Removing the regulation of sphingolipid biosynthesis by completely knocking out Orosomucoid-like protein (ORM) genes results in ceramide hyperaccumulation and nonviable seeds with strongly reduced oil content.
Abstract
Orosomucoid-like proteins (ORMs) interact with serine palmitoyltransferase (SPT) to negatively regulate sphingolipid biosynthesis, a reversible process critical for balancing the intracellular sphingolipid levels needed for growth and programmed cell death. Here, we show that ORM1 and ORM2 are essential for life cycle completion in Arabidopsis (Arabidopsis thaliana). Seeds from orm1−/− orm2−/− mutants, generated by crossing CRISPR/Cas9 knockout mutants for each gene, accumulated high levels of ceramide, indicative of unregulated sphingolipid biosynthesis. orm1−/− orm2−/− seeds were nonviable, displayed aberrant embryo development, and had >80% reduced oil content versus wild-type seeds. This phenotype was mimicked in Arabidopsis seeds expressing the SPT subunit LCB1 lacking its first transmembrane domain, which is critical for ORM-mediated regulation of SPT. We identified a mutant for ORM1 lacking one amino acid (Met-51) near its second transmembrane domain that retained its membrane topology. Expressing this allele in the orm2 background yielded plants that did not advance beyond the seedling stage, hyperaccumulated ceramides, and showed altered organellar structures and increased senescence- and pathogenesis-related gene expression. These seedlings also showed upregulated expression of genes for sphingolipid catabolic enzymes, pointing to additional mechanisms for maintaining sphingolipid homeostasis. ORM1 lacking Met-51 had strongly impaired interactions with LCB1 in a yeast (Saccharomyces cerevisiae) model, providing structural clues about regulatory interactions between ORM and SPT.
INTRODUCTION
Sphingolipids are essential, abundant endomembrane and plasma membrane lipids that contribute to membrane function, vesicular trafficking, and the mediation of cellular processes in eukaryotes (Coursol et al., 2003; Liang et al., 2003; Chen et al., 2006; Markham et al., 2011). The unique and defining structural feature of sphingolipids is the long-chain base (LCB) or sphingoid base. The simplest LCB, sphinganine (d18:0), derives from the condensation of Ser and palmitoyl-CoA catalyzed by serine palmitoyltransferase (SPT) and subsequent reduction of the 3-ketosphinganine product. LCBs can be further modified by hydroxylation, desaturation, and phosphorylation to yield a range of structural variants (Markham et al., 2006; Chen et al., 2009). Free LCBs and their phosphorylated forms typically occur in low concentrations in eukaryotic cells. LCBs exert signaling functions such as modulating cell proliferation and apoptosis in mammalian cells and serve as a trigger of programmed cell death (PCD) and associated pathogen defense responses in plant cells (Alden et al., 2011; Zheng et al., 2018; Huby et al., 2020). The majority of LCBs occur in ceramides. These N-acylated LCBs are synthesized by ceramide synthase–mediated condensation of an LCB and a fatty acyl-CoA. Ceramide synthases have defined substrate specificities that result in ceramides with distinct pairings of structurally diverse LCBs and fatty acids (Chen et al., 2009; Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2015a). In mammalian cells, ceramides function as regulators of apoptotic processes, and perturbations in their levels are associated with inflammation, obesity, diabetes, and cancer. In plants, ceramide accumulation has been shown to initiate PCD (Liang et al., 2003; Bi et al., 2014; Dadsena et al., 2019). Ceramides provide the hydrophobic backbone for more complex sphingolipids, including glucosylceramides (GlcCer) and glycosylinositolphosphoceramides (GIPCs), the principal glycosphingolipids of plant cells.

SPT activity is highly regulated in eukaryotes to modulate the requirement of sphingolipids for growth and membrane function while limiting the accumulation of LCBs and ceramides until needed to trigger specific cellular functions, such as PCD-mediated pathogen defense in plants (Peer et al., 2010). SPT is composed of the subunits LCB1 and LCB2 and the accessory protein known as small subunit of SPT (ssSPT) or TSC3 in yeast (Saccharomyces cerevisiae; Gable et al., 2000; Kimberlin et al., 2013). SPT is primarily regulated by posttranslational mechanisms in order to rapidly respond to perturbations in intracellular sphingolipid concentrations. ORMs or orosomucoid-like proteins (or ORMDLs in mammals) are now recognized as noncatalytic proteins that negatively regulate SPT (Breslow et al., 2010; Han et al., 2010). In Arabidopsis (Arabidopsis thaliana), two ORM genes, ORM1 (At1g01230) and ORM2 (At5g42000), were previously identified by Kimberlin et al. (2016). In S. cerevisiae, Orm1p and Orm2p suppress SPT activity in response to elevated sphingolipid levels through a physical interaction that requires the first transmembrane domain of LCB1 (Han et al., 2019). Sphingolipid-responsive regulation of the ORM-SPT interaction in S. cerevisiae is mediated by phosphorylation/dephosphorylation of an N-terminal domain of the ORMs (Breslow et al., 2010). This domain is absent from ORM/ORMDL of multicellular eukaryotes, suggesting that an alternative mechanism regulates the ORM-SPT interaction, such as a recently demonstrated mechanism of direct binding of a ceramide molecule to mammalian ORMDL and S. cerevisiae ORM to confer negative SPT regulation (Davis et al., 2019). In addition, ORMDL expression levels vary with sphingolipid availability in mammalian cells (Gupta et al., 2015).
S. cerevisiae cells are viable after knockout of the two ORM genes, but they accumulate increased amounts of LCBs and ceramides and are sensitive to tunicamycin, an inducer of endoplasmic reticulum (ER) stress (Breslow et al., 2010). However, a full understanding of the biochemical and physiological functions of ORM or ORMDL proteins in multicellular eukaryotes is only beginning to emerge. A recent report showed that ORMDL proteins are critical for nerve myelination and for suppressing the accumulation of toxic sphingolipid biosynthetic intermediates in mice (Clarke et al., 2019). Downregulation of ORM2 using an artificial microRNA in an Arabidopsis ORM1 T-DNA mutant yielded fertile plants with increased accumulation of LCBs and ceramides and early senescence (Li et al., 2016). In addition, RNA interference (RNAi)–induced suppression of Arabidopsis ORM1 and ORM2 resulted in plants with a normal appearance but with increased sensitivity to the ceramide synthase inhibitor fumonisin B1 and increased LOH2 ceramide synthase activity (Kimberlin et al., 2016). Beyond Arabidopsis, RNAi of ORM genes in rice (Oryza sativa) was linked to reduced pollen viability (Chueasiri et al., 2014). However, the lack of complete ORM knockout mutants in Arabidopsis or other plants has precluded assessment of SPT regulation in the absence of ORM proteins.
In this study, to advance our understanding of ORM-mediated sphingolipid biosynthesis, we generated orm1 orm2 double mutants using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. Loss of SPT regulation resulted in nonviable seeds with low oil content that accumulated high levels of ceramides. We mimicked this phenotype by removing the first transmembrane domain of LCB1, which is known to interact with ORM for SPT regulation (Han et al., 2019). These studies also uncovered a single amino acid deletion mutant of ORM1 that had severely altered membrane and organellar structures and that also hyperaccumulated ceramides. Using a S. cerevisiae model, we showed that the deleted amino acid, which occurs in a position preceding the second membrane-spanning domain of ORM, strongly reduced the ORM-LCB1 interaction. This finding provides important information about the structural features of ORM and ORMDL proteins that are associated with their regulatory interaction with the LCB1 subunit of SPT.
RESULTS
ORMs Are Essential for Plant Development
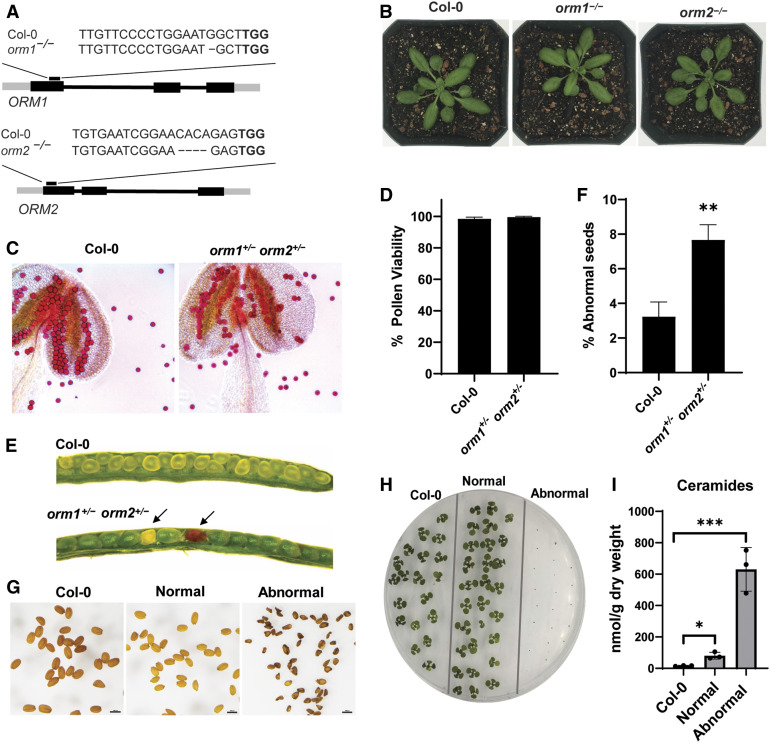
We designed two single guide RNAs to target regions in the coding sequence of each of the two Arabidopsis ORM genes (Figure 1A). We introduced these constructs into Arabidopsis via Agrobacterium tumefaciens–mediated transformation to generate CRISPR/Cas9-induced knockouts of the ORM1 and ORM2 genes. We screened T1 and T2 transformants by restriction enzyme digestion of the PCR amplicons encompassing the ORM1 and ORM2 target sites to obtain homozygous lines with mutations in each gene. These lines were also verified by PCR to lack Cas9 transgenes. These homozygous single mutants were visually indistinguishable from wild-type plants under optimal growth conditions (Figure 1B). The population of mutants obtained contained nucleotide deletions resulting in frameshifts and premature stop codons, as determined by PCR–restriction enzyme digestion and sequencing (Supplemental Figure 1). To obtain double knockout mutants, we crossed the orm1−/− and orm2−/− single mutants. No progeny with homozygous knockout mutations in both genes were obtained after analyzing 155 plants from the F2 generation and 60 plants from the F3 generation. To gain more insight into the basis for the apparent lethality associated with the double mutant, we performed viability staining on pollen from plants genotyped as orm1+/− orm2+/− (Supplemental Figure 2A). Nearly all of the pollen from these mutants was viable, similar to pollen from the wild-type plants (Figures 1C and 1D), rather than 25% nonviability that would be expected for pollen lethality in this mutant.
