Abstract
Glioma is one of the most prevalent types of malignancy in the central nervous system worldwide, and the prognosis of patients with late stage glioma remains poor. Thus, the development of promising therapeutic strategies against glioma is essential. Long non-coding RNAs (lncRNAs) are functional RNA molecules involved in the initiation and progression of tumors, including glioma. Investigation on the regulatory roles of lncRNAs may facilitate the development of effective treatments. lncRNA NBAT1 is associated with the growth and metastasis of cancer; however, its underlying molecular mechanisms remain unknown. Thus, the present study aimed to investigate the effects of NBAT1 in glioma. The expression levels of NBAT1, microRNA (miRNA/miR)-21 and SOX7 in patients with glioma, and healthy donors using reverse transcription-quantitative PCR analysis. Human glioma cells (A172 and AM138) and normal astrocytes were used to establish the NBAT1-knockdown and overexpression models. Cell Counting Kit-8 and Transwell assays were performed to determine whether NBAT1 exerted effects on cell proliferation, migration and invasion. The results demonstrated that NBAT1 expression decreased in glioma tissues compared to normal samples. Additionally, downregulation of NBAT1 was detected in human glioma cells compared with normal astrocytes. Overexpression of NBAT1 inhibited glioma cell proliferation, migration and invasion. In addition, miR-21 was identified as a potential target of NBAT1, and the effects of miR-21-induced cell proliferation and metastasis were reversed following overexpression of NBAT1. Furthermore, SOX7 was predicted as the potential target of miR-21, and its expression was upregulated in glioma cells by overexpression of NBAT1 compared with the vehicle only control. Taken together, the results of the present study provide novel insight into the functions of NBAT1 in glioma, suggesting that the NBAT1/miR-21/SOX7 axis may act as a potential therapeutic target for the treatment of patients with glioma.
Keywords: glioma, long non-coding RNA NBAT1, proliferation, migration, invasion
Introduction
Glioma is one of the most common and aggressive primary malignancies of the brain, and the global incidence was 5% in 2013; it was reported to account for ~80% of malignant neoplasia and ~30-40% of primary tumors in the central nervous system, worldwide (1). Despite recent advances in adjuvant radiotherapy, hormone therapy, chemotherapy and targeted biologic therapy, the overall survival outcomes of most patients with glioma remain poor (2,3). Furthermore, the aforementioned treatments exhibit side effects, such as anemia and constipation (3); thus, the development of novel therapeutic strategies against glioma remains essential.
Long non-coding RNAs (lncRNAs) are a novel class of non-protein coding RNAs, >200 nt long (4–6). lncRNAs are involved in a number of biological processes, including cell differentiation and gene expression (7). Increasing evidence suggests that lncRNAs are associated with the initiation and progression of different types of tumor, including glioma (8–15). Furthermore, aberrant expression levels of lncRNAs have been reported in tumor cells, indicating the potential role of lncRNAs in tumorigenesis (16–18). Thus, investigation of the regulatory functions of lncRNAs in cancer may help facilitate the development of effective therapies.
lncRNA NBAT1 is involved in carcinogenesis, and previous studies have reported the dysregulation of NBAT1 in different types of cancer (19,20). NBAT1 promotes the initiation and development of tumors via several pathways, including PRC2 signaling (21,22), and thus is considered a promising biomarker and prognostic indicator of glioma (20,23). However, the potential molecular targets of NBAT1 in glioma have not yet been fully investigated.
MicroRNAs (miRNAs/miRs) are endogenously expressed non-coding RNAs that are ~22 nt in length and are considered potential targets of lncRNAs (24,25). Increasing evidence suggests that the levels of miRNAs are dysregulated in cancer, which may contribute to tumor development (26,27). In addition, miRNAs such as miR-140 and miR-152 are able to interact with lncRNAs in glioma (28,29). However, the specific functions of miRNAs in glioma remain unclear.
The present study aimed to investigate the functions of the NBAT1-mediated miR-21/SOX7 signaling pathway in glioma cell proliferation and metastasis, which may provide novel insight for the effective treatment of patients with glioma.
Materials and methods
Patient samples
A total of 40 pairs of primary tumor and paracarcinoma tissue samples (>5 cm from tumor margin) were collected from patients with glioma (16 men and 24 women; age range, 45–65 years; mean age, 51 years) undergoing surgical resection at The First Affiliated Hospital of Jinzhou Medical University (Jinzhou, China) between June 2015 and August 2017. Prior to surgery, none of the patients have received chemotherapy or radiotherapy. Metastasis was observed in 14 and absent in 26 patients, and 22 cases were diagnosed with grade I or II glioma, while 18 were diagnosed with grade III or IV glioma, according to the World Health Organization grading system (https://www.cancer.net/cancer-types/brain-tumor/grades-and-prognostic-factors). The biopsies were examined by two independent pathologists from the Department of Histopathology, The First Affiliated Hospital of Jinzhou Medical University (Jinzhou, China). All samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until further experimentation. The present study was approved by the Institutional Review Board of The First Affiliated Hospital of Jinzhou Medical University (approval no. JYD159302; Jinzhou, China). Written informed consent was provided by all patients prior to the study start.
Cell culture
The human glioma cell lines (AM38, Gli-6, GSC11 and A172) and normal human astrocyte cell line (A735) were purchased from the American Type Culture Collection. Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (all purchased from HyClone; Cytiva) at 37°C in a humidified incubator containing 5% CO2.
Cell transfection
To establish the NBAT1 knockdown model, short hairpin (sh)RNA sequences targeting NBAT1 (sh-NBAT1), as well as the negative control (sh-NC) were synthesized by Shanghai GenePharm Co., Ltd. Following annealing, the shRNAs were integrated into the lentiviral pU6-Luc-Puro vector (Shanghai GenePharm Co., Ltd.). To establish the NBAT1 overexpression model, wild-type (WT; o/e-NBAT1) or mutant (o/e-NC) NBAT1 fragments were amplified by PCR using Taq polymerase (Thermo Fisher Scientific, Inc.) and subcloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). The primer sequences used were as follows: Forward, 5′-TCAGCAGAAACGGCACGAT-3′ and reverse 5′-AGATGACCCAGGCACCTCC-3′. The following thermocycling conditions were used: 95°C for 30 sec, followed by 30 cycles at 95°C for 15 sec, 60°C for 20 sec, 68°C for 10 sec and a final extension at 68°C for 5 min.
