Abstract
X-linked ubiquitin-specific peptidase 9 (USP9X) serves important roles in the development and progression of various human cancers. However, its role and molecular mechanism in liver cancer require further elucidation. In the present study, USP9X was found to be upregulated in liver cancer tissues. At the same time, overexpression of USP9X promoted the proliferation, invasiveness and migration of liver cancer cells, which were subsequently suppressed by USP9X silencing. On a molecular level, the results revealed that USP9X knockdown suppressed elements of the Janus kinase 2 (JAK2)/STAT3 signaling pathway, including JAK2, STAT3, matrix metalloproteinase-2 and c-Myc. By contrast, overexpression of USP9X had the opposite effect. In conclusion, the results of the present study suggest that USP9X is upregulated in patients with liver cancer, which may accelerate the proliferation, invasiveness and migration of liver cancer cells by regulating the JAK2/STAT3 signaling pathway.
Keywords: liver cancer, X-linked ubiquitin-specific peptidase 9, Janus kinase 2, STAT3
Introduction
Liver cancer is a common malignant tumor, which due to its high metastatic potential, has become the primary cause of cancer-related death (1,2). although great progress has been made in the diagnosis and treatment of liver cancer, patient prognosis is still unsatisfactory (3–5). Thus, exploring its potential pathological mechanism is particularly important to find new strategies that can improve treatment of patients with liver cancer.
X-linked ubiquitin-specific peptidase 9 (USP9X) is a member of the deubiquitinase family that has been shown to be involved in a variety of biological pathways, such as regulating cell transformation and survivalf6,7). Biochemical studies have identified that USP9X is widely expressed in tissues, which has been confirmed in TCGA database, and it can regulate the protein function by the deubiquitination pathway (8–14). USP9X plays important roles in the development and progression of several human cancers (15). For example, the USP9X protein is significantly increased in non-small cell lung cancer, breast cancer, leukemia, cervical cancer, follicular lymphoma, colon cancer and esophageal squamous cell carcinoma (8–14). Overexpression of USP9X in cancer activates multiple important pathways, such as the Ras/mitogen-activated protein kinase, PI3K/AKT, Rho/ Rho-associated protein kinase, Jagged/Notch, NK-κB and Wnt/β-catenin pathways (16). In addition, USP9X plays an important role in promoting liver cancer cell proliferation, but low USP9X expression can inhibit the bioactivity of these cells (17). This indicates that the protein stability of USP9X plays an important role in regulating the proliferation and cell activity of liver cancer cells. However, the molecular mechanism by which USP9X regulates the proliferation, invasiveness and migration of liver cancer cells is remains to be clarified.
The Janus kinase 2 (JAK2)/STAT3 pathway plays an important part in several cellular activities, including processes involved with the cytoplasmic domains of several cytokine receptors and mediating signals triggered by several hematopoietic growth factors, including erythropoietin (Epo), thrombopoietin (Tpo), and granulocyte colony-stimulating factor (G-CSF) (18,19). This pathway can regulate cytokines, adhesion molecules and other inflammatory mediators (18,20,21), and transduce signals from activated receptors or intracellular kinases to the nucleus to activate and regulate gene transcription (22). JAK2/STAT3 signaling also serves a critical role in tumor cell proliferation, invasiveness and migration (23,24), where the oncogene STAT3 is a key factor (25). Several studies have shown that STAT3 activation can improve the expression of c-Myc, which promotes cellular proliferation (26). On the other hand, the STAT3 activation is associated with the upregulation of matrix metalloproteinase-2 (MMP2), and further promote tumor invasive growth (27–31).
In the present study, the expression of USP9X in liver cancer tissues, and its role in the proliferation, invasiveness and migration of liver cancer cells were investigated to provide novel insight into the role of USP9X in liver cancer.
Materials and methods
Tissue samples
A total of 14 patient samples were collected from seven liver cancer patients and seven patients without liver cancer at the No. 1 People's Hospital of Xuzhou, (Xuzhou, China). Part of the surgically removed tissues were used for histological diagnosis, and remainder was immediately frozen in liquid nitrogen and stored at −80°C. All specimens were pathologically diagnosed. The present study was approved by the Research Ethics Committee of the No. 1 People's Hospital of Xuzhou and written informed consent was obtained from each patient.
Cell culture
The 293T cell line and the HepG2 liver cancer cell line were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai. All cells were identified by STR profiling. Cells were cultured in DMEM (Hyclone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Invigentech) in a 5% CO2 incubator at 37°C. The inhibitor of JAK2 (Fedratinib, http://www.medchemexpress.cn/tg-101348.html?src=bd-product) was added 24 h post-transfection at a concentration of 10 µg/ml, and the cells were collected 6 h later.
