RELATED TO ABI3 AND VP1 (RAV) gene function is conserved in the regulation of flowering time in monocots and dicots, although via different mechanisms, and reveal roles in rice gynoecium development.
Abstract
In plants, correct formation of reproductive organs is critical for successful seedset and perpetuation of the species. Plants have evolved different molecular mechanisms to coordinate flower and seed development at the proper time of the year. Among the plant-specific RELATED TO ABI3 AND VP1 (RAV) family of transcription factors, only TEMPRANILLO1 (TEM1) and TEM2 have been shown to affect reproductive development in Arabidopsis (Arabidopsis thaliana). They negatively regulate the floral transition through direct repression of FLOWERING LOCUS T and GIBBERELLIN 3-OXIDASE1/2, encoding major components of the florigen. Here we identify RAV genes from rice (Oryza sativa), and unravel their regulatory roles in key steps of reproductive development. Our data strongly suggest that, like TEMs, OsRAV9/OsTEM1 has a conserved function as a repressor of photoperiodic flowering upstream of the floral activators OsMADS14 and Hd3a, through a mechanism reminiscent of that one underlying floral transition in temperate cereals. Furthermore, OsRAV11 and OsRAV12 may have acquired a new function in the differentiation of the carpel and the control of seed size, acting downstream of floral homeotic factors. Alternatively, this function may have been lost in Arabidopsis. Our data reveal conservation of RAV gene function in the regulation of flowering time in monocotyledonous and dicotyledonous plants, but also unveil roles in the development of rice gynoecium.
In plants, the correct formation of reproductive organs is critical not only for successful seedset but also for the perpetuation of the species. Accordingly, floral evocation must take place at a favorable time of the year to guarantee pollination and maximum survival possibilities for the offspring. Plants that are affected in their flowering time often have a lower amount of seeds resulting in yield losses. Indeed, precocious flowering is frequently associated with reduced photosynthetic capacity due to a shortened vegetative phase (Endo-Higashi and Izawa, 2011). Conversely, delayed flowering can affect seed maturation due to exposure to unfavorable conditions. A negative correlation also exists between grain size and grain number (Guo et al., 2018; Li et al., 2018), two important agronomic traits which are controlled by both genetic determinants and environmental conditions.
Plants have evolved different molecular mechanisms to coordinate flower and seed development at the proper time of the year. Actually, the switch from vegetative to reproductive growth is controlled by multiple genetic determinants that integrate the responses to environmental and physiological conditions of the plant. Ultimately, the regulatory pathways underlying the floral transition converge on floral integrators that are able to activate genes in the shoot apical meristem (SAM) that control the initiation and development of the inflorescence meristem (IM), and then of floral meristems (FM) from which floral organs differentiate. Upon fertilization, the carpel transforms into a fruit in which the seeds develop.
In the last decade, the molecular basis of the floral transition has been unveiled in different plant species: the florigen, a long distance signaling molecule, is first produced in leaves under favorable conditions, and then transported to the apical meristem to initiate reproductive development (Andrés and Coupland, 2012). In the model species Arabidopsis (Arabidopsis thaliana), photoperiodic flowering is triggered by FLOWERING LOCUS T (FT), a small globular protein of 21 kD (Kardailsky et al., 1999; Kobayashi et al., 1999). The expression of FT is activated under inductive long days (LD) in vascular tissues of leaves by the positive regulator CONSTANS (CO; Suárez-López et al., 2001; An et al., 2004), and precocious flowering is prevented by the counteraction of the RELATED TO ABI3 AND VP1 (RAV) transcription repressors TEMPRANILLO1 (TEM1) and TEM2 (Castillejo and Pelaz, 2008). Since prevention of precocious flowering is important for reproductive fitness other repressors such as FLOWERING LOCUS C (FLC), SCHLAFMUTZE (SMZ), SCHNARCHZAPFENZ (SNZ), TARGET OF EARLY ACTIVATION TAGGED 1 (TOE1), TOE2, FLOWERING LOCUS M (FLM) or SHORT VEGETATIVE PHASE (SVP) play key roles in the control of FT expression in response to vernalization, age, photoperiod or ambient temperature (Hartmann et al., 2000; Scortecci et al., 2001, 2003; Aukerman and Sakai, 2003; Schmid et al., 2003; Jung et al., 2007; Lee et al., 2007; Li et al., 2008;; Mathieu et al., 2009; Lee et al., 2013; Posé et al., 2013). Once flowering is induced, FM identity genes, such as the MADS-box genes SUPPRESSOR OF CONSTANS1 (SOC1) and APETALA1 (AP1), are induced which in turn repress TEMs expression (Kaufmann et al., 2010; Tao et al., 2012). Under noninductive short days (SD), CO is not active and there is no CO-dependent FT induction. In this light regime, the accumulation of the plant hormones gibberellins (GA) triggers floral transition by inducing SOC1 and LEAFY (LFY; Wilson et al., 1992; Blázquez et al., 1997; Moon et al., 2003; Eriksson et al., 2006; Hisamatsu and King, 2008). Interestingly, TEMs also regulate GA accumulation by repressing GA-3-OXIDASE 1 (GA3OX1) and GA3OX2 genes (Osnato et al., 2012).
In the crop species rice (Oryza sativa), two closely related genes have been described as FT orthologs: Heading date 3a (Hd3a), which promotes flowering under inductive SD, and Rice Flowering locus T 1 (RFT1), which does it under noninductive LD (Kojima et al., 2002; Komiya et al., 2009). An evolutionarily conserved module defined by the rice orthologs of Arabidopsis CO and FT controls photoperiodic flowering (Shrestha et al., 2014). The rice homolog of CO, Hd1, functions as activator of Hd3a in SD, and contrarily as repressor in LD (Yano et al., 2000; Nemoto et al., 2016). Nevertheless, additional rice-specific regulators have been discovered: the floral activator Early Heading date 1 (Ehd1, Doi et al., 2004), and its repressor Grain number, plant height and heading date 7 (Ghd7; Itoh et al., 2010). The Ehd1-Ghd7 pathway determines the critical day-length necessary for the induction of the florigen through a double gating mechanism dependent on the circadian clock and phytochrome-mediated light perception (Itoh et al., 2010). Additional studies on phylogenetic reconstructions revealed the presence of FLC homologs in monocots genomes (Ruelens et al., 2013). These FLC-like factors repress the floral transition in response to cold in temperate cereals (Winfield et al., 2009; Greenup et al., 2010), but appeared to have acquired an opposite function in tropical cereals. Indeed, the rice FLC homolog OsMADS51 activates the expression of the floral promoter Ehd1 (Kim et al., 2007).
Upon FT/Hd3a activation, the resulting gene product moves through the phloem from the leaf to the SAM (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007; Notaguchi et al., 2008; Komiya et al., 2009) where it triggers transcriptional reprogramming able to confer the competence to form flowers (Corbesier et al., 2007; Torti et al., 2012). Intriguingly, FT-like proteins do not have DNA binding activity per se and must interact with bZIP transcription factors (TFs), i.e. FLOWERING LOCUS D (FD) in Arabidopsis and OsFD1 in rice (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011; Brambilla et al., 2017; Collani et al., 2019), to activate the expression of downstream genes. The interaction of FT and FD proteins require the 14-3-3 adaptor proteins for the formation of an active Floral Activation Complex (FAC; Taoka et al., 2011; Collani et al., 2019), which in Arabidopsis directly activates the FM identity gene AP1 (Wigge et al., 2005), whereas the rice FAC complex induces the expression of the AP1-like genes OsMADS14/15/18 and the SEPALLATA-like gene OsMADS34 that are required for the specification of IM identity (Kobayashi et al., 2012; Gómez-Ariza et al., 2019) The florigens also mediate the transcriptional repression of PINE, a gene encoding a Zinc Finger type TF involved in the negative regulation of stem elongation. Therefore, the FAC coordinates the formation of reproductive structures (by activating IM identity genes) and internode elongation (by repressing PINE), guaranteeing the emergence of the panicle from the flag leaf, known as heading, which occurs when rice inflorescence development is completed (Gómez-Ariza et al., 2019).
In the rice inflorescence, primary and secondary branches form on the flanks of the IM (rachis) and terminate in spikelet meristems that develop floret meristems from which the palea, the lemma, two lodicules, six stamens and one central carpel differentiate. The identity of different floral organs is specified by the interaction of MADS domain TFs belonging to the SEPALLATA (OsMADS1), APETALA3 (OsMADS16) and AGAMOUS (OsMADS3–OsMADS58) subfamilies (Jeon et al., 2000; Kyozuka et al., 2000; Nagasawa et al., 2003; Dreni et al., 2011). Eventually, the FM is consumed during gynoecium development, when a single ovule primordium forms inside the carpel (Dreni et al., 2007). Recently, OsMADS1 was also shown to be essential during seed development, specifically in the regulation of grain size and shape (Liu et al., 2018).
In this study, we identified four members of the RAV family of TF in rice, which share sequence similarity with Arabidopsis TEMs. In silico/coexpression analyses based on available transcriptomics data revealed that OsRAVs interact with genes belonging to the MADS-box superfamily at different developmental stages, suggesting that RAVs are unknown players in the gene regulatory network underlying reproductive development in rice. Specifically, OsRAV8 and OsRAV9 negatively correlate with IM identity genes of the AP1 subfamily, whereas OsRAV11 and OsRAV12 act downstream of MADS-domain floral homeotic factors of the SEP and AG subfamilies. Molecular and functional studies using knock-down and knock-out mutant lines indicate a conserved function for OsRAV9/OsTEM1 as repressor of photoperiodic flowering upstream of the floral activators OsMADS14 and Hd3a, and reveal a role for OsRAV11 and OsRAV12 in the correct development of female reproductive organs, downstream of OsMADS1 and OsMADS13.
RESULTS
The Rice Genome Contains Four RAV Genes
Genes belonging to the RAV subfamily are present in all land plant species and encode for putative TFs, which are characterized by two DNA binding domains, an APETALA2-type at the N terminus and a B3-type at the C terminus (Kagaya et al., 1999). In the model species, Arabidopsis, the subfamily of RAV genes is composed of six members (Riechmann, 2002); in addition to the TEMs, four other genes belong to this family: RAV1 and RAV1-like, which are phylogenetically close to TEMs, and RAV3 and RAV3-like, which are the most divergent and nothing has been reported about their function. The functional characterization of the closest four indicate their regulatory role in different stages of plant development (Hu et al., 2004; Feng et al., 2005; Castillejo and Pelaz, 2008; Osnato et al., 2012; Matías-Hernández et al., 2014, 2016; Aguilar-Jaramillo et al., 2019) and in the response to abiotic stresses (Fu et al., 2014).
