Abstract
Seven alkaloidal compounds (2–8) and one polyketide (1) were isolated from a semisolid rice culture of the marine-derived fungus Aspergillus sp. F452. Structures of the isolated compounds were elucidated based on spectroscopic data and comparisons with previously reported data. The alkaloidal compounds (2–8) displayed weak to moderate inhibitory activities against Staphylococcus aureus-derived sortase A (SrtA) without affecting cell viability. Aspermytin A (1) strongly inhibited SrtA activity, with an IC50 value of 146.0 μM, and significantly reduced bacterial adherence to fibronectin-coated surfaces. The present results indicate that the underlying mechanism of action of compound 1 is associated with the inhibition of SrtA-mediated S. aureus adhesion to fibronectin, thus potentially serving as an SrtA inhibitor.
Keywords: marine-derived fungus, Aspergillus sp., metabolites, sortase A, fibronectin
1. Introduction
Antibiotic-resistant bacteria are the most prominent limitation in conventional antimicrobial treatment [1]. Microorganisms can acquire antibiotic resistance when their survival is at risk. Whereas antibiotics have a long-standing history of success in treatment of bacterial infections, recent increasing antimicrobial resistance has stimulated the search for anti-virulence drugs as an alternative to conventional antibiotics, despite their high importance, for counteracting bacterial pathogens [2,3].
The pathogenesis of bacterial infections is initiated with bacterial adhesion to host tissue surfaces mediated via specific interactions between host ligands and bacterial surface proteins [4]. In particular, in Gram-positive bacteria including Staphylococcus aureus, this fundamental stage of infection proceeds through sortase-mediated anchoring of surface proteins in host cells to the bacterial cell wall envelope [5]. In S. aureus, sortase A (SrtA) cleaves surface proteins between threonine and glycine residues in LPXTG sorting signals at their C-termini and is subsequently incorporated into the bacterial cell wall envelope via a transpeptidation reaction [6,7]. Numerous knockout studies have revealed that SrtA plays a critical role in the pathogenesis of Gram-positive bacterial infections by modulating bacterial adhesion to host tissues [8,9,10]. SrtA decorates the surfaces of Gram-positive bacteria with a diverse array of proteins that enable each microbe to effectively interact with its environment and is not required for bacterial growth or viability [2,3]. It is thus considered a promising target for the development of anti-virulence drugs that aim to interfere with important virulence mechanisms, such as adhesion to host tissues.
The secondary metabolites of marine fungi, including polyketides, alkaloids, terpenes, lactones, and peptides, are a rich source of bioactive natural products [11]. Many bioactive compounds with varying degrees of action, such as antibiotic, antiviral, antimicrobial, and anticancer properties, have been isolated from marine fungal sources. Recent investigations of marine fungal metabolites looking for bioactive compounds indicate their potential as a source of new medicines [12,13]. Previously, we reported several novel natural products isolated from marine-derived fungi; polyaromatic metabolites from Penicillium sp. exhibited moderate cytotoxicity and significant inhibitory activity against S. aureus SrtA [14], asperphenins from Aspergillus sp. induced significant cytotoxicity in diverse cancer cells [15], and peptides from A. allahabadii and A. ochraceopetaliformis displayed SrtA inhibitory activity [16].
In further study, chemical investigation of Aspergillus sp. F452 was performed [15], whose crude extract inhibited S. aureus-derived SrtA (63% inhibition at 100 μg/mL). Bioassay-guided separation of the extract yielded seven alkaloidal (2–8) and one polyketide (1) compound, whose structures were analyzed through combined spectroscopic methods. This study describes the structures and biological activities of these compounds. Among them, compound 1 (aspermytin A) significantly inhibited S. aureus-derived SrtA. The in vivo bioactivity and underlying mechanism of action were also found to be associated with the inhibition of SrtA-mediated S. aureus adhesion to the eukaryotic cell matrix protein fibronectin.
