Differences in outputs between Fls3 and Fls2immunity receptors suggest they use distinct mechanisms to activate immunity in tomato.
Abstract
Plants mount defense responses by recognizing indicators of pathogen invasion, including microbe-associated molecular patterns (MAMPs). Flagellin, from the bacterial pathogen Pseudomonas syringae pv. tomato (Pst), contains two MAMPs, flg22 and flgII-28, that are recognized by tomato (Solanum lycopersicum) receptors Flagellin sensing2 (Fls2) and Fls3, respectively, but to what degree each receptor contributes to immunity and whether they promote immune responses using the same molecular mechanisms are unknown. Here, we characterized CRISPR/Cas9-generated Fls2 and Fls3 tomato mutants and found that the two receptors contribute equally to disease resistance both on the leaf surface and in the apoplast. However, we observed striking differences in certain host responses mediated by the two receptors. Compared to Fls2, Fls3 mediated a more sustained production of reactive oxygen species and an increase in transcript abundance of 44 tomato genes, with two genes serving as specific reporters for the Fls3 pathway. Fls3 had greater in vitro kinase activity than Fls2 and could transphosphorylate a substrate. Using chimeric Fls2/Fls3 proteins, we found no evidence that a single receptor domain is responsible for the Fls3-sustained reactive oxygen species, suggesting involvement of multiple structural features or a nullified function of the chimeric construct. This work reveals differences in certain immunity outputs between Fls2 and Fls3, suggesting that they might use distinct molecular mechanisms to activate pattern-triggered immunity in response to flagellin-derived MAMPs.
Pseudomonas syringae pv. tomato (Pst) causes bacterial speck disease on tomato (Solanum lycopersicum), manifesting as small, necrotic lesions on leaves, stems, fruit, and flowers (Jones, 1991). The tomato-Pst pathosystem serves as a model for studying bacterial pathogenesis and plant immunity (Pedley and Martin, 2003; Oh and Martin, 2011). The virulence of Pst is primarily determined by a suite of 36 type III effectors, which are translocated into the plant cell during the infection process. Two effectors, AvrPto and AvrPtoB, play major roles in interfering with plant immune responses and promote enhanced multiplication of Pst in the leaf apoplast (Buell et al., 2003; Lin and Martin, 2005; Kvitko et al., 2009; Cunnac et al., 2011; Martin, 2012).
Plants detect pathogens by recognizing conserved microbe-associated molecular patterns (MAMPs) via pattern recognition receptor (PRR)-triggered immunity (PTI) or by recognizing pathogen effectors through nucleotide-binding leucine-rich repeat (LRR) receptor (NLR)-triggered immunity (NTI). For Pst, the motility-associated protein flagellin contains two MAMPs, flg22 and flgII-28, that are recognized by the PRRs Flagellin sensing2 (Fls2) and Fls3, respectively (Gómez-Gómez and Boller, 2000; Robatzek et al., 2007; Hind et al., 2016). Upon recognition of these and other MAMPs, a suite of molecular events occurs to promote defense, including the production of reactive oxygen species (ROS), activation of a mitogen-activated protein kinase (MAPK) cascade, transcriptional reprogramming, callose deposition at the cell wall, stomatal closure, and calcium fluxes (Couto and Zipfel, 2016; Li et al., 2016). While PTI has generally been associated with a moderate inhibition of bacterial pathogen growth in Arabidopsis (Arabidopsis thaliana) and tomato (∼10-fold; Zipfel et al., 2004; Lacombe et al., 2010; Schwizer et al., 2017), a recent study shows that flagellin-mediated PTI plays a major role in immunity on the leaf surface for some tomato accessions, decreasing bacterial populations by ∼150-fold (Roberts et al., 2019b).
Fls2 and Fls3 bind flg22 and flgII-28 through their extracellular LRR domain. In Arabidopsis, upon binding flg22, FLS2 associates with the coreceptor BRI1-ASSOCIATED RECEPTOR KINASE (BAK1), and both FLS2 and BAK1 are transphosphorylated to initiate downstream signaling (Sun et al., 2013; Couto and Zipfel, 2016; Saijo et al., 2018). Tomato has two Fls2 genes, Fls2.1 (Solyc02g070890) and Fls2.2 (a paralog of Fls2.1 located 3.8 kb away from Fls2.1; Solyc02g070910). Fls2.1 appears to encode the only functional Fls2 in tomato, as a mutation in Fls2.1 causes a complete loss of flg22 recognition (Jacobs et al., 2017). Not all solanaceous species have Fls3. While tomato, potato (Solanum tuberosum), and pepper (Capsicum annuum) respond to flgII-28, Nicotiana benthamiana and petunia (Petunia axillaris) do not (Cai et al., 2011; Clarke et al., 2013; Hind et al., 2016). Previous studies have examined MAPK activation and gene expression as immunity outputs of Fls2 and Fls3 and have found some similarities between the components involved in Fls2 and Fls3 signaling in tomato (Rosli et al., 2013; Hind et al., 2016). Similar to Fls2.1, Fls3 gene expression is induced in leaves after treatment with flg22, flgII-28, or DC3000∆avrPto∆avrPtoB (Rosli et al., 2013; Hind et al., 2016). After flgII-28 treatment, there is an increase in the phosphorylation of MAPKs in protoplasts transfected with Fls3 (Hind et al., 2016). Additionally, N. benthamiana plants silenced for Bak1 have a reduced flgII-28 ROS response compared to control plants, and Fls3 and Arabidopsis BAK1 interact upon flgII-28 treatment when they are coexpressed in N. benthamiana leaves (Hind et al., 2016). Together, these observations suggest that there may be some similar factors involved in Arabidopsis FLS2 and tomato Fls3 signaling, but further analysis is needed to determine the molecular mechanisms of Fls3.
Fls2 and Fls3 belong to a family of non-RD kinases since they lack conserved Arg and Asp residues in the activation loop. In Arabidopsis, FLS2 has weak autophosphorylation activity that requires the presence of the entire FLS2 intracellular domain, including the inner juxtamembrane domain (iJM) and the kinase domain (KD; Gómez-Gómez et al., 2001; Xiang et al., 2008; Lu et al., 2010; Cao et al., 2013). Chimeric constructs containing different domains of PRRs have been made to study which domains are important for receptor functions (Albert and Felix, 2010; Albert et al., 2010; Brutus et al., 2010; Mueller et al., 2012; Kouzai et al., 2013; Holton et al., 2015; Hohmann et al., 2018; Wu et al., 2019; Zhou et al., 2020). Chimeric constructs combining the ectodomain of Arabidopsis receptor-like kinase (RLK) EF-Tu receptor (EFR), which detects the bacterial MAMP Ef-Tu, with the intracellular domain of cell wall-associated kinase AtWAK1, which recognizes oligogalacturonides released from the cell wall, results in a functional chimeric protein that recognizes Ef-Tu and activates AtWAK1-specific plant defenses. However, combining the AtWAK1 ectodomain with the EFR intracellular domain does not result in effective defense, as it does not cause a significant reduction in bacterial growth compared to the negative control (Brutus et al., 2010). Combining the FLS2 ectodomain with the EFR intracellular domain also results in a functional protein (Brutus et al., 2010). A chimeric cross-species protein combining the Arabidopsis EFR extracellular domain with the transmembrane and intracellular domains of rice (Oryza sativa) Xa21 (an LRR-RLK that recognizes the sulfated protein RaxX of Xanthamonas oryzae pv. oryzae and encodes resistance) was also functional (Holton et al., 2015). Chimeric constructs swapping Arabidopsis FLS2 and tomato Fls2.1 LRRs aided the authors in finding specific LRR repeats in tomato that are responsible for recognizing the flg15 peptide, which is not recognized by Arabidopsis FLS2 (Robatzek et al., 2007; Mueller et al., 2012).
NLR-triggered immunity is activated upon the recognition of pathogen effectors by NLRs. In the case of bacterial-plant interactions, this causes a suite of molecular events including activation of a MAPK cascade, production of ROS, transcriptional reprogramming, and localized, controlled cell death (hypersensitive response), that is typically associated with a significant inhibition of growth (∼100- to 1,000-fold; Chandra et al., 1996; Jia and Martin, 1999; Abramovitch et al., 2003; Pedley and Martin, 2004; Lolle et al., 2020; Thomas et al., 2020)). The Pst effectors AvrPto and AvrPtoB are recognized by the cytoplasmic kinase Pto from tomato, which acts with the NLR Prf to activate the immune response and results in cell death (Salmeron et al., 1996; Lin and Martin, 2007; Xing et al., 2007; Dong et al., 2009; Mucyn et al., 2009; Gutierrez et al., 2010; Martin, 2012; Mathieu et al., 2014). AvrPto and AvrPtoB are unrelated effectors, but both are bound by Pto.
