Abstract
Shank-associated RH domain interactor (SHARPIN) is a component of the linear ubiquitin chain activation complex, which is essential for p53 signaling and inflammation. Previous studies have demonstrated that SHARPIN functions in tumor cell survival, growth, invasion and tumorigenesis. These functions include the regulation of p53 proteins via poly-ubiquitination, interaction with a type II protein arginine methyltransferase 5 in melanoma cells, modulating ras-associated protein-1 through p38 and c-Jun N-terminal kinases/c-Jun signaling, and mediating phosphoinositide 3-kinase/AKT signaling via phosphatase and tensin homologue deleted on chromosome 10. Hence, SHARPIN not only participates in the inflammatory response but also serves a critical role in tumor cells. The present review summarizes the biological functions of the absence or presence of SHARPIN with regard to activating the canonical NF-κB signaling pathway and the effects on p53 and other signaling pathways for the modulation of tumorigenesis. Therefore, this review provides insight into the underlying role and mechanisms of SHARPIN in tumorigenesis, as well as its potential application in cancer therapy.
Keywords: SHARPIN, tumorigenesis, LUBAC, p53, PRMT5, PTEN
1. Introduction
Cancer is a major global public health burden, with ~21.6 million new cancer cases predicted for 2030 (1). In general, tumorigenesis is the process that promotes the transformation of normal cells into invasive cells, overcoming the constraints that usually limit proliferation and survival. These alterations can give rise to a number of potentially deleterious circumstances or vulnerabilities that can be lethal to patients if left unchecked (2). Despite extensive research investigating tumorigenesis, the precise underlying molecular mechanisms remain unclear.
Shank-associated RH domain interactor (SHARPIN) is an ~40-kDa multifunctional adaptor protein that is amplified and overexpressed in a number of human cancer types. Studies have shown that the SHARPIN promotes cancer cell proliferation, tumor formation and metastasis (3–6). SHARPIN was first identified in C57BL/KaLawRij mice, functioning as a shank binding protein at the postsynaptic density of excitatory synapses in the central nervous system (7). In addition, SHARPIN has important physiological functions in several organisms (7) and is ubiquitously expressed in various types of cells and tissues (8). SHARPIN is an autosomal gene that is conserved among a number of mammalian species, including humans, chimpanzees, dogs, rats and mice (8). SHARPIN is located on cell membranes and in the nuclei, and primarily functions in immune and inflammatory responses (8). A previous study has reported that mutations in this gene contribute to chronic proliferative dermatitis, which is accompanied by immune system malfunction and multi-organ inflammation (7). Moreover, SHARPIN deficiency results in an autoinflammatory phenotype in an inflammatory mouse model (7). There is evidence demonstrating that SHARPIN is a component of the linear ubiquitin chain assembly complex (LUBAC), which participants in a range of complex biological functions (9,10) (Fig. 1). Moreover, the LUBAC is also involved in various molecular and cellular processes, such as embryogenesis (11) and apoptosis (12). SHARPIN has been well characterized as a crucial regulator of canonical NF-κB signaling during the inflammatory response (10,13) and T-cell differentiation (14,15). A recent study reported that SHARPIN serves an important role in promoting breast cancer progression (16). A previous study also showed that SHARPIN regulates p53 protein levels in tumor cell lines through the mouse double minute 2 homolog (MDM2)-dependent pathway (17). Additionally, it has been reported that SHARPIN interaction with protein arginine methyltransferase 5 (PRMT5) mediates tumor cell growth (3). Notably, Zhou et al (18) demonstrated that SHARPIN upregulates ras-associated protein-1 (Rap1), which promotes melanoma development through p38 and c-Jun N-terminal kinases (JNK)/c-Jun signaling pathways. A previous study also revealed that SHARPIN mediates the phosphatase and tensin homologue deleted on chromosome 10 (PTEN) signaling pathway, which promotes tumorigenesis by phosphoinositide 3-kinase (PI3K)/AKT signaling (19). Therefore, research investigating SHARPIN has improved our understanding of its functions in humans; however, a comprehensive summary of mechanisms by which SHARPIN regulates tumorigenesis has not been available until now. Hence, the following sections summarize the current data on the possible functions of the aforementioned proteins and SHARPIN in tumorigenesis.
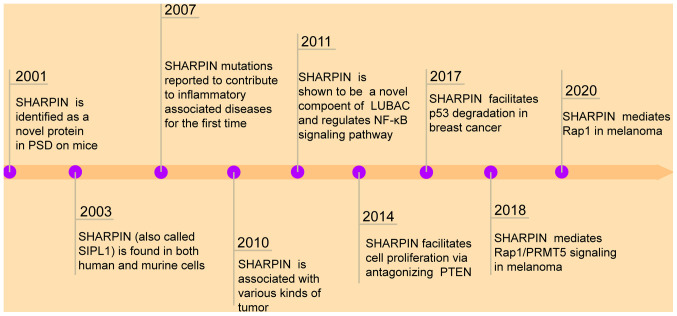
Figure 1.
