A Borealin homolog in Arabidopsis functions as a part of the chromosome passenger complex.
Abstract
The Aurora B kinase, encoded by the AURORA 3 (AUR3) gene in Arabidopsis (Arabidopsis thaliana), is a key regulator of cell division in all eukaryotes. Aurora B has at least two central functions during cell division; it is essential for the correct, i.e. balanced, segregation of chromosomes in mitosis and meiosis by controlling kinetochore function, and it acts at the division plane, where it is necessary to complete cytokinesis. To accomplish these two spatially distinct functions, Aurora B in animals is guided to its sites of action by Borealin, inner centromere protein (INCENP), and Survivin, which, together with Aurora B, form the chromosome passenger complex (CPC). However, besides Aurora homologs, only a candidate gene with restricted homology to INCENP has been described in Arabidopsis, raising the question of whether a full complement of the CPC exists in plants and how Aurora homologs are targeted subcellularly. Here, we have identified and functionally characterized a Borealin homolog, BOREALIN RELATED (BORR), in Arabidopsis. Together with detailed localization studies including the putative Arabidopsis INCENP homolog, these results support the existence of a CPC in plants.
Equal chromosome segregation during cell division is crucial for the survival, growth, and reproduction of every organism. Chromosome segregation is assured by the M-phase checkpoint, which involves two regulatory units, the spindle assembly checkpoint (SAC) and the chromosomal passenger complex (CPC; Carmena et al., 2012).
Chromatids are held together by Cohesin, a proteinaceous ring-like structure, which needs to be cleaved by Separase to allow chromatid separation in anaphase. The SAC inhibits the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase, which mediates the degradation of Securin, an inhibitor of Separase, until all kinetochores are properly attached to microtubules of opposing poles (London and Biggins, 2014; Komaki and Schnittger, 2016).
The CPC has been well characterized in yeast and mammals, where it has been found to consist of four proteins: Aurora kinase B (Aurora B), Borealin, inner centromere protein (INCENP), and Survivin. The CPC fulfills several functions during mitosis. In particular, it is involved in ensuring that all kinetochores are attached to microtubules emanating from opposing poles (Kitagawa and Lee, 2015). The CPC localizes to the inner centromere where it activates Aurora B in response to low interkinetochore tension. Active Aurora B phosphorylates kinetochore proteins, leading to the destabilization of erroneous microtubule attachments. Once proper kinetochore-microtubule attachments are established from opposing poles, which gives high interkinetochore tension, Aurora B is spatially separated from kinetochores, resulting in proper bistable spindle formation.
Aurora B belongs to the Aurora kinase family of Ser/Thr kinases that are highly conserved in the eukaryotic kingdom (van der Waal et al., 2012; Weimer et al., 2016). Whereas yeast possesses a single Aurora homolog, mammals have three Aurora kinases, Aurora A, Aurora B, and Aurora C, among which only Aurora C acts in meiosis (Goldenson and Crispino, 2015). Since these three kinases share a common consensus phosphorylation motif, it is thought that interacting proteins are important for their localization and substrate specificity.
Aurora A interacts with the spindle assembly factor TPX2, and localizes to spindle microtubules to regulate spindle assembly (Gruss and Vernos, 2004). By contrast, Aurora B and Aurora C, the catalytic subunit of the CPC, are involved in the correction of erroneous kinetochore-microtubule attachments, activation of the SAC, and cytokinesis in mitosis and meiosis. These diverse functions are based on dynamic localization patterns controlled by the three noncatalytic subunits of the CPC (van der Horst and Lens, 2014).
INCENP, the largest noncatalytic subunit of the CPC, directly binds to all other components of the complex in animals and yeast. Borealin and Survivin interact with the conserved N-terminal region of INCENP, whereas Aurora B binds to the C-terminal domain of INCENP, called the IN-box (Carmena et al., 2012). The IN-box is required for interaction with and activation of Aurora B (Honda et al., 2003).
Whereas the N terminus of Borealin acts as the INCENP-binding region, its C terminus contains a homodimerization domain that is involved in a stable CPC localization at centromeres (Bekier et al., 2015). In addition, the phosphorylation status of the central part affects the centromere localization and steady-state level of Borealin itself (Kaur et al., 2010; Date et al., 2012).
Aurora kinases in plants are categorized into two groups, α-Aurora and β-Aurora, based on localization pattern and sequences (Weimer et al., 2016). Interestingly, these two groups have mixed features of the animal Aurora A and Aurora B/C groups. For instance, AUR1 and AUR2 in Arabidopsis, which both belong to the α class, localize to spindle microtubules, which is reminiscent of Aurora A, but they also localize to the central region of phragmoplasts paralleling Aurora B/C at the cell cleavage site. AUR3, a member of the β-Aurora group, localizes to kinetochores, similar to Aurora B (Komaki and Schnittger, 2017); however, in contrast to Aurora B, AUR3 does not accumulate at the division plane (Demidov et al., 2005).
Whereas aur1 and aur2 single mutants do not show any obvious growth alterations, the aur1 aur2 double null mutant is gametophytic lethal (Van Damme et al., 2011). A weak loss-of-function aur1 aur2 double mutant exhibits altered division plane orientation, reduced pollen viability, and enhanced vascular cell differentiation. These defects can be rescued by expressing either AUR1 or AUR2, but not AUR3, indicating that α-Auroras and β-Auroras have distinct functions (Van Damme et al., 2011; Demidov et al., 2014; Lee et al., 2019).
Given the sparse information about the noncatalytic subunits of the plant CPC, it remains unclear whether there is a conserved CPC function in plants. The putative homolog of INCENP in Arabidopsis has a long N-terminal region of unknown function, which is conserved only in plants. Although Arabidopsis incenp mutants, also known as wyrd (wyr), show defects in gametophytic cell division (Kirioukhova et al., 2011), it remains unclear whether Arabidopsis INCENP acts as part of a putative plant CPC because of missing information about its localization and binding partners.
Here, we present the identification and functional characterization of an Arabidopsis Borealin homolog, which colocalizes with the INCENP homolog to the inner centromere and the central domain of the phragmoplast. We also observed that only AUR3 acts as the catalytic subunit of the plant CPC. These data underscore the mixed features of plant Aurora kinases.
RESULTS
Identification of a Putative Borealin Homolog in Plants
As a first step to determine whether plants have a functional CPC, we searched for Borealin and Survivin homologs in the Arabidopsis genome. Given that the putative INCENP homolog of Arabidopsis, WYR, shares only very weak similarities with its animal counterpart (Kirioukhova et al., 2011), we expected a similar situation for Borealin and Survivin. Indeed, standard BLAST searches did not result in the identification of likely candidates. Therefore, we made use of the fact that Borealin is transcriptionally controlled by the tumor suppressor protein Retinoblastoma in animals (Cam et al., 2004; Date et al., 2007). Mining a dataset of the genome-wide binding sites of the Arabidopsis Retinoblastoma homolog RETINOBLASTOMA-RELATED (RBR1) revealed several unknown genes likely involved in cell division control (Supplemental Fig. S1; Bouyer et al., 2018). One of these genes, AT4G39630, showed a weak similarity to Borealin and will be referred to as BOREALIN-RELATED (BORR; Supplemental Fig. S1A).
