Analysis of a unique tomato epiallele reveals an epigenetic pathway that, along with the ripening process, modulates vivipary in tomato fruits.
Abstract
Vivipary, wherein seeds germinate prior to dispersal while still associated with the maternal plant, is an adaptation to extreme environments. It is normally inhibited by the establishment of dormancy. The genetic framework of vivipary has been well studied; however, the role of epigenetics in vivipary remains unknown. Here, we report that silencing of METHYLTRANSFERASE1 (SlMET1) promoted precocious seed germination and seedling growth within the tomato (Solanum lycopersicum) epimutant Colorless non-ripening (Cnr) fruits. This was associated with decreases in abscisic acid concentration and levels of mRNA encoding 9-cis-epoxycarotenoid-dioxygenase (SlNCED), which is involved in abscisic acid biosynthesis. Differentially methylated regions were identified in promoters of differentially expressed genes, including SlNCED. SlNCED knockdown also induced viviparous seedling growth in Cnr fruits. Strikingly, Cnr ripening reversion suppressed vivipary. Moreover, neither SlMET1/SlNCED-virus-induced gene silencing nor transgenic SlMET1-RNA interference produced vivipary in wild-type tomatoes; the latter affected leaf architecture, arrested flowering, and repressed seed development. Thus, a dual pathway in ripening and SlMET1-mediated epigenetics coordinates the blockage of seed vivipary.
Fruits are developmental structures unique to flowering plants. They also play a central role in seed development and dispersal (Giovannoni, 2004; Lozano et al., 2009). In tomato (Solanum lycopersicum), the onset of fruit ripening occurs after the cell expansion in the developing ovary has completed and seed has matured. Ripening is characterized by higher respiration and the autocatalytic synthesis of ethylene (Lin et al., 2009), and it leads to fruit softening, alternation in texture and colors, enrichment of organic acids, nutrients, and pigments, and the development of aroma and flavor (Rose et al., 2004; Tieman et al., 2006, 2017, Seymour et al., 2008, 2013; Uluisik et al., 2016; Wang and Seymour, 2017; Zhu et al., 2018). This process is modulated by a complex genetic network comprising master transcription factors (TFs), including MADS-RIN (an MADS-box TF RIPENING INHIBITOR; Vrebalov et al., 2002), HB1 (a class I homeodomain Leu zipper TF; Lin et al., 2008), TAGL1 (an AGAMOUS clade MADS-box TF; Itkin et al., 2009; Vrebalov et al., 2009), and an APETALA2/ERF (ethylene response factor) SlAP2a TF (Chung et al., 2010). These TFs appear to be involved in modulation of fruit ripening through either transcriptional up- or down-regulation of gene expression for ethylene synthesis and other ripening-related physiological processes (Zhou et al., 2012; Karlova et al., 2014; Wang et al., 2020).
Another key player in the regulatory network controlling tomato fruit ripening is the gene that resides at the Colorless non-ripening (Cnr) locus. Cnr fruit cannot ripen and remains colorless. Its texture alters due to loss of cell-to-cell adhesion in fruit tissues (Eriksson et al., 2004). The CNR gene encodes the SQUAMOSA promoter-binding protein-box TF SPL-CNR (Manning et al., 2006). The expression of CNR is developmentally controlled, being mainly expressed in ripening fruits, and is fine-tuned by microRNAs (Manning et al., 2006; Chen et al., 2015a). However, using the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein9) gene-editing technique, Gao et al. (2019) recently produced CNR-knockout (KO) mutants and found that CNR-KO failed to phenocopy nonripening as seen in the naturally occurring Cnr mutant or in KD (knockdown by RNA interference [RNAi] or virus-induced gene silencing [VIGS]) tomato fruits (Manning et al., 2006; Chen et al., 2015a, 2015b). Such phenotypic discrepancies raise an intriguing issue about the precise functionality of CNR in tomato fruit ripening. However, CRISPR/Cas9-mediated KO mutants of genes essential for development do not often show any obvious phenotype as in naturally occurring mutants or in silencing/RNAi-KD lines. This phenomenon could be explained by noncoding RNA-mediated transcriptional adaptation (i.e. genetic compensation), reinforcing that CNR and the other ripening genes NON-RIPENING and FRUITFULL1/2 may play essential roles not only in ripening but also in other developmental and physiological processes (Lai et al., 2020; Wang et al., 2020).
Cnr results from a spontaneous epimutation that causes hypermethylation in the promoter of CNR (Manning et al., 2006). This epimutation occurs naturally, and differential methylation has been found in the region 2.4 kb upstream from the CNR translational start site (Manning et al., 2006). The Cnr mutant also possesses a hypermethylated epigenome (Zhong et al., 2013; Chen et al., 2015b, 2018a), most likely due to the lack of induction of the SlDML2 gene that encodes a DEMETER-like DNA demethylase governing tomato fruit ripening (Liu et al., 2015). DOMAINS REARRANGED METHYLTRANSFERASE7 (SlDRM7), METHYLTRANSFERASE1 (SlMET1), and CHROMOMETHYLASE2 (SlCMT2) and SlCMT3, which are essential for RNA-directed DNA methylation (RdDM) and for methylation maintenance, are essential to maintain the Cnr epiallele; and virus-induced silencing of these genes causes Cnr fruits to ripen (Chen et al., 2015b). These recent discoveries reveal that both genetic and epigenetic mechanisms are involved in the modulation of fruit ripening in tomato (Seymour et al., 2013).
In tomato and other flowering plants, accompanying fruit development and ripening, seed develops, matures, and becomes dormant within ripe fruit. However, under certain physiological conditions, seed can germinate within fruits, a phenomenon called vivipary (VP). VP has evolutionary and ecological advantages for some plant species in surviving extreme environments (Eyster, 1931; Dintu et al., 2015). However, in the form of preharvest sprouting, VP can substantially reduce yield and product quality in vegetables, grain, and fruit crops, thus posing a threat to global food security (Gubler et al., 2005; Shu et al., 2016b). The genetics and genes responsible for VP have been extensively studied in plants, including cereal and fruit crops such as barley (Hordeum vulgare), maize (Zea mays), sorghum (Sorghum bicolor), wheat (Triticum aestivum), rice (Oryza sativa), tomato, and gourd (Lagenaria siceraria; Robertson, 1952; McCarty et al., 1991; Hable et al., 1998; Burbidge et al., 1999; Agrawal et al., 2001; McKibbin et al., 2002; Porch et al., 2006; Suzuki et al., 2006, 2008; Fang et al., 2008; Benech-Arnold and Rodríguez, 2018; Nakamura, 2018; N’Gaza et al., 2019). In particular in maize, there are at least 25 independent Viviparous (VP) mutants, and these VP mutants are grouped into three classes (Durantini et al., 2008). Class 1 includes mutants such as VP1. Maize VP1, the first characterized viviparous gene, encodes a TF belonging to the AFL (ABI3/FUS3/LEC2) subfamily of the B3 TFs (Carbonero et al., 2017). The Arabidopsis (Arabidopsis thaliana) putative ortholog gene to VP1 is the ABSCISIC ACID-INSENSITIVE3 (ABI3), and these genes are not impaired in abscisic acid (ABA) biosynthesis. However, VP mutations in class 2 block early steps prior to the branching point that separates ABA and ζ-carotenoid biosynthesis, resulting in reduction of carotenoid accumulation in both endosperm and vegetative tissues (Singh et al., 2003). Class 3 mutants either affect later steps of ABA biosynthesis (Schwartz et al., 1997) or regulate the synthesis of the molybdenum cofactor required for the last step in ABA biosynthesis (Porch et al., 2006; Suzuki et al., 2006). These mutants are frequently unable to complete seed maturation but lead to precocious germination (Carbonero et al., 2017). On the other hand, emerging evidence suggests that there may be an epigenetic layer of controls involved in VP and the associated processes of seed dormancy and germination. Dynamic seed-seedling epigenomes have been reported in Arabidopsis and soybean (Glycine max; An et al., 2017; Bouyer et al., 2017; Kawakatsu et al., 2017; Narsai et al., 2017). Cis-acting noncoding antisense RNA, microRNA, and genes that affect DNA methylation (Singh et al., 2013; Yamauchi et al., 2014; Fedak et al., 2016; Huo et al., 2016; Chen et al., 2017) may affect seed germination and VP, although the underlying mechanisms remain unknown.
While our original goal was to reveal how RdDM and/or DNA methylation maintenance determine the nonripening epiphenotype in Cnr, we observed, by serendipity, that deficiency in DNA methylation affected not only fruit ripening but also the growth of viviparous seedlings within the Cnr fruits. These findings suggested that that epigenetic mechanism may involve seed VP in tomato. In this article, we exploit further the Cnr epiallele and report that an epigenetic pathway along with the ripening process controls VP in tomato.
RESULTS
Silencing of SlMET1 Induces VP in Cnr Fruits
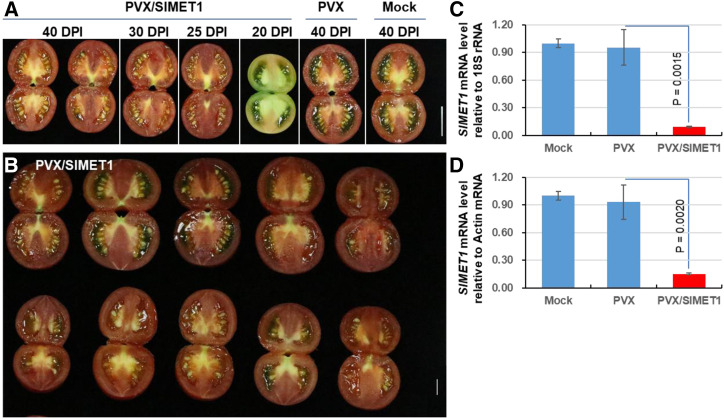
The Cnr mutant possesses a hypermethylated epigenome (Manning et al., 2006; Chen et al., 2018a), and several genes such as SlDRM7, SlMET1, SlCMT2, and SlCMT3 that are essential for RdDM and for methylation maintenance are required to maintain the Cnr epiallele (Chen et al., 2015b). KD of these genes, SlCMT3 in particular by Potato virus X (PVX)-VIGS, results in ripening reversion of Cnr fruits (Chen et al., 2015b). SlMET1-KD led to a partial reversion of tomato ripening in approximately 30% of Cnr fruits, while SlCMT4-KD did not have any effect on the colorless nonripening phenotype (Fig. 1, A and B). These results were consistent with our previous reports (Chen et al., 2015b). Reversion of fruit ripening was illustrated by the development of red color on the treated Cnr tomato. It is noteworthy that color alterations represent a valid indicator of ripening, consistent with changes in tomato physical, physiological, agrochemical, biochemical, and molecular characteristics, as described in our previous VIGS studies (Manning et al., 2006; Lin et al., 2008; Zhou et al., 2012; Kong et al., 2013; Chen et al., 2015a, 2015b, 2018a; Lai et al., 2015; Lai et al., 2020).
