Abstract
The powerful ability of human amnion-derived mesenchymal stem cells (hAMSCs) to promote angiogenesis suggests that they may facilitate angiogenesis-associated therapeutic strategies. However, the molecular mechanisms underlying hAMSC-induced angiogenesis remain largely unknown. The present study results suggested that enhanced migration and tube formation in human umbilical vein endothelial cells (HUVECs) was induced by conditioned medium from hAMSCs (hAMSC-CM). In addition, culture with this conditioned medium resulted in the increased expression of circular RNA ATP binding cassette subfamily B member 10 (circ-ABCB10) and vascular endothelial growth factor A (VEGFA). In the present study genes related to thecirc-ABCB10/microRNA (miR)-29b-3p/VEGFA pathway were predicted using bioinformatics software, and further investigated using in vitro luciferase reporter assays. Loss-of-function assays were performed using small interfering RNAs (siRNAs). The results suggested that siRNA-silencing of circ-ABCB10 in HUVECs weakened migration and tube formation of HUVECs following hAMSC-CM treatment and reduced the levels of VEGFA expression. Treatment with an miR-29b-3p inhibitor could largely rescue these effects in HUVECs, following circ-ABCB10 silencing. The present study results suggest that the circ-ABCB10/miR-29b-3p/VEGFA pathway may be involved in the pro-angiogenic role of hAMSC-CM in HUVECs.
Keywords: circ-ABCB10, hAMSCs, angiogenesis, HUVECs, VEGFA
Introduction
Angiogenesis, the formation of new blood vessels, plays an important role in bone regeneration and osteoblast differentiation (1) and provides essential nutrients and oxygen during bone formation (2). Previous studies have indicated an association between angiogenesis and osteogenesis during bone healing and regeneration (2,3). Compared with vasculogenesis, angiogenesis occurs more frequently and plays a major role during the process of bone healing (4). Thus, treatments that improve angiogenesis maybe a beneficial for bone regeneration (4).
Angiogenesis is a complex process that can be manipulated by interactions between vascular cells and the microenvironment (3). Previous studies have suggested that mesenchymal stem cells (MSCs) may have a function in cell-based regulation of angiogenesis (5,6). MSCs can promote tube formation and enhance the proliferation of endothelial cells via a paracrine pathway (7). Zuo et al (8) found that conditioned medium (CM) from MSCs could enhance angiogenesis in a post-infarction model. MSCs can also be obtained from multiple tissues including bone marrow, adipose tissue, peripheral blood and synovial tissue (9,10). However, the wide use of MSCs in therapeutic angiogenesis is limited by the low number of cells that can be isolated from these tissues and their instability that results from donor age-dependent differentiation (11). Therefore, alternative seed cells are required for the pursuit of this type of therapeutic strategy.
Human amnion-derived mesenchymal stem cells (hAMSCs) show a powerful ability to promote angiogenesis (12) and exhibit stable viability and low level anti-inflammatory properties (13). Abundant amounts of amniotic tissuecan be donated with fewer ethical issues compared with other tissue types commonly donated for the isolation of seed cells (14). A previous study we carried out showed that hAMSCs significantly enhanced the angiogenesis of human umbilical vein endothelial cells (HUVECs) both in vivo and in vitro (15), however, the molecular mechanism underlying this process remain largely unknown.
Circular RNA (circRNA) is a subclass of endogenous non-coding RNA with a loop structure generated by back splicing (16). Recent studies have found that circRNAs are widely expressed in various human cell types and have multiple biological functions in the process of development and disease (17,18). The role of circRNAs in angiogenesis may involve competing endogenous RNAs (ceRNA) (19). Thus, the present study aimed to investigate whether circRNAs play a role in hAMSC-induced angiogenesis.
The present study investigated the pro-angiogenic capacity of CM from hAMSCs on HUVECs in a co-culture system. The results suggested that circ-ATP binding cassette subfamily B member 10 (ABCB10) may play an important role in the promotion of angiogenesis induced by hAMSC-CM via the circ-ABCB10/microRNA (miR)-29b-3p/vascular endothelial growth factor A (VEGFA) axis.
Materials and methods
Ethics statement
Study protocols were approved by The Ethics Committee of The School of Stomatology, Nanjing Medical University (approval no. PJ2013-037-001). The methodologies used in the present study were compliant with the Declaration of Helsinki and written informed consent was obtained from each participant.
Cell culture
hAMSCs were isolated from donated human placentas from full term pregnancies (from healthy pregnant females; length of pregnancy range 38-41 weeks) and identified, following a previously-described method (15). A total of three donors from the Obstetrics Department of Nanjing Maternal and Child Health Hospital Affiliated to Nanjing Medical University (Nanjing, Jiangsu, China), aged 24-27 (mean age, 25.3), were enrolled between February 2018 and March 2018 in this study. The cells were cultured in α-MEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/l penicillin and 100 mg/l streptomycin at 37˚C and 5% CO2. Cells at passages 3-5 were used in all experiments. HUVECs were purchased from China Infrastructure of Cell Line Resources and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/l penicillin and 100 mg/l streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C and 5% CO2.
Collection of CM
When hAMSCs had grown to 70-80% confluence in 15 cm plates, cells were washed three times with PBS and incubated at 37˚C and 5% CO2 with 15 ml serum-free α-MEM containing penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin) and 15 ml serum-free DMEM containing penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin) for 24 h. A 30 ml quantity of medium was collected after 24 h, centrifuged at 3,000 x g and 4˚C for 3 min followed by a further centrifugation for 5 min at 1,500 x g and 4˚C. Medium was then filtered through 0.45-µm filters (Merck KGaA) and stored at -80˚C for use as CM. For the collection of CM, cells from the three different donors were collected separately. Subsequently, three replicate experiments using CM from each donor were conducted (15), unless otherwise stated. Serum-free medium (containing 50% v/v DMEM, 50% v/v α-MEM, 100 U/ml penicillin and 100 µg/ml streptomycin) was used as control unconditioned medium.
