Diacylglycerol kinases DGK2 and DGK4 produce phosphatidic acid at the ER for signaling in pollen tube growth and for the biosynthesis of phosphatidylglycerol in leaves of Arabidopsis.
Abstract
Phosphatidic acid (PA) is a key phospholipid in glycerolipid metabolism and signaling. Diacylglycerol kinase (DGK) produces PA by phosphorylating diacylglycerol, a crucial step in PA metabolism. Although DGK activity is known to be involved in plant development and stress response, how specific DGK isoforms function in development and phospholipid metabolism remains elusive. Here, we showed that Arabidopsis (Arabidopsis thaliana) DGK2 and DGK4 are crucial for gametogenesis and biosynthesis of phosphatidylglycerol and phosphatidylinositol in the endoplasmic reticulum (ER). With comprehensive transcriptomic data of seven DGKs and genetic crossing, we found that dgk2-1/– dgk4-1/– plants were gametophyte lethal, although parental single homozygous plants were viable. The dgk2-1/+ dgk4-1/+ double heterozygote showed defective pollen tube growth and seed development because of nonviable mutant gametes. DGK2 and DGK4 were localized to the ER and were involved in PA production for pollen tube growth. Transgenic knockdown lines of DGK2 and DGK4 confirmed the gametophyte defect and also revealed defective leaf and root growth. Glycerolipid analysis in the knockdown lines showed that phosphatidylglycerol and phosphatidylinositol metabolism was affected differently in floral buds and leaves. These results suggest that DGK2 and DGK4 are essential during gametogenesis and are required for ER-localized phospholipid metabolism in vegetative and reproductive growth.
INTRODUCTION
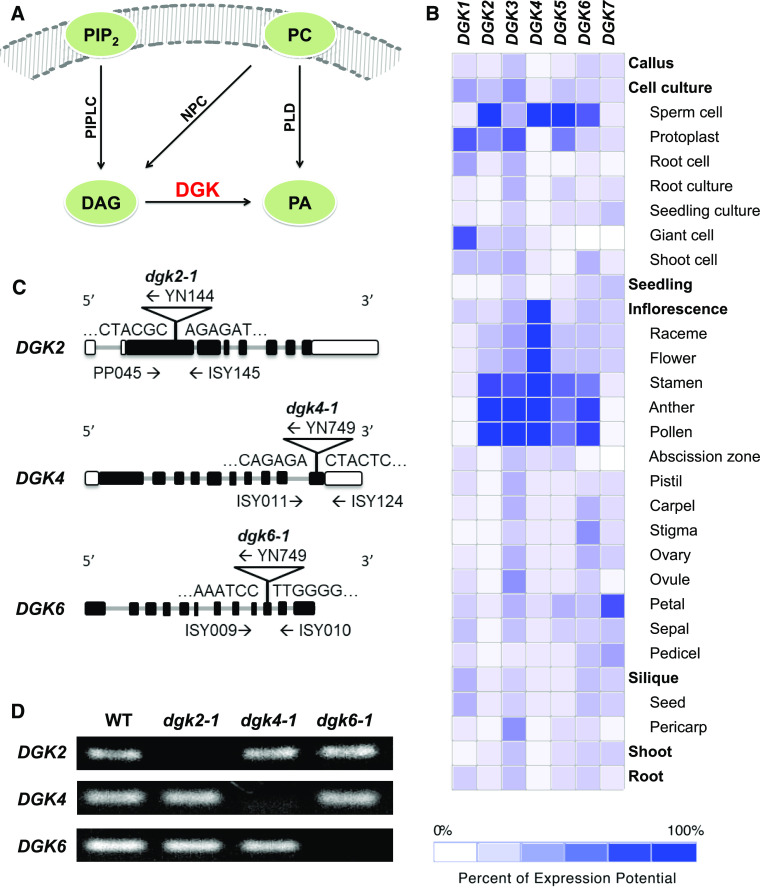
Phospholipid signaling plays a crucial role in plant development and stress response (Heilmann, 2016; Hong et al., 2016). In Arabidopsis (Arabidopsis thaliana) and other seed plants, phosphatidic acid (PA) is a representative class of phospholipid signals (Munnik, 2001). PA can be mainly produced from major membrane phospholipid classes by phospholipase D (PLD) or from diacylglycerol (DAG) by diacylglycerol kinase (DGK). In lipid signaling, DAG can be produced from major membrane phospholipid classes by nonspecific phospholipase C or from minor phosphoinositides by phosphoinositide-specific phospholipase C (Figure 1A; Testerink and Munnik, 2005). Compared with the extensively studied PLD enzyme family (Hong et al., 2016), the physiological function of the DGK family has been less investigated in Arabidopsis and other seed plants.
Figure 1.
Characterization of Arabidopsis DGK2, DGK4, and DGK6. NPC, nonspecific phospholipase C; PC, nonspecific phospholipase; PIP2, phosphatidylinositol 4,5-bisphosphate; PIPLC, phosphoinositide-specific phospholipase C.
(A) Schematic representation of DGK function in plant PA signaling.
(B) Heatmap of tissue-specific expression pattern of DGK genes. Data were analyzed with GENEVESTIGATOR.
(C) Positions of T-DNA and primers used for PCR-based genotyping. Black bars, exons; gray bars, introns; white bars, untranslated regions.
(D) RT-PCR analysis to detect transcripts of DGK2, DGK4, and DGK6 in homozygous dgk2-1, dgk4-1, and dgk6-1 mutants. WT, wild type.
DGK is a highly conserved family of lipid kinases among most unicellular and multicellular organisms. These lipid kinases phosphorylate DAG to produce PA. Regulation of DAG and PA homeostasis is critical in membrane lipid homeostasis, because both DAG and PA are central mediators in phospholipid biosynthesis and signaling in animals and plants. In animals, DAG has been identified as a lipid second messenger that modulates the function of the protein kinase C family (Asaoka et al., 1992; Martelli et al., 2002) and is involved in regulating proteins that affect various biological processes such as carcinogenesis, metastasis, cell growth, development, survival, and apoptosis (Asaoka et al., 1992; Ebinu et al., 1998; Tognon et al., 1998; Ron and Kazanietz, 1999; Sakane et al., 2002). By contrast, in plants, the signaling role of DAG has been an open question because neither members of the protein kinase C family nor putative effectors of DAG signaling have been revealed yet. Instead, plants use PA as a lipid signal to trigger downstream effector responses to overcome diverse stress events including wounding, osmotic pressure, salinity, cold, pathogen attack, and drought (Munnik, 2001; Testerink and Munnik, 2005). Thus, DGK represents a primary enzyme family in phospholipid signaling that may contribute critically to the response to biotic and abiotic stresses (Arisz et al., 2009).

The activity of plant DGKs has been reported in different plant species, including tomato (Lycopersicum esculentum), Arabidopsis, maize (Zea mays), wheat (Triticum aestivum), rice (Oryza sativa), and apple (Malus domestica). From a protein database search, at least 47 DGK proteins are present in 22 plant species, including crop plants such as grape (Vitis vinifera), sweet orange (Citrus × sinensis), and cotton (Gossipium hirsutum; Escobar-Sepúlveda et al., 2017). However, a limited number of reports are available for functional studies of DGK proteins. In tomato, two different cDNAs, named CALMODULIN BINDING DGK (LeCBDGK) and LeDGK1, have been isolated. LeCBDGK contains a calmodulin binding domain that is located close to the C terminus of the protein, but this domain is absent in LeDGK1. Both proteins were demonstrated to be active in vitro. For LeCBDGK, Ca2+ is an important factor for its membrane association (Snedden and Blumwald, 2000). Overexpression of a rice DGK, BTH-INDUCED DGK (OsBIDK1), demonstrated improved disease resistance in transgenic tobacco (Nicotiana tabacum) against Tobacco mosaic virus and Phytophthora parasitica var nicotianae (Zhang et al., 2008).
In Arabidopsis, DGKs are encoded by seven genes (DGK1 to DGK7; Katagiri et al., 1996; Gómez-Merino et al., 2004, 2005). DGK2 is ubiquitously expressed in the whole plant except for stems, and its transcript level is induced by cold exposure (4°C) and wounding (Gómez-Merino et al., 2004). DGK7 is expressed in most tissues, and the protein is primarily found in flowers and young tissues (Gómez-Merino et al., 2005). As reported recently, single knockout mutants of DGK2, DGK3, or DGK5 attenuated PA production and improved tolerance to freezing temperatures (Tan et al., 2018). In addition, different single and double mutants of DGKs showed altered sensitivity to 24-epibrassinolide treatment under optimal and high salinity conditions (Derevyanchuk et al., 2019). Thus, cumulative evidence highlights the role of DGKs in the plant PA signaling response to biotic and abiotic stresses (Gómez-Merino et al., 2004, 2005; Mérida et al., 2008; Arisz et al., 2009; Tan et al., 2018).
In contrast to the relatively well-studied role of DGKs in stress response, their involvement in plant growth and development is an emerging issue. Root elongation is greatly affected by inhibition of DGK function (Gómez-Merino et al., 2005). Recently, dgk4 knockout mutants were found to be defective in pollen tube growth (Vaz Dias et al., 2019). Both DAG and PA are precursors for the biosynthesis of most glycerolipid classes, which are the major components of cellular membranes and are thus associated with plant growth and development. Although an increasing body of evidence revealed the involvement of DGK in PA signaling under stress conditions, a remaining question is whether specific DGK isoforms are essential for plant growth and housekeeping glycerolipid metabolism in Arabidopsis under normal growth conditions.
In this work, we focused on the role of DGKs in reproductive development. We selected DGK2, DGK4, and DGK6 for their characteristic gene expression patterns in reproductive organs. From unbiased genetic crossing between these three DGKs, we discovered that plants doubly heterozygous for DGK2 and DGK4 (dgk2-1/+ dgk4-1/+) showed impaired development in both the male and female gametophytes. Suppressing DGK2 expression in DGK4 knockout plants (35Spro:amiDGK2 dgk4-1/–) or vice versa (35Spro:amiDGK4 dgk2-1/–) conferred gametophyte defects similar to dgk2-1/+ dgk4-1/+ plants, as well as adverse effects to vegetative growth, including shorter roots and smaller seedlings. The lipid profile of the flower buds and rosette leaves of these transgenic 35Spro:amiDGK2 dgk4-1/– plants indicated altered biosynthesis of phosphatidylglycerol (PG) and phosphatidylinositol (PI), two phospholipid classes derived from PA in the endoplasmic reticulum (ER). We propose that DGK2 and DGK4 are ER-localized DGKs required for vegetative and gametophyte development as well as glycerolipid metabolism.
RESULTS
Gene Expression Profile of Seven Arabidopsis DGKs in Different Tissues
To identify which DGK genes are involved in reproductive development, we first analyzed the tissue-specific expression profiles of seven DGK genes by using the bioinformatics search tool GENEVESTIGATOR (www.genevestigator.com). Each DGK gene showed a distinct tissue expression pattern (Figure 1B). DGK1, DGK3, and DGK7 showed rather ubiquitous expression in various tissues or cell types, whereas DGK2, DGK4, and DGK6 were primarily expressed in floral organs. In particular, these genes were preferentially expressed in male reproductive organs, which suggests a potential role in reproductive processes. We therefore focused on DGK2, DGK4, and DGK6 for further functional characterization.
Isolation of dgk2, dgk4, and dgk6 Mutants in Arabidopsis
To investigate the role of DGK2, DGK4, and DGK6 in reproductive development in vivo, we isolated T-DNA insertion mutants for DGK2, DGK4, and DGK6, named dgk2-1, dgk4-1, and dgk6-1, respectively. Each single homozygous mutant dgk2-1, dgk4-1, and dgk6-1 was isolated by PCR-based genotyping. DNA sequencing confirmed that the T-DNA insertions were located within the exons (Figure 1C). No transcript for DGK2, DGK4, and DGK6 was detected in dgk2-1, dgk4-1, and dgk6-1, respectively, by RT-PCR analysis (Figure 1D), and they are thus considered null mutants for their respective target gene. Under normal growth conditions, individual single homozygous mutants showed no visible growth defects, which suggests that the function of these genes may be redundant in vivo.
