Barley calmodulin HvCaM1 regulates salt tolerance by mediating the expression of Na+ transport-related HKT1 genes and the transcriptional regulation by a CaM-binding transcription activator, HvCAMTA4.
Abstract
Calcium (Ca2+) signaling modulates sodium (Na+) transport in plants; however, the role of the Ca2+ sensor calmodulin (CaM) in salt tolerance is elusive. We previously identified a salt-responsive calmodulin (HvCaM1) in a proteome study of barley (Hordeum vulgare) roots. Here, we employed bioinformatic, physiological, molecular, and biochemical approaches to determine the role of HvCaM1 in barley salt tolerance. CaM1s are highly conserved in green plants and probably originated from ancestors of green algae of the Chlamydomonadales order. HvCaM1 was mainly expressed in roots and was significantly up-regulated in response to long-term salt stress. Localization analyses revealed that HvCaM1 is an intracellular signaling protein that localizes to the root stele and vascular systems of barley. After treatment with 200 mm NaCl for 4 weeks, HvCaM1 knockdown (RNA interference) lines showed significantly larger biomass but lower Na+ concentration, xylem Na+ loading, and Na+ transportation rates in shoots compared with overexpression lines and wild-type plants. Thus, we propose that HvCaM1 is involved in regulating Na+ transport, probably via certain class I high-affinity potassium transporter (HvHKT1;5 and HvHKT1;1)-mediated Na+ translocation in roots. Moreover, we demonstrated that HvCaM1 interacted with a CaM-binding transcription activator (HvCAMTA4), which may be a critical factor in the regulation of HKT1s in barley. We conclude that HvCaM1 negatively regulates salt tolerance, probably via interaction with HvCAMTA4 to modulate the down-regulation of HvHKT1;5 and/or the up-regulation of HvHKT1;1 to reduce shoot Na+ accumulation under salt stress in barley.
Salt stress is a major abiotic factor restricting crop growth and productivity, which is exacerbated by global climate change and human activities (Munns and Tester, 2008; Munns et al., 2020). Excess sodium (Na+) causes ion toxicity and also ion imbalance through competitively inhibiting the uptake of some mineral nutrients such as potassium (K+). Maintaining lower Na+ accumulation in shoots is crucial for salt-tolerant plant species and genotypes under salt stress (Munns, 2005; Munns and Tester, 2008; Horie et al., 2012; Deinlein et al., 2014; Shen et al., 2016, 2017). Lower shoot Na+ accumulation in plants is attributed to either shoot Na+ exclusion or vacuolar sequestration, both of which are regulated by membrane transporters, such as some members of high-affinity potassium transporters (HKTs), Na+/H+ exchangers (NHXs), and Salt Overly Sensitive1 (SOS1; Apse et al., 1999; Shi et al., 2000; Zhang and Blumwald, 2001; Davenport et al., 2007; Barragán et al., 2012; Huang et al., 2020). Activation of membrane transport systems in roots plays a key role in reducing shoot Na+ accumulation and enhancing salt tolerance in many plant species (Zhu, 2002; Shabala and Cuin, 2008; Ismail and Horie, 2017). However, the mechanisms underlying Na+ exclusion and translocation and the link to calcium (Ca2+) signaling require further investigation.
Ca2+-mediated signal transduction plays a regulatory role in plant salt tolerance (Zhu, 2002, 2016; Demidchik et al., 2018). For instance, SOS3 (calcineurin-like protein [CBL4]) is a Ca2+ sensor, which perceives the Na+-induced cytosolic Ca2+ rise via the interaction with SOS2 (calcineurin-interacting protein kinase [CIPK24]), causing phosphorylation and activation of the plasma membrane SOS1/NHX7 (Shi et al., 2000; Zhu, 2002; El Mahi et al., 2019). The activation of SOS1 in root epidermal cells and xylem parenchyma cells leads to the extrusion of Na+ from roots to rhizosphere (Shi et al., 2002; Zhu, 2016). Meanwhile, a rapid elevation of cytosolic Ca2+ under Na+ stress can also be captured by other Ca2+ sensors, including calmodulin (CaM), calmodulin-like proteins, and calcium-dependent protein kinases (Galon et al., 2010; Cho et al., 2016). Multiomics techniques have revealed that CaM exhibits salt-induced expression changes at the transcriptional and translational levels in seedlings or roots, suggesting that these Ca2+ sensors participate in salt stress signaling (Zhang et al., 2012; Wu et al., 2014; Zhu et al., 2015; Shen et al., 2018; El Mahi et al., 2019). However, regulation of the complex Ca2+ signaling by CaMs to function in plant salt tolerance is still elusive.
CaMs usually encode a single peptide protein consisting of a pair of unique Ca2+-binding EF-hands and have no other functional domains or motifs (Reddy et al., 2011). Meanwhile, CaM protein sequences are highly conserved in green plants, with an average 90% identity of 65 members detected in 15 representative plant species from Chlorophyceae to angiosperms (Zhu et al., 2015). Moreover, CaMs have less variation and fewer family members than other Ca2+ sensors (Xu et al., 2015; Zhu et al., 2015). However, little attention has been paid to the origin and evolution of CaMs in green plants. The recent advancement of assembled genomes (Kersey, 2019) and transcriptome databases (Leebens-Mack et al., 2019) will likely unlock the evolution of CaMs, providing insights into their roles in stress tolerance of plants.
CaM may function through posttranscriptional modification similar to that in SOS3 via the regulation of downstream receptors, such as CaM-binding proteins (CBPs; Galon et al., 2010; Batistič and Kudla, 2012; El Mahi et al., 2019). In different plants, CaM can adjust the affinity of the Ca2+/CaM complex for the selection of special receptors (e.g. phosphodiesterase) and targets (e.g. Cyclic Nucleotide-Gated Channel12) after recognizing the cytosolic Ca2+ change for a specific stress response (Gifford et al., 2013; DeFalco et al., 2016). In Arabidopsis (Arabidopsis thaliana), AtCaM1/4 shows an up-regulated expression under salt stress, resulting in enhanced salt tolerance via binding with S-nitrosoglutathione reductase in nitric oxide signaling (Zhou et al., 2016). CaM-binding protein kinases (AtCaM2/3/5) modulate the activity of heat shock factors through phosphorylation of a CaM-binding protein kinase (AtCBK3) in heat stress signaling (Zhang et al., 2009), while AtCaM6 and AtCaM7 are involved in the light-dependent stress pathway and with reactive oxygen species signaling (Kushwaha et al., 2008; Al-Quraan et al., 2011). In rice (Oryza sativa), OsCaM1-1 also shows expression patterns similar to AtCaM1/4 (Chinpongpanich et al., 2012), and its overexpression mitigates salt-induced oxidative damage (Kaewneramit et al., 2019). In addition, overexpression of GmCaM4 enhances salt tolerance by regulating the expression of a series of salt-responsive genes, including a key Pro biosynthetic enzyme, P5CS1, in soybean (Glycine max; Yoo et al., 2005; Rao et al., 2014). Moreover, many transcription factors, including CaM-binding transcription activators (CAMTAs), CBP60s, MYBs, WRKYs, bZIPs, bHLHs, NACs, and GTs, have been identified as CBPs, suggesting that CaM might play important roles in the transcriptional regulation of salt stress tolerance in plants (Galon et al., 2010; Xi et al., 2012; Shkolnik et al., 2019). For example, AtCAMTA6 regulates the expression of AtHKT1 via the binding sites of CGCG-core or CGTGT-core, contributing to Na+ homeostasis during seed germination (Shkolnik et al., 2019). Therefore, the roles of CaMs in the salt stress response have been reported, but their function in the regulation of Na+ transport remains to be revealed.
As one of the most salt-tolerant diploid crop species with a large and complex genome, barley (Hordeum vulgare) has been used as a model plant to decipher the physiological and molecular mechanisms of salinity tolerance in plants (Chen et al., 2005; Munns and Tester, 2008; Nevo and Chen, 2010; Mascher et al., 2017; Dai et al., 2018). However, there are limited reports on CaM members and their functions in barley. We have previously identified one salt-responding CaM protein (protein identifier A0MMD0) in the root proteome of wild barley genotypes (Wu et al., 2014; Shen et al., 2018).
We hypothesized that HvCaM1 plays a critical role in the establishment of higher salt tolerance in barley via the regulation of Ca2+ signaling transduction, transcriptional factors, and membrane transport. Here, we cloned HvCaM1 (gene identifier HORVU0Hr1G001270) and comprehensively analyzed its function through overexpression (OE) and RNA interference (RNAi) in barley with a range of physiological and molecular techniques. We found that HvCaM1 negatively regulates salt tolerance in barley and participates in Na+ transport from roots to shoots. Furthermore, we also detected that HvCaM1 regulates two HvHKT1 members via the differential interaction with the transcriptional activator HvCAMTA4.