Endothelial cell proliferation assay
The proliferation of HUVECs was determined using a Cell Counting Kit 8 (CCK-8) assay (Donjindo Molecular Technologies, Inc.), according to the manufacturer's instructions. HUVECs were initially seeded into 96-well plates at a density of 1x103 cells/well and incubated at 37˚C for 12 h. After synchronization with DMEM containing 2% FBS for 24 hat 37˚Cand 5%CO2, cells were co-cultured with a dilution of 50 or 80% v/v or 100% v/v CM in fresh control medium (containing 50% v/v DMEM, 50% v/vα-MEM, 100 U/ml penicillin and 100 µg/ml streptomycin) for 24 h. Cell proliferation was then measured at 450 nm using a microplate reader (BioTek Instruments, Inc.). All experimental results are presented as the means of three replicates performed under the same conditions.
Tube-formation assay
HUVECs were seeded onto 96-well plates coated with Matrigel reduced factor (50 µl per well at 37˚C for 1 h; Becton, Dickinson and Company), at a density of 3,000 cells/well and cultured for 4-6 h at 37˚C and 5% CO2 with hAMSC-CM (80% v/v CM) or control unconditioned medium, both supplemented with 1% FBS. Images of tube formation were obtained using an optical light microscope at 100X (Carl Zeiss, Inc.) and then five fields were randomly selected from each well for quantitative analysis of the total tube length with Image J software (version 1.8.0; http://imagej.nih.gov/ij/).
Wound healing assay and transwell migration
Prior to the wound healing assay, HUVECs cultured in DMEM containing 10% FBS were plated (1x105 cells per well in a 6-well plate) at the logarithmic growth phase until a monolayer was formed. A straight line was scraped with a 200 µl pipette tip to create a gap across the cell monolayer. The cell debris was then removed by washing with PBS three times. hAMSC-CM (80% v/v CM) containing 2% FBS or control unconditioned medium containing 2% FBS was added and HUVECs were allowed to proliferate for 16 h (use of 2% FBS is a potential limitation of this study, but this was necessary to maintain cell viability). Microscopic images (5 per well at labeled locations) were obtained at 0 and 16 h using an optical light microscope 40X objective lens (Carl Zeiss, Inc.). For each image, the sizes of the gaps were measured using Image J software (version 1.8.0; National Institutes of Health). For each group, five random locations within each gap were measured. Cell mobility was calculated as [width at 0 h-width at 16 h]/width at 0 h] x100.
For the Transwell migration assay, 8.0-µm pore culture inserts (24 well; Corning, Inc.) and HUVECs in log phase were prepared. A total of 1.5x104 cells/well were suspended in 200 µl control unconditioned medium without FBS and loaded into the upper chambers of the insert, and 600 µl hAMSC-CM or control unconditioned medium containing 10% FBS was added into the lower chambers. After incubation at 37˚C for 12 h, migrated cells that had attached to the lower surface of the filter were fixed at 25˚C in 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet at 25˚C for 1 h. Cells were counted and photographed in five random fields under an optical light microscope using a 100X objective lens (Carl Zeiss, Inc.). The migration assay was performed independently three times.
RNA interference, miRNA inhibitor and transfection
Small interfering RNAs (siRNAs) specific for circ-ABCB10 and miR-29b-3p inhibitor were purchased from Guangzhou RiboBio Co., Ltd. siRNA (50 nM) or inhibitor (100 nM) was transfected with Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After transfection for 48 h, circ-ABCB10 and VEGFA expression levels were assessed by reverse transcription-quantitative PCR (RT-qPCR). siRNA and inhibitor sequences are shown in Table SI.
Luciferase reporter assay
The potential binding sites of circ-ABCB10 and miR-29b-3p were determined using TargetScan version 7.1 (http://www.targetscan.org/vert_71) and the miRanda database (August 2010 release; http://www.microrna.org/microrna/home.do/). Vectors containing circ-ABCB10 sequences with the predicted miR-29b-3p binding sites or sequences mutated were purchased from Guangzhou RiboBio Co., Ltd. and named ‘circ-wild-type (WT)’ and ‘circ-mutant (Mut)’. Vectors were then co-infected with miR-29b-3p mimic or mimic negative control (NC) in 293T cells (purchased from China Infrastructure of Cell Line Resources). A dual-luciferase assay was then performedwith the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's instructions 24 h after the co-transfection. The activity of Renilla luciferase was used as an internal control, and the relative luciferase activity was calculated and presented as the ratio of firefly luciferase activity to Renilla luciferase activity. The predicted sequences are shown in Table SI.
Western blot analysis
CM or control unconditioned medium was removed when the cells had reached 80% confluence and the cells were washed three times with pre-chilled 1X PBS, before 300 µl RIPA buffer (Beyotime Institute of Biotechnology) was added to the cells to extract cellular protein. After incubation for 30 min on ice, lysed cells were transferred into a 1.5-ml centrifuge tube and centrifuged at 4˚C for 15 min at 10,656 x g. Protein quantification was performed using the bicinchoninic acid method and then loaded at 40 µg per lane. Protein was separated using 10% SDS-PAGE and electrotransferred to PVDF membranes (Merck KGaA). The membranes were incubated with primary antibody against VEGFA (1:200; cat. no. ab171828; Abcam) at room temperature for 1 h, or GAPDH (1:1,000; cat. no. ab8245; Abcam), AKT [1:1,000; cat. no. 4691; Cell Signaling Technology, Inc. (CST)], phosphorylated (p)-AKT (1:1,000; cat. no. 4060; CST), ERK1/2 (1:1,000; cat. no. 4695; CST) and p-ERK1/2 (1:1,000; cat. no. 4377; CST) primary antibodies at 4˚C overnight. Then, membranes were washed with 0.1% Tween-20 in TBS, and incubated with a secondary antibody (1:10,000; cat. no. ab150081; Abcam) conjugated to horseradish peroxidase at room temperature for 1 h. ECL detection was performed using Pierce ECL reagent (Thermo Fisher Scientific, Inc) and the densitometry carried out using Image Lab software (version 3.0; Bio-Rad Laboratories, Inc.).