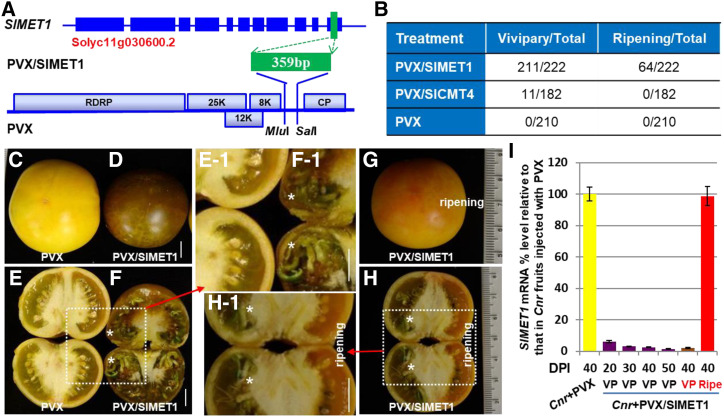
Figure 1.
Induction of tomato vivipary by SlMET1 silencing. A, Schematic of the SlMET1 gene and the PVX/SlMET1 VIGS construct. A 359-bp fragment corresponding to the last exon of SlMET1 was cloned into the PVX vector to generate PVX/SlMET1. The genome organization of PVX and two cloning sites are indicated. RDRP is viral RNA-dependent RNA polymerase. The triple-gene block encodes three viral movement proteins of 25, 12, and 8 kD (K). CP is viral coat protein. B, Summary of VP and ripening reversion in Cnr fruits. The numerator represents the number of Cnr fruits showing either VP or ripening reversion. The denominator represents the total number of fruits that were inoculated with needle injection of PVX/SlMET1, PVX/SlCMT4 (Chen et al., 2015b), or the empty VIGS vector PVX. C to F, VP within Cnr fruits. Cnr fruits injected with PVX produced mature seeds (C and E). Precocious seed germination and viviparous seedling growth were present in PVX/SlMET1-injected Cnr fruits (PVX/SlMET1; D and F). G and H, Fruit ripening suppresses VP. A PVX/SlMET1-injected Cnr fruit (PVX/SlMET1) showed ripening reversion. Seeds matured in the ripening sector of the fruit shown in G, but seeds in the nonripe sectors became viviparous (H). The dotted boxes in E, F, and H are enlarged, as indicated by the red arrows, to show closeup images in E-1, F-1, and H-1, respectively. Examples of viviparous seedlings are indicated by asterisks. Fruits were photographed at 40 DPI (G and H) or 50 DPI (C–F). Bars = 1 cm. I, Suppression of endogenous SlMET1 expression in Cnr fruits by VIGS. RT-qPCR assays were performed on three biological duplicate samples collected from control (Cnr+PVX) or VP (Cnr+PVX/SlMET1) fruits at 20, 30, 40, and 50 DPI. Red-highlighted VP and Ripe signify viviparous seeds/seedlings or mature seeds collected from nonripening or ripening sectors of PVX/SlMET1-injected fruits. Data are shown as means ± sd (n = 3). Reduction of SlMET1 expression was found to be statistically different (one-way ANOVA, P < 0.05) in all VP samples compared with the Cnr+PVX control (purple versus yellow bars). However, the mRNA level was not obviously reduced in the Ripe sectors (G and H), likely due to mixtures of such sectors showing limited and sporadic ripening reversion with surrounding nonripe tissues for RT-qPCR analyses.
Apart from ripening reversion, we observed unexpected but predominant VP from almost all seeds in more than 95% of PVX/SlMET1-injected Cnr fruits (Fig. 1, B–F). Viviparous seedling growth was also found in about 6% of the PVX/SlCMT4-injected Cnr fruits (Fig. 1B; Supplemental Fig. S1, A–C). There was no VP at any developmental stage in control Ailsa Craig (AC) or Cnr fruits injected with PVX alone or Cnr fruits infected with PVX/SlDRM7, PVX/SlCMT2, or PVX/SlCMT3 (Supplemental Fig. S2, A–G). Within the SlMET1-KD Cnr fruits, VP appeared at around 20 d post inoculation (DPI), equivalent to 36 DPA, and seedling growth was evident at 30, 40, and 50 DPI (Fig. 1, D and F; Supplemental Fig. S3, A–D). We also observed that all seeds in sectors of PVX/SlMET1-injected Cnr fruits that showed partial ripening reversion remained dormant, whereas those seeds in the nonripe sectors of the same fruits became viviparous (Fig. 1, G and H). Moreover, in fruits or sectors of fruits where VP was occurring, there was a marked decrease in abundance of the SlMET1 transcripts in viviparous seeds/seedlings (Fig. 1, G and I; Supplemental Fig. S4). However, the SlMET1 level in sectors with limited and sporadic ripening reversion was not significantly reduced (Fig. 1I; Supplemental Fig. S4A), likely because tomato samples used for such analyses might have coexisted with normal Cnr fruit tissues. Nonetheless, a close link of reduction in SlMET1 expression with more pronounced and even ripening reversion was evident in Cnr fruits that were VIGSed by PVX/SlMET1 (Supplemental Fig. S4B; Chen et al., 2015b).
In addition, PVX infection alone was shown to have no influence on SlMET1 expression in seed and surrounding tissues at different stages of Cnr fruit development (i.e. at different DPI; Supplemental Fig. S5, A and B). It should be noted that our VIGS experiments were performed on fruits at 5 to 20 DPA on the same trusses or various trusses of Cnr plants. Thus, fruits that were collected at a definite DPI for molecular analysis might be at different stages of DPA, but they had undergone the same length of VIGS treatment. Collectively, these data indicate that suppression of SlMET1 triggers, but ripening reversion inhibits, VP in Cnr fruits.
Reduction of ABA Biosynthesis in SlMET1-KD Cnr Fruits
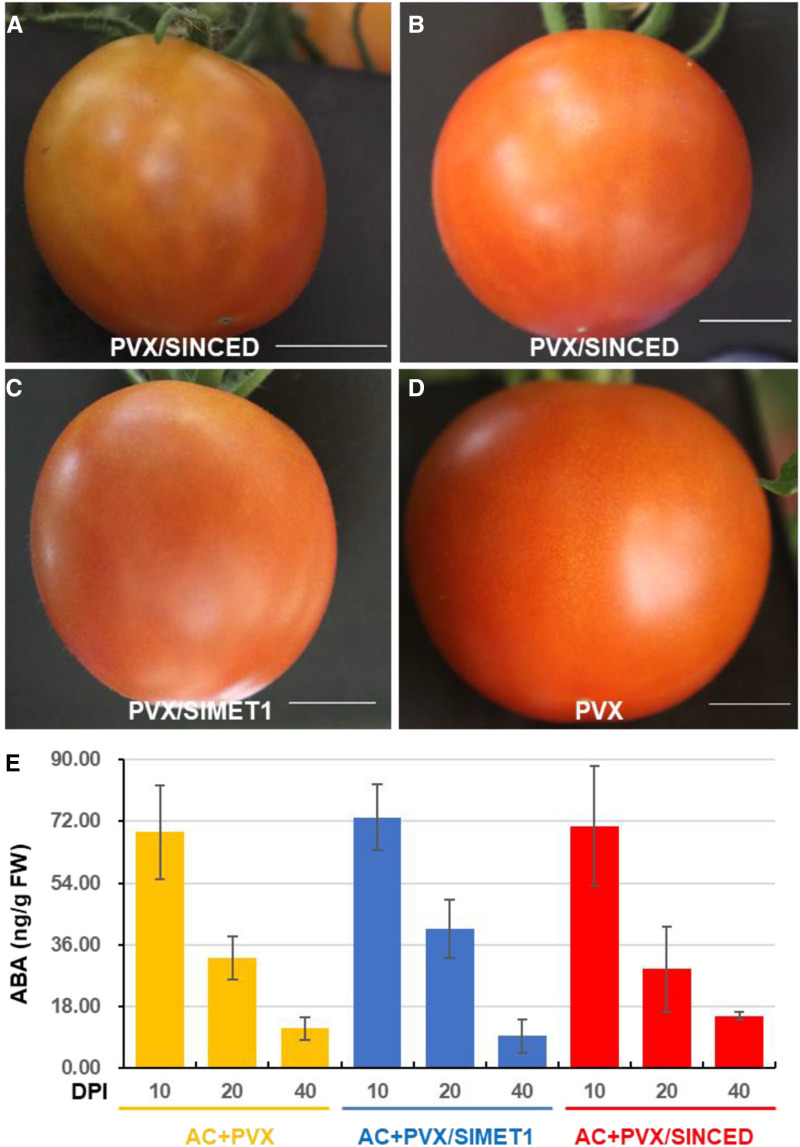
Our observation of VP induced by VIGS of SlMET1 or SlCMT4, but not other RdDM and methylation maintenance genes (Chen et al., 2015b), suggests that these two genes could be involved in establishing and/or maintaining dormancy during seed development within fruits. We investigated the mechanism by which SlMET1 could influence VP during fruit development and ripening because of the much stronger effect that SlMET1-KD had on VP compared with SlCMT4-KD. ABA is well documented to be required to maintain seed dormancy and inhibit germination in many plants, including tomato (Shu et al., 2016b). Using [2H6](+)-cis,trans-ABA as an internal standard, we performed HPLC-electrospray-mass spectrometry to measure ABA concentrations in mature seeds collected from wild-type AC and Cnr fruits at three developmental and ripening stages: mature green (30 DPA), breaker (36 DPA on average), and 10 d after breaker (47 DPA on average). These analyses revealed a gradual reduction in ABA (Fig. 2A), suggesting that ABA synthesis is developmentally modulated in AC and Cnr. We further determined ABA accumulation in both viviparous seeds and nongerminating mature seeds collected from the AC and Cnr fruits that were injected with PVX/SlMET1 or PVX at 30 DPI (Fig. 2B). In Cnr, PVX/SlMET1 treatment led to an approximately 63% reduction in ABA levels (1.89 ± 0.5 ng g−1 fresh tissue weight) compared with the PVX control (5.15 ± 1.09 ng g−1 fresh tissue weight; Fig. 2B). Interestingly, a nearly 41% reduction in ABA levels (9.35 ± 4.85 ng g−1 fresh tissue weight) in AC-treated PVX/SlMET1 compared with the PVX-injected AC control (15.75 ± 2.12 ng g−1 fresh tissue weight) was also observed (Fig. 2B), although no VP was induced in wild-type ripe AC fruits (see below). These findings suggest that SlMET1 may act via ABA synthesis or metabolism to epigenetically regulate VP in Cnr fruits, although reduction in ABA alone cannot trigger VP in AC fruits.