A172 and AM38 cells were seeded into 6-well plates at a density of 2×105 and subsequently transfected with the recombinant lentiviral vectors (50 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 8 h at 37°C. The culture medium was replaced with fresh DMEM supplemented with 10% FBS 8 h post-transfection. Knockdown and overexpression of NBAT1 were confirmed by reverse transcription-quantitative (RT-q)PCR analysis.
The miR-21 mimics or inhibitors and the corresponding NCs (all 40 pg/µl) were purchased from Shanghai GenePharm Co., Ltd. and transfected into glioma cells (2×105) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 8 h at 37°C. The culture medium was replaced with fresh DMEM supplemented with 10% FBS at 8 h post-transfection. Subsequent experiments were performed 24 h post-transfection.
RT-qPCR
miRNA was extracted from tissues or cell lines using the miRNeasy Mini kit (Qiagen China Co., Ltd.), and miR-21 expression was determined using the TaqMan MicroRNA Assay kit (cat. no. 4427975; Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was subsequently performed using an Applied Biosystem 7500 PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. U6 small nuclear RNA was used as the internal control.
Total RNA was extracted from clinical samples or cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.) and subsequently reverse-transcribed into cDNA using the PrimeScript™ RT kit (Takara Biotechnology Co., Ltd.). The temperature protocol was as follows: 42°C for 45 min, 99°C for 5 min and 5°C for 5 min. qPCR was performed using the SYBR® Green PCR Master Mix (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The following primer sequences were used for qPCR: NBAT1 forward, 5′-ATTTCTGCTCCTGGGTCTTAC-3′ and reverse, 5′-GCAAGAGCACAAGAGGAAGA-3′; SOX7 forward, 5′-CAAGGACGAGAGGAAACGGC-3′ and reverse, 5′-TACGGCCTCTTCTGGGACAG-3′; and GAPDH forward, 5′-GCAAGAGCACAAGAGGAAGA-3′ and reverse, 5′-ACTGTGAGGAGGGGAGATTC-3′. The following thermocycling conditions were used for qPCR: 95°C for 5 min, followed by 45 cycles at 95°C for 15 sec, 60°C for 20 sec and 72°C for 10 sec. Relative mRNA expression levels were calculated using the 2−∆∆Cq method (30) and normalized to the internal reference gene GAPDH.
Western blotting
Total protein was extracted from tissues or cells using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology). Protein concentration was determined using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology), and 40 µg protein/lane was separated via 10% SDS-PAGE. The separated proteins were subsequently transferred onto nitrocellulose membranes (EMD Millipore) and blocked with tris-buffered saline (TBS) containing 5% skimmed milk for 2 h at room temperature. The membranes were incubated with primary antibodies against SOX7 (1:1,000; cat. no. MAB2766; Novus Biologicals, Ltd.) and GAPDH (1:1,000; cat. no. sc-47724; Santa Cruz Biotechnology Inc.) overnight at 4°C. Membranes were washed three with TBS + Tween-20 (0.05%) and subsequently incubated with horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (1:5,000; cat. no. sc-2371; Santa Cruz Biotechnology Inc.) for 1 h at 37°C. Protein bands were visualized using an enhanced chemiluminescence protein detection kit (Pierce; Thermo Fisher Scientific, Inc.) and quantified via densitometric analysis using ImageJ software (version 1.48; National Institutes of Health).
Cell counting kit-8 (CCK-8) assay
Transfected A172/AM38 cells were harvested 24 h post-transfection and seeded into 96-well plates at a density of 1×104 cells/well. Briefly, 10 µl of CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to the wells on days 1, 2, 3 and 4 post-inoculation, according to the manufacturer's protocol. Following incubation at 37°C for an additional 2 h, cell proliferation was subsequently analyzed at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Transwell assay
For the migration assay, a total of 1×105 A172/AM38 cells were plated in the upper chamber of Transwell plates with a pore size of 8 µm (BD Biosciences) in FBS-free DMEM medium. For the invasion assay, Transwell membranes were precoated with Matrigel® (Sigma-Aldrich; Merck KGaA) at room temperature for 48 h. DMEM medium (500 µl) supplemented with 10% FBS was added to the lower chambers. Following overnight incubation at 37°C, non-migratory/invasive cells were removed using a cotton swab, whereas the migratory/invasive cells in the lower chambers were fixed with 4% paraformaldehyde for 15 min and stained with 0.5% crystal violet for 20 min at room temperature. Stained cells were counted in five randomly selected fields using an inverted light microscope (magnification, ×100; Olympus Corporation).
Northern blot analysis
Total RNA was extracted from tissues or cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). An equal amount of RNA (25 µg) was loaded onto 15% TBE-urea gels and separated using a 15% urea-PAGE gel. The separated proteins were subsequently transferred onto positively charged nylon membranes (Cytiva) and cross-linked by UV irradiation. The blots were hybridized by DIG-labelled probe for miR-21 (cat. no. 339546; Qiagen AB) overnight at 42°C. The membranes were washed three times with a low-stringency buffer (2×SSC containing 0.1% SDS; Thermo Fisher Scientific, Inc.), and the RNA levels were determined using the DIG Luminescent Detection kit [Roche Diagnostics (Shanghai) Co., Ltd] and semi-quantified using Multi Gauge software (version 3.0; Fujifilm Wako Pure Chemical Corporation). U6 (cat. no. 339508; Qiagen) RNA was used as the loading control.