Enzyme-linked immunosorbent assay (ELISA)
CM from hAMSCs was collected. HUVECs at 80% confluence were incubated at 37˚C with CM (80% v/v of hAMSC-CM in fresh control unconditioned medium) for 24 h, and then the medium was collected, which was referred to as ‘CM-aft’. To measure the changes in VEGFA protein expression levels in CM and CM-aft, an ELISA assay was performed with the human VEGF Quantikine ELISA kit (cat. no. DVE00; R&D Systems, Inc.) according to the manufacturer's protocol. Absorbance was detected at 450 nm. Both CM and CM-aft were transferred into ultra-filtration conical tubes (EMD Millipore) and concentrated to 100X. Then, both 100X CM and 100X CM-aft were used to extract total RNA.
RT-qPCR
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The quantity and quality of RNA was measured using Nanodrop 2000 spectrophotometry (Thermo Fisher Scientific, Inc.). Total RNA (1,000 ng) was reverse-transcribed to cDNA at 42˚C for 30 min using Prime Script RT master mix (Takara Bio, Inc.) and the TaqMan microRNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for the RT of miRNA with specific primers at 42˚C for 60 min and then 70˚C for 30 min. The level of RNA expression was detected using SYBR Green Master mix (Takara Bio, Inc.) using an ABI 7500 system (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The thermocycling conditionswere as follows: 95˚C for 5 min, before 40 cycles of 95˚C for 10 sec and 60˚C for 45 sec. Relative fold changes of RNA expression were calculated using the 2-ΔΔCq method (20). GAPDH or U6 were used as the internal standard controls. RT-qPCR analysis was independently performed three times. Primer sequences are shown in Table SI.
Statistical analysis
All data were collected and analyzed from three independent repeats. All image analysis was performed by a blinded observer. Data are presented as the mean ± SD, unless otherwise indicated. Data were analyzed using SPSS 20.0 software (IBM Corp.) to perform one-way ANOVA followed by Tukey's test, or a two-tailed Student's t-test (unpaired). P<0.05 was considered to indicate a statistically significant difference.
Results
hAMSC-CM enhances HUVEC proliferation, migration and tube formation
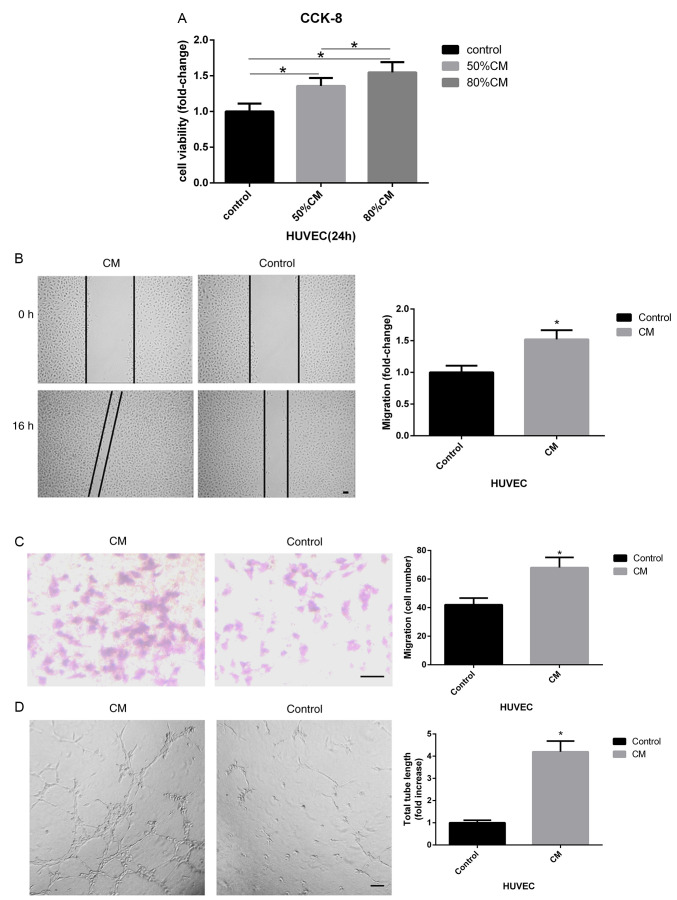
Dilutions of 50 and 80% v/v hAMSC-CM in fresh medium, referred to as ‘50% CM’ and ‘80% CM’, were used to investigate the effect of CM on HUVEC proliferation using a CCK-8 assay. The results suggested that 80% CM promoted the viability of HUVECs to a higher level compared with 50% CM (Fig. 1A). Therefore, 80% CM was used for all further experiments. The present study investigated the effects of hAMSC-CM on the migration of HUVECs during wound healing and in Transwell assays. The results indicated that significantly increased migratory ability of HUVECs was induced by hAMSC-CM (Fig. 1B and C). In addition, tube-formation assays were performed to investigate the pro-angiogenic role of hAMSC-CM on HUVECs. It was found that the total tube length formed by HUVECs was significantly higher when treated with hAMSC-CM compared with the control (Fig. 1D).
