Multiparametric in vivo analyses using dual-reporting genetically encoded fluorescent indicators reveal the spatiotemporal coordination of signaling compound dynamics in Arabidopsis roots.
Abstract
Deciphering signal transduction processes is crucial for understanding how plants sense and respond to environmental changes. Various chemical compounds function as central messengers within deeply intertwined signaling networks. How such compounds act in concert remains to be elucidated. We have developed dual-reporting transcriptionally linked genetically encoded fluorescent indicators (2-in-1-GEFIs) for multiparametric in vivo analyses of the phytohormone abscisic acid (ABA), Ca2+, protons (H+), chloride (anions), the glutathione redox potential, and H2O2. Simultaneous analyses of two signaling compounds in Arabidopsis (Arabidopsis thaliana) roots revealed that ABA treatment and uptake did not trigger rapid cytosolic Ca2+ or H+ dynamics. Glutamate, ATP, Arabidopsis PLANT ELICITOR PEPTIDE, and glutathione disulfide (GSSG) treatments induced rapid spatiotemporally overlapping cytosolic Ca2+, H+, and anion dynamics, but except for GSSG, only weakly affected the cytosolic redox state. Overall, 2-in-1-GEFIs enable complementary, high-resolution in vivo analyses of signaling compound dynamics and facilitate an advanced understanding of the spatiotemporal coordination of signal transduction processes in Arabidopsis.
INTRODUCTION
Understanding how plants sense and respond to environmental and extracellular fluctuations is key for our strategic progressions to limit the consequences of climate change on plant growth and crop productivity. Plants have evolved complex signal transduction networks that enable the sensing and integration of extracellular signals, and the processing and transduction of the underlying information into physiological, growth, and developmental responses. Within such signaling networks, spatiotemporal concentration changes of hormones, the divalent cation Ca2+, and reactive oxygen species (ROS) mediate various downstream responses (Dodd et al., 2010; Kudla et al., 2010; Shan et al., 2012; Vanstraelen and Benková, 2012; Mittler, 2017; Waszczak et al., 2018; Smirnoff and Arnaud, 2019). Among the plant hormones, abscisic acid (ABA) functions as a central regulator of the plant water status (Cutler et al., 2010; Finkelstein, 2013; Yoshida et al., 2019). Dynamic concentration changes of signaling compounds require inter- and intracellular transport, including long-distance transport and signaling, that often depend on proton (H+) and electrochemical gradients across membranes (Schumacher, 2014; Choi et al., 2016; Sze and Chanroj, 2018). In addition, environmental and cellular H+ concentration (pH) can affect plant growth, development, and molecular properties (Shavrukov and Hirai, 2016). Therefore, H+ may also function in signaling (Sze and Chanroj, 2018).
In plants, hormonal, Ca2+, ROS, and pH signaling processes are intertwined on several levels (Hauser et al., 2011; Vanstraelen and Benková, 2012; Gilroy et al., 2014; Steinhorst and Kudla, 2014; Edel and Kudla, 2016), for example, to regulate stomatal movements or root hair and pollen tube growth (Munemasa et al., 2015; Mangano et al., 2016; Hauser et al., 2017; Michard et al., 2017). In response to the growth hormone auxin, extracellular ATP, touch, or wounding, Ca2+ signals are accompanied by an apoplastic alkalization and/or cytosolic acidification (Monshausen et al., 2009, 2011; Behera et al., 2018). However, except for the auxin response (Shih et al., 2015; Dindas et al., 2018), the underlying mechanisms are not well understood. Current models suggest that cytosolic Ca2+ and extracellular ROS signals are important for cell-to-cell communication and long-distance signaling (Gilroy et al., 2014; Steinhorst and Kudla, 2014). Although some components, such as the ROS-producing Arabidopsis (Arabidopsis thaliana) RESPIRATORY BURST OXIDASE HOMOLOG D (AtRBOHD), the ion channel TWO PORE CHANNEL1 (TPC1), GLUTAMATE RECEPTOR-LIKE CHANNELS (GLRs), Ca2+-DEPENDENT PROTEIN KINASES (CDPKs/CPKs), and CALCINEURIN B-LIKE (CBL) proteins together with CBL-INTERACTING PROTEIN KINASES (CIPKs), have been linked to such processes (Choi et al., 2016; Waszczak et al., 2018), the underlying mechanisms remain unclear.
To decipher the coordination and interdependence of signaling processes, it is important to monitor the spatiotemporal dynamics of signaling compounds. Genetically encoded fluorescent indicators (GEFIs) are currently the state-of-the-art technology for high-resolution in vivo monitoring of biological processes (Grossmann et al., 2018; Hilleary et al., 2018; Walia et al., 2018). In general, GEFIs consist of a ligand binding sensory domain (SD) that is fused to fluorescent proteins (FPs). When binding to their ligand, structural changes in the SD affect the absorption and emission properties of attached FPs. Thus, ligand binding can affect (1) the fluorescence emission of a single FP, (2) the emission ratio of a single FP after excitation at two wavelengths, or (3) the emission ratio of two attached FPs. In addition, single FPs have been engineered for responsiveness to changes in H+, anion, and metal concentrations or redox conditions (Okumoto et al., 2012; Walia et al., 2018). Although the number of GEFIs is steadily increasing, only a fraction has been introduced into plants, and fewer have been used in simultaneous multiparametric analyses (Okumoto et al., 2012; Kostyuk et al., 2018; Walia et al., 2018). To facilitate the use of GEFIs in multiparametric analyses, we introduce here the concept of dual-reporting transcriptionally linked GEFIs (2-in-1-GEFIs) that enable the in vivo monitoring of at least two signaling compounds simultaneously. Through the genetic fusion of two distinct indicators, seven 2-in-1-GEFIs were generated that enable time-efficient and complementary in vivo analyses of ABA, Ca2+, H+, Cl–, H2O2, and the glutathione redox potential (EGSH) at unpreceded spatiotemporal resolution. Microscopic analyses of these 2-in-1-GEFIs in Arabidopsis roots confirmed their functionality and revealed that extracellularly applied ABA was rapidly taken up, but without discernible effect on cytosolic Ca2+ and pH levels. By contrast, treatments with glutamate, ATP, AtPEP1, and glutathione disulfide (GSSG) induced spatiotemporally overlapping cytosolic concentration dynamics of Ca2+, H+, and Cl–, without noticeable rapid effects on the cytosolic redox state.
RESULTS
Approaches to Optimize FRET-Based ABA Indicators
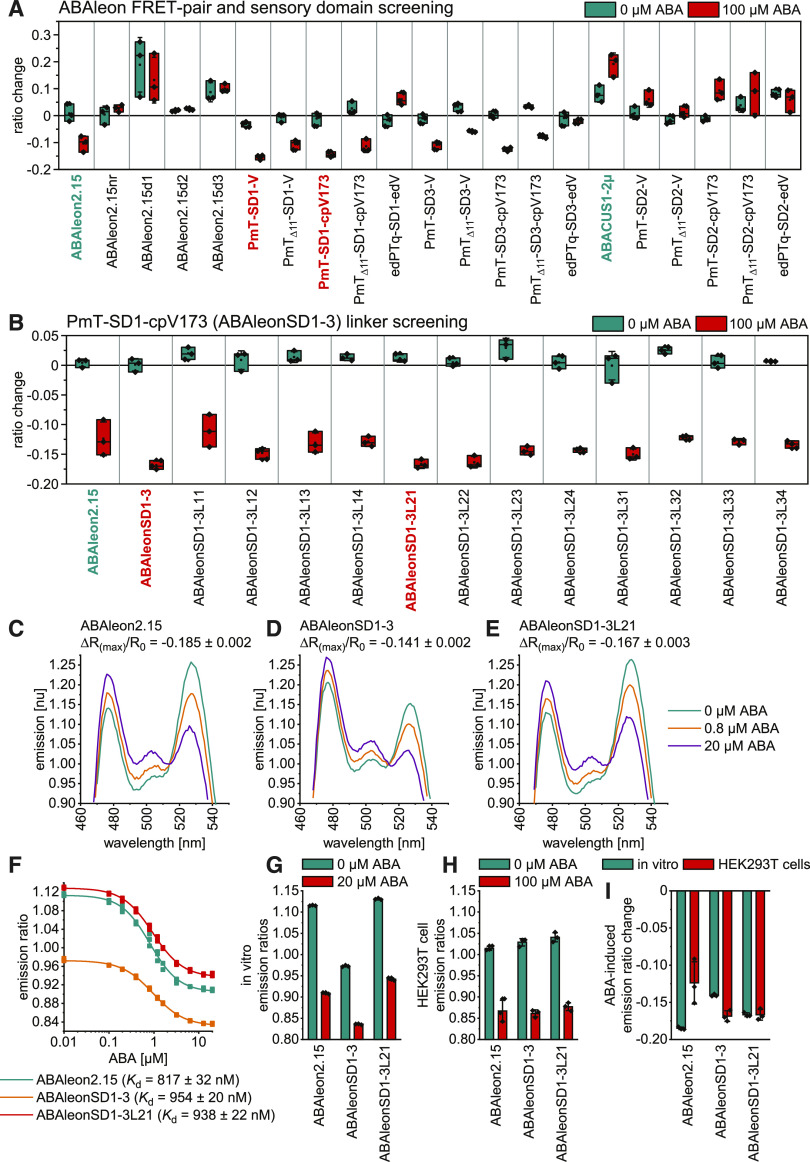
Currently available ABA indicators are ABACUS and ABAleon (Jones et al., 2014; Waadt et al., 2014). Because their expression in plants affects ABA signaling and because they exhibit a relatively small signal-to-noise ratio (Waadt et al., 2015), we aimed to optimize these indicators before utilizing them for multiparametric analyses. As a testing system, we chose human embryonic kidney (HEK293T) cells that allow for efficient medium-throughput plate reader–based screens. Compared to ABAleon2.15 and ABACUS1-2µ used as positive controls and nonresponsive ABAleon2.15nr as negative control, the initial screening aimed to evaluate deletion variants of ABAleon2.15 and various combinations of five FP Förster resonance energy transfer (FRET)-pairs and three SDs (Figure 1A; Supplemental Figures 1A to 1D). Sixty minutes after application of 0 or 100 µM ABA, ABAleon2.15 deletion variants (d1, d2, and d3), as well as ABACUS1-2µ and SD2 variants, exhibited emission ratio changes even in control conditions. Compared to ABAleon2.15, two indicators (PmTurquoise-SD1-Venus and PmTurquoise-SD1-cpVenus173) exhibited negligible responses to control treatments but increased negative emission ratio changes in response to ABA (Figure 1A). Because PmTurquoise-SD1-cpVenus173 (ABAleonSD1-3) differed from ABAleon2.15 only in the sequences that link the SD with the attached FPs (Supplemental Figures 1A and 1D), an additional ABAleonSD1-3 linker screening was performed (Figure 1B). This led to the identification of ABAleonSD1-3L21 (linkers LD and T; Supplemental Figure 1E) with similar properties as ABAleonSD1-3 (Figure 1B).
Figure 1.
Development of ABAleonSD1-3L21.
(A) and (B) FRET-pair and SD (A) and linker screening (B) of ABA indicator variants after expression in HEK293T cells. Shown are emission ratio changes in response to 60-min treatments with 0 and 100 µM ABA. Central lines in boxes, median; boxes, 25th and 75th percentiles; diamonds, data points; square dots, mean; whiskers, ±sd. Reference indicators are shown in cyan and new candidates in red.
(C) to (E) Representative ABA-dependent normalized (nu) in vitro emission spectra of ABAleon2.15 (C), ABAleonSD1-3 (D), and ABAleonSD1-3L21 (E) with indicated ΔR(max)/R0.
(F) ABA-dependent in vitro emission ratios and apparent ABA Kd of ABAleons.
(G) and (H) Comparison of ABA-dependent ABAleon emission ratios (mean ± sd, n = 3; diamonds, data points): in vitro (G) and in HEK293T cells (H).
(I) In vitro and HEK293T cell comparison of ABA-induced maximum emission ratio changes (mean ± sd, n = 3; diamonds, data points).
Information on ABA indicator topologies is given in Supplemental Figure 1.
To corroborate these findings, ABAleon2.15, ABAleonSD1-3, and ABAleonSD1-3L21 proteins were purified from Escherichia coli and characterized in vitro. Although all three ABAleons were functional (Figures 1C to 1G), their properties in vitro were markedly different from results obtained in HEK293T cells (Figures 1G to 1I). Basal emission ratios (at 0 µM ABA) were not the same in both assay systems (Figures 1G and 1H). ABAleon2.15 exhibited a higher ABA-induced emission ratio change in vitro, while ABAleonSD1-3 responses were larger in the HEK293T cell system (Figure 1I). Notably, ABAleonSD1-3L21 responded similarly in vitro and in HEK293T cells (Figure 1I). In vitro calibrations revealed that ABAleonSD1-3 responded more weakly to ABA [maximum emission ratio change ΔR(max)/R0 = –0.141] than ABAleon2.15 and ABAleonSD1-3L21 [ΔR(max)/R0 = –0.185 and –0.167, respectively; Figures 1C to 1E]. Apparent ABA Kd of ABAleonSD1-3 (954 nM) and ABAleonSD1-3L21 (938 nM) were slightly lower compared with ABAleon2.15 (817 nM; Figure 1F). Altogether, the ABA indicator screening led to the identification of ABAleonSD1-3L21 as a candidate for further evaluation in planta.