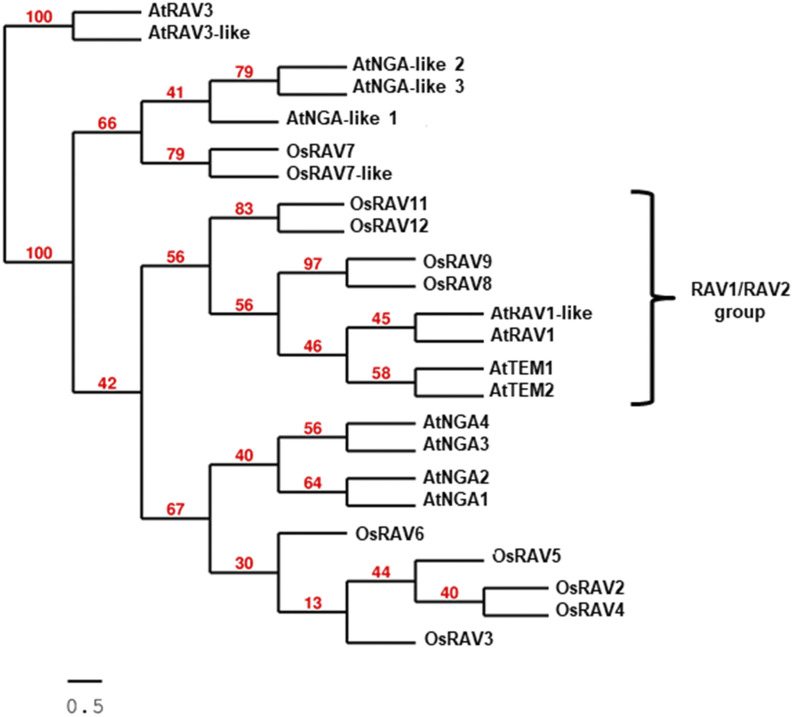
A preliminary phylogenomics analysis indicates a clear separation between AP2-B3 coding genes (RAV) and those encoding only the B3 domain (ARF, NGA, VAL), as reported in Supplemental Figure S1. Specifically in rice, although twelve genes were named RAV (Supplemental Table S1; Swaminathan et al., 2008), only four encode putative proteins containing both DNA binding domains as predicted by gene orthology and paralogy (OsRAV8, OsRAV9, OsRAV11, and OsRAV12; Supplemental Fig. S2A). Further analysis carried out by searching the SALAD (Surveyed conserved motif Alignment diagram and the Associating Dendrogram) database (Mihara et al., 2010) revealed the absence of RAV proteins in green and red algae, and the presence of multiple conserved motifs in addition to the AP2 and B3 domains (Supplemental Fig. S2B), including the bipartite nuclear localization signal and the B3 repression domain (Supplemental Fig. S3) together with features associated to posttranslational modifications (Supplemental Fig. S4). We also performed a phylogenetic analysis based on the deduced full-length protein sequences retrieved by a BLAST-P search against the proteomes of the two model species (Fig. 1; Supplemental Fig. S3). Although the four OsRAV proteins clearly clustered together with AtRAV1/AtRAV1-like and TEM1/TEM2, OsRAV8 and OsRAV9 showed the highest similarity with these Arabidopsis RAV factors (Riechmann, 2002).
Figure 1.
Phylogenetic analysis of RAV proteins in model species. Tree represents the most related RAV-like proteins from Arabidopsis (At) and Oryza sativa (Os) retrieved using TEM1 as query in a BLAST-P search, alignment with MUSCLE, and selected with G-blocks. TEM1 and TEM2 are RAV2-like and RAV2, respectively. Phylogenetic analysis with bootstrapping procedure (n = 100) as statistical test for branch support was carried out with PhyML, and tree visualization with TreeDyn. Scale bar indicates the number of substitutions per site. Values in red indicate percentage branch support values.
In the rice genome, the regions corresponding to the OsRAV8 and OsRAV9 loci may have originated recently, likely due to tandem duplication events after speciation. Indeed, these genes are physically linked (having a distance of about 50 Kb) at the tip of the short arm of chromosome 1 in a region that is enriched in sequences related to retro-transposons (Supplemental Fig. S5, A and B). Furthermore, when we searched the Plant Genome Duplication Database (Lee et al., 2013) using the OsRAV8-OsRAV9 locus identifiers, we found intragenome syntenic relationships with a region on the long arm of chromosome 1 containing OsRAV11 (Supplemental Fig. S5C), and a region on chromosome 5 containing OsRAV12 (Supplemental Fig. S5D). As a result of these phylogenomics and phylogenetic analyses, OsRAV11 and the related gene OsRAV12 appeared as within-species paralogs of OsRAV8-OsRAV9 with similar genomic structures.
Expression Patterns of RAV Genes and Floral MADS-box Genes Are Correlated
To gain insights into the possible function of OsRAV genes in various biological processes and metabolic pathways, we carried out an in silico coexpression analysis by using the Rice Functionally Related gene Expression Network Database (RiceFREND) platform (Sato et al., 2013) and constructed a coexpressed gene list for OsRAV9 as guide gene (Supplemental Dataset S1). Among the top 474 genes displaying a positive correlation with OsRAV9 (Pearson’s Correlation Coefficient higher than 0.3), we found enrichment for biological processes gene ontology (GO) categories related to reproductive processes, response to chemical stimuli, and response to oxidative stress (Supplemental Fig. S6, A and B). Supporting this, we also found overrepresentation for molecular function of the GO terms oxidoreductase and iron-binding activities (Supplemental Fig. S6C). Taken together, these findings suggest a possible role for RAV in reproductive development as well as abiotic stress response (similarly to Arabidopsis RAVs). In particular, peroxidases and oxidoreductases are detoxification enzymes activated upon accumulation of reactive oxygen species (Choudhury et al., 2013), and ion-binding proteins are able to sequester excess iron to avoid reaction with oxygen and the formation of damaging reactive oxygen species (Selote et al., 2015). On the other hand, GO analysis of the top 176 genes displaying a negative correlation with OsRAV9 suggested a possible function in signal transduction pathways, due to the enrichment of the terms related to protein dephosphorylation and regulation of transcription for biological processes (Supplemental Fig. S7A). In particular, we found a significant negative Pearson’s Correlation Coefficient between OsRAV9 and genes involved in reproductive development including the florigen Hd3a and the IM identity genes such as OsMADS14, OsMADS15, and OsMADS34 (Table 1). Furthermore, a recent transcriptomics analysis of apical meristems revealed a negative correlation between the expression of the closely related gene OsRAV8 and IM identity genes OsMADS14, OsMADS15, OsMADS18, and OsMADS34 at the transition from vegetative to reproductive growth (Gómez-Ariza et al., 2019; Supplemental Fig. S7). Taken together, coexpression and available transcriptomics datasets may suggest a possible role for at least OsRAV8 and OsRAV9 in the negative regulation of the transition from vegetative to reproductive phase, similarly to RAV genes in Arabidopsis.
Table 1. List of the top 20 genes negatively correlated with OsRAV9.
Genes listed in bold indicate involvement in the regulation of reproductive development. ABA, Abscisic acid; PCC, Pearson’s Correlation Coefficient.
| Weighted PCC | Locus Identifier | Gene name | Description |
|---|---|---|---|
| −0.568 | LOC_Os03g54160 | OsMADS14 | MADS-domain containing Transcription Factor |
| −0.542 | LOC_Os03g54170 | OsMADS34 | MADS-domain containing Transcription Factor |
| −0.526 | LOC_Os07g01820 | OsMADS15 | MADS-domain containing Transcription Factor |
| −0.497 | LOC_Os04g13150 | Cyclin-like F-box domain containing protein | |
| −0.491 | LOC_Os10g06560 | Cyclin-dependent kinase G-1 | |
| −0.477 | LOC_Os09g19500 | Protein kinase-like domain containing protein | |
| −0.476 | LOC_Os01g11940 | OsFT-like1 | Similar to SP3D |
| −0.464 | LOC_Os01g19880 | Conserved hypothetical protein | |
| −0.463 | LOC_Os10g06510 | Protein kinase-like domain containing protein | |
| −0.458 | LOC_Os02g55990 | Longin-like domain containing protein. | |
| −0.451 | LOC_Os07g17230 | OsWRKY123 | WRKY-domain containing Transcription Factor |
| −0.446 | LOC_Os05g43930 | OMT | Similar to O-methyltransferase ZRP4 (EC 2.1.1.-) |
| −0.434 | LOC_Os11g31770 | Conserved hypothetical protein | |
| −0.433 | LOC_Os05g38290 | Similar to Protein phosphatase 2C (PP2C) | |
| −0.4315 | LOC_Os02g52780 | bZIP | Similar to ABA-responsive element binding protein 2 (AREB2) |
| −0.423 | LOC_Os03g04990 | Conserved hypothetical protein | |
| −0.424 | LOC_Os06g06320 | Hd3a | HEADING DATE 3A |
| −0.422 | LOC_Os12g13910 | Conserved hypothetical protein | |
| −0.411 | LOC_Os09g27680 | Conserved hypothetical protein | |
| −0.407 | LOC_Os08g36340 | HAK4 | Similar to HIGH-AFFINITY POTASSIUM TRANSPORTER 4 |
OsRAV Genes Are Differentially Expressed during Plant Development
Because a role for OsRAV genes in rice plant development is yet to be elucidated, we first inferred their expression profiles by searching publicly available collections of transcriptomics data (Sato et al., 2011). OsRAV9 and OsRAV12 are expressed in vegetative tissues (i.e. leaves and roots), the former at early stages of plant development (Supplemental Fig. S8A), and the latter at maturity (Supplemental Fig. S8B), whereas OsRAV11 is widely expressed in different organs with the exception of anthers (Supplemental Fig. S8C). Moreover, the expression of OsRAV9 seems to follow a diurnal oscillation: its transcript levels are almost undetectable during the day, then increase at dusk and reach a peak in the middle of the night (Supplemental Fig. S8D). Likewise, the expression of OsRAV11 and OsRAV12 appeared to oscillate during the day with a peak after dusk (Supplemental Fig. S8, E and F), although with a smaller amplitude compared to OsRAV9.
To validate these expression data, we designed specific primers for each of the four OsRAV genes (Supplemental Fig. S8G). With respect to OsRAV8, we performed standard reverse transcription (RT)-PCR reactions, and even if a clear band was amplified using genomic DNA as control template, no amplification was observed when using cDNA obtained from different vegetative and reproductive tissues (Supplemental Fig. S8H), confirming that this gene is not transcribed at detectable levels in the samples examined. Only very recently OsRAV8 was found to be expressed; it was absent in all previous studies likely because of its very specific expression in the apical meristem only at the time of floral transition (Gómez-Ariza et al., 2019).