Figure 1.
ORM Double Knockout Mutant Is Seed Lethal.
(A) Schematic representation of CRISPR/Cas9-induced mutations in ORM genes. Gene structures of ORM1 and ORM2; black boxes represent exons. The CRISPR/Cas9 target site is indicated as well as the nucleotide deletions for each gene in the single mutants.
(B) Representative images of 25-d-old wild-type Col-0, orm1−/−, and orm2−/− plants.
(C) Representative images of pollen and anthers (treated with Alexander stain) collected from wild-type Col-0 and orm1+/− orm2+/− plants.
(D) Viability of pollen determined by counts of ∼100 pollen grains from five randomly selected flowers from independent Col-0 and orm1+/− orm2+/− plants. Shown are the mean ± sd.
(E) Developing seeds in siliques from wild-type Col-0 and orm1+/− orm2+/− plants. Shriveled, brown (abnormal) seeds are indicated by arrows.
(F) Percentage of shriveled and brown (abnormal) seeds in siliques determined by counts of an average of 200 developing seeds from 10 randomly selected siliques of the independent wild-type Col-0 and orm1+/− orm2+/− plants. Shown are the mean ± sd. Asterisks denote significant differences as determined by two-tailed Student’s t test, with a significance of P ≤ 0.01.
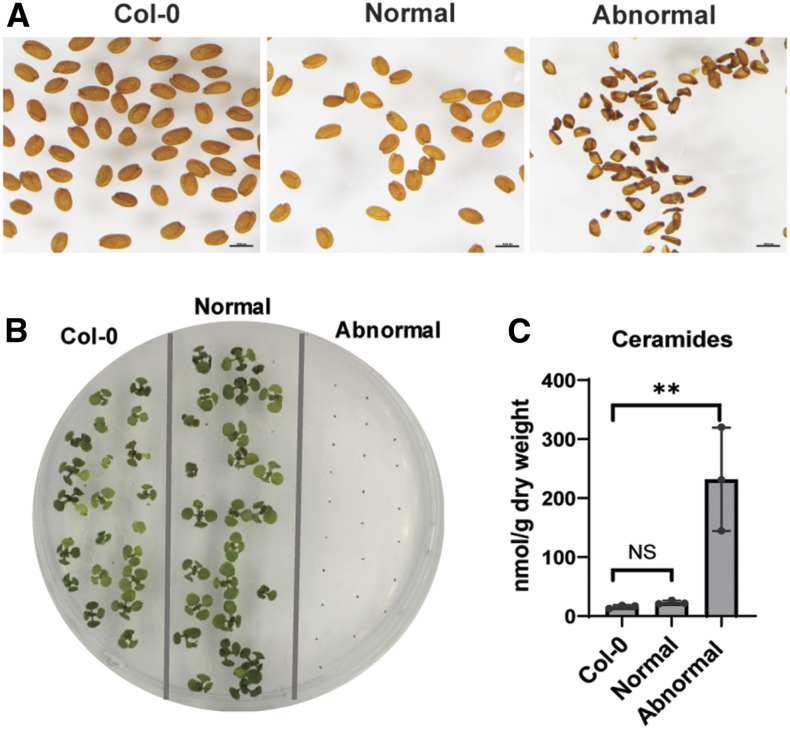
(G) Seeds from wild-type Col-0; seeds from orm1+/− orm2+/− were separated and classified into normal and the darker, shriveled seeds as abnormal. Bars = 1 mm.
(H) Phenotypes of 10-d-old seedlings from wild-type Col-0 seeds and normal and abnormal seeds from orm1+/− orm2+/−. Abnormal seeds did not germinate.
(I) Ceramide content in seeds from wild-type Col-0 and normal and abnormal seeds from orm1+/− orm2+/−. Shown are the mean ± sd, n = 3. Asterisks indicate significant differences based on one-way ANOVA followed by Tukey’s multiple comparisons test, with a significance of *, P ≤ 0.05 and ***, P ≤ 0.001.
Instead, a population of seeds from these plants had dark-colored seed coats and were severely wrinkled. This phenotype was observed for ∼7% of seeds collected from the F2 orm1+/− orm2+/− plants of orm1−/− and orm2−/− crosses, which is consistent with the expected 6.25% Mendelian ratio for the occurrence of homozygous double mutants. The remaining seeds were visually indistinguishable from wild-type seeds (Figures 1E and 1F). Of the seeds in these two populations, dark, wrinkled seeds did not germinate, whereas seeds with normal appearance showed no impairment in germination on solid Suc-containing medium (Figures 1G and 1H) and soil. Strikingly, free ceramide concentrations in pooled abnormal seeds were ∼40-fold higher than those in wild-type seeds and ∼8-fold higher than in the normal-appearing seed segregants from orm1+/−orm2+/− plants (Figure 1I). We also observed a similar seed phenotype in Atlcb1+/− plants expressing a version of the LCB1 subunit of SPT lacking its first transmembrane domain (LCB1ΔTMD1) that is required for SPT-ORM regulatory interactions (Han et al., 2019). In these experiments, the segregating seeds from Atlcb1+/− plants expressing LCB1ΔTMD1 included a population of shrunken, nonviable seeds with a 14-fold increase in ceramide levels compared with wild-type seeds (Figure 2).
Figure 2.
Atlcb1+/− Plants Expressing LCB1ΔTMD1 Phenocopy the ORM Double Knockout Mutant.
(A) Seeds from wild-type Col-0; seeds from Atlcb1+/− plants expressing LCB1ΔTMD1 were separated and classified into normal and abnormal darker and shriveled seeds. Bars = 1 mm.
(B) Phenotypes of 10-d-old seedlings from wild-type Col-0 seeds and normal and abnormal seeds from Atlcb1+/− expressing LCB1ΔTMD1. Abnormal seeds did not germinate.
(C) Ceramide content in seeds from wild-type Col-0 and normal and abnormal seeds from LCB1ΔTMD1. Shown are the mean ± sd, n = 3. Asterisks indicate significant difference based on one-way ANOVA followed by Tukey’s multiple comparisons test, with a significance of **, P ≤ 0.01. NS, not significant.
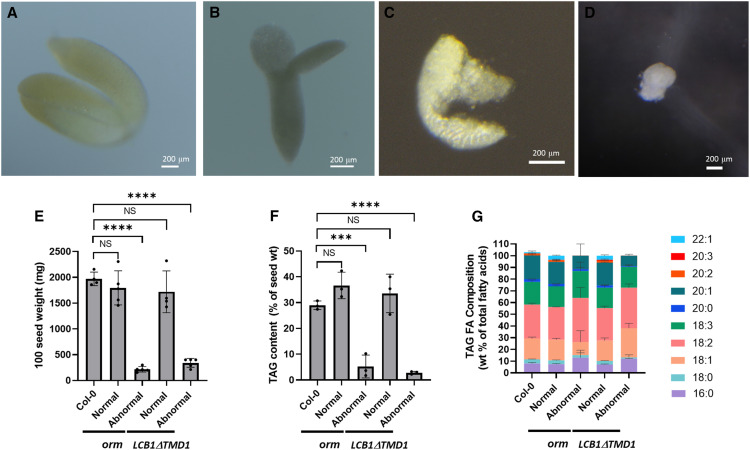
We examined seeds from the orm1−/− and orm2−/− crosses and LCB1ΔTMD1 in more detail to understand the basis for the loss of viability. The weight of mature nonviable, abnormal seeds was 80 to 90% lower than that of normal seed segregants from these lines (Figure 3E). Embryos dissected from the abnormal seeds had variable appearance ranging from cell clusters with undifferentiated appearance to embryo-like structures that were up to one-third the size of those from normal seeds (Figures 3A to 3D). Underlying this phenotype, oil content of the abnormal seeds, as measured by the fatty acid content of purified triacylglycerols (TAG), was 85 to 90% lower than that of normal seed segregants (Figure 3F).
Figure 3.
Abnormal Seeds from ORM and LCB1ΔTMD1 Mutant Plants Have Altered Embryo Morphology and Reduced TAG Concentrations.
(A) to (D) Morphology of embryos from the wild-type seeds (A) and abnormal seeds from orm1+/− orm2+/− plants (see [B] to [D]) showing that the embryo is not fully developed. Embryos were dissected from mature seeds.
(E) The 100 seed weight. Values are the mean ± sd of seeds harvested from four independent plants. Asterisks indicate significant difference based on one-way ANOVA followed by Tukey’s multiple comparisons test, with a significance of ****, P ≤ 0.0001. NS, not significant; TMD, transmembrane domain.
(F) TAG content in seeds from wild-type Col-0 and normal and abnormal seeds from orm1+/− orm2+/− and Atlcb1+/− expressing LCB1ΔTMD1. Values are the mean ± sd of three independent lipid extractions. Asterisks indicate significant difference based on one-way ANOVA followed by Tukey’s multiple comparisons test, with a significance of ***, P ≤ 0.001 and ****, P ≤ 0.0001. NS, not significant; TMD, transmembrane domain.
(G) Composition of TAG as weight percent of fatty acid in seeds from wild-type Col-0 and normal and abnormal seeds from orm1+/− orm2+/− and Atlcb1+/− expressing LCB1ΔTMD1. Values are the mean ± sd of three independent samples. TMD, transmembrane domain.
The most striking difference in fatty acid composition of TAG from the abnormal seeds was a reduction in the overall content of C20 and C22 very long chain fatty acids derived from ER-localized elongation reactions. Notably, the fatty acids 20:2, 20:3, and 22:1 were not detectable in TAG from the abnormal seeds (Figure 3G).
Overall, these results indicate that ORMs are essential for the completion of a full life cycle in Arabidopsis. Lethality due to the absence of ORM proteins is associated with the recovery of nonviable seeds with undeveloped embryos that accumulate excessive ceramide concentrations and have strongly reduced TAG levels. This was phenocopied in plants with deregulated SPT activity due to the loss of the transmembrane domain of LCB1 that abolishes ORM regulation of SPT (Han et al., 2019). The identification of nearly the same phenotype in ORM-null mutants and LCB1-ΔTMD1 lines also indicated that the loss of seed viability is associated with the role of ORM proteins in sphingolipid metabolism, rather than other reported functions of ORM in Arabidopsis (Yang et al., 2019).
