Papaver rhoeasS-determinants, which specify self-incompatibility and rejection of self-pollen, trigger growth arrest and programmed cell death in vegetative Arabidopsis tissues when expressed ectopically.
Abstract
Self-incompatibility (SI) is used by many angiosperms to reject self-pollen and avoid inbreeding. In field poppy (Papaver rhoeas), SI recognition and rejection of self-pollen is facilitated by a female S-determinant, PrsS, and a male S-determinant, PrpS. PrsS belongs to the cysteine-rich peptide family, whose members activate diverse signaling networks involved in plant growth, defense, and reproduction. PrsS and PrpS are tightly regulated and expressed solely in pistil and pollen cells, respectively. Interaction of cognate PrsS and PrpS triggers pollen tube growth inhibition and programmed cell death (PCD) of self-pollen. We previously demonstrated functional intergeneric transfer of PrpS and PrsS to Arabidopsis (Arabidopsis thaliana) pollen and pistil. Here, we show that PrpS and PrsS, when expressed ectopically, act as a bipartite module to trigger a self-recognition:self-destruct response in Arabidopsis independently of its reproductive context in vegetative cells. The addition of recombinant PrsS to seedling roots expressing the cognate PrpS resulted in hallmark features of the P. rhoeas SI response, including S-specific growth inhibition and PCD of root cells. Moreover, inducible expression of PrsS in PrpS-expressing seedlings resulted in rapid death of the entire seedling. This demonstrates that, besides specifying SI, the bipartite PrpS-PrsS module can trigger growth arrest and cell death in vegetative cells. Heterologous, ectopic expression of a plant bipartite signaling module in plants has not been shown previously and, by extrapolation, our findings suggest that cysteine-rich peptides diversified for a variety of specialized functions, including the regulation of growth and PCD.
Pollen-pistil interactions are complex, crucial events in plant reproductive biology, involving bidirectional signaling between the pistil and the pollen landing on it. Many of the responses regulating pollination take place within the pollen grains, which constitute the highly reduced haploid male gametophyte. The pollen grain is composed of the specialized vegetative cell that contains within itself two sperm cells, complete with cell walls and plasma membranes. The pollen’s role is to deliver two sperm cells to the embryo sac so that double fertilization can take place. Thus, pollen represents a unique gametophytic structure; for example, serial analysis of gene expression studies have revealed that 83% of the pollen-expressed gene tags are pollen specific and thus thought to be critical for pollen function (da Costa-Nunes and Grossniklaus, 2003; Honys and Twell, 2004; Mergner et al., 2020).
Self-incompatibility (SI) is an important mechanism used by flowering plants to prevent selfing. It is controlled by a multiallelic S-locus allowing self/nonself recognition between pistil and pollen. In several SI systems, when male and female S-determinant allelic specificities match, self (incompatible) pollen is recognized and rejected before fertilization can occur. A key characteristic of SI determinants is that they are extremely tightly regulated, both in a developmental and a tissue-specific manner, being expressed solely in pistil and pollen cells during a narrow developmental window, as the tissues approach maturity (Takayama and Isogai, 2005). SI in poppy (Papaver rhoeas) is controlled and specified by S-determinants expressed specifically in the stigma (PrsS; Foote et al., 1994) and pollen (PrpS; Wheeler et al., 2009). PrpS encodes a novel integral membrane protein with several predicted transmembrane domains; PrsS encodes a small, secreted protein and is the founding member of the large family of S-protein homologs (SPHs), which are found in most dicotyledonous plants, some fungi, and metazoa (Rajasekar et al., 2019). This family of small, secreted proteins have features similar to cysteine-rich peptides (CRPs), which include ligands known to be involved in diverse signaling pathways (Wheeler et al., 2010; Marshall et al., 2011; Bircheneder and Dresselhaus, 2016; Liu et al., 2017). However, aside from PrsS, the functional roles of SPHs in plants remain to be established (Rajasekar et al., 2019).
A long-standing model for SI in P. rhoeas is that PrsS acts as a signaling ligand to trigger SI in incompatible pollen. While PrpS is distinct from typical plant receptors (e.g. receptor-like kinases), its allele-specific interaction with PrsS activates a network of intracellular signals in incompatible pollen that result in the rapid inhibition of pollen tube growth and, ultimately, programmed cell death (PCD). Key hallmark features of the P. rhoeas SI response include a rapid increase of cytosolic free Ca2+ ([Ca2+]cyt; Franklin-Tong et al., 1993), a dramatic drop in cytoplasmic pH ([pH]cyt; Wilkins et al., 2015), and distinctive alterations of the actin cytoskeleton (Geitmann et al., 2000; for review, see Wang et al., 2019). We previously demonstrated that a cognate PrpS-PrsS interaction in Arabidopsis (Arabidopsis thaliana) pollen growing in vitro triggered hallmark features of the P. rhoeas SI response (de Graaf et al., 2012; Wang et al., 2020) and showed that PrpS and PrsS, when expressed in pollen and pistil, respectively, in Arabidopsis, function to prevent self-seed set, effectively rendering Arabidopsis self-incompatible (Lin et al., 2015). These findings demonstrated that the P. rhoeas S-determinants can be functionally transferred between highly diverged plant species (Bell et al., 2010). However, as the SI response is triggered within the unique, highly specialized context of the pollen, it was unclear whether the PrpS-PrsS module triggers a pollen-specific pathway or whether this pair of proteins can trigger growth arrest and cell death pathways in other parts of the plant.
Cellular responses in plants require an integrated signal perception and signal transduction network; such networks are responsible for orchestrating and coordinating a plethora of diverse processes including growth and development. As such, signaling processes allow tissues and organs to communicate with each other efficiently. A major class of proteins involved in cellular communication are those involved in short-range peptide signaling, utilizing small secreted proteins or peptides that act as ligands that interact with some sort of receptors (Sparks et al., 2013). Many signaling peptides are perceived by receptor-like kinases, and it is thought that much of the specificity of responses is due to the localized expression of ligands and their receptors (for review, see Breiden and Simon, 2016). For example, although CLAVATA3 CLE family peptides act in both roots and shoots (Fletcher et al., 1999), they nevertheless function in both organs specifically in apical meristematic tissues.
Heterologous expression of plant genes in other plant species has often been used to identify function phenotypically by dominant gene activity (Diener and Hirschi, 2000). Ectopic expression has also been used to demonstrate function; for example, Boutilier et al. (2002) showed that constitutive expression of the BABY BOOM transcription factor promotes cell proliferation and morphogenesis during embryogenesis. However, transfer of two genes encoding a receptor-ligand pair that are normally specifically expressed in certain tissues for a specific function to a completely different cellular context, to our knowledge, has not previously been explored. Thus far, examples of the ectopic expression of single genes in plant cells has typically been restricted to reiterate their function in other cell types to show functional relatedness or to recapitulate an evolutionarily divergent event using similar genes from different species. One of the best known examples is perhaps the expression of chimeric RNase genes in anthers of transformed tobacco (Nicotiana tabacum) and oilseed rape (Brassica napus) plants, which specifically destroyed the tapetal cells of developing pollen, resulting in male sterility (Mariani et al., 1990).
Here, we have examined the effect of the ectopic expression of PrpS and PrsS from P. rhoeas in vegetative cells of Arabidopsis, using characteristic markers of P. rhoeas SI-PCD to examine function. We show that the heterologous, ectopic expression of these genes, which specify a tightly controlled reproductive trait in the male gametophyte, can trigger a self-recognition:self-destruct response, resulting in growth arrest and PCD in vegetative sporophytic cells. Ectopic expression of PrpS and PrsS in Arabidopsis recapitulates major cellular aspects of the P. rhoeas SI response in vegetative cells, providing evidence that this heterologous, bipartite module can signal to similar cellular targets in different cell types.
RESULTS
PrsS Treatment Results in S-Specific Root Growth Inhibition of PrpS-Expressing Seedlings
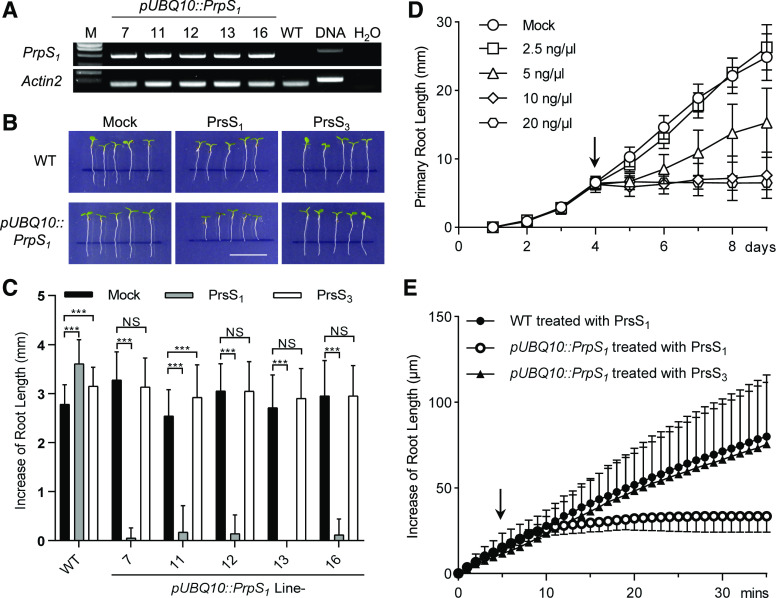
In P. rhoeas, the interaction of cognate PrpS and PrsS triggers a Ca2+-dependent signaling network in pollen, resulting in a rapid growth arrest followed by PCD of incompatible pollen after SI induction (Franklin-Tong et al., 1993, 1995, 1997). To examine if the PrpS-PrsS module might also work outside the specific context of pollen-pistil interactions, we examined if growth inhibition and PCD caused by the PrpS-PrsS module also could be triggered in other tissues. We therefore expressed PrpS1 under the control of a constitutive UBQ10 promoter in Arabidopsis plants and established five independent single T-DNA insertion lines (pUBQ10::PrpS1 lines 7, 11, 12, 13, and 16). Reverse transcription quantitative PCR (RT-qPCR) showed that PrpS1 mRNA was substantially expressed in these transgenic Arabidopsis seedlings (Fig. 1A; Supplemental Fig. S1). Focusing first on root growth, we did not observe differences in the root length between Arabidopsis Columbia-0 (Col-0) wild-type and pUBQ10::PrpS1 seedlings (Fig. 1B), demonstrating that PrpS1 expression alone did not alter seedling development. Next, we applied recombinant PrsS1 protein to 4-d-old root tips of wild-type and pUBQ10::PrpS1 seedlings. Exposure to PrsS1 protein did not show any inhibition effect of normal development and growth of Arabidopsis wild-type seedlings (Supplemental Fig. S2). However, PrsS1 treatment of pUBQ10::PrpS1 seedlings resulted in a rapid and complete inhibition of root growth (Fig. 1, B and C). The growth of pUBQ10::PrpS1 seedling roots was inhibited by recombinant PrsS1 protein in a dose-dependent manner (Fig. 1D). Treatment of pUBQ10::PrpS1 roots with 5 ng μL−1 PrsS1 significantly inhibited their growth rate, while 10 ng μL−1 or more completely blocked root elongation. This provides evidence that the PrpS-PrsS module, although its constitutive components are normally only expressed in pollen and pistil, respectively, and triggers a response in pollen specifically, can also act to trigger the inhibition of growth of vegetative, sporophytic cells.
