Abstract
The antitumor enzyme L-asparaginase (L-Asp) has commonly been used for the treatment of acute lymphoblastic leukemia. However, the effects of L-Asp on acute myeloid leukemia (AML) and their underlying mechanisms have not been fully elucidated. In the present study, the effects of L-Asp on cell proliferation and apoptosis were investigated using the AML cell lines U937, HL-60 and KG-1a. The effects of combining L-Asp with mitoxantrone (MIT) and cytarabine (Ara-c) were also analyzed. The combination of MIT and Ara-C is known as MA therapy, and is a widely used therapeutic regimen for the treatment of elderly patients with refractory AML. When applied alone, L-Asp inhibited cell proliferation and induced apoptosis in each of the cell lines tested. Furthermore, the combined use of L-Asp with MA therapy further potentiated the inhibition of cell proliferation while increasing the induction of apoptosis. These findings provide evidence for the potential antitumor effect of L-Asp in AML, and indicate that improved efficacy maybe achieved by combining L-Asp with MIT and Ara-c. This combination may provide a promising new therapeutic strategy for the treatment of AML.
Keywords: acute myeloid leukemia, antitumor, apoptosis, cytarabine, L-asparaginase
Introduction
Acute myeloid leukemia (AML) is a biologically and clinically heterogeneous disorder that is characterized by clonal expansion of myeloid progenitors in the bone marrow and peripheral blood (1). The incidence of AML increases with age, and >60% of patients are at least 60 years old at the time of diagnosis (2). The 5-year overall survival (OS) rate of AML is 40% for patients younger than 60, but only 10-20% for older patients (3,4). The majority of patients who relapse after complete remission (CR) do not survive for more than 5 years (5). Even though several studies have reported improvements in the outcomes of younger AML patients over the last 40 years (6-8), the prognosis of patients aged ≥60 is poor and has remained virtually unchanged (6).
Combination chemotherapy remains the primary choice of AML treatment, and the achievement of CR is crucial for the long-term survival of patients with AML. For more than four decades, induction chemotherapy with cytarabine and an anthracycline (‘7+3’ therapy) has been applied as the standard chemotherapy regimen for patients newly diagnosed with classical AML (9). However, the remission rate of ‘7+3’ therapy is not satisfactory. Although CR is achieved in 50-75% of adult AML patients treated with the deoxycytidine analog cytarabine (Ara-c) and an anthracycline antibiotic [including daunorubicin or idarubicin, or anthracenedione mitoxantrone (MIT)], only 20-30% of patients experience long-term disease-free survival (DFS) (10).
MA therapy, which comprises MIT combined with Ara-c, is now widely used for the treatment of elderly patients with AML (11-13). Ahmed et al (14) previously reported the use of L-asparaginase (L-Asp) in combination with other conventional chemotherapeutic drugs in the treatment of elderly patients with refractory AML. The findings suggest that L-Asp may prove effective in the treatment of AML, and that L-Asp plus conventional chemotherapeutic drugs may offer beneficial effects in elderly patients with refractory AML.
L-Asp has been widely used in the treatment of pediatric leukemia since its antitumor activity was first discovered, which significantly improves the survival rate of children with leukemia (15). Nonetheless, the antitumor mechanism of L-Asp remains unclear. It is generally considered that leukemia cells lack sufficient activity of asparagine synthetase (16). Whereas, L-Asp can potentially decompose and deplete asparagine present in the blood surrounding the leukemia cells, leading to the dysfunction of protein synthesis and eventual cell death (17,18).
Autophagy is a biological process in which large molecules and damaged organelles in the cytoplasm are degraded. Despite being a recycling mechanism that assists cells in all eviating nutrient stress, autophagy has been demonstrated to regulate cell differentiation, cell death and the cell cycle (19-21).
Microtubule-associated protein 1 light chain 3 [LC3; a homolog of autophagy-related protein 8 (Atg8)] is a protein normally localized to the autophagosome membrane (22). LC3 was originally identified as a subunit of the neuronal microtubule-associated proteins (MAPs) MAPlA and MAPlB, and its expression can regulate the microtubule binding activity of MAPlA and MAPlB (23). There are three human LC3 isoforms (LC3A, LC3B and LC3C) that undergo post-translational modifications during autophagy (24). The C-terminus of newly synthesized LC3 is normally cleaved by Atg4 protease to form the cytosolic soluble LC3-I (25). Following autophagosome formation, LC3-I is modified by shearing and ubiquitination, before being coupled with phosphatidylethanolamine on the surface of the autophagosome membrane to form a membrane-bound LC3-II, located on the inner and outer membranes of the autophagosome (24,26). Unlike other Atg family of proteins located on autophagosome membranes, LC3-II remains stable on autophagosome membranes until it fuses with lysosomes, and is therefore usually used as a marker for autophagy detection (27-29).
In a previous study, Song et al (30) reported asparaginase-induced autophagic properties in chronic myeloid leukemia (CML) cell lines K562 and KU812. Therefore, it was hypothesized that L-Asp may exert antiproliferative effects on AML cells by inducing autophagy. In the present study, the effects of L-Asp on cell proliferation, apoptosis, morphological changes and the autophagic activity of AML cell lines U937, HL-60 and KG-1a were investigated. In addition, the effects of combining L-Asp priming with MA therapy were also assessed. The aim of this study was to elucidate of the mechanism of L-Asp efficacy and to investigate whether the combination of L-Asp priming with MA therapy could be beneficial in AML treatment.