Lentivirus production
The short hairpin (sh) RNA USP9X plasmid and the associated shControl (which contained a non-targeting oligomers and doesn't affect the wild-type USP9X) were purchased from Shanghai GenePharma Co., Ltd. shUSP9X sequence: 5′-CCGGCCACCTCAAACCAAGGATCAACTCGAGTTGATCCTTGGTTTGAGGTGGTTTTT-3′; shControl sequence: 5′-GATCTTCTCCGAACGTGT- CACGTTTCAAGAGAACGTGACACGTTCGGAGAAT TTTTTG-3′. Transfection was performed with PolyJet transfection reagent (SignaGen Laboratories) according to the manufacturer's instructions. The lentiviruses were produced by co-transfecting 293T cells with both the core and the packaging plasmids. The lentivirus was collected after incubation at 37°C for 72 h.
Establishment of stable cell lines
The plasmids of control and shUSP9X were GFP-tagged. Moderate amounts (1 ml) of lentiviruses were used to infect HepG2 cells (5×105 cells/well in 6-well plate) and then cultured at 37°C for 72 h. Cells were continuously cultured in the DMEM medium containing 2.5 µg/ml puromycin (Sigma Aldrich; Merck KGaA). The surviving cells were cultured into cell lines stably expressing control and shUSP9X. Cell lines stably expressing the control or shUSP9X constructs were confirmed by western blot analysis.
Transient overexpression of USP9X
The Myc-USP9X was generated by cloning the human USP9X cDNA into the expression vector pcDNA3.1-Myc at the BamHI and XhoI restriction sites (Youbio, http://www.youbio.cn). An empty carrier containing the Myc tag (Myc-Vector) was used as a negative control (Youbio). The final concentration of all plasmids was ~1 mg/ml. Transfection of USP9X was performed using PolyJet transfection reagent (SignaGen Laboratories, http://signagen.com) according to the manufacturer's instructions and then incubated at 37°C for 24 h. Subsequent experiments were performed 24 h post-transfection. The efficiency of overexpression was verified by western blot analysis.
5-ethynyl-20-deoxyuridine (EdU) incorporation assay
The effect of USP9X on the proliferation of HepG2 cells was measured by EdU incorporation using an EdU assay kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's instructions. Briefly, a total of 8×104 cells/well were cultured in 48-well plates for 24 h, followed by a 2-h incubation with 50 µmol/l EdU at 37°C. Cells were then washed with PBS and fixed with 4% paraformaldehyde for 30 min, followed by permeabilization with 0.5% Triton X-100 for a further 30 min. Afterwards, the cells were washed five times with PBS and incubated with 100 µl 1X Apollo® reaction cocktail (apart of the EdU kit) for 30 min at room temperature. Finally, the cell nuclei were dyed with 100 μl of Hoechst solution (5 µg/ml) for 30 min and visualized with a fluorescent microscope (IX71; Olympus, Corporation). The results are analyzed through Image J v1.8.0.
Cell viability assay
Cell viability was assessed with the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.). A total of 1×104 cells/well were cultured in 96-well plates and at the designated time point, 10 µl of CCK-8 reagent was added into the medium. After 4 h at 37°C, the absorbances were measured at 450 nm using a Synergy Mx Multi-Mode Microplate Reader. The cell viability was calculated according to the absorbances. The absorbances were normalized against a DMSO control.
Colony formation assay
A total of 300 cells were seeded into 6-cm dishes and cultured for 15 days at 37°C. Cells were fixed with methanol at room temperature for 1 h and stained with 0.1% crystal violet at room temperature for 30 min to assess the number of colonies. Cells were washed twice with PBS and the plate images were captured using a digital camera (Alpha 7 II, SONY, http://www.sonystyle.com.cn/products/ilc/ilce_7m2/ilce_7m2.html). Positive colony formation, defined as colonies with >50 cells, was confirmed by manual counting.
Transwell invasion assay
Transwell assays were performed with a polycarbonate filter membrane with a diameter of 6.5 mm and pore size of 8 µm (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. To assess invasion ability, the filters were pre-coated with 10 µg Matrigel. A cell suspension of 3×104 cells (in each group) in serum-free culture media was added into the inserts, and each insert was placed into the lower chamber filled with culture media containing 10% FBS as a chemoattractant. After 48 h in culture at 37°C, the non-invasive cells were removed from the upper chamber with cotton-tipped swabs, and the filters were fixed with methanol for 30 min and stained with a 0.1% crystal violet solution for a further 30 min at room temperature. Finally, five fields of adherent cells in each well were randomly selected and counted with an inverted microscope and counted.
Wound healing assay
Cells (5×106) cultured in DMEM supplemented with 10% fetal bovine serum were seeded into 6-well plates and incubated in a 5% CO2 incubator at 37°C for 12 h. Then, scratches were made with a pipette tip in the middle of the wells, and the cells were washed with PBS to remove cell debris before incubation in serum-free media for 50 h. At the designated time, five randomly selected fields were imaged with an inverted microscope. The number of cells across the wound was normalized to that of the control group. The results were analyzed using Image J v1.8.0 (National Institutes of Health).