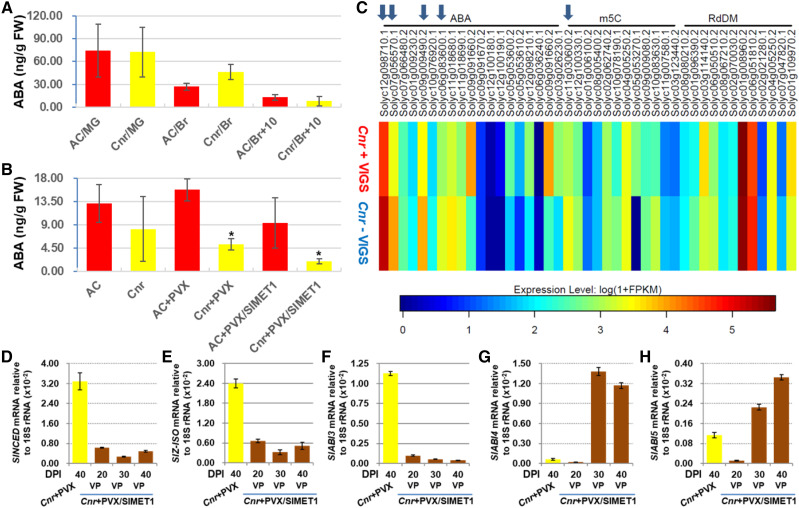
Figure 2.
Influences of SlMET1 silencing on ABA biosynthesis and ABA-related gene expression. A, ABA at different fruit developmental stages. Detection was carried out on four biological fruit samples collected at mature green (30 DPA), breaker (36 DPA on average), and 10 d after breaker (47 DPA on average) stages. Data are shown as means ± sd (n = 4). Red bars represent AC and yellow bars represent Cnr. FW, Fresh weight. B, SlMET1 knocked down by VIGS affects ABA accumulation. ABA assays were on four different AC or Cnr fruits mock inoculated or injected with PVX or PVX/SlMET1 at 30 DPI (equivalent to 47 DPA on average). Significant reduction of ABA levels was seen in viviparous seeds/seedlings from SlMET1-KD (Cnr+PVX/SlMET1) Cnr fruits when compared with Cnr fruits injected with PVX (Cnr+PVX; one-way ANOVA, P < 0.0016, as indicated by asterisks). Data are shown as means ± sd (n = 4). C, Identification of epigenetically regulated differentially expressed genes (epiDEGs) by comparative RNA-seq and comparative WGBS. Arrows from left to right indicate four epiDEGs (SlZ-ISO, SlNCED, SlABI5, and SlABI3) in addition to SlMET1, which was knocked down by VIGS. The four epiDEGs along with SlABI4 were chosen for further investigation due to their pivotal role in ABA biosynthesis and response. Names and SOL identifiers for these epiDEGs are detailed in Supplemental Table S2. D to H, Effects of SlMET1-KD on the expression of ABA biosynthesis and ABA-responsive genes. The expression of SlNCED (D), SlZ-ISO (E), and SlABI3 (F) was significantly down-regulated (one-way ANOVA, P < 0.05) at 20, 30, or 40 DPI, while that of SlABI4 (G) and SlABI5 (H) was significantly up-regulated (one-way ANOVA, P < 0.05) at 30 or 40 DPI in viviparous seeds/seedlings from PVX/SlMET1-injected Cnr fruits, compared with mature seeds from PVX-injected control Cnr fruits. RT-qPCR assays were performed on three different samples collected at different DPI as indicated. RNA transcript level was normalized against 18S rRNA. Data are shown as means ± sd (n = 3).
Influences of SlMET1 on the Expression of ABA-Related Genes
To elucidate how SlMET1 silencing reduced ABA levels, we conducted comparative transcriptomic RNA-sequencing (RNA-seq; Fig. 2C; Supplemental Fig. S6, A–C) and whole-genome bisulfite sequencing (WGBS; Supplemental Fig. S7, A and B) on viviparous seeds/seedlings and nonviviparous seeds from Cnr or the SlMET1-KD Cnr fruits that were collected at 40 DPI and reverse transcription quantitative PCR (RT-qPCR; Fig. 2, D–H). We were aware that these comparisons were not strict between equivalent materials (i.e. mature dormant seeds versus fully germinated seedlings). Indeed, once VP took places, seeds would become developmentally different from properly matured seeds no matter whether they were collected at earlier or later stages of fruit development. On the other hand, if non-VP mature seeds were collected from normal and VIGSed Cnr fruits, no differentially expressed genes (DEGs) and/or differentially methylated regions (DMRs) would be expected to be identified. Nevertheless, through mining DEGs along with DMRs in our experiments, we identified two ABA biosynthesis genes, SlNCED and SlZ-ISO, that encode 9-cis-epoxycarotenoid dioxygenase and ζ-carotene isomerase, respectively, and two ABA-responsive genes, ABI3 and ABI5 (Fig. 2C; Supplemental Fig. S6C). We also included ABI4 for subsequent analysis. SlNCED is the key enzyme in ABA biosynthesis and SlZ-ISO is involved in enzymatic production of ABA precursors, while ABI3, ABI4, and ABI5 are known to regulate genes that either inhibit dormancy release or suppress seed germination (Giraudat et al., 1992; Albertos et al., 2015; Shu et al., 2016a, 2016b). Consistent with seedling growth, the expression of SlNCED, SlZ-ISO, or SlABI3 was down-regulated in viviparous tissues of the SlMET1-KD Cnr fruits at 20, 30, and 40 DPI (Fig. 2, D–F). However, the levels of SlABI4 and SlABI5 transcripts were found to be reduced at 20 DPI but increased by 30 and 40 DPI when viviparous seeds/seedlings became apparent (Fig. 2, G and H).
Silencing of SlNCED Induces VP in Cnr Fruits and Epigenetic Regulation of SlNCED and ABA-Related Gene Expression
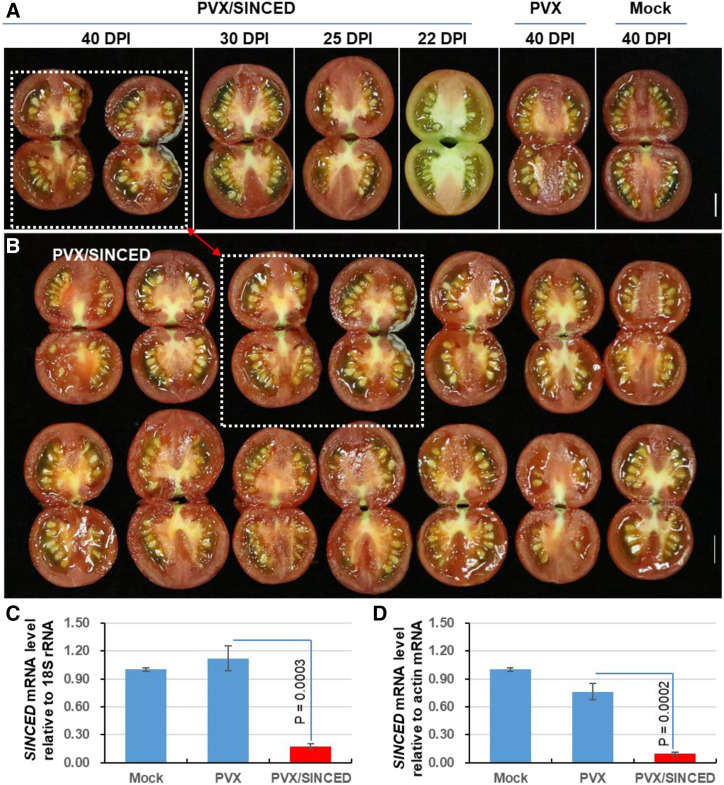
To determine whether SlNCED, SlZ-ISO, SlABI3, and other ABA-related genes were indeed involved in the regulation of VP, we attempted to silence or overexpress them in transgenic Cnr plants, but this was unsuccessful due to the recalcitrance of this epimutant to transformation. We thus decided to employ VIGS to silence SlNCED, SlZ-ISO, and SlABI3 and examined their role in Cnr seed development. To achieve this, we first cloned a 215-bp fragment of the SlNCED gene into the PVX vector to produce PVX/SlNCED (Fig. 3A). In mock- or PVX-injected fruits, seeds developed and matured normally (Fig. 3, B–D). However, in 92 of the 97 Cnr fruits injected with PVX/SlNCED, all seeds exhibited VP at about 20 DPI when these fruits approached the color-turning/breaker stage (Fig. 3E), and subsequently, viviparous seedling growth was evident at 30 and 40 DPI (Fig. 3, F and G). Consistent with this, VIGS knocked down the endogenous SlNCED expression by approximately 80% compared with expression in mock or PVX controls (Fig. 3H). No ripening reversion was observed in any of these SlNCED-VIGS, SlZ-ISO-VIGS, or SlABI3-VIGS Cnr fruits (Fig. 3B).