Evaluation of FRET-Based ABA Indicators in Arabidopsis
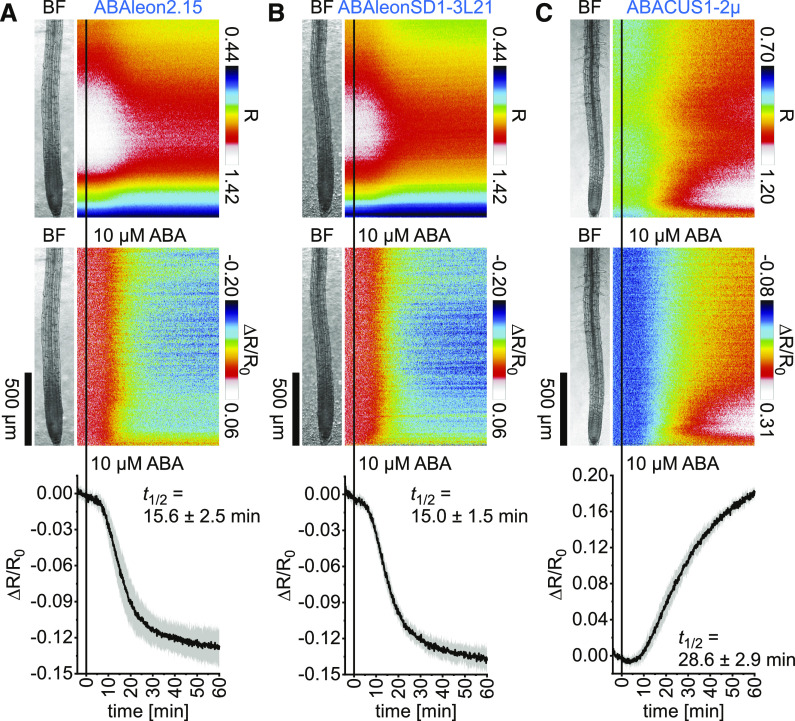
Five-day-old Arabidopsis seedlings expressing ABAleon2.15, ABAleonSD1-3L21, or ABACUS1-2µ in the cytosol and nucleus were compared for their ABA responses in roots. Therefore, spatiotemporal vertical response profiles of R (Figure 2, top) or emission ratio changes normalized to 4-min average baseline recordings (ΔR/R0; Figure 2, middle) and overall emission ratio changes (Figure 2, bottom) were acquired in response to 10 µM ABA treatments. ABAleon2.15 and ABAleonSD1-3L21 responded similarly to ABA, with a sigmoidal emission ratio decrease at a half response time (t1/2) of ∼15 min (Figures 2A and 2B). Note that the root cap exhibited a much weaker response to ABA than the other tissues (Figures 2A and 2B, middle). Compared to ABAleons, ABACUS1-2µ responded slower to ABA (t1/2 ∼ 29 min), but with larger increasing emission ratio changes that were more pronounced in the meristematic and early elongation zone (Figure 2C; Supplemental Movie 1). None of the indicators exhibited emission ratio changes in response to control treatments (Supplemental Figure 2). Because both ABAleons exhibited similar ABA response patterns with faster response kinetics than ABACUS1-2µ, we decided to use ABAleonSD1-3L21 for our multiparameter imaging approach.
Figure 2.
Comparison of ABA Indicator ABA Responses in Arabidopsis.
(A) to (C) Roots of 5-d-old Arabidopsis expressing ABAleon2.15 (n = 9; see [A]), ABAleonSD1-3L21 (n = 8; see [B]), and ABACUS1-2µ (n = 6; see [C]) were imaged for 64 min at a frame rate of 10 min−1 and treated with 10 µM ABA at t = 0 min. Shown are average vertical response profiles of emission ratios (R; top) and emission ratio changes (ΔR/R0; middle) normalized to 4-min average baseline recordings. An adjacent representative bright field (BF) root image is shown for orientation. (Bottom) Full image average emission ratio changes (mean ± sd) with indicated t1/2.
A representative experiment is provided as Supplemental Movie 1. Data of 0 µM ABA control experiments are shown in Supplemental Figure 2.
Concept and Design of 2-in-1-GEFIs
To establish multiparametric in vivo analyses of signaling compounds in Arabidopsis, six GEFIs were chosen: ABAleonSD1-3L21 (ABA), R-GECO1 (Ca2+), Arabidopsis codon-optimized red-fluorescing (P)A-17 (H+), E2GFP (H+ and Cl–), Grx1-roGFP2 (EGSH), and roGFP2-Orp1 (H2O2). Their properties have been well characterized in vitro (Figure 1; Bizzarri et al., 2006; Gutscher et al., 2008, 2009; Arosio et al., 2010; Zhao et al., 2011; Shen et al., 2014). In addition, except for PA-17, these indicators have also been applied and characterized in Arabidopsis (Figure 2; Meyer et al., 2007; Marty et al., 2009; Keinath et al., 2015; Waadt et al., 2017; Demes et al., 2019; Nietzel et al., 2019).
Multiparametric analyses of signaling compounds require the generation of transgenic plants that express several GEFIs simultaneously. Because the generation of transgenic plants is time-consuming and the insertion of multiple transgenes into the Arabidopsis genome could result in epigenetic silencing effects (Behera et al., 2015), we aimed to express two GEFIs from one mRNA. Therefore, two GEFIs were genetically fused via sequences encoding a 14–amino acid ASGGSGGTSGGGGS-linker (GSL), or the self-cleaving 22–amino acid P2A linker that enables the expression of two separate polypeptides (Kim et al., 2011). Overall, seven 2-in-1-GEFIs have been generated: ABAleonSD1-3L21-P2A-R-GECO1 (ABA and Ca2+), PA-17-P2A-ABAleonSD1-3L21 (H+ and ABA), R-GECO1-GSL-E2GFP (Ca2+, H+, and Cl–), PA-17-P2A-Grx1-roGFP2 (H+ and EGSH), PA-17-P2A-roGFP2-Orp1 (H+ and H2O2), R-GECO1-P2A-Grx1-roGFP2 (Ca2+ and EGSH), and R-GECO1-P2A-roGFP2-Orp1 (Ca2+ and H2O2). See Supplemental Data Sets 1B and 1C for information about constructs and transgenic Arabidopsis plants. Note that the GEFI lines used for microscope analyses exhibited rather normal growth phenotypes when grown vertically on half-strength Murashige and Skoog (0.5 MS) medium for 5 d (Supplemental Figure 3A). However, 25- to 28-d-old, soil-grown GEFI lines expressing ABA indicators or R-GECO1-E2GFP variants were markedly impaired in growth (Supplemental Figures 3B to 3D; Supplemental Data Set 2). Therefore, data should be interpreted with caution when obtained using such transgenic GEFI lines at an age when growth phenotypes become apparent. In the following, we describe the application the 2-in-1-GEFIs in Arabidopsis and highlight the resulting biological findings.
ABA Does Not Trigger Rapid Ca2+ or pH Changes in Roots
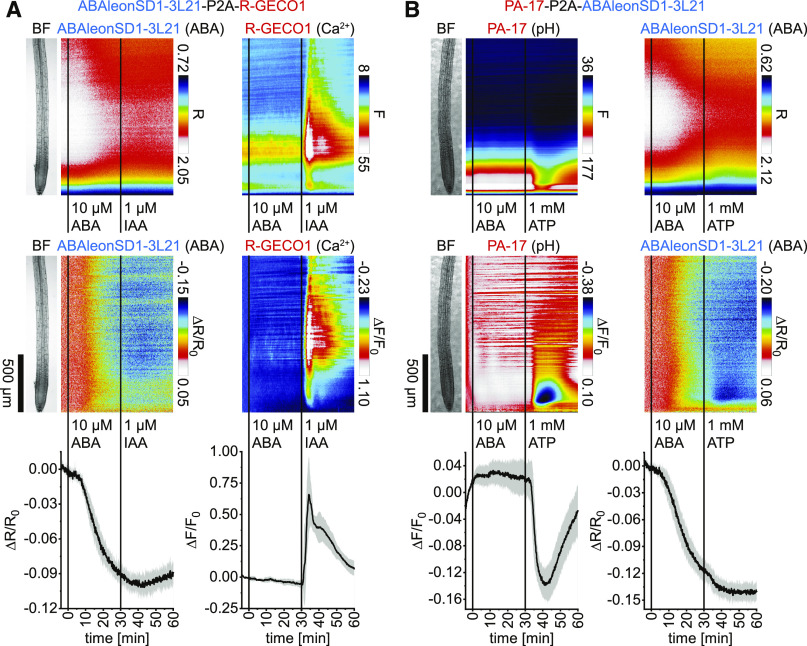
Interrelations of cytosolic concentration changes of ABA, Ca2+, and H+ are well known for stomatal aperture regulation (Munemasa et al., 2015; Eisenach and De Angeli, 2017; Jezek and Blatt, 2017) but less understood in roots. To investigate the interrelation of ABA with Ca2+ in roots, ABAleonSD1-3L21-P2A-R-GECO1 seedlings were monitored in response to 10 µM ABA, which induced a typical ABAleonSD1-3L21 emission ratio decrease (Figure 3A, left). The Ca2+ indicator R-GECO1 did not respond to this treatment (Figure 3A, right). However, subsequent 1 µM indole-3-acetic-acid (IAA; auxin) treatment at 30 min induced a biphasic Ca2+ signal that initiated in the root elongation zone and spread from there to neighboring regions (Figure 3A, right; Supplemental Movie 2), as observed previously by Waadt et al. (2017).
Figure 3.
Application of ABA Does Not Trigger Rapid Changes of Cytosolic Ca2+ or pH Levels in Arabidopsis Roots.
(A) and (B) Analyses of roots of 5-d-old Arabidopsis expressing ABAleonSD1-3L21-P2A-R-GECO1 (ABA and Ca2+; n = 5; see [A]) in response to 10 µM ABA (t = 0 min) and 1 µM IAA (t = 30 min) and PA-17-P2A-ABAleonSD1-3L21 (pH and ABA; n = 8; see [B]) in response to 10 µM ABA (t = 0 min) and 1 mM ATP (t = 30 min). Images were acquired for 64 min at a frame rate of 10 min−1. (Top) Shown are average vertical response profiles of emission ratios (R) or fluorescence emissions (F). (Middle) Signal changes (ΔR/R0 or ΔF/F0) normalized to 4-min average baseline recordings. An adjacent representative bright field (BF) root image is shown for orientation. (Bottom) Full image signal changes (mean ± sd).
Representative experiments are provided as Supplemental Movies 2 and 3.
To investigate the effect of ABA on cytosolic pH in roots, PA-17-P2A-ABAleonSD1-3L21 seedlings were treated with 10 µM ABA. From 0 to 30 min after ABA treatment PA-17 fluorescence remained unchanged (Figure 3B, left). However, in response to 1 mM ATP PA-17 fluorescence emission decreased, indicating a rapid and transient cytosolic acidification with a maximum pH drop in the root meristematic zone that also spread to the elongation zone (Figure 3B, left). In this experiment, ABAleonSD1-3L21 exhibited a typical ABA response pattern that was not affected by ATP (Figure 3B, right; Supplemental Movie 3). These experiments established the functionality of both 2-in-1-GEFIs and revealed that ABA does not trigger rapid cytosolic Ca2+ or pH changes in roots.
Auxin, ATP, and Glutamate Treatments Trigger Spatiotemporally Overlapping Dynamics of Ca2+, H+, and Cl–
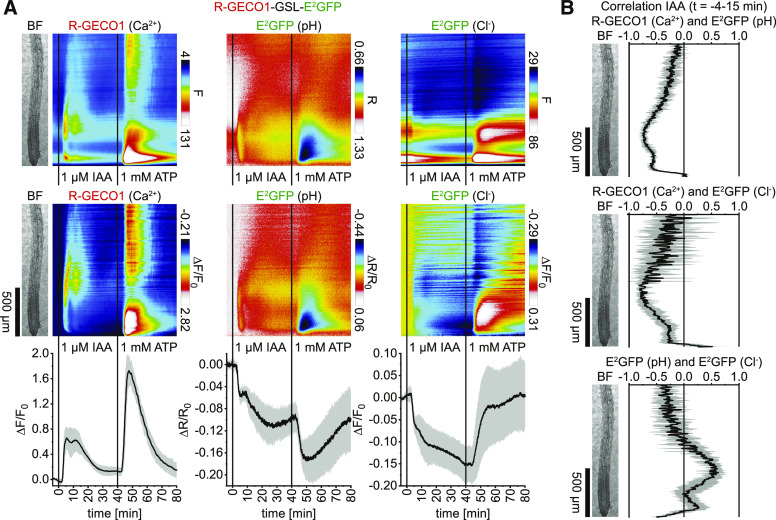
Next, we used R-GECO1-GSL-E2GFP to simultaneously monitor Ca2+, H+, and Cl–. Because anions, such as Cl–, quench the fluorescence of E2GFP and because its excitation ratiometric pH readout is Cl– independent, E2GFP provides a means to simultaneously assess cytosolic H+ and Cl– changes (Bizzarri et al., 2006; Arosio et al., 2010). In response to 1 µM IAA, R-GECO1 reported biphasic Ca2+ signals in the root elongation zone that traveled to neighboring regions. Subsequent 1 mM ATP treatments triggered Ca2+ signals in the root cap and meristematic zone that proceeded shootward (Figure 4A, left). Interestingly, both Ca2+ signals coincided with a cytosolic acidification reported by E2GFP (Figure 4A, middle; Supplemental Movie 4). IAA also induced a cytosolic Cl– influx, indicated by a E2GFP fluorescence emission decrease in the entire imaged root, with a maximum decrease in the meristematic zone (Figure 4A, right). ATP however induced cytosolic Cl– influx in the upper elongation zone and above but Cl– efflux in the lower elongation and meristematic zone (Figure 4A, right). Correlation analyses of the initial 15 min during the IAA response indicated a remarkable spatiotemporal overlap of cytosolic Ca2+, H+, and Cl– dynamics in the meristematic and elongation zone (Figure 4B).