Historical process of resolving SHARPIN function. SHARPIN was originally identified in the PSD of mice, functioning in the nervous system. SHARPIN is reported to participate in inflammatory-associated diseases and tumors. Researchers gradually realized that SHARPIN is a novel component of the LUBAC and mediates the NF-κB signaling pathway. Subsequently, SHARPIN-mediated regulation of p53, PRMT5, Rap1 and PTEN signaling in tumors has been reported. SHARPIN, shank-associated RH domain interactor; PSD, postsynaptic density; LUBAC, linear ubiquitin chain assembly complex; PRMT5, protein arginine methyltransferase 5; PTEN, phosphatase and tensin homolog, Rap1, ras-associated protein-1.
2. SHARPIN is a key member of the LUBAC family of proteins
Numerous studies have demonstrated that LUBAC consists of three structurally related proteins: Heme-oxidized IRP2 ligase 1L (HOIL-1L), HOIL-1 interacting protein (HOIP) and SHARPIN, with molecular weights of 120, 58 and 40 kD, respectively (20). LUBAC is involved in post-translational modifications that regulate a multitude of cellular processes, including cell death, development, carcinogenesis and autoimmune diseases (21,22). The HOIL-1L subunit is an accessory molecule that is involved in the stabilization of LUBAC. For example, cells lacking HOIL-1L have significantly decreased linear ubiquitylation (23), and the primary function of SHARPIN is to maintain the linear ubiquitylation activity in LUBAC. HOIP is a catalytic subunit that is associated with regulatory proteins, such as HOIL-1L and SHARPIN (24). A recent study demonstrated that the aforementioned subunits interact with each other in the trimeric core of LUBAC, contributing to the overall stabilization of the complex (25).
A recent study demonstrated that deficiency in SHARPIN or HOIP in mice causes severe inflammation in adulthood or embryonic lethality, respectively. By contrast, HOIL-1 deficiency contributes no overt phenotype (11). Previously, the LUBAC was shown to serve a pivotal role in complete activation of the canonical NF-κB signaling pathway, whereas the absence of one subunit resulted in attenuated activation of this pathway, particularly when SHARPIN was absent (26). These data suggest that HOIP, HOIL-1L and SHARPIN are all necessary for efficient activation of the NF-κB signaling pathway. Notably, Rodgers et al (27) suggested that linear ubiquitination is required for activation of the NACHT, LRR and PYD domains-containing protein 3 inflammasome, and this finding further expands the role of LUBAC as an innate immune regulator.
3. SHARPIN regulates the canonical NF-κB signaling pathway
In brief, the canonical NF-κB signaling pathway can be triggered by different stimulators, such as tumor necrosis factor (TNF)-α, interleukin (IL)-1 and pathogen-associated molecular patterns (28,29). When stimulators combine with the TNF receptor (TNFR), TNFR type 1-associated death domain protein, FAS-associated death domain protein and TNFR-associated factor 2 proteins and protein kinases are recruited in the cytoplasm as a result of phosphorylation and activation of the inhibition of the κB kinase (IKK) complex (30). The classical IKK complex consists of two catalytic subunits and a regulatory subunit, IKKα, IKKβ and the NF-κB essential modulator (NEMO), respectively. Furthermore, the activated IKK complex facilitates the phosphorylation of IκB, which releases NF-κB dimers that freely translocate into the nucleus and bind with DNA, promoting the transcription of relevant target genes (30,31). Recent studies have demonstrated that the LUBAC mediates linear ubiquitylation, which is involved in the canonical NF-κB signaling pathway (Fig. 2A).
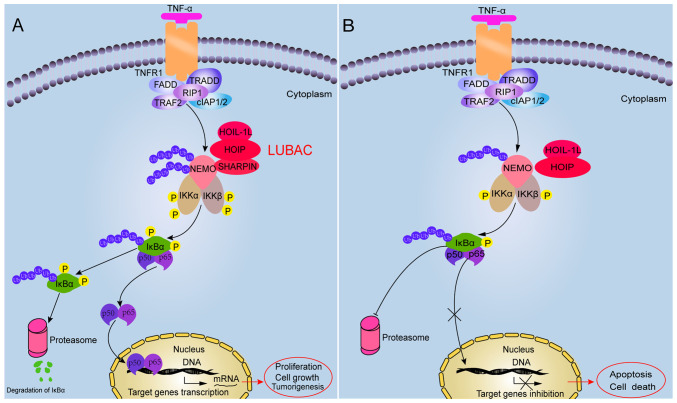
Figure 2.
