Chloroplast autophagy and chloroplast-associated ubiquitination independently contribute to chloroplast degradation, the management of ROS damage, and the adaptation to starvation.
Abstract
Autophagy and the ubiquitin-proteasome system are the major degradation processes for intracellular components in eukaryotes. Although ubiquitination acts as a signal inducing organelle-targeting autophagy, the interaction between ubiquitination and autophagy in chloroplast turnover has not been addressed. In this study, we found that two chloroplast-associated E3 enzymes, SUPPRESSOR OF PPI1 LOCUS1 and PLANT U-BOX4 (PUB4), are not necessary for the induction of either piecemeal autophagy of chloroplast stroma or chlorophagy of whole damaged chloroplasts in Arabidopsis (Arabidopsis thaliana). Double mutations of an autophagy gene and PUB4 caused synergistic phenotypes relative to single mutations. The double mutants developed accelerated leaf chlorosis linked to the overaccumulation of reactive oxygen species during senescence and had reduced seed production. Biochemical detection of ubiquitinated proteins indicated that both autophagy and PUB4-associated ubiquitination contributed to protein degradation in the senescing leaves. Furthermore, the double mutants had enhanced susceptibility to carbon or nitrogen starvation relative to single mutants. Together, these results indicate that autophagy and chloroplast-associated E3s cooperate for protein turnover, management of reactive oxygen species accumulation, and adaptation to starvation.
Dysfunctional proteins and damaged organelles must be degraded to maintain cellular homeostasis in eukaryotes. The digestion of intracellular components also allows the reuse of the resulting small molecules, such as amino acids and lipids (Araújo et al., 2011; Izumi et al., 2019). Accordingly, in vivo turnover of intracellular macromolecules is important in the adaptation to stresses that increase cellular damage and in the efficient use of nutrients once assimilated. Autophagy and the ubiquitin-proteasome system (UPS) are evolutionarily conserved systems for the degradation of proteins and organelles (Dikic, 2017).
In the UPS, a small polypeptide, ubiquitin, acts as a tag for proteins that should be degraded by the 26S proteasome complex (Finley, 2009). Protein ubiquitination begins with the ATP-consuming activation of ubiquitin by E1 enzymes. The activated ubiquitin is transferred from E1 to ubiquitin-conjugating E2 enzymes. Ubiquitin ligases (E3) then shift the ubiquitin from E2 to specific target proteins. The buildup of polyubiquitin chains via the ubiquitination of ubiquitin’s own Lys-48 residue typically acts as the signal inducing proteasome-mediated breakdown (Chau et al., 1989). Generally, eukaryotic genomes encode numerous E3s; in Arabidopsis (Arabidopsis thaliana), for example, genomics studies indicate that more than 1,500 E3s are expressed (Hua and Vierstra, 2011; Metzger et al., 2014; Shu and Yang, 2017). Individual E3s may have specific targets, thereby allowing highly controlled, selective protein degradation in response to changing circumstances (Dikic, 2017).
By contrast, during the major type of autophagy, known as macroautophagy, nascent, double membrane-bound vesicles (termed autophagosomes) engulf a portion of the cytoplasm including proteins and organelles, which allows the bulk digestion of a subset of intracellular components (Mizushima and Komatsu, 2011). This degradation system is vital in enabling cells to adapt to starvation through the efficient recycling of cytoplasmic components. A family of autophagy-related (ATG) proteins have been identified in the budding yeast Saccharomyces cerevisiae (Nakatogawa et al., 2009). The production of autophagosomal membranes requires the activation of the ubiquitin-like proteins ATG8 and ATG12 via protein conjugation cascades, in a process similar to ubiquitin activation and handling by E1s, E2s, and E3s in the UPS (Mizushima et al., 1998; Ichimura et al., 2000). First, ATG12 is activated by the E1-like protein ATG7 and transferred to the E2-like ATG10, initiating the formation of a protein complex comprising ATG12, ATG5, and ATG16. Meanwhile, ATG8 is processed by the protease ATG4, activated by ATG7, and then transferred to the E2-like ATG3. Subsequently, ATG8 becomes conjugated to the lipid phosphatidylethanolamine, a building block of the autophagosomal membrane, with the aid of the autophagy-related E3 ATG12-ATG5-ATG16 complex. A mutant analysis of core ATG orthologs in Arabidopsis revealed that the functions of those proteins are largely conserved in plants (Marshall and Vierstra, 2018; Soto-Burgos et al., 2018; Yoshimoto and Ohsumi, 2018).
Microautophagy represents another type of membrane dynamic whereby cytoplasmic components are incorporated into the vacuolar lumen, during which the vacuolar membrane itself supports target sequestration (Oku and Sakai, 2018). One well-characterized microautophagy process is the degradation of peroxisomes in the methylotrophic yeast Komagataella phaffii (previously known as Pichia pastoris). In this micropexophagy process, core ATG proteins are required for the formation of a specific membrane structure to complete the sequestration of targeted peroxisomes (Mukaiyama et al., 2004).
Recent advances in the study of autophagy, particularly in yeasts and mammals, have established a role of autophagy in the selective degradation of dysfunctional components, such as very large protein complexes and organelles (Anding and Baehrecke, 2017). Notably, in mammals, ubiquitination acts as the signal inducing such selective autophagy processes. For instance, p62/Sequestosome-1 (SQSTM1) and Next to BRCA1 Gene1 (NBR1), which are autophagic receptors that bind the mammalian homologs of ATG8, also interact with ubiquitinated proteins to degrade them through autophagosomal transport (Komatsu et al., 2007; Kirkin et al., 2009). The involvement of ubiquitination in the turnover of organelles was established through research on a type of mitophagy: the protein kinase PTEN-Induced Putative Kinase1 accumulates on depolarized mitochondria, allowing a cytosolic E3, parkin, to ubiquitinate the surfaces of dysfunctional mitochondria (Pickles et al., 2018). The ubiquitinated mitochondria then become targets of autophagosomal transport to the lysosomes for degradation.
Ubiquitination-triggered selective autophagy also occurs in plants. Arabidopsis and tobacco (Nicotiana tabacum) orthologs of NBR1 play roles in the degradation of ubiquitinated protein aggregates similar to that of NBR1 in mammals (Svenning et al., 2011; Zientara-Rytter et al., 2011; Zhou et al., 2013). When the function of the 26S proteasome complex is impaired by the presence of inhibitors, the proteasome itself is ubiquitinated and then degraded by autophagy (Marshall et al., 2015). During this proteaphagy process, 26S proteasome Regulatory Particle Non-ATPase10 acts as a receptor through binding to both ubiquitin and ATG8. These findings reveal that plants have selective autophagy processes that recognize ubiquitinated forms of target proteins.
Plant cells are unique in containing photosynthetic organelles, the chloroplasts. Since chloroplasts accumulate considerable damage during photosynthesis and contain the majority of nutrients in green plant tissues, chloroplast turnover is critical for the management of oxidative damage and the recycling of assimilated nutrients (Ishida and Makino, 2018; Nakamura and Izumi, 2018). Our previous studies established that there are two types of autophagy that degrade chloroplasts. During leaf senescence or sugar starvation, autophagosomes transport a portion of the chloroplast stroma in a specific form enclosed in small bodies containing stromal proteins, termed Rubisco-containing bodies (RCBs; Ishida et al., 2008; Izumi et al., 2015). This RCB-mediated, piecemeal-type autophagy is important in amino acid recycling during developmental growth or adaptation to sugar starvation (Izumi et al., 2013; Ono et al., 2013; Hirota et al., 2018). By contrast, chloroplast damage due to strong visible light or ultraviolet B, termed photoinhibition, induces vacuolar transport of entire chloroplasts, in a process known as chlorophagy (Izumi et al., 2017). Chlorophagy is a selective autophagic process that eliminates specific chloroplasts that have accumulated envelope-associated damage (Nakamura et al., 2018). Since the vacuolar membrane assists in sequestering damaged chloroplasts, chlorophagy is likely a microautophagy pathway that requires some core ATG proteins including ATG5 and ATG7 (Nakamura et al., 2018).
Recent studies have also revealed that ubiquitination is involved in the degradation of chloroplast-associated proteins. SUPPRESSOR OF PPI1 LOCUS1 (SP1) is a chloroplast outer envelope-anchored E3 (Ling et al., 2012). The main targets of SP1 are the subunits of a protein import complex in the chloroplast outer envelope known as TOC (translocon at the outer envelope membrane of chloroplasts). SP1-mediated turnover of TOC proteins is mediated by the 26S proteasome, in a process termed chloroplast-associated protein degradation (CHLORAD), and is important in chloroplast development and in the control of reactive oxygen species (ROS) production during abiotic stresses (Ling et al., 2012, 2019; Ling and Jarvis, 2015). Notably, another recent study demonstrated that the plastid ferrochelatase2 (fc2) mutant can conditionally accumulate the ROS singlet oxygen (1O2) in chloroplasts, which subsequently leads to the degradation and removal of photodamaged chloroplasts (Woodson et al., 2015). A genetic suppressor screen of fc2 mutants identified the cytosolic E3 PLANT U-BOX4 (PUB4) as playing a role in this pathway. It was hypothesized that PUB4 may directly (or indirectly) ubiquitinate photodamaged chloroplasts to mark them for removal.