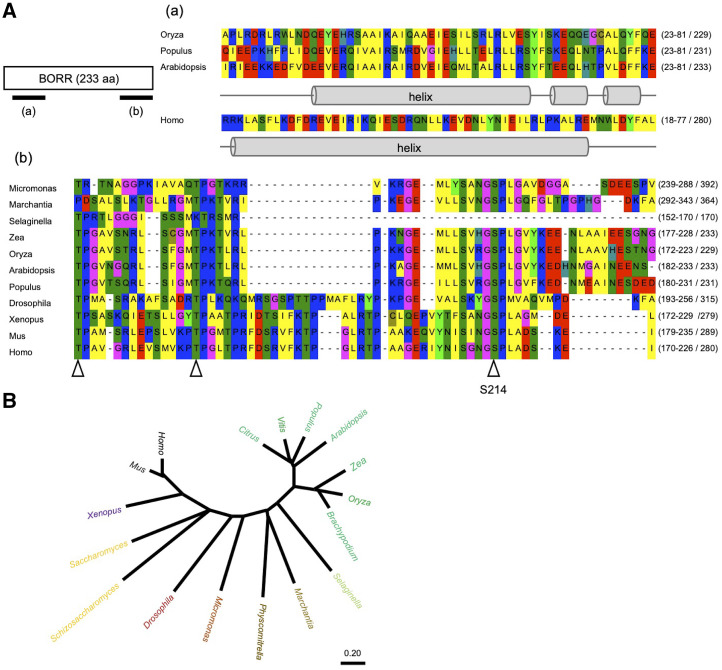
The C-terminal region of the corresponding protein shows 37% similarity with the central region of human Borealin, which includes highly conserved consensus sites for CDK phosphorylation (Fig. 1A). A second stretch of homology can be found in the N terminus, which is predicted to adopt an α-helical structure. In animals and yeast, this N-terminal α-helix forms a three-helical bundle with INCENP and Survivin (Fig. 1A; Jeyaprakash et al., 2007). Notably, the Arabidopsis BORR is considerably shorter than the human homolog (233 versus 280 amino acids), partly as the result of a truncated C-terminal domain. Using BORR as a template, we found putative Borealin genes in all branches of the plant kingdom, including angiosperms, gymnosperms, pteridophytes, bryophytes, and algae (Fig. 1B).
Figure 1.
BORR gene structure in plants. A, Protein sequence of BORR in Arabidopsis. Lowercase letters indicate predictions of the N-terminal helix in BORR (a) and alignment of the most conserved sequence region of the protein (b). Arrowheads in b indicate the conserved CDK consensus sites. B, Phylogenetic analysis of the Borealin family in yeast, animals, and plants. The tree was constructed using MEGA X by the neighbor-joining method.
Phenotypic Analysis of borr Mutants
Since no mutants for BORR were available in the public mutant collections of Arabidopsis, we generated a mutant by CRISPR/Cas9. The resulting borr-1 allele has a T insertion in the second exon, i.e. between nucleotides 280 and 281 downstream of the start codon, leading to a stop codon and a predicted truncated protein of 67 amino acids (Supplemental Fig. S1A). Whereas the heterozygous borr-1 plants grew as the wildtype, we were unable to obtain homozygous mutant plants. Consistently, we observed aborted seeds and undeveloped ovules in siliques of borr-1 −/+ (Fig. 2, A and B).
Figure 2.
BORR is required for seed development. A, Wild-type or heterozygous borr mutant plants pollinated with pollen from wild-type or heterozygous borr mutants. Black and gray arrowheads indicate tiny white ovules (unfertilized ovules/early arrested seeds) and collapsed brown seeds or seeds without green embryo (late aborted seeds), respectively. Scale bar = 1 mm. B, Proportions of the seed phenotypes shown in A. Total number (n) of seeds and ovules from five hand-pollinated siliques are shown in each cross. C, Seeds and ovules from the heterozygous borr siliques cleared in chloral hydrate solution. At the globular embryo stage, developing seeds with abnormal embryos, developing seeds with no embryo, and unfertilized ovules were observed. Scale bar = 50 μm. D, Classification of the embryo phenotype for each cross. Total number (n) of developing seeds and unfertilized ovules from six to eight hand-pollinated pistils are shown.
To assess whether this lethal phenotype was indeed due to the frameshift mutation in the BORR gene, we constructed reporter lines in which the genomic region of BORR was fused to an open reading frame (ORF) encoding for GFP. Expression of either an N-terminal or C-terminal GFP fusion to BORR (GFP:BORR or BORR:GFP, respectively) complemented the lethal phenotype of borr-1 mutant plants, corroborating that loss of BORR affects plant reproduction.
To address the nature of the lack of homozygous borr-1 mutants, we conducted reciprocal crosses of heterozygous mutants with the wild type. When we used borr-1 −/+ as the male parent, the transmission rate was determined to be 44%. Consistently, 96% of borr-1 −/+ pollen resembled wild-type pollen and, based on 4′,6-diamino-phenylindole staining, contained two sperm cells.
By contrast, when we used borr-1 −/+ as the female parent, the transmission rate was reduced to 29%, indicating that BORR is especially needed for the development and/or function of the female gametophyte. However, when analyzing the mature female gametophyte, no obvious developmental defects were observed, suggesting that the reduced transmission of borr-1 through the female gametophyte originated shortly before or after/during fertilization. In accordance with this, we found 12% unfertilized ovules/early arrested seeds and 19% late-aborted seeds when we used borr-1 −/+ as the female (n = 250) in contrast to 3% unfertilized ovules/early-arrested seeds and 0.3% late-aborted seeds in control crosses when we used the wild type as the female and male parent (n = 299; Fig. 2, A and B).
In addition, we observed abnormal embryo development, including delayed and distorted embryos or embryo-like structures in ∼10% of all ovules/seeds analyzed when borr-1 +/− was used as the female parent in crosses with the wild type as male versus 4% in the control crosses supporting a female gametophytic effect of borr (n = 313 and n = 408; Fig. 2, C and D). One likely explanation for this is that the divisions leading to the development of the embryo sac cause aneuploidy in borr that precludes and/or severely interferes with embryo development.
In addition, BORR is needed during embryo development, since the number of seeds with embryonic defects almost doubled in self-fertilized borr-1 mutants (Fig. 2D). Thus, BORR is an essential gene needed for cell proliferation and development during early stages of the plant life cycle.