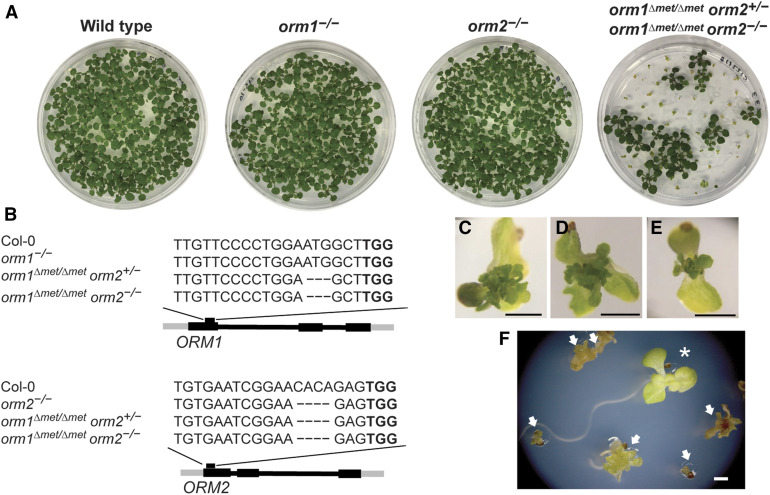
The availability of progeny from orm1−/− and orm2−/− crosses also allowed us to assess the contributions of each ORM gene to the viability and growth of Arabidopsis plants. In addition to our inability to obtain homozygous double mutants for these genes, we observed that orm1−/− orm2+/− mutants were strongly dwarfed, had yellow leaves, and senesced prior to flowering (Figure 4A). By contrast, orm1+/−orm2−/− mutants had a distinct bushy phenotype, with increased leaf number compared to wild-type plants and delayed flowering time (Figures 4B and 4C). Overall, these results revealed stronger growth phenotypes for the homozygous ORM1 knockout in the orm2+/− background than the homozygous ORM2 knockout in the orm1+/− background.
Figure 4.
orm1−/− orm2+/− and orm1+/− orm2−/− Plants Have Distinct Growth Phenotypes.
(A) Representative image of 35-d-old orm1+/− orm2+/− and orm1−/− orm2+/− plants. The orm1−/− orm2+/− plants showed reduced size and yellow regions corresponding to cell death.
(B) Representative image of 18-d-old wild-type Col-0 and orm1+/− orm2−/− plants. Mutants showed reduced size, abnormal leaf shape, and a bushy phenotype.
(C) Representative image of 50-d-old orm1+/− orm2−/− and orm1+/− orm2+/− plants. The orm1+/− orm2−/− plants showed a bushy phonotype and delayed flowering.
(D) Representative image of 80-d-old orm1+/− orm2−/− plant.
orm1Δmet/Δmet orm2−/− Mutant Does Not Survive beyond the Seedling Stage
Screening of gene-edited lines also revealed a mutant with an in-frame deletion of a single codon that resulted in a deletion of the Met residue at amino acid 51 relative to the wild-type ORM1 (Figure 5B). This line also carried nucleotide deletions in ORM2 that led to a frameshift and premature stop codon (Supplemental Figures 1 and 2B). Seedlings with the genotype orm1Δmet/Δmet orm2+/− showed a phenotype like the wild type and the single mutants under normal growth conditions (Figure 5A).
Figure 5.
orm1Δmet/Δmet orm2−/− Plants Exhibit Developmental Defects and Do Not Progress beyond the Seedling Stage.
(A) and (C) to (E) Representative images (A) of the 12-d-old wild-type Col-0, orm1−/−, orm2−/−, orm1Δmet/Δmet orm2+/−, and orm1Δmet/Δmet orm2−/− seedlings. Seedlings with the same phenotype as the wild type correspond to orm1Δmet/Δmet orm2+/−; small seedlings showing developmental defects correspond to orm1Δmet/Δmet orm2−/−; enlarged images are shown in (C) to (E). Bars = 1 mm.
(B) CRISPR/Cas9-induced mutations in ORM1 and ORM2. Structures of the ORM genes; black boxes represent exons. The position of the CRISPR target site is marked as well as the nucleotide deletions in each mutant.
(F) Phenotypes of 18-d-old seedlings; arrows indicate orm1Δmet/Δmet orm2−/− and asterisk indicates orm1Δmet/Δmet orm2+/−. Bar = 1 mm.
However, we could only recover plants of the genotype orm1Δmet/Δmet orm2−/− in solid medium supplemented with Suc. The resulting seedlings were severely dwarfed and had a proliferation of small, deformed chlorotic leaves. These plants persisted in a visually viable state for 20 to 25 d after planting, but did not progress beyond the seedling stage, indicating that the orm1Δmet/Δmet orm2−/− mutation is seedling lethal (Figures 5A and 5C to 5F). Complementation of this mutant with the Arabidopsis ORM1 cDNA under the control of its native promoter was sufficient to rescue the seedling lethality and recover fertile plants, although many of the independent complemented mutant lines were smaller than wild-type plants, which is similar to the phenotype of orm1+/−orm2−/− plants, as described above (Supplemental Figure 3).
orm1Δmet/Δmet orm2−/− Mutant Hyperaccumulates Selected Sphingolipids
Based on the finding that downregulating ORM expression triggers sphingolipid accumulation (Breslow et al., 2010; Kimberlin et al., 2016; Li et al., 2016), we conducted extensive sphingolipidomic profiling of our gene-edited mutants from seedlings grown on Suc medium at 12 to 15 d after planting. The orm1Δmet/Δmet orm2−/− mutant accumulated 3.7-fold more sphingolipids than wild-type seedlings (Figure 6A). No significant differences in the levels of free LCBs, ceramides with nonhydroxylated fatty acids (Cer), or other sphingolipid classes were detected in the orm1−/−, orm2−/−, or orm1Δmet/Δmet orm2+/− mutants compared to wild-type plants (Figures 6B to 6E and 6G to 6I). In strong contrast, orm1Δmet/Δmet orm2−/− seedlings showed heightened accumulation of LCB (5-fold), Cer (90-fold), and ceramides with hydroxylated fatty acids (hCer; 12-fold) compared to wild-type seedlings of similar age (Figures 6B and 6D; Supplemental Figure 4).
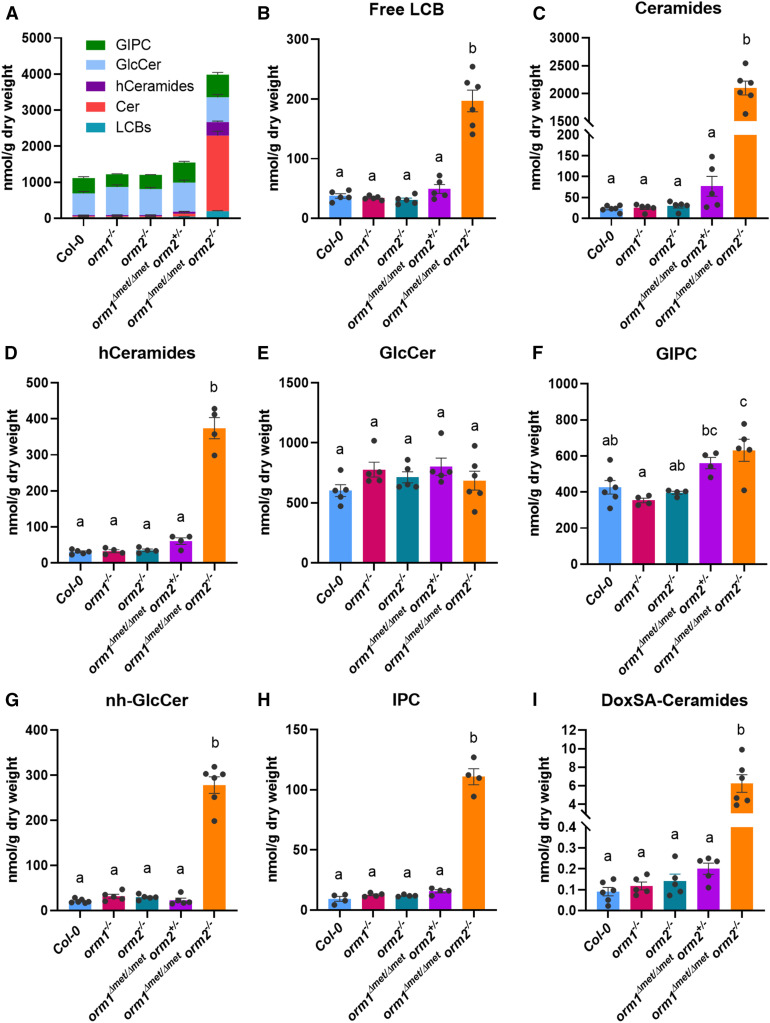
Figure 6.
Selected Sphingolipid Classes Highly Accumulate in the orm1Δmet/Δmet orm2−/− Mutant.
(A) to (G) Total sphingolipid content (A) in wild-type, orm1−/−, orm2−/−, orm1Δmet/Δmet orm2+/−, and orm1Δmet/Δmet orm2−/−. Content of the following sphingolipid classes in the mutants: free LCBs (B), Cer (Ceramides; see [C]), hCer (hCeramides; see [D]), GlcCer (E), and GIPCs (F). Content of atypical sphingolipids nh-GlcCer (G) and IPCs (H).
(I) Content of atypical deoxy-LCB m18:0 in ceramides. Normally, SPT condenses Ser with palmitoyl-CoA to form d18:0. However, the unusual condensation of Ala gives rise to a deoxy-LCB, DoxSA m18:0. Measurements are the average of four to six replicates consisting of pooled 12- to 15-d-old seedlings grown on different plates. Bars represent se of the mean. Different letters indicate significant difference based on one-way ANOVA followed by Tukey’s multiple comparisons test (P ≤ 0.05).