As mentioned above, ubiquitination acts as a signal that induces autophagic degradation as well as proteasomal breakdown; however, the interaction between autophagy and chloroplast-associated E3s has not been addressed. In this study, we focused on the requirement of chloroplast-associated E3s for the induction of chloroplast-targeting autophagy in Arabidopsis and showed that PUB4 and SP1 are dispensable for the induction of both chlorophagy and RCB-mediated autophagy. We further assessed the relationship between PUB4-related degradation and autophagy through the phenotypic analysis of single or double mutant Arabidopsis lines with mutations affecting the two pathways. During developmental growth or under starvation conditions, single mutants of autophagy genes or PUB4 showed distinct phenotypes; conversely, double mutants with mutations affecting both systems showed synergistic phenotypes. These results further support the notion that autophagy and PUB-related ubiquitination independently contribute to protein turnover to control oxidative damage and nutrient recycling.
RESULTS
PUB4 Mutation Does Not Affect the Induction of Chlorophagy
Ubiquitination acts as a direct signal to induce mitochondrion-targeting autophagy in mammals (Pickles et al., 2018). In Arabidopsis, both chlorophagy and PUB4-related ubiquitination serve to remove damaged chloroplasts during photooxidative conditions (Woodson et al., 2015; Nakamura et al., 2018). Therefore, we began our assessment of the requirement for PUB4 in the induction of chlorophagy by using a mutant allele of Arabidopsis PUB4, pub4-6 (Supplemental Fig. S1A), in which an amino acid substitution in PUB4 compromises the ubiquitination of ROS-overaccumulating chloroplasts (Woodson et al., 2015).
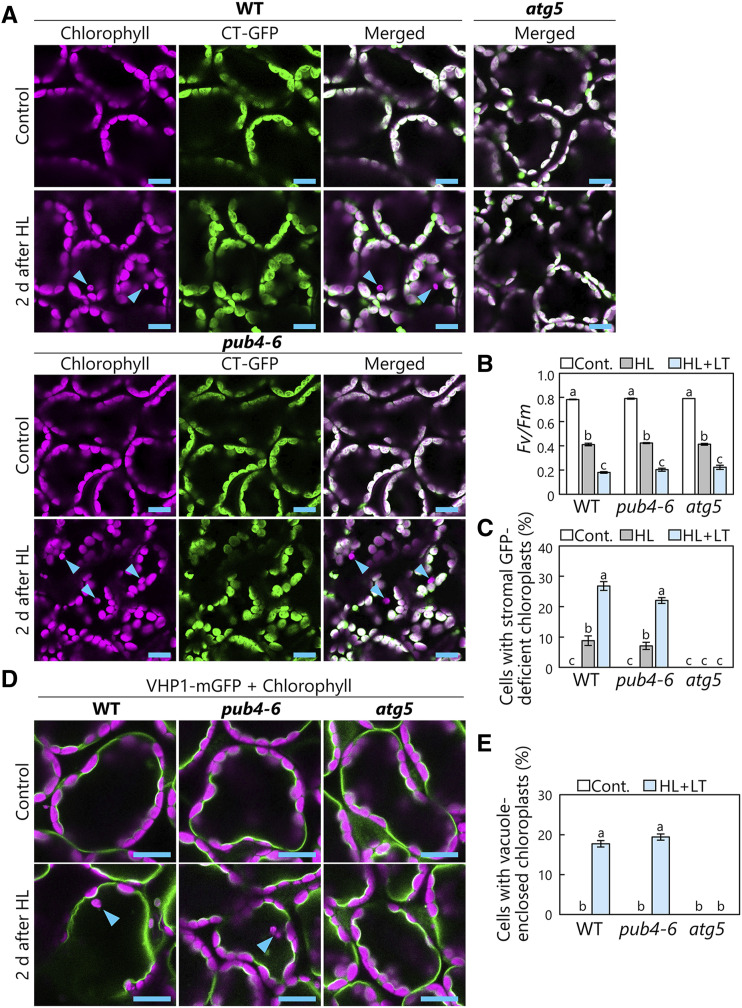
When chloroplasts are incorporated into the vacuole via chlorophagy, vacuole-enclosed chloroplasts lose their envelopes and stromal components earlier than they lose thylakoid structures and chlorophyll signals, as evidenced by numerous observations of chlorophagy via confocal microscopy or transmission electron microscopy in previous studies (Izumi et al., 2017; Nakamura and Izumi, 2018). Therefore, in leaves expressing fluorescent proteins targeted to the chloroplast stroma, vacuole-enclosed chloroplasts exhibit a chlorophyll signal without a stromal marker signal, and the number of such stromal marker-deficient chloroplasts reflects the degree of chlorophagy activity (Izumi et al., 2017; Nakamura et al., 2018; Nakamura and Izumi, 2018). Thus, we prepared wild-type, pub4-6, and atg5-1 plants expressing a GFP fused to a transit peptide of a chloroplast protein, RecA, under the control of the cauliflower mosaic virus 35S promoter (Pro-35S:CT-GFP; Köhler et al., 1997), and subjected them to treatments with 2 h of high light (HL; 2,000 µmol m−2 s−1) at either 23°C or low temperature (LT; 10°C). Chloroplasts deficient in this stromally targeted GFP fusion were visible in wild-type plants but not in atg5-1 plants (Fig. 1A, arrowheads), confirming the occurrence of core ATG-dependent chlorophagy in HL-damaged leaves. Stromal marker-deficient chloroplasts also appeared in HL-damaged pub4-6 leaves (Fig. 1A, arrowheads). Since the maximum quantum yield of PSII (Fv/Fm) decreases in response to the level of photoinhibition, the reduction of the Fv/Fm ratio in this assay (Fig. 1B) reflects the extent of HL-induced chloroplast damage. In wild-type and pub4-6 plants, similar levels of PSII damage led to similar levels of induction of stromal GFP-deficient chloroplasts (Fig. 1, B and C). Additionally, the greater declines of Fv/Fm seen at 10°C as compared with 23°C corresponded to the higher induction levels of stromal GFP-deficient chloroplasts in both wild-type and pub4-6 plants at 10°C (Fig. 1, B and C). These observations confirm the occurrence of chlorophagy in pub4-6 plants.
Figure 1.
Chlorophagy occurs in pub4-6 mutant leaves. A, Confocal images of wild-type (WT), pub4-6, and atg5 mesophyll cells expressing chloroplast stroma-targeted (CT)-GFP from either nontreated control plants or plants 2 d after HL (2,000 µmol m−2 s−1) exposure for 2 h at 10°C. Chlorophyll (magenta) and CT-GFP (green) signals are shown. In the merged images, overlapping regions of chlorophyll and GFP appear white. Only merged images are shown for atg5 plants. Arrowheads indicate CT-GFP-deficient chloroplasts. Bars = 10 µm. B, Fv/Fm ratios measured before (Cont.) and immediately after HL treatment for 2 h at 23°C (HL) or at LT (HL+LT; 10°C). Error bars indicate se (n = 4). C, Proportions of cells with stromal CT-GFP-deficient chloroplasts in a fixed region, measured 2 d after HL treatment. Error bars indicate se (n = 4). D, Confocal images of wild-type, pub4-6, and atg5 mesophyll cells expressing the tonoplast marker VHP1-mGFP from either nontreated control plants or plants 2 d after HL treatment for 2 h at 10°C. Chlorophyll (magenta) and VHP1-mGFP (green) signals are shown. Only merged images are shown. Arrowheads indicate vacuole-enclosed chloroplasts. Bars = 10 µm. E, Proportions of cells with vacuole-enclosed chloroplasts in the fixed region, measured 2 d after HL treatment for 2 h at 10°C. Error bars indicate se (n = 4). In B, C, and E, different letters denote significant differences based on Tukey’s test (P < 0.05).
To directly observe chloroplast accumulation in the vacuolar lumen, we exposed transgenic lines expressing the vacuolar membrane marker VACUOLAR H+-PYROPHOSPHATASE1 (VHP1)-monomeric GFP (ProVHP1:VHP1-mGFP; Segami et al., 2014) to HL (Fig. 1D). A confocal microscopy assay confirmed that similar amounts of chloroplasts were present inside the vacuole in wild-type and pub4-6 plants, but not in atg5-1 plants, after HL damage (Fig. 1, D and E). Additionally, we confirmed that chloroplasts accumulated in the vacuole in T-DNA insertional knockout mutants of PUB4 (pub4-1 and pub4-2; Supplemental Fig. S1, B and C; Wang et al., 2013), supporting the proposal that PUB4 itself is dispensable in the induction of chlorophagy.