2. Results and Discussion
2.1. Isolation and Structure Elucidation of Compounds 1–8
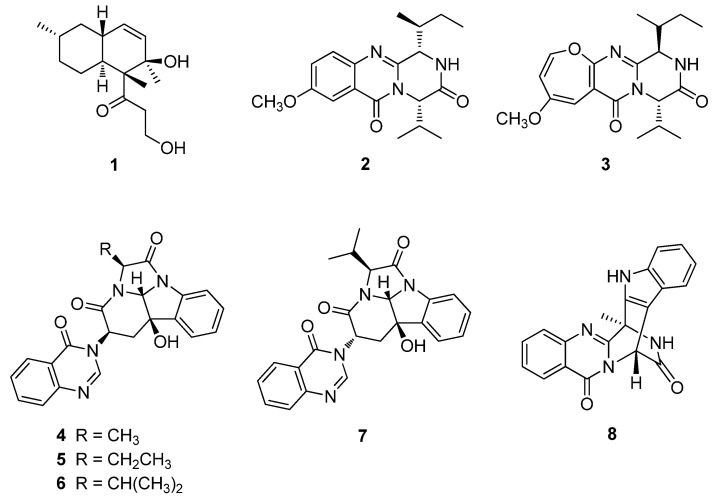
The fungal strain F452 [15] was cultured in semisolid rice medium and extracted with MeOH and CH2Cl2. Following solvent evaporation, the combined extract was separated by solvent partitioning followed by reversed-phase C18 vacuum flash chromatography and semi-preparative high performance liquid chromatography (HPLC) to yield eight compounds. Compounds 1–8 were identified as aspermytin A (1) [17], versicomide A (2) [18], versicoloid A (3) [19], isochaetominines A–C (4–6) [20,21], 14-epi-isochaetominine C (7) [21], and fumiquinazoline K (8) [22], respectively, via combined spectroscopic analyses, including high-resolution fast atom bombardment mass spectroscopy, 1H- and 13C-nuclear magnetic resonance (NMR), 2-D NMR, and UV spectroscopy (Figure 1). The spectroscopic data for these compounds were in good agreement with those in the literature.
Figure 1.
Structures of compounds 1–8 from Aspergillus sp. F452.
2.2. SrtA Inhibitory Activity of Compounds 1–8
Recombinant SrtA derived from S. aureus ATCC6538p was purified from Escherichia coli extracts using metal chelate-affinity chromatography [23]. The enzyme activity was determined from the fluorescence intensity upon cleavage of a peptide substrate containing the LPETG motif [24]. Throughout the separation process, the crude extract and chromatographic fractions containing fungal metabolites of strain F452 inhibited the activity of SrtA. Accordingly, the same bioassay was performed using pure compounds. The inhibitory potencies of the pure compounds against recombinant SrtA, expressed as IC50 values, are shown in Table 1 and are compared to those of the known SrtA inhibitors berberine chloride (IC50 = 85.9 μM) and para-hydroxymercuribenzoic acid (pHMB) (IC50 = 112.5 μM). The pure compounds 1–8 displayed weak to significant SrtA inhibition (IC50 values of 269.4–146.0 μM). Among them, compound 1 exhibited the most potent inhibitory activity.
Table 1.
Inhibitory activity of compounds 1–8 toward the activity of the SrtA enzyme and bacterial growth of S. aureus ATCC6538p.
| Compounds | SrtA IC50 μM (μg/mL) | MIC μM (μg/mL) 1 |
|---|---|---|
| 1 | 146.0 ± 2.3 (38.9 ± 0.6) | >480.5 (>128) |
| 2 | 269.4 ± 3.9 (92.5 ± 1.4) | >372.7 (>128) |
| 3 | 193.5 ± 2.5 (69.5 ± 0.9) | >356.1 (>128) |
| 4 | 267.9 ± 4.1 (107.8 ± 1.5) | >318.1 (>128) |
| 5 | 232.5 ± 3.7 (96.8 ± 1.4) | >307.4 (>128) |
| 6 | 216.4 ± 2.5 (93.1 ± 1.1) | >297.4 (>128) |
| 7 | 237.1 ± 3.8 (102.1 ± 1.2) | >297.4 (>128) |
| 8 | 235.1 ± 2.6 (83.8 ± 0.9) | >359.2 (>128) |
| Berberine chloride | 85.9 ± 1.2 (31.9 ± 0.4) | >332.2 (>128) |
| pHMB | 112.5 ± 1.7 (33.1 ± 0.5) | ND 2 |
| Ampicillin | ND | 0.4 (0.1) |
1 MIC means minimum inhibitory concentration. 2 ND means not determined. pHMB (para-hydroxymercuribenzoic acid) and berberine chloride were used as reference inhibitors of SrtA. Ampicillin was used as a standard antibacterial drug.
