The pathogen-responsive protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulate camalexin biosynthesis upon pathogen infection via differential phosphorylation of the transcription factor WRKY33.
Abstract
Camalexin is a major phytoalexin that plays a crucial role in disease resistance in Arabidopsis (Arabidopsis thaliana). We previously characterized the regulation of camalexin biosynthesis by the mitogen-activated protein kinases MPK3 and MPK6 and their downstream transcription factor WRKY33. Here, we report that the pathogen-responsive CALCIUM-DEPENDENT PROTEIN KINASE5 (CPK5) and CPK6 also regulate camalexin biosynthesis in Arabidopsis. Chemically induced expression of constitutively active CPK5 or CPK6 variants was sufficient to induce camalexin biosynthesis in transgenic Arabidopsis plants. Consistently, the simultaneous mutation of CPK5 and CPK6 compromised camalexin production in Arabidopsis induced by the fungal pathogen Botrytis cinerea. Moreover, we identified that WRKY33 functions downstream of CPK5/CPK6 to activate camalexin biosynthetic genes, thereby inducing camalexin biosynthesis. CPK5 and CPK6 interact with WRKY33 and phosphorylate its Thr-229 residue, leading to an increase in the DNA binding ability of WRKY33. By contrast, the MPK3/MPK6-mediated phosphorylation of WRKY33 on its N-terminal Ser residues enhances the transactivation activity of WRKY33. Furthermore, both gain- and loss-of-function genetic analyses demonstrated the cooperative regulation of camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6. Taken together, these findings indicate that WRKY33 functions as a convergent substrate of CPK5/CPK6 and MPK3/MPK6, which cooperatively regulate camalexin biosynthesis via the differential phospho-regulation of WRKY33 activity.
INTRODUCTION
Plants have developed a multilayer surveillance system to recognize invading pathogens. Plant perception of pathogen-associated molecular patterns (PAMPs) and pathogen-derived effectors triggers several early signaling events, including the influx of calcium ions (Ca2+), activation of mitogen-activated protein kinases (MAPKs), and induction of reactive oxygen species (ROS) and ethylene production (Jones and Dangl, 2006; Bigeard et al., 2015; Cui et al., 2015; Couto and Zipfel, 2016; Tang et al., 2017; Peng et al., 2018). These early events are involved in signaling the late-stage defense responses, including defense gene activation, phytoalexin production, cell wall strengthening, and the hypersensitive response, to confer disease resistance (Meng and Zhang, 2013; Seybold et al., 2014; Cui et al., 2015; Qi et al., 2017).
Calcium-dependent protein kinases (CDPKs) perceive the rapid Ca2+ influx triggered after pathogen perception. CDPKs, together with MAPKs, play pivotal roles in regulating plant defense responses through the phosphorylation of downstream substrate proteins (Boudsocq and Sheen, 2013; Meng and Zhang, 2013; Romeis and Herde, 2014; Seybold et al., 2014; Bigeard et al., 2015). Much progress has been made in understanding how MAPK signaling functions in plant immune responses, whereas the roles of CDPKs in plant immunity are less well characterized. Arabidopsis (Arabidopsis thaliana) contains 34 putative genes encoding CDPKs, several of which were shown to be activated upon plant perception of PAMPs or pathogen effectors (Boudsocq et al., 2010; Boudsocq and Sheen, 2013; Dubiella et al., 2013; Gao et al., 2013). Independent studies have demonstrated that CPK5 and CPK1/CPK2/CPK4/CPK11 are all involved in phosphorylating the respiratory burst oxidase homolog D (RBOHD) to promote ROS production (Boudsocq et al., 2010; Dubiella et al., 2013; Gao et al., 2013). Functional genomics analysis has identified CPK4, CPK5, CPK6, and CPK11 as overlapping regulators of transcriptional reprogramming triggered by bacterial PAMPs or effectors (Boudsocq et al., 2010; Gao et al., 2013). In addition, CPK5 and CPK6 are also involved in pathogen-induced ethylene production and hypersensitive cell death (Dubiella et al., 2013; Gravino et al., 2015; Liu et al., 2017). However, how these pathogen-responsive CDPKs regulate downstream defense responses remains largely unclear. In addition, the crosstalk between CDPKs and MAPKs has been implicated in plant immune responses (Ludwig et al., 2005; Boudsocq et al., 2010), but the underlying mechanisms are currently unknown.
Phytoalexins are low-molecular-weight antimicrobial compounds produced by plants after exposure to pathogens, which play crucial roles in plant disease resistance (Piasecka et al., 2015). Camalexin (3-thiazol-2′-yl-indole), a major phytoalexin in Arabidopsis, accumulates to high levels in response to a variety of pathogens (Tsuji et al., 1992; Thomma et al., 1999; Bednarek et al., 2009; Schlaeppi et al., 2010; Stotz et al., 2011; Hiruma et al., 2013) and is then exported to the extracellular space by the transporters PLEIOTROPIC DRUG RESISTANCE12 (PDR12) and PENETRATION3 (PEN3)/PDR8 to defend against pathogens (He et al., 2019). Arabidopsis mutants defective in camalexin biosynthesis exhibit enhanced susceptibility to a number of fungal and oomycetic pathogens, highlighting the importance of camalexin in plant resistance to these pathogens (Glazebrook and Ausubel, 1994; Thomma et al., 1999; Bednarek et al., 2009; Schlaeppi et al., 2010; Stotz et al., 2011; Hiruma et al., 2013).
Camalexin is an indolic compound derived from Trp metabolism. The first step in camalexin biosynthesis in Arabidopsis is catalyzed by two homologous cytochrome P450 enzymes, CYP79B2 and CYP79B3, which convert Trp into indole-3-acetaldoxime (IAOx; Zhao et al., 2002). From IAOx, several branches of indolic metabolism diverge, leading to the formation of camalexin, indole glucosinolates, auxin (indole-3-acetic acid), and several other indolics (Zhao et al., 2002; Nafisi et al., 2007; Sønderby et al., 2010). In the camalexin biosynthetic pathway, IAOx is later converted to indole-3-acetonitrile (IAN) by the P450 enzyme CYP71A13 (Nafisi et al., 2007). Thereafter, IAN is conjugated with glutathione by glutathione S-transferases to form GSH(IAN) (Su et al., 2011), which is further processed by γ-glutamyl peptidases to generate Cys(IAN) (Geu-Flores et al., 2011). Finally, camalexin is synthesized from Cys(IAN) via a two-step reaction catalyzed by the P450 enzyme CYP71B15/PAD3 (Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009).
In Arabidopsis, camalexin biosynthetic genes, including CYP79B2, CYP71A13, and PAD3, are expressed at very low levels in the absence of environmental stress and are highly induced by pathogen infection to activate camalexin biosynthesis (Schuhegger et al., 2006; Nafisi et al., 2007). The involvement of the transcription factor WRKY33 in regulating camalexin biosynthesis has been well characterized (Qiu et al., 2008; Mao et al., 2011). The induction of camalexin biosynthesis is largely blocked in wrky33 mutants after infection by the fungal pathogen Botrytis cinerea or the bacterial pathogen Pseudomonas syringae (Qiu et al., 2008; Mao et al., 2011). WRKY33 binds to the promoters of CYP71A13 and PAD3 in vivo, suggesting that WRKY33 directly activates these camalexin biosynthetic genes (Qiu et al., 2008; Mao et al., 2011; Birkenbihl et al., 2017). We previously showed that the pathogen-responsive MAPKs MPK3 and MPK6 positively regulate camalexin biosynthesis by phosphorylating WRKY33 (Ren et al., 2008; Mao et al., 2011). However, how MPK3/MPK6-mediated phosphorylation regulates WRKY33 activity remains unknown. WRKY33 also regulates camalexin biosynthesis by interacting with another immune-related MAPK, MPK4 (Qiu et al., 2008). WRKY33 forms a nuclear complex with MPK4 and its substrate MKS1. It is thought that the phosphorylation of MKS1 by active MPK4 results in the release of WRKY33 from the complex to activate camalexin biosynthesis (Qiu et al., 2008).
In this study, we report that the Arabidopsis CDPKs CPK5 and CPK6 positively regulate camalexin biosynthesis in response to B. cinerea infection. In the absence of pathogen attack, conditional expression of a constitutively active CPK5 or CPK6 variant in transgenic Arabidopsis plants is sufficient to trigger the activation of camalexin biosynthetic genes and therefore the production of camalexin. Consistently, the induction of camalexin biosynthesis by B. cinerea is compromised in the cpk5 cpk6 double mutant due to decreased induction of camalexin biosynthetic genes. Moreover, we demonstrate that WRKY33 functions downstream of CPK5/CPK6 to activate camalexin synthetic genes and therefore induce camalexin biosynthesis. WRKY33 is further identified as a substrate of CPK5/CPK6, which phosphorylate the Thr-229 residue of WRKY33 and thus enhance its DNA binding ability. By contrast, phosphorylation of WRKY33 by MPK3/MPK6 on the N-terminal Ser residues of WRKY33 increases its transactivation activity. These results, together with further gain- and loss-of-function genetic analyses, indicate that the differential phosphorylation of WRKY33 by CPK5/CPK6 and MPK3/MPK6 cooperatively upregulates camalexin biosynthesis in Arabidopsis upon pathogen infection.
RESULTS
Conditional Expression of a Constitutively Active CPK5 Variant Induces Camalexin Biosynthesis in the Absence of Pathogen Attack
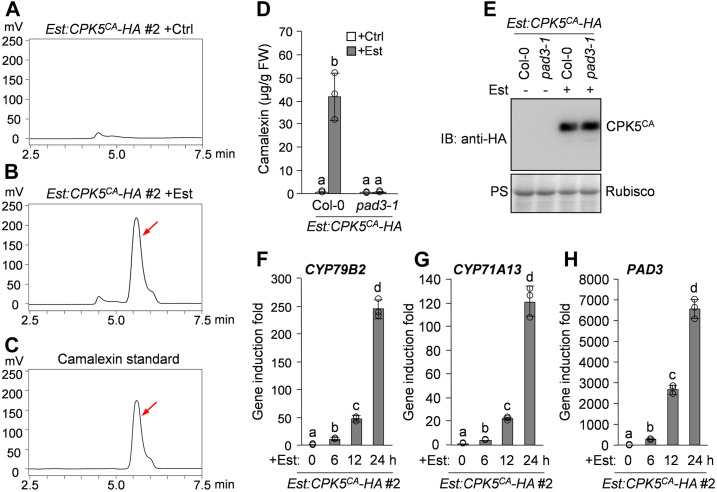
In plants, host responses to an attempted pathogen infection include the activation of secondary metabolic reprogramming (Piasecka et al., 2015). To investigate whether pathogen-responsive CDPKs regulate the biosynthesis of antimicrobial metabolites in Arabidopsis, we generated a conditional gain-of-function transgenic plant (named Est:CPK5CA-HA), in which a hemagglutinin (HA)-tagged constitutively active CPK5 variant (CPK5CA) that lacks the C-terminal autoinhibitory and Ca2+ binding domains of CPK5 was expressed under the control of an estradiol (Est)-inducible promoter (Supplemental Figure 1B). We then profiled the metabolites secreted into the liquid culture medium by the Est:CPK5CA-HA transgenic seedlings after treatment with Est. HPLC analysis with fluorescence detection revealed an induced metabolite in the seedling culture after Est treatment (Figures 1A and 1B). This Est-induced metabolite had the same retention time as the synthetic camalexin standard (Figures 1B and 1C), suggesting that CPK5CA expression induces camalexin biosynthesis. Mutation of the camalexin biosynthetic gene PAD3 in the Est:CPK5CA-HA plant by genetic crossing blocked the production of the Est-induced metabolite (Figures 1D and 1E), confirming that the metabolite induced by CPK5CA in our HPLC analysis is camalexin. The Est-induced protein levels of CPK5CA in different transgenic lines were closely correlated with the levels of camalexin production (Supplemental Figure 1), further confirming that conditional expression of CPK5CA induces camalexin biosynthesis.
Figure 1.
Conditional Activation of CPK5 Induces Camalexin Biosynthesis in Arabidopsis.