The dgk2 dgk4 Double Mutant Impairs the Male Gametophyte
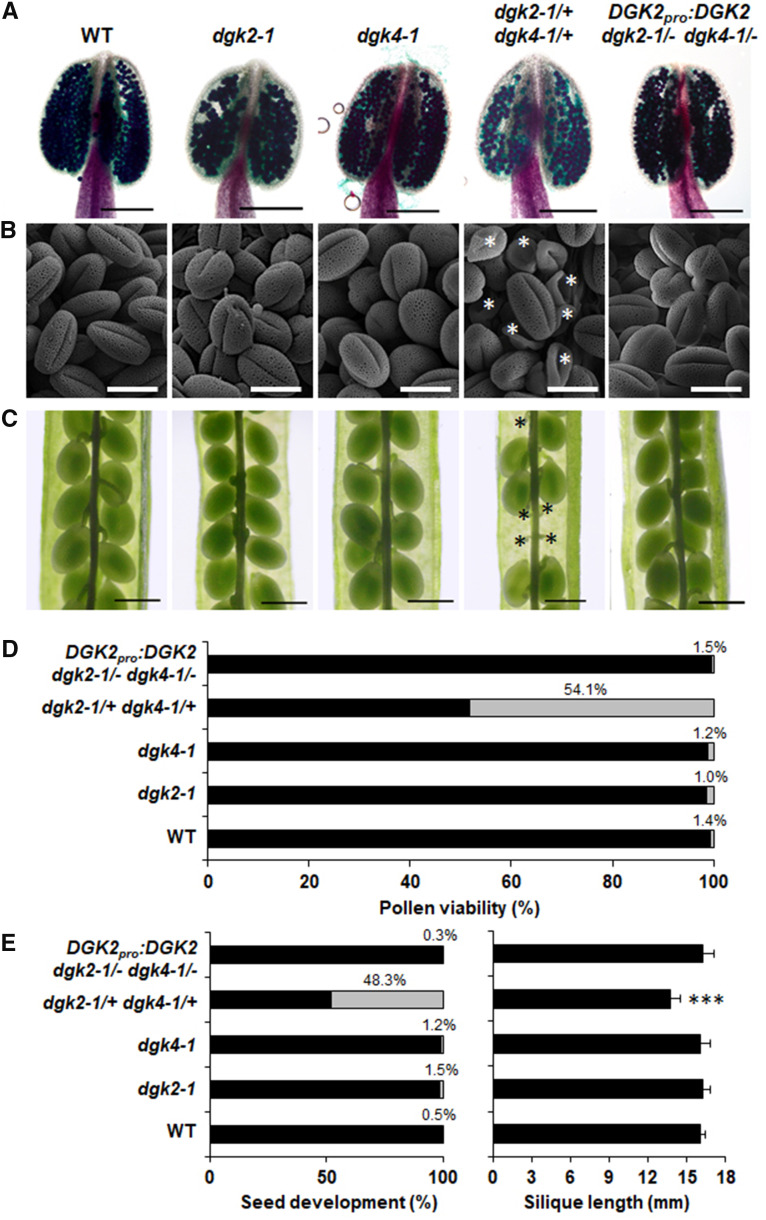
To further investigate a possible overlapping function between DGK2, DGK4, and DGK6, we crossed dgk2-1, dgk4-1, and dgk6-1 to generate double mutants of three different combinations. We isolated viable dgk2-1 dgk6-1 and dgk4-1 dgk6-1 double mutants without any visible growth phenotypes, but we failed to identify a single dgk2-1 dgk4-1 double homozygous plant after genotyping more than 116 offspring seedlings of a dgk2-1/+ dgk4-1/+ plant, implying that the dgk2-1/– dgk4-1/– may be gametophyte, embryo, or seed lethal. To investigate whether the dgk2-1/+ dgk4-1/+ affects the male reproductive organs, we scored pollen viability by Alexander staining (Alexander, 1969). Overall, 54.1% of pollen grains in dgk2-1/+ dgk4-1/+ stained green, which indicates reduced viability (Figures 2A and 2D). In agreement with this observation, scanning electron microscopy revealed that ∼50% of pollen grains were shrunken in dgk2-1/+ dgk4-1/+ (Figure 2B). Furthermore, mature siliques of dgk2-1/+ dgk4-1/+ frequently produced aborted seeds, as indicated by a significant number of empty slots (48.3%) in siliques (Figure 2C, black asterisks; Figure 2E, left). In addition, mature siliques were shorter on dgk2-1/+ dgk4-1/+ double heterozygous plants than on the wild type (Figure 2E, right). The percentage of empty slots in siliques was much higher than the expected percentage of double homozygous mutant embryos (6.3%) generated from self-crossing, which suggests that embryo lethality is not sufficient to explain these numbers.
Figure 2.
Defective Gametogenesis in the Double Heterozygous Mutant of dgk2-1 and dgk4-1 (dgk2-1/+ dgk4-1/+).
(A) and (B) Pollen viability in 5-week-old plants by Alexander staining (A) and morphology of pollen grains by scanning electron microscopy imaging (B). Bars in (A) = 200 μm and bars in (B) = 20 μm. White asterisks indicate aborted pollen. The first flower was avoided for observation. WT, wild type.
(C) Developing seeds in a mature silique. Shrunken seeds were marked with a black asterisk. Bars = 500 μm.
(D) Percentage of viable (black bars) or nonviable (grey bars) pollen observed in (A). More than 800 pollen grains were counted for each line.
(E) Percentage of normal (black bars) or shrunken (grey bars) seeds (left) and silique length (right). More than 500 seeds were counted, and at least eight siliques were measured for each line. Statistical significance was analyzed by Student’s t test: ***, P < 0.001.
Next, to investigate whether these morphological phenotypes are present in parental single mutants, we looked at dgk2-1 and dgk4-1. A recent publication reported that dgk4 mutants exhibit smaller vegetative tissue and reduced pollen tube growth (Vaz Dias et al., 2019), and we included one of these previously published mutants (GK-709G04, hereafter dgk4-2) as a control to compare phenotypes. However, under our growth conditions, we did not observe any effect on plant size (Supplemental Figure 1A) or rosette leaf size (Supplemental Figure 1B) in dgk2-1, dgk4-1, or dgk4-2. We further investigated the reproductive growth phenotype in mature flowers (Supplemental Figure 1C) and siliques (Supplemental Figure 1D) as well as the male gametophyte by Alexander staining (Supplemental Figure 1E) and pollen morphology by scanning electron microscopy (Supplemental Figure 1F). We observed no remarkable mutant phenotype in any of the single mutants. Because of the lack of a second insertion allele for dgk2, we were unable to test whether an independent dgk2 dgk4 double mutant could reproduce the same morphological phenotypes as dgk2-1/+ dgk4-1/+.
To examine whether the defects observed in pollen grains and seeds were due to the combined loss of DGK2 and DGK4, we performed genetic complementation by introducing the genomic sequence of DGK2 (DGK2pro:DGK2) or DGK4 (DGK4pro:DGK4) into dgk2-1/+ dgk4-1/+ plants. We isolated a number of DGK2pro:DGK2 dgk2-1/– dgk4-1/– plants by PCR-based genotyping, which showed no evidence of reduced pollen viability (Figures 2A, 2B, and 2D), aborted seed development, or shorter siliques (Figures 2C and 2E). We also attempted to generate DGK4pro:DGK4 dgk2-1/– dgk4-1/– but were unable to isolate this line after genotyping hundreds of seedlings. These results indicate that transduction of the DGK2 genomic sequence can fully complement the reproductive defect caused by disruption of DGK2 and DGK4. Thus, the male gametophyte defects may be caused by the combined loss of DGK2 and DGK4.
Tissue Localization of DGK2 and DGK4
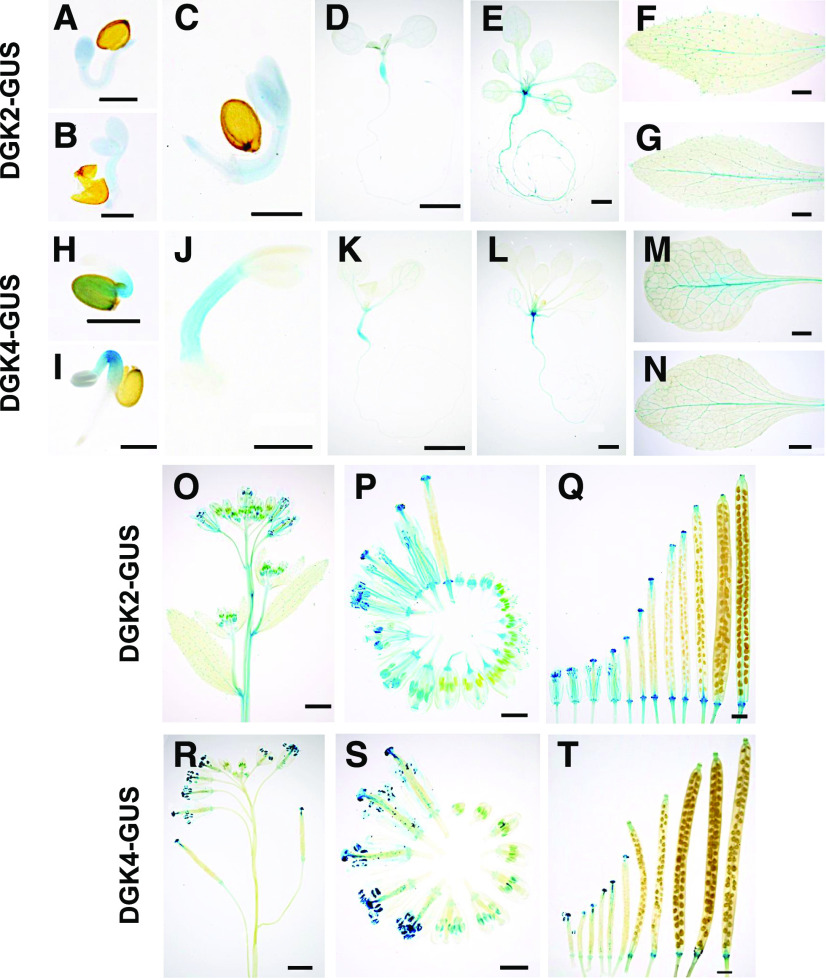
To study the tissue-specific localization pattern of DGK2 and DGK4, we generated transgenic plants harboring DGK2pro:DGK2-GUS or DGK4pro:DGK4-GUS and performed histochemical β-glucuronidase (GUS) staining assays. In 1- to 3-d-old seedlings, DGK2-GUS and DGK4-GUS lines showed different staining patterns; DGK2-GUS displayed a stronger signal in cotyledons than in hypocotyls (Figures 3A to 3C), whereas DGK4-GUS exhibited the opposite pattern (Figures 3H to 3J). This staining pattern was confirmed with an additional independent transgenic line (Supplemental Figures 2A and 2F). In 7- and 14-d-old seedlings, GUS staining was observed in hypocotyls and roots of DGK2-GUS (Figures 3D and 3E) and DGK4-GUS (Figures 3K and 3L). In vegetative tissues, GUS staining was barely detectable, except for a faint staining in the leaf veins of DGK2-GUS (Figures 3F and 3G) and DGK4-GUS (Figures 3M and 3N).
Figure 3.
Tissue-Specific Localization of DGK2 and DGK4.
DGK2pro:DGK2-GUS line #23 (see [A] to [G] and [O] to [Q]) and DGK4pro:DGK4-GUS line #12 (see [H] to [N] and [R] to [T]) were subjected to histochemical GUS staining.
(A) to (E) and (H) to (L) Time-course GUS staining profile of germinating seedlings. Seeds were stratified in sterile water for 1 d and placed on MS agar plates. Seedlings on day 1 (see [A] and [H]), day 2 (see [B] and [I]), day 3 (see [C] and [J]), day 7 (see [D] and [K]), and day 14 (see [E] and [L]) after plating.
(F) and (M) Cauline leaves.
(G) and (N) Rosette leaves.
(O) and (R) Inflorescence stems.
(P) and (S) Flowers at different developmental stages.
(Q) and (T) Developing siliques.
Bars in (A) to (C), (H), (I), and (J) = 500 μm; bars in (P), (Q), (S), and (T) = 1 mm; and bars in (D) to (G), (K) to (O), and (R) = 2 mm.
In reproductive tissues, DGK2-GUS inflorescence stems were stained slightly, but this was not the case for DGK4-GUS (Figures 3O and 3R). During flower development, GUS staining was observed specifically in developing anthers and stigmas (Figures 3P and 3S). In developing siliques, GUS staining was limited to receptacles for both DGK2-GUS and DGK4-GUS (Figures 3Q and 3T). These staining patterns were also confirmed with an additional independent transgenic line (Supplemental Figures 2B to 2E and 2G to 2J). Thus, DGK2-GUS and DGK4-GUS showed stronger staining in developing anthers, with a distinct pattern in germinating seedlings and inflorescence stems.
