Abstract
Long non-coding RNA (lncRNA) MEG3 is a key biomarker and therapeutic target in lung cancer; however, its underlying molecular mechanism in lung cancer progression remains unclear. The present study demonstrated a novel regulatory axis in lung cancer, lncRNA MEG3/dyskeratosis congenita 1 (DKC1), and further investigated the effects and molecular mechanism of lncRNA MEG3/DKC1 in lung cancer. RT-qPCR and western blot analysis were performed to determine gene and protein expression levels. The RNA immunoprecipitation assay was performed to verify binding between lncRNA MEG3 and DKC1. Flow cytometry analysis was performed to assess cell apoptosis, while the Cell Counting Kit-8 assay was performed to determine cell viability. Transwell and wound healing assays were performed to assess cell invasion and migration, respectively. Telomerase activity was measured using the quantitative TeloTAGGG Telomerase PCR-ELISA kit. The results demonstrated that lncRNA MEG3 was downregulated, while its binding protein, DKC1, was upregulated in lung cancer cells. Furthermore, lncRNA MEG3 inhibited cell proliferation, migration, invasion and telomerase activity in A549 cells by downregulating DKC1. lncRNA MEG3 inhibited non-small cell lung cancer progression by inhibiting telomere function, cell proliferation, telomerase activity, cell migration and invasion via regulation of the DKC1 protein expression. LncRNA MEG3/DKC1 was identified as a novel dual-directional regulatory axis in the present study, acting as a promising target for the treatment of lung cancer.
Keywords: lncRNA MEG3, DKC1, lung cancer, telomerase activity, cell apoptosis, migration, invasion
Introduction
Lung cancer is considered the most common type of tumor and is a predominant cause of cancer-associated mortality in China (1–3). Currently, more than 1.5 million cases of mortality are reported from lung carcinomas annually, and the 5-year survival rate is only 17% (1,4). Early stage symptoms of lung cancer are atypical and the majority of the different types of lung cancer are not detected until advanced stage. The survival rate for lung cancer at early stage is approximately 50% (5). Timely screening and effective treatment are fundamental for improving the clinical outcomes of lung cancer (6,7). Biopsy is used as the gold standard for the diagnosis of lung cancer due to its minimal invasive risks; however, this technique poses difficulties regarding accessibility to lung lesions, thus resulting in potential misdiagnosis and subsequent complications (8,9). Therefore, it is critical to identify and develop novel therapeutic approaches for the diagnosis of lung cancer, with minimal invasiveness. Furthermore, the current gold standard treatment for lung cancer exhibits poor prognostic values (10). In summary, the development of novel therapeutic approaches for diagnosis, and screening novel therapy targets are essential for improving clinical outcomes in lung cancer.
Non-small cell lung cancer (NSCLC) accounts for 85% of all lung cancer cases, and is considered the predominant subtype of lung cancer (11). It is beneficial to understand the pathogenesis of NSCLC, in order to develop novel therapy targets and biomarkers to improve treatment and diagnosis of lung cancer, respectively. Long non-coding RNAs (lncRNAs) are RNA molecules with lengths exceeding 200 nucleotides, which are transcribed from 90% of the genome and are involved in a number of cellular processes (12). LncRNAs are considered to act as pivotal players in the progression of several types of disease, and have been reported to be beneficial in the diagnosis and treatment of lung cancer. Thus, aberrant expression of lncRNAs may be used as indicators for the diagnosis of lung cancer, whereby the degree of lncRNA aberration is considered more significant compared with miRNA expression (13). Furthermore, understanding the regulatory mechanisms of lncRNAs in lung cancer is beneficial to the identification of underlying pathogenic molecular mechanisms, and thus development of effective therapeutic strategies.
Regarded as a notable regulator, lncRNA MEG3 possesses inhibitory effects on several types of cancer (14–17). Furthermore, lncRNA MEG3 has been reported to act as a critical regulatory player in lung cancer. Previous studies have demonstrated that lncRNA MEG3 expression significantly decreases in NSCLC, and low lncRNA MEG3 expression levels have been associated with a poor prognosis in patients with lung cancer (18,19). LncRNA MEG3 is also considered a key epigenetic regulator of epithelial-to-mesenchymal transition (20). Given that lncRNA MEG3 is a vital regulator in lung cancer, clarifying its underlying molecular mechanism in lung cancer progression is beneficial in understanding the pathological process, and thus identifying effective therapeutic targets in lung cancer. A previous study indicated an association between lncRNAs and proteins, which plays a vital role in several types of physiological processes (21). Thus, the present study investigated whether particular proteins may be involved in lung cancer progression via interaction with lncRNA MEG3.
