Silencing of petunia tRNA-specific methyltransferase 61A decreases mRNA m1A levels, leading to abnormal leaf development.
Abstract
N1-methyladenosine is a unique type of base methylation in that it blocks Watson-Crick base pairing and introduces a positive charge. m1A is prevalent in yeast and mammalian mRNA and plays a functional role. However, little is known about the abundance, dynamics, and topology of this modification in plant mRNA. Dot blotting and liquid chromatography tandem mass spectrometry analyses revealed a dynamic pattern of m1A mRNA modification in various tissues and at different developmental stages in petunia (Petunia hybrida), a model system for plant growth and development. We performed transcriptome-wide profiling of m1A in petunia mRNA by m1A mRNA immunoprecipitation followed by a deep-sequencing approach (m1A-seq, using an m1A-specific antibody). m1A-seq analysis identified 4,993 m1A peaks in 3,231 genes expressed in petunia corollas; there were 251 m1A peaks in which A residues were partly replaced by thymine and/or reverse transcription stopped at an adenine site. m1A was enriched in coding sequences, with single peaks located immediately after start codons. Ethylene treatment upregulated 400 m1A peaks in 375 mRNAs and downregulated 603 m1A peaks in 530 mRNAs in petunia corollas; 975 m1A peaks in mRNA were only detected in corollas treated with air and 430 were only detected in corollas treated with ethylene. Silencing of petunia tRNA-specific methyltransferase 61A (PhTRMT61A) reduced the m1A level in mRNA in vivo and in vitro. In addition, PhTRMT61A silencing caused abnormal leaf development, and the PhTRMT61A protein was localized to the nucleus. Thus, m1A in mRNA is an important epitranscriptome marker and plays a role in plant growth and development.
More than 100 types of chemical modifications have been identified in cellular RNAs, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), inosine, and so on (Frye et al., 2016; Li et al., 2016a; Zhao et al., 2017). These modifications may play important roles in mRNA metabolism (Cui et al., 2017).
m6A, an abundant type of RNA modification in eukaryotic cells, has been identified in humans (Homo sapiens), Saccharomyces cerevisiae, Arabidopsis (Arabidopsis thaliana), bacteria, viruses, and other species (Chen et al., 2015; Deng et al., 2015; Huang et al., 2015; Wan et al., 2015; Wei et al., 2017). m6A primarily appears near the stop codons and 3′ untranslated regions (UTRs) of mRNAs (Dominissini et al., 2012; Deng et al., 2015). Dynamic m6A regulation is found in various growth and development processes, indicating a possible epigenetic regulation role during RNA processing and exerting biological functions (Hastings, 2013; Shen et al., 2015; Daoud et al., 2016; Yang et al., 2016). Deficiency of the m6A modification leads to various kinds of diseases in humans (Wei et al., 2017). In Arabidopsis, reduced m6A levels in mRNA resulted in developmental defects, including reduction of apical dominance, and defects in floral organ number, size, and identity (Bodi et al., 2012). In Arabidopsis, MTA (ortholog of human METTL3), MTB (METTL14), FKBP12 INTERACTING PROTEIN37, VIRILIZER, and the E3 ubiquitin ligase HAKAI are required for m6A formation (Růžička et al., 2017). Downregulation of these proteins led to reduced relative m6A levels and shared pleiotropic phenotypes, which included aberrant vascular formation in the root (Růžička et al., 2017).
m5C has been found to occur on tRNAs, ribosomal RNAs (rRNAs), mRNAs, and long noncoding RNAs (lncRNAs) in many organisms (Edelheit et al., 2013; Burgess et al., 2015; Squires et al., 2012; Hussain et al., 2013; Khoddami and Cairns, 2013). m5C sites play important roles in rRNA processing, structure, and translation (Hong et al., 1997; Sharma et al., 2013; Gigova et al., 2014). Similarly, m5C sites in tRNAs are required for tRNA stability and efficient translation (Schaefer et al., 2009); Tuorto et al., 2012, 2015) and are induced under oxidative stress conditions (Chan et al., 2010, 2012). Transcriptome-wide profiling of RNA m5C in Arabidopsis has been reported, including mRNAs, long noncoding RNAs, and other noncoding RNAs (David et al., 2017; Cui et al., 2017). A dynamic pattern of m5C mRNA modification in various tissues and at different developmental stages was revealed by liquid chromatography tandem mass spectrometry (LC-MS/MS) and dot blot analyses (Cui et al., 2017). In addition, Arabidopsis tRNA-specific methyltransferase 4B (TRM4B) exhibits m5C RNA methyltransferase activity (David et al., 2017; Cui et al., 2017).
m1A, which is also abundant in human mRNAs (Li et al., 2016a), is a unique type of base methylation in that it blocks Watson-Crick base pairing, introduces a positive charge and can dramatically alter protein-RNA interactions and RNA secondary structures through electrostatic effects (Roundtree et al., 2017). A dynamic m1A methylome has been revealed in the eukaryotic mRNA of mammals. m1A is responsive to many kinds of cellular stress (Dominissini et al., 2016; Li et al., 2016a). The mapping of this type of modification shows that it is uniquely located near the first splice site and translation start site in coding sequences (CDS), and m1A is generally correlated with the upregulation of translation (Dominissini et al., 2016; Li et al., 2016a). In mRNAs, m1A exists within highly structured 5′ UTRs and may affect translation (Dominissini et al., 2016; Li et al., 2016a Cenik et al., 2017). The technique of m1A-ID-seq was developed for the transcriptome-wide characterization of m1A (Li et al., 2016a). However, the abundance, dynamics, and topology of m1A in plant mRNA are not known.
The gaseous phytohormone ethylene modulates various plant developmental processes, including organ growth, flowering, fruit ripening, senescence, and abscission (Abeles et al., 1992; (O’Neill et al., 1993; Clark et al., 1997) Wang et al., 2002). In flower development, ethylene has been reported to play important roles in sexual determination (Yamasaki et al., 2003) and petal senescence (Woltering and van Doorn, 1988; van Doorn and van Meeteren, 2003; van Doorn and Woltering, 2008). Several reports suggested that ethylene was also involved in floral organ abscission (Patterson and Bleecker, 2004). Ethylene could promote or inhibit petal growth, and ethylene treatment significantly reduced petal size, inhibited expansion of petal abaxial subepidermal cells, and decreased petal water content in rose (Rosa spp.; Reid et al., 1989a, 1989b; Ma et al., 2008). In addition, besides the transcriptome, proteome, and metabolome profiles, ethylene treatment changed the phosphoproteome profile in higher plants (Slade et al., 2012; Wang et al., 2013; Yin et al., 2014; Zhang et al., 2015; Du et al., 2016; Gupta et al., 2018; Der Agopian et al., 2020). Our previous study showed that ethylene treatment changed the proteome and ubiquitylome profiles in petunia (Petunia hybrida) corollas (Guo et al., 2017). However, the effect of ethylene treatment on the m1A methylome of mRNA remains unknown in plants.
Petunia is a model system for plant growth and development, especially in flower development, flower senescence, anthocyanin synthesis, and synthesis of floral volatiles (Gerats and Vandenbussche, 2005; Chen et al., 2017; Liu et al., 2017). In petunia, ethylene is involved in flower senescence, the synthesis of floral volatiles, and regulation of the timing of nucleic acid and protein degradation during petal senescence (Wilkinson et al., 1997; Langston et al., 2005; Chapin and Jones, 2009; Guo et al., 2017). Methylated RNA immunoprecipitation (IP) sequencing, referred to in this study as m1A-seq, was applied using an m1A-specific antibody to perform transcriptome-wide profiling of m1A sites in petunia. A total of 4,993 m1A peaks were identified in 3,231 genes expressed in corollas. Several features of m1A in petunia mRNA were revealed through further analyses of m1A-seq data. Ethylene treatment reduced transcriptome-wide m1A levels in petunia corollas. We found that the petunia tRNA-specific methyltransferase 61A (PhTRMT61A) silencing resulted in a decrease in mRNA m1A peaks in petunia corollas and abnormal leaf development. The results of this study suggest that m1A in mRNA is an important epitranscriptome marker and could play a regulatory role in plant development in petunia.
RESULTS
m1A Dot Blotting and MS Analysis
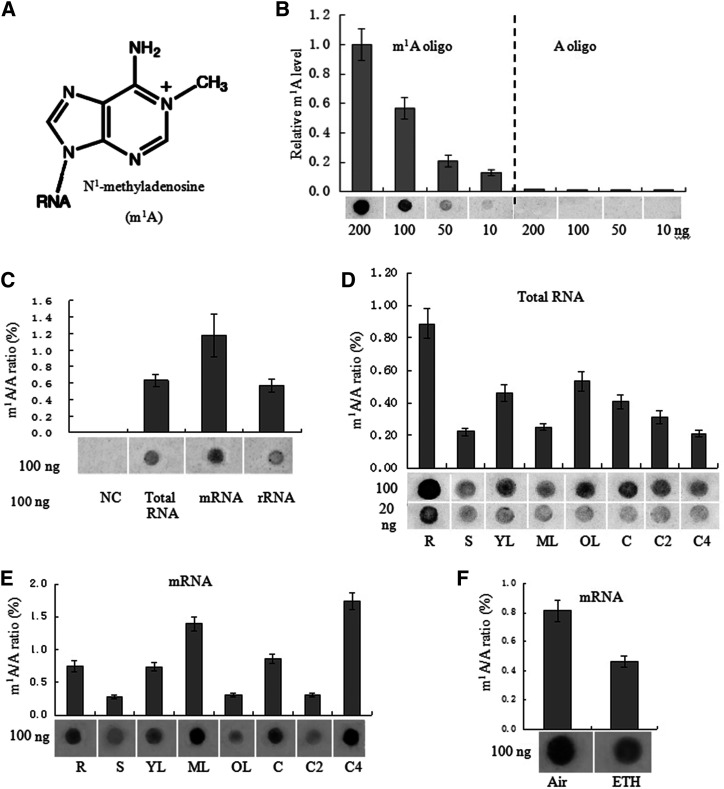
To determine whether m1A modifications are present in plant RNA, dot blotting assays were performed with an m1A antibody that has been confirmed to specifically bind m1A in mammalian mRNA (Dominissini et al., 2016; Li et al., 2016a). Here, the results of dot blotting further showed the specificity of the m1A antibody for detecting m1A modification (Fig. 1, A and B). m1A was detected in the total RNA, mRNA, and rRNA of the tested tissues of petunia ‘Mitchell’ (Fig. 1C). Among all petunia RNA species tested, mRNA showed the strongest signal (Fig. 1C). m1A was present at different levels in total RNA and mRNA isolated from roots, stems, leaves, and corollas of different senescent stages in petunia (Fig. 1, D and E). The m1A level is dynamic in petunia, and mature leaves and senescent corollas exhibit strong m1A signals in mRNA (Fig. 1E). In addition, m1A was present at various levels in the total RNA of different plant species including petunia, Brunfelsia latifolia, Solanum lycopersicum, Capsicum annuum, Arabidopsis, Phalaenopsis aphrodite, and Oncidium hybridum, and the strongest signal was detected in B. latifolia (Supplemental Fig. S1).