Although no changes were detected in GlcCer concentrations, the levels of GlcCer containing nonhydroxylated fatty acids (nhGlcCer), not typically found in abundance in Arabidopsis, were 13-fold higher in orm1Δmet/Δmet orm2−/− seedlings than in wild-type seedlings (Figures 6E and 6G; Supplemental Figures 5 and 6). GIPC levels increased by 48% in the orm1Δmet/Δmet orm2−/− mutant compared to wild-type seedlings (Figure 6F; Supplemental Figure 7). The LCB composition of the single mutants and orm1Δmet/Δmet orm2+/− did not change significantly compared to wild type (Figures 7A and 7B). However, in orm1Δmet/Δmet orm2−/−, the levels of free and phosphorylated forms of d18:0 were the most strongly increased, with lesser increases in the amounts of t18:0 and t18:1 free and phosphorylated species (Figures 7A and 7B).
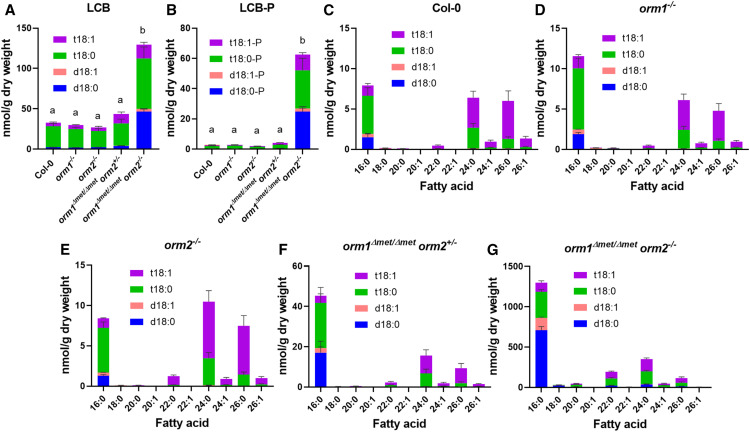
Figure 7.
Free LCB and Ceramide Compositions and Concentrations Are Strongly Affected in the orm1Δmet/Δmet orm2−/− Mutant.
(A) to (G) Free LCB composition (d18:0, d18:1, t18:0, t18:1, [A]) and free LCB-phosphate (LCB-P) composition in the wild-type, orm1−/−, orm2−/−, orm1Δmet/Δmet orm2+/−, and orm1Δmet/Δmet orm2−/− (B). Bars show averages of four to six replicates consisting of 12- to 15-d-old pooled seedlings grown on different plates. Error bars represent the se of the mean. Different letters indicate significant difference, for each LCB, based on one-way ANOVA followed by Tukey’s multiple comparisons test (P ≤ 0.05). Ceramide molecular species compositions representing the exact pairings of LCB and fatty acid for the wild-type (C), orm1−/− (D), orm2−/− (E), orm1Δmet/Δmet orm2+/− (F), and orm1Δmet/Δmet orm2−/− (G) plants. Measurements for all panels are the average of four to six replicates consisting of 12- to 15-d-old pooled seedlings grown on different plates. Bars represent se of the mean.
Cer profiles of the single mutants were similar to those of the wild type (Figures 7C to 7E). By contrast, the orm1Δmet/Δmet orm2+/− mutant had increased amounts of Cer with C16 fatty acids relative to wild-type and single mutant plants (Figure 7F). This phenotype was more accentuated in orm1Δmet/Δmet orm2−/− seedlings, which primarily accumulated Cer species with C16 fatty acids linked to the dihydroxy LCB d18:0 and d18:1 (Figure 7G). Increased amounts of Cer with C22, C24, and C26 fatty acids as well as atypical C18 and C20 fatty acid–containing species were also detected in orm1Δmet/Δmet orm2−/− seedlings relative to wild-type plants and mutants of either ORM gene (Figure 7G). Overall, the primary change in the composition of all sphingolipid classes, especially Cer, hCer, and nhGlcCer, in the orm1Δmet/Δmet orm2−/− seedlings was the change in the total and/or relative amounts of those containing C16 fatty acids bound to dihydroxy LCB, which are derived from the LOH2 ceramide synthase (Figure 7G; Supplemental Figures 4 and 6; Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2015a). The orm1Δmet/Δmet orm2−/− plants also contained aberrant forms of hCer and GIPCs with currently undefined structures based on liquid chromatography–mass spectrometry ionization as well as Cer with the LCB deoxy-sphinganine (DoxSA), which is derived from the condensation of Ala, rather than Ser, to palmitoyl-CoA by SPT (Figure 6I). In addition, the concentration of inositolphosphorylceramides (IPCs), the precursors of GIPCs, increased nearly 12-fold in small orm1Δmet/Δmet orm2−/− seedlings versus the wild type (Figure 6H).
Overall, these findings are consistent with the notion that SPT regulation by the orm1Δmet-encoded polypeptide is deficient and that the flux of excess LCB occurs through the LOH2 ceramide synthase to produce Cer backbones with C16 fatty acids and dihydroxy LCB, a portion of which are channeled to GIPCs but accumulate as IPC intermediates.
Integrity of Cellular Component Is Compromised in the orm1Δmet/Δmet orm2−/− Mutant
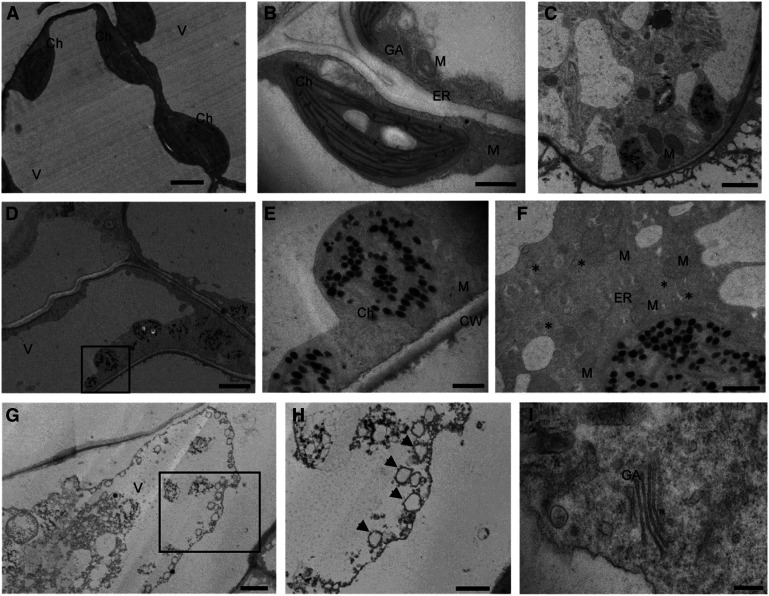
Given that sphingolipids are abundant endomembrane and plasma membrane components that contribute to vesicular trafficking, we used transmission electron microscopy (TEM) to evaluate the subcellular phenotypes associated with enhanced sphingolipid accumulation in 10-d-old orm1Δmet/Δmet orm2−/− seedlings relative to the wild-type seedlings of the same age. Mesophyll cells from the wild-type seedlings showed large vacuoles with turgor pressure pushing organelles to the periphery (Figure 8A). Chloroplasts of the wild-type cells had the typical oval shape and well-defined thylakoid membranes (Figures 8A and 8B). By contrast, the orm1Δmet/Δmet orm2−/− mutant cells displayed a lack of vacuolar turgor (Figure 8D). In addition, chloroplasts of orm1Δmet/Δmet orm2−/− cells were round and showed marked disintegration of thylakoids and highly abundant osmiophilic structures that resemble plastoglobuli (Figures 8C to 8F).
Figure 8.
Subcellular Features are Strongly Altered in the orm1Δmet/Δmet orm2−/− Mutant.
(A) to (H) Representative TEM images of the wild-type seedlings (see [A] and [B]) and orm1Δmet/Δmet orm2−/− (see [C] to [I]). Longitudinal sections of leaves from 10-d-old seedlings were prepared for TEM analysis. Boxes represent sections enlarged in (E) and (H). Asterisks indicate vesicles and arrows autophagosomes. Bar = 200 nm in (I); bars = 800 nm in (E) and (F); bar = 1 μm in (B); bars = 2 μm in (A) and (C); and bars = 4 μm in (D) and (G). Ch, Chloroplast; CW, cell wall; GA, Golgi apparatus; M, mitochondrion; V, vacuole.
Notably, increased vesicle numbers were observed around the ER network in orm1Δmet/Δmet orm2−/− cells (Figure 8F). Furthermore, electrodense material and double membrane vesicles consistent with autophagosomes were detected inside the vacuoles of these cells. Moreover, entire chloroplasts were engulfed and appeared to be in the process of degradation (Figures 8G and 8H). Despite these large defects, Golgi stacks were detectable in orm1Δmet/Δmet orm2−/− cells (Figure 8I).
Genes for Ceramide Synthases, LCB Kinase, and LCB-Phosphate Lyase Are Upregulated in the orm1Δmet/Δmet orm2−/− Mutant
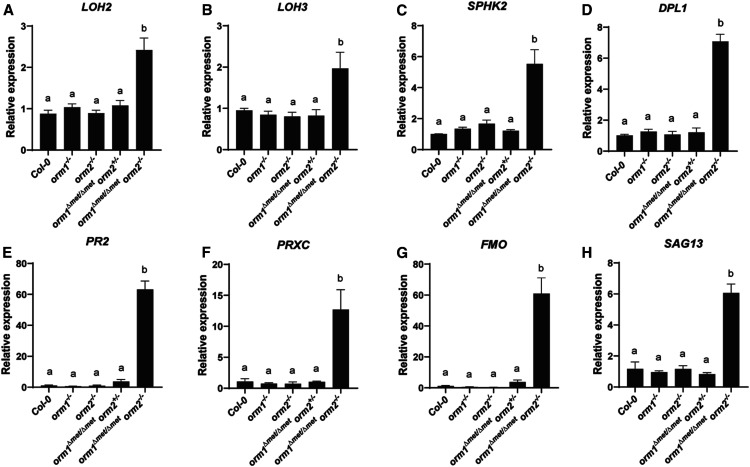
Given the increased concentrations of most sphingolipid classes in orm1Δmet/Δmet orm2−/−, we examined the expression of genes in 12-d-old seedlings for key sphingolipid biosynthetic and catabolic enzymes, including the SPT-associated polypeptides LCB1 and ssSPTa, ceramide synthases (LOH1, LOH2, and LOH3), sphingosine kinases (SPHK1 and SPHK2), and the LCB catabolic enzyme LCB-phosphate lyase (or DPL1). No significant differences were detected in the expression of genes for LCB1, ssSPTa, or LOH1 in any mutant analyzed (Supplemental Figures 8A to 8C). However, consistent with the increased amounts of ceramides in orm1Δmet/Δmet orm2−/−, the ceramide synthase gene LOH2 showed an ∼2.5-fold increase in expression and the ceramide synthase gene LOH3 showed an ∼2-fold increase in orm1Δmet/Δmet orm2−/− plants compared to the wild type and the other mutants examined (Figures 9A and 9B). Most notably, the expression of the key sphingolipid catabolism-associated genes SPHK2 and DPL1 increased by approximately six- to sevenfold, respectively, in orm1Δmet/Δmet orm2−/− plants relative to the wild type and other ORM mutants (Figures 9C and 9D). This result is consistent with the notion that the induction of LCB catabolism is one route (in addition to ceramide biosynthesis) for the mitigation of unregulated LCB production in the orm1Δmet/Δmet orm2−/− mutant.