Figure 1.
Expression of PrpS in transgenic Arabidopsis triggers root growth inhibition after cognate PrsS treatment. A, RT-PCR shows the expression of PrpS1 mRNA in pUBQ10::PrpS1 transgenic seedlings. Actin2 was used as a housekeeping gene control. Quantification of the relative expression levels is shown in Supplemental Figure S1. B and C, S-specific inhibition of root growth of pUBQ10::PrpS1 seedlings after PrsS1 treatment. B, Images of 4-d-old seedlings 24 h after treatment with PrsS proteins (10 ng µL−1). Black lines indicate the positions of root tips when treated. Only pUBQ10::PrpS1 seedlings (line 12) treated with PrsS1 (bottom, center) display inhibited root growth. This line was used for all the other experiments if not specified. Bar = 1 cm. C, Quantitation of increases in seedling root length from different transgenic lines (see A) treated with PrsS proteins (10 ng µL−1) 24 h after treatment (means ± sd; n = 20–25 seedlings). All five lines had root growth significantly inhibited by PrsS1 when comparisons were made with either PrsS3 or mock treatment for each line (two-way ANOVA multiple comparison: NS, not significant; ***P < 0.001). D, Root growth of pUBQ10::PrpS1 seedlings was inhibited by PrsS1 in a dose-dependent manner. The x axis indicates time (days) after transfer of plates to the growth chamber. The arrow indicates when the treatment was added. Data shown are means ± sd (n = 20–25 seedlings). E, PrsS1 treatment induces rapid root growth inhibition of pUBQ10::PrpS1 seedlings in an S-specific manner. The arrow indicates the time point of PrsS addition (10 ng µL−1). Two-way ANOVA shows that PrsS1 treatment significantly inhibited root growth (P < 0.001) while PrsS3 did not (P = 0.29), in comparison with wild-type (WT) seedlings treated with PrsS1. Data shown are means ± sd (n = 6).
The S-allele-specific inhibition is a key feature of the P. rhoeas SI response. To test this, we treated pUBQ10::PrpS1 and wild-type seedlings with either PrsS1 or PrsS3 recombinant protein. pUBQ10::PrpS1 seedling roots were strongly inhibited by the PrsS1 protein, while the PrsS3 protein had no effect; wild-type seedling roots were not inhibited by any treatment (Fig. 1, B and C). As only a cognate PrpS-PrsS combination caused growth inhibition, this shows that the S-determinants maintain their S-specificity in Arabidopsis roots.
As the SI response in pollen triggers rapid inhibition of incompatible pollen tube growth, we examined the timing of inhibition of growth of the roots in more detail, using a perfusion chamber system in combination with confocal microscopy (Krebs and Schumacher, 2013). Under these conditions, wild-type seedling roots elongated at a rate of ∼2.3 μm min−1, and the addition of PrsS proteins did not affect this (Fig. 1E; Supplemental Fig. S3, A and D). However, the addition of PrsS1 protein resulted in a rapid reduction of root growth of pUBQ10::PrpS1 seedlings (P < 0.001, two-way ANOVA; Fig. 1E). Growth was completely inhibited within 5 to 20 min after the addition of PrsS1 (Supplemental Fig. S3, B, E, and G). This inhibition was only observed with a cognate PrpS-PrsS interaction; in a compatible interaction, using the noncognate recombinant PrsS3 protein, roots of pUBQ10::PrpS1 seedlings grew at a similar rate to wild-type roots (P = 0.29, two-way ANOVA; Fig. 1E; Supplemental Fig. S3, C and F). Taken together, these data demonstrate that PrpS and PrsS interaction in roots rapidly elicits the inhibition of growth. This response is strikingly similar to what was observed in P. rhoeas pollen tubes during the SI response (Thomas and Franklin-Tong, 2004). However, a key difference is that root is a multicellular organ that increases its length by diffuse growth, whereas the pollen tube is a single cell elongating by tip growth. Together, these data demonstrate that the PrpS-PrsS bipartite signaling module can operate ectopically to inhibit the growth of vegetative cells.
PrsS Triggers Cell Death and DEVDase Activation in PrpS-Expressing Seedlings
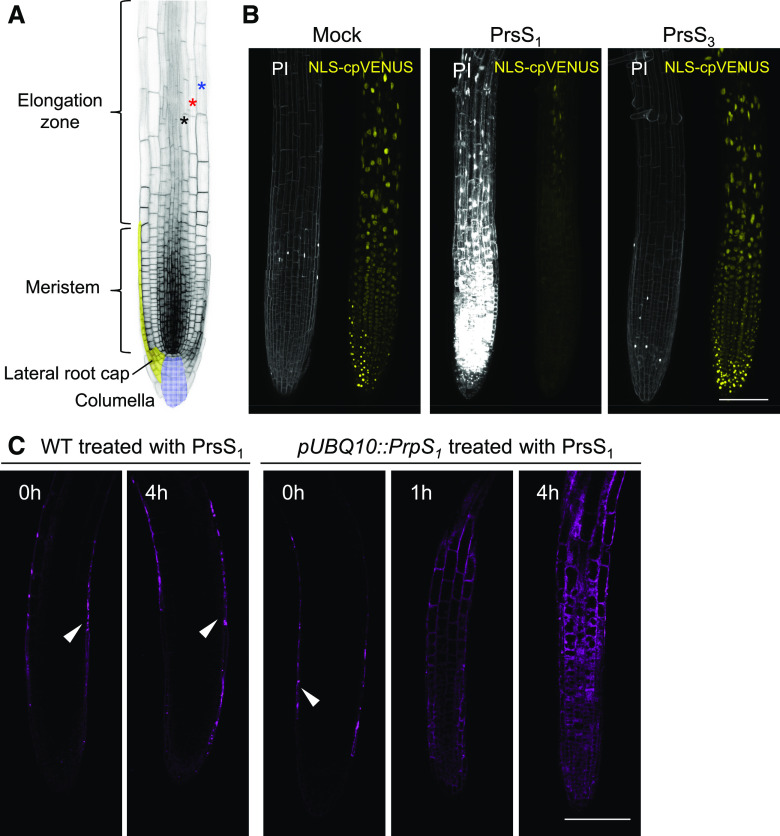
In P. rhoeas pollen, downstream of PrpS and PrsS interaction, after inhibition of growth, a distinctive PCD program is triggered. To investigate if this aspect of the SI response could be recapitulated in PrpS-expressing Arabidopsis roots, we examined root cells for evidence of death after PrsS treatment. We first examined plasma membrane permeability using propidium iodide (PI) staining and nuclear integrity using a nucleus-localized fluorescent protein marker line (pUBQ10::NLS-YC3.6 [Nagai et al., 2004] containing both nucleus-localized eCFP and cpVENUS). Twenty-four hours after the addition of PrsS1 to pUBQ10::PrpS1 seedling root tips, we found that many cells showed plasma membrane permeabilization to PI and loss of nuclear integrity, providing evidence of death (Fig. 2, A and B). This occurred in the whole root tip region, including the different cell types in the root cap, the root meristem, transition zone, and elongation zone (Fig. 2, A and B). Examining temporal changes to the root after PrsS1 treatment, we observed a gradual increase in the number of dead cells (Supplemental Fig. S4). A significant increase in PI staining was initially observed in the lateral root cap 1 h after PrsS1 treatment. At 2 h, cell death was observed in the columella root cap region. Cell death in the meristem was observed 4 h after PrsS1 treatment, and the number of cells affected increased over time. In contrast, in the controls (mock treated and treated with PrsS3), only a few PI-positive cells were observed in the root cap (Fig. 2B; Supplemental Fig. S4), which undergoes PCD as part of its regular developmental program (Fendrych et al., 2014). These results provide good evidence that cognate combinations of PrpS-PrsS, besides specifying SI, can operate to trigger cell death in vegetative cells.
Figure 2.
PrsS treatment results in cell death of PrpS-expressing seedling root cells. A, Image of a root illustrating different regions of the root tip. Cell files of epidermis, cortex, and endodermis are indicated by blue, red, and black stars, respectively. B, PrsS treatment results in the S-specific cell death of pUBQ10::PrpS1 roots. Representative images are shown for pUBQ10::PrpS1 roots expressing NLS-YC3.6 stained with PI 24 h after treatment (n > 6). No death was observed in roots mock treated with buffer (Mock), as shown by the absence of PI staining (PI; left images). Cognate PrsS1 treatment (10 ng µL−1) resulted in high levels of PI staining (white) in pUBQ10::PrpS1 seedling roots, but those treated with compatible PrsS3 (10 ng µL−1) did not. The NLS-cpVENUS signal (yellow) also reveals evidence of cell death, as it is lost after cognate PrsS1 addition. Images were taken 24 h after treatment. C, PrsS treatment activates a DEVDase activity in pUBQ10::PrpS1 seedling roots. DEVDase activity was monitored using the CR(DEVD)2 probe (purple). Besides endogenous DEVDase activity detected in the lateral root cap (indicated by white arrowheads), no DEVDase activity was observed in the root tip of wild-type (WT) seedlings before or after PrsS1 (10 ng µL−1) treatment. For the PrpS1-expressing root, DEVDase activity was observed within 1 h of PrsS1 addition in different cell types, including epidermis, cortex, and endodermis, of both the meristem and elongation zone of the root tip, and activity subsequently increased further. Representative images (n = 5) of single Z-optical sections are shown here; a full projection image is shown in Supplemental Figure S5. Bars = 100 µm.