(A) to (C) Est-induced expression of CPK5CA in Est:CPK5CA-HA transgenic plants induces camalexin production. Two-week-old Est:CPK5CA-HA seedlings were treated with 10 μM Est (B) or the solvent control (ethanol [Ctrl]; [A]) for 24 h in liquid medium. The metabolites in the medium were analyzed by HPLC with fluorescence detection. Pure camalexin was used as the reference standard (C). Arrows indicate the peak of camalexin.
(D) and (E) CPK5CA-induced camalexin production is abolished in the pad3-1 mutant background. Two-week-old Est:CPK5CA-HA and Est:CPK5CA-HA/pad3-1 seedlings were treated with 10 μM Est or solvent control for 24 h, and camalexin production was quantified (D). The expression levels of CPK5CA-HA were analyzed by immunoblotting (IB) with anti-HA antibody, and total protein loading was assessed by Ponceau S staining (PS; [E]). FW, fresh weight.
(F) to (H) Est-induced CPK5CA expression in Est:CPK5CA-HA seedlings leads to the activation of camalexin biosynthetic genes. Two-week-old Est:CPK5CA-HA seedlings were treated with 10 μM Est. Transcript levels of camalexin biosynthetic genes CYP79B2 (F), CYP71A13 (G), and PAD3 (H) at the indicated time points were analyzed by RT-qPCR.
In (D), (F), (G), and (H), error bars indicate sd (n = 3 biological repeats), small circles represent individual data points, and different letters above the columns indicate significant differences (P < 0.05), as determined by one-way ANOVA.
The induction of camalexin biosynthesis in Est:CPK5CA-HA plants after Est treatment was associated with the activation of camalexin biosynthetic genes. As shown in Figures 1F to 1H, the P450 genes involved in pathogen-induced camalexin biosynthesis, including CYP79B2, CYP71A13, and PAD3, were highly activated by the Est-induced expression of CPK5CA. A luciferase (LUC) reporter-aided analysis of the promoter activities of CYP79B2, CYP71A13, and PAD3 in Arabidopsis protoplasts also revealed the activation of these camalexin biosynthetic genes upon coexpression of CPK5CA (Supplemental Figures 2A to 2C). Additionally, a previous transcriptional profiling analysis also showed the CPK5CA-mediated activation of CYP79B2 and CYP71A13 in Arabidopsis protoplasts (Boudsocq et al., 2010). Therefore, these results indicate that CPK5CA expression (i.e., CPK5 activation) induces camalexin biosynthesis through the transcriptional activation of camalexin biosynthetic genes.
CPK5 and CPK6 Function Redundantly in Regulating B. cinerea-Induced Camalexin Biosynthesis
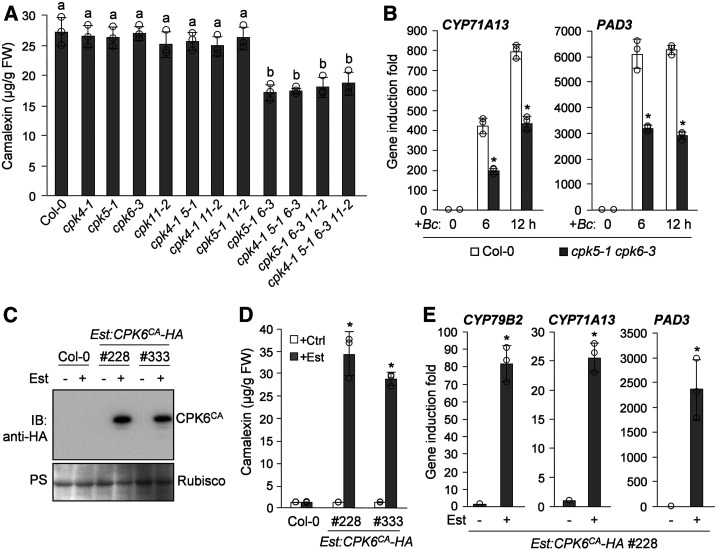
To investigate the role of CPK5 in pathogen-induced camalexin biosynthesis, we examined the induction of camalexin biosynthesis in the cpk5-1 knockout mutant after infection by B. cinerea. We did not observe reduced camalexin production in cpk5-1 compared with the Arabidopsis wild-type Columbia (Col-0; Figure 2A). CPK4, CPK5, CPK6, and CPK11 play overlapping roles in Arabidopsis immunity (Boudsocq et al., 2010; Gao et al., 2013). To reveal the potential functional redundancy of these CPKs in regulating camalexin biosynthesis, we isolated the cpk4-1, cpk6-3, and cpk11-2 knockout mutants and generated different cpk double, triple, and quadruple mutants. As shown in Figure 2A, like cpk5-1, the cpk4-1, cpk6-3, and cpk11-2 single mutants also exhibited normal induction of camalexin biosynthesis after B. cinerea infection. Notably, compromised induction of camalexin biosynthesis was observed in the cpk5-1 cpk6-3 but not the cpk4-1 cpk5-1, cpk5-1 cpk11-2, or cpk4-1 cpk11-2 double mutants, indicating that CPK5 and CPK6, but not CPK4 or CPK11, function redundantly to positively regulate B. cinerea-induced camalexin biosynthesis, which is consistent with the close phylogenetic relationship of CPK5 to CPK6 (Boudsocq and Sheen, 2013). Moreover, as observed in the cpk triple and quadruple mutants, mutation of CPK4 and/or CPK11 in the cpk5-1 cpk6-3 double mutant background also did not affect the induction of camalexin production in cpk5-1 cpk6-3 (Figure 2A), further indicating that CPK4 and CPK11 are not required for B. cinerea-induced camalexin biosynthesis. The compromised camalexin production in cpk5-1 cpk6-3 after B. cinerea infection is associated with the greatly reduced induction of camalexin biosynthetic genes, including CYP71A13 and PAD3 (Figure 2B), which is consistent with the involvement of the activation of these genes in CPK5CA-induced camalexin biosynthesis. Together, these results indicate that CPK5 and CPK6 play a redundant and positive role in regulating B. cinerea-induced camalexin biosynthesis through the activation of camalexin biosynthetic genes. Additionally, CPK5 and CPK6 expression was also induced by B. cinerea infection, with similar induction patterns (Supplemental Figure 3), further supporting the positive and redundant role of CPK5 and CPK6 in inducing camalexin biosynthesis.
Figure 2.
CPK5 and CPK6 Redundantly Regulate Camalexin Biosynthesis.
(A) CPK5 and CPK6 play redundant roles in B. cinerea-induced camalexin biosynthesis. Camalexin produced from 2-week-old Col-0 and cpk mutant seedlings was measured at 24 h postinoculation with B. cinerea spores. Error bars indicate sd (n = 3 biological repeats), small circles represent individual data points, and different letters above the columns indicate significant differences (P < 0.05), as determined by one-way ANOVA. FW, fresh weight.
(B) Requirement of CPK5 and CPK6 in B. cinerea-induced expression of camalexin biosynthetic genes. Two-week-old Col-0 and cpk5-1 cpk6-3 seedlings were inoculated with B. cinerea spores. Transcript levels of CYP71A13 and PAD3 at the indicated time points were analyzed by RT-qPCR. Asterisks above the columns indicate significant differences (P < 0.05) compared with Col-0 at specific time points, as determined by Student’s t test. Error bars indicate sd (n = 3 biological repeats).
(C) to (E) Est-induced expression of CPK6CA in Est:CPK6CA-HA transgenic plants induces camalexin production and activates camalexin biosynthetic gene expression. Two-week-old Est:CPK6CA-HA transgenic seedlings were treated with 10 μM Est or the solvent control (Ctrl) for 48 h. The expression of CPK6CA-HA was analyzed by immunoblotting (IB) with anti-HA antibody, and total protein loading was assessed by Ponceau S staining (PS; [C]). Meanwhile, camalexin production was quantified (D), and the transcript levels of CYP79B2, CYP71A13, and PAD3 in one Est:CPK6CA-HA line were analyzed by RT-qPCR (E). Asterisks above the columns indicate significant differences (P < 0.05) compared with the Est-treated Col-0 (D) or the Est:CPK6CA-HA transgenic line without Est treatment (E), as determined by Student’s t test. Error bars indicate sd (n = 3 biological repeats).
To provide gain-of-function evidence supporting the role of CPK6 in regulating camalexin biosynthesis, we generated Est:CPK6CA-HA transgenic Arabidopsis plants expressing an HA-tagged constitutively active CPK6 variant (CPK6CA) under the control of an Est-inducible promoter (Figure 2C). As shown in Figure 2D, induction of camalexin production was observed in Est:CPK6CA-HA plants after Est treatment, which was also associated with the activation of camalexin synthetic genes (Figure 2E). A LUC reporter assay in Arabidopsis protoplasts also showed the activation of the CYP79B2, CYP71A13, and PAD3 promoters in response to the coexpression of CPK6CA (Supplemental Figures 2A, 2D, and 2E). These results indicate that, similar to CPK5, conditional activation of CPK6 also can induce camalexin biosynthesis via the transcriptional activation of camalexin synthetic genes, further confirming the positive role of CPK6 in regulating camalexin biosynthesis.
WRKY33 Acts Downstream of CPK5/CPK6 in Regulating Camalexin Biosynthesis
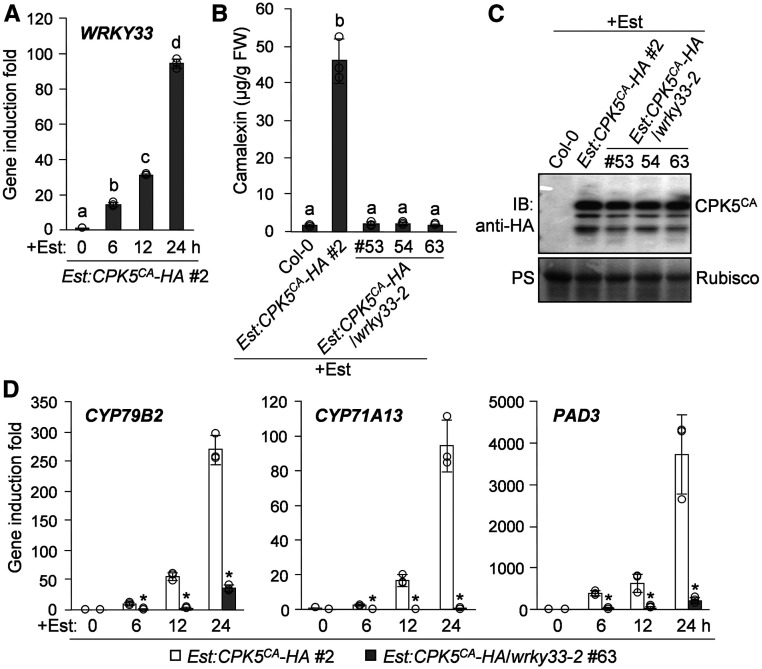
In search for the transcription factor(s) that regulate camalexin biosynthesis downstream of CPK5/CPK6, we found that the induction of CPK5CA expression in Est:CPK5CA-HA plants was sufficient to highly activate the expression of WRKY33 (Figure 3A), which encodes a transcription factor essential for pathogen-induced camalexin biosynthetic gene activation and camalexin production (Qiu et al., 2008; Mao et al., 2011). To determine whether WRKY33 functions downstream of CPK5/CPK6 in regulating camalexin biosynthesis, we transformed the Est:CPK5CA-HA construct into the wrky33-2 mutant background. As shown in Figures 3B and 3C, CPK5CA-induced camalexin production was blocked in Est:CPK5CA-HA/wrky33-2 plants, in which the Est-induced CPK5CA expression levels were comparable to that in Est:CPK5CA-HA plants. The blockage of camalexin biosynthesis in Est:CPK5CA-HA/wrky33-2 after Est treatment was associated with the loss of activation of the camalexin biosynthetic genes CYP79B2, CYP71A13, and PAD3 (Figure 3D). Consistently, a LUC reporter assay in Arabidopsis protoplasts showed that the activation of the CYP71A13 and PAD3 promoters by CPK5CA is also dependent on WRKY33 (Supplemental Figure 4), which directly regulates CYP71A13 and PAD3 expression (Qiu et al., 2008; Mao et al., 2011; Birkenbihl et al., 2017). These results indicate that WRKY33 functions downstream of CPK5/CPK6 to activate camalexin biosynthetic genes, thus inducing camalexin biosynthesis.