Figure 1.
Detection of m1A in petunia ‘Mitchell’. A, Chemical structure of m1A. B, Dot blot analysis shows that the anti-m1A antibody specifically recognizes an m1A-modified RNA oligo. The graph shows quantification of the dot blot results with three biological replicates by ImageJ, with representative blots shown below the graph. C to F, Dot blot (shown below the graphs) and LC-MS/MS assays were performed for the detection and quantification of m1A levels in P. hybrida among different RNA species of 20-d-old wild-type (without any specific treatment) whole seedlings (C) in total RNA (D) and mRNA (E) from various tissues of wild-type plants, including roots (R), stems (S), young leaves (2 cm; YL), mature leaves (4 cm; ML), old leaves (OL), corollas of open flowers (C), corollas of flowers open for 2 d (C2), corollas of flowers open for 4 d (C4), and in mRNA from corollas treated with air or 2 μm ethylene (ETH) for 16 h (F). Error bars indicate the mean ± sd; n = 3. NC, Normal control (no RNA was added).
Since the m1A level in mRNA is dynamically regulated by senescence in corollas and ethylene plays an important role in flower senescence, we further examined the effect of ethylene treatment on the m1A level of mRNA in corollas. Our previous study showed that ethylene treatment accelerated petunia flower senescence (Guo et al., 2017). In this study, similar results were observed. Petunia flowers exhibited the first visible symptom of senescence at ∼16 h after 2 μL L−1 ethylene treatment: the margins of the corollas began to involute, and a few translucent dots appeared in the corollas (Supplemental Fig. S2). At 48 h after ethylene treatment, the petunia flowers exhibited obvious symptoms of senescence (Supplemental Fig. S2). Air-treated petunia corollas were fully turgid 0 to 48 h after flower opening, exhibited no symptoms of senescence, and were visually indistinguishable from flowers at anthesis (Supplemental Fig. S2). The effect of ethylene treatment on m1A level in mRNA in petunia corollas was examined by dot blotting assays with the m1A antibody and the results showed that 16 h of 2 μL L−1 ethylene treatment reduced the m1A level in mRNA in petunia corollas (Fig. 1F).
To further confirm the results of the dot blotting assays and quantify m1A in RNA, quantitative LC-MS/MS of pure RNA preparations was performed. Mass spectrograms of adenine and methyladenosine are shown in Supplemental Figure S3. Previously, an RNA purification procedure was adopted to minimize contamination by these abundant RNAs (Li et al., 2016a). Since m1A can be rearranged as m6A under alkaline conditions (Dimroth rearrangement; Macon and Wolfenden, 1968), RNA is digested at neutral pH to avoid underestimation of m1A abundance. mRNA consistently showed the highest m1A abundance (1.17% m1A/A) among the three RNA species examined (Fig. 1C). Among the various tissues examined, stems showed the lowest (0.28%) and senescent corollas the highest (1.82%) abundance of m1A in their mRNA (Fig. 1E).
Transcriptome-Wide Profile of m1A in Petunia
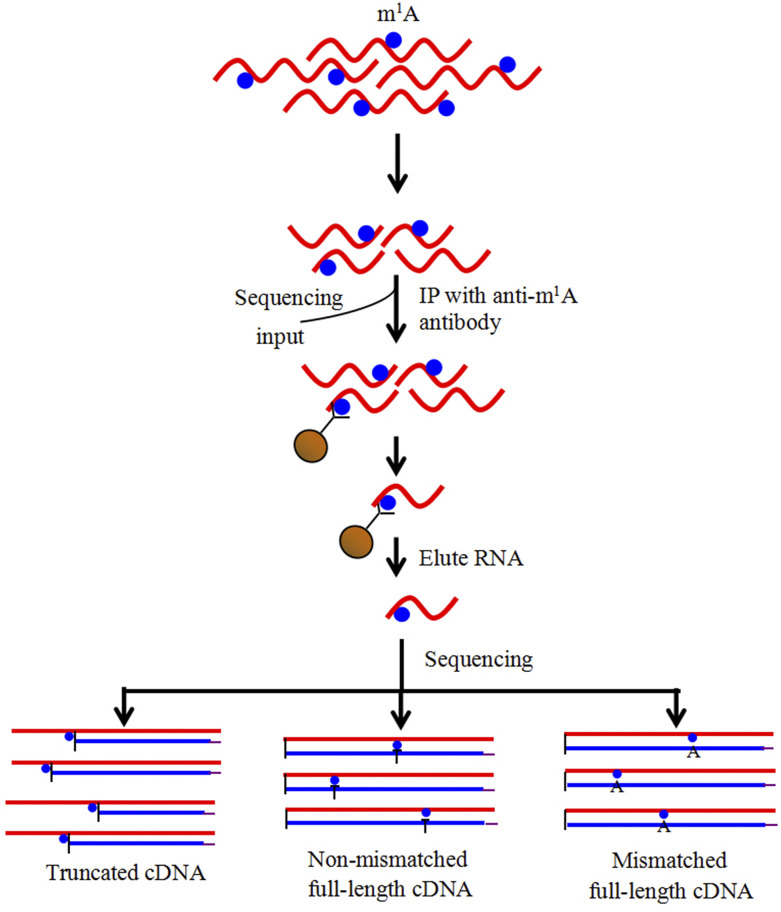
To identify and localize m1A sites at the transcriptome-wide level, we analyzed m1A methylation in mRNA extracted from petunia ‘Mitchell’ corollas treated with air or ethylene for 16 h using the antibody-based m1A-seq approach. We applied m1A-seq to mRNA from petunia corollas in two replicates and determined methylated positions (m1A peaks) using the MACS peak-calling algorithm (Zhang et al., 2008; Dominissini et al., 2016). RNA was fragmented into ∼50-nucleotide-long oligonucleotides (input) and immunoprecipitated using the anti-m1A antibody (Fig. 2).
Figure 2.
m1A-seq using an m1A antibody to enrich and identify m1A sites. RNA is fragmented and subjected to IP using an anti-m1A antibody. Eluted RNA fragments are converted to cDNA and sequenced. m1A causes both reverse transcription stops and read-through accompanied by mismatches, to produce typical peaks with a central trough.
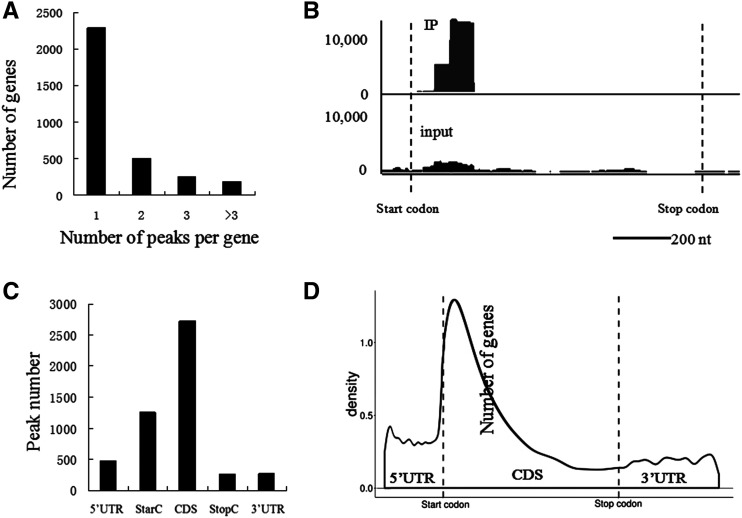
We identified 4,993 putative m1A peaks originating from 3,231 expressed genes from the transcriptome of petunia corollas after combining the libraries of two replicates using strict peak-calling criteria (P < 10−10, fold change >2; Supplemental Data Set 1). Each methylated gene contained an average of 1.55 peaks, and 72% of these genes were methylated once (Fig. 3A; Supplemental Data Set 1). One representative view (PhADPRM, Peaxi162Scf00089g01220) of typical m1A peaks in mRNA is shown in Figure 3B.
Figure 3.
The m1A methylome of petunia ‘Mitchell’ corollas treated with air and ethylene for 16 h. A, Percentages of methylated genes exhibiting 1, 2, 3, or 3+ peaks per gene in corollas. B, Representative views of typical m1A peaks in mRNA (PhADPRM, Peaxi162Scf00089g01220). Start codons and stop codons are shown. C, Distribution of m1A peaks across mRNA segments. Each segment was normalized according to its average length in the RefSeq annotation. The total normalized transcripts were binned into regions of 1% total length, and the percentage of m1A peaks belonging to each bin was calculated. D, Metagene profiles of the m1A peak distribution along a normalized transcript composed of three rescaled nonoverlapping segments illustrated below.