Western blotting
At the designated times, the cells were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology). The protein concentration was determined using the BCA kit (cat. no. P0010; Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Equal amounts of protein lysate (80 µg) were resolved using 10% SDS-PAGE. The proteins were then transferred onto PVDF membranes and blocked with 3% BSA at room temperature for 2 h. Membrane were incubated with primary antibodies at 4°C overnight, and subsequently incubated with HRP goat anti mouse/rabbit IgG secondar antibody (1:4,000; cat. nos. sa00001-1 and sa00001-2; ProteinTech Group, Inc.) at room temperature for 2 h. 1X TBST was used for washing. The antibodies used were against: USP9X [1:1,000; cat. no. 14898; Cell Signaling Technology (CST), Inc.], phosphorylated (Tyr1007/1008)-JAK2 (1:1,000; cat. no. 3771, CST, Inc.), p-STAT3 (Tyr705; 1:1,000; cat. no. 9145; CST, Inc.), JAK2 (1:1,000; cat. no. 17670-1-AP; ProteinTech Group, Inc.), STAT3 (1:1,000, 10253-2-AP, ProteinTech Group, Inc.), MMP2 (1:1,000; cat. no. 10373-2-AP; ProteinTech Group, Inc.), c-Myc (1:1,000; cat. no. 10828-1-AP; ProteinTech Group, Inc.), β-actin (1:5,000; cat. no. 60008-1-Ig; ProteinTech Group, Inc.) and Myc-tag (1:1,000; cat. no. 16286-1-AP; ProteinTech Group, Inc.). Bound antibodies were detected with ECL Plus Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.) and exposed to X-ray films. Band densities were quantified using ImageJ software (version 1.8.0; National Institutes of Health). The relative amount of protein was determined by normalizing the densitometry value of interest to that of the loading control. Relative protein expression was determined by normalizing the densitometry value of target protein bands and corresponding internal parameters.
Immunohistochemistry
Immunohistochemical staining was performed using the procedures provided by the S-P immunohistochemical kit (OriGene Technologies, Inc.). The sections were fixed with 4% paraformaldehyde for 3 days and sealed with 10% goat serum (Vicmed, http://www.vicmed.cn/a/dbyjxgcp/myzhxg/399.html) for 30 min at room temperature. The sections were then incubated with USP9X antibody (1:100; cat. no. 14898; CST, Inc.) at 4°C overnight, followed by incubation with biotin-conjugated goat anti-rabbit IgG (original concentration; cat. no. PV-9001) and HRP-conjugated streptavidin (1:20; cat. no. ZLI-9018; both from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.). The reaction was developed using 3,3′diaminobenzidine reagent (OriGene Technologies, Inc.). Sections were stained with hematoxylin to stain the nuclei for 2 min and dehydrated by incubation in increasing concentrations of ethanol (50, 75, 80, 90 and 100%) followed by 100% xylene for 5 min. All steps were performed at room temperature. The slides were observed under the Olympus BX-53 light microscope (magnification, ×40; Olympus, Corporation), and images processed using Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Survival analysis
The association between USP9X and survival of patients with liver cancer was assessed using the GEPIA database (http://gepia.cancer-pku.cn/) in order to understand the clinical significance of USP9X.
Statistical analysis
The results are representative of ≥3 experimental repeats. All quantitative data are presented as the mean ± SEM. Statistical analysis was performed using SPSS (version 16.0; SPSS, Inc.). One-way analysis of variance followed by Tukey's post hoc test were used to compare differences between multiple groups. Paired Student's t-test was used to compare differences between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
USP9X is highly expressed in human liver cancer tissues
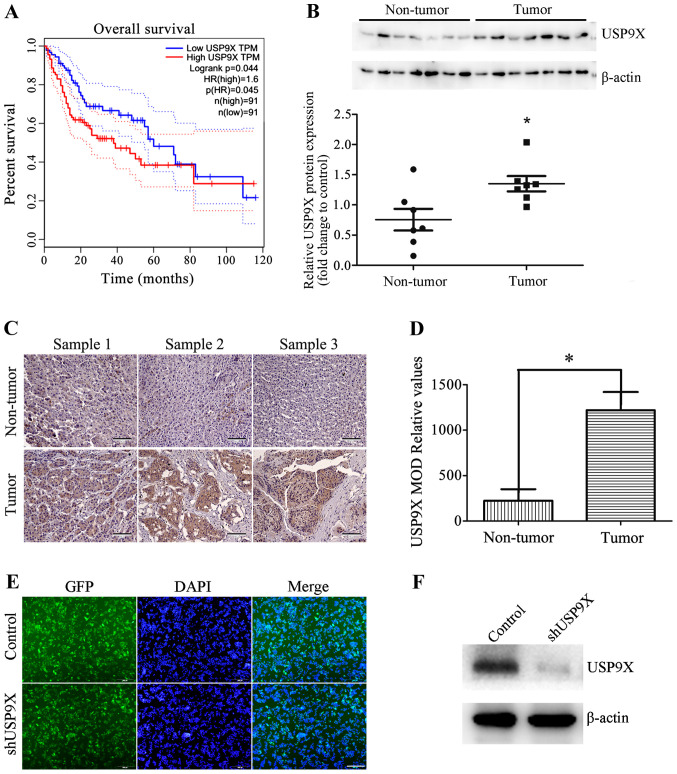
To understand the clinical significance of USP9X, the association between USP9X protein expression levels and the survival of patients with liver cancer was analyzed using The Cancer Genome Atlas database. The results show that high levels of USP9X is associated with poor prognosis in patients with liver cancer (Fig. 1A). The USP9X protein expression level of seven non-tumorous liver tissues and seven liver cancer tissues were also examined by western blot analysis. The results indicate that USP9X is expressed to a greater degree in liver cancer tissues than non-tumorous tissues (Fig. 1B). These results were also consistent with immunohistochemical analysis (Fig. 1C and D). These results revealed that USP9X may play an important role in the development of liver cancer.