To investigate if a similar pathway to that triggered in P. rhoeas pollen was utilized in the death of the root cells, as a DEVDase is implicated as a key PCD executor (Bosch and Franklin-Tong, 2007) in P. rhoeas pollen SI-PCD, we examined this protease activity in the pUBQ10::PrpS1 seedling roots. The chemically synthesized probe CR(DEVD)2 was employed to detect DEVDase activity in roots in vivo. In wild-type roots, consistent with the occurrence of normal, constitutive root cap PCD (Fendrych et al., 2014), DEVDase activity was detected in the outermost layer of the root in the root cap prior to treatment (Fig. 2C; Supplemental Fig. S5A). The addition of PrsS proteins to wild-type seedling roots did not affect DEVDase activity even after 4 h (Fig. 2C; Supplemental Fig. S5A). However, treatment of pUBQ10::PrpS1 roots with PrsS1 induced the activation of DEVDase activity in several different zones and cell types, including the root cap, meristem, and elongation zone (Fig. 2C; Supplemental Fig. S5B). When pUBQ10::PrpS1 roots were treated with PrsS3 protein, no major differences in DEVDase activity were observed compared with that in untreated roots (Supplemental Fig. S5, C and D). This demonstrates that DEVDase activation is induced by PrpS-PrsS interaction in these PrpS1-expressing Arabidopsis seedling roots and that DEVDase activation is S-allele specific in these vegetative tissues. This suggests that a similar pathway is reconstituted in these vegetative cells by this bipartite module.
PrsS Treatment Triggers an S-Specific Ca2+ Signature in PrpS-Expressing Roots
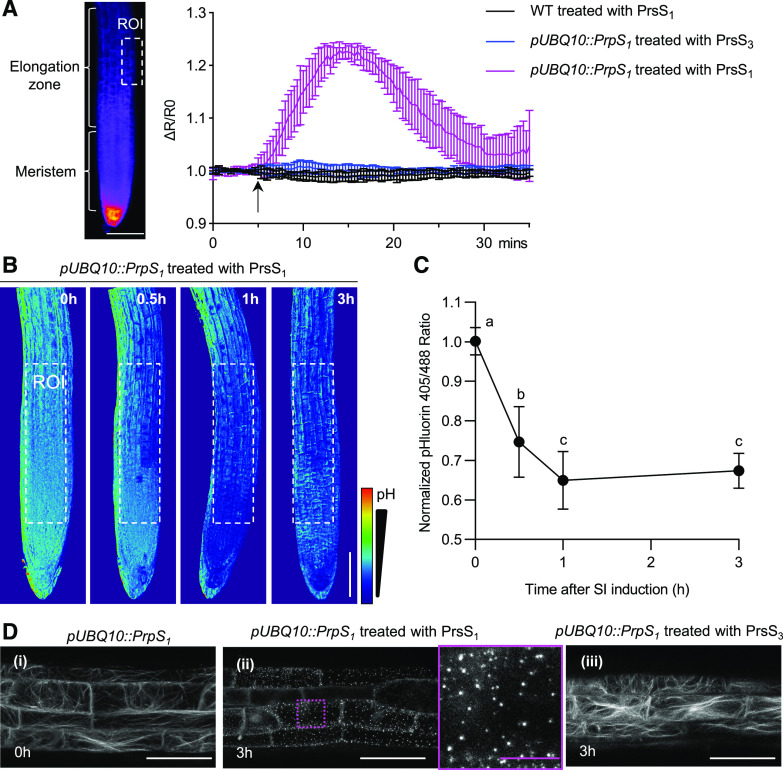
We next investigated whether other hallmark downstream features of the P. rhoeas SI response were triggered in the PrpS-expressing roots after the addition of cognate PrsS proteins. To monitor the [Ca2+]cyt spatiotemporally, the genetically encoded calcium indicator YC3.6 (Nagai et al., 2004; Krebs et al., 2012) was coexpressed with PrpS1 in Arabidopsis seedlings. We observed no obvious change in the [Ca2+]cyt when wild-type seedlings were treated with PrsS1 protein (Fig. 3A; Supplemental Fig. S6A). However, when PrsS1 protein was added to PrpS1-expressing seedlings, we detected transient [Ca2+]cyt increases in their roots. The increase was first observed in the elongation zone of the root, peaking ∼10 min after PrsS protein addition, and subsequently gradually decreased back to the approximate resting level within ∼25 min (Fig. 3A; Supplemental Fig. S6B). An increase in [Ca2+]cyt in the meristem and columella regions was also observed (Supplemental Fig. S6B). These [Ca2+]cyt dynamics were not observed in PrpS1-expressing seedlings treated with PrsS3 protein (Fig. 3A; Supplemental Fig. S6C), demonstrating that this [Ca2+]cyt response was S-specific. We also examined roots for increases in nuclear Ca2+ ([Ca2+]nuc) after the addition of PrsS by introducing an NLS-YC3.6 construct into the pUBQ10::PrpS1 transgenic seedlings. We observed increases in [Ca2+]nuc at the root tip, including columella, meristem, and elongation zone (Supplemental Fig. S7), which were spatiotemporally similar to the [Ca2+]cyt response. Our observation of unsynchronized Ca2+ signatures in different parts of the root hints at a possible transmission of Ca2+ signaling between neighboring tissues in the pUBQ10::PrpS1 root triggered by PrsS1. As increases in [Ca2+]cyt are a key feature of the SI response, our data suggest that we may be observing an SI-like response in vegetative tissues.
Figure 3.
Key hallmarks of the P. rhoeas SI response are observed in the Arabidopsis PrpS-expressing roots after PrsS treatment. A, PrsS induces transient increases in [Ca2+]cyt in cognate PrpS-expressing Arabidopsis seedling roots. The quantitation of changes in [Ca2+]cyt was measured in Arabidopsis seedling roots in the elongation zone (region of interest [ROI]; left image, dotted box) using the Ca2+ marker YC3.6 signal expressed as fractional ratio changes (ΔR/R0; mean ± sd; n = 6). After PrsS addition (10 ng µL−1; indicated by the arrow), an increase in [Ca2+]cyt was observed in the elongation zone of pUBQ10::PrpS1 roots treated with PrsS1 (magenta); controls (black and blue) did not display this response. WT, Wild type. B and C, PrsS triggers acidification in cognate PrpS-expressing Arabidopsis seedling root. B, Ratiometric (405:488 nm) imaging of pUBQ10::PrpS1 roots expressing the pH sensor pHluorin after PrsS1 (10 ng µL−1) addition revealed that the signal ratio decreased, indicating cytosolic acidification. Bar = 100 µm. C, Quantification of the pHluorin ratio measured in the region of interest (white dotted boxes in B) of these roots shows a significant decrease in [pH]cyt after SI induction (means ± sd; n = 12; one-way ANOVA with multiple comparison test, between A and B and between B and C, P < 0.001; C versus C is not significant). The pHluorin ratio at time 0 h was normalized to 1. D, PrsS treatment triggers S-specific loss of actin filaments and the formation of actin foci in roots. Representative images (n > 6) of confocal imaging of pUBQ10::PrpS1 roots (elongation zone) expressing LifeAct-mRuby2 are shown. i, Prior to treatment, typical longitudinal actin filament bundles were observed. ii, At 3 h after treatment with PrsS1 (10 ng µL−1), actin foci were observed (magenta dotted box and magnification of this region to the right). iii, The same line at 3 h after the addition of PrsS3 proteins (10 ng µL−1) displayed normal longitudinal actin filament bundles. Images are full projections. White bars = 50 µm, and magenta bar = 10 µm.
PrsS Induces S-Specific Cytoplasmic Acidification in PrpS-Expressing Roots
Another hallmark feature of the P. rhoeas SI is cytosolic acidification. We examined PrpS1-expressing roots treated with PrsS proteins for alterations in [pH]cyt using the genetically encoded pH-sensitive GFP variant, pHluorin (Moseyko and Feldman, 2001). After 30 min of PrsS1 treatment, PrpS1-expressing roots displayed a significant drop in [pH]cyt (P < 0.0001, one-way ANOVA; Fig. 3, B and C). Further cytoplasmic acidification continued until ∼1 h, and levels remained low, as the pHluorin 405:488 ratio at 3 h was not significantly different to that at 1 h (P = 0.7975, one-way ANOVA; Fig. 3, B and C). This rapid drop in [pH]cyt was only observed in PrpS1-expressing roots treated with cognate PrsS1 proteins and not in wild-type seedlings treated with PrsS1/3 proteins or PrpS1-expressing roots treated with PrsS3 proteins (Supplemental Fig. S8). The temporal pH dynamics after PrpS-PrsS interaction in Arabidopsis root was similar to that observed in P. rhoeas pollen after SI induction. These data demonstrate that the cognate PrpS-PrsS interaction in Arabidopsis roots induces cytoplasmic acidification and further support the idea that a P. rhoeas SI-like signaling pathway is triggered in Arabidopsis roots after the interaction of cognate PrpS and PrsS.