Figure 3.
WRKY33 Is Essential for CPK5-Mediated Induction of Camalexin Biosynthesis.
(A) Est-induced CPK5CA-HA expression in Est:CPK5CA-HA plants leads to the transcriptional activation of WRKY33. Two-week-old Est:CPK5CA-HA seedlings were treated with 10 μM Est. WRKY33 transcript levels at the indicated time points were analyzed by RT-qPCR.
(B) to (D) WRKY33 is required for CPK5CA-induced production of camalexin and the activation of camalexin biosynthetic genes. Two-week-old Est:CPK5CA-HA and Est:CPK5CA-HA/wrky33-2 seedlings were treated with 10 μM Est. Camalexin production was quantified at 24 h posttreatment (B). The expression of CPK5CA-HA was analyzed by immunoblotting (IB) with anti-HA antibody, and total protein loading was assessed by Ponceau S staining (PS; [C]). The transcript levels of CYP79B2, CYP71A13, and PAD3 at the indicated time points were analyzed by RT-qPCR (D). FW, fresh weight.
Different letters above the columns in (A) and (B) indicate significant differences (P < 0.05), as determined by one-way ANOVA. Significant differences at specific time points in (D) are indicated by asterisks (*, P < 0.05), as determined by Student’s t test. Error bars in (A), (B), and (D) indicate sd (n = 3 biological repeats), and small circles represent individual data points.
Phosphorylation of WRKY33 by CPK5/CPK6 in Vitro and in Vivo
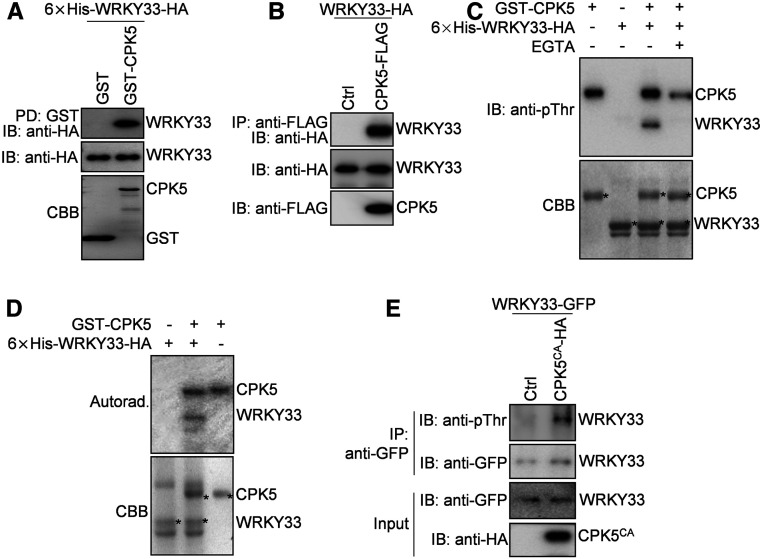
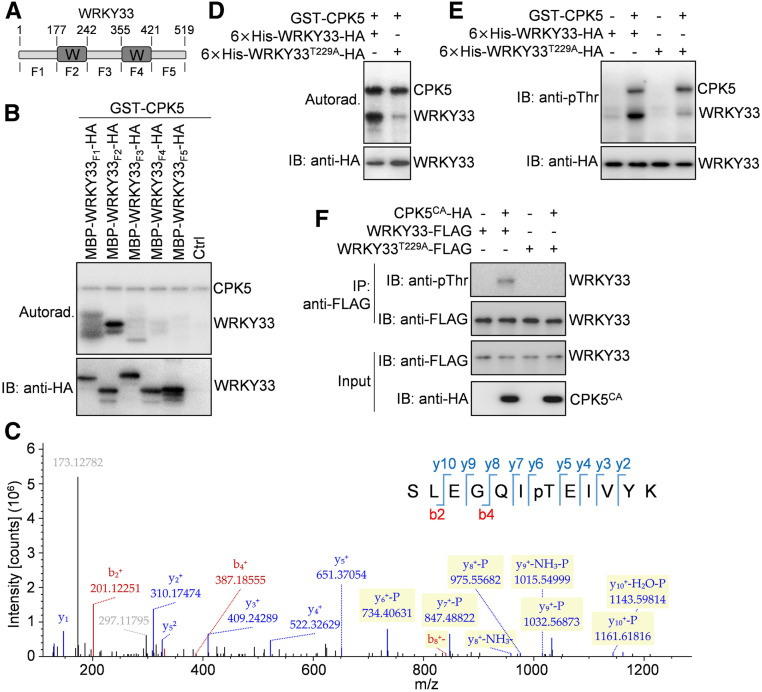
We previously demonstrated that WRKY33 binds to its own promoter, suggesting the self-activation of WRKY33 expression (Mao et al., 2011). Thus, we speculated that the positive regulation of WRKY33 expression by CPK5/CPK6 might result from CPK5/CPK6-mediated phosphorylation and activation of WRKY33, which in turn activates the expression of its own and camalexin biosynthetic genes. To test this hypothesis, we examined whether CPK5 interacts with WRKY33. As shown in an in vitro pull-down assay, the hexa-histidine (6×His)-tagged WRKY33 was pulled down by glutathione S-transferase (GST)-tagged CPK5 but not by GST itself (Figure 4A). Consistently, a coimmunoprecipitation (co-IP) assay using coexpressed FLAG-tagged CPK5 and HA-tagged WRKY33 in Arabidopsis protoplasts showed that CPK5 coimmunoprecipitated with WRKY33 (Figure 4B). These results indicate that CPK5 interacts with WRKY33 in vitro and in vivo.
Figure 4.
CPK5 Interacts with and Phosphorylates WRKY33 in Vitro and in Vivo.
(A) CPK5 interacts with WRKY33 in an in vitro pull-down assay. GST- or GST-CPK5-bound glutathione beads were incubated with 6×His-WRKY33-HA proteins, and the pulled-down (PD) proteins were analyzed by immunoblotting with anti-HA antibody (IB: anti-HA; top panel). The protein inputs were assessed by immunoblotting (middle panel) and Coomassie Brilliant Blue (CBB) staining (bottom panel).
(B) CPK5 associates with WRKY33 in Arabidopsis protoplasts. CPK5-FLAG and WRKY33-HA proteins were coexpressed in protoplasts. The proteins immunoprecipitated from protoplast extracts using anti-FLAG agarose beads (IP: anti-FLAG) were analyzed by immunoblotting with anti-HA antibody (IB: anti-HA; top panel). The protein inputs were assessed by immunoblotting (bottom two panels). Ctrl, vector control.
(C) and (D) CPK5 phosphorylates WRKY33 in vitro. The phosphorylation reactions without (C) or with (D) the addition of [γ-32P]ATP were performed using GST-CPK5 as the kinase and 6×His-WRKY33-HA as the substrate. A negative control reaction with the addition of 5 mM EGTA was included in (C). After separation by SDS-PAGE, the phosphorylated proteins were detected by immunoblotting with anti-pThr antibody (C) or by autoradiography (Autorad. [D]; top panels). The protein inputs were assessed by Coomassie Brilliant Blue staining (bottom panels), and the target protein bands are marked by asterisks.
(E) CPK5 phosphorylates WRKY33 in Arabidopsis protoplasts. CPK5CA-HA and WRKY33-GFP proteins were coexpressed in protoplasts. The WRKY33-GFP proteins immunoprecipitated from protoplast extracts using anti-GFP agarose beads (IP: anti-GFP) were analyzed by immunoblotting with anti-pThr (IB: anti-pThr) or anti-GFP (IB: anti-GFP; top two panels) antibody. The protein inputs were assessed by immunoblotting (bottom two panels).
To further explore whether CPK5 phosphorylates WRKY33, we performed in vitro phosphorylation assays using an anti-phospho-threonine (anti-pThr) or anti-phospho-serine (anti-pSer) antibody. As shown in Figure 4C, in the presence of Ca2+, GST-CPK5 strongly phosphorylated 6×His-WRKY33-HA, which was detected by anti-pThr antibody but not by anti-pSer antibody (Supplemental Figure 5). The phosphorylation of 6×His-WRKY33-HA by GST-CPK5 was blocked in the presence of EGTA (a Ca2+ chelator; Figure 4C), highlighting the specificity of CPK5-mediated phosphorylation. Another phosphorylation assay using γ-32P-labeled ATP confirmed the in vitro phosphorylation of 6×His-WRKY33-HA by GST-CPK5 (Figure 4D), which also indicates that the anti-pThr antibody can be successfully used to detect the CPK5-mediated phosphorylation of WRKY33. Therefore, we used this antibody to test whether CPK5 could phosphorylate WRKY33 in Arabidopsis protoplasts, in which HA-tagged CPK5CA was coexpressed with GFP-tagged WRKY33. As shown in Figure 4E, WRKY33-GFP was phosphorylated by the coexpressed CPK5CA-HA in Arabidopsis protoplasts. Together, these results demonstrate that CPK5 phosphorylates WRKY33 in vitro and in vivo. In addition to CPK5, CPK6 also interacts with and phosphorylates WRKY33 based on co-IP, pull-down, and phosphorylation assays (Supplemental Figure 6), which is consistent with the redundant role of CPK5 and CPK6 in regulating camalexin biosynthesis.
CDPKs are known as Ser/Thr protein kinases. There are several Ser and Thr residues in WRKY33 protein. To map the WRKY33 residue(s) phosphorylated by CPK5/CPK6, we split WRKY33 protein into five fragments, designated WRKY33F1 (amino acids 1 to 177), WRKY33F2 (amino acids 178 to 242), WRKY33F3 (amino acids 243 to 355), WRKY33F4 (amino acids 356 to 421), and WRKY33F5 (amino acids 422 to 519; Figure 5A). The two WRKY domains of WRKY33 are located in the WRKY33F2 and WRKY33F4 fragments, respectively. An in vitro phosphorylation assay showed that GST-CPK5 strongly phosphorylated the maltose binding protein (MBP)/HA double-tagged WRKY33F2, while the recombinant WRKY33F1 but not the three other WRKY33 fragments was phosphorylated very weakly by GST-CPK5 (Figure 5B), suggesting that the major WRKY33 residue(s) phosphorylated by CPK5 is located in WRKY33F2. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of a phosphorylation reaction containing GST-CPK5 and MBP-WRKY33F2-HA proteins revealed the phosphorylation of Thr-229 in WRKY33F2 by CPK5 (Figure 5C). Mutating Thr-229 to Ala (T229A) in the full-length WRKY33 largely blocked its phosphorylation by CPK5 in the in vitro phosphorylation assays using either γ-32P-labeled ATP or anti-pThr antibodies (Figures 5D and 5E), whereas the T229A mutation in WRKY33 did not affect its interaction with CPK5 in the in vitro pull-down assay (Supplemental Figure 7A). Consistently, phosphorylation of WRKY33 by CPK5CA in Arabidopsis protoplasts was lost when Thr-229 was mutated to Ala in WRRY33 (Figure 5F), while the interaction between WRKY33 and CPK5 in protoplasts was also not affected by the T229A mutation in WRKY33, as indicated by a co-IP assay (Supplemental Figure 7B). Taken together, these data indicate that the Thr-229 residue in WRKY33 is the major CPK5 phosphorylation site in vitro and in vivo. Additionally, the very weak phosphorylation of WRKY33T229A and WRKY33F1 by CPK5 observed in the in vitro phosphorylation assays (Figures 5B, 5D, and 5E) suggests that there may be other minor CPK5 phosphorylation site(s) located in WRKY33F1.
Figure 5.
CPK5 Phosphorylates the Thr-229 Residue of WRKY33.
(A) Schematic diagram showing the domain structure of WRKY33 protein. WRKY33 contains two WRKY domains (W) and three other regions of unknown function. The amino acid positions are labeled on the top. The fragments of WRKY33 used for the following experiments are indicated at the bottom.