To address BORR function after embryo development, we generated BORR knockdown plants by expressing two artificial microRNAs (amiRNAs) targeted against the second (amiBORR1) and third (amiBORR2) exons, respectively, of BORR (Fig. 3A). Most transgenic plants expressing amiBORR1 (19 of 25) and amiBORR2 (15 of 20) showed a dwarf phenotype. For the following analyses, we selected two transgenic plants for each construct (amiBORR1-1, amiBORR1-2, amiBORR2-1, and amiBORR2-2). Each of the four knockdown plants had a similar level of BORR transcript reduction (Fig. 3B) and displayed a dwarf phenotype with curled leaves during the vegetative stage (Supplemental Fig. S2A). At the flowering stage, all BORR knockdown plants were bushy and exhibited a typical bonsai phenotype (sometimes also called broom stick phenotype), which is commonly observed in mutants with low APC/C activity. The bonsai phenotype is characterized by short inflorescences that are often curled at the very end, with only a few developing siliques (Supplemental Fig. S2, B and C; Saze and Kakutani, 2007; Zheng et al., 2011).
Figure 3.
Phenotypical analysis of amiRNA-mediated BORR knockdown plants. A, Sequence alignments of amiRNAs and their target sites on BORR mRNA. B, Relative expression level of BORR in the amiBORR plants was confirmed by RT-qPCR analysis with three biological replicates. C, Nine-day-old wild-type and amiBORR seedlings. Scale bar = 1 cm. D, Root growth measurements of wild-type and amiBORR plants. Four-day-old seedlings were transferred to a Murashige and Skoog plate and root lengths were measured for 5 d. Asterisks indicated significance using Student's t test (**P < 0.01). Error bars indicate the SD (n = 10). E, Confocal images of 7-d-old wild-type and amiBORR1-1 roots stained with propidium iodide. amiBORR1-1 root mutations were categorized as Mild and Severe based on the phenotype. Arrowheads indicate the boundary between the dividing region and the elongation region of the root. Regions marked by white and yellow dotted boxes are shown in close-up under Columella region and Dividing region, respectively. Scale bars =50 μm. F and G, Root meristem size (F) and number of meristematic cortex cells (G). Asterisks indicated significance using Student's t test (**P < 0.01). Error bars indicate the SD (n = 15). H, Representative images of normally distributed and lagging chromosomes in 7-d-old wild-type and amiBORR1-1 root cells. Microtubules and centromeres were visualized by TagRFP:TUA5 and GFP:CENH3, respectively. The arrowhead indicates a lagging chromosome. Scale bar = 5 μm. I, Frequency of lagging chromosomes in anaphase in H. Error bars indicate the SD (n = 50). J, Representative images of AUR3 accumulation levels at kinetochores in 7-d-old wild-type and amiBORR#1-1 root cells. Microtubules and AUR3 were visualized by TagRFP:TUA5 and AUR3:GFP, respectively. Scale bar = 5 μm. K, AUR3 signal intensity at kinetochores in J. Forty AUR3-GFP signals at kinetochores from 10 cells were measured. The center line indicates the median, the box represents the interquartile range, error bars were determined as 1.5× the interquartile range, and the circle represents an outlier. Asterisks indicated significance using Student's t test (**P < 0.01). Error bars indicate the SD. a.u., Arbitrary units.
Primary root growth was also compromised in all BORR knockdown plants (Fig. 3, C and D). Microscopic analyses of amiBORR1-1 revealed that the root meristem size was reduced (Fig. 3, E–G). Moreover, an aberrant pattern of cell divisions was found in the columella of all roots analyzed (25 of 25). In 40% of all seedlings (10 of 25), altered division patterns were also present in the epidermal, cortex, and endodermal layers, underlining that BORR is required for proper cell division (Fig. 3E).
To assess whether Arabidopsis BORR has a function in chromosome segregation as well, amiBORR1-1 was introgressed into a transgenic line expressing both a microtubule (RFP:TUA5) and a centromere (GFP:CENH3) marker (Komaki and Schnittger, 2017). Indeed, we could frequently observe lagging chromosomes in amiBORR1-1 plant cells (9 of 50), a phenotype that hardly occurred in the wild-type control plant cells (1 of 50; Fig. 3, H and I). To examine whether the lagging chromosomes are related to a compromised AUR3 localization, we introduced a previously generated AUR3:GFP reporter into amiBORR#1-1 plants (Komaki and Schnittger, 2017). Whereas some AUR3:GFP signal could be still detected at the kinetochores in amiBORR1-1 plants, the signal intensity was much weaker than in wild-type plants, suggesting that Arabidopsis BORR ensures chromosome segregation through AUR3 localization (Fig. 3, J and K). Taken together, corresponding to Borealin function in animals, these results indicate that BORR is required for proper chromosome segregation and cell division in Arabidopsis.
Interaction Scheme of CPC Components in Arabidopsis
To reveal the molecular network of the Arabidopsis CPC, we investigated the interaction of BORR with INCENP and the three Aurora kinases of Arabidopsis. Although Borealin has a conserved coiled-coil domain, which is known as an INCENP binding site in other organisms (Jeyaprakash et al., 2007), an interaction between BORR and INCENP was not detected by a yeast two-hybrid (Y2H) interaction assay (Fig. 4A). We next performed an in vivo coimmunoprecipitation (IP) assay. To this end, anti-GFP immunoprecipitates from total protein extracts of plants expressing both BORR:RFP and GFP:INCENP, or from plants expressing BORR:RFP and free GFP as a negative control, were probed with an anti-RFP antibody. BORR:RFP was only detected in the extract of plants coexpressing GFP-INCENP, indicating that Arabidopsis BORR could be part of a CPC in vivo (Fig. 4C).
Figure 4.
Interaction among the CPC components. A, Interaction among the CPC components as revealed by Y2H assays. B, Interaction between AUR3 and various regions of INCENP or IN-box-mutated INCENP. Monomeric GFP (mGFP) was used as a negative control. Each strain was spotted on SD plates without Trp and Leu (−TL; control media) or without Trp, Leu, and His (−TLH; selection media) and photographed after incubation at 30°C for 2 d. AD, GAL4-activation domain; BD, GAL4-DNA binding domain. C, IP of INCENP with BORR. 7-d-old Arabidopsis seedlings expressing BORR:RFP and mGFP:INCENP or BORR:RFP and mGFP were used for IP with an anti-GFP antibody. Both input and IP fraction were subjected to immunoblotting with an anti-RFP antibody. The asterisk indicates a nonspecific band.
Consistent with the topology of the CPC in animal and yeast, we further found that INCENP interacts specifically with AUR3 through its C-terminal region whereas no interaction was detected with AUR1 or AUR2 (Fig. 4, A and B). The C-terminal region of INCENP contains an Aurora B binding domain, called IN-box, which is conserved from yeast to mammals (Adams et al., 2000). Since Arabidopsis INCENP also possesses a putative IN-box in its C terminus, we exchanged Trp-1723 with Gly and Phe-1745 with Ala. These residues are conserved in yeast and animals, and the Trp-1723 mutation leads to loss of interaction with Aurora B in in vitro assays (Sessa et al., 2005), as well as in chicken culture cells (Xu et al., 2009). Consistent with a key role of the putative IN-box, Arabidopsis INCENPF1745A partially failed, and INCENPW1723G completely failed, to interact with AUR3 (Fig. 4B).