RESULTS
Evolutionary Bioinformatics and Phylogenetic Analysis of HvCaM1
We first cloned the full-length cDNA and genome DNA of HvCaM1 from the barley 'Golden Promise'. HvCaM1 contains two exons and one intron encoding a peptide consisting of 149 amino acids, with two typical EF-hands (Supplemental Fig. S1A). No difference in coding sequences (CDSs) and protein sequences was found between the Tibetan wild barley accessions and barley cultivars (Supplemental Table S1). Sequence similarity analysis showed that HvCaM1 exhibits, on average, 98.8% identity with the CaM members in rice and Arabidopsis with the four conserved Ca2+-binding sites (Supplemental Fig. S1B). Phylogenetic analysis showed that HvCaM1 is relatively close to OsCaM1-1 and AtCaM1 (Supplemental Fig. S1C).
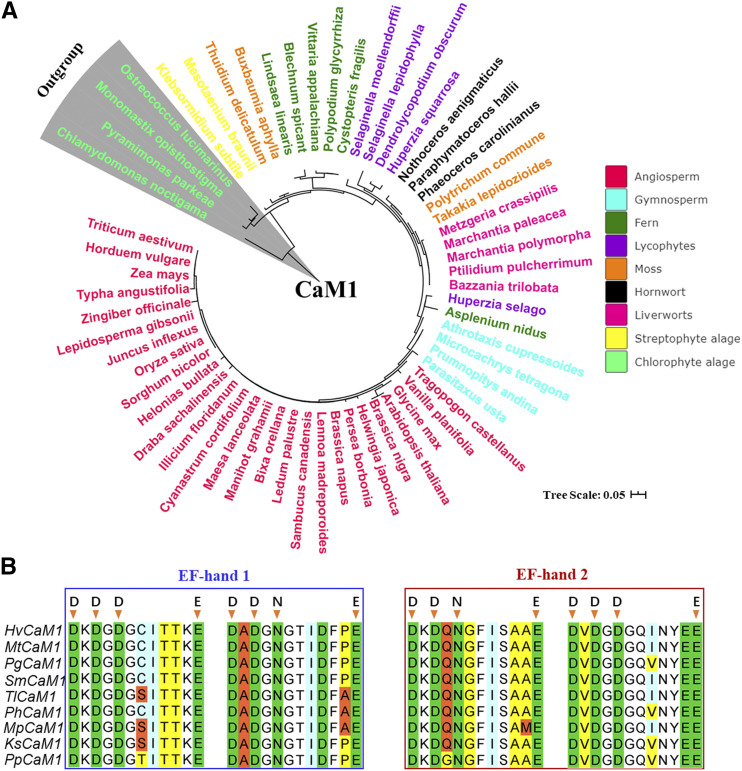
Evolutionary analysis further indicated that CaM1 likely originates from the ancestors of the Volvocales order of chlorophyte algae (Chlamydomonadaceae; Fig. 1A). CaM1s are particularly conserved in angiosperms, with up to 87% identity detected in the CaM1 proteins of 60 representative plant species (Supplemental Table S2). Moreover, sequence analysis showed that each CaM1 protein contains two conserved EF-hand motifs and the ubiquitous presence of a consensus sequence of DDGDGE and DXDXNE in the protein structure of Ca2+-binding sites in major lineages (Fig. 1B). However, the difference of a couple of amino acids in the protein sequences of different chlorophyte algae indicated that CaM1s likely evolved from Chlamydomonadaceae but are conserved from streptophyte algae (e.g. Klebsormidium subtile) and then diversified in their function during the adaptation of land plants (Fig. 1; Supplemental Fig. S1).
Figure 1.
Phylogenetic tree and sequence analysis of HvCaM1. A, Evolution analysis of predicted CaM1 candidates in 60 representative species of the major lineage of green plants. All sequences were downloaded from the 1,000 Plant Transcriptome and EnsemblPlants databases. B, Sequence alignment of the two conserved EF-hand domains among nine representative species of major lineages. Hv, Hordeum vulgare; Ks, Klebsormidium subtile; Mp, Marchantia polymorpha; Mt, Microcachrys tetragona; Pg, Polypodium glycyrrhiza; Ph, Paraphymatoceros hallii; Pp, Pyramimonas parkeae; Sm, Selaginella moellendorffii; Tl, Takakia lepidozioides. Arrowheads indicate conserved amino acids in the four Ca2+-binding sites.
HvCaM1 Is Up-Regulated in Roots under Salt Stress and Mainly Localized in Stele and Vascular Cells of Roots
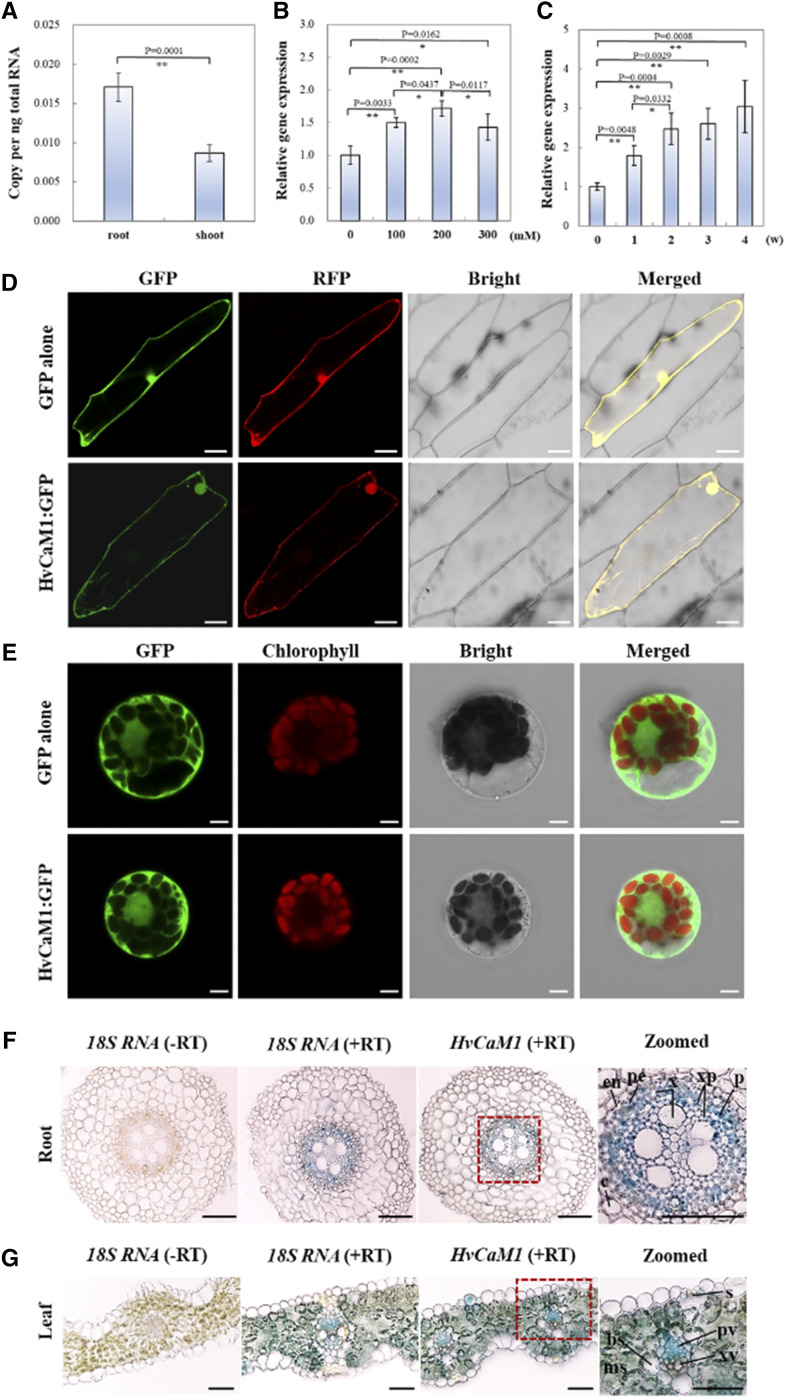
We investigated the expression of HvCaM1 in roots and shoots of barley seedlings. Quantitative PCR (qPCR) analysis showed that HvCaM1 is expressed in both roots and shoots, with significantly higher expression in roots (Fig. 2A). Moreover, the expression of HvCaM1 in roots was greatly affected by salt concentration, with the highest up-regulated value occurring at 200 mm NaCl treatment (Fig. 2B). Interestingly, the expression of HvCaM1 was significantly up-regulated by 3.04-fold after 4 weeks of long-term salt stress compared with the control value (Fig. 2C).
Figure 2.
Expression pattern analysis of HvCaM1. A, Absolute quantification of HvCaM1 transcript in roots and shoots under the normal condition. B, Relative expression of HvCaM1 in roots exposed to salt concentration gradients within 2 h. C, Relative HvCaM1 expression within 4 weeks of salt exposure (200 mm). Asterisks represent highly significant and significant differences determined by independent Student’s t test (*P < 0.05 and **P < 0.01; n = 4; error bars indicate se). D and E, Subcellular localization of HvCaM1 in onion epidermal cells (D) and barley leaf protoplasts (E). F and G, In situ PCR analysis of HvCaM1 transcript in root (F) and leaf (G) cross sections of 14-d-old seedlings. bs, Bundle sheath; c, cortex; en, endodermis; ms, mesophyll; p, phloem; pe, pericycle; pv, phloem vessel; s, stomatal guard cell; xp, xylem parenchyma; xv, xylem vessel. Bars = 50 μm (D), 5 μm (E), and 100 μm (F and G).
