Abstract
Purpose
While the association between the gut microbiome and the immune system has been studied in autoimmune disorders, little is known about ocular disease. Previously we reported that IRT5, a mixture of five probiotic strains, could suppress autoimmune dry eye. In this study, we investigated the mechanism by which IRT5 performs its immunomodulatory function in a mouse model of autoimmune dry eye.
Methods
NOD.B10.H2b mice were used as an autoimmune dry eye model. Either IRT5 or PBS was gavaged orally for 3 weeks, with or without 5 days of antibiotic pretreatment. The effects on clinical features, extraorbital lacrimal gland and spleen proteins, and fecal microbiota were analyzed.
Results
The ocular staining score was lower, and tear secretion was higher, in the IRT5-treated groups than in the PBS-treated groups. After IRT5 treatment, the downregulated lacrimal gland proteins were enriched in the biological processes of defense response and immune system process. The relative abundances of 33 operational taxonomic units were higher, and 53 were lower, in the feces of the IRT5-treated groups than in those of the PBS-treated groups. IRT5 administration without antibiotic pretreatment also showed immunomodulatory functions with increases in the Lactobacillus helveticus group and Lactobacillus hamsteri. Additional proteomic assays revealed a decrease of proteins related to antigen-presenting processes in the CD11b+ and CD11c+ cells of spleen in the IRT5-treated groups.
Conclusions
Changes in the gut microbiome after IRT5 treatment improved clinical manifestations in the autoimmune dry eye model via the downregulation of antigen-presenting processes in immune networks.
Keywords: IRT5, probiotics, autoimmune dry eye, gut microbiome, proteomics, antigen presentation, cornea
The human microbiome project and advancement in metagenomic analysis have revealed that the gut microbiome modulates human diseases by affecting the metabolism and both innate and adaptive immunity.1–5 Dry eye disease associated with Sjögren syndrome is a well-known autoimmune disease.6 Many autoimmune disease or metabolic syndromes, including Sjögren syndrome, may be affected by aberrant interaction between the immune system and gut microbiome.3–5,7,8 For example, a dysbiotic intestinal microbiome has been shown to affect autoimmune uveitis and dry eye.8–13
Whether probiotics exert beneficial effects on inflammatory or metabolic diseases has long been debated.14,15 Recent meta-analytic human studies reported favorable microbiome effects on clinical manifestations in diabetes, necrotizing enterocolitis, inflammatory bowel disease, and eczema but not in rheumatoid arthritis.16–20 There has also been a clinical trial reporting that administration of Bifidobacterium mixture may attenuate dry eye syndrome.21 IRT5 is a probiotic mixture of five strains that includes Bifidobacterium bifidum, Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus reuteri, and Streptococcus thermophilus.22 IRT5 exerts an anti-inflammatory effect in experimental autoimmune models of myasthenia gravis, colitis, and encephalomyelitis.22–24 Previously, we demonstrated the beneficial effect of IRT5 on the clinical manifestations of autoimmune uveitis and dry eye models but not on those of a corneal allotransplantation model.25
Although IRT5, especially B. bifidum, has been studied for its anti-inflammatory effects, the precise mechanism by which IRT5 improves dry eye is not known.26,27 A recent human study reported protein changes in the lacrimal and tear fluid of patients with dry eye.28 Therefore, to understand the pathophysiologic changes of the lacrimal gland and its role as a possible target organ during crosstalk between the gut microbiome and the immune system, we investigated the mechanism by which IRT5 probiotics alter the gut microbiota and the proteome of the extraorbital lacrimal glands in a mouse model of Sjögren syndrome.
Methods
Animals
Twelve-week-old male NOD.B10.H2b mice (from The Jackson Laboratory, Bar Harbor, ME, USA) were used as the autoimmune dry eye model (n = 35). Due to the poor breeding properties of NOD.B10.H2b mice, we conducted four repeated experiments focused on different analysis. The number of mice used in each experiment is as follows.
-
•
Experiment 1: Five mice in the PBS group and five mice in the IRT5 group were used for the clinical evaluation and proteomic analysis of the extraorbital lacrimal glands after antibiotic pretreatment.
-
•
Experiment 2: Four mice in the PBS group and five mice in the IRT5 group were used for the clinical evaluation and proteomic analysis of the lymph nodes and spleen after antibiotic pretreatment.