PrsS Triggers Actin Cytoskeletal Remodeling in PrpS-Expressing Seedling Roots
As highly characteristic alterations to the actin cytoskeleton are a key feature of P. rhoeas SI, we examined the dynamics of the actin cytoskeleton to see if this characteristic response also took place in roots. We added recombinant PrsS to pUBQ10::PrpS1 transgenic seedling roots that also expressed the genetically encoded actin marker, LifeAct-mRuby2 (Dyachok et al., 2014; Bascom et al., 2018). Wild-type roots displayed typical actin filament bundles before and after PrsS1 application (Supplemental Fig. S9A). PrpS1-expressing roots showed a similar actin organization prior to the addition of recombinant PrsS1 (Fig. 3D). However, by 60 min after PrsS1 application, the mRuby2 signal in the PrpS1-expressing seedling roots was much reduced, fragmented actin filaments were detected, and small punctate actin foci had formed (Supplemental Fig. S9B). At 3 h, the actin foci were brighter and larger (Fig. 3D; Supplemental Fig. S9B). These distinctive actin alterations are very similar to what has been described for incompatible pollen in the P. rhoeas SI response (Snowman et al., 2002). In roots, we also observed abnormally thick actin bundles and actin aggregation around the nucleus at 3 h after cognate PrsS treatment (Supplemental Fig. S9B). PrpS1-expressing roots did not undergo any actin remodeling after treatment with recombinant PrsS3 protein (Fig. 3D), demonstrating that actin remodeling is an S-specific event. Together, these observations demonstrate that interaction of PrpS and PrsS in Arabidopsis roots triggers a signaling network involving hallmark features observed in incompatible pollen in the P. rhoeas SI response (Snowman et al., 2002), suggesting that they can recapitulate an SI-like response in vegetative tissues.
PrsS Treatment Results in an S-Specific Cell Death of PrpS-Expressing Leaf Protoplasts
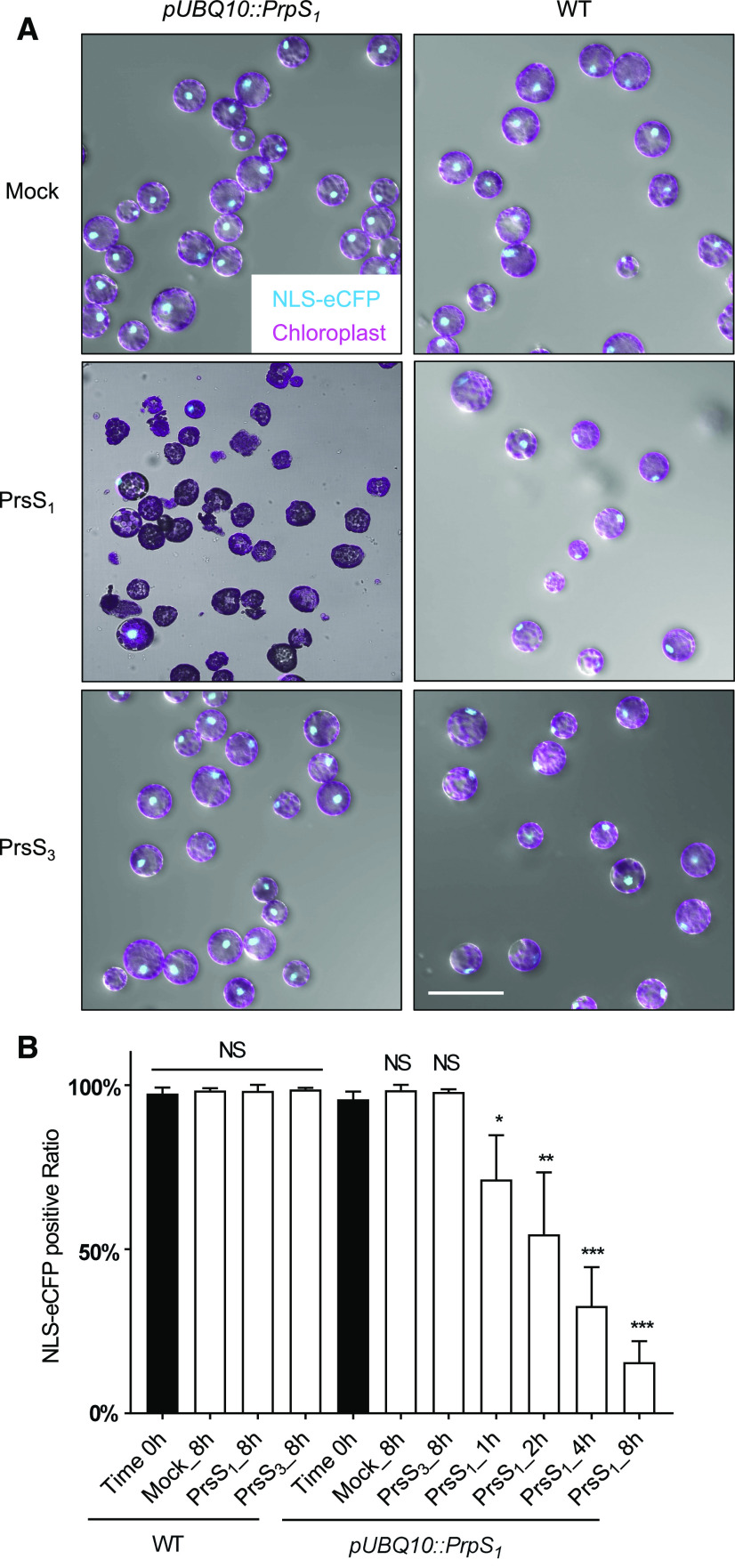
As our data suggested that a P. rhoeas SI-PCD-like signaling pathway could be triggered in Arabidopsis root cells, we wondered whether this response might also be observed in other somatic cell types. We therefore examined whether the viability of leaf protoplasts derived from PrpS1-expressing Arabidopsis plants might also be affected by PrsS1 protein treatment. We utilized a nucleus-localized eCFP (NLS-eCFP) signal as a cell viability marker for leaf protoplasts. After 8 h of incubation with PrsS1 protein, only PrpS1-expressing protoplasts showed a loss of the NLS-eCFP signal, together with abnormal cell shape and leakage of cellular contents (Fig. 4A). In contrast, treatment with PrsS3 protein or mock treatment with buffer had no effect; these control protoplasts appeared viable and intact and the same as untreated wild-type protoplasts (Fig. 4A). Quantitative, temporal analysis showed a gradual and significant decrease in the ratio of protoplasts displaying a positive NLS-eCFP signal. Prior to treatment, this was 95.7%, and it decreased to 71.2% at 1 h (P < 0.05, one-way ANOVA; Fig. 4B), progressively decreasing down to 15.6% after 8 h (P < 0.001, one-way ANOVA; Fig. 4B). This was not observed in the wild-type protoplasts or PrpS1-expressing protoplast incubated with PrsS3 proteins, which displayed NLS-eCFP signals not significantly different from the untreated controls (P = 0.7532, one-way ANOVA; Fig. 4B). These data demonstrate that the interaction between PrpS and PrsS in leaf protoplasts is sufficient to induce cell death in an S-specific manner. This provides further evidence that the S-determinants can operate ectopically in totipotent protoplasts.
Figure 4.
PrpS-expressing leaf protoplasts treated with PrsS undergo S-specific cell death. A, Representative images of pUBQ10::PrpS1/pUBQ10::NLS-YC3.6 (line 11) leaf protoplasts after PrsS treatment (10 ng µL−1) for 8 h showing bright-field images combined with autofluorescent chloroplast signals (magenta) and fluorescent NLS-eCFP signals (turquoise), indicating nuclear integrity. Only PrpS1-expressing protoplasts treated with cognate PrsS1 (center, left) showed loss of the nuclear signal, abnormal cell shape, and leakage of cellular content. This provides evidence for S-specific cell death triggered by cognate PrsS1 in undifferentiated cells. Bars = 100 µm. B, Quantification of the loss of nuclear integrity in pUBQ10::PrpS1/pUBQ10::NLS-YC3.6 (line 11) leaf protoplasts over time by counting NLS-eCFP signals (turquoise). PrsS1 or PrsS3 treatment (10 ng µL−1) did not affect the nuclear integrity of wild-type (WT) protoplasts. The percentage of PrpS1-expressing protoplasts with an NLS-eCFP signal was significantly reduced by PrsS1 treatment, from ∼96% at 0 h to ∼16% at 8 h, but no significant difference was observed with PrsS3 after 8 h. Data show means ± sd; 100 to 150 cells were counted in each treatment for each time point, n = 3 experiments. One-way ANOVA with multiple comparisons of time 0 h with each of the other treatments at each time point (NS, not significant; *P < 0.05; **P < 0.01; and ***P < 0.001).
Coexpression of PrsS and PrpS Triggers S-Specific Cell Death in Whole Plants
Finally, we investigated whether PrsS, when expressed in planta, was able to exert the same effect as treatment with recombinant PrsS protein in whole plants. We introduced PrsS into the pUBQ10::PrpS1 background line under the control of an estradiol-inducible promoter (pH3.3::XVE::PrsS1/3/pUBQ10::PrpS1, referred to as XVE::PrsS1/3/PrpS1 hereafter). XVE::PrsS1/PrpS1 seeds completely failed to germinate on medium containing estradiol. In contrast, no significant difference in the germination rate (95.5%–97.5%) of the background pUBQ10::PrpS1 line and the XVE::PrsS3/PrpS1 line was observed before and after estradiol induction (Table 1). This effect on seed germination demonstrated that simultaneous expression of cognate PrpS and PrsS in seeds induces cell death in planta.
Table 1. Coexpression of cognate PrsS and PrpS completely abolishes Arabidopsis seed germination.