The subcellular localization detected by GFP showed that HvCaM1 appears to be expressed in the nucleus and plasma membrane but not in the cytosol of onion (Allium cepa) epidermal cells, while it is expressed in the nucleus, cytosol, and plasma membrane in barley leaf mesophyll protoplasts, suggesting that it may be an intracellular signal protein (Fig. 2, D and E). We performed in situ PCR to determine the cellular localization of HvCaM1 transcripts in cross sections of roots and leaves. In roots, the HvCaM1 transcripts were mainly detected in phloem and xylem parenchyma cells of the stele (Fig. 2F). In leaves, the transcripts were found predominantly in phloem vessel and mesophyll cells and to a lesser extent in stomatal cells (Fig. 2G). Additionally, observation of fluorescence signals from GFP fused with the 2.1-kb promoter of HvCaM1 in the transformed barley seedlings also confirmed that HvCaM1 mainly localizes in the stele of roots (Supplemental Fig. S2A) and in stomatal guard cells, subsidiary cells, and leaf mesophyll cells (Supplemental Fig. S2B).
HvCaM1 Negatively Regulates Salt Tolerance via the Modulation of Na+ Transport in Barley
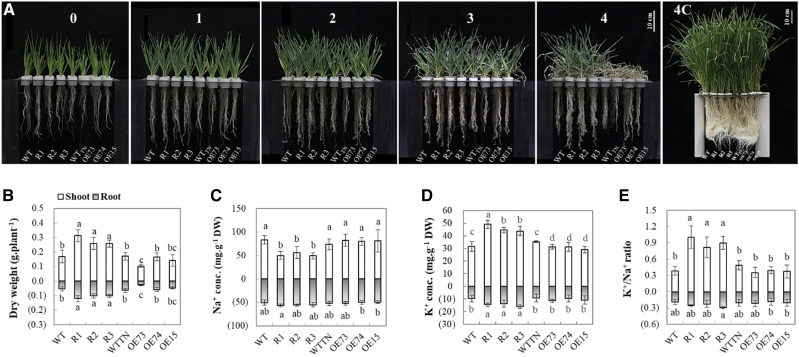
To examine the function of HvCaM1, we generated 46 and 52 independent knockdown (RNAi) and OE barley lines, respectively. Three RNAi lines and three OE lines were examined by PCR verification via specific primers (Supplemental Fig. S3A), showing the significant average decrease and increase in the expression levels of HvCaM1 by 59% and 2.41-fold, respectively, in comparison with the wild-type plants (Supplemental Fig. S3B). The phenotypic difference between the genetically transformed lines and the wild-type plant was evident 3 weeks after 200 mm salt exposure (Fig. 3A). RNAi lines showed significantly higher salt tolerance than OE and wild-type seedlings 4 weeks after salt stress, as reflected by shoot and root dry weight (Fig. 3B). Only the dry weight of OE73 was significantly lower than that of the wild type in response to salt stress. Meanwhile, shoots of RNAi lines had higher relative water content than those of OE lines and wild-type plants at 4 weeks of salt stress (Supplemental Fig. S4A). When planted in the saline soil, these RNAi lines were less affected in grain yield traits (spikes per plant, seeds per spike, setting rate, and kernel weight) in comparison with wild-type plants (Supplemental Fig. S5). These results indicate that HvCaM1 may function as a negative regulator of salt tolerance in barley.
Figure 3.
Phenotypic differences and ion responses of wild-type (WT), RNAi, and OE seedlings under salt stress. A, Phenotypes under 200 mm salt treatment and the control condition. Observation was conducted every week from the 14-d-old seedling (referred as the 0 time point) until 4 weeks. Bars = 10 cm. B to E, Dry weight (B), Na+ concentration (C), K+ concentration (D), and K+/Na+ ratio (E) of barley genotypes under salt stress for 4 weeks. RNAi lines, R1, R2, and R3; OE lines, OE73, OE74, and OE15; WTTN, transgenically negative line; 4C, 4 weeks under the control condition from 14-d-old seedlings. Data are means of six replicates ± se (n = 6). Different lowercase letters above the columns represent significant differences at P < 0.05 determined by one-way ANOVA. DW, Dry weight.
After 4 weeks of salt treatment, RNAi lines had a significantly lower shoot Na+ concentration (52 mg g−1 dry weight) than OE (81.4 mg g−1 dry weight) and wild-type (78.6 mg g−1 dry weight) plants, but such differences were not found in roots (Fig. 3C). RNAi lines showed a significantly higher K+ concentration in both roots and shoots (1.52- and 1.45-fold higher) than in wild-type plants, respectively. The OE lines showed significant NaCl-induced reduction of shoot K+ in comparison with the wild type (Fig. 3D). Consistently, RNAi lines showed a significant 144% and 109% higher shoot K+/Na+ ratio than the wild-type and OE lines under salt stress, respectively (Fig. 3E). In addition, RNAi lines had a significantly lower Na+ transport rate from roots to shoots than both OE and wild-type plants, but there was no difference in K+ transport rate or Ca2+ concentration among these genetic lines (Supplemental Fig. S4, B–D). Thus, we hypothesized that HvCaM1 negatively regulates salt tolerance in barley, probably due to root-to-shoot Na+ translocation.
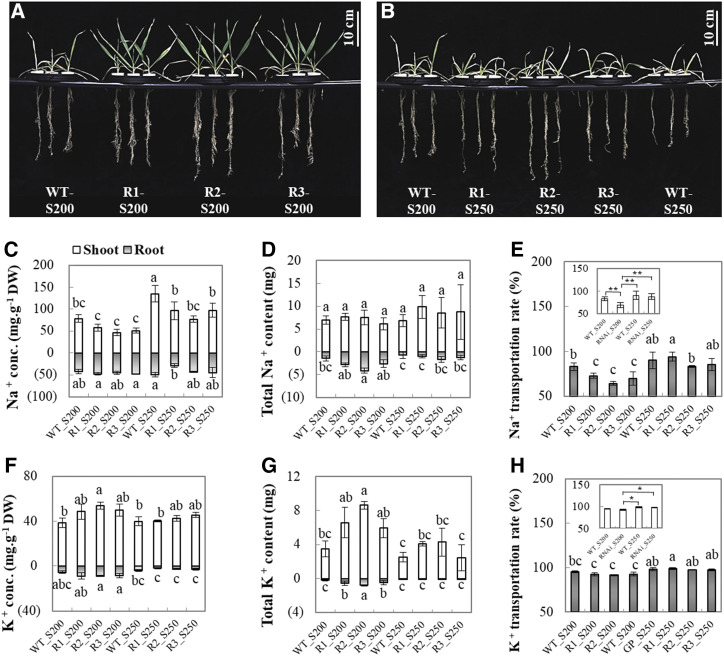
To validate the hypothesis that the involvement of HvCaM1 in the regulation of salt tolerance is related to Na+ transport, we examined the responses of RNAi lines (R1, R2, and R3) and wild-type seedlings to 200 mm (S200) and 250 mm (S250) salt stress solutions (Fig. 4). After 4 weeks of salt stress, three RNAi lines had significantly larger root and shoot dry weight than the wild-type plant under both S200 and S250 treatments, but there was no difference under S300 treatment (Fig. 4, A and B; Supplemental Table S3). Furthermore, wild-type plants had significantly higher shoot Na+ concentrations than RNAi lines under S250 treatment. Such a difference was not detected in roots (Fig. 4C). Meanwhile, three RNAi lines and wild-type plants showed little difference in the total Na+ accumulation per plant (Fig. 4D), but the RNAi lines had much lower Na+ transport rates than the wild-type plants at S200 treatment, while differences at S250 were not significant (Fig. 4E). Clearly, the higher salt stress tolerance of RNAi lines is closely associated with less Na+ transport from roots to shoots, which was mediated by HvCaM1. On the other hand, RNAi lines had higher K+ concentration and accumulation in plant tissues in comparison with wild-type plants at S200 treatment, but no difference was found in the root-to-shoot K+ transport rate (Fig. 4, F–H). Therefore, our results indicate that HvCaM1 negatively regulates salt tolerance, indirectly resulting in reduced Na+ transport from roots to shoots under salt stress.
Figure 4.
Na and K element analysis of wild-type (WT) and RNAi plants under salt conditions. A and B, Wild-type and RNAi seedlings exposed to 200 mm (S200) and 250 mm (S250) salt solutions for 4 weeks. C to H, Na+ concentration (C), Na+ content per plant (D), Na+ transportation rate (E), K+ concentration (F), K+ content per plant (G), and K+ transportation rate (H) of wild-type and RNAi plants under salt stress. RNAi lines, R1, R2, and R3. Data are means of four replicates ± se (n = 4). Different lowercase letters above the columns represent significant differences at P < 0.05 determined by one-way ANOVA. Asterisks in the insets of E and H represent significant and highly significant differences as determined by independent Student’s t test (*P < 0.05 and **P < 0.01). DW, Dry weight.