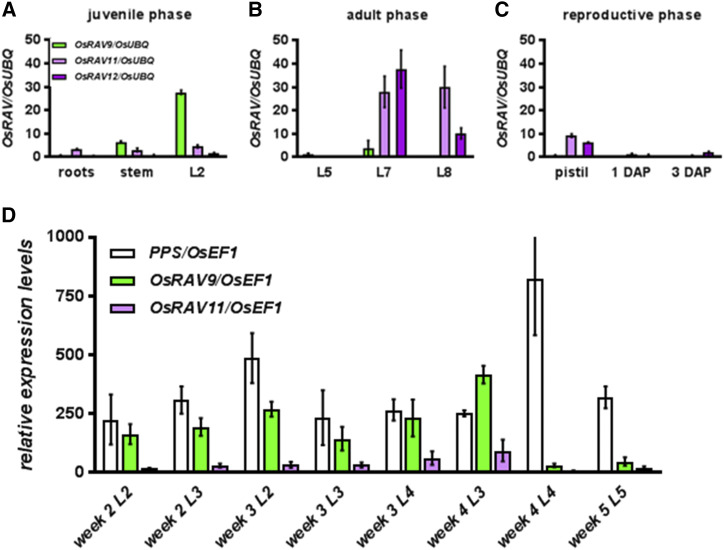
The expression profiles of the other three OsRAVs were investigated by RT-quantitative PCR (qPCR) in wild-type plants at different developmental stages. During the juvenile phase, the mRNAs of OsRAV9 and OsRAV11 were detected in roots, basal region of the stem comprising the SAM, and young leaves (Fig. 2A), although the abundance was much higher for OsRAV9 than OsRAV11. During the adult phase, the expression of OsRAV9 became almost undetectable in vegetative tissues resembling Arabidopsis TEM genes expression, whereas OsRAV11 and OsRAV12 displayed high transcript levels in mature leaves (Fig. 2B), resembling the expression behavior of the positive regulator of leaf senescence AtRAV1 in Arabidopsis (Woo et al., 2010). OsRAVs were also expressed in female reproductive organs (Fig. 2C), and their transcript levels decreased after pollination, suggesting that their activities might be restricted to the gynoecium before anthesis.
Figure 2.
Patterns of OsRAV genes expression during plant development. A to C, Transcript levels of OsRAVs, relative to OsUBQ, in vegetative and reproductive tissues of wild-type plants. A, RT-qPCR in roots, stems, and L2 dissected from 1-week-old seedlings. B, RT-qPCR in mature leaves (L5, L6, L7) dissected from 10-week-old adult plants. C, RT-qPCR in mature pistils and fertilized ovaries 1 and 3 d after pollination (DAP). D, Transcript abundance of PPS (white), OsRAV9 and OsRAV11, relative to OsUBQ, in juvenile (L2) and adult (L3-L4-L5) leaves. At week 2, L3 is formed and L4 is emerging. At week 3, L4 expands and L5 is emerging. At week 4, L5 is fully expanded. Expression data are mean values of three biological replicates, and error bars represent sd.
To investigate in detail a possible role of OsRAVs in phase changes during vegetative growth, we dissected differentiating leaves from wild-type plants to monitor their transcript levels at different developmental stages. Precisely, in rice, the juvenile phase is limited to the second leaf (L2), because the transition to the adult phase occurs during the development of the third to fifth leaf (L3–L5) when the midrib differentiates (Itoh et al., 2005). We also analyzed the expression of the Peter Pan Syndrome (PPS) gene (Tanaka et al., 2011), the rice ortholog of the Arabidopsis COP1 (Liu et al., 2008), as a marker for the transition from the juvenile to adult phase. In accordance with its function, PPS displayed a peak of expression in the fourth leaf four weeks after germination. OsRAV9 exhibited the highest transcript levels in L3 and then decreased in L4 and L5 (Fig. 2D), indicating that its transcription was drastically reduced at the transition to the adult phase. Also the expression of OsRAV11 was detected in young leaves, although at very low levels as compared with PPS and OsRAV9 (Fig. 2D).
In summary, these molecular analyses suggest diversification of expression patterns for OsRAV9 and OsRAV11, the former being transcribed at higher levels in the vegetative phase and specifically in juvenile leaves, and the latter at maturity, in particular in old leaves and female reproductive structures.
OsRAV9 Negatively Regulates Floral Transition
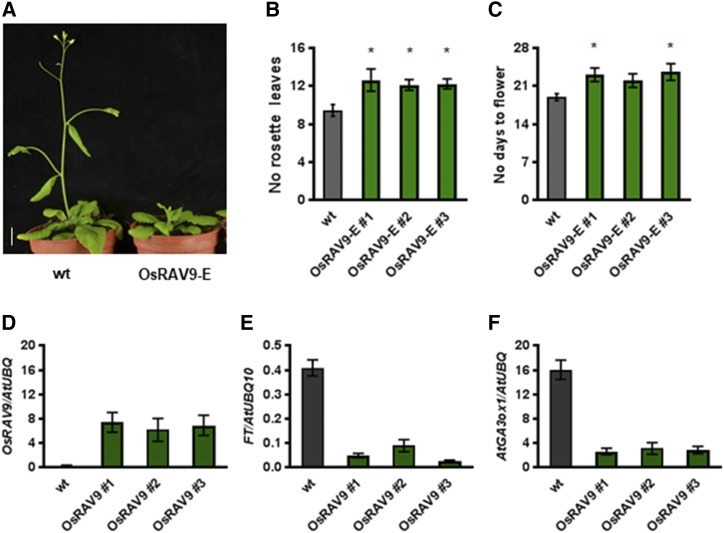
To investigate the functional conservation between rice and Arabidopsis RAV genes, we used the Pro-35S:OsRAV9 and Pro-35S:OsRAV11 constructs to transform Arabidopsis plants (Supplemental Fig. S9A). After selection of several independent transgenic lines (Supplemental Fig. S9B), phenotypic analyses of selected T2 generations revealed a mild late flowering phenotype of transgenic plants expressing OsRAV9 (OsRAV9-E), but no phenotypic alterations in plants expressing OsRAV11 (Supplemental Fig. S9C). Furthermore, molecular analyses of three representative T3 lines revealed also a correlation between the late flowering phenotype of OsRAV9-E plants (Fig. 3, A to D) and the down-regulation of FT and GA3oxidase1 (Fig. 3, E and F), two downstream targets of the TEM factors in Arabidopsis (Castillejo and Pelaz, 2008; Osnato et al., 2012). Taken together, these findings suggest that rice OsRAV9 and the Arabidopsis TEM genes have an at least partial conserved function as repressor of photoperiodic flowering in Arabidopsis.
Figure 3.
Late flowering phenotype of Arabidopsis plants expressing OsRAV9. A, Representative images of wild-type (wt; left) and OsRAV9-E transgenic (right) plants grown for 4 weeks under LD. Scale bar = 1 cm. B and C, Flowering time scored as number of rosette leaves and number of days to flower of wild-type (gray) and OsRAV9-E lines (green) grown under LD. D to F, Relative expression levels of OsRAV9 and TEM1 downstream targets in wild-type and representative T3 OsRAV9-E lines grown for 1 week under LD. D, Ectopic expression of OsRAV9 in transgenic Arabidopsis lines. E and F, Down-regulation of FT and AtGA3ox1 in OsRAV9-E lines compared with wild-type. Flowering time data are the average of 25 plants each genotype, with sem. Three biological replicates gave similar results, and one was chosen as representative. Data were analyzed by one-way ANOVA followed by Dunnett's multiple comparisons test between wild-type and transgenic lines. Asterisks indicate statistical significance (*P < 0,05). Expression data are reported as mean values of three biological replicates; error bar represents the sem. AtUBQ10 was used for normalization.
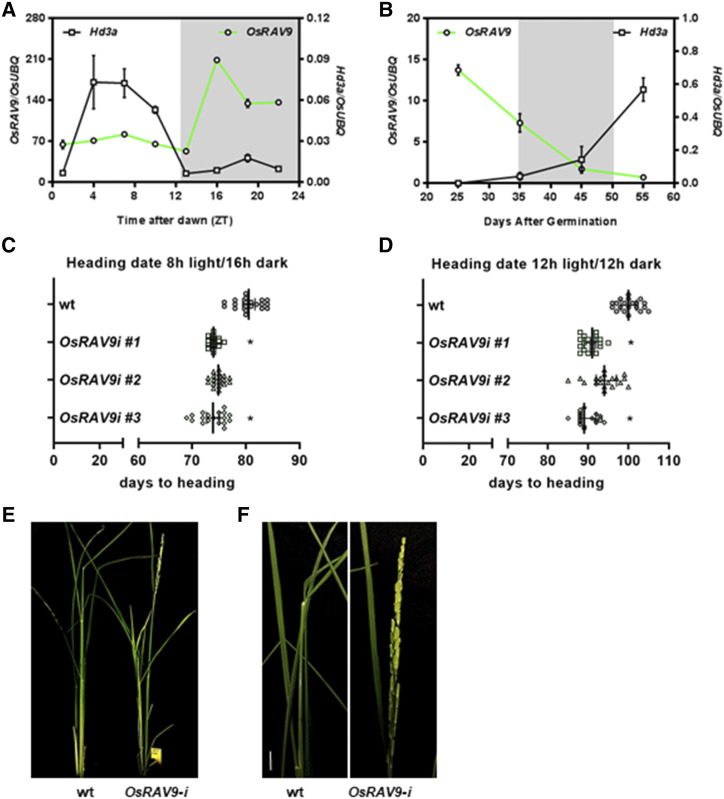
Besides the high sequence identity between OsRAV9 and AtTEMs, their expression patterns were similar as both displayed high transcripts levels in the juvenile phase that decreased as the plant aged shortly before the transition from vegetative to reproductive growth (Fig. 2; Castillejo and Pelaz, 2008). Consistently, expression analyses in wild-type plants showed a mutual exclusive pattern for OsRAV9 and the florigen Hd3a, not only during the day (Fig. 4A) but also throughout plant development (Fig. 4B), similarly to the opposite expression patterns of TEM and FT in Arabidopsis (Castillejo and Pelaz, 2008). Actually, the expression of OsRAV9 was high at early stages and dropped at around the transition to the adult phase. Conversely, the expression of Hd3a is almost undetectable at early stages of vegetative growth, and increases in the adult phase (Fig. 4B; Kojima et al., 2002; Komiya et al., 2009). The florigen begins to be produced in adult leaves 4 weeks after germination under inductive conditions, and triggers floral transition in adult plants when it reaches its maximum accumulation around 6 weeks .