Dyskeratosis congenita 1 (DKC1) protein is a key component of the telomerase complex and an essential structural subunit of the telomerase ribonucleoprotein (RNP). DKC1 has the ability to activate telomerase RNP activity, resulting in telomere shortening, as well as promoting effects of lung cancer (22). DKC1 protein, which is reported to play key roles in several types of cancer, is also aberrantly expressed in lung cancer, whereby high DKC1 expression is considered an unfavorable indicator of prognosis (23). Furthermore, DKC1 is speculated to be the binding protein of lncRNA MEG3, via StarBase analysis. Thus, the present study aimed to investigate the associations between lncRNA MEG3 and DKC1 protein in lung cancer, and determine the potential molecular mechanism involved in this interaction.
Materials and methods
Cell culture and transfection
HBE, A549, NCI-H1975 and SK-MES-1 cell lines were purchased from the American Type Culture Collection, while the PLA-801D cell line was purchased from Mingzhou Biotechnology Co., Ltd. All the cells were cultured in DMEM medium (Gibco) supplemented with 10% FBS at 37°C in 5% CO2. Overexpression-MEG3, overexpression-negative control (NC), short hairpin (sh)RNA-DKC1 and shRNA-NC were purchased from Shanghai GenePharma Co., Ltd. Transfection was performed using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After transfection for 48 h, the subsequent experimentations were performed.
Western blotting
Total protein (5 mg/ml) was extracted from A549 cells using RIPA lysis buffer and the lysates were centrifuged at 15,000 × g for 20 min at 4°C. Total protein was quantified using the bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.) and the samples (30 µg) were separated via SDS-PAGE on a 15% gel. The separated proteins were subsequently transferred onto polyvinylidene difluoride membranes and blocked with 5% skim milk overnight at 4°C. The membranes were incubated with primary antibodies against DKC1 (cat. no. ab156877; Abcam; 1/1,000), Bax (cat. no. ab32503; Abcam; 1: 150) and Bcl-2 (cat. no. ab185002; Abcam; 1:500) overnight at 4°C. Following the primary incubation, the membranes were incubated with a IgG-HRP secondary antibody (cat. no. ab ab7090, Abcam, 1:3,000) for 1 h at room temperature. GAPDH (cat. no. ab181602) served as the internal control. Protein bands were visualized using an enhanced chemiluminescence detection system (Thermo Fisher Scientific, Inc.) and quantified using ImageQuant LAS500 (GE Healthcare).
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from cells using TRIzol® reagent and reverse transcribed into cDNA using the QuantiNova Reverse Transcription kit (Qiagen GmbH). qPCR was subsequently performed using a CFX96 Real-Time PCR Detection system (Bio-Rad Laboratories, Inc.). The following thermocycling conditions were used for RT-qPCR: Denaturation at 95°C for 5 sec; 45 cycles of 95°C for 5 sec, 60°C for 1 min, 95°C for 5 sec, 60°C for 1 min. Relative mRNA levels were quantified using the 2−ΔΔCq method (24). The primer sequences used for qPCR were as follows: DKC1 forward, 5′-CTCGGAAGTGGGGTTTAGGT-3′ and reverse, 5′-GAGGTGGTTGCTGAAGTGGT-3′; lncRNA MEG3 forward, 5′-GCTATGCTCATACTTTGACTC-3′ and reverse, 5′-CATCATAAGGGTGATGACAG-3′; and β-actin forward, 5′-CATGTACGTTGCTATC-CAGGC-3′ and reverse, 5′-CCGTGCTGGGATTGGTCATA-3′. β-actin was used as the internal control.
RNA immunoprecipitation (RIP) assay
The binding between lncRNA MEG3 and DKC1 was predicted using StarBase database (http://starbase.sysu.edu.cn/index.php). The RIP assay was performed to verify binding between lncRNA MEG3 and DKC1, using the EZ-Magna RIP kit (EMD Millipore), according to the manufacturer's protocol. Briefly, the lysates of A549 cells were incubated with the DKC1 antibody (cat. no. ab93777, Abcam) or IgG-treated magnetic beads, following cell lysis in RIPA lysis buffer overnight at 4°C. Following RNA purification, the levels of lncRNA MEG3 in the magnetic beads treated with DKC1 antibody or IgG were determined via RT-qPCR analysis.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to assess cell viability within different groups. A549 cells were seeded into 96-well plates at a density of 1×105. Following treatment for 24, 48 and 72 h, 10 µl CCK-8 reagent was added to each well and incubated in the dark for 2 h, according to the manufacturer's protocol. Cell viability was subsequently analyzed at a wavelength of 450 nm, using a microplate reader.
Migration assay
A total of 1×105 A549 cells were plated in the upper chambers of Transwell plates in serum-free medium for 24 h. Transwell membranes were precoated with Matrigel (Becton) at 37°C for 2 h. Medium supplemented with 20% FBS was plated in the lower chambers. Cells in the upper chambers were discarded using cotton swabs, while the migratory cells in the lower chambers were fixed with 4% methanol, prior to staining with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA). Stained cells were counted in randomly selected fields using a fluorescent microscope (Thermo Fisher Scientific, Inc.; magnification, ×100).