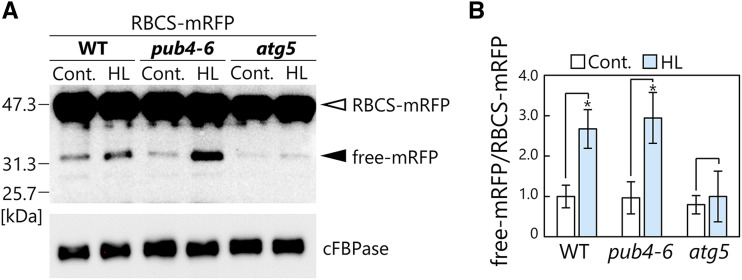
To corroborate the confocal microscopy results, we assessed the activity of chloroplast autophagy via a cleavage assay based on immunoblot detection (Fig. 2). When fluorescent protein-tagged chimeric proteins are transported into the lytic organelles through autophagy, fluorescent polypeptide tags are released and accumulate in the vacuole/lysosomes because of their tolerance for lytic degradation activity (Mizushima et al., 2010). Therefore, the biochemical detection of fluorescent tags derived from autophagic degradation of target proteins is widely used to assess the activity of various types of autophagy pathways. We subjected plants expressing the stroma-localized Rubisco small subunit (RBCS) fused to monomeric red fluorescent protein (mRFP; ProRBCS:RBCS-mRFP; Ono et al., 2013) to HL treatment; this construct was selected because RFP is more tolerant of lytic activity compared with GFP (Kimura et al., 2007). We observed faint RFP signals spread out in the vacuolar lumen, in addition to RBCS-mRFP-deficient chloroplasts, in the HL-damaged leaves of wild type and pub4-6 but not atg5 plants (Supplemental Fig. S2). Immunoblot detection of RFP in protein extracts showed that free mRFP was released in the leaves of wild-type and pub4-6 plants but only minimally in atg5 plants (Fig. 2A). Free mRFP increased in response to HL treatment in wild-type and pub4-6 plants but not in atg5 plants (Fig. 2). This biochemical evidence supports the notion that the pub4-6 mutation does not affect the activity of chlorophagy.
Figure 2.
Biochemical assay for chloroplast autophagy activity in leaves damaged by HL. A, RFP and cFBPase (loading control) detected by immunoblotting of soluble protein extracts of wild-type (WT), pub4-6, and atg5 leaves expressing RBCS-mRFP from either nontreated control plants or plants 2 d after HL (2,000 µmol m−2 s−1) exposure for 2 h at 10°C. The open arrowhead indicates RBCS-mRFP fusion, and the closed arrowhead indicates free mRFP derived from autophagic degradation of RBCS-mRFP. Confocal images of leaves in the same treatment groups are shown in Supplemental Figure S2. B, Quantification of free mRFP/RBCS-mRFP ratio shown relative to that of nontreated wild-type plants, which was set to 1. Error bars indicate se (n = 4). Asterisks denote significant differences due to HL treatment based on Student’s t test (*P < 0.05).
Piecemeal-Type Chloroplast Autophagy via RCBs during Sugar Starvation Occurs in pub4-6 Plants
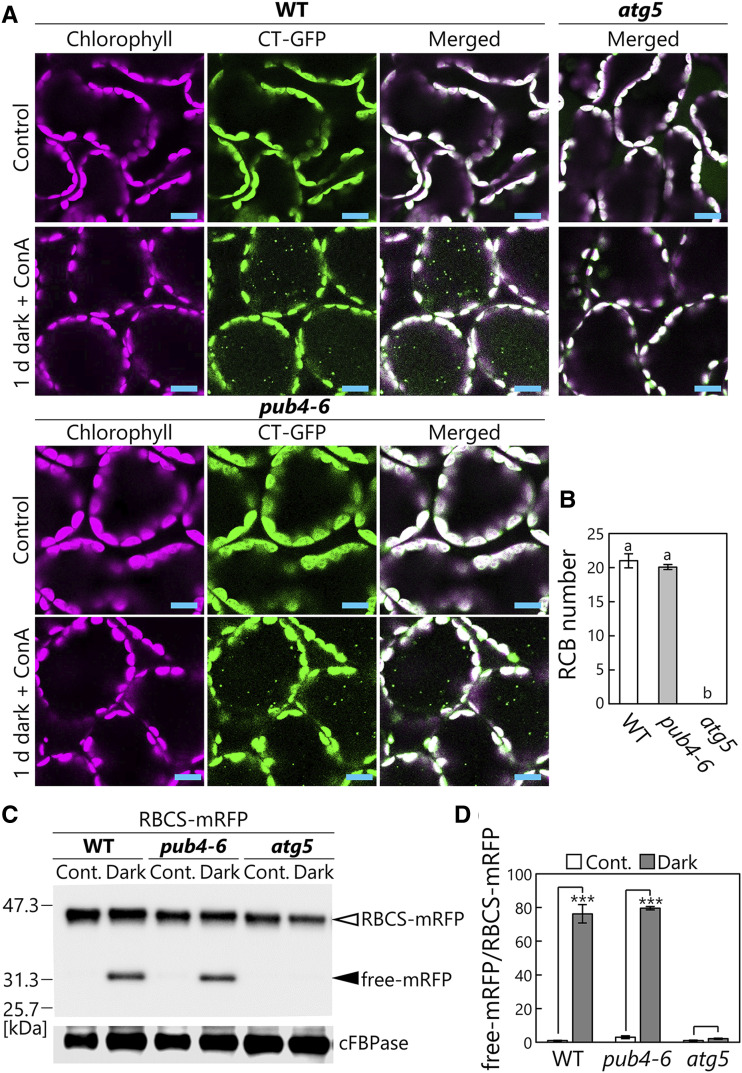
We next assessed the occurrence of another type of chloroplast-targeting autophagy, RCB-mediated autophagy, that partially transports the chloroplast stroma as a type of autophagic cargo, termed RCBs, especially in response to sugar starvation (Ishida et al., 2008; Izumi et al., 2010). To induce this pathway, we incubated detached leaves expressing Pro-35S:CT-GFP in darkness as a sugar-starvation condition. Additionally, we supplied concanamycin A, which inhibits vacuolar H+-ATPase and consequently reduces vacuolar lytic activity, to stabilize the resulting accumulation of RCBs. After 1 d of this treatment, similar amounts of RCBs appeared in wild-type and pub4-6 plants, but no RCBs were seen in atg5 mutant plants (Fig. 3, A and B). To biochemically evaluate the activity of RCB-mediated autophagy via the cleavage assay, we incubated RBCS-mRFP-expressing leaves in darkness without concanamycin A, since we postulated that RCBs are digested by lytic activity, and mRFP signals derived from lytic cleavage of RBCS-mRFP accumulate in the vacuole in a fashion similar to their accumulation after the induction of chlorophagy (Fig. 2; Supplemental Fig. S2). We observed the dispersed RFP signals in the vacuolar lumen in the leaves of wild-type and pub4-6 but not atg5 plants after the 2-d incubation in darkness without concanamycin A (Supplemental Fig. S3). Consistent with this observation, a cleavage assay with anti-RFP antibody in dark-treated leaves revealed the autophagy-dependent release of similar amounts of free mRFP in wild-type and pub4-6 plants (Fig. 3, C and D). These results indicate that comparable piecemeal chloroplast autophagy activity occurs in wild-type and pub4-6 plants. Thus, we concluded that PUB4 function is not required for the induction of both full organelle-type and partial-type chloroplast-targeting autophagy.
Figure 3.
Comparable activity of piecemeal-type chloroplast autophagy between wild-type (WT) and pub4-6 leaves. A, Confocal images of wild-type, pub4-6, and atg5 mesophyll cells expressing CT-GFP from either nontreated control leaves or leaves incubated in darkness for 1 d with concanamycin A (ConA) added. Chlorophyll (magenta) and CT-GFP (green) signals are shown. In the merged images, overlapping regions of chlorophyll and GFP appear white. Only merged images are shown for atg5 plants. Small GFP dots represent RCBs in the vacuole. Bars = 10 µm. B, Number of RCBs in a fixed area after 1 d of incubation in darkness with concanamycin A. Error bars indicate se (n = 3). Different letters denote significant differences based on Tukey’s test (P < 0.05). C, RFP and cFBPase (loading control) detected by immunoblotting of soluble protein extracts from wild-type, pub4-6, and atg5 leaves expressing RBCS-mRFP. Proteins extracted from either nontreated control leaves (Cont.) or leaves after 2 d of incubation in darkness were used. The open arrowhead indicates RBCS-mRFP fusion, and the closed arrowhead indicates free mRFP derived from autophagic degradation of RBCS-mRFP. The confocal images for leaves in the same treatment groups are shown in Supplemental Figure S3. D, Quantification of free mRFP/RBCS-mRFP ratio shown relative to that of nontreated wild-type leaves, which was set to 1. Error bars indicate se (n = 3). Asterisks denote significant differences due to the dark treatment based on Student’s t test (***P < 0.001).