Subcellular Localization of DGK2 and DGK4
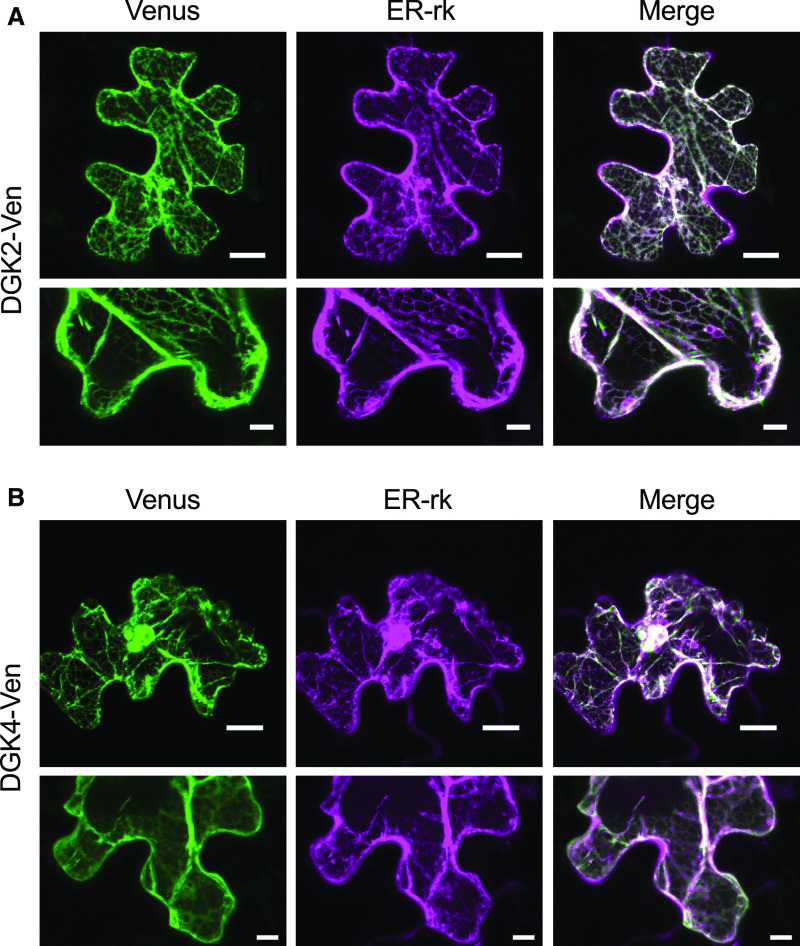
To investigate the subcellular localization of DGK2 and DGK4, we expressed DGK2pro:DGK2-Ven or DGK4pro:DGK4-Ven (fusions between DGK2/4 and the Venus [Ven] version of the green fluorescent protein [GFP]) transiently in Nicotiana benthamiana leaves and observed the subcellular location of the Ven fluorescent signal. We used three copies of the Ven reporter because a single copy did not give a clear fluorescent signal, possibly due to weak promoter activity from DGK2pro and DGK4pro. Both DGK2-Ven and DGK4-Ven showed a clear reticulated signal pattern (Figures 4A and 4B), which is typically observed with ER-localized proteins. We cobombarded leaves with a known ER marker (ER-rk; Nelson et al., 2007), which clearly overlapped with the Ven signal from DGK2-Ven and DGK4-Ven (Figures 4A and 4B). To test whether transiently produced DGK2-Ven and DGK4-Ven fusion proteins were intact in terms of protein size, we transfected DGK2pro:DGK2-Ven or DGK4pro:DGK4-Ven into protoplasts produced from Arabidopsis leaves for clearer protein immunoblot detection than from N. benthamiana leaves. We first confirmed that DGK2-Ven and DGK4-Ven still localized to the ER in protoplasts (Supplemental Figure 3A), which agreed with our results in N. benthamiana leaves (Figure 4). Next, immunoblot analysis of total protein from protoplasts producing DGK2-Ven or DGK4-Ven with an anti-GFP antibody indicated that both DGK2-Ven and DGK4-Ven appeared to accumulate at their predicted respective molecular weights, indicating that the proteins are intact and the Ven moiety was not cleaved off (Supplemental Figure 3B). Thus, DGK2 and DGK4 may be localized mainly at the ER.
Figure 4.
ER Localization of DGK2-Ven and DGK4-Ven in N. benthamiana Leaf Epidermal Cells.
(A) and (B) N. benthamiana leaves were bombarded with particles carrying the constructs DGK2pro:DGK2-Ven (A) or DGK4pro:DGK4-Ven (B). Fluorescence signals for Ven (green) and ER marker (ER-rk, magenta) are merged. Bars = 20 μm.
Defective Pollen Germination and Tube Elongation in dgk2-1/+ dgk4-1/+
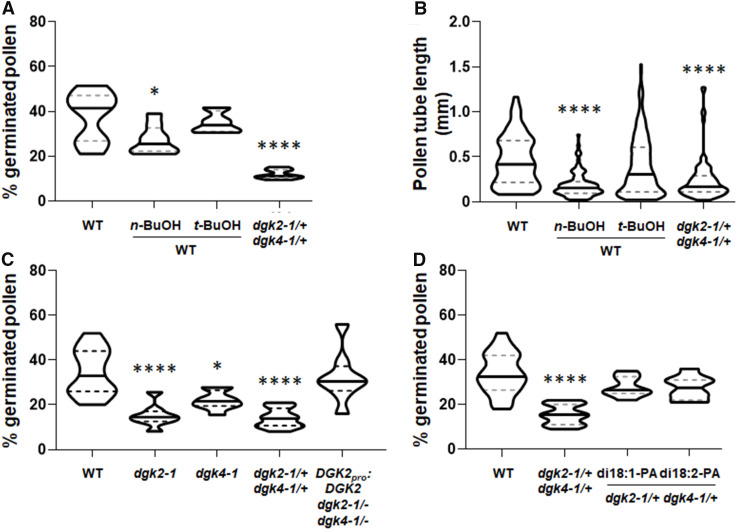
To elucidate the role of DGK2 and DGK4 in pollen tube growth, we investigated the germination (Figure 5A) and elongation (Figure 5B) of pollen in wild-type (Columbia-0 [Col-0]) and dgk2-1/+ dgk4-1/+ heterozygous plants. PA is involved in pollen germination and elongation of the pollen tube in tobacco and is produced in part by PLD (Potocký et al., 2003). PLD activity prefers primary alcohols such as n-butanol over water in hydrolyzing phospholipids (Yang et al., 1967). Thus, PLD activity in the presence of n-butanol produces phosphatidylbutanol and hence reduces PA production. Consistent with the observations in tobacco (Potocký et al., 2003), germination and elongation of the Arabidopsis wild-type pollen were affected by treatment with n-butanol, but not tert-butanol, which is a tertiary alcohol and not a preferred substrate for PLD (Figures 5A and 5B). Because n-butanol treatment may also produce butylaldehyde by endogenous alcohol dehydrogenase activity (Tadege and Kuhlemeier, 1997), we treated the Arabidopsis wild-type pollen with butylaldehyde and observed an effect on germination that was similar to that with n-butanol (Supplemental Figure 4). Compared with the wild type treated with n-butanol, dgk2-1/+ dgk4-1/+ pollen tubes had similar lengths (Figure 5B) but a more severely reduced germination rate (Figure 5A), which suggests that DGK-derived PA signal plays a more predominant role in germination than elongation during pollen tube growth. Next, we tested whether DGK2 and DGK4 were redundant in pollen germination by investigating pollen germination in the single mutants dgk2-1 and dgk4-1. In agreement with a recent report for DGK4 (Vaz Dias et al., 2019), pollen germination was reduced in dgk4-1 (Figure 5C). However, we found that this phenotype was more pronounced in dgk2-1 than in dgk4-1, suggesting that DGK2 may possess a predominant function over DGK4 in pollen germination.
Figure 5.
Effect of DGK2 and DGK4 on Pollen Tube Growth.
(A) and (B) Percentage of germinated pollen (A) and length of pollen tube (B) of the wild type (WT; Col-0), dgk2-1/+ dgk4-1/+, and WT treated with n-butanol (n-BuOH) or tert-butanol (t-BuOH).
(C) Percentage of germinated pollen in the wild type (WT), dgk2-1/–, dgk4-1/–, dgk2-1/+ dgk4-1/+, and DGK2pro:DGK2 dgk2-1 dgk4-1.
(D) Chemical complementation of defective pollen germination in dgk2-1/+ dgk4-1/+ by dioleoyl PA (di18:1-PA) and dilinoleoyl PA (di18:2-PA) in vitro. More than 50 pollen grains were counted in each assay.
Statistical significance was analyzed by Student’s t test: *, P < 0.05; ****, P < 0.0001.
To test whether defective pollen tube growth was caused by the mutation, we analyzed the germination rate of pollen in DGK2pro:DGK2 dgk2-1/– dgk4-1/–. The germination rate of DGK2pro:DGK2 dgk2-1/– dgk4-1/– pollen was indistinguishable from that of the wild type, indicating that the decreased germination rate in dgk2-1/+ dgk4-1/+ was complemented by the introduction of the DGK2pro:DGK2 construct (Figure 5C). These results suggest that reduced pollen germination rates are due to the loss of DGK2 and DGK4. To further examine whether the reduced germination rate of dgk2-1/+ dgk4-1/+ pollen was due to defective PA production, we performed a chemical complementation assay by supplementing ER-derived (eukaryotic) molecular species of PA (dioleoyl PA and dilinoleoyl PA). The addition of both PA species largely reversed the defective pollen germination in dgk2-1/+ dgk4-1/+, which was indistinguishable from the germination rate in the wild-type pollen (Figure 5D). These results suggest that DGK2 and DGK4 are involved in PA production for pollen tube growth.
Reciprocal Genetic Crossing
To investigate viability of the male and female gametophytes in the dgk2-1 and dgk4-1 single mutants, we performed reciprocal genetic crosses and examined the segregation of genotypes in the resulting F1 progeny by PCR-based genotyping (Supplemental Table 1). Crossing dgk2-1/DGK2 pollen with dgk2-1/dgk2-1 stigma produced F1 seeds with a segregation ratio of 23:25 for dgk2-1/DGK2:dgk2-1/dgk2-1, very close to the theoretical ratio of 24:24. Conversely, crossing dgk2-1/dgk2-1 pollen with dgk2-1/DGK2 stigma resulted in a segregation ratio of 24:24 for dgk2-1/DGK2:dgk2-1/dgk2-1, which agrees with the theoretical ratio of 24:24. Next, crossing dgk2-1/DGK2 pollen with DGK2/DGK2 stigma produced F1 seeds with a segregation ratio of 51:49 for DGK2/DGK2:dgk2-1/DGK2, and crossing DGK2/DGK2 pollen with dgk2-1/DGK2 stigma resulted in a segregation ratio of 50:50 for DGK2/DGK2:dgk2-1/DGK2. Likewise, crossing dgk4-1/DGK4 pollen on a dgk4-1/dgk4-1 stigma produced F1 seeds with a segregation ratio of 24:24 for dgk4-1/DGK4:dgk4-1/dgk4-1, which agrees with the theoretical ratio of 24:24. Conversely, crossing dgk4-1/dgk4-1 pollen with dgk4-1/+ stigma resulted in a segregation ratio of 45:49 for dgk4-1/DGK4:dgk4-1/dgk4-1, close to the theoretical ratio of 47:47. Finally, crossing dgk4-1/DGK4 pollen with DGK4/DGK4 stigma produced F1 seeds with a segregation ratio of 51:49 for wild type:dgk4-1/DGK4, and crossing DGK4/DGK4 pollen with dgk4-1/DGK4 stigma resulted in a segregation ratio of 52:48 for wild type:dgk4-1/DGK4. In all cases, statistical analysis indicated no significant defect in the penetrance of mutant allele between male and female gametophytes. These data suggest that both male and female gametophytes from dgk2-1 and dgk4-1 were viable, albeit with a reduced germination rate during in vitro culture of dgk2-1 and dgk4-1 pollen (Figure 5C).