Wound healing assay
Cells were seeded in 6-well plates until they reached 90% confluency, and a pipette tip was used to scratch a line across the cell monolayers. The cells were subsequently washed with PBS. Following transfection, the cells were observed under a microscope and the wound healing images were captured. The cells were serum-starved during the wound healing assay. After incubation for 24 h, cell migration was observed under a fluorescence microscope (Thermo Fisher Scientific, Inc.; magnification, ×100).
Flow cytometric analysis of apoptosis
Following transfection, the cells were centrifuged at 200 × g for 5 min at room temperature and subsequently resuspended in binding buffer (eBioscience; Thermo Fisher Scientific, Inc.), and subsequently stained with annexin V-fluorescein isothiocyanate (FITC) for 5 min at room temperature and propidium iodide for 15 min at room temperature in the dark, using the FITC Annexin V/PI Apoptosis Detection kit I (Guangzhou RiboBio Co., Ltd.). Apoptotic cells were subsequently analyzed using the FACScan flow cytometer.
Measurement of telomerase activity
Telomerase activity was assessed using the Quantitative TeloTAGGG Telomerase PCR-ELISA kit (Roche Diagnostics GmbH), according to the manufacturer's protocol. Following transfection, the cells were lysed and total protein was collected. Subsequently, biotin-labeled primers were incubated with the cell extracts. The telomere repeat sequences were loaded onto the primer ends and submitted to PCR amplification. Following specific probes detection, the PCR products were plated onto 96-wells plates coated with streptavidin (5 pmol biotin/well; Thermo Fisher Scientific, Inc.). Telomerase activity was subsequently analyzed at a wavelength of 450 nm, using the STAT-FAX 3200 ELISA reader (Awareness Technology, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS software (version 17.0) and GraphPad Prism software (version 5.0; GraphPad Software, Inc.). Data are presented as the mean ± standard deviation and experiments were performed in triplicate. One-way analysis of variance followed by Tukey's post-hoc test was used to compare differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA MEG3 is downregulated and DKC1 is upregulated in lung cancer cells
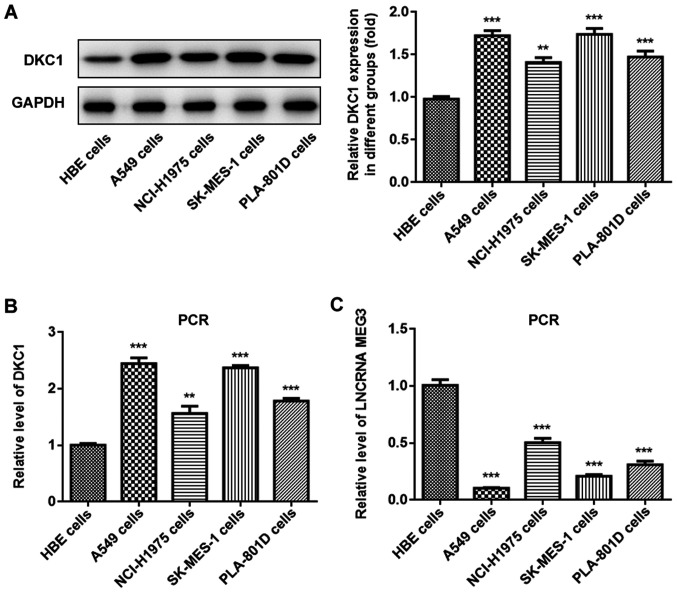
The expression levels of lncRNA MEG3 and DKC1 were assessed in different lung cancer cell lines and normal lung cell lines, in order to determine their role in lung cancer progression. Western blot and RT-qPCR analyses demonstrated that DKC1 expression significantly increased in A549 cells compared with human bronchial epithelial cells (Fig. 1A and B). Furthermore, lncRNA MEG3 expression decreased in all the lung cell lines, particularly in A549 cells (Fig. 1C); thus this cell line was selected for further experimentation.
Figure 1.
LncRNA MEG3 is downregulated and DKC1 is upregulated in lung cancer cells. The level of DKC1 was detected in the study groups via (A) western blot and (B) PCR analyses. The lncRNA MEG3 level was detected via (C) PCR assay in the study groups. ***P<0.001 and **P<0.01 vs. HBE group. LncRNA, long non-coding RNA; DKC1, dyskeratosis congenita 1; HBE, human bronchial epithelioid cells.
Verification of the binding between lncRNA MEG3 and DKC1 protein
As predicted by the StarBase database, the results of the RIP assay verified that DKC1 is the binding protein of lncRNA MEG3. The results of the present study demonstrated that lncRNA MEG3 expression increased in the DCK1 group compared with the IgG group, confirming that lncRNA MEG3 binds to DK1 protein in lung cancer cells (Fig. 2).
Figure 2.