-
•
Experiment 3: Three mice in the PBS group and four mice in the IRT5 group were used for clinical evaluation and gut microbiome analysis after antibiotic pretreatment.
-
•
Experiment 4: Four mice in the PBS group and five mice in the IRT5 group were used for clinical evaluation and gut microbiome analysis without antibiotic pretreatment.
Mice were bred in a specific pathogen-free facility at the Biomedical Research Institute of Seoul National University Hospital (Seoul, Korea) and maintained at 22–24°C, relative humidity 55% ± 5%, with free access to food and water. All mice were managed in accordance with the Association for Research in Vision and Ophthalmology guidelines for the Use of Animals in Ophthalmic and Vision Research. The study was approved by the Institutional Animal Care and Use Committee of the Seoul National University Biomedical Research Institute (IAUCUC No. 17-0093-C1A0 and 18-0129-S1A0).
Antibiotics Pretreatment and IRT5 Treatment
Of the 35 mice, antibiotic pretreatment was performed on 26 and the remaining 9 were not pretreated.
In the pretreatment group, a cocktail of 1 g l−1 ampicillin, 500 mg l−1 vancomycin, and 1 g l−1 metronidazole (all from Sigma-Aldrich, St. Louis, MO, USA) was administered in drinking water for 5 days before the start of the treatment. The IRT5 probiotic powder containing 2 × 108 CFU g−1 of five strains (L. casei, L. acidophilus, L. reuteri, B. bifidum, and S. thermophilus) was kindly provided by Young-Tae Ahn (Korea Yakult Co., Giheung, Korea). Either PBS alone (n = 12) or 1 × 109 IRT5 probiotics (n = 14) in 300 µL PBS was gavaged orally once a day for 3 weeks.
In the non-pretreatment group, either PBS (n = 4) or IRT5 probiotics (n = 5) was gavaged orally once a day for 3 weeks.
Clinical Evaluation
Phenol red–impregnated cotton threads (FCI Ophthalmics, Pembroke, MA, USA) were inserted for 60 seconds into the lateral canthus of mice under anesthesia (anesthetized using a mixture of zoletil and xylazine at a ratio of 1:3). The amount of tear secreted was determined by measuring the wet length of the wet thread in millimeters. After instilling one drop of 3% Lissamine Green B (Sigma-Aldrich) to the lower lateral conjunctival sac, corneal epithelial defect was scored in a blinded manner as follows: 0 if no punctuate staining was observed, 1 if less than one-third of the cornea was staining, 2 if two-thirds or less was stained, and 3 if more than two-thirds of the cornea was stained.25,29
Proteomics Sample Preparation
Lacrimal gland samples from NOD mice treated with PBS (control, n = 5) or IRT5 (n = 5) after antibiotic pretreatment were individually pulverized using a Cryoprep device (CP02; Covaris, Inc., Woburn, MA, USA). The pulverized tissue powder was sonicated in lysis buffer (4% SDS, 0.1M Tris-HCl pH 7.6, 1× Halt protease inhibitor cocktail [Hoffmann-La Roche AG, Basel, Switzerland] in 10 mL). The homogenate was centrifuged at 16,000 × g and 20°C for 10 minutes, and the supernatant was collected for protein digestion. The protein concentration was measured using a bicinchoninic acid (BCA) protein assay (BCA Protein Assay Kit; Thermo Fisher Scientific, San Jose, CA, USA). One hundred micrograms of proteins from each tissue was digested using a filter-aided sample preparation (FASP) method following published instructions.30 The peptides were labeled with TMT 10-plex reagent (Thermo Fisher, Waltham, MA, USA), according to the manufacturer's protocol. The five peptide samples from the PBS-treated mice were labeled as 126, 127N, 127C, 128N, and 128C, and the five from the IRT5 mice were labeled as 129N, 129C, 130N, 130C, and 131. The labeled peptides were pooled and fractionated into 12 fractions using high-pH reversed-phase fractionation. The peptides from the 12 fractions were dried and desalted using a C18 spin column (Thermo Fisher).