Here, we investigated some of the similarities and differences in the immunity outputs between Fls2 and Fls3 in tomato, including ROS production, transcript abundance of pathway-specific reporter genes, kinase activity, and whole-plant responses to Pst infection, and we present data suggesting there may be some differences in the molecular signaling between these two flagellin-sensing receptors in tomato.
RESULTS
Fls3 and Fls2 Each Contribute to Disease Resistance in Tomato
We used the CRISPR/Cas9 gene-editing system to develop tomato lines that were insensitive to the peptides flgII-28, flg22, or both flgII-28 and flg22. In the background of Rio Grande-prf3 (RG-prf3), which has a 1.1-kb deletion in Prf that inactivates the defense responses associated with bacterial effectors AvrPto and AvrPtoB (Salmeron et al., 1994), we generated mutations in Fls3 alone (∆Fls3), Fls2.1 alone (∆Fls2.1), both Fls2.1 and its paralog Fls2.2 together (∆Fls2.1/2.2), and Fls2.1, Fls2.2, and Fls3 together (∆Fls2.1/2.2/3) and verified the presence of mutations using Sanger sequencing (Supplemental Fig. S1A; Supplemental Table S1; Supplementary Methods). The CRISPR/Cas9-generated mutations resulted in a 1-bp insertion (+1 bp) in the guide RNA region of Fls3 for ∆Fls3, a 7-bp deletion (−7 bp) in Fls2.1 for ∆Fls2.1, a 7-bp deletion in Fls2.1 and a 1-bp insertion (+1 bp) in Fls2.2 for ∆Fls2.1/2.2, and a 1-bp insertion in each of the three Fls genes for ∆Fls2.1/2.2/3. Loss of flgII-28 or flg22 peptide recognition was confirmed in each mutant line using ROS assays (Supplemental Fig. S1B; Supplemental Table S1).
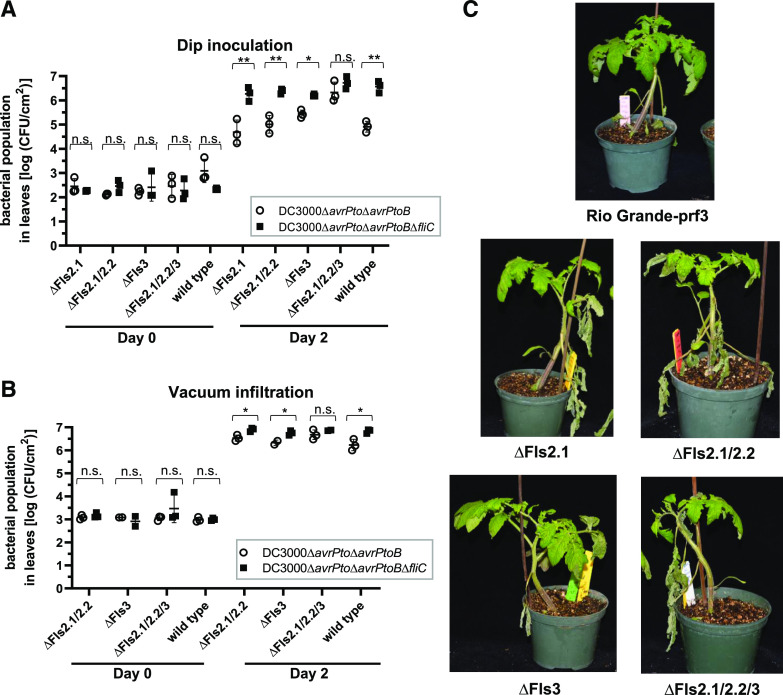
To test the contribution of Fls3 and Fls2 to flagellin recognition on the leaf surface and mounting of defense responses, the four mutant lines and wild-type RG-prf3 were dip inoculated with Pst DC3000 strains deleted for the effector genes avrPto and avrPtoB (DC3000∆avrPto∆avrPtoB) or avrPto, avrPtoB, and fliC, the latter of which encodes the flagellin protein that makes up the bacterial flagellum (DC3000∆avrPto∆avrPtoB∆fliC). Bacterial growth in leaves was measured 2 d after inoculation. In the wild-type, ∆Fls2.1, ∆Fls2.1/2.2, and ∆Fls3 lines, we observed significant differences in bacterial populations between the two strains, suggesting that all these lines are able to recognize flagellin. However, in the ∆Fls2.1/2.2/3 line there was no difference in bacterial growth between DC3000∆avrPto∆avrPtoB and DC3000∆avrPto∆avrPtoB∆fliC (Fig. 1A; Supplemental Fig. S2A).
Figure 1.
Both Fls2 and Fls3 contribute to disease resistance on the leaf surface and in the leaf apoplast in tomato. A and B, Bacterial populations in tomato leaves of CRISPR/Cas9-generated mutants of Fls2.1 (∆Fls2.1), Fls3 (∆Fls3), Fls2.1 and Fls2.2 (∆Fls2.1/2.2), or Fls2.1, Fls2.2, and Fls3 (∆Fls2.1/2.2/3), or of RG-prf3 (wild type) were measured 0 and 2 d after dip-inoculating plants in bacterial suspensions (1 × 108 CFU mL−1; A), or vacuum-infiltrating plants with bacterial suspensions (1 × 104 CFU mL−1; B). The means of three individual plants are indicated as separate points. For each population, the top and bottom horizontal lines show the ± sd, and the middle horizontal line is the mean of the three plants. Statistical significance was determined by pairwise t test (*P < 0.05, and **P < 0.01 or P > 0.05; ns, not significant). C, Photos of CRISPR/Cas9-generated mutant plants vacuum infiltrated with DC3000∆avrPto∆avrPtoB at 1 × 104 CFU mL−1, taken 1 week postinoculation. Experiments were repeated three times with similar results, and data are representative of a single replicate. (See also Supplemental Figs. S1 and S2; Supplemental Tables S1 and S2.)
We next tested whether Fls3 and Fls2 contribute to disease resistance in the leaf apoplast by vacuum infiltrating DC3000∆avrPto∆avrPtoB and DC3000∆avrPto∆avrPtoB∆fliC into ∆Fls2.1/2.2, ∆Fls3, and ∆Fls2.1/2.2/3 plants and the wild-type line (as a control) and measuring bacterial growth 2 d later. Similar to dip-inoculated plants, we observed differences in bacterial populations between the two strains for all of the lines except ∆Fls2.1/2.2/3. However, the differences in the populations observed for the vacuum-inoculated ∆Fls2.1/2.2, ∆Fls3, and RG-prf3 were much smaller compared to the dip-inoculated plants, with about 3-fold differences in bacterial populations when vacuum infiltrated versus 10- to 40-fold differences when dip inoculated (Fig. 1B; Supplemental Fig. S2B). Along with the observations in Figure 1A, this suggests that Fls3 and Fls2 act both on the leaf surface and in the apoplast, but their effect is greater on the leaf surface. This agrees with observations that bacterial entry through stomata elicits a strong plant defense response (Melotto et al., 2008; Roberts et al., 2019b). We also observed that, compared to RG-prf3, speck symptoms were more severe on the plants that are unable to recognize flg22 and/or flgII-28 (∆Fls2.1, ∆Fls2.1/2.2, ∆Fls3, and ∆Fls2.1/2.2/3). Symptoms were similar on ∆Fls2.1, ∆Fls2.1/2.2, and ∆Fls3 plants, and were slightly more severe on ∆Fls2.1/2.2/3 plants (Fig. 1C). Together, these observations suggest that Fls3 and Fls2 both make important contributions to Pst resistance in tomato.