The m1A peak distribution within the transcriptome regions was investigated next. Most of the m1A peaks were apparently positioned in CDSs (Fig. 3C), showing that the m1A distribution was not random. Then, the m1A peaks were normalized using the relative fraction of the segment occupied in the transcriptome, and the results showed that m1A was markedly enriched in CDSs with a single peak positioned closely after the start codon (Fig. 3D).
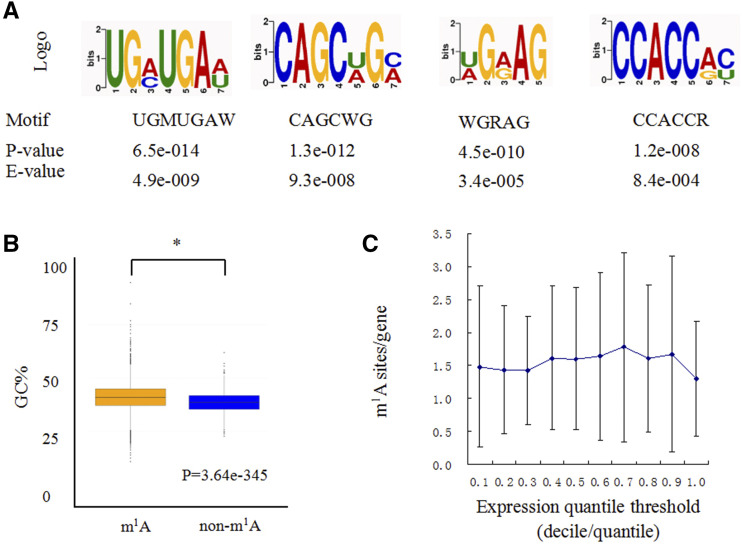
The sequence patterns of m1A peaks may define or be associated with their locations. An unbiased search for common motifs enriched in segments around m1A peak summits was performed (Fig. 4A). The most significantly enriched motifs (P < 0.05) were UGMUGAW (E = 4.9e−009), CAGCWGM (E = 9.3e−008), WGRAG (E = 3.4e−005) and CCACCRY (E=8.4e−004), which were present in >70% of the methylated peak summits (Fig. 4A). In addition, GC content is a key determinant of RNA structure. We found that the m1A peak sequences exhibited a higher GC content than those of the non-m1A controls (P = 3.64e−345; Fig. 4B), which showed that the combination of m1A sequence and structure features may specify methylation locations (Ozanick et al., 2005; Takuma et al., 2015).
Figure 4.
Sequence and structure traits of m1A peaks. A, Three representative sequence motifs enriched with m1A. B, GC content comparisons between m1A peaks and control sequences. GC% is the percentage of G + C content within the m1A peaks. Statistical analysis was performed using one-way ANOVA followed by Duncan’s NMRT. Asterisks indicate significant differences at the P = 0.05 level. The data points (whiskers) represents the GC content ratio of the modified m1A sequence and the corresponding nonmodified m1A sequence. The blocks are drawn based on these data and are the characteristics of the box graph. The bars are the main shaft of the box. C, Transcripts with different expression levels are divided into 10 groups, and the m1A density of each group is calculated separately. The error bars represent the se of m1A sites/gene of two biological repeats.
To examine the correlation between the m1A peaks and transcript abundance, RNA-seq was performed for the same petunia corollas and generated 29.3 to 37.3 million reads that uniquely mapped to the petunia reference genome, with three highly reproducible biological replicates (R > 0.88; Supplemental Data Set 2). We detected 23,769 mRNA sequences. Treatment with ethylene resulted in 685 downregulated and 1,226 upregulated unigenes (absolute log2-fold change >1 and P < 0.05; Supplemental Data Set 3). The coverage of m1A in the expressed genes was 13.6%. The distribution of transcripts containing m1A sites was compared with the general distribution of transcript abundance. The results showed that there is no significant correlation between m1A level and gene expression (Fig. 4C).
Next, Gene Ontology (GO) analyses of the methylated genes were performed, and the results showed that the top 10 P < 0.05 categories of methylated genes significantly enriched were those involved in small molecule binding, nucleotide binding, nucleoside phosphate binding, purine ribonucleoside triphosphate binding, ATP binding, pyrophosphatase activity, hydrolase activity, action on acid anhydrides and phosphorus-containing anhydrides, purine nucleotide binding, nucleoside-triphosphatase activity, ribonucleotide binding, and others (Supplemental Fig. S4; Supplemental Data Set 4). The most dramatic results for molecular function were observed in binding, especially binding of nucleotide and small molecule, suggesting that m1A mRNA modification might play an important role in the nucleus, including in RNA slicing and transcription regulation in petunia corollas.
Since m1A can lead to reverse transcription stoppage and read-through accompanied by mismatches, we examined the peaks in which an A was replaced by thymine (T) and/or reverse transcription stopped at the A site. The results showed that among the 4993 m1A peaks originating from 3231 expressed genes, there were 251 m1A peaks in 199 expressed genes in which the A residues were partially replaced by thymine (T) and/or the reverse transcription stopped at the A site (Supplemental Data Set 5).
To obtain the m1A data in noncoding RNA species like lncRNA and circular RNA (circRNA) of petunia corollas, we used StringTie and Coding Potential Calculator (CPC) software to identify lncRNAs and used the find_circ tool (Memczak et al., 2013) to identify the circRNAs in the transcriptome of petunia corollas. After careful screening and bioinformatic analysis, a total of 14,017 lncRNAs (Supplemental Data Set 6) and 1,079 circRNAs (Supplemental Data Set 7) were obtained in this study. Further, 485 and 175 putative m1A peaks originating from 476 lncRNAs and 134 circRNAs were identified from the transcriptome of petunia corollas, respectively (Supplemental Data Sets 8 and 9, respectively), using MACS default parameters to identify methylation sites. Each methylated lncRNA and circRNA contained an average of 1.02 and 1.31 peaks, respectively, and 98% of lncRNAs and 76% of circRNA were methylated once. For lncRNAs modified by m1A, two types of m1A pattern were observed: modification of one m1A site and methylation of two m1A sites. For circRNAs modified by m1A, 3, 7, 18, and 147 circRNAs were modified by 4, 3, 2, and 1 m1A sites, respectively.
Effects of Ethylene Treatment on the m1A Level in mRNA in Petunia Corollas
The results of dot blotting showed that ethylene treatment greatly reduced the m1A level in the mRNA of petunia ‘Mitchell’ corollas. We further analyzed the m1A patterns of corollas under 16 h of ethylene and air treatment. We found that 400 and 603 peaks in 375 and 530 mRNAs were up- and downregulated, respectively, under ethylene treatment. In addition, 975 m1A peaks in mRNA were not detected, and 430 novel m1A sites were identified in corollas treated with ethylene (Supplemental Figs. S5 and S6; Supplemental Data Set 10). These results, together with the results of dot blotting, showed that ethylene treatment results in a slight reduction in m1A levels in the petunia corolla. In addition, we found that 55 and 59 peaks in lncRNAs (Supplemental Data Set 11), and 15 and 17 peaks in circRNAs (Supplemental Data Set 12), were up- and downregulated, respectively, by ethylene.
To determine the functional differences in genes associated with upregulated versus downregulated m1A levels, we analyzed the genes whose m1A levels were quantified for GO enrichment in a clustering analysis (Supplemental Fig. S7; Supplemental Data Sets 13 and 14). The analyses of molecular functions indicated that many genes with upregulated m1A levels were highly enriched in the categories of ubiquitin-like protein transferase activity, inorganic diphosphatase activity, protein Tyr/Ser/Thr phosphatase activity, ubiquitin-protein transferase activity, protein Tyr phosphatase activity, carbohydrate transmembrane transporter activity, sugar transmembrane transporter activity, carbohydrate transporter activity, ER retention sequence binding, 1,3-β-d-glucan synthase activity and others (Supplemental Fig. S7A; Supplemental Data Set 13). Many genes with downregulated m1A levels were enriched in the categories of 1,3-β-d-glucan synthase activity, nucleotide binding, nucleoside phosphate binding, small molecule binding, nucleoside-triphosphatase activity, helicase activity, pyrophosphatase activity, RNA binding, purine ribonucleoside triphosphate binding, ATPase activity, and others (Supplemental Fig. S7B; Supplemental Data Set 14). These results showed that m1A could play important roles in protein degradation, nutritional transport, transcription or translation regulation, and so on during flower senescence.
Association between the m1A Methylome and Transcriptome after Ethylene Treatment
The correlation between the whole m1A methylome and the mRNA transcriptome in corollas treated with ethylene and air was analyzed according to the quantitative results. Among 809 mRNAs whose abundance was detected, there were 956 m1A peaks with significantly changed methylation levels after ethylene treatment (P < 0.00001, fold change >2; Supplemental Data Set 15). Among the 956 m1A peaks, 384 were upregulated and 572 were downregulated. Quantitative ratios from the m1A methylome and transcriptome were compared upon ethylene treatment, as shown in Supplemental Figure S8. The Pearson's correlation coefficient was 0.30 when all significantly changed m1A peaks were considered based on their mRNA abundance. Therefore, the m1A methylome and transcriptome were slightly positively correlated.