The spleens of the NOD mice treated with PBS (control, n = 4) or IRT5 (n = 5) after antibiotics pretreatment were cut in half. The CD11b+ cells were sorted in one half and the CD11c+ cells were sorted in the other half. The cervical and mesenteric lymph nodes of each mouse were pooled and only CD3+ cells were obtained. After cell sorting, the cells from each mouse were pooled and proteomic sample preparation was carried out. The cells were lysed using the same lysis buffer mentioned above and centrifuged for 10 minutes at 16,000 × g in order to acquire the protein supernatant. The protein concentration of each cell was measured using BCA and 50 µg protein was digested using FASP digestion. The digested peptides were labeled with TMT 6-plex reagent (Thermo Fisher) and pooled into one sample (126: IRT5 lymph node CD3+, 127: IRT5 spleen CD11b+, 128: IRT5 spleen CD11c+, 129: PBS lymph node CD3+, 130: PBS spleen CD11b+, and 131: PBS spleen CD11c+). The pooled sample was fractionated into 12 fractions and each fraction was desalted and analyzed using liquid chromatography–mass spectrometry (LC-MS) analysis.
LC-MS/MS Analysis
The desalted peptides were resuspended in 0.1% formic acid in water and injected into a Q Exactive orbitrap hybrid mass spectrometer (Thermo Fisher) coupled with an Easy-nLC 1000. The peptides (1 µg) were loaded onto a trap column (2 cm × 75 µm i.d. packed with 2 µm C18) and an analytical column (70 cm × 75 µm i.d. packed with 3 µm C18). A 180-minute gradient with a flow rate of 0.45 µL/min separated the peptides depending on linear acetonitrile (ACN) gradient (changing from 5% to 40% solvent B in 150 minutes from 40% to 80% solvent B in 5 minutes, holding at 80% solvent B for 10 minutes, and equilibrating the column with 5% solvent B for 15 minutes). A data-dependent scan was used, and the top 12 peaks were selected and isolated for fragmentation. The resolution of the complete MS was 70,000 and that of the MS/MS was 17,500 for the TMT 6-plex and 35,000 to distinguish TMT 10-plex mass. Precursor ions were fragmented using a normalized collision energy of 30. The dynamic exclusion was set to 30 seconds.
Proteomics Data Analysis
Raw data from the MS were processed using postexperiment monoisotopic mass refinement to increase sensitivity in peptide identification by selecting unique mass class. Refined data were analyzed using Proteome Discoverer 2.2 (Thermo Fisher). Seqeust HT and the Uniprot mouse reference were used (released in August 2018). For strict peptide identification, 0.01 false discovery rate (FDR) was applied as the peptide level. In addition, more than one unique peptide was always used to identify a protein. To quantify the protein ratio, only proteins that had more than two unique peptides were selected and quantified using reporter ion intensities. As the sum of the ion intensities of the reporter ions from each plex should be identical, all the peptide intensities were normalized to the total reporter ion intensity. Gene ontology analysis of differentially expressed proteins (DEPs) was performed to understand the biological functions of the DEPs in the mouse model. Biological process, cellular components, molecular function, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were validated using DAVID bioinformatics resources. A cutoff of ≤0.05 was applied, and the STRING database was used for interpreting protein interactions. Expression changes were weighed based on degree and betweenness centrality, which reflects the amount of control that a node exerts over the interactions of other nodes in the network.