DGK2 Knockdown in the dgk4 Mutant Confers a Reproductive and Vegetative Growth Phenotype
Our genetic evidence suggests that the loss of both DGK2 and DGK4 may affect gamete viability during gametogenesis. However, whether these DGKs are essential specifically during gametogenesis or ubiquitously throughout the plant life cycle remains uncertain. For this reason, we created weaker loss-of-function lines for DGK2 and DGK4 by using artificial microRNA (amiRNA)–mediated gene suppression (Schwab et al., 2006; Ossowski et al., 2008). We produced two transgenic plant lines that overexpress one of two specific miRNA sequences designed to target DGK2 (amiDGK2-I and amiDGK2-II; Figure 6A) in the dgk4-1 mutant background (35Spro:amiDGK2-I dgk4-1 and 35Spro:amiDGK2-II dgk4-1). Among 24 transgenic plants each, we obtained four ami-DGK2 transgenic lines (35Spro:amiDGK2-I dgk4-1 lines #1 and #7 and 35Spro:amiDGK2-II dgk4-1 lines #2 and #4), with much lower DGK2 expression (Figure 6B). Compared with the wild type, DGK2 expression in 7-d-old seedlings decreased by 79, 93, 85, and 85% in 35Spro:amiDGK2-I dgk4-1 lines #1 and #7 and 35Spro:amiDGK2-II dgk4-1 lines #2 and #4, respectively. To confirm the gametophyte phenotypes observed in dgk2-1/+ dgk4-1/+ double heterozygotes, we first analyzed pollen viability of transgenic plants (Figure 6C). Compared with dgk2-1/+ dgk4-1/+ with 50.5% nonviable pollen, 35Spro:amiDGK2-I dgk4-1 lines #1 and #7 produced 4.4 and 3.5% nonviable pollen, respectively. However, in 35Spro:amiDGK2-II dgk4-1 lines #2 and #4, 28.9 and 68.4% of pollen grains were nonviable. Next, observation of seeds showed a similar trend; the proportion of aborted seeds in 35Spro:amiDGK2-I dgk4-1 lines #1 and #7 was 13.8 and 17.9% but that of 35Spro:amiDGK2-II dgk4-1 lines #2 and #4 was 45.9 and 68.1%, respectively (Figure 6D, left). All transgenic lines had shorter siliques than dgk2-1/+ dgk4-1/+ or the wild type (Figure 6D, right; Supplemental Figure 5). These results confirmed the reproductive phenotypes in dgk2-1/+ dgk4-1/+ and suggest that the 35Spro:amiDGK2-II dgk4-1 line #4 is a stronger knockdown allele, whereas the others are weaker alleles than dgk2-1/+ dgk4-1/+ in terms of the strength of their gametophytic phenotypes.
Figure 6.
Construction and Screening of the 35Spro:amiDGK2 dgk4-1 Plants.
(A) Schematic representation of the 35Spro:amiDGK2-I and 35Spro:amiDGK2-II constructions. Two target sites of amiRNAs designed are indicated. LB, left border; RB, right border; ter, transcriptional terminator.
(B) Relative transcript levels of DGK2 in 7-d-old seedlings of the wild type (WT) and four independent transgenic lines: 35Spro:amiDGK2-I dgk4-1 (lines #1 and #7) and 35Spro:amiDGK2-II dgk4-1 (lines #2 and #4). Values are mean ± sd of three biological replicates and with three technical replicates.
(C) Percentage of viable (black bars) or nonviable (grey bars) pollen by Alexander staining. More than 800 pollen grains were counted for each line.
(D) Percentage of normal (black bars) or aborted (grey bars) seeds (left) and length of silique (right). More than 500 seeds were counted, and at least eight siliques were measured for each line.
Statistical significance was analyzed by Student’s t test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To rule out the possibility that these morphological phenotypes were due to a possible other mutation in dgk4-1 that is not related to the DGK4 insertion but somehow produces these phenotypes when DGK2 transcript levels are reduced, we produced two transgenic plant lines that overexpress one of two specific miRNA sequences designed to target DGK4 (amiDGK4-I and amiDGK4-II) (Supplemental Figure 6) in the dgk2-1 mutant background (35Spro:amiDGK4-I dgk2-1 and 35Spro:amiDGK4-II dgk2-1). Consistent with our observations in 35Spro:amiDGK2-I dgk4-1 and 35Spro:amiDGK2-II dgk4-1 (Figure 6; Supplemental Figure 5), two representative lines of 24 screened lines for each transgenic plant (35Spro:amiDGK4-I dgk2-1 #2 and #20 and 35Spro:amiDGK4-II dgk2-1 #9 and #22) showed similar aborted seeds and shorter siliques (Supplemental Figures 6A and C) as well as reduced pollen viability (Supplemental Figure 6B). These results suggest that the reproductive growth phenotypes observed in these lines are due to suppression of DGK2 and DGK4.
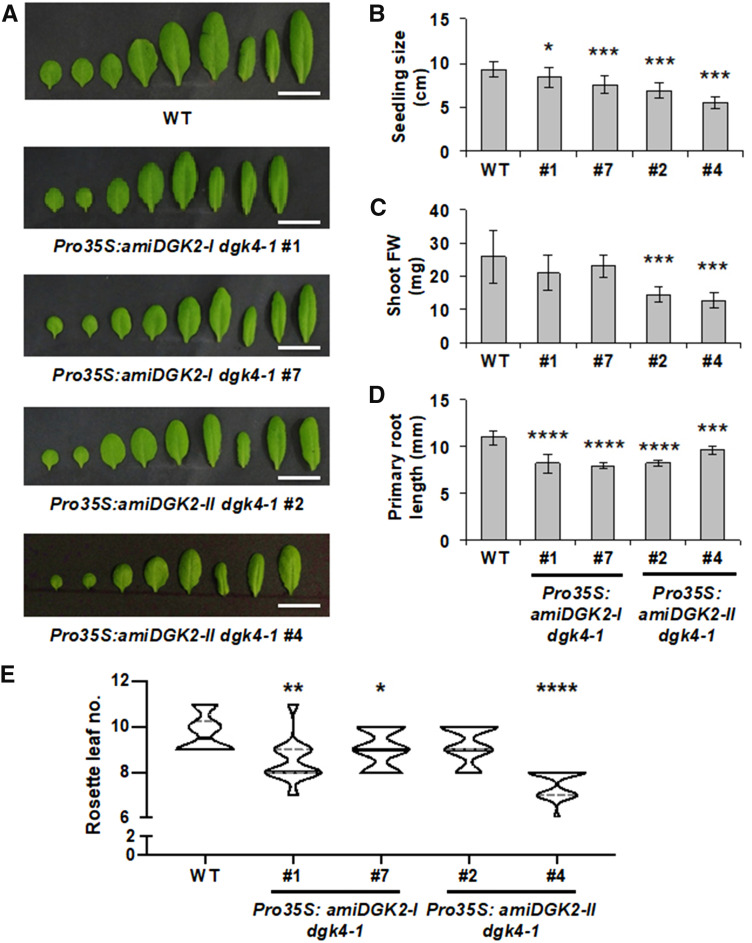
We then characterized the vegetative growth of these 35Spro:amiDGK2 dgk4-1 transgenic plants in soil and in Murashige and Skoog (MS) medium (Figure 7). In 24-d-old, soil-grown plants under long-day conditions, the rosette leaves of transgenic lines were round shaped (Figure 7A) and significantly smaller (Figure 7B), in particular for the stronger knockdown allele (35Spro:amiDGK2-II dgk4-1 line #4). Also, compared to the wild type, shoot fresh weight was significantly reduced in 35Spro:amiDGK2-II dgk4-1 lines #2 and #4 (Figure 7C). In 14-d-old seedlings grown on an MS plate, primary root length was significantly reduced in all transgenic lines (Figure 7D). Moreover, a slight but significant early flowering phenotype was observed in the transgenic lines: although the wild-type plants started bolting 9.7 leaves on average, all transgenic lines showed significantly reduced average leaf numbers (8.5, 9.0, and 7.3 for 35Spro:amiDGK2-I dgk4-1 lines #1 and #7 and 35Spro:amiDGK2-II dgk4-1 line #4, respectively; Figure 7E). These results suggest that function of DGK2 and DGK4 is required for both vegetative and reproductive growth.
Figure 7.
Vegetative Growth of the Wild Type, 35Spro:amiDGK2-I dgk4-1, and 35Spro:amiDGK2-II dgk4-1.
(A) Detached rosette leaves of 24-d-old plants. Bars = 2 cm. WT, wild type.
(B) and (C) Seedling size (B) and fresh weight (FW) of the shoot part (C) of 24-d-old, soil-grown wild-type (WT) and 35Spro:amiDGK2 plants. Values are mean ± sd of 14 plants for each.
(D) Primary root length of 14-d-old wild-type (WT) and 35Spro:amiDGK2 seedlings grown on MS medium. Values in (A) to (D) are mean ± sd of eight plants.
(E) Flowering time under long-day (16-h-light/8-h-dark cycle) conditions. The number of rosette leaves was counted when plants were bolting (n = 14). WT, wild type.
Statistical significance was analyzed by Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Enzyme Activity of Recombinant DGK4
DGK activity of DGK4 was not examined previously, whereas that of DGK2 has been reported (Gómez-Merino et al., 2004). To test whether DGK4 encodes a functional DGK, we conducted in vitro enzyme activity assays with recombinant DGK4 protein. We cloned the DGK4 coding sequence into an Escherichia coli vector (pMAL5x) to produce a protein fused with maltose binding protein (MBP). We determined enzymatic activity from total protein extracts (after induction of protein expression) by quantifying the amount of fluorescence-labeled PA produced after incubation of recombinant DGK4 with fluorescence-labeled DAG (1-NBD-decanoyl-2-decanoyl-sn-glycerol [NBD-DAG]) and nonlabeled ATP as substrates. Significant DGK activity was detected for extracts obtained from cells producing MBP-fused DGK4, compared to that harboring the empty vector (pMAL5x; Supplemental Figure 7). Thus, DGK4 is a functional DGK that phosphorylates DAG to produce PA in vitro.
Polar Glycerolipid Profiles in Rosette Leaves and Floral Buds of 35Spro:amiDGK2-II dgk4-1
To investigate whether the changes in polar glycerolipid contents are associated with the phenotype expression of the knockdown lines, we chose 35Spro:amiDGK2-II dgk4-1 lines #2 and #4 for lipid analysis because of their stronger phenotype among four knockdown lines. Because obvious phenotypes were observed in gametophytes and rosette leaf size, we analyzed polar glycerolipid composition in floral buds and 24-d-old rosette leaves.
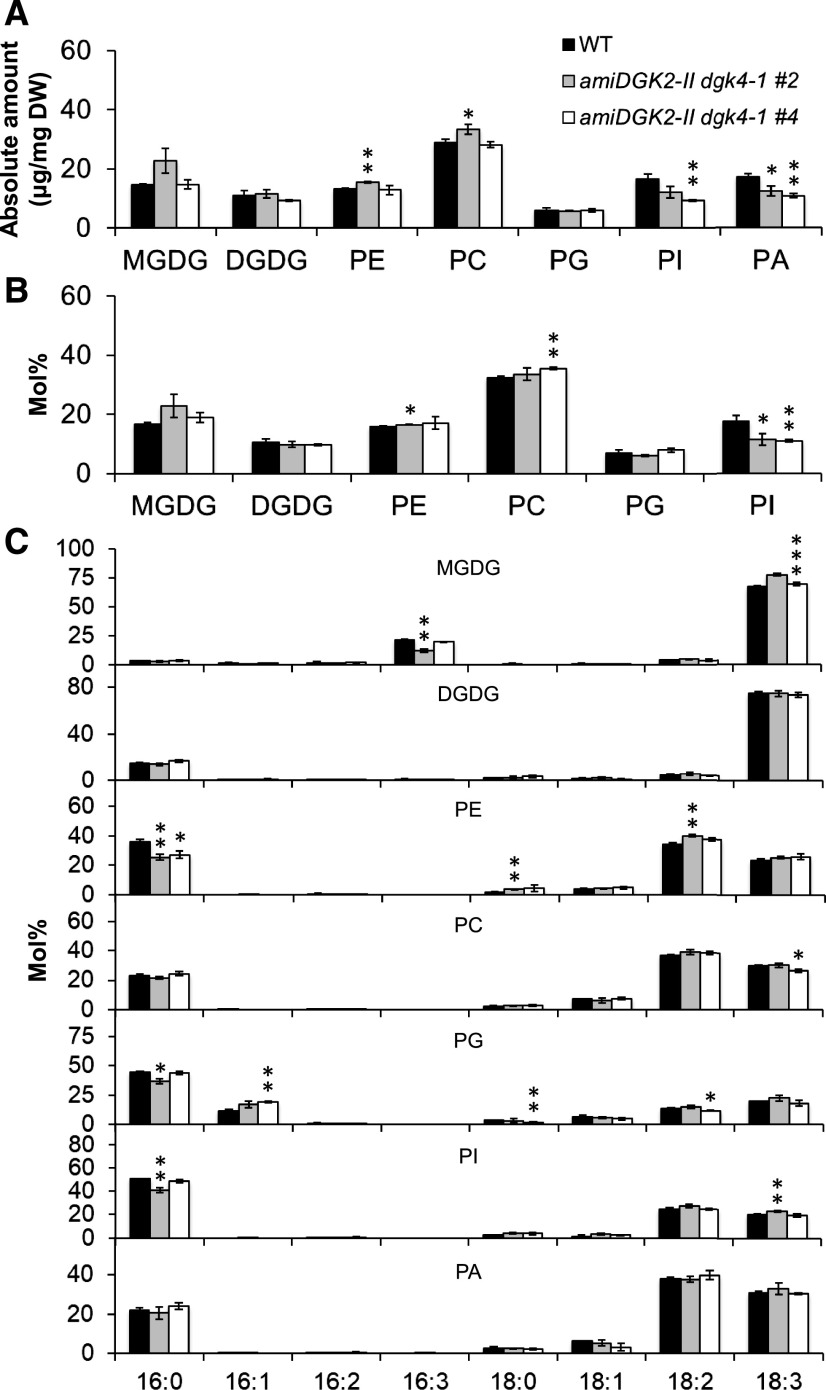
In floral buds, we first quantified the absolute amount of glycerolipid classes normalized by tissue dry weight, including PA, a reaction product of DGK. Consistent with our previous report (Nakamura et al., 2014), PA content was high in floral buds (Figure 8A). The amount of PA was reduced by 28% in 35Spro:amiDGK2-II dgk4-1 lines #2 and #4 compared to the wild type, demonstrating that DGK2 and DGK4 are involved in PA production in vivo (Figure 8A). No other glycerolipid class showed significant changes consistently between the two knockdown lines. Next, we analyzed the composition (mol %) of major polar glycerolipid classes. Most lipid classes showed no consistent change between the two lines, but we observed a significant and consistent reduction in PI content in the two knockdown lines relative to the wild type (Figure 8B). We further analyzed the fatty acid composition of these lipid classes (Figure 8C). The 16:0 content of phosphatidylethanolamine (PE) was significantly and consistently reduced in the two knockdown lines compared with the wild type. Thus, in flower buds, DGK2 and DGK4 may affect PA and PI significantly, but not other glycerolipid classes.