Silencing of HvCaM1 Modulates HKT1 Expression in Roots
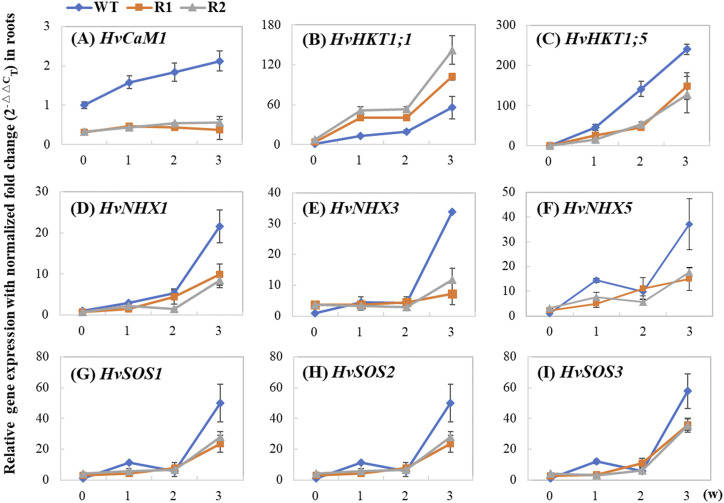
To determine whether HvCaM1 regulates Na+ transport, qPCR analysis of the well-known genes associated with Na+ and K+ transport (HKT1;1–HKT1;5 except HKT1;2, HKT2;1–HKT2;3, NHX1–NHX6, and SOS1–SOS3 members) was performed for roots of the wild type and RNAi lines under the normal condition and salt treatment (Fig. 5). The relative expression (salt stress/control) of all examined Na+ and K+ transporter genes showed the dramatic difference between the two RNAi lines and wild-type plants at 3 weeks after salt treatment (Fig. 5; Supplemental Fig. S6). Moreover, all eight genes were greatly down-regulated in the two RNAi lines in comparison with the wild-type plants, except for HvHKT1;1, which was clearly up-regulated in the RNAi lines. Interestingly, over the 3-week salt treatment, relative expression of HvHKT1;5 showed a similar pattern to that of HvCaM1, while HvHKT1;1 showed a completely opposite response, suggesting the involvement of HvCaM1 in the regulation of both HvHKT1;1 and HvHKT1;5 expression (Fig. 5, A–C). On the contrary, four NHX genes (HvSOS1/HvNHX7, HvNHX1, HvNHX3, and HvNHX5) showed different patterns of relative expression from that of HvCaM1, although these genes were also greatly up-regulated in the wild-type plants under salt treatment (Fig. 5, D–G). HvSOS2 and HvSOS3 showed a similar expression pattern to that of the NHX genes (Fig. 5, H and I). Therefore, the involvement of HvCaM1 in regulating salt stress tolerance may influence the expression of HvHKT1;1 and HvHKT1;5 responsible for Na+ transport.
Figure 5.
Expression analysis of Na+ and K+ transporters under salt stress. The expression levels were investigated in wild-type (WT) and RNAi (R1 and R2) roots when exposed to 200 mm NaCl for 0, 1, 2, and 3 weeks (w). A, HvCaM1. B, HvHKT1;1. C, HvHKT1;5. D, HvNHX1. E, HvNHX3. F, HvNHX5. G, HvSOS1. H, HvSOS2. I, HvSOS3. Data are means of four replicates ± sd.
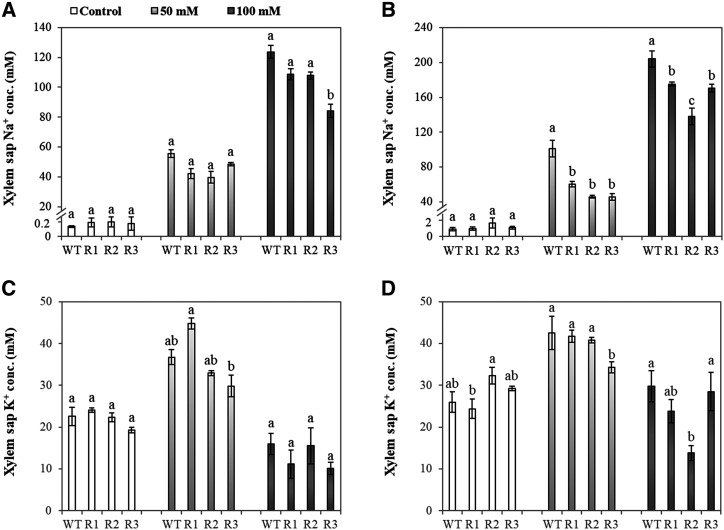
Furthermore, the physiological role of Na+ transporters, mainly for HvHKT1s in Na+ transportation from roots to shoots, was validated by xylem sap analysis (Fig. 6). Na+ concentrations in the xylem sap of HvCaM1 RNAi lines were significantly lower than in the wild-type plants after 7 d of 50 and 100 mm NaCl treatments (Fig. 6B). There was a slight difference in K+ concentration in the xylem sap among these barley lines (Fig. 6, C and D).
Figure 6.
Na+ and K+ concentrations in xylem sap of wild-type (WT) and RNAi plants under control and salt conditions. Na+ (A and B) and K+ (C and D) concentrations of xylem sap are shown in 4-week-old wild-type and RNAi seedlings exposed to control, 50 mm, and 100 mm NaCl solutions for 2 d (A and C) and 7 d (B and D). Data are means ± se (n = 4–6). RNAi lines, R1, R2, and R3. Different lowercase letters represent significant differences at P < 0.05 as determined by one-way ANOVA.
HvCaM1 Interacts with HvCAMTA4 to Regulate the Preferential Expression of HKT1s
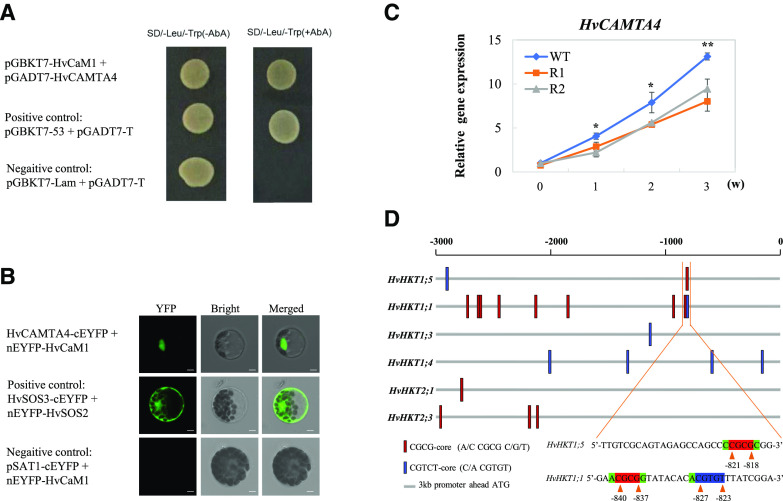
To understand how HvCaM1 affects HKT1 expression (i.e. HvHKT1;1 and HvHKT1;5), we performed a yeast two-hybrid assay using the library constructed from barley roots under salt stress. We identified nine candidate proteins that interact with HvCaM1 in the assay (Supplemental Table S4). HvCAMTA4 (HORVU2Hr1G069950) was among the candidate proteins and contains an IQ domain for CaM binding and a CG-1 domain with specific DNA-binding activity (specific CGCG-core and corecognized CGTCT-core with an abscisic acid response element) to function as a coactivator of transcription.
A bimolecular fluorescence complementation (BiFC) assay confirmed the interaction between HvCaM1 and HvCAMTA4 in both the yeast (Saccharomyces cerevisiae) system and barley protoplasts (Fig. 7, A and B). Compared with the sites for protein-protein interaction between HvSOS2 and HvSOS3 in the nucleus, cytoplasm, and plasma membrane, the interaction between HvCaM1 and HvCAMTA4 was limited to the nucleus, indicating a role for HvCaM1 in the regulation of HvCAMTA4-mediated transcriptional activation (Fig. 7B). Furthermore, qPCR analysis showed consistent patterns of expression for HvCAMTA4 and HvHKT1;5, with a significant correlation of r = 0.97 (P < 0.01), in the roots of wild-type and RNAi lines under salt stress (Figs. 5C and 7C).
Figure 7.
HvCaM1 interacts with HvCAMTA4 via specific binding sites in the promoters of HKT1 genes. A, Yeast interaction assay between HvCaM1 and HvCAMTA4. B, BiFC assay between HvCaM1 and HvCAMTA4 in barley protoplasts. Bars = 5 μm. C, Expression levels of HvCAMTA4 in roots of wild-type (WT) and RNAi plants under 200 mm NaCl for 3 weeks (w). Asterisks represent significant and highly significant differences as determined by independent Student’s t test in roots of wild-type and RNAi plants (*P < 0.05 and **P < 0.01). Data are means of four replicates ± sd. D, The cis-regulatory elements of CGCG-core and CGTGT-core along 3-kb promoter regions of HKT genes, marked as red and blue, respectively. The specific sequences are enlarged in the promoters of HvHKT1;5 and HvHKT1;1.