(B) The N-terminal WRKY domain of WRKY33 is phosphorylated by CPK5. Phosphorylation of the MBP/HA-tagged WRKY33 fragments by GST-CPK5 in the presence of [γ-32P]ATP was detected by autoradiography (Autorad.; top panel). The protein inputs were assessed by immunoblotting (IB; bottom panel).
(C) The Thr-229 residue of WRKY33 is phosphorylated by CPK5. An LC-MS/MS analysis of the phosphorylation reaction containing GST-CPK5 and MBP-WRKY33F2-HA proteins revealed the phosphorylation of Thr-229 in WRKY33F2 by CPK5.
(D) and (E) The Thr-229 residue is the major CPK5 phosphorylation site in WRKY33 in vitro. Phosphorylation of the wild-type and T229A mutant forms of 6×His-WRKY33-HA proteins by GST-CPK5 in the presence (D) or absence (E) of [γ-32P]ATP was detected by autoradiography (D) or immunoblotting (E), respectively (top panels). The protein inputs were assessed by immunoblotting (bottom panels).
(F) Mutation of Thr-229 in WRKY33 blocked its phosphorylation by CPK5CA in Arabidopsis protoplasts. The wild-type and T229A mutant forms of WRKY33-FLAG proteins were coexpressed with CPK5CA-HA in protoplasts. The WRKY33-FLAG proteins immunoprecipitated from protoplast extracts using anti-FLAG agarose beads (IP: anti-FLAG) were analyzed by immunoblotting using anti-pThr (IB: anti-pThr) or anti-FLAG (IB: anti-FLAG) antibody (top two panels). The protein inputs were assessed by immunoblotting (bottom two panels).
The CPK5 Phosphorylation Site in WRKY33 Is Essential for Its Full Activity in Vivo
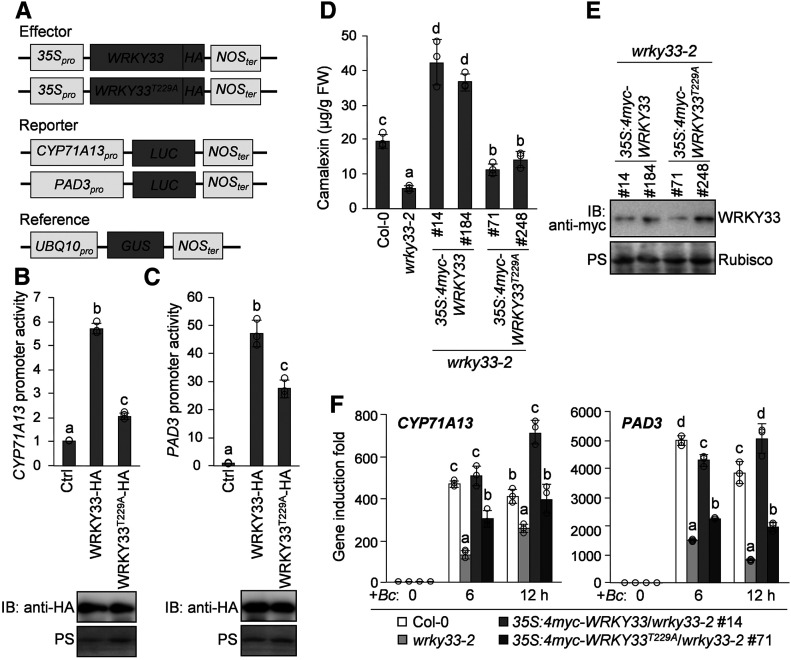
To determine the importance of WRKY33 phosphorylation by CPK5/CPK6 in vivo, we investigated the transcriptional activity of WRKY33T229A using a reporter construct containing the LUC gene driven by the promoter of CYP71A13 or PAD3, both of which are direct targets of WRKY33 (Qiu et al., 2008; Mao et al., 2011; Birkenbihl et al., 2017). We performed transactivation assays in Arabidopsis protoplasts by coexpressing the CYP71A13Pro:LUC or PAD3Pro:LUC reporter with the wild-type or T229A mutant version of WRKY33 driven by the 35S promoter (Figure 6A). As shown in Figures 6B and 6C, the protein expression levels of WRKY33 and WRKY33T229A were comparable in protoplasts, whereas the abilities of WRKY33T229A to transactivate the CYP71A13Pro:LUC and PAD3Pro:LUC reporters were significantly reduced compared with that of WRKY33. These results suggest that the Thr-229 residue is essential for the full activity of WRKY33 in transcriptionally activating the expression of camalexin biosynthetic genes.
Figure 6.
The Thr-229 Residue of WRKY33 Is Essential for Its Full Transcriptional Activity in Vivo.
(A) Schematic diagrams of the effector, reporter, and reference constructs used in the LUC reporter-aided analysis of WRKY33 activities.
(B) and (C) The WRKY33T229A mutant shows significantly reduced transcriptional activity compared with WRKY33 in Arabidopsis protoplasts. The reporter construct CYP71A13pro:LUC (B) or PAD3pro:LUC (C) was cotransfected with the effector construct 35S:WRKY33-HA or 35S:WRKY33T229-HA or the vector control (Ctrl) into protoplasts. The reference construct UBQ10pro:GUS was included in all transfections and served as an internal transfection control. The LUC activities were normalized to GUS activities, and the data are shown as relative fold increases over the vector control. Protein levels of WRKY33-HA and WRKY33T229-HA were assessed by immunoblotting (IB), and total protein loading was assessed by Ponceau S staining (PS).
(D) to (F) The WRKY33T229A mutant only partially complemented the defects of wrky33-2 mutant in the B. cinerea-induced production of camalexin and the activation of camalexin biosynthetic genes. Two-week-old Col-0, wrky33-2, 35S:4myc-WRKY33/wrky33-2, and 35S:4myc-WRKY33T229A/wrky33-2 seedlings were inoculated with B. cinerea spores. Camalexin production was measured at 24 h postinoculation (D). Protein levels of 4myc-WRKY33 and 4myc-WRKY33T229 in transgenic lines were assessed by immunoblotting, and total protein loading was assessed by Ponceau S staining (E). The transcript levels of CYP71A13 and PAD3 at the indicated time points postinoculation were analyzed by RT-qPCR (F). FW, fresh weight.
In (B), (C), (D), and (F), error bars indicate sd (n = 3 biological repeats), small circles represent individual data points, and different letters above the columns indicate significant differences (P < 0.05) among the data from a single treatment ([B] to[D]) or a single time point (F), as determined by one-way ANOVA.
Furthermore, we investigated the ability of WRKY33T229A to complement the camalexin induction defect of the wrky33-2 mutant after infection by B. cinerea. To do this, we transformed the wrky33-2 mutant with 4myc-tagged WRKY33 or WRKY33T229A driven by the 35S promoter. More than 10 independent lines of each construct were analyzed to determine the level of camalexin production after B. cinerea inoculation. As shown in Figures 6D and 6E, among the transgenic lines with similar levels of WRKY33 protein, the 35S:4myc-WRKY33T229A/wrky33-2 lines produced much lower levels of camalexin than the 35S:4myc-WRKY33/wrky33-2 lines after B. cinerea infection. Notably, B. cinerea-induced camalexin levels in the 35S:4myc-WRKY33/wrky33-2 lines were much higher than those in Col-0 plants, whereas none of the 35S:4myc-WRKY33T229A/wrky33-2 lines showed full rescue of the camalexin production defect in wrky33-2. The 35S:4myc-WRKY33/wrky33-2 lines also exhibited much stronger B. cinerea resistance than the 35S:4myc-WRKY33T229A/wrky33-2 lines (Supplemental Figure 8). These results reveal that WRKY33T229A is much less efficient in complementing the wrky33 mutant phenotype than wild-type WRKY33. Consistently, the induction of CYP71A13 and PAD3 expression after B. cinerea inoculation was much lower in 35S:4myc-WRKY33T229A/wrky33-2 lines compared with 35S:4myc-WRKY33/wrky33-2 lines (Figure 6F). Taken together, these data indicate that WRKY33T229A, a loss-of-phosphorylation mutant, lacks the full activity of WRKY33 in activating the expression of camalexin biosynthetic genes, highlighting the importance of WRKY33 phosphorylation by CPK5/CPK6 in pathogen-induced camalexin biosynthesis.
The CPK5-Mediated Phosphorylation of WRKY33 Increases Its DNA Binding Ability
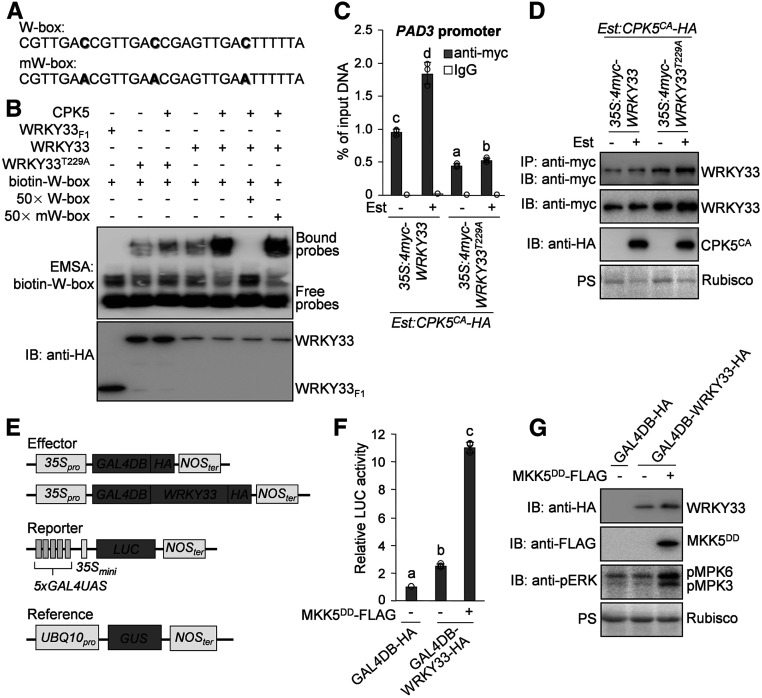
WRKY33 is a type I WRKY transcription factor containing two WRKY domains (Eulgem et al., 2000). The WRKY domain is known as a DNA binding domain, which recognizes the TTGAC(C/T) cis-element, also known as the W-box element, in its target genes (Eulgem et al., 2000). Early studies on type I WRKYs including sweet potato (Ipomoea batatas) SPF1, parsley (Petroselinum crispum) WRKY1, and Arabidopsis WRKY1 suggested that the DNA binding activity of these three type I WRKYs is primarily mediated by their C-terminal WRKY domains, while their N-terminal WRKY domains contribute little to DNA binding (Ishiguro and Nakamura, 1994; de Pater et al., 1996; Eulgem et al., 1999). However, recent structural studies showed that both the C- and N-terminal WRKY domains of Arabidopsis WRKY33 are capable of binding to the W-box element (Brand et al., 2013; Xu et al., 2020). We therefore tested the DNA binding ability of the two WRKY domains of WRKY33 by performing an electrophoresis mobility shift assay (EMSA). As shown in Supplemental Figure 9, consistent with the results of structural studies (Brand et al., 2013; Xu et al., 2020), both the C- and N-terminal WRKY domains of WRKY33 showed W-box binding activity. Notably, the WRKY33 N-terminal WRKY domain displayed even higher DNA binding affinity than its C-terminal WRKY domain (Supplemental Figure 9).
The CPK5 phosphorylation site (Thr-229) is located in the N-terminal WRKY domain of WRKY33 (Supplemental Figure 10). Therefore, we speculated that the phosphorylation of WRKY33 on Thr-229 by CPK5 regulates the DNA binding activity of WRKY33. We therefore performed an EMSA to test this hypothesis. To determine whether CPK5-mediated phosphorylation of WRKY33 alters its DNA binding activity in the EMSA, we phosphorylated WRKY33 using Ca2+-activated CPK5, and a control reaction without the addition of CPK5 was used as unphosphorylated WRKY33. As shown in Figure 7B, the EMSA revealed that the W-box binding activity of the phosphorylated WRKY33 is much higher than that of the unphosphorylated WRKY33, indicating that the CPK5-mediated phosphorylation of WRKY33 increases its DNA binding ability. Consistently, the T229A mutation blocked the enhancement of the W-box binding activity of WRKY33 by CPK5-mediated phosphorylation in the EMSA (Figure 7B).