Figure 8.
Glycerolipid Contents in Flower Buds of the Wild Type and 35Spro:amiDGK2-II dgk4-1.
(A) Amount of major polar glycerolipid classes normalized by tissue dry weight (DW). WT, wild type.
(B) Contents of major polar glycerolipid classes shown in mol % fractions. WT, wild type.
(C) Fatty acid composition (mol%) of the polar glycerolipid classes. WT, wild type.
Data are mean ± sd from at least three biological replicates. The asterisks indicate significance by Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
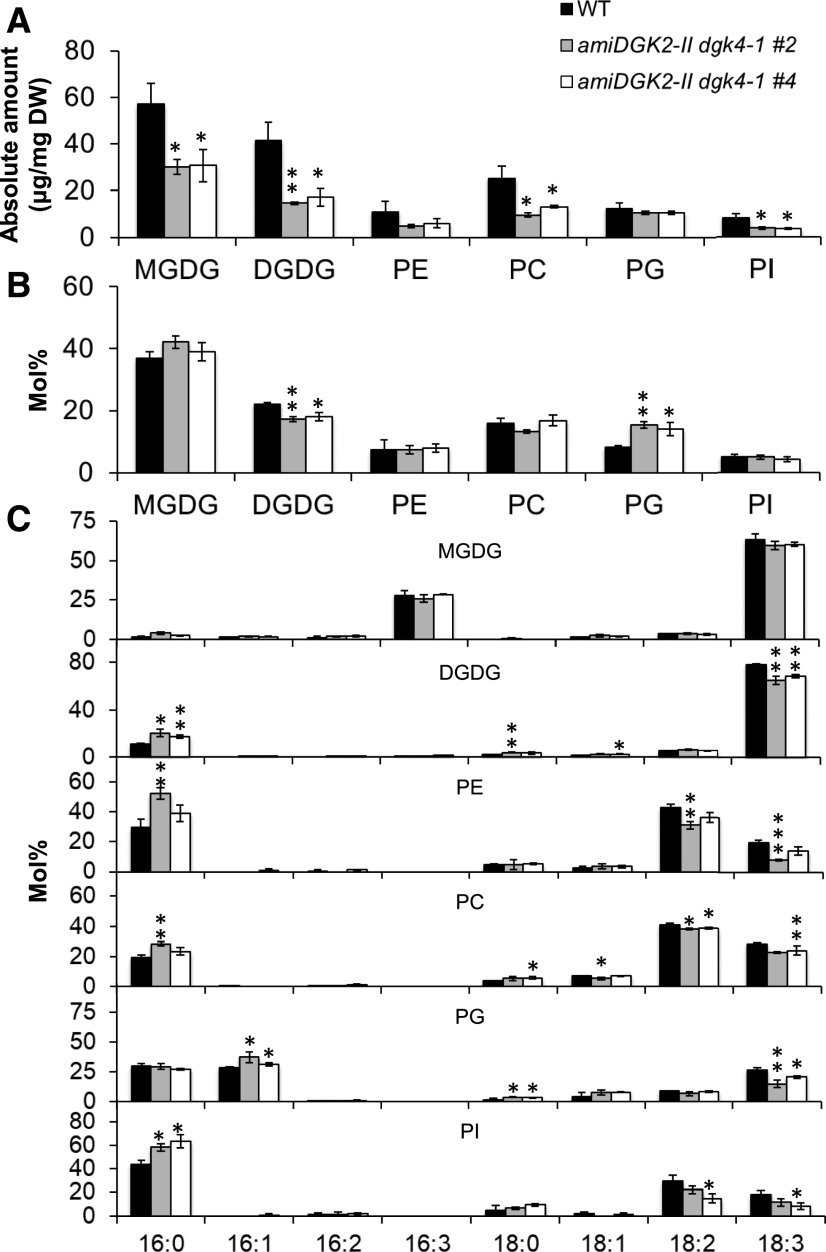
In rosette leaves, we first analyzed the absolute amount of lipid content normalized by tissue dry weight. Levels of most lipid class analyzed decreased in the knockdown lines; monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), phosphatidylcholine (PC), and PI levels decreased by 47, 61, 55, and 56%, respectively, compared to the wild type (Figure 9), presumably because leaf size and fresh weight were significantly reduced in the knockdown lines (Figures 7A to 7C). However, we saw no significant differences for PE or PG, which may alter relative membrane lipid composition. Thus, we next analyzed polar glycerolipid composition in mol % (Figure 9B). In the knockdown lines, only the PG fraction, and not the PE fraction, consistently increased at the expense of DGDG. Analysis of fatty acid composition in these glycerolipid classes showed an increase of 16:0 content at the expense of polyunsaturated fatty acids in DGDG, PC, PE, and PI (Figure 9C). MGDG showed no significant changes in fatty acid composition, but PG increased significantly in t16:1 content but decreased in 18:3 content. Because t16:1 is a signature fatty acid in plastidic PG, this change suggests a specific decrease in extraplastidic PG biosynthesis.
Figure 9.
Glycerolipid Contents in Rosette Leaves of the 24-d-Old Wild Type and 35Spro:amiDGK2-II dgk4-1.
(A) Amount of major polar glycerolipid classes normalized by tissue dry weight (DW). WT, wild type.
(B) Contents of major polar glycerolipid classes shown in mol % fractions. WT, wild type.
(C) Fatty acid composition (mol %) of the polar glycerolipid classes. WT, wild type.
Data are mean ± sd from at least three biological replicates. The asterisks indicate significance by Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
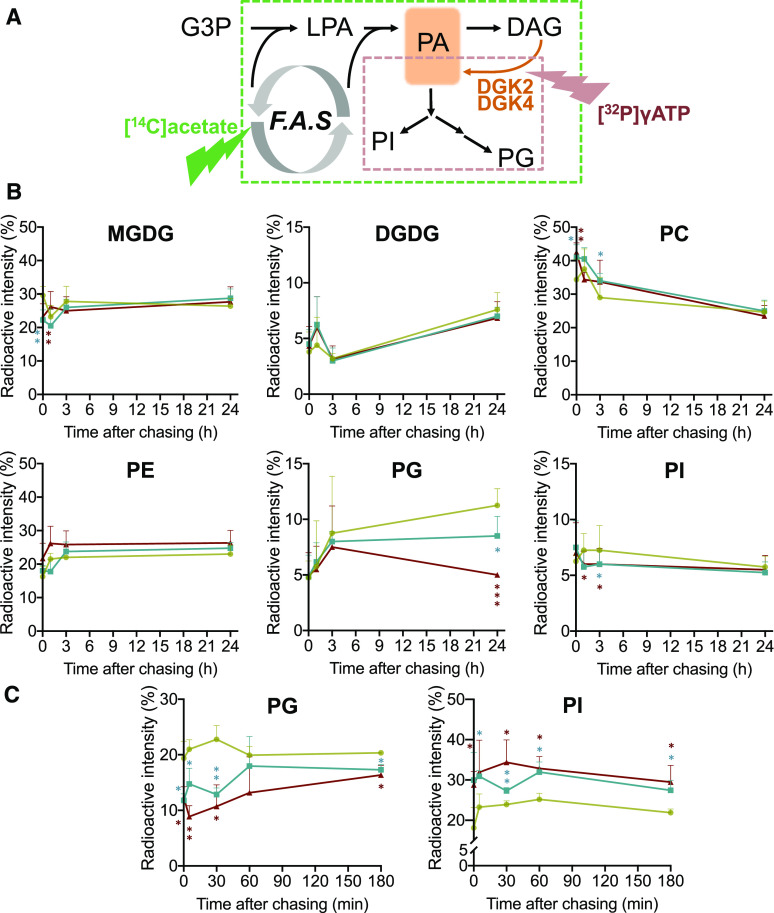
To further investigate whether extraplastidic PG biosynthesis was compromised in the knockdown lines, we performed a pulse-chase radiolabeling experiment using [14C]acetate or [γ-32P]ATP. With [14C]acetate, neo-synthesized glycerolipids are labeled, and thus de novo synthesis of these lipid classes can be monitored (Figure 10A, green dashed square; Bates et al., 2017). With [γ-32P]ATP, PA produced from DAG by DGK activity is labeled, which allows the monitoring of the metabolic fate of DGK-derived PA (Figure 10A, red dashed square). With [14C]acetate pulse-chase labeling, the amount of 14C-labeled PG, but not the other major glycerolipid classes, significantly decreased in both knockdown lines compared to the wild type (Figure 10B), which suggests that DGK2 and DGK4 may be involved in de novo biosynthesis of PG. Next, [γ-32P]ATP pulse-chase radiolabeling revealed that production of labeled PG considerably decreased in the knockdown lines, which caused a relative increase of labeled PI because these two phospholipid classes are produced from PA in the ER-localized glycerolipid metabolic pathway (Figure 10C). These results suggest that ER-localized PG biosynthesis is compromised in the knockdown mutant lines and that PA produced by DGK2 and DGK4 is mainly used as a precursor for the biosynthesis of PG.
Figure 10.
Pulse-Chase Radiolabeling Experiments in Rosette Leaves of the 24-d-Old Wild Type and 35Spro:amiDGK2-II dgk4-1.
(A) Schematic illustration of two distinct radiolabeling methods used in this experiment. [14C]Acetate is incorporated into the fatty acid synthesis pathway (F.A.S), which produces 14C-labeled 18:1-CoA to be incorporated into glycerol backbone and thus radiolabel lipid metabolites indicated in the territory of the green dashed square. [γ-32P]ATP is a substrate of DGK; hence, immediately radiolabeled compound is a product PA. Since PG and PI are derived from PA in ER-localized phospholipid metabolism, metabolites within the territory of the red dashed square will be radiolabeled. G3P, glycerol 3-phosphate; LPA, lysophosphatidic acid.