SP1 Is Also Dispensable for the Induction of Chlorophagy and RCB-Mediated Autophagy
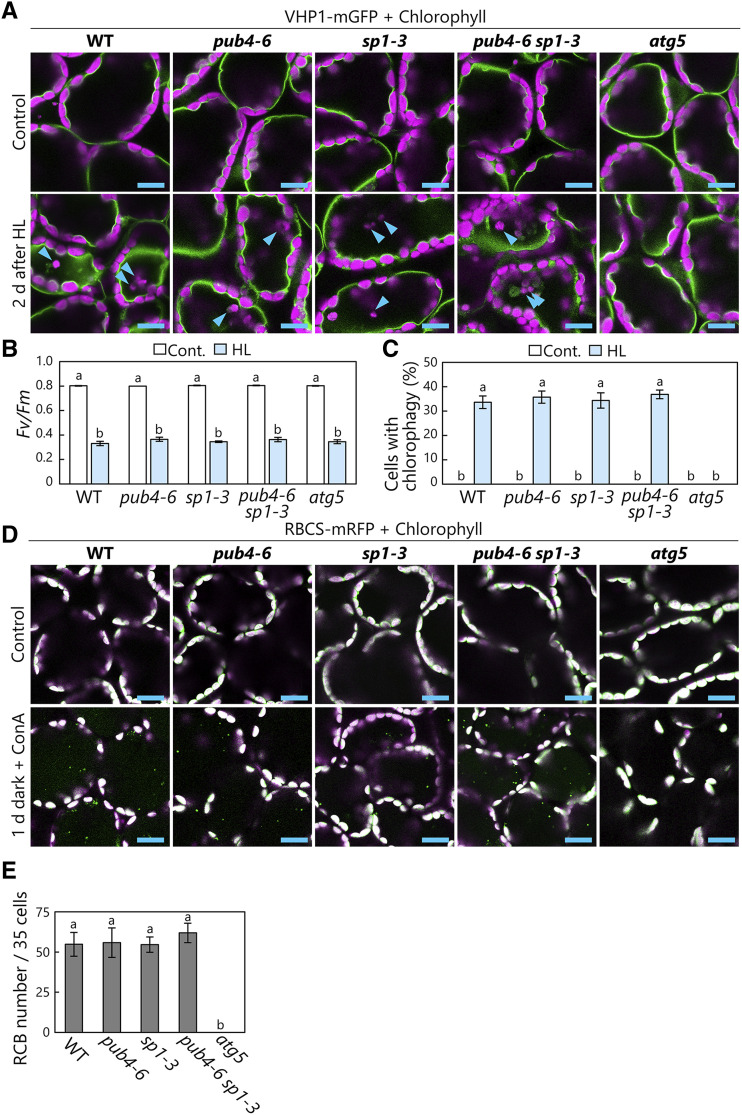
We next investigated the involvement of SP1, a chloroplast envelope-anchored E3 that controls the degradation of TOC proteins (Ling et al., 2012), in the induction of the two main types of chloroplast autophagy. After constructing transgenic lines expressing VHP1-mGFP in the wild-type and sp1-3 mutant backgrounds, we observed that vacuole-enclosed chloroplasts had a similar appearance in both backgrounds (Fig. 4A). HL treatment caused similar PSII damage in the transgenic wild-type and sp1-3 plants, and their levels of chlorophagy induction were also comparable (Fig. 4B). Upon subjecting further transgenic lines expressing stromal RBCS-mRFP in the wild-type and sp1-3 backgrounds to sugar starvation, we observed similar numbers of RCBs accumulating in each genotype (Fig. 4, D and E). These results revealed that chloroplast autophagy can occur without SP1 function.
Figure 4.
SP1 is dispensable for the induction of chloroplast autophagy. A, Confocal images of wild-type (WT), pub4-6, sp1-3, pub4-6 sp1-3, and atg5 mesophyll cells expressing the tonoplast marker VHP1-mGFP from either nontreated control leaves or leaves 2 d after HL (2,000 µmol m-2s−1) treatment for 2 h at 10°C. Arrowheads indicate vacuole-enclosed chloroplasts. Chlorophyll (magenta) and VHP1-mGFP (green) signals are shown. Only merged images are shown. B, Fv/Fm ratios measured before (Cont.) and immediately after HL treatment. Error bars indicate se (n = 6). C, Proportions of cells with vacuole-enclosed chloroplasts in a fixed region, measured 2 d after HL treatment. Error bars indicate se (n = 4). D, Confocal images of wild-type, pub4-6, sp1-3, pub4-6 sp1-3, and atg5 mesophyll cells expressing stromal RBCS-mRFP from either nontreated control leaves or leaves incubated in darkness for 1 d with concanamycin A (ConA) added. Chlorophyll (magenta) and RBCS-mRFP (green) signals are shown. In the merged images, overlapping regions of chlorophyll and RFP appear white. Only merged images are shown. Small RFP dots represent RCBs in the vacuole. E, Number of RCBs in a fixed number of cells (35) after 1 d of incubation in darkness with concanamycin A. Error bars indicate se (n = 6). In B, C, and E, different letters denote significant differences based on Tukey’s test (P < 0.05). Bars = 10 µm (A and D).
Here, to examine whether there is any potential redundancy between PUB4 and SP1 in the induction of chloroplast autophagy, we generated pub4-6 sp1-3 double mutants and additionally checked the chlorophagy activity and RCB production (Fig. 4). Chlorophagy after HL damage (Fig. 4A) and RCB production during sugar starvation (Fig. 4D) occurred even in pub4-6 sp1-3 double mutants. The induction levels of both autophagy pathways were the same among wild-type, pub4-6, sp1-3, and pub4-6 sp1-3 plants (Fig. 4, C and E), indicating that PUB4 and SP1 do not have redundant roles in chloroplast autophagy. Thus, it is unlikely that these previously identified chloroplast-associated E3s act as triggers of chloroplast autophagy.
Disrupted ROS Management and Enhanced Chlorosis in the Double Mutants of Autophagy and PUB4
An independent role of SP1, the turnover of chloroplast outer envelope-anchored translocon proteins by the 26S proteasome termed CLORAD, was well established (Ling et al., 2012, 2019; Ling and Jarvis, 2015). By contrast, previous studies have proposed that both autophagy proteins and PUB4 contribute to the removal of damaged chloroplasts under photooxidative stress (Woodson et al., 2015; Nakamura et al., 2018). Our data indicate that the two systems act independently (Figs. 1–3). Therefore, to further define their relationship genetically, we generated double mutants of PUB4 and two autophagy genes: pub4-6 atg5-1 and pub4-6 atg7-2.
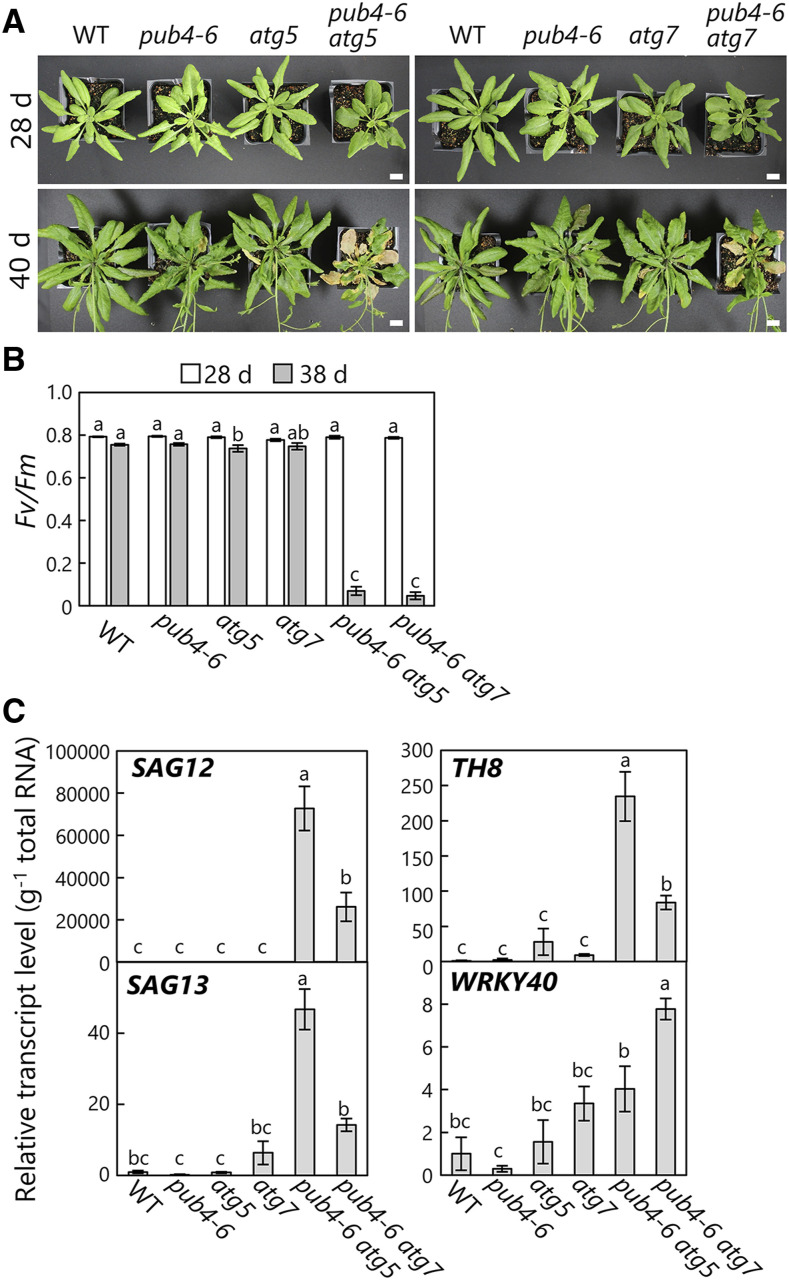
Interestingly, the double mutants had more extreme phenotypes than the respective single mutants. At 28 d after sowing, the plants did not exhibit clear phenotypic differences between genotypes (Fig. 5A, top), but after 40 d, pub4-6 atg5-1 or pub4-6 atg7-2 plants showed accelerated chlorosis of rosette leaves (Fig. 5A, bottom). This visible chlorosis was consistent with the reduction of leaf photosynthetic capacity: the Fv/Fm ratio was lower in the leaves of the double mutants than in the wild type and the respective single mutants at 38 d after sowing (Fig. 5B). We further evaluated the transcript levels of senescence-associated genes (SAGs) and observed that the transcript abundances of SAG12 and SAG13 were elevated in the leaves of 37-d-old pub4-6 atg5-1 and pub4-6 atg7-2 plants relative to the respective single mutants (Fig. 5C, left). These results indicate an acceleration of leaf senescence symptoms in the double mutants. Numerous studies have demonstrated that atg single mutants display early-senescence phenotypes compared with wild-type plants (Hanaoka et al., 2002; Yoshimoto et al., 2009; Guiboileau et al., 2012); therefore, our data show that the pub4-6 mutation synergistically enhances such phenotypes.