Bioinformatics prediction and dual-luciferase reporter assay
TargetScan (www.targetscan.org), miRanda (www.microrna.org/microrna) and LncBase Predicted v2 (http://carolina.imis.athenainnovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) databases were used to predict the potential binding sites of NBAT1 and SOX7 transcripts on miR-21.
WT fragments of the 3′-untranslated region (3′UTR) of NBAT1 and SOX7 containing the potential binding sites of miR-21 were purchased from Shanghai GenePharma Co., Ltd. and cloned into the pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega Corporation) according to the manufacturer's protocol. The QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) was used to generate the NBAT1- and SOX7-3′UTR-MUT reporter containing mutant miR-21 binding sites. The constructed luciferase vectors were co-transfected with miR-21 mimics or miR-NC into DH5α competent cells (Thermo Fisher Scientific, Inc.). Following incubation at 37°C for 48 h, firefly and Renilla luciferase activities were detected using a Dual Luciferase Reporter assay system (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS software 17.0 (SPSS, Inc.), and data are presented as the mean ± standard deviation. Student's t-test (paired and unpaired) was used to compare differences between two groups. One-way analysis of variance followed by Student-Newman-Keuls or Tukey's post-hoc test were performed to compare differences among multiple groups. Pearson's correlation analysis was performed to determine the correlations between mRNA expression levels. All experiments were performed in triplicate. P<0.05 was considered to indicate a statistically significant difference.
Results
NBAT1 is downregulated in glioma tissues and cells and associated with poor prognosis
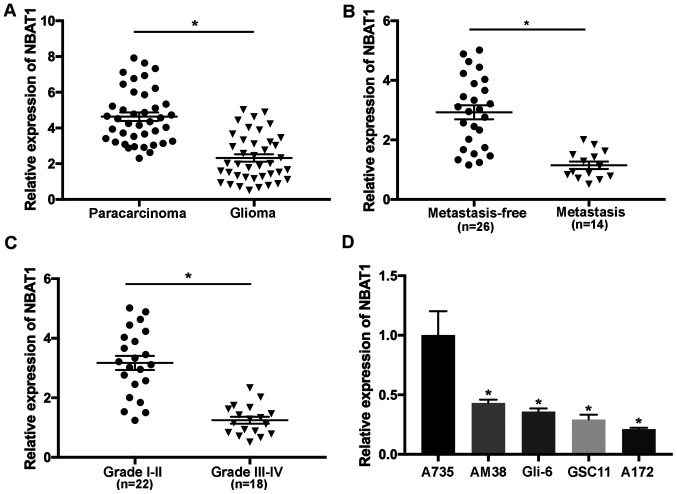
The expression levels of NBAT1 in 40 paired glioma and paracarcinoma samples were determined using RT-qPCR analysis. The results demonstrated that NBAT1 expression was significantly downregulated in glioma tissues compared with that in the paracarcinoma tissues (P<0.05; Fig. 1A). The effects of NBAT1 on metastasis of patients with glioma were also investigated. The results revealed that NBAT1 expression was significantly decreased in patients with metastatic glioma compared with the controls (patients with non-metastatic glioma; P<0.05; Fig. 1B). In addition, NBAT1 expression was significantly decreased in aggressive (grade III or IV) compared with low-grade (I or II) glioma, suggesting that downregulation of NBAT1 was associated with the development of glioma (P<0.05; Fig. 1C). NBAT1 expression was significantly downregulated in glioma cells compared with A735 cells (P<0.05; Fig. 1D). Taken together, these results suggested that NBAT1 expression was downregulated in glioma, which may also be associated with metastasis.
Figure 1.
NBAT1 is downregulated in glioma tissues and cells. (A) The expression levels of NBAT1 were determined in 40 glioma and paired paracarcinoma samples via RT-qPCR analysis. (B) NBAT1 expression was determined in patients with metastatic glioma compared with those without metastasis. (C) NBAT1 mRNA expression was assessed in glioma tissues from patients with different tumor stages via RT-qPCR analysis. (D) The expression levels of NBAT1 were determined in glioma cells (A172, GSC11, Gli-6 and AM38) and normal human astrocytes (A735). *P<0.05 vs. corresponding control. RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of NBAT1 suppresses glioma cell proliferation, migration and invasion
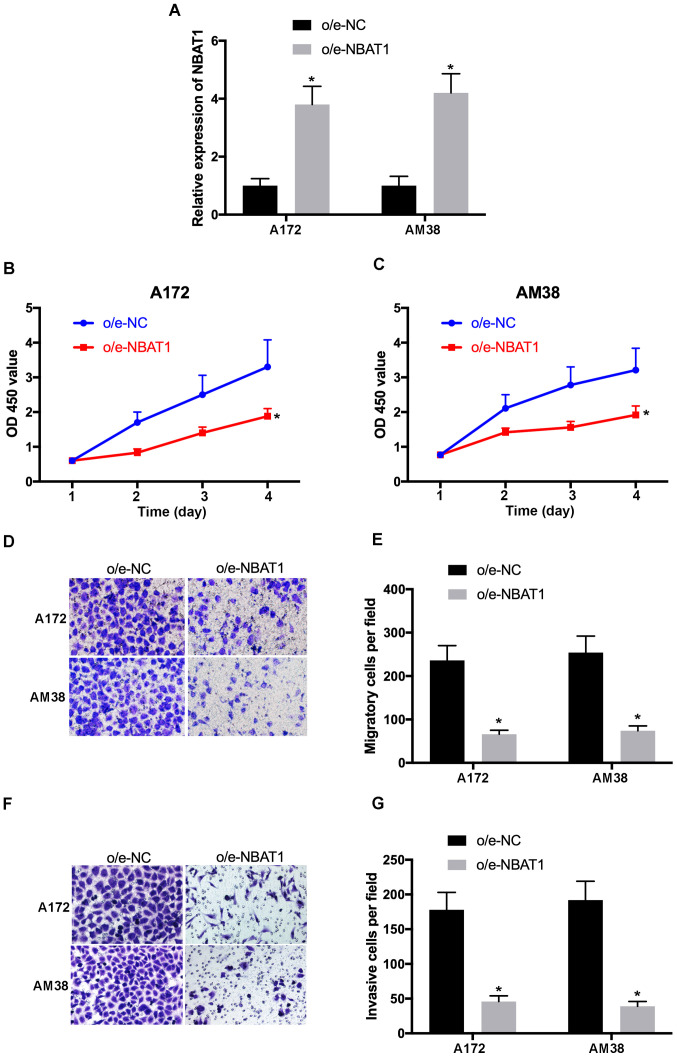
To determine the effects of NBAT1 on glioma cell proliferation, migration and invasion, NBAT1 was overexpressed in A172 and AM38 cells, and transfection efficiency was determined by RT-qPCR (Fig. 2A). The CCK-8 assay demonstrated that the proliferative ability of o/e-NBAT1-transfected A172 and AM38 cells significantly decreased compared with that of the control (P<0.05; Fig. 2B and C). In addition, the Transwell assay results revealed that the migratory and invasive abilities of o/e-NBAT1-transfected A172 and AM38 cells were significantly inhibited compared with those of the control groups (P<0.05; Fig. 2D-G). Collectively, these results suggested that overexpression of NBAT1 may suppress the proliferation, migration and invasion of glioma cells.