(B) and (C) Pulse-chase radiolabeling analysis of polar glycerolipid metabolism using [14C]acetate (B) or [γ-32P]ATP (C). Rosette leaves of the wild type (khaki) and lines 35Spro:amiDGK2-II dgk4-1 #2 (blue) and #4 (dark brown) were labeled for 1 h with [14C]acetate or 30 min with [γ-32P]ATP, and radiolabeled polar glycerolipids were analyzed at different times after chasing. Data are mean ± sd from three biological replicates. The asterisks indicate significance by Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Role of DGK2 and DGK4 in Growth and Development
In this work, we focused on DGK2 and DGK4 because of the gametophyte-lethality phenotype seen in the double mutant (Figure 2). We showed that DGK2 and DGK4 had redundant but essential functions: the single mutant of either gene was fully viable, but a double homozygous mutant was not obtained unless we introduced the genomic sequence of DGK2 (DGK2pro:DGK2; Figure 2). Although the genetic complementation by DGK4pro:DGK4 was unsuccessful, possibly because of a requirement of distantly located cis-element not included in the cloned DGK4pro:DGK4 fragment, similar phenotypes were observed in 35Spro:amiDGK2 dgk4-1 (Figure 6) and 35Spro:amiDGK4 dgk2-1 (Supplemental Figure 6) transgenic knockdown plants. dgk2-1/+ dgk4-1/+ double heterozygous plants produced shrunken and nonviable pollen in anthers (Figures 2A, 2B, and 2D), siliques with empty seed slots (Figures 2C and 2E, left) and of reduced length (Figure 2E, right), and reduced pollen tube growth (Figure 5). Functional reporter assays revealed that DGK2 and DGK4 have similar distribution at the tissue and subcellular levels. GUS reporter assays revealed that the staining of DGK2-GUS and DGK4-GUS was predominant in developing anthers (Figure 3; Supplemental Figure 2). In addition, fluorescent reporter assays of DGK2-Ven and DGK4-Ven in tobacco leaves and Arabidopsis leaf protoplasts revealed an ER localization (Figure 4; Supplemental Figure 3). The similarity in tissue and subcellular distribution between DGK2 and DGK4 may account for their functional redundancy. However, we noted that the single mutants showed defective pollen germination (Figure 5C), which agrees with a previous report for DGK4 (Vaz Dias et al., 2019) but is newly observed for DGK2. The pollen germination phenotype was stronger in dgk2-1 than dgk4-1 (Figure 5C). The result of reciprocal genetic crossing between dgk2-1 and dgk4-1 indicated no defective penetrance of mutant phenotypes between the male and female gametophytes (Supplemental Table 1), although Vaz Dias et al. (2019) reported that transmission of their dgk4 mutant allele was significantly impaired through the male, but not female, gametophyte. Since Alexander staining of dgk2-1, dgk4-1, and dgk4-2 single mutants did not reveal evidence of nonviable pollen (Supplemental Figure 1E), our results suggest that DGK2 and DGK4 have nonoverlapping function in pollen tube growth but are redundant in gametophytic viability.
It is well established that pollen germination and elongation of the pollen tube require PA signaling, which is produced by PLD activity. In tobacco pollen, n-butanol treatment competes with PLD-derived PA production and affects pollen tube growth, and exogenous supplementation of PA recovers the pollen tube growth defect (Potocký et al., 2003). Because PA content decreased consistently in floral buds of two transgenic knockdown lines (Figure 8A) and supplementation of eukaryotic PA species to pollen rescued the tube growth defect (Figure 5D), DGK2 and DGK4 may be involved in PA production during pollen tube growth. In pollen tube elongation, the defect in dgk2-1/+ dgk4-1/+ was as severe as that in the wild type treated with n-butanol (Figure 5B), which suggests that PA production from the DGK and PLD pathways is equally important in pollen tube elongation. For pollen germination, dgk2-1/+ dgk4-1/+ showed more severe defects than the n-butanol–treated wild type (Figure 5A) that was however rescued by exogenous supplementation of eukaryotic PA species (Figure 5D). Thus, DGK-derived PA may play a more prominent role than PLD-derived PA in pollen germination, but both pathways contribute equally to pollen tube elongation. Thus, our results suggest that DGK activity may represent an alternative pathway for PA signaling during pollen tube growth, in which both DGK2 and DGK4 play a major role.
In addition to the reproductive organs, clear GUS staining was observed for both DGK2-GUS and DGK4-GUS in roots, hypocotyls, and leaf veins of seedlings (Figure 3; Supplemental Figure 2). Although this finding suggests a possible role of DGK2 and DGK4 in these vegetative tissues, the gametophyte lethality of the double mutant hampered further investigation to address whether DGK2 and DGK4 are essential only for gametogenesis or ubiquitously in any subsequent developmental stage after gametogenesis. To investigate a possible role of DGK2 and DGK4 in vegetative growth, we took an alternative approach that produces amiRNA-based knockdown lines for DGK2 in a dgk4-1 background and DGK4 in a dgk2-1 background. As expected, the knockdown lines not only confirmed the gametophyte phenotypes (Figure 6; Supplemental Figure 6) but also revealed significant defects in vegetative growth (Figure 7). In agreement with the results of our GUS staining in roots, root length was significantly reduced in the knockdown lines (Figure 7D). These knockdown lines also showed round-shaped rosette leaves (Figure 7A) and reduced seedling size (Figure 7B) and fresh weight (Figure 7C), which may be associated with a possible role of DGK2 and DGK4 in the vasculature, where intense GUS staining was observed for both DGK2 and DGK4 (Figure 3; Supplemental Figure 2). Although a viable vegetative phenotype in the knockdown lines indicates a somewhat minor contribution of DGK2 and DGK4, or that residual activity in the lines may be sufficient to meet most needs of total DGK activity in vegetative growth, these growth phenotypes may be associated with altered phospholipid metabolism in these tissues.
Role of DGK2 and DGK4 in Phospholipid Metabolism
Results of in vitro pollen tube growth assays provided an important implication regarding an alternative pathway for PA signaling. The effect of DGK2 and DGK4 suppression on pollen germination and pollen tube elongation suggests that these DGKs are involved in pollen tube growth, which is supported by a previous report on tobacco pollen tubes indicating that both PLD and DGK activities play distinct roles in PA production for pollen tube growth (Pleskot et al., 2012). Tip growth in the elongating pollen tube is depolarized when phosphoinositide-specific phospholipase C hydrolyzes phosphatidylinositol 4,5-bisphosphate to produce DAG (Helling et al., 2006), which may be the substrate for DGK to produce PA. Unlike Arabidopsis PLD activity, which mainly localizes at the plasma membrane (Fan et al., 1999), our data demonstrate that both DGK2 and DGK4 localized to the ER (Figure 4; Supplemental Figure 3A). Two concurrent pathways for PA production by PLD and DGK may have distinct subcellular localization, which might be associated with the differential function between PLD and DGK in PA signaling and modulating the downstream effectors.
The ER localization of DGK2 and DGK4 and vegetative growth phenotypes in the knockdown lines prompted us to explore whether DGK2 and DGK4 may have a role in basal phospholipid metabolism (not in the context of metabolism for signaling). This has been an open question because the reaction catalyzed by DGK follows the direction opposite to the major flux of de novo phospholipid biosynthesis (Kornberg and Pricer, 1953). Thus, DGK has drawn less attention in the context of phospholipid biosynthesis (anabolism) than PA signaling, which involves catabolic reactions. Here, an analysis of polar glycerolipids in our knockdown lines revealed intriguing changes in lipid contents. In floral buds, which contain developing male and female gametophytes, the absolute amount of PA per tissue dry weight was reduced significantly, which indicates that DGK2 and DGK4 may play a major role in PA production (Figure 8A). The analysis of major polar glycerolipid composition (in mol %) showed reduced PI content (Figure 8B), which suggests relatively reduced PI biosynthesis. Because PA is converted to CDP-DAG, which serves as a common precursor for the biosynthesis of PI and PG in the ER phospholipid metabolism (Zhou et al., 2013), this result suggests that PA, synthesized by DGK2 and DGK4, may serve as a precursor for the biosynthesis of PI and PG in the ER. Regarding PG content in the knockdown lines, the molecular profiles were distinct between floral buds and rosette leaves. PG content in mol % fraction remained unchanged in floral buds (Figure 8B) but significantly increased in rosette leaves (Figure 9B). Although this result appears to be inconsistent with the proposal that PG biosynthesis may be impaired due to knockdown of DGK2 and DGK4, we noted a significant increase in t16:1-containing PG, a signature fatty acid species in chloroplastic PG (Browse et al., 1985). Indeed, pulse-chase radiolabeling of rosette leaves with [14C]acetate showed that PG biosynthesis was specifically affected in the knockdown lines (Figure 10B). Moreover, radiolabeling of DGK activity-derived PA with [γ-32P]ATP and chasing its metabolism to PG and PI indicated a reduced flux toward PG biosynthesis versus PI (Figure 10C). Hence, increased t16:1 fatty acid composition in the PG fraction of knockdown lines may be due to reduced PG synthesis in the ER that does not contain t16:1, which was observed more in rosette leaves than floral buds perhaps because of the abundance of chloroplasts in rosette leaves as a photosynthetic tissue. Floral organs contain higher amount of PI than leaves (Nakamura and Ohta, 2007), so the effect of DGK2 and DGK4 suppression on PI content may be more pronounced in floral buds than in rosette leaves. In addition to the changes in phospholipid contents, we also noted a significant reduction of DGDG content in rosette leaves (Figure 9), but not floral buds (Figure 8). Since galactolipid biosynthesis occurs exclusively in the plastids, this change implies a possible effect of ER-located DGK2 and DGK4 on plastid galactolipid biosynthesis. Galactolipid biosynthesis pathways are present both at the outer and inner envelopes of chloroplasts (Benning and Ohta, 2005). Whereas the inner envelope–located pathway determines MGDG content and is essential for photosynthetic function (Kobayashi et al., 2007), the outer envelope–located pathway plays a minor role and may affect only DGDG content under normal growth conditions (Kelly et al., 2003). Considering that rosette leaves of 35Spro:amiDGK2-II dgk4-1 had reduced a DGDG content, but not MGDG (Figure 9), and showed no obvious photosynthetic defect judging from the leaf color (Figure 7A), DGK2 and DGK4 might also be involved in the outer envelope–located galactolipid biosynthesis, possibly at the ER–chloroplast contact site. How ER-located enzymes affect chloroplastic glycerolipid biosynthesis remains open for investigation.
In conclusion, we propose that DGK2 and DGK4 play a tissue-specific role to produce PA: in pollen, they generate PA for signaling in pollen tube growth, whereas in leaves they provide a precursor for the biosynthesis of PG. Our results offer a new pathway for PA signaling in pollen tube growth and for PG biosynthesis in ER phospholipid metabolism.
METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col-0 accession) was germinated and grown on half-strength MS medium containing 1% (w/v) Suc and 0.8% (w/v) agarose (Murashige and Skoog, 1962) under 16-h-light/8-h-dark photoperiod conditions with white-light illumination (75 μmol m−2 s−1, GC-560H; Firstek) at 22°C. For plant growth past 2 weeks after germination, seedlings were transplanted to soil and grown under the same light and temperature conditions.
Plant Materials
Seeds for the following mutants were obtained from the Nottingham Arabidopsis Stock Centre: dgk2-1 (SAIL_289E03), dgk4-1 (SALK_069158C), dgk4-2 (GK-709G04), and dgk6-1 (SALK_011187C). Homozygous plants were isolated by PCR-based genotyping using gene-specific primers and T-DNA–specific primers as illustrated (Figure 1C). The primers were for dgk2-1 (PP045/ISY145 and PP045/YN144), dgk4-1 (ISY011/ISY124 and ISY011/YN749), and dgk6-1 (ISY009/ISY010 and ISY009/YN749). See Supplemental Table 2 for oligonucleotide sequences. For biological replicates in transcript and lipid analyses, planting, harvesting, preparation, and analysis of samples were conducted separately on separate days to ensure the most rigorous reproducibility. Statistical analysis data are shown in the Supplemental Data Set.
Plasmid Vector Construction and Transgenic Plants Production
DGK2pro:DGK2
We amplified 5800 bp of the genomic sequence for DGK2 by PCR from the Arabidopsis wild-type (Col-0) genomic DNA using primers PP010 and PP011. The PCR product was cloned into plasmid pENTR/D–TOPO (Invitrogen, Thermo Fisher Scientific) to obtain pAA001 (pENTR_ProDGK2:DGK2).
DGK2pro:DGK2-GUS
To create the GUS reporter construct translationally fused to the C terminus of the open reading frame (ORF) of DGK2, a SfoI site was introduced 5′ to the stop codon of pAA001 by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000) with primer ISY193 to obtain pAA005. The GUS cassette was inserted into the SfoI site of pAA005 to obtain pAA017.
DGK2pro:DGK2-Ven
The triple (3x) repeat of a Ven fluorescent reporter construct was obtained from pCC102 (Lin et al., 2015) and inserted into the SfoI site of pAA005 to obtain pAA018.
DGK4pro:DGK4-GUS
To create the GUS reporter construct translationally fused to the C terminus of the ORF of DGK4, we amplified 3057 bp of the genomic sequence for DGK4 by PCR from the Arabidopsis wild-type (Col-0) genomic DNA using primers ISY172 and ISY124. The PCR product was cloned into the plasmid pENTR/D–TOPO (Invitrogen, Thermo Fisher Scientific) to obtain pAA047. The SfoI site was added 5′ to the stop codon of pAA047 by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000) with primer ISY194 to obtain pAA049. The GUS cassette was inserted into the SfoI site of pAA049 to obtain pAA052.