Figure 1.
Expression of USP9X in patients with liver cancer. (A) The Cancer Genome Atlas database analysis showed that high expression levels of USP9X were associated with poor prognosis in patients with liver cancer. (B) Representative western blots and quantification data show that the protein expression levels of USP9X were higher in liver cancer tissues than in non-tumor liver tissues. (C) Representative images of USP9X expression innormal liver tissues and liver cancer tissues determined by immunohistochemistry. Scale bar, 100 µm. (D) Histogram showing the quantification of the relative levels of USP9X from immunohistochemistry analysis. (E) GFP images show that shUSP9X and the control were successfully transfected into HepG2 cells, generating stable USP9X-knockdown cell lines. Scale bar, 500 µm. (F) Silencing efficiency of shUSP9X in HepG2 cells was examined by western blot. *P<0.05. GFP, green-fluorescent protein; MOD, mean optical density; sh, short hairpin; USP9X, X-linked ubiquitin-specific peptidase 9; TPM, total productive maintenance; p(HR), proportion of high resistance; n, number.
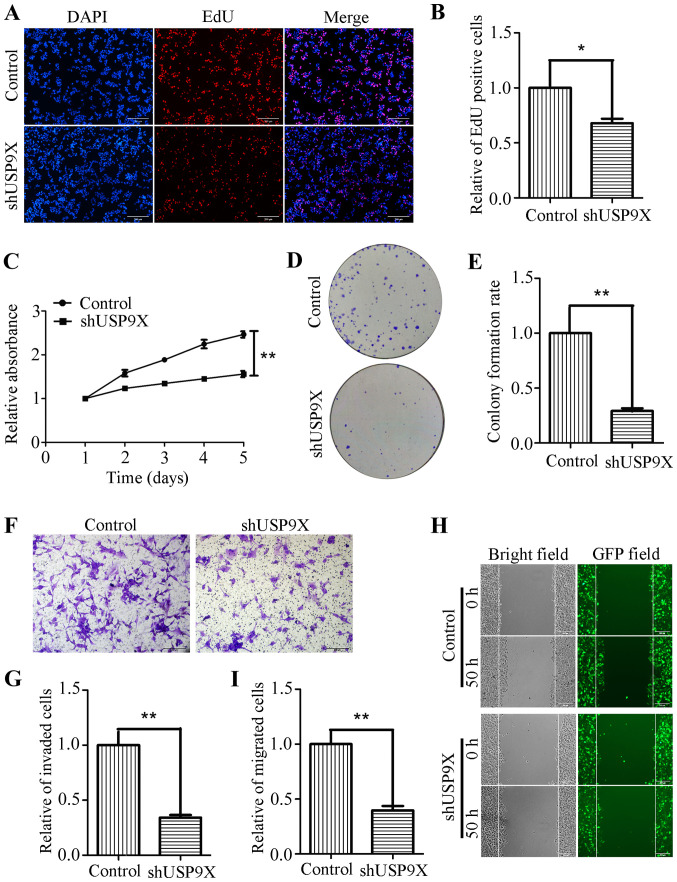
Silencing of USP9X inhibits the proliferation, invasiveness and migration of human liver cancer cells
Due to the increased expression of USP9X in liver cancer tissues, it was hypothesized that USP9X might be associated the proliferation, invasiveness and migration of liver cancer cells. To address this problem, USP9X expression was silenced using shRNA. shUSP9X and the respective control lentiviruses were packaged into 293T cells before being transfection into the HepG2 cell line to successfully generate stable cell lines with loss of USP9X (Fig. 1E and F). To determine whether USP9X-knockdown could affect the proliferation, invasion and migration of liver cancer cells, a number of functional experiments were performed. The EdU incorporation assay, showed that the number of EdU-positive cells was markedly reduced in HepG2 cells transfected with shUSP9X, compared with the control (Fig. 2A and B). In addition, the CCK-8 assay revealed that the proliferation rate of the shUSP9X group was also significantly decreased compared with the control (Fig. 2C). The ability of HepG2 cells to form colonies was also significantly decreased upon silencing of USP9X compared to the control group (Fig. 2D and E). Furthermore, Transwell assays (with Matrigel coating) showed that cell invasiveness was inhibited in HepG2 cells transfected with shUSP9X (Fig. 2F and G). The wound healing assay indicated that, compared with the control group, the number of migrated cells at 50 h was significantly decreased in shUSP9X HepG2 cells compared to the control group (Fig. 2H and I). These results show that silencing of USP9X significantly inhibits the proliferation, invasiveness and migration of liver cancer cells.