Figure 2.
Overexpression of NBAT1 suppresses glioma cell proliferation, migration and invasion. (A) Transfection efficiency of o/e-NBAT1 was confirmed via RT-qPCR analysis. The proliferative ability of (B) A172 and (C) AM38 cells transfected with o/e-NBAT1 or o/e-NC was assessed using the Cell Counting Kit-8 assay. (D) Cell migration was assessed using Transwell assays (magnification, ×100). (E) The migratory ability of A172 and AM38 cells transfected with o/e-NBAT1 or o/e-NC. (F) Cell invasion was assessed using Transwell assays (magnification, ×100). (G) The invasive ability of A172 and AM38 cells transfected with o/e-NBAT1 or o/e-NC. *P<0.05 vs. o/e-NC. o/e, overexpression vector; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; OD, optical density.
miR-21 is the potential target of NBAT1 in glioma
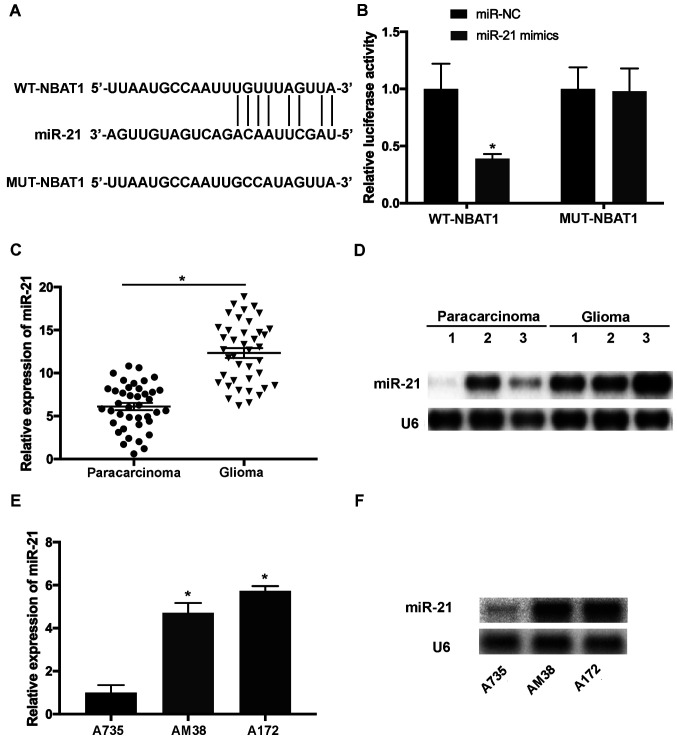
To determine whether NBAT1 functions by suppressing its target miRNAs in glioma, the potential binding sites of miR-21 in NBAT1 transcripts were predicted using LncBase Predicted v2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) (Fig. 3A). Luciferase reporter vectors containing WT-NBAT1 and MUT-NBAT1 sequences of predicted miR-21 binding sites were constructed. The results demonstrated that miR-21 mimics significantly attenuated the activity of the luciferase plasmid containing the WT binding sites compared with that of the MUT control (P<0.05; Fig. 3B). RT-qPCR and northern blot analyses indicated that miR-21 expression was upregulated in glioma tissues compared with that in paracarcinoma tissues (P<0.05; Fig. 3C and D). In addition, upregulated miR-21 expression was observed in A172 and AM38 cells compared with that in A735 cells (P<0.05; Fig. 3E and F).
Figure 3.
miR-21 is a downstream target of NBAT1 in glioma. (A) The putative binding sites of miR-21 on NBAT1 transcripts. (B) Overexpression of miR-21 significantly decreased the luciferase activity of WT-NBAT1, whereas no significant changes were observed in MUT-NBAT1. (C) The RNA levels of miR-21 were increased in glioma compared with the paracarcinoma tissues. (D) The levels of miR-21 were evaluated in glioma samples using northern blotting. (E) The expression levels of miR-21 were increased in A172 and AM38 cells compared with those in A735 cells. (F) The expression of miR-21 in cell lines was assessed using northern blotting. *P<0.05. miR, microRNA; WT, wild-type; MUT, mutant; NC, negative control.