Figure 4.
Application of Auxin and ATP Triggers Spatiotemporally Overlapping Cytosolic Concentration Changes of Ca2+, H+, and Cl–.
Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1-GSL-E2GFP (Ca2+, pH, and Cl–) in response to 1 µM IAA (t = 0 min) and 1 mM ATP (t = 40 min; n = 6). Images were acquired for 84 min at a frame rate of 10 min−1.
(A) Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R). (Middle) Signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. (Bottom) Full image signal changes (mean ± sd).
(B) Spatiotemporal Pearson correlation analyses (mean ± sd) of indicated GEFI responses during the application of IAA (t = –4 to 15 min). An adjacent representative bright field (BF) root image is shown for orientation.
A representative experiment is provided as Supplemental Movie 4.
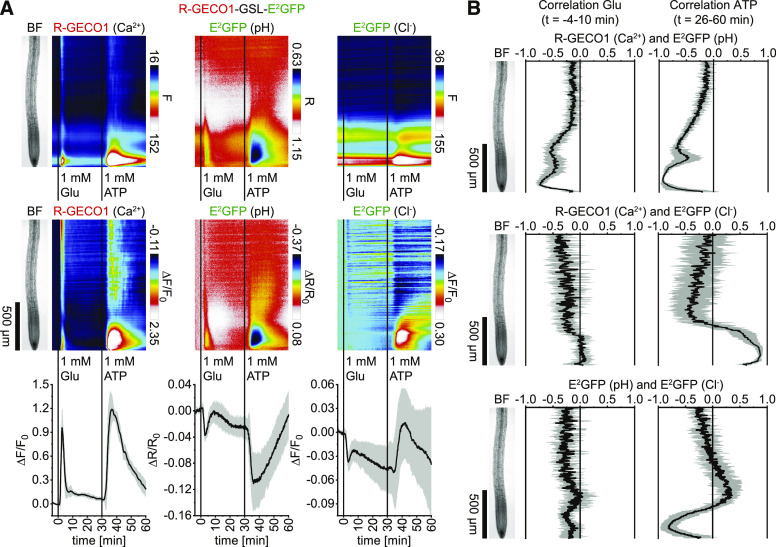
In additional experiments, the effect of 1 mM glutamate was assessed. Compared to IAA, glutamate treatments triggered a more rapid Ca2+ transient (Figure 5A, left) that arrived in a wave-like shape from upper root regions (Supplemental Movie 5). H+ and Cl– also displayed a rapid and transient initial cytosolic influx with a maximum acidification in the meristematic zone, followed by a weak transient alkalization in the early elongation zone and a prolonged cytosolic H+ influx (Figure 5A, middle and right; see signal change data sets). During the initial 10 min of the glutamate response, Ca2+ and H+ dynamics exhibited a noticeable spatiotemporal overlap/correlation in the meristematic and early elongation zone (Figure 5B, left). Subsequent responses to ATP, used as positive control, were as observed before (Figures 4A and 5A). Correlation analyses indicated a remarkable coincidence of cytosolic Ca2+ and H+ influx and Cl– efflux in the meristematic zone (Figure 5B, right).
Figure 5.
Application of Glutamate Triggers Spatiotemporally Overlapping Cytosolic Concentration Changes of Ca2+, H+, and Cl–.
Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1-GSL-E2GFP (Ca2+, pH, and Cl–) in response to 1 mM glutamate (Glu; t = 0 min) and 1 mM ATP (t = 30 min; n = 7). Images were acquired for 64 min at a frame rate of 10 min−1.
(A) Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R). (Middle) Signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. (Bottom) Full image signal changes (mean ± sd).
(B) Spatiotemporal Pearson correlation analyses (mean ± sd) of indicated GEFI responses during the application of glutamate (left; t = –4 to 10 min) or ATP (right; t = 26 to 60 min). An adjacent representative bright field (BF) root image is shown for orientation.
A representative experiment is provided as Supplemental Movie 5. See also Supplemental Figures 4 and 5 for related experiments.
To increase the spatial resolution for pH measurements, R-GECO1-P2A-E2GFP was fused to the N terminus of LOW TEMPERATURE INDUCED PROTEIN 6B (LTI6b) or VESICLE TRANSPORT V-SNARE11 (VTI11). This enabled the targeting of E2GFP to the cytosolic side of the plasma membrane (LTI6b; Cutler et al., 2010) or the tonoplast (VTI11; Takemoto et al., 2018), while R-GECO1 remained in the cytosol and the nucleus (Supplemental Figures 4A and 5A). Compared to previous analyses (Figure 5), these indicators reported very similar Ca2+ and pH response patterns, irrespective of the subcellular localization of E2GFP. However, it appeared that Cl– responses were more variable with noticeably higher Cl– dynamics at the tonoplast (Supplemental Figures 4B and 5B). Note that R-GECO1-P2A-E2GFP-LTI6b expression induced more severe growth defects in Arabidopsis than in the other R-GECO1-E2GFP variants (Supplemental Figure 3D). Therefore, results obtained with this GEFI should be interpreted with caution. Taken together, our results demonstrate that R-GECO1-GSL-E2GFP enables the simultaneous monitoring of cytosolic Ca2+, H+, and Cl– dynamics, that in response to IAA, ATP and glutamate exhibited a remarkably high spatiotemporal overlap.
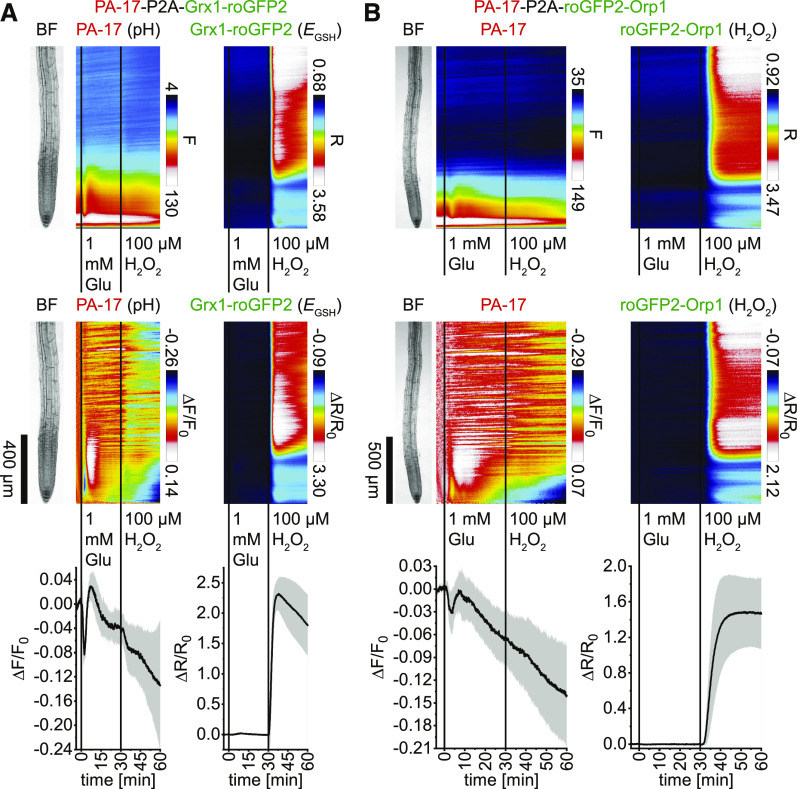
Glutamate Treatment Induces Cytosolic Acidification without Noticeable Effects on the Cytosolic Redox State
To test whether glutamate has an effect on the cytosolic redox state, Arabidopsis seedlings expressing PA-17-P2A-Grx1-roGFP2 (pH and EGSH) or PA-17-P2A-roGFP2-Orp1 (pH and H2O2) were exposed to 1 mM glutamate and 100 µM H2O2 treatments as positive control. As observed before, glutamate triggered a biphasic cytosolic acidification that prolonged during the 100 µM H2O2 response (Figures 6A and 6B, left; Supplemental Movies 6 and 7). Application of glutamate did not induce cytosolic redox changes, whereas 100 µM H2O2 treatments triggered a steep cytosolic oxidation that remained high for longer than 30 min (Figures 6A and 6B, right). During this response, the roGFPs indicated a cytosolic oxidation predominantly in epidermis and cortex cells of the elongation zone and above, with faster responses in upper root regions. Except for the epidermis, cells of the meristematic zone only slightly increased their redox state in response to H2O2 (Supplemental Movies 6 and 7). Together, these data indicate that 100 µM H2O2 treatments rapidly induce cytosolic oxidation and that the root meristematic zone is less sensitive to this oxidative stress.
Figure 6.
Application of Glutamate Triggers a Rapid Cytosolic Acidification without Noticeable Effects on the Cytosolic Redox State.
(A) and (B) Analyses of roots of 5-d-old Arabidopsis expressing PA-17-P2A-Grx1-roGFP2 (pH and EGSH; n = 6; see [A]) and PA-17-P2A-roGFP2-Orp1 (pH and H2O2; n = 8; see [B]) in response to 1 mM glutamate (Glu; t = 0 min) and 100 µM H2O2 (t = 30 min). Images were acquired for 64 min at a frame rate of 10 min−1. Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R). (Middle) Signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. An adjacent representative bright field (BF) root image is shown for orientation. (Bottom) Full image signal changes (mean ± sd). Note that experiments in (A) and (B) were acquired at different magnifications.
Representative experiments are provided as Supplemental Movies 6 and 7.
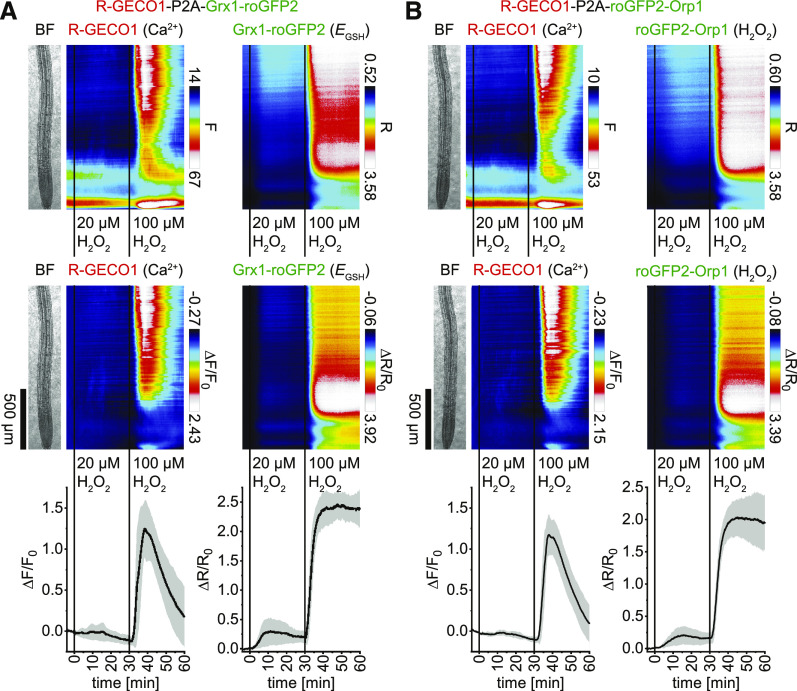
H2O2 Treatment Triggers Spatiotemporally Overlapping but Also Distinct Patterns of Cytosolic Oxidation and Ca2+ Dynamics
Current models propose an interdependence of Ca2+ and ROS signaling (Gilroy et al., 2014; Steinhorst and Kudla, 2014). To investigate the spatiotemporal relationships of cytosolic Ca2+ and ROS signals, we first treated Arabidopsis seedlings expressing R-GECO1-P2A-Grx1-roGFP2 (Ca2+ and EGSH) or R-GECO1-P2A-roGFP2-Orp1 (Ca2+ and H2O2) with 20 and 100 µM H2O2. The EGSH and H2O2 indicators responded to both treatments with similar patterns, albeit with increased signal changes in response to 100 µM H2O2 (Figures 7A and 7B, right; Supplemental Movies 8 and 9). Although 20 µM H2O2 treatments induced a detectable cytosolic oxidation, discernible Ca2+ signals were not observed (Figure 7; Supplemental Movies 8 and 9). In response to 100 µM H2O2 treatments, cytosolic oxidation preceded detectable Ca2+ signals. Although both signals appeared to arrive from upper root regions, Ca2+ signals in the elongation zone exhibited a maximum response in the vasculature, whereas cytosolic oxidation was more pronounced in epidermis and cortex cells. Both signals exhibited a minimum response in the meristematic zone (Figure 7; Supplemental Movies 8 and 9). We conclude that these 2-in-1-GEFIs exhibit sufficient sensitivity for resolving the interrelation of cytosolic Ca2+ and ROS signals, which, in response to 100 µM H2O2 treatments, exhibit overlapping, but not similar, spatiotemporal response patterns. In addition, the roGFPs facilitate the detection of cytosolic oxidation in response to H2O2 below the threshold of Ca2+ channel activation.