Newly harvested seeds of different lines were sown on LRC2 plates containing estradiol (10 µm) or solvent (ethanol; mock control), and germination rate was recorded 4 d after being placed into a growth chamber. The numbers in parentheses indicate the number of germinated seeds/the total number of seeds being examined. After mock treatment, the seed germination rates of the lines pUBQ10::PrpS1, XVE-PrsS1/PrpS1, and XVE-PrsS3/PrpS1 are comparable to each other (∼95%–97%). Estradiol induction does not affect the germination rate of pUBQ10::PrpS1 or XVE-PrsS3/PrpS1, whereas induced expression of PrsS1 completely abolishes XVE-PrsS1/PrpS1 germination.
| Treatment | pUBQ10::PrpS1 | XVE-PrsS1/PrpS1 | XVE-PrsS3/PrpS1 |
|---|---|---|---|
| Mock | 95.5% (128/134) | 96.1% (147/153) | 97.5% (197/202) |
| Estradiol | 96.6% (113/117) | 0.0% (0/108) | 97.0% (131/135) |
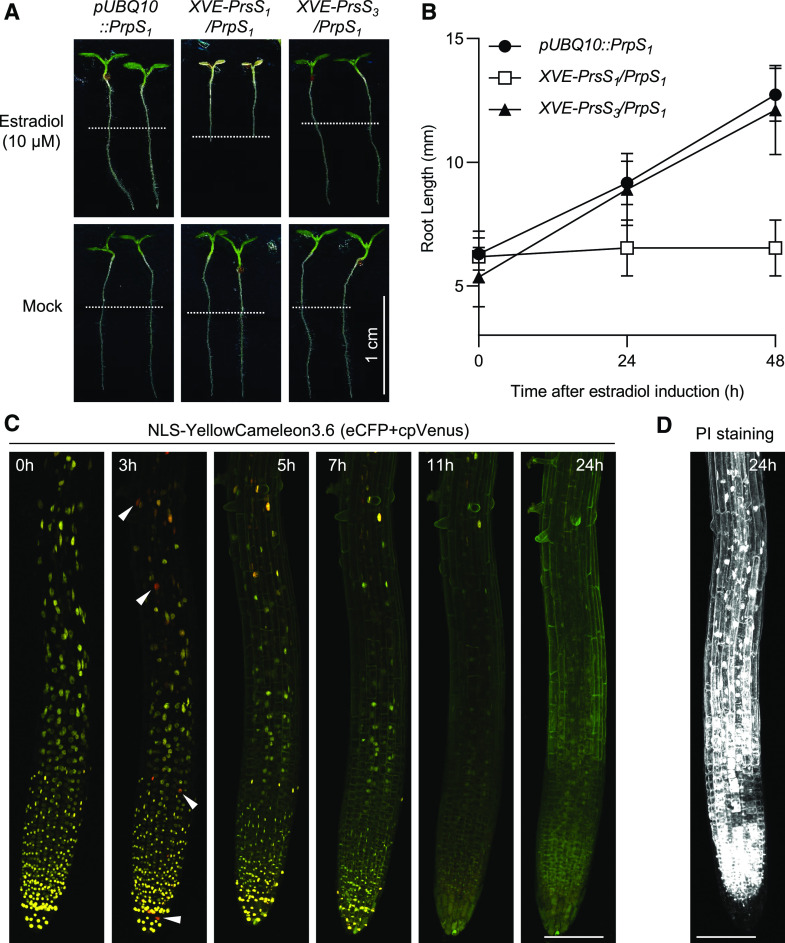
To test this hypothesis and examine cell viability after estradiol induction further, we induced PrsS1/3 expression by transferring XVE::PrsS1/3/PrpS1 seedlings to medium containing estradiol. Root growth was rapidly inhibited after transfer to estradiol, whereas pUBQ10::PrpS1 and XVE::PrsS3/PrpS1 seedlings were not affected (Fig. 5, A and B; Supplemental Fig. S10). Strikingly, the XVE::PrsS1/PrpS1 seedlings were stunted and cotyledons were white after 48 h on estradiol (Fig. 5A). These data show that the estradiol-induced expression of PrsS1 (Supplemental Fig. S11) is sufficient to cause S-specific root growth inhibition and subsequent systemic PCD of the entire pUBQ10::PrpS1 seedling. Moreover, time-lapse examination of XVE::PrsS1/PrpS1 seedling roots expressing NLS-YC3.6 after estradiol treatment revealed localized increases in [Ca2+]nuc 3 h after estradiol induction (Fig. 5C), providing evidence for estradiol-induced expression of PrsS and subsequent PrpS-PrsS interaction. At 5 h, a dramatic decrease in nuclear integrity was observed in root tips, and this continued for up to 11 h, when almost no cells with intact nuclei were observed in root tips (Fig. 5C). PI staining showed that besides the loss of nuclear integrity, plasma membrane permeability was also affected (Fig. 5D). Thus, cell death triggered by the coexpression of cognate PrpS and PrsS was observed in whole root tissues (Supplemental Fig. S12). Control plants that expressed noncognate PrsS3 and PrpS1 exhibited no major changes in nuclear integrity after estradiol induction (Supplemental Fig. S12). Together, these data demonstrate that the coexpression of cognate PrpS and PrsS induces the death of the whole plant in Arabidopsis. This suggests that this two-component system is capable of triggering cell death when they are expressed together, regardless of tissue or cell type.
Figure 5.
Ectopic expression of PrpS and PrsS in Arabidopsis triggers cell death in whole seedlings in an S-specific manner. A and B, Root growth of XVE-PrsS1/PrpS1 seedlings was inhibited after estradiol induction in an S-specific manner. A, Four-day-old seedlings were transferred to new medium containing 10 µm estradiol. Images were taken 48 h after treatment. White dashed lines indicate the positions of the root tips at the time of transfer. Estradiol-induced expression of PrsS1 resulted in the death of the whole seedling (top, center), whereas no obvious effect was observed when PrsS3 was expressed (top, right). B, Quantification of root length at 24 and 48 h after estradiol induction reveals the inhibition of root growth in XVE-PrsS1/PrpS1 lines upon transfer to estradiol plates, whereas the growth of roots of XVE-PrsS3/PrpS1 and pUBQ10::PrpS1 seedlings was not affected (means ± sd; n = 20 seedlings). C, Estradiol induction resulted in nuclear disintegration and cell death of XVE-PrsS1/PrpS1 seedlings. NLS-YC3.6 was used to monitor the nuclear integrity after estradiol induction over time. Confocal images of merged eCFP (green) and cpVENUS (red) channels are shown. The yellow signal (green-red overlap) shows intact nuclei; extensive nuclear disintegration (loss of yellow signal) was observed as early as 5 h after estradiol induction and was almost complete by 11 h. The fluorescence signal was so weak at 5 h that the confocal laser power was increased from 1.5% (0 and 3 h) to 3.5% (5, 7, 11, and 24 h) to allow visualization of the seedling. NLS-YC3.6 monitors [Ca2+]nuc and reveals that increases (red signal; indicated by white arrowheads) could be observed 3 h after induction. Bar = 100 µm. D, PI staining of a representative root at 24 h after estradiol induction reveals that virtually all the cells are dead (white signal). Bar = 100 µm.
DISCUSSION
The S-locus in P. rhoeasr encodes a pair of S-determinants, PrpS and PrsS. Their tissue- and development-specific expression, solely in pollen and pistil, respectively, is tightly regulated, and they interact in an allele-specific manner to specify and mediate the SI response within the male gametophyte pollen during early pollination. We previously demonstrated functional intergeneric transfer of the P. rhoeas S-determinants to the reproductive system of Arabidopsis (de Graaf et al., 2012; Lin et al., 2015). Here, we show that PrpS and PrsS do not just function as S-determinants to specify SI but that they can operate beyond their usual reproductive context. We demonstrate that the effect of this self-recognition:self-destruct mechanism is not confined to the male gametophyte but that PrpS and PrsS can also act as a heterologous bipartite module to trigger a canonical SI-like response, resulting in growth inhibition and PCD independent of the reproductive context when ectopically expressed in sporophytic tissues of Arabidopsis.
Pollen is a highly specialized gametophytic organism with very specific, precise functions related to reproduction. As such, pollen displays a distinct molecular profile that is distinct from all other plant tissues (da Costa-Nunes and Grossniklaus, 2003; Honys and Twell, 2004; Mergner et al., 2020). The finding here that PrpS and PrsS can act outside of this reproductive context to trigger an SI-like growth arrest and PCD response in vegetative cells of the sporophyte when expressed ectopically is surprising, exciting, and not predicted by our earlier studies. To our knowledge, ectopic transfer of a two-component module from the reproductive context into the vegetative sporophytic one has not previously been reported in plants. This is a milestone, as it demonstrates that these two genes, which are normally responsible for controlling a reproductive trait, are sufficient to trigger signaling to growth arrest and cell death in numerous cell types, independent of their particular tissue-specific and developmental context.
Our data showing that the PrpS-PrsS module can act ectopically provide potential new clues to the possible origin and evolution of bipartite genetic modules that act in cell-cell signaling networks. PrsS has homologs in a large family named after them, the SPHs (also known as plant self-incompatibility protein S1 homologs in the databases), comprising more than 1,800 homologous sequences in more than 70 plant species as well as in fungi and metazoa (Ride et al., 1999; Rajasekar et al., 2019). Over 90 SPHs have been identified in Arabidopsis (Rajasekar et al., 2019). Based on the large number of SPH family members, all encoding proteins with signal peptides, together with their wide distribution, it has previously been proposed that they may be ligands involved in a wide range of signaling pathways (Ride et al., 1999). It has been suggested that this family of proteins may have evolved to act as a versatile and stable scaffold to display a variety of peptides in the predicted extracellular loops, each interacting with a different receptor (Rajasekar et al., 2019). Our findings here, showing that PrsS can trigger responses in vegetative tissues, provide further hints that (depending on how they have evolved) perhaps other SPHs may be involved in signaling in different tissues.
PrsS and SPHs are members of the CRPs, which include the Brassica spp. pollen S-determinant SCR/SP11 (Schopfer et al., 1999; Takayama et al., 2000), defensins (Bircheneder and Dresselhaus, 2016), LUREs (Okuda et al., 2009; Takeuchi and Higashiyama, 2016), and rapid alkanization factors (RALFs; Pearce et al., 2001; Li and Yang, 2018), which are known to interact with receptors to activate diverse signaling networks involved in plant growth, defense, and reproduction (Wheeler et al., 2010; Marshall et al., 2011; Takeuchi and Higashiyama, 2016; Liu et al., 2017). Although comparatively few secreted peptides have been shown to interact with receptors in plants, genome analysis has revealed the existence of hundreds of predicted secreted proteins that may act as ligands (Lease and Walker, 2006). It has been suggested that CRPs have diversified for a huge variety of specialized functions (Manners, 2007; Silverstein et al., 2007; Bircheneder and Dresselhaus, 2016); rapid evolution from an origin in plant defense to regulate plant reproduction has been proposed (Bircheneder and Dresselhaus, 2016). Analysis of Arabidopsis SPH genes in the available databases reveal that they are mainly, but not exclusively, expressed in reproductive tissues (Supplemental Figs. S13 and S14; Mergner et al., 2020). Notably, several SPHs are expressed in silique septum, silique valves, flower pedicles, and senescent leaves, which all undergo PCD in various cellular/developmental contexts (Beers, 1997; Gómez et al., 2014). This hints that this family may have evolved a general function in several diverse tissues to signal to growth and PCD, as we have found for PrsS in this study.