PhTRMT61A Is a Methyltransferase for m1A of mRNA, and Its Suppression Changes the Development of Leaves in Petunia
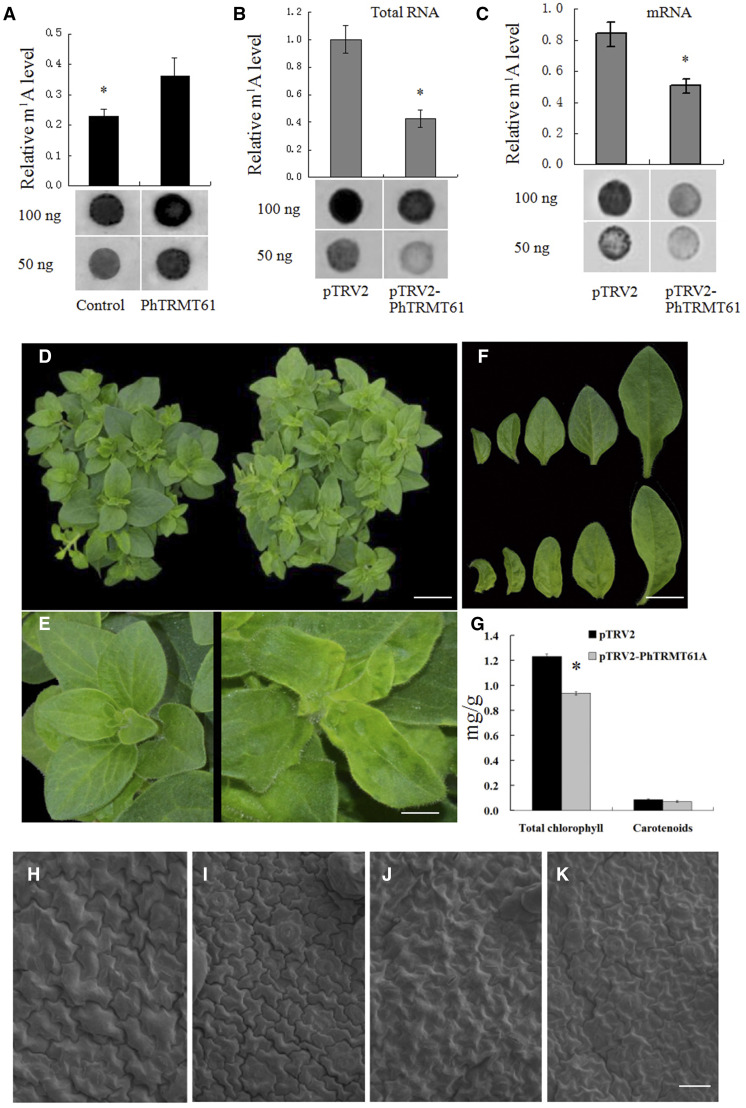
The tRNA methyltransferase responsible for the formation of m1A58 identified in mammals and yeast is a two-subunit enzyme encoded by TRMT6 and TRMT61 (Anderson et al., 1998, 2000; Ozanick et al., 2005, 2007; Saikia et al., 2010). In mammals, the TRMT61 subfamily includes two members, TRMT61A and TRMT61B. However, using the complementary DNA (cDNA) sequences of human TRMT61A and TRMT61B (locus NP_689520.2 and NP_060380.3) as queries in BLAST searches of the NCBI database and the Sol Genomics Network, we found that the TRMT61 subfamily only includes one member in the genomes of Arabidopsis, S. lycopersicum, Oryza sativa, and petunia, and the encoded proteins are named AtTRMT61A (NP_568302.1), SlTRMT61A (XP_010316290.1), OsTRMT61A (XP_015636972.1), and PhTRMT61A, respectively (Supplemental Fig. S9). PhTRMT61A showed 72.7% and 92.9% identity to AtTRMT61A and SlTRMT61A, respectively (Supplemental Table S1). The full-length cDNA of PhTRMT61A was recombinantly expressed in Escherichia coli (His-PhTRMT61A), and recombinant His-PhTRMT61A proteins were obtained. We incubated His-PhTRMT61A proteins with the total RNA or mRNA of petunia ‘Mitchell’ seedlings. Dot blotting revealed a marked increase in the level of m1A in both the total RNA and mRNA incubated with His-PhTRMT61A proteins (Fig. 5A), indicating that PhTRMT61A could be a methyltransferase for m1A in mRNAs in petunia.
Figure 5.
PhTRMT61A is a methyltransferase for m1A in mRNA, and silencing PhTRMT61A causes phenotypic alterations. A, Dot blot analysis of total RNA nucleosides from petunia ‘Ultra’ corollas incubated with or without the recombinant PhTRMT61A protein. B and C, Dot blotting analysis of total RNA (B) and mRNA (C) nucleosides from the corollas of pTRV2- and pTRV2-PhTRMT61A-treated plants. D and E, Five-week-old VIGS-mediated PhTRMT61A-silenced plants (right) compared to control plants (left). The plants were digitally extracted for comparison. F, Leaves of PhTRMT61A-silenced plants (bottom) compared with those of control plants (top). The leaves were digitally extracted for comparison. G, Effects of PhTRMT61A silencing on the contents of total chlorophyll and carotenoids in young leaves (2 cm). H to K, Scanning electron micrographs of adaxial (H and I) and abaxial (J and K) epidermal cells of leaves of PhTRMT61A-silenced plants (I and K) compared with those of control plants (J and K). Statistical analysis was performed using one-way ANOVA followed by Duncan's NMRT with three biological replicates. Asterisks indicate significant differences at the P = 0.05 level and data are presented as the means ± sd (A–C and G). Scale bars = 3.0 cm (D), 0.5 cm (E), 1.3 cm (F), and 40 μm (H–K).
We further investigated the effects of PhTRMT61A suppression on RNA m1A methylation in petunia. We established an efficient virus-induced gene silencing (VIGS) system in petunia ‘Ultra’ (Tan et al., 2014; Chen et al., 2017). The pTRV2-PhTRMT61A vector was constructed to suppress the expression of PhTRMT61A. Thirty to 35 petunia ‘Ultra’ plants were subjected to infection by pTRV2-PhTRMT61A and pTRV2 vectors. Approximately 30 d after petunia seedlings were infected according to our previous protocol (Chen et al., 2017), we extracted the total RNA of the young leaves of plants treated with pTRV2-PhTRMT61A and pTRV2 and purified the mRNA. The results of dot blotting with the m1A antibody showed that PhTRMT61A silencing significantly reduced the m1A level in both the total RNA and mRNA (Fig. 5, B and C).
No significant difference in the height of the stems, plant size or leaf number was observed in PhTRMT61A-silenced plants compared with the control. However, a chlorotic and wrinkled leaf phenotype was found in pTRV2-PhTRMT61A-infected plants (Fig. 5, D–F). We further measured chlorophyll and carotenoid contents in leaves from PhTRMT61A-silenced and control plants. PhTRMT61A silencing significantly decreased the total leaf chlorophyll content compared with the control but did not significantly change the carotenoid content (Fig. 5G). As expected, pTRV2-PhTRMT61A treatment significantly reduced the expression of PhTRMT61A in petunia leaves (Supplemental Fig. S10). In addition, scanning electron micrographs of adaxial and abaxial epidermal cells in leaves showed small cells in the plants subjected to PhTRMT61A silencing (Fig. 5, H–K; Table 1).
Table 1. Effects of PhTRMT61A silencing on the cell size in leaves.
Data are means ± se from 15 to 20 samples. Statistical analysis was performed using one-way ANOVA followed by Duncan's NMRT. The asterisk indicates significant difference at *P = 0. 05.
| Trait | pTRV2 | pTRV2-PhTRMT61A | pTRV2-PhTRMT61A/pTRV2 |
|---|---|---|---|
| % | |||
| Diameter of adaxial epidermal cells of leaf (µm) | 39.24 ± 3.22 | 26.23 ± 3.49* | 66.8 |
| Diameter of abaxial epidermal cells of leaves (µm) | 34.62 ± 3.86 | 26.37 ± 7.78* | 76.1 |
| Cell density of adaxial epidermal cells of leaves (cells 0.1 mm−2) | 56.32 ± 7.72 | 128.31 ± 9.61 | 227.7 |
| Cell density of abaxial epidermal cells of leaves (cells 0.1 mm−2) | 69.59 ± 8.64 | 134.83 ± 11.5 | 193.7 |
We investigated PhTRMT61A expression by quantitative PCR (qPCR). PhTRMT61A expression was high in the leaves and roots and low in the stems (Supplemental Fig. S11A). In addition, treatment with 2 μL L−1 ethylene significantly decreased PhTRMT61A mRNA levels in corollas (Supplemental Fig. S11B).
The PhTRMT61A Protein Is Localized to the Nucleus
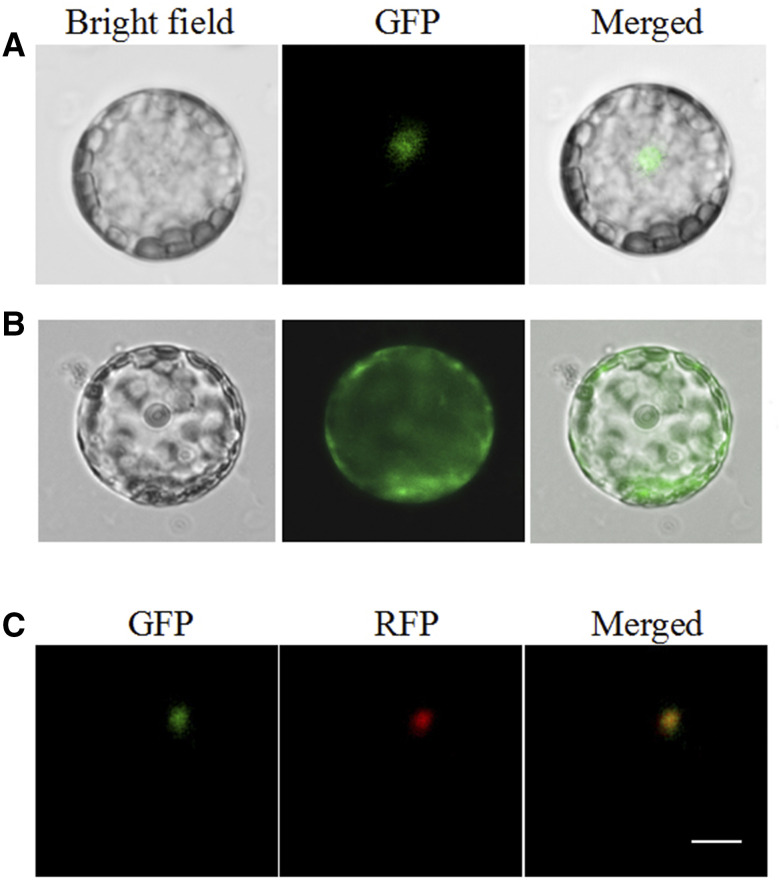
We further investigated the subcellular localization of PhTRMT61A in petunia ‘Mitchell’ cells. The pSAT-35S:GFP-TRMT61A vector, which included the full ORF sequence of PhTRMT61A, was constructed. The pSAT-35S:GFP-TRMT61A and pSAT-35S:GFP vectors were transiently expressed in petunia leaf protoplasts. After ∼20 h, the fluorescence signal of the PhTRMT61A-GFP fusion protein was detected in the nucleus of protoplasts (Fig. 6, A and B). To further confirm the results, EOBI-red fluorescent protein, which is a nuclear localized MYB factor (Spitzer-Rimon et al., 2012; Liu et al., 2017), was used as a nuclear marker. The results further confirmed that PhTRMT61A localizes to the nucleus (Fig. 6C).