Figure 4.
Role of OsRAV9 in heading date. A, Diurnal oscillation of Hd3a and OsRAV9 expression in leaves of 4-week-old wild-type (wt) plants. Gray block indicates night. B, Mutually exclusive expression patterns of Hd3a and OsRAV9 in leaves throughout wild-type (wt) plant development. Gray block indicates floral transition. C and D, Scatter plots representing heading date as number of days to flower of wild-type (in gray) and OsRAV9-i plants (in light green) grown under inductive photoperiods (8-h light/16-h dark, 12-h light/12-h dark). Lines represent the median with 95% of confidence interval. E, Early flowering phenotype of a selected OsRAV9 silencing line (RAV9-i 3, right) compared with wild-type (left) 100 d after germination (DAG). F, Close-up view showing wild-type panicle at the booting stage and OsRAV9-i panicle at anthesis. For molecular analyses, plants were grown under 12-h light/12-h dark at 28°C, whereas for heading date plants were grown under different daylengths. Bars = 10 cm. Expression data are reported as mean values of three biological replicates; error bar represents the sem. Flowering time data are the average of 18 to 20 plants each genotype. Three biological replicates gave similar results, and one was chosen as representative. Data were analyzed by one-way ANOVA followed by Dunnett's multiple comparisons test between wild-type and transgenic lines. Asterisks indicate statistical significance ( *P < 0.05 and**P < 0.033).
Because the dynamics of OsRAV9 expression strikingly resembled those of TEM genes, we decided to investigate if the function was also conserved in rice. We used an RNA interference (RNAi) silencing strategy due to the absence of insertion mutants in public collections for OsRAV9. Wild-type rice calli were transformed with an RNAi construct carrying a Gene Sequence Tag specific for the 3′ end of the gene under the control of a constitutive promoter (Supplemental Fig. S10A). We obtained eleven independent transgenic lines for the silencing construct, and selected T1 OsRAV9-RNAi transformants by monitoring its transcript levels at the peak of expression (Supplemental Fig. S10B). T2 generation lines were obtained by self-pollination, and three lines with the highest silencing of OsRAV9 were characterized. The RNAi lines (reported as OsRAV9-i) flowered slightly earlier than the wild-type under inductive photoperiods, more clearly under SD than under 12 h of light (Fig. 4, C and D; Supplemental Fig. S10C), but not under noninductive LD (Supplemental Fig. S10C). Under SD, the down-regulation of OsRAV9 resulted in significantly early flowering plants (Fig. 4C). Under 12-h light/12-h dark regime, transgenic lines flowered on average 90 DAG and underwent anthesis 1 week later, whereas wild-type plants were still at the booting stage (Fig. 4, E and F). These findings suggested that OsRAV9 functions as floral repressor in rice like TEMs do in Arabidopsis, likely upstream of the florigen Hd3a under inductive conditions. Therefore, hereafter we refer to OsRAV9 as OsTEM1.
OsTEM1 Regulates Floral Activators OsMADS14 and Hd3a
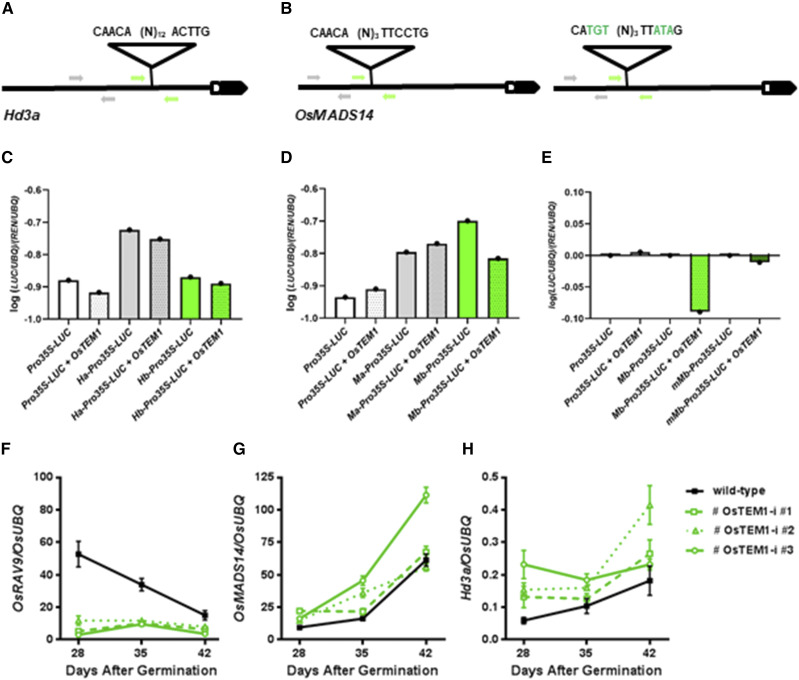
Based on the results obtained, it was tempting to speculate a role for OsTEM1 in the direct repression of Hd3a as a noncanonical RAV binding site is present in its regulatory region (Fig. 5A). However, we could not exclude an indirect effect on Hd3a via additional transcriptional regulators. Actually, a negative correlation also exists between OsTEM1 and additional floral activators including OsFT-like1 and IM identity genes (Table 1), previously shown to act in a regulatory loop with the florigen, upstream of Hd3a in the leaf and downstream of the Hd3a/14-3-3/OsFD1 complex in reproductive meristems (Kobayashi et al., 2012). OsMADS14 is also expressed in adult leaves, and a perfect RAV binding site was found in its promoter (Fig. 5B). Therefore, we used a transient expression system based on a dual Renilla-Luciferase assay to investigate the direct interaction between OsTEM1 and potential target genes. The effector vector Pro-35S:OsTEM1 was transiently coexpressed with reporter vectors containing different regulatory regions of Hd3a and OsMADS14 (Fig. 5, C and D; Supplemental Fig. S11). We evaluated transactivation ability of the floral repressor on target promoters by measuring the relative expression of the LUCIFERASE reporter genes from firefly(LUC) and from renilla (REN) in cell lysates. Despite the biological variability between independent replicates, a clear reduction of LUC/REN relative transcript levels was always observed when OsTEM1 was cotransformed with a reporter vector carrying promoter sequences of OsMADS14 containing the RAV binding site but not Hd3a (Fig. 5, C and D; Supplemental Fig. S11, B and C). However, this reduction was abolished when we cotransfected the effector vector with a mutated version of the reporter vector carrying the RAV binding site of the OsMADS14 promoter. For statistical analyses purposes, the values of these three replicates including intact and mutated RAV binding sites are shown as logarithmic values (Fig. 5E; Supplemental Fig. S11D). Therefore, the transient cotransformation assays soundly suggest that transcription repression mediated by OsTEM1 is stronger on DNA regulatory sequences of OsMADS14 and is likely mediated by the RAV binding site.
Figure 5.
Interaction between OsRAV9/OsTEM1 and floral activators. A, Noncanonical RAV binding site in the promoter of Hd3a, 785 bp upstream of the transcription starting site. B, Perfect RAV binding site in the promoter of OsMADS14, 2250 bp upstream of the transcription starting site, and mutated version (right). Arrows represent oligonucleotides used to amplify fragments of ProHd3a (H) and ProOsMADS14 (M) without RAV binding sites (Ha and Ma, in gray) and with RAV binding sites (Hb and Mb, in green). C to E, Transactivation activity of OsTEM1 in transiently transformed protoplasts, reported as ratio of transcript levels of LUC and REN reporter genes relative to UBQ. C, RT-qPCR of protoplasts cotransformed with Pro-35S:OsTEM1 and reporter vectors containing sequences of ProHd3a (Ha in gray, Hb in green). D, RT-qPCR of protoplasts cotransformed with Pro-35S:OsTEM1 and reporter vectors containing sequences of ProOsMADS14 (Ma in gray, Mb in green). E, Analysis of protoplasts cotransformed with Pro-35S-OsTEM1 and reporter vectors containing sequences of ProOsMADS14 (Mb in green, mutated Mb in dark green). Values are the mean of three independent replicates. F to H, Expression analysis of genes involved in heading date in independent T2 lines (green) compared with wild-type plants grown for 28, 35, and 42 d under inductive conditions. F, Down-regulation of OsRAV9/OsTEM1 in silencing lines (green). G and H, Up-regulation of the floral activators OsMADS14 and Hd3a in silencing lines (green lines) compared with wild-type (black line). Expression data are mean value of three biological replicates with three technical replicates each, and error bars represent sd.
Finally, we monitored the expression levels of OsTEM1 and floral activators shortly before and around the floral transition in wild-type and OsTEM1-i lines grown under inductive conditions. As expected, transcripts levels of OsTEM1 were confirmed to be higher at week 4 in wild-type plants, and strong down-regulation was observed in the transgenic lines (Fig. 5F). Conversely, the expression of OsMADS14 increased from week 4 to 6 in wild-type plants, and a clear up-regulation was detected in OsTEM1-i lines 2 and 3 at 35 DAG (Fig. 5G). The related IM genes OsMADS15 and OsMADS34 were expressed at extremely low levels in leaves at floral transition (Supplemental Fig. S12A); however, OsMADS18 was transcribed at higher levels in vegetative tissues, and alteration of its mRNA abundance was found in the transgenic line with highest OsRAV9 silencing (Supplemental Fig. S12B). Furthermore, considerable increase in Hd3a mRNA levels was also detected in silencing lines around floral transition (Fig. 5H). Taken together, transient cotransformation of protoplasts and comparative expression analysis of wild-type and transgenic RNAi lines suggest that OsTEM1 controls heading date via direct repression of OsMADS14, although it also modulates Hd3a expression.
We also tested the effect of the down-regulation of OsTEM1 on other genetic pathways involved in the control of heading date (Supplemental Fig. S12C), and, although variable, we found up-regulation of the flowering inductors OsMADS50, OsMADS51, and Ehd1 in silencing lines (Supplemental Fig. S12, D and E), perhaps suggesting additional roles in parallel pathways. As a consequence, down-regulation of the OsTEM1 repressor and up-regulation of different floral activators in transgenic lines resulted in early flowering phenotype.
OsRAV11 and OsRAV12 Regulate Carpel Development
The fact that OsRAV11/OsRAV12 have diversified from OsTEM1 in their expression patterns and coding sequences prompted us to hypothesize that these genes might have acquired different roles in plant development, likely after floral induction based on their expression pattern in the reproductive phase (Fig. 2C). A preliminary spatio-temporal expression analysis indicated that OsRAV11 was expressed at early stages of flower development before organ primordia differentiation because its mRNA was detected in the spikelet meristem (Supplemental Fig. S13A). Later on, transcripts became first restricted to the carpel primordia similar to carpel identity genes, and afterward specifically limited to the apical part of the developing gynoecium (Supplemental Fig. S13B).