Figure 9.
Expression of Genes Associated with Sphingolipid Homeostasis, Plant Defense Responses, and Senescence are Upregulated in the orm1Δmet/Δmet orm2−/− Mutant.
(A) to (H) Wild-type, orm1−/−, orm2−/− orm1Δmet/Δmet orm2+/−, and orm1Δmet/Δmet orm2−/− seedlings of 12-d-old plants were used to examine gene expression by qPCR to monitor genes encoding enzymes in sphingolipid biosynthetic and catabolic pathways: ceramide synthase gene LOH2 (A), ceramide synthase gene LOH3 (B), sphingosine kinase 2 gene SPHK2 (C), and LCB-phosphate lyase gene DPL1 (D) and selected pathogenesis- and senescence-related genes: β-1,3-glucanase gene PR2 (E), class III peroxidase gene PRXC (F), flavin monooxygenase gene FMO (G), and senescence-related 13 gene SAG13 (H). PP2AA3 transcript levels were used as a control for the sphingolipid genes and UBQ for the pathogenesis- and senescence-related genes. Specific primers used for this analysis are shown in the Supplemental Table. Gene expression levels are normalized to those in wild-type seedlings. Values are the mean ± sd (n = 6 to 12). Different letters indicate significant difference based on one-way ANOVA followed by Tukey’s multiple comparisons test (P ≤ 0.05).
Defense and Senescence Genes Are Upregulated in the orm1Δmet/Δmet orm2−/− Mutant
The accumulation of ceramides has been linked to the activation of signaling pathways that lead to PCD (Liang et al., 2003; Bi et al., 2014). To examine whether the high amounts of ceramides in orm1Δmet/Δmet orm2−/− activate PCD, we performed qPCR of marker genes using RNA extracted from 12-d-old seedlings. The expression of the pathogenesis-related genes (PR-2, PRXC, FMO, PR3) was significantly higher in orm1Δmet/Δmet orm2−/− than in the wild type and the other mutants (Figures 9E to 9G; Supplemental Figure 8E). A similar expression pattern was also observed for the senescence-related gene SAG13 (Figure 9H).
ORM1ΔMet51 Fails to Interact with LCB1 to Suppress SPT Activity
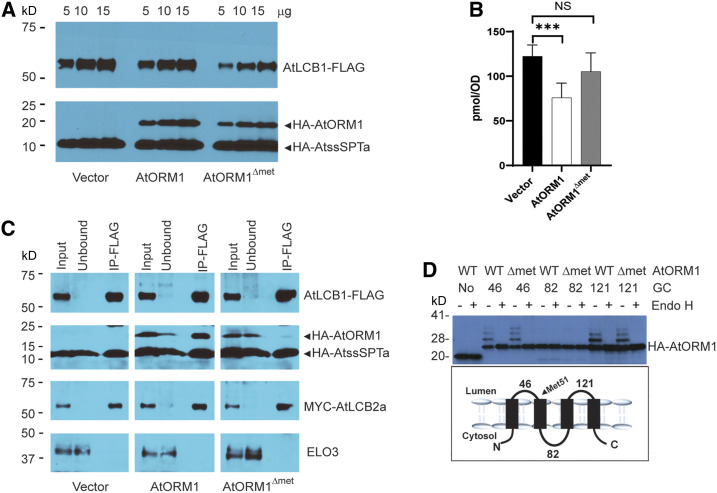
Our results clearly show that ORM1 lacking Met-51 is strongly impaired in repressing SPT activity. This amino acid is located in the ER luminal domain immediately adjacent to the second transmembrane domain of ORM1 (Supplemental Figure 9). We hypothesized that, without this amino acid, the conformation of the second transmembrane domain of ORM1 is altered such that the interaction with LCB1 for the repression of SPT activity is disrupted. To better understand this regulatory mechanism, we stably expressed the Arabidopsis ORM1ΔMet51 mutant protein in an S. cerevisiae mutant background in which AtLCB1, AtLCB2, and AtssSPTa replaced the corresponding S. cerevisiae SPT-associated polypeptides, as confirmed by immunoblotting (Figure 10A). We assessed in vivo SPT activity by measuring the DoxSA produced when expressing AtLCB1C144W (Figure 10B). Deoxy-LCBs cannot be phosphorylated/degraded and are used as a readout for in situ SPT activity (Gable et al., 2010; Kimberlin et al., 2016). When expressed in this S. cerevisiae background, the wild-type Arabidopsis ORM1 was able to suppress DoxSA production, which is consistent with its function as a negative regulator of SPT activity. By contrast, DoxSA concentrations in AtORM1ΔMet51-expressing cells were similar to those in vector control cells lacking ORM1, which is consistent with a lack of repressed SPT activity.
Figure 10.
AtORM1ΔMet51 Fails to Regulate SPT Activity and Does Not Interact with LCB1.
(A) AtORM1ΔMet51 was stably expressed in S. cerevisiae with the native SPT complex replaced by the Arabidopsis SPT complex (see Methods). AtLCB1-FLAG, MYC-AtLCB2a, and HA-AtssSPTa without or with HA-AtORM1 or HA-AtORM1ΔMet51 were expressed in S. cerevisiae strain lcb1 tsc3. Five, 10, and 15 µg of microsomal proteins was loaded and analyzed by SDS-PAGE (4 to 12%; Invitrogen) and detected with anti-LCB1 (1:3000) and anti-HA (Covance) antibodies.
(B) DoxSA levels were determined from cells expressing AtLCB1C144W and AtLCB2a, HA-AtssSPTa along with vector, HA-AtORM1 wild-type, or HA-AtORM1ΔMet51. Shown are the mean ± sd of DoxSA levels from six independent colonies for each strain. Asterisks denote significant differences, as determined by two-tailed Student’s t test with a significance of P ≤ 0.001; NS, not significant, n = 6.
(C) Coimmunoprecipitation of FLAG-tagged AtLCB1 in S. cerevisiae expressing AtLCB1-FLAG, MYC-AtLCB2a, HA-AtssSPTa, and either HA-AtORM1 or HA-AtORM1ΔMet51. Solubilized S. cerevisiae microsomes were incubated with anti-FLAG beads, and protein was eluted with FLAG peptide. Solubilized microsomes (Input), unbound and bound (IP-FLAG) were analyzed by immunoblotting. ELO3, an integral ER membrane protein, was used as a negative control.
(D) Topology mapping of AtORM1ΔMet51. GCs were inserted after the indicated amino acids, and the GC-tagged proteins were expressed in S. cerevisiae. Increased mobility following treatment of microsomes with endoglycosidase H (Endo H) revealed that the GCs at residues 46 and 121 are glycosylated and therefore reside in the lumen of the ER. However, the GC at residue 82 is not glycosylated, indicating that residue 82 is located in the cytosol. AtORM1ΔMet51 retains the topology of wild-type (WT) ORM1.
ORMs interact with the first transmembrane domain of LCB1 to repress SPT activity in S. cerevisiae (Han et al., 2019), although the structural components of ORM associated with this interaction have not been defined. To test whether AtORM1ΔMet51 physically interacts with AtLCB1, as does the wild-type ORM1, we performed coimmunoprecipitation of FLAG-tagged AtLCB1 with solubilized microsomes from S. cerevisiae cells expressing Myc-AtLCB2a, hemagglutinin (HA)-AtssSPTa, and HA-AtORM1 or HA-AtORM1ΔMet51. Pull-downs of AtLCB1 resulted in coimmunoprecipitation of AtLCB2a and AtORM1, but not ELO3, an ER protein that does not interact with SPT. By contrast, only trace amounts of HA-AtORM1ΔMet51 were detected in the AtLCB1 pull-downs (Figure 10C).
This finding indicates that Met-51 is critical for the ORM-LCB1 physical interaction to regulate SPT activity. To determine whether the impaired ORM-LCB1 interaction is due to gross or subtle alterations in the secondary structure of ORM induced by the Met-51 deletion, we compared the membrane topology of AtORM1 and AtORM1ΔMet51. We inserted glycosylation cassettes into the two predicted ER luminal loops (at amino acids 46 and 121) and into the cytosolic loop between the second and third transmembrane domains (at amino acid 82) and expressed the proteins in S. cerevisiae along with reconstituted Arabidopsis SPT. The analysis showed that the cassettes in the predicted luminal domains were glycosylated, while the cassette in the predicted cytosolic domain was not (Figure 10D). Thus, we conclude that ORM1 with the Met-51 deletion retains the topology of wild-type ORM1.
DISCUSSION
Our findings identified the essential role of sphingolipid biosynthetic regulation at the level of SPT for seed viability, which was previously unclear due to the lack of complete knockout mutants for ORM genes in plants. We showed that orm1−/− orm2−/− seeds have impaired embryo development accompanied by hyperaccumulation of the cytotoxic sphingolipid biosynthetic intermediates ceramides. Strongly enhanced ceramide accumulation was also observed in the S. cerevisiae orm1Δ/orm2Δ mutant (Breslow et al., 2010; Han et al., 2010) and recently in Ormdl1/3 mutant mice (Clarke et al., 2019). We also confirmed that impaired seed viability in the mutant is due solely to the function of ORMs in SPT regulation, rather than other ascribed ORM functions (Yang et al., 2019). This was achieved by mimicking this phenotype by removing the first transmembrane domain of LCB1, which is required for ORM binding to SPT (Han et al., 2019). Furthermore, through gene editing, we recovered the orm1Δmet/Δmet orm2−/− mutant, which expresses an ORM1 structural variant that is strongly compromised in the regulation of SPT activity. This mutant provided valuable insight into cellular responses to unchecked sphingolipid biosynthesis. These responses include compromised organellar structures, the induction of catabolic genes to maintain sphingolipid homeostasis, and clues about the structural requirements of ORM for interaction with LCB1.