Difference between SHARPIN presence and absence in regulating the NF-κB signaling pathway. (A) Presence of SHARPIN promotes canonical NF-κB signaling activation. Once stimulators bind with the receptors, associated molecules are recruited to activate the IKK complex. The complete LUBAC is involved in this process that serves a role in ubiquitination and fully activates the IKK complex. Subsequently, the phosphorylation and ubiquitination of IKBα contributes to the degradation of IKBα, which generates dimers. The dimers translocate into the nucleus and mediate target gene transcription. (B) Absence of SHARPIN attenuates canonical NF-κB signaling activation. When SHARPIN is absent, the LUBAC attenuates the phosphorylation and ubiquitination of IKBα, resulting in reduced IKBα degradation and inhibiting dimer translocation into the nucleus. Therefore, transcription of the target genes is inhibited. SHARPIN, shank-associated RH domain interactor; IKK, inhibition of κB kinase; LUBAC, linear ubiquitin chain assembly complex; TNF, tumor necrosis factor; TNFR1, TNF receptor 1; TRADD, TNFR type 1-associated DEATH domain protein; TRAF2, TNFR-associated factor 2; RIP1, receptor-interacting serine/threonine protein kinase 1; cIAP1/2, cellular inhibitor of apoptosis protein-1/2; FADD, FAS-associated death domain protein; HOIL-1L, heme-oxidized IRP2 ligase 1L; HOIP, HOIL-1 interacting protein; NEMO, NF-κB essential modulator; p, phosphorylated; Ub, ubiquitinated.
Numerous studies have suggested that NEMO possesses a specific ubiquitin-binding region that interacts with the LUBAC (23,32–34). The NF-κB activated state is influenced by the process of NEMO conjugating with a linear poly-ubiquitin chain (35). Furthermore, NEMO deficiency leads to decreased interaction with LUBAC, preventing SHARPIN-mediated linear ubiquitination and NF-κB activation (14,36). Consistent with this, SHAPRIN deficiency leads to inhibition of LUBAC-mediated linear poly-ubiquitination of endogenous NEMO and attenuates the activation of the NF-κB signaling pathway (37). Therefore, p65/p50 cannot translocate into the nucleus to induce target gene expression due to decreased phosphorylation and degradation of NF-κB inhibitor α (IKBα) (28) (Fig. 2B). These findings suggest that the NEMO-SHARPIN interaction is essential to mediate canonical NF-κB signaling. Hence, these results suggest that SHARPIN is important in regulating canonical NF-κB signaling and that this disruption may impact downstream physiological functions.
4. SHARPIN is a mediator of p53
It is well known that p53 is a vital tumor suppressor and its presence was initially reported in response to various types of stress >40 years ago (38). The p53 protein acts as a transcription factor in cells and is functionally inactivated in the majority of cancer types (39). Several studies have demonstrated that activated p53 is associated with numerous downstream responses and is involved in important physiological and pathological processes, including cell cycle arrest, DNA repair, apoptosis, metabolism, invasion, metastasis and tumorigenesis (40–42). p53 is maintained at very low cellular levels in normal cells due to binding with E3 ubiquitin ligase mouse double minute 2 homolog (MDM2), which contributes to p53 ubiquitination and rapid degradation by the proteasome (43). SHARPIN may be involved in promoting p53 ubiquitination and maintaining cellular levels of p53 (44). Mechanistically, MDM2 recognizes the N-terminal trans-activation domain of p53 and mediates p53 transcription as an inhibitor of p53 transcriptional activation (45). Nevertheless, in response to cellular stress signals, p53 is rapidly activated by phosphorylation, which releases MDM2 and promotes p53 stabilization (46). MDM2 regulates p53 through direct binding, mono-ubiquitylation, polyubiquitylation (44) or negative feedback regulation (47), which maintains appropriate cellular levels of p53.
A recent study demonstrated that SHARPIN may be upstream of p53 signaling in breast cancer cells, as depleted SHARPIN resulted in decreased cell proliferation and increased expression of p53 (17). Furthermore, the study reported that SHARPIN modulates p53 protein levels through poly-ubiquitination and degradation in a MDM2-dependent manner, which determines the fate of p53 in tumor cells (Fig. 3). Therefore, p53 protein levels in cells are indirectly regulated by SHARPIN. Nevertheless, whether other accessory molecules or signaling pathways are involved in SHARPIN-mediated regulation of the p53/MDM2 complex needs further clarification.