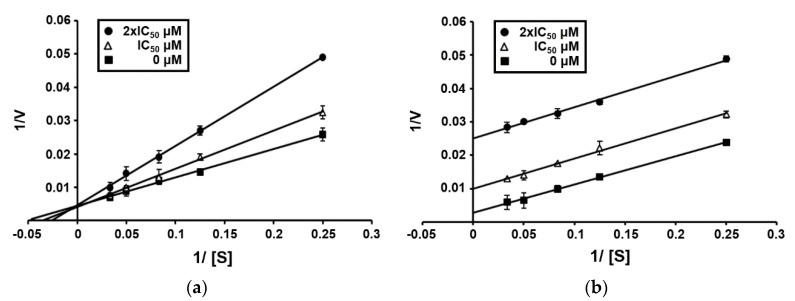
To determine the type of inhibition, kinetic studies were performed with compounds 1 and 3 at IC50 or twofold IC50 based on a Lineweaver and Burk plot [25] (Figure 2). Inhibitor constants were obtained by a Dixon plot. Inhibitory kinetics show that compound 1 behaved as a mixed inhibitor (Ki = 265.0 μM). In contrast, compound 3 behaved as an uncompetitive inhibitor (Ki = 83.0 μM). Moreover, the binding of compounds 1 and 3 to SrtA was reversible because the enzyme activity was indeed recovered by dialysis within 2 h, excluding the possible existence of a covalent bond between inhibitor and enzyme.
Figure 2.
Lineweaver–Burk plot of SrtA inhibition by compounds 1 (a) and 3 (b). [S], substrate concentration [μM]; V, reaction velocity (Δabsorbance unit/min). Each data point represents the mean of three experiments.
2.3. Antibacterial Activity and Cytotoxicity of Compounds 1–8
Because SrtA inhibitors are expected to serve as anti-infective agents and inhibit bacterial pathogenesis without affecting cell viability [8], the minimum inhibitory concentrations (MICs) of these compounds were also measured to exclude the possible effects of test compounds on S. aureus cell adhesion to the eukaryotic cell matrix protein fibronectin owing to the inhibition of cell growth. The compounds did not exhibit inhibitory activity against S. aureus ATCC6538p (MIC > 128 μg/mL) (Table 1). In the cytotoxicity assay against A549 (lung cancer) and K562 (leukemia) cell lines, compounds 1–8 displayed weak (2–7: IC50 > 13–50 μM) to no inhibitory activity (1 and 8: IC50 > 100 μM), comparable to etoposide (IC50 = 0.5 μM).