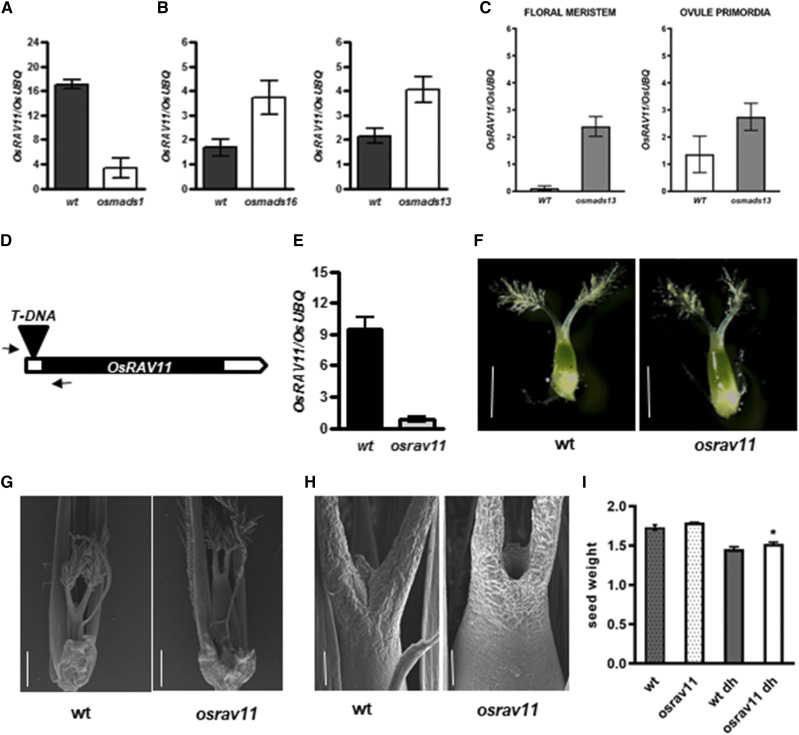
The analysis of available transcriptomics data sets performed on floral homeotic mutants that revealed OsRAV11 down-regulation in OsMADS1-RNAi inflorescences (Supplemental Table S2; Khanday et al., 2013), and conversely up-regulation in osmads13 mutant (M. Osnato and M.M. Kater, unpublished data), suggested an interaction between RAV and MADS-box genes controlling floral organ development. This could be a direct effect since we found CArG-boxes, consensus sequences recognized and bound by MADS-domain TFs, in OsRAV11 and also in OsRAV12 regulatory sequences (Supplemental Fig. S13D). The regulation by OsMADS1 and OsMADS13 is further supported by the fact that both appear as regulators of OsRAV genes in the Environmental Gene Regulatory Influence Networks (Wilkins et al., 2016). Therefore, OsRAV11 might have a role in the development of the pistil, likely downstream of class-d and class-E MADS-domain floral homeotic factors.
Consistently, we found down-regulation of OsRAV11 in developing panicles of the osmads1 mutant (Fig. 6A), characterized by the conversion of floral organs into glume-like structures (Agrawal et al., 2005; Khanday et al., 2013), and up-regulation in young panicles of the floral homeotic mutants osmads16 and osmads13 (Fig. 6B), in which stamens and ovules are homeotically converted into extra carpels respectively (Nagasawa et al., 2003; Dreni et al., 2007) perhaps because the ectopic activatio of OsRAV11. Accordingly, we observed strong upregulation of OsRAV11 specifically in osmads13 ovule primordia (Fig. 6C; M. Osnato and M.M. Kater, unpublished data), which later develop as carpels.
Figure 6.
Molecular and functional characterization of OsRAV11. A to C, Misregulation of OsRAV11 in floral homeotic mutants. A, Down-regulation of OsRAV11 in osmads1 developing inflorescences. B, Up-regulation of OsRAV11 in osmads16 and osmads13 developing inflorescences. C, Strong activation of OsRAV11 in specific cell types (floral meristem and ovule primordia) isolated from osmads13 mutant flowers. D, Schematic representation of the T-DNA insertion in the 5′ untranslated region of OsRAV11. Arrows indicate primers used for genotyping. E, Down-regulation of OsRAV11 in pistils dissected from osrav11 mutant flowers at maturity. F to H, Morphological analyses of female reproductive structures at maturity. F, Representative images of wild-type and osrav11 carpels dissected from mature flowers at anthesis obtained by optical microscopy. G and H, Representative images of reproductive structures obtained by scanning electron microscopy (sem). G, Wild-type and osrav11 carpels upon fertilization. Glumes were partly removed to show female reproductive organs. H, Apical tissues of wild-type and osrav11 gynoecia after pollination. Bars = 1 mm (F), 500 μm (G), and 100 μm (H). I, Bar plots representing the weight of seed produced by wild-type (in gray) and osrav11 (in white) plants grown in the greenhouse under inductive conditions. Expression data are reported as mean value of three biological replicates; error bar represents the sem. Phenotypic data are the average of three biological replicates of the weight of 100 seeds each genotype, with sem. Statistical significance was examined by Student’s two-tailed unpaired t test (*P < 0.05).
Therefore, we explored the function of OsRAV11 using a knock-out mutant characterized by the insertion of T-DNA in the 5′ untranslated region of the gene (Fig. 6, D and E). The loss of OsRAV11 function did not cause alteration of heading date, but elongated carpels (Fig. 6F). A detailed phenotypic analysis by sem upon fertilization indicated alteration in size and shape of the pistil, with an enlarged ovary compared to wild-type pistils (Fig. 6G). Specifically, differentiation of apical tissues of the gynoecium were altered in the osrav11 mutant as carpel tissues did not fuse (Fig. 6H). Likewise, we observed alteration of seed morphology (Supplemental Fig. S13): mutant plants produced seeds with increased seed length and decreased seed width (Supplemental Fig. S3G), resulting in statistically significant alterations of length-to-width ratio and circularity (Supplemental Fig. S3H). Interestingly, we observed a slight increase in seed weight after de-husking (Fig. 6I).
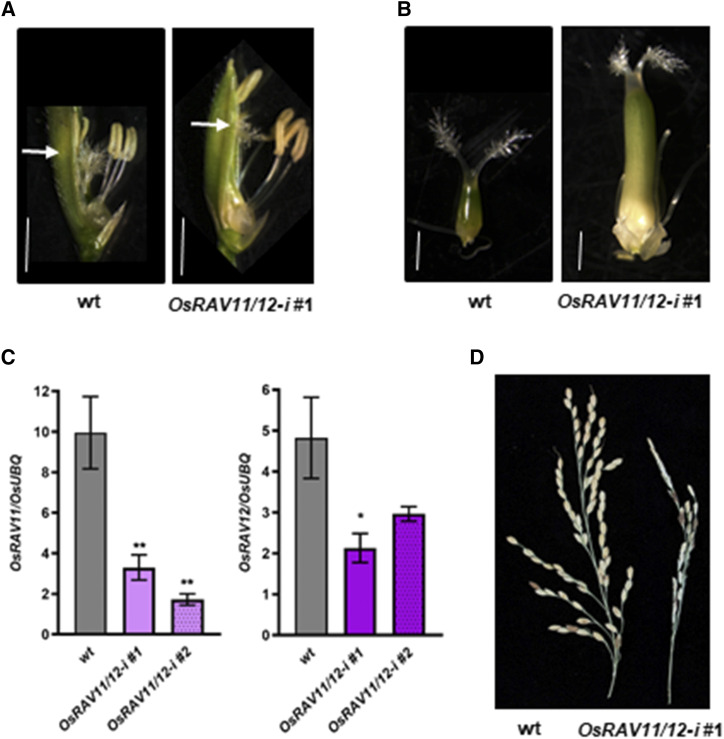
Ultimately, to investigate the possible redundancy between OsRAV11 and the closely related OsRAV12 gene in the formation of female reproductive organs, we generated transgenic rice plants in which the expression levels of both genes were reduced by RNA interference (Supplemental Fig. S14). Vegetative growth of knock-down lines (from now on OsRAV11/12-i) was normal, whereas flowers showed alterations caused by abnormal morphology of female reproductive organs (Fig. 7A). Indeed, elongated cylindrical pistils with highly reduced stigmas and enlarged ovaries (Fig. 7B) were observed in transgenic plants with reduced levels of both OsRAV11 and OsRAV12 in the gynoecium (Fig. 7C), indicating that these two genes redundantly regulate the basal-apical patterning of the gynoecium. Interestingly, flowers of these transgenic lines produced viable pollen, but most of the ovules were not fertilized, resulting in very poor seedset. Consequently, most of the transgenic lines displayed severe fertility defects (Fig. 7D).
Figure 7.
Morphogenetic effects of OsRAV11/OsRAV12 silencing in reproductive phase. A, Representative images of mature flowers dissected from wild-type (wt) and OsRAV11/12-i inflorescences. Arrows indicate the position of the stigmas. B, Representative images of mature carpels dissected from wild-type and one T3 OsRAV11/12-I flowers obtained by optical microscopy. Bars =1 mm. C, Down-regulation of OsRAV11 and OsRAV12 in mature pistils of two representative T3 OsRAV11/12-i lines as compared with wild-type. Expression data are reported as mean values of three biological replicates with sem. Data were analyzed by ordinary one-way ANOVA followed by Dunnett's multiple comparisons test between wild-type and transgenic lines. Asterisks indicate statistical significance (*P < 0.05 and **P < 0.033). D, Fertility defects of mature OsRAV11/12-i panicles compared with wild-type.
To conclude, molecular and functional characterization of OsRAV11 and OsRAV12 suggest that these two genes might redundantly regulate the differentiation of the female reproductive structures. Intriguingly, a decreased activity of OsRAV11 correlated with an increase in seed weight, whereas the loss of OsRAV11 and OsRAV12 activity associates with sterility problems.
DISCUSSION
Arabidopsis and Rice RAV Genes Are Closely Related
RAV proteins belong to the plant-specific B3 superfamily of TFs (Swaminathan et al., 2008), and are characterized by an additional AP2 DNA binding domain at the N terminus. AP2-B3 type proteins were not found in the green algae Chlamydomonas reinhardtii, but they appeared early in the evolution of land plant species. The presence of two DNA binding domains suggests that these TFs achieve high affinity by specifically binding bipartite sequences in regulatory regions of downstream targets (Kagaya et al., 1999; Castillejo and Pelaz, 2008; Osnato et al., 2012; Matías-Hernández et al., 2016).