Materials and methods
Cell culture
Human AML cell lines U937, HL-60, and KG-1a were purchased from China Infrastructure of Cell Line Resources (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences). U937 cells were cultured in RPMI-1640 medium (SH30809, Hyclone; GE Healthcare Life Sciences) supplemented with 10% heat-inactivated fetal bovine serum (FBS; 10099-141; Gibco; Thermo Fisher Scientific, Inc.), whereas HL-60 and KG-1a cells were maintained in IMDM medium (SH30228.01B; Hyclone; GE Healthcare Life Sciences) supplemented with 20% FBS. All cells were cultured at 37˚C under 5% CO2.
Materials and reagents
Ara-c (trade name, Cytosar) was purchased from Pfizer Inc. and diluted in the provided water for injection with benzyl alcohol according to the manufacturer's protocol. MIT (trade name, Militant) and L-Asp (derived from Escherichia coli) were purchased from Sichuan Shenghe Pharmaceutical Co., Ltd. and Changzhou Qianhong Bio-Pharma Co., Ltd., respectively, and were diluted in sterile 0.9% sodium chloride solution when used.
Chloroquine (CQ; cat. no. 14774), primary antibodies for western blotting: Rabbit LC-3A/B (D3U4C; cat. no. 12741) and rabbit glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 14C10; Rabbit; cat. no. 2118), were all purchased from Cell Signaling Technology (Cell Signaling Technology, Inc.).
Treatment of AML cells U937, HL-60, and KG-1a
The cells were cultured at a density of 5,000 cells/well in the 96-well plates (100 µl/well) at 37˚C. AML cell lines U937, HL-60 and KG-1a were treated with Ara-c, MIT and/or L-Asp following the attainment of steady-state exponential growth. For all cells, when determining the half maximal inhibitory concentrations (IC50), Ara-c and MIT were applied at concentrations of 0-8 µg/ml for 24 h, and L-Asp was applied at concentrations of 0-6.4 U/ml for 48 h. In subsequent experiments, U937, HL-60 and KG-1a cells were treated with Ara-c + MIT or Ara-c + MIT + L-Asp at their respective IC50 for 24 and 48 h. The IC50 values were calculated using GraphPad Prism version 6.0 (GraphPad Software, Inc.).
Cell proliferation analysis
Following each drug treatment regimen, cell viability was measured using Cell Counting kit-8 (CCK-8; CK04; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocols. Cell growth inhibition rates were calculated as follows: Growth inhibition rate (%)=[(Acontrol-Ablank of control)-(Asample-Ablank of sample)]/(Acontrol-Ablank of control). A indicates the absorbance at 460 nm.
Flow cytometric analysis
AML cells (U937, HL-60 and KG-1a) cultured at a density of 4x105 cells/well in 6 cm dishes were separated into four different treatment groups: Control, Ara-c + MIT, L-Asp and Ara-c + MIT + L-Asp. The control group was cultured in media supplemented with 10% FBS for 48 h without drug treatment. The Ara-c + MIT group was cultured in media with 10% FBS for 24 h then treated with Ara-c and MIT for the next 24 h. The L-Asp group was treated with L-Asp for 48 h. The Ara-c + MIT + L-Asp group was treated with L-Asp alone for 24 h and then together with Ara-c and MIT for the next 24 h. At the end of their respective treatment regimen, cells were subsequently stained with Annexin V/FITC and propidium iodide (PI) using an Annexin V-FITC Apoptosis Detection kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol, and analyzed using the ACEA NovoCyte Flow Cytometer (ACEA Biosciences; Agilent Technologies, Inc.). Each experiment was repeated three times, and the mean values were applied to calculate the rates of apoptosis induction for each treatment group. FlowJo software (version 7.6.1, FlowJo LLC) was used for analysis.
Cell imaging
AML cells from the different treatment groups were seeded into 10-cm dishes at the density of 1x106 cells/well. Cell morphology was assessed using Olympus BX63 (Olympus Corporation) under x100 and x400 magnification.
Protein extraction for autophagy detection
In the present study, U937, HL-60 and KG-1a cells were seeded into 6 cm dishes at a density of 4x105 cells/well, and divided into two groups: CQ+ and CQ-. The CQ+ group was treated with 10 µM CQ for 30 min, followed by L-Asp for 0, 3, 6, 12, 24 and 48 h. The CQ-group was directly treated with L-Asp for 0, 3, 6, 12, 24 and 48 h without CQ pre-treatment. For the CQ+ and CQ-groups, the concentration of L-Asp used was determined on the basis of its IC50 value in each cell line. At each time point, protein samples of the AML cells from each treatment group were extracted for western blot analysis.