The ROS Response Induced by flgII-28 Is More Sustained Compared to That Induced by flg22
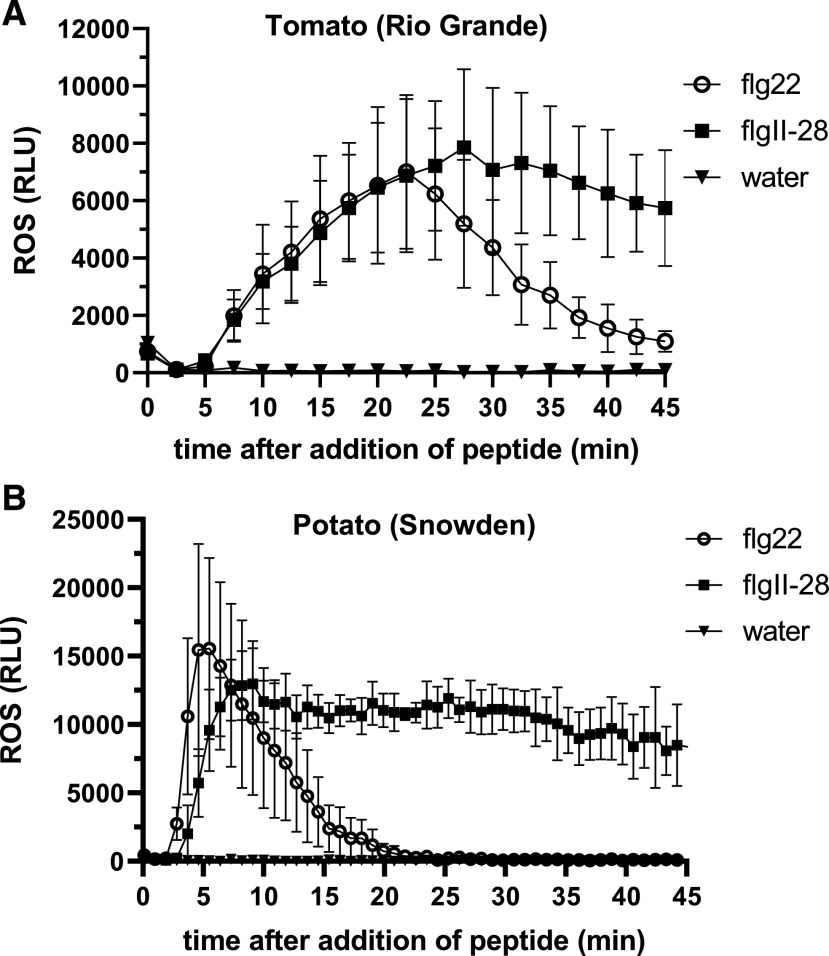
We previously observed that a more sustained ROS response was mediated by Fls3 compared to Fls2 in an accession of the wild species Solanum pimpinellifolium and hypothesized that this might indicate that the two PRRs use different molecular mechanisms (Hind et al., 2016). To follow up on this observation, we tested whether tomato (RG-prf3) had a sustained ROS response to flgII-28 compared to flg22. As previously observed, we saw a sustained ROS response for flgII-28 compared to flg22 (Fig. 2A; Supplemental Fig. S3, A and B). While the amplitude of the flgII-28 and flg22 responses was similar, the flgII-28 ROS response continued for up to 100 min versus 60 min for flg22 (Fig. 2A). To see the generality of this sustained response, we tested another tomato accession (CR293), a tomato heirloom (Galina), and a tomato breeding line (OH05-8144) and found that the flgII-28 response was sustained in all of these lines compared to flg22 (Supplemental Fig. S3B).
Figure 2.
The flgII-28 ROS response is sustained in both tomato and potato. Oxidative burst produced over 45 min in response to 100 nm flgII-28 or flg22 peptide or water in tomato variety RG-prf3 (A) or potato variety ‘Snowden’ (B). ROS was measured in relative light units (RLU), and results shown are means ± sd (n = 4 for tomato, n = 3 for potato) and are representative of three independent experiments. See also Supplemental Figures S3 and S4.
To investigate whether this sustained ROS response was specific to tomato, we also tested the flgII-28 ROS response in another solanaceous species, potato, for which flgII-28 is a major MAMP (Clarke et al., 2013; Moroz and Tanaka, 2019). We found that two potato varieties, ‘Snowden’ and ‘Dakota Crisp’, both had a sustained ROS response for flgII-28 compared to flg22, though the amplitude of the response to both MAMPs was similar (Fig. 2B; Supplemental Fig. S3C). Our observation of a sustained ROS response in potato is in agreement with previous studies (Clarke et al., 2013; Moroz and Tanaka, 2019). Because only one variety of eggplant (Solanum melongena MM643) was previously tested and shown nonresponsive to flgII-28 (Clarke et al., 2013), we tested the eggplant variety ‘Shikou’ for its flgII-28 response and found that it did not respond to flgII-28 (Supplemental Fig. S4). A previous study observed a sustained ROS response for pepper (‘Jalapeno Early’; Clarke et al., 2013). Together, our data and previous reports suggest that the flgII-28 extended ROS response is conserved in all solanaceous species that respond to flgII-28.
Fls3 Specifically Regulates Expression of a Set of Defense-Related Genes in Tomato Leaves
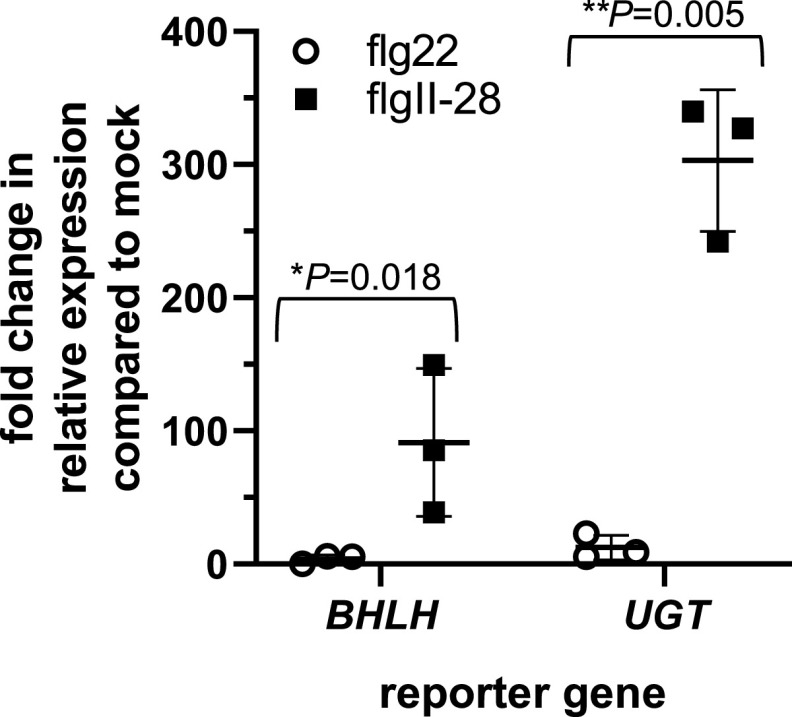
Reporter genes are useful for monitoring the activity of specific signaling pathways. From data generated in a previous RNA sequencing (RNA-Seq)analysis of flagellin-induced genes in tomato (Rosli et al., 2013; Pombo et al., 2017) we identified 44 genes whose transcript abundance was significantly increased 6 h after applying flgII-28 but not after flg22 application (Supplemental Table S2). Gene ontology term enrichment analysis revealed that many of these genes were defense related, including genes related to proteolysis, peptidase and hydrolase activity, and metabolism (Supplemental Table S3). Among these genes, the two most highly induced encode a basic helix-loop-helix (bHLH) transcription factor (bHLH; Solyc03g114230) and a UDP-glucosyltransferase (UGT; Solyc09g098080). To test whether these genes might serve as specific reporters for the Fls3 pathway, we used reverse transcription quantitative (RT-qPCR) to estimate gene expression in leaves of bHLH and UGT, after infiltrating with 1 μm flgII-28, flg22, or water as a control. Transcript abundance of both bHLH and UGT significantly increased 6 h after flgII-28 treatment compared to flg22 treatment (Fig. 3). After flgII-28 treatment, bHLH gene expression increased by ∼110-fold and UGT by ∼300-fold relative to water treatment. Comparatively, flg22 induced expression of bHLH by just 3-fold and UGT by 12-fold. These data suggest that bHLH and UGT can serve as specific reporters for the Fls3 response to flgII-28.
Figure 3.
Expression of two tomato genes is induced by the Fls3 pathway but not the Fls2 pathway. RT-qPCR of the two most highly induced genes (bHLH transcription factor [bHLH, Solyc03g114230] and a UDP-glucosyltransferase [UGT, Solyc09g098080]) was performed 6 h after infiltrating with 1 μm flg22 or flgII-28 peptide or water and normalized to an internal control (ARD2). Data shown are the average fold change in expression compared to the water control (mock) for three individual plants from a single experimental replicate shown as separate points. The top and bottom horizontal lines show the ± sd, and the middle horizontal lines are the means of the three plants. Significance was determined by pairwise t test and P values are shown above each pairwise comparison. Asterisks represent significance determined by pairwise t test (*P < 0.05 and **P < 0.01). Results are representative of three independent experiments.