Expression Pattern and Subcellular Localization of the CPC Components in Mitosis
To investigate the spatial and temporal expression pattern of BORR, we generated transgenic plants harboring a genomic fragment of the BORR gene fused to a GUS gene before the STOP codon. As expected, strong GUS activity was observed in both the shoot and root meristems of seedlings, indicating that BORR is expressed in proliferating cells (Fig. 5, A–C). In addition, BORR was also found to be strongly expressed in flowering tissues including both the male and female reproductive organs (Fig. 5, D–F).
Figure 5.
Expression and subcellular localization of CPC components. A to F, Expression patterns of BORR:GUS in the shoot (A), root (B), lateral roots (C), inflorescence (D), ovules (E), and young flower buds (F). Plants used in A to C were 10 d old and those in D to F were 4 weeks old. Three independent transgenic lines were analyzed and representative images are shown. Scale bars = 500 μm. G, Subcellular localization of GFP:INCENP, BORR:GFP, and AUR3:GFP during the cell cycle. Each reporter line was crossed with TagRFP:TUA5-expressing plants to visualize microtubule structures. Scale bar = 10 μm. H, Colocalization of the CPC components during the cell cycle. Scale bar = 10 μm. For live imaging in G and H, root tips of 5-d-old seedlings were used.
To reveal the subcellular localization of the CPC components in plants, we made use of the functional BORR reporter line used for the complementation studies above. In addition, we constructed reporter lines in which the genomic region of INCENP was fused to an ORF encoding for GFP. In contrast to BORR, only the N-terminal GFP fusion of INCENP (GFP:INCENP) could complement the lethal phenotype of incenp homozygous mutants (Supplemental Fig. S1, D and E). For AUR3, we used a previously published reporter line (AUR3:GFP) in the wild-type background (Komaki and Schnittger, 2017).
First, we crossed the three CPC reporter lines with RFP:TUA5-expressing plants to check the localization of CPC components during mitosis. All three CPC components showed the same localization pattern (Fig. 5G; Supplemental Movies S1–S3). In interphase, they localized to the nucleus. Before nuclear envelope breakdown (NEB), the CPC components strongly accumulated at the kinetochores until anaphase onset. Once chromosomes moved toward the spindle poles, they localized to the middle part of the phragmoplast. Interestingly, in early telophase, the CPC components moved back to the nucleus even though there still was an expanding phragmoplast.
To corroborate that all three CPC components colocalize, we created transgenic plants that expressed BORR:RFP together with GFP:INCENP, BORR:RFP with AUR3:GFP, and RFP:INCENP with AUR3:GFP (Table 1). Microscopical analyses revealed a tight colocalization pattern through the cell cycle in all three lines, suggesting that BORR, INCENP, and AUR3 work together as a plant CPC (Fig. 5H; Supplemental Figs. S4 and S5; Supplemental Movies S4–S6).
Table 1. Genetic material generated in this study.
FP, Fluorescent protein.
| Construct | Genetic Background |
|---|---|
| FP Expression Lines | |
| PROBORR:mGFP:BORR | borr-1 |
| PRORPS5A:TagRFP:TUA5 | |
| PROBORR:BORR:mGFP | borr-1 |
| PRORPS5A:TagRFP:TUA5 | |
| PROCENH3:TagRFP:CENH3 | |
| PROBORR:BORRS214A:mGFP | borr-1 |
| PRORPS5A:TagRFP:TUA5 | |
| PROBORR:BORRS214D:mGFP | borr-1 |
| PRORPS5A:TagRFP:TUA5 | |
| PROINCENP:mGFP:INCENP | wyr-2 |
| PRORPS5A:TagRFP:TUA5 | |
| PROBORR:BORR:TagRFP | |
| PROINCENP:INCENP:mGFP | wyr-2 |
| PROAUR3:AUR3:mGFP | PRORPS5A:TagRFP:TUA5 |
| PROBORR:BORR:TagRFP | |
| PROINCENP:mRUBY3:INCENP | |
| GUS Expression Line | |
| PROBORR:BORR:GUS | Wild type |
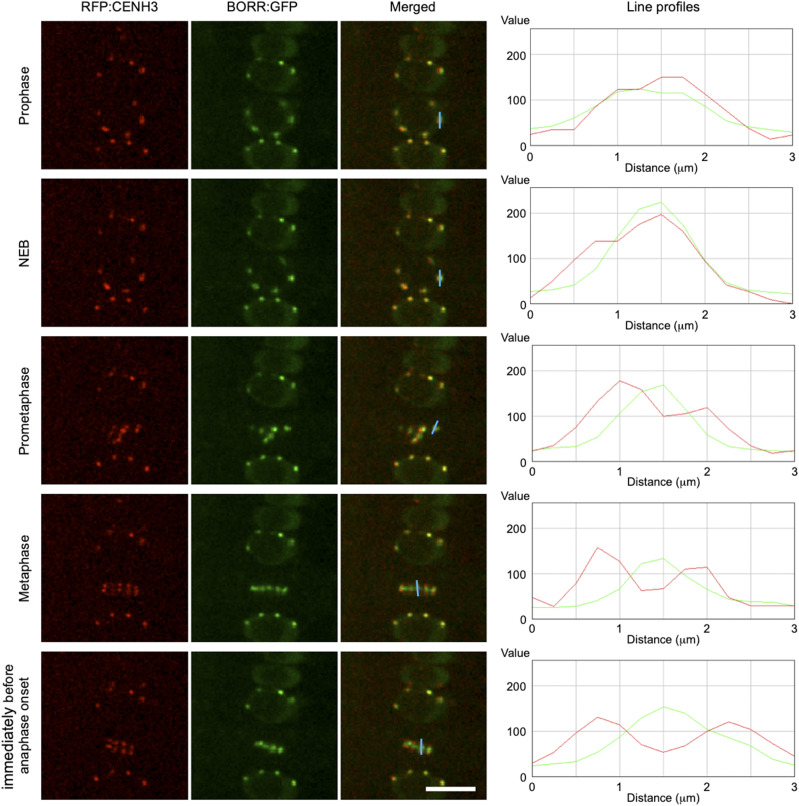
In other organisms, it has been reported that the CPC localizes to the inner centromere region to monitor kinetochore-microtubule attachments (Hindriksen et al., 2017). To reveal the localization of the CPC precisely, we crossed the BORR:GFP line with the inner kinetochore marker RFP:CENH3. Just after NEB, these two fluorescent reporters showed a high level of overlapping signal. After prometaphase, BORR:GFP localized closer to the inner region of the kinetochore than RFP:CENH3. Yet, both proteins still overlapped in their localization pattern. Shortly before anaphase onset, RFP:CENH3 formed two lines along the metaphase plate, and BORR:GFP localized between these two lines with no overlap (Fig. 6; Supplemental Movie S7). This result demonstrated that the CPC localizes to the inner centromere region in plants.