2.4. Inhibition of SrtA-mediated S. aureus Adhesion to Fibronectin by Compound 1
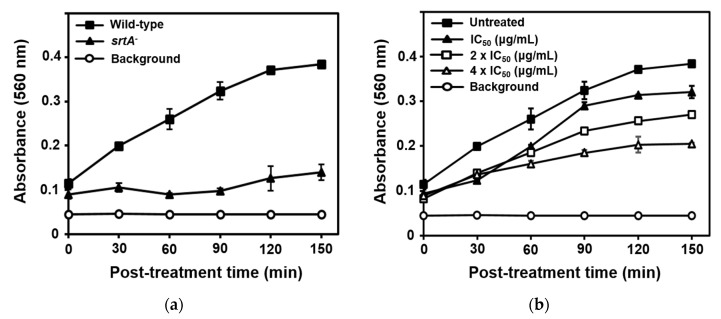
An active SrtA enzyme is required for the attachment of S. aureus to eukaryotic cell matrix proteins, such as fibronectin and fibrinogen, thus accelerating bacterial adhesion to host tissues and subsequent invasion [26]. The srtA− mutant strain cannot bind these proteins. Thus, SrtA inhibitors should inhibit SrtA activity in vivo and in turn reduce fibronectin-binding protein surface display. Initially, the SrtA-mediated fibronectin-binding capacities of S. aureus strain Newman (wild-type) and its isogenic srtA knockout mutant (srtA−) to fibronectin were evaluated. As shown in Figure 3a, the fibronectin-binding activity of the srtA knockout mutant was significantly reduced compared to that of the wild type. Based on SrtA inhibition intensity, compound 1 was selected. The results of the inhibition of S. aureus adhesion to fibronectin via fibronectin-binding protein by compound 1 are shown in Figure 3b. As expected, treatment of strain Newman with 0-, 1-, 2-, or 4-fold the SrtA IC50 of compound 1 significantly reduced bacterial adherence to fibronectin-coated surfaces. The onset and magnitude of inhibition of Newman strain adhesion to fibronectin by compound 1 with 4× the SrtA IC50 value was comparable to the behavior of the untreated srtA knockout mutant, as shown in Figure 3a. The results of the fibronectin-binding assay suggested the potential of this compound in treating S. aureus infections through inhibition of SrtA activity.
Figure 3.
Adhesion of S. aureus strain Newman (wild-type) and the isogenic srtA knockout mutant (srtA−) to fibronectin (a), and inhibition of Newman strain adhesion to fibronectin by compound 1 (b) with 0×, 1×, 2×, or 4× the SrtA IC50 value. The results are presented as the mean ± standard deviation of three replicates.
3. Materials and Methods
3.1. General Experimental Procedures
The UV spectra were acquired with a Hitachi U-3010 spectrophotometer (Tokyo, Japan). The NMR spectra were recorded in DMSO-d6 solution using Bruker Avance (400, 500, or 600) instruments (Billerica, MA, USA). IR spectra were recorded on a JASCO 4200 FT-IR spectrometer (Easton, MD, USA) using a ZnSe cell. High-resolution FABMS data were acquired using a JEOL JMS 700 mass spectrometer (Tokyo, Japan) with 6 keV-energy, emission current 5.0 mA, xenon as inert gas, and meta-nitrobenzyl alcohol (NBA) as the matrix at the Korea Basic Science Institute (Daegu, Korea). Low-resolution ESIMS data were recorded on an Agilent Technologies 6130 quadrupole mass spectrometer with an Agilent Technologies 1200 series HPLC system (Santa Clara, CA, USA). HPLC analyses were performed on a Spectrasystem p2000 equipped with a Spectrasystem RI-150 refractive index detector (Waltham, MA, USA). All of the solvents used were spectroscopic grade or distilled from glass prior to use.
3.2. Fungal Material
The isolation and identification of Aspergillus sp. (strain number F452) have previously been reported [15]. The fungal strain was isolated from submerged, decaying wood off the shore of Jeju Island, Korea, and identified using standard molecular biological protocols by DNA amplification and sequencing of the ITS region. The nucleotide sequence of F452 has been deposited in the GenBank database under accession number KF384188.
3.3. Extraction and Isolation
The isolated strain was cultivated on a YPG agar plate (5 g yeast extract, 5 g peptone, 10 g glucose, and 16 g agar in 1 L artificial seawater) for 4 days. The agar plugs (1 cm × 1 cm, 5 pieces each) were inoculated into 100 mL YPG media in a 250 mL Erlenmeyer flask for 5 days, then separately transferred to 2.8 L glass Fernbach flasks with rice media (200 g rice, 2 g peptone, and 2 g yeast extract with 200 mL artificial seawater in each flask, boiled in an autoclave for 20 min at 120 °C; 50 flasks in total).