Fls3 and Fls2 Have Different Kinase Activities In Vitro
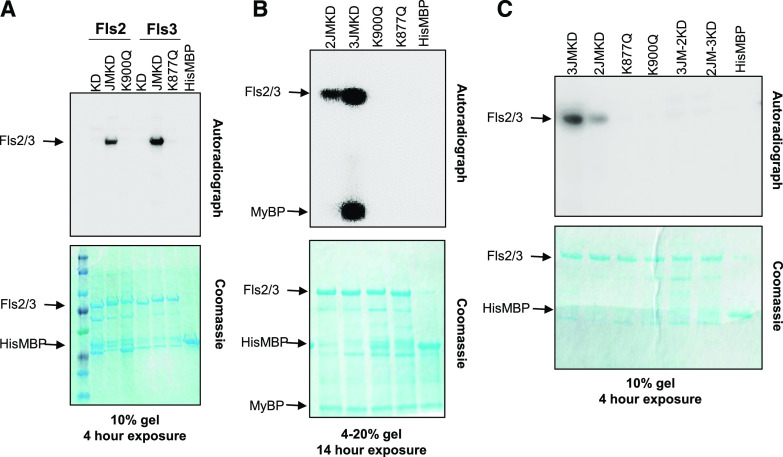
It is possible that different kinase activities of Fls3 and Fls2 account for their differences in immunity output. It was previously reported that Arabidopsis Fls2 has weak kinase activity in vitro that requires the entire intracellular domain for activity (both the iJM and KD; Gómez-Gómez et al., 2001; Xiang et al., 2008; Lu et al., 2010; Cao et al., 2013). To test whether tomato Fls3 and Fls2 have in vitro kinase activity and whether their activity is dependent on the presence of the iJM, we made recombinant proteins in BL21 Escherichia coli that contained the Fls3 or Fls2 KD alone, or the entire cytoplasmic domain containing the JM plus the KD (JMKD), expressed in the pDEST-HisMBP vector (which has an N-terminal 6×His-MBP tag), and assayed their kinase activity. As negative controls, we generated variants with substitutions in their ATP binding sites, Fls2-JMKD(K900Q) and Fls3-JMKD(K877Q), and a control that expressed a short E. coli sequence to allow expression of the His-MBP protein in the pDEST-HisMBP vector. We found that both tomato Fls2 and Fls3 have kinase activity that depended on the presence of the iJM (Fig. 4A), and the in vitro kinase activity for Fls3 was greater than that for Fls2 (Fig. 4). We added myelin basic protein (MyBP) to the in vitro kinase assays to test whether Fls2 and Fls3 could transphosphorylate this generic substrate. We observed that Fls3, but not Fls2, could transphosphorylate MyBP (Fig. 4B). Because Fls2 and Fls3 both require the iJM for in vitro kinase activity, we wanted to know whether they required their own cognate iJM for activity. We swapped the JM domain between Fls3 and Fls2 (2JM-3KD and 3JM-2KD) and tested them in in vitro kinase assays. We found that swapping the iJMs between Fls2 and Fls3 completely abolished in vitro kinase activity for both proteins (Fig. 4C). These data suggest that Fls3 and Fls2 have differences in their kinase activity that require their corresponding iJMs.
Figure 4.
Fls3 and Fls2 vary in their kinase activity in vitro. The KD alone or the iJM plus the KD (JMKD) of Fls3 and Fls2 were expressed as recombinant proteins in the pDEST-HisMBP vector and tested for in vitro kinase activity. The kinase-inactive mutants for Fls3(K877Q) and Fls2(K900Q) were generated in the background of the JMKD constructs. HisMBP protein was used as a negative control. A, Fls3 or Fls2 KDs or JMKDs were tested in in vitro kinase assays. B, MyBP was added to the in vitro kinase assays to test the ability of the kinases to transphosphorylate the MyBP substrate. C, Kinase assays swapping the iJMs of Fls3 and Fls2 to test the requirement for their cognate iJM. 3JM-2KD expresses the iJM from Fls3 and the KD from Fls2, and 2JM-3KD expresses the iJM from Fls2 and the KD from Fls3. Data shown are representative of at least three independent experiments. Exposure length to the phosphor-screen (4 or 14 h) and the SDS-PAGE gel percentage are indicated.
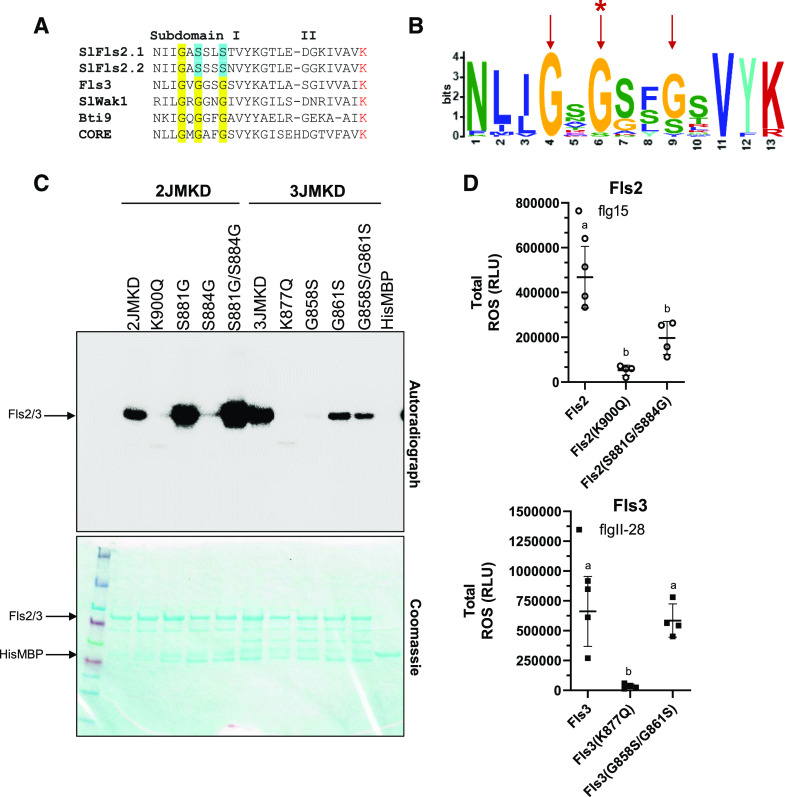
Subdomain I Contributes to the in Vitro Differences in Kinase Activity for Fls3 and Fls2
We hypothesized that subdomain I of the KD, which is involved in correct positioning of the ATP substrate in the active site, contributes to the differences in kinase activity we observed between Fls2 and Fls3. We aligned the Fls2 (Fls2.1 and Fls2.2) and Fls3 subdomain I sequences with other immunity-associated tomato kinases and found that Fls2 deviates from the glycine-rich motif (GxGxxG) present in other kinases (Fig. 5A). Fls2 has the conserved, first position Gly, but the second and third glycines are replaced by serines (GxSxxS). Fls3 has the standard GxGxxG motif. We compared the sequence motif in all 42 tomato RLKs in the Fls2/Fls3 class (class IX-a; Wei et al., 2015). We aligned the subdomain I KD regions of the 42 RLKs to determine the conservation and/or consensus of each of the residues in the GxGxxG motif. Two genes in this class, Solyc10g085110 and Solyc03g118330, likely do not encode active kinases because they lack the subdomain I region and the Lys responsible for binding of ATP (Supplemental Fig. S5A). We found that of the 40 RLKs in this class that have a subdomain I, only Fls2 deviates from the GxGxxG motif at the second-position Gly (Fig. 5B; Supplemental Fig. S5A). The third-position Gly is more flexible in the class, with 28 RLKs harboring a Gly, 10 RLKs a Ser, 1 RLK a Val, and 1 RLK an Ile. The first position Gly is conserved in all 40 RLKs. To test whether the serines in the Fls2 subdomain I motif impact Fls2 kinase activity, we made substitutions of each Ser individually or simultaneously to glycines (GxSxxS to GxGxxS [S881G], GxSxxG [S884G], or GxGxxG [S881G/S884G]), effectively swapping the motifs between Fls2 and Fls3. We found that the S881G mutation alone increased the kinase activity of Fls2 (Fig. 5C). In contrast, S884G resulted in a nearly complete loss of kinase activity. However, mutating both residues (S881G/S884G) led to an even stronger increase of kinase activity than S881G alone (Fig. 5C). To see whether substituting the glycines to serines in the Fls3 subdomain I motif would impact kinase activity, we mutated the glycines to serines either individually or simultaneously (GxGxxG to GxSxxG [G858S], GxGxxS [G861S], or GxSxxS [G858S/G861S]). We found that the G858S mutation alone caused a near complete loss of in vitro kinase activity. The G861S mutation resulted in a strong reduction of kinase activity compared to the wild-type 3JMKD. Mutating both residues (G858S/G861S) also resulted in a reduction of kinase activity that was slightly less than that for G861S alone (Fig. 5C). These data support the hypothesis that the subdomain I motif contributes to the increased kinase activity of Fls3 compared to Fls2 in vitro.
Figure 5.
Subdomain I contributes to the differences in kinase activity between Fls3 and Fls2 in vitro. A, Subdomain I in tomato showing the GxGxxG motif. The Lys involved in ATP binding is shown in red. Yellow and blue highlighting indicate conserved glycines in the motif and the deviant serines in Fls2, respectively. B, MEME LOGO showing the prevalence of the GxGxxG motif in class IXa of the tomato RLKs, which includes Fls2 and Fls3. Red arrows indicate the three Gly residues in the GxGxxG motif. Only Fls2 has a different residue in the second Gly position (red asterisk). C, In vitro kinase activity of Fls2JMKD and Fls3JMKD containing mutations in subdomain I. Data are representative of three independent replicates. D, Total ROS accumulated over 45 min after applying 100 nm flg15 (Fls2 proteins) or flgII-28 peptide (Fls3 proteins) on leaf discs collected after transient expression of the various proteins in N. benthamiana. Protein overexpression was driven by the CaMV 35S promoter. Shown are the means of four individual plants indicated as separate points. The top and bottom horizontal lines show the ± sd, and the middle horizontal line is the mean of the three plants. Data are from a single experiment and are representative of three independent replicates. Significance was determined via ANOVA followed by Tukey’s posttest with a significance cutoff of P < 0.05 (shown as lowercase letters). See also Supplemental Figure S5. RLU, Relative light unit.