Figure 6.
CPC localizes to inner centromeres. Localization of the CPC from prophase to immediately before anaphase onset. Inner kinetochores and CPC complexes are visualized by TagRFP:CENH3 and BORR:GFP, respectively. For live cell imaging, root tips of 5-d-old seedlings were used. The blue bar indicates the position where the line profiles were obtained. Scale bar = 10 μm.
In humans, Borealin function is regulated by phosphorylation. In particular, the phosphorylation of S219, a putative Cdk1 target residue, in the central region of the protein affects its stability and centromere localization (Kaur et al., 2010; Date et al., 2012). Since this Cdk1 phosphorylation site, residing in the region most conserved between human and plant Borealin, is conserved in BORR (Fig. 1B), we generated a phospho-mimic (BORRS214D:GFP) and a dephospho variant (BORRS214A:GFP; Dissmeyer and Schnittger, 2011) and transformed them into the heterozygous borr-1 mutants to evaluate their functionality. Notably, both constructs could complement the lethal phenotype of borr-1 mutant plants and showed a normal BORR localization pattern (Supplemental Fig. S1, A–C). Thus, the physiological importance of this conserved CDK target motif has yet to be resolved in plants.
Expression Pattern and Subcellular Localization of the CPC Components in Meiosis
Since the plant meiotic spindle checkpoint seems to be less stringent (Wijnker and Schnittger, 2013; Komaki and Schnittger, 2016), we asked whether BORR and INCENP are present in meiosis. Analyzing male meiosis, we found that both proteins localized to kinetochores until the onset of anaphase I (Fig. 7; Supplemental Movies S8 and S9). Interestingly, although no cytokinesis occurs after meiosis I in Arabidopsis meiocytes, both proteins localized to the division plane shortly after anaphase I, representing either a late anaphase midzone or a phragmoplast midzone-like structure (Fig. 7; Supplemental Movies S8 and S9). The transition between the two phases is currently only poorly defined; however, the localization of BORR and INCENP could possibly contribute to the understanding of the composition and dynamics of these structures.
Figure 7.
Subcellular localization of CPC components in meiosis. A and B, Subcellular localization of BORR:GFP (A) and GFP:INCENP (B) during meiosis. Each reporter line was crossed with TagRFP:TUA5-expressing plants to visualize the microtubule structures. Scale bars = 10 μm. For live imaging of A and B, flower buds of 1-month-old plants were used.
The localization of BORR and INCENP in the second meiotic division resembled the localization of both proteins in mitosis, i.e. at the kinetochores and subsequently at the phragmoplast (Fig. 7; Supplemental Movies S8 and S9).
DISCUSSION
The CPC is essential for proper cell division in animals and yeast. Whether CPC activity exists in plants and how the potential complex is composed was poorly understood prior to the presented work. Here, we have identified and characterized BORR, a Borealin homolog in Arabidopsis. Whereas the presence of a functional homolog of Survivin in plants is currently still unclear, the existence of BORR together with localization data for AUR3 and an INCENP homolog demonstrates that a CPC is present in plants and of equal importance as in animals.
BORR and INCENP localize to kinetochores in mitosis and meiosis. Both proteins could also be coprecipitated from seedlings, indicating that BORR and INCENP indeed work in one complex. Consistent with previous reports, we observed that AUR3 accumulates at mitotic kinetochores (Fig. 5G; Supplemental Movie S3; Demidov et al., 2005), whereas AUR1 and AUR2 localize to the mitotic spindle (Van Damme et al., 2011). In addition, out of the three AUR proteins in Arabidopsis, only AUR3 interacted with INCENP in our Y2H assays (Fig. 4). These data suggest that only AUR3 acts as the catalytic subunit of the CPC at the inner centromere in plants.
In animals, the CPC localizes to the inner centromere to monitor interkinetochore tension. Since proper kinetochore-microtubule attachments are not established during prophase, the distance between the inner centromere and kinetochores is very small, allowing the CPC-dependent centromere-localized Aurora B activity to act on kinetochores and to destabilize erroneous attachments of microtubules.
Once proper attachments are formed and interkinetochore tension is built up, Aurora B localized to the inner centromere is spatially separated from kinetochores leading to the formation of a stable bipolar spindle. Since we found that the kinetochore signals moved away from BORR signals during cell-cycle progression, we propose that the plant CPC also acts as a tension sensor.
At anaphase onset, the animal CPC translocates from kinetochores to the cell division plane. Although plant cell division is strikingly different (Müller and Jürgens, 2016), we demonstrated that Arabidopsis BORR, INCENP, and AUR3 also accumulate at the division plane at the beginning of cell division (Fig. 5G; Supplemental Movies S1–S3). Interestingly, they accumulate at the reforming nuclei at telophase before the expanding phragmoplast is completely disassembled, i.e. cytokinesis is finished, whereas in animals the CPC stays at the cleavage site until the two daughter cells are formed (Carmena et al., 2012).
Remarkably, previous studies revealed that the two α-Aurora members, AUR1 and AUR2, localize to the division plane until the end of cytokinesis (Demidov et al., 2005; Kawabe et al., 2005). Therefore, the plant CPC might be needed for the initiation of cell division but not be necessary for later steps. In animals, TPX2 recruits Aurora A to the division plane (Kufer et al., 2002). However, TPX2 does not localize to the division plane in plants, although it and its homologs also interact with α-Aurora members in Arabidopsis (Petrovská et al., 2012; Boruc et al., 2019). Thus, it is still not clear how α-Aurora localization to the division plane is controlled.
It seems likely that there is a yet unidentified interaction partner of the CPC in plants that has affinity to the plus-end of microtubules and causes localization of the CPC, i.e. AUR3, to kinetochores. After separation of the chromosomes in anaphase, this or a different factor promotes the accumulation in the spindle midzone/early phragmoplast.
Since each component of the CPC is needed for its activity, loss of any CPC component leads to the same mutant phenotypes as seen in Aurora B mutants (Honda et al., 2003; Vader et al., 2006). CPC mutants typically exhibit cell division defects and lagging chromosomes resulting from incorrect microtubule-kinetochore attachments. These phenotypes are frequently coupled with ploidy changes causing cancer in mammals (Tang et al., 2017). Notably, a complete loss of function of any of the CPC components leads to lethality (Cutts et al., 1999; Uren et al., 2000; Lu et al., 2008; Yamanaka et al., 2008). The sporophyte of borr-1 heterozygous knockout plants grew like the wild type. However, no homozygous borr mutants could be recovered and the heterozygous mutants harbor undeveloped ovules, aborted seeds, and embryonic defects in the forming siliques. Similar effects were observed in the INCENP mutant wyr (Kirioukhova et al., 2011). However, in contrast to borr, the loss of INCENP causes an arrest in development of both the female and the male gametophytes. We can currently not rule out whether INCENP has a specific and BORR-independent function during the gametophyte life phase. It is also possible that INCENP has a shorter half-life than BORR, given that it is a large protein and hence the gametophytes run out of sporophytically inherited protein levels much earlier than is the case for the much smaller BORR protein.