Fermentation in rice media was conducted under static conditions for 6 weeks followed by extraction of each flask with MeOH (1 L × 3) and CH2Cl2 (1 L × 3). The solvent was combined and evaporated to obtain an organic extract. The combined extracts (247.33 g) were successively partitioned between n-BuOH (177.12 g) and H2O (70.05 g); the former fraction was repartitioned using H2O-MeOH (15:85) (83.31 g) and n-hexane (93.18 g). The H2O-MeOH fraction was separated by C18 reversed-phase vacuum flash chromatography using a sequential mixture of MeOH and H2O as eluents (five fractions in the gradient, H2O-MeOH, from 60:40 to 0:100), acetone, and finally EtOAc.
Based on the results of 1H NMR analyses and bioactivity tests, the 30:70 H2O-MeOH fraction (9.20 g) was separated by semi-preparative reversed-phase HPLC (YMC-ODS-A column, 10 × 250 mm; H2O-MeOH, 50:50; 1.7 mL/min) and yielded compounds 1 (tR = 46.1 min), 2 (tR = 22.2 min), 3 (tR = 13.5 min), 4 (tR = 37.2 min), 5 (tR = 40.5 min), 6 (tR = 66.9 min), 7 (tR = 74.1 min), and 8 (tR = 15.4 min). Compounds 1, 4, and 5 were further purified by analytical HPLC (YMC-ODS-A column, 4.6 × 250 mm; H2O-MeCN, 65:35; 0.7 mL/min; tR = 18.2, 11.9, and 14.5 min, respectively). The purified metabolites were isolated at the following yields: 5.5, 3.4, 4.4, 2.9, 5.1, 9.3, 1.9, and 7.9 mg for 1–8, respectively.
3.4. SrtA Inhibition Assay
The srtA gene from S. aureus ATCC6538p was expressed and recombinant SrtA was purified as previously described [23]. The SrtA inhibition test was carried out by analyzing the increased fluorescence intensity resulting from the cleavage of synthetic peptide substrate dabcyl-LPETG-edans (AnaSpec, Inc., Fremont, CA, USA) [9,24] with slight modification. The reaction was carried out with 100 μL buffer (50 mM Tris-HCl, 5 mM CaCl2, and 150 mM NaCl, pH 7.5), 7.5 µM synthetic peptide, 7.5 µM purified SrtA, and test samples at various concentrations. Each sample was dissolved in dimethyl sulfoxide (DMSO) and diluted with reaction buffer to obtain a final concentration of 1% DMSO, which did not influence enzyme activity. The SrtA inhibition assay was conducted at 37 °C for 1 h, and inhibition was quantified fluorometrically using a microplate reader (FLx800, BioTek Instruments, Winooski, VT, USA) at excitation and emission wavelengths of 350 and 495 nm, respectively. pHMB and berberine chloride were used as reference inhibitors of SrtA.
3.5. Enzyme Kinetics
All sortase (SrtA) assays were performed at 37 °C in SrtA enzyme buffer as described above. The inhibitors 1 and 3 were dissolved in DMSO and immediately diluted to the desired working concentration with the same SrtA buffer. The enzymatic inhibition measurements were carried out at different substrate concentrations in the presence and absence of a given concentration of inhibitor, and their kinetics were evaluated by the Lineweaver and Burk plot method [25]. For the dialysis kinetic studies, a solution of enzyme (0.96 mL, 75 μM) and fixed inhibitor concentration (0.04 mL) was prepared and dialyzed against 100 mL buffer at 37 °C for 2 h using regenerated cellulose dialysis membranes SPECTRAPOR® (Rancho Dominguez, CA, USA). Aliquots of 100 μL of the enzyme-inhibitor mixture were taken in time intervals of 0, 30, 60, 90, and 120 min and added to 0.020 mL of substrate (7.5 μM), and after an hour of incubation the enzyme activity was measured. A control solution prepared with enzyme and buffer (SrtA buffer, 100 μL; SrtA, 7.5 μM) was treated similarly.