Figure 1.
hAMSC-CM increases the proliferation, migration and tube formation of HUVECs. (A) CCK-8 assay of HUVECs treated with different dilutions of hAMSC-CM. (B) Influence of hAMSC-CM on HUVEC migration was measured by wound healing assay. Scale bar, 100 µm. (C) Effect of hAMSC-CM on HUVEC migration was measured by Transwell assay. Scale bar, 100 µm. (D) Influence of hAMSC-CM on HUVEC angiogenesis was measured by tube formation assay. Scale bar, 200 µm. (E) Changes in mRNA expression levels that may be involved in the process of angiogenesis in HUVECs were measured by reverse transcription-quantitative PCR. (F) Protein expression levels of VEGFA, p-ERK1/2, ERK1/2, p-AKT and AKT in HUVECs were detected by western blotting. (G) VEGFA concentration in hAMSC-CM or hAMSC-CM treated with HUVECs for 24 h was assessed by ELISA. *P<0.05 vs. control unless otherwise indicated. HUVECs, human umbilical vein endothelial cells; hAMSCs, human amnion-derived mesenchymal stem cells; CCK-8, Cell Counting Kit-8;CM, conditioned medium; CM, hAMSC-CM; CM-aft, hAMSC-CM incubated with HUVECs for 24 h; VEGF, vascular endothelial growth factor; p-, phosphorylated; MSCs, mesenchymal stem cells; Ang-1, angiopoietin; HIF-1α, hypoxia-inducible factor 1-α; KDR, kinase insert domain receptor.
To investigate the potential molecular mechanism of hAMSC-CM-induced angiogenesis in HUVECs, the mRNA expression level of a number of genes that may be involved in angiogenesis was determined using RT-qPCR. After incubation with hAMSC-CM for 24 h, HUVECs showed a significantly increased mRNA expression level of VEGFA compared with that of the control group (Fig. 1E). This change in VEGFA expression was also identified at the protein level using western blotting. In addition, western blotting results identified increased levels of p-ERK1/2 but not p-AKT (Fig. 1F). MSCs can enhance tube formation in endothelial cells by secreting proteins such as VEGFA (21). Therefore, the present study hypothesized that a similar pattern could occur during tube formation in HUVECs treated with hAMSC-CM. However, ELISA results suggested that the concentration of VEGFA in hAMSC-CM was low, while the concentration of VEGFA in hAMSC-CM significantly increased after treating HUVECs for 24 h (Fig. 1G). It was demonstrated that the increased levels of VEGFA may have been secreted from HUVECs treated with hAMSC-CM and not from hAMSC-CM directly.
Collectively, the present results suggested that hAMSC-CM enhanced HUVEC proliferation, migration and tube formation, which may result from the induction of the VEGF signaling pathway.
circ-ABCB10 involvement in the pro-angiogenic role of hAMSC-CM
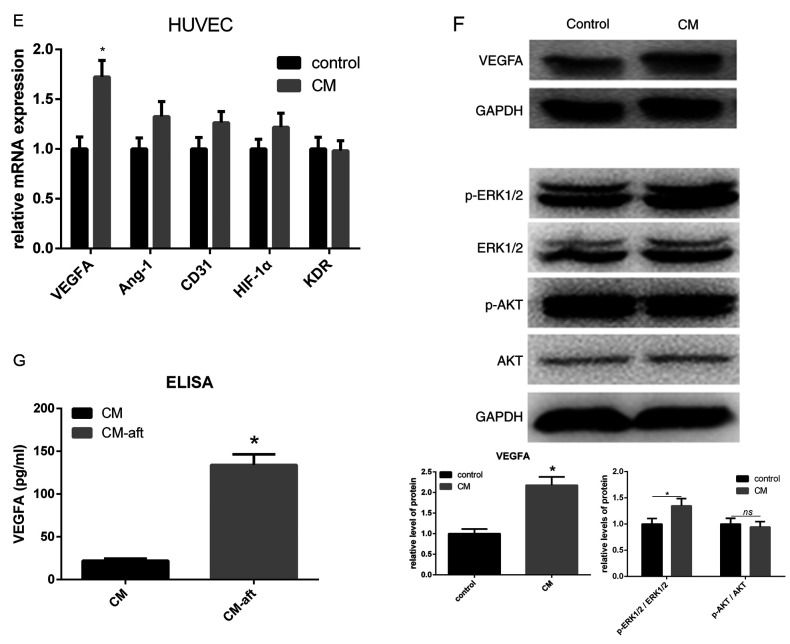
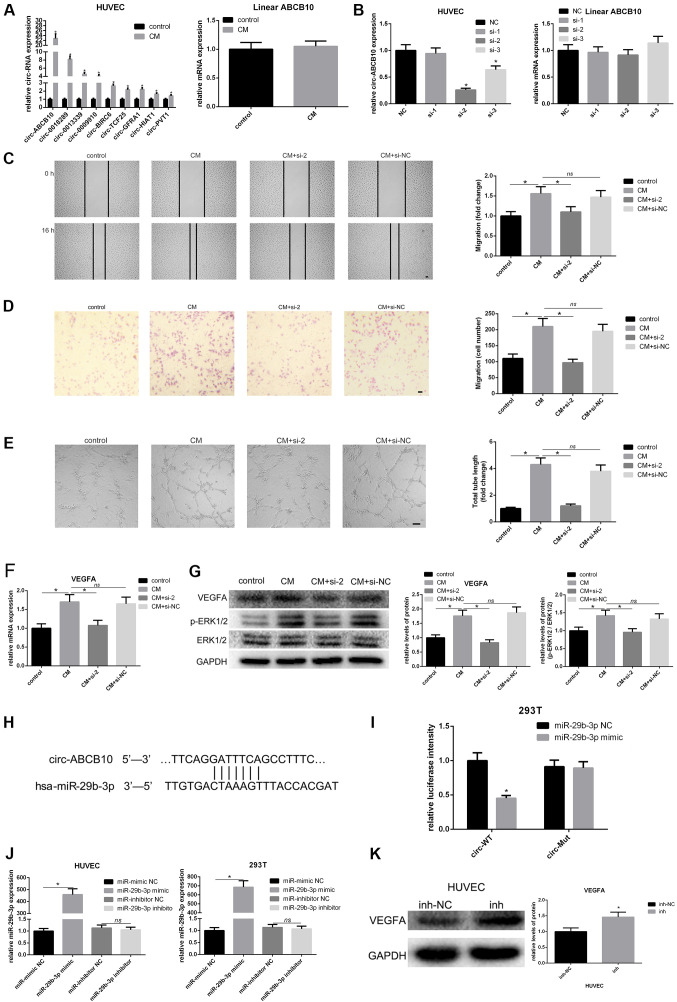
Previous studies showed that circRNAs may be involved in the process of angiogenesis during development or disease (19,22-26), including circ_002136, which plays an important role in regulating angiogenesis in glioma via the miR-138-5p/SOX13 axis (27). The present study investigated several circRNAs that may potentially regulate the process of angiogenesis. The present results suggested a number of circRNAs were upregulated in HUVECs after treatment with hAMSC-CM. Among them, circ-ABCB10 had the highest fold-change, while linear ABCB10 expression level was unchanged (Fig. 2A). To investigate the potential function of hAMSC-CM-induced circ-ABCB10 expression in HUVECs, three siRNAs, si-1, si-2 and si-3, were designed to specifically target the junction site of circ-ABCB10 in HUVECs. The present study examined the silencing efficiency of these three siRNAs and found si-2 had the best silencing capacity. Therefore, si-2 was used in further investigations (Fig. 2B). As expected, both scratch test and transwell assays in HUVECs treated with hAMSC-CM revealed a reduced migration in the si-2 group compared with the NC group (Fig. 2C and D). A tube formation assay was performed to investigate the impact on angiogenesis caused by silencing of circ-ABCB10 in HUVECs incubated with hAMSC-CM. Similarly, it was found that the si-2 group showed a significantly decreased total tube length compared with the NC group (Fig. 2E).
