipients contain mRNA transcripts which predict development of antibody-mediated rejection. Transplantation. 2017;101:2419–2428. doi: 10.1097/TP.0000000000001834. [DOI] [PubMed] [Google Scholar]
- 133.Korutla, L. et al. Noninvasive diagnosis of recurrent autoimmune type 1 diabetes after islet cell transplantation. Am. J. Transplant.19, 1852–1858 (2019). [DOI] [PMC free article] [PubMed]
- 134.Gardiner C, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brennan K, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020;10:1039. doi: 10.1038/s41598-020-57497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mateescu B, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Webber, J. & Clayton, A. How pure are your vesicles? J Extracell Vesicles2, 19861 (2013). [DOI] [PMC free article] [PubMed]
- 138.Tian Y, et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles. 2020;9:1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Van Deun J, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 140.Simonsen JB. What are we looking at? extracellular vesicles, lipoproteins, or both? Circ. Res. 2017;121:920–922. doi: 10.1161/CIRCRESAHA.117.311767. [DOI] [PubMed] [Google Scholar]
- 141.Flaherty SE, III, et al. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science. 2019;363:989–993. doi: 10.1126/science.aaw2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takov K, Yellon DM, Davidson SM. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J. Extracell. Vesicles. 2019;8:1560809. doi: 10.1080/20013078.2018.1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lobb RJ, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sódar BW, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016;6:24316. doi: 10.1038/srep24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zheng X, et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J. Extracell. Vesicles. 2020;9:1750202. doi: 10.1080/20013078.2020.1750202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmelter C, et al. Comparison of two solid-phase extraction (SPE) methods for the identification and quantification of porcine retinal protein markers by LC-MS/MS. Int. J. Mol. Sci. 2018;19:3847. doi: 10.3390/ijms19123847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fang S, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE. 2017;12:e0175050. doi: 10.1371/journal.pone.0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dorayappan, K. D. P. et al. A microfluidic chip enables isolation of exosomes and establishment of their protein profiles and associated signaling pathways in ovarian cancer. Cancer Res.79, 3503–3513 (2019). [DOI] [PMC free article] [PubMed]
- 149.Zarovni N, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods (San. Diego, Calif.) 2015;87:46–58. doi: 10.1016/j.ymeth.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 150.Woo HK, et al. Urine-based liquid biopsy: non-invasive and sensitive AR-V7 detection in urinary EVs from patients with prostate cancer. Lab Chip. 2018;19:87–97. doi: 10.1039/c8lc01185k. [DOI] [PubMed] [Google Scholar]
- 151.Ghosh A, et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE. 2014;9:e110443. doi: 10.1371/journal.pone.0110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bijnsdorp IV, et al. Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method. J. Extracell. Vesicles. 2017;6:1313091. doi: 10.1080/20013078.2017.1313091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu F, et al. The exosome total isolation chip. ACS Nano. 2017;11:10712–10723. doi: 10.1021/acsnano.7b04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen J, Xu Y, Lu Y, Xing W. Isolation and visible detection of tumor-derived exosomes from plasma. Anal. Chem. 2018;90:14207–14215. doi: 10.1021/acs.analchem.8b03031. [DOI] [PubMed] [Google Scholar]
- 155.Alegre E, et al. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 2014;138:828–832. doi: 10.5858/arpa.2013-0134-OA. [DOI] [PubMed] [Google Scholar]
- 156.Sáenz-Cuesta M, et al. Methods for extracellular vesicles isolation in a hospital setting. Front. Immunol. 2015;6:50. doi: 10.3389/fimmu.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Knol JC, et al. Peptide-mediated ‘miniprep’ isolation of extracellular vesicles is suitable for high-throughput proteomics. EuPA Open Proteom. 2016;11:11–15. doi: 10.1016/j.euprot.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Welton JL, Webber JP, Botos LA, Jones M, Clayton A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles. 2015;4:27269. doi: 10.3402/jev.v4.27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rim KT, Kim SJ. Quantitative analysis of exosomes from murine lung cancer cells by flow cytometry. J. Cancer Prev. 2016;21:194–200. doi: 10.15430/JCP.2016.21.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao L, et al. Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients. J. Cancer. 2017;8:1145–1152. doi: 10.7150/jca.18026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Soares Martins T, Catita J, Martins Rosa I, da Cruz E Silva OAB, Henriques AG. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE. 2018;13:e0198820. doi: 10.1371/journal.pone.0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kevin Dooley, S. C. J. et al. Developing an Engineered Exosome Therapeutic Platform for Immuno-Oncology. Keystone Symposium on Cancer Immunotherapy, Whistler, Canada (2019).
