Abstract
Age-related macular degeneration (AMD) is a common cause of blindness worldwide. While recent studies have revealed that the loss of choroidal endothelial cells (ChECs) is critical to the disease pathogenesis of dry AMD, in vitro studies are needed to fully elucidate the disease mechanism. However, these studies remain hindered due to the lack of publically available human ChEC lines. To address this need, ChECs were harvested form donor tissue and enriched for by using magnetic cell separation using anti-CD31 conjugated microbeads. Next, lenti-viral vectors with endothelial-specific promoters driving genes necessary for immortalization, CDH5p-hTERT and CDH5p TAg, were generated. Stable integration of both gene cassettes allowed cells to maintain their proliferative state and yielded an immortalized cell line (iChEC-1). Immunocytochemical analysis of iChEC-1 confirmed the expression of important ChEC markers such as CA4, a marker of choriocapillaris endothelial cells, CDH5, and CD34, pan-endothelial cell markers. qRT-PCR analysis of expanded clones from iChEC-1 further showed that the line maintained expression of other important endothelial markers, vWF, PECAM1, and PLVAP, similar to primary cells. Functional responses were characterized by tube-forming assays and repopulation of decellularized choroid with the immortalized cell line. In conclusion, the iChEC-1 line presents a suitable immortalized human ChEC line for future in vitro studies of AMD.
Keywords: choroid, endothelial cells, immortalization, macular degeneration, eye
Introduction
Age-related macular degeneration (AMD) is a leading cause of blindness, which is expected to affect up to 196 million individuals worldwide by the year 2020 (Friedman et al., 2004; Wong et al., 2014). The disease is characterized by degeneration of the central retina due to a combination of environmental and genetic factors. While the underlying etiology of AMD has historically centered on degeneration of the retinal-pigmented epithelial cells, recent studies have shown that the death of choroidal endothelial cells (ChECs) play a pivotal role in both development and progression of atrophic “dry AMD” (McLeod et al., 2009; Bhutto et al., 2012; Gelfand and Ambati, 2016; Mullins et al., 2011; Whitmore et al., 2013). Moreover, in the neovascular (“wet”) end stage of AMD, which affects as many as 5-9% of older adults (Friedman et al., 2004), ChECs become activated to form pathologic vascular complexes that invade the retina and ultimately lead to death of photoreceptor cells (Green and Enger, 2005).
Although additional research is needed to more fully understand the role that ChECs play in AMD pathogenesis and progression, several barriers that impede scientific progress exist. First, commercial sources of human choroidal endothelial cells are not readily available, and choroidal ECs in tissue culture have distinctive features compared to EC from other vascular sources (Palanisamy et al., 2018). Second, human donor tissue, with a sufficiently short death-to-preservation interval to allow for reliable isolation and expansion endothelial cell cultures, is not readily accessible for most researchers. In addition, following isolation, primary ChECs can be passaged a limited number of times before cellular senescence occurs. The difficulty in acquiring and maintain human ChECs leaves researchers with limited options for the study of disease pathology in vitro.
In this study we sought to generate an immortalized cell line from human donor tissue to extend the utility of harvested ChECs. We hypothesized that an endothelial cell-specific promoter driving expression of the SV40 TAg and hTERT would be sufficient to immortalize ChECs while maintaining appropriate gene expression. Furthermore, limiting the expression of immortalization genes by the use of a cell-type specific promoter would restrict immortalization to only cells that continue to express endothelial cell specific genes following transduction and expansion. To accomplish this, we isolated primary human ChECs which were subsequently transduced with immortalization factors driven by the endothelial cell specific promoter, CDH5p (Prandini et al., 2005). The resulting immortalized human ChEC line (iChEC-1) was characterized and determined to retain choroidal endothelial cell phenotypes.
Materials and Methods
Isolating Primary ChECs:
Human donor eyes were obtained by the Iowa Lions Eye Bank (Iowa City, IA). For the generation of this cell line, eyes from a one month old, Caucasian female who passed away due to pulmonary hypertension and cardiac failure, were obtained with full consent of the next of kin. All experiments were conducted in accordance with the Declaration of Helsinki. Isolation techniques were performed essentially as previously described (Skeie and Mullins, 2008; Songstad et al., 2017).
Briefly, we made a circumferential cut just anterior to the equator and removed the cornea, ciliary body, iris and lens. Next, we removed the vitreous and the neural retina, followed by removal of the RPE leaving behind choroid/scleral tissue. The choroid was peeled from the sclera and cut into approximately 2mm squares with a razor blade followed by incubation with 2mg/mL collagenase (Life Technologies) for 1 hr at 37°C and the resulting suspension was passed through a 100μm filter followed by a 40μm nylon filter. After ~36 hours of culture in Endothelial Cell Growth Media (R&D Systems; Cat. No. CCM07), non-adherent cells (presumably including CD31-positive hematopoietic cells) were discarded. Adherent cells were liberated using TrypLE (Thermo Fisher Scientific; Cat. No. 12604013) and, to obtain a homogenous population, we performed magnetic cell separation according to the manufacturer’s instructions using anti-CD31 conjugated microbeads (Miltenyi Biotec cat# 130-091-935) on an automated separator system (autoMACS, Miltenyi Biotec). We maintained harvested ChECs by feeding with Endothelial Cell Growth Media and incubating at 37°C with 5% CO2. Confluent cells were passaged using TrypLE.
Generating Lentiviral ChEC Immortalization Vector:
Lentiviral packaging vectors were constructed in a stepwise fashion. First, the promoter for endothelial cell specific Cadherin 5 (CDH5) (Prandini et al., 2005), which is expressed by choroidal endothelial cells (Schubert et al., 2014), was cloned into the pENTR5’-TOPO plasmid (Thermo Fisher Scientific; Carlsbad, CA; Cat. No K59120). Next the human Telomerase gene (hTERT) and SV40 large T (TAg) were amplified from either the pLOX-Ttag-iresTK or pLOX-TERT-iresTK, a gift from Didier Trono (Addgene plasmid # 12246 and # 12245) (Salmon et al.,2000), and individually cloned into the pENTR/D-topo plasmid upstream of a blasticidin resistance cassette (Thermo Fisher Scientific; Cat. No. K240020).Finally the pENTR5’CDH5p and pENTR/D-hTERT or pENTR/D-TAg were cloned into the pDEST R4R3 Vector II (Thermo Fisher Scientific; Cat. No. A11145) using Gateway™ LR Clonase™ II Enzyme mix (Thermo Fisher Scientific; Cat. No. 11791100). The construct maps are depicted in Figure 1.
Figure 1. Generation of the Human iChEC-1 Cell line.