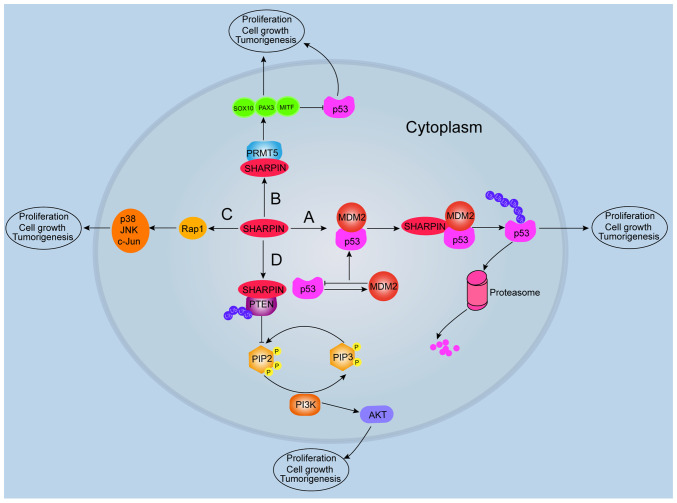
Figure 3.
SHARPIN regulates multiple signaling pathways in tumorigenesis. A: SHARPIN mediates p53 ubiquitination and rapid degradation by the proteasome, which promotes tumorigenesis. B: SHARPIN interacts with PRMT5 and activates SOX10, PAX3 and MITF, which promotes tumorigenesis. PRMT5 also regulates PAX3 and MITF and inhibits p53 in tumorigenesis. C: SHARPIN upregulates Rap, which promotes melanoma development through p38 and JNK/c-Jun signaling. D: SHARPIN can bind with PTEN, which mediates tumorigenesis through the PI3K/AKT signaling pathway. SOX10, SRY-box transcription factor 10; MITF, melanocyte inducing transcription factor; PAX3, paired box 3; PRMT5, protein arginine methyltransferase 5; SHARPIN, shank-associated RH domain interactor; Rap1, ras-associated protein 1; JNK, c-Jun N-terminal kinases; MDM2, mouse double minute 2 homolog; PTEN, phosphatase and tensin homologue deleted on chromosome 10; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PIP2, phosphatidylinositol (4,5)-bisphosphate; PI3K, phosphoinositide 3-kinases; p, phosphorylated; Ub, ubiquitinated.
SHARPIN interacts with PRMT5
PRMT5 is a member of the PRMT family of proteins and can catalyze symmetric methylation of histone and non-histone proteins. PRMT5 catalyzes the transfer of a methyl group from S-adenosylmethionine to the guanidino nitrogen atoms of arginine (48). PRMT5 is commonly activated in cancer, which is mediated in part by the PRMT5 co-factor methylosome protein 50 (also known as p44) (49). Researchers have demonstrated that PRMT5 serves an essential role in lung cancer, leukemia (49), tumorigenesis (50), cell survival and human embryonic stem cell proliferation (51). Stopa et al (52) provides an overall literature review of PRMT5 overexpression, which appears to be an important factor in the tumorigenicity of a large number of cancer types, such as gastric (53) and breast cancer (54). In addition, PRMT5 mediates the methylation of Arg1175 of epidermal growth factor receptor, which controls extracellular signal-regulated kinase activation (55). Similarly, E2F-1 may be directly methylated by PRMT5, which influences the stability of E2F-1. Depleting PRMT5 using small interfering RNA resulted in decreased arginine methylation of E2F-1 but increased E2F-1 levels in one study (56).
Recent reports have shown that the interaction of SHARPIN with PRMT5 contributes to regulating the transcription of cancer-associated genes (3,56). For example, Fu et al (57) reported that SHARPIN activates PRMT5 to specifically target histone H3R2 for mono-methylation, which is responsible for the subsequent activation of cancer-associated genes and mediates metastasis in invasive lung cancer cells. Tamiya et al (3) also demonstrated that PRMT5 activity is increased after SHARPIN binding. This study observed that SHARPIN activates PRMT5 by regulating SRY-box transcription factor 10, paired box gene 3 (PAX3) and microphthalmia-associated (MITF) transcription factors in melanoma development (Fig. 3). These results demonstrate that both SHARPIN and PRMT5 are involved in tumorigenesis.
PRMT5 regulates p53 signaling
Scoumanne et al (42) reported that PRMT5-knockdown prevented p53 stabilization and reduced p53 expression in response to DNA damage and cell cycle arrest. Subsequently, expression of both the MDM2 and p21 target genes was inhibited. The study also demonstrated that PRMT5 is required for p53 protein synthesis and suggested that PRMT5-mediated regulation of p53 serves a critical role in colon carcinoma cell survival. Furthermore, another study revealed that PRMT5 influences the methylation of p53 in response to DNA damage (58). Similarly, PRMT5 functions in a negative regulatory mechanism that underlies p53-dependent apoptosis in Caenorhabditis elegans (59). PRMT5 regulates Mdm4 (p53 regulator) expression, which influences p53-induced cell cycle arrest (60,61). PRMT5 is also important for regulating the p53-dependent mechanism of apoptosis (13). Furthermore, PRMT5 modulates p53 function in cell proliferation (42) and tumorigenesis (62), and p53 signaling has important functions in tumorigenesis (63,64). Overall, these results demonstrate that PRMT5 is directly involved in regulating p53 function in cell apoptosis, the cell cycle, survival and proliferation.