Figure 2.
Downregulation of USP9X inhibits liver cancer cell proliferation, invasiveness and migration. (A) Images and (B) statistical analysis of the EdU incorporation assay while silencing USP9X in HepG2 cells. Scale bar, 200 µm. (C) CCK-8 assay results. (D) Images and (E) statistical analysis of colony formation assays upon USP9X silencing in HepG2 cells. (F) Images and (G) statistical analysis of Transwell assay results (with Matrigel coating) after shUSP9X-knockdown in HepG2 cells. Scale bar, 200 µm. (H) Images and (I) statistical analysis of wound healing assays while silencing USP9X in HepG2 cells. Scale bar, 200 µm. *P<0.05 and **P<0.01. EdU, 5-ethynyl-20-deoxyuridine; GFP, green-fluorescent protein; sh, short hairpin; USP9X, X-linked ubiquitin-specific peptidase 9.
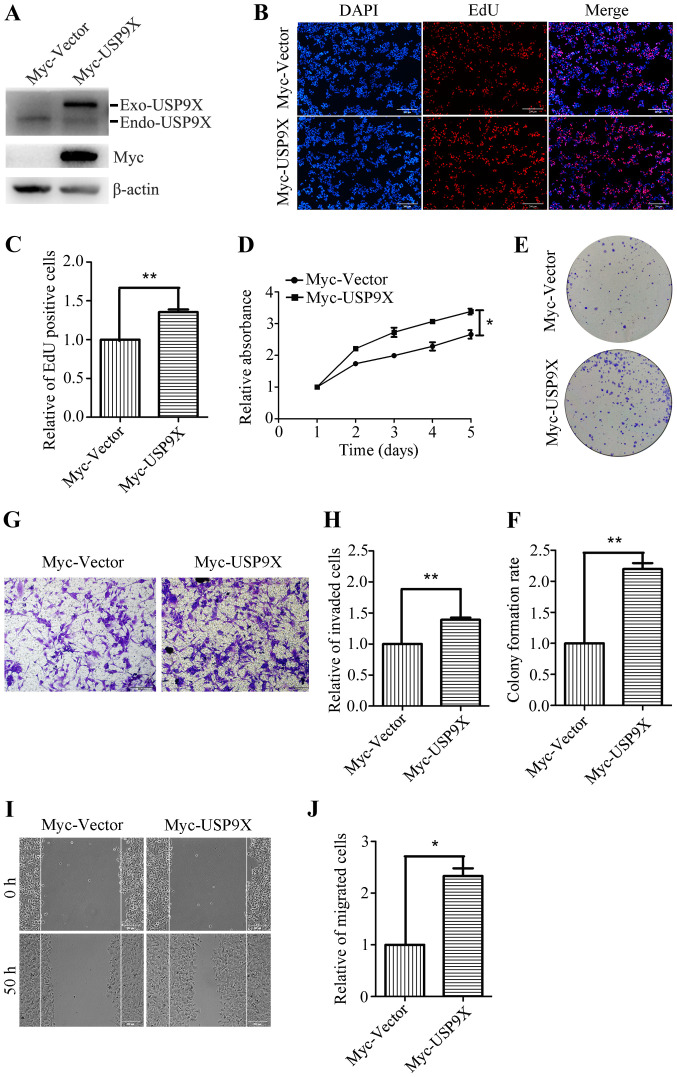
Overexpression of USP9X promotes the proliferation, invasion and migration of human liver cancer cells
To further investigate the role of USP9X in the proliferation and metastatic potential of human liver cancer cells, HepG2 cells were transiently transfected with Myc-Vector and Myc-USP9X plasmids. Transfection efficiency was confirmed by western blotting, and the Myc-tag antibody was used to detect the exogenous USP9X. (Fig. 3A). After 24 h of transfection, the effects of overexpression of USP9X on cell proliferation and metastasis were investigated. The EdU incorporation assay showed that the number of EdU positive cells was significantly increased in the USP9X overexpressing cells (Fig. 3B and C). The CCK-8 assay also revealed that the proliferation rate of the USP9X overexpressing cells was increased compared with the control (Fig. 3D). Moreover, the ability of USP9X overexpressing cells to form colonies was also significantly increased compared with the control cells (Fig. 3E and F). The Transwell assay (with Matrigel coating) showed that the number of invasive cells was increased by ~40% in USP9X overexpressing cells (Fig. 3G and H). Furthermore, the wound healing assay indicated that the number of migratory cells was notably increased upon Myc-USP9X transfection, compared with the control (Fig. 3I and J). These results reveal that USP9X overexpression significantly promotes the proliferation, invasiveness and migration of liver cancer cells.
Figure 3.