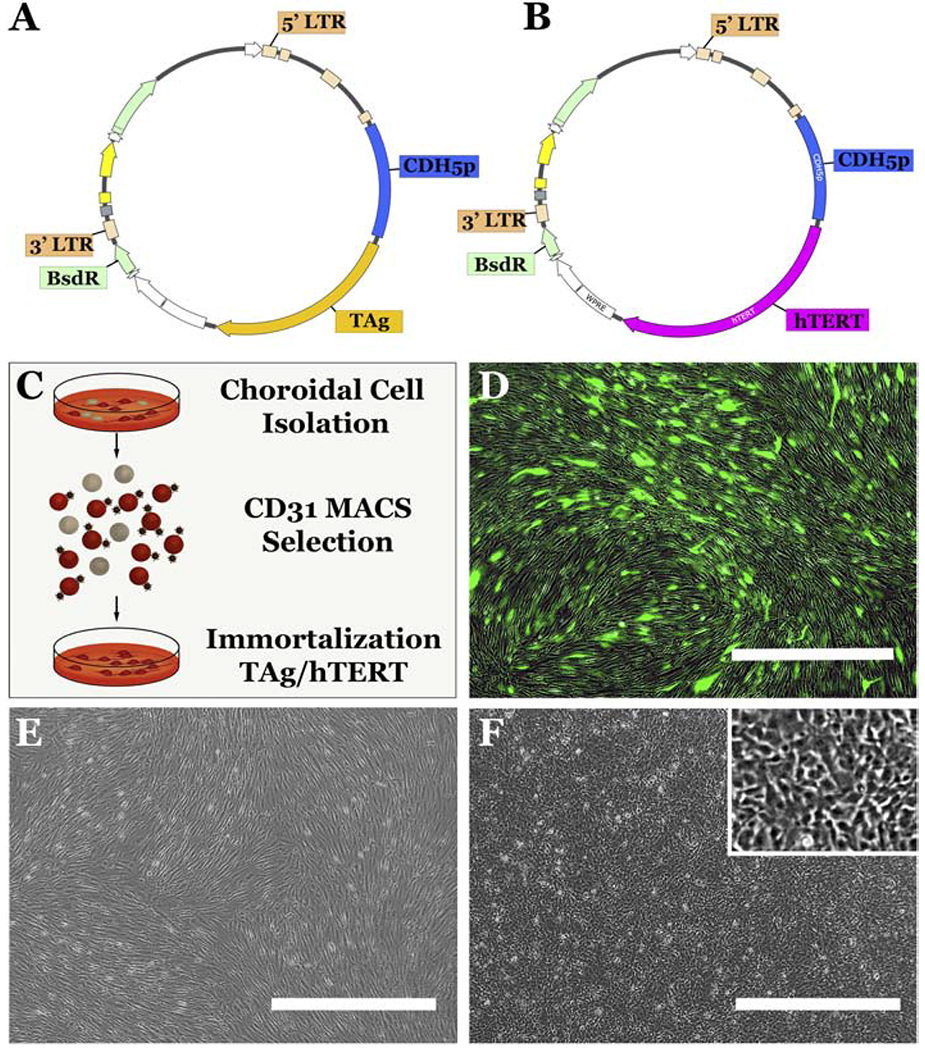
A-B: Schematics depicting HIV lenti viral constructs containing CDH5p-TAg (A) and CDH5p-hTERT(B) cassettes. C: Schematic depicting the protocol used to generate the iChEC-1 cell line. Primary cultures of human ChECs were obtained post-mortem and adherent CD31-positive endothelial cells were enriched using CD31 magnetic beads (MACS). Following isolation, CD31 positive ChECs were transduced with HIV based lenti viral vectors containing CDH5p-TAg (A) and/or CDH5p-hTERT(B). D: HIV-CMVp-GFP transduction of primary human ChECs at an MOI of 10 demonstrating high transduction efficiency. E-F: phase micrographs of primary ChECs and iChEC-1 cells. Scalebar = 1,000μm. CDH5p=CDH5 promoter; TAg= SV40 T-antigen; LTR = long terminal repeat; BsdR=blasticidin resistance ; ChEC= choroidal endothelial cells; MACS= Magnetic-activated Cell Sorting
Producing CDH5p-hTERT and CDH5p Ttag Lentivirus:
Virus production was performed as previously described (Songstad et al., 2017). Briefly, 293FT cells (Thermo Fisher Scientific; Cat. No. R700-07) were transfected with pLP1 (Thermo Fisher Scientific; Cat. No. K4975-00), pLP2 (Thermo Fisher Scientific, Cat. No. K4975-00), pLP/VSVG (Thermo Fisher Scientific; Cat. No. K4975-00), and the transgene cassette. The cell culture media was collected for three days, filtered and centrifuged overnight. The supernatant was subsequently removed and the viral pellet was resuspended in lentiviral suspension buffer (40 mg/ml lactose/PBS buffer).
Transduction of Human ChECs with CDH5p-hTERT and/or CDH5p-TtAG Lentivirus and Selection of ChEC-Like Cells:
Human ChECs were passaged onto 6-well tissue culture plates at a density of 100,000 cells per well and subsequently transduced at a multiplicity of infection (MOI) of 10 with both CDH5-hTERT and CDH5-TAg lentivirus in Endothelial Cell Growth Media. After 48 hours, the cells were fed with fresh Endothelial Cell Media containing 10 μg/mL Blasticidin to eliminate cells that were not transduced. Selection was maintained for one week post-lentiviral transduction.
Culture of immortalized iChEC-1 cell line:
We maintained immortalized ChECs, now referred to as iChEC-1, using the conditions described for primary ChECs above (i.e., by feeding with Endothelial Cell Growth Media (R&D Systems; Cat. No. CCM07) and incubating at 37°C with 5% CO2). Confluent cells were passaged using TrypLE (Thermo Fisher Scientific; Cat. No. 12604013).