Our findings emphasize that the full significance of ORMs to plant viability can only be assessed by complete knockout of the corresponding genes. By contrast, Arabidopsis ORM-suppressed plants previously generated by RNAi or artificial microRNA methods were fully viable, although the response to bacterial pathogens was altered in these plants and early senescence was observed with the most extreme suppression of ORM expression (Kimberlin et al., 2016; Li et al., 2016). Similar to our findings, a recent report revealed the inability to recover mice lacking all three ORMDL genes (Clarke et al., 2019). However, we were able to more precisely determine that lethality occurs during seed development rather than during gametogenesis. This finding contrasts with those from previous studies of plants with strongly reduced sphingolipid biosynthetic capacity due to impaired SPT activity (Dietrich et al., 2008; Teng et al., 2008; Kimberlin et al., 2013). In these mutants, pollen is defective in endomembrane formation and is unable to complete maturation. Sphingolipids accumulate to exceptionally high levels in Arabidopsis pollen relative to leaves (Luttgeharm et al., 2015b; Ischebeck, 2016). As such, it is likely that pollen is able to tolerate unregulated sphingolipid synthesis that results from complete ORM knockout.
The mechanism underlying the loss of seed viability from unregulated SPT activity in orm1−/− orm2−/− and orm1Δmet/Δmet orm2−/− mutants likely involves a combination of the functions of sphingolipids as major structural components of the endomembrane and as bioactive mediators of cellular activities such as PCD that lead to aberrant embryo development. As shown in orm1Δmet/Δmet orm2−/− seedlings, strong upregulation of sphingolipid biosynthesis results in large alterations in membrane and organellar structures in plant cells (Figure 8). These seedlings appear to have defects in ER function, as indicated by the relative reduction in the total content of very long chain fatty acids in the abnormal seeds from the progeny of orm1−/− and orm2−/− crosses and LCB1ΔTMD1 transgenic lines (Figure 3G). These fatty acids are formed by ER-localized enzymes including the FAE1-encoded β-ketoacyl-CoA synthase. The hyperaccumulation of ceramides in these seeds also likely triggers PCD in embryonic cells, as indicated by the enhanced expression of PCD-related genes in orm1Δmet/Δmet orm2−/− seedlings (Figures 9E to 9G; Supplemental Figure 8E).
Among the gene-edited ORM variants identified in our studies was a mutant that contained an in-frame deletion of Met-51 combined with a homozygous knockout of ORM2 (orm1Δmet/Δmet orm2−/−). Seeds from this mutant were viable, in contrast to orm1−/− orm2−/−; however, the plants did not advance beyond the seedling stage and had strong developmental defects. Like the orm1−/− orm2−/− seeds, the orm1Δmet/Δmet orm2−/− seedlings hyperaccumulated ceramides with C16 fatty acids. These seedlings also accumulated aberrant sphingolipids including DoxSA-containing ceramides, GlcCer containing nonhydroxylated fatty acids, and IPCs, all of which were nearly absent from wild-type seedlings. Cells from the orm1Δmet/Δmet orm2−/− seedlings displayed gross defects in membrane and organellar structures as well as apparent autophagosome-like structures. The early cell death displayed by the orm1Δmet/Δmet orm2−/− seedlings can be attributed to the activation of PCD pathways, as indicated by the high transcript levels of pathogenesis- and senescence-related genes that have been shown to be activated by the accumulation of LCB and ceramides.
Notably, Met-51 is predicted to occur at a position that is adjacent to the second transmembrane domain of ORMs, but is not a conserved residue across eukaryotic ORM or ORMDL proteins (Supplemental Figure 9). Using S. cerevisiae mutants containing the Arabidopsis SPT complex, we determined that the ORM1 Met-51 mutant has greatly reduced interaction with Arabidopsis LCB1, which is required for ORM-induced suppression of SPT activity. Given that Met-51 is not conserved in eukaryotes, it is likely that LCB1 does not directly interact with this residue. Instead, the lack of this amino acid likely produces a conformational change at the second transmembrane domain of ORM that impedes its regulatory interaction with the first transmembrane domain of LCB1. The maintenance of the topology of ORM1ΔMet51 in microsomal membranes was verified by endoglycosidase H digestion studies using the mutant ORM1 protein carrying glycosylation cassettes. To date, no residues or structural features in ORMs have been identified that are associated with their interaction with the LCB1/LCB2 heterodimer of SPT. Our findings point to the possible interaction of the first transmembrane domain of LCB1 with the second transmembrane domain of ORM as the basis for SPT regulation. Additional structural studies are required to fully elucidate these potential regulatory interactions between ORM and LCB1.
The use of gene editing also allowed us to assess the redundancy of ORM1 and ORM2. Notably, single mutants and progeny from the crosses that genotype as orm1+/− orm2+/− had an appearance similar to the wild-type plants under normal conditions. However, orm1−/− orm2+/− seedlings displayed early senescence and did not flower (Figure 4A). By comparison, orm1+/− orm2−/− plants were fertile but were strongly dwarfed and had delayed flowering compared to the wild type and orm1+/− orm2+/− plants (Figures 4B and 4C). Perhaps the stronger phenotype associated with the complete ORM1 knockout in the ORM2 heterozygous background reflects the finding that ORM1 is more highly expressed than ORM2 throughout the plant except in pollen (Kimberlin et al., 2016). The normal appearance of mutants genotyped as ORM1/orm2−/− and orm1−/−/ORM2 suggests that ORM1 and ORM2 are functionally redundant, despite the phenotypic differences observed in orm1−/− orm2+/− and orm1+/− orm2−/− seedlings. However, we did observe that orm1+/− orm2−/− plants have a highly bushed appearance and are strongly delayed in flowering (>80 d to flowering; Figure 4D), pointing to a meristem defect (Tantikanjana et al. 2001). This phenotype requires further investigation, but it suggests that ORM2 contributes more strongly to meristem function than ORM1, perhaps due to cell type–specific differences in the expression of the ORM genes or to a nonsphingolipid-related function of ORM proteins.
Our results also revealed transcriptional mechanisms for maintaining sphingolipid homeostasis upon the enhanced production of LCBs in the orm1Δmet/Δmet orm2−/− mutant. LOH2 and LOH3 (encoding the functionally distinct ceramide synthases), SPHK2 (enconding a LCB kinase) and DPL1 (encoding the last step in LCB degradation) were transcriptionally upregulated in the mutant. Notably, upregulating LOH2 expression was associated with the preponderance of ceramides containing C16 fatty acids and dihydroxy LCBs (the principal products of LOH2 ceramide synthase activity) in free ceramides and GlcCer, including nhGlcCer, which accumulated in orm1Δmet/Δmet orm2−/− seedlings but were detected at only low concentrations in the wild type and ORM1 and ORM2 single mutants. These findings are consistent with our previous report that LOH2 activity is upregulated in Arabidopsis ORM RNAi plants, presumably as a pathway for reducing cytotoxicity of free LCBs and ceramides (which are metabolized to GlcCer; Kimberlin et al., 2016). No changes were detected in LCB1 or ssSPTa transcript levels in the orm1Δmet/Δmet orm2−/− mutant, indicating that the transcriptional regulation of genes for SPT complex proteins is not a pathway for maintaining sphingolipid homeostasis in response to deregulated LCB biosynthesis. Instead, the expression of genes involved in the catabolism of LCBs increased approximately six- to sevenfold (SPHK2 and DPL1) in this mutant, suggesting that an unknown mechanism is activated in response to increased ceramide and/or LCB levels.
Our overall findings about the metabolic and developmental defects associated with the deregulation of SPT by disrupting ORM genes or removing the first transmembrane domain of LCB1 are schematically summarized in Figure 11. These sphingolipid-related regulatory processes identified in Arabidopsis are likely found in other plant species due to the conservation of sphingolipid metabolic enzymes in the plant kingdom. Still unanswered is how ORM interactions with LCB1 are regulated in response to perturbations in intracellular sphingolipid levels or abiotic and biotic stresses (e.g., bacterial and fungal pathogenesis). Similar to mammalian ORMDLs, plant ORMs lack the Ser-rich N-terminal extension found in S. cerevisiae ORMs, which is phosphorylated or dephosphorylated in response to intracellular sphingolipid levels to mediate ORM-LCB1 interactions (Breslow et al., 2010; Han et al., 2010). Mammalian ORMDLs have recently been shown to bind ceramides directly, which affects the interactions of ORMs with LCB1 (Davis et al., 2019). A similar regulatory mechanism might occur in plants. In this regard, we previously speculated that LOH2-derived ceramides or glycosphingolipids enriched in dihydroxy LCBs and C16 fatty acids likely provide minimal SPT regulation relative to those containing trihydroxy LCBs and very long chain fatty acids based on the hyperaccumulation of sphingolipids found in sbh1 sbh2 mutants and LOH2-overexpressing plants (Chen et al., 2008; Luttgeharm et al., 2015a). Still, how ORMs reversibly regulate SPT activity in response to cellular sphingolipid requirements remains an outstanding question in plants.
Figure 11.
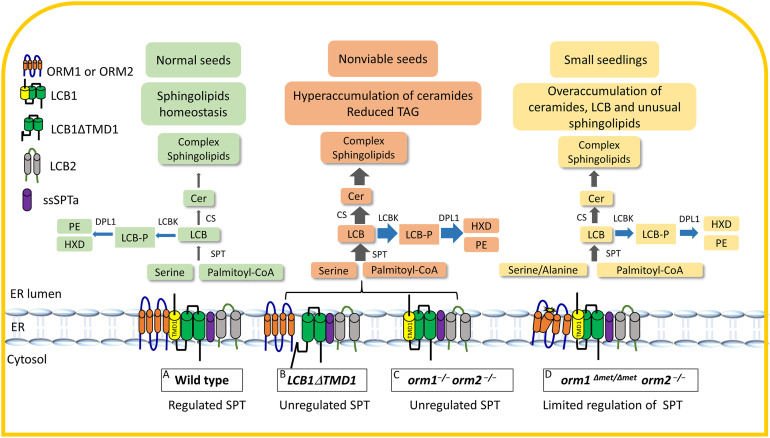
Model of ORM-Mediated Sphingolipid Biosynthesis in Wild-Type Plants and ORM and LCB1 Mutants.