Western blot analysis
Confluent AML cells grown on 6 cm dishes were washed with ice-cold PBS. Following centrifugation at 1,500 x g for 5 min to discard the medium, 10 ml ice-cold PBS was added in and mixed well with the cells in 10 ml centrifuge tubes. After a period of 5 min, centrifugation was performed at 1,500 x g to discard the PBS, twice before subsequent lysisusing 2X SDS-PAGE sample buffer (100 mM 1M Tris-HCl pH 6.8, 2% 2-mercaptoethanol, 20% glycerol and 4% SDS). Bicinchoninic acid assay (P0010S; Beyotime Institute of Biotechnology) was used for protein quantification. The lysates were subjected to separation by SDS-PAGE in 12% gels at 15 µg per lane, before the separated proteins were transferred onto Immobilon®-P PVDF (Millipore; EMD Millipore). Following blocking in PBS containing 5% non-fat dried milk and 0.1% Tween-20 for 1 h at room temperature, the membranes were incubated overnight at 4˚C with the LC3A/B antibody (14/16 kDa. #4108S; CST) using a dilution of 1:1,000. On the same membranes, incubation with the LC3A/B antibody was followed by 2 h incubation at room temperature with primary GAPDH antibody (GAPDH; 14C10; Rabbit; cat. no. 2118) using a dilution of 1:4,000. The membranes were subsequently incubated for 2 h with goat anti-rabbit (cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) at a dilution of 1:5,000 at room temperature. The protein bands were visualized using an ECL-Plus kit (00161543; Thermo Fisher Scientific, Inc.) coupled with X-ray radio graphs (038401501; Carestream Health, Inc.).
Statistical analysis
All statistical analysis was performed in GraphPad Prism version 6.0 (GraphPad Software, Inc.). All data are presented as the mean ± standard deviation (SD). One-way ANOVA followed by Tukey's multiple comparison test was used to determine statistical significance. P<0.05 was considered to indicate a statistically significant difference.
Results
Effects of L-Asp, Ara-c and MIT alone on AML cell viability
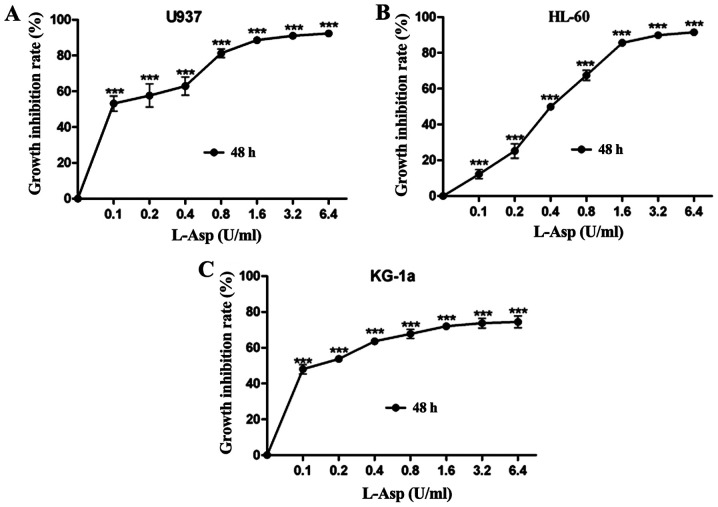
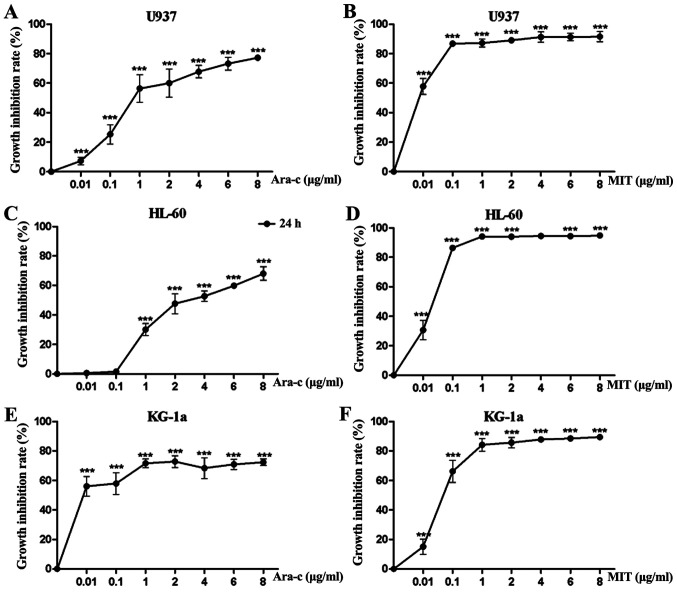
To determine the growth inhibitory effect of L-Asp on the AML cell lines U937, HL-60 and KG-1a, the cells were treated with ascending concentrations for 48 h prior to analysis. L-Asp significantly inhibited cell proliferation in all three AML cell lines in a dose-dependent manner, and was even effective at 0.1 U/ml, which was the lowest concentration of tested (Fig. 1).The 48 h IC50 values of L-Asp were calculated to be 0.106, 0.44 and 0.098 U/ml for U937, HL-60 and KG-1a, respectively. Additionally, the concentration-growth inhibition rate curves of Ara-c and MIT treatments alone were produced for each of the three AML cell lines. As presented in Fig. 2, Ara-c inhibited cell proliferation in all three AML cell lines in a dose-dependent manner, while the inhibition effect of MIT would reach a threshold at certain concentrations (0.1-1 µg/ml). For U937, HL-60 and KG-1a cells, the 24 h IC50 values of Ara-c and MIT were calculated to be 0.809, 3.07, 0.002 µg/ml and 0.002, 0.021 and 0.068 µg/ml, respectively.