3.6. Antibacterial Activity Assay
The MICs of test compounds were determined as previously described [27]. S. aureus ATCC6538p (5 mL) was cultured in tryptic soy broth to saturation at 37 °C and diluted to an OD600 of 0.01. The culture was incubated for an additional 2 h and diluted to an OD600 of 0.005. In each well of a 96-well plate, 180 µL of cells was mixed with 20 µL of a concentrated test compound solution in 10% DMSO (final concentration, 1% DMSO). Culture plates were incubated overnight at 37 °C, and the OD600 was measured using a Multiskan Spectrum spectrophotometer (Thermo Labsystems Inc., Beverly, MA, USA). MIC values were defined as the lowest concentration of the test compounds inhibiting cell growth. Ampicillin was used as a positive control.
3.7. Cytotoxicity Assay
The effect of compounds (1–8) on cell proliferation was measured by the sulforhodamine B (SRB) cellular protein-staining method [28]. In brief, A549 (lung cancer) and K562 (leukemia) cells (1 × 104 cells in 190 μL of complete DMEM) were seeded in 96-well plates with various concentrations of compounds (1–8) and incubated at 37 °C in a humidified atmosphere with 5% CO2. After 72 h of compound (1–8) treatment, the cells were fixed with 10% TCA solution for 1 h, and cellular proteins were stained with a solution of 0.4% SRB in 1% acetic acid. The stained cells were dissolved in 10 mM Tris buffer (pH 10.0). The effect of compounds (1–8) on cell viability was calculated as a percentage relative to a solvent-treated control, and the IC50 values were calculated using a nonlinear regression analysis (percent survival versus concentration). Etoposide was used as a positive control.
3.8. Fibronectin-Binding Assay
S. aureus strains used were Newman (wild-type) and the isogenic srtA knockout mutant (srtA−) [9]. These strains were cultured in tryptic soy broth at 37 °C at 200 rpm up to mid-log phase (OD600 = 0.5). The fibronectin-binding assay was performed as described previously [24,29]. Cells were treated with test compounds at their indicated concentrations. Every 30 min for 2.5 h, a 0.65 mL cell suspension was centrifuged at 10,000× g for 10 min, and the supernatant was eliminated. Following incubation overnight at −20 °C, pellets were resuspended in 0.65 mL phosphate-buffered saline (PBS) and distributed as a 100 µL scale in fibronectin-coated flat-bottomed 96-well plates (Corning Life Sciences, Tewksbury, MA, USA). The cell suspension was removed and washed with PBS following incubation at 37 °C for 2 h. Bound cells were fixed via incubation with 2% (v/v) glutaraldehyde for 30 min. After a second wash with PBS, cells were stained for 15 min with 100 μL crystal violet dye (12.5 g/L). Each well was washed with PBS and covered with aluminum foil. Plates were dried overnight and absorbance was measured at 560 nm using a microplate reader.
4. Conclusions
Seven alkaloidal compounds (2–8) and one polyketide (1) were isolated from a semisolid rice culture of the marine-derived fungus Aspergillus sp. F452. The structures of these compounds were obtained through a combination of spectroscopic analyses, and their data were in good agreement with previous reports. Bioactivity studies have revealed that compound 1 from a marine-derived fungus Aspergillus sp., separated from the mussel Mytilus edulis, shows significant neurotrophic effects on PC-12 cells [17]. Compound 3 from the deep sea-derived A. versicolor SCSIO 05,879 exhibits antifungal activity against the phytopathogenic fungus Colletotrichum acutatum (minimum inhibitory concentration (MIC) of 1.6 μg/mL) [19]. Compounds 4–7 from a marine-derived fungus Aspergillus sp. F452 exhibit weak inhibition against Na+/K+-ATPase (IC50 values of 20–78 μM) [20]. Compound 6 also displays weak inhibition against Bacillus subtilis, and compound 7 from A. fumigatus, an endophytic fungus, shows weak cytotoxic activity against the human prostate cancer cell line PC3 [21]. In our measurement of SrtA enzyme activity, compounds 1–8 displayed moderate to significant SrtA inhibition, comparable to berberine chloride and pHMB, against S. aureus-derived SrtA, a transpeptidase responsible for anchoring surface proteins to the peptidoglycan cell wall in Gram-positive bacteria. Further bioassays of compound 1 indicated that the underlying mechanism of action was associated with the inhibition of adhesion of S. aureus to fibronectin via fibronectin-binding protein. Our results demonstrate the potential of these metabolites for the development of new agents to treat Gram-positive bacterial infections by inhibiting SrtA activity.