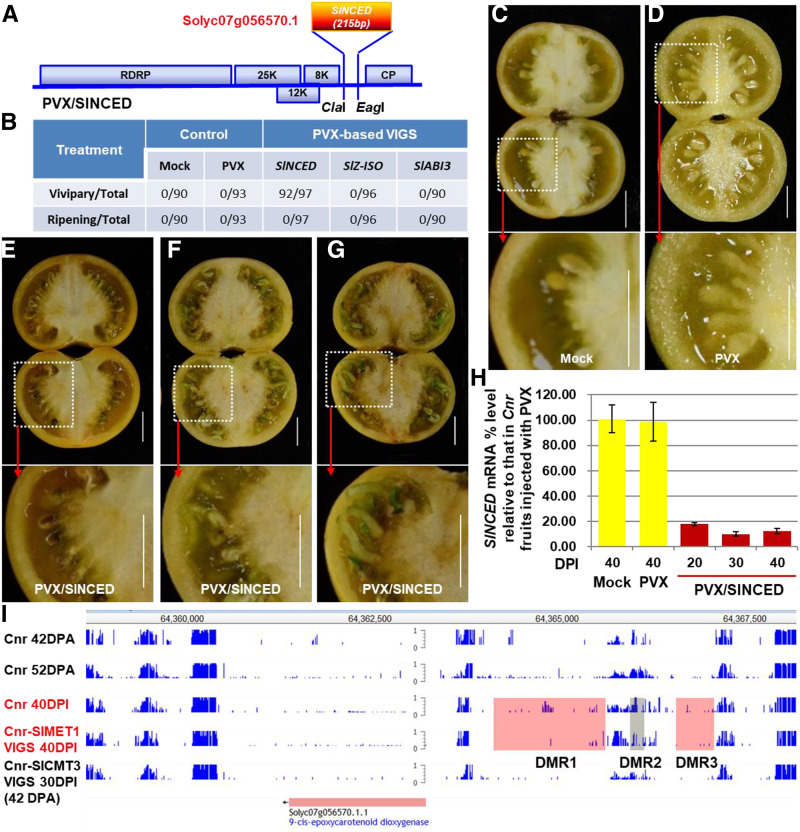
Figure 3.
Induction of tomato VP by SlNCED silencing. A, Schematic of VIGS construct PVX/SlNCED. The genome organization of PVX and two cloning sites are indicated. RDRP is the viral RNA-dependent RNA polymerase. The triple-gene block encodes three viral movement proteins of 25, 12, and 8 kD (K). CP is the viral coat protein. B, Summary of VP and ripening reversion in Cnr fruits. The numerator represents the number of viviparous or ripening reversion fruits. The denominator represents the total number of fruits that were injected with PVX/SlNCED, PVX/SlZ-ISO, PVX/SlABI3, the empty VIGS vector PVX, or Tris-EDTA buffer (Mock). C to G, Mock-treated Cnr fruits (C) or those injected with PVX alone (D) produced mature seeds, while precocious seed germination and viviparous seedling growth were present in Cnr fruits injected with PVX/SlNCED (E–G). The enlarged dotted boxes below each image, as indicated by the red arrows, clearly show mature seeds (C and D), germinating seeds (E and F), and viviparous seedlings (F and G). Fruits were photographed at 20 DPI (E), 30 DPI (F), and 40 DPI (C, D, and G). Bars = 1 cm. H, Suppression of endogenous SlNCED expression by VIGS. RT-qPCR assays were performed on three biological duplicate samples collected from mock-, Cnr+PVX control (PVX)-, or VP (Cnr+PVX/SlMET1)-treated fruits at various DPI as indicated. Data are shown as means ± sd (n = 3). Reduction of SlNCED expression in viviparous seeds/seedlings was found to be statistically significant compared with mock or PVX control seeds (one-way ANOVA, P < 0.05). I, DMRs in the promoter of the SlNCED gene. The level of DNA methylation in the specific promoter regions DMR1 and DMR3 (highlighted red) decreased but increased in DMR2 (highlighted gray) of the SlNCED gene in the SlMET1-silenced Cnr viviparous seeds. WGBS data sets for pericarps of healthy Cnr fruits at 42 or 52 DPA (Zhong et al., 2013) and pericarps of SlCMT3-silenced Cnr fruits at 30 DPI (Chen et al., 2015b) were also included as extra controls for comparative bioinformatics analysis. The gene identifier and its coordinates on the tomato chromosome are indicated. The 0-to-1 scale bar indicates the level of DNA methylation from 0% to 100% methylated cytosine.
To investigate how SlMET1 regulated SlNCED expression, we identified DMRs in the SlNCED promoter and found that the levels of DNA methylation were reduced in DRM1 and DRM3 but were increased in DMR2 in the SlMET1-KD Cnr fruits (Fig. 3I; Supplemental Figs. S8–S10). To our surprise, the whole methylome in CG, CHG, or CHH context, where H is A, T, or C, increased (Fig. 4, A–E; Supplemental Fig. S7A), likely due to up-regulation of SlCMT or SlDRM gene expression in the SlMET1-KD Cnr seed tissues (Fig. 4, F and G).
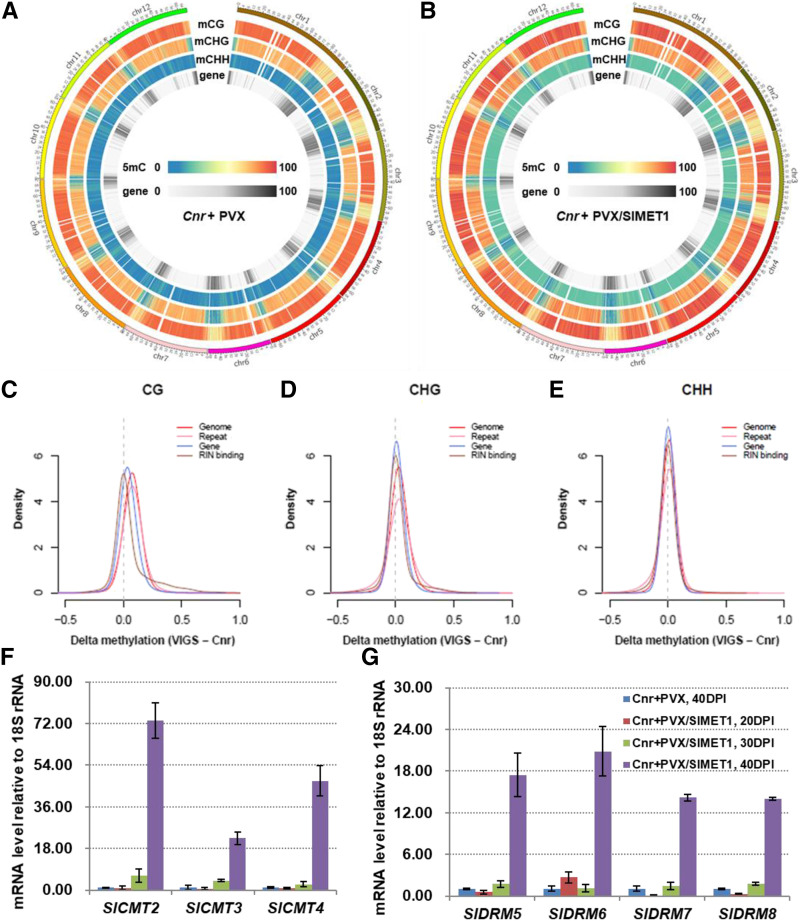
Figure 4.
Influence of SlMET1 silencing on DNA methylation level of the Cnr methylome and on the expression of SlCMT and SlDRM genes. A to E, Influence of SlMET1-KD on global DNA methylation profiles of the Cnr methylome. WGBS profiles of 12 tomato chromosomes for mature seeds or viviparous seeds/seedlings from Cnr fruits injected with the empty VIGS vector PVX (A; Cnr+PVX) or PVX/SlMET1 (B; Cnr+PVX/SlMET1) at 40 DPI are illustrated in the methylated CG (mCG), mCHG, and mCHH contexts (where H is A, T, or C) and methylated cytosine (mC) for genes. DNA methylation level of the Cnr methylone in the CG (C), CHG (D), and CHH (E) contexts for the whole genome, repeat, gene, and RIN-binding sequences are further indicated. Delta methylation (VIGS – Cnr) shows the changed methylation levels between SlMET1-KD Cnr fruits (Cnr+PVX/SlMET1) and control Cnr fruits (Cnr+PVX). The overall DNA methylation levels in the methylome increased in viviparous seeds/seedlings in the SlMET1-KD Cnr fruits. F and G, Regulation of SlCMT (F) and SlDRM (G) gene expression by SlMET1 silencing. RT-qPCR assays were performed on seed or viviparous seed/seedling samples collected from Cnr fruits injected with PVX (Cnr+PVX) or PVX/SlMET1 (Cnr+PVX/SlMET1), respectively, at various DPI as indicated in G. Data are shown as means ± sd (n = 3). Statistical analysis of gene expression between the treatments of Cnr+PVX and Cnr+PVX/SlMET1 at 40 DPI shows a significant difference (one-way ANOVA, P < 0.05).