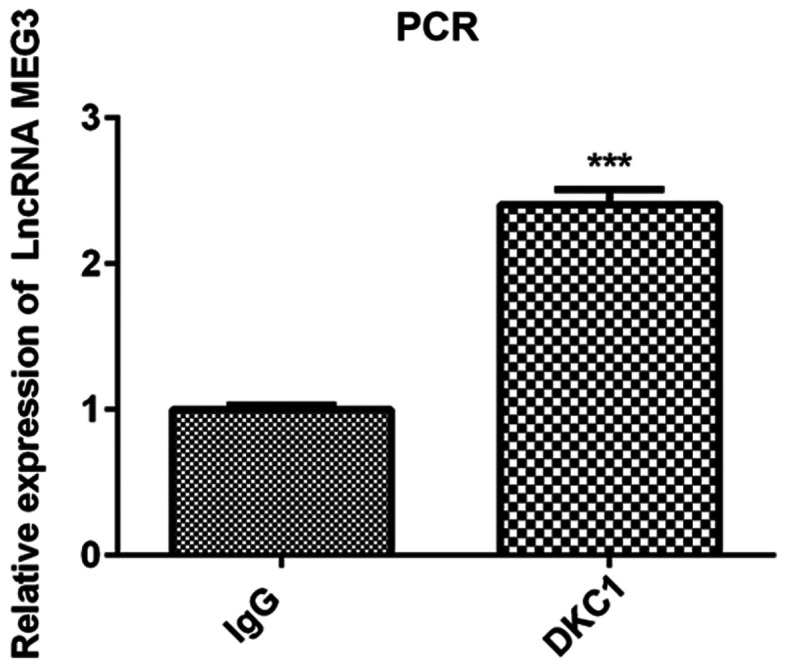
Verification of the binding between lncRNA MEG3 and DKC1. The expression of MEG3, which was detected by PCR, increased in the DKC1 vs. IgG group. ***P<0.001 vs. IgG group. LncRNA, long non-coding RNA; DKC1, dyskeratosis congenita 1.
LncRNA MEG3 inhibits cell proliferation by downregulating DKC1 in A549 cells
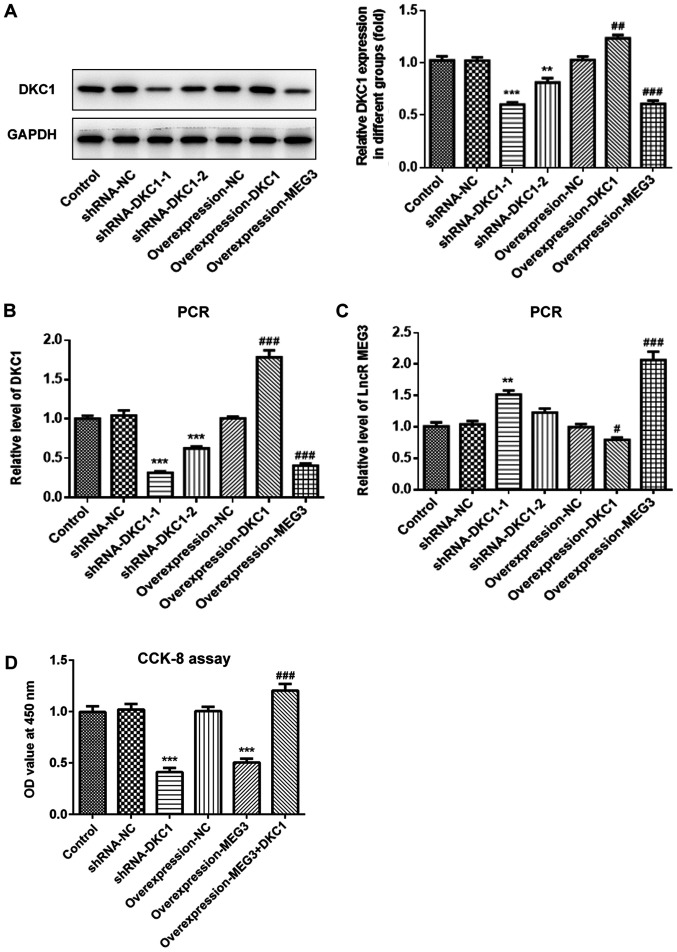
Due to the low expression of lncRNA MEG3 in A549 cells, the present study subsequently analyzed the effects of overexpressing lncRNA MEG3, as well as DKC1 knockdown or overexpression in lung cancer cells, in order to investigate the effects of lncRNA MEG3/DKC1 in lung cancer progression. Both RT-qPCR and western blot analyses demonstrated successful overexpression of lncRNA MEG3, DKC1 knockdown and overexpression of DKC1 (Fig. 3A-C). The shRNA-DKC1-1, which demonstrated higher knockdown efficiency, was selected for silencing of DKC1. The results demonstrated that overexpression of lncRNA MEG3 or DKC1 knockdown significantly decreased cell proliferation. Furthermore, overexpression of DKC1 reversed the antiproliferative effects exhibited by overexpressing lncRNA MEG3 (Fig. 3D).
Figure 3.