Figure 7.
Application of H2O2 Triggers Overlapping but Also Distinct Patterns of Cytosolic Ca2+ Concentration Changes and Oxidation.
(A) and (B) Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1-P2A-Grx1-roGFP2 (Ca2+ and EGSH; n = 8; see [A]) and R-GECO1-P2A-roGFP2-Orp1 (Ca2+ and H2O2; n = 8; see [B]) in response to 20 µM H2O2 (t = 0 min) and 100 µM H2O2 (t = 30 min). Images were acquired for 64 min at a frame rate of 10 min−1. (Top) Average vertical response profiles of fluorescence emissions (F) or emission ratios (R). (Middle) Signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. An adjacent representative bright field (BF) root image is shown for orientation. (Bottom) Full image signal changes (mean ± sd).
Representative experiments are provided as Supplemental Movies 8 and 9.
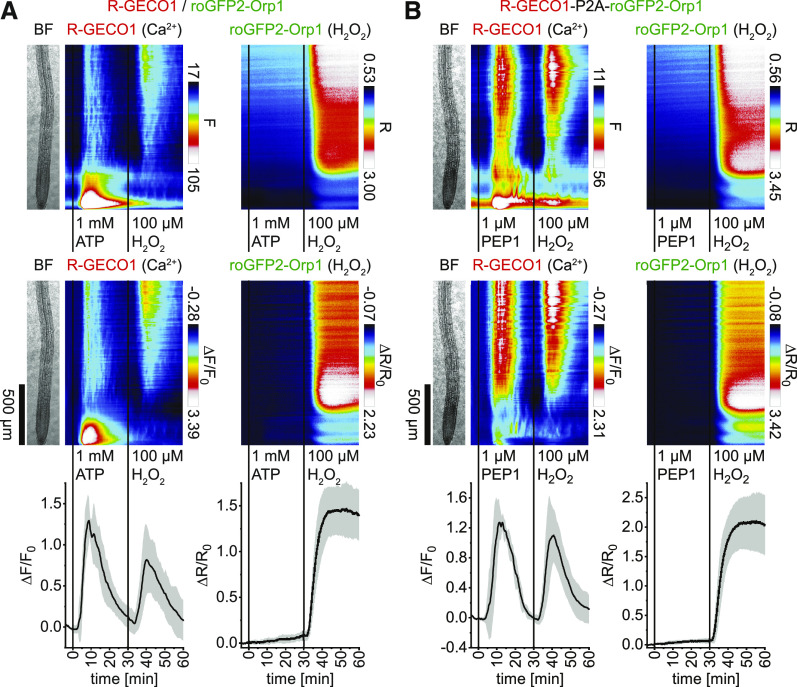
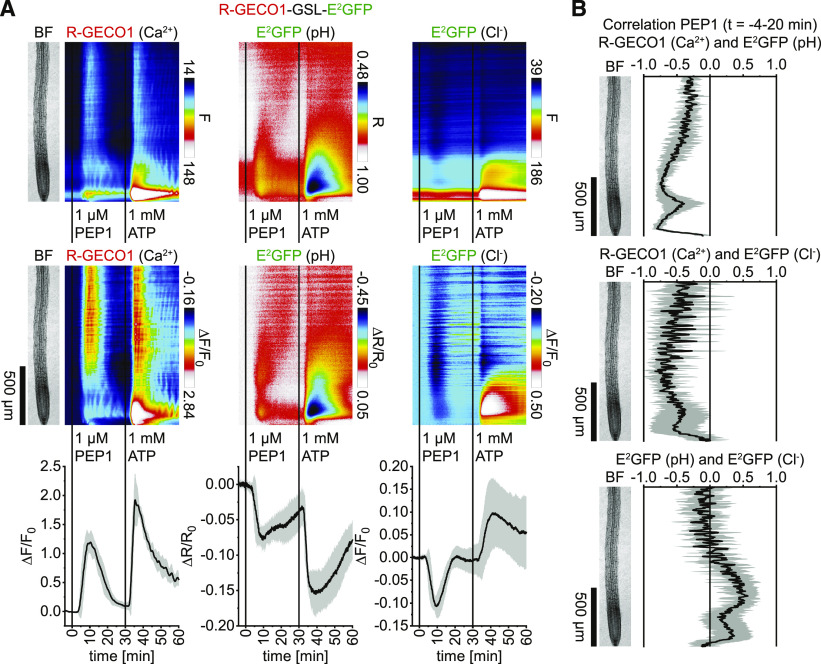
ATP and AtPEP1 Treatments Trigger Cytosolic Ca2+, H+, and Cl– Dynamics and a Weak Cytosolic Oxidation
Extracellular ATP and the signaling peptide AtPEP1 function as damage-associated elicitors that trigger Ca2+ signals and ROS production (Song et al., 2006; Demidchik, 2009; Tanaka et al., 2010; Ma et al., 2014). To investigate the spatiotemporal relationships of these processes, Arabidopsis seedlings expressing R-GECO1 and roGFP2-Orp1 or Grx1-roGFP2 from individual expression cassettes located on one T-DNA were subjected to 1 mM ATP and consecutive 100 µM H2O2 treatments as a positive control. ATP triggered typical Ca2+ responses, but its effect on the cytosolic redox state was rather weak (Figure 8A; Supplemental Figure 6A). H2O2 (100 µM) treatments induced cytosolic Ca2+ signals and oxidation as observed before (Supplemental Movies 10 and 11). Experiments using R-GECO1-P2A-roGFP2-Orp1 and R-GECO1-P2A-Grx1-roGFP2 revealed that 1 µM (At)PEP1 treatments induced Ca2+ signals that initiated in epidermis cells, followed by an overall Ca2+ burst, after which Ca2+ oscillations in the meristem appeared that proceeded to the vasculature and further shootward. However, PEP1 treatments had only weak effects on the cytosolic redox state (Figure 8B; Supplemental Figure 6B; Supplemental Movies 12 and 13). To better resolve the roGFP responses, the initial 30-min signal change vertical response profiles were extracted from original data sets and calibrated to the same color scale. The data indicate a detectable cytosolic oxidation in response to glutamate, ATP, and PEP1 that was however low compared to the 20 µM H2O2 response (Supplemental Figure 7). We also investigated the effect of PEP1 using R-GECO1-GSL-E2GFP. PEP1 (1 µM) triggered cytosolic Ca2+, H+, and Cl– dynamics that, during the initial 20 min of the response, exhibited a spatiotemporal overlap/correlation mainly in the meristematic and elongation zone (Figures 9A and 9B; Supplemental Movie 14). Together, these experiments established that PEP1 triggers spatiotemporally overlapping Ca2+, H+, and Cl– dynamics in roots. However, the effect of PEP1 and ATP on the cytosolic redox state was below the threshold of ROS-induced Ca2+ signaling (Supplemental Figure 7).
Figure 8.
Cytosolic Oxidation Is Only Weakly Affected by ATP and PEP1.
(A) and (B) Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1 and roGFP2-Orp1 (Ca2+ and H2O2; n = 7; see [A]) in response to 1 mM ATP and 100 µM H2O2 and R-GECO1-P2A-roGFP2-Orp1 (Ca2+ and H2O2; n = 7; see [B]) in response to 1 µM PEP1 and 100 µM H2O2. Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R) and (middle) signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. An adjacent representative bright field (BF) root image is shown for orientation. (Bottom) Full image signal changes (mean ± sd).
Representative experiments are provided as Supplemental Movies 10 and 12. See also Supplemental Figures 6 and 7 for related data and experiments.
Figure 9.
Application of PEP1 Triggers Spatiotemporally Overlapping Cytosolic Concentration Changes of Ca2+, H+, and Cl–.
Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1-GSL-E2GFP (Ca2+, pH, and Cl–; n = 6) in response to 1 µM PEP1 (t = 0 min) and 1 mM ATP (t = 30 min). Images were acquired for 64 min at a frame rate of 10 min−1.
(A) Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R) and (middle) signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. (Bottom) Full image signal changes (mean ± sd).
(B) Spatiotemporal Pearson correlation analyses (mean ± sd) of indicated GEFI responses during the application of PEP1 (t = –4 to 20 min). An adjacent representative bright field (BF) root image is shown for orientation.
A representative experiment is provided as Supplemental Movie 14.
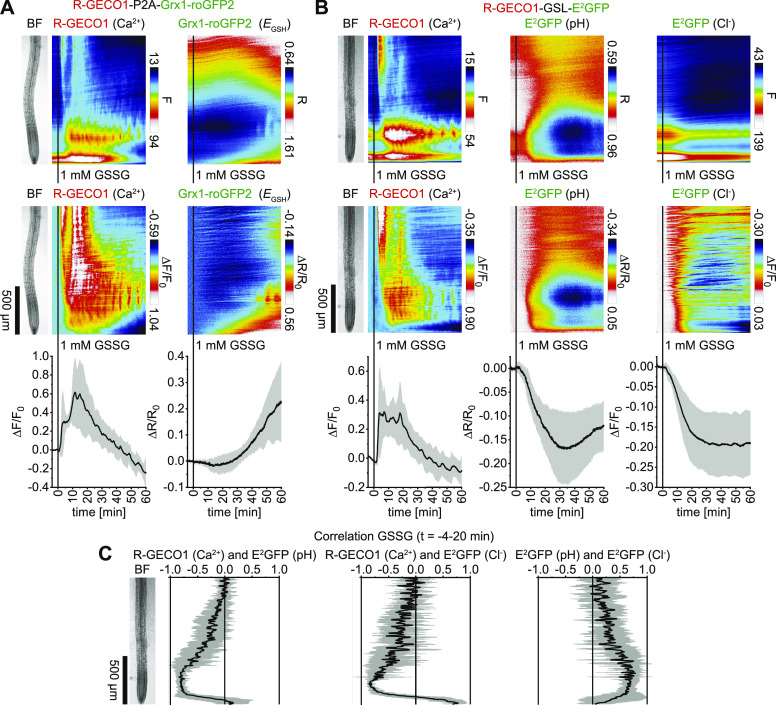
GSSG Treatments Trigger Rapid Cytosolic Ca2+, H+, and Cl– Dynamics That Precede a Slow-Progressing Cytosolic Oxidation
GSSG triggers cytosolic Ca2+ signals (Gomez et al., 2004) and directly oxidizes Grx1-roGFP2 (Gutscher et al., 2008). We sought to resolve the spatiotemporal relationships of these responses in Arabidopsis seedlings expressing R-GECO1-P2A-Grx1-roGFP2. Although 1 mM GSSG-induced Ca2+ signals were variable, they appeared to arrive from upper root regions and traveled toward the root tip, followed by a second Ca2+ burst in the vasculature and oscillations in the meristematic and elongation zone (Figure 10A, left; Supplemental Movie 15). After the initial Ca2+ signal reached the root tip, in this region a cytosolic oxidation was observed that slowly progressed toward the elongation zone, where oscillation became visible (Figure 10A, right; Supplemental Movie 15). Note that the Ca2+ and EGSH oscillations were shifted in phase (Supplemental Movie 15). Additional experiments using R-GECO1-GSL-E2GFP revealed that GSSG treatments also induced cytosolic H+ and Cl– influx, exhibiting the most noticeable spatiotemporal overlap with Ca2+ signals in the meristematic and early elongation zone during the initial 20 min of the GSSG response (Figures 10B and 10C; Supplemental Movie 16). In summary, GSSG treatments trigger Ca2+ signals, cytosolic acidification, and Cl– influx that precede a cytosolic oxidation.
Figure 10.
GSSG-Triggered Cytosolic Concentration Changes of Ca2+, H+, and Cl– Precede a Cytosolic Oxidation.
(A) and (B) Analyses of roots of 5-d-old Arabidopsis expressing R-GECO1-P2A-Grx1-roGFP2 (Ca2+ and EGSH; n = 5; see [A]) and R-GECO1-GSL-E2GFP (Ca2+, pH and Cl–; n = 6; see [B]) in response to 1 mM GSSG (t = 0 min). Images were acquired for 64 min at a frame rate of 10 min−1. Average vertical response profiles of (top) fluorescence emissions (F) or emission ratios (R) and (middle) signal changes (ΔF/F0 or ΔR/R0) normalized to 4-min average baseline recordings. (Bottom) Full image signal changes (mean ± sd).