Fecal Microbiota 16S Ribosomal RNA Analysis
Fecal pellets were collected directly from the anus of each mouse by holding the mouse in one hand while allowing it to defecate into a sterile tube. The collected feces were immediately stored at –80°C. For the analysis of bacteria in the feces, PCR amplification was performed using extracted DNA and the bacterial PCR primers 341F (5′-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3′; underlined sequence indicates the target region primer) and 805R (5′-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGACAG-GACTACHVGGGTATCTAATCC-3′) targeting the V3 to V4 regions of the 16S ribosomal RNA (rRNA) gene. The reaction conditions were as follows: 3 minutes of initial denaturation at 95°C, 25 cycles of 30-second denaturation at 95°C, 30-second primer annealing at 55°C, 30-second elongation at 72°C, and a final extension at 72°C for 5 minutes. Secondary amplification was performed using the i5 forward primer (5′-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXXX-TCGTCGGCAGCGTC-3′; X indicates the barcode region) and i7 reverse primer (5′-CAAGCAGAAGACGGCATACGAGAT-XXXXXXXX-GTCTCGTGGGCTCGG-3′) for attaching the Illumina NexTera barcode. The secondary amplification conditions were identical to those of the first amplification except that the number of amplification cycles was set to eight. The amplified products were confirmed using gel electrophoresis on 1.0% agarose gel and visualized using a Gel Doc system (BioRad, Hercules, CA, USA). CleanPCR (CleanNA) was used for purifying the amplified products. The same concentrations of purified product were pooled and short fragments were removed using CleanPCR (nontarget product). The product size and quality were evaluated on a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) using a DNA 7500 chip. Mixed amplicons were pooled and sequenced using an Illumina MiSeq Sequencing system (Illumina, Inc., San Diego, CA, USA) at ChunLab, Inc. (Seoul, Korea) according to the manufacturer's instruction. Raw read processing began with quality checking and filtering of low-quality (<Q25) reads by Trimmomatic 0.32.31 Paired-end sequence data were merged using PANDAseq after passing the quality control.32 Then, the primers were trimmed with ChunLab's in-house program at a similarity cutoff of 0.8. HMMER's hmmsearch program was used to detect nonspecific amplicons that do not encode 16S rRNA.33 The sequence was denoised using DUDE-Seq and nonredundant reads extracted with UCLUST-clustering.34,35 After the EzBioCloud database was searched for taxonomic assignment using USEARCH (8.1.1861_i86linux32), more precise pairwise alignment was performed.35,36 To detect chimeras in reads with <97% best hit similarity rates, UCHIME and the nonchimeric 16S rRNA database from EzBioCloud were used.37 Reads that were not identified to the species level (with <97% similarity) in the EzBioCloud database were compiled and UCLUST was used for de novo clustering to generate additional Operational Taxonomic Unit (OTUs).35 OTUs with single reads were omitted from further analysis.
Sequence Analysis
We clustered the sequences and grouped them as OTUs. BIOiPLUG software (ChunLab) was used to summarize the OTU data and to calculate the microbial alpha and beta diversity, and a linear discriminant analysis (LDA) of the effect size (LEfSe) was determined for each sample. Alpha diversity is an analysis of the diversity within a community, including the Chao1 index, Shannon index, and phylogenetic diversity index. The microbial beta diversity was compared using Bray-Curtis and UniFrac. An LEfSe and the Kruskal-Wallis test were used to estimate the effect of abundance in each sample on the effect of differences and to identify the bacterial taxa that showed significant differences in their demarcation. Only those taxa that showed a P value <0.05 and a log LDA score ≥2 were ultimately considered.38 The multiple test corrections were based on the FDR. An FDR of 0.05 was used as a statistically significant cutoff.
Statistical Analyses
GraphPad Prism software, Version 7.04 (GraphPad Prism, Inc., San Diego, CA, USA) was used. For comparing the two groups, independent t-tests or nonparametric Mann-Whitney U tests were used. To compare changes over time, Wilcoxon matched-pairs signed rank tests were used. The Shapiro-Wilk test was used for testing the normality of the data. Differences were considered statistically significant at P < 0.05.
Results
IRT5 Treatment Improves Clinical Manifestations in an Autoimmune Dry Eye Model
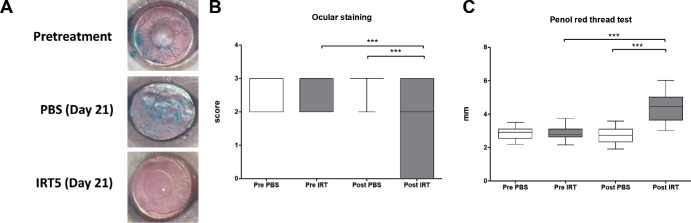
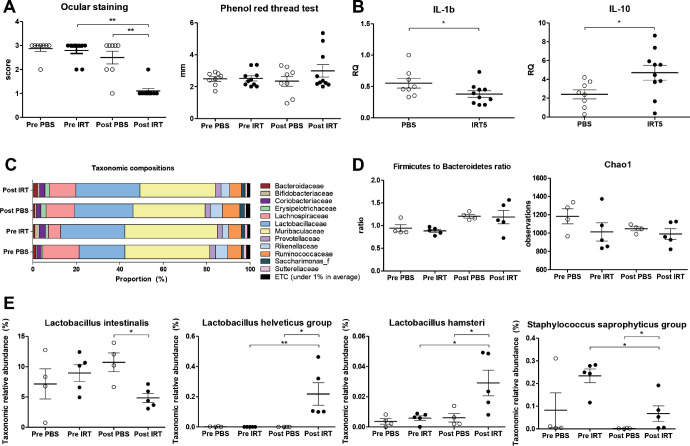
Compared to the pretreatment group (P < 0.0001, Wilcoxon matched-pairs signed rank test) and the PBS treatment group (P < 0.0001, Mann-Whitney U test), 3 weeks of treatment with IRT5 significantly decreased the ocular staining score (Figs. 1A, 1B). Tear secretion also increased after IRT5 treatment compared with the pretreatment group (P < 0.0001, Wilcoxon matched-pairs signed rank test) or PBS treatment group (P < 0.0001, Mann-Whitney U test, Fig. 1C).