(A) to (D) ORM proteins and LCB1 are integral ER membrane proteins with multiple transmembrane domains (TMDs). The ORM proteins contain four TMDs, with both termini located in the cytosol, while LCB1 has three TMDs, with its N terminus located in the ER lumen and C terminus located in the cytosol. LCB1, along with LCB2 and ssSPTa, comprise SPT, which catalyzes the first step in sphingolipid biosynthesis. TMD1 of LCB1 is required for ORM binding to SPT (A). Expression of LCB1 without its first transmembrane domain (B) or the complete knockout of ORM1 and ORM2 (C) results in the loss of SPT regulation. This is characterized by strongly enhanced accumulation of ceramides and selected complex sphingolipids and the loss of seed viability marked by a strong reduction in TAG content. The lack of Met-51 before the TMD2 of ORM1 it thought to causes a conformational change that dramatically decreases its interaction with LCB1 for SPT regulation (D). CER, ceramide; CS, ceramide synthase; DPL1, LCB phosphate lyase; HXD, hexadecenal; LCBK, long chain base kinase; LCB-P, long chain bases-phosphate; PE, phosphoethanolamine. Grey arrows indicate de novo sphingolipid biosynthesis and blue arrows indicate catabolic reactions. Thickness of arrows represents relative metabolic flux levels.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) was used as the wild-type reference in this study. Arabidopsis seedlings were grown on Murashige and Skoog (MS) medium supplemented with 1% (w/v) Suc and 0.8% (w/v) agar, pH 5.7, with 16-h-light (100 μmol/ m−2 s−1)/8-h-dark conditions at 22°C. The light source for growth chamber–grown seedlings was supplied by standard wide-spectrum fluorescent bulbs type F32/841/ECO 32 W (maximum intensity, 480 to 570 nm). For Arabidopsis plants in soil, seeds were sown, and after 2 d of stratification at 4°C, plants were grown at 22°C with 16-h-light (100 μmol/ m−2 s−1)/8-h-dark conditions. The light source for these plants came from wide-spectrum fluorescent bulbs of type F32/841/ECO 32 W and/or F72/T12/CW/VHO 160 W and F96/T12/CW/VHO215 215 W (maximum intensity, 480 to 570 nm).
Generation of CRISPR/Cas9 ORM Mutants
For CRISPR/Cas9-mediated gene editing of ORM1 and ORM2, designed target sites (Figure 1A) were fused with a single guide RNA and expressed under the control of the U6 promoter. The egg cell–specific EC1 promoter was used to drive Cas9 expression as previously reported by Wang et al. (2015). In short, BsaI sites were incorporated by PCR into the ORM target sequences (primers P1 to P4; Supplemental Table). The purified PCR products were digested with BsaI and ligated to the BsaI-linearized binary vector pHEE401E. The final CRISPR/Cas9 binary vector was electroporated into Agrobacterium tumefaciens strain GV3101 and then transformed into the Arabidopsis Col-0 wild-type plants via the floral dip method (Clough and Bent, 1998). The seeds were screened for hygromycin resistance on MS plates containing 25 mg/L hygromycin. For genotyping, fragments including the target regions of ORM1 and ORM2 were amplified by PCR from the genomic DNA of transgenic plants (primers P5 to P8; Supplemental Table). Amplicons were digested with the restriction enzymes BslI (ORM1) and DraIII (ORM2). The specific indels were identified by DNA sequencing. To analyze for nontransgenic plants, progeny of hygromycin-selected and confirmed homozygous (CRISPR/Cas9 mutation) T1 plants were sown directly on soil without hygromycin selection. These plants were then screened by PCR (P9+P10; Supplemental Table) for the lack of the Cas9 gene with the presence of the CRISPR mutation, in the T2 generation. The plants lacking Cas9 but containing the CRISPR mutation were kept and used for further studies as mutated but nontransgenic lines.
Genetic Complementation of orm1Δmet/Δmet orm2−/−
For genetic complementation of the mutant orm1Δmet/Δmet orm2−/−, ORM1 cDNA was synthesized with included silent mutations of the ORM1 gRNA target sequence to mitigate possible editing of the transgene. The cDNA was amplified by overlapping PCR and cloned into the EcoRI and XbaI sites of binary vector pBinGlyRed3 under the control of the native ORM1 promoter 600-bp region upstream of the ORM1 start codon (primers P11 to P16; Supplemental Table). orm1Δmet/Δmet orm2+/− plants were transformed with the pBinGlyRed3-ORM1 construct by the floral dip method (Clough and Bent, 1998). Transformants were selected based on Discosoma red fluorescent protein fluorescence and genotyped. Mutation was confirmed by sequencing.
Generation of the LCB1ΔTMD1 Mutant
LCB1ΔTMD1 was generated by deleting coding sequence for 17 amino acids corresponding to the first transmembrane domain of AtLCB1 (amino acids 35 to 51). LCB1ΔTMD1 under the control of the LCB1 native promoter was cloned into the pBinGlyRed3 binary vector, which was transformed into A. tumefaciens GV3101 by electroporation. Heterozygous LCB1/lcb1-knockout mutants (SALK_077745) were transformed by the floral dip method (Clough and Bent, 1998).
Pollen Staining
Anthers of mature plants were isolated and smeared on a glass slide. The pollen was stained using Alexander staining method (Alexander, 1969) for 1 h at 25°C. Pollen imaging was performed using the EVOS FL Auto Cell Imaging System.
Sphingolipid Extraction and Analysis
Sphingolipids were extracted as described in Markham and Jaworski (2007). Briefly, 12- to 15-d-old Arabidopsis seedlings grown on solid medium were collected from independent plates for each biological replicate. The seedlings were lyophilized, and 10 to 30 mg of tissue was homogenized and extracted with isopropanol:heptane:water (55:20:25, v/v/v). We used 1 to 4 mg of plant material for each biological replicate for sphingolipid analysis from seeds. Internal standards for the different sphingolipid classes were added. The supernatants were dried and de-esterified with methylamine in ethanol:water (70:30, v/v). The lipid extract was re-suspended in tetrahydrofuran:methanol:water (5:2:5, v/v/v) containing 0.1% (v/v) formic acid. The sphingolipid species were analyzed using a Shimadzu Prominence ultra-performance liquid chromatography system and a 4000 QTRAP mass spectrometer (AB SCIEX). Data analysis and quantification were performed using the software Analyst 1.5 and MultiQuant 2.1 as described by Markham and Jaworski (2007), Kimberlin et al. (2013), and Davis et al. (2020).
Lipid Extraction Analysis
To quantify the TAG content, lipids were extracted from ∼1 mg of seeds using a method based on that of Bligh and Dyer (1959). Seeds were ground using a glass rod in 13 × 100-mm glass screw cap tubes with 3 mL of methanol:chloroform (2:1, v/v). Triheptadecanoin (17:0-TAG) was added to the seeds as an internal standard prior to extraction. After 1 h of incubation at 25°C, 1 mL of chloroform and 1.9 mL of water were added. The solution was mixed thoroughly and centrifuged at 400g for 10 min. The lower organic phase containing total lipids was transferred to a new glass tube and solvent evaporated under a N2 stream with heating at 40°C. The sample was redissolved in 1 mL of heptane and loaded onto a solid phase extraction column (Supelco Supelclean LC-Si SPE column; Sigma-Aldrich) pre-equilibrated with heptane. A purified TAG fraction was eluted from the column and converted to fatty acid methyl esters, which were analyzed by gas chromatography as previously described (Zhu et al., 2016). TAG fatty acid content was quantified relative to 17:0 fatty acid methyl ester from the internal standard.
RNA Isolation and Quantitative RT-PCR
RNA was extracted from 12- to 15-d-old Arabidopsis seedlings grown on solid MS medium. Each replicate corresponds to pooled seedlings from independent plates. RNA extraction was performed using an RNeasy Kit (Qiagen) according to the manufacturer’s protocol. The isolated RNA (1 μg) was treated with DNase I (Invitrogen). cDNA conversion was performed with a RevertAid cDNA synthesis kit (Thermo Fisher Scientific). SYBR Green was used as the fluorophore in a qPCR supermix (Qiagen). PP2AA3 and UBIQUITIN (UBQ) were used as internal reference genes. qPCR was performed using a Bio-Rad MyiQ iCycler qPCR instrument. The thermal cycling conditions were an initial step of 95°C for 10 min followed by 45 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Primers used in this study are listed in Supplemental Table.
Electron Microscopy
Ten-day-old wild-type and orm1Δmet/Δmet orm2−/− seedlings were used for TEM. The samples were cut and fixed with 2.5% (v/v) glutaraldehyde and 2.0% (v/v) paraformaldehyde in 0.1 M cacodylate buffer. The samples were subjected to postfixation with 1% (w/v) osmium tetroxide in 0.1 M cacodylate buffer, dehydrated with ethanol and acetone, and embedded with a Spurr’s Embedding Kit. Ultrathin sections (100 nm) were cut and stained with uranyl acetate and lead citrate. Samples were imaged on a Hitachi H7500 transmission electron microscope at an accelerating voltage of 80 kV.
Saccharomyces cerevisiae Cell Growth and Expression Plasmids
Saccharomyces cerevisiae strain TDY9113 (Mata tsc3Δ:NATlcb1Δ:KAN ura3 leu2 lys2 trp1Δ) lacking endogenous SPT was used for the expression of Arabidopsis SPT subunits and ORM proteins as described by Kimberlin et al. (2016). For DoxSA quantification, S. cerevisiae strain TDY9113 expressing AtLCB1C144W was grown in 1.5% (w/v) Gal and 0.5% (w/v) Glc supplemented with 40 mM Ala. Plasmids for the expression of AtLCB1-FLAG, Myc-AtLCB2a, and HA-AtssSPTa in S. cerevisiae were as described by Kimberlin et al. (2013) and for HA-AtORM1 as described by Kimberlin et al. (2016). AtLCB1C144W was generated by QuikChange mutagenesis (Agilent Technologies) and confirmed by sequencing. The open reading frame of AtORM1ΔMet51 was amplified by PCR and inserted into pPR3-N (Dualsystems Biotech) for expression with an N-terminal HA tag. LCB and DoxSA quantifications were performed as previously described by Kimberlin et al. (2016).