Immunocytochemistry:
Cells were fixed in a 4% paraformaldehyde (Alfa Aesar, Haverhill, MA; Cat. No. 43368) solution for 5 minutes and washed using 1× Dulbecco’s phosphate-buffered saline (Thermo Fisher Scientific; Cat. No. 14190250). Subsequently, the cells were incubated with Super Block T20 (TBS) Blocking Buffer (Thermo Fisher Scientific; Cat. No. 37536) for 30 minutes. Next, cells were incubated for 16 hours with a solution of primary antibody and Super Block. The following primary antibodies were used: anti-CA4 (R&D Systems; Cat. No. MAB21861), anti-CD34 (Thermo Fisher Scientific, Cat. No. MA1-10202), anti-CDH5/VE-Cadherin (Abeam; Cat. No. ab33168), anti-CFH (Abeam; Cat. No. ab8842), anti-hTERT (Novus Biological; Cat. No. NB100-317), and anti-SV40 TAg (Abeam; Cat. No. abl6879). To detect primary antibodies, Alexa Fluor 488 donkey anti-mouse (Thermo Fisher Scientific; Cat. No. A21202), Alexa Fluor 488 donkey anti-rabbit (Thermo Fisher Scientific; Cat. No. R37118), or Alexa Fluor 568 Donkey anti-Sheep (Thermo Scientific; Cat. No. A-21099) secondary antibodies were used. Cell nuclei were counterstained using DAPI (Thermo Fisher Scientific; Cat. No. 62248). Samples were imaged using a Leica DM 2500 SPE confocal microscope (Leica Microsystems).
Tube forming assay:
In order to assess the endothelial cell phenotype of tube formation in the iChEC-1 line, Matrigel (Corning Incorporated, Life Sciences, MA, USA, 354234) was thawed at 4C and diluted in EC medium to a concentration of 10 mg/mL. A volume of 290 μL was dispensed with cooled pipet tip to a 24-well flat-bottom tissue culture plate (Nunc Cat. No. 142475, Thermo Scientific). The plate was subsequently incubated for 1 hr at 37°C, followed by addition of ChECs (trypsinized at approximately 80% confluency). A total of 300 μL of the cell suspension was added to each well, at densities ranging from 3.7×l04 to 3×l05. The plate was incubated for 18 to 24 hours at 37°C and 5% CO2. Following incubation, medium was carefully removed and the wells were washed twice with HBSS. Cells were labeled by adding 300 μL of Calcein AM dye per well (Corning, 8 μg/mL in HBSS) and incubating for 1 h at 37°C, 5% CO2, followed by photomicrography on an inverted fluorescence microscope (1X81, Olympus, Tokyo, Japan).
Genotyping:
Single nucleotide polymorphisms associated with age-related macular degeneration were selected from the NHGRI-EBI Catalog of published genome-wide association studies (https://www.ebi.ac.uk/gwas/). SNPs within 3 most significantly AMD-associated genomic regions (1q31.3, 6q21.3, and 10q26.13) were genotyped. The genomic coordinates of these SNPs were visualized using the UCSC Genome Browser (http://genome.ucsc.edu/) and primers were designed manually or using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Genomic DNA was isolated using the NucleoSpin Tissue kit (Macherey-Nagel Cat. No. 740952) and regions between 300-1236 bp were individually PCR amplified. The Carver Nonprofit Genetic Testing Laboratory at the University of Iowa performed Sanger sequencing with an ABI 3730×1 sequencer (Carlsbad, CA).
Recellularization of Choroid Assay:
In order to evaluate the capacity of iChEC-1 cells to recellularize choroidal scaffolds, human choroid were decellularized as previously described(Chirco et al., 2017). Briefly, flash frozen, archived RPE/Choroid tissue was thawed and washed in dH2O followed by a solution of 1% Triton X-100. Then the tissue was rinsed with 1x PBS and then treated with a solution containing 0.1% SDS + 0.1M EDTA. Another 1×PBS wash was applied followed by treatment with a 10 U/mL DNase I solution. The tissue was again washed in 1×PBS followed by incubation in Endothelial Cell Growth Media for 7 days. Immortalized endothelial cells were seeded outside of the tissue. After 7 days, recellularized punches were fixed in 4% paraformaldehyde and embedded for cryostat sectioning. Sections were labeled with DAPI and antibodies directed against collagen IV (Abcam, ab6586) to visualize ChECs and the extracellular matrix scaffold, respectively, as described previously(Chirco et al., 2017).
Gene expression analysis
RNA was collected from samples using the NucleoSpin RNA kit (Macherey-Nagel; Cat. No. 740955) and cDNA was generated using SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific; Cat. No. 11754). To obtain copy numbers, qRT-PCR was performed on the QuantStudio 6 Flex Real-Time PCR System (Thermo Fischer Scientific; Cat. No. 4485697) using Power SYBR Green Master Mix (Thermo Fisher Scientific; Cat. No. 4368577). The following primers were used: vWF (Forward:CCATCGAGGTGAAGCACAGT; Reverse:CCATGTTCCCACCCACGTAA), PECAM1 (Forward:CTGATGCCGTGGAAAGCAGA; Reverse:GGAGCAGGGCAGGTTCATAA), PLVAP (Forward:CAGCTAGCCTGGAGGAGTTC; Reverse:TATCCCTGCATCCTCCGCAA) and 18s rRNA (Forward:CGGCTACCACATCCAAGGAAG; Reverse:GCTGGAATTACCGCGGCTGCT).
For RT-PCR, targeted genes were amplified using BIOLASE DNA Polymerase (Bioline; Cat. No. BIO-21042) and the following primers: TJP1 (Forward:AGCCATTCCCGAAGGAGTTG; Reverse:ATCACAGTGTGGTAAGCGCA), VEGFR2 (Forward:CGGGGCATGTACTGACGATT; Reverse: TACCAGTGGATGTGATGCGG), FLT1 (Forward:CAAATAAGCACACCACGCCC; Reverse:CGCCTTACGGAAGCTCTCTT), ENG (Forward:ATACCACAGCCTTCATCTGCG; Reverse:ATATGTCACCTCGCCCCTCT), CCR3 (Forward: TTCAGGAGTGGTGACGCCTA, Reverse: CACTTCTCCCTGGCTTGTGT), POLR2A (Forward:TAACCTGCTGCGGATGATCTGGAA; Reverse:GATGTTGAAGAGCAGCGTGGCATT)
For RNA-sequencing, cells were grown in EC growth media in a 12 well tissue culture plate (Costar #3513). When the cells were at approximately 95% confluency, RNA was extracted from 3 replicate wells using a commercial kit (RNeasy, Qiagen). Quality control and RNA sequencing were performed at the Genomics Division of the Iowa Institute of Human Genetics at The University of Iowa as described previously using 151 base pair paired end reads on an Illumina HiSeq4000 (Zeng et al. 2014). Reads were mapped to human genome build hg38 and the GENCODE comprehensive gene annotation for chromosomes (ver. 28; Harrow et al., 2012) using STAR (ver. 2.5.3a modified; Dobin et al., 2013). Reads were counted using featureCounts from Rsubread (ver. 1.20.6; Liao, Smyth & Shi, 2014), and counts were normalized to counts per million (CPM) using edgeR (ver. 3.20.9; Robinson, McCarthy, & Smyth, 2010). Genes were ranked by median CPM across the three replicates using R (ver. 3.4.3; R Core Team, 2017).