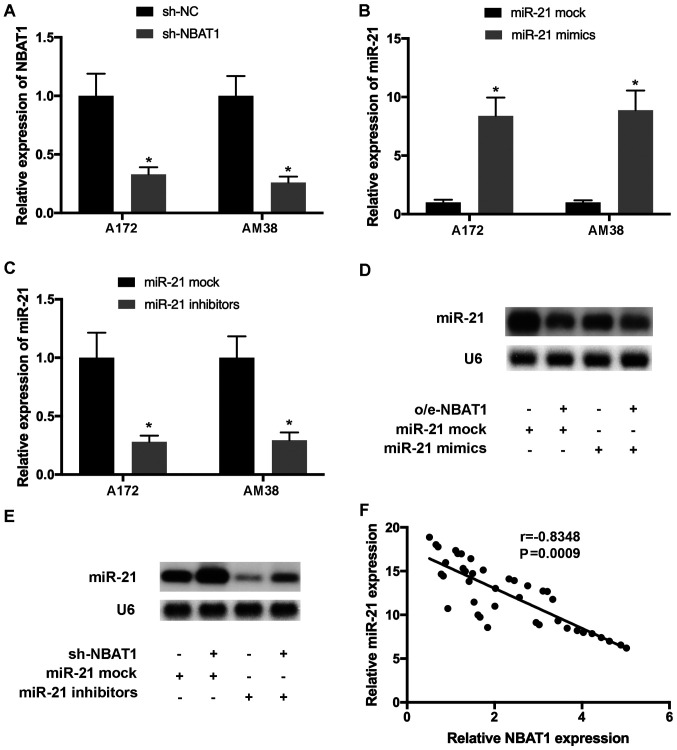
To further investigate the effects of NBAT1 on miR-21 expression, glioma cells were transfected with o/e-NBAT1 and miR-21 mimics, or with sh-NBAT1 and miR-21 inhibitors, and transfection efficiency was determined by RT-qPCR (Fig. 4A-C). Northern blot analysis demonstrated that upregulation of miR-21 expression by transfection with miR-21 mimics was reversed following the addition of o/e-NBAT1 (Fig. 4D). Similarly, downregulation of miR-21 expression by transfection with miR-21 inhibitors was counteracted following co-transfection with sh-NBAT1 (Fig. 4E). Pearson's correlation analysis demonstrated that the levels of miR-21 and NBAT1 were negatively correlated in glioma tissues (Fig. 4F).
Figure 4.
miR-21 expression is downregulated by NBAT1. A172 and AM38 cells were transfected with (A) sh-NBAT1, (B) miR-21 mimics or (C) miR-21 inhibitors. (D) Upregulated miR-21 expression in glioma cells transfected with miR-21 mimics was reversed following transfection with o/e-NBAT1. (E) Downregulated miR-21 expression in glioma cells transfected with miR-21 inhibitors was counteracted following transfection with sh-NBAT1. (F) Pearson's correlation analysis demonstrated that the levels of miR-21 and NBAT1 were inversely correlated in glioma tissues (r=−0.8348; P=0.0009). *P<0.05 vs. mock control. miR, microRNA; sh, short hairpin; NC, negative control; o/e, overexpression vector.
miR-21 serves important roles in the proliferation and metastasis of glioma cells
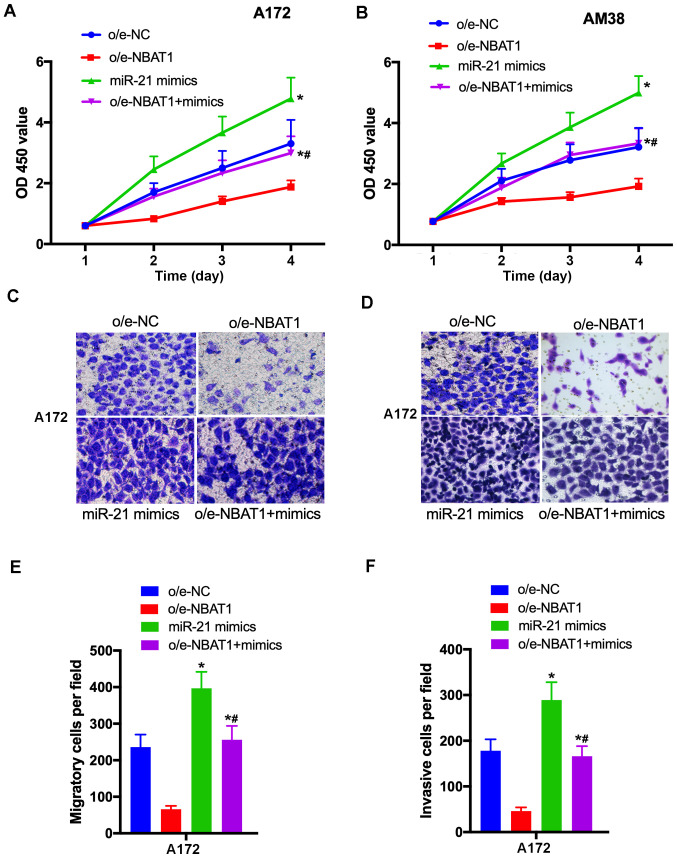
The effects of miR-21 on the progression of glioma were further assessed. The CCK-8 assay demonstrated that the proliferative ability of glioma cells significantly increased following transfection with the miR-21 mimics compared with the vehicle only control (P<0.05; Fig. 5A and B). In addition, the migratory and invasive abilities of glioma cells were enhanced following transfection with the miR-21 mimics compared with the vehicle only control (P<0.05; Fig. 5C-F). Taken together, these results suggest that miR-21 promoted glioma cell proliferation, migration and invasion.
Figure 5.
miR-21 promotes glioma cell proliferation, migration and invasion. (A and B) The proliferative ability of A172 and AM38 cells transfected with o/e-NC, o/e-NBAT1, miR-21 mimics or co-transfected with o/e-NBAT1 and miR-21 mimics was assessed via the Cell Counting Kit-8 assay. (C and E) The migratory ability of glioma cells transfected with o/e-NC, o/e-NBAT1 or the miR-21 mimics or co-transfected with o/e-NBAT1 and the miR-21 mimics was determined by the Transwell assay. (D and F) The invasive ability of A172 cells increased following transfection with the miR-21 mimics. *P<0.05 vs. o/e-NC; #P<0.05 vs. o/e-NBAT1. miR, microRNA; o/e, overexpression vector; NC, negative control; OD, optical density.