Figure 1.
L-Asp inhibits the proliferation of acute myeloid leukemia cells. (A) U937, (B) HL-60 and (C) KG-1a cells were treated with ascending concentrations of L-Asp for 48 h. Each experiment was conducted three times (n=3). ***P<0.001 vs. control (0 U/ml). L-Asp, L-asparaginase.
Figure 2.
Ara-c and MIT inhibit acute myeloid leukemia cell proliferation. (A) U937, (C) HL-60 and (E) KG-1a cells were treated with ascending concentrations of Ara-c for 24 h. (B) U937, (D) HL-60 and (F) KG-1a were treated with ascending concentrations of MIT for 24 h. Each experiment was conducted three times (n=3). ***P<0.001 vs. control (0 µg/ml). Ara-c, cytarabine; MIT, mitoxantrone.
Effects of Ara-c + MIT and L-Asp + Ara-c + MIT treatments as inhibitors of AML cell proliferation
AML cell lines U937, HL-60 and KG-1a were first pretreated with L-Asp for 24 h and subsequently with Ara-c and MIT additively for the next 24 h. Each of the three cell lines were treated with drugs at concentrations corresponding to their respective IC50 values (Table I). The combined effects of L-Asp pretreatment and subsequent Ara-c + MIT treatment were analyzed in terms of growth inhibition rates, calculated using the aforementioned formula.
Table I.
IC50 values of Ara-c and MIT for 24 h treatment and L-Asp for 48 h treatment in the AML cell lines U937, HL-60 and KG-1a.
| Cell line | IC50 value | ||
|---|---|---|---|
| Ara-c (µg/ml) | MIT (µg/ml) | L-Asp (U/ml) | |
| U937 | 0.809 | 0.002 | 0.106 |
| HL-60 | 3.07 | 0.021 | 0.44 |
| KG-1a | 0.002 | 0.068 | 0.098 |
IC50, half maximal inhibitory concentration; Ara-c, cytarabine; MIT, mitoxantrone; AML, acute myeloid leukemia.
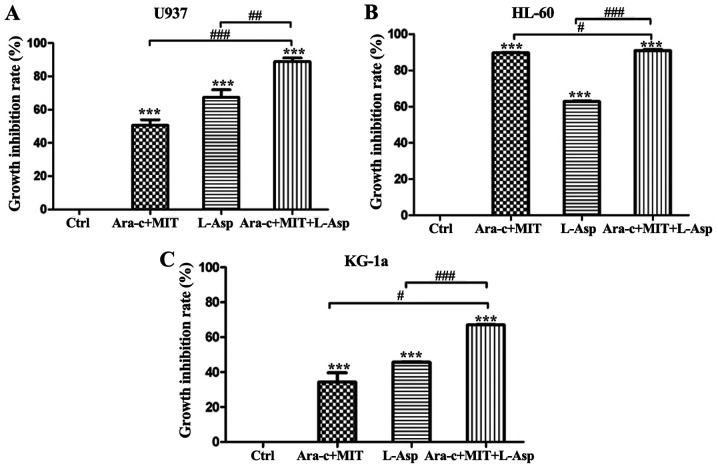
Treatment with Ara-c + MIT and L-Asp alone inhibited the proliferation of U937 cells by 50.67 and 67.34%, respectively, while the inhibition of proliferation induced by treatment with L-Asp and Ara-c + MIT was significantly greater at 88.84% (Fig. 3A). Anti-proliferative effects were also observed in KG-1a cells, with the inhibition rates of 34.39, 45.70 and 67.04% for Ara-c + MIT, L-Asp alone and Ara-c + MIT + L-Asp treatment, respectively (Fig. 3C). Finally, treatment with Ara-c + MIT or L-Asp alone exerted suppressive effects on HL-60 cell proliferation, with inhibition rates of 89.54 and 62.78% respectively; while combined Ara-c + MIT + L-Asp treatment exhibited the highest inhibitory effect in this cell line, with an inhibition rate of 90.78% (Fig. 3B).
Figure 3.
Comparison of the inhibitory effects of Ara-c + MIT, L-Asp alone and Ara-c + MIT + L-Asp on acute myeloid leukemia cell proliferation. Growth inhibition rates of (A) U937, (B) HL-60 and (C) KG-1a cells were evaluated using a Cell Counting kit-8 assay. In each cell line, the inhibitory effect of Ara-c + MIT + L-Asp was significantly greater compared with that of Ara-c + MIT or L-Asp alone. Each experiment was repeated three times (n=3). ***P<0.001 vs. control. #P<0.05, ##P<0.005 and ###P<0.001 as indicated. Ctrl, control; Ara-c, cytarabine; MIT, mitoxantrone; L-Asp, L-asparaginase.