Figure 2.
hAMSC-CM-induces loss of function of circ-ABCB10 in HUVECs. (A) Changes in levels of circular RNAs that may be involved in angiogenesis in HUVECs were measured by RT-qPCR. circ-ABCB10 expression showed the highest increase, while linear ABCB10 remained unchanged. (B) Specific siRNAs targeting circ-ABCB10 were transfected into HUVECs, and the efficiency and expression of linear ABCB10 were measured by RT-qPCR. (C) Scratch test and (D) Transwell assays showed the effect of downregulation of circ-ABCB10 on HUVEC migration induced by hAMSC-CM. Scale bar, 100 µm. (E) Tube formation assay results showed that the effect of circ-ABCB10 downregulation on HUVEC angiogenesis was induced by hAMSC-CM. Scale bar, 200 µm. (F) RT-qPCR results showed changes in VEGFA mRNA expression levels in HUVECs induced by hAMSC-CM when circ-ABCB10 was downregulated (G) Western blotting results showed the corresponding changes in protein expression levels of VEGFA and p-ERK1/2. (H) Potential miR-29b-3p binding site in the 5' to 3' sequence of circ-ABCB10. (I) Dual-luciferase assay. HEK293T cells were co-transfected with hsa-miR-29b-3p or miR-NC and plasmid with wild-type or mutant circ-ABCB10. (J) The efficiency of transfection of miR-29b-3p. (K) The expression of VEGFA potentially targeted by miR-29b-3p in HUVEC transfected with inhibitor-NC or miR-29b-3p inhibitor was detected by western blot. *P<0.05 vs. control or NC, unless otherwise indicated. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; HUVECs, human umbilical vein endothelial cells; hAMSCs, human amnion-derived mesenchymal stem cells; CM, conditioned medium; VEGF, vascular endothelial growth factor; p-, phosphorylated; MSCs, mesenchymal stem cells; WT, wild-type; Mut, mutant; circ, circular RNA; NS, not significant.
The present study investigated whether circ-ABCB10 positively regulated VEGFA. RT-qPCR and western blotting results suggested that silencing of circ-ABCB10 significantly reduced the expression of VEGFA at both the mRNA and protein level in hAMSC-CM-treated HUVECs. In addition, activation of p-ERK1/2 showed the same effect (Fig. 2F and G). There might be a potential association between circ-ABCB10 and miR-29b-3p, which matched the hypothesis of ceRNAs predicted by the TargetScan and miRanda databases (Fig. 2H). A luciferase assay was performed to investigate whether circ-ABCB10 could sponge miR-29b-3p in vitro, and it was demonstrated that luciferase intensity was significantly decreased when miR-29b-3p mimic was co-transfected with WT circ-ABCB10 (Fig. 2I). In addition, the efficiency of transfection of miR-29b-3p mimic was detected (Fig. 2J). In order to confirm the transfection efficiency of miR-29b-3p mimic and inhibitor, qPCR was performed. As shown in Fig. 2J, miR-29b-3p mimic significantly increased miR-29b-3p expression, while miR-29b-3p inhibitor had no effect. Several studies have shown that the miRNA inhibitor, which binds to the mature miRNA and blocks its function in RNA-induced silencing complex (RISC), may not directly degrade the target miRNA (28-30). Functional experiments and detection of target genes are still reliable to evaluate the role of miRNA inhibitor (28-30). Notably, miR-29b-3p has been reported to target VEGF gene and inhibit its expression (31). Furthermore, it has been directly confirmed that miR-29b-3p could bind the mRNA of VEGFA in RISC by a recent study (32). Thus, the protein expression of VEGFA was determined by western blotting, following transfection with miR-29b-3p inhibitor. The results showed the miR-29b-3p inhibitor significantly increased the VEGFA expression (Fig. 2K). Thus, the circ-ABCB10/miR-29b-3p/VEGFA axis may be involved in the pro-angiogenic effects of hAMSC-CM on HUVECs.