Figure 6.
Subcellular localization of PhTRMT61. Petunia protoplasts were transformed with a construct carrying GFP or PhTRMT61-GFP under the control of the CaMV 35S promoter to assess subcellular localization. A, Expression of the PhTRMT61-GFP fusion protein. B, Expression of GFP protein. C, Subcellular localization of PhTRMT61-GFP with EOBI-red fluorescent protein as markers. All images were captured with a confocal laser scanning system. Scale bar = 5 µm.
DISCUSSION
Recent studies have shown that m1A affects many aspects of RNA metabolism in mammals (Dominissini et al., 2016; Li et al., 2016a). However, the abundance, function, and distribution of m1A in plant RNA remain unknown. Here, we performed transcriptome-wide profiling of m1A sites in petunia corolla RNAs through m1A-seq using an m1A-specific antibody.
Our dot blot and LC-MS/MS assays indicated that m1A shows differences in abundance in different RNA species and presents the highest abundance in mRNA and the lowest abundance in rRNA (Fig. 1C); m5C levels are also highest in mRNA in Arabidopsis (Cui et al., 2017). Similar to m6A in Arabidopsis (Bodi et al., 2012), m1A is an abundant mRNA modification in plants, in which the m1A/A ratio is as high as 1.82%, and the m1A levels in mRNA are spatially and temporally regulated (Fig. 1E). In addition, m1A presents differences in abundance in the total RNA of different plant species and exhibits the highest abundance in the woody plant B. latifolia (Supplemental Fig. S1).
When m1A-seq was applied, 4,993 m1A peaks were identified in 3,231 genes expressed in petunia, and each methylated gene presented an average of 1.55 peaks, showing 13.6% coverage of m1A in expressed genes (Supplemental Data Set 1). In mammalian cells, Dominissini et al. (2016) identified 7,154 peaks in 4,151 gene transcripts, and each methylated gene exhibited an average of 1.72 peaks. Li et al. (2016) identified 901 m1A peaks in 600 genes in HEK293T cells. These results showed that the number of m1A-methylated genes in petunia corollas can be compared with that in mammalian cells.
It is worth noting that analysis of the m1A peaks identified in this study showed that m1A is preferentially positioned in CDSs and that there is one peak of m1A enrichment within CDSs (Fig. 3, C and D), which is located closely after the start codon. Together with the introduction of a positive charge on m1A, these distribution features imply that m1A modifications in CDSs could play a regulatory role in transcriptional and translational processes in petunia. However, in human mRNA, m1A is enriched near start codons upstream of the first splice site and presents a positive correlation with transcription and translation (Dominissini et al., 2016; Li et al., 2016a). These results suggest potential divergence of the m1A methylation machinery and its functions in different eukaryotic species. In addition, m6A is preferentially located at the 3′ end of transcripts in Arabidopsis (Bodi et al., 2012). The obtained m6A-IP reads are markedly enriched near start codons and stop codons and within 3′ UTRs in Arabidopsis (Luo et al., 2014). In Arabidopsis, the greatest number of m5C sites was identified within the CDS. While m5C sites were evenly distributed within 5′ UTRs and CDSs, m5C sites were most abundant in the first quarter of 3′ UTRs (David et al., 2017). In another study, m5C was enriched in CDSs, with two peaks positioned closely before stop codons and after start codons in Arabidopsis (Cui et al., 2017). These results indicated the existence of differences in the methylation machinery of different RNA methylation modifications in plants.
By applying m1A-seq, 4,993 m1A peaks were identified in 3,231 genes expressed in petunia corollas, among which were identified only 251 m1A peaks in which the A residues were partly replaced by T residues and/or reverse transcription stopped at A sites, showing that read-through accompanied by mismatches and reverse transcription stops did not occur in most m1A peaks identified in this study. However, Dominissini et al. (2016) showed a high rate of mismatches and reverse transcription stops. This disparity may be due to the different reverse transcriptases used in these experiments.
In humans, m1A is regulated by stress, and most peaks appear to be stress specific (Li et al., 2016a). Ethylene treatment reduces the total m1A level in mRNA (Fig. 1F; Supplemental Data Set 10). In addition, dot blotting showed that the m1A level in mRNA is dynamically regulated by senescence in corollas (Fig. 1E). These results showed that mRNA m1A could be associated with flower senescence.
The correlation between the whole m1A methylome and mRNA transcriptome in corollas treated with ethylene and air was analyzed, and the obtained Pearson's correlation coefficient, denoted by r, which is a statistical measure of the strength of a linear relationship between paired data and is, by design, constrained between −1 and 1 (Amdisen, 1987), was 0.30 (Supplemental Fig. S8). This suggested that the m1A methylome and transcriptome were slightly positively correlated, implying that the changing pattern of the m1A methylome was coincident with that of the transcriptome after ethylene treatment to a certain extent.
In S. cerevisiae, TRMT61, a Rossman fold methyltransferase superfamily member, has been identified as a methyltransferase responsible for tRNA m1A58 formation, which uses S-adenosyl-Met as a methyl donor (Anderson et al., 2000; Kozbial and Mushegian, 2005). In humans, TRMT61B localizes to the mitochondria and has been identified as a mitochondria-specific tRNA methyltransferase responsible for tRNA m1A58 formation (Chujo and Suzuki, 2012; Bar-Yaacov et al., 2016). In fact, there is only one member in the TRMT family in many plant species genomes. In this study, we showed that PhTRMT61A could be an m1A methyltransferase for mRNA. First, an in vitro methylation assay showed that His-PhTRMT61A increased the m1A modification level in the mRNA from petunia corollas. Second, the total RNA of E. coli containing His-PhTRMT61A presented a higher m1A modification level than that of wild-type E. coli. Finally, PhTRMT61A silencing resulted in a significant reduction of m1A modification levels in both total RNA and mRNA in petunia corollas, implying that PhTRMT61A indeed affects m1A modification in vivo.
In yeast, TRMT61 plays an important role in the cellular stabilization of initiator tRNAMet (Anderson et al., 1998; Kadaba et al., 2004). TRMT61A silencing in human cells causes a slow growth phenotype, indicating a role of TRMT61A in cell proliferation (Saikia et al., 2010). In this study, in addition to decreasing the m1A level in mRNA, PhTRMT61A silencing affected the development of leaves in petunia; in particular, PhTRMT61A silencing caused a chlorotic leaf phenotype and reduced chlorophyll contents (Fig. 5, D–G). Additionally, PhTRMT61A silencing resulted in a reduced size of leaf epidermal cells (Fig. 5, H–K), while the size of the leaves was not significantly changed, showing that PhTRMT61A silencing could promote cell division. Analysis of qPCR assay results showed that the transcription of PhTRMT61A was high in the leaves (Supplemental Fig. S11A), supporting its function in leaf development. However, the direct connection between the phenotypic change and the reduction in mRNA m1A levels mediated by PhTRMT61A silencing requires further study. In Arabidopsis, mutations in m6A methyltransferase result in an embryo-lethal phenotype (Zhong et al., 2008), and reduced m6A levels cause developmental defects in floral organs (Bodi et al., 2012). In Arabidopsis, mutations in TRM4B, the mRNA m5C methyltransferase, cause a decrease m5C peaks and defects in root development due to reduced cell division in the root apical meristem (Cui et al., 2017). In addition, trm4b mutants show increased sensitivity to oxidative stress (David et al., 2017). These results reveal the important role of RNA modifications in plant growth and development.
In addition, ethylene treatment significantly decreased PhTRMT61A expression in corollas (Supplemental Fig. S11B). The possibility that PhTRMT61A plays a role in the decrease in m1A levels in response to ethylene cannot be ruled out, and the underlying mechanism requires further study.
In S. cerevisiae, TRMT61A and TRMT61B are responsible for the methylation of nuclear and mitochondrial RNA, respectively (Chujo and Suzuki, 2012; Bar-Yaacov et al., 2016). In this study, nuclear localization of PhTRMT61A indicated that PhTRMT61A is responsible for the methylation of nuclear RNA rather than of chloroplast or mitochondrial RNA, and that PhTRMT61A-mediated methylation of RNA occurs prior to nuclear export of RNA.
MATERIALS AND METHODS
Plant Material
Petunia (Petunia hybrida) ‘Mitchell’ and ‘Ultra’ plants were grown in a greenhouse (23 ± 2°C; Liu et al., 2011). To avoid self-pollination, which will increase ethylene production and accelerate petal senescence, we emasculated the flowers 1 d before they were fully opened and harvested them at the anthesis stage (corollas 90° reflexed). Roots, stems, and leaves were collected at the vegetative stage when the plants were ∼10 cm in height. Three biological repeats, each including three technical repeats, were performed for each experiment.