Then we identified binding sites along the 3-kb promoter region of HKT genes using the two CAMTA-recognized motifs (CGCG-core and CGTCT-core; Supplemental Table S5). Within a 2-kb region, the cis-regulatory elements were detected in all HvHKT1s but not in HvHKT2s of barley (Fig. 7D). Among the members of the barley HKT1 subfamily, the CGCG-core was only found in the promoters of HvHKT1;5 and HvHKT1;1, while HvHKT1;3 and HvHKT1;4 specifically had only CGTCT-cores. Moreover, only one CGCG-core site was found in the promoter of HvHKT1;5 (−818 to −821 bp), which is quite close to the regions in HvHKT1;1 (−837 to −840 bp for CGCG-core and −823 to −827 bp for CGTCT-core) and might be considered an important cis-regulatory region (Fig. 7D). This indicated that HvCaM1 likely regulates the expression of HvHKT1;5 and HvHKT1;1 for Na+ transport through coordinated interaction with HvCAMTA4.
DISCUSSION
Down-Regulation of HvCaM1 Enhances Salt Tolerance in Barley
In plants, CaMs are Ca2+ sensors responding rapidly to early signaling transduction under salt stress (Zhang et al., 2012; Rao et al., 2014; Zhou et al., 2016). However, the roles of CaM in the long-term response to salt stress remain elusive. Previously, we discovered a salt-responding HvCaM1 in the analysis of the barley root proteome (Wu et al., 2014; Shen et al., 2018). This study shows that HvCaM1 is involved in the regulation of salt tolerance via the modulation of gene expression of Na+ transporters HvHKT1;5 and HvHKT1;1 and HvCAMTA4-mediated transcriptional regulation. In Arabidopsis, the knockout mutants Atcam1 and Atcam4 show hypersensitivity to salt stress (Zhou et al., 2016). Similarly, higher expression of both rice OsCaM1 and soybean GmCaM4 enhances salt tolerance (Chinpongpanich et al., 2012; Rao et al., 2014). However, we found that the knockdown (RNAi) lines of HvCaM1 showed a large increase of salt tolerance, as reflected by their larger biomass than wild-type and OE lines under 200 and 250 mm salt treatments (Figs. 3 and 4). There are many reports showing that gene silencing is beneficial for enhanced resistance and production (e.g. AtWRKY11, OsMADS26, and TaDA1), but overexpressing the same gene has negative effects in transgenic plants (Journot-Catalino et al., 2006; Khong et al., 2015; Liu et al., 2020). One possible explanation is that constitutive overexpression of a regulator gene can modulate the expression of downstream stress-associated genes, which could impose a major energy cost to the plants (Munns et al., 2020; Shabala et al., 2020). For example, drought-hypersensitive OsMADS26-overexpressing rice lines modulated 412 differentially expressed genes as compared with 95 differentially expressed genes in the RNAi rice lines, which displayed an enhanced drought tolerance (Khong et al., 2015).
HvCaM1 appears to be highly conserved in comparison with its closest homologous genes, AtCaM1 and OsCaM1 (Fig. 1; Supplemental Fig. S1). Why does it show a different function in salt tolerance in barley? Our previous studies on the root proteome found that HvCaM1 is significantly down-regulated in the salt-tolerant barley genotypes (XZ16, XZ26, and CM72) under salt stress (Wu et al., 2014; Shen et al., 2018). A plausible explanation is that distinct functions of CaMs among the plants may be driven by natural selection or adaptation favoring a complex survival system in responding to external environments (Zhu et al., 2015; Zhao et al., 2019). Barley originated from the Middle East, where drought and high salinity are natural environmental conditions (Nevo and Chen, 2010), in contrast to the natural habitats of Arabidopsis, rice, and soybean (Hyten et al., 2006; Kovach et al., 2009; Beilstein et al., 2010).
Moreover, the expression level of HvCaM1 varied greatly with NaCl concentrations and the exposed time in salt treatments (Fig. 2, B and C). As intracellular Ca2+ sensors, the expression level of CaMs could be changed with cytosolic Ca2+ oscillation elicited by Na+ stress (Reddy et al., 2011; Ismail and Horie, 2017). In comparison with barley, Arabidopsis, rice, and soybean are less tolerant to salt (Munns and Tester, 2008; Horie et al., 2012), and all CaM1s in these four species are linked to salt tolerance/sensitivity. In addition, there is no difference in the protein sequence of HvCaM1 among those genotypes (Supplemental Table S1) and high similarity to CaM1s in Arabidopsis, rice, and soybean, suggesting that its function is probably related to posttranscriptional modification or translational regulatory networks (Batistič and Kudla, 2012; El Mahi et al., 2019). Therefore, unlike AtCaM1 and other CaM1s, HvCaM1 may negatively regulate salt tolerance with the function of long-term adaptation to salinity in barley, which has not been previously identified.
HvCaM1 May Be Involved in the Modulation of the Long Distance of Na+ Transport from Roots to Shoots
In general, roots have higher salt tolerance than shoots, and lower shoot Na+ concentration is mainly associated with less Na+ transport from roots, which is an important characteristic for higher salt tolerance in most plants, including barley (Shabala et al., 2010; Zahra et al., 2014; Shen et al., 2016). Here, it is obvious that HvCaM1 knockdown lines had significantly lower shoot Na+ concentration than the wild-type or OE lines under both 200 and 250 mm NaCl (Figs. 3B and 4C). Lower shoot Na+ concentration likely results from either the increase of root Na+ exclusion or the decrease of root-to-shoot Na+ transport (Wu et al., 2019; Huang et al., 2020). Interestingly, we confirmed that the lower shoot Na+ concentration in RNAi lines is closely associated with a lower Na+ transportation rate from roots to shoots (Fig. 4E; Supplemental Fig. S4B) and a lower xylem sap Na+ concentration (Fig. 6) but with unchanged Na+ accumulation per plant (Fig. 4D). These results indicated that HvCaM1 is involved in root-to-shoot Na+ transportation, which is further supported by the preferential expression of HvCaM1 in root stele and vascular systems (Fig. 2, A and E). Another important factor that affects the amount of Na+ transport to the shoot is the rate of xylem Na+ loading. The xylem Na+ loading is thermodynamically active (Shabala, 2013) and is also mediated by either SOS1 or CCC transporters operating at the xylem parenchyma interface (Colmenero-Flores et al., 2007; Ishikawa et al., 2018). Here, we showed that transcript levels of SOS genes are significantly down-regulated in RNAi lines (Fig. 5G), which may restrict the amounts of xylem Na+ loading, thus reducing Na+ transport between roots and shoots. However, this requires a detailed investigation in the future.
High K+ accumulation and K+/Na+ ratios are beneficial for plants to cope with salt stress (Chen et al., 2007; Shabala et al., 2010). In this study, RNAi lines had higher K+ concentration and K+/ Na+ ratios than wild-type and OE plants (Figs. 3, D and E, and 4, F and G), suggesting that HvCaM1 might affect K+ homeostasis. However, there was no difference in K+ transportation rate among these barley genotypes under salt stress (Fig. 4H; Supplemental Fig. S4B), indicating that HvCaM1 is not involved in the regulation of K+ transportation. In short, we propose that the preferential expression and cellular localization of HvCaM1 makes it possible for barley to modulate long-distance Na+ transport from roots to shoots (Zhu, 2016).
HvCaM1 Regulates Na+ Transport via Preferential Transcriptional Regulation of HvHKT1s
CaMs may bind with quantitatively changed Ca2+ signals across membrane systems (Gifford et al., 2013; DeFalco et al., 2016; Demidchik et al., 2018). The major membrane transporters conferring cellular and whole-plant Na+ homeostasis are HKTs and NHXs/SOS1 (Zhu, 2002; El Mahi et al., 2019; Munns et al., 2020; Shabala et al., 2020). Here, only some HKT1 members, in particular HvHKT1;5 and HvHKT1;1, were significantly affected by the knockdown of HvCaM1 (Fig. 5, A–C; Supplemental Fig. S6, A and B). HKTs regulate long-distance Na+ delivery, depending on root-to-shoot Na+ translocation and Na+ exclusion from the reproductive organ (Cao et al., 2020). HvHKT1;5 negatively regulates salt tolerance in barley (Huang et al., 2020). Thus, lower expression of HvHKT1;5 in RNAi roots potentially indicates less Na+ transport from roots to shoots, which is consistent with the function of HvCaM1 in Na+ transport. Correspondingly, lower Na+ loading from roots to shoots via the xylem was confirmed with higher salt tolerance by the down-regulation of HvCaM1 and HvHKT1;5 (Fig. 6, A and B; Huang et al., 2020). By contrast, another Na+ transporter, HvHKT1;1, plays a role in Na+ retrieval from shoots to roots, and overexpression of HvHKT1;1 in Arabidopsis reduces Na+ accumulation (Han et al., 2018). However, there is no report on the overexpression and silencing of HvHKT1;1 in barley, and the level of salt tolerance is very different between barley and Arabidopsis, which requires further investigation.