Effects of lncRNA MEG3 on cell proliferation in the lung cells via downregulation of DKC1. Level of DKC1 was assessed in the study groups via (A) western blotting and (B) PCR. (C) lncRNA MEG3 was detected in the study groups via PCR. (D) Cell survival rate in the different groups was evaluated via CCK-8 assay. (A-C) ***P<0.001, **P<0.01 vs. shRNA-NC and ###P<0.001, ##P<0.01, #P<0.05 vs. OverExp-NC group. (D) ***P<0.001 vs. control and ###P<0.001 vs. OverExp-MEG3 group. LncRNA, long non-coding RNA; DKC1, dyskeratosis congenita 1.
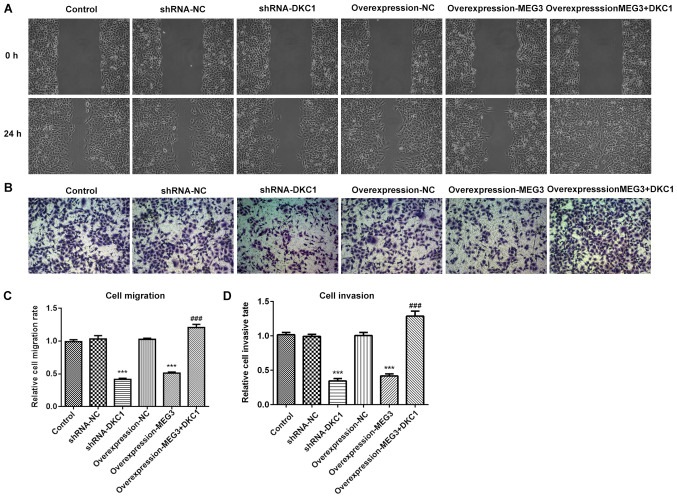
LncRNA MEG3 inhibits cell migration and invasion by downregulating DKC1 in A549 cells
Both cell migration and invasion decreased following overexpression of lncRNA MEG3 or DKC1 knockdown, compared with the control group (Fig. 4A-D). Notably, overexpression of DKC1 reversed the inhibitory effects exhibited by overexpressing lncRNA MEG3. Taken together, these results suggest that A549 cell migration and invasion may be inhibited by lncRNA MEG3 via the downregulation of DKC1.
Figure 4.
Effects of lncRNA MEG3 on cell migration as well as invasion in the lung cells via downregulation of DKC1. Images for (A) cell migration evaluated by wound healing assay and (B) cell invasion evaluated by Transwell assay in the study groups (magnification, ×100). Quantitative histogram for (C) cell migration and (D) invasion. ***P<0.001 vs. control and ###P<0.01 vs. OverExp-MEG3 group. DKC1, dyskeratosis congenita 1.
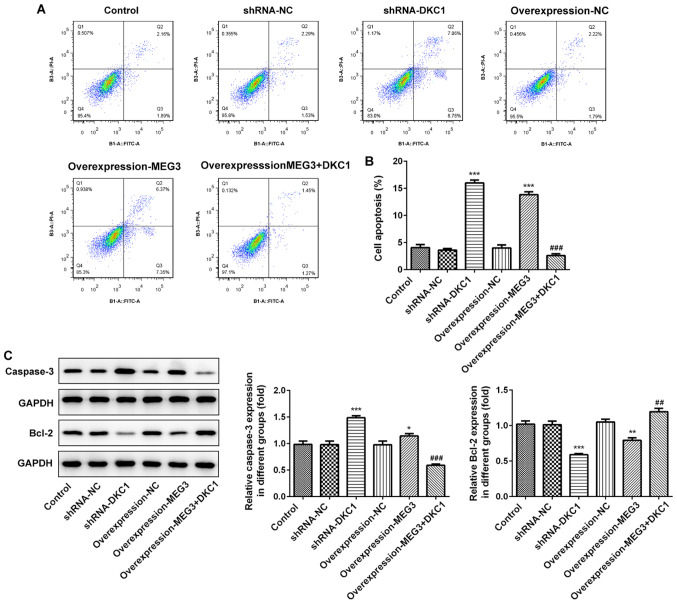
LncRNA MEG3 promotes cell apoptosis by downregulating DKC1 in A549 cells
Overexpression of lncRNA MEG3 or DKC1 knockdown increased cell apoptosis (Fig. 5A and B). Assessment of the apoptosis-associated proteins demonstrated that the expression level of pro-apoptosis protein-caspase protein increased, while the expression level of anti-apoptosis protein-bcl-2 decreased, following overexpression of lncRNA MEG3 or DKC1 knockdown (Fig. 5C). Notably, overexpression of DKC1 reversed the proapoptotic effects exhibited by overexpressing lncRNA MEG3, suggesting that lncRNA MEG3 promotes cell apoptosis by downregulating DKC1.
Figure 5.
Effects of lncRNA MEG3 on cell apoptosis and apoptosis-related proteins in the lung cells via downregulating DKC1. (A) Images for cell apoptosis and (B) quantitative histogram for cell apoptosis percent in the study groups. (C) The levels of apoptosis-related proteins in the study groups. ***P<0.001, **P<0.01, *P<0.05 vs. control and ###P<0.001, ##P<0.01 vs. OverExp-MEG3 group. DKC1, dyskeratosis congenita 1.