- 163.McKiernan J, et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2-10ng/ml at initial biopsy. Eur. Urol. 2018;74:731–738. doi: 10.1016/j.eururo.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 164.McKiernan J, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2:882–889. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 165.Avalon GloboCare and its Subsidiary Genexosome Technologies Announce Discovery and Development of World’s First Saliva-Based Exosomal Biomarker “miR-185” as Dual Diagnostic and Therapeutic Target for Oral Cancer. GLOBE NEWSWIRE. https://www.globenewswire.com/news-release/2019/01/28/1706078/0/en/Avalon-GloboCare-and-its-Subsidiary-Genexosome-Technologies-Announce-Discovery-and-Development-of-World-s-First-Saliva-Based-Exosomal-Biomarker-miR-185-as-Dual-Diagnostic-and-Thera.html (Accessed 28 Jan 2019).
- 166.Avalon GloboCare Announces Breakthrough in Identifying Human Angiogenic Exosomes/Extracellular Vesicles (EV) Derived from Endothelial Cells. GLOBE NEWSWIRE. https://www.globenewswire.com/news-release/2019/02/11/1716606/0/en/Avalon-GloboCare-Announces-Breakthrough-in-Identifying-Human-Angiogenic-Exosomes-Extracellular-Vesicles-EV-Derived-from-Endothelial-Cells.html (Accessed 11 Feb 2019).
- 167.PureTech Health Announces Collaboration with Roche to Advance Technology for Oral Administration of Antisense Oligonucleotides. PureTech News. http://puretechhealth.com/news/23-press-releases/1029-puretech-health-announces-collaboration-with-roche-to-advance-technology-for-oral-administration-of-antisense-oligonucleotides (Accessed 20 Jul 2018).
- 168.Agrawal AK, et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017;13:1627–1636. doi: 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 169.Figueroa JM, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19:1494–1502. doi: 10.1093/neuonc/nox085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kimura, H. et al. CKAP4, a DKK1 Receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin. Cancer Res. 25, 1936–1947 (2019). [DOI] [PubMed]
- 171.Yokoyama S, et al. Clinical implications of carcinoembryonic antigen distribution in serum exosomal fraction-Measurement by ELISA. PLoS ONE. 2017;12:e0183337. doi: 10.1371/journal.pone.0183337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rodriguez M, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer. 2014;53:713–724. doi: 10.1002/gcc.22181. [DOI] [PubMed] [Google Scholar]
- 173.Niu L, et al. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2019;110:433–442. doi: 10.1111/cas.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hannafon BN, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang, X. et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol carcinogen, 58, 674–685 (2019). [DOI] [PubMed]
- 177.Arbelaiz A, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66:1125–1143. doi: 10.1002/hep.29291. [DOI] [PubMed] [Google Scholar]
- 178.Matsumura T, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer. 2015;113:275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sandfeld-Paulsen B, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016;10:1595–1602. doi: 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Taverna S, et al. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017;7:3170. doi: 10.1038/s41598-017-03460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rodríguez-Martínez A, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019;21:21. doi: 10.1186/s13058-019-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Alegre E, et al. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta. 2016;454:28–32. doi: 10.1016/j.cca.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 183.Lin, Y. et al. Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin. Cancer Res.25, 3035–3045 (2019). [DOI] [PubMed]
- 184.Stokman MF, et al. Changes in the urinary extracellular vesicle proteome are associated with nephronophthisis-related ciliopathies. J. Proteom. 2019;192:27–36. doi: 10.1016/j.jprot.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 185.Hisey CL, Dorayappan KDP, Cohn DE, Selvendiran K, Hansford DJ. Microfluidic affinity separation chip for selective capture and release of label-free ovarian cancer exosomes. Lab Chip. 2018;18:3144–3153. doi: 10.1039/c8lc00834e. [DOI] [PubMed] [Google Scholar]