Acknowledgments
We thank the Basic Science Research Institute in Daegu, Korea, for providing the mass data and the National Center for Inter-University Research Facilities (NCIRF), Seoul National University, for providing the NMR data.
Author Contributions
S.C.P. and J.-Y.H. carried out the isolation and structural elucidation; B.C. performed the fibronectin-binding assays; J.L. and E.C. contributed to the SrtA inhibition and antibacterial activity assays; D.-C.O. reviewed and evaluated all data; J.S. and K.-B.O. supervised the research work and prepared the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation (NRF, grant No. 2018R1D1A1B07043375) of Korea funded by the Ministry of Education, Science and Technology and the NRF (grant No. 2018R1A4A1021703) of Korea funded by the Ministry of Science, ICT and Future Planning. This work was also supported by the BK21 Plus Program of the Department of Agricultural Biotechnology, Seoul National University, Seoul, Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gould I.M. Antibiotic resistance: The perfect storm. Int. J. Antimicrob Agents. 2009;34:S2–S5. doi: 10.1016/S0924-8579(09)70549-7. [DOI] [PubMed] [Google Scholar]
- 2.Maresso A.W., Schneewind O. Sortase as a target of anti-infective therapy. Pharm. Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 3.Cascioferro S., Totsika M., Schillaci D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014;77:105–112. doi: 10.1016/j.micpath.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Rasko D.A., Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug. Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickx A.P., Budzik J.M., Oh S.Y., Schneewind O. Architects at the bacterial surface-sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian S.K., Skaar E.P., Gaspar A.H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D.M., Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 7.Clancy K.W., Melvin J.A., McCafferty D.G. Sortase transpeptidases: Insights into mechanism, substrate specificity and inhibition. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazmanian S.K., Liu G., Jensen E.R., Lenoy E., Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian S.K., Ton-That H., Su K., Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss W.J., Lenoy E., Murphy T., Tardio L., Burgio P., Projan S.J., Schneewind O., Alksne L. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J. Antimicrob. Chemother. 2004;53:480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- 11.Rateb M.E., Ebel R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011;28:290–344. doi: 10.1039/c0np00061b. [DOI] [PubMed] [Google Scholar]
- 12.Jin L., Quan C., Hou X., Fan S. Potential pharmacological resources: Natural bioactive compounds from marine-derived fungi. Mar. Drugs. 2016;14:76. doi: 10.3390/md14040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youssef F.S., Ashour M.L., Singab A.N.B., Wink M. A comprehensive review of bioactive peptides from marine fungi and their biological significance. Mar. Drugs. 2019;17:559. doi: 10.3390/md17100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julianti E., Lee J.-H., Liao L., Park W., Park S., Oh D.-C., Oh K.-B., Shin J. New polyaromatic metabolites from a marine-derived fungus Penicillium sp. Org. Lett. 2013;15:1286–1289. doi: 10.1021/ol4002174. [DOI] [PubMed] [Google Scholar]
- 15.Liao L., Bae S.Y., Won T.H., You M., Kim S.-H., Oh D.-C., Lee S.K., Oh K.-B., Shin J. Asperphenins A and B, lipopeptidyl benzophenones from a marine-derived Aspergillus sp. fungus. Org. Lett. 2017;19:2066–2069. doi: 10.1021/acs.orglett.7b00661. [DOI] [PubMed] [Google Scholar]