The regulation of HvHKT1s by HvCaM1 probably occurs at the transcriptional or posttranscriptional level (Reddy et al., 2011; Wang et al., 2015). In general, CaM functions in transcriptional regulation through specific CBP transcription factors, such as CAMTAs (Galon et al., 2010). CAMTA binds to Ca2+/CaM and participates in transcriptional regulation by recognizing and binding to a specific cis-element, (G/A/C)CGCG(C/G/T) (Yang and Poovaiah, 2002; Shen et al., 2015). AtCAMTA6 negatively regulates salt tolerance via HKT1, probably with the specific CGCG-core or abscisic acid response element-cooperating CGTCT-core motif (Shkolnik et al., 2019). Here, we identified an interacting partner of HvCaM1, HvCAMTA4, through yeast two-hybrid screening, showing transcriptional function in the nucleus (Fig. 7, A and B; Supplemental Table S4). Meanwhile, the expression pattern of HvCAMTA4 was similar to those of HvHKT1;5 and HvCaM1 (Fig. 7C), indicating its possible role as a linker protein for HvCaM1 to transcriptionally regulate the expression of HvHKT1;5. Interestingly both HvHKT1;1 and HvHKT1;5 contain regulatory elements (Fig. 7D) for HvCAMT4 (Shkolnik et al., 2019), and there are different cis-regulatory element sites in the promoter regions of HvHKT1;5 and HvHKT1;1. Therefore, it may be concluded that HvCAMTA4 regulates HvHKT1;5 independently while it modulates HvHKT1;1 together with regulatory proteins for abscisic acid signaling in response to salt stress (Whalley and Knight, 2013). Moreover, HvHKT1;1 is an important Na+ transporter that confers salt tolerance (Han et al., 2018), but HvHKT1;5 is a negative regulator of salt tolerance in barley (Huang et al., 2020). Overall expression of the key salt-responsive genes (HKTs, NHXs, and SOSs) is down-regulated in the HvCaM1 RNAi lines with the exception of HvHKT1;1. Interestingly, HvHKT1;1 and HvHKT1;5 in the R1 and R2 RNAi lines are similarly highly expressed after salt-induced up-regulation and down-regulation at week 3, respectively (Fig. 5). This could be the explanation for the differential expression of HvHKT1;1 and HvHKT1;5 in HvCaM1 knockdown lines. However, the possibility of other HvCaM1-interacting proteins, such as CBP60b, participating in the regulation of HKT1 transportation activity by posttranscriptional modification requires future experimentation (Lin et al., 2014).
In summary, we found that a transcriptional activator, HvCAMTA4, interacts with HvCaM1 under salt stress to regulate the expression of HvHKT1;5 and HvHKT1;1 at different sites of cis-regulatory elements of the promoter regions. However, the inherent interaction and signal transduction between HvCaM1 and HvCAMTA4 in mediating HvHKT1 activity remain unknown. Meanwhile, the protein sequence of HvCaM1 is quite conserved among plant species. Thus, RNAi or gene editing of CaM1s may be used to improve the salt tolerance of different crops.
MATERIALS AND METHODS
Growth Conditions and Sampling
Barley (Hordeum vulgare) ‘Golden Promise’ (cv GP) was used for the experiments. Barley germination and growth conditions were performed as described in previous studies (Wu et al., 2014; Shen et al., 2016). In the hydroponic experiments, salt treatment began in 14-d-old seedlings by adding 100 mm NaCl per day to reach a final salt concentration of 100, 200, 250, or 300 mm, according to experimental purposes. For a screening assay, the seedlings were exposed to the 200 mm salt condition and photographed every week. For gene expression analysis, roots and shoots were sampled under normal conditions. Moreover, roots were sampled under 0, 100, 200, and 300 mm salt treatments and at 0, 1, 2, 3, and 4 weeks under the 200 mm salt level. The normal condition without salt addition was used as the control. The modified one-fifth-strength Hoagland solution was renewed every 3 d. In the soil experiment, seedlings were transplanted into 10-L containers with mixed artificial soil, supplied with the same conditions as described by Shen et al. (2018). Soil salt treatment began at the heading stage by applying 1 L of 200 mm NaCl solution to the bottom of the pots every 3 d to a final content of 2% (w/w) NaCl. Seedlings supplied with tap water were used as the control. Yield traits, including spikes per plant, seeds per spike, setting rate, and kernel weight, were recorded. There were six biological replicates in both hydroponic and soil experiments.
Gene Cloning and Bioinformatics Analysis
HvCaM1 (HORVU0Hr1G001270) was cloned from cv GP. Single-nucleotide polymorphism and amino acid variation of HvCaM1 were validated among barley genotypes (‘Morex’, ‘CM72’, and ‘GP’ as well as Tibetan wild barley accessions XZ16, XZ26, and XZ169; Wu et al., 2014; Shen et al., 2018). Evolutionary bioinformatics were performed according to Zhao et al. (2019) with some modifications as described by Feng et al. (2020). The candidate sequences of CaM1 members among 60 representative plants were selected from the 1,000 Plant Transcriptome and EnsemblPlants databases by BLAST-P (Zhu et al., 2015; Leebens-Mack et al., 2019; Supplemental Table S2). Multiple sequence alignment and phylogenetic analysis were performed by MAFFT and FastTree via the maximum likelihood method, respectively. The phylogenetic tree was annotated by the Interactive Tree of Life resource (http://itol.embl.de). Jalview was used to perform the alignment of protein sequences. The primers used are listed in Supplemental Table S6.
Plant Transformation and Verification
Genetic transformation was mediated by Agrobacterium tumefaciens strain AGL1 according to Harwood (2014) with some modification. To generate the RNAi construct, a 388-bp specific fragment (primers RNAi_HvCaM1_F/R) of HvCaM1 transcript was cloned into pDONR-Zeo cloning vector by the Gateway BP Clonase II enzyme mix kit (11789; Invitrogen), according to Miki and Shimamoto (2004) and Miki et al. (2005). The positive cloning plasmid was then recombined with pANDA vector by Gateway LR reaction (11791; Invitrogen). To generate the OE plasmid, a 450-bp complete coding region (primers OE_HvCaM1_F/R) of HvCaM1 was amplified and then constructed into pBract214 vector with the similar method of RNAi construction. There were more than 10 independent transgenic events of RNAi and OE. Among them, three RNAi lines (R1, R2, and R3), three OE lines, and one transgenically negative line were used as the materials. Only PCR-positive plants with specific test primers (R_Test_F/R for RNAi and OE_Test_F/R for OE) were used in the following experiments.
Gene Expression Analysis
The transcript levels of HvCaM1 and transporter-associated genes were detected by qPCR. Briefly, total RNA of samples was extracted by the MiniBEST kit and reversed by the PrimeScript RT reagent kit (Takara). Then, qPCR was performed with SYBR Green Supermix (Bio-Rad) on a Roche LightCycler 480 instrument. For absolute qPCR analysis, a pDONR plasmid containing the complete CDS region of HvCaM1 was used to quantify the transcript concentration by diluting to 100, 10−1, 10−2, 10−3, 10−4, and 10−5 ng μg−1 (a standard curve of R2 > 0.999), as described by Wu et al. (2016). For relative gene expression analysis, the comparative 2−△△Ct method was performed using the reference gene α-Tubulin (Wu et al., 2014). The transcripts of wild-type (cv GP) roots under the normal condition were regarded as the controls. The primers of HKT, NHX, and SOS genes were designed following Fu et al. (2018) and Huang et al. (2020). There were four biological replicates and three technical repeats for gene expression analysis.
Element Determination and Analysis
After 4 weeks of salt exposure (200 or 250 mm), the fresh weight (FW) of roots and shoots was determined, and then tissues were dried in an oven at 80°C for 2 d to obtain dry weight (DW). Relative water content (RWC) was calculated using the following formula: RWC (%) = (FW − DW)/FW × 100%. Dried samples were digested by HNO3 using microwave digestion equipment (Multiwave 3000; Anton Paar) according to Shen et al. (2018). For element analysis, the concentrations of Na+, K+, and Ca2+ were determined by an inductively coupled plasma-optical emission spectrometer (iCAP 6000 series; Thermo Fisher Scientific). In addition, Na+ or K+ content was calculated based on concentration and dry weight, while the transportation rates were also calculated as follows: (shoot content)/(total content per plant) × 100%. There were four biological replicates for element analysis.