SHARPIN regulates PRMT5 activity, which induces the transcriptional activity of PAX3 and MITF in melanoma growth; in turn, PAX3 and MITF can inhibit p53 activation (3). Consistent with these findings, a recent study demonstrated that PAX3 specifically binds to the promoter of p53 leading to repressed p53 expression in glioblastoma (65). In addition, MITF binds p53 to regulate cyclin-dependent kinase inhibitor 1A in melanoma cells (66). Overall, these data suggest that PRMT5 is capable of regulating p53 indirectly, and SHARPIN mediates p53 through PRMT5-dependent signaling with PAX3 and MITF.
5. SHARPIN interacts with Rap1 in tumorigenesis
A study by Lilja et al (67) demonstrated that SHARPIN is indispensable for regulating integrin inactivation by Shanks interaction with Rap1 in cells. This finding suggests that SHARPIN and Rap1 functions may be closely associated. Furthermore, a recent study also supported this viewpoint, demonstrating that SHARPIN promotes melanoma progression through Rap1 via the p38 and JNK/Jun signaling pathways (18) (Fig. 3). However, the specific mechanism by which SHARPIN mediates Rap1 remains unclear, and whether this interaction is direct or indirect needs to be further explored.
6. SHARPIN functions in the PTEN signaling pathway
Studies have shown that PTEN is a pivotal tumor suppressor gene, and that it is frequently deleted in late-stage human cancer types and has important functions in crosstalk with the PI3K/ATK signaling pathway, mediating several fundamental cellular processes under different circumstances via multiple downstream targets (68–70). For example, a study has suggested that PI3K/AKT signaling is attenuated in the brains of patients with Alzheimer's disease (71). Moreover, PI3K/AKT/PTEN is mediated by intracellular ROS production (72). The main function of PTEN is to catalyze the conversion of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to phosphatidylinositol (4,5)-bisphosphate (73). PTEN is an antagonist of PI3K and, when dephosphorylated, serves as a negative regulator of PI3K/AKT signaling in normal cells, while loss or inactivation of PTEN contributes to hyperactivation of PI3K/AKT in primary T-cell leukemia (74) and adult B-cell acute lymphoblastic leukemia (75). Once activated, the PI3K/AKT pathway promotes cell survival by blocking the function of pro-apoptotic proteins and promoting protein synthesis and cell proliferation. Consequently, these results indicate that inactivation of PTEN is a critical step during tumorigenesis.
There has been much interest in the transcriptional and post-translational modulation of PTEN expression, protein stability and activity mediated by miRNA (76), phosphorylation (77), acetylation (78) and ubiquitination (79). A recent study reports that PTEN knockdown could significantly promote mouse neuronal cell proliferation and differentiation in vitro (80). Moreover, a previous study demonstrated that SHARPIN is a negative regulator of PTEN in human tumor cell lines and human primary cervical cancer cells both in vitro and in vivo (81). Furthermore, it has been reported that SHARPIN interacts with PTEN through its ubiquitin-like domain (81). It is well known that the major function of PTEN is inhibition of the PI3K signaling pathway, while loss of PTEN activates the PI3K/AKT pathway (82). Additionally, attenuation of PTEN function activates PI3K/AKT signaling and elicits tumorigenesis (82). Hence, reducing PTEN-induced PIP3 phosphatase activity and enhancing the activity of the PI3K/AKT signaling pathway promotes tumorigenesis (83). In accordance with this, De Melo et al (84) suggested that SHARPIN facilitated PTEN poly-ubiquitination via lysine63 and formation of the SHARPIN/PTEN complex, which did not lead to PTEN degradation. These biochemical processes alter the affinity of the SHARPIN/PTEN complex and reduce the phosphatase activity of PTEN (Fig. 3). The study proposed that SHARPIN promotes poly-ubiquitination of PTEN in a manner that does not alter PTEN stability, and that SHARPIN/PTEN binding is enhanced by poly-ubiquitination of PTEN.
7. Conclusion
Overall, there has been a marked increase in the understanding of the function of SHARPIN in recent years. The present review summarizes our understanding of how SHARPIN mediates canonical NF-κB signaling and crosstalk with various mechanisms, regulating tumorigenesis. Furthermore, the function of SHARPIN in the NF-κB, p53, PRMT5, Rap1 and PTEN signaling pathways was explored. The aforementioned results shed light on possible functions of these proteins in tumor cells, and may provide novel targets for cancer treatment in the future. However, the detailed molecular function of associated co-activators or co-repressors contributing to these pathways needs further investigation.
Acknowledgements
The authors would like to thank Professor Shaogang Qu (Nanfang Hospital, Southern Medical University, Guangzhou, China) for assistance with the manuscript revision.