Figure 5.
Accelerated leaf senescence in pub4-6 atg double mutant plants. A, Photographs of 28-d-old or 40-d-old wild-type (WT), pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants. Bars = 10 mm. B, Fv/Fm ratios measured in leaves of 28- or 38-d-old plants. Error bars indicate se (n = 4). C, Transcript levels of the senescence-associated genes SAG12 and SAG13 and the ROS-responsive genes TH8 and WRKY40 in leaves of 35-d-old plants. The respective transcript levels are normalized to the level of 18S rRNA and are shown relative to the wild-type expression, which was set to 1. Error bars indicate se (n = 3). In B and C, different letters denote significant differences based on Tukey’s test (P < 0.05).
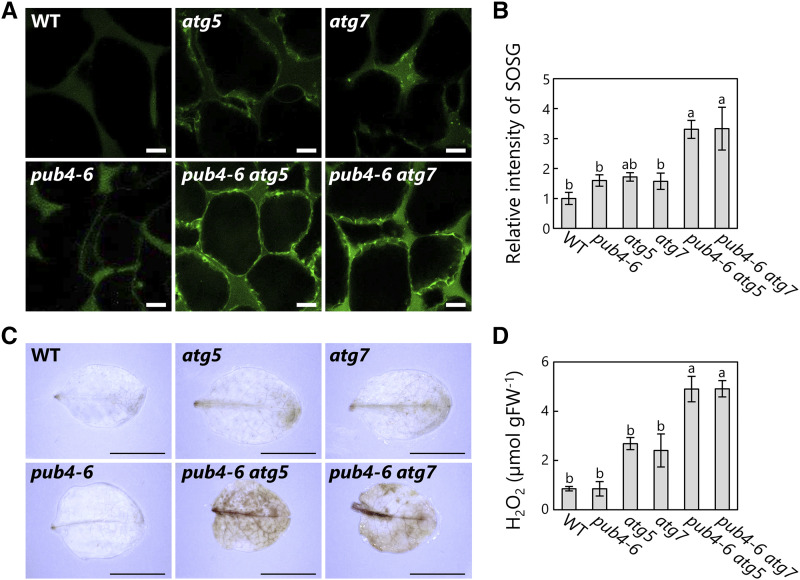
We additionally checked the transcript levels of marker genes of ROS accumulation and identified elevated expression of two ROS-responsive genes, THIOREDOXIN H-TYPE8 (TH8) and WRKY40 (Laloi et al., 2007; Shao et al., 2013), in the double mutants (Fig. 5C, right). Accordingly, we directly assessed the accumulation of ROS in senescent rosette leaves. 1O2 staining with the fluorescent probe singlet oxygen sensor green (SOSG) showed greater 1O2 production in the double mutants than in the respective single mutants (Fig. 6, A and B). Furthermore, the leaves of the double mutants displayed a stronger signal for 3,3′-diaminobenzidine, which stains hydrogen peroxide (H2O2; Fig. 6C). Consistent with this cytological observation, quantification of the H2O2 contents of homogenized leaf lysates showed elevated H2O2 in senescing leaves of the double mutants (Fig. 6D). These results reveal increased accumulation of ROS in the pub4-6 atg double mutants during senescence, associated with the early-chlorosis phenotype.
Figure 6.
Enhanced ROS production in the leaves of pub4-6 atg double mutant plants. A, Confocal images of 1O2 stained by SOSG in mesophyll cells from 35-d-old wild-type (WT), pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants. Bars = 10 µm. B, Relative intensities of SOSG in a fixed area of mesophyll cells relative to the wild-type intensity, which was set to 1. Error bars indicate se (n = 4). C, H2O2 staining by 3,3′-diaminobenzidine in leaves from 35-d-old plants. Bars = 10 mm. D, Amounts of H2O2 in leaf extracts from 35-d-old plants, in micromoles per gram fresh weight (FW). Error bars indicate se (n = 3). In B and D, different letters denote significant differences based on Tukey’s test (P < 0.05).
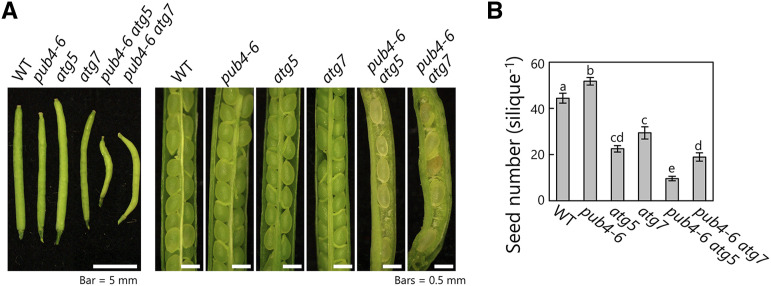
Through phenotypic analysis, we further observed that the pub4-6 atg double mutants had abnormal siliques containing defective seeds (Fig. 7A). We therefore counted the number of normal seeds per silique and observed that the number was lower in the atg single mutants than in the wild type and even lower in the double mutant (Fig. 7B). Thus, the synergistic effects of the autophagy deficiency and PUB4 mutations visibly influenced seed production, which is a critical event in the final stages of the plant life cycle.
Figure 7.
Impaired seed production in pub4-6 atg double mutant plants. A, Photographs of siliques from main stems in wild-type (WT), pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants. Bars = 5 mm (left) and 0.5 mm (right). B, Number of normal mature seeds per silique of main stem. Error bars indicate se (n = 17–19). Different letters denote significant differences based on Tukey’s test (P < 0.05).
Effects of Autophagy Deficiency and the pub4-6 Mutation on the Accumulation of Ubiquitinated Proteins
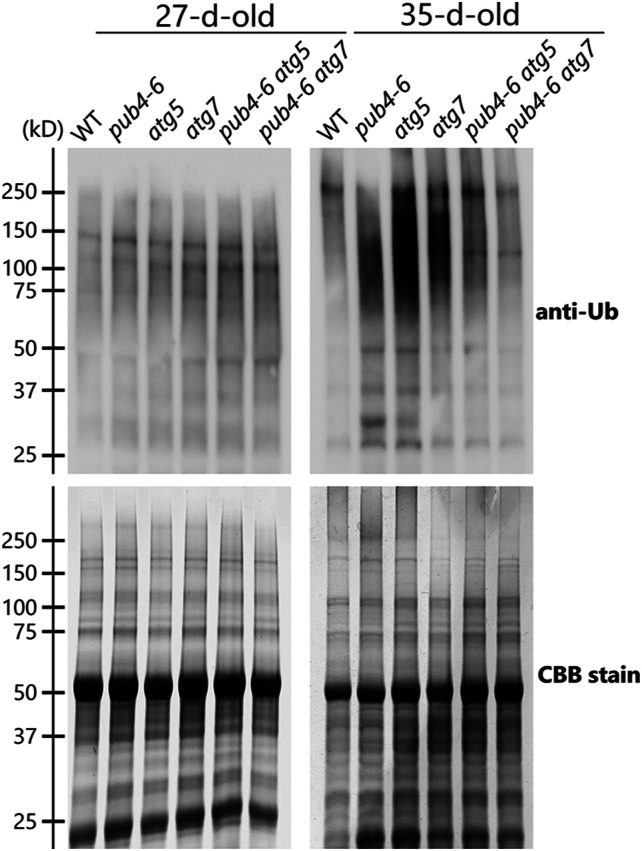
Since PUB4 is an E3 ubiquitin ligase, we evaluated the effects of pub4-6 mutation (either alone or in combination with the autophagy mutations) with regard to the appearance of ubiquitinated proteins through immunoblot analysis of total leaf protein extracts (Fig. 8). In the leaves of 27-d-old plants, the ubiquitination state of total proteins did not show clear differences among the analyzed genotypes (Fig. 8, left). By contrast, in the leaves of 35-d-old plants, smear bands representing polyubiquitinated proteins were more prominent in the total protein extracts of atg5 or atg7 single mutants than in those of the wild type (Fig. 8, right), indicating that active autophagy helps prevent the accumulation of ubiquitinated proteins. In the presence of the pub4-6 mutation, decreased levels of polyubiquitinated proteins were observed in both the wild-type and atg backgrounds (Fig. 8, right); thus, PUB4-related ubiquitination also functions in senescing leaves. These results reveal the contributions of autophagy and PUB4-related ubiquitination to protein degradation in senescing leaves.
Figure 8.
Accumulation of ubiquitinated proteins in response to leaf senescence. Ubiquitinated proteins were detected by immunoblotting of total protein extracts from leaves in 27- or 35-d-old wild-type (WT), pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants (top). Total proteins were stained with Coomassie Brilliant Blue (CBB) as a loading control (bottom). Representative results from one of three independent experiments are shown.