SOX7 is a downstream target of miR-21
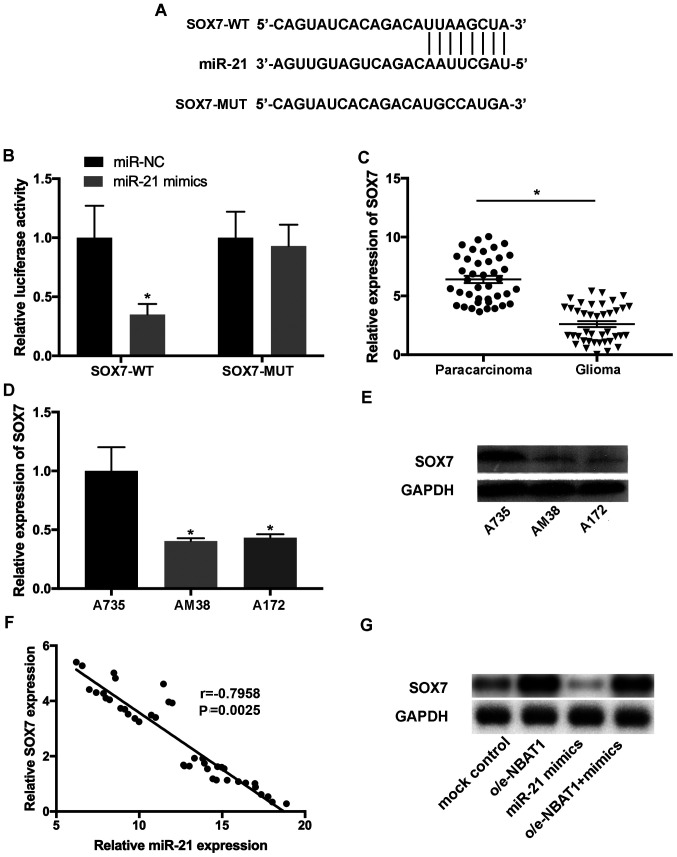
To identify the putative targets of miR-21, the complementary sequence of miR-21 in SOX7 transcripts was predicted (Fig. 6A). The interaction between SOX7 and miR-21 was confirmed using a dual-luciferase assay. Luciferase reporter plasmids containing SOX7-WT) and SOX7-MUT fragments of the predicted miR-21 binding sites were constructed. The results demonstrated that the miR-21 mimics significantly decreased the activity of the luciferase vector containing the SOX7-WT sequence, but not the mutant control (Fig. 6B). RT-qPCR analysis indicated that SOX7 expression was downregulated in glioma tissues compared with that in the paracarcinoma tissues (Fig. 6C). In addition, downregulated SOX7 expression was observed in A172 and AM38 cells compared with that in A735 cells (P<0.05; Fig. 6D and E). Pearson's correlation analysis demonstrated that the levels of SOX7 and miR-21 were inversely correlated in glioma samples (Fig. 6F).
Figure 6.
SOX7 is a potential target of miR-21 in glioma. (A) The complementary sequence of miR-21 in SOX7 transcripts was predicted. (B) The interaction between SOX7 and miR-21 was validated using the dual-luciferase reporter assay. (C) SOX7 expression was downregulated in glioma compared with paracarcinoma tissues. (D) The mRNA levels of SOX7 decreased in A172 and AM38 cells compared with those in A735 normal astrocytes. (E) The protein levels of SOX7 decreased in glioma cell lines. (F) Pearson's correlation analysis demonstrated that the levels of SOX7 and miR-21 were inversely correlated in glioma tissues (r=−0.7958; P=0.0025). (G) SOX7 expression was downregulated following transfection with miR-21 mimics and upregulated following overexpression of NBAT1 in glioma cells. *P<0.05 vs. normal control. miR, microRNA; WT, wild-type; MUT, mutant; NC, negative control; o/e, overexpression vector.
To further investigate the effects of SOX7 on the progression of glioma, the expression levels of SOX7 were assessed in glioma cells transfected with control, o/e-NBAT1 or the miR-21 mimics, or co-transfected with o/e-NBAT1 and the miR-21 mimics. SOX7 expression significantly decreased following transfection with the miR-21 mimics compared with that in the control group, which was reversed following overexpression of NBAT1 (Fig. 6G). Collectively, these results suggested that SOX7 may be a novel target of miR-21, and that the NBAT1/miR-21/SOX7 signaling pathway may be involved in the progression of glioma.
Discussion
lncRNAs are a novel class of non-protein coding RNAs that are >200 nt in length (4). Previous studies on lncRNAs have reported their importance on the pathogenesis of cancer, and increasing evidence indicates that lncRNAs act as key factors in the regulation of cancer cells (25–29). lncRNAs function as oncogenes or tumor suppressors in cancer, and aberrant expression levels of lncRNAs are associated with tumor development (31). For example, lncRNA PVT1 modulates the cell proliferation, invasion and tumor growth in orthotopic xenografts of breast cancer (32). In addition, upregulated LINC01116 expression is associated with unfavorable overall survival and metastasis of patients with breast cancer (33). The expression levels of lncRNA PlncRNA-1 are downregulated in cancer tissues; however, restored PlncRNA-1 expression inhibits proliferation and promotes apoptosis of breast cancer cells via the TGF-β1/phosphoglycerate dehydrogenase axis (34). Furthermore, downregulation of lncRNA SNHG5 suppresses proliferation and migration of gastric cancer cells via the miR-32/Kruppel-like factor 4 axis (35). These findings suggest that lncRNAs are key regulators during the initiation and progression of cancer. Consistent with previous findings, the results of the present study demonstrated the key roles of NBAT1 on the proliferation and metastasis of glioma cells.