Glossary
Abbreviations
- SHARPIN
shank-associated RH domain interactor
- LUBAC
linear ubiquitin chain assembly complex
- MDM2
mouse double minute 2 homolog
- PRMT5
protein arginine methyltransferase 5
- PI3K
phosphoinositide 3-kinases
- PTEN
phosphatase and tensin homologue deleted on chromosome ten
- IKK
inhibition of κB kinase
- NEMO
NF-κB essential modulator
- HOIL-1L
heme-oxidized IRP2 ligase 1L
- HOIP
HOIL-1 interacting protein
- PAX3
paired box gene 3
- MITF
microphthalmia-associated transcription factor
- PIP3
- PI3K
phosphoinositide 3-kinase
- Rap1
ras-associated protein-1
Funding
The present study was supported by grants from the Science and Technology Program of Foshan (grant no. 1920001000694), the Scientific Research Startup of Shunde Hospital (grant no. SRSP2019012) and the Medical Scientific Research Foundation of Guangdong Province (grant no. A2020175).
Availability of data and materials
Not applicable.
Authors' contributions
CZ, DX, KZ and JY jointly conceived and designed the review, researched the literature and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human development index (2008–2030): A population-based study. Lancet Oncol. 2012;13:790–801. doi: 10.1016/S1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamiya H, Kim H, Klymenko O, Kim H, Feng Y, Zhang T, Han JY, Murao A, Snipas SJ, Jilaveanu L, et al. SHARPIN-mediated regulation of protein arginine methyltransferase 5 controls melanoma growth. J Clin Invest. 2018;128:517–530. doi: 10.1172/JCI95410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung J, Kim JM, Park B, Cheon Y, Lee B, Choo SH, Koh SS, Lee S. Newly identified tumor-associated role of human Sharpin. Mol Cell Biochem. 2010;340:161–167. doi: 10.1007/s11010-010-0413-x. [DOI] [PubMed] [Google Scholar]
- 5.Ojo D, Wu Y, Bane A, Tang D. A role of SIPL1/SHARPIN in promoting resistance to hormone therapy in breast cancer. Biochim Biophys Acta Mol Basis Dis. 2018;1864:735–745. doi: 10.1016/j.bbadis.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Bii VM, Rae DT, Trobridge GD. A novel gammaretroviral shuttle vector insertional mutagenesis screen identifies SHARPIN as a breast cancer metastasis gene and prognostic biomarker. Oncotarget. 2015;6:39507–39520. doi: 10.18632/oncotarget.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Potter CS, Sundberg JP, Hogenesch H. SHARPIN is a key regulator of immune and inflammatory responses. J Cell Mol Med. 2012;16:2271–2279. doi: 10.1111/j.1582-4934.2012.01574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rittinger K, Ikeda F. Linear ubiquitin chains: Enzymes, mechanisms and biology. Open Biol. 2017;7:170026. doi: 10.1098/rsob.170026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 11.Peltzer N, Darding M, Montinaro A, Draber P, Draberova H, Kupka S, Rieser E, Fisher A, Hutchinson C, Taraborrelli L, et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature. 2018;557:112–117. doi: 10.1038/s41586-018-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Y, Joo D, Liu G, Tu H, You J, Jin J, Zhao X, Hung MC, Lin X. Linear ubiquitination of cFLIP induced by LUBAC contributes to TNFα-induced apoptosis. J Biol Chem. 2018;293:20062–20072. doi: 10.1074/jbc.RA118.005449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teh CE, Lalaoui N, Jain R, Policheni AN, Heinlein M, Alvarez-Diaz S, Sheridan JM, Rieser E, Deuser S, Darding M, et al. Linear ubiquitin chain assembly complex coordinates late thymic T-cell differentiation and regulatory T-cell homeostasis. Nat Commun. 2016;7:13353. doi: 10.1038/ncomms13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redecke V, Chaturvedi V, Kuriakose J, Hacker H. SHARPIN controls the development of regulatory T cells. Immunology. 2016;148:216–226. doi: 10.1111/imm.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Z, Tang J, Yang Q, Li X, Zhu J, Wu G. Atypical ubiquitin-binding protein SHARPIN promotes breast cancer progression. Biomed Pharmacother. 2019;119:109414. doi: 10.1016/j.biopha.2019.109414. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Yu S, Wang W, Li X, Hou Y, Liu Z, Shi Y, Mu K, Niu G, Xu J, et al. SHARPIN facilitates p53 degradation in breast cancer cells. Neoplasia. 2017;19:84–92. doi: 10.1016/j.neo.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou S, Liang Y, Zhang X, Liao L, Yang Y, Ouyang W, Xu H. SHARPIN promotes melanoma progression via Rap1 signaling pathway. J Invest Dermatol. 2020;140:395–403.e6. doi: 10.1016/j.jid.2019.07.696. [DOI] [PubMed] [Google Scholar]
- 19.De Melo J, Wu V, He L, Yan J, Tang D. SIPL1 enhances the proliferation, attachment, and migration of CHO cells by inhibiting PTEN function. Int J Mol Med. 2014;34:835–841. doi: 10.3892/ijmm.2014.1840. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki K, Iwai K. Roles of linear ubiquitinylation, a crucial regulator of NF-κB and cell death, in the immune system. Immunol Rev. 2015;266:175–189. doi: 10.1111/imr.12308. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda F. Linear ubiquitination signals in adaptive immune responses. Immunol Rev. 2015;266:222–236. doi: 10.1111/imr.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwai K, Fujita H, Sasaki Y. Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat Rev Mol Cell Biol. 2014;15:503–508. doi: 10.1038/nrm3836. [DOI] [PubMed] [Google Scholar]