Figure 1.
Clinical manifestations in the mouse cornea. (A) Representative images of mouse cornea before and after the PBS and IRT5 treatment. Lissamine Green B was used to visualize the wounded areas of the mouse corneas. (B) The ocular staining score of the IRT5-treated group (n = 14, 28 eyes) was significantly lower than that of the PBS-treated group (n = 12, 24 eyes). (C) Tear secretion in the IRT5-treated group was significantly higher than that of the PBS-treated group. ****P < 0.0001 (data presented as box-and-whisker plot with median).
IRT5 Probiotics Change Immunomodulatory or Ionic Transport-Related Protein Expression in the Extraorbital Glands
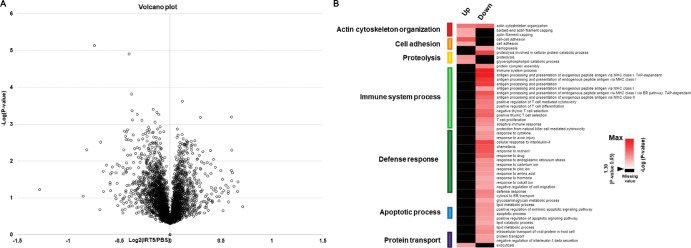
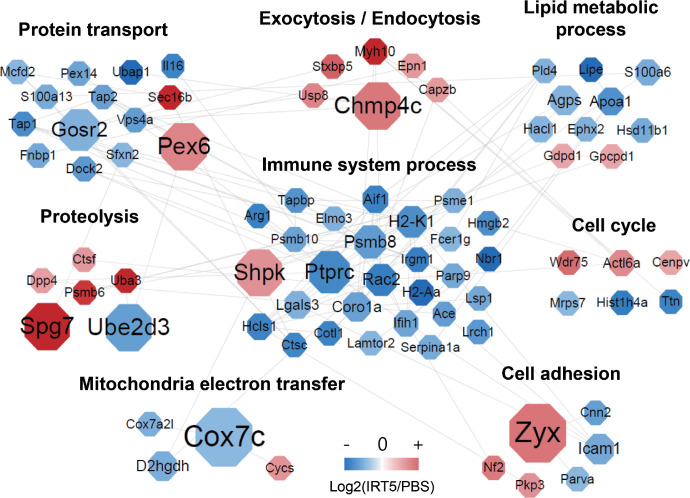
Quantitative proteome analysis identified a total of 5379 proteins in the extraorbital glands with a FDR of less than 1% at peptide spectrum match levels (Supplementary Table S1). Among the identified proteins, 202 proteins were selected as DEPs (changes in expression ≥1.15-fold of the reporter ion intensity using Student's t-test; P < 0.05) (Fig. 2A, Supplementary Table S2). To gain insight into the functional roles of DEPs in the effects of ITR5 treatment, a comparison of the gene ontology (GO) of the biological processes (BPs) of the DEPs was performed. A heatmap of GOBP enrichment analysis showed significantly represented GOBP terms (P < 0.05) for DEPs in the IRT5-treated group compared with the PBS-treated group. Proteins related to immune system process and defense response were significantly downregulated and proteins associated with actin cytoskeleton organization, cell adhesion, and proteolysis were significantly upregulated (Fig. 2B). To understand the changes in the maps of the cellular networks in the lacrimal glands after IRT5 treatment, we constructed network models using the DEPs from the lacrimal glands. We grouped the network proteins into eight modules, including protein transport, exocytosis/endocytosis, lipid metabolic process, proteolysis, immune system process, cell cycle, mitochondria electron transfer, and cell adhesion based on the GO biological processes and KEGG pathways (Fig. 3).