We next tested whether substitutions in subdomain I of Fls3 and Fls2 affect the ROS output in transient in planta assays in the context of the full-length proteins. We cloned the double mutants Fls2(S881G/S885G) and Fls3(G858S/G861S) into the pGWB417 plant protein expression vector that has a C-terminal myc-tagged protein, effectively swapping the subdomain I motif between Fls2 and Fls3. We also cloned the kinase inactive pGWB417::Fls2(K900Q) and pGWB417::Fls3(K877Q) as negative controls, and transiently expressed the proteins in N. benthamiana leaves. ROS responses to 100 nm flgII-28 or flg15 peptide were measured and compared to the wild-type Fls2 or Fls3 proteins. Surprisingly, there was a statistically significant reduction in total ROS production of Fls2(S881G/S884G), which effectively has the Fls3 subdomain I motif, compared to wild-type Fls2. There was no statistically significant difference in total ROS production of Fls3(G858S/G861S), which effectively has the Fls2 subdomain I motif, compared to wild-type Fls3 (Fig. 5D). We confirmed that all proteins were transiently expressed via immunoblotting (Supplemental Fig. S5B). These results suggest that although the subdomain I sequence influences in vitro kinase activity, the differences in kinase activity alone do not explain the differences in ROS output between Fls2 and Fls3.
No Single Domain Appears to Explain the Sustained ROS Response of Fls3
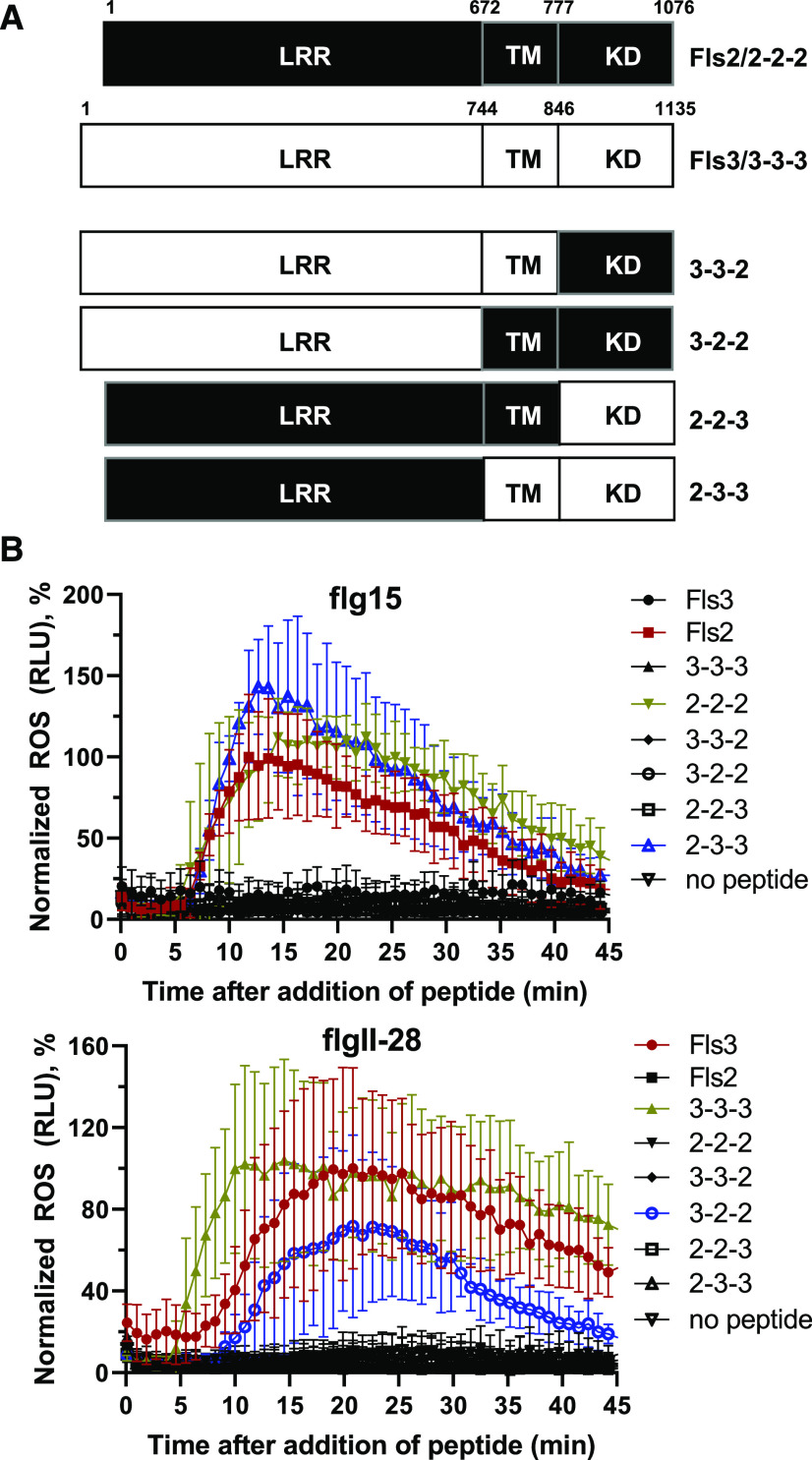
It is also possible that differences in the Fls2 and Fls3 LRR domains, which are the extracellular domains that bind the flg22 or flgII-28 peptides, are responsible for the different readouts of these two PRRs. To test this possibility, we generated chimeric constructs that swapped the LRR domain, transmembrane domain (TM), and KD between Fls2 and Fls3 for transient expression in N. benthamiana. We included the presumed signal peptide and N terminus within the LRR domain, the iJM and outer JM within the TM region, and the C terminus within the KD (Fig. 6A). We used a low concentration of peptide in the ROS assays (10 nm flgII-28 or flg15) to allow detection of subtle differences in the ROS responses of the chimeric constructs. As a control, we also generated the reconstituted wild-type constructs, which have the same sequence as the wild-type Fls3 or Fls2 constructs but were generated in the same manner as the chimeric constructs (labeled 3-3-3 or 2-2-2, respectively).
Figure 6.
No single domain governs the sustained ROS response. A, Schematic diagram showing the design of the chimeric Fls2 and Fls3 constructs. Amino acid numbers corresponding to the domains used to generate the chimeric proteins are shown above each schematic. In the labels for each chimeric construct, numbers 2 and 3 indicate whether the domain was from Fls2 or Fls3 and correspond to the LRR, TM, and KD, respectively. Reconstituted Fls2 (2-2-2) and Fls3 (3-3-3) were included as controls and are identical in sequence to wild-type Fls2 and Fls3, respectively. B, ROS curves comparing the chimeric constructs for the sustained response. Constructs were transiently expressed in N. benthamiana leaves under control of the CaMV 35S promoter, and ROS activity was measured over 45 min after the addition of 10 nm flg15 or flgII-28 peptide. Data are the means of four plants, and error bars represent the ± sd. Data are from a single experiment and are representative of three independent experiments. The two chimeric constructs that responded to peptide are shown in blue, the unadulterated controls in red, and the reconstituted controls in gold. See also Supplemental Figure S6.
In our transient assays, only the chimeric constructs with the TM and KD from the same receptor responded to peptide (2-3-3 and 3-2-2; Fig. 6B). Specifically, the response of the chimeric construct with the LRR from Fls2 and the TM and KD from Fls3 (2-3-3) to the flg15 peptide was similar to those of the wild-type Fls2 and reconstituted wild-type construct (2-2-2) in both amplitude and duration of response (Fig. 6B; Supplemental Fig. S6, A and B). The time at which the maximum ROS response to flg15 peptide occurred was also similar between 2-3-3, Fls2, and 2-2-2 (∼12 min after peptide treatment). The normalized ROS responses at the maximum amplitudes were not statistically different between 2-3-3, Fls2, and 2-2-2 (Supplemental Fig. S6A). Additionally, 45 min after peptide treatment there was no significant difference between the ROS responses of 2-3-3, Fls2, and 2-2-2 (Supplemental Fig. S6B).