(C) Spatiotemporal Pearson correlation analyses (mean ± sd) of indicated GEFI responses during the application of GSSG (t = –4 to 20 min; data from [B]). An adjacent representative bright field (BF) root image is shown for orientation.
Representative experiments are provided as Supplemental Movies 15 and 16.
DISCUSSION
Optimization of ABA Indicators in HEK293T Cells
Optimization procedures of FRET-based indicators usually comprise the testing of FRET-pair-, SD-, and linker variants (Okumoto et al., 2012; Hochreiter et al., 2015). Although such testing has been performed on ABACUS, the linkers between the SD and attached FPs remained invariant (Jones et al., 2014). Early optimizations of ABAleon focused on SD modifications that led to the development of ABAleon2.15 with improved stereospecificity for (+)-ABA (Waadt et al., 2014). Here, we have developed ABAleonSD1-3L21 that, compared to ABAleon2.15, exhibits an improved signal-to-noise ratio in the HEK293T cell testing system (Figure 1B). HEK293T cells are a convenient system for GEFI screenings because they can be easily transfected, cultivated, and analyzed in a plate reader (Tian, 2009). HEK293T cells contain neglectable ABA levels and are therefore well suited for ABA indicator screenings with a potential to facilitate the heterologous characterization of ABA transporters using ABA indicators. Successful characterizations of plasma membrane proteins in HEK293T cells have been demonstrated for RBOHs and CYCLIC NUCLEOTIDE-GATED ION CHANNEL (CNGC)–type Ca2+ channels (Gao et al., 2016; Han et al., 2019). The differences of ABAleon characteristics between HEK293T cell and in vitro analyses might be due to a lower stability of ABAleon2.15 in HEK293T cells. Similar issues have been reported for Ca2+ indicators (Tian, 2009). However, in vitro characterizations of ABA indicators are time consuming, and screenings using E. coli are not practical due to a likely even lower protein stability in this system (Jones et al., 2014; Waadt et al., 2014). Note that previously measured properties of ABAleon2.15 [ΔR(max)/R0 ∼ –0.10 and Kd ∼500 nM; Waadt et al., 2014] were different compared to results in Figures 1C and 1F. Here, we used a faster sandwich-tag purification procedure with subsequent characterization of freshly purified proteins that might give more reliable results.
ABAleon2.15 and ABAleonSD1-3L21 exhibited similar ABA responses in Arabidopsis. ABACUS1-2µ responded slower than ABAleons to ABA (Figure 2), probably due to the lower ABA affinity (Jones et al., 2014). However, this indicator might have advantages for the analyses of ABA dynamics in the root tip, the root–hypocotyl junction and in guard cells, where ABAleons are close to saturation (Figure 2; Waadt et al., 2015). In the future, optimization of ABA indicators will require the development of alternative SDs and the investigation of alternative biosensor designs to improve their dynamic range and minimize their influence on plant growth.
2-in-1-GEFIs Facilitate Simultaneous Multiparametric Analyses
Multiplexed live imaging in plants has been performed via the combination of GEFIs with fluorescent dyes, the use of GEFIs in parallel experiments, or through dual expression of Ca2+ indicators (Monshausen et al., 2007, 2009, 2011; Loro et al., 2012; Schwarzländer et al., 2012; Ngo et al., 2014; Keinath et al., 2015; Behera et al., 2018; Kelner et al., 2018; Wagner et al., 2019). However, GEFI-based simultaneous analyses of two signaling compounds have been established in Arabidopsis only for Ca2+ and ABA (Waadt et al., 2017). Because most GEFIs are FRET- or green FP-based, simultaneous multiparametric analyses have become possible through the development of red FP-based indicators for Ca2+, redox/H2O2, and pH (Bilan and Belousov, 2017; Martynov et al., 2018; Walia et al., 2018). Yet, except for R-GECO1, their application in plants is rare. Here, we introduced the intensiometric red fluorescing pH indicator (P)A-17 (Shen et al., 2014), which is well suited for pH analyses in Arabidopsis with similar responsiveness compared to the ratiometric E2GFP (Figure 3B, left; Figure 4A, middle; Figure 5A, middle).
As the generation of stable transgenic organisms is time consuming, simultaneous expression of GEFIs, or the generation of dual-reporting GEFIs, is advantageous. Moreover, the latter approach minimizes epigenetic silencing effects, often observed in lines carrying multiple transgenes (Behera et al., 2015). Dual-reporting GEFIs have been developed for pH and Cl– (ClopHensor; Arosio et al., 2010) and for phosphatidylinositol 3,4,5-trisphosphate localization and H2O2 concentration (PIP-SHOW; Mishina et al., 2012). For the generation of 2-in-1-GEFIs, we were inspired by ClopHensor and the incorporated E2GFP that we fused with R-GECO1 in analogy to R-GECO1-GSL-mTurquoise (Waadt et al., 2017). Note that recent studies indicated that ClopHensor/E2GFP also responds to NO3– (Demes et al., 2019). Because our imaging buffer contained 5 mM Cl– and the microscope-dish agarose medium contained 4 mM NO3–, the observed E2GFP responses likely depended on both anion species. In contrast to R-GECO1-GSL-E2GFP, the other 2-in-1-GEFIs have been linked via the self-cleaving P2A-peptide, which enables efficient cleavage in Arabidopsis (Supplemental Figures 4 and 5; Burén et al., 2012). In addition, P2A-based 2-in-1-GEFIs exhibit similar responses compared to indicators expressed from separate expression cassettes (Figure 8; Supplemental Figure 6). However, because only one expression cassette is used, P2A-linked GEFIs can be more easily screened at the microscope. Our work established several 2-in-1-GEFIs based on the P2A-linkage, which is applicable to any eukaryotic system (Kim et al., 2011).
Cytosolic Ca2+, H+, and Anion Dynamics Exhibit a High Spatiotemporal Overlap
Previous work established that mechanical stimulation, wounding, ATP, and auxin simultaneously induce Ca2+ and pH dynamics (Monshausen et al., 2009, 2011; Behera et al., 2018). We found that, in addition to auxin and ATP, also glutamate, PEP1, and GSSG trigger cytosolic Ca2+, H+, and anion dynamics with high spatiotemporal overlap (Supplemental Figure 8). The linkage of Ca2+ and H+ dynamics may depend on H+ pumps and Ca2+/H+-coupled transport via CATION/PROTON EXCHANGERS (CAXs) or Ca2+-ATPases (Bonza and De Michelis, 2011; Pittman and Hirschi, 2016). However, knowledge about their role in Ca2+ signaling is fragmentary, probably due to functional overlap of gene family members (Behera et al., 2018). Simultaneous Ca2+ and H+ dynamics in response to auxin are mediated by the auxin/H+-symporter AUXIN RESISTANT1 (AUX1) and the Ca2+ channel CNGC14 that are functionally coupled (Shih et al., 2015; Dindas et al., 2018). Since the activation of plasma membrane H+-ATPases is coupled to AUX1 (Inoue et al., 2016), this could explain the subsequent cytosolic H+ efflux.
Mechanical stimulation-induced Ca2+ and H+ dynamics depend on the RECEPTOR-LIKE KINASE (RLK) FERONIA, which acts as a receptor for RAPID ALKALIZATION FACTOR (RALF) peptides (Haruta et al., 2014; Shih et al., 2014; Stegmann et al., 2017). Several RLKs, including the ATP receptor DOES NOT RESPOND TO NUCLEOTIDES1 (DORN1) and PEP RECEPTORS (PEPRs), can induce Ca2+ signals, apoplastic alkalization (coupled to cytosolic acidification), and ROS production (Qi et al., 2010; Choi et al., 2014; Ma et al., 2014; Seybold et al., 2014; Haruta et al., 2015; Chen et al., 2017; Kimura et al., 2017). The effect of DORN1 and PEPRs on cytosolic anion efflux was observed in guard cells during stomatal closure (Chen et al., 2017; Zheng et al., 2018). Our analyses revealed that PEP1 induced a transient cytosolic anion influx along the entire imaged root axis. Extracellular ATP triggered cytosolic anion influx in the differentiation and elongation zone but efflux in the meristematic zone (Figure 9). Early research revealed that cytosolic, but not extracellular, ATP is required for adenine nucleotide activation of R-type anion channels (Hedrich et al., 1990) and protein kinase–mediated activation of S-type anion channels (Schmidt et al., 1995). It is likely that ATP-triggered Ca2+ signals activate Ca2+-dependent protein kinases required for the activation of anion channels (Mori et al., 2006). The differences in PEP1- and ATP-induced cytosolic anion dynamics in roots might be due to the distinct Ca2+ signatures observed in the meristematic zone (Figure 9).
Several transporters and channels are known to be permeable for anions such as Cl– and NO3– (Hedrich, 2012; Li et al., 2017; Colmenero-Flores et al., 2019). Among them, members of the NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER (NPF) family contribute to the import of these anions into the cytosol via active anion (A–)/H+ symport. Passive export of Cl– and NO3– from the cytosol is facilitated by SLOW-TYPE ANION CHANNEL1 HOMOLOGS (SLAC1/SLAH), ALUMINUM-ACTIVATED MALATE TRANSPORTERS (ALMTs), and members of the NPF-type NITRATE EXCRETION TRANSPORTER (NAXT) subfamily. In addition, Cl– and NO3– sequestration into the vacuole is facilitated by members of the CHLORIDE CHANNEL (CLC) family, the DETOXIFICATION EFFLUX CARRIERS DTX33 and DTX35, and by AtALMT9 (Li et al., 2017; Colmenero-Flores et al., 2019). Interestingly, NPFs, SLAC1/SLAHs, and Brachypodium distachyon BdALMT12 are subject to Ca2+-dependent posttranslational regulation (Kudla et al., 2018; Luu et al., 2019). In the future, it will be interesting to identify the transporters and channels that contribute to the observed anion dynamics in Arabidopsis roots and to elucidate their potential interconnection with Ca2+ signaling mechanisms (Supplemental Figure 8).
On the Interdependence of Ca2+ and ROS Signaling
The interdependence of Ca2+ and ROS signaling has been extensively discussed by Gilroy et al. (2014) and Steinhorst and Kudla (2014). In the context of long-distance and systemic signaling, current models propose that Ca2+ signals trigger the ion channel TPC1 for signal amplification. Ca2+ signal propagation occurs via plasmodesmata or Ca2+-dependent activation of RBOHs. RBOH-derived extracellular ROS propagate to adjacent cells to activate plasma membrane-localized Ca2+ channels (Choi et al., 2016; Evans et al., 2016). Ca2+-dependent activation of RBOHs is well established (Kadota et al., 2015; Han et al., 2019). However, whether RBOH-dependent ROS contribute to Ca2+ channel activation, has only been inferred from pharmacological and genetic analyses (Kwak et al., 2003; Evans et al., 2016). In Arabidopsis guard cells, hyperpolarization-activated Ca2+-permeable channels can be activated by 50 µM H2O2 (Pei et al., 2000). In Vicia faba guard cells such channels exhibit a half response at 76 µM H2O2 (Köhler et al., 2003). Analyses in root epidermis cells revealed a Ca2+ channel activation by 10 µM H2O2 in the elongation zone and by 1 mM H2O2 in the maturation zone (Demidchik et al., 2007). The threshold concentrations of ROS required to activate Ca2+ channels may depend on the cell type, the location (apoplast or cytosol), and the chemical nature of ROS (Demidchik et al., 2007).
Our analyses revealed that in Arabidopsis roots 20 µM extracellular H2O2 triggered a detectable cytosolic oxidation, but no considerable Ca2+ signals (Figure 7). By contrast, glutamate, ATP, and PEP1, which efficiently trigger Ca2+ signals, induced a cytosolic oxidation rather below this threshold (Supplemental Figure 7). These data are consistent with a slow progressing cytosolic oxidation in response to the pathogen-associated molecular pattern flagellin fragment flg22 (Nietzel et al., 2019). Whether such cytosolic oxidation is Ca2+ dependent requires further experimentation. However, compared to 20 µM H2O2 responses, our data suggest that glutamate-, ATP-, and PEP1-induced ROS production, inferred from their effects on the cytosolic redox status, is not sufficient to trigger root Ca2+ channels (Supplemental Figure 7). We hypothesize that the observed ROS dependence of Ca2+ signaling may be indirectly linked to the effects of ROS on the cell wall, which binds considerable amounts of Ca2+ in Ca2+-pectate cross-linked complexes (Hepler and Winship, 2010; Peaucelle et al., 2012; Kärkönen and Kuchitsu, 2015). Such a model would be consistent with a rather slow H2O2 activation of Ca2+ channels in patch-clamp analyses (20 to 60 min; Demidchik et al., 2007). Receptor kinases of the gene family of HYDROGEN-PEROXIDE-INDUCED Ca2+ INCREASES1 (HPCA1) also likely contribute to H2O2-induced Ca2+ signaling (Wu et al., 2020). Another possibility would be that a signaling component triggers both Ca2+ and ROS signaling. The BOTRYTIS-INDUCED KINASE1 (BIK1) could be a good candidate for such a mechanism (Liu et al., 2013; Kadota et al., 2014; Li et al., 2014; Kimura et al., 2017; Tian et al., 2019).