circ-ABCB10/miR-29b-3p/VEGFA axis may be involved in the pro-angiogenic effects of hAMSC-CM on HUVECs
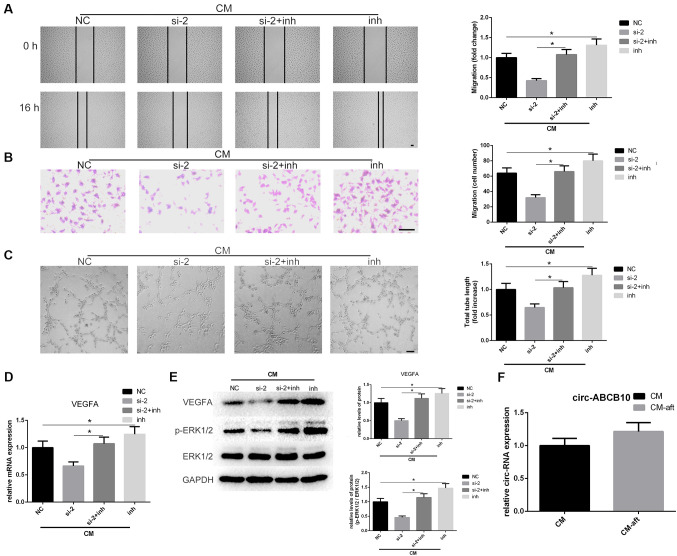
To investigate whether inhibiting miR-29b-3p can attenuate the effect caused by silencing circ-ABCB10 in hAMSC-CM-treated HUVECs, a series of functional experiments were performed. Wound healing and Transwell assays results suggested that co-transfection of si-2 and miR-29b-3p inhibitor significantly reversed the inhibition of migration caused by knockdown of circ-ABCB10 (Fig. 3A and B). Tube-formation assay results identified that the suppression of angiogenesis in hAMSC-CM-treated HUVECs, resulting from knockdown of circ-ABCB10, was rescued by co-transfection of si-2 and miR-29b-3p inhibitor (Fig. 3C). To understand the association between circ-ABCB10, miR-29b-3p and VEGFA, the present study investigated whether reduced expression of VEGFA caused by knockdown of circ-ABCB10 could be rescued by inhibiting miR-29b-3p in HUVECs. RT-qPCR and western blotting results suggested that the reduction of VEGFA caused by knockdown of circ-ABCB10 could be inhibited by co-transfecting si-2 and miR-29b-3p inhibitor. In addition, the activation of p-ERK1/2 showed a similar effect (Fig. 3D and E).
Figure 3.
Loss of miR-29b-3p rescues the effects caused by circ-ABCB10 downregulation in hAMSC-CM-treated HUVECs. (A) Scratch test and (B) Transwell assays results indicated that circ-ABCB10 downregulation on hAMSC-CM-induced HUVEC migration was largely rescued by a miR-29b-3p inhibitor. Scale bar, 100 µm. (C) Tube-formation assay results showed the effect of circ-ABCB10 downregulation on hAMSC-CM-induced HUVEC angiogenesis was significantly rescued by the miR-29b-3p inhibitor. Scale bar, 200 µm. (D) VEGFA mRNA expression levels were recovered when HUVECs were co-transfected with miR-29b-3p inhibitor. (E) VEGFA protein expression levels and ERK1/2 activation were rescued by miR-29b-3p inhibitor. (F) circ-ABCB10 expression levels in hAMSC-CM were unchanged after incubation with HUVECs for 24 h. *P<0.05. HUVECs, human umbilical vein endothelial cells; hAMSCs, human amnion-derived mesenchymal stem cells; CM, conditioned medium; CM, hAMSC-CM; CM-aft, hAMSC-CM incubated with HUVECs for 24 h; VEGF, vascular endothelial growth factor; p-, phosphorylated; MSCs, mesenchymal stem cells; siRNA, small interfering RNA; circ, circular RNA; inh, inhibitor.
The present study investigated whether increased circ-ABCB10 in HUVECs originated from hAMSC-CM. The expression levels of circ-ABCB10 in hAMSC-CM and hAMSC-CM incubated with HUVECs for 24 h, referred to as hAMSC-CM-aft, were analyzed. It was demonstrated that unlike the changes of expression of VEGFA, circ-ABCB10 expression levels between hAMSC-CM and hAMSC-CM-aft exhibited no statistically significant change (Fig. 3F). Thus, the present results suggested that hAMSC-CM may partially promote HUVEC migration and tube formation via the circ-ABCB10/miR-29b-3p/VEGFA axis.
Discussion
Angiogenesis is important in the process of therapeutic bone regeneration, as bone healing or remodeling requires oxygen and nutrients from blood vessels to maintain normal metabolism (2). Therapeutic angiogenesis induced by stem cells has been considered to be a potential solution to provide a more beneficial vascular niche during bone regeneration (33). Kim et al (12), found that hAMSCs exhibit angiogenesis, but the details of the molecular mechanisms involved in the pro-angiogenic process remain unknown. Previous studies have focused on the significant effect on angiogenesis in various stem cell types (5-8,34). MSCs can secrete multiple types of growth factors and cytokines such as VEGF, insulin-like growth factor 1 and hepatocyte growth factor in a paracrine manner (7,35). A previous study revealed promising findings regarding angiogenesis in a 3D contact co-culturing model of hAMSCs with HUVECs, which may be involved in hAMSC secretion of matrix metallopeptidases (15). The present study hypothesized that hAMSC-CM may promote angiogenesis of HUVECs in a non-contact co-culture system in a similar paracrine manner. The present results suggested a pro-angiogenic capacity of CM from hAMSCs on HUVECs, using a non-contact co-culture model in vitro. The present study investigated the molecular mechanism involved in this process. It was found that hAMSC-CM significantly promoted tube formation and migration of HUVECs with significant increases in circ-ABCB10 and VEGFA expression levels. In addition, the present results suggested silencing circ-ABCB10 in HUVECs in the non-contact co-culture model led to weakened induction of angiogenesis and reduced expression of VEGFA, following hAMSC-CM treatment of HUVECs. It was demonstrated that the transfection of miR-29b-3p inhibitor could largely rescue the weakened angiogenesis induced by hAMSC-CM treatment of HUVECs and decreased the expression of VEGFA caused by the knockdown of circ-ABCB10.