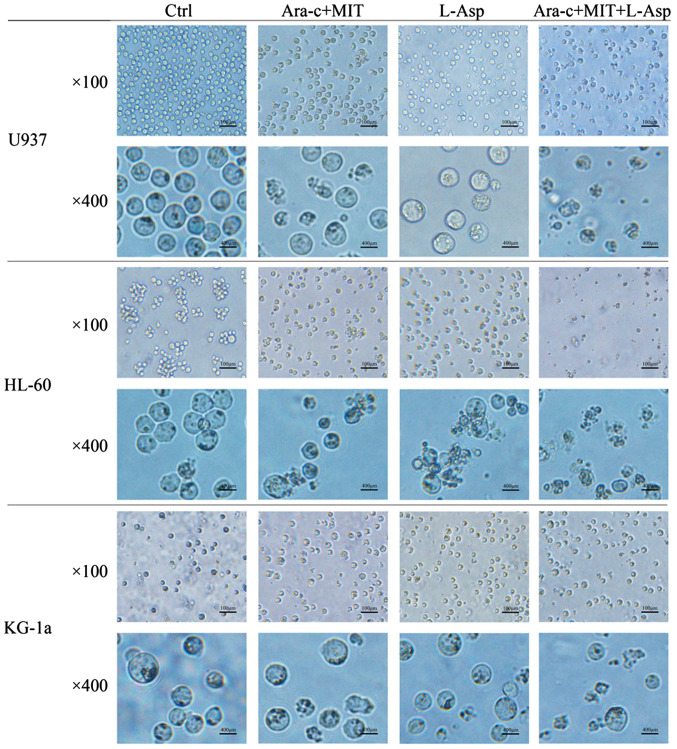
The morphological changes of AML cells from the different treatment groups were examined using microscopy. In the control group, U937, HL-60 and KG-1a cells all appeared circular and bright with good refraction, whereas some of the cells that were treated with Ara-c + MIT or L-Asp began to exhibit irregular shapes and some granulation (Fig. 4). Notably, normally shaped living cells could barely be observed in the medium in the Ara-c + MIT + L-Asp treatment group (Fig. 4).
Figure 4.
Comparison of cell morphological changes in the Ara-c + MIT, L-Asp alone and Ara-c + MIT + L-Asp treatment groups. Results are shown for U937, HL-60, and KG-1a acute myeloid leukemia cells imaged under x100 and x400 magnification. Ctrl, control; Ara-c, cytarabine; MIT, mitoxantrone; L-Asp, L-asparaginase.
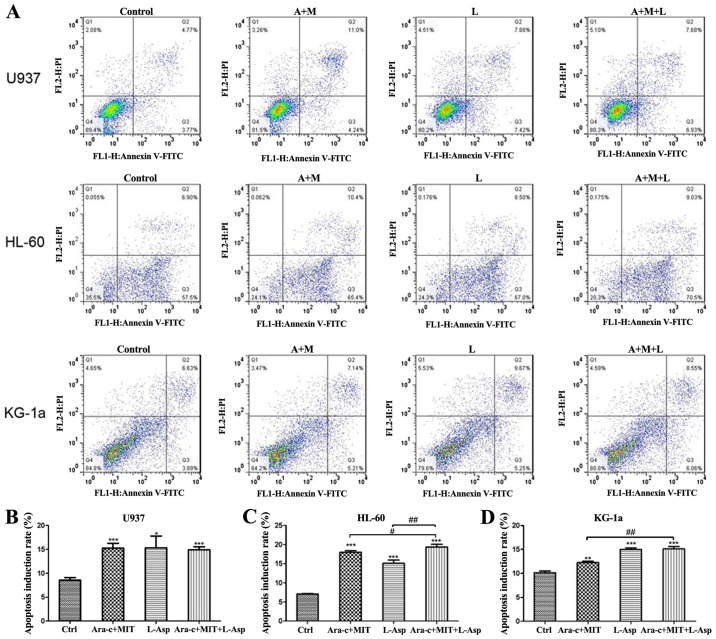
Effects of Ara-c + MIT and L-Asp + Ara-c + MIT on the apoptosis of AML cells
Cell apoptosis was detected by flow cytometry using Annexin V-FITC/PI dual staining. The results indicate that all three treatment (Ara-c + MIT, L-Asp alone and Ara-c + MIT + L-Asp) induced significant cell apoptosis in U937, HL-60 and KG-1a cells compared with the control (Fig. 5). However, cell apoptosis in the Ara-c + MIT + L-Asp treatment group did not differ significantly from that of the Ara-c + MIT treatment group or the L-Asp treatment group in U937 cells (Fig. 5A and B). Ara-c + MIT + L-Asp demonstrated significantly increased cell apoptosis compared with Ara-c + MIT or L-Asp alone in HL-60 cells (Fig. 5A and C); whereas Ara-c + MIT + L-Asp treatment appeared to have potentiated cell apoptosis compared with Ara-c + MIT treatment, but no statistical significance was observed between Ara-c + MIT + L-Asp and L-Asp alone in KG-1a cells (Fig. 5A and D).
Figure 5.