We then analyzed potential DMRs of SlZ-ISO and SlABI3 and performed similar VIGS experiments to silence both genes (Fig. 3B; Supplemental Figs. S11, A–F, and S12, A–F). In contrast to SlNCED, VIGS of SlZ-ISO or SlABI3 was unable to induce VP, although DMRs were identified in the promoters of both genes (Supplemental Figs. S11G and S12G). Seeds of these silenced lines showed normal development and maturation (Supplemental Figs. S11, D–F, and S12, D–F). To test other genes that are directly or indirectly involved in ABA biosynthesis (Zang et al., 2016), we also examined the effect of Phytoene Synthase1 (SlPSY1) and Deetiolated1 (SlDET1) on the induction of VP in Cnr fruits. Silencing of SlPSY1 or SlDET1 intensified brown pigmentation in the outer epidermis of Cnr fruits (Supplemental Figs. S13, A and B, and S14, A and B). However, seeds matured and did not show VP in these fruits (Supplemental Figs. S13C and S14C). DRMs were identified in the SlPSY1 promoter, but only limited changes in DNA methylation were observed in the SlDET1 promoter (Supplemental Figs. S13D and S14, D and E). These results suggest that all the tested genes are likely to be under SlMET1-mediated epigenetic modulation, but these genes may have different functions in other fruit physiological processes rather than seed VP.
VIGS of SlMET1 or SlNCED Cannot Induce VP in AC fruits
Ripening reversion blocked VP in Cnr fruits (Fig. 1), suggesting that, in addition to SlMET1-mediated epicontrol of VP, ripening per se may also play a role in the prevention of seeds from VP in normal fruits. To test this idea, we performed VIGS on SlMET1 and SlNCED in wild-type AC fruits. Silencing of both genes occasionally resulted in slight hydration on AC fruits (Fig. 5, A–D), similar, but to a much lesser extent, to the heavy hydration observed on SlMET1-KD Cnr fruits (Fig. 1, D and G). During the time course for each treatment, ABA was also reduced in these fruits (Fig. 5E). However, neither SlNCED-KD (Fig. 6) nor SlMET1-KD (Fig. 7) induced viviparous seedling growth within these treated AC fruits.
Figure 5.
Hydration phenotypes in AC fruits. A and B, Hydration on AC fruits injected with PVX/SlNCED. C, Hydration on AC fruits injected with PVX/SlMET1. D, AC fruit injected with PVX. Photographs were taken at 25 DPI (A, B, and D) or 19 DPI (C). Bars = 1 cm. E, Measurement of ABA in AC fruits. Fruits were injected with PVX, PVX/SlMET1, or PVX/SlNCED and collected at different stages (DPI) of VIGS treatment for ABA measurement. Data are shown as means ± sd (n = 3). FW, Fresh weight.
Figure 6.
Silencing of SlMET1 induces no VP in AC fruits. A, AC fruits with mock treatment or injected with PVX or PVX/SlMET1. Photographs were taken at 20, 25, 30, or 40 DPI. Bar = 2 cm. B, Ten extra AC fruits injected with PVX/SlMET1 at 40 DPI. Bar = 1 cm. C and D, Suppression of endogenous SlMET1 expression in AC fruits by VIGS. RT-qPCR assays were performed on different samples collected from three mock AC control fruits or three AC fruits infected with the VIGS empty vector PVX or PVX/SlMET1 at 40 DPI. Data are shown as means ± sd (n = 3). Student’s t test indicated that reduction of SlMET1 gene expression in PVX/SlMET1-VIGS AC fruits compared with that in PVX-treated fruits was statistically significant. P values are indicated.
Figure 7.
Silencing of SlNCED induces no VP in AC fruits. A, AC fruits with mock treatment or injected with PVX or PVX/SlNCED. Photographs were taken at 22, 25, 30, and 40 DPI. Bar = 1 cm. B, Twelve extra AC fruits injected with PVX/SlNCED at 40 DPI. Boxed fruits in A and B are the same. Bar = 1 cm. C and D, Suppression of endogenous SlNCED expression in AC fruits by VIGS. RT-qPCR assays were performed on different samples collected from three mock AC control fruits or three AC fruits infected with the VIGS empty vector PVX or PVX/SlNCED at 40 DPI. Data are shown as means ± sd (n = 3). Student’s t test indicated that reduction of SlNCED gene expression in PVX/SlNCED-VIGS AC fruits compared with that in PVX-treated fruits was statistically significant. P values are indicated.
Pleiotropic Effects of SlMET1 on Tomato Development and Seed Production
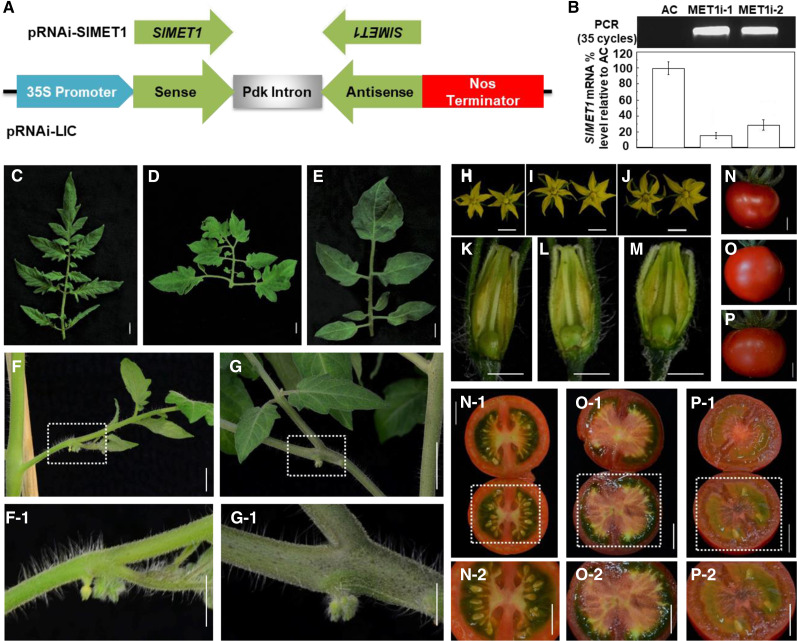
To further define the role of SlMET1 in tomato fruit and seed development, we generated two independent SlMET1-RNAi lines by transforming AC tomato with pRNAi-SlMET1 (Fig. 8A). RNAi was sufficient to silence endogenous SlMET1 expression in MET1i-1 and MET1i-2 transgenic lines (Fig. 8B). Both lines produced various abnormal phenotypes, including changes of compound leaf architectures (Fig. 8, C–E) and early abortion of floral development (Fig. 8, F, F-1, G, and G-1). Nevertheless, these RNAi AC plants still produced some morphologically normal but slightly large flowers (Fig. 8, H–J) that appeared to undergo proper anthesis and fruit set (Fig. 8, K–M). Compared with wild-type AC, there were very few fruits that went through normal development and ripened in the MET1i-1 and MET1i-2 transgenic plants (Fig. 8, N–P), and the very few ripe fruits from the MET1i-1 and MET1i-2 transgenic plants produced almost no seeds (Fig. 8, N-1, N-2, O-1, O-2, P-1, and P-2). Consistent with CRISPR/Cas9-mediated SlMET1-KO (Yang et al., 2019), these findings demonstrate that RNAi of SlMET1 has pleiotropic effects on leaf, flower, and seed development in the transgenic AC tomato lines.
Figure 8.
Influence of SlMET1 RNAi on tomato development and seed production. A, Schematic of the pRNAi-SlMET1 construct. B, Detection of the RNAi transgene (top) and RNAi-mediated suppression of endogenous SlMET1 expression (bottom). Transgene insertion was detected by PCR of genomic DNA in two independent RNAi lines, MET1i-1 and MET1i-2, but not in the wild-type AC tomato. RT-qPCR analysis shows that expression of endogenous SlMET1 was markedly reduced in both RNAi lines compared with the AC control (one-way ANOVA, P < 0.05). Data are shown as means ± sd (n = 3). C to E, SlMET1 RNAi affects tomato leaf development. A normal compound leaf from AC (C) and typical abnormal leaves from RNAi lines MET1i-1 (D) and MET1i-2 (E) are shown. F and G, SlMET1 RNAi influences floral development in the two RNAi lines MET1i-1 (F) and MET1i-2 (G). The dotted boxes were enlarged to show clearer phenotypes (F-1 and G-1). H to J, Fully developed flowers from AC (H) and the two RNAi lines MET1i-1 (I) and MET1i-2 (J). K to M, Floral development and fruit setting in AC (K) and the two RNAi lines MET1i-1 (L) and MET1i-2 (M). N to P, Ripe fruits from AC (N) and the two RNAi lines MET1i-1 (O) and MET1i-2 (P). Opened fruits are shown in N-1, O-1, and P-1, respectively. The dotted boxes are enlarged to show closeups of the seed in AC (N-2) and the two RNAi lines MET1i-1 (O-2) and MET1i-2 (P-2) fruits. Almost no seed was seen in the MET1i-1 fruit (O, O-1, and O-2), and only three seeds were visible in the MET1i-2 fruit (P, P-1, and P-2). These surviving seeds matured and showed no VP, although they were able to germinate after being sown into compost. Bars = 1 cm in C to J, N to P, N-1, N-2, O-1, O-2, P-1, and P-2; bars = 0.5 cm in F-1 and K to M; and bar = 0.25 cm in G-1.