Ethylene Treatment
Petunia ‘Mitchell’ flowers were treated with ethylene according to our previous protocols (Guo et al., 2017). After being harvested at anthesis, the flower stems were placed in flasks with distilled water and then treated with air or 2 μL L–1 ethylene for 16 h. Corollas were collected and frozen in liquid nitrogen and stored at −80°C. Three biological repeats, each including three technical repeats, were performed for each experiment.
RNA Extraction, Library Construction, and Sequencing
RNA extraction, library construction, and sequencing were performed according to previously described protocols (Guo et al., 2017). The total RNA of the samples was isolated using a TRIzol Kit (Promega) and treated with RNase-free DNase I (Takara Bio) for 30 min at 37°C to remove residual DNA. All mRNA was broken up into ∼50-bp fragments by adding fragmentation buffer. First-strand cDNA was generated using random hexamer-primed reverse transcription with Thermo-X reverse transcriptase, followed by the synthesis of second-strand cDNA using RNase H and DNA polymerase I. The cDNA fragments were purified using a QIAquick PCR extraction kit. These purified fragments were then washed with EB buffer for end reparation poly (A) addition and ligated to sequencing adapters. Following agarose gel electrophoresis and extraction of the cDNA from gels, the cDNA fragments were purified and enriched by PCR to construct the final cDNA library. The cDNA library was sequenced on the Illumina sequencing platform (HiSeq 2000) using paired-end technology by Gene Denovo. A Perl program was written to select clean reads by removing low-quality sequences (with >50% of bases having a quality of <20 in one sequence), reads with >5% N bases (bases unknown), and reads containing adaptor sequences. Three biological repeats, each including three technical repeats, were performed for each experiment.
Read Alignment and Normalization of Gene Expression Levels
Read alignment was performed according to previously described protocols (Guo et al., 2017). We mapped the sequencing reads to the reference sequence with SOAPaligner/soap2 (Li et al., 2009), a tool designed for short sequence alignment. The coverage of reads in a single gene was used to calculate the expression level of that gene. We obtained the expression levels of all genes detected using this method.
Reads that could be uniquely mapped to a gene were used to calculate expression levels. The gene expression level was measured as the number of uniquely mapped reads per kilobase of exon region per million mappable reads (Guo et al., 2017). This value can be directly used for comparing the differences in gene expression between samples. All expression data statistics and visualization were conducted with the R package (http://www.r-project.org/).
lncRNA Prediction
StringTie software (v 1.2.2; http://ccb.jhu.edu/software/stringtie/) was used to assemble and merge the reads on the petunia reference genome. The combined transcripts were compared with the known reference transcripts using cuffcompare software v2.2.1 (Trapnell et al., 2010) and the candidate lncRNAs were obtained. Among the candidate lncRNAs, the transcripts with a length <200 bp and an ORF length >300 bp were discarded. Any potential coding of the remaining transcripts was evaluated using Pfamscan (http://xfam.org/; Punta et al., 2012) and CPC (Kong et al., 2007). Only transcripts determined to be noncoding by both CPC and Pfamscan were considered lncRNAs.
Identification of circRNAs
After RNA sequencing, all the high-quality clean data were used for identification of circRNAs according to the methods of Ye et al. (2015). Clean reads were mapped to the petunia reference genome (https://solgenomics.net/organism/Petunia_axillaris/genome) using bowtie2 software (v2.2.9; http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml). The reads that could not be mapped to the genomes were obtained. For these unmapped reads, the 20-nucleotide anchors were first extracted from both ends and aligned independently to the petunia reference genome to identify the unique anchor positions by the widely used tool find_circ described previously (Memczak et al., 2013). The reversed orientation of the aligned anchors suggested circRNA splicing. Then, the anchor alignments were extended to generate the GU/AG splice sites flanking the complete read alignments and breakpoints. Finally, a circRNA was identified as a candidate if it had at least two distinct back-spliced reads.
Quantification of m1A by LC-MS/MS
Quantification analysis of m1A was performed as described previously (Li et al., 2016a), with slight modifications. A 150 ng sample of purified RNA was digested into nucleosides with 0.5 U of P1 nuclease (N8630, Sigma-Aldrich) in 20 μL of 10 mm ammonium acetate buffer (pH 5.3) at 42°C for 6 h, followed by digestion with 0.5 U of alkaline phosphatase (P4252, Sigma-Aldrich) with 2.5 μL of 0.5 m MES buffer (pH 6.5) added to remove the phosphate groups of the nucleosides. The mixture was incubated at 37°C for 6 h and diluted to 50 μL for LC-MS/MS.
After being separated by ultra-performance liquid chromatography on a C18 column, the nucleosides were detected by triple-quadrupole MS (6200 series TOF/6500 series Q-TOF vB.06.01 [B6157], Agilent) in positive-ion multiple reaction-monitoring mode. The mass transitions of mass-to-charge ratios (m/z) 282.0 to 150.1 (m1A), m/z 282.0 to 150.1 (m6A), and m/z 268.0 to 136.0 (A) were monitored and recorded. We assessed a series of concentrations of pure authentic nucleoside standards for every batch of experiments to obtain the corresponding standard curves, since we can deduce the concentrations of nucleosides in mRNA samples by fitting the signal intensities to the standard curves. Then, the m1A/A ratios were calculated.
Dot Blot Assays
Dot blot analysis was performed as described previously (Shen et al., 2016; Cui et al., 2017; Nagarajan et al., 2019). Purified RNA was first denatured and spotted onto a Magna Nylon Transfer membrane, followed by UV crosslinking (UVP Crosslinker, CL-1000, Analytik Jena). The membrane was washed with 1× Tris-buffered saline with Tween 20 (PBST), blocked with 5 g nonfat milk in 100 mL PBST, and incubated with an anti-m1A antibody (1:1,000; D345-3, MBL) for 1 h at 25°C. The blots were washed three times with 5 g Tween-20 in 100 mL PBS buffer. Then, the membranes were incubated with a goat anti-mouse IgG-HRP secondary antibody (1:5,000; M21002, Abmart) for 1 h at 25°C. The blots were washed four times with 1× PBST and visualized using the High-sig ECL western Blotting Substrate (Tanon). Three biological repeats, each including three technical repeats, were performed for each experiment.
m1A-Seq
m1A-seq was performed by CloudSeq Biotech according to the published procedure (Li et al., 2016a), with slight modifications. Briefly, fragmented RNA was incubated with an anti-m1A polyclonal antibody (202003, Synaptic Systems) in immunoprecipitation buffer (10 mm Tris-Cl [pH 7.4], 150 mm NaCl, and 0.1% NP-40) for 2 h at 4°C. The mixture was then immunoprecipitated by incubation with protein-A beads (Thermo Fisher) at 4°C for an additional 2 h. Then, bound RNA was eluted from the beads with N1-methyladenosine (PR3732, Berry & Associates) in IPP buffer and extracted with TRIzol reagent (Thermo Fisher) according to the manufacturer’s instructions. The purified RNA was used for RNA-seq library generation with the NEBNext Ultra RNA Library Prep Kit (New England Biolab). Both the input sample without IP and the m1A IP samples were subjected to 150-bp paired-end sequencing on an Illumina HiSeq sequencer. Two biological repeats were performed for each experiment.
Peak Calling
Adaptors and low-quality bases were trimmed from raw sequencing reads using Cutadapt software (v1.9.3; Martin, 2011). The reads were aligned to the petunia genome (https://solgenomics.net/organism/Petunia_axillaris/genome) using Tophat2 (v2.0.12; Kim et al., 2013). Peaks enriched via IP over input experiments were identified using MACS2 (v2.1.0.20140616; Zhang et al., 2008). MACS2-identified peaks were intersected with a database of exons from the petunia genome (RefSeq annotation). Peaks were allocated to the feature containing the segment with which they shared the largest overlap. Peaks falling in intergenic sequences or exhibiting overlap shorter than 50 nucleotides were excluded from further analyses. For each cell type, only peaks identified (fold change ≥2 or ≥4, as indicated, false discovery rate ≤0.05) in replicates were considered. Common peaks were defined as peaks that were independently identified in both air- and ethylene-treated RNA. Negative peaks were identified by switching the IP and input samples (Zhang et al., 2008).
m1A-Seq Data Analysis
Genome sequences and annotations were downloaded from https://solgenomics.net/organism/Petunia_axillaris/genome (v1.6.2). Paired-end reads were harvested from an Illumina HiSeq 4000 sequencer and were subjected to quality control according to Q30 values. Thereafter, the reads were subjected to 3′ adaptor trimming using Cutadapt software (v1.9.3) to remove low-quality reads and generate clean reads (Martin, 2011). First, the clean reads of input libraries were aligned to the genome using STAR software (Dobin et al., 2013). Second, the clean reads of the input libraries were mapped to the genome with Hisat2 software (v2.04; Kim et al., 2015) and assembled with StringTie software (v1.2.2). The assembled transcripts were merged and compared with known transcripts using Cuffcompare software (v2.2.1). Thereafter, the clean reads of all libraries were aligned to the reference genome with Hisat2 software. Methylated sites on RNAs (peaks) were identified with MACS software (Liu, 2014). Differentially methylated sites were identified with diffReps (Shen et al., 2013). The peaks identified by both software platforms overlapping with exons in the mRNA were identified and chosen with homemade scripts. GO enrichment analysis was performed using information extracted from the reference annotation with homemade software. Pathway enrichment analysis was performed using blast2GO software (Conesa et al.,2005).
Annotation of mRNA Segments
Nonoverlapping mRNA regions (“5 UTR,” “start codon (startC),” “CDS,” “stop codon (stopC),” and “3 UTR”) were used for m1A peak analysis. The “startC” and “stopC” segments were defined as the sequences 100 nucleotides upstream and downstream, respectively, of the start and stop codons. The location of the mRNA segments was determined using BEDTools (Li et al. 2016b).