Figure 2.
Volcano plot of total protein identified in the extraorbital lacrimal glands after the PBS (n = 5) and IRT5 (n = 5) treatment and gene ontology analysis of 202 DEPs. (A) Volcano plot displaying all proteins that were identified by at least two unique peptides. (B) The heat map of the GOBP enrichment analysis showed significantly represented GOBP terms (P < 0.05) in the IRT5-treated group (only biological processes that satisfy P < 0.05 were included). Proteins associated with immune system process were significantly downregulated and proteins related to actin cytoskeleton organization, cell adhesion, and proteolysis were upregulated.
Figure 3.
Protein network showing the proteomic changes after the PBS (n = 5) and IRT5 (n = 5) treatment. Protein interaction was schematized as a network using STRING and the Cytoscape program.
Among these DEPs, Table 1 lists the proteins that exhibited high betweenness centrality (the top 10% of protein-protein interactions). The proteins that possessed the high betweenness centrality (>0.5, the top 7% of protein-protein interactions in decreasing order) were mitochondrial cytochrome c oxidase subunit 7C (Cox7c), zyxin (Zyx), charged multivesicular body protein 4c (Chmp4c), peroxisome assembly factor 2 (Pex2), paraplegin (Spg7), ubiquitin-conjugating enzyme E2 D3 (Ube2d3), sedoheptulokinase (Shpk), receptor-type tyrosine-protein phosphatase C (PTPRC), and Golgi SNAP receptor complex member 2 (Gosr2). Among these, Shpk and Ptprc are enriched in immune system process with high betweenness centrality in this network model. Additionally, Pex6, Gosr2, protein transport protein Sec16B (Sec16b), antigen peptide transporter 2 (TAP2), and antigen peptide transporter 1 (TAP1) are grouped in protein transport forming a close network with immune system process within the protein-protein network model (Fig. 3).
Table 1.
Differentially Expressed Proteins that Have High Betweenness Centrality
| Gene | Protein | Betweenness | ||
|---|---|---|---|---|
| Name | Name | Centrality | Fold Change | P Value |
| Cox7c | Cytochrome c oxidase subunit 7C, mitochondrial | 1.000 | 0.870 | 0.001 |
| Zyx | Zyxin | 1.000 | 1.269 | 0.022 |
| Chmp4c | Charged multivesicular body protein 4c | 0.667 | 1.261 | 0.012 |
| Pex6 | Peroxisome assembly factor 2 | 0.667 | 1.229 | 0.016 |
| Spg7 | Paraplegin | 0.667 | 1.544 | 0.049 |
| Ube2d3 | Ubiquitin-conjugating enzyme E2 D3 | 0.667 | 0.813 | 0.045 |
| Shpk | Sedoheptulokinase | 0.650 | 1.197 | 0.041 |
| Ptprc | Receptor-type tyrosine-protein phosphatase C | 0.521 | 0.757 | 0.000 |
| Gosr2 | Golgi SNAP receptor complex member 2 | 0.500 | 0.855 | 0.003 |
| Psmb8 | Proteasome subunit beta type-8 | 0.254 | 0.805 | 0.008 |
| Rac2 | Ras-related C3 botulinum toxin substrate 2 | 0.221 | 0.728 | 0.005 |
| H2-K1 | H-2 class I histocompatibility antigen, K-B alpha chain | 0.215 | 0.772 | 0.000 |
| Icam1 | Intercellular adhesion molecule 1 | 0.171 | 0.835 | 0.025 |
| Agps | Alkyldihydroxyacetonephosphate synthase, peroxisomal | 0.150 | 0.858 | 0.009 |
| D2hgdh | D-2-hydroxyglutarate dehydrogenase, mitochondrial | 0.150 | 0.845 | 0.047 |
*> 0.05 P value, >1.2- or <0.83-fold change, >0.1 betweenness centrality cutoff was applied.