Deficiency of Autophagy and PUB4 Enhances Susceptibility to Starvation
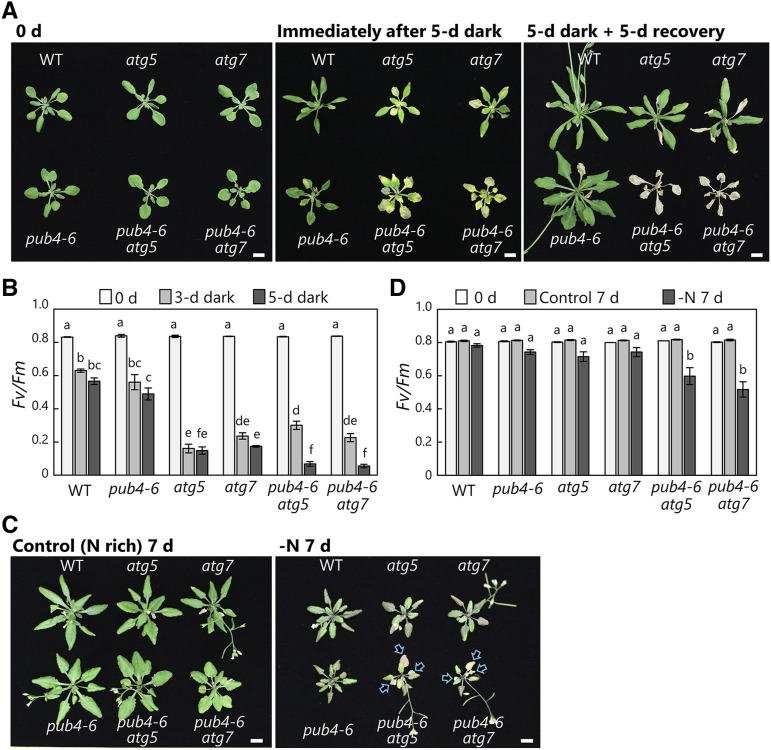
Plants must reutilize previously assimilated nutrients to survive starvation conditions; thus, such conditions emphasize the physiological importance of intracellular protein degradation systems. In fact, it is widely recognized that the growth defects and cell death phenotypes of atg mutants are enhanced under nitrogen- or carbon-starvation conditions (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Hirota et al., 2018). Therefore, we subjected the autophagy gene and pub4-6 double mutant plants, along with the wild type and the respective single mutants, to nitrogen or carbon starvation (Fig. 9).
Figure 9.
Enhanced susceptibility to starvation in pub4-6 atg double mutant plants. A, Wild-type (WT), pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants grown for 20 d (0 d; before treatment) under long-day conditions (14 h of light/10 h of dark), subjected to an extended dark treatment for 5 d (Immediately after 5-d dark), and allowed to recover under the long-day condition for 5 d (5-d dark + 5-d recovery). B, Fv/Fm ratios measured in leaves from 20-d-old plants (0 d of treatment) and then from plants after 3 and 5 d of dark treatment. Error bars indicate se (n = 3–4). C, Wild type, pub4-6, atg5, atg7, pub4-6 atg5, and pub4-6 atg7 plants grown for 20 d (0 d; before treatment) in nutrient-rich solution and then for a further 7 d either in the same nutrient-rich solution (Control) or in nitrogen-deficient solution (−N). Arrows indicate dead leaves by chlorosis. D, Fv/Fm ratios measured in leaves from 20-d-old plants (0 d of the treatment) and from plants after 7 d of growth in the control or −N conditions. Error bars indicate se (n = 4). In B and D, different letters denote significant differences based on Tukey’s test (P < 0.05). Bars = 10 mm (A and C).
When the plants were exposed to 5 d of darkness to induce sugar starvation, leaf chlorosis appeared in the atg5 or atg7 single mutants and pub4-6 atg double mutants (Fig. 9A, center). After this treatment, the atg single mutants started to grow again, whereas the double mutants died (Fig. 9A, right). This susceptibility was consistent with declines in photosynthetic capacity in the various genotypes: the Fv/Fm ratio was lower in atg mutant and pub4-6 atg double mutant leaves than in wild-type and pub4-6 plants after 3 d of darkness, and the double mutants showed further reductions after 5 d of darkness (Fig. 9B).
We further exposed the plants to nitrogen-deficiency treatments (Fig. 9, C and D). The double mutants showed enhanced leaf chlorosis phenotypes in the nitrogen-deficient condition (Fig. 9C, arrows), which is consistent with the greater declines of Fv/Fm seen in the double mutant leaves after 7 d of growth in nitrogen-deficient solution (Fig. 9D). The pub4-6 atg double mutants consistently displayed enhanced phenotypes relative to the respective single mutants during natural senescence (Figs. 5–7) and under carbon or nitrogen starvation (Fig. 9). These results suggest a parallel cooperation of autophagy and PUB4 in protein degradation in response to senescence or starvation.
DISCUSSION
Ubiquitination acts as a signal inducing various types of selective autophagy. Recent studies have revealed the contributions of autophagy and ubiquitination to chloroplast turnover (Woodson et al., 2015; Izumi et al., 2017; Nakamura et al., 2018; Nakamura and Izumi, 2018). Thus, we assessed the involvement of chloroplast-associated E3s in the induction of chloroplast-targeting autophagy. Our microscopy observations, biochemical assays of autophagy flux, and phenotypic analyses of the mutants all indicated that autophagy does not depend on the chloroplast-associated E3s PUB4 and SP1. Genetic analyses further suggested that autophagic processes and those mediated by PUB4 act in parallel.
Relationship between PUBs and Autophagy
PUBs, U-box-type E3s in plants, constitute a large gene family—PUB1 to PUB64 were annotated in the Arabidopsis genome (Azevedo et al., 2001)—and are involved in multiple processes (Trujillo, 2018). Interestingly, the pub4-6 mutant allele leads to a single amino acid substitution (Gly-255 to Arg) and a semidominant phenotype. This suggests that the pub4-6 allele may have a dominant-negative effect on closely related members of the PUB family (Woodson et al., 2015), although this has not yet been tested directly. This study clearly indicates that chlorophagy occurs normally in pub4-6 mutant plants; therefore, the function of PUB4 is not directly linked to the induction of chlorophagy (Figs. 1–3). Instead, PUB4 may be involved in a parallel, but distinct, pathway of chloroplast degradation and removal. Unlike chlorophagy, the damaged chloroplasts removed via the PUB4 pathway become degraded in the cytoplasm before interacting with the tonoplast. Furthermore, these chloroplasts do not retain recognizable thylakoid structures or chlorophyll once transported into the central vacuole (Woodson et al., 2015). The identification of direct ubiquitination targets of PUB4 may help to further elucidate the precise mechanism of PUB4-mediated chloroplast degradation in response to oxidative damage.
Autophagy plays an important role in the degradation of cytoplasmic ubiquitinated protein aggregates. In mammals, the autophagic receptors p62/SQSTM1 and NBR1 bind ubiquitinated proteins to transport them to the lysosomes as autophagosomal cargos (Komatsu et al., 2007; Kirkin et al., 2009). Plant orthologs of NBR1 (NBR1 in Arabidopsis and Joka2 in tobacco) serve similar functions (Svenning et al., 2011; Zientara-Rytter et al., 2011; Zhou et al., 2013). The enhanced accumulation of ubiquitinated proteins has been observed in Arabidopsis atg mutant lines and nbr1 mutants during senescence or abiotic stresses (Zhou et al., 2013; Munch et al., 2014). In this study, we observed the hyperaccumulation of ubiquitinated proteins in senescent leaves of atg5 or atg7 mutants as compared with the wild type (Fig. 8). By contrast, the pub4-6 mutation reduced the level of ubiquitinated proteins (Fig. 8). If PUB4-mediated ubiquitination is required to induce the autophagic degradation of ubiquitinated proteins during leaf senescence, atg single mutants and pub4-6 atg double mutants should show similar phenotypes. However, we observed enhanced phenotypic severity in the double mutants, including accelerated chlorosis and an increase in ROS contents in senescent leaves (Figs. 5 and 6). In addition, pub4-6 single mutants did not show similar phenotypes to atg single mutants (Figs. 4–9; Woodson et al., 2015). These results support the notion that autophagy and PUB4 cooperate to carry out protein degradation in parallel, and thus the impairment of both systems in the double mutants leads to synergistic phenotypes.
The remobilization of nitrogen or carbon sources from vegetative tissues to seeds is impaired in atg mutant plants (Guiboileau et al., 2012; Barros et al., 2017), resulting in reduced seed production. Although the pub4-6 single mutation does not by itself affect seed production, the introduction of the pub4-6 mutation into atg mutant backgrounds further reduced seed production as compared with the atg single mutants (Fig. 7). These results suggest that autophagy is the major pathway supporting nutrient recycling and that the additional defects brought about by the loss of PUB4 activity in atg plants is due to aggravation of the autophagy phenotypes.
Relationship between SP1 and Autophagy
Previous studies have demonstrated that the major TOC subunits TOC33, TOC75, and TOC159 are the specific targets of the chloroplast outer envelope-anchored E3 SP1 (Ling et al., 2012). A recent study further identified additional players in the extraction of polyubiquitinated TOC proteins to the cytosol: the chloroplast outer envelope channel protein SP2 and cytoplasmic AAA+ chaperone (CELL DIVISION CYCLE48) form complexes with SP1 to mediate the release of polyubiquitinated TOCs and their digestion by 26S proteasomes (Ling et al., 2019). This system, termed CHLORAD, is important for the regulation of protein import via TOC complexes in response to plastid biogenesis or abiotic stresses (Ling and Jarvis, 2015; Ling et al., 2019). Accumulating evidence has established an essential role for SP1 in the degradation of TOCs by cytoplasmic 26S proteasomes. However, this study does not support a direct connection between SP1 and autophagy-mediated degradation.