- 23.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 24.Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G, et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498:318–324. doi: 10.1038/nature12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita H, Tokunaga A, Shimizu S, Whiting AL, Aguilar-Alonso F, Takagi K, Walinda E, Sasaki Y, Shimokawa T, Mizushima T, et al. Cooperative domain formation by homologous motifs in HOIL-1L and SHARPIN plays A crucial role in LUBAC stabilization. Cell Rep. 2018;23:1192–1204. doi: 10.1016/j.celrep.2018.03.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsunaga Y, Nakatsu Y, Fukushima T, Okubo H, Iwashita M, Sakoda H, Fujishiro M, Yamamotoya T, Kushiyama A, Takahashi S, et al. LUBAC formation is impaired in the livers of mice with MCD-dependent nonalcoholic steatohepatitis. Mediators Inflamm. 2015;2015:125380. doi: 10.1155/2015/125380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, Amatya R, Kelly TJ, Iwai K, Ting J, et al. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J Exp Med. 2014;211:1333–1347. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokunaga F. Linear ubiquitination-mediated NF-κB regulation and its related disorders. J Biochem. 2013;154:313–323. doi: 10.1093/jb/mvt079. [DOI] [PubMed] [Google Scholar]
- 29.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 30.D'Ignazio L, Batie M, Rocha S. Hypoxia and inflammation in cancer, focus on HIF and NF-κB. Biomedicines. 2017;5:21. doi: 10.3390/biomedicines5020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J Biol Chem. 2013;288:31728–31737. doi: 10.1074/jbc.M113.495846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokunaga F, Iwai K. Involvement of LUBAC-mediated linear polyubiquitination of NEMO in NF-kappaB activation. Tanpakushitsu Kakusan Koso. 2009;54:635–642. (In Japanese) [PubMed] [Google Scholar]
- 34.Iwai K, Tokunaga F. Linear polyubiquitination: A new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Bal E, Laplantine E, Hamel Y, Dubosclard V, Boisson B, Pescatore A, Picard C, Hadj-Rabia S, Royer G, Steffann J, et al. Lack of interaction between NEMO and SHARPIN impairs linear ubiquitination and NF-κB activation and leads to incontinentia pigmenti. J Allergy Clin Immunol. 2017;140:1671–1682.e2. doi: 10.1016/j.jaci.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 37.Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 2011;30:3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane D, Levine A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozaki T, Nakagawara A. Role of p53 in cell death and human cancers. Cancers (Basel) 2011;3:994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37:4965–4976. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 2007;420:25–27. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 44.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lessel D, Wu D, Trujillo C, Ramezani T, Lessel I, Alwasiyah MK, Saha B, Hisama FM, Rading K, Goebel I, et al. Dysfunction of the MDM2/p53 axis is linked to premature aging. J Clin Invest. 2007;127:3598–3608. doi: 10.1172/JCI92171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Lin J, Levine AJ. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–152. doi: 10.1007/BF03401562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1999;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 48.Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y, Du X, Li J, Li P, Ren R, Pan J. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126:3961–3980. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehman I, Basu SM, Das SK, Bhattacharjee S, Ghosh A, Pommier Y, Das BB. PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res. 2018;46:5601–5617. doi: 10.1093/nar/gky291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durant ST, Cho EC, La Thangue NB. p53 methylation-the Arg-ument is clear. Cell Cycle. 2008;8:801–802. doi: 10.4161/cc.8.6.7850. [DOI] [PubMed] [Google Scholar]
- 51.Gkountela S, Li Z, Chin CJ, Lee SA, Clark AT. PRMT5 is required for human embryonic stem cell proliferation but not pluripotency. Stem Cell Rev. 2014;10:230–239. doi: 10.1007/s12015-013-9490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M, Yao B, Gui T, Guo C, Wu X, Li J, Ma L, Deng Y, Xu P, Wang Y, et al. PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics. 2020;10:4437–4452. doi: 10.7150/thno.42047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vinet M, Suresh S, Maire V, Monchecourt C, Nemati F, Lesage L, Pierre F, Ye M, Lescure A, Brisson A, et al. Protein arginine methyltransferase 5: A novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019;8:2414–2428. doi: 10.1002/cam4.