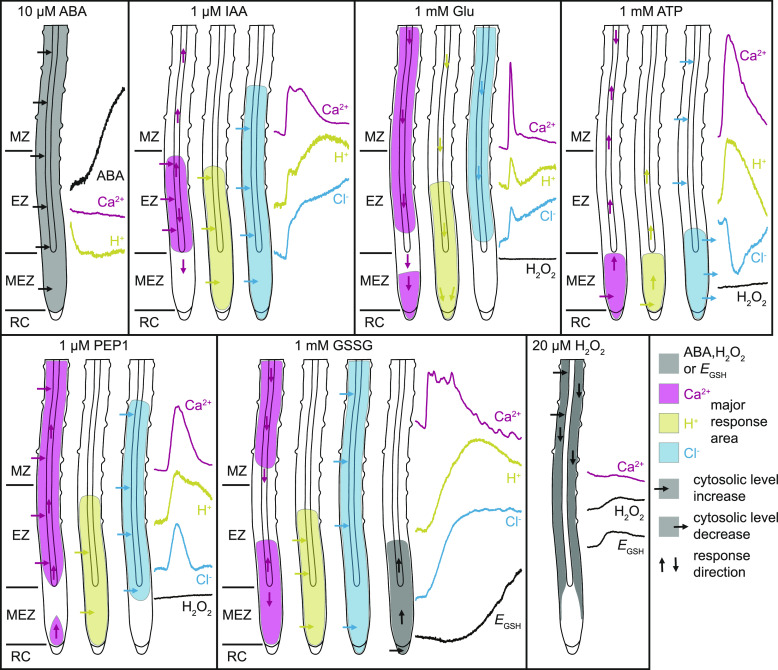
Conclusions
Our work established 2-in-1-GEFI–based simultaneous multiparametric in vivo analyses of signaling compounds in Arabidopsis. Using the 2-in-1-GEFIs, we observed that in roots ABA does not trigger rapid cytosolic Ca2+ or pH changes. However, auxin, glutamate, ATP, PEP1, and GSSG induce cytosolic Ca2+, H+, and anion dynamics with high spatiotemporal overlap (Figure 11; Supplemental Figure 8). These results suggest an interdependence and coordination of ion fluxes that need to be dissected in future research. Findings that glutamate-, ATP-, and PEP1-induced cytosolic oxidation is below the threshold required for triggering Ca2+ channels argue against the current model of a ROS-assisted Ca2+ signal propagation mechanism (Evans et al., 2016). We hypothesize that ROS may have an indirect effect on Ca2+ signaling. Overall, 2-in-1-GEFI–based imaging will allow for high-resolution in vivo analyses of signaling processes beyond the model plant Arabidopsis.
Figure 11.
Schematic Model of Observed Cytosolic ABA, Ca2+, H+, Cl–, and Redox Changes in Arabidopsis Roots.
ABA treatment and uptake did not induce rapid cytosolic Ca2+ or H+ concentration changes. By contrast, IAA, glutamate (Glu), ATP, PEP1, and GSSG triggered cytosolic Ca2+, H+, and Cl– concentration changes with high spatiotemporal overlap. For comparison of the obtained data see also Supplemental Figure 8 and Supplemental Movies 4, 5, 14, and 16. Compared to 20 µM H2O2 and 1 mM GSSG, redox changes in response to Glu, ATP, and PEP1 were very low and below the threshold required to trigger ROS-induced Ca2+ signaling. Regions with highest response are color coded according to the adjacent response curves that were taken from the corresponding main figures (ABA, H2O2, and EGSH, black; Ca2+, magenta; H+, yellow; Cl–, cyan). For a better illustration of cytosolic H+ and Cl– concentration increases, the PA-17 and E2GFP response curves were inverted. Horizontal arrows indicate cytosolic concentration increase or decrease and vertical arrows the direction of the response. EZ, elongation zone; MEZ, meristematic zone; MZ, maturation zone; RC, root cap.
METHODS
Generation of Plasmids
Oligonucleotides used for cloning procedures (Supplemental Data Set 1A) were obtained from Eurofinsgenomics. Plasmids (Supplemental Data Set 1B) were constructed using classical cloning procedures and the GreenGate system (Lampropoulos et al., 2013) utilizing enzymes from Thermo Fisher Scientific. Arabidopsis codon-optimized DNA fragments of PmTurquoise and PA-17 were designed using GeneArt gene synthesis (Thermo Fisher Scientific). Expression of GEFIs in Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) was performed utilizing the promoter of a ubiquitous and highly expressed reference gene ASPARTIC PROTEASE A1 (APA1, AT1G11910) that was chosen based on searches using Genevestigator (Hruz et al., 2008). The expression cassette also included the terminator of the HEAT SHOCK PROTEIN18.2 (HSP18.2) gene (AT5G59720; Nagaya et al., 2010; Waadt et al., 2014).
Optimization of ABA Indicators in HEK293T Cells
Transformation and cultivation of HEK293T cells were performed as described previously (Ogasawara et al., 2008; Zhang et al., 2018). Spectral characteristics of ABA indicators were recorded in flat-bottomed 96-well plates (Greiner BIO-ONE) using a Safire plate reader (TECAN) operated by XFLUOR4.51 software with the following parameters: fluorescence emission scan, bottom mode; excitation, wavelength 440 nm, bandwidth, 12.5 nm; emission wavelength scan from 460 to 600 nm, bandwidth, 10 nm; gain, 100 to 115; flashes, 10; integration time, 40 to 60 µs; and temperature, 37°C. HEK293T cells were kept in Hank’s balanced salt solution (HBSS; Thermo Fisher Scientific), and fluorescence emission spectra were recorded before (t0) and 60 min (t60) after exchange of solution to either HBSS with 100 µM (±)-ABA (Merck) and 0.1% (v/v) ethanol (treatment) or HBSS with 0.1% (v/v) ethanol (solvent control). ABA indicator emission ratios were calculated as average emission at 518 to 538 nm divided by average emission at 470 to 490 nm after subtraction of the nontransfected HEK293T cell background emission spectrum. ΔR/R0 was calculated as [R(t60) – R(t0)]/R(t0). Experiments were performed in triplicates.
Purification and In Vitro Characterization of ABAleons
BL21-CodonPlus (DE3)-RIL cells transformed with pET28-6xHis-ABAleon-(P)StrepII constructs were shaken at 150 rpm and 37°C in 2× 1 liter of Luria-Bertani medium supplemented with 50 µg mL−1 kanamycin and 30 µg mL−1 chloramphenicol. At an OD600 of 0.5, 1 mM isopropyl β-d-1-thiogalactopyranoside (Carl Roth) was added, and protein expression was conducted in a shaking incubator at 24°C for 6 h. Cultured cells were harvested by several centrifugation steps at 4°C and 5000g, and the bacterial pellet was flash frozen in liquid N2 and stored at –80°C.
The bacterial pellet was thawed on ice and resuspended in 30 mL of lysis buffer (1× PBS [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4], 10 mM imidazole [Merck], 1× Roche protease inhibitor EDTA-free, 1 mM phenylmethylsulfonyl fluoride [Carl Roth], and 1 mg mL−1 lysozym [VWR], pH 7.4). After 40 to 60 min of incubation on ice, cells were disrupted through microtip-based sonication (25% amplitude, 21× 20 s), and cell debris were removed by centrifugation (2× 30 min; 20,000g at 4°C) and filtering through 0.45-µm syringe filters (Merck).
The 6×-His purification was conducted in 20-mL gravity columns (VWR) loaded with 4 mL of HisPur nickel-nitrilotriacetic acid resin (Thermo Fisher Scientific). After binding of proteins to the nickel-nitrilotriacetic acid resin, columns were washed five times with 10 mL of His-wash buffer (1× PBS and 25 mM imidazole, pH 7.4), and proteins were eluted in 3× 2 mL of His-elution buffer (1× PBS and 250 mM imidazole, pH 7.4). Purified proteins were then loaded onto a 20-mL gravity column supplemented with 3 mL of 50% (v/v) Strep Tactin Superflow (IBA). After washing four times with 7.5 mL of SII-wash buffer 1 (30 mM Tris-HCl, pH 7.4, and 250 mM NaCl) and washing three times with 7.5 mL of SII-wash buffer 2 (30 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10 mM MgCl2, 1 mM MnCl2), proteins were eluted in 3x 1.5 mL of SII-elution buffer (30 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10 mM MgCl2, 1 mM MnCl2, and 2.5 mM desthiobiotin; IBA) and concentrated to a final volume of ∼1 mL using Amicon Ultra-4 30 K filters (Merck). Purity of proteins was analyzed by SDS-PAGE using 10% Mini-PROTEAN TGX Precast Gels (Bio-Rad) and InstantBlue staining (Expedeon). In a similar procedure, protein yield was calculated according to a BSA standard curve.
For vitro calibration, a 100 mM (+)-ABA (TCI) stock solution dissolved in 100% (v/v) ethanol was used for an ABA dilution series in SII-wash buffer 2 and 0.2% (v/v) ethanol. Ten microliters of each ABA dilution were added to three wells of black flat-bottomed µCLEAR 96-well plates (Greiner BIO-ONE) containing 90 μL of ∼1.1 µM ABAleon protein, diluted in SII-wash buffer 2, or to 90 μL SII-wash buffer 2 alone as background control. Fluorescence emission spectra were recorded after 20 min of incubation at room temperature in the dark using a TECAN Infinite M1000 plate reader (TECAN) operated by the i-control 1.10.4.0 software with the following parameters: fluorescence emission scan, bottom mode; excitation wavelength, 440 nm; bandwidth, 10 nm; emission wavelength scan from 460 to 650 nm; bandwidth, 10 nm; gain, 98 to 104; flashes, 10 at 100 Hz; integration time, 60 µs; temperature, 21 to 22°C. ABA-dependent ABAleon emission ratios were calculated as described above. ΔR(max)/R0 was calculated as [R(at 20 µM ABA) – R(at 0 µM ABA)]/R(at 0 µM ABA). Apparent ABA Kd of ABAleons were calculated by fitting the emission ratio values of all three replicates to a four-parameter logistic function using OriginPro 2018 (OriginLab).
Generation of Transgenic Arabidopsis Plants Expressing GEFIs
Agrobacterium strain ASE containing the pSOUP helper plasmid and the respective plant expression vectors (Supplemental Data Set 1B) were used for transformation of Arabidopsis ecotype Col-0 by floral dip (Clough and Bent, 1998) to generate the transgenic lines listed in Supplemental Data Set 1C. Seeds of transformed plants were surface sterilized for 10 to 15 min in 70% (v/v) ethanol, washed three times with 100% (v/v) ethanol, and sowed on 0.5 MS medium (Duchefa) supplemented with 5 mM MES-KOH, pH 5.8, 0.8% (w/v) phytoagar (Duchefa), and 10 µg mL−1 glufosinate-ammonium or 25 µg mL−1 hygromycin B (Merck) for herbicide selection. After 3 to 6 d of stratification in the dark at 4°C, transgenic plants were grown for 6 d in a growth room (16-h-light/8-h-dark cycle; 22°C; 65% RH; photon fluence rate, 100 µmol m−2 s−1, Philips Green Power LED deep red/blue 120 LO). Positive transformants were then transferred to herbicide-free 0.5 MS medium–containing Petri dishes. After 1 d of recovery, GEFI expression was confirmed by visual inspection using a Discovery.V20 fluorescence stereo microscope (Zeiss) equipped with GFP, yellow fluorescent protein, and red fluorescent protein filters and a Plan S 1.0× FWD 81-mm lens. Approximately 40 herbicide-resistant and fluorescing seedlings were then transferred to round 7-cm pots containing classic soil (Einheitserde) and grown until seed ripening in the growth room. ABAleon-expressing plants were covered with a plastic lid and grown in a Conviron CMP6010 growth cabinet (16-h-light/8-h-dark cycle; 20°C; 65% RH; photon fluence rate, 150 µmol m−2 s−1, Philips Alto F39T5/841 light bulbs). To confirm proper GEFI expression, compare GEFI fluorescence emissions and avoid silencing effects in next generations, one leaf of each individual ∼3-week-old plant was examined using an SP5 II confocal laser scanning microscope (Leica) equipped with a HCX PL APO CS 20.0 × 0.7 IMM UV light objective (Leica Microsystems) using emission and excitation settings listed in Supplemental Data Set 1D. For each construct, at least two transgenic lines with highest GEFI expression, proper 3:1 segregation in the second generation, and least silencing were selected for further propagation. One line, indicated with “(microscope)” in Supplemental Data Set 1C, was used for microscopic experiments.