Xylem Sap Analysis
Na+ and K+ concentrations in the xylem sap were measured according to Huang et al. (2020). Four-week-old seedlings of the wild type and RNAi lines (R1–R3) were exposed to 0 (control), 50, and 100 mm NaCl. After 2 and 7 d, the stems were cut 1 to 2 cm above the root and shoot junction. The cut surface was cleaned using soft filter papers to avoid contamination. Xylem sap exudates were collected within 1 h with a micropipette and stored in ice-chilled Eppendorf tubes. About 10 µL of sap was collected for each replicate pooled from two to six plants and then diluted with 3 to 10 mL of 2% (v/v) HNO3 for ion measurement. The concentrations of Na+ and K+ in the solution were determined by an inductively coupled plasma-mass spectrometer (ELAN DRC-e; Perkin-Elmer). Four to six biological replicates for each treatment and time point were used.
Tissue-Specific and Subcellular Localization
To generate the stable expression plasmid for tissue-specific localization, approximately 2.1-kb upstream fragments of the HvCaM1 start were amplified with the primers Pro_HvCaM1_F/R and then cloned into pCAMBIA1300 (deleted 35S promoter) fused with the GFP coding region. The recombinant plasmid was transferred into AGL1 to obtain genetically modified barley materials. Fresh root and leaf samples of 14-d-old seedlings (verified by PCR with Pro_Test_F/R) were used to observe GFP fluorescence with the confocal laser microscope at 488/490 to 540 nm.
To generate the transiently expressing plasmid for subcellular localization, the 447-bp coding region of HvCaM1 without the stop code was amplified and fused with the 5′ terminus of the sGFP gene into pCAMBIA1300 vector after linearization digestion using the homologous recombination cloning kit (C112; Vazyme). The recombinant and empty vectors driven by the 35S promoter were transformed separately into onion (Allium cepa) epidermis cells by a biolistic PDS-1000/He particle device (Bio-Rad) along with a cell-localized RFP marker, as described previously (Huang et al., 2020). GFP and RFP fluorescence signals were measured using a confocal laser microscope (LSM780; Zeiss) at 488/490 to 540 nm and 561/580 to 630 nm, respectively.
In Situ PCR
For in situ PCR analysis, a 388-bp sequence of the HvCaM1 transcript, the same as in RNA silencing, was amplified and performed according to Athman et al. (2014) with some modification, as described by Long et al. (2018). In brief, 1-cm-long barley leaf and root sections from 14-d-old seedlings were immersed into fixative solution (63% [v/v] ethanol, 5% [v/v] acetic acid, and 2% [v/v] formaldehyde) for 4 h at 4°C. The samples were then embedded in 5% (w/v) low-melting-temperature agarose and cut into 60- to 70-μm cross sections. After DNase I treatment (Takara), reverse transcription was performed with gene-specific reverse primers using digoxigenin (DIG)-labeled dUTP and KOD FX enzyme (Toyobo) for PCR amplification. The tested samples were then incubated with anti-DIG-alkaline phosphate antibody, stained with BM purple substrate (Roche), and mounted in 40% (v/v) glycerol after washing. The ribosomal 18S transcript was amplified by in situ PCR as the positive control, while only water without primer addition was used as a negative control. A Leica microscope was used for observation and imaging, according to the color reaction between alkaline phosphate and DIG-labeled antibody.
Yeast Two-Hybrid Interaction Assay
The candidate gene HvCAMTA4 (HORVU2Hr1G069950) was originally screened from a yeast (Saccharomyces cerevisiae) library constructed with the total cDNA of salt-treated barley roots by the CloneMiner II kit (A11180; Invitrogen). To verify the interaction between HvCaM1 and HvCAMTA4, the cDNA of HvCAMTA4 was amplified into pGADT7 as prey, while the coding region of HvCaM1 cloned into pGBKT7 was used as a bait according to Cho et al. (2016). The combination of pGBKT7-53 and pGADT7-T was used as the positive control, while empty vector of pGBKT7-Lam and pGADT7-T was used as the negative control. The yeast two-hybrid interaction assay was performed after cotransformation of both the bait and prey constructs into Y2HGold yeast competent cells (Clontech). The transformed yeast cells were then screened on synthetic dextrose double dropout medium (lacking Leu and Trp) with or without aureobasidin A addition.
BiFC Assay
For the BiFC interaction assay, the coding regions of HvCaM1 and HvCAMTA4 were separately cloned into pSAT1-cEYFP/nEYFP vectors to generate HvCaM1-cEYFP and nEYFP-HvCAMTA4 vectors with the Vazyme homologous recombination kit. Barley protoplast isolation and transformation were performed as described by Ye et al. (2019). Seven-day-old barley seedlings were used to isolate the protoplast. The genes HvSOS3 (HORVU1Hr1G080820; CBL4) and HvSOS2 (HORVU7Hr1G090260; CIPK24) involved in the SOS pathway were used to generate HvCBL4-cEYFP and nEYFP-HvCIPK24 vectors as the positive control, while HvCaM1-cEYFP and nEYFP were used as the negative control. A total of 10 mg of plasmid DNA of each construct was transformed by polyethylene glycol into the barley protoplast. The YFP fluorescence signals were detected by a confocal laser microscope at 514/520 to 550 nm.
Statistical Analysis
The significant differences of dry weight, relative water content, element content, yield, and gene expression among genotypes, treatments, or tissues were analyzed by one-way ANOVA or independent Student’s t test using SPSS 20.0 software (IBM SPSS Statistics). The significance levels at P < 0.05 and P < 0.01 were defined as significant and highly significant, respectively.
Accession Numbers
All accession numbers and species for sequence alignment and gene expression are listed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cloning and sequence analysis of HvCaM1.
Supplemental Figure S2. Tissue-specific localization of HvCaM1.
Supplemental Figure S3. PCR verification in RNAi and OE lines.
Supplemental Figure S4. Physiological indexes of wild-type, RNAi, and OE seedlings under salt stress.
Supplemental Figure S5. Growth performance and yield traits of wild-type and RNAi plants in the saline soil (n = 6).
Supplemental Figure S6. Expression analysis of Na+ and K+ transporters under salt stress (remaining).
Supplemental Table S1. Sequence variation in the HvCaM1 CDS region of barley genotypes.
Supplemental Table S2. Accession numbers for sequence alignment and gene expression.
Supplemental Table S3. Dry weight of wild-type and RNAi seedlings under salt conditions for 4 weeks (n = 4).
Supplemental Table S4. Positive interaction candidates with HvCaM1 by yeast two-hybrid screening assay.
Supplemental Table S5. CAMTA cis-regulatory elements in the 3-kb promoters of HKT genes.
Supplemental Table S6. Primers used in this study.
Acknowledgments
We thank Dr. Peter Dominay (University of Glasgow) and Dr. Hiroyuki Tsuji (Nara Institute of Science and Technology) for providing the vectors pBract214 and pANDA, respectively. We also thank Dr. Yong Han (Murdoch University) and Drs. Zhonchang Wu and Jiming Xu (Zhejiang University) for technical support.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31901429, 31771685, and 3161001022), the China Postdoctoral Science Foundation (grant nos. 2019T120520 and 2018M640561), the China Agriculture Research System (grant no. CARS–05), and the Jiangsu Collaborative Innovation Center for Modern Crop Production.
Articles can be viewed without a subscription.