Motif Discovery and GO Analysis
Motif discovery was performed according to the previously described methods of Li et al. (2016b). We used the MEME algorithm (http://meme-suite.org/) to perform de novo searches for enriched motifs in demethylase-sensitive regions. To ensure the reliability of the identified motifs, we used two sequence sets as controls. The motif search was performed with demethylase-sensitive regions. The P-value was used to denote the significance of motif enrichment: the lower the P-value, the more significant the motifs (P < 0.05).
All GO term association and enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/; Wu et al., 2015; Xie et al., 2015).
Sequence Analysis
Alignments were performed, and a phylogenetic tree was generated using the DNAMAN software (Lynnon). Identity search for nucleotides and translated amino acids was performed using the National Center for Biotechnology Information BLAST network server (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Expression and Purification of the Recombinant PhTRMT61A Protein
Expression and purification of the recombinant PhTRMT61A protein were according to a previous protocol (Chujo and Suzuki, 2012). The full-length cDNA of PhTRMT61A was amplified by RT-PCR of total RNA from petunia corollas using the primers listed in Supplemental Table S2. The PCR product was cloned into the corresponding sites of the pCZN1 vector (Novagen) to obtain the expression vector pCZN1-PhTRMT61A, which produced His-fused PhTRMT61A (His-PhTRMT61A). The Escherichia coli Top10 strain was transformed with pCZN1-PhTRMT61A and cultured in LB media containing 50 mg mL−1 ampicillin. When the bacteria reached an optical density of 0.5, protein expression was induced by the addition of 50 mm isopropylthio-β-galactoside, and the cells were grown for 5 h at 20°C. Cells were harvested and suspended in buffer A (20 mm Tris-HCl containing 1 mm phenylmethylsulfonyl fluoride and bacteria protease inhibitor cocktail [pH 8.0]; Sigma-Aldrich), followed by sonication on ice. Cell lysates were cleared by ultracentrifugation at 100,000g for 20 min at 4°C. The supernatant was loaded onto a Ni-IDA -Sepharose Cl-6B chelating column (Smart Life Sciences). After washing off unbound proteins with buffer A, recombinant proteins were eluted with a 50-mL linear gradient from 0 to 500 mm imidazole in buffer A. Fractions containing the recombinant proteins were pooled and dialyzed overnight against buffer B (20 mm Tris-HCl, 250 mm imidazole, and 0.15 m NaCl [pH8.0]). The concentration of purified His-PhTRMT61A was determined by the SDS-PAGE protein assay and western-blot analysis.
Construction of VIGS Vectors and Transformation
VIGS vectors were constructed according to our previously reported protocols (Liu et al., 2019). Specific primers (Supplemental Table S2) for PhTRMT61A were designed to clone 416 bp of the CDSs of this gene, and the pTRV2-PhTRMT61A vector was constructed. Agrobacterium tumefaciens (strain AGLO) was transformed with the pTRV2 or pTRV2-PhTRMT61A and pTRV1 vectors. The A. tumefaciens culture was grown overnight at 28°C in Luria-Bertani medium with 200 mm acetosyringone and 50 mg L−1 kanamycin. The A. tumefaciens cells were harvested and resuspended in inoculation buffer (10 mm MgCl2, 10 mm MES [pH 5.5], and 200 mm acetosyringone to OD600 = 10). After an additional 3 h of incubation at 28°C, A. tumefaciens containing pTRV1 was mixed with A. tumefaciens containing the pTRV2 derivatives in a 1:1 ratio. Then, ∼450 μL of this mixture was applied to the cut surface of 30-d-old petunia ‘Ultra’ plantlets. Thirty-six petunia plants with each vector were subjected to infection with pTRV2-PhTRMT61A and pTRV2 vectors, respectively.
qPCR Assays
qPCR assays were performed on a LightCycler 480 Real-Time PCR system (Roche) according to our previously described methods (Liu et al., 2011). Specific primers (Supplemental Table S3) were designed using the sequence of PhTRMT61A (Peaxi162Scf00129g01325.1, https://solgenomics.net/organism/Petunia_axillaris/genome). The samples were subjected to thermal cycling as follows: DNA polymerase activation at 95°C for 4 min; 36 cycles of 45 s at 95°C, 45 s at 53°C or 55°C, 45 s at 72°C, and 45 s at 80°C, with a final elongation step of 8 min at 72°C. The amplicon was analyzed by electrophoresis and sequenced once for identity confirmation. Quantification was based on the analysis of the threshold cycle (Ct) value, as described previously (Pfaffl, 2001). Petunia Actin served as an internal reference gene (accession no. FN014209) and was subjected to quantitative PCR to quantify cDNA abundance. Three biological repeats, each including three technical repeats, were performed for each experiment.
Pigment Profiling
Total chlorophyll and total carotenoids were measured according to previously described methods (Liu et al., 2019). Four leaves were harvested and freeze dried, and three replicate methanol extractions were prepared. The absorbance of the solution was read at 646.8, 663.2, and 470 nm against the solvent (acetone) blank. The individual concentrations of total chlorophyll and total carotenoids were measured by spectrophotometry and calculated using previously described methods (Lichtenthaler, 1987). Three biological repeats, each including three technical repeats, were performed for each experiment.
Scanning Electron Micrographs
Samples for scanning electron micrography were prepared according to previously described protocols (Yang et al., 2017). Leaves from PhTRMT61A-silenced plants and control plants were cut into ∼4-mm2 pieces. The samples were fixed in 4 mL glutaraldehyde in 100 mL 0.1 mol L−1 phosphate-buffered saline for 4 h at 4°C and then washed three times in the same buffer. The samples were then fixed in 1% osmium tetroxide for 2 h at 25°C and rinsed 3 times using the same buffer. The samples were dehydrated in increasing grades of ethanol and then dried with a critical point drier (CPD 030, Bal-Tec). The dried samples were fixed on the sample stage and coated with gold with ion-sputtering equipment. The samples were observed with a scanning electron microscope (XL-30-ESEM, FEI) at a 10 kV acceleration and photographed.
Subcellular Localization
Subcellular localization was performed according to previously described protocols (Liu et al., 2019). For protein subcellular localization, the full-length form of PhTRMT61A was fused to GFP at the C-terminal end of the pSAT-1403TZ plasmid (Tzfira et al., 2005) to form the pSAT-35S:GFP-TRMT61A vector. The control vector (35S:GFP) and the fusion vector were separately transformed into petunia ‘Mitchell’ protoplasts. Digital images were obtained using a confocal laser scanning system (ECLIPSE TE2000-E, Nikon). The sequences of the primers are described in Supplemental Table S2. Three biological repeats were performed for each experiment.
Statistical Analyses
Statistical analysis was performed using one-way ANOVA followed by Duncan’s new multiple range test (NMRT) with three replicates. P ≤ 0.05 was considered significant.
Accession Numbers
Sequence data from this manuscript can be found in GenBank (the following secure token has been created to allow review of record GSE136921 while it remains in private status: yxifsqkyxxstbmp). The raw data of RNA sequencing of GSE136921 have been submitted to the National Center for Biotechnology Information.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Detection and quantification of m1A levels in different plant species.
Supplemental Figure S2. Effect of ethylene on flowers of petunia ‘Mitchell’.
Supplemental Figure S3. Mass spectrogram of A and methyladenosine.
Supplemental Figure S4. GO-based enrichment analysis of mRNA with m1A.
Supplemental Figure S5. m1A is dynamically regulated by ethylene.
Supplemental Figure S6. Venn diagram of m1A peaks and m1A-methylated genes under ethylene and air treatment.
Supplemental Figure S7. GO-based enrichment analysis of mRNA with up- and downregulated m1A levels.
Supplemental Figure S8. Concordance between changes in the m1A methylome and those in the mRNA transcriptome after air and ethylene treatment.
Supplemental Figure S9. Predicted amino acid sequence alignment and neighbor-joining tree of TRMT61A.
Supplemental Figure S10. Effects of pTRV2-PhTRMT61A treatment on the expression of PhTRMT61A in leaves as determined by quantitative PCR.
Supplemental Figure S11. Expression of PhTRMT61A determined by quantitative PCR in different organs and developmental stages and in response to 16 h of 2 μL L–1 exogenous ethylene.
Supplemental Table S1. Comparative analysis of PhTRMT61A amino acid sequences with those of its closest homologs in Arabidopsis, S. lycopersicum, O. sativa, and H. sapiens.
Supplemental Table S2. Primer sequences of PhTRMT61A used in vector construction for recombinant protein production, VIGS, and subcellular localization.
Supplemental Table S3. Primer sequences of PhTRMT61A and PhACTIN used in quantitative real-time PCR.
Supplemental Data Set 1. Methylated mRNA sites in petunia corollas treated with air and ethylene.
Supplemental Data Set 2. List of all gene reads per kilobase of exon region per million mappable reads for RNA sequences in petunia corollas treated with air and ethylene.
Supplemental Data Set 3. Differentially expressed gene list of RNA sequences in petunia corollas treated with air and ethylene.
Supplemental Data Set 4. GO analyses of molecular function for all methylated genes.
Supplemental Data Set 5. m1A mismatch replaced by T and reverse transcription stopped at the A site in mRNA.
Supplemental Data Set 6. lncRNA profiling of petunia corollas.
Supplemental Data Set 7. circRNA profiling of petunia corollas.
Supplemental Data Set 8. All methylated lncRNAs.
Supplemental Data Set 9. All methylated circRNAs.
Supplemental Data Set 10. Differentially methylated sites in mRNA in petunia corollas.
Supplemental Data Set 11. Differentially methylated sites in lncRNA in petunia corollas.
Supplemental Data Set 12. Differentially methylated sites in circRNA in petunia corollas.