Immunoprecipitation
Microsomal membrane proteins were prepared from S. cerevisiae cells expressing FLAG-tagged AtLCB1, Myc-tagged AtLCB2a, HA-tagged AtssSPTa, and HA-tagged AtORM1 or AtORM1ΔMet51. Microsomal membrane proteins were solubilized in 1.5% digitonin at 4°C for 2.5 h and incubated with FLAG-beads (Sigma-Aldrich) overnight. The bound proteins were eluted in immunoprecipitation buffer (50 mM Hepes-KOH, pH 6.8, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM calcium chloride, and 15% [v/v] glycerol) containing 0.25% (w/v) digitonin and 200 μg/mL FLAG peptide, resolved on a 4 to 12% (w/v) Bis-Tris NuPAGE gel (Invitrogen), and detected by immunoblotting with anti-HA (1:5000 dilution; Covance), anti-Myc (1:3000 dilution; Sigma-Aldrich), and anti-FLAG (1:5000 dilution; GenScript) antibodies.
Membrane Topology Mapping of AtORM1ΔMet51
AtORM1 or AtORM1ΔMet51-encoding synthetic cDNAs with an in-frame glycosylation cassette (GC) inserted after codon 46, 82 or 121 were synthesized by GenScript and ligated into pPR3-N for expression with an N-terminal HA tag. The HA-ORM1-GC–tagged proteins were expressed (along with AtLCB1-FLAG, MYC-AtLCB2a, and HA-AtssSPTa) in S. cerevisiae strain TDY9113. Isolation of microsomal proteins, digestion with endoglycosidase H, and immunodetection of the AtORM1 proteins were performed as previously described (Kimberlin et al., 2016).
Statistical Analyses
Two-tailed Student’s t test was performed to evaluate statistically significant differences compared to the control (wild type). One-way ANOVA followed by Tukey’s test was used to determine the differences among the five genotypes for a given variable. Values of P < 0.05 were considered statistically significant. The statistical analyses were done using GraphPad Prism 8.3.0. t test and ANOVA results are shown in Supplemental Data Set.
Accession Numbers
Accession numbers for the genes studied in this work are as follows: ORM1 (At1G01230); ORM2 (At5g42000); LCB1 (At4g36480); ssSPTa (At1g06515); LOH1 (At3g25540); LOH2 (At3g19260); LOH3 (At1g13580); SPHK1 (At4g21540); SPHK2 (At2g46090); DPL1 (At1g27980); PP2AA3 (At1g13320); PRXC (At3g49120); PR2 (At3g57260); PR3 (At3g12500); FMO (At1g19250); SAG13 (At2g29350); UBQ (At5g25760).
Supplemental Data
Supplemental Figure 1. Predicted protein sequences of ORMs in the CRISPR/Cas9 mutants.
Supplemental Figure 2. PCR/digestion-based genotyping of CRISPR/Cas9 ORM mutants.
Supplemental Figure 3. Complementation of orm1Δmet/Δmet orm2−/−.
Supplemental Figure 4. Ceramide compositions with hydroxylated fatty acids in ORM mutants.
Supplemental Figure 5. Glucosylceramide compositions in ORM mutants.
Supplemental Figure 6. Composition of glucosylceramides containing non-hydroxylated fatty acids in ORM mutants.
Supplemental Figure 7. Glycosylinositolphosphoceramide compositions in ORM mutants.
Supplemental Figure 8. Expression of genes associated with sphingolipid biosynthetic and catabolic pathways and pathogenesis.
Supplemental Figure 9. Amino acid sequence alignment of ORM proteins.
Supplemental Table. Primer sequences used for cloning, RT-PCR, qPCR, and genotyping.
Supplemental Data Set. Result of t tests and ANOVA.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jaydeeo Kolape for microscopy technical assistance and Jules Russ for the TEM sample preparation at the Microscopy Core Facility in the Center for Biotechnology at the University of Nebraska–Lincoln. This work was supported by the National Science Foundation (grant MCB 1818297 to E.B.C., T.M.D., and J.E.M.) and the Mexican National Council of Science and Technology (Consejo Nacional de Ciencia y Tecnologia; to A.G.-S.).
AUTHOR CONTRIBUTIONS
A.G.-S., G.H., T.M.D., and E.B.C. designed the study; A.G.-S., G.H., L.G., Y.L., R.E.C., and J.E.M. performed the experiments and analyzed the data, along with G.H., T.M.D., and E.B.C.; and A.G.-S., G.H., T.M.D., and E.B.C. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Alden K.P., Dhondt-Cordelier S., McDonald K.L., Reape T.J., Ng C.K., McCabe P.F., Leaver C.J.(2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410: 574–580. [DOI] [PubMed] [Google Scholar]
- Alexander M.P.(1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Bi F.C., et al. (2014). Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26: 3449–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J.(1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., Weissman J.S.(2010). Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Cahoon E.B., Saucedo-García M., Plasencia J., Gavilanes-Ruíz M.(2009). Plant Sphingolipids: Structure, Synthesis and Function In Lipids in Photosynthesis: Essential and Regulatory Functions, Wada H., and Murata N., eds (Dordrecht: Springer Netherlands; ), pp. 77–115. [Google Scholar]
- Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B.(2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Markham J.E., Dietrich C.R., Jaworski J.G., Cahoon E.B.(2008). Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20: 1862–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueasiri C., Chunthong K., Pitnjam K., Chakhonkaen S., Sangarwut N., Sangsawang K., Suksangpanomrung M., Michaelson L.V., Napier J.A., Muangprom A.(2014). Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development. PloS One 5: e106386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B.A., et al. (2019). The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. eLife 8: e51067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coursol S., Fan L.M., Le Stunff H., Spiegel S., Gilroy S., Assmann S.M.(2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654. [DOI] [PubMed] [Google Scholar]
- Dadsena S., et al. (2019). Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nat. Commun. 10: 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.L., Gable K., Suemitsu J., Dunn T.M., Wattenberg B.W.(2019). The ORMDL/Orm-serine palmitoyltransferase (SPT) complex is directly regulated by ceramide: Reconstitution of SPT regulation in isolated membranes. J. Biol. Chem. 294: 5146–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.A., et al. (2020). The lipid flippases ALA4 and ALA5 play critical roles in cell expansion and plant growth. Plant Physiol. 182: 2111–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C.R., Han G., Chen M., Berg R.H., Dunn T.M., Cahoon E.B.(2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54: 284–298. [DOI] [PubMed] [Google Scholar]
- Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, Dunn TM(2010). A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J Biol Chem 285: 22846–22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K., Slife H., Bacikova D., Monaghan E., Dunn T.M.(2000). Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275: 7597–7603. [DOI] [PubMed] [Google Scholar]
- Gupta S.D., Gable K., Alexaki A., Chandris P., Proia R.L., Dunn T.M., Harmon J.M.(2015). Expression of the ORMDLS, modulators of serine palmitoyltransferase, is regulated by sphingolipids in mammalian cells. J. Biol. Chem. 290: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Gupta S.D., Gable K., Bacikova D., Sengupta N., Somashekarappa N., Proia R.L., Harmon J.M., Dunn T.M.(2019). The ORMs interact with transmembrane domain 1 of Lcb1 and regulate serine palmitoyltransferase oligomerization, activity and localization. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864: 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lone M.A., Schneiter R., Chang A.(2010). Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huby E., Napier J.A., Baillieul F., Michaelson L.V., Dhondt-Cordelier S.(2020). Sphingolipids: Towards an integrated view of metabolism during the plant stress response. New Phytol. 225: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T.(2016). Lipids in pollen - They are different. Biochim. Biophys. Acta 1861 (9 Pt B): 1315–1328. [DOI] [PubMed] [Google Scholar]
- Kimberlin A.N., Han G., Luttgeharm K.D., Chen M., Cahoon R.E., Stone J.M., Markham J.E., Dunn T.M., Cahoon E.B.(2016). ORM expression alters sphingolipid homeostasis and differentially affects ceramide synthase activity. Plant Physiol. 172: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin A.N., Majumder S., Han G., Chen M., Cahoon R.E., Stone J.M., Dunn T.M., Cahoon E.B.(2013). Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 25: 4627–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yin J., Rong C., Li K.E., Wu J.X., Huang L.Q., Zeng H.Y., Sahu S.K., Yao N.(2016). Orosomucoid proteins interact with the small subunit of serine palmitoyltransferase and contribute to sphingolipid homeostasis and stress responses in Arabidopsis. Plant Cell 28: 3038–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T.(2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17: 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgeharm K.D., Chen M., Mehra A., Cahoon R.E., Markham J.E., Cahoon E.B.(2015a). Overexpression of Arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death, and mycotoxin resistance. Plant Physiol. 169: 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgeharm K.D., Kimberlin A.N., Cahoon R.E., Cerny R.L., Napier J.A., Markham J.E., Cahoon E.B.(2015b). Sphingolipid metabolism is strikingly different between pollen and leaf in Arabidopsis as revealed by compositional and gene expression profiling. Phytochemistry 115: 121–129. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G.(2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Li J., Cahoon E.B., Jaworski J.G.(2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281: 22684–22694. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Molino D., Gissot L., Bellec Y., Hématy K., Marion J., Belcram K., Palauqui J.C., Satiat-Jeunemaître B., Faure J.D.(2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer M., Stegmann M., Mueller M.J., Waller F.(2010). Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 584: 4053–4056. [DOI] [PubMed] [Google Scholar]
- Tantikanjana T., Yong J.W.H., Letham D.S., Griffith M., Hussain M., Ljung K., Sandberg G., Sundaresan V.(2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 15: 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C., Dong H., Shi L., Deng Y., Mu J., Zhang J., Yang X., Zuo J.(2008). Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 146: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., Riezman H., Feussner I.(2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192: 841–854. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J.(2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Kimberlin A.N., Elowsky C.G., Liu Y., Gonzalez-Solis A., Cahoon E.B., Alfano J.R.(2019). A plant immune receptor degraded by selective autophagy. Mol. Plant 12: 113–123. [DOI] [PubMed] [Google Scholar]
- Zheng P., Wu J.X., Sahu S.K., Zeng H.Y., Huang L.Q., Liu Z., Xiao S., Yao N.(2018). Loss of alkaline ceramidase inhibits autophagy in Arabidopsis and plays an important role during environmental stress response. Plant Cell Environ. 41: 837–849. [DOI] [PubMed] [Google Scholar]
- Zhu L.H., et al. (2016). Dedicated industrial oilseed crops as metabolic engineering platforms for sustainable industrial feedstock production. Sci. Rep. 6: 22181. [DOI] [PMC free article] [PubMed] [Google Scholar]