In this study, we focused on the four rice RAV genes, which display striking similarities with four RAV genes of Arabidopsis (Supplemental Fig. S15) that were already described as regulators of different aspects of plant development and stress responses. Specifically, AtRAV1 and AtRAV1-like redundantly control leaf senescence (Woo et al., 2010), AtRAV1 and TEM2 modulate sensitivity to drought and salinity (Fu et al., 2014), TEM1 and TEM2 repress trichome formation (Matías-Hernández et al., 2016), and floral transition under inductive and noninductive photoperiods (Castillejo and Pelaz, 2008; Osnato et al., 2012) as well as in response to low temperatures (Marín-González et al., 2015) and to plant age (Aguilar-Jaramillo et al., 2019). Interestingly, preliminary coexpression analyses suggested that OsRAV9 and OsRAV11 could act in signal transduction pathways activated in response to abiotic stresses. Nevertheless, OsRAV9 and the closely related OsRAV8 could also act in Gene Regulatory Networks controlling the transition from vegetative to reproductive growth in the leaf and in the apical meristem, respectively.
OsRAV9/OsTEM1 Is a New Player in Flowering
Although phylogenomics and phylogenetic analyses revealed that OsRAV genes might have originated from duplication events from a common ancestor after speciation and separation of monocots and dicots, it is difficult to draw conclusions from orthology with AtRAVs. Furthermore, the expression domains of the four OsRAV paralogous genes appear to have diversified likely due to polymorphisms in their regulatory regions. Regardless of the remarkable similarities in the gene structures and coding sequences, OsRAV8 and OsRAV9 are expressed in different tissues. Indeed, OsRAV9 mRNA is abundant in juvenile leaves, and its reduction marks the transition to the adult phase when plants acquire the competence to flower, whereas the transcripts of OsRAV8 are detected in the apical meristem and its levels also decrease at floral transition when IM identity genes are activated (Gómez-Ariza et al., 2019). Therefore, at least OsRAV9 displayed an expression pattern that resembles that of TEM genes in Arabidopsis. Supporting the functional conservation, the ectopic expression of OsRAV9 in Arabidopsis plants correlated with the repression of the TEM targets FT and AtGA30X1 and delayed flowering time. In addition, silencing of OsRAV9/OsTEM1 in transgenic rice lines resulted in early flowering due to up-regulation of the floral activators OsMADS14 and Hd3a.
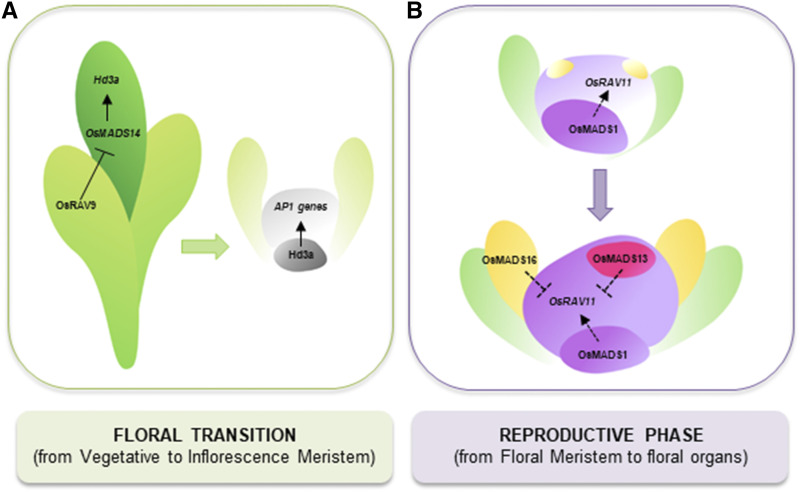
The mechanism of action seems to be different in the two species. To avoid precocious flowering, Arabidopsis TEMs directly target the florigens FT (Castillejo and Pelaz, 2008) and AtGA3ox1/2 (Osnato et al., 2012), whereas OsTEM1 might regulate Hd3a indirectly via repression of OsMADS14 as proposed in Figure 8A. Although this AP1/FUL-like gene is expressed at high levels in reproductive tissues, its mRNA accumulates also during the vegetative phase. Accordingly, an additional role for OsMADS14 as activator of the florigen in the leaf has been previously proposed, likely acting via a positive regulatory loop with Hd3a (Kobayashi et al., 2012). Actually, knock-down lines silencing OsMADS14, OsMADS15, OsMADS18, and OsMADS34 display delayed flowering (Kobayashi et al., 2012), and conversely transgenic rice ectopically expressing OsMADS14 and OsMADS18 are early flowering (Jeon et al., 2000; Fornara et al., 2004). A very recent study also indicates that AP1-like genes could regulate drought-escape; indeed, the early flowering phenotype under drought conditions correlates with increased expression of OsMADS18 (Groen et al., 2020).
Figure 8.
Model for interaction between RAV and MADS factors during reproductive growth. A, OsRAV9 represses the transcription of OsMADS14, a positive regulator of the florigen Hd3a, in the leaf. Upon floral transition, the complex Hd3a-OsFD activates the expression of IM identity genes OsMADS14-15-18-34 in the apical meristem. B, The expression of OsMADS1 in developing flowers marks the formation of lemma/palea (in green) and central carpel (in pink) from the FM, whereas the presence of OsMADS16 and OsMADS13 prevents the expression of genes involved in carpel development in stamen primordia (in yellow) and in ovule primordium (in violet).
Upon floral transition, the FAC activates the expression of OsMADS14, together with OsMADS15, OsMADS18, and OsMADS34, which specify IM identity and trigger the development of reproductive structures (Kobayashi et al., 2010; Kobayashi et al., 2012). Based on transcriptomics analysis, we speculate that OsRAV8 could play a role in the maintenance of the vegetative state of the apical meristem, thus preventing the formation of the rachis under noninductive conditions.
Mechanisms Controlling Floral Transition in Cereals
It has long been known that AP1/FUL-like proteins control seasonal flowering in cereals growing in temperate regions (Fjellheim et al., 2014). Precisely, the floral transition is triggered by the activation of VERNALIZATION 1-like (VRN1-like) and FT-like genes in wheat leaves in response to increasing day-length and prolonged exposure to low temperatures (Danyluk et al., 2003; Shimada et al., 2009; Winfield et al., 2009). At least in wheat, VRN1 regulates flowering by directly binding the promoter of the downstream target FT-like1 (Deng et al., 2015; Tanaka et al., 2018). Surprisingly, the function of AP1/FUL-like proteins as floral activators acting in leaves seems to be well conserved in rice (Jeon et al., 2000; Kobayashi et al., 2012), despite the fact that it is a cereal crop of tropical origin that does not require vernalization. In this study, we propose a new mechanism that is largely independent from previously described molecular networks determining heading date in rice (Tsuji et al., 2013). Interestingly, the presence of TEM orthologs in the Poacea family (Supplemental Fig. S1) opens up the intriguing possibility that the RAV-AP1-FT regulatory module could be conserved among tropical (Oryza spp. tribe) and temperate (Triticeae tribe) cereals.
Moreover, as mentioned above, AtRAV1 and TEM2 seem to modulate sensitivity to drought and salinity (Fu et al., 2014). OsTEM1 has been recently shown to play a role in response to abiotic stresses (OsRAV2; Duan et al., 2016), and OsTEM1 expression was reduced in plants growing under drought conditions (Plessis et al., 2015) where the transcription of OsMADS18 was increased (Groen et al., 2020). Therefore, RAV genes could be involved in adaptive growth by modulating heading date in response to environmental limitations and fluctuations, by integrating external and internal physiological conditions.
Mechanisms Controlling Floral Organ Development in Rice
In the last decade, genetic and functional genomics analyses in different plant species revealed that flower development is governed by a complex framework based on MADS-domain TFs. Rice members of the grass-specific LOF-SEP clade sequentially regulate different steps of flower development upon the vegetative to reproductive phase change. First, OsMADS34 forms tetrameric complex with the AP1-like factors OsMADS14 and OsMADS15 in the IM to initiate inflorescence branch meristem primordia development from which secondary branches and spikelets differentiate (Kobayashi et al., 2012). After the formation of rudimentary glumes and sterile lemmas, the spikelet meristem is converted into floret meristem, which produces different floral organs. Another grass-specific LOF-SEP factor, OsMADS1, plays a central role in the determination of floral organ identity, as it interacts physically and genetically with AP3-like and AG-like factors, which are involved in the development of male and female reproductive organs (Prasad et al., 2001; Li et al., 2011; Khanday et al., 2013, 2016). Recently, transcriptomics analysis performed on OsMADS1 knock-down panicles at very early stages of flower development (Khanday et al., 2013) unveiled misregulation of floral homeotic MADS-box genes as well as down-regulation of genes encoding B3-type TFs, including OsARFs and OsRAV11 (Supplemental Table S2). Precisely, the ARF-type TFs OsETTIN1 and - OsETTIN2 control the differentiation of apical tissues of the carpel, and at least OsETTIN2 is directly regulated by OsMADS1 (Khanday et al., 2013). Intriguingly, the aberrant carpel morphology of loss of OsETTINs (Khanday et al., 2013) is similar to that of transgenic lines with reduced level of OsRAV11 and OsRAV12 (Fig. 7B).
OsRAV11 and OsRAV12 Regulate the Development of Gynoecium
In Arabidopsis, the early flowering TEM loss of function mutants do not display evident alterations of flower development. Nonetheless, the late flowering TEM overexpressing lines show fertility defects and produce shorter siliques containing fewer seeds. This phenotype could be related to decreased content of GA, or alternatively to misregulation of genes involved in the regulation of later organ development. In rice, further molecular analyses suggest that RAV genes might have acquired additional functions at later stages of vegetative and reproductive growth. Indeed, OsRAV11 and OsRAV12 are highly expressed not only in mature leaves at ripening, indicating a possible regulatory role in leaf senescence similarly to AtRAV1, but also in the gynoecium before fertilization. Actually, functional characterization of knock-down and knock-out mutants points at a new function for OsRAV11 and OsRAV12 in the correct formation of female reproductive organs. Indeed, the whole basal-apical pattern was distorted; the ovary was enlarged in osrav11 and misshapen in OsRAV11/12-i plants. Down-regulation of both genes resulted in reduced stigmas and larger carpels than in single osrav11 mutants, which may suggest a redundant function of these genes in carpel development and differentiation. Therefore, we hypothesize a role for OsRAV11 and OsRAV12 in the determination of the basal-apical pattern of the pistil, perhaps in parallel with ARFs. Furthermore, we propose that at least OsRAV11 may control the differentiation of the gynoecium downstream of MADS-domain TFs (Fig. 8B), due to its mis-regulation in the floral homeotic mutants osmads1, osmads13 and osmads16 (Fig. 6, A and C).