Examination of the literature and databases reveals that no functional data are currently available for any Arabidopsis SPHs. Nevertheless, association networks for one of the SPH genes, AT1G51250, using STRING analysis (Szklarczyk et al., 2019), for example, reveals associations/putative interactions with several proteins. These include APPB1 and AT4G02250, which are plant pectin methylesterase inhibitor proteins, implicated in mediating growth; RALFL8, RALFL15, and RALFL26 (RALF-like cell signaling peptides), implicated in regulating plant stress, growth, and development; and LCR72, a Cys-rich peptide, predicted to encode a PR protein, that itself interacts with other defensins. These interactions hint that this SPH may signal to regulate growth and stress response. As both RALFs and pectin methylesterase inhibitors are broadly expressed (Supplemental Figs. S15 and S16; Mergner et al., 2020), this suggests that some SPHs may also potentially interact with these proteins to mediate these responses in various tissues. However, to our knowledge, no studies to date have identified a function for any SPH in another tissue, and as no other partners for SPHs have been identified to date, we cannot speculate much further about the possible functions of putative homologs of SPHs or their putative interactors, which is currently a black box. Although PrpS, being a small transmembrane protein with no known homologs, is not a receptor in the classic sense, our findings here, showing that the PrpS-PrsS module can act as a receptor-ligand-like module outside its usual reproductive context in vegetative tissues, provide a rare example of a specialized bipartite gene module that can act in a cell-autonomous manner. Thus, our finding that PrpS-PrsS can function in vegetative tissues, together with information on SPH homologs and their possible interactors, may provide clues about how the SPHs might potentially have coevolved to function in different cell types, an interesting avenue to be explored in the future.
Although the downstream cellular responses observed here in Arabidopsis roots in response to PrsS are strikingly similar to what was observed in P. rhoeas pollen tubes during the SI response (Wilkins et al., 2014; Wang et al., 2019), a key difference is that roots utilize diffuse growth, whereas a pollen tube elongates by tip growth. Diffuse growth is used by most plant cells and is often contrasted to tip growth. However, despite differences in spatial patterning, there may be considerable overlap in the regulatory processes involved in these two types of growth (Yang, 2008; Cosgrove, 2018). Our evidence that the PrpS-PrsS module can also inhibit diffuse growth and does not apparently distinguish between these two types of growth supports this concept. Moreover, it is of interest that the peptides of several other CRP members function to regulate different types of growth. For example, RALFs are involved in the arrest of root growth and development (Pearce et al., 2001; Haruta et al., 2014; Blackburn et al., 2020), LUREs (specifically expressed in synergid cells) act to control directional growth of pollen tubes to the embryo sac (Okuda et al., 2009), SCR/SP11 act as the male S-determinant in Brassica spp. to inhibit self pollen (Schopfer et al., 1999; Takayama et al., 2000), and ZmES4 induces pollen tube growth arrest and bursting to release sperm cells during fertilization (Amien et al., 2010; for review, see Kanaoka and Higashiyama, 2015; Higashiyama and Yang, 2017; Blackburn et al., 2020). Further studies are needed to determine if there is a common growth-arrest mechanism triggered by these different CRP-mediated signaling pathways. Moreover, it would be of considerable interest to investigate if PrsS interacts with RALFs as a putative candidate player in the SI signaling pathway in pollen, as they are involved in signaling via reactive oxygen species to inhibit primary root elongation (Haruta et al., 2014) and it has been established that reactive oxygen species are involved in the SI-PCD response in pollen (Wilkins et al., 2011).
We previously showed that PrpS and PrsS could function to mediate SI and PCD in Arabidopsis pollen, despite the fact that this species is self-compatible (de Graaf et al., 2012; Lin et al., 2015). We proposed that the P. rhoeas SI system worked in Arabidopsis pollen because it could recruit existing proteins to form new signaling networks, by multitasking of endogenous components that can act in signaling networks that they do not normally operate in, to provide a specific, predictable physiological outcome. This successful transfer between species suggested that the signaling network and cellular targets downstream of the PrpS-PrsS interaction might be present in a wide range of angiosperm species (de Graaf et al., 2012), as this was the simplest explanation of why these genes work in such an evolutionarily diverged (more than 100 million years; Bell et al., 2010) species. However, we did not explore whether this might extend beyond the particular context of pollen involved in the SI response. Here, we have extended our studies to show that this pair of genes can also act in other cell types in Arabidopsis. Our findings here, showing that this module can trigger growth arrest and PCD in sporophytic vegetative cells, provide firm evidence for this idea of plug and play and extends it, by showing that PrpS-PrsS can also act in an ectopic situation to trigger a signaling network and response that appears to be common and ubiquitously expressed and not just restricted to pollen. As key components can be harnessed in different cell types to reconstitute key P. rhoeas SI-PCD-like phenomena in vegetative cells, this provides hints about the functional diversification and recruitment of preexisting components and the plasticity of cell signaling downstream of the PrpS-PrsS interaction leading to growth arrest and PCD in plant cells.
Our study has substantially extended previous studies (de Graaf et al., 2012; Lin et al., 2015) and reveals that the events downstream of the P. rhoeas PrpS-PrsS interaction can be triggered in different cell types of Arabidopsis. This lays the foundations for new opportunities to elucidate key mechanisms triggered by cognate PrpS-PrsS interactions. Although the P. rhoeas SI system has provided an excellent model system to investigate cell-cell recognition, intracellular signaling, and PCD at the molecular level, the extremely limited genetic resources in this system have provided an obstacle to progress, as certain approaches were not possible. Our findings here suggest that Arabidopsis plants express an SI-like response in vegetative tissues with all the key features of P. rhoeas SI, opening up new opportunities to genetically dissect the signaling networks involved. Expression of the bipartite PrpS-PrsS module in different tissues has the potential to be applied to devise biochemical or genetic approaches to search for downstream components. Using this system in vegetative tissue or whole plants has the advantage that it overcomes the bottleneck that many reproductive researchers are faced with (i.e. that of limited material), as collecting sufficient pollen at the correct developmental stage is laborious, time-consuming, and difficult to scale up. Being able to perform experiments on bulk plant tissue or on whole plants could allow us to identify new genes/proteins involved in the downstream pathway; these could then be examined and validated in pollen to establish if they authentically play a role in the SI response. For example, root growth assays could provide a simple assay for screening large sets of T-DNA mutants or chemical library screening. Biochemical approaches, such as purification of candidate proteins or profiling of PrpS-PrsS-induced metabolomic changes using pollen, are generally impossible due to the small amount of tissue available. Using a heterologous expression system to enable a bulk purification, from leaves or roots, for example, of putative proteases with caspase-like activities or actin-binding proteins implicated in the actin remodeling might be possible. In conclusion, this ectopic Arabidopsis self-recognition:self-destruct system will allow us to test new hypotheses about the cellular mechanisms and genetic components involved in the SI-PCD response and tip growth of plant cells in the future.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were gas sterilized, sown out on LRC2 plates (2.15 g L−1 Murashige and Skoog medium basal salts [Duchefa Biochemie], 0.1 g L−1 MES, pH adjusted to 5.7 with KOH, and 1% [w/v] Plant Tissue Culture Agar [Neogen]), and stored in a cold room (4°C) for 3 d before being moved to a growth chamber for vertical growth with continuous light emitted by white fluorescent lamps (intensity of 120 µmol m−2 s−1) at 22°C. Unless specifically stated, 4-d-old seedlings after being placed in the growth chamber were used for experiments. When necessary, seedlings were transferred to Jiffy pots in soil and grown under glasshouse conditions under a 16-h-light/8-h-dark regime at 22°C. Plants were protected by Arasystem to stop the pollen spreading when flowering and keep the seed stocks pure. Seeds were collected when the plants were completely dry and kept at room temperature or 4°C for long-term storage.
Cloning and Transgenic Lines
All the expression vectors were generated using either Gateway cloning (Invitrogen) or GreenGate cloning (Lampropoulos et al., 2013). High-fidelity Phusion DNA polymerase (New England Biolabs) was used for all the DNA fragment amplification.
The expression clones pUBQ10::PrpS1 were obtained using Gateway cloning (Invitrogen). PrpS1 gDNA was amplified using primers F-attB1-PrpS1/R-attB2-PrpS1 with gDNA of line BG16 (de Graaf et al., 2012) as template. The resulting PCR fragments were cloned into pDONR221 using BP clonase (Invitrogen) to obtain pEN-L1-PrpS1-L2. The entry vector pEN-L4-pUBQ10-R1 was obtained from the PSB Gateway Vector collection (Fendrych et al., 2014). These entry clones were recombined into Gateway destination vector pB7m24GW (Karimi et al., 2002) using LR Clonase II plus enzyme (Invitrogen) to obtain the expression clone pUBQ10::PrpS1.
The expression clones pH3.3::XVE::PrsS1/3 were generated using Gateway cloning (Invitrogen). The DNA fragment of the H3.3 promoter was amplified using K1H3-F and X1H3-R primers. PCR products were digested using KpnI and XhoI restriction enzymes followed by DNA gel purification. Plasmid pEN-L4-pRPS5A::XVE-R1 (Huysmans et al., 2018) was digested using the same restriction enzymes, followed by DNA gel electrophoresis. The vector backbone without the RPS5A promoter was cut out and purified. The KpnI-pH3.3-XhoI DNA fragment was ligated into the linearized vector backbone to generate pEN-L4-pH3.3::XVE-R1. PrsS1/3 gDNA was amplified using primer sets F-attB1-PrsS1/3/R-attB2-PrsS1/3 with plasmid pSLR1::PrsS1/3 (Lin et al., 2015) as template. The resulting PCR fragments were cloned into pDONR221 using BP clonase (Invitrogen) to obtain pEN-L1-PrsS1/3-L2. These entry clones were recombined into Gateway destination vectors pB7m24GW-FAST-Green using LR Clonase II plus enzyme (Invitrogen) to obtain the expression clone pH3.3::XVE::PrsS1/3.