- 16.Hwang J.-Y., Lee J.-H., Park S.C., Lee J., Oh D.-C., Oh K.-B., Shin J. New peptides from the marine-derived fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Mar. Drugs. 2019;17:488. doi: 10.3390/md17090488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukamoto S., Miura S., Yamashita Y., Ohta T. Aspermytin A: A new neurotrophic polyketide isolated from a marine-derived fungus of the genus Aspergillus. Bioorg. Med. Chem. Lett. 2004;14:417–420. doi: 10.1016/j.bmcl.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 18.Pan C., Shi Y., Chen X., Chen C.-T.A., Tao X., Wu B. New compounds from a hydrothermal vent crab-associated fungus Aspergillus versicolor XZ-4. Org. Biomol. Chem. 2017;15:1155–1163. doi: 10.1039/C6OB02374F. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., He W., Huang X., Tian X., Liao S., Yang B., Wang F., Zhou X., Liu Y. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 2016;64:2910–2916. doi: 10.1021/acs.jafc.6b00527. [DOI] [PubMed] [Google Scholar]
- 20.Liao L., You M., Chung B.K., Oh D.-C., Oh K.-B., Shin J. Alkaloidal metabolites from a marine-derived Aspergillus sp. fungus. J. Nat. Prod. 2015;78:349–354. doi: 10.1021/np500683u. [DOI] [PubMed] [Google Scholar]
- 21.Xie F., Li X.-B., Zhou J.-C., Xu Q.-Q., Wang X.-N., Yuan H.-Q., Lou H.-X. Secondary metabolites from Aspergillus fumigatus, an endophytic fungus from the liverwort Heteroscyphus tener (Steph) Schiffn. Chem. Biodivers. 2015;12:1313–1321. doi: 10.1002/cbdv.201400317. [DOI] [PubMed] [Google Scholar]
- 22.Heredia M.L., de la Cuesta E., Avendano C. Acid-promoted reactions in 1-hydroxy, 1-dimethylaminomethyl and 1-methylene-4-arylmethyl-2,4-dihydro-1H-pyrazino[2,1-b]-quinazoline-3,6-diones. Tetrahedron. 2002;58:6163–6170. doi: 10.1016/S0040-4020(02)00619-1. [DOI] [Google Scholar]
- 23.Oh K.-B., Kim S.-H., Lee J., Cho W.-J., Lee T., Kim S. Discovery of diarylacrylonitriles as a novel series of small molecule sortase A inhibitors. J. Med. Chem. 2004;47:2418–2421. doi: 10.1021/jm0498708. [DOI] [PubMed] [Google Scholar]
- 24.Oh K.-B., Oh M.-N., Kim J.-G., Shin D.-S., Shin J. Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. Appl. Microbiol. Biotechnol. 2006;70:102–106. doi: 10.1007/s00253-005-0040-8. [DOI] [PubMed] [Google Scholar]
- 25.Lineweaver H., Burk D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934;56:658–666. doi: 10.1021/ja01318a036. [DOI] [Google Scholar]
- 26.Alksne L.E., Projan S.J. Bacterial virulence as a target for antimicrobial chemotherapy. Curr. Opin. Biotechnol. 2000;11:625–636. doi: 10.1016/S0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- 27.Frankel B.A., Bentley M., Kruger R.G., McCafferty D.G. Vinyl sulfones: Inhibitors of srtA, a transpeptidase required for cell wall protein anchoring and virulence in Staphylococcus aureus. J. Am. Chem. Soc. 2004;126:3404–3405. doi: 10.1021/ja0390294. [DOI] [PubMed] [Google Scholar]
- 28.Kim T.S., Shin Y.-H., Lee H.-M., Kim J.K., Choe J.H., Jang J.-C., Um S., Jin H.S., Komatsu M., Cha G.-H., et al. Ohmyungsamycins promote antimicrobial responses through autophagy activation via AMP-activated protein kinase pathway. Sci. Rep. 2017;7:3431. doi: 10.1038/s41598-017-03477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgalai I., Foster H.A. Comparison of adhesion of wound isolates of Staphylococcus aureus to immobilized proteins. J. Appl. Micrbiol. 2003;94:413–420. doi: 10.1046/j.1365-2672.2003.01858.x. [DOI] [PubMed] [Google Scholar]