Besides the specification of the identity of different floral organs, OsMADS1 has been proposed as a key trait of agronomical interest. During spikelet development, OsMADS1 also interacts with Gγ subunits of GS3 and DEP1 (Fan et al., 2006; Huang et al., 2009; Liu et al., 2018), which regulate its transcriptional activity on a common set of target genes involved in the determination of seed size and shape. The dominant negative mutation osmads1(lgy3) causes an alternatively spliced protein variant and correlates with more slender grain, as the mutated protein promotes cell proliferation in longitudinal direction (Liu et al., 2018). Pyramiding of lgy3 and dep1-1 alleles in a japonica cultivar resulted not only in a 10% increase in grain yield, but also improved grain length-to-width ratio and grain chalkiness (Liu et al., 2018). We can hypothesize that the elongated seed phenotype associated to down-regulation of OsMADS1 (Liu et al., 2018) could be mediated through the down-regulation of B3 genes including OsRAV11 since a similar slender phenotype is observed in osrav11 and OsRAV11/12-i carpels. Further analyses are required to understand the interactions with putative upstream regulators, interacting proteins and downstream targets constituting the molecular network that regulates the development of the gynoecium in rice.
MATERIALS AND METHODS
RAV Sequence Analyses
OsRAV8 and AtTEM1 were used as queries in phylogenomics analyses in sequenced land plants species (eudicotyledons, monocotyledones, Amborellales) using gene tree tool of Pan-taxonomic Compara (http://www.gramene.org/). TEM1 protein sequence was used as query in a BLAST-P (https://blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE=BlastSearch&PROGRAM=blastp) search against the proteomes of Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa).
TF binding sites (CArG box for MADS-domain proteins, consensus sequence composed of CAACA and CCTG elements at a distance of 3-9 nucleotides for RAV proteins) were searched in the regulatory regions of genes of interest by using the Promoter Analysis tool of Plant PAN3.0 (http://plantpan.itps.ncku.edu.tw/index.html).
Gene and protein sequences were retrieved from The Arabidopsis Information Resource (www.arabidopsis.org) and GRAMENE (www.gramene.org).
Phylogenetic Analyses
Analysis of phylogenetic relationships between 24 RAV-related full-length protein sequences and construction of phylogenetic tree were performed using tools available at http://www.phylogeny.fr/ (Dereeper at al., 2008). MUSCLE was used for protein sequences alignment, and G-blocks for a more stringent selection (https://www.ebi.ac.uk/Tools/msa/muscle). PhyML was used for phylogenetic analysis with bootstrapping procedure (n = 100) as statistical test for branch support (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=phyml), and TreeDyn for tree visualization (http://www.phylogeny.fr/one_task.cgi?task_type=treedyn). Sequence Diversity Diagram (SeDD; http://vda-lab.github.io/sedd.html) was used to compare two sets of RAV protein sequences from Arabidopsis and Oryza sativa and visualize conserved versus diversified positions in the two species. Motif clustering analysis was carried out using SALAD version 3 (https://salad.dna.affrc.go.jp/salad/en/) with OsRAV9 sequence, and the prediction of functional and structural motifs in RAV protein sequences via web-based tools of ExPAsY (https://prosite.expasy.org/scanprosite; de Castro et al., 2006).
Expression Profiles
Expression profiles of rice RAV genes were inferred from a large collection of microarray data derived from different tissues at different developmental stages under natural field conditions (http://ricexpro.dna.affrc.go.jp/). Coexpression analyses were carried out by using OsRAV9 as single guide genes and searching RiceFREND (http://ricefrend.dna.affrc.go.jp/), the Plant Coexpression Database (PLANEX; Yim et al., 2013), and the Rice Oligonucleotide Array Database (Cao et al., 2012).
Plant Material and Growth Conditions
Wild-type and transgenic Arabidopsis (Col-0 background) seeds were sown on soil pots, and plants were grown under LD (16-h light/8-h dark at 22°C) until maturity. Wild-type and transgenic rice (O. sativa ssp. japonica) seeds were surface sterilized and sown on Murashige Skoog medium with 30 g L−1 Suc. After germination, seedlings were transferred to soil pots and grown under controlled conditions until maturity (SD: 8-h light at 28°C, 16-h dark at 24°C; 12/12: 12-h light at 28°C, 12-h dark at 24°C or LD; LD:16-h light at 28°C, 8-h dark at 24°C). For functional characterization of OsRAV genes, transgenic lines (cv Nipponbare) and insertion mutant (PFG_2A-10680, cv Hwayoung) were used. For expression analysis, segregating progenies of floral homeotic mutants (cv Dongjin) were genotyped (Supplemental Table S3), grown for 10 weeks in LD and then transferred to inductive conditions. Pools of developing inflorescences were harvested 3 weeks after floral transition from homozygous mutants (osmads1, osmads13, osmads16) and wild-type plants.
Cloning and Generation of Arabidopsis and Rice Transgenic Plants
The coding sequence of OsRAV9/OsTEM1 was amplified using primer sets SPp538-SPp541, subcloned in pCRII and pENTR-3C by restriction/ligation, and introduced in pALLIGATOR2 vector downstream of Pro-35S by Gateway technology (Supplemental Table S4). Arabidopsis plants (Col-0, tem1-tem2) were transformed with the Pro-35S:OsTEM1 construct by floral dip, and GFP-positive seeds were selected by fluorescence microscopy. For the generation of the RNAi constructs, the Gene Sequence Tags specific for the 3′ ends of OsRAV9/OsTEM1 and OsRAV11 were amplified using primer sets SPp516-SPp539 and SPp527-SPp537, respectively, then subcloned in pCRII, and afterward cloned in pENTR-3C by restriction/ligation (as KpnI-EcoRV fragments). The resulting constructs were digested with PvuI before LR recombination to pBios-378 plant expression vector. Scutellum-derived rice calli (‘Nipponbare’) were transformed by Agrobacterium spp. cocultivation. Independent transformation events were selected, 11 for OsRAV9 and 7 for OsRAV11 constructs, regenerated and propagated. Plants that underwent regeneration but did not contain the transgenic cassette were used as transformation control. The primers used for genotyping and cloning are listed in Supplemental Tables S3 and S4.
Direct Binding of OsTEM1 to Downstream Genes
To generate the set of reporter vectors, different promoter regions of OsMADS14 and Hd3a were cloned as SalI-PstI fragments in a modified pGreenII 0800-LUC carrying Pro-35S:LUC and Pro-35S:REN (as internal control to estimate the proportion of transformed protoplasts). Primers SPp1764- SPp1765 were designed to introduce mutations at the RAV binding site contained in ProOsMADS14 by PCR-based method. The corresponding reporter vector was used as template for site-directed mutagenesis (15 cycles: 10´´ at 98°C, 20´´ at 66°C, 20´´ at 66°C), and the resulting vector used in transactivation assays. Protoplasts were isolated from calli by digesting the cell wall with Macerozyme R-10 and Cellulase (Yakult Pharmaceuticals), and transfected with different combinations of reporter Pro-35S:OsTEM1 and effector constructs using polyethylene glycol. After 18 h incubation in darkness at 24°C, transformed protoplasts were pelleted and resuspended in homogenization buffer for RNA extraction. Transactivation activity of OsTEM1, based on the relative ratio of mRNA abundance of LUC and REN reporter genes, was assessed by RT-qPCR. The primers used for cloning and expression analyses are listed in Supplemental Tables S3 and S4.
RNA Extraction and Expression Analyses
For expression analyses in Arabidopsis, pools of 20 seedlings grown for 1 week under LD were collected at ZT12. For expression analyses in rice, pools of 10 to 15 samples from different tissues and/or developmental stages were collected at ZT13 unless otherwise stated. RNA was extracted with PureLink RNA mini kit (Ambion) and treated with DNaseI RNase free (Ambion). For large-scale experiments, RNA was extracted with Maxwell RSC Plant RNA kit (Promega), and DNase treatment was performed on-column. Then 1 μg of DNase-treated RNA was retro-transcribed with SuperScript III (Invitrogen), and cDNA was used for RT-qPCR with Light Cycler 480 SYBR Green I master on Light Cycler 480 II (Roche). Three biological replicates and three technical replicates were performed. In situ hybridization was performed as previously reported (Dreni et al., 2007). The primers used for expression analyses are listed in Supplemental Table S5.
Phenotypic analyses
Morphological analysis of reproductive structures was performed by Optical Microscopy (Olympus DP71) and SEM. For SEM, flowers at anthesis were fixed in 2.5% (v/v) glutaraldehyde in 0.1 m P-buffer (pH 7.4) overnight at 4°C, washed 4 times for 10 min in 0.1 m P-buffer, postfixed in 1% osmium tetraoxide with 0.7% ferrocyanide in P-buffer, washed in water, dehydrated in an ascending ethanol series (50%, 70%, 80%, 90%, and 95% for 10 min each and twice with 100% ethanol), and dried by critical-point drying with CO2. Alteration on the morphology of the seed were analyzed by using the Smart-grain software (Tanabata et al., 2012). At least 100 seeds per genotype (for three independent biological replicates) were spread uniformly on the glass with a black background, and images were captured with HP Scanner at 300 dpi. The software determined seed shape parameters such as seed length, width, perimeter, and area, and also calculated length-to-width ratio and circularity.
Statistical Analyses
All statistical analyses are shown in Supplemental Table S6. Statistical significance of each experiment was determined by using GraphPad Prism 7 (https://www.graphpad.com/scientific-software/prism). For flowering time phenotype, we chose one-way ANOVA; multiple comparisons were then corrected with Dunnet's or Dunn's tests. For other kind of data, we compared two columns by using unpaired Student’s two-tailed t test, with confidence interval at 95%.
Following instructions of GraphPad Prism, we first transformed ratios in log of ratios (Y = log(Y)), for Figure 5 and Supplemental Figure S11. Then we created a column data table and entered two columns of data (control – OsTEM1, +OsTEM1) with matched values on the same row. We chose t tests from the list of column analyses and selected conditions as follows: Experimental design: Paired; Assume Gaussian distribution: Yes; Choose test: Ratio paired t test. On the second tab of the t test dialog, we chose to compute +OsTEM1 versus −OsTEM1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers. AtRAV locus identifiers: AT1G13260 (AtRAV1), AT3G25730 (AtRAV1-like), At1g25560 (AtRAV2-like/TEM1), AT1G68840 (RAV2/TEM2), At1g50680 (AtRAV3), At1g51120 (AtRAV3-like). OsRAV locus identifiers: LOC_Os10g39190 (OsRAV2), LOC_Os08g06120 (OsRAV3), LOC_Os03g02900 (OsRAV4), LOC_Os04g49230 (OsRAV5), LOC_Os02g45850 (OsRAV6), LOC_Os11g05740 (OsRAV7), LOC_Os01g04750 (OsRAV8), LOC_Os01g04800 (OsRAV9/OsTEM1), LOC_Os01g49830 (OsRAV11), LOC_Os05g47650 (OsRAV12). OsMADS locus identifiers: LOC_Os03g54160 (OsMADS14), LOC_Os07g01820 (OsMADS15), LOC_Os07g41370 (OsMADS18), LOC_Os03g54170 (OsMADS34), LOC_Os03g03100 (OsMADS50/OsSOC1), LOC_Os01g69850 (OsMADS51).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Phylogenomic analysis of TEM homologs in sequenced land plants.