The dual-expression clones pUBQ10::PrpS1_pUBQ10::NLS-YC3.6, pUBQ10::PrpS1_pUBQ10::YC3.6, pUBQ10::PrpS1_pUBQ10::pHGFP, and pUBQ10::PrpS1_pUBQ10::LifeAct-mRuby2 were generated using GreenGate cloning. Promoter UBQ10 was amplified using primer sets F-A-pUBQ10/R-B-pUBQ10 and F-D-pUBQ10/R-E-pUBQ10 with entry vector pEN-L4-pUBQ10-R1 as template. The resulting PCR fragments were cloned into pJET1.2 using the CloneJET PCR Cloning Kit (Thermo Fisher) to obtain the entry vectors pEN-A-pUBQ10-B and pEN-D-pUBQ10-E. Similarly, pEN-B-PrpS1-C was generated by cloning of the PrpS1 DNA fragment amplified using primers F-B-PrpS1/R-C-PrpS1 into pJET1.2. To create entry vectors for terminator RBCS (tRBCS), NLS-YC3.6, YC3.6, and tMAS, the corresponding DNA fragments were amplified using primer sets F-C-tRBCS/R-D-tRBCS, F-E-NLS/R-F-YC3.6, F-E-YC3.6/R-F-YC3.6, and F-F-tMAS/R-G-tMAS with expression vector pUBQ10::NLS-YC3.6 (Krebs et al., 2012) as template. The resulting PCR fragments were cloned into pJET1.2 to obtain entry vectors pEN-C-tRBCS-D, pEN-E-NLS-YC3.6-F, pEN-E-YC3.6-F, and pEN-F-tMAS-G. pEN-E-pHGFP-F and pEN-E-LifeAct-mRuby2-F was generated by cloning of the DNA fragment pHGFP and LifeAct-mRuby2 into pJET1.2, respectively. pHGFP was amplified using primer set F-E-pHGFP/R-F-pHGFP with the genomic DNA of transgenic line pUBQ10::pHGFP (Fendrych et al., 2014) as template. LifeAct-mRuby2 was amplified using primers F-E-LifeAct/R-F-mRuby2 with the genomic DNA of transgenic line pNTP303::LifeAct-mRuby2 as template. These entry clones were cloned into GreenGate destination vector pFAST-RK-AG (Decaestecker et al., 2019) to obtain the dual-expression vectors pUBQ10::PrpS1_pUBQ10::NLS-YC3.6, pUBQ10::PrpS1_pUBQ10::YC3.6, pUBQ10::PrpS1_pUBQ10::pHGFP, and pUBQ10::PrpS1_pUBQ10::LifeAct-mRuby2. Detailed primer information can be found in Supplemental Table S1.
The expression vectors were transformed into GV3101 Agrobacterium tumefaciens competent cells. The floral dipping method was adopted to stably transform Col-0 Arabidopsis plants as described previously (Fendrych et al., 2014).
T1 transgenic seeds were screened with LRC2 plates with corresponding antibiotics or using a fluorescence stereomicroscope by checking the fluorescence exhibited by the seeds. Lines with a single T-DNA insertion were obtained by selecting a 3:1 segregation ratio with T2 seeds. T3 homozygous seeds were used for all the experiments, if not specified.
RNA Extraction, cDNA Synthesis, and RT-PCR
To examine the PrpS1 mRNA expression in the transgenic line, 10 4-d-old seedlings from each line were collected, with wild-type seedlings as the control. To examine the PrsS1/3 mRNA expression in the pH3.3::XVE::PrsS1/3/pUBQ10::PrpS1 transgenic line before and after estradiol treatment, 4-d-old seedlings were transferred onto LRC2 plates containing 10 μm estradiol (LRC2 plates containing 0.1% [v/v] ethanol as a mock treatment control) for 6 h before being collected for RNA extraction. Total RNA was extracted using the RNeasy Mini Kit (Qiagen). cDNA was synthesized using 500 ng of total RNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. RT-qPCR was performed with the LightCycler 480 (Roche) using SYBR Green followed by data analysis using qBase. PrpS1 and PrsS1/3 mRNA expression were examined using primer sets F-PrpS1-Q/R-PrpS1-Q and F-PrsS1/3-Q/R-PrsS1/3-Q, respectively, with Actin2 as the housekeeping control (F-Actin2-Q/R-Actin2-Q). Detailed primer information can be found in Supplemental Table S1.
PrsS Protein Treatment
Recombinant PrsS proteins were produced as described (Foote et al., 1994) and stored in −70°C. PrsS proteins were dialyzed in one-fifth strength LRC2 liquid medium overnight in 4°C before use. The concentration of PrsS proteins was determined using the Bradford assay (Bio-Rad), during which the standard curve was generated using BSA (Sigma-Aldrich). To examine the effect of PrsS proteins on seedling growth, 10 μL of PrsS proteins with the desired concentration (the PrsS protein concentration used in all the experiments was 10 ng µL−1, unless specified) needed for different experiments was added to the root tip of each seedling using a pipette on LRC2 plates. The plates were kept horizontally for 30 min to allow PrsS proteins to dry before being placed back in the growth chamber vertically. When the PrsS protein treatment was needed during live-cell imaging, a perfusion chamber system was adopted. Samples were mounted and treated as described (Krebs and Schumacher, 2013) with minor modifications: instead of cotton, glass wool was used, and one-half-strength Murashige and Skoog solution was replaced with one-fifth-strength LRC2 solution. The procedure and concentration of PrsS proteins we used here were similar to what were used to induce the SI-PCD response in pollen growing in vitro (de Graaf et al., 2012; Wilkins et al., 2015), apart from the composition of the medium, where, instead of liquid pollen germination medium, one-fifth-strength LRC2 liquid medium was used here.
Protoplast Preparation
Leaves from 4-week-old plants were harvested and the lower epidermis was removed using double-sided tape as described (Wu et al., 2009). These leaf samples were immediately transferred into a petri dish containing protoplast enzymes (1% [w/v] cellulose R10 Yakult and 0.25% [w/v] macerozyme R10 Yakult) in protoplast washing solution (0.4 m mannitol, 10 mm CaCl2, 20 mm KCl, 0.1% [w/v] BSA, and 20 mm MES, pH 5.7 adjusted using KOH). Samples were incubated at room temperature with light on an orbital shaker set to 40 rpm for up to 2 h, followed by gentle filtration using a 70-μm cell strainer into a 50-mL tube. Protoplasts were washed three times with the protoplast washing solution by centrifuging at 100g for 3 min and aspirating off the supernatant, followed by suspension in protoplast washing solution, before being subjected to PrsS treatment. PrsS proteins used for protoplast treatment were dialyzed in protoplast washing solution without BSA overnight in 4°C. BSA was added back to the protoplast washing solution after PrsS protein concentration determination using the Bradford assay (Bio-Rad). PrsS protein treatment for protoplast was carried out on a 12-well tissue culture plate. PrsS proteins were added to the protoplast directly to a final concentration 10 ng µL−1. Protoplast washing solution was added as a mock control. During treatments, plates were placed in the Arabidopsis growth chamber with continuous light emitted by white fluorescent lamps (intensity of 120 µmol m−2 s−1) at 22°C. Fifty microliters of protoplast samples was taken from the plate at 0 h (before treatment) and at 1, 2, 4, and 8 h for viability examination and confocal imaging.
Estradiol Induction
β-Estradiol (Sigma-Aldrich) was dissolved in pure ethanol, and a 10 mm stock solution was prepared. The stock solution was stored in −20°C for up to 1 month. Four-day-old seedlings grown on LRC2 plates were transferred onto LRC2 plates containing estradiol (10 μm) or 0.1% (v/v) ethanol (mock control) for specified periods of time according to different experiments.
DEVDase Activity Assay
The CV-Caspase3&7 detection Kit (Enzo Life Science) was used for measuring the DEVDase activities of seedlings after PrsS protein treatment. DEVDase activity probe CR(DEVD)2 powder was reconstituted using 100 μL of dimethyl sulfoxide to obtain CR(DEVD)2 stock solution and kept in −20°C if not utilized immediately. Before use, the stock solution was diluted 1:5 in MilliQ water to make the staining solution. The working solution was made by further diluting the staining solution 1:20 in one-fifth-strength LRC2 solution. Samples were incubated in the working solution for 1 h at room temperature before imaging.
Imaging, Image Analysis, and Figure Preparation
Imaging of the root calcium signature was performed using a Zeiss LSM710 microscope using a PlanApochromat 20× objective (numerical aperture 0.8). YC3.6 or NLS-YC3.6 was excited with 405 nm, and fluorescence emissions of 460 to 515 nm and 515 to 570 nm were collected for eCFP and cpVenus, respectively. When PI staining was performed in conjunction with NLS-YC3.6 signal acquisition, seedling samples were mounted with one-fifth-strength LRC2 medium containing 5 μg mL−1 PI. A new imaging track was set up for PI signal acquisition. PI was excited with 561 nm, and fluorescence emissions between 580 and 700 nm were collected.
Imaging of the root pHGFP signal was performed using a Zeiss LSM710 microscope using a PlanApochromat 20× objective (numerical aperture 0.8). pHGFP was excited with 405 and 488 nm, and fluorescence emissions between 495 and 545 nm were collected.
LifeAct visualization was acquired using a Leica SP8 confocal laser scanning system with HCPL APO CS2 40×/1.10 (water) objective and HyD detector. LifeAct-mRuby2 was excited with 559 nm, and fluorescence emissions between 570 and 700 nm were collected.
DEVDase activities were visualized using a Leica SP8 confocal laser scanning system with Fluostar VISIR 25×/0.95 (water) objective and HyD detector. Samples were excited with 592 nm, and fluorescence emissions between 610 and 690 nm were collected.
All the images were processed and analyzed using Fiji. To quantify the calcium signal from NLS-YC3.6 or YC3.6 images, fluorescence intensities of the selected regions of interest were extracted using Fiji for both the eCFP and cpVenus channels. Fractional ratio changes (ΔR/R) were calculated as (R − R0)/R0, where R0 is the average ratio of the first 5 min (15 frames) of each measurement.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 8.0 for Windows.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Actin2 (At3g18780), UBQ10 (At4g05320), and H3.3 (At4g40040).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR shows that expression of PrpS1 mRNA varies in different pUBQ10::PrpS1 lines.