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, Lee HJ, Wang YN, Liu M, Liao HW, et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011;13:174–181. doi: 10.1038/ncb2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho EC, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, Lu YC, Stimson L, Khan O, Konietzny R, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31:1785–1797. doi: 10.1038/emboj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu T, Lv X, Kong Q, Yuan C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget. 2017;8:54809–54820. doi: 10.18632/oncotarget.18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 59.Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009;5:e1000514. doi: 10.1371/journal.pgen.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerhart SV, Kellner WA, Thompson C, Pappalardi MB, Zhang XP, Montes de Oca R, Penebre E, Duncan K, Boriack-Sjodin A, Le B, et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep. 2018;8:9711. doi: 10.1038/s41598-018-28002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Diehl JA. PRMT5-dependent p53 escape in tumorigenesis. Oncoscience. 2015;2:700–702. doi: 10.18632/oncoscience.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scaglione A, Patzig J, Liang J, Frawley R, Bok J, Mela A, Yattah C, Zhang J, Teo SX, Zhou T, et al. PRMT5-mediated regulation of developmental myelination. Nat Commun. 2018;9:2840. doi: 10.1038/s41467-018-04863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu F, Cheng G, Hamard PJ, Greenblatt S, Wang L, Man N, Perna F, Xu H, Tadi M, Luciani L, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J Clin Invest. 2015;125:3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H, Wang H, Huang Q, Liu Q, Guo Y, Lu J, Li X, Xue C, Han Q. Transcriptional repression of p53 by PAX3 contributes to gliomagenesis and differentiation of glioma stem cells. Front Mol Neurosci. 2018;11:187. doi: 10.3389/fnmol.2018.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C, Zhao L, Su Q, Fan X, Wang Y, Gao S, Wang H, Chen H, Chan CB, Liu Z. Phosphorylation of MITF by AKT affects its downstream targets and causes TP53-dependent cell senescence. Int J Biochem Cell Biol. 2016;80:132–142. doi: 10.1016/j.biocel.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 67.Lilja J, Zacharchenko T, Georgiadou M, Jacquemet G, De Franceschi N, Peuhu E, Hamidi H, Pouwels J, Martens V, Nia FH, et al. SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras. Nat Cell Biol. 2017;19:292–305. doi: 10.1038/ncb3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JT, Shan J, Zhong J, Li M, Zhou B, Zhou A, Parsons R, Gu W. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell Res. 2013;23:552–564. doi: 10.1038/cr.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 70.Worby CA, Dixon JE. Pten. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Wu C, Han B, Xu F, Mao M, Guo X, Wang J. Dexmedetomidine attenuates repeated propofol exposure-induced hippocampal apoptosis, PI3K/Akt/Gsk-3β signaling disruption, and juvenile cognitive deficits in neonatal rats. Mol Med Rep. 2016;14:769–775. doi: 10.3892/mmr.2016.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakanishi A, Wada Y, Kitagishi Y, Matsuda S. Link between PI3K/AKT/PTEN pathway and NOX proteinin diseases. Aging Dis. 2014;5:203–211. doi: 10.14336/AD.2014.0500203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 74.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA, Barata JT. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118:3762–3774. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gomes AM, Soares MV, Ribeiro P, Caldas J, Povoa V, Martins LR, Melao A, Serra-Caetano A, de Sousa AB, Lacerda JF, et al. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica. 2014;99:1062–1068. doi: 10.3324/haematol.2013.096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J Biol Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 78.Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2018;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- 79.Yang JM, Schiapparelli P, Nguyen HN, Igarashi A, Zhang Q, Abbadi S, Amzel LM, Sesaki H, Quinones-Hinojosa A, Iijima M. Characterization of PTEN mutations in brain cancer reveals that pten mono-ubiquitination promotes protein stability and nuclear localization. Oncogene. 2017;36:3673–3685. doi: 10.1038/onc.2016.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Z, Han X, Shen L, Zou H, Zhang B, Liu J, Gong A. PTEN silencing enhances neuronal proliferation and differentiation by activating PI3K/Akt/GSK3β pathway in vitro. Exp Cell Res. 2018;363:179–187. doi: 10.1016/j.yexcr.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: The long and the short of it. Trends Biochem Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He L, Ingram A, Rybak AP, Tang D. Shank-interacting protein-like 1 promotes tumorigenesis via PTEN inhibition in human tumor cells. J Clin Invest. 2012;120:2094–2108. doi: 10.1172/JCI40778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Melo J, Lin X, He L, Wei F, Major P, Tang D. SIPL1-facilitated PTEN ubiquitination contributes to its association with PTEN. Cell Signal. 2014;26:2749–2756. doi: 10.1016/j.cellsig.2014.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.