Results
Immortalization of human ChECs
Primary human ChECs were isolated from donor tissue and enriched using CD31 bound magnetic beads as described above (Figure 1C). To immortalize freshly isolated human ChECs, CDH5p-hTERT and CDH5p-SV40 TAg plasmids were constructed and packaged into lentiviral vectors. By expressing the immortalization gene under control of the endothelial cell specific CDH5 promoter and including a blasticidin resistance cassette, these vectors were capable of specifically immortalizing endothelial cells that conferred blasticidin resistance (Figure 1A&B). As a positive control for HIV mediated transduction, we also transduced a group of human ChECs with HIV-CMVp-GFP, which demonstrated a transduction efficiency of approximately 71 percent after 72 hours (Figure 1D).
To generate immortalized cell lines, primary human ChECs were passaged to a 6-well culture plate at a density of 100,000 cells per well. Cells were subsequently transduced with either HIV-CDH5p-hTERT at a multiplicity of infection (MOI) of 10 or with HIV-CDH5p-hTERT and HIV-CDH5p-SV40 TAg both at an MOI of 10 (i.e. total viral load, MOI = 20). Starting at 72 hours post-transduction, cells underwent positive selection with blasticidin (10μg/mL) for one week. Afterwards, the untreated cells, CDH5p-hTERT+ ChECs and CDH5p-hTERT+/CDH5p-SV40 TAg+ were continuously passaged as per the methods section above. Only cells transduced with CDH5p-hTERT+ /CDH5p-SV40 TAg+ continued to proliferate after 10 expansions (the proliferative capacity of cells transduced with CDH5p-hTERT+ only was not significantly different than untransduced control cells). Notably, a phenotypic difference occurred between the untreated (Figure 1E) and iChEC-1 cells (Figure 1F) by passage 4. Specifically, as compared to untransduced ChECs, iChEC-1 cells tended to adopt a less elaborate morphology, with a smaller portion of its body habitus manifested as long spindly projections and/or elaborate lamellipodia (i.e., iChEC-1 cells exhibited a more proliferative cellular morphology). However, the iChEC-1 cells maintained a cobble-stone like phenotype at confluency (Figure 1F).
Characterization of immortalized Human ChECs
We performed immunocytochemistry to assess the expression of both endothelial cell markers and the immortalization factors hTERT and SV40 TAg (Figure 2). The ChEC-1 cell line was positive for both the pan-endothelial cell markers CD34 (Figure 2A), vWF (Figure 2E) and the choriocapillaris enriched marker CA4 (Figure 2C) (“Maturation of the fetal human choriocapillaris.,” 2009; Songstad et al., 2017). As expected, iChEC-1 cells also expressed hTERT and SV40 TAg, the latter of which is localized to the nucleus (Figure 2I). Interestingly,hTERT displayed a nuclear and cytoplasmic phase, which has been previously reported (Iannilli, et al., 2013; Kyo et al., 2003; Lepreux et al., 2008). The cells were positive for the CFH protein, which harbors genetic variants that contribute to the risk of age-related macular degeneration (Figure 2G) (Whitmore et al., 2014). Lastly, we performed RT-PCR to examine the expression of TJP1, VEGFR2, FLT1, ENG, and CCR3 (Figure 6). POLR2A, a house keeping gene, served as a control. Whereas it may be upregulated under disease conditions(Takeda et al., 2009), basal expression of CCR3 was undetectable.
Figure 2. Characterization of Human iChEC-1 Cells.
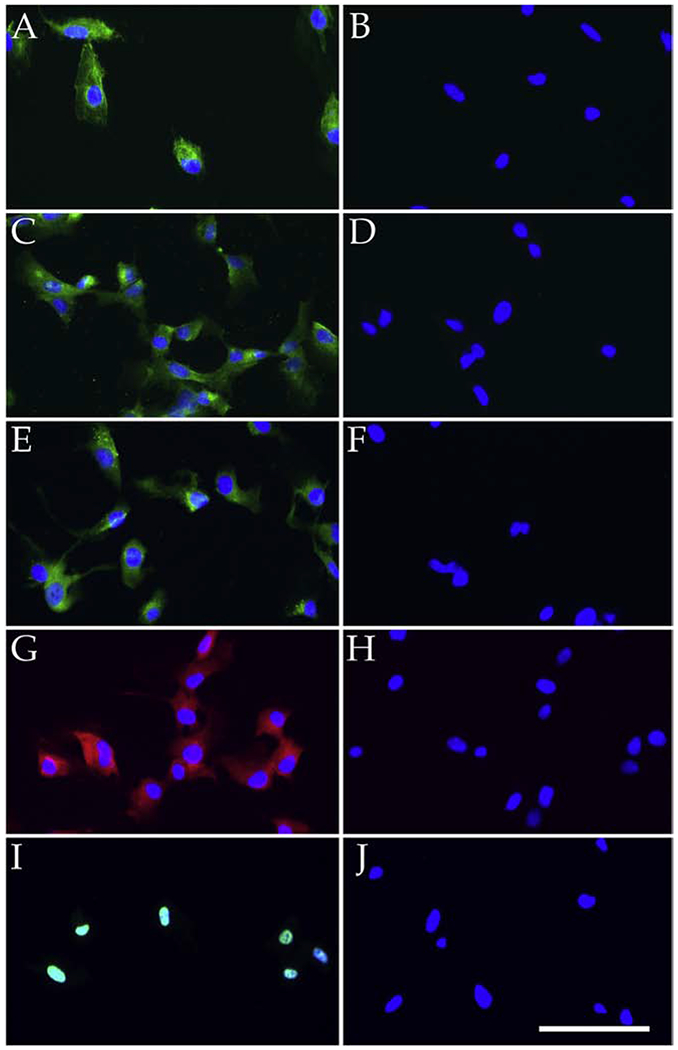
Cells were immunolabeled with antibodies directed against endothelial cell markers, CD34 (A; green), CA4 (C; green), von Willebrand factor (E; green), factor H (G, red) as well as TAg (I; green). Panels B, D, F, H, and J show labeling with secondary antibody only. All sections were counterstained with DAPI (A-J). Scalebar indicates 100μm.
Figure 6. RT-PCR analysis of iChEC.