Comparison of cell apoptosis induction rates in the Ara-c + MIT, L-Asp alone and Ara-c + MIT + L-Asp treatment groups. (A) Flow cytometry dot blots show the numbers of apoptotic cells as evaluated using Annexin V-FITC and PI in U937, HL-60 and KG-1a cells. (B) Ara-c + MIT + L-Asp treatment did not induce a difference in cell apoptosis compared with the Ara-c + MIT or L-Asp alone groups in U937 cells. (C) Ara-c + MIT + L-Asp treatment significantly increased apoptosis compared with that in the Ara-c + MIT and L-Asp treatment groups in HL-60 cells. (D) Ara-c + MIT + L-Asp significantly increased cell apoptosis compared with that in the Ara-c + MIT group, but not with the L-Asp group. Each experiment was conducted three times (n=3). *P<0.05, **P<0.005, ***P<0.001 vs. Ctrl. #P<0.01, ##P<0.001 as indicated. Ctrl, control; Ara-c, cytarabine; MIT, mitoxantrone; L-Asp, L-asparaginase; FITC, fluorescein isothiocyante; PI, propidium iodide.
Autophagy detection
To evaluate the effects of L-Asp on autophagy on the U937, HL-60 and KG-1a cell lines, western blot analysis was used to assess the ratio of autophagy-associated protein LC3A/B expression levels in the cells (Fig. 6A). Each cell line was divided into two groups, namely a CQ+ group that was treated with 10 µM CQ for 3 h prior to protein extraction, and a CQ-group that was not treated with CQ. In both treatment groups, cells were treated with L-Asp at a concentration of 0.1 U/ml for 0, 3, 6, 12, 24 and 48 h. A time-dependent increase in LC3A/B-II/I ratio following L-Asp treatment was only observed in U937 cells, and not in HL-60 and KG-1a cells (Fig. 6B-D).
Figure 6.
Measurement of LC3A/B expression levels in each treatment group by western blot analysis. (A) Representative western blot images for each group. Quantified relative protein expression ratios for LC3A/B-II/I in (B) U937, (C) HL-60 and (D) KG-1a cells, with each experiment conducted three times (n=3). *P<0.05, **P<0.01, ***P<0.001 vs. the corresponding 0 h group. CQ, chloroquine; LC3A/B, light chain 3A/B.
Discussion
AML is a malignant myeloproliferative disease that is derived from hematopoietic stem cells (HSCs). It accounts for ~70% of acute leukemia cases and has an incidence of 2-4/100,000(31). Although chemotherapy, targeted therapy, and HSC transplantation (32-35) have all conferred improvements in CR and long-term DFS, the majority of AML patients ultimately relapse and develop refractory leukemia due to primary/secondary drug resistance, leading to treatment failure (4,5). The etiology of AML is complex, and its pathogenesis is not completely understood. In light of the high mortality associated with AML and the lack of significant improvements in prognosis over the last 30 years (36,37), novel therapeutic approaches for AML are required.
In the present study, it was observed that L-Asp treatment alone inhibited cell proliferation and induced apoptosis in the AML cell lines U937, HL-60 and KG-1a. The effects of L-Asp + Ara-c + MIT were compared those of L-Asp alone and Ara-c+ MIT. Combining L-Asp with Ara-c + MIT in the treatment procedure potentiated the inhibition of AML cell proliferation in each of the three cell lines investigated, and resulted in greater cell apoptosis in HL-60 and KG-1a cells. The present study investigated the effects of L-Asp on AML cell lines, and the results support the suggestion that the combination of L-Asp, MIT and Ara-c could be a promising new therapeutic strategy for AML.
Autophagy is an evolutionary conserved process that occurs throughout the eukaryotic phylogenetic tree and is essential for cellular survival and development (38). Aside from the maintenance of homeostasis, this catabolic process protects organisms from conditions including neurodegenerative diseases, heart disease, liver disease and aging (39).
It is generally recognized that autophagy can have favorable and unfavorable outcomes in tumorigenesis and metastasis (40). Previous studies demonstrated that basal autophagy activation can modulate downstream signaling pathways that are involved in maintaining adequate HSC proliferation and differentiation (41). Impaired autophagy can result in the transformation of HSCs into a preleukemic state, increasing the likelihood of malignant leukemic transformation (42). Previous studies by Karvela et al (42) and Rothe et al (43) illustrated that autophagy may serve a carcinogenic role in CML. Therefore, autophagy may serve diverse functions in leukemia physiology.
L-Asp may serve a role autophagy in AML cells. Several studies have implicated autophagic effects of L-Asp in a number of malignancies including leukemia. Yu et al (44) and Song et al (45) proposed that L-Asp could induce autophagy in ovarian cancer and CML, potentially contributing to antitumor effects. Additionally, other studies have demonstrated that suppressing autophagy may improve the antineoplastic effect of L-Asp in glioblastoma, whereas activating this process could overcome the L-Asp-induced immune suppression in macrophages (39); suggesting that L-Asp treatment may induce autophagy by promoting apoptosis, and inhibiting cell growth in AML cells.
LC3-II content or the LC3-II/LC3-I ratio is positively correlated with the number of autophagosomes and the autophagic activity of cells to some extent (46-48). In the present study, the LC3-II/I ratio was used to evaluate the autophagic activity of cells.