Supplemental Data Set 13. Molecular function GO analysis of upregulated methylated sites.
Supplemental Data Set 14. Molecular function GO analysis of downregulated methylated sites.
Supplemental Data Set 15. Association between m1A methylome and transcriptome.
Acknowledgments
We thank Cloud-Seq Biotech and BioMarker Technologies for the m1A-seq service.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31870692, 31770737, and 31661143047).
References
- Abeles FB, Morgan PW, Saltveit ME Jr.(1992) Ethylene in Plant Biology. Academic Press, San Diego [Google Scholar]
- Amdisen A.(1987) Pearson’s correlation coefficient, p-value, and lithium therapy. Biol Psychiatry 22: 926–928 [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG(2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Björk GR, Tamame M, Hinnebusch AG(1998) The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B, et al. (2016) Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol 14: e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG(2012) Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front Plant Sci 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess AL, David R, Searle IR(2015) Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol 15: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik C, Chua HN, Singh G, Akef A, Snyder MP, Palazzo AF, Moore MJ, Roth FP(2017) A common class of transcripts with 5′-intron depletion, distinct early coding sequence features, and N1-methyladenosine modification. RNA 23: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC(2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3: 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ(2010) A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin LJ, Jones ML(2009) Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. J Exp Bot 60: 2179–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liu H, Wei Q, Zhao H, Liu J, Yu Y(2017) The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. J Exp Bot 68: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Tran H, Liang Z, Lin H, Zhang L(2015) Identification and analysis of the N6-methyladenosine in the Saccharomyces cerevisiae transcriptome. Sci Rep 5: 13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Suzuki T(2012) Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18: 2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DG, Richards C, Hilioti Z, Lind-Iversen S, Brown K(1997) Effect of pollination on accumulation of ACC synthase and ACC oxidase transcripts, ethylene production and flower petal abscission in geranium (Pelargonium × hortorum L.H. Bailey). Plant Mol Biol 34: 855–865 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M(2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y, Zhang B, Leo V, Vardy LA, Lu T, et al. (2017) 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol Plant 10: 1387–1399 [DOI] [PubMed] [Google Scholar]
- Daoud H, Zhang D, McMurray F, Yu A, Luco SM, Vanstone J, Jarinova O, Carson N, Wickens J, Shishodia S, et al. (2016) Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. J Med Genet 53: 200–207 [DOI] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR(2017) Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C(2015) Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res 43: 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Agopian RG, Fabi JP, Cordenunsi-Lysenko BR(2020) Metabolome and proteome of ethylene-treated papayas reveal different pathways to volatile compounds biosynthesis. Food Res Int 131: 108975. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR(2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. (2016) The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Song J, Forney C, Palmer LC, Fillmore S, Zhang Z(2016) Proteome changes in banana fruit peel tissue in response to ethylene and high-temperature treatments. Hortic Res 3: 16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R(2013) Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9: e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T(2016) RNA modifications: What have we learned and where are we headed? Nat Rev Genet 17: 365–372 [DOI] [PubMed] [Google Scholar]
- Gerats T, Vandenbussche M(2005) A model system for comparative research: Petunia. Trends Plant Sci 10: 251–256 [DOI] [PubMed] [Google Scholar]
- Gigova A, Duggimpudi S, Pollex T, Schaefer M, Koš M(2014) A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 20: 1632–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liu J, Wei Q, Wang R, Yang W, Ma Y, Chen G, Yu Y(2017) Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiol 173: 668–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Min CW, Meng Q, Agrawal GK, Rakwal R, Kim ST(2018) Comparative phosphoproteome analysis upon ethylene and abscisic acid treatment in Glycine max leaves. Plant Physiol Biochem 130: 173–180 [DOI] [PubMed] [Google Scholar]
- Hastings MH.(2013) m6A mRNA methylation: A new circadian pacesetter. Cell 155: 740–741 [DOI] [PubMed] [Google Scholar]
- Hong B, Brockenbrough JS, Wu P, Aris JP(1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol 17: 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Xiong J, Yang Y, Liu S, Yuan B, Feng Y(2015) Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. RSC Advances 5: 64046–64054 [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. (2013) NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J(2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR(2013) Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL(2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL(2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G(2007) CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozbial PZ, Mushegian AR(2005) Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston BJ, Bai S, Jones ML(2005) Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. J Exp Bot 56: 15–23 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J(2009) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C(2016a) Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat Chem Biol 12: 311–316 [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Yi C(2016b) Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat Methods 14: 23–31 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK.(1987) Chlorophylls and carotenoids: Pigments of photosynthesis. Methods Enzymol 148: 350–352 [Google Scholar]
- Liu F, Xiao Z, Yang L, Chen Q, Shao L, Liu J, Yu Y(2017) PhERF6, interacting with EOBI, negatively regulates fragrance biosynthesis in petunia flowers. New Phytol 215: 1490–1502 [DOI] [PubMed] [Google Scholar]
- Liu J, Chang X, Ding B, Zhong S, Peng L, Wei Q, Meng J, Yu Y(2019) PhDHS is involved in chloroplast development in petunia. Front Plant Sci 10: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y(2011) Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J Exp Bot 62: 825–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. (2014) Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. In Kidder BL, ed, Stem Cell Transcriptional Networks, Methods in Molecular Biology, volume 1150, Humana Press, Totowa, NJ, pp 81–95 [DOI] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. (2014) Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J(2008) Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol 148: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macon JB, Wolfenden R(1968) 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 7: 3453–3458 [DOI] [PubMed] [Google Scholar]
- Martin M.(2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338 [DOI] [PubMed] [Google Scholar]
- Nagarajan A, Janostiak R, Wajapeyee N(2019) Dot blot analysis for measuring global N6-methyladenosine modification of RNA. Methods Mol Biol 1870: 263–271 [DOI] [PubMed] [Google Scholar]
- O’Neill SD, Nadeau JA, Zhang XS, Bui AQ, Halevy AH(1993) Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell 5: 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT(2005) The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Bujnicki JM, Sem DS, Anderson JT(2007) Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res 35: 6808–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB(2004) Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW.(2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MS, Dodge LL, Mor Y, Evans RY(1989a) Effects of ethylene on rose opening. Acta Hortic (261): 215–220 [Google Scholar]
- Reid MS, Evans RY, Dodge LL, Mor Y(1989b) Ethylene and silver thiosulfate influence opening of cut rose flowers. J Am Soc Hortic Sci 114: 436–440 [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C(2017) Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. (2017) Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 215: 157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T(2010) Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Lyko F(2009) RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kötter P, Entian KD(2013) Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 41: 9062–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, Jia GF, Chen J, Feng YQ, Yuan BF, Liu SM(2015) Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab 100: E148–E154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H(2016) N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell 38: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ(2013) diffReps: Detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 8: e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade WO, Ray WK, Williams PM, Winkel BS, Helm RF(2012) Effects of exogenous auxin and ethylene on the Arabidopsis root proteome. Phytochemistry 84: 18–23 [DOI] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, Cna’ani A, Ben Zvi MM, Masci T, Edelbaum O, Yu Y, Shklarman E, Ovadis M, et al. (2012) The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24: 5089–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T(2012) Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuma H, Ushio N, Minoji M, Kazayama A, Shigi N, Hirata A, Tomikawa C, Ochi A, Hori H(2015) Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI). J Biol Chem 290: 5912–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Liu J, Huang F, Guan J, Zhong S, Tang N, Zhao J, Yang W, Yu Y(2014) PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Mol Plant 7: 1384–1387 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L(2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne HJ, et al. (2015) The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34: 2350–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F(2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, van Meeteren U(2003) Flower opening and closure: A review. J Exp Bot 54: 1801–1812 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ(2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V(2005) pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, Lang Z(2015) Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol 16: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Stier G, Lin J, Liu G, Zhang Z, Chang Y, Reid MS, Jiang CZ(2013) Transcriptome changes associated with delayed flower senescence on transgenic petunia by inducing expression of etr1-1, a mutant ethylene receptor. PLoS One 8: e65800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR(2002) Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ji X, Guo X, Ji S(2017) Regulatory role of N6 -methyladenosine (m6A) methylation in RNA processing and human diseases. J Cell Biochem 118: 2534–2543 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ(1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15: 444–447 [DOI] [PubMed] [Google Scholar]
- Woltering EJ, van Doorn WG(1988) Role of ethylene and senescence of petals: Morphological and taxonomical relationships. J Exp Bot 39: 1605–1616 [Google Scholar]
- Wu Q, Cheng Z, Zhu J, Xu W, Peng X, Chen C, Li W, Wang F, Cao L, Yi X, et al. (2015) Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci Rep 5: 9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kang H, Liu W, Wang GL(2015) Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J Proteome Res 14: 2017–2025 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Fujii N, Takahashi H(2003) Characterization of ethylene effects on sex determination in cucumber plants. Sex Plant Reprod 16: 103–111 [Google Scholar]
- Yang W, Cai Y, Hu L, Wei Q, Chen G, Bai M, Wu H, Liu J, Yu Y(2017) PhCESA3 silencing inhibits elongation and stimulates radial expansion in petunia. Sci Rep 7: 41471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Huang W, Huang JT, Shen F, Xiong J, Yuan EF, Qin SS, Zhang M, Feng YQ, Yuan BF, et al. (2016) Increased N6-methyladenosine in human sperm RNA as a risk factor for asthenozoospermia. Sci Rep 6: 24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L(2015) Widespread noncoding circular RNAs in plants. New Phytol 208: 88–95 [DOI] [PubMed] [Google Scholar]
- Yin X, Sakata K, Komatsu S(2014) Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J Proteome Res 13: 5618–5634 [DOI] [PubMed] [Google Scholar]
- Zhang XL, Qi MF, Xu T, Lu XJ, Li TL(2015) Proteomics profiling of ethylene-induced tomato flower pedicel abscission. J Proteomics 121: 67–87 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C(2017) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG(2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]