Supplemental Figure S2. Treatment with PrsS1 proteins does not inhibit the growth of wild-type seedlings.
Supplemental Figure S3. Recombinant PrsS protein treatment triggers rapid root growth inhibition of PrpS-expressing seedlings in an S-specific manner.
Supplemental Figure S4. PrsS treatment results in S-specific cell death of PrpS-expressing seedling root cells.
Supplemental Figure S5. PrsS treatment results in S-specific activation of DEVDase in PrpS-expressing seedling roots.
Supplemental Figure S6. PrsS treatment triggers S-specific alterations in [Ca2+]cyt.
Supplemental Figure S7. PrsS treatment triggers nucleus-localized Ca2+ changes in an S-specific manner.
Supplemental Figure S8. PrsS treatment triggers S-specific cytosolic pH decreases in PrpS-expressing roots.
Supplemental Figure S9. PrsS treatment triggers S-specific formation of actin foci in PrpS-expressing roots.
Supplemental Figure S10. Coexpression of PrpS and PrsS in Arabidopsis triggers root growth inhibition in an S-specific manner.
Supplemental Figure S11. Estradiol treatment induces the expression of PrsS mRNA transcript in XVE-PrsS/PrpS1 lines.
Supplemental Figure S12. Coexpression of PrpS and PrsS in whole Arabidopsis plants using estradiol triggers S-specific cell death in whole seedling roots.
Supplemental Figure S13. Transcript expression patterns of the SPH genes in Arabidopsis tissues.
Supplemental Figure S14. Protein expression patterns of the SPH genes in Arabidopsis tissues.
Supplemental Figure S15. Expression patterns of RALFs in Arabidopsis tissues.
Supplemental Figure S16. Expression patterns of pectin methylesterase inhibitors in Arabidopsis tissues.
Supplemental Table S1. Primers for vector construction and mRNA detection.
Acknowledgments
We thank members of the PCD laboratory for critical comments on the article, Dr. Melanie Krebs (University of Heidelberg) for sharing YC3.6-related vectors and seeds, and Dr. Rafael Andrade Buono (Gent University) for providing the root image for Figure 2A.
Footnotes
This work was supported by the European Research Council (grant no. PROCELLDEATH 749 639234 to M.K.N.), the Biotechnology and Biological Sciences Research Council (grant no. BB/P005489/1 to V.E.F.-T. and M.B.), the Fonds Wetenschappelijk Onderzoek (grant nos. G011215N and 12I7417N to M.T. and Z.L., respectively), and the China Scholarship Council (grant no. 201806760049 to F.X.).
Articles can be viewed without a subscription.
References
- Amien S, Kliwer I, Márton ML, Debener T, Geiger D, Becker D, Dresselhaus T(2010) Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom CS Jr., Winship LJ, Bezanilla M(2018) Simultaneous imaging and functional studies reveal a tight correlation between calcium and actin networks. Proc Natl Acad Sci USA 115: E2869–E2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers EP.(1997) Programmed cell death during plant growth and development. Cell Death Differ 4: 649–661 [DOI] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS(2010) The age and diversification of the angiosperms re-revisited. Am J Bot 97: 1296–1303 [DOI] [PubMed] [Google Scholar]
- Bircheneder S, Dresselhaus T(2016) Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J Exp Bot 67: 4849–4861 [DOI] [PubMed] [Google Scholar]
- Blackburn MR, Haruta M, Moura DS(2020) Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol 182: 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Franklin-Tong VE(2007) Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proc Natl Acad Sci USA 104: 18327–18332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiden M, Simon R(2016) Q&A: How does peptide signaling direct plant development? BMC Biol 14: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ.(2018) Diffuse growth of plant cell walls. Plant Physiol 176: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes JA, Grossniklaus U(2003) Unveiling the gene-expression profile of pollen. Genome Biol 5: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker W, Andrade Buono R, Pfeiffer M, Vangheluwe N, Jourquin J, Karimi M, van Isterdael G, Beeckman T, Nowack MK, Jacobs TB(2019) CRISPR-TSKO: A technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31: 2868–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BHJ, Vatovec S, Juárez-Díaz JA, Chai L, Kooblall K, Wilkins KA, Zou H, Forbes T, Franklin FCH, Franklin-Tong VE(2012) The Papaver self-incompatibility pollen S-determinant, PrpS, functions in Arabidopsis thaliana. Curr Biol 22: 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A, Hirschi H(2000) Heterologous expression for dominant-like gene activity. Trends Plant Sci 5: 10–11 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Sparks JA, Liao F, Wang YS, Blancaflor EB(2014) Fluorescent protein-based reporters of the actin cytoskeleton in living plant cells: Fluorophore variant, actin binding domain, and promoter considerations. Cytoskeleton (Hoboken) 71: 311–327 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM(1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Foote HCC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FCH(1994) Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc Natl Acad Sci USA 91: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong VE, Hackett G, Hepler PK(1997) Ratio-imaging of Ca2+ in the self-incompatibility response in pollen tubes of Papaver rhoeas. Plant J 12: 1375–1386 [Google Scholar]
- Franklin-Tong VE, Ride JP, Franklin FCH(1995) Recombinant stigmatic self-incompatibility (S-) protein elicits a Ca2+ transient in pollen of Papaver rhoeas. Plant J 8: 299–307 [Google Scholar]
- Franklin-Tong VE, Ride JP, Read ND, Trewavas AJ, Franklin FCH(1993) The self-incompatibility response in Papaver rhoeas is mediated by cytosolic-free calcium. Plant J 4: 163–177 [Google Scholar]
- Geitmann A, Snowman BN, Emons AMC, Franklin-Tong VE(2000) Alterations in the actin cytoskeleton of pollen tubes are induced by the self-incompatibility reaction in Papaver rhoeas. Plant Cell 12: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez MD, Vera-Sirera F, Pérez-Amador MA(2014) Molecular programme of senescence in dry and fleshy fruits. J Exp Bot 65: 4515–4526 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR(2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T, Yang WC(2017) Gametophytic pollen tube guidance: Attractant peptides, gametic controls, and receptors. Plant Physiol 173: 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D(2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Buono RA, Skorzinski N, Radio MC, De Winter F, Parizot B, Mertens J, Karimi M, Fendrych M, Nowack MK(2018) NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 30: 2197–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Higashiyama T(2015) Peptide signaling in pollen tube guidance. Curr Opin Plant Biol 28: 127–136 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A(2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K(2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Krebs M, Schumacher K(2013) Live cell imaging of cytoplasmic and nuclear Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb Protoc 2013: pdb.prot073031. [DOI] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J(2013) GreenGate: A novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Walker JC(2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Yang WC(2018) Ligands switch model for pollen-tube integrity and burst. Trends Plant Sci 23: 369–372 [DOI] [PubMed] [Google Scholar]
- Lin Z, Eaves DJ, Sanchez-Moran E, Franklin FCH, Franklin-Tong VE(2015) The Papaver rhoeas S determinants confer self-incompatibility to Arabidopsis thaliana in planta. Science 350: 684–687 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Jiao H, Li L, Qiao X, Fabrice MR, Wu J, Zhang S(2017) Expansion and evolutionary patterns of cysteine-rich peptides in plants. BMC Genomics 18: 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM.(2007) Hidden weapons of microbial destruction in plant genomes. Genome Biol 8: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani C, Beuckeleer MD, Truettner J, Leemans J, Goldberg RB(1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347: 737–741 [Google Scholar]
- Marshall E, Costa LM, Gutierrez-Marcos J(2011) Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J Exp Bot 62: 1677–1686 [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, Papacek M, Chen X, Chaudhary A, Samaras P, Richter S, Shikata H, Messerer M, et al. (2020) Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579: 409–414 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ(2001) Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ 24: 557–563 [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A(2004) Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA 101: 10554–10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr.(2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekar KV, Ji S, Coulthard RJ, Ride JP, Reynolds GL, Winn PJ, Wheeler MJ, Hyde EI, Smith LJ(2019) Structure of SPH (self-incompatibility protein homologue) proteins: A widespread family of small, highly stable, secreted proteins. Biochem J 476: 809–826 [DOI] [PubMed] [Google Scholar]
- Ride JP, Davies EM, Franklin FCH, Marshall DF(1999) Analysis of Arabidopsis genome sequence reveals a large new gene family in plants. Plant Mol Biol 39: 927–932 [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB(1999) The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Silverstein KAT, Moskal WA Jr., Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA(2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ(2002) Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E, Wachsman G, Benfey PN(2013) Spatiotemporal signalling in plant development. Nat Rev Genet 14: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019) STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Isogai A(2005) Self-incompatibility in plants. Annu Rev Plant Biol 56: 467–489 [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A(2000) The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA 97: 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Higashiyama T(2016) Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531: 245–248 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Franklin-Tong VE(2004) Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 429: 305–309 [DOI] [PubMed] [Google Scholar]
- Wang L, Lin Z, Triviño M, Nowack MK, Franklin-Tong VE, Bosch M(2019) Self-incompatibility in Papaver pollen: Programmed cell death in an acidic environment. J Exp Bot 70: 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Triviño M, Lin Z, Carli J, Eaves DJ, Van Damme D, Nowack MK, Franklin-Tong VE, Bosch M(2020) New opportunities and insights into Papaver self-incompatibility by imaging engineered Arabidopsis pollen. J Exp Bot 71: 2451–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BH, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FC, Franklin-Tong VE(2009) Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 459: 992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, Vatovec S, Franklin-Tong VE(2010) The pollen S-determinant in Papaver: Comparisons with known plant receptors and protein ligand partners. J Exp Bot 61: 2015–2025 [DOI] [PubMed] [Google Scholar]
- Wilkins KA, Bancroft J, Bosch M, Ings J, Smirnoff N, Franklin-Tong VE(2011) Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of Papaver. Plant Physiol 156: 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KA, Bosch M, Haque T, Teng N, Poulter NS, Franklin-Tong VE(2015) Self-incompatibility-induced programmed cell death in field poppy pollen involves dramatic acidification of the incompatible pollen tube cytosol. Plant Physiol 167: 766–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KA, Poulter NS, Franklin-Tong VE(2014) Taking one for the team: Self-recognition and cell suicide in pollen. J Exp Bot 65: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS(2009) Tape-Arabidopsis sandwich: A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.(2008) Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]