DISCUSSION
In this article, we report that SlMET1 controls VP in Cnr fruits and can have a profound effect on different developmental events in AC tomato. In Cnr fruits, SlMET1 participates in regulating the expression of ABA-biosynthesis/response genes, likely through changing methylation levels in their promoter sequences (Fig. 9). SlNCED is the key enzyme in ABA biosynthesis, and SlZ-ISO, SlPSY1, and SlPDS are also involved in enzymatic production of ABA precursors, while SlDET1 plays an important role in regulating genes that are required for ABA precursor biosynthesis (Fig. 9A). All these genes may be modulated by the SlMET1-mediated epigenetic mechanism in Cnr. SlMET1 is a facilitator for ABA production as well as for SlNCED and SlZ-ISO expression, since SlMET1-KD leads to reduction of ABA and down-regulation of SlNCED and SlZ-ISO (Fig. 9A). Such escalation of SlNCED transcripts is probably via a transcriptional repressor that may fail its binding to hypermethylated DMR1 and DMR3 along with an activator that may bind to hypomethylated DMR2 in the SlNCED promoter in Cnr fruits. Methylation levels in the three DMRs are directly affected by SlMET1, although changes in methylation were subtle for some cytosines (Fig. 3I; Supplemental Figs. S8–S10). This phenomenon may not be particularly surprising, since slight methylation alternations have been reported in the Cnr epimutant (Manning et al., 2006). There are only 18 differentially methylated cytosines within the DMR of the CNR gene promoter, of which only eight were essential for the nonripening phenotype, and reduction of methylation levels of the eight cytosines caused Cnr fruits to ripen (Chen et al., 2015b). Nevertheless, the precise role of the three DRMs in the regulation of SlNCED expression remains to be elucidated.
Figure 9.
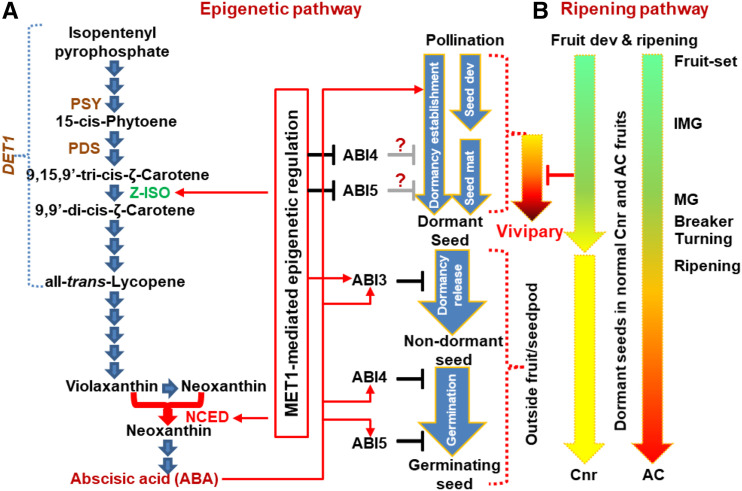
Models of the SlMET1-directed epipathway and the ripening-related pathway in VP. A, Epipathway in VP. SlMET1 directs the epiregulation of expression of NCED and Z-ISO genes. A simplified ABA biosynthesis pathway is shown (PDS, phytoene desaturase). ABA and SlMET1-directed epiregulation of ABA-responsive genes are involved in VP. Silencing of SlMET1 or SlNCED, but not SlZ-ISO, SlPSY1, and SlDET1, resulted in VP in tomato fruits, suggesting an SlMET1→SlNCED→ABA⊣VP epipathway to prevent VP (→ and ⊣ indicate positive and negative regulation, respectively). SlMET1-KD-mediated reduction of ABA may result in a decrease in ABI3 gene expression, which is known to suppress dormancy release. However, ABI3-KD cannot induce viviparous seeds, suggesting that the SlMET1→SlNCED→ABA⊣VP epipathway is not via the influence of ABI3 on dormancy release. Again, SlMET1-KD-mediated increase of ABI4/ABI5 expression contradicts the well-documented roles of ABI4/ABI5 in inhibiting germination, suggesting that an extra SlMET1⊣ABAI4/ABI5→VP control to suppress dormancy establishment may also contribute to the epigenetic modulation of VP in tomato fruits. It should be noted that dormancy in developing fruit or mature seeds, the latter extracted from fruits and put to germinate, may not have exactly the same regulation mechanism although both involve ABA. B, Impact of ripening on VP. The genetic and molecular framework for this pathway remains to be elucidated. It should be pointed out that any specific ripening-associated genes or ripening hormone ethylene do not necessarily involve tomato VP and that gene(s) influencing ripening in Cnr would not necessarily lead to the VP effect directly. However, that ripening reversion suppresses VP supports the existence of a ripening-associated pathway. This idea is also consistent with our finding that silencing of SlMET1 or SlNCED as well as transgenic SlMET1 RNAi all fail to induce VP in AC fruits. Taken together, we propose that an epigenetic pathway involved in SlMET1→SlNCED→ABA⊣VP along with SlMET1⊣ABAI4/ABI5→VP may occur in the course of dormancy establishment during the early seed development (Seed dev) and/or late seed maturation (Seed mat) before the breaker/tuning stages. In addition to the epipathway (A), a ripening-associated genetic pathway may also act independently to block viviparous seedling growth during tomato development and fruit ripening (B). Such dual reassurances would warranty proper maturation of robust seeds for growing progeny. This model explains the unanticipated findings reported in this article, although it does not exclude the involvement of other SlMET1-triggered specific or nonspecific epimodulations in tomato VP. Fruit dev, Fruit development; IMG, immature green; MG, mature green.
Tomato seeds mature in fruits when fruits reach mature green/breaker/ripening stages, normally at 35 to 40 DPA (Downie et al., 2003). Suppression of SlNCED by direct or indirect SlMET1-mediated epiregulation reduces ABA biosynthesis and induces VP in Cnr fruits. These events may occur during early stages of fruit development at which seeds have not established or at least reduced dormancy. This view is supported by the fact that ABI3 silencing had no effect on VP in fruits (Supplemental Fig. S6) and that ABI3 is known to inhibit dormancy release (Fig. 9A; Shu et al., 2016b). On the other hand, the increase of ABI4 or ABI5 along with the reduced ABA levels in the SlMET1-KD Cnr fruits (Fig. 2) appear to contradict their roles in suppressing seed germination outside fruits or seed pods (Fig. 9A; Gubler et al., 2005; Shu et al., 2016b). However, our finding may suggest that when seeds still reside within fruit tissues, ABI4 and ABI5 might have a different activity in the blockage of dormancy establishment, thus promoting viviparous seedling growth (Fig. 9A). We speculate that SlMET1→SlNCED→ABA (likely along with SlMET1⊣ABI4/ABI5) may affect the establishment of seed dormancy in the course of early seed development and/or late maturation during tomato fruit development and ripening (Fig. 9A).
Strikingly, VP is completely suppressed if ripening occurred in PVX/SlMET1-treated Cnr fruits (Fig. 1, G and H). This would suggest that the SlMET1→SlNCED→ABA pathway is not the exclusive mechanism responsible for withholding VP in tomato. Consistent with this hypothesis, ABA production peaks at mature green stages and is reduced at later stages of fruit development and ripening in wild-type AC and Cnr fruits (Figs. 2, A and B, and 5E; Ji et al., 2014). Most significantly, SlMET1-KD or SlNCED-KD by VIGS failed to induce VP in wild-type AC fruits in which ABA production was also reduced (Figs. 2B and 5–7). Moreover, RNAi of SlMET1 blocks seed production without developing VP in transgenic tomato (Fig. 8) and other plants (Hu et al., 2014; Yamauchi et al., 2014; Chen et al., 2017), likely due to extremely early germination, parthenocarpy, or pollen deficiencies that would be lethal to seed development. Furthermore, overexpression of SlNCED increases the level of ABA that inhibits seed germination, while a mutation in SlNCED is the cause of ABA deficiency without showing a genuine VP phenotype in tomato (Burbidge et al., 1999; Thompson et al., 2000). Taken together, our results along with published data support that ripening de facto contributes to inhibiting VP in fruits (Fig. 9B). A more thorough analysis of the revertant sectors through gene expression studies will be needed in order to provide some insight as to factors that might specially regulate the observed phenomena (Fig. 1, G and H). Nevertheless, there may exist an epigenetic SlMET1→SlNCED→ABA pathway and a ripening-associated genetic pathway that independently functions to prevent the occurrence of VP in order to guarantee the production of mature seeds during tomato development and fruit ripening. Disruption of either pathway would not cause VP; this explains why VP was only induced in SlMET1- or SlNCED-KD nonripe Cnr fruits but not in SlMET1-/SlNCED-KD ripening-reoccurring Cnr or AC fruits (Fig. 9). This model is further supported by a recent finding that seeds of a ripening-inhibitor mutant accession exhibit limited precocious germination and viviparous seedling growth in developing fruits (Wang et al., 2016).
MATERIALS AND METHODS
Plant Materials and Growth
Wild-type tomato (Solanum lycopersicum) line AC and the Cnr epimutant (AC background) were grown in insect-free growth rooms or greenhouses at 25°C under a 16-h-light/8-h-dark cycle with a humidity of 60% to 80%. AC seeds were also sown and grown in growth chambers under conditions as described by Manning et al. (2006) and Chen et al. (2015a, 2015b) to generate cotyledons for tomato transformation.
VIGS and RNAi Constructs
VIGS constructs were generated as previously described by van Wezel et al. (2002) and Chen et al. (2015b). Briefly, a nontranslatable 200- to 400-bp fragment corresponding to the coding region of each gene was PCR amplified and cloned into MluI/SalI or ClaI/EagI sites of the PVX vector (Bruce et al., 2011) to produce PVX/SlMET1, PVX/SlCMT4, PVX/SlNCED, PVX/SlZ-ISO, PVX/SlABI3, PVX/SlPSY1, and PVX/SlDET1. To generate the SlMET1 RNAi construct pRNAi-SlMET1, a 250-bp SlMET1 fragment was PCR amplified using tomato cDNA as template and cloned in the sense and antisense orientations into the pRNAi-LIC vector (Chen et al., 2018b). Primers used for making these constructs are listed in Supplemental Table S1. All constructs were verified by DNA sequencing.
VIGS
PVX-based VIGS in Cnr and AC fruits at various developmental stages on different trusses on the same plants and on different plants was carried out in repeated experiments as described by Chen et al. (2015b). In each experiment, carpopodium of at least 40 fruits at 5 to 20 DPA were mock injected with Tris-EDTA buffer or injected with recombinant viral RNAs generated by in vitro transcription from each of the VIGS constructs. Tomato plants were grown and maintained in growth rooms at 25°C with supplementary lighting to give a 16-h photoperiod. Fruits were examined daily and photographed with a Coolpix 995 digital camera (Nikon).