When we applied 10 nm of the flgII-28 peptide, only the chimeric construct with the LRR from Fls3 and the TM and KD from Fls2 (3-2-2) activated a ROS response (Fig. 6B). However, while the time at which the maximum response occurred was similar between 3-2-2, wild-type Fls3, and the reconstituted wild-type construct (3-3-3; ∼20 min after peptide treatment), and the normalized maximum amplitudes of the ROS responses were not statistically significant (Fig. 6B; Supplemental Fig. S6A), the ROS response for 3-2-2 was not sustained like those of Fls3 and 3-3-3 (Fig. 6B; Supplemental Fig. S6B). At 45 min after flgII-28 treatment, the ROS response for 3-2-2 was significantly less than those for Fls3 and 3-3-3 and was not significantly different from those for the flgII-28 nonresponders (Supplemental Fig. S6B). All proteins were confirmed to be expressed via immunoblotting, though there were some differences in levels of expression between the chimeric constructs (Supplemental Fig. S6C). However, expression of 3-2-2 was similar to that of Fls3 and 3-3-3, so expression alone does not explain the differences in sustained ROS response between Fls3, 3-3-3, and 3-2-2. Additionally, the expression of 2-3-3 was reduced compared to that of Fls2 and 2-2-2 (to levels similar to those observed for Fls3 and 3-3-3), but the response of 2-3-3 to peptide was similar to those of Fls2 and 2-2-2, therefore supporting that expression levels alone do not account for the differences observed between the chimeric constructs. The data in Figure 6 and Supplemental Figure S6 were normalized to the in-plate Fls3 or Fls2 controls to also account for differences in expression between constructs, plants, and technical replicates. In conclusion, although we cannot rule out that our domain swapping might have nullified some of the functions of the receptors, as reported for other receptors (Albert et al., 2010; Mueller et al., 2012), these data suggest that no single domain is responsible for the sustained ROS response.
DISCUSSION
While both Fls2 and Fls3 recognize flagellin-derived MAMPs, their individual contributions to disease resistance are unknown. We used the CRISPR/Cas9 gene-editing system to generate mutations in the Fls3 and two Fls2 genes in tomato and used the resulting lines to examine the contributions of the two PRRs to disease resistance. Both Fls3 and Fls2.1 had similar effects on inhibition of Pst growth on the leaf surface, and in the leaf apoplast, with the greatest effects occurring on the leaf surface (Fig. 1; Supplemental Fig. S2). Stomata play a large role in the bacterial infection process and subsequent plant defense response (Melotto et al., 2006, 2017). A recent study shows that surface-based responses and PTI play a large role in defense for diverse tomato accessions (Roberts et al., 2019b). Our observations with Fls2 and Fls3 are consistent with previous reports showing that PTI and recognition of pathogens on leaf surfaces are important to plant defense. Additionally, we found that a 7-bp deletion in Fls2.1 alone completely abolished flg22 recognition in tomato, further supporting a previous report that Fls2.1 is the only functional receptor for flg22 recognition in tomato leaves (Jacobs et al., 2017).
Fls2 and Fls3 appear to contribute equally to PTI, which suggests that they may use some similar mechanisms and/or signaling pathways. In fact, we reported recently that activation of the Fls2 or Fls3 pathways induces the expression of the wall-associated kinase Wak1 gene, and both PRRs coimmunoprecipitate with the Wak1 protein, which appears to act independently of ROS production and at a later stage of PTI (Zhang et al., 2020a). However, the differences in several readouts from the Fls2 and Fls3 pathways also suggest underlying differences in some aspect of their functions. These could be mechanistic differences, variances in the amount of each receptor protein, distribution of the receptors in plant tissues, efficiency and/or duration of receptor activation, and/or stability of the flg15, flg22, and flgII-28 peptides used. While our results do not address all of these possibilities, we discuss below the evidence we have found for differences in the immune pathways between Fls2 and Fls3.
One observation that suggests there may be differences in the molecular mechanisms between Fls2 and Fls3 is that the ROS response to flgII-28 is sustained compared to its response to flg22 (Fig. 2; Supplemental Fig. S3). We observed this sustained ROS response in both tomato and potato leaves, and data from another study support a sustained ROS response for flgII-28 in pepper (Clarke et al., 2013). Although Moroz and Tanaka (2019) reported differences in the amplitude of flgII-28 and flg22 ROS responses in potato, we did not observe these differences under our conditions. This may be attributed to the use of different potato varieties or peptide concentrations in the assays (100 nm in our study versus 1 μm in Moroz and Tanaka [2019]). Additionally, although N. benthamiana does not have an endogenous Fls3, transient overexpression of Fls3 in N. benthamiana also results in a sustained response compared to Fls2 (Hind et al., 2016; this study). Previous efforts to express Fls3 in Arabidopsis have been unsuccessful (Hind et al., 2016), so it is currently unknown whether Fls3 can function in species outside the Solanaceae, but future efforts to express Fls3 in more diverse species could determine whether solanaceous-specific protein partners are involved in the Fls3 ROS response.
We analyzed RNA-Seq data from previous studies (Rosli et al., 2013; Pombo et al., 2017) and identified 44 genes whose transcript abundance in leaves increased upon application of flgII-28, and not flg22, peptide. Two of these genes, bHLH and UGT, were developed as reporter genes, and their transcript abundance was confirmed to increase only in response to flgII-28. This suggests that these two genes are regulated by an Fls3-specific signaling pathway that is independent of Fls2 and supports the hypothesis that there are differences in molecular signaling pathways between Fls3 and Fls2. We previously identified three genes whose expression is induced specifically during PTI (Pombo et al., 2014). These genes were not identified as specific Fls3 reporters in the RNA-Seq data, indicating that they may be induced by both flg22 and flgII-28. The possible role of any of the 44 genes in Fls3-mediated immunity is unknown. While we have yet to find an Fls2-specific reporter gene, future studies using pathway-specific reporter genes may help dissect the differences in the signaling components activated by Fls2 and Fls3.
Arabidopsis FLS2 has weak autophosphorylation activity in vitro and in vivo that requires the presence of the iJM (Gómez-Gómez et al., 2001; Xiang et al., 2008; Lu et al., 2010; Cao et al., 2013). We found that tomato Fls2 and Fls3 both have relatively strong autophosphorylation activity in vitro that can be detected after only 4 h of exposure to a phosphor-screen. This activity is dependent on the presence of the iJM, which may be due to specific important residues within the domain. For example, a previous study examined the requirement of the iJM for rice PRR Xa21 function and found that the C-terminal region of the iJM was required for autophosphorylation (Chen et al., 2010). The authors found a conserved Thr residue (T705) within the JM at the C-terminal end of the domain that was conserved among plant RLKs, and mutation of this residue (T705A or T705E) resulted in increased susceptibility to X. oryzae pv oryzae. It is still unknown what molecular role this residue and the iJM as a whole play in kinase activity, but the authors propose that the Thr may be serving a dual role in receiving and donating a phosphoryl group (Chen et al., 2010). Tomato Fls2 and Fls3 both have this conserved Thr residue, and future research is needed to determine the molecular role of their iJMs. Some other plant RLK chimeras require the cognate iJM to function. For example, a chimeric construct containing the extracellular domain of the rice chitin elicitor receptor CEBiP and the KD from the rice blast resistance protein Pi-d2 is only functional if the TM originated from Pi-d2 (Kouzai et al., 2013). However, not all RLKs require their cognate iJM to function. Swapping the iJMs of the Arabidopsis RLK CERK1 with BAK1 and Fls2 still resulted in a functional CERK1 protein (Zhou et al., 2020). Our future efforts will focus on understanding why there is a cognate iJM requirement for Fls2 and Fls3.
Subdomain I of the Fls3 KD contributes to the stronger in vitro kinase activity of this protein (Figs. 4 and 5). A previous study speculated that substituting the second Gly for a Ser, as seen in Arabidopsis FLS2 (S879), would lead to a major reduction in kinase activity for FLS2, which supports the finding in this study that Arabidopsis has weak autophosphorylation ability (Schwessinger et al., 2011). Therefore, one would predict that changing of S881 to S881G in tomato Fls2 would result in a dramatic increase in kinase activity (Fig. 5C). In fact, when we made this substitution we did observe a dramatic increase in kinase activity, whereas the S884G substitution resulted in a complete abolishment of kinase activity. However, mutation of both residues (S881G/S884G) caused an even greater increase in kinase activity than S881G alone. Conversely, the G858S substitution in Fls3 caused a complete abolishment of kinase activity, and the G861S and G858S/G861S mutations resulted in a dramatic reduction, but not abolition, of Fls3 kinase activity (Fig. 5C). Further studies are needed to determine why Fls2 requires a Ser at residue 884 in the context of GxSxxS or GxGxxS for kinase activity, but these differences between Fls3 and Fls2 suggest that there may be differences in the molecular mechanisms of kinase activation between Fls2 and Fls3. Kinase activity alone, however, does not explain the sustained ROS response in planta. When we made the Fls2(S881G/S884G) and Fls3(G858S/G861S) mutations in the context of the full-length protein and overexpressed them in N. benthamiana leaves (which effectively swap the subdomain I motifs between Fls2 and Fls3), the Fls2(S881G/S884G) mutation resulted in a decrease of total ROS production rather than the predicted increase. For the Fls3(G858S/G861S) substitution, we observed no statistically significant effect of the mutation on total ROS production. It is currently unknown why the in vitro results do not translate to the in planta ROS assays, but future experiments studying the kinase activity in vivo will help uncover the mechanisms of kinase activation in Fls3 and Fls2.