Using knockdown plants, we could further reveal that reduction of BORR results in mitotic defects, including lagging chromosomes and abnormal cell division, which appear to be likely caused by a compromised localization of AUR3. A previous study reported that hesperadin treatment, which inhibits the AUR3 kinase activity in vitro, induces lagging chromosomes in tobacco BY-2 cells (Kurihara et al., 2006), suggesting that the AUR3 function in chromosome segregation is conserved in the plant lineage.
Interestingly, the BORR knockdown plants showed a typical bonsai phenotype, which is characterized by inhibition of internode elongation and premature termination of the shoot apical meristem. Although the molecular mechanism is still not known, the bonsai phenotype is associated with a reduction of APC/C activity (Saze and Kakutani, 2007; Zheng et al., 2011).
The SAC is another M-phase checkpoint, which works together with the CPC to ensure faithful chromosome segregation. The primary role of the SAC is delaying APC/C activity until all kinetochores are properly attached to the spindle microtubules. Therefore, one possible explanation of the bonsai phenotype is that the SAC is over-activated in BORR knockdown plants. Indeed, these two M-phase checkpoints are directly or indirectly connected in other organisms (Trivedi and Stukenberg, 2016). Further studies are needed to understand the relationship between the SAC and the CPC in plants.
The final noncatalytic CPC subunit, Survivin, remains elusive in plants. Survivin localizes to the inner centromere upon phosphorylation of histone H3 at Thr-3, which in animals is catalyzed by the Haspin kinase (Kelly et al., 2010). Survivin localization is required for recruitment of the entire CPC to the inner centromere. Therefore, inhibition of Haspin kinase activity leads to dissociation of the CPC from the inner centromere in mammals. Recently, it was shown that inhibition of Haspin kinase activity with 5-iodotubercidin induces disruption of AUR3 localization at the inner centromere in BY-2 tobacco culture cells as well (Kozgunova et al., 2016). Although this result indicates that Haspin kinase activity is important for proper AUR3 localization at the inner centromere, no Survivin homolog could be identified in plants on a sequence level. However, a functional homolog might still exist. Alternatively, plants might employ a different mechanism for CPC localization.
The phosphorylation status of its different components is of key importance for the regulation of the CPC in animals and yeast. For instance, yeast INCENP is cooperatively phosphorylated by Cdk1 and Aurora, which prevents CPC binding to the spindle before anaphase (Goto et al., 2006; Nakajima et al., 2011). In addition, Casein kinase2 phosphorylates the human Survivin, which leads to its exclusion from the nucleus in interphase (Barrett et al., 2011). Borealin is also phosphorylated by many kinases, including Cdk1, which is required for targeting of the CPC to kinetochores (Kaur et al., 2010; Date et al., 2012). However, the mutation of a conserved CDK phosphorylation site neither obviously altered BORR localization nor reduced the activity of the protein in a way that would result in a mutant phenotype. Hence, further work is needed to shed light on the regulation of the plant CPC, with this study opening the door for an in-depth analysis of plant CPC function focusing on kingdom-specific aspects of its regulation and activity.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) accession Columbia (Col-0) was used as the wild type in this study. All mutants are in the Col-0 background. Plants were grown on a solid medium containing one-half strength Arabidopsis nutrient solution (Haughn and Somerville, 1986), 1% (w/v) Suc, and 1.5% (w/v) agar in a growth chamber (16 h of light at 21°C/8 h of dark at 18°C). The transfer DNA insertion line GABI_65B09 (wyr-2) was obtained from the Nottingham Arabidopsis Stock Center. The borr-1 line was generated by CRISPR/CAS9 (Fauser et al., 2014). Primer pairs for genotyping are described in Supplemental Table S1 and Supplemental Figure S1.
Plasmid Construction and Transgenic Plants
The plasmid construction for the CRISPR/CAS9 system was performed as described in Fauser et al. (2014). To obtain the borr knockout plants, 20-bp gene-specific spacer sequences of the BORR gene (Supplemental Table S1) were cloned into the pEn-Chimera, followed by LR recombination reactions with the destination vector pDe-CAS9. The plasmid construction for the amiRNA system was performed as described in Carbonell et al. (Carbonell et al., 2014). To obtain the BORR knockdown plants, 75-bp gene-specific sequences of BORR gene with the AtMIR390a backbone (Supplemental Table S2; Carbonell et al., 2014) were synthesized and cloned into pDONR221, followed by LR recombination reactions with the destination vector pGWB602. To create the PROBORR:BORR:GUS construct, 2 kb upstream of the start codon and 1 kb downstream of the stop codon of the BORR gene were amplified by PCR and cloned into pENTR2B by SLiCE. The SmaI site was inserted in front of the stop codon of the BORR construct. The resulting construct was linearized by SmaI digestion and was ligated to the GUS gene, followed by LR recombination reactions with the destination vector pGWB501 (Nakagawa et al., 2007). To create PROBORR:BORR:FPs constructs, the GUS gene in the PROBORR:BORR:GUS construct was replaced by the ORF for monomeric GFP (mGFP) or TagRFP-T. To create the PROBORR:GFP:BORR construct, the SmaI site was inserted in front of the start codon of the BORR construct. To create PROINCENP:FPs:INCENP, the genomic fragment of INCENP gene was amplified by PCR and cloned into pENTR2B by the SLiCE method. The SmaI site was inserted in front of the start codon of INCENP. The resulting construct was linearized by SmaI digestion and ligated to the mGFP or mRUBY3 gene followed by LR recombination reactions with the destination vector pGWB501. Primer pairs for plasmid construction are described in Supplemental Table S1. Transgenic plants were generated by the floral dip method. The Agrobacterium tumefaciens strain GV3101 (pMP90), harboring the gene of interest on a binary plasmid, was grown in 3 mL of Luria-Bertani medium at 28°C. Agrobacteria were resuspended in 3 mL of transformation buffer containing 5% (w/v) Suc and 0.05% (v/v) Silwet l-77 (Momentive Performance Materials), and used for plant transformation.
Expression Analysis by RT-qPCR
Total RNA was isolated from 7-d-old seedlings with the RNeasy Plant Mini Kit (Qiagen). Then, 300 mg of total RNA were reverse transcribed with ReverTra Ace reverse transcription quantitative PCR (RT-qPCR) Master Mix with gDNA Remover (TOYOBO) according to the manufacturer’s instructions. qPCR was performed using the Roche LightCycler 480 and the TB Green Premix Ex Taq (TaKaRa). PP2AA3 (AT1G13320) was used as the reference gene (Czechowski et al., 2005). Primer pairs for BORR and PP2AA3 are described in Supplemental Table S1. All experiments were performed in three biological replicates.