Figure 7.
Differential Regulation of WRKY33 Activity by CPK5 and MPK3/MPK6-Mediated Phosphorylation.
(A) Oligonucleotide sequences of the W-box probe and its mutated form (mW-box). The mutated nucleotides in the mW-box probe are highlighted in boldface.
(B) Phosphorylation of WRKY33 by CPK5 enhances the W-box binding activity of WRKY33 in vitro. Freshly prepared 6×His-WRKY33-HA and 6×His-WRKY33T229A-HA proteins were subjected to phosphorylation by GST-CPK5, and control reactions without GST-CPK5 were processed side-by-side. The DNA binding activities of recombinant WRKY33 and WRKY33T229A proteins in the above reactions were determined by EMSA using the biotin-labeled W-box probes (top panel). The truncated 6×His-WRKY33F1-HA protein was used in the EMSA as a negative control. The specificity of W-box binding activity was demonstrated by the competition assays using a 50-fold excess of unlabeled W-box or mW-box probes. The protein inputs were assessed by immunoblotting (IB; bottom panel).
(C) and (D) CPK5-mediated phosphorylation of WRKY33 enhances its DNA binding ability in vivo. ChIP-qPCR analysis was performed using 35S:4myc-WRKY33/Est:CPK5CA-HA and 35S:4myc-WRKY33T229A/Est:CPK5CA-HA transgenic plants. Input chromatin was isolated from 2-week-old seedlings treated with 10 μM Est or the solvent control for 24 h. The WRKY33-chromatin complex was immunoprecipitated using anti-myc antibody and protein G-agarose. A control reaction was processed side-by-side using mouse IgG. ChIP- and input-DNA samples were quantified by qPCR using primers specific to the W-box-containing region in the PAD3 promoter. The ChIP results are presented as percentage of input DNA (C). The input and immunoprecipitated proteins in the ChIP assay were assessed by immunoblotting, and total protein loading was assessed by Ponceau S staining (PS [D]).
(E) Schematic diagrams of the effector, reporter, and reference constructs used in the transactivation assay. The reporter construct contains the LUC reporter gene driven by five repeats of GAL4UAS and the 35S minimal promoter (35Smini). In the effector constructs, GAL4DB-HA alone or fused with WRKY33 was expressed under the control of the 35S promoter.
(F) and (G) MPK3/MPK6-mediated phosphorylation of WRKY33 enhances its transactivation activity. Arabidopsis protoplasts were cotransfected with the reporter construct and one of the effector constructs as well as the 35S:MKK5DD-FLAG construct or vector control. The reference construct UBQ10pro:GUS was included in all transfections and served as an internal transfection control. The LUC activities were normalized to the GUS activities, and the data are shown as relative fold increases over the background from the transfection with the 35S:GAL4DB-HA construct (F). The expression of GAL4DB-WRKY33-HA and MKK5DD-FLAG and the activation of MPK3/MPK6 by MKK5DD were analyzed by immunoblotting using anti-HA, anti-FLAG, and anti-phospho-ERK1/2 (anti-pERK) antibodies, respectively, and total protein loading was assessed by Ponceau S staining (G).
In (C) and (F), error bars indicate sd (n = 3 biological repeats), small circles represent individual data points, and different letters above the columns indicate significant differences (P < 0.05), as determined by one-way ANOVA.
Furthermore, we transformed 4myc-tagged WRKY33 or WRKY33T229A driven by the 35S promoter into Est:CPK5CA-HA plants and subjected these transgenic plants to chromatin immunoprecipitation (ChIP)-qPCR analysis. As shown in Figure 7C, Est-induced expression of CPK5CA enhanced the ability of WRKY33 to bind to the PAD3 promoter in 35S:4myc-WRKY33/Est:CPK5CA-HA plants, whereas the WRKY33T229A mutant was compromised not only in the basal PAD3 promoter binding activity but also in the CPK5CA-enhanced PAD3 promoter binding ability in 35S:4myc-WRKY33T229A/Est:CPK5CA-HA plants. Therefore, both in vitro EMSA and in vivo ChIP-qPCR analyses demonstrated that phosphorylation of WRKY33 by CPK5 enhances the DNA binding activity of WRKY33. Additionally, Est-induced expression of CPK5CA did not alter the WRKY33 protein levels in either 35S:4myc-WRKY33/Est:CPK5CA-HA or 35S:4myc-WRKY33T229A/Est:CPK5CA-HA plants (Figure 7D), suggesting that the CPK5-mediated phosphorylation does not regulate the WRKY33 protein stability.
The MPK3/MPK6-Mediated Phosphorylation of WRKY33 Enhances Its Transactivation Activity
We previously reported that WRKY33 is also phosphorylated by the MAPKs MPK3 and MPK6, which also play an essential role in pathogen-induced camalexin production (Ren et al., 2008; Mao et al., 2011). Unlike CPK5/CPK6, MPK3/MPK6 phosphorylate WRKY33 on its N-terminal Ser residues followed by Pro (Supplemental Figure 10; Mao et al., 2011). We demonstrated that the phosphorylation of WRKY33 by MPK3/MPK6 does not alter the DNA binding activity of WRKY33 (Mao et al., 2011), but how MPK3/MPK6 regulate WRKY33 activity remains unknown. To investigate whether MPK3/MPK6-mediated phosphorylation regulates the protein stability of WRKY33, we transformed 4myc-tagged WRKY33 under the control of the 35S promoter into the conditional gain-of-function Est:MKK5DD-HA transgenic Arabidopsis plants that were generated previously (He et al., 2019). As shown in Supplemental Figure 11, after treatment with Est, expression of the constitutively active MKK5DD activated the downstream MPK3 and MPK6 in the 35S:4myc-WRKY33/Est:MKK5DD-HA transgenic lines, but the 4myc-WRKY33 protein levels were not altered after MPK3/MPK6 activation in these lines, indicating that MPK3/MPK6-mediated phosphorylation does not affect WRKY33 protein stability in vivo.
To explore whether the phosphorylation of WRKY33 by MPK3/MPK6 regulates the transactivation activity of WRKY33, we generated a 35S promoter-driven effector construct expressing WRKY33 fused with the DNA binding domain of the yeast transcription factor GAL4 (GAL4DB-WRKY33-HA) and a reporter construct containing the LUC gene driven by the GAL4-specific upstream activation sequence (GAL4UAS) and the 35S minimal promoter (Figure 7E). We performed transactivation assays by cotransforming the effector and reporter constructs with or without the 35S:MKK5DD-FLAG construct into Arabidopsis protoplasts. As shown in Figures 7F and 7G, the expression of GAL4DB-WRKY33-HA in protoplasts resulted in a significant increase in LUC activity from the reporter construct, confirming that WRKY33 functions as a transcriptional activator. In response to MPK3/MPK6 activation by MKK5DD-FLAG, the transactivation activity of GAL4DB-WRKY33-HA, as indicated by LUC activity, was dramatically enhanced in protoplasts, whereas GAL4DB-WRKY33-HA protein levels were unchanged (Figures 7F and 7G). These results indicate that MPK3/MPK6-mediated phosphorylation enhances the transactivation activity of WRKY33 but does not affect its protein stability. We also performed transactivation assays in Arabidopsis protoplasts to test whether the CPK5-mediated phosphorylation of WRKY33 affects its transactivation activity. As shown in Supplemental Figure 12, upon coexpression of CPK5CA-FLAG, the transactivation activity of GAL4DB-WRKY33-HA was not altered in protoplasts, indicating that the CPK5-mediated phosphorylation of WRKY33 does not regulate its transactivation activity.
Taken together, these results suggest that WRKY33 functions as a convergent substrate of CPK5/CPK6 and MPK3/MPK6, which cooperatively regulate WRKY33 activity through differential phosphorylation. Specifically, phosphorylation of WRKY33 on the Thr-229 residue by CPK5 increases the DNA binding ability of WRKY33, while the MPK3/MPK6-mediated phosphorylation of WRKY33 on its N-terminal Ser residues enhances the transactivation activity of WRKY33.
CPK5/CPK6 and MPK3/MPK6 Cooperatively Regulate Camalexin Biosynthesis and B. cinerea Resistance
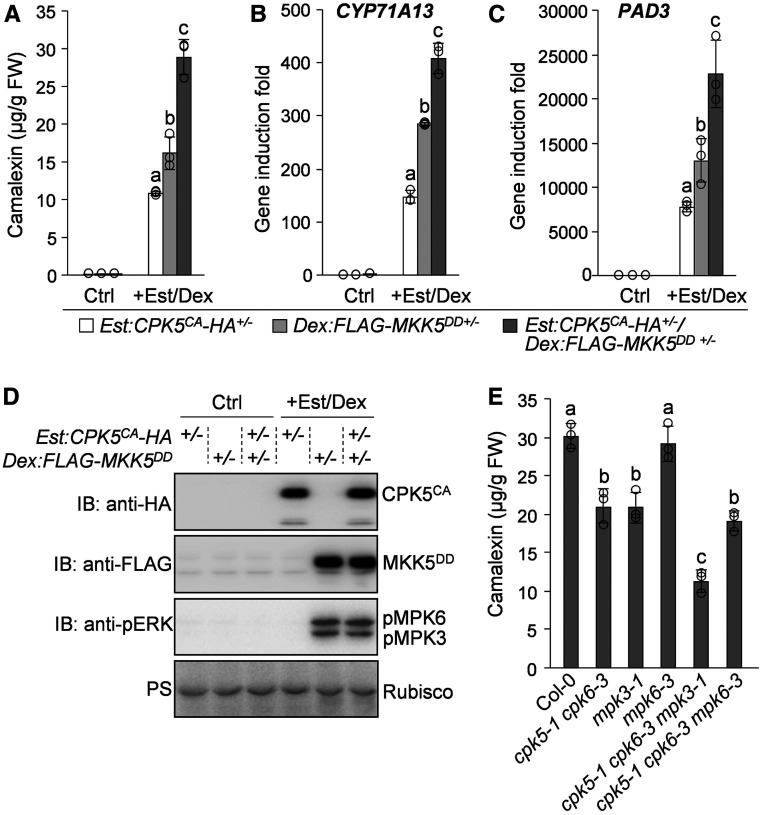
The finding that CPK5/CPK6 and MPK3/MPK6 differentially phospho-regulate WRKY33 activity prompted us to investigate whether these two types of kinases function cooperatively to regulate camalexin biosynthesis and disease resistance. First, we generated the Dex:FLAG-MKK5DD transgenic Arabidopsis plants expressing FLAG-tagged constitutively active MKK5DD under the control of a dexamethasone (Dex)-inducible promoter. We then crossed Est:CPK5CA-HA, Dex:FLAG-MKK5DD, and Col-0 plants with each other to generate the F1 seeds of Est:CPK5CA-HA+/−, Dex:FLAG-MKK5DD+/−, and Est:CPK5CA-HA+/−/Dex:FLAG-MKK5DD+/−. Following treatment with Est and Dex to induce the expression of CPK5CA and/or MKK5DD, we compared the levels of camalexin production in these F1 plants. As shown in Figures 8A and 8D, similar to CPK5CA-induced camalexin biosynthesis in Est:CPK5CA-HA+/− plants, MPK3/MPK6 activation by MKK5DD in Dex:FLAG-MKK5DD+/− plants after Dex treatment also highly induced camalexin production, which is consistent with previous reports (Ren et al., 2008; Mao et al., 2011). Notably, coexpression of CPK5CA and MKK5DD in Est:CPK5CA-HA+/−/Dex:FLAG-MKK5DD+/− plants had an additive effect on the induction of camalexin biosynthesis (Figures 8A and 8D), which was associated with the higher induction of CYP71A13 and PAD3 expression in this plant compared with Est:CPK5CA-HA+/− and Dex:FLAG-MKK5DD+/− plants (Figures 8B and 8C). These results indicate that the cooperative action of CPK5 and MPK3/MPK6 positively regulates camalexin biosynthetic gene expression and camalexin biosynthesis.