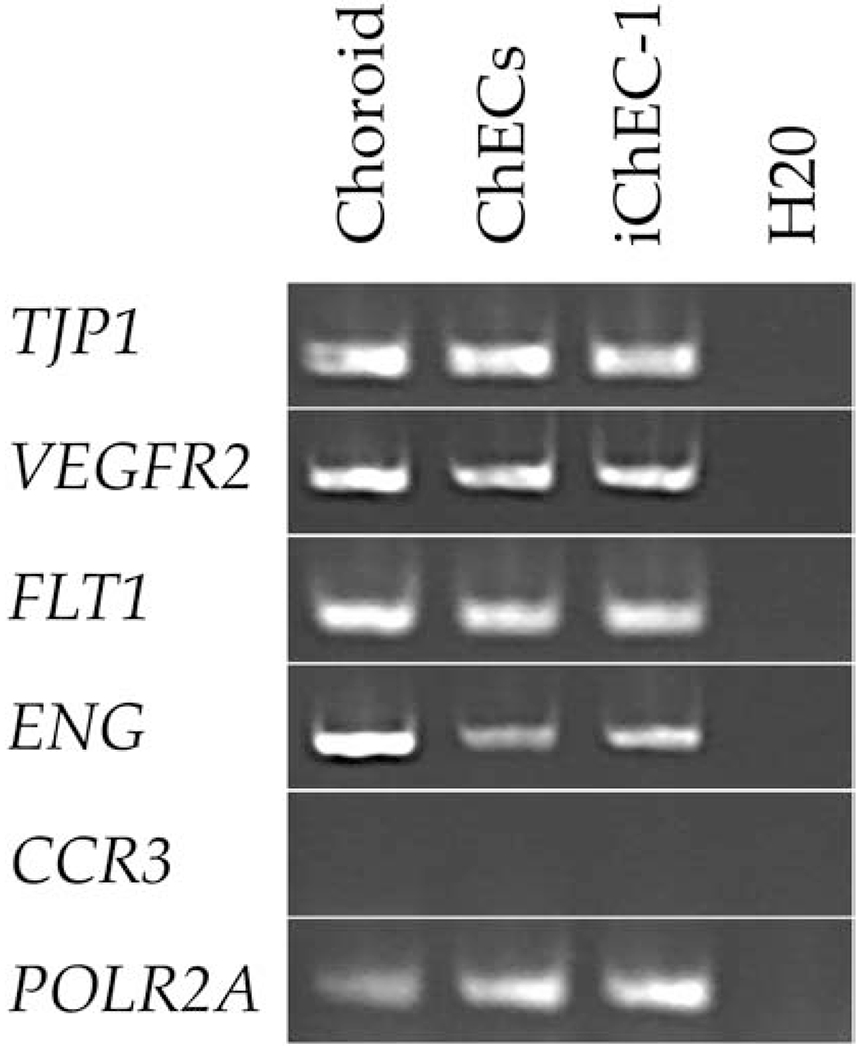
Expression analysis of EC associated genes of TJP1, VEGFR2, FLT1, ENG, CCR3, and POLR2A in uncultured choroid, primary ChECs, and iChEC-1. Choroid = non-cultured, human RPE/choroid mRNA; ChECs= primary culture of choroidal endothelial cells; iChEC-1=immortalized choroidal endothelial cell-1; H2O= Water control
To demonstrate phenotypic stability with passage, we next performed a limiting dilution to isolate monoclonal cell populations. Cells were plated at an average dilution of 0.5 cells per well in a 96 well tissue culture plate and nine clones were obtained. qRT-PCR analysis demonstrated that clones maintained expression of the pan-endothelial cell markers vWF and PECAM1, and the endothelial cell fenestrae associated marker PLVAP (Figure 3).
Figure 3.
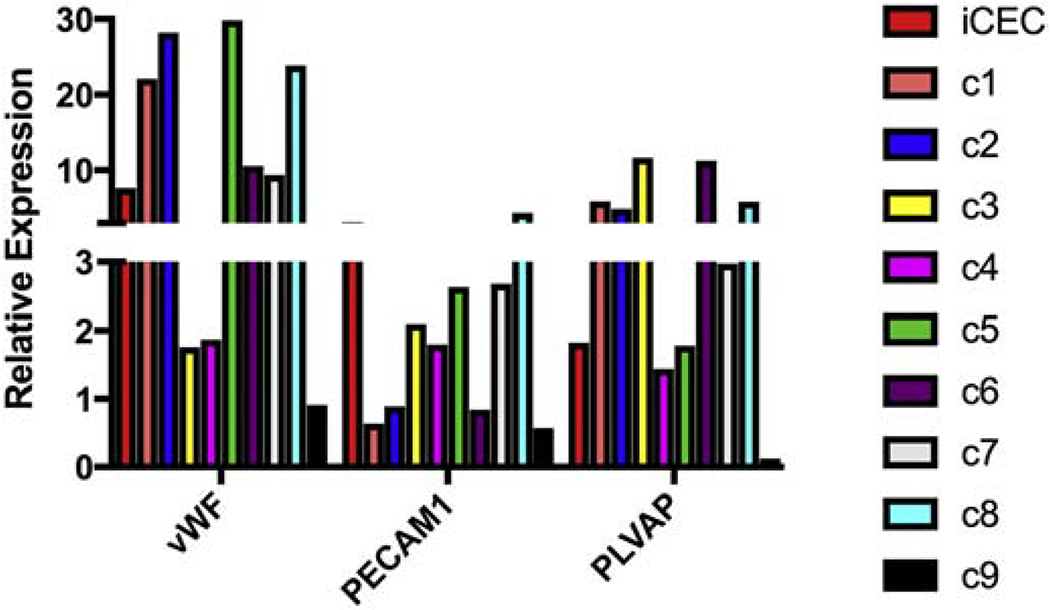
Quantitative RT-PCR analysis of clonally expanded iChEC-1. Clones of iChEC-1 were obtained using a limiting dilution. Comparison of iChEC-1 clones to the parent line demonstrated intact expression of endothelial cell markers across all clones. Relative expression = [(absolute copy number of the gene / absolute copies of 18s rRNA)/(copies of gene in hChEC/copies of 18s rRNA in hChEC)] . TAg = SV40 T-antigen; iChEC = immortalized choroidal endothelial cell; hChEC= primary human choroidal endothelial cells; and C= clone .
Cell-line specific genotyping of AMD-associated risk loci
In order to further characterize and facilitate the use of iChEC-1 cells in AMD disease modeling, we genotyped the cell line with respect to SNPs from published genome-wide association studies at the 3 most heavily implicated loci in AMD (Figure 4). Of note, this line is homozygous for protein-coding missense risk alleles of CFH (rs800292) and SLC44A4 (rs12661281). It is also homozygous for non-protective alleles at C2 (rs9332739) and CFB (rs641153). Heterozygosity was observed for missense risk alleles of CFH (rs1061147, rs1061170) and ARMS2 (rs10490924). The majority of AMD-associated SNPs lie within non-protein coding regions and, as expected, a combination of risk and non-risk alleles of unknown significance exists at these loci. Furthermore, only a subset of genes was expressed at each locus, as determined by RNA-seq (Figure 4).