Confocal Microscopy and Image Analysis
Root tips of 5-d-old seedlings were used for live cell imaging. Samples were put on glass-bottom dishes and covered with a solid medium containing one-half strength Arabidopsis nutrient solution, 1% (w/v) Suc, and 1.5% (w/v) agar. Confocal images of mitotic cells were acquired by an inverted Nikon ECLIPSE Ti-U microscope equipped with a YOKOGAWA CSU-X spinning disc detector unit connected to an EM-CCD camera (iXon3 DU897; Andor) and a laser combiner system (500 series; Andor), using a Plan Apo 60×/1.20 water immersion objective. GFP was excited at 488 nm with a 520/35 emission filter and TagRFP-T and mRUBY3 at 561 nm with a 617/73 emission filter. Images were obtained at 20-s intervals and corrected for sample drift using the StackReg plugin (version 1.49; ImageJ). To obtain line profile data, images were analyzed by the RGB Profiler plugin (version 1.49; ImageJ).
For meiotic live cell imaging, flower buds from 1-month-old plants were used. Sample preparation was performed as described in Prusicki et al. (2018).
Images were obtained every 1 min for BORR:GFP and every 3 min for GFP:INCENP. Images were corrected for the sample drift by the StackReg plugin (version 1.49; ImageJ).
GUS Histochemical Analysis
Samples were fixed in 90% (v/v) acetone for 15 min and were washed in 50 mm sodium phosphate buffer. The fixed samples were incubated in GUS solution [50 mm sodium phosphate buffer (pH 7.0), 0.5% (v/v) Triton X-100, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, and 0.5 mg mL−1 X-gluc] for 1 h at 37°C. After staining, the samples were cleared in chloral hydrate solution (8 g chloral hydrate, 1 mL 100% [v/v] glycerol, and 2 mL distilled water).
Y2H Assay
Y2H assays were performed as described in Komaki and Schnittger (2017). All of the complementary DNAs (cDNAs) tested were amplified by PCR using gene-specific primers from cDNA made from total RNA of wild-type Arabidopsis, followed by PCR with universal attB primers, and cloned into pDONR221. The subcloned BORR cDNA was recombined into pGBT9-C (DNA-BD), in which GAL4-BD is fused with the C terminus of BORR by LR recombination reactions. The other subcloned cDNAs were recombined into either the conventional vector pGBT9 (DNA-BD) or pGAD424 (AD). Primer pairs for plasmid construction are described in Supplemental Table S1.
Protein Extraction and IP Assay
For IP, 1 g of 7-d-old seedlings expressing BORR:RFP with GFP:INCENP or GFP was ground to a fine powder in liquid nitrogen using a mortar. Total protein was extracted in 2 mL of extraction buffer containing 50 mm Tris-HCl (pH8.0), 150 mm NaCl, 1% (v/v) IGEPAL CA-630, and an EDTA-Free Protease Inhibitor Cocktail (Roche) for 30 min on ice and centrifuged for 10 min at 20,000g at 4°C. The supernatant was incubated for 1 h on ice with anti-GFP magnetic beads (Miltenyi Biotec), and beads were washed four times with extraction buffer on the magnetic field. Then, the beads were boiled in SDS sample buffer to release the proteins. Protein samples were detected with 1:2,000 diluted anti-GFP (A6455; Thermo Scientific) and 1:1,000 diluted anti-RFP (AB233; Evrogen) as primary antibodies and subsequently with 1:10,000 diluted anti-rabbit IgG, HRP-linked antibody (NA934; GE Healthcare) as secondary antibody.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource database under the following accession numbers: AT4G32830 (AUR1), AT2G25880 (AUR2), AT2G45490 (AUR3), AT4G39630 (BORR), AT5G55820 (INCENP/WYRD). Borealin protein sequence data for other organisms from this article can be found in the National Center for Biotechnological Information data libraries under the following accession numbers: XP_003570304.1 (Brachypodium distachyon), XP_006428073.1 (Citrus clementina), NP_609279.1 (Drosophila melanogaster), NP_001243804.1 (Homo sapiens), PTQ34788.1 (Marchantia polymorpha), XP_002503544.1 (Micromonas commoda), NP_080836.3 (Mus musculus), XP_015650359.1 (Oryza sativa), XP_024357151.1 (Physcomitrella patens), XP_024458072.1 (Populus trichocarpa), AJV31725.1 (Saccharomyces cerevisiae), CAA22184.2 (Schizosaccharomyces pombe), XP_024537708.1 (Selaginella moellendorffii), XP_002280684.1 (Vitis vinifera), NP_001002902.1 (Xenopus tropicalis), NP_001142076.1 (Zea mays).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Complementation test of BORR and INCENP mutants.
Supplemental Figure S2. Both BORR and AUR3 localize to inner centromeres.
Supplemental Figure S3. Plant growth phenotypes of wild-type and BORR knockdown plants.
Supplemental Figure S4. Colocalization of CPC components.
Supplemental Figure S5. Both BORR and AUR3 localize to inner centromeres.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Synthesized oligonucleotides used in this study.
Supplemental Movie S1. Subcellular localization of GFP:INCENP during mitosis.
Supplemental Movie S2. Subcellular localization of BORR:GFP during mitosis.
Supplemental Movie S3. Subcellular localization of AUR3:GFP during mitosis.
Supplemental Movie S4. Colocalization of AUR3:GFP and BORR:RFP.
Supplemental Movie S5. Co-localization of AUR3:GFP and RFP:INCENP.
Supplemental Movie S6. Colocalization of GFP:INCENP and BORR:RFP.
Supplemental Movie S7. Comparison of subcellular localization between BORR:GFP and RFP:CENH3.
Supplemental Movie S8. Subcellular localization of BORR:GFP during meiosis.
Supplemental Movie S9. Subcellular localization of GFP:INCENP during meiosis.
Acknowledgments
We thank Konstantinos Lampou for critical reading of and helpful comments on the manuscript.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SCHN 736/8–1 to A.S.), the Japan Society for the Promotion of Science (KAKENHI grant no. JP18K45678 to S.K.), the Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas no. JP19H04864 to S.K.), and the University of Hamburg (core funding to A.S.).
Articles can be viewed without a subscription.