Overexpression of USP9X promotes liver cancer cell proliferation, invasiveness and migration. (A) Overexpression efficiency of Myc-USP9X in HepG2 cells was verified by western blot analysis. (B) Images and (C) statistical analysis of EdU incorporation assay in USP9X overexpressing HepG2 cells. Scale bar, 200 µm. (D) CCK-8 assay results. (E) Images and (F) statistical analysis of plate colony formation while upregulating USP9X in HepG2 cells. (G) Images and (H) statistical analysis of Transwell assay result (with Matrigel coating) while upregulating USP9X in HepG2 cells. Scale bar, 200 µm. (I) Representative images and (J) statistical analysis of wound healing assay in USP9X overexpressing HepG2 cells. *P<0.05 and **P<0.01. EdU, 5-ethynyl-20-deoxyuridine; Endo, endonuclease; Exo, exonuclease; USP9X, X-linked ubiquitin-specific peptidase 9.
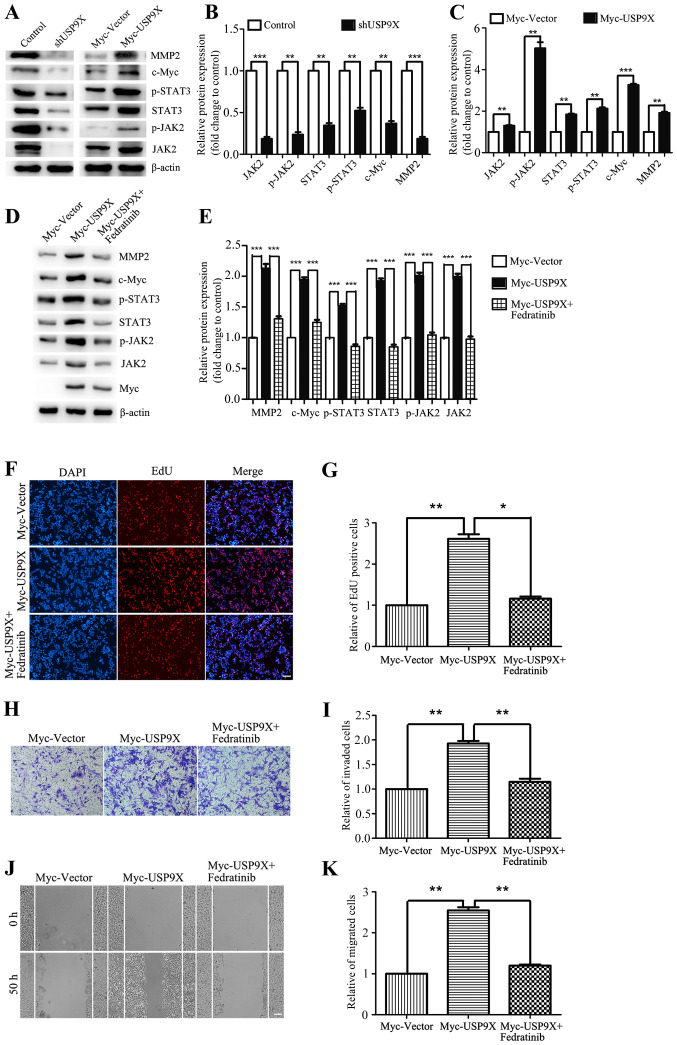
USP9X positively regulates the JAK2/STAT3 pathway to promote the proliferation, invasiveness and migration of liver cancer cells
The aforementioned results have shown that USP9X is important for the proliferation, invasivenessand migration of liver cancer cells. However, the underlying mechanisms were yet to be addressed. By western blot analysis, USP9X silencing was found to block downstream effectors of the JAK2/STAT3 pathway, thereby inhibiting the expression of c-Myc and MMP2 (Fig. 4A and B). By contrast, overexpression of USP9X significantly upregulated the protein expression levels of the JAK2/STAT3 signaling pathway thus increasing the expression of c-Myc and MMP2. The relative ratios of the experimental group and the control group to their respective internal parameters were used for statistical analysis (Fig. 4A and C). When evaluating the effect of the JAK2 inhibitor Fedratinib on USP9X overexpressing cells, it was found that JAK2 inhibition was able to effectively rescue the expression of JAK2/STAT3 pathway components, c-Myc and MMP2 (Fig. 4D and E), and also restore the cellular proliferation, invasiveness and migration induced by USP9X overexpression (Fig. 4F-K). In conclusion, these results suggest that USP9X positively regulates the JAK2/STAT3 pathway, thereby promoting the proliferation, invasiveness and migration of liver cancer cells.
Figure 4.