Figure 4.
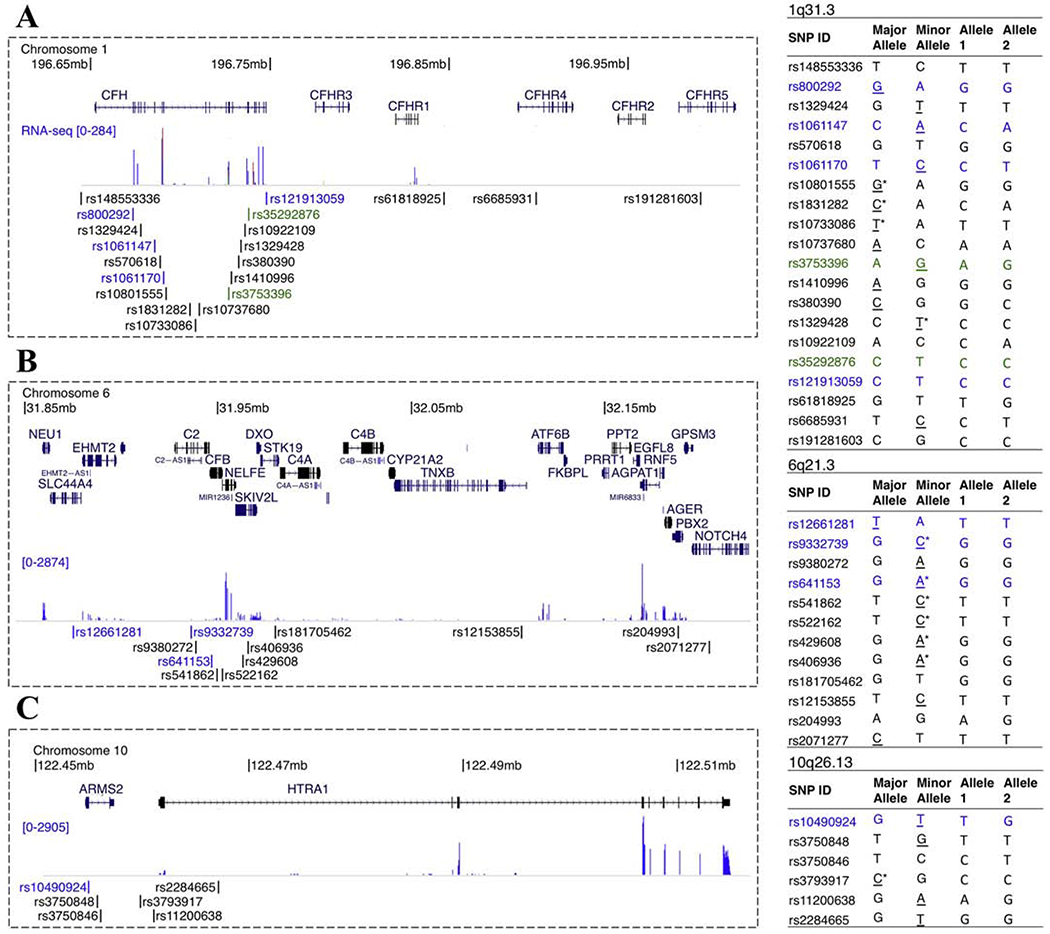
Cell-line specific characterization of AMD-associated risk loci. SNPs significantly associated with AMD at chromosomes 1q31.3 (A), 6q21.3 (B) and 10q26.13 (C) were PCR amplified and sequenced. Genomic coordinates are marked at the top of each image and gene names are shown above each locus for coding genes and to the left in small text for non-coding genes. RNA-seq signal and SNP locations are shown below the gene annotations. Missense coding SNPs are marked in blue and synonymous coding SNPs are marked in green. Tables to the right of each locus indicate the major and minor alleles on the plus strand based on dbSNP build 150. The risk modifying allele (if reported) at each SNP is underlined with protective alleles marked by asterisks. Alleles 1 and 2 represent the cell-line specific genotype. All bases are reported with respect to the plus strand of hg38.
Functional validation of Human iChEC-1 cells
To characterize the ability of iChEC-1 cells to continue to perform endothelial cell functions we evaluated their ability to form capillary-like networks and repopulate a decellularized choroid as previously described (Chirco et al., 2017). To determine if iChEC-1 cells retain the ability to form a capillary-like network, a matrigel tube forming assay was performed. At 18-24 hours post-seeding, iChEC-1 cells formed reticulated vascular networks characteristic of endothelial cells (Figure 5A & B). To determine if iChEC-1 cells are capable of migrating into and taking up residence within a decellularized choroidal extracellular matrix, we next performed an in vitro transplantation experiment. As described above, human choroidal punches were decellularized and plated in a 6 well culture dish. IChEC-1 cells were plated around the punch and allowed to migrate into the punch over the course of 7 days. As hypothesized, iChEC-1 cells reintegrated into the choroid scaffolds (Figure 5C). Interestingly, the iChEC-1 cells appeared to migrate specifically into choriocapillary lumens (Figure 5C).
Figure 5. Assessment of iChEC-1 cell function.