Figure 8.
Cooperative Regulation of Camalexin Biosynthesis by CPK5/CPK6 and MPK3/MPK6.
(A) to (D) Coexpression of CPK5CA and MKK5DD has additive effects on the induction of camalexin biosynthesis and the activation of camalexin biosynthetic genes. Col-0 and homozygous lines of Est:CPK5CA-HA and Dex:FLAG-MKK5DD were pairwise crossed to generate Est:CPK5CA-HA+/−, Dex:FLAG-MKK5DD+/−, and Est:CPK5CA-HA+/−/Dex:FLAG-MKK5DD+/− F1 plants. Camalexin produced from 2-week-old seedlings was measured after treatment with 10 μM Est for 24 h and 5 μM Dex for 16 h (A). The transcript levels of CYP71A13 (B) and PAD3 (C) after treatment with Est and Dex were analyzed by RT-qPCR. The expression of CPK5CA-HA and FLAG-MKK5DD and the activation of MPK3/MPK6 by FLAG-MKK5DD were analyzed by immunoblotting (IB), and total protein loading was assessed by Ponceau S staining (PS [D]). FW, fresh weight.
(E) CPK5/CPK6 and MPK3 contribute additively to B. cinerea-induced camalexin production. Camalexin produced from 2-week-old Col-0 and mutant seedlings indicated was measured at 24 h postinoculation with B. cinerea spores.
In (A), (B), (C), and (E), error bars indicate sd (n = 3 biological repeats), small circles represent individual data points, and different letters above the columns indicate significant differences (P < 0.05), as determined by one-way ANOVA.
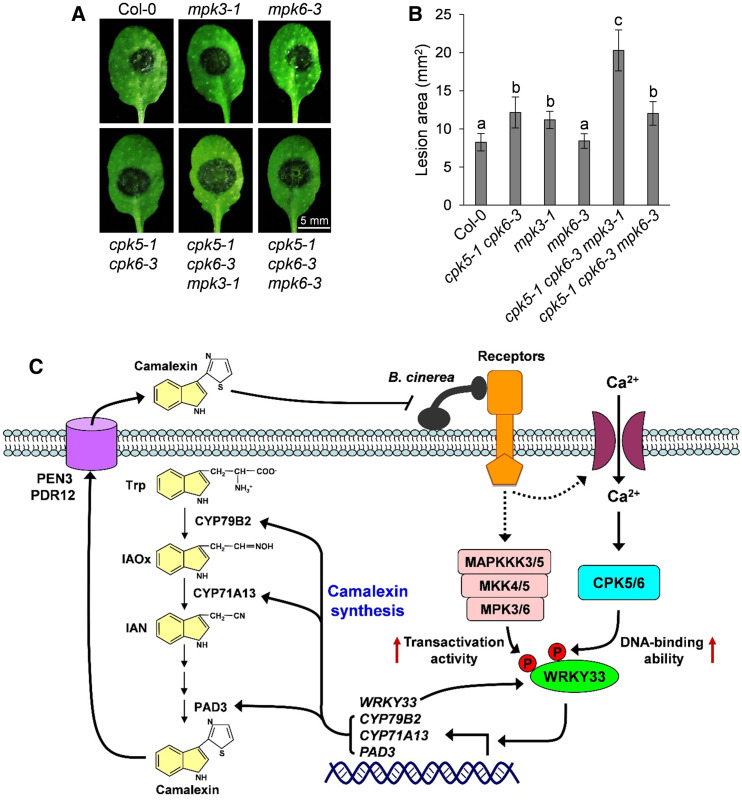
Furthermore, to provide loss-of-function evidence supporting the cooperative regulation of camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6, we generated the cpk5 cpk6 mpk3 and cpk5 cpk6 mpk6 triple mutants and compared the camalexin production levels in these triple mutants with those in the parental single and double mutants following B. cinerea inoculation (Figure 8E). Again, compromised camalexin production was observed in cpk5 cpk6 (Figure 8E). The B. cinerea-induced camalexin production was also compromised in mpk3, but not in mpk6, and was further reduced in cpk5 cpk6 mpk3, but not in cpk5 cpk6 mpk6, compared with cpk5 cpk6 (Figure 8E). These results indicate that CPK5/CPK6 function cooperatively with MPK3 to positively regulate pathogen-induced camalexin biosynthesis. Based on our previous report (Ren et al., 2008), the absence of a defect in the induction of camalexin biosynthesis when MPK6 was mutated in different backgrounds is likely due to the redundancy of MPK3/MPK6 and the relatively minor contribution of MPK6 in regulating camalexin biosynthesis.
Consistent with the finding that CPK5/CPK6 and MPK3 cooperatively regulate camalexin biosynthesis, we also observed an additive contribution of these kinase genes to B. cinerea resistance in Arabidopsis. As shown in Figures 9A and 9B, compared with Col-0 plants, the cpk5 cpk6 and mpk3 mutants displayed increased susceptibility to B. cinerea, while the cpk5 cpk6 mpk3 triple mutant showed even stronger susceptibility. The increased B. cinerea susceptibility in these mutants was closely correlated with their reduced camalexin production, highlighting the importance of camalexin production for Arabidopsis resistance to B. cinerea. Therefore, both gain- and loss-function evidence indicates that CPK5/CPK6 function cooperatively with MPK3/MPK6 to positively regulate camalexin biosynthesis and B. cinerea resistance, which is consistent with the observation that these two types of kinases cooperatively regulate WRKY33 activity via the differential phosphorylation of this transcription factor.
Figure 9.
CPK5/CPK6 and MPK3/MPK6 Cooperatively Regulate Arabidopsis Resistance to B. cinerea.
(A) and (B) Additive contribution of CPK5/CPK6 and MPK3/MPK6 to B. cinerea resistance. Fully developed leaves of 4-week-old Col-0 and mutant plants indicated were inoculated with droplets of B. cinerea spore suspension. Leaf images were taken at 48 h postinoculation (A). Meanwhile, lesion size was quantified (B). Error bars indicate sd (n = 20 leaves from different plants of each genotype), and different letters above the columns in (B) indicate significant differences (P < 0.05), as determined by one-way ANOVA. The experiments were independently repeated three times with similar results.
(C) A model depicting the cooperative regulation of camalexin biosynthesis and B. cinerea resistance by CPK5/CPK6 and MPK3/MPK6. B. cinerea infection triggers the activation of CPK5/CPK6 and MPK3/MPK6, which in turn phosphorylate their convergent substrate WRKY33. CPK5/CPK6- and MPK3/MPK6-mediated phosphorylation of WRKY33 enhances its DNA binding and transactivation activities, respectively. WRKY33 autoactivates its own transcription to amplify the immune signal. The activated WRKY33 further upregulates the expression of camalexin biosynthetic genes including CYP79B2, CYP71A13, and PAD3 to drive the synthesis of camalexin, which is then exported to the extracellular space by PEN3 and PDR12 transporters to defend against B. cinerea.
DISCUSSION
Pathogen-triggered phytoalexin production is an integral part of plant defense responses that plays a critical role in disease resistance (Piasecka et al., 2015). The biosynthetic pathways of several phytoalexins have been elucidated (Sønderby et al., 2010; Geu-Flores et al., 2011; Piasecka et al., 2015), but the signaling pathways regulating their biosynthesis are mostly unknown. We previously characterized the regulation of camalexin synthesis by MPK3/MPK6 and their downstream transcription factor WRKY33 (Ren et al., 2008; Mao et al., 2011). In this study, we demonstrated that CPK5/CPK6 function cooperatively with MPK3/MPK6 to regulate camalexin biosynthesis based on both gain- and loss-of-function genetic analyses (Figures 1, 2, and 8). We also identified that WRKY33 acts downstream of CPK5/CPK6 to activate camalexin synthetic genes, thereby inducing camalexin biosynthesis (Figure 3; Supplemental Figure 4). In vitro and in vivo phosphorylation assays demonstrated that WRKY33 is a substrate of CPK5/CPK6 (Figure 4; Supplemental Figure 6), which phosphorylate the Thr-229 residue of WRKY33 and thus increase its DNA binding ability (Figures 5 and 7A to 7D). By contrast, the MPK3/MPK6-mediated phosphorylation of WRKY33 on its N-terminal Ser residues enhances the transactivation activity of WRKY33 (Figures 7E to 7G). Taken together, we conclude that WRKY33 functions as a convergent substrate of CPK5/CPK6 and MPK3/MPK6, which cooperatively regulate camalexin biosynthesis through the differential phospho-regulation of WRKY33 activity (Figure 9C).
Dual-Level Regulation of WRKY33 by CPK5/CPK6
In addition to the phospho-regulation of WRKY33 protein by CPK5/CPK6, the activation of CPK5 in Est:CPK5CA-HA plants after Est treatment also highly induced the transcription of WRKY33 (Figure 3A), suggesting a dual-level regulation of WRKY33 by CPK5/CPK6. Our previous ChIP-qPCR assay revealed the direct binding of WRKY33 to the W-boxes in its own promoter (Mao et al., 2011), indicating that WRKY33 can autoactivate its own expression. In this study, we demonstrated that WRKY33 acts as a substrate of CPK5/CPK6 (Figure 4; Supplemental Figure 6), which phosphorylate WRKY33 to enhance its W-box binding activity (Figures 5 and 7A to 7D). Therefore, in Est:CPK5CA-HA plants after Est treatment, the CPK5CA-mediated phosphorylation of the basal-level WRKY33 protein is likely involved in turning on WRKY33 expression, followed by the amplification of WRKY33 expression via a positive feedback regulation loop mediated by CPK5 and WRKY33, ultimately leading to high-level WRKY33 induction (Figure 9C).
WRKY33 Is a Key Regulator of Arabidopsis Immunity
WRKY transcription factors are involved in regulating plant responses to pathogen infection (Pandey and Somssich, 2009). Accumulating evidence indicates that WRKY33 is a key regulator of Arabidopsis immunity. This study, together with our previous work (Mao et al., 2011), demonstrates that WRKY33 plays an essential role in pathogen-induced camalexin production via the direct activation of camalexin biosynthetic genes. WRKY33 also positively regulates PAMP/pathogen-triggered ROS production and ethylene biosynthesis through transcriptionally activating the expression of RBOHD and the 1-aminocyclopropane-1-carboxylic acid synthase genes ACS2 and ACS6, respectively (Li et al., 2012; Zhao et al., 2020). Moreover, ChIP sequencing analysis indicated that WRKY33 directly upregulates the transcription of PROPEP2 and PROPEP3, encoding plant elicitor peptides, thereby amplifying immune responses (Birkenbihl et al., 2017). WRKY33 is also involved in the establishment of systemic acquired resistance via the direct transcriptional activation of AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) to induce the production of pipecolic acid, a mobile metabolite that induces systemic acquired resistance (Wang et al., 2018). Therefore, within the large WRKY transcription factor family in Arabidopsis (Eulgem et al., 2000), WRKY33 is likely a master regulator of immune responses, whose homologs in crop plants could be promising candidates for engineering disease resistance.
Multitasking of CPK5/CPK6 in Arabidopsis Immunity
The perception of pathogens triggers an influx of Ca2+ that activates CDPKs to transduce and propagate immune signals for the establishment of disease resistance (Boudsocq and Sheen, 2013; Romeis and Herde, 2014; Seybold et al., 2014). In this study, we found that the pathogen-responsive CPK5 and CPK6 phosphorylate WRKY33 and thus enhance its DNA binding ability to induce camalexin biosynthesis. CPK5/CPK6, together with CPK4 and CPK11, also phosphorylate another subgroup of WRKY transcription factors, including WRKY8, WRKY28, and WRKY48, to regulate nucleotide binding domain Leucine-rich repeat protein-mediated transcriptional reprogramming (Gao et al., 2013). CPK5/CPK6 also positively regulate pathogen-induced ethylene production (Gravino et al., 2015). Since WRKY33 directly activates ACS2/ACS6 expression to induce ethylene biosynthesis (Li et al., 2012), it is tempting to speculate that CPK5/CPK6 regulate ethylene induction via the phosphorylation of WRKY33. In addition to WRKYs, CPK5 also phosphorylates RBOHD to increase its enzymatic activity for ROS production (Dubiella et al., 2013). CPK5/CPK6 have also been implicated in regulating hypersensitive cell death and PAMP-triggered transcriptional reprogramming (Boudsocq et al., 2010; Dubiella et al., 2013; Liu et al., 2017). Further identification of the related substrates of CPK5/CPK6 will reveal the molecular mechanisms underlying the CPK5/CPK6-mediated regulation of these defense responses.