In addition to the effects on kinase activity due to the subdomain I motif, we also observed differences in transphosphorylation in vitro. Fls3 could transphosphorylate MyBP, whereas Fls2 could not, which further supports differences in kinase activity between Fls3 and Fls2. While it is currently unknown what may be the in planta transphosphorylation target(s) of Fls3, in Arabidopsis, upon binding flg22, FLS2 associates with the coreceptor BAK1 and both FLS2 and BAK1 are transphosphorylated to initiate downstream signaling (Sun et al., 2013; Couto and Zipfel, 2016; Saijo et al., 2018). The presumed orthologs of BAK1 in tomato are Serk3A/Serk3B, but this has not been experimentally investigated. Additionally, Fls2 and Fls3 both coimmunoprecipitate with Wak1 independently of flg22/flgII-28 or Arabidopsis BAK1 (Zhang et al., 2020a). It is possible that Fls3 and Fls2 may interact differently with the Serks, and that will be a focus of future studies.
We hypothesized that either the kinase domain or the LRR domain may be responsible for the sustained ROS response observed for Fls3. Therefore, we generated chimeric Fls2 and Fls3 constructs to test whether either of these domains was solely responsible for the sustained ROS response (Fig. 6; Supplemental Fig. S6). When we overexpressed the chimeric constructs in N. benthamiana leaves and measured the ROS responses to flgII-28 or flg15, we discovered that there is a requirement for the TM (which includes the outer JM and the iJM) and the KD to be from the same receptor, as only the constructs with a TM and KD originating from the same receptor responded to peptide (Fig. 6; Supplemental Fig. S6). Because the TM region included the iJM, this observation agrees with the in vitro kinase results showing that kinase activity requires the cognate iJM and suggests that the lack of ROS responses in the chimeric constructs with a TM and KD from different receptors may be due to a lack of kinase activity (Fig. 4C). We also found that neither of the two chimeric constructs that responded to peptide (2-3-3 to flg15 or 3-2-2 to flgII-28) had a sustained ROS response (Fig. 6B; Supplemental Fig. S5, A and B). While the shape of the curve and the time at maximum amplitude resembled the receptor matching their LRR domain (12 min for Fls2 and 2-3-3, and 20 min for Fls3 and 3-2-2), both chimeric constructs were statistically indistinguishable from their negative controls at 45 min after peptide treatment, while Fls3 and 3-3-3 maintained the ROS levels at ∼50% to 60% of their maximum amplitude (Fig. 6; Supplemental Fig. S6).
While we cannot rule out the possibility that swapping large regions of these proteins may nullify their original function, our data collectively suggest that the biological function of the receptors could be more complicated than a single domain being responsible for the sustained response. Rather, it may be a combination of factors or structural features unique to Fls3 that additively lead to a difference in the immune outputs. While previous data show that Fls3 and Fls2 have some similar molecular characteristics (Hind et al., 2016), our data suggest that Fls3 may act in a different signaling pathway from that of Fls2. Determining the possible different components of the Fls3 and Fls2 pathways may shed light on how Fls3 evolved as a solanaceous-specific flagellin receptor and help us better understand the molecular mechanisms of plant immunity.
MATERIALS AND METHODS
Plant Growth Conditions, Inoculations, and Bacterial Growth Assays
Tomato (Solanum lycopersicum) seedlings were grown under the conditions described previously and inoculated as described in Roberts et al. (2019b; see Supplemental Methods for more information). For the dip inoculations, 3-week-old seedlings were placed in a 100% relative humidity chamber for 14 h prior to inoculation, then dipped into the bacterial suspension of 1 × 108 colony-forming units (CFUs) mL−1 for 10 s. Bacterial populations were quantified on Day 0 and Day 2 or 3, as described previously (Roberts et al., 2019b; see Supplemental Methods for more information; for a list of bacterial strains used in this study, see Supplemental Table S4).
Generation of CRISPR/Cas9-Mediated Knockout Lines
Guide RNAs were designed to target Fls3, Fls2.1, or Fls2.1/Fls2.2 as described previously (Jacobs et al., 2017; Zhang et al., 2020b) using the tomato genome version SL2.5 (Tomato Genome Consortium, 2012). To induce mutations in both Fls3 and Fls2.1/2.2 in the same plant (∆Fls2.1/2.2/3), the constructs used to induce the individual mutations were transformed into the Agrobacterium tumefaciens strain LBA4404 and the cultures were mixed 1:1 prior to tomato transformation. Tomato transformations were performed at the Boyce Thompson Institute transformation facility (Gupta and Van Eck, 2016; Van Eck et al., 2019). More information is available in Supplemental Figure S1, Supplemental Tables S1, S4, and S5, and Supplemental Methods, as well as on the Plant CRISPR database at plantcrispr.org.
ROS Bioassays
ROS production in leaf discs (30 mm2, cork borer size 2) was measured in response to flgII-28, flg22, or flg15 peptides as described in Roberts et al. (2019b) using 100-nm concentrations for all assays except for the chimeric constructs (10 nm flgII-28 and flg15; Fig. 6; Supplemental Fig. S6). Every experiment was repeated at least three times, with four plants per experiment; values represent the means of the four plants ± sd as determined using the Prism 8 program. Shown is one representative replicate for each experiment. See Supplemental Methods for information about the peptides used in this study.
Reporter Genes
Three 5-week-old RG-prf3 plants were syringe-infiltrated with 1 μm flgII-28 or flg22 peptide or water and sampled 6 h postinfiltration. Biological replicates were taken from each of the three plants infiltrated with peptide or water. RNA extraction, complementary DNA synthesis, and RT-qPCR were performed as described previously (Pombo et al., 2014), and significance was determined using a pairwise t test with the Prism 8 program. Primers used for RT-qPCR are available in Supplemental Table S4. Gene ontology terms were determined using the Plant Transcriptional Regulatory Map software (http://plantregmap.cbi.pku.edu.cn/go.php).
Cloning
The chimeric constructs were generated via overlap extension PCR. The 2KD and 2JMKD PCR products were inserted into the entry vector pJLSMART (Mathieu et al., 2014). Chimeric construct open reading frames were then recombined into the Gateway vector pGWB417 (Nakagawa et al., 2007, 2009) using LR Clonase II following the manufacturer’s instructions (Thermo Fisher Scientific; https://www.thermofisher.com/us/en/home.html). Mutagenesis of the 2JMKD, 3JMKD, Fls2, and Fls3 clones was performed in the entry vectors using the Q5 Site-Directed Mutagenesis kit following the manufacturer’s instructions (New England Biolabs; www.neb.com). For a list of all primers and constructs used in this study, see Supplemental Tables S4 and S5, and for additional information, see Supplementary Methods.
Agroinfiltrations
Agroinfiltrations of binary vectors into Nicotiana benthamiana were performed as previously described (Hind et al., 2016). All cultures were prepared to a final OD600 of 0.2. The Fls3- and Fls2- containing bacterial cultures were mixed 1:1 with a construct expressing the p19 viral suppressor of silencing. See Supplemental Methods for more information.
In Vitro Kinase Assays
HisMBP-tagged proteins were transformed into BL21 (DE3) pLys Rosetta cells and grown in culture at 37°C until the OD600 reached 0.6 to 0.8. Protein expression was induced using 1 mm isopropylthio-β-galactoside at 28°C for 3 to 4 h. Cell pellets were suspended in column buffer (20 mm Tris-HCl [pH 7.5], 200 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and 10% [v/v] glycerol) supplemented with Complete Easy protease inhibitor cocktail (Millipore Sigma; https://www.sigmaaldrich.com/united-states.html), lysed by sonication, mixed with amylose resin (New England BioLabs), and eluted with 10 mm maltose. In vitro kinase assays were performed using 5 μg of each of the various kinase proteins and/or 3 μg of myelin basic proteins and conducted as described previously (Roberts et al., 2019a).
Immunoblotting
For the transiently expressed proteins in N. benthamiana, total protein was extracted from agroinfiltrated leaves, and 5 to 10 μg was run on SDS-PAGE, blotted on polyvinylidene difluoride membrane, and detected with anti-Myc antibodies (A00704, Genscript; www.genscript.com) and ECL Plus chemiluminescent substrate (Thermo Fisher Scientific; www.thermofisher.com) as described in Roberts et al. (2019a).
Accession Numbers
See Supplemental Table S4 for accession information.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Development and characterization of CRISPR/Cas9-generated mutations in Fls2.1, Fls2.2, and Fls3.
Supplemental Figure S2. Additional replicates of bacterial growth in the CRISPR/Cas9 knockout lines.