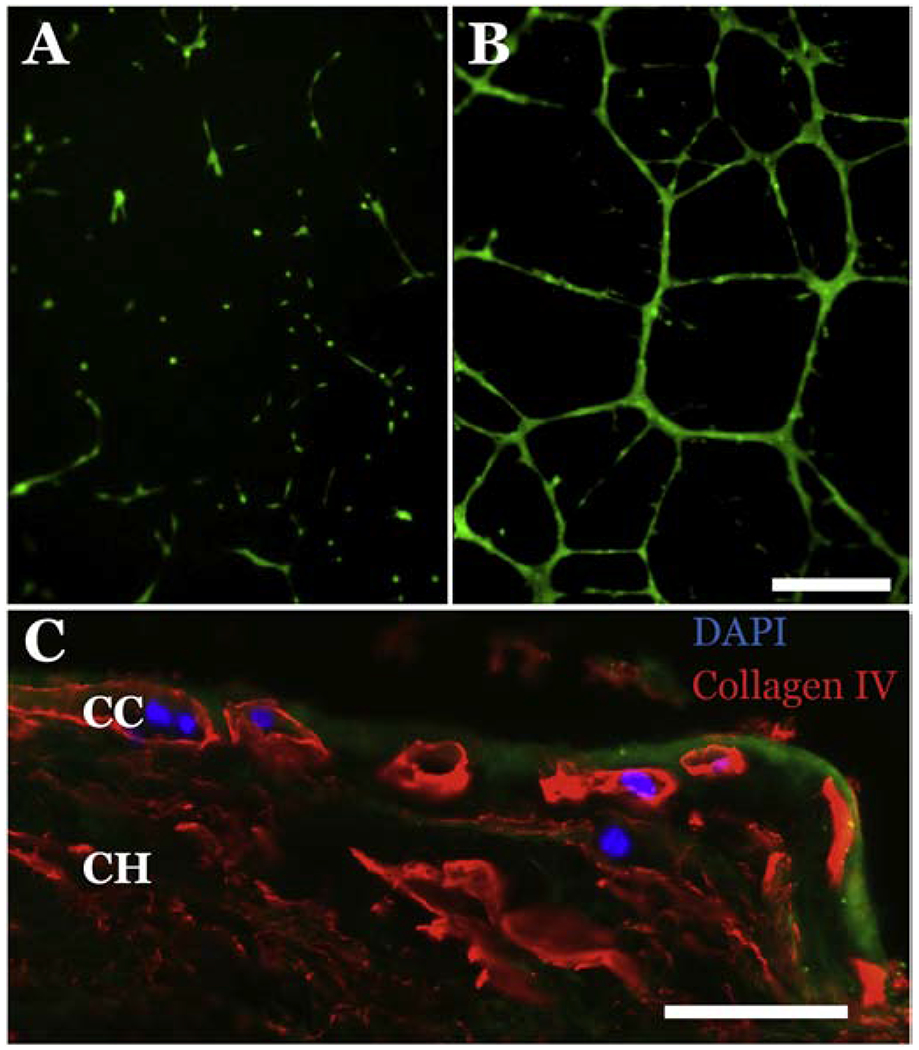
A-B: Tube formation assay demonstrating that iChEC-1 cells form tubes in matrigel (A, 5×104 cells; B, 1×105 cells), note calcein-labeled (green) cells in vascular networks. Scalebar =250μm. C: iChEC-1 cells (nuclei, DAPI) home to empty capillary tubes (Collagen IV, red) in decellularized human choroid explants. Green = Phalloidin ; CC = choriocapillaris; CH= choroid, Scalebar = 50 μm.
Discussion
In this study, we show that cell specific promoters are sufficient to drive immortalization of a purified population of human choroidal endothelial cells. By co-opting the transcriptional machinery to maintain cell proliferation we have been able to successfully maintain cellular identity. That is, the continued expansion of the line depends on maintaining an endothelial cell specific transcriptional program and in turn an endothelial cell specific phenotype as confirmed by immunocytochemistry and quantitative RT-PCR. Similar to primary cells, iChEC-1 cells maintain the ability to form vascular tubes and repopulate choroidal extracellular matrix scaffolds in vitro. Further studies will be necessary to determine possible consequences of long-term culture, such as loss of eNOS expression by dedifferentiation, which is common to culturing cells in vitro (Lacorre et al., 2004; Thum et al., 2000). Furthermore, while human ChECs likely have many phenotypic similarities to non-human choroidal endothelial cells (Uhlmann et al., 2001), access to a human cell line offers an opportunity to understand human-specific genetic loci (e.g., ARMS2).
A recent study reported the use of a constitutive promoter coupled to a temperature sensitive SV40 TAg to control the proliferation of endothelial cells post-immortalization (Loeven et al., 2018). Using this strategy, the authors demonstrate that by increasing the temperature under which the cells are cultured they could inactivate SV40 TAg, slow cell cycle progression, and promote endothelial cell maturation. Although a promising approach, we opted to use cell type specific promoters in our immortalization process in an attempt to avoid the possibility of immortalizing contaminating non-endothelial cell types such as choroidal fibroblasts, which are often present post-isolation. Interestingly, as shown above when using an endothelial cell specific promoter to drive immortalization endothelial cell fate can be maintained. That said, as indicated in Figure 1, immortalization does induce a change in cellular morphology, specifically cells adopt a highly proliferative less complex structure. As such, a combination of the two approaches may ultimately be ideal. Access to a human choroidal endothelial cell line that retains the phenotypic and functional qualities of the primary parent cell addresses a major bottleneck associated with in vitro studies of AMD. Furthermore, genetic variation at AMD-associated SNPs, especially those with unclear functional relevance (i.e. synonymous coding mutations or variants within non-protein coding sequences), can contribute to phenotypic variation between cell lines in unpredictable ways.
Availability of this newly developed resource will allow us and others to perform experiments designed to examine protective pathways necessary to regulate complement activation or evaluate genetic predispositions to choroidal neovascularization. Extending our understanding of AMD pathogenesis will ultimately provide opportunities for treatment of this common cause of blindness.
Highlights:
Choroidal endothelial cells (ChECs) were isolated from a human donor eye
Transduction with CDH5p-hTERT/CDH5p-TAg yielded an immortalized cell line (iChEC-1).
iChEC-1 retain phenotypic and functional properties of choroidal endothelial cells.
iChEC-1 are a suitable immortalized human ChEC line for future studies
Acknowledgements
Supported in part by: NIH grants EY024605, EY026087, P30 EY025580, Research to Prevent Blindness, and. the Elmer and Sylvia Sramek Charitable Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors have submitted an invention disclosure form for the use of the CDH5 immortalization construct through the University of Iowa Research Foundation.