Diverse Roles of MPK3/MPK6 in Arabidopsis Immunity
Similar to CPK5/CPK6, MPK3/MPK6 are also involved in regulating a range of immune responses. The multifunctionality of MPK3/MPK6 can be largely attributed to their ability to phosphorylate distinct substrates. In this study and our previous work (Mao et al., 2011), we revealed that MPK3/MPK6 phosphorylate WRKY33 to enhance its transactivation activity and further induce camalexin biosynthesis. The MPK3/MPK6-mediated phosphorylation of WRKY33 is also involved in upregulating the expression of ACS2/ACS6 and ALD1 to induce the production of ethylene and pipecolic acid, respectively (Li et al., 2012; Wang et al., 2018). MPK3/MPK6 also directly phosphorylate ACS2/ACS6 to stabilize these ACS proteins and induce ethylene production (Liu and Zhang, 2004; Han et al., 2010). Besides WRKY33, several other pathogen-responsive WRKYs have also been identified as potential MPK3/MPK6 substrates in protein microarray- and in vitro phosphorylation-based screenings (Popescu et al., 2009; Sheikh et al., 2016). In addition to WRKYs, MPK3/MPK6 also phosphorylate the transcription factors ERF6 and ERF104, leading to the stabilization of ERF6 and the release of ERF104, respectively, to activate defensin gene expression (Bethke et al., 2009; Meng et al., 2013). High-throughput genomic and phosphoproteomic approaches have enabled the proteome-wide identification of potential MPK3/MPK6 substrates, especially WRKYs (Popescu et al., 2009; Hoehenwarter et al., 2013; Sheikh et al., 2016; Rayapuram et al., 2018). Further plant-based analyses will reveal details of their involvement in immune signaling in Arabidopsis.
Crosstalk between CPK5/CPK6 and MPK3/MPK6 in Arabidopsis Immunity
Transcriptome profiling analysis revealed that CPK5/CPK6 and MPK3/MPK6 synergistically regulate a subset of defense response genes (Boudsocq et al., 2010), implying coregulation of common downstream factors by these two types of kinases. In this study, we demonstrated that CPK5/CPK6 and MPK3/MPK6 differentially phosphorylate and thus cooperatively activate their common substrate WRKY33 to induce camalexin production, uncovering a molecular mechanism of crosstalk between CPK5/CPK6 and MPK3/MPK6 in regulating Arabidopsis immunity. Therefore, we further propose that CPK5/CPK6 and MPK3/MPK6 likely also cooperatively regulate other WRKY33-associated defense responses, including the pathogen-induced expression of PROPEPs and the production of ethylene, ROS, and pipecolic acid (Li et al., 2012; Birkenbihl et al., 2017; Wang et al., 2018; Zhao et al., 2020), via cophosphorylation of WRKY33. In addition, the WRKY8, WRKY28, and WRKY48 transcription factors, which have been identified as the substrates of CPK5/CPK6 (Gao et al., 2013), are also phosphorylated by MPK3/MPK6, as revealed by protein microarray and/or in vitro phosphorylation assays (Popescu et al., 2009; Sheikh et al., 2016). Further study is needed to identify whether these WRKYs serve as the convergent substrates to mediate the cooperative regulation of pathogen/PAMP-induced transcriptional reprogramming by CPK5/CPK6 and MPK3/MPK6.
METHODS
Plant Materials, Growth Conditions, and Treatments
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild-type control. All mutants and transgenic plants used in this study are in the Col-0 background. The knockout mutants cpk4-1 (SALK_081860; Zhu et al., 2007), cpk5-1 (SAIL_657C06; Boudsocq et al., 2010), cpk6-3 (SALK_025460; Boudsocq et al., 2010), cpk11-2 (SALK_054495; Zhu et al., 2007; Boudsocq et al., 2010), mpk3-1 (SALK_151594; Ren et al., 2008), mpk6-3 (SALK_127507; Liu and Zhang, 2004), pad3-1 (Glazebrook and Ausubel, 1994), and wrky33-2 (GABI_324B11; Mao et al., 2011) were described previously. The double, triple, and quadruple mutants were generated by genetic crossing, and homozygous lines were used for the experiments.
Arabidopsis seedlings were grown on half-strength Murashige and Skoog plates for 7 d at 22°C in a growth chamber with a 12-h-fluorescent light (60 μE m−2 s−1)/12-h-dark cycle, then transferred to 6 mL of liquid half-strength Murashige and Skoog medium in 20-mL gas chromatography vials (10 seedlings per vial), and cultured for another 7 d under continuous fluorescent light (60 μE m−2 s−1) to avoid the effect of the light/dark cycle on metabolite biosynthesis and gene expression. Thereafter, the seedlings in gas chromatography vials were inoculated with Botrytis cinerea (strain T4) spores (8 × 104 spores/mL) or treated with 10 μM Est, 5 μM Dex, or the solvent control, and samples of liquid medium and seedlings were collected at the indicated time points after treatment to analyze metabolite production and gene expression.
Plasmid Construction and Generation of Transgenic Plants
The protein-coding regions of CPK5, CPK6, MKK5, and WRKY33 and the CPK5CA, CPK6CA, and WRKY33F1-WRKY33F5 gene fragments were PCR amplified from Arabidopsis Col-0 cDNA and cloned into plant expression vectors, protoplast transfection vectors, or Escherichia coli expression vectors to generate the recombinant constructs used in this study (Supplemental Table 1). The T229A and T215D/S221D (DD) mutations were introduced into WRKY33 and MKK5, respectively, by site-directed mutagenesis. The effector, reporter, and reference vectors used for the LUC reporter-aided analysis of promoter activity or transactivation activity in Arabidopsis protoplasts were described previously (Li et al., 2015). For the promoter activity assay, the promoters of CYP79B2, CYP71A13, and PAD3 were PCR amplified from Col-0 genomic DNA and used to substitute the 35S promoter in the pHBT-LUC vector (Gao et al., 2013) to generate the CYP79B2pro:LUC, CYP71A13pro:LUC, and PAD3pro:LUC reporter constructs (Supplemental Table 1). For the transactivation assay, WRKY33 was subcloned into the effector vector and fused to GAL4DB (Supplemental Table 1). The primers used to generate the constructs are listed in Supplemental Table 2.
Transgenic Arabidopsis plants were generated by Agrobacterium tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998). For all transgenic plants, 30 to 50 T1 plants per construct were screened for transgene expression using immunoblotting, and T3 homozygous lines with a single transgene insertion were used for phenotypic characterization or genetic crossing. The transgenic Arabidopsis plants used in this study are listed in Supplemental Tables 3 and 4.
Metabolic Analysis
Samples of liquid medium were collected at the indicated time points after different treatments for metabolic analyses. The liquid medium samples were diluted with methanol and subjected to HPLC analyses on an LC-6AD system with an RF-10AXL fluorescence detector (Shimadzu). Samples were analyzed using an Inertsil ODS-SP column (250 × 4.6 mm, 5 µm, GL Sciences) with 5% (v/v) methanol as solvent A and 100% methanol as solvent B at a flow rate of 0.7 mL/min (gradient of solvent B: 60% for 2 min, followed by a 14-min linear gradient to 100%, then ended after 4 min at 100%). The camalexin peak was identified by referring to the standard (Sigma-Aldrich). The camalexin concentrations were calculated based on the comparison of peak areas (excitation at 310 nm and emission at 390 nm) in samples with those of known amounts of the standard.
Gene Expression Analysis
Total RNA was extracted from seedling samples using TRIzol reagent (Invitrogen). After DNase treatment, 1 μg of total RNA was used for RT. Expression of CYP79B2, CYP71A13, PAD3, and WRKY33 at different time points after inoculation with B. cinerea or treatment with Est or Dex was quantified by qPCR using a real-time qPCR machine (CFX Connect, Bio-Rad) as previously described (He et al., 2019). The transcript of elongation factor 1α (EF1α) was used as a reference. The levels of CYP79B2, CYP71A13, PAD3, and WRKY33 expression were calculated as fold induction relative to the basal levels before treatment. The primer pairs used for qPCR are listed in Supplemental Table 2.
B. cinerea Resistance Assay
Arabidopsis seeds were sown in soil and grown under a 12-h-light/12-h-dark cycle in a growth chamber at 22°C and 65% humidity for 4 weeks. To quantify disease resistance, mature rosette leaves were detached and inoculated with 10-μL drops of B. cinerea spore suspension (2 × 105 spores/mL). The inoculated leaves were kept in Petri dishes on wet filter paper. The lesion size was measured 48 h after inoculation with B. cinerea.
Immunoprecipitation and co-IP Assays
Arabidopsis protoplasts were transfected with the indicated constructs and cultured for 12 h. Total proteins were isolated with 0.5 mL of extraction buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, 0.5% [v/v] Triton X-100, and 1× protease inhibitor cocktail from Roche). After vortexing vigorously for 30 s, the samples were centrifuged at 16,000g for 10 min at 4°C, and the supernatant was incubated with anti-FLAG (Sigma-Aldrich, catalog no. A2220) or anti-GFP (ChromoTek, catalog no. gta-20) agarose beads for 2 h at 4°C with gentle shaking. The beads were collected and washed three times with washing buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, and 0.1% [v/v] Triton X-100) and once more with 50 mM Tris-HCl, pH 7.5. The immunoprecipitated proteins were released from the beads by boiling in SDS-PAGE sample loading buffer and analyzed by immunoblotting with anti-HA (Roche, catalog no. 12013819001) or anti-pThr (Cell Signaling Technology, catalog no. 9381) antibody.
In Vitro Pull-Down and Phosphorylation Assays
The 6×His-, GST-, and MBP-fused recombinant proteins were expressed in E. coli strain BL21(DE3) and purified using Ni-NTA agarose (Qiagen), Pierce glutathione agarose (Thermo Fisher Scientific), and amylose resin (New England Biolabs), respectively, according to each manufacturer’s instructions. Approximately 10 μg of GST or GST-fused proteins was incubated with 5 μL of prewashed glutathione agarose beads in 1 mL of pull-down buffer (20 mM Tris-HCl, pH 7.5, 1 mM β-mercaptoethanol, 3 mM EDTA, 150 mM NaCl, and 1% [v/v] Nonidet P-40) for 30 min at 4°C with gentle shaking. The beads were harvested by centrifugation at 1000g for 1 min and inoculated with 10 μg of 6×His-fused proteins in 1 mL of pull-down buffer for 1 h at 4°C with gentle shaking. Finally, the beads were harvested and washed three times with 1 mL of pull-down buffer and once more with 1 mL of 50 mM Tris-HCl, pH 7.5. The pulled-down proteins were released from beads by boiling in SDS-PAGE sample loading buffer and analyzed by immunoblotting with anti-HA antibody (Roche, catalog no. 12013819001).
The in vitro phosphorylation reactions were performed at 25°C for 2 h in 30 μL of kinase buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.1 mM CaCl2, and 1 mM DTT) containing 10 μg of substrate proteins, 1 μg of kinases, and 0.1 mM ATP with or without 5 μCi of [γ-32P]ATP. Negative control reactions with the addition of 5 mM EGTA were included to confirm the specificity of CPK5/CPK6-mediated phosphorylation. The reactions were stopped by adding 4× SDS loading buffer, and the phosphorylation of recombinant proteins was analyzed by autoradiography or by immunoblotting with anti-pThr (Cell Signaling Technology, catalog no. 9381) or anti-pSer (Santa Cruz Biotechnology, catalog no. sc-81514) antibody after protein separation by SDS-PAGE.