Stem cell-based therapy is an important strategy in the treatment of various diseases, especially angiogenesis-boosting therapies (5,6). The present results suggested increased proliferation, migration and tube formation of HUVECs following hAMSC-CM treatment. VEGFA expression level was increased in HUVECs cultured with hAMSC-CM. The present study examined whether hAMSC-CM delivered hAMSC-secreted VEGFA to HUVECs. ELISA assay results suggested that the protein expression level of VEGFA in hAMSC-CM was lower compared with hAMSC-CM incubated with HUVECs for 24 h. This effect may be attributed to a different manner of secretion in diverse stem cell types (35) or may be caused by mediation of upstream molecules regulating VEGFA signaling.
circRNAs are not just byproducts of transcription, but can also mediate cell proliferation, migration, invasion and apoptosis in both disease and development (16). circRNAs may have an important role in angiogenesis by acting as a miRNA sponge (36). In a recent study, circ-SHKBP1 was found to regulate the angiogenesis of glioma-exposed endothelial cells via interactions with miR-379 and miR-544a (19). Another previous study indicated that has_circ_0054633 protects HUVECs from the damage caused by high glucose by sponging miR-218 and upregulating the expression of roundabout homolog 1 and heme oxygenase-1(36). Therefore, the present study selected a number of circRNAs that may be involved in angiogenesis and detected expression changes in co-cultured HUVECs. Among them, circ-ABCB10 showed the greatest increase in expression and was predicted to sponge up miR-29b-3p targeting VEGFA. The present luciferase reporter assays and loss-of-function and rescue assay results demonstrated thatcirc-ABCB10 may increase the angiogenesis of HUVECs via the miR-29b-3p/VEGFA axis. The present results supported the hypothesis that hAMSC-CM may promote the angiogenesis of HUVECs by regulating molecules upstream of the VEGFA pathway.
Several circRNAs involved in biofunctions including proliferation and angiogenesis were found in cancer exosomes (37,38). A recent study indicated exosomal circ-IARS secreted from pancreatic cancer cells could mediate the permeability of HUVEC monolayers to enhance tumor metastasis (39). The present study investigated whether circ-ABCB10, which enhanced the angiogenesis of HUVECs, originates from exosomes released by hAMSCs. We thereby measured circ-ABCB10 expression levels in hAMSC-CM and hAMSC-CM incubated with HUVECs for 24 h. However, the expression levels of circ-ABCB10 in hAMSC-CM showed no statistically significant difference in levels in hAMSC-CM incubated with HUVECs for 24 h. Hence, it is possible that the increased expression of circ-ABCB10 was indirectly induced by hAMSC-CM, possibly by the promotion of circ-ABCB10 transcription rather than direct delivery by exosomes in hAMSC-CM. Whilst extensive study on the biofunction of circRNAs has been undertaken, the molecular mechanism associated with the transcription of circRNAs remains elusive. Previous studies focused on transcription factors regulating the transcription of circRNAs (40,41). Liang et al (40), found that hypoxia-inducible factor 1-α regulates the hypoxia-induced expression of circ-DENND4C in breast cancer cells. Another study showed that Twist1 promotes the expression of circular cullin 2 during epithelial-mesenchymal transition (41). Therefore, hAMSC-CM may contain secretory proteins that can interact with transcription factors to mediate the transcription of circ-ABCB10 in HUVECs. However, further studies are required to investigate this effect.
In summary, the present results suggested hAMSC-CM enhanced migration and tube formation of HUVECs via the circ-ABCB10/miR-29b-3p/VEGFA pathway. Thus, the present results could help to elucidate the molecular mechanism of hAMSC-induced angiogenesis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by The National Natural Science Foundation of China (grant no. 81670966) and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-87).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZT, MS and NC contributed to the study design and conception. ZT, JT, ZY and QZ performed the experiments. ZT and XY analyzed the data. ZT wrote the manuscript. NC and ZY revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Study protocols were approved by The Ethics Committee of the School of Stomatology, Nanjing Medical University (approval no. PJ2013-037-001). Written informed consent was obtained from each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hu N, Jiang D, Huang E, Liu X, Li R, Liang X, Kim SH, Chen X, Gao JL, Zhang H, et al. BMP9-regulated angiogenic signalling plays an important role in the osteogenic differentiation of mesenchymal progenitor cells. J Cell Sci. 2013;126:532–541. doi: 10.1242/jcs.114231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusumbe AP, Ramasamy SK, Adams RH. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507:376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanczler JM, Oreffo RO. Osteogenesis and angiogenesis: The potential for engineering bone. Eur Cell Mater. 2008;15:100–114. doi: 10.22203/ecm.v015a08. [DOI] [PubMed] [Google Scholar]
- 5.Daley GQ, Scadden DT. Prospects for stem cell-based therapy. Cell. 2008;132:544–548. doi: 10.1016/j.cell.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Satija NK, Singh VK, Verma YK, Gupta P, Sharma S, Afrin F, Sharma M, Sharma P, Tripathi RP, Gurudutta GU. Mesenchymal stem cell-based therapy: A new paradigm in regenerative medicine. J Cell Mol Med. 2009;13:4385–4402. doi: 10.1111/j.1582-4934.2009.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinnaird T, Stabile E, Burnett MS, Lee CW, Barr S, Fuchs S, Epstein SE. Marrow-derived stromal cells express genes encoding a broad spectrum of arteriogenic cytokines and promote in vitro and in vivo arteriogenesis through paracrine mechanisms. Circ Res. 2004;94:678–685. doi: 10.1161/01.RES.0000118601.37875.AC. [DOI] [PubMed] [Google Scholar]