References
- Al-Quraan NA, Locy RD, Singh NK(2011) Implications of paraquat and hydrogen peroxide-induced oxidative stress treatments on the GABA shunt pathway in Arabidopsis thaliana calmodulin mutants. Plant Biotechnol Rep 5: 225–234 [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E(1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Athman A, Tanz SK, Conn VM, Jordans C, Mayo GM, Ng WW, Burton RA, Conn SJ, Gilliham M(2014) Protocol: A fast and simple in situ PCR method for localising gene expression in plant tissue. Plant Methods 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM(2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Kudla J(2012) Analysis of calcium signaling pathways in plants. Biochim Biophys Acta 1820: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S(2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhang M, Liang X, Li F, Shi Y, Yang X, Jiang C(2020) Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline-alkaline tolerance in maize. Nat Commun 11: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Newman I, Zhou M, Mendham N, Zhang G, Shabala S(2005) Screening plants for salt tolerance by measuring K+ flux: A case study for barley. Plant Cell Environ 28: 1230–1246 [Google Scholar]
- Chen Z, Pottosin II, Cuin TA, Fuglsang AT, Tester M, Jha D, Zepeda-Jazo I, Zhou M, Palmgren MG, Newman IA, et al. (2007) Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol 145: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinpongpanich A, Limruengroj K, Phean-O-Pas S, Limpaseni T, Buaboocha T(2012) Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res Notes 5: 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KM, Nguyen HTK, Kim SY, Shin JS, Cho DH, Hong SB, Shin JS, Ok SH(2016) CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol 209: 664–678 [DOI] [PubMed] [Google Scholar]
- Colmenero-Flores JM, Martínez G, Gamba G, Vázquez N, Iglesias DJ, Brumós J, Talón M(2007) Identification and functional characterization of cation-chloride cotransporters in plants. Plant J 50: 278–292 [DOI] [PubMed] [Google Scholar]
- Dai F, Wang X, Zhang XQ, Chen Z, Nevo E, Jin G, Wu D, Li C, Zhang G(2018) Assembly and analysis of a qingke reference genome demonstrate its close genetic relation to modern cultivated barley. Plant Biotechnol J 16: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M(2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30: 497–507 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Marshall CB, Munro K, Kang HG, Moeder W, Ikura M, Snedden WA, Yoshioka K(2016) Multiple calmodulin-binding sites positively and negatively regulate Arabidopsis CYCLIC NUCLEOTIDE-GATED CHANNEL12. Plant Cell 28: 1738–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI(2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I(2018) Calcium transport across plant membranes: Mechanisms and functions. New Phytol 220: 49–69 [DOI] [PubMed] [Google Scholar]
- El Mahi H, Pérez-Hormaeche J, De Luca A, Villalta I, Espartero J, Gámez-Arjona F, Fernández JL, Bundó M, Mendoza I, Mieulet D, et al. (2019) A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol 180: 1046–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Liu W, Qiu CW, Zeng F, Wang Y, Zhang G, Chen ZH, Wu F(2020) HvAKT2 and HvHAK1 confer drought tolerance in barley through enhanced leaf mesophyll H+ homoeostasis. Plant Biotechnol J 18: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Shen Q, Kuang L, Yu J, Wu D, Zhang G(2018) Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol Biochem 130: 248–257 [DOI] [PubMed] [Google Scholar]
- Galon Y, Finkler A, Fromm H(2010) Calcium-regulated transcription in plants. Mol Plant 3: 653–669 [DOI] [PubMed] [Google Scholar]
- Gifford JL, Jamshidiha M, Mo J, Ishida H, Vogel HJ(2013) Comparing the calcium binding abilities of two soybean calmodulins: Towards understanding the divergent nature of plant calmodulins. Plant Cell 25: 4512–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Yin S, Huang L, Wu X, Zeng J, Liu X, Qiu L, Munns R, Chen ZH, Zhang G(2018) A sodium transporter HvHKT1;1 confers salt tolerance in barley via regulating tissue and cell ion homeostasis. Plant Cell Physiol 59: 1976–1989 [DOI] [PubMed] [Google Scholar]
- Harwood WA.(2014) A protocol for high-throughput Agrobacterium-mediated barley transformation In Henry R, and Furtado A, eds, Cereal Genomics: Methods and Protocols. Humana Press, Totowa, NJ, pp 251–260 [DOI] [PubMed] [Google Scholar]
- Horie T, Karahara I, Katsuhara M(2012) Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice (N Y) 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Kuang L, Wu L, Shen Q, Han Y, Jiang L, Wu D, Zhang G(2020) The HKT transporter HvHKT1;5 negatively regulates salt tolerance. Plant Physiol 182: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB(2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103: 16666–16671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Cuin TA, Bazihizina N, Shabala S(2018) Xylem ion loading and its implications for plant abiotic stress tolerance. Adv Bot Res 87: 267–301 [Google Scholar]
- Ismail AM, Horie T(2017) Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol 68: 405–434 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T(2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewneramit T, Buaboocha T, Sangchai P, Wutipraditkul N(2019) OsCaM1-1 overexpression in the transgenic rice mitigated salt-induced oxidative damage. Biol Plant 63: 335 [Google Scholar]
- Kersey PJ.(2019) Plant genome sequences: Past, present, future. Curr Opin Plant Biol 48: 1–8 [DOI] [PubMed] [Google Scholar]
- Khong GN, Pati PK, Richaud F, Parizot B, Bidzinski P, Mai CD, Bès M, Bourrié I, Meynard D, Beeckman T, et al. (2015) OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol 169: 2935–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR(2009) The origin and evolution of fragrance in rice (Oryza sativa L.). Proc Natl Acad Sci USA 106: 14444–14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha R, Singh A, Chattopadhyay S(2008) Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S, et al. (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Du W, Yang Y, Schumaker KS, Guo Y(2014) A calcium-independent activation of the Arabidopsis SOS2-like protein kinase24 by its interacting SOS3-like calcium binding protein1. Plant Physiol 164: 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li H, Hao C, Wang K, Wang Y, Qin L, An D, Li T, Zhang X(2020) TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol J 18: 1330–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L, Persson DP, Duan F, Jørgensen K, Yuan L, Schjoerring JK, Pedas PR(2018) The iron-regulated transporter 1 plays an essential role in uptake, translocation and grain-loading of manganese, but not iron, in barley. New Phytol 217: 1640–1653 [DOI] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K(2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K(2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Munns R.(2005) Genes and salt tolerance: Bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M(2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen ZH, Foster KJ, et al. (2020) Energy costs of salt tolerance in crop plants. New Phytol 225: 1072–1090 [DOI] [PubMed] [Google Scholar]
- Nevo E, Chen G(2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ 33: 670–685 [DOI] [PubMed] [Google Scholar]
- Rao SS, El-Habbak MH, Havens WM, Singh A, Zheng D, Vaughn L, Haudenshield JS, Hartman GL, Korban SS, Ghabrial SA(2014) Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol Plant Pathol 15: 145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Ali GS, Celesnik H, Day IS(2011) Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S.(2013) Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot 112: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Chen G, Chen ZH, Pottosin I(2020) The energy cost of the tonoplast futile sodium leak. New Phytol 225: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA(2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala S, Cuin TA, Pang J, Percey W, Chen Z, Conn S, Eing C, Wegner LH(2010) Xylem ionic relations and salinity tolerance in barley. Plant J 61: 839–853 [DOI] [PubMed] [Google Scholar]
- Shen C, Yang Y, Du L, Wang H(2015) Calmodulin-binding transcription activators and perspectives for applications in biotechnology. Appl Microbiol Biotechnol 99: 10379–10385 [DOI] [PubMed] [Google Scholar]
- Shen Q, Fu L, Dai F, Jiang L, Zhang G, Wu D(2016) Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genomics 17: 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yu J, Fu L, Wu L, Dai F, Jiang L, Wu D, Zhang G(2018) Ionomic, metabolomic and proteomic analyses reveal molecular mechanisms of root adaption to salt stress in Tibetan wild barley. Plant Physiol Biochem 123: 319–330 [DOI] [PubMed] [Google Scholar]
- Shen QF, Fu LB, Qiu L, Feng X, Zhang GP, Wu DZ(2017) Time-course of ionic responses and proteomic analysis of a Tibetan wild barley at early stage under salt stress. Plant Growth Regul 81: 11–21 [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK(2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK(2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik D, Finkler A, Pasmanik-Chor M, Fromm H(2019) CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A key regulator of Na+ homeostasis during germination. Plant Physiol 180: 1101–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W(2015) The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HJ, Knight MR(2013) Calcium signatures are decoded by plants to give specific gene responses. New Phytol 197: 690–693 [DOI] [PubMed] [Google Scholar]
- Wu D, Shen Q, Qiu L, Han Y, Ye L, Jabeen Z, Shu Q, Zhang G(2014) Identification of proteins associated with ion homeostasis and salt tolerance in barley. Proteomics 14: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Wu D, Yamaji N, Yamane M, Kashino-Fujii M, Sato K, Feng Ma J(2016) The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol 172: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shabala L, Zhou M, Su N, Wu Q, Ul-Haq T, Zhu J, Mancuso S, Azzarello E, Shabala S(2019) Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J 100: 55–67 [DOI] [PubMed] [Google Scholar]
- Xi J, Qiu Y, Du L, Poovaiah BW(2012) Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci 185–186: 274–280 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu M, Lu L, He M, Qu W, Xu Q, Qi X, Chen X(2015) Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol Genet Genomics 290: 1403–1414 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW(2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Ye L, Wang Y, Long L, Luo H, Shen Q, Broughton S, Wu D, Shu X, Dai F, Li C, et al. (2019) A trypsin family protein gene controls tillering and leaf shape in barley. Plant Physiol 181: 701–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, Heo WD, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, et al. (2005) Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Zahra J, Nazim H, Cai SG, Han Y, Wu DZ, Zhang BL, Haider SI, Zhang GP(2014) The influence of salinity on cell ultrastructures and photosynthetic apparatus of barley genotypes differing in salt stress tolerance. Acta Physiol Plant 36: 1261–1269 [Google Scholar]
- Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S(2012) Mechanisms of plant salt response: Insights from proteomics. J Proteome Res 11: 49–67 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E(2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhou RG, Gao YJ, Zheng SZ, Xu P, Zhang SQ, Sun DY(2009) Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol 149: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wang Y, Chan KX, Marchant DB, Franks PJ, Randall D, Tee EE, Chen G, Ramesh S, Phua SY, et al. (2019) Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc Natl Acad Sci USA 116: 5015–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Jia L, Chu H, Wu D, Peng X, Liu X, Zhang J, Zhao J, Chen K, Zhao L(2016) Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting S-nitrosoglutathione reductase via direct binding. PLoS Genet 12: e1006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK.(2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK.(2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Dunand C, Snedden W, Galaud JP(2015) CaM and CML emergence in the green lineage. Trends Plant Sci 20: 483–489 [DOI] [PubMed] [Google Scholar]