References
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC(2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol 10: 1075–1078 [DOI] [PubMed] [Google Scholar]
- Barrett RM, Colnaghi R, Wheatley SP(2011) Threonine 48 in the BIR domain of survivin is critical to its mitotic and anti-apoptotic activities and can be phosphorylated by CK2 in vitro. Cell Cycle 10: 538–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekier ME, Mazur T, Rashid MS, Taylor WR(2015) Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores. Nat Commun 6: 6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Deng X, Mylle E, Besbrugge N, Van Durme M, Demidov D, Tomaštíková ED, Tan TC, Vandorpe M, Eeckhout D, et al. (2019) TPX2-LIKE PROTEIN3 is the primary activator of α-Aurora kinases and is essential for embryogenesis. Plant Physiol 180: 1389–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Heese M, Chen P, Harashima H, Roudier F, Grüttner C, Schnittger A(2018) Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet 14: e1007797. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD(2004) A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 16: 399–411 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cuperus JT, Carrington JC(2014) New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165: 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC(2012) The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13: 789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts SM, Fowler KJ, Kile BT, Hii LL, O’Dowd RA, Hudson DF, Saffery R, Kalitsis P, Earle E, Choo KH(1999) Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum Mol Genet 8: 1145–1155 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR(2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date D, Dreier MR, Borton MT, Bekier ME II, Taylor WR(2012) Effects of phosphatase and proteasome inhibitors on Borealin phosphorylation and degradation. J Biochem 151: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date DA, Jacob CJ, Bekier ME, Stiff AC, Jackson MW, Taylor WR(2007) Borealin is repressed in response to p53/Rb signaling. Cell Biol Int 31: 1470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D, Lermontova I, Weiss O, Fuchs J, Rutten T, Kumke K, Sharbel TF, Van Damme D, De Storme N, Geelen D, et al. (2014) Altered expression of Aurora kinases in Arabidopsis results in aneu- and polyploidization. Plant J 80: 449–461 [DOI] [PubMed] [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A(2005) Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Schnittger A(2011) Use of phospho-site substitutions to analyze the biological relevance of phosphorylation events in regulatory networks. Methods Mol Biol 779: 93–138 [DOI] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H(2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Goldenson B, Crispino JD(2015) The aurora kinases in cell cycle and leukemia. Oncogene 34: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kiyono T, Tomono Y, Kawajiri A, Urano T, Furukawa K, Nigg EA, Inagaki M(2006) Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat Cell Biol 8: 180–187 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Vernos I(2004) The mechanism of spindle assembly: Functions of Ran and its target TPX2. J Cell Biol 166: 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Somerville C(1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hindriksen S, Lens SMA, Hadders MA(2017) The ins and outs of Aurora B inner centromere localization. Front Cell Dev Biol 5: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Körner R, Nigg EA(2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 14: 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E(2007) Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131: 271–285 [DOI] [PubMed] [Google Scholar]
- Kaur H, Bekier ME, Taylor WR(2010) Regulation of Borealin by phosphorylation at serine 219. J Cell Biochem 111: 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Matsunaga S, Nakagawa K, Kurihara D, Yoneda A, Hasezawa S, Uchiyama S, Fukui K(2005) Characterization of plant Aurora kinases during mitosis. Plant Mol Biol 58: 1–13 [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H(2010) Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirioukhova O, Johnston AJ, Kleen D, Kägi C, Baskar R, Moore JM, Bäumlein H, Gross-Hardt R, Grossniklaus U(2011) Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development 138: 3409–3420 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Lee SH(2015) The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Schnittger A(2016) The spindle checkpoint in plants—a green variation over a conserved theme? Curr Opin Plant Biol 34: 84–91 [DOI] [PubMed] [Google Scholar]
- Komaki S, Schnittger A(2017) The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev Cell 43: 172–185 [DOI] [PubMed] [Google Scholar]
- Kozgunova E, Suzuki T, Ito M, Higashiyama T, Kurihara D(2016) Haspin has multiple functions in the plant cell division regulatory network. Plant Cell Physiol 57: 848–861 [DOI] [PubMed] [Google Scholar]
- Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA(2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol 158: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Matsunaga S, Kawabe A, Fujimoto S, Noda M, Uchiyama S, Fukui K(2006) Aurora kinase is required for chromosome segregation in tobacco BY-2 cells. Plant J 48: 572–580 [DOI] [PubMed] [Google Scholar]
- Lee KH, Avci U, Qi L, Wang H(2019) The α-Aurora kinases function in vascular development in Arabidopsis. Plant Cell Physiol 60: 188–201 [DOI] [PubMed] [Google Scholar]
- London N, Biggins S(2014) Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 15: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LY, Wood JL, Ye L, Minter-Dykhouse K, Saunders TL, Yu X, Chen J(2008) Aurora A is essential for early embryonic development and tumor suppression. J Biol Chem 283: 31785–31790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Jürgens G(2016) Plant cytokinesis—No ring, no constriction but centrifugal construction of the partitioning membrane. Semin Cell Dev Biol 53: 10–18 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Cormier A, Tyers RG, Pigula A, Peng Y, Drubin DG, Barnes G(2011) Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol 194: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovská B, Cenklová V, Pochylová Z, Kourová H, Doskočilová A, Plíhal O, Binarová L, Binarová P(2012) Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol 193: 590–604 [DOI] [PubMed] [Google Scholar]
- Prusicki MA, Keizer EM, van Rosmalen RP, Komaki S, Seifert F, Müller K, Wijnker E, Fleck C, Schnittger A(2018) Live cell imaging of meiosis in Arabidopsis thaliana. eLife 8: e42834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T(2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A(2005) Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell 18: 379–391 [DOI] [PubMed] [Google Scholar]
- Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J(2017) Aurora kinases: Novel therapy targets in cancers. Oncotarget 8: 23937–23954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, Stukenberg PT(2016) A centromere-signaling network underlies the coordination among mitotic events. Trends Biochem Sci 41: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH(2000) Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 10: 1319–1328 [DOI] [PubMed] [Google Scholar]
- Vader G, Kauw JJ, Medema RH, Lens SM(2006) Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep 7: 85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E(2011) Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23: 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Lens SM(2014) Cell division: Control of the chromosomal passenger complex in time and space. Chromosoma 123: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal MS, Hengeveld RC, van der Horst A, Lens SM(2012) Cell division control by the Chromosomal Passenger Complex. Exp Cell Res 318: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Demidov D, Lermontova I, Beeckman T, Van Damme D(2016) Aurora kinases throughout plant development. Trends Plant Sci 21: 69–79 [DOI] [PubMed] [Google Scholar]
- Wijnker E, Schnittger A(2013) Control of the meiotic cell division program in plants. Plant Reprod 26: 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Ogawa H, Vagnarelli P, Bergmann JH, Hudson DF, Ruchaud S, Fukagawa T, Earnshaw WC, Samejima K(2009) INCENP-aurora B interactions modulate kinase activity and chromosome passenger complex localization. J Cell Biol 187: 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Heike T, Kumada T, Shibata M, Takaoka Y, Kitano A, Shiraishi K, Kato T, Nagato M, Okawa K, et al. (2008) Loss of Borealin/DasraB leads to defective cell proliferation, p53 accumulation and early embryonic lethality. Mech Dev 125: 441–450 [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen X, McCormick S(2011) The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]