USP9X positively regulates the JAK2/STAT3 signaling pathway. (A-C) Western blot assay and quantification showed that in HepG2 cells, the protein levels of JAK2/STAT3 pathway components, c-Myc and MMP2 were reduced in the context of USP9X loss by shRNA, while the results were contrary when USP9X increased in expression. (D and E) Western blot assay showed that the JAK2 inhibitor Fedratinib effectively rescued the expression of JAK2/STAT3 pathway components, c-Myc and MMP2. The inhibitor was added 24 h after transfection at a concentration of 10 µg/ml, and the cells were collected 6 h later. (F-K) EdU incorporation assay, Transwell invasion assay and wound healing assay showed that Fedratinib effectively restored pcell roliferation, invasiveness and migration induced by overexpressing USP9X. *P<0.05, **P<0.01 and ***P<0.001. EdU, 5-ethynyl-20-deoxyuridine; JAK2, Janus kinase 2; MMP2, matrix metalloproteinase-2; p-, phosphorylated; USP9X, X-linkedubiquitin-specific peptidase 9.
Discussion
USP9X is involved in the regulation of numerous key biological processes, such as proteasome activity, organ production, tumor production and transcriptional regulation (6,7). It can simultaneously affect the ubiquitination process to regulate apoptosis, proliferation and adhesion of cells (8). Previous research has found that USP9X is significantly increased in human non-small cell lung cancer, breast cancer, leukemia, cervical cancer, follicular lymphoma, colon cancer and esophageal squamous cell carcinoma, which indicates that USP9X could be a potential biomarker in human cancers (8–14). However, the functions of USP9X in liver cancer have not yet been clarified. In the present study, the protein expression of USP9X in normal liver tissues and liver cancer tissues was detected. The results show that USP9X was unregulated in patients with liver cancer, compared with those in normal liver tissues. When analyzing the effects of USP9X on the proliferation, invasiveness and migration of liver cancer cells, the results indicated that silencing USP9X inhibited cell proliferation, invasiveness and migration, whereas USP9X overexpression had the opposite effect. These findings suggest that USP9X is a oncogene in liver cancer, as well as being a potential biomarker.
It is well known that inhibition of tumor-suppressor genes or activation of oncogenes is a key event for the development and malignant progression of liver cancer (32–35). USP9X plays an important role in the proliferation of liver cancer cells (17). In the present study, USP9X was found to be involved in the regulation of the JAK2/STAT3 pathway. Western blot analyses confirmed that USP9X overexpression could increase the levels of JAK2 and STAT3, along with c-Myc and MMP2. A number of studies have previously indicated that STAT3 activation improves the expression of c-Myc, which subsequently promotes cellular proliferation (26). Conversely, STAT3 activation has been associated with the upregulation of MMP2, and further promotion of tumor invasive (27–31). At the same time, USP9X-knockdown inhibits the expression of c-Myc and MMP2, thereby inhibiting the proliferation, invasiveness and migration of liver cancer cells. Overexpressing USP9X exerted the opposite effect, which was rescued by the JAK2 inhibitor Fedratinib. Therefore, upregulation of USP9X may be an important event in the development and malignant progression of liver cancer.
To the best of our knowledge, the present study shows for the first time evidence linking USP9X to the proliferation and malignant progression of human liver cancer by regulating the JAK2/STAT3 pathway. However, the specific mechanism by which USP9X regulates the JAK2/STAT3 pathway requires further investigation. These results lay the foundation for further study into the role of USP9X in liver cancer, and exploring the diagnosis and treatment of liver cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
All authors participated in the design, interpretation and analysis of the data and reviewed the manuscript. XS and WY performed the experiments, WY and YL designed the experiments, CW drafted the initial manuscript, and YH and YL critically revised the manuscript for important intellectual content.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of the No. 1 People's Hospital of Xuzhou (Xuzhou, China; approval no. xyy11[2020]-XJSXZ-010). Written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. CONCORD Working Group Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Parkin DM, Li LD, Chen YD, Bray F. Estimation and projection of the national profile of cancer mortality in China: 1991-2005. Br J Cancer. 2004;90:2157–2166. doi: 10.1038/sj.bjc.6601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulik L, Heimbach JK, Zaiem F, Almasri J, Prokop LJ, Wang Z, Murad MH, Mohammed K. Therapies for patients with hepatocellular carcinoma awaiting liver transplantation: A systematic review and meta-analysis. Hepatology. 2018;67:381–400. doi: 10.1002/hep.29485. [DOI] [PubMed] [Google Scholar]