References
- Baba T, Grebe R, Hasegawa T, Bhutto I, Merges C, McLeod DS, & Lutty GA (2009) Maturation of the fetal human choriocapillaris. Investigative Ophthalmology & Visual Science, 50(7), 3503–3511. doi: 10.1167/iovs.08-2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutto I, & Lutty G (2012). Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Molecular Aspects of Medicine, 33(4), 295–317. http://doi.Org/10.1016/j.mam.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirco KR, Worthington KS, Flamme-Wiese MJ, Riker MJ, Andrade JD, Ueberheide BM, Stone EM, Tucker BA, Mullins RF, 2017. Preparation and evaluation of human choroid extracellular matrix scaffolds for the study of cell replacement strategies. Acta Biomater 57, 293–303. doi: 10.1016/j.actbio.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner., 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PTVM, Nemesure B, Mitchell P, Kempen J, Eye Diseases Prevalence Research Group, 2004. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol 122, 564–572. doi: 10.1001/archopht.122.4.564 [DOI] [PubMed] [Google Scholar]
- Gelfand BD, Ambati J, 2016. A Revised Hemodynamic Theory of Age-Related Macular Degeneration. Trends Mol Med 22, 656–670. doi: 10.1016/j.molmed.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green WR, Enger C, 2005. Age-related macular degeneration histopathologic studies: the 1992 Lorenz E. Zimmerman Lecture. 1992., Retina; (Philadelphia, Pa.). [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project., 22(9), 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannilli F, Zalfa F, Gartner A, Bagni C, & Doth CG (2013). Cytoplasmic TERT Associates to RNA Granules in Fully Mature Neurons: Role in the Translational Control of the Cell Cycle Inhibitor pl5INK4B. PLoS ONE, 8(6), e66602 10.1371/journal.pone.0066602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo S, Masutomi K, Maida Y, Kanaya T, Yatabe N, Nakamura M, et al. (2003). Significance of immunological detection of human telomerase reverse transcriptase: re-evaluation of expression and localization of human telomerase reverse transcriptase. The American Journal of Pathology, 163(3), 859–867. 10.1016/S0002-9440(10)63446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacorre D-A, Baekkevold ES, Garrido I, Brandtzaeg P, Haraldsen G, Amalric F, Girard J-P, 2004. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood 103, 4164–4172. doi: 10.1182/blood-2003-10-3537 [DOI] [PubMed] [Google Scholar]
- Lepreux S, Doudnikoff E, Aubert I, Bioulac-Sage P, Bloch B, & Martin-Negrier M-L (2008). Cytoplasmic expression of human telomerase catalytic protein (hTERT) in neutrophils: an immunoelectron microscopy study. Ultrastructural Pathology, 32(5), 178–183. 10.1080/01913120802034504 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Loeven MA, van Gemst JJ, Schophuizen CMS, Tilakaratna V, van den Heuvel LP, Day AJ, et al. (2018). A Novel Choroidal Endothelial Cell Line Has a Decreased Affinity for the Age-Related Macular Degeneration-Associated Complement Factor H Variant 402H. Investigative Ophthalmology & Visual Science, 59(2), 722–730. 10.1167/iovs.IOVS-17-22893 [DOI] [PubMed] [Google Scholar]
- McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, & Lutty GA (2009). Relationship between RPE and choriocapillaris in age-related macular degeneration. Investigative Ophthalmology & Visual Science, 50(10), 4982–4991. 10.1167/iovs.09-3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Johnson MN, Faidley EA, Skeie JM, Huang J, 2011. Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration. Invest. Ophthalmol. Vis. Sci 52, 1606–1612. doi: 10.1167/iovs.10-6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy K, Nareshkumar RN, Sivagurunathan S, Raman R, Sulochana KN, Chidambaram S, 2018. Anti-angiogenic effect of adiponectin in human primary microvascular and macrovascular endothelial cells. Microvasc. Res doi: 10.1016/j.mvr.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Prandini M-H, Dreher I, Bouillot S, Benkerri S, Moll T, & Huber P (2005). The human VE-cadherin promoter is subjected to organ-specific regulation and is activated in tumour angiogenesis. Oncogene, 24(18), 2992–3001. 10.1038/sj.onc.1208483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL https://www.R-project.org/. [Google Scholar]
- Robinson MD, McCarthy DJ, & Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, & Trono D (2000). Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Molecular Therapy: the Journal of the American Society of Gene Therapy, 2(4), 404–414. 10.1006/mthe.2000.0141 [DOI] [PubMed] [Google Scholar]
- Schubert C, Pryds A, Zeng S, Xie Y, Freund KB, Spaide RF, Merriam JC, Barbazetto I, Slakter JS, Chang S, Munch IC, Drack AV, Hernandez J, Yzer S, Merriam JE, Linneberg A, Larsen M, Yannuzzi LA, Mullins RF, Allikmets R, 2014. Cadherin 5 is regulated by corticosteroids and associated with central serous chorioretinopathy. Hum. Mutat 35, 859–867. doi: 10.1002/humu.22551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeie JM, Mullins RF, 2008. Elastin-mediated choroidal endothelial cell migration: possible role in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci 49, 5574–5580. doi: 10.1167/iovs.08-1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad AE, Worthington KS, Chirco KR, Giacalone JC, Whitmore SS, Anfinson KR, Ochoa D, Cranston CM, Riker MJ, Neiman M, Stone EM, Mullins RF, Tucker BA, 2017. Connective Tissue Growth Factor Promotes Efficient Generation of Human Induced Pluripotent Stem Cell- Derived Choroidal Endothelium. Stem Cells Transl Med 6, 1533–1546. doi: 10.1002/sctm.16-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Baffi JZ, Kleinman ME, Cho WG, Nozaki M, Yamada K, Kaneko H, Albuquerque RJC, Dridi S, Saito K, Raisler BJ, Budd SJ, Geisen P, Munitz A, Ambati BK, Green MG, Ishibashi T, Wright JD, Humbles AA, Gerard CJ, Ogura Y, Pan Y, Smith JR, Grisanti S, Hartnett ME, Rothenberg ME, Ambati J, 2009. CCR3 is a target for age-related macular degeneration diagnosis and therapy. Nature 460, 225–230. doi: 10.1038/nature08151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Haverich A, Borlak J, 2000. Cellular dedifferentiation of endothelium is linked to activation and silencing of certain nuclear transcription factors: implications for endothelial dysfunction and vascular biology. FASEB J. 14, 740–751. [DOI] [PubMed] [Google Scholar]
- Uhlmann S, Friedrichs U, Eichler W, Hoffmann S, Wiedemann P, 2001. Direct measurement of VEGF-induced nitric oxide production by choroidal endothelial cells. Microvasc. Res 62, 179–189. doi: 10.1006/mvre.2001.2334 [DOI] [PubMed] [Google Scholar]
- Whitmore SS, Braun TA, Skeie JM, Haas CM, Sohn EH, Stone EM, Scheetz TE, Mullins RF, 2013. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Mol. Vis 19, 2274–2297. [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Sohn EH, Chirco KR, Drack AV, Stone EM, Tucker BA, Mullins RF, 2014. Complement activation and choriocapillaris loss in early AMD: Implications for pathophysiology and therapy. Prog Retin Eye Res. doi: 10.1016/j.preteyeres.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng C-Y, Wong TY, 2014. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2, el06–16. doi: 10.1016/S2214-109X(13)70145-l [DOI] [PubMed] [Google Scholar]
- Zeng S, Whitmore SS, Sohn EH, Riker MJ, Wiley LA, Scheetz TE, et al. (2016). Molecular response of chorioretinal endothelial cells to complement injury: implications for macular degeneration. The Journal of Pathology, 238(3), 446–456. 10.1002/path.4669 [DOI] [PMC free article] [PubMed] [Google Scholar]