Supplemental Figure S3. Additional tomato and potato accessions with sustained flgII-28 ROS response.
Supplemental Figure S4. Eggplant variety ‘Shikou’ does not respond to flgII-28.
Supplemental Figure S5. Supplemental information related to Figure 5.
Supplemental Figure S6. Only the chimeric constructs with a TM and KD originating from the same receptor respond to flgII-28 or flg15 peptide.
Supplemental Table S1. Generation of CRISPR/Cas9-mediated knockouts of the flagellin-sensing genes.
Supplemental Table S2. List of genes induced upon flgII-28 treatment by RNA-Seq (Pombo et al., 2017; Rosli et al., 2013).
Supplemental Table S3. Gene ontology term analysis of flgII-28-induced genes from RNASeq (all terms, q < 0.05; Pombo et al., 2017; Rosli et al., 2013).
Supplemental Table S4. Constructs and strains used in this study.
Supplemental Table S5. Primers used in this study.
Supplemental Methods. Plant growth conditions, inoculations, and bacterial growth assays; generation of CRISPR/Cas9-mediated knockout lines; peptides used in the ROS bioassays; cloning; and agroinfiltration.
Acknowledgments
We thank Ning Zhang for helpful comments on the manuscript; Jing Zhang, Fabian Giska, and Ning Zhang for performing supporting experiments; Kevin Chen and Ben Carter for experimental assistance; Brian Bell, Jay Miller, and Nick Vail for greenhouse assistance; and Jason Ingram for providing potato tubers used in this study.
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1546625 to G.B.M.) and the USDA-Binational Agriculture Research Fund (BARD; grant no. IS–4931–16C).
Articles can be viewed without a subscription.
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB(2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Felix G(2010) Chimeric receptors of the Arabidopsis thaliana pattern recognition receptors EFR and FLS2. Plant Signal Behav 5: 1430–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Jehle AK, Mueller K, Eisele C, Lipschis M, Felix G(2010) Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J Biol Chem 285: 19035–19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G(2010) A domain swap approach reveals a role of the plant wall-associated kinase1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, et al. (2011) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7: e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Aceti DJ, Sabat G, Song J, Makino S, Fox BG, Bent AF(2013) Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog 9: e1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Martin GB, Low PS(1996) The Pto kinase mediates a signaling pathway leading to the oxidative burst in tomato. Proc Natl Acad Sci USA 93: 13393–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chern M, Canlas PE, Jiang C, Ruan D, Cao P, Ronald PC(2010) A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. J Biol Chem 285: 10454–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA(2013) Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C(2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A(2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA 108: 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Xiao F, Fan F, Gu L, Cang H, Martin GB, Chai J(2009) Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21: 1846–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Bauer Z, Boller T(2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gupta S, Van Eck J(2016) Modification of plant regeneration medium decreases the time for recovery of Solanum lycopersicum cultivar M82 stable transgenic lines. Plant Cell Tissue Organ Cult 127: 417–423 [Google Scholar]
- Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, Rathjen JP(2010) Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J 61: 507–518 [DOI] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O’Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al. (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Hohmann U, Santiago J, Nicolet J, Olsson V, Spiga FM, Hothorn LA, Butenko MA, Hothorn M(2018) Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc Natl Acad Sci USA 115: 3488–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton N, Nekrasov V, Ronald PC, Zipfel C(2015) The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog 11: e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, Zhang N, Patel D, Martin GB(2017) Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol 174: 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Martin GB(1999) Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol Biol 40: 455–465 [DOI] [PubMed] [Google Scholar]
- Jones JB.(1991) Bacterial speck In Jones JB, Jones JP, Stall RE, and Zitter TA, eds, Compendium of Tomato Diseases. APS Press, St. Paul, MN, pp 26–27 [Google Scholar]
- Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y(2013) Expression of the chimeric receptor between the chitin elicitor receptor CEBiP and the receptor-like protein kinase Pi-d2 leads to enhanced responses to the chitin elicitor and disease resistance against Magnaporthe oryzae in rice. Plant Mol Biol 81: 287–295 [DOI] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei C-F, Russell AB, Martin GB, Schneider DJ, Collmer A(2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P(2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N-C, Martin GB(2007) Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse pseudomonas syringae pathovars to infect tomato. Mol Plant Microbe Interact 20: 806–815 [DOI] [PubMed] [Google Scholar]
- Lin N-C, Martin GB(2005) An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact 18: 43–51 [DOI] [PubMed] [Google Scholar]
- Lolle S, Stevens D, Coaker G(2020) Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr Opin Immunol 62: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P(2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB.(2012) Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB In Martin F, and Kamoun S, eds, Effectors in Plant-Microbe Interactions. Wiley-Blackwell, Ames, IA, pp 123–154 [Google Scholar]
- Mathieu J, Schwizer S, Martin GB(2014) Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog 10: e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, He SY(2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY(2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY(2017) Stomatal defense a decade later. Plant Physiol 174: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz N, Tanaka K(2019) FlgII-28 is a major flagellin-derived defense elicitor in potato. Mol Plant Microbe Interact 33: 247–255 [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Wu AJ, Balmuth AL, Arasteh JM, Rathjen JP(2009) Regulation of tomato Prf by Pto-like protein kinases. Mol Plant Microbe Interact 22: 391–401 [DOI] [PubMed] [Google Scholar]
- Mueller K, Bittel P, Chinchilla D, Jehle AK, Albert M, Boller T, Felix G(2012) Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ishiguro S, Kimura T(2009) Gateway vectors for plant transformation. Plant Biotechnol (Tokyo) 26: 275–284 [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Oh C-S, Martin GB(2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16: 132–140 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB(2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB(2004) Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J Biol Chem 279: 49229–49235 [DOI] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fei Z, Martin GB, Rosli HG(2017) Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem. Sci Rep 7: 44905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fernandez-Pozo N, Dunham DM, Fei Z, Martin GB(2014) Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biol 15: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Bittel P, Chinchilla D, Köchner P, Felix G, Shiu SH, Boller T(2007) Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol 64: 539–547 [DOI] [PubMed] [Google Scholar]
- Roberts R, Hind SR, Pedley KF, Diner BA, Szarzanowicz MJ, Luciano-Rosario D, Majhi BB, Popov G, Sessa G, Oh CS, et al. (2019a) Mai1 protein acts between host recognition of pathogen effectors and mitogen-activated protein kinase signaling. Mol Plant Microbe Interact 32: 1496–1507 [DOI] [PubMed] [Google Scholar]
- Roberts R, Mainiero S, Powell AF, Liu AE, Shi K, Hind SR, Strickler SR, Collmer A, Martin GB(2019b) Natural variation for unusual host responses and flagellin-mediated immunity against Pseudomonas syringae in genetically diverse tomato accessions. New Phytol 223: 447–461 [DOI] [PubMed] [Google Scholar]
- Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin GB(2013) Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol 14: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S(2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ(1994) Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell 6: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ(1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C(2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwizer S, Kraus CM, Dunham DM, Zheng Y, Fernandez-Pozo N, Pombo MA, Fei Z, Chakravarthy S, Martin GB(2017) The tomato kinase Pti1 contributes to production of reactive oxygen species in response to two flagellin-derived peptides and promotes resistance to Pseudomonas syringae infection. Mol Plant Microbe Interact 30: 725–738 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J(2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Thomas NC, Hendrich CG, Gill US, Allen C, Hutton SF, Schultink A(2020) The immune receptor Roq1 confers resistance to the bacterial pathogens Xanthomonas, Pseudomonas syringae, and Ralstonia in tomato. Front Plant Sci 11: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck J, Keen P, Tjahjadi M(2019) Agrobacterium tumefaciens-mediated transformation of tomato. Methods Mol Biol 1864: 225–234 [DOI] [PubMed] [Google Scholar]
- Wei Z, Wang J, Yang S, Song Y(2015) Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 58: 121–134 [DOI] [PubMed] [Google Scholar]
- Wu J, Reca IB, Spinelli F, Lironi D, De Lorenzo G, Poltronieri P, Cervone F, Joosten MHAJ, Ferrari S, Brutus A(2019) An EFR-Cf-9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1- and BAK1-dependent recognition of elf18. Mol Plant Pathol 20: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xing W, Zou Y, Liu Q, Liu J, Luo X, Huang Q, Chen S, Zhu L, Bi R, Hao Q, et al. (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449: 243–247 [DOI] [PubMed] [Google Scholar]
- Zhang N, Pombo MA, Rosli HG, Martin GB(2020a) Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol 183: 1869–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Roberts HM, Van Eck J, Martin GB(2020b) Generation and molecular characterization of CRISPR/Cas9-induced mutations in 63 immunity-associated genes in tomato reveals specificity and a range of gene modifications. Front Plant Sci 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Liu J, Wang J, Chen S, Chen L, Wang J, Wang HB, Liu B(2020) The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J Integr Plant Biol 62: 556–562 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T(2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]