DGK4pro:DGK4-Ven
The 3x repeat of a Ven fluorescent reporter construct was obtained from pCC102 (Lin et al., 2015) and inserted into the SfoI site of pAA049 to obtain pAA053. The obtained entry vector plasmids pAA001, pAA017, and pAA052 were recombined into the pKGW destination vector with the use of LR Clonase (Thermo Fisher Scientific; Karimi et al., 2002) to obtain pAA008, pAA033, and pAA054, respectively. These plant binary vectors were introduced into dgk2-1/+ dgk4-1/+ double heterozygotes via Agrobacterium (Agrobacterium tumefaciens)–mediated transformation. In total, 24 T1 plants were genotyped, and T2 seeds from those carrying transgene were harvested individually. To distinguish the transgene from endogenous DGK2 or DGK4, the following primers were designed and used for PCR: DGK2pro:DGK2 (PP025/KK097), DGK2pro:DGK2-GUS (PP023/KK098), and DGK4pro:DGK4-GUS (ISY011/KK098). Lines used for the observations were DGK2pro:DGK2 line #5; DGK2pro:DGK2-GUS lines #11 and #23, and DGK4pro:DGK4-GUS lines #4 and #12.
35Spro:amiDGK2 dgk4-1
Two specific miRNA sequences targeting DGK2 (amiDGK2-I and amiDGK2-II) were designed (miR-sense, 5′-TTGTTAAGTACTATTAGCCCC-3′ and 5′-TACCATTTAAAGATTGGCCCT-3′, respectively) according to WMD3 software (http://wmd3.weigelworld.org/; Schwab et al., 2006; Ossowski et al., 2008). The assembled artificial miRNA precursor fragment was cloned into the XhoI and XbaI sites of pYN2047 (Lin et al., 2015) to obtain pAA056 and pAA057, respectively (Figure 6A) and then recombined into the pKGW destination vector by using LR Clonase (Karimi et al., 2002) to obtain pAA060 and pAA061, respectively. These plasmids were introduced into dgk4-1 via Agrobacterium-mediated transformation. In total, 24 T1 plants were genotyped for the transgene (35Spro:amiDGK2-I, PP074/CH072; 35Spro:amiDGK2-II, PP078/CH072), and the T2 seeds from those carrying transgene were harvested individually. 35Spro:amiDGK2-I dgk4-1 lines #1 and #7 and 35Spro:amiDGK2-II dgk4-1 lines #2 and #4 were used for further observation.
35Spro:amiDGK4 dgk2-1
Two specific miRNA sequences targeting DGK4 (amiDGK4-I and amiDGK4-II) were designed (miR-sense, 5′-TGTTTTATCGCAGATTTCCGC-3′ and 5′-TGTTTTATCGCAGACTTCCAC-3′, respectively) according to WMD3 software (Schwab et al., 2006; Ossowski et al., 2008). The assembled artificial miRNA precursor fragment was cloned into the XhoI and XbaI sites of pYN2047 (Lin et al., 2015) to obtain pAA058 and pAA059, respectively, and then recombined into the pBGW destination vector by using LR Clonase (Karimi et al., 2002) to obtain pAA062 and pAA063, respectively. These plasmids were introduced into dgk2-1 via Agrobacterium-mediated transformation. In total, 24 T1 plants were genotyped for the transgene (35Spro:amiDGK4-I, PP082/CH072; 35Spro:amiDGK4-II, PP086/CH072), and the T2 seeds from those carrying transgene were harvested individually. 35Spro:amiDGK4-I dgk2-1 lines #2 and #20 and 35Spro:amiDGK4-II dgk2-1 lines #9 and #22 were used for further observation.
Observation of Growth Phenotypes
Flowering time was scored as the number of rosette leaves when bolting inflorescences were ∼1 cm in height when grown in long-day (16-h-light/8-h-dark) conditions. To determine the extent of vegetative growth, we measured the fresh weight and diameter of the aerial part of 24-d-old seedlings (defined as seedling size).
GUS Staining
GUS staining was observed at various growth stages and in different organs with histochemical GUS staining. Freshly harvested tissues were immersed in 90% (v/v) acetone on ice for 15 min and transferred to GUS staining solution for overnight incubation at 37°C (Lin et al., 2015). Samples were transferred to 70% (v/v) ethanol to stop staining and then to distaining solvent (ethanol:acetic acid, 6:1 in volume) to remove pigments.
Alexander Staining of Pollen
Alexander staining was performed by staining anthers in a drop of Alexander staining buffer (Alexander, 1969; Lin et al., 2015), and images were taken by using an upright fluorescent microscope (Axio Imager Z1; Zeiss) equipped with a camera (AxioCam ERc5s; Zeiss).
In Vitro Germination and Tube Elongation of Pollen
For in vitro germination and tube elongation of pollen, pollen grains from 20 mature flowers were collected in liquid medium and germinated in vitro in solid medium as described by He et al. (2018) in the absence of 10 μM myoinositol.
Cryo-Scanning Electron Microscopy
Fresh flower samples were frozen in a liquid nitrogen slush and transferred to a preparation chamber at –160°C for 5 min. Sublimation was performed at –85°C for 15 min. Subsequently, the samples were coated with platinum (Pt) at –130°C, transferred to a scanning electron microscopy chamber, and observed at –160°C using cryo-scanning electron microscopy (FEI Quanta 200 scanning electron microscope; Thermo Fisher Scientific) with cryo-scanning electron microscopy preparation system (PP2000TR; Quorum Technologies) at 20 kV.
Transient Gene Expression Assay by Particle Bombardment
To perform transient gene expression assays in tobacco leaves, 5 μg of plasmid DNA suspended with 1.25 mg of tungsten particles in water was mixed with 50 μL of 2.5 M CaCl2 and 20 μL of 0.1 M spermidine, precipitated, and suspended in ethanol. Leaves (3 to 5 cm in length) excised from plants were placed with the abaxial side upward on MS agarose medium in a Petri dish. Bombardment was performed with a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) with helium gas pressure of 450 psi. The ER marker plasmid (ER-rk) was as reported by Nelson et al. (2007). More than 40 cells randomly selected from more than five different leaves were observed for DGK2-Ven and DGK4-Ven each, all of which showed results to the images in Figure 4 and supported ER location of DGK2-Ven and DGK4-Ven.
Confocal Laser Scanning Microscopy
Fluorescence of 3xVen and monomeric red fluorescent protein was observed under a microscope (LSM 510 Meta; Zeiss) equipped with objectives (Plan-Apochromat 20×/0.8-NA and Plan-Apochromat 10×/0.45-NA). Images were captured under a microscope (LSM 510 v3.2; Zeiss) with filters for Ven (514 nm laser, 520 to 555 nm band-pass) and monomeric red fluorescent protein (561 nm laser, 590 to 630 nm band-pass).
Protoplast Transformation and Immunoblot Analysis
Protoplasts were isolated from mesophyll cells of well-expanded leaves of 3- to 5-week old Arabidopsis according to Wu et al. (2009). Protoplast transfection was performed as reported previously (Yoo et al., 2007) with 20 μg of plasmid DNA stored at 4°C and ∼4 × 104 protoplasts in 200 μL of MMg solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7) at room temperature. Plasmid was transformed into protoplasts by adding 220 μL of 40% (w/v) polyethylene glycol solution, followed by incubation at room temperature for 6 min. The transformed protoplasts were washed with W5 buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM Glc, and 2 mM MES, pH 5.7) and then incubated in W5 buffer containing 1% (w/v) BSA at room temperature for 16 h under white light (75 μmol m−2 s−1, GC-560H) at 22°C. The transfected protoplasts were collected and suspended with 2× SDS-PAGE loading buffer, and 20 µg was run on SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. The blotted membrane was rinsed twice in TBS buffer with 0.1% (w/v) Tween 20 (TBST) and blocked in 5% (w/v) nonfat milk powder in TBST for 1 h. The membrane was washed in TBST and probed with a primary rabbit anti-GFP antibody (A-11122; Invitrogen) at a dilution of 1:2500 overnight at 4°C, washed three times in TBST, and then incubated with secondary antibody (goat anti-rabbit IgG–horseradish peroxidase, ab6720; Invitrogen) at a dilution of 1:10,000 at room temperature for 1 h. Signals were visualized using chemiluminescent substrate (Lumigen ECL; GE Healthcare) and image analyzer (Image Quant LAS 4000; GE Healthcare). More than 30 cells were observed for DGK2-Ven and DGK4-Ven each, all of which showed a clear overlap with ER marker signal.
In Vitro DGK4 Enzyme Activity
To produce recombinant DGK4 N-terminally fused with maltose-binding protein (MBP) in Escherichia coli, 1464 bp of the ORF for DGK4 was amplified with primers PP069 and PP058. The fragment was cloned into the SalI and XbaI sites of the plasmid pMAL-c5x (New England Biolabs) to obtain pMAL5x-DGK4 (pAA048), which was introduced into E. coli C41 (DE3) strain (Lucigen). A 50-mL culture in M9 medium at an A600 of 0.5 to 0.6 was treated with isopropyl-d-thiogalactopyranoside (2 mM at final concentration) to induce production of MBP-DGK4 fusion protein at 23°C for 20 h. The cells were collected by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 15% [w/v] glycerol), and lysed by a few repeats of freeze-thaw cycles followed by sonication. The crude enzyme suspension was obtained by centrifugation at 3000g for 20 min at 4°C. Protein concentration in the supernatant was determined by the Bradford method (Bradford, 1976) with BSA as a standard.
For the enzyme assay, reaction conditions and total lipid recovery procedures were as described previously (Gómez-Merino et al., 2004), with minor modification. Briefly, the standard assay mixture was in a total volume of 250 μL that contained 40 mM Tricine, 1 mM CHAPS, 0.02% (v/v) Triton X-100, 5 mM MgCl2, 1 mM ATP, 50 µM NBD-DAG (Cayman Chemical), 450 µM 1-2-dioleoyl-sn-glycerol (Avanti Polar Lipids), and ∼10 µg of crude enzyme. DAG substrate mixture was prepared by mixing 50 µM NBD-DAG and 450 µM 1-2-dioleoyl-sn-glycerol, dried under nitrogen stream, and resuspended and sonicated in the assay buffer. After the reaction, total lipid was separated by one-dimensional silica gel thin layer chromatography (TLC) with a solvent system of ethyl acetate:iso-octane:formic acid/water (52:8:12:40, by volume) for separation of PA (Nakamura et al., 2005). Fluorescent bands of the products on TLC plates were determined, and the intensities were quantified by using an image analyzer (ImageQuant LAS 4000).
RNA Extraction and Transcript Analysis
RNA extraction and RT-qPCR were performed as described previously (Lin et al. 2015). Briefly, total RNA was isolated from 7-d-old seedlings using TRI reagent (Ambion) including DNase treatment, and cDNA was synthesized using the SuperScript III first-strand synthesis kit (Invitrogen, Thermo Fisher Scientific).
RT-qPCR was performed with the 7500 Real-Time PCR System (Applied Biosystems). Reactions (10 μL of total volume) were performed in a 96-well plate containing 5 μL of 2× SYBR Green (Ambion), 2 ng of cDNA, and 200 nM of each gene-specific primer under the following standard PCR program: 95°C for 10 min; 40 cycles of 95°C for 15 s and 60°C for 1 min. The comparative threshold cycle method was used to determine relative gene expression, with the expression of ACTIN2 (At3g18780; KK129/KK130) as an internal control. Data are mean ± sd from three biological replicates, each with three technical replicates. The primer sets for quantitative RT-qPCR were as reported by Nakamura et al. (2014).
For detection of transcripts, the full-length protein coding sequence of DGK2 (2139 bp), DGK4 (1464 bp), and DGK6 (1401 bp) was amplified with the cDNA synthesized from the total RNA of 7-d old seedlings (wild type, dgk2-1, dgk4-1, and dgk6-1) under the standard PCR program (94°C for 5 min; 38 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 3 min; 72°C for 10 min) with the following primer sets: DGK2 (PP007/PP008), DGK4 (PP069/PP058), and DGK6 (PP150/PP151).
Glycerolipid Analysis
Polar glycerolipids were analyzed as described by Nakamura et al. (2003, 2014). Briefly, total lipid was extracted from samples with chloroform-methanol solvent system as described by Bligh and Dyer (1959). Each lipid class was separated by two-dimensional silica-gel TLC with the solvent system as described by Nakamura et al. (2003), scraped off, and immersed for 3 h at 80°C in HCl-methanol, including pentadecanoic acid (1 mM) as an internal standard. The reaction products were extracted with hexane, concentrated, and analyzed with gas chromatography equipped with a flame ionization detector (GC-2010, Shimadzu) and ULBON HR-SS-10 column (Shinwa Chemical Industries).