- 6.Khut PY, Tucker B, Lardelli M, Wood SA. Evolutionary and expression analysis of the zebrafish deubiquitylating enzyme, usp9. Zebrafish. 2007;4:95–101. doi: 10.1089/zeb.2006.0502. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grützmann R, Aust D, Rümmele P, et al. Australian Pancreatic Cancer Genome Initiative The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 9.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Liu Y, Yang B, Cao H, Yang CX, Ouyang W, Zhang SM, Yang GF, Zhou FX, Zhou YF, et al. Elevated expression of USP9X correlates with poor prognosis in human non-small cell lung cancer. J Thorac Dis. 2015;7:672–679. doi: 10.3978/j.issn.2072-1439.2015.04.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng J, Hu Q, Liu W, He X, Cui L, Chen X, Yang M, Liu H, Wei W, Liu S, et al. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn Pathol. 2013;8:177. doi: 10.1186/1746-1596-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 14.Rolén U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, Masucci MG. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- 15.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: Therapeutic implications. Eur J Cancer. 2004;40:2217–2229. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Pardali K, Moustakas A. Actions of TGF-β as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Hu H, Tang C, Jiang Q, Luo W, Liu J, Wei X, Liu R, Wu Z. Reduced ubiquitin-specific protease 9X expression induced by RNA interference inhibits the bioactivity of hepatocellular carcinoma cells. Oncol Lett. 2015;10:268–272. doi: 10.3892/ol.2015.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, Harr MW, Levine RL, Xu H, Tefferi A, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19:283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu JH, Kim KH, Kim H. SOCS 3 and PPAR-γ ligands inhibit the expression of IL-6 and TGF-β1 by regulating JAK2/STAT3 signaling in pancreas. Int J Biochem Cell Biol. 2008;40:677–688. doi: 10.1016/j.biocel.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths DS, Li J, Dawson MA, Trotter MW, Cheng YH, Smith AM, Mansfield W, Liu P, Kouzarides T, Nichols J, et al. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13:13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q, Wang F, Wang Y, Wang X. m6A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019;10:171. doi: 10.1038/s41419-019-1417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong E, Sucic S, Monje FJ, Savalli G, Diao W, Khan D, Ronovsky M, Cabatic M, Koban F, Freissmuth M, et al. STAT3 controls IL6-dependent regulation of serotonin transporter function and depression-like behavior. Sci Rep. 2015;5:9009. doi: 10.1038/srep09009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chua CY, Liu Y, Granberg KJ, Hu L, Haapasalo H, Annala MJ, Cogdell DE, Verploegen M, Moore LM, Fuller GN, et al. IGFBP2 potentiates nuclear EGFR-STAT3 signaling. Oncogene. 2015;35:738–747. doi: 10.1038/onc.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen MF, Greibe E, Skovbjerg S, Rohde S, Kristensen AC, Jensen TR, Stentoft C, Kjær KH, Kronborg CS, Martensen PM. Folic acid mediates activation of the pro-oncogene STAT3 via the Folate Receptor alpha. Cell Signal. 2015;27:1356–1368. doi: 10.1016/j.cellsig.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 26.Shirogane T, Fukada T, Muller JMM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–719. doi: 10.1016/S1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 27.Kasza A. Signal-dependent Elk-1 target genes involved in transcript processing and cell migration. Biochim Biophys Acta. 2013;1829:1026–1033. doi: 10.1016/j.bbagrm.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Ou Y, Liu L, Xue L, Zhou W, Zhao Z, Xu B, Song Y, Zhan Q. TRAP1 shows clinical significance and promotes cellular migration and invasion through STAT3/MMP2 pathway in human esophageal squamous cell cancer. J Genet Genomics. 2014;41:529–537. doi: 10.1016/j.jgg.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Feng J, Yu SY, Li CZ, Li ZY, Zhang YZ. Integrative proteomics and transcriptomics revealed that activation of the IL-6R/JAK2/STAT3/MMP9 signaling pathway is correlated with invasion of pituitary null cell adenomas. Mol Cell Endocrinol. 2016;436:195–203. doi: 10.1016/j.mce.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu B, Zhang K, Li S, Li H, Yan Z, Huang L, Wu J, Han X, Jiang W, Mulatibieke T, et al. HIC1 attenuates invasion and metastasis by inhibiting the IL-6/STAT3 signalling pathway in human pancreatic cancer. Cancer Lett. 2016;376:387–398. doi: 10.1016/j.canlet.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Cao K, Gong H, Qiu Z, Wen Q, Zhang B, Tang T, Zhou X, Cao T, Wang B, Shi H, et al. Hepatitis B virus X protein reduces the stability of Nrdp1 to up-regulate ErbB3 in hepatocellular carcinoma cells. Tumour Biol. 2016;37:10375–10382. doi: 10.1007/s13277-016-4936-y. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Zheng C, Shao J, Chen L, Liu X, Shao J. Overexpression of eEF1A1 regulates G1-phase progression to promote HCC proliferation through the STAT1-cyclin D1 pathway. Biochem Biophys Res Commun. 2017;494:542–549. doi: 10.1016/j.bbrc.2017.10.116. [DOI] [PubMed] [Google Scholar]
- 34.Wang C, Yao B, Xu M, Zheng X. RIP1 upregulation promoted tumor progression by activating AKT/Bcl-2/BAX signaling and predicted poor postsurgical prognosis in HCC. Tumour Biol. 2016;37:15305–15313. doi: 10.1007/s13277-016-5342-1. [DOI] [PubMed] [Google Scholar]
- 35.Xia W, Zhuang J, Wang G, Ni J, Wang J, Ye Y. P4HB promotes HCC tumorigenesis through downregulation of GRP78 and subsequent upregulation of epithelial-to-mesenchymal transition. Oncotarget. 2017;8:8512–8521. doi: 10.18632/oncotarget.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.