- 8.Zuo S, Jones WK, Li H, He Z, Pasha Z, Yang Y, Wang Y, Fan GC, Ashraf M, Xu M. Paracrine effect of wnt11-overexpressing mesenchymal stem cells on ischemic injury. Stem Cells Dev. 2012;21:598–608. doi: 10.1089/scd.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soncini M, Vertua E, Gibelli L, Zorzi F, Denegri M, Albertini A, Wengler GS, Parolini O. Isolation and characterization of mesenchymal cells from human fetal membranes. J Tissue Eng Regen Med. 2007;1:296–305. doi: 10.1002/term.40. [DOI] [PubMed] [Google Scholar]
- 10.Santhagunam A, Dos Santos F, Madeira C, Salgueiro JB, Cabral JM. Isolation and ex vivo, expansion of synovial mesenchymal stromal cells for cartilage repair. Cytotherapy. 2014;16:440–453. doi: 10.1016/j.jcyt.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Ilancheran S, Moodley Y, Manuelpillai U. Human fetal membranes: A source of stem cells for tissue regeneration and repair? Placenta. 2009;30:2–10. doi: 10.1016/j.placenta.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Kim SW, Zhang HZ, Kim CE, An HS, Kim JM, Kim MH. Amniotic mesenchymal stem cells have robust angiogenic properties and are effective in treating hindlimb ischaemia. Cardiovasc Res. 2012;93:525–534. doi: 10.1093/cvr/cvr328. [DOI] [PubMed] [Google Scholar]
- 13.Parolini O, Alviano F, Bagnara GP, Bilic G, Bühring HJ, Evangelista M, Hennerbichler S, Liu B, Magatti M, Mao N, et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international workshop on placenta derived stem cells. Stem Cells. 2008;26:300–311. doi: 10.1634/stemcells.2007-0594. [DOI] [PubMed] [Google Scholar]
- 14.Leyva-Leyva M, Barrera L, López-Camarillo C, Arriaga-Pizano L, Orozco-Hoyuela G, Carrillo-Casas EM, Calderón-Pérez J, López-Díaz A, Hernandez-Aguilar F, González-Ramírez R, et al. Characterization of mesenchymal stem cell subpopulations from human amniotic membrane with dissimilar osteoblastic potential. Stem Cells Dev. 2012;22:1275–1287. doi: 10.1089/scd.2012.0359. [DOI] [PubMed] [Google Scholar]
- 15.Jiang F, Ma J, Liang Y, Niu Y, Chen N, Shen M. Amniotic mesenchymal stem cells can enhance angiogenic capacity via MMPs in vitro and in vivo. Biomed Res Int. 2015;2015(324014) doi: 10.1155/2015/324014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao T, Chen Q, Fu L, Guo J. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47:497–504. doi: 10.1111/hepr.12871. [DOI] [PubMed] [Google Scholar]
- 17.Hou LD, Zhang J. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30:1–6. doi: 10.1177/0394632016686985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang W, Ji M, He G, Yang L, Niu Z, Jian M, Wei Y, Ren L, Xu J. Silencing CDR1as inhibits colorectal cancer progression through regulating microrna-7. Onco Targets Ther. 2017;10:2045–2056. doi: 10.2147/OTT.S131597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, Cai H, Ma J, Liu L, Wang P, et al. Circ-SHKBP1 regulates the angiogenesis of u87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2008;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Fierro FA, O'Neal AJ, Beegle JR, Chávez MN, Peavy TR, Isseroff RR, Egaña JT. Hypoxic pre-conditioning increases the infiltration of endothelial cells into scaffolds for dermal regeneration pre-seeded with mesenchymal stem cells. Front Cell Dev Biol. 2015;3(68) doi: 10.3389/fcell.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 23.Gao YL, Zhang MY, Xu B, Han LJ, Lan SF, Chen J, Dong YJ, Cao LL. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8:86625–86633. doi: 10.18632/oncotarget.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, Liu Y. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495:189–196. doi: 10.1016/j.bbrc.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(11215) doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6(30919) doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, Ma J, Shao L, Wang D, Shen S, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. J Exp Clin Cancer Res. 2019;38(65) doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson DW, Bracken CP, Szubert JM, Goodall GJ. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One. 2013;8(e55214) doi: 10.1371/journal.pone.0055214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2(E98) doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W, Guo J, Gu R, Lei B, Ding X, Ma J, Xu G. MicroRNA-29b-3p inhibits cell proliferation and angiogenesis by targeting VEGFA and PDGFB in retinal microvascular endothelial cells. Mol Vis. 2020;26:64–75. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655–667. doi: 10.1111/jcmm.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren L, Ma D, Liu B, Li J, Chen J, Yang D, Gao P. Preparation of three-dimensional vascularized MSC cell sheet constructs for tissue regeneration. Biomed Res Int. 2014;2014(301279) doi: 10.1155/2014/301279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards SS, Zavala G, Prieto CP, Elliott M, Martínez S, Egaña JT, Egaña JT, Bono MR, Palma V. Functional analysis reveals angiogenic potential of human mesenchymal stem cells from Wharton's jelly in dermal regeneration. Angiogenesis. 2014;17:851–866. doi: 10.1007/s10456-014-9432-7. [DOI] [PubMed] [Google Scholar]
- 35.Ratajczak MZ, Kucia M, Jadczyk T, Greco NJ, Wojakowski W, Tendera M, Ratajczak J. Pivotal role of paracrine effects in stem cell therapies in regenerative medicine: Can we translate stem cell-secreted paracrine factors and microvesicles into better therapeutic strategies? Leukemia. 2012;26:1166–1173. doi: 10.1038/leu.2011.389. [DOI] [PubMed] [Google Scholar]
- 36.Pan L, Lian W, Zhang X, Han S, Cao C, Li X, Li M. Human circular RNA-0054633 regulates high glucose-induced vascular endothelial cell dysfunction through the microrna-218/roundabout 1 and microrna-218/heme oxygenase-1 axes. Int J Mol Med. 2018;42:597–606. doi: 10.3892/ijmm.2018.3625. [DOI] [PubMed] [Google Scholar]
- 37.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 38.Steinbichler TB, Dudás J, Riechelmann H, Skvortsova II. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–181. doi: 10.1016/j.semcancer.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumour metastasis. J Exp Clin Cancer Res. 2018;37(177) doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37:4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 41.Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, Qin Y, Zhang H, Gao WF, Lei YY, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78:4150–4162. doi: 10.1158/0008-5472.CAN-17-3009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.