Supplemental Figure S2. Analysis of genes encoding AP2-B3 (RAV) proteins.
Supplemental Figure S3. Similarity clustering based on distribution patterns of known conserved motifs in RAV proteins.
Supplemental Figure S4. Similarity clustering based on distribution patterns of conserved features related to posttranslational modification.
Supplemental Figure S5. Analysis of RAV genes in O. sativa ssp. japonica.
Supplemental Figure S6. GO analysis of genes strongly coexpressed with OsRAV9.
Supplemental Figure S7. GO test of genes negatively correlated with OsRAV9 and expression analysis of genes acting in apical meristems at floral transition in rice.
Supplemental Figure S8. Inferred expression profiles of OsRAV genes during plant development.
Supplemental Figure S9. Analysis of T1 and T2 transgenic lines ectopically expressing OsRAV9 and OsRAV11 in Arabidopsis.
Supplemental Figure S10. Molecular and phenotypic analysis of T1 transgenic rice lines silencing OsRAV9.
Supplemental Figure S11. Transactivation activity of OsTEM1 on floral activators in rice protoplasts.
Supplemental Figure S12. Expression analyses of genes involved in the floral transition in OsRAV9-i rice lines.
Supplemental Figure S13. Analyses of OsRAV11 and OsRAV12.
Supplemental Figure S14. Molecular characterization of OsRAV11 and OsRAV12.
Supplemental Figure S15. Analysis of RAV proteins from Arabidopsis and O. sativa.
Supplemental Table S1. List of RAV genes in O. sativa reported by Swaminathan et al. (2008).
Supplemental Table S2. List of genes encoding B3-domain TFs down-regulated in OsMADS1 knock-down panicles (modified from Khanday et al., 2013).
Supplemental Table S3. List of primers used for genotyping.
Supplemental Table S4. List of primers used for cloning.
Supplemental Table S5. List of primers used for expression analyses.
Supplemental Table S6. Statistical analyses.
Supplemental Dataset S1. Coexpression analyses
Acknowledgments
We thank Dr. Marti Bernardo, Dr. Jordi Morata, and Dr. Victor Gonzalez for advice on bioinformatic analyses. Dr. Sebastien Santini for the Phylogeny.fr tool. Elia Lacchini for technical help with set-up of genotyping of osmads1 mutants. Dr. Ludovico Dreni for help with rice transgenic plants. Pilar Fontanet for advice and technical help on protoplast co-transformation assays. Dr. Paula Suárez-López for critical reading of the manuscript. We also thank the Servei de Microscopia at the Universitat Autònoma de Barcelona for help with SEM. A.E.A.-J. performed this work within the frame of a Ph.D. Program of the Universitat Autònoma de Barcelona.
Footnotes
This work was supported by the Ministerio de Ciencia, Tecnología e Innovación Productiva (grant no. PGC2018–095804–B–I00) the Ministerio de Economía y Competitividad (grant no. BFU2015–64409–P), Government of Catalonia | Agència de Gestió d'Ajuts Universitaris i de Recerca (CERCA Programme, Consolidated Research Group no. 2014 SGR 1406 and Investigator Training Program PhD fellowship to A.E.A.-J.), Flower Power (to M.M.K. and S.P.), EvoRepRice (FIRST; to M.M.K.), the EMBO short-term fellowship (to M.O.), and the Ministerio de Economía y Competitividad through the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016–2019 (grant no. SEV–2015 0533).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T(2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H(2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Aguilar-Jaramillo AE, Marín-González E, Matías-Hernández L, Osnato M, Pelaz S, Suárez-López P(2019) TEMPRANILLO is a direct repressor of the microRNA miR172. Plant J 100: 522–535 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G(2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H(2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D(1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, et al. (2017) Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 29: 2801–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Jung K-H, Choi D, Hwang D, Zhu J, Ronald PC(2012) The rice oligonucleotide array database: An atlas of rice gene expression. Rice (N Y) 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S(2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Choudhury S, Panda P, Sahoo L, Panda SK(2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav 8: e23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collani S, Neumann M, Yant L, Schmid M(2019) FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol 180: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F(2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N(2006) ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B(2015) Direct links between the vernalization response and other key traits of cereal crops. Nat Commun 6: 5882. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A(2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PBF, An G, Colombo L, Kater MM(2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM(2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan YB, Li J, Qin RY, Xu RF, Li H, Yang YC, Ma H, Li L, Wei PC, Yang JB(2016) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol Biol 90: 49–62 [DOI] [PubMed] [Google Scholar]
- Endo-Higashi N, Izawa T(2011) Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol 52: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O(2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q(2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Feng J-X, Liu D, Pan Y, Gong W, Li-Geng M, Luo J-C, Deng X W, Zhu Y-X(2005) An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol 59: 853–868 [DOI] [PubMed] [Google Scholar]
- Fjellheim S, Boden S, Trevaskis B(2014) The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front Plant Sci 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM(2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Kang HK, Son SH, Kim SK, Nam KH(2014) A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol 55: 1892–1904 [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J, Brambilla V, Vicentini G, Landini M, Cerise M, Carrera E, Shrestha R, Chiozzotto R, Galbiati F, Caporali E, et al. (2019) A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat Plants 5: 358–362 [DOI] [PubMed] [Google Scholar]
- Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B(2010) ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol 153: 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen SC, Ćalić I, Joly-Lopez Z, Platts AE, Choi JY, Natividad M, Dorph K, Mauck WM III, Bracken B, Cabral CLU, et al. (2020) The strength and pattern of natural selection on gene expression in rice. Nature 578: 572–576 [DOI] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX(2018) GRAIN SIZE AND NUMBER1 negatively regulates the OSMKKK10-OSMKK4-OSMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P(2000) Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T, King RW(2008) The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J Exp Bot 59: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY(2004) Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res 14: 8–15 [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X(2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T(2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Itoh J-I, Nonomura K-I, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y(2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 2347. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA(2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G(2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua N-H, Park C-M(2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T(1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D(1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Khanday I, Das S, Chongloi GL, Bansal M, Grossniklaus U, Vijayraghavan U(2016) Genome-wide targets regulated by the OsMADS1 transcription factor reveals its DNA recognition properties. Plant Physiol 172: 372–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanday I, Yadav SR, Vijayraghavan U(2013) Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol 161: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G(2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J(2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J(2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T(1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M(2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K(2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K(2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH(2013) Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH(2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H(2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D(2011) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xu R, Duan P, Li Y(2018) Control of grain size in rice. Plant Reprod 31: 237–251 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ(2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X, et al. (2018) G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat Commun 9: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-González E, Matías-Hernández L, Aguilar-Jaramillo AE, Lee JH, Ahn JH, Suárez-López P, Pelaz S(2015) SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant Physiol 169: 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M(2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M(2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernández L, Aguilar-Jaramillo AE, Marín-González E, Suárez-López P, Pelaz S(2014) RAV genes: Regulation of floral induction and beyond. Ann Bot 114: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matías-Hernández L, Aguilar-Jaramillo AE, Osnato M, Weinstain R, Shani E, Suárez-López P, Pelaz S(2016) TEMPRANILLO reveals the mesophyll as crucial for epidermal trichome formation. Plant Physiol 170: 1624–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T(2010) SALAD database: A motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res 38: D835–D842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I(2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y(2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Nonoue Y, Yano M, Izawa T(2016) Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J 86: 221–233 [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T(2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S(2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Plessis A, Hafemeister C, Wilkins O, Gonzaga ZJ, Meyer RS, Pires I, Müller C, Septiningsih EM, Bonneau R, Purugganan M(2015) Multiple abiotic stimuli are integrated in the regulation of rice gene expression under field conditions. eLife 4: e08411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M(2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U(2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Riechmann JL. (2002) Transcriptional regulation: A genomic overview. The Arabidopsis Book 1: e0085, doi:10.1199/tab.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K(2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: 2280. [DOI] [PubMed] [Google Scholar]
- Sato Y, Antonio B, Namiki N, Motoyama R, Sugimoto K, Takehisa H, Minami H, Kamatsuki K, Kusaba M, Hirochika H, et al. (2011) Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H, Ohyanagi H, Sugimoto K, Itoh J, Antonio BA, et al. (2013) RiceFREND: A platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res 41: D1214–D1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA(2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU(2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Scortecci K, Michaels SD, Amasino RM(2003) Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol 52: 915–922 [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM(2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26: 229–236 [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, et al. (2009) A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J 58: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R, Gómez-Ariza J, Brambilla V, Fornara F(2014) Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann Bot 114: 1445–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G(2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T(2008) The plant B3 superfamily. Trends Plant Sci 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K(2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tanabata T, Shibaya T, Ebana K, Yano M(2012) SmartGrain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol 160: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka C, Itoh T, Iwasaki Y, Mizuno N, Nasuda S, Murai K(2018) Direct interaction between VRN1 protein and the promoter region of the wheat FT gene. Genes Genet Syst 93: 25–29 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Itoh H, Sentoku N, Kojima M, Sakakibara H, Izawa T, Itoh J, Nagato Y(2011) The COP1 ortholog PPS regulates the juvenile-adult and vegetative-reproductive phase changes in rice. Plant Cell 23: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H(2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plan J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 Proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G(2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K(2013) Florigen in rice: Complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol 16: 228–235 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D(2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wilkins O, Hafemeister C, Plessis A, Holloway-Phillips MM, Pham GM, Nicotra AB, Gregorio GB, Jagadish SV, Septiningsih EM, Bonneau R, et al. (2016) EGRINs (Environmental Gene Regulatory Influence Networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell 28: 2365–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR(1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ(2009) Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song IJ, Kim JH, Lee HY, Nam HG, Lim PO(2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61: 3947–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim WC, Yu Y, Song K, Jang CS, Lee B-M(2013) PLANEX: The plant co-expression database. BMC Plant Biol 13: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]