Phenotypic Characterization of GEFI Lines
Seeds were surface sterilized and sown on 0.5 MS medium supplemented with 5 mM MES-KOH, pH 5.8, and 0.8% (w/v) phytoagar. After stratification for 6 d, plants were grown in the growth room (16-h-light/8-h-dark cycle; 22°C; 65% RH; photon fluence rate, 100 µmol m−2 s−1, Philips Green Power LED deep red/blue 120 LO), and images were acquired from 5-d-old vertically grown seedlings. Six- to 7-d-old seedlings were then transferred to soil in single pots and further grown in the growth room until 25 to 28 d old. ABAleon and respective control plants were grown as described above. Pictures from 7 to 12 plants per genotype were acquired from the top, and rosette areas were extracted using the magnetic lasso tool in Photoshop CS6 (Adobe) or the Rosette Tracker Fiji plugin (De Vylder et al., 2012) and quantified using Fiji (Schindelin et al., 2012). Raw data of rosette area quantifications are presented in Supplemental Data Set 2.
Microscopic Analyses
Seeds of GEFI-expressing lines were surface sterilized and sown in four horizontal rows on square Petri dishes containing Long Ashton mineral solution modified to contain 1 mM K+ [LAK medium; 1 mM KH2PO4, 2 mM Ca(NO3)2, 1 mM MgSO4, 30 µM H3BO3, 10 µM MnSO4, 1 µM ZnSO4, 1 µM CuSO4, 0.03 µM (NH4)6Mo7O24, and 50 µM FeNaEDTA; Barragán et al., 2012] supplemented with 10 mM MES-Tris, pH 5.6, and 0.8% (w/v) phytoagar. After 6 d of stratification in the dark at 4°C, seedlings were grown vertically in a Conviron CMP 6010 growth cabinet (16-h-light/8-h-dark cycle; 22°C; 65% RH; photon fluence rate, 150 µmol m−2 s−1, Philips Alto F39T5/841 light bulbs). After 4 d, seedlings were transferred to microscope dishes (MatTek) containing 200 μL of polymerized LAK medium supplemented with 10 mM MES-Tris, pH 5.6, and 0.7% (w/v) low melting point agarose (Carl Roth). Seedlings were incubated vertically overnight in the growth cabinet until the microscopic experiments were conducted.
Before microscopic analyses, seedlings on microscope dishes were placed horizontally and topped with 90 μL of imaging buffer (5 mM KCl, 50 µM CaCl2, and 10 mM MES-Tris, pH 5.6; Allen et al., 2001). Using a 200-μL pipette tip, seedlings were gently attached back to the LAK medium-low melting point agarose bed and incubated for 10 to 50 min for recovery, until the GEFI fluorescence emission baseline was stable. Microscopic analyses were performed with Leica SP5 II and Leica SP8 confocal laser scanning microscopes using a 10× air objective and photomultiplier tube detectors (Leica Microsystems). Microscope settings were as follows: image format, 1024 × 178 pixels (1536 × 256 pixels for RW300 experiment); bidirectional scanning at 400 Hz; zoom, 0.75 (SP8) or 1 (SP5 II and RW300 experiment at SP8); pinhole, 5 airy units; line accumulation, 2 (SP5 II) and 1 (SP8); line average, 1 (SP5 II) and 2 (SP8); offset, –0.4% for blue, cyan, green, and yellow emissions and –0.2% for red emissions; and frame rate, 6 s. Laser intensities and gain settings were optimized for each GEFI and kept stable for all experimental replicates. Emission and excitation settings for each GEFI are listed in Supplemental Data Set 1D. After 4-min baseline recordings, chemical treatments were performed by dropping 10 μL of 10-fold concentrated stock solutions (Supplemental Data Set 1E) close to the imaged region.
Image processing and analyses were conducted using Fiji (Schindelin et al., 2012). Image processing included background subtraction (2-4), gaussian blur (1), median (1), 32-bit conversion, thresholding of background noise (2-5), and ratio image calculation for ratiometric GEFIs. Normalized fluorescence intensity (ΔF/F0), ΔR/R0, Pearson correlation, and root tip–localized, time-dependent vertical response profile analyses were conducted using a custom-built Fiji plugin (https://github.com/RainerWaadt/GEFI_Analyzer). Fluorescence emissions (F) and emission ratios (R) were measured as average values from each entire processed movie frame, and signal changes (ΔF/F0 and ΔR/R0) were calculated relative to the average value of a 4-min baseline recording as [F(t) – F(baseline)]/F(baseline)] and [R(t) – R(baseline)]/R(baseline)]. Means and sd of experimental replicates were calculated using Excel (Microsoft). For time-dependent vertical response profiles, root tips were detected within each movie frame, and regions of interest were drawn to cover the entire x axis and a defined area above the root tip. Vertical response profiles were calculated from each movie frame region of interest as average of all x axis pixel values within each y axis pixel line (similar to the Plot Profile command in Fiji) and plotted in a time-dependent manner. Time-dependent signal change vertical response profiles were calculated using the raw response profiles as a basis and applying the signal change formulas to each y axis pixel line. Average time-dependent vertical response profiles of multiple experimental replicates were generated using the average Z projection command in Fiji. Pearson correlation analyses were conducted on selected time windows of respective vertical response profiles and correlation coefficients were calculated along the y axis. Means and sd of experimental replicates were calculated using Excel (Microsoft).
Statistical Analysis
For phenotypic analyses presented in Supplemental Figure 3 and Supplemental Data Set 2, box plot and statistical analyses using pairwise one-way -ANOVA Tukey test comparisons relative to Col-0 wild type were conducted using OriginPro (OriginLab).
Accession Numbers
The Arabidopsis Genome Initiative locus numbers for the genes used in this article are as follows: ABI1 (AT4G26080); APA1 (AT1G11910); AtPEP1 (AT5G64900); HSP18.2 (AT5G59720); LTI6b (AT3G05890); PYL1 (AT5G46790); PYR1 (AT4G17870); VTI11 (AT5G39510).
Supplemental Data
Supplemental Figure 1. Topologies of ABA indicators.
Supplemental Figure 2. Solvent control experiments of ABA indicators in Arabidopsis.
Supplemental Figure 3. Plant growth phenotype analyses of transgenic GEFI lines.
Supplemental Figure 4. Targeting of E2GFP to the plasma membrane.
Supplemental Figure 5. Targeting of E2GFP to the tonoplast.
Supplemental Figure 6. Cytosolic EGSH is only weakly affected by ATP and PEP1 treatments.
Supplemental Figure 7. Glutamate-, ATP- and PEP1-dependent cytosolic oxidation is below the threshold of ROS-induced Ca2+ signaling.
Supplemental Figure 8. Cytosolic Ca2+, H+ and anion concentration changes exhibit a high spatiotemporal overlap.
Supplemental Data Set 1. Lists of materials, imaging settings and chemicals used in this work.
Supplemental Data Set 2. Raw data of rosette area quantifications.
Supplemental Movie 1. ABA indicator ABA responses in Arabidopsis.
Supplemental Movie 2. ABAleonSD1-3L21-P2A-R-GECO1 in response to ABA and IAA.
Supplemental Movie 3. PA-17-P2A-ABAleonSD1-3L21 in response to ABA and ATP.
Supplemental Movie 4. R-GECO1-GSL-E2GFP in response to IAA and ATP.
Supplemental Movie 5. R-GECO1-GSL-E2GFP in response to glutamate and ATP.
Supplemental Movie 6. PA-17-P2A-Grx1-roGFP2 in response to glutamate and H2O2.
Supplemental Movie 7. PA-17-P2A-roGFP2-Orp1 in response to glutamate and H2O2.
Supplemental Movie 8. R-GECO1-P2A-Grx1-roGFP2 in response to H2O2.
Supplemental Movie 9. R-GECO1-P2A-roGFP2-Orp1 in response to H2O2.
Supplemental Movie 10. R-GECO1 and roGFP2-Orp1 in response to ATP and H2O2.
Supplemental Movie 11. R-GECO1 and Grx1-roGFP2 in response to ATP and H2O2.
Supplemental Movie 12. R-GECO1-P2A-roGFP2-Orp1 in response to PEP1 and H2O2.
Supplemental Movie 13. R-GECO1-P2A-Grx1-roGFP2 in response to PEP1 and H2O2.
Supplemental Movie 14. R-GECO1-GSL-E2GFP in response to PEP1 and ATP.
Supplemental Movie 15. R-GECO1-P2A-Grx1-roGFP2 in response to GSSG.
Supplemental Movie 16. R-GECO1-GSL-E2GFP in response to GSSG.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the groups at Centre for Organismal Studies (Heidelberg) for generous access to equipment and GreenGate modules, Jana Hakenjos for initial help with ABAleon purifications, Andreas Meyer (University of Bonn) for roGFP2 PCR templates, Eugenia Russinova (Vlaams Instituut voor Biotechnologie Gent) for providing the AtPEP1 peptide, and Shintaro Munemasa (Okayama University) for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (grant WA 3768/1-1 to R.W., AN 1323/1-1 to Z.A., and KU 931/14-1 to J.K.).
AUTHOR CONTRIBUTIONS
R.W. conceived the project, generated most of the plasmids and transgenic plants, conducted the in vitro characterization of ABAleons, contributed to the phenotype analyses in Supplemental Figure 3, performed all microscope analyses, and wrote the article. P.K. conducted the characterization of ABA indicators in HEK293T cells and revised the article. Z.A. generated the transgenic lines RW307 and RW308, contributed to the phenotype analyses in Supplemental Figure 3, and revised the article. C.W. developed the GEFI Analyzer Fiji plugin and revised the article. G.B. generated the plasmids indicated with GB and conducted preliminary ABA indicator characterizations. K.L. introduced A-17 and generated PA-17 GreenGate modules. J.K. supervised the ABA indicator characterization in HEK293T cells and revised the article. K.S. supervised and hosted the project.