Previous studies showed the importance of autophagy in the response to abiotic stresses such as drought or salt stress (Avin-Wittenberg, 2019). Since sp1 mutants also show susceptibility to salt or osmotic stresses (Ling and Jarvis, 2015), we would expect that cooperation between the autophagy and CHLORAD systems helps plants adapt to such stress conditions. The generation and analysis of autophagy and SP1 double mutants could provide evidence for such cooperation in Arabidopsis plants.
Mechanisms for Chloroplast Autophagy
The molecular mechanism conferring selectivity in chloroplast autophagy remains largely unknown. Chlorophagy selectively removes membrane-damaged chloroplasts (Nakamura et al., 2018). Although this study does not show that ubiquitination is involved in the recognition of damaged chloroplasts (Figs. 1, 2, and 4), unidentified E3s might function in chlorophagy. In budding yeast, the anchoring of the transmembrane protein ATG32 into the mitochondrial outer envelope is an initiation signal for mitophagy (Kanki et al., 2009; Okamoto et al., 2009). The phosphorylation of ATG32 allows its interaction with ATG11, inducing the recognition of ATG32 by ATG8 for autophagosomal sequestering (Kanki et al., 2013; Furukawa et al., 2018). Therefore, the accumulation of specific proteins on the chloroplast outer envelope and their interaction with ATG8 might act as an induction signal for chlorophagy. This study also showed that some chloroplast-associated E3s are dispensable for the induction of piecemeal-type chloroplast autophagy that delivers a portion of stroma in response to sugar starvation (Figs. 3 and 4). During this process, Endosomal Sorting Complexes Required for Transport III proteins mediate the sorting of autophagosomes containing chloroplast stroma to the vacuole (Spitzer et al., 2015). However, how autophagosomes interact with the chloroplast envelope to degrade a portion of chloroplasts also remains uncertain.
Here, we conclude that previously identified chloroplast-related E3s are not required for the induction of chloroplast-targeting autophagy. Our data also indicate that autophagy and ubiquitination independently support plant growth and stress adaptation. Plants have multiple systems for protein degradation; for example, previous studies have revealed that diverse intrachloroplastic proteases and autophagy-independent vesicular systems contribute to chloroplast protein degradation in addition to autophagy- and UPS-dependent systems (Martínez et al., 2008; Ling et al., 2012; Wang and Blumwald, 2014; Nishimura et al., 2016; Izumi and Nakamura, 2018). The existence of this multiplicity of complex systems for organelle and protein turnover may provide a means for plants to optimally orchestrate chloroplast turnover to ensure their health throughout the life cycle and overcome periods of nutrient insufficiency.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for all experiments. Plants were grown in soil or hydroponic solution in chambers at 23°C under a 14-h-light/10-h-dark photoperiod using fluorescent lamps or light-emitting diode lamps (120 µmol m−2 s−1). The Arabidopsis T-DNA insertion lines of atg5 (atg5-1; SAIL_129_B07), atg7 (atg7-2; GABI_655B06), sp1 (sp1-3; SALK_002099), and pub4 (pub4-1; SALK_108269 and pub4-2; SALK_054373) were described previously (Thompson et al., 2005; Hofius et al., 2009; Ling et al., 2012; Wang et al., 2013). The Arabidopsis PUB4 point mutation line (pub4-6) was described previously (Woodson et al., 2015). Transgenic Arabidopsis expressing chloroplast stroma-targeted GFP driven by the cauliflower mosaic virus 35S promoter (Pro-35S:CT-GFP), VHP1-mGFP driven by the VHP1 promoter (ProVHP1:VHP1-mGFP), δTIP-YFP driven by the δTIP promoter (ProδTIP:δTIP-YFP), and RBCS2B-mRFP driven by the RBCS2B promoter (ProRBCS:RBCS-mRFP) were described previously (Köhler et al., 1997; Ono et al., 2013; Segami et al., 2014; Nakamura et al., 2018).
Light Treatment
The HL exposure for 2 h was provided by a xenon light source (MAX303; Asahi Spectra) equipped with a mirror module (MAX-VIS; Asahi Spectra) to extract visible light (wavelengths between 385 and 740 nm) and a rod lens (MDRLQ1B; Asahi Spectra) to emit light with uniform intensity in a chamber at 23°C or 10°C. Plants were subjected to the light exposure at 14 d after sowing, and the second rosette leaves were observed 2 d later by confocal microscopy. After the HL treatments, plants were cultivated in the indicated growth conditions until microscopic observation.
Starvation Treatment
For the sugar-starvation treatment of detached leaves, second rosette leaves of 21-d-old plants were excised and incubated in MES-NaOH (pH 5.5) buffer for 2 d in darkness. For the treatment with concanamycin A, which is an inhibitor of vacuolar H+-ATPase, MES-NaOH (pH 5.5) containing 1 µm concanamycin A was infiltrated into the leaves with a 1-mL syringe and then the leaves were incubated in darkness for 1 d. Concanamycin A inhibits the degradation of RCBs in the vacuolar lumen, resulting in the vacuolar accumulation of RCBs (Ishida et al., 2008). For the sugar-starvation treatment of whole plants, 20-d-old plants in soil were put in a growth chamber without light supply at 23°C for the indicated days.
For the nitrogen-starvation treatment of whole plants, plants were hydroponically grown in rock-wool blocks containing nutrient-rich solution as previously described (Izumi et al., 2010), with slight modification. The rock-wool blocks with 20-d-old plants were washed with distilled water and then soaked with either nitrogen-deficient solution for −N treatment or nutrient-rich solution as a control. The nutrient-rich solution contained 4 mm KNO3, 2.5 mm potassium phosphate buffer (pH 5.5), 2 mm Ca(NO3)2, 2 mm MgSO4, 50 µm Fe-EDTA, 70 µm H3BO4, 14 µm MnCl2, 0.2 µm Na2MoO4, 10 µm NaCl, 0.5 µm CuSo4, 1 µm ZnSO4, and 0.01 µm CoCl2. The nitrogen-deficient solution was prepared by replacing KNO3 and Ca(NO3)2 with KCl and CaCl2, respectively.
Measurement of the Quantum Yield of PSII
The Fv/Fm was calculated using a pulse-modulated fluorometer (FluorCam FC 800; Photon Systems Instruments) at room temperature. Before measurement of fluorescence emission, the plants were incubated in darkness for 30 min. Then, initial (minimum) and subsequent maximum PSII fluorescence in the dark-adapted state were measured with a measuring light and a saturating pulse.
Imaging Analysis with Laser Scanning Confocal Microscopy
Laser scanning confocal microscopy (LSCM) was performed with an inverted Carl Zeiss LSM800 system equipped with a C-Apochromat LD63× water-immersion objective lens (numerical aperture, 1.15; Carl Zeiss) or an inverted Nikon C2 system equipped with a CFI Apochromat LWD λS 40×C WI (numerical aperture, 1.15). For quantitative evaluation of the frequency of chlorophagy, four different regions (204 × 204 × 40 μm each) per plant were monitored through LSCM by adjusting the focus to calculate the proportion of cells with stromal GFP-deficient chloroplasts or vacuole-enclosed chloroplasts.
For quantitative evaluation of RCB number, images of four different areas (204 × 204 μm each) per plant were obtained by LSCM and the number of RCBs was counted. For quantitative evaluation of vacuolar RFP intensity, the RFP intensity of four different vacuolar areas (25 × 25 μm) per plant was measured. The intensity of vacuolar RFP in nontreated control leaves of wild-type plants was set to 1.
Immunoblotting
Immunoblotting was performed as previously described (Ishida et al., 2008; Izumi et al., 2015), with slight modification. For the cleavage assay of RBCS-mRFP, leaves were homogenized in HEPES-NaOH (pH 7.5) containing 14 mm 2-mercaptoethanol, 10% (v/v) glycerol, and protease inhibitor cocktail (Roche) and then centrifuged at 10,000g for 10 min. The supernatants (soluble protein extract) were mixed with an equal volume of SDS sample buffer consisting of 200 mm Tris-HCl (pH 8.5), 2% (w/v) SDS, 0.7 m 2-mercaptoethanol, and 20% (v/v) glycerol and incubated for 5 min at 95°C. For the detection of ubiquitinated proteins, leaves were homogenized in HEPES-NaOH (pH 7.5) containing 14 mm 2-mercaptoethanol, 10% (v/v) glycerol, 2% (w/v) SDS, and protease inhibitor cocktail (Roche) and then centrifuged at 10,000g for 5 min. The supernatants were incubated for 5 min at 95°C as total protein extract. Equal amounts of protein were analyzed by SDS-PAGE using TGX FastCast acrylamide gels (Bio-Rad). The protein amounts were quantified by Lowry assay (Bio-Rad). Immunoblotting was performed with an anti-RFP 1G9 clone antibody (MBL), an anti-cFBPase antibody (Agrisera), or an anti-ubiquitin P4D1 clone antibody (Cytoskeleton). Goat anti-mouse IgG (H+L) secondary antibody DyLight 800 4X PEG (Invitrogen), goat anti-rabbit IgG (H+L) secondary antibody DyLight 800 4X PEG (Invitrogen), or goat anti-mouse HRP secondary antibody (Dako) was used as secondary antibody. Their fluorescence signals, or chemiluminescence signals developed with SuperSignal West Dura Extended Duration Substrate (Pierce), were detected using a ChemiDoc MP system (Bio-Rad).