Since autophagy is a dynamic process, the static level of LC-3II/I alone is not sufficient to accurately measure changes in intracellular autophagic flux. Chloroquine (CQ), which is a lysosome inhibitor that inhibits autophagosome-lysosome fusion (31), is commonly used for studying autophagy and autophagy flux. To this end, changes in autophagy, as indicated by the LC3 A/B expression ratio, were evaluated in AML cells treated with L-Asp with and without CQ. However, inconsistent and in conclusive autophagic effects were observed in the three AML cell lines following L-Asp treatment, indicating that the L-Asp-induced effects on AML cells may be attributed to alternative mechanisms. Also, as this time-dependent response occurred in CQ- and CQ+ groups, the increase in LC3A/B II/I ratio presented in Fig. 6B-D may be due to factors other than autophagy. Therefore, whether L-Asp treatment produced any notable changes in autophagy in the three AML cell lines warrants further investigation. And additional experimental methods should be performed to unravel the signaling pathways mediating the anti-proliferative and pro-apoptotic effects of L-Asp on AML cells.
In conclusion, data from the present study demonstrated that L-Asp significantly suppressed cell growth while increasing apoptosis in the AML cell lines U937, HL-60 and KG-1a. When L-Asp was combined with Ara-c and MIT, these physiological effects were potentiated further. These results support the suggestion that a combination of L-Asp and MA may be useful as an AML therapy.
Acknowledgements
The authors would like to thank the China-America Institute of Neuroscience, Beijing Luhe Hospital, Capital Medical University (Beijing, China) for their valuable technical assistance with the experiments.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
TC performed most of the experiments in the present study and analyzed the obtained data. JZ and HuZ interpreted the data. YuZ, YoZ and XZ helped TC to perform the CCK8 experiments. HeZ was a major contributor to conceiving of the study and participated in writing the manuscript.
Ethics approval and consent to participate
The current study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Beijing Luhe Hospital, Capital Medical University (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Saultz JN, Garzon R. Acute myeloid, leukemia: A concise review. J Clin Med. 2016;5(pii: E33) doi: 10.3390/jcm5030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartwright RA, Staines A. Acute leukaemias. Baillieres Clin Haematol. 1992;5:1–26. doi: 10.1016/j.emc.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Dohner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European Leukemia Net. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.3390/jcm5030033. [DOI] [PubMed] [Google Scholar]
- 5.Stein EM, Tallman MS. Emerging therapeutic drugs for AML. Blood. 2016;127:71–78. doi: 10.1182/blood-2015-07-604538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Davies SM, Xiang Y, Robison LL, Ross JA. Trends in leukemia incidence and survival in the United States (1973-1998) Cancer. 2003;97:2229–2235. doi: 10.1002/cncr.11316. [DOI] [PubMed] [Google Scholar]
- 7.Luke C, Nguyen AM, To B, Seshadri R, Hughes T, Bardy P, Colbeck M, Buranyi-Trevarton D, McMellon M, Roder D. Myeloid leukaemia treatment and survival-the South Australian experience, 1977 to 2002. Asian Pac J Cancer Prev. 2006;7:227–233. [PubMed] [Google Scholar]
- 8.Norsworthy KJ, DeZern AE, Tsai HL, Hand WA, Varadhan R, Gore SD, Gojo I, Pratz K, Carraway HE, Showel M, et al. Timed sequential therapy for acute myelogenous leukemia: Results of a retrospective study of 301 patients and review of the literature. Leuk Res. 2017;61:25–32. doi: 10.1016/j.leukres.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaled S, Al-Malki M, Marcucci G. Acute myeloid leukemia: Biologic, prognostic, and therapeutic insights. Oncology (Williston Park) 2016;30:318–329. [PubMed] [Google Scholar]
- 10.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106:1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 11.Zeidner JF, Foster MC, Blackford AL, Litzow MR, Morris LE, Strickland SA, Lancet JE, Bose P, Levy MY, Tibes R, et al. Final results of a randomized multicenter phase II study of alvocidib, cytarabine and mitoxantrone versus cytarabine and daunorubicin (7 + 3) in newly diagnosed high-risk acute myeloid leukemia (AML) Leuk Res. 2018;72:92–95. doi: 10.1016/j.leukres.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini NY, Cerny J, Furtado VF, Desmond A, Zhou Z, Raffel G, Puthawala I, Bednarik J, Shanahan L, Miron PM, et al. Elderly do benefit from induction chemotherapy: High dose mitoxantrone-based (‘5 + 1’) induction chemotherapy regimen in newly diagnosed acute myeloid leukemia. Am J Hematol. 2018;94:209–215. doi: 10.1002/ajh.25347. [DOI] [PubMed] [Google Scholar]