References
- Allen G.J., Chu S.P., Harrington C.L., Schumacher K., Hoffmann T., Tang Y.Y., Grill E., Schroeder J.I.(2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Arosio D., Ricci F., Marchetti L., Gualdani R., Albertazzi L., Beltram F.(2010). Simultaneous intracellular chloride and pH measurements using a GFP-based sensor. Nat. Methods 7: 516–518. [DOI] [PubMed] [Google Scholar]
- Barragán V., Leidi E.O., Andrés Z., Rubio L., De Luca A., Fernández J.A., Cubero B., Pardo J.M.(2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S., Wang N., Zhang C., Schmitz-Thom I., Strohkamp S., Schültke S., Hashimoto K., Xiong L., Kudla J.(2015). Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytol. 206: 751–760. [DOI] [PubMed] [Google Scholar]
- Behera S., Zhaolong X., Luoni L., Bonza M.C., Doccula F.G., De Michelis M.I., Morris R.J., Schwarzländer M., Costa A.(2018). Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilan D.S., Belousov V.V.(2017). New tools for redox biology: From imaging to manipulation. Free Radic. Biol. Med. 109: 167–188. [DOI] [PubMed] [Google Scholar]
- Bizzarri R., Arcangeli C., Arosio D., Ricci F., Faraci P., Cardarelli F., Beltram F.(2006). Development of a novel GFP-based ratiometric excitation and emission pH indicator for intracellular studies. Biophys. J. 90: 3300–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza M.C., De Michelis M.I. (2011). The plant Ca2+-ATPase repertoire: Biochemical features and physiological functions. Plant Biol. (Stuttg.) 13: 421–430. [DOI] [PubMed] [Google Scholar]
- Burén S., Ortega-Villasante C., Otvös K., Samuelsson G., Bakó L., Villarejo A.(2012). Use of the foot-and-mouth disease virus 2A peptide co-expression system to study intracellular protein trafficking in Arabidopsis. PLoS One 7: e51973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Cao Y., Li H., Kim D., Ahsan N., Thelen J., Stacey G.(2017). Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., Lee S.Y., Stacey G.(2014). Identification of a plant receptor for extracellular ATP. Science 343: 290–294. [DOI] [PubMed] [Google Scholar]
- Choi W.G., Hilleary R., Swanson S.J., Kim S.H., Gilroy S.(2016). Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 67: 287–307. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colmenero-Flores J.M., Franco-Navarro J.D., Cubero-Font P., Peinado-Torrubia P., Rosales M.A.(2019). Chloride as a beneficial macronutrient in higher plants: New roles and regulation. Int. J. Mol. Sci. 20: 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R.(2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- De Vylder J., Vandenbussche F., Hu Y., Philips W., Van Der Straeten D.(2012). Rosette tracker: An open source image analysis tool for automatic quantification of genotype effects. Plant Physiol. 160: 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., Shabala S.N., Davies J.M.(2007). Spatial variation in H2O2 response of Arabidopsis thaliana root epidermal Ca2+ flux and plasma membrane Ca2+ channels. Plant J. 49: 377–386. [DOI] [PubMed] [Google Scholar]
- Demidchik V., et al. (2009). Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca2+ channels. Plant J. 58: 903–913. [DOI] [PubMed] [Google Scholar]
- Dindas J., Scherzer S., Roelfsema M.R.G., von Meyer K., Müller H.M., Al-Rasheid K.A.S., Palme K., Dietrich P., Becker D., Bennett M.J., Hedrich R.(2018). AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 9: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D.(2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620. [DOI] [PubMed] [Google Scholar]
- Demes E., Besse L., Satiat-Jeunemaitre B., Thomine S., De Angeli A.(2019). Dynamic measurement of cytosolic pH and [NO3–] uncovers the role of the vacuolar transporter AtCLCa in the control of cytosolic pH. bioRxiv 716050, doi:10.1101/716050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel K.H., Kudla J.(2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33: 83–91. [DOI] [PubMed] [Google Scholar]
- Eisenach C., De Angeli A.(2017). Ion transport at the vacuole during stomatal movements. Plant Physiol. 174: 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Choi W.G., Gilroy S., Morris R.J.(2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171: 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.(2013). Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.F., Gu L.L., Wang H.Q., Fei C.F., Fang X., Hussain J., Sun S.J., Dong J.Y., Liu H., Wang Y.F.(2016). Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 3096–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S., Suzuki N., Miller G., Choi W.G., Toyota M., Devireddy A.R., Mittler R.(2014). A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19: 623–630. [DOI] [PubMed] [Google Scholar]
- Gomez L.D., Noctor G., Knight M.R., Foyer C.H.(2004). Regulation of calcium signalling and gene expression by glutathione. J. Exp. Bot. 55: 1851–1859. [DOI] [PubMed] [Google Scholar]
- Grossmann G., Krebs M., Maizel A., Stahl Y., Vermeer J.E.M., Ott T.(2018). Green light for quantitative live-cell imaging in plants. J. Cell Sci. 131: 131. [DOI] [PubMed] [Google Scholar]
- Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P.(2008). Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5: 553–559. [DOI] [PubMed] [Google Scholar]
- Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P.(2009). Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 284: 31532–31540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.P., Köster P., Drerup M.M., Scholz M., Li S., Edel K.H., Hashimoto K., Kuchitsu K., Hippler M., Kudla J.(2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221: 1935–1949. [DOI] [PubMed] [Google Scholar]
- Haruta M., Gray W.M., Sussman M.R.(2015). Regulation of the plasma membrane proton pump (H(+)-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 28: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R.(2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Li Z., Waadt R., Schroeder J.I.(2017). SnapShot: Abscisic acid signaling. Cell 171: 1708–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Waadt R., Schroeder J.I.(2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21: R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R.(2012). Ion channels in plants. Physiol. Rev. 92: 1777–1811. [DOI] [PubMed] [Google Scholar]
- Hedrich R., Busch H., Raschke K.(1990). Ca2+ and nucleotide dependent regulation of voltage dependent anion channels in the plasma membrane of guard cells. EMBO J. 9: 3889–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Winship L.J.(2010). Calcium at the cell wall-cytoplast interface. J. Integr. Plant Biol. 52: 147–160. [DOI] [PubMed] [Google Scholar]
- Hilleary R., Choi W.G., Kim S.H., Lim S.D., Gilroy S.(2018). Sense and sensibility: The use of fluorescent protein-based genetically encoded biosensors in plants. Curr. Opin. Plant Biol. 46: 32–38. [DOI] [PubMed] [Google Scholar]
- Hochreiter B., Garcia A.P., Schmid J.A.(2015). Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors (Basel) 15: 26281–26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P.(2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S.I., Takahashi K., Okumura-Noda H., Kinoshita T.(2016). Auxin influx carrier AUX1 confers acid resistance for arabidopsis root elongation through the regulation of plasma membrane H+-ATPase. Plant Cell Physiol. 57: 2194–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Danielson J.A., Manojkumar S.N., Lanquar V., Grossmann G., Frommer W.B.(2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek M., Blatt M.R.(2017). The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol. 174: 487–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C.(2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56: 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C.(2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kärkönen A., Kuchitsu K.(2015). Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 112: 22–32. [DOI] [PubMed] [Google Scholar]
- Keinath N.F., Waadt R., Brugman R., Schroeder J.I., Grossmann G., Schumacher K., Krebs M.(2015). Live cell imaging with R-GECO1 sheds light on flg22- and chitin-induced transient [Ca(2+)]cyt patterns in Arabidopsis. Mol. Plant 8: 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A., Leitão N., Chabaud M., Charpentier M., de Carvalho-Niebel F.(2018). Dual color sensors for simultaneous analysis of calcium signal dynamics in the nuclear and cytoplasmic compartments of plant cells. Front. Plant Sci. 9: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y.(2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Waszczak C., Hunter K., Wrzaczek M.(2017). Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 29: 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler B., Hills A., Blatt M.R.(2003). Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol. 131: 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuk A.I., Panova A.S., Bilan D.S., Belousov V.V.(2018). Redox biosensors in a context of multiparameter imaging. Free Radic. Biol. Med. 128: 23–39. [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K.(2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Becker D., Grill E., Hedrich R., Hippler M., Kummer U., Parniske M., Romeis T., Schumacher K.(2018). Advances and current challenges in calcium signaling. New Phytol. 218: 414–431. [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I.(2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A., Sutikovic Z., Wenzl C., Maegele I., Lohmann J.U., Forner J.(2013). GreenGate---A novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Tester M., Gilliham M.(2017). Chloride on the move. Trends Plant Sci. 22: 236–248. [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M.(2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X., Zhou J.M.(2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110: 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro G., Drago I., Pozzan T., Schiavo F.L., Zottini M., Costa A.(2012). Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J. 71: 1–13. [DOI] [PubMed] [Google Scholar]
- Luu K., Rajagopalan N., Ching J.C.H., Loewen M.C., Loewen M.E.(2019). The malate-activated ALMT12 anion channel in the grass Brachypodium distachyon is co-activated by Ca2+/calmodulin. J. Biol. Chem. 294: 6142–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Guo J., Kang Y., Doman K., Bryan A.C., Tax F.E., Yamaguchi Y., Qi Z.(2014). AtPEPTIDE RECEPTOR2 mediates the AtPEPTIDE1-induced cytosolic Ca(2+) rise, which is required for the suppression of Glutamine Dumper gene expression in Arabidopsis roots. J. Integr. Plant Biol. 56: 684–694. [DOI] [PubMed] [Google Scholar]
- Mangano S., Juárez S.P., Estevez J.M.(2016). ROS regulation of polar growth in plant cells. Plant Physiol. 171: 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L., Siala W., Schwarzländer M., Fricker M.D., Wirtz M., Sweetlove L.J., Meyer Y., Meyer A.J., Reichheld J.P., Hell R.(2009). The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 9109–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynov V.I., Pakhomov A.A., Deyev I.E., Petrenko A.G.(2018). Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta, Gen. Subj. 1862: 2924–2939. [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Brach T., Marty L., Kreye S., Rouhier N., Jacquot J.P., Hell R.(2007). Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52: 973–986. [DOI] [PubMed] [Google Scholar]
- Michard E., Simon A.A., Tavares B., Wudick M.M., Feijó J.A.(2017). Signaling with ions: The keystone for apical cell growth and morphogenesis in pollen tubes. Plant Physiol. 173: 91–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina N.M., Bogeski I., Bolotin D.A., Hoth M., Niemeyer B.A., Schultz C., Zagaynova E.V., Lukyanov S., Belousov V.V.(2012). Can we see PIP(3) and hydrogen peroxide with a single probe? Antioxid. Redox Signal. 17: 505–512. [DOI] [PubMed] [Google Scholar]
- Mittler R.(2017). ROS are good. Trends Plant Sci. 22: 11–19. [DOI] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Messerli M.A., Shi C., Gilroy S.(2007). Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA 104: 20996–21001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Weisenseel M.H., Gilroy S.(2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Miller N.D., Murphy A.S., Gilroy S.(2011). Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65: 309–318. [DOI] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I.(2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S., Hauser F., Park J., Waadt R., Brandt B., Schroeder J.I.(2015). Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K.(2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51: 328–332. [DOI] [PubMed] [Google Scholar]
- Ngo Q.A., Vogler H., Lituiev D.S., Nestorova A., Grossniklaus U.(2014). A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev. Cell 29: 491–500. [DOI] [PubMed] [Google Scholar]
- Nietzel T., et al. (2019). The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2 O2 and thiol redox integration and elucidates intracellular H2 O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol. 221: 1649–1664. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283: 8885–8892. [DOI] [PubMed] [Google Scholar]
- Okumoto S., Jones A., Frommer W.B.(2012). Quantitative imaging with fluorescent biosensors. Annu. Rev. Plant Biol. 63: 663–706. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S., Höfte H.(2012). Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I.(2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734. [DOI] [PubMed] [Google Scholar]
- Pittman J.K., Hirschi K.D.(2016). CAX-ing a wide net: Cation/H(+) transporters in metal remediation and abiotic stress signalling. Plant Biol. (Stuttg.) 18: 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C.A., Berkowitz G.A. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 107: 21193–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Schelle I., Liao Y.J., Schroeder J.I.(1995). Strong regulation of slow anion channels and abscisic acid signaling in guard cells by phosphorylation and dephosphorylation events. Proc. Natl. Acad. Sci. USA 92: 9535–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K.(2014). pH in the plant endomembrane system-an import and export business. Curr. Opin. Plant Biol. 22: 71–76. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Logan D.C., Johnston I.G., Jones N.S., Meyer A.J., Fricker M.D., Sweetlove L.J.(2012). Pulsing of membrane potential in individual mitochondria: A stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold H., Trempel F., Ranf S., Scheel D., Romeis T., Lee J.(2014). Ca2+ signalling in plant immune response: From pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 204: 782–790. [DOI] [PubMed] [Google Scholar]
- Shan X., Yan J., Xie D.(2012). Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 15: 84–91. [DOI] [PubMed] [Google Scholar]
- Shavrukov Y., Hirai Y.(2016). Good and bad protons: Genetic aspects of acidity stress responses in plants. J. Exp. Bot. 67: 15–30. [DOI] [PubMed] [Google Scholar]
- Shen Y., Rosendale M., Campbell R.E., Perrais D.(2014). pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 207: 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H.W., DePew C.L., Miller N.D., Monshausen G.B.(2015). The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr. Biol. 25: 3119–3125. [DOI] [PubMed] [Google Scholar]
- Shih H.W., Miller N.D., Dai C., Spalding E.P., Monshausen G.B.(2014). The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 24: 1887–1892. [DOI] [PubMed] [Google Scholar]
- Smirnoff N., Arnaud D.(2019). Hydrogen peroxide metabolism and functions in plants. New Phytol. 221: 1197–1214. [DOI] [PubMed] [Google Scholar]
- Song C.J., Steinebrunner I., Wang X., Stout S.C., Roux S.J.(2006). Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 140: 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N., Belkhadir Y., Zipfel C.(2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. [DOI] [PubMed] [Google Scholar]
- Steinhorst L., Kudla J.(2014). Signaling in cells and organisms - Calcium holds the line. Curr. Opin. Plant Biol. 22: 14–21. [DOI] [PubMed] [Google Scholar]
- Sze H., Chanroj S.(2018). Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 177: 875–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K., Ebine K., Askani J.C., Krüger F., Gonzalez Z.A., Ito E., Goh T., Schumacher K., Nakano A., Ueda T.(2018). Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E2457–E2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Swanson S.J., Gilroy S., Stacey G.(2010). Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol. 154: 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Hou C., Ren Z., Wang C., Zhao F., Dahlbeck D., Hu S., Zhang L., Niu Q., Li L., Staskawicz B.J., Luan S.(2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572: 131–135. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Benková E.(2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28: 463–487. [DOI] [PubMed] [Google Scholar]
- Waadt R., Hitomi K., Nishimura N., Hitomi C., Adams S.R., Getzoff E.D., Schroeder J.I.(2014). FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Hsu P.K., Schroeder J.I.(2015). Abscisic acid and other plant hormones: Methods to visualize distribution and signaling. BioEssays 37: 1338–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Krebs M., Kudla J., Schumacher K.(2017). Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis. New Phytol. 216: 303–320. [DOI] [PubMed] [Google Scholar]
- Wagner S., et al. (2019). Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol. 224: 1668–1684. [DOI] [PubMed] [Google Scholar]
- Walia A., Waadt R., Jones A.M.(2018). Genetically encoded biosensors in plants: Pathways to discovery. Annu. Rev. Plant Biol. 69: 497–524. [DOI] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J.(2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69: 209–236. [DOI] [PubMed] [Google Scholar]
- Wu F., et al. (2020). Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578: 577–581. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Christmann A., Yamaguchi-Shinozaki K., Grill E., Fernie A.R.(2019). Revisiting the basal role of ABA - Roles outside of stress. Trends Plant Sci. 24: 625–635. [DOI] [PubMed] [Google Scholar]
- Zhang X., Köster P., Schlücking K., Balcerowicz D., Hashimoto K., Kuchitsu K., Vissenberg K., Kudla J.(2018). CBL1-CIPK26-mediated phosphorylation enhances activity of the NADPH oxidase RBOHC, but is dispensable for root hair growth. FEBS Lett. 592: 2582–2593. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Araki S., Wu J., Teramoto T., Chang Y.F., Nakano M., Abdelfattah A.S., Fujiwara M., Ishihara T., Nagai T., Campbell R.E.(2011). An expanded palette of genetically encoded Ca2+ indicators. Science 333: 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Kang S., Jing Y., Ren Z., Li L., Zhou J.M., Berkowitz G., Shi J., Fu A., Lan W., Zhao F., Luan S.(2018). Danger-associated peptides close stomata by OST1-independent activation of anion channels in guard cells. Plant Cell 30: 1132–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]