ABA Assay
To analyze the ABA concentration in viviparous seeds/seedlings and mature nongerminating seeds, approximately 0.5-g samples were collected from PVX/SlMET1- or PVX-infected Cnr fruits. After adding the internal standard [2H6](+)-cis,trans-ABA, organic compounds were extracted and purified from four biological duplicates of each treatment. The relative amount of ABA was then quantified using HPLC-electrospray-mass spectrometry and calculated as described by Dobrev and Kamínek (2002).
RNA Extraction and RT-qPCR
Total RNA was extracted from viviparous seeds/seedlings or mature nongerminating seeds using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized using a FastQuant RT Kit with gDNA Eraser (Tiangen). Real-time PCR was performed using a CFX96/384 real-time system (Bio-Rad) with the UltraSYBR Mixture (CoWin Bioscience) and gene-specific primers (Supplemental Table S1). 18S rRNA or actin mRNA was used as an internal control, and at least two biological duplicates and four technical duplicates were used for each repeated experiment. The relative level of specific gene expression was calculated using the ∆∆Ct method as described by Chen et al. (2015b).
Transcriptome RNA-Seq
For the RNA-seq experiment, viviparous seeds/seedlings or mature nongerminating seeds were harvested from PVX/SlMET1 or PVX-injected Cnr fruits, respectively. Five micrograms of pooled RNA extracted from samples collected from three different fruits was used to construct the library for RNA-seq using the Illumina Genome Analyzer (Solexa). Library construction and RNA-seq were done by OneGene. Low-quality reads were removed from the RNA-seq raw data, and high-quality reads were aligned to the tomato genome (version SL2.50; https://solgenomics.net). Functional annotation of sequences, Gene Ontology enrichment analysis, and bioinformatics analyses were performed by BGI as described by Zouari et al. (2014). Furthermore, qualified reads were aligned to the tomato reference genome using STAR v2.5.3a (Dobin et al., 2013) with key parameters –runThreadN 10 \–genomeDir $gnomedir–readFilesIn $nfqR1 $nfqR2 \–outSAMstrandField intronMotif–limitBAMsortRAM 41000000000 \–outFileNamePrefix $nbam–outSAMtype BAM SortedByCoordinate \–outBAMsortingThreadN 8–outFilterMultimapNmax 5–winAnchorMultimapNmax 5–alignIntronMax 1000 \–alignEndsType EndToEnd. Expression values were computed using Cufflinks v2.2.1.
Tomato Transformation
AC seeds were surface sterilized and germinated on plates containing one-quarter-strength Murashige and Skoog medium (pH 5.6–6) in the growth chamber as described by Gonzalez et al. (2007). After 5 to 7 d, cotyledons of explants were collected and cocultivated with Agrobacterium tumefaciens strain GV3101 carrying the binary vector pRNAi-SlMET1 to induce shoots under the selection of kanamycin resistance as described by Dobrev and Kamínek (2002). Regenerated shoots with 3 to 4 cm length were cut off from independent calli and transferred to rooting medium for root development as described by How Kit et al. (2010). To confirm the stable transformation event, putatively transformed plantlets with well-developed roots were subjected to molecular analyses through PCR and RT-qPCR. When compared with nontransformed AC, nine independent lines were confirmed to be transformed with the RNAi construct and transferred to compost, but only two survived to maturity in the greenhouse. These two lines were named MET1i-1 and MET1i-2 and used in this study.
DNA Preparation and WGBS
Genomic DNA was isolated from viviparous seeds/seedlings or mature nongerminating seeds that were harvested from PVX/SlMET1 or PVX-injected Cnr fruits using the DNeasy Plant Mini Kit (Qiagen). The integrity and purity of genomic DNA were checked using gel electrophoresis and NanoPhotometer (Implen). The concentration of genomic DNA was measured using the Qubit DNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies). To construct the WGBS library, genomic DNA was first fragmented by sonication to an average size of approximately 250 bp, followed by blunting of DNA ends, deoxyadenosine addition to the 3′ end, and adaptor ligation, according to the manufacturer’s instructions (Illumina). Ligated DNA fragments were bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research) and followed by direct PCR sequencing using HighSeq4000. After collecting the raw data, data filtering was first done to remove those low-quality reads, and the clean data sets were mapped to the reference tomato genome (https://solgenomics.net) by BSMAP. The uniquely mapped reads were used to determine the genomic DNA methylation status. These initial WGBS and related standard bioinformatics analysis were performed by BGI. The cytosine methylation information was used for further in-house bioinformatics analyses (Zhong et al., 2013; Chen et al., 2015b, 2017). Briefly, the tomato genome fasta and annotation files were obtained from EnsemblePlants (ftp://ftp.ensemblgenomes.org/pub/plants/release-38/fasta/solanum_lycopersicum/dna/). Raw fastq sequencing files were processed by removing the adaptor sequence. After adaptor trimming, we did quality checking using FastQC analysis (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). After preprocessing, clean sequencing reads were aligned to the tomato genomes using the pipleilne methylpy v0.1.0. Unmethylated chloroplast DNA was used as a control to calculate the sodium bisulfite reaction nonconversion rate of unmodified cytosines. The reference genome was indexed by methylpy. The reads were aligned using methylpy with the following key parameters: methylpy paired-end-pipeline \–merge-by-max-mapq True \–binom-test True \–unmethylated-control chloroplast \–min_cov 3 \–num-procs 28–sort-mem 45000000000 \–remove-clonal True \–aligner-options “-p 28” \–trim-read False. Illustrative figures were created using the R environment (Li et al., 2009; Schultz et al., 2015; R Core Team, 2017). The genome-wide DNA methylation profile figures were created using the software package Circos (http://circos.ca/) with key parameters angle_offset* = -82\ chromosomes_units = 40000.
Identification of epiDEGs
To identify epiDEGs by comparative RNA-seq and WGBS, reads were aligned to the tomato genome (Solanum lycopersicum.SL2.50; ftp://ftp.ensemblgenomes.org/pub/plants/release-37/fasta/solanum_lycopersicum/dna/Solanum_lycopersicum.SL2.50.dna.toplevel.fa.gz) using software Methylpy 1.3 as described by Schultz et al. (2015). Chloroplast DNA was used as a control to calculate the sodium bisulfite reaction nonconversion rate of unmodified cytosines. A binomial test was used to determine the methylation status of cytosines with a minimum coverage of three reads (Schultz et al., 2015). After finding the DMR, we used the software bedtools2 (https://bedtools.readthedocs.io/en/latest/) to find the closest upstream gene in the GFF file (Solanum_lycopersicum.SL2.50.37) within 1 kb. The fragments per kilobase of exon model per million reads mapped of genes is calculated using the cuffnorm from the cufflink suite as described by Trapnell et al. (2010).
Statistical Analysis
Experiments such as VIGS, tomato transformation, and RT-qPCR and ABA assays were repeated at least twice, and the resulting data are represented as means ± sd wherever appropriate. Duncan’s multiple range tests in a one-way ANOVA procedure or Student’s t test were performed to analyze whether there were significant differences between different treatments wherever appropriate.
Accession Numbers
Accession numbers for all relevant sequences are listed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Induction of tomato VP by SlCMT4 silencing.
Supplemental Figure S2. Ripening reversion but no VP in Cnr fruits by VIGS of SlCMT2, SlCMT3, or SlDRM7.
Supplemental Figure S3. Induction of tomato VP by SlMET1 silencing.
Supplemental Figure S4. Correlation of SlMET1 expression with ripening reversion and VP in Cnr fruits.
Supplemental Figure S5. No influence of PVX infection on endogenous SlMET1 expression in Cnr fruits.
Supplemental Figure S6. Differential gene expression in viviparous seeds/seedlings versus mature seeds.
Supplemental Figure S7. Influence of SlMET1 silencing on DNA methylation level of the Cnr methylome.
Supplemental Figure S8. Differential DNA methylation in CC, CHG, and CHH contexts in DMR1 within the SlNCED gene promoter
Supplemental Figure S9. Differential DNA methylation in CC, CHG, and CHH contexts in DMR2 within the SlNCED gene promoter.
Supplemental Figure S10. Differential DNA methylation in CC, CHG, and CHH contexts in DMR3 within the SlNCED gene promoter.
Supplemental Figure S11. SlZ-ISO-KD cannot induce VP in Cnr fruits.
Supplemental Figure S12. SlABI3 silencing cannot induce VP in Cnr fruits.
Supplemental Figure S13. SlPSY1 silencing enhances pigmentation but cannot induce VP in Cnr fruits.
Supplemental Figure S14. SlDET1 silencing enhances pigmentation but cannot induce VP in Cnr fruits.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Names and SOL identifiers of epiDEGs.
Acknowledgments
We thank David Baulcombe for the kind gift of the original PVX vector. We thank Graham Seymour for revising the article and William Finch-Savage, Jiankang Zhu, Leon V. Kochian, and Huizhong Wang for critical reading of the article. We also thank Kenneth Manning and Paul Hunter for their initial participation in this project and Hui Wang, Chaoqun Wu, and Pengcheng Zhang for their technical assistance.
Footnotes
This work was supported by the Zhejiang Provincial Natural Science Foundation (grant no. LY19C150006), the Ministry of Agriculture of the People’s Republic of China (grant no. 2016ZX08009001–004), the National Natural Science Foundation of China (grant no. 31872636), the Ministry of Science and Technology of the People’s Republic of China (grant no. 2017YFE0110900), Hangzhou Normal University (grant nos. 9995C5021841101 and 201108), the Hangzhou City Innovative Program for Science Excellence (grant no. 20131028), the U.S. National Science Foundation (grant nos. DMS–1903226 and DMS–1925066), the U.S. National Institutes of Health (grant nos. R01GM113242 and R01GM122080), the U.K. Biotechnology and Biological Sciences Research Council (grant no. BBS/E/H/00YH0271), and the U.K. Royal Society (grant no. RG072176).