LC-MS/MS Analysis
The LC-MS/MS analysis was performed as reported previously (Li et al., 2014). Briefly, the phosphorylation reaction using GST-CPK5 as the kinase and MBP-WRKY33F2-HA as the substrate was performed at 25°C for 2 h. The proteins in the reaction were then digested using trypsin (1:50 trypsin-to-protein ratio, w/w) at 37°C overnight. Phosphopeptides were enriched for LC-MS/MS analysis with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). The MS/MS spectra were analyzed with Thermo Proteome Discoverer (Thermo Fisher Scientific, version 2.2), and the identified phosphorylated peptides were manually inspected to ensure confidence in the assignment of phosphorylation sites.
EMSA
The DNA oligonucleotide probe (5′-CGTTGACCGTTGACCGAGTTGACTTTTTA-3′) with three W-boxes (underlined) was synthesized and labeled with biotin at the 5′ end (Sangon). The gel mobility shift assays were performed using a LightShift Chemiluminescent EMSA Kit (Thermo Scientific) according to the manufacturer’s instructions. Briefly, freshly prepared recombinant WRKY33, WRKY33T229A, WRKY33F2, or WRKY33F4 protein (1 μg) was incubated with biotin-labeled DNA probes (10 pmol) for 30 min at 25°C in the presence or absence of unlabeled competitor DNA. The resulting protein-DNA complexes were separated on a 6% (w/v) native polyacrylamide gel and transferred to a nylon membrane. After cross-linking to the membrane by UV light radiation, the biotin-labeled DNA probes were detected using a streptavidin-conjugated horseradish peroxidase. To test the effect of CPK5-mediated phosphorylation on the DNA binding activity of WRKY33, recombinant WRKY33 and WRKY33T229A proteins were incubated with GST-CPK5 in kinase reaction buffer containing 0.1 mM ATP for 1 h at 25°C before performing the EMSA.
ChIP-qPCR Analysis
Two-week-old 35S:4myc-WRKY33/Est:CPK5CA-HA and 35S:4myc-WRKY33T229A/Est:CPK5CA-HA transgenic seedlings treated with 10 μM Est for 24 h were processed as described previously (Yamaguchi et al., 2014). Chromatin was isolated from 0.3 g of frozen tissues and sonicated with a Covaris M220 sonicator (peak incident power of 75 W, duty factor of 10%) for 5 min. Immunoprecipitation was performed by incubating the sheared chromatin samples with 4 μg of anti-myc antibody (Millipore, catalog no. 05-724) or mouse IgG (negative control) for 1 h at 4°C. The protein-chromatin immunocomplexes were captured using herring sperm DNA- and BSA-blocked protein G-agarose (Invitrogen). The immunoprecipitated DNA was purified using a ChIP DNA Clean and Concentrator kit (Zymo Research). The immunoprecipitated and input DNA samples were analyzed by qPCR using primers specific for the W-box-containing regions of PAD3. The ChIP-qPCR results are presented as percentages of the input DNA.
Analysis of Promoter Activity and Transactivation Activity
Arabidopsis protoplasts were cotransfected with the indicated reporter and effector constructs. The reference construct UBQ10pro:GUS was included in all transfections and served as an internal transfection control. After culturing for 8 h, the transfected protoplasts were collected and resuspended in cell lysis buffer (25 mM Tris-phosphate, pH 7.8, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% [v/v] glycerol, 1% [v/v] Triton X-100, and 2 mM DTT). The LUC activities in the protoplast lysates were analyzed using a LUC assay kit (GoldBio) and a multimode plate reader (SpectraMax iD5) according to each manufacturer’s instructions. For the GUS activity assay, aliquots of protoplast lysate were reacted with a fourfold volume of GUS assay solution (10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, and 1 mM methylumbelliferyl-β-d-glucuronide) at 37°C for 1 h, and the fluorescent signals were measured using a SpectraMax iD5 reader. The promoter and transactivation activities were calculated as the ratio of LUC activity to GUS activity, and the data are shown as relative fold increases over the control.
Statistical Analyses
At least three independent repetitions were performed for the experiments in this study. Results from one of the independent repeats that gave similar results are shown. Student’s t test was used to determine whether the difference between the two groups of data is statistically significant (Supplemental Data Set). Asterisks above the columns indicate differences that are statistically significant (P < 0.05). When more than two groups of data at a specific time point or from a specific treatment are compared, one-way ANOVA with Tukey’s posthoc test was performed (P < 0.05; Supplemental Data Set). Different letters above the data points are used to indicate differences that are statistically significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CPK4 (AT4G09570), CPK5 (AT4G35310), CPK6 (AT2G17290), CPK11 (AT1G35670), MPK3 (AT3G45640), MPK6 (AT2G43790), MKK5 (AT3G21220), EF1α (AT5G60390), CYP79B2 (AT4G39950), CYP71A13 (AT2G30770), PAD3 (AT3G26830), and WRKY33 (AT2G38470).
Supplemental Data
Supplemental Figure 1. Induction levels of camalexin production in Est:CPK5CA-HA transgenic lines are correlated with Est-induced protein levels of CPK5CA.
Supplemental Figure 2. Activation of CPK5 or CPK6 induces the expression of camalexin biosynthetic genes in Arabidopsis protoplasts.
Supplemental Figure 3. CPK5 and CPK6 expression in Arabidopsis is induced by B. cinerea infection.
Supplemental Figure 4. WRKY33 is required for CPK5-mediated activation of the promoters of camalexin biosynthetic genes in Arabidopsis protoplasts.
Supplemental Figure 5. The phosphorylation of WRKY33 by CPK5 is not detected by an anti-pSer antibody.
Supplemental Figure 6. CPK6 interacts with and phosphorylates WRKY33.
Supplemental Figure 7. The T229A mutation in WRKY33 does not affect its interaction with CPK5.
Supplemental Figure 8. The Thr-229 residue of WRKY33 is required for its function in Arabidopsis resistance to B. cinerea.
Supplemental Figure 9. Both the N- and C-terminal WRKY domains of WRKY33 are capable of binding to the W-box element.
Supplemental Figure 10. The amino acid sequence of WRKY33 showing the CPK5/CPK6 and MPK3/MPK6 phosphorylation sites.
Supplemental Figure 11. MPK3/MPK6-mediated phosphorylation of WRKY33 does not alter its protein stability.
Supplemental Figure 12. CPK5-mediated phosphorylation of WRKY33 does not alter its transactivation activity.
Supplemental Table 1. Recombinant constructs generated in this study.
Supplemental Table 2. Primers used in this study.
Supplemental Table 3. Transgenic Arabidopsis plants generated by Agrobacterium-mediated transformation in this study.
Supplemental Table 4. Transgenic Arabidopsis plants generated by genetic crossing in this study.
Supplemental Data Set. Results of ANOVAs and t tests for the data presented in each figure.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the ABRC for providing mutant seeds. We also thank Ping He, Libo Shan, and Bo Li for providing protoplast transfection vectors. This work was supported by the National Natural Science Foundation of China (grant 31970282 to X.M. and grant 31800215 to J.Z.).
AUTHOR CONTRIBUTIONS
J.Z. and X.M. designed the experiments; J.Z., X.W., Y.H., and T.S. performed the experiments and analyzed the data; P.W., S.D., and S.Z. contributed new analytical tools; X.M. and J.Z. wrote the article.
References
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P.(2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106. [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J.(2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H.(2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Kracher B., Roccaro M., Somssich I.E.(2017). Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29: 20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E.(2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J.(2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J.(2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L.H., Fischer N.M., Harter K., Kohlbacher O., Wanke D.(2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41: 9764–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Couto D., Zipfel C.(2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16: 537–552. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E.(2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. [DOI] [PubMed] [Google Scholar]
- de Pater S., Greco V., Pham K., Memelink J., Kijne J.(1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24: 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.P., Schulze W.X., Romeis T.(2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E.(2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Schmelzer E., Hahlbrock K., Somssich I.E.(1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18: 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P.(2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F., Møldrup M.E., Böttcher C., Olsen C.E., Scheel D., Halkier B.A.(2011). Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23: 2456–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F.M.(1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91: 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravino M., Savatin D.V., Macone A., De Lorenzo G.(2015). Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 84: 1073–1086. [DOI] [PubMed] [Google Scholar]
- Han L., Li G.J., Yang K.Y., Mao G., Wang R., Liu Y., Zhang S.(2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64: 114–127. [DOI] [PubMed] [Google Scholar]
- He Y., Xu J., Wang X., He X., Wang Y., Zhou J., Zhang S., Meng X.(2019). The Arabidopsis pleiotropic drug resistance transporters PEN3 and PDR12 mediate camalexin secretion for resistance to Botrytis cinerea. Plant Cell 31: 2206–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Fukunaga S., Bednarek P., Pislewska-Bednarek M., Watanabe S., Narusaka Y., Shirasu K., Takano Y.(2013). Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc. Natl. Acad. Sci. USA 110: 9589–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W., Thomas M., Nukarinen E., Egelhofer V., Röhrig H., Weckwerth W., Conrath U., Beckers G.J.(2013). Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell. Proteomics 12: 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S., Nakamura K.(1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244: 563–571. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L.(2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Li B., Jiang S., Yu X., Cheng C., Chen S., Cheng Y., Yuan J.S., Jiang D., He P., Shan L.(2015). Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27: 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., et al. (2014). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S.(2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Hake K., Wang W., Zhao T., Romeis T., Tang D.(2017). CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1-mediated immunity. Plant Cell 29: 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S.(2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A.A., Saitoh H., Felix G., Freymark G., Miersch O., Wasternack C., Boller T., Jones J.D., Romeis T.(2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102: 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S.(2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.Y., Mordorski B., Liu Y., Zhang S.(2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S.(2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J.(2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E.(2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., van Wersch R., Zhang Y.(2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 31: 403–409. [DOI] [PubMed] [Google Scholar]
- Piasecka A., Jedrzejczak-Rey N., Bednarek P.(2015). Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 206: 948–964. [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P.(2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wang J., Gong Z., Zhou J.M.(2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38: 92–100. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N., Bigeard J., Alhoraibi H., Bonhomme L., Hesse A.M., Vinh J., Hirt H., Pflieger D.(2018). Quantitative phosphoproteomic analysis reveals shared and specific targets of Arabidopsis mitogen-activated protein kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteomics 17: 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S.(2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Herde M.(2014). From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 20: 1–10. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F.(2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62: 840–851. [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatos A., Halkier B.A., Glawischnig E.(2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold H., Trempel F., Ranf S., Scheel D., Romeis T., Lee J.(2014). Ca2+ signalling in plant immune response: From pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 204: 782–790. [DOI] [PubMed] [Google Scholar]
- Sheikh A.H., Eschen-Lippold L., Pecher P., Hoehenwarter W., Sinha A.K., Scheel D., Lee J.(2016). Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana. Front. Plant Sci. 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby I.E., Geu-Flores F., Halkier B.A.(2010). Biosynthesis of glucosinolates: Gene discovery and beyond. Trends Plant Sci. 15: 283–290. [DOI] [PubMed] [Google Scholar]
- Stotz H.U., Sawada Y., Shimada Y., Hirai M.Y., Sasaki E., Krischke M., Brown P.D., Saito K., Kamiya Y.(2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 67: 81–93. [DOI] [PubMed] [Google Scholar]
- Su T., Xu J., Li Y., Lei L., Zhao L., Yang H., Feng J., Liu G., Ren D.(2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23: 364–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J.M.(2017). Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29: 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Nelissen I., Eggermont K., Broekaert W.F.(1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171. [DOI] [PubMed] [Google Scholar]
- Tsuji J., Jackson E.P., Gage D.A., Hammerschmidt R., Somerville S.C.(1992). Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 98: 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Schuck S., Wu J., Yang P., Döring A.C., Zeier J., Tsuda K.(2018). A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30: 2480–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.P., Xu H., Wang B., Su X.D.(2020). Crystal structures of N-terminal WRKY transcription factors and DNA complexes. Protein Cell 11: 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.F., Kwon C.S., William D.A., Wagner D.(2014). PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. The Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen Q., Zhou S., Sun Y., Li X., Li Y.(2020). H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to Verticillium dahliae toxins. Plant Physiol. 182: 640–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L.(2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Glazebrook J.(1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]