A number of lncRNAs can regulate gene expression by interacting with their target miRNAs directly. For example, lncRNA BC032469 binds to miR-1207-5p and human telomerase reverse transcriptase, and induces cancer cell proliferation (36). Furthermore, lncRNA H19 regulates the proliferation, migration and invasion of gastric cancer cells through its target miRNAs, such as miR-675 (37,38). However, the functions and underlying molecular mechanisms of lncRNAs remain largely unknown and require further investigation. Similarly, the present study demonstrated the regulatory function of miR-21 during the proliferation, migration and invasion of glioma cells, and its underlying molecular mechanisms were elucidated.
Increasing evidence suggests that aberrant NBAT1 expression is associated with various types of cancer, such as ovarian cancer, breast cancer and osteosarcoma (19,21,22). NBAT1 inhibits the proliferation and promotes apoptosis of ovarian cancer cells (19). NBAT1 regulates the migration and invasion of breast cancer cells by targeting Dickkopf Wnt signaling pathway inhibitor 1/polycomb repressive complex 2 signaling (21). A previous study has reported that miR-21 is a potential target of NBAT1 in osteosarcoma cells (22). NBAT1 has also been demonstrated to be associated with the proliferation and metastasis of glioma cells (20). Consistent with previous findings, the results of the present study demonstrated that NBAT1 was downregulated in glioma tissues and cells. Furthermore, overexpression of NBAT1 suppressed glioma cell proliferation, migration and invasion. However, the effects of sh-NBAT1 on the biological behavior of glioma cells were not investigated in the present study.
miRNAs may function as oncogenes or tumor suppressors by modulating gene expression, and are considered to be targets of lncRNAs (24,25). Previous studies have reported that the expression levels of miRNAs are impaired in several types of cancer, including glioma, and miR-140 and miR-152 interact with lncRNAs in glioma cells (26–29). The present study aimed to identify novel targets of NBAT1 in glioma. The results of the present study suggested that NBAT1 was able to bind with miR-21, the expression levels of which were notably upregulated in glioma tissues and cells. Additionally, the expression of miR-21 was reduced by NBAT1 in glioma cells, and their expression levels were inversely correlated in glioma tissues. Inhibition of miR-21 suppressed the proliferation, migration and invasion of glioma cells. The results of the present study were consistent with a previous study, which suggested that NBAT1 inhibited the progression of osteosarcoma via miR-21 signaling (22).
SOX7 is a member of the SOX transcription factors (39) and is considered to be an essential regulator in cardiovascular development (40). SOX7 functions as a tumor suppressor in endometrial cancer and as an oncogene in gastric cancer (41–44). The positive association between SOX7 expression and distant metastasis-free survival has been reported in prostate cancer and breast cancer (45,46). In the present study, SOX7 was identified as a downstream target of miR-21, and the results demonstrated that SOX7 expression levels were decreased in glioma tissues and cells compared with those in paracarcinoma tissues and normal astrocytes, respectively. Of note, downregulated SOX7 expression induced by transfection with miR-21 mimics was reversed following overexpression of NBAT1. Collectively, these results highlighted the key functions of NBAT1/miR-21/SOX7 signaling on the progression of glioma. A major limitation of the present study is that in vivo analysis was not performed, thus prospective studies will focus on performing in vivo experiments to validate these findings. Furthermore, the expression levels of proliferation- and invasion-associated molecules should be investigated to confirm the existing findings.
In conclusion, the results of the present study suggested that NBAT1 may act as a potential tumor suppressor by upregulating SOX7 via suppressing miR-21, thus inhibiting glioma cell proliferation, migration and invasion. These results indicate the essential roles of NBAT1 and its underlying molecular mechanisms in the proliferation, migration and invasion of glioma cells, providing evidence on the potential functions of NBAT1 in tumorigenesis, and suggest that the NBAT1/miR-21/SOX7 axis may be a promising therapeutic target for the treatment of patients with glioma.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Outstanding Talent Cultivation Foundation of Liaoning Province (grant no. 2015020349), the President Foundation of Jinzhou Medical University (grant no. XZJJ20130206) and the Natural Science Foundation of Liaoning Province (grant no. 2019-ZD-0824).
Availability of data and materials
The datasets generated or analyzed during the present study are included in this published article.
Authors' contributions
WG conceived and designed the present study. NG, RW, XF and CL performed the experiments and interpreted the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of The First Affiliated Hospital of Jinzhou Medical University (approval no. JYD159302; Jinzhou, China). Written informed consent was provided by all patients for the use of clinical tissue prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112:1241–1246. doi: 10.1038/bjc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansouri A, Mansouri S, Hachem LD, Klironomos G, Vogelbaum MA, Bernstein M, Zadeh G. The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: A systematic review. Cancer. 2016;122:2469–2478. doi: 10.1002/cncr.30088. [DOI] [PubMed] [Google Scholar]
- 3.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: A review on the treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 4.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills JD, Chen J, Kim WS, Waters PD, Prabowo AS, Aronica E, Halliday GM, Janitz M. Long intervening non-coding RNA 00320 is human brain-specific and highly expressed in the cortical white matter. Neurogenetics. 2015;16:201–213. doi: 10.1007/s10048-015-0445-1. [DOI] [PubMed] [Google Scholar]