Reverse Transcription Quantitative PCR
Total RNA was isolated from rosette leaves using the RNeasy kit (Qiagen) and used for cDNA synthesis with the PrimeScript RT reagent kit with gDNA eraser (Takara). An aliquot of the synthesized cDNA derived from an equal amount of total RNA was subjected to reverse transcription quantitative PCR analysis using the KAPA SYBR FAST qPCR Kit (KAPA Bio-systems) using a real-time PCR system (CFX96; Bio-Rad). The level of 18S rRNA was measured as an internal control (Izumi et al., 2012). The gene-specific primers were described previously (Laloi et al., 2007; Carbonell-Bejerano et al., 2010; Shao et al., 2013; Supplemental Fig. S4).
Detection of H2O2 and 1O2
H2O2 staining was performed as previously described (Orozco-Cardenas and Ryan, 1999), with slight modification. Leaves were infiltrated with an H2O2 staining solution consisting of 1 mg mL−1 3,3′-diaminobenzidine (pH 3.8) using a 50-mL syringe and incubated for 8 h in darkness with gentle shaking. The samples were then boiled in bleaching solution consisting of 60% (v/v) ethanol, 20% (v/v) acetic acid, and 20% (v/v) glycerol for 10 min. The quantification of H2O2 amount in leaf lysates was performed as previously described (Chakraborty et al., 2016) by using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen) and the plate reader SPARK (Tecan).
1O2 staining was performed using SOSG (Invitrogen). Leaves were infiltrated with 100 µm SOSG using a 50-mL syringe and incubated for 10 min in darkness. The leaves were washed briefly by distilled water, and their mesophyll cells were observed by confocal microscopy. For quantitative evaluation of SOSG intensity, the intensity of SOSG signal in a region of fixed size (204 × 204 μm) for each plant was measured. The intensity of the SOSG signal in nontreated control leaves of wild-type plants was set to 1.
Statistical Analysis
For statistical analysis, Student’s t test was used for the comparison of paired samples, and Tukey’s test or Dunnett’s test was used for the comparison of multiple samples, as indicated in the figure legends.
Accession Numbers
Genetic information from this article can be found in the Arabidopsis Information Resource under the following accession numbers: ATG5, AT5G17290; ATG7, AT5G45900; PUB4, AT2G23140; SP1, AT1G63900; SAG12, AT5G45890; SAG13, AT2G29350; TH8, AT1G69880; and WRKY40, AT1G80840.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chlorophagy occurs in T-DNA insertional knockout mutants of PUB4.
Supplemental Figure S2. Vacuolar accumulation of chloroplast marker in leaves damaged by HL.
Supplemental Figure S3. Vacuolar accumulation of chloroplast marker in sugar-starved leaves.
Supplemental Figure S4. Primer sequences used in this study.
Acknowledgments
We thank Dr. Kohki Yoshimoto and Dr. Yoshinori Ohsumi for the use of atg mutant plants, Dr. Maureen R. Hanson for the use of Pro35S:CT-GFP, and Dr. Shoji Segami for the use of VHP1pro:VHP1-mGFP. We thank Motoko Chiba for technical assistance.
Footnotes
This work was supported by the Japan Society for the Promotion of Science (grant nos. JP17H05050, JP18H04852, JP19H04712, and JP20H04916 to M.I., JP19J01681, JP20K15501, and JP20H05352 to S.N., and JP17H06350 to S.H., and Research Fellowship for Young Scientists to S.N.), the Japan Science and Technology Agency (grant no. JPMJPR16Q1 to M.I.), the U.S. Department of Energy (grant no. DE–SC0019573 to J.D.W.), and the Biotechnology and Biological Sciences Research Council (grant nos. BB/K018442/1, BB/N006372/1, BB/R009333/1, and BB/R016984/1 to R.P.J.).
Articles can be viewed without a subscription.
References
- Anding AL, Baehrecke EH(2017) Cleaning house: Selective autophagy of organelles. Dev Cell 41: 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR(2011) Protein degradation: An alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T.(2019) Autophagy and its role in plant abiotic stress management. Plant Cell Environ 42: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K(2001) The U-box protein family in plants. Trends Plant Sci 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Barros JAS, Cavalcanti JHF, Medeiros DB, Nunes-Nesi A, Avin-Wittenberg T, Fernie AR, Araújo WL(2017) Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol 175: 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA(2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Hill AL, Shirsekar G, Afzal AJ, Wang GL, Mackey D, Bonello P(2016) Quantification of hydrogen peroxide in plant tissues using Amplex Red. Methods 109: 105–113 [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A(1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Dikic I.(2017) Proteasomal and autophagic degradation systems. Annu Rev Biochem 86: 193–224 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD(2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Finley D.(2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fukuda T, Yamashita SI, Saigusa T, Kurihara Y, Yoshida Y, Kirisako H, Nakatogawa H, Kanki T(2018) The PP2A-like protein phosphatase Ppg1 and the far complex cooperatively counteract CK2-mediated phosphorylation of Atg32 to inhibit mitophagy. Cell Rep 23: 3579–3590 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C(2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y(2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Izumi M, Wada S, Makino A, Ishida H(2018) Vacuolar protein degradation via autophagy provides substrates to amino acid catabolic pathways as an adaptive response to sugar starvation in Arabidopsis thaliana. Plant Cell Physiol 59: 1363–1376 [DOI] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NHT, Mattsson O, Jørgensen LB, Jones JDG, Mundy J, Petersen M(2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD(2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Ishida H, Makino A(2018) Impacts of autophagy on nitrogen use efficiency in plants. Soil Sci Plant Nutr 64: 100–105 [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T(2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Makino A, Ishida H(2013) Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Wada S, Kondo E, Kurusu T, Kuchitsu K, Makino A, Ishida H(2015) Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol 167: 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Ishida H, Nakamura S, Hidema J(2017) Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29: 377–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Nakamura S(2018) Chloroplast protein turnover: The influence of extraplastidic processes, including autophagy. Int J Mol Sci 19: 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Nakamura S, Li N(2019) Autophagic turnover of chloroplasts: Its roles and regulatory mechanisms in response to sugar starvation. Front Plant Sci 10: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H(2012) RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J Exp Bot 63: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H(2010) The autophagic degradation of chloroplasts via Rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, Hirota Y, Saigusa T, Aoki Y, Uchiumi T, et al. (2013) Casein kinase 2 is essential for mitophagy. EMBO Rep 14: 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ(2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T(2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR(1997) Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K(2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Broad W, Trösch R, Töpel M, Demiral Sert T, Lymperopoulos P, Baldwin A, Jarvis RP(2019) Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363: 836. [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P(2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Ling Q, Jarvis P(2015) Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr Biol 25: 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD(2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD(2018) Autophagy: The master of bulk and selective recycling. Annu Rev Plant Biol 69: 173–208 [DOI] [PubMed] [Google Scholar]
- Martínez DE, Costa ML, Gomez FM, Otegui MS, Guiamet JJ(2008) ‘Senescence-associated vacuoles’ are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J 56: 196–206 [DOI] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM(2014) RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta 1843: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M(2011) Autophagy: Renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y(1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B(2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H, Baba M, Osumi M, Aoyagi S, Kato N, Ohsumi Y, Sakai Y(2004) Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol Biol Cell 15: 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D, Rodriguez E, Bressendorff S, Park OK, Hofius D, Petersen M(2014) Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Hidema J, Sakamoto W, Ishida H, Izumi M(2018) Selective elimination of membrane-damaged chloroplasts via microautophagy. Plant Physiol 177: 1007–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Izumi M(2018) Regulation of chlorophagy during photoinhibition and senescence: Lessons from mitophagy. Plant Cell Physiol 59: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y(2009) Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W(2016) Chloroplast proteases: Updates on proteolysis within and across suborganellar compartments. Plant Physiol 171: 2280–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y(2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Oku M, Sakai Y(2018) Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40: e1800008. [DOI] [PubMed] [Google Scholar]
- Ono Y, Wada S, Izumi M, Makino A, Ishida H(2013) Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ 36: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M, Ryan CA(1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD(2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Vigié P, Youle RJ(2018) Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 28: R170–R185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S, Makino S, Miyake A, Asaoka M, Maeshima M(2014) Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: Artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26: 3416–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Duan GY, Bock R(2013) A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell 25: 4209–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Yang W(2017) E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol 58: 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Burgos J, Zhuang X, Jiang L, Bassham DC(2018) Dynamics of autophagosome formation. Plant Physiol 176: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Li F, Buono R, Roschzttardtz H, Chung T, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS(2015) The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T(2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD(2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M.(2018) News from the PUB: Plant U-box type E3 ubiquitin ligases. J Exp Bot 69: 371–384 [DOI] [PubMed] [Google Scholar]
- Wang H, Lu Y, Jiang T, Berg H, Li C, Xia Y(2013) The Arabidopsis U-box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J 74: 511–523 [DOI] [PubMed] [Google Scholar]
- Wang S, Blumwald E(2014) Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles. Plant Cell 26: 4875–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J(2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y(2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K(2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Ohsumi Y(2018) Unveiling the molecular mechanisms of plant autophagy: From autophagosomes to vacuoles in plants. Plant Cell Physiol 59: 1337–1344 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z(2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzyńska A, Sirko A(2011) Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]