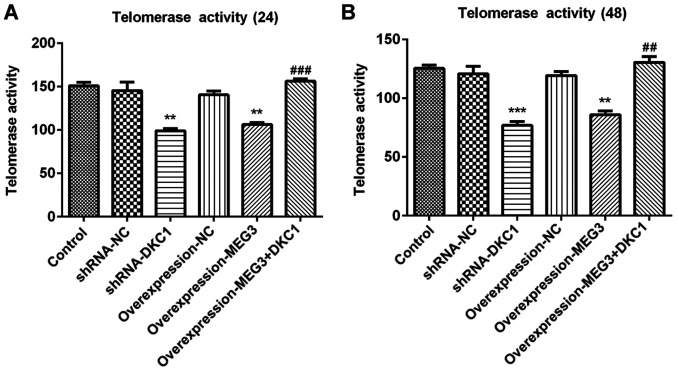
lncRNA MEG3 inhibits telomerase activity by downregulating DKC1 in A549 cells
Regarded as a well-known tumor biomarker (25), telomerase activity was assessed in several types of cancer cells. The results of the present study demonstrated that overexpression of lncRNA MEG3 or DKC1 knockdown remarkably decreased telomerase activity at 24 and 48 h, compared with the control group (Fig. 6A and B). Furthermore, the inhibitory effects of overexpressing lncRNA MEG3 on cell apoptosis were abolished following overexpression of DKC1, suggesting that the inhibitory effects of lncRNA MEG3 on telomerase activity were achieved by downregulating DKC1.
Figure 6.
Effects of lncRNA MEG3 on telomerase activity in the lung cells via down-regulating of DKC1. Telomerase activity at (A) 24 and (B) 48 h in the study groups. ***P<0.001, **P<0.01 vs. control and ###P<0.001, ##P<0.01 vs. OverExp-MEG3 group. DKC1, dyskeratosis congenita 1; sh, short hairpin; NC, negative control.
Discussion
Lung cancer is considered the leading cause of mortality, thus critical measures are required for its quick and efficient treatment. Dysregulation of lncRNAs have been confirmed to play a key role in the progression of lung cancer. Several types of lncRNAs are aberrantly expressed in lung cancer, including lncRNA MEG3, which is considered a key RNA in lung cancer (13,26–28). LncRNA MEG3 expression is significantly decreased in lung cancer, which is associated with a poor prognosis for patients with lung cancer (19). lncRNA MEG3 has been reported to enhance the anticancer effects of vincristine and cisplatin (19,29). Furthermore, lncRNA MEG3 has been demonstrated to promote cell apoptosis, inhibit cell migration and invasion, and induce cell apoptosis in lung cancer (30). However, the underlying molecular mechanisms of lncRNA MEG3 in the progression of lung cancer remain unclear.
Increasing evidence suggests that lncRNAs play a regulatory role by binding to certain types of protein (21). The results of the present study demonstrated that lncRNA MEG3 inhibited progression of NSCLC by suppressing telomere function, by targeting DKC1 protein. Furthermore, DKC1 was verified as a novel binding protein of lncRNA MEG3 in the present study.
DKC1 has been reported to be upregulated in renal cell carcinoma, resulting in enhanced cell proliferation, migration and invasion (31). Furthermore, overexpression of DKC1 is a prognostic indicator in liver cancer (32). However, the effects of DKC1 expression in lung cancer remain to be determined. The results of the present study indicated that DKC1 was upregulated in lung cancer, suggesting its potential role in the progression of lung cancer.
The present study revealed that DKC1 is the binding protein of lncRNA MEG3. The role of lncRNA MEG3 binding to DKC1 in lung cancer was further investigated. Vital cell processes, including cell proliferation, apoptosis, invasion and migration were investigated. The results demonstrated that cell proliferation, invasion and migration were all inhibited, while cell apoptosis increased following DKC1 knockdown. The effects of DKC1 knockdown were in accordance with the effects of overexpressing lncRNA MEG3 in lung cancer. Overexpression of DKC1 reversed the effects of overexpressing lncRNA MEG3 on cell apoptosis, proliferation, invasion and migration, suggesting that the effects of lncRNA MEG3 were achieved by downregulating DKC1.
DKC1 serves as the encoding gene of the telomerase subunit, dyskerin, and has been reported to be elevated in several types of cancer cells (33). Furthermore, DKC1 has the ability to induce telomerase activity, resulting in an increase in length of the telomere (34). Its ability to enhance cell proliferation in cancer has made telomerase the focus of research. Maintaining the molecular mechanisms of telomere DNA is vital for cell proliferation, without limitation in cancer. Targeting telomerase activity is considered to be key in easing tumor progression (35–37). Currently, DKC1 is highly expressed in different types of cancer, including glioma and renal cell carcinoma (31,38,39). In the present study, telomerase activity was significantly inhibited following DKC1 knockdown and overexpression of lncRNA MEG3, while the effects of overexpressing lncRNA MEG3 on telomerase activity were abolished by overexpressing DKC1, suggesting that the effects of lncRNA MEG3 on telomerase activity was achieved by targeting and silencing DKC1. Taken together, the results of the present study support the hypothesis that the lncRNA MEG3/DKC1 regulatory axis is a promising therapeutic target for lung cancer.