- 13.Pardee TS, Anderson RG, Pladna KM, Isom S, Ghiraldeli LP, Miller LD, Chou JW, Jin G, Zhang W, Ellis LR, et al. A Phase I study of CPI-613 in combination with high-dose cytarabine and mitoxantrone for relapsed or refractory acute myeloid leukemia. Clin Cancer Res. 2018;24:2060–2073. doi: 10.1158/1078-0432.CCR-17-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed T, Holwerda S, Klepin HD, Isom S, Ellis LR, Lyerly S, Manuel M, Dralle S, Berenzon D, Powell BL, Pardee TS. High dose cytarabine, mitoxantrone and l-asparaginase (HAMA) salvage for relapsed or refractory acute myeloid leukemia (AML) in the elderly. Leuk Res. 2015;39:945–949. doi: 10.1016/j.leukres.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia LT, Chen SY, Yang AG. Cancer gene therapy targeting cellular apoptosis machinery. Cancer Treat Rev. 2012;38:868–876. doi: 10.1016/j.ctrv.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Dübbers A, Würthwein G, Müller HJ, Schulze-Westhoff P, Winkelhorst M, Kurzknabe E, Lanvers C, Pieters R, Kaspers GJ, Creutzig U, et al. Asparagine synthetase activity in paediatric acute leukaemias: AML-M5 subtype shows lowest activity. Br J Haematol. 2000;109:427–429. doi: 10.1046/j.1365-2141.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- 17.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11:372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 18.Shen HM, Pervaiz S. TNF receptor superfamily-induced cell death: Redox-dependent execution. FASEB J. 2006;20:1589–1598. doi: 10.1096/fj.05-5603rev. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puissant A, Robert G, Auberger P. Targeting autophagy to fight hematopoietic malignancies. Cell Cycle. 2010;9:3470–3478. doi: 10.4161/cc.9.17.13048. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Baehrecke EH. Autophagy, cell death, and cancer. Mol Cell Oncol. 2015;2(e985913) doi: 10.4161/23723556.2014.985913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 23.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- 24.Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y, Pei Y, Wan B, Guo J, Yu L. Molecular cloning and characterization of rat LC3A and LC3B-two novel markers of autophagosome. Biochem Biophys Res Commun. 2006;339:437–442. doi: 10.1016/j.bbrc.2005.10.211. [DOI] [PubMed] [Google Scholar]
- 25.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12(226) doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 27.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I. Autophagy basics. Microbiol Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- 30.Song P, Ye L, Fan J, Li Y, Zeng X, Wang Z, Wang S, Zhang G, Yang P, Cao Z, Ju D. Asparaginase induces apoptosis and cytoprotective autophagy in chronic myeloid leukemia cells. Oncotarget. 2015;6:3861–3873. doi: 10.18632/oncotarget.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18(pii: E1865) doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gollner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, Pabst C, Sauer T, Lerdrup M, Tavor S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23:69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hourigan CS, Gale RP, Gormley NJ, Ossenkoppele GJ, Walter RB. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31:1482–1490. doi: 10.1038/leu.2017.113. [DOI] [PubMed] [Google Scholar]
- 34.Xu B, Zhao Y, Wang X, Gong P, Ge W. MZH29 is a novel potent inhibitor that overcomes drug resistance FLT3 mutations in acute myeloid leukemia. Leukemia. 2017;31:913–921. doi: 10.1038/leu.2016.297. [DOI] [PubMed] [Google Scholar]
- 35.Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. 2017;129:1627–1635. doi: 10.1182/blood-2016-10-696039. [DOI] [PubMed] [Google Scholar]
- 36.Estey EH. Treatment of acute myeloid leukemia. Haematologica. 2009;94:10–16. doi: 10.1097/CCO.0b013e328358f62d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian S, Kartha RV. MicroRNA-mediated gene regulations in human sarcomas. Cell Mol Life Sci. 2012;69:3571–3585. doi: 10.1007/s00018-012-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabat AM, Pott J, Maloy KJ. The mucosal immune system and its regulation by autophagy. Front Immunol. 2016;7(240) doi: 10.3389/fimmu.2016.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ndoye A, Weeraratna AT. Autophagy-An emerging target for melanoma therapy. F1000Res. 2016;5(pii: F1000) doi: 10.12688/f1000research.8347.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li CJ, Liao WT, Wu MY, Chu PY. New insights into the role of autophagy in tumor immune microenvironment. Int J Mol Sci. 2017;18(pii: E1566) doi: 10.3390/ijms18071566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang WX, Zhou JC, Peng D, Hua F, Li K, Yu JJ, Lv XX, Cui B, Liu SS, Yu JM, et al. Disrupting the TRIB3-SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome-mediated HSC activation. Autophagy. 2019:1–15. doi: 10.1080/15548627.2019.1635383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karvela M, Baquero P, Kuntz EM, Mukhopadhyay A, Mitchell R, Allan EK, Chan E, Kranc KR, Calabretta B, Salomoni P, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy. 2016;12:936–948. doi: 10.1080/15548627.2016.1162359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothe K, Lin H, Lin KB, Leung A, Wang HM, Malekesmaeili M, Brinkman RR, Forrest DL, Gorski SM, Jiang X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood. 2014;123:3622–3634. doi: 10.1182/blood-2013-07-516807. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Zhang X, Tian H, Zhang Z, Tian Y. Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J BUON. 2018;23:1396–1401. [PubMed] [Google Scholar]
- 45.Song X, Liu L, Chang M, Geng X, Wang X, Wang W, Chen TC, Xie L, Song X. NEO212 induces mitochondrial apoptosis and impairs autophagy flux in ovarian cancer. J Exp Clin Cancer. 2019;38(239) doi: 10.1186/s13046-019-1249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32(2272e.1-11):2261–2672. doi: 10.1038/onc.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitter SK, Song C, Qi X, Mao H, Rao H, Akin D, Lewin A, Grant M, Dunn W Jr, Ding J, et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014;10:1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Z, Chen C, Wu Z, Miao Y, Muhammad I, Ding L, Tian E, Hu W, Ni H, Li R, et al. A dual role of P53 in regulating colistin-induced autophagy in PC-12 cells. Front Pharmacol. 2017;8(768) doi: 10.3389/fphar.2017.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.