Articles can be viewed without a subscription.
References
- Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H(2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol 125: 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sánchez-Vicente I, Nambara E, Lorenzo O(2015) S-Nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun 6: 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An YC, Goettel W, Han Q, Bartels A, Liu Z, Xiao W(2017) Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci Rep 7: 12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benech-Arnold RL, Rodríguez MV(2018) Pre-harvest sprouting and grain dormancy in Sorghum bicolor: What have we learned? Front Plant Sci 9: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V(2017) DNA methylation dynamics during early plant life. Genome Biol 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce G, Gu M, Shi N, Liu Y, Hong Y(2011) Influence of retinoblastoma-related gene silencing on the initiation of DNA replication by African cassava mosaic virus Rep in cells of mature leaves in Nicotiana benthamiana plants. Virol J 8: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB(1999) Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J 17: 427–431 [DOI] [PubMed] [Google Scholar]
- Carbonero P, Iglesias-Fernández R, Vicente-Carbajosa J(2017) The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J Exp Bot 68: 871–880 [DOI] [PubMed] [Google Scholar]
- Chen M, Xie S, Ouyang Y, Yao J(2017) Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1. Plant Sci 260: 80–89 [DOI] [PubMed] [Google Scholar]
- Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, et al. (2015a) Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep 5: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kong J, Qin C, Yu S, Tan J, Chen YR, Wu C, Wang H, Shi Y, Li C, et al. (2015b) Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep 5: 9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yu Z, Kong J, Wang H, Li Y, Zhao M, Wang X, Zheng Q, Shi N, Zhang P, et al. (2018a) Comparative WGBS identifies genes that influence non-ripe phenotype in tomato epimutant Colourless non-ripening. Sci China Life Sci 61: 244–252 [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang X, Fan Y, Li B, Ryabov E, Shi N, Zhao M, Yu Z, Qin C, Zheng Q, et al. (2018b) A genetic network for systemic RNA silencing in plants. Plant Physiol 176: 2700–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J(2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Dintu KP, Sibi CV, Ravichandran P, Satheeshkumar K(2015) Vivipary in Ophiorrhiza mungos L.: A rare phenomenon in angiosperms. Plant Biol (Stuttg) 17: 294–295 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR(2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Kamínek M(2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Downie B, Gurusinghe S, Dahal P, Thacker RR, Snyder JC, Nonogaki H, Yim K, Fukanaga K, Alvarado V, Bradford KJ(2003) Expression of a GALACTINOL SYNTHASE gene in tomato seeds is up-regulated before maturation desiccation and again after imbibition whenever radicle protrusion is prevented. Plant Physiol 131: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durantini D, Giulini A, Malgioglio A, Pilu R, Tuberosa R, Sanguineti C, Gavazzi G(2008) Vivipary as a tool to analyze late embryogenic events in maize. Heredity 101: 465–470 [DOI] [PubMed] [Google Scholar]
- Eriksson EM, Bovy A, Manning K, Harrison L, Andrews J, De Silva J, Tucker GA, Seymour GB(2004) Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol 136: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyster WH.(1931) Vivipary in maize. Genetics 16: 574–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedak H, Palusinska M, Krzyczmonik K, Brzezniak L, Yatusevich R, Pietras Z, Kaczanowski S, Swiezewski S(2016) Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc Natl Acad Sci USA 113: E7846–E7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhu N, Zhu X, Wu M, Jiang CZ, Grierson D, Luo Y, Shen W, Zhong S, Fu DQ, et al. (2019) Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic Res 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ.(2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM(1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A(2007) The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51: 642–655 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV(2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8: 183–187 [DOI] [PubMed] [Google Scholar]
- Hable WE, Oishi KK, Schumaker KS(1998) Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol Gen Genet 257: 167–176 [DOI] [PubMed] [Google Scholar]
- How Kit A, Boureau L, Stammitti-Bert L, Rolin D, Teyssier E, Gallusci P(2010) Functional analysis of SlEZ1 a tomato enhancer of zeste (E(z)) gene demonstrates a role in flower development. Plant Mol Biol 74: 201–213 [DOI] [PubMed] [Google Scholar]
- Hu L, Li N, Xu C, Zhong S, Lin X, Yang J, Zhou T, Yuliang A, Wu Y, Chen YR, et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci USA 111: 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H, Wei S, Bradford KJ(2016) DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci USA 113: E2199–E2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A(2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ji K, Kai W, Zhao B, Sun Y, Yuan B, Dai S, Li Q, Chen P, Wang Y, Pei Y, et al. (2014) SlNCED1 and SlCYP707A2: Key genes involved in ABA metabolism during tomato fruit ripening. J Exp Bot 65: 5243–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA(2014) Transcriptional control of fleshy fruit development and ripening. J Exp Bot 65: 4527–4541 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR(2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Chen W, Shen J, Qin C, Lai T, Zhang P, Wang Y, Wu C, Yang X, Hong Y(2013) Virus-induced gene complementation in tomato. Plant Signal Behav 8: e27142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Wang X, Ye B, Jin M, Chen W, Wang Y, Zhou Y, Blank A, Gu M, Zhang P, et al. (2020) Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J Exp Bot 71: 2995–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Wang Y, Zhou T, Mei F, Zhang P, Zhou Y, Shi N, Hong Y(2015) Virus-induced LeSPL-CNR silencing inhibits fruit ripening in tomato. J Agric Sci 7: 184 [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R(2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D(2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D(2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, et al. (2015) A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA 112: 10804–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Giménez E, Cara B, Capel J, Angosto T(2009) Genetic analysis of reproductive development in tomato. Int J Dev Biol 53: 1635–1648 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB(2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK(1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM, Lazzeri PA, Gale MD, Lenton JR, Holdsworth MJ(2002) Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA 99: 10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S.(2018) Grain dormancy genes responsible for preventing pre-harvest sprouting in barley and wheat. Breed Sci 68: 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J(2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Gaza ALF, Kouassi KI, Koffi KK, Kouakou KL, Baudoin JP, Zoro BIA(2019) Prevalence and variation of viviparous germination with respect to fruit maturation in the bottle gourd Lagenaria siceraria (Molina) Standley (Cucurbitaceae). Heliyon 5: e02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porch TG, Tseung CW, Schmelz EA, Settles AM(2006) The maize Viviparous10/Viviparous13 locus encodes the Cnx1 gene required for molybdenum cofactor biosynthesis. Plant J 45: 250–263 [DOI] [PubMed] [Google Scholar]
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Robertson DS.(1952) The genotype of the endosperm and embryo as it influences vivipary in maize. Proc Natl Acad Sci USA 38: 580–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Saladié M, Catalá C(2004) The plot thickens: New perspectives of primary cell wall modification. Curr Opin Plant Biol 7: 296–301 [DOI] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. (2015) Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR(1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Seymour G, Poole M, Manning K, King GJ(2008) Genetics and epigenetics of fruit development and ripening. Curr Opin Plant Biol 11: 58–63 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JKC(2013) Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J 11: 269–278 [DOI] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang P, Li Y, Wang S, Tang S, Liu C, et al. (2016a) ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J 85: 348–361 [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH(2016b) Two faces of one seed: Hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JK, Mazourek M, Chomet P, Brutnell TP(2003) Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Singh S, Randhawa H, Singh J(2013) Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in wheat (Triticum aestivum L.). PLoS ONE 8: e77009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR(2008) The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol 146: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Settles AM, Tseung CW, Li QB, Latshaw S, Wu S, Porch TG, Schmelz EA, James MG, McCarty DR(2006) The maize viviparous15 locus encodes the molybdopterin synthase small subunit. Plant J 45: 264–274 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB(2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23: 363–374 [DOI] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MF Jr., Lin T, Nguyen C, Bies D, Rambla JL, Beltran KS, Taylor M, Zhang B, et al. (2017) A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ(2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L(2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik S, Chapman NH, Smith R, Poole M, Adams G, Gillis RB, Besong TM, Sheldon J, Stiegelmeyer S, Perez L, et al. (2016) Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol 34: 950–952 [DOI] [PubMed] [Google Scholar]
- van Wezel R, Dong X, Liu H, Tien P, Stanley J, Hong Y(2002) Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol Plant Microbe Interact 15: 203–208 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J(2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang D, Seymour GB(2017) Tomato flavor: Lost and found? Mol Plant 10: 782–784 [DOI] [PubMed] [Google Scholar]
- Wang R, Angenent GC, Seymour G, de Maagd RA(2020) Revisiting the role of master regulators in tomato ripening. Trends Plant Sci 25: 291–301 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang L, Xu X, Qu W, Li J, Xu X, Wang A(2016) Seed development and viviparous germination in one accession of a tomato rin mutant. Breed Sci 66: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Johzuka-Hisatomi Y, Terada R, Nakamura I, Iida S(2014) The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol Biol 85: 219–232 [DOI] [PubMed] [Google Scholar]
- Yang Y, Tang K, Datsenka TU, Liu W, Lv S, Lang Z, Wang X, Gao J, Wang W, Nie W, et al. (2019) Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J Integr Plant Biol 61: 1224–1242 [DOI] [PubMed] [Google Scholar]
- Zang G, Zou H, Zhang Y, Xiang Z, Huang J, Luo L, Wang C, Lei K, Li X, Song D, et al. (2016) The De-Etiolated 1 homolog of Arabidopsis modulates the ABA signaling pathway and ABA biosynthesis in rice. Plant Physiol 171: 1259–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhou T, Zhang H, Lai T, Qin C, Shi N, Wang H, Jin M, Zhong S, Fan Z, Liu Y, et al. (2012) Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening. Sci Rep 2: 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C, et al. (2018) Rewiring of the fruit metabolome in tomato breeding. Cell 172: 249–261.e12 [DOI] [PubMed] [Google Scholar]
- Zouari I, Salvioli A, Chialva M, Novero M, Miozzi L, Tenore GC, Bagnaresi P, Bonfante P(2014) From root to fruit: RNA-seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genomics 15: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]