Radioisotope Pulse-Chase Labeling Assay
Radiolabeling analysis of polar glycerolipids in different suppression lines of 35Spro:amiDGK2-II dgk4-1 of Arabidopsis was as follows: the rosette leaf samples were labeled with 0.15 mM sodium [14C]acetate (52 mCi/mmol PerkinElmer) in half-strength MS liquid medium. After 1 h of incubation with the radioisotope-containing solution, the tissue samples were rinsed twice with water and harvested at the indicated times: 0, 1, 3, and 24 h. Total lipids were extracted from tissue samples as described by Lin et al. (2019). Each polar glycerolipid class was separated on an HPTLC plate (Silica gel 60G, Merck) with the first development solvent system of methyl acetate/isopropanol/chloroform/methanol/0.45% KCl (25:25:25:12:4, by volume) and second development solvent system of hexane/diethylether/acetic acid (70:30:2, by volume). Lipid standards used were as described by Lin et al. (2018).
To perform radioisotope labeling assay with Arabidopsis rosette leaves, tissue samples were labeled with 1 mM [γ-32P]ATP (1 Ci mmol−1, PerkinElmer) in half-strength MS liquid medium. After 30 min of incubation with radioisotope-containing solution, tissue samples were rinsed twice with water, and the labeled compounds were extracted at the indicated times: 0, 5, 30, 60, and 180 min. Tissue was ground to a fine powder under liquid N2, and lipids were extracted as described by Lin et al. (2019). Each lipid class was separated on a TLC plate (Silica gel 60G) with the solvent system of chloroform:methanol:acetic acid (65:25:15, by volume). Lipid standards used were described in Lin et al. (2018). Radioactive spots were visualized by using an imaging plate (BAS-MS 2040; Fuji Film) in the hypercassette (10239344; GE Healthcare), and the intensities were quantified with an image analyzer (Typhoon FLA 7000; GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: DGK2 (At5g63770), DGK4 (At5g57690), DGK6 (At4g28130), ACTIN2 (At3g18780).
Supplemental Data
Supplemental Figure 1. Morphology of single mutants for DGK2 and DGK4.
Supplemental Figure 2. Tissue-specific localization of DGK2 and DGK4.
Supplemental Figure 3. Detection of DGK2-Ven and DGK4-Ven from protein extract of Arabidopsis leaf protoplasts transiently producing these fusion proteins.
Supplemental Figure 4. Effect of n-butanol and butyraldehyde treatment on wild-type pollen tube growth.
Supplemental Figure 5. A representative image of siliques from the wild type and four independent transgenic lines.
Supplemental Figure 6. Reproductive growth phenotype of 35Spro:amiDGK4 dgk2-1 plants.
Supplemental Figure 7. In vitro DGK enzyme activity assay of recombinant DGK4 fused with maltose binding protein (MBP) and produced in Escherichia coli.
Supplemental Table 1. Reciprocal genetic crossing of Arabidopsis dgk2-1 and dgk4-1.
Supplemental Table 2. List of oligonucleotide primer sequences used in this study.
Supplemental Data Set. Statistical analysis data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ian Sofian Yunus (Institute of Plant and Microbial Biology, Academia Sinica; currently at Imperial College London) for isolation of T-DNA mutants (dgk2-1, dgk4-1, and dgk6-1) and technical support with molecular cloning. This research was supported by Academia Sinica (Career Development Award grant AS-CDA-107-L02 to Y.N.).
AUTHOR CONTRIBUTIONS
A.E.A., V.C.N., and F.G. performed experiments, analyzed data, and wrote the article; Y.N. conceived research, supervised experiments and data analysis, and wrote the article. All authors commented on the article and approved the content.
References
- Alexander M.P.(1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Arisz S.A., Testerink C., Munnik T.(2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791: 869–875. [DOI] [PubMed] [Google Scholar]
- Asaoka Y., Nakamura S., Yoshida K., Nishizuka Y.(1992). Protein kinase C, calcium and phospholipid degradation. Trends Biochem. Sci. 17: 414–417. [DOI] [PubMed] [Google Scholar]
- Bates P.J., Reyes-Reyes E.M., Malik M.T., Murphy E.M., O’Toole M.G., Trent J.O.(2017). G-Quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 1861 (5 Pt B): 1414–1428. [DOI] [PubMed] [Google Scholar]
- Benning C., Ohta H.(2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280: 2397–2400. [DOI] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J.(1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Bradford M.M.(1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Browse J., McCourt P., Somerville C.R.(1985). A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science 227: 763–765. [DOI] [PubMed] [Google Scholar]
- Derevyanchuk M., Kretynin S., Kolesnikov Y., Litvinovskaya R., Martinec J., Khripach V., Kravets V.(2019). Seed germination, respiratory processes and phosphatidic acid accumulation in Arabidopsis diacylglycerol kinase knockouts – The effect of brassinosteroid, brassinazole and salinity. Steroids 147: 28–36. [DOI] [PubMed] [Google Scholar]
- Ebinu J.O., Bottorff D.A., Chan E.Y.W., Stang S.L., Dunn R.J., Stone J.C.(1998). RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science 280: 1082–1086. [DOI] [PubMed] [Google Scholar]
- Escobar-Sepúlveda H.F., Trejo-Téllez L.I., Pérez-Rodríguez P., Hidalgo-Contreras J.V., Gómez-Merino F.C.(2017). Diacylglycerol kinases are widespread in higher plants and display inducible gene expression in response to beneficial elements, metal, and metalloid ions. Front. Plant Sci. 8: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Zheng S., Cui D., Wang X.(1999). Subcellular distribution and tissue expression of phospholipase Dalpha, Dbeta, and Dgamma in Arabidopsis. Plant Physiol. 119: 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Merino F.C., Arana-Ceballos F.A., Trejo-Téllez L.I., Skirycz A., Brearley C.A., Dörmann P., Mueller-Roeber B.(2005). Arabidopsis AtDGK7, the smallest member of plant diacylglycerol kinases (DGKs), displays unique biochemical features and saturates at low substrate concentration: The DGK inhibitor R59022 differentially affects AtDGK2 and AtDGK7 activity in vitro and alters plant growth and development. J. Biol. Chem. 280: 34888–34899. [DOI] [PubMed] [Google Scholar]
- Gómez-Merino F.C., Brearley C.A., Ornatowska M., Abdel-Haliem M.E.F., Zanor M.-I., Mueller-Roeber B.(2004). AtDGK2, a novel diacylglycerol kinase from Arabidopsis thaliana, phosphorylates 1-stearoyl-2-arachidonoyl-sn-glycerol and 1,2-dioleoyl-sn-glycerol and exhibits cold-inducible gene expression. J. Biol. Chem. 279: 8230–8241. [DOI] [PubMed] [Google Scholar]
- He S.-L., Hsieh H.-L., Jauh G.-Y.(2018). SMALL AUXIN UP RNA62/75 are required for the translation of transcripts essential for pollen tube growth. Plant Physiol. 178: 626–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I.(2016). Plant phosphoinositide signaling - dynamics on demand. Biochim. Biophys. Acta 1861 (9 Pt B): 1345–1351. [DOI] [PubMed] [Google Scholar]
- Helling D., Possart A., Cottier S., Klahre U., Kost B.(2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18: 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhao J., Guo L., Kim S.-C., Deng X., Wang G., Zhang G., Li M., Wang X.(2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62: 55–74. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A.(2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Katagiri T., Mizoguchi T., Shinozaki K.(1996). Molecular cloning of a cDNA encoding diacylglycerol kinase (DGK) in Arabidopsis thaliana. Plant Mol. Biol. 30: 647–653. [DOI] [PubMed] [Google Scholar]
- Kelly A.A., Froehlich J.E., Dörmann P.(2003). Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H.(2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA 104: 17216–17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Pricer W.E. Jr.(1953). Enzymatic esterification of alpha-glycerophosphate by long chain fatty acids. J. Biol. Chem. 204: 345–357. [PubMed] [Google Scholar]
- Lin Y.C., Kanehara K., Nakamura Y.(2019). Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol. 223: 1904–1917. [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Kobayashi K., Wada H., Nakamura Y.(2018). Phosphatidylglycerophosphate phosphatase is required for root growth in Arabidopsis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863: 563–575. [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Liu Y.C., Nakamura Y.(2015). The choline/ethanolamine kinase family in Arabidopsis: Essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27: 1497–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A.M., Bortul R., Tabellini G., Bareggi R., Manzoli L., Narducci P., Cocco L.(2002). Diacylglycerol kinases in nuclear lipid-dependent signal transduction pathways. Cell. Mol. Life Sci. 59: 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérida I., Ávila-Flores A., Merino E.(2008). Diacylglycerol kinases: At the hub of cell signalling. Biochem. J. 409: 1–18. [DOI] [PubMed] [Google Scholar]
- Munnik T.(2001). Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6: 227–233. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F.(1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nakamura Y., Arimitsu H., Yamaryo Y., Awai K., Masuda T., Shimada H., Takamiya K., Ohta H.(2003). Digalactosyldiacylglycerol is a major glycolipid in floral organs of Petunia hybrida. Lipids 38: 1107–1112. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Awai K., Masuda T., Yoshioka Y., Takamiya K., Ohta H.(2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280: 7469–7476. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ohta H.(2007). The diacylglycerol forming pathways differ among floral organs of Petunia hybrida. FEBS Lett. 581: 5475–5479. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Teo N.Z., Shui G., Chua C.H., Cheong W.F., Parameswaran S., Koizumi R., Ohta H., Wenk M.R., Ito T.(2014). Transcriptomic and lipidomic profiles of glycerolipids during Arabidopsis flower development. New Phytol. 203: 310–322. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A.(2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D.(2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Pleskot R., Pejchar P., Bezvoda R., Lichtscheidl I.K., Wolters-Arts M., Marc J., Zárský V., Potocký M.(2012). Turnover of phosphatidic acid through distinct signaling pathways affects multiple aspects of pollen tube growth in tobacco. Front. Plant Sci. 3: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocký M., Eliás M., Profotová B., Novotná Z., Valentová O., Zárský V.(2003). Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130. [DOI] [PubMed] [Google Scholar]
- Ron D., Kazanietz M.G.(1999). New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 13: 1658–1676. [PubMed] [Google Scholar]
- Sakane F., Imai S., Yamada K., Murakami T., Tsushima S., Kanoh H.(2002). Alternative splicing of the human diacylglycerol kinase δ gene generates two isoforms differing in their expression patterns and in regulatory functions. J. Biol. Chem. 277: 43519–43526. [DOI] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A.(2000). Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D.(2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden W.A., Blumwald E.(2000). Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J. 24: 317–326. [DOI] [PubMed] [Google Scholar]
- Tadege M., Kuhlemeier C.(1997). Aerobic fermentation during tobacco pollen development. Plant Mol. Biol. 35: 343–354. [DOI] [PubMed] [Google Scholar]
- Tan W.-J., Yang Y.-C., Zhou Y., Huang L.-P., Xu L., Chen Q.-F., Yu L.-J., Xiao S.(2018). DIACYLGLYCEROL ACYLTRANSFERASE and DIACYLGLYCEROL KINASE modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol. 177: 1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C., Munnik T.(2005). Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 10: 368–375. [DOI] [PubMed] [Google Scholar]
- Tognon C.E., Kirk H.E., Passmore L.A., Whitehead I.P., Der C.J., Kay R.J.(1998). Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol. Cell. Biol. 18: 6995–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz Dias F., Serrazina S., Vitorino M., Marchese D., Heilmann I., Godinho M., Rodrigues M., Malhó R.(2019). A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tube growth. New Phytol 222: 1434–1446. [DOI] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S.(2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.F., Freer S., Benson A.A.(1967). Transphosphatidylation by phospholipase D. J. Biol. Chem. 242: 477–484. [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang W., Chen J., Zhang H., Song F.(2008). Overexpression of a rice diacylglycerol kinase gene OsBIDK1 enhances disease resistance in transgenic tobacco. Mol. Cells 26: 258–264. [PubMed] [Google Scholar]
- Zhou Y., Peisker H., Weth A., Baumgartner W., Dörmann P., Frentzen M.(2013). Extraplastidial cytidinediphosphate diacylglycerol synthase activity is required for vegetative development in Arabidopsis thaliana. Plant J. 75: 867–879. [DOI] [PubMed] [Google Scholar]