- 6.Hsiao J, Yuan TY, Tsai MS, Lu CY, Lin YC, Lee ML, Lin SW, Chang FC, Liu Pimentel H, Olive C, et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of dravet syndrome. EBioMedicine. 2016;9:257–277. doi: 10.1016/j.ebiom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Dong M, Fan D, Hou P, Li H, Liu L, Lin C, Liu J, Su L, Wu L, et al. lncRNA ANCR down-regulation promotes TGF-β-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329–67343. doi: 10.18632/oncotarget.18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Grzmil M, Morin P, Jr, Lino MM, Merlo A, Frank S, Wang Y, Moncayo G, Hemmings BA. MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human glioblastoma. Cancer Res. 2011;71:2392–2402. doi: 10.1158/0008-5472.CAN-10-3112. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 11.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, et al. lncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, Lui WM, Fung CF, Wong TS, Leung GK. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Zhi F, Wang Q, Xue L, Shao N, Wang R, Deng D, Wang S, Xia X, Yang Y. The use of three long non-coding RNAs as potential prognostic indicators of astrocytoma. PLoS One. 2015;10:e0135242. doi: 10.1371/journal.pone.0135242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vital AL, Tabernero MD, Castrillo A, Rebelo O, Tão H, Gomes F, Nieto AB, Resende Oliveira C, Lopes MC, Orfao A. Gene expression profiles of human glioblastomas are associated with both tumor cytogenetics and histopathology. Neuro Oncol. 2010;12:991–1003. doi: 10.1093/neuonc/noq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang X, Huang Y, Wang Y, Lu Y, Fu D, Chen J. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1) PLoS One. 2013;8:e68469. doi: 10.1371/journal.pone.0068469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6:537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amit D, Matouk IJ, Lavon I, Birman T, Galula J, Abu-Lail R, Schneider T, Siegal T, Hochberg A, Fellig Y. Transcriptional targeting of glioblastoma by diphtheria toxin-A driven by both H19 and IGF2-P4 promoters. Int J Clin Exp Med. 2012;5:124–135. [PMC free article] [PubMed] [Google Scholar]
- 18.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Yan C, Jiang Y, Wan Y, Zhang L, Liu J, Zhou S, Cheng W. Long noncoding RNA NBAT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer. Onco Targets Ther. 2017;10:1993–2002. doi: 10.2147/OTT.S124645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li J, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–32425. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, Wang G, Yang J, Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am J Cancer Res. 2017;7:2009–2019. [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey GK, Kanduri C. Fighting neuroblastomas with NBAT1. Oncoscience. 2015;2:79–80. doi: 10.18632/oncoscience.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie H, Fu JL, Xie C. miR-138-5p is downregulated in patients with atrial fibrillation and reverses cardiac fibrotic remodeling via repressing CYP11B2. Eur Rev Med Pharmacol Sci. 2018;22:4642–4647. doi: 10.26355/eurrev_201807_15523. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Sun J, Chen Y, Su W, Shan H, Li Y, Wang Y, Zheng N, Shan H, Liang H. lncRNA PFAR promotes lung fibroblast activation and fibrosis by targeting miR-138 to regulate the YAP1-Twist axis. Mol Ther. 2018;26:2206–2217. doi: 10.1016/j.ymthe.2018.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan TB, Robert LC, Teebagy PA, Morgan SE, Beatty EW, Cicuto BJ, Nowd PK, Rieger-Christ KM, Bryan DJ. Spatiotemporal microRNA profile in peripheral nerve regeneration: miR-138 targets vimentin and inhibits Schwann cell migration and proliferation. Neural Regen Res. 2018;13:1253–1262. doi: 10.4103/1673-5374.235073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Z, Feng Q, Xu L, Li S, Zhou L. CREPT regulated by miR-138 promotes breast cancer progression. Biochem Biophys Res Commun. 2017;493:263–269. doi: 10.1016/j.bbrc.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, Peng G, Liao Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. doi: 10.1016/j.abb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018 Feb 8; doi: 10.3727/096504018X15178768577951. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, Guan GH, Liu Q, Qian YH, Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 32.Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W, Feng H. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723–4734. doi: 10.1038/s41388-018-0310-4. [DOI] [PubMed] [Google Scholar]
- 33.Hu HB, Chen Q, Ding SQ. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1987–1993. doi: 10.26355/eurrev_201804_14726. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Gao H, Zhou S, Liao Y. lncRNA PlncRNA-1 overexpression inhibits the growth of breast cancer by upregulating TGF-β1 and downregulating PHGDH. Breast Cancer. 2018;25:619–625. doi: 10.1007/s12282-018-0858-4. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 36.Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 37.Yan J, Zhang Y, She Q, Li X, Peng L, Wang X, Liu S, Shen X, Zhang W, Dong Y, et al. Long noncoding RNA H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase 3 signaling pathway. Cell Physiol Biochem. 2017;42:2364–2376. doi: 10.1159/000480028. [DOI] [PubMed] [Google Scholar]
- 38.Li P, Tong L, Song Y, Sun J, Shi J, Wu Z, Diao Y, Li Y, Wang Z. Long noncoding RNA H19 participates in metformin-mediated inhibition of gastric cancer cell invasion. J Cell Physiol. 2019;234:4515–4527. doi: 10.1002/jcp.27269. [DOI] [PubMed] [Google Scholar]
- 39.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 40.Francois M, Koopman P, Beltrame M. Soxf genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol. 2010;42:445–448. doi: 10.1016/j.biocel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Katoh M. Expression of human SOX7 in normal tissues and tumors. Int J Mol Med. 2002;9:363–368. [PubMed] [Google Scholar]
- 42.Stovall DB, Cao P, Sui G. SOX7: From a developmental regulator to an emerging tumor suppressor. Histol Histopathol. 2014;29:439–445. doi: 10.14670/hh-29.10.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3:1546–1556. doi: 10.18632/oncotarget.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui J, Xi H, Cai A, Bian S, Wei B, Chen L. Decreased expression of Sox7 correlates with the upregulation of the Wnt/β-catenin signaling pathway and the poor survival of gastric cancer patients. Int J Mol Med. 2014;34:197–204. doi: 10.3892/ijmm.2014.1759. [DOI] [PubMed] [Google Scholar]
- 45.Zhong WD, Qin GQ, Dai QS, Han ZD, Chen SM, Ling XH, Fu X, Cai C, Chen JH, Chen XB, et al. SOXs in human prostate cancer: Implication as progression and prognosis factors. BMC Cancer. 2012;12:248. doi: 10.1186/1471-2407-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stovall DB, Wan M, Miller LD, Cao P, Maglic D, Zhang Q, Stampfer MR, Liu W, Xu J, Sui G. The regulation of SOX7 and its tumor suppressive role in breast cancer. Am J Pathol. 2013;183:1645–1653. doi: 10.1016/j.ajpath.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated or analyzed during the present study are included in this published article.