The present study is not without limitations. First, in vitro analysis was performed only in A549 cells; thus, prospective studies should focus on investigating the effects of the lncRNA MEG3/DKC1 dual regulatory axis, in other lung cancer cells or in vivo. DKC1 and lncRNA MEG3 are well-established prognostic indicators in lung cancer, and the results of the present study provided vital evidence for further study on DKC1, as well as lncRNA MEG3 in the progression of lung cancer.
Overall, the present study identified a novel dual- directional regulatory axis, the lncRNA MEG3/DKC1 axis. Overexpression of lncRNA MEG3 inhibited cell proliferation, migration, invasion and telomerase activity by sponging DKC1, thus the lncRNA MEG3/DKC1 axis is a promising therapeutic target for patients with lung cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the study are available from the corresponding author upon reasonable request.
Authors' contributions
ZY designed the present study and drafted the initial manuscript. ZY and ZW performed the experiments. YD analyzed the experimental data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Shen M, Xu Z, Jiang K, Xu W, Chen Y, Xu Z. Long noncoding nature brain-derived neurotrophic factor antisense is associated with poor prognosis and functional regulation in non-small cell lung caner. Tumor Biol. 2017;39:1010428317695948. doi: 10.1177/1010428317695948. [DOI] [PubMed] [Google Scholar]
- 3.Hong QY, Wu GM, Qian GS, Hu CP, Zhou JY, Chen LA, Li WM, Li SY, Wang K, Wang Q, et al. Prevention and management of lung cancer in China. Cancer. 2015;121(Suppl 17):S3080–S3088. doi: 10.1002/cncr.29584. [DOI] [PubMed] [Google Scholar]
- 4.Shu J, Wang L, Han F, Chen Y, Wang S, Luo F. BTBD7 Downregulates E-cadherin and promotes epithelial-mesenchymal transition in lung cancer. Biomed Res Int. 2019;2019:5937635. doi: 10.1155/2019/5937635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman RM, Sanchez R. Lung cancer screening. Med Clin North Am. 2017;101:769–785. doi: 10.1016/j.mcna.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Dowd EL, Baldwin DR. Early diagnosis pivotal to survival in lung cancer. Practitioner. 2014;258:21–24, 2-3. [PubMed] [Google Scholar]
- 7.Jacobsen MM, Silverstein SC, Quinn M, Waterston LB, Thomas CA, Benneyan JC, Han PKJ. Timeliness of access to lung cancer diagnosis and treatment: A scoping literature review. Lung Cancer. 2017;112:156–164. doi: 10.1016/j.lungcan.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Guibert N, Tsukada H, Hwang DH, Chambers E, Cibas ES, Bale T, Supplee J, Ulrich B, Sholl LM, Paweletz CP, Oxnard GR. Liquid biopsy of fine-needle aspiration supernatant for lung cancer genotyping. Lung Cancer. 2018;122:72–75. doi: 10.1016/j.lungcan.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Heerink WJ, de Bock GH, de Jonge GJ, Groen HJ, Vliegenthart R, Oudkerk M. Complication rates of CT-guided transthoracic lung biopsy: Meta-analysis. Eur Radiol. 2017;27:138–148. doi: 10.1007/s00330-016-4357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodard GA, Jones KD, Jablons DM. Lung cancer staging and prognosis. Cancer Treat Res. 2016;170:47–75. doi: 10.1007/978-3-319-40389-2_3. [DOI] [PubMed] [Google Scholar]
- 11.Shi YX, Sheng DQ, Cheng L, Song XY. Current landscape of epigenetics in lung cancer: Focus on the mechanism and application. J Oncol. 2019;2019:8107318. doi: 10.1155/2019/8107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323. doi: 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang AX, Hu JL, Wang YM, Gao Y. Comprehensive analysis of lncRNA and mRNA expression profiles in lung cancer. Clin Lab. 2017;63:313–320. doi: 10.7754/Clin.Lab.2016.160812. [DOI] [PubMed] [Google Scholar]
- 14.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 15.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: A tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Yang H, Xia W, Cui L, Xu R, Lu H, Xue D, Tian Z, Ding T, Cao Y, et al. LncRNA MEG3 inhibits the progression of prostate cancer by facilitating H3K27 trimethylation of EN2 through binding to EZH2. J Biochem. 2019;167:295–301. doi: 10.1093/jb/mvz097. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Zhang HW, Chen HN, Deng XJ, Tu YX, Jackson AO, Qing JN, Wang AP, Patel V, Yin K. Perivascular adipose tissue dysfunction aggravates adventitial remodeling in obese mini pigs via NLRP3 inflammasome/IL-1 signaling pathway. Acta Pharmacol Sin. 2019;40:46–54. doi: 10.1038/s41401-018-0068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu JL, Meng FM, Li HJ. High expression of lncRNA MEG3 participates in non-small cell lung cancer by regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci. 2018;22:5938–5945. doi: 10.26355/eurrev_201809_15923. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Liu T, Wang K, Qu X, Pang Z, Liu S, Liu Q, Du J. Down-regulation of long non-coding RNA MEG3 indicates an unfavorable prognosis in non-small cell lung cancer: Evidence from the GEO database. Gene. 2017;630:49–58. doi: 10.1016/j.gene.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292:82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Garcia I, Marcos T, Muñoz-Barrutia A, Serrano D, Pio R, Montuenga LM, Ortiz-de-Solorzano C. Multiscale in situ analysis of the role of dyskerin in lung cancer cells. Integr Biol (Camb) 2013;5:402–413. doi: 10.1039/c2ib20219k. [DOI] [PubMed] [Google Scholar]
- 23.Penzo M, Ludovini V, Treré D, Siggillino A, Vannucci J, Bellezza G, Crinò L, Montanaro L. Dyskerin and TERC expression may condition survival in lung cancer patients. Oncotarget. 2015;6:21755–21760. doi: 10.18632/oncotarget.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Kulić A, Plavetić ND, Gamulin S, Jakić-Razumović J, Vrbanec D, Sirotković-Skerlev M. Telomerase activity in breast cancer patients: Association with poor prognosis and more aggressive phenotype. Med Oncol. 2016;33:23. doi: 10.1007/s12032-016-0736-x. [DOI] [PubMed] [Google Scholar]
- 26.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan BS, Yang MC, Singh S, Chou YC, Chen HY, Wang MY, Wang YC, Chen RH. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene. 2019;38:5612–5626. doi: 10.1038/s41388-019-0812-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Feng C, Li Y, Ma Y, Cai R. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460:1–8. doi: 10.1007/s11010-019-03564-1. [DOI] [PubMed] [Google Scholar]
- 29.Xia H, Qu XL, Liu LY, Qian DH, Jing HY. LncRNA MEG3 promotes the sensitivity of vincristine by inhibiting autophagy in lung cancer chemotherapy. Eur Rev Med Pharmacol Sci. 2018;22:1020–1027. doi: 10.26355/eurrev_201802_14384. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Zhu Z, Shi S, Wang J, Li N. Long non-coding RNA MEG3 regulates migration and invasion of lung cancer stem cells via miR-650/SLC34A2 axis. Biomed Pharmacother. 2019;120:109457. doi: 10.1016/j.biopha.2019.109457. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Pan Y, Jiang R, Hou P, Shan H, Chen F, Jiang T, Bai J, Zheng J. DKC1 serves as a potential prognostic biomarker for human clear cell renal cell carcinoma and promotes its proliferation, migration and invasion via the NF-κB pathway. Oncol Rep. 2018;40:968–978. doi: 10.3892/or.2018.6484. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Zhang J, Huang C, Liu H. Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS One. 2012;7:e43147. doi: 10.1371/journal.pone.0052675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nersisyan L, Hopp L, Loeffler-Wirth H, Galle J, Loeffler M, Arakelyan A, Binder H. Telomere length maintenance and its transcriptional regulation in lynch syndrome and sporadic colorectal carcinoma. Front Oncol. 2019;9:1172. doi: 10.3389/fonc.2019.01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards LA, Kumari A, Knezevic K, Thoms JA, von Jonquieres G, Napier CE, Ali Z, O'Brien R, Marks-Bluth J, Maritz MF, et al. DKC1 is a transcriptional target of GATA1 and drives upregulation of telomerase activity in normal human erythroblasts. Haematologica. 2020;105:1517–1526. doi: 10.3324/haematol.2018.215699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardes de Jesus B, Blasco MA. Telomerase at the intersection of cancer and aging. Trends Gene. 2013;29:513–520. doi: 10.1016/j.tig.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizukoshi E, Kaneko S. Telomerase-targeted cancer immunotherapy. Int J Mol Sci. 2019;20:1823. doi: 10.3390/ijms20081823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene. 2019;38:6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao FA, Chu K, Chen HR, Zhang M, Shi PC, Bai J, You YP. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Invest New Drugs. 2019;37:1177–1186. doi: 10.1007/s10637-019-00748-w. [DOI] [PubMed] [Google Scholar]
- 39.Penzo M, Casoli L, Ceccarelli C, Treré D, Ludovini V, Crinò L, Montanaro L. DKC1 gene mutations in human sporadic cancer. Histol Histopathol. 2013;28:365–372. doi: 10.14670/HH-28.365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the study are available from the corresponding author upon reasonable request.