Among the DEPs, proteins that have been reported to be associated with dry eye syndrome, Sjögren syndrome, or autoimmune disease through modulating immunity are shown in Table 2. Ptprc, which plays an important role in T-cell activation and B-cell proliferation in the MRL/MpJ-lpr mouse model, was downregulated and clustered in immune system process after IRT5 treatment. High mobility group protein B2 (Hmgb2), which is reportedly highly expressed in Sjögren syndrome, was downregulated after IRT5 treatment. These proteins are mostly involved in immune system process and may be significant in restoring the functioning of the lacrimal gland after IRT5 treatment. Although its relationship with dry eye is not clear, Cox 7c, one of the terminal oxidases of mitochondrial electron transfer, was also downregulated after IRT5 treatment.
Table 2.
DEPs Related to Dry Eye, Sjögren Syndrome, or Autoimmune Disease Through Immune Modulation
| Biological Process | Protein Name | Gene Name | Fold Changes |
|---|---|---|---|
| Immune response | Receptor-type tyrosine-protein phosphatase C | Ptprc | 0.757 |
| High mobility group protein B2 | Hmgb2 | 0.774 | |
| Proteasome subunit beta type-8 | Psmb8 | 0.805 | |
| H-2 class II histocompatibility antigen, A-B alpha chain | H2-Aa | 0.666 | |
| H-2 class I histocompatibility antigen, K-B alpha chain | H2-K1 | 0.772 | |
| Proteasome activator complex subunit 1 | Psme1 | 0.853 | |
| Antigen peptide transporter 1 | Tap1 | 0.765 | |
| Antigen peptide transporter 2 | Tap2 | 0.815 | |
| Proteasome subunit beta type 9 | Psmb9 | 0.770 | |
| Cell adhesion | Intercellular adhesion molecule 1 | Icam1 | 0.835 |
*> 0.05 P value, >1.2- or <0.83-fold change was applied for enriching the DEPs.
Treatment with IRT5 Probiotics Changes Gut Microbiome Composition
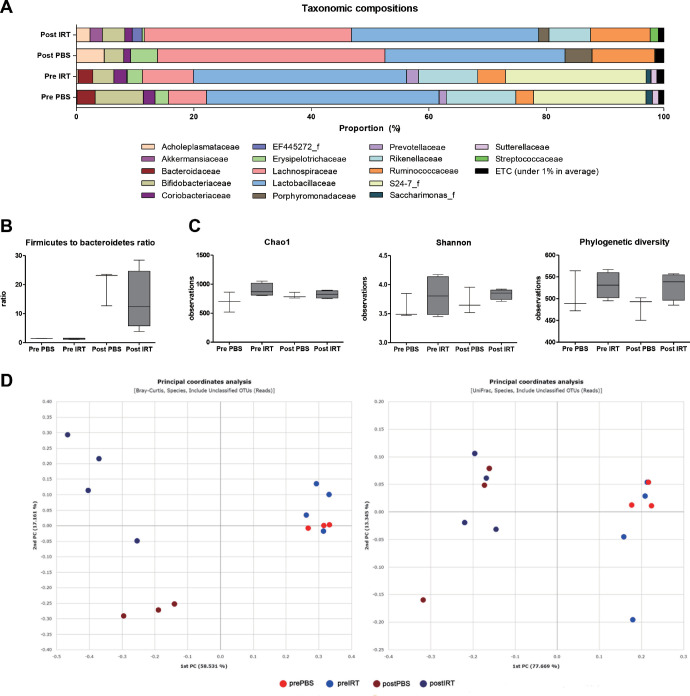
To promote the survival of IRT5 probiotics in the gut, oral antibiotics were applied for 5 days prior to treatment in 26 mice in both the PBS (n = 12) and IRT5 (n = 14) groups. Of the 26 mice, 3 mice from the PBS group and 4 from the IRT5 group underwent gut microbiome analysis. The average taxonomic composition changed in both groups (Fig. 4A, Supplementary Table S3). The Firmicutes to Bacteroidetes ratio was not significantly different between the two groups, and there was also no difference between pre- and posttreatment samples (Fig. 4B). There were no significant differences in the alpha diversity analyzed using the Chao1 index, Shannon index, or phylogenetic diversity index (Fig. 4C). The separation between the IRT5 and PBS groups was observed using a Bray-Curtis principal coordinate analysis (Fig. 4D).
Figure 4.
Gut microbiome changes after the PBS (n = 3) and IRT5 (n = 4) treatment with antibiotic pretreatment. (A) Average taxonomic composition at the family level before and after the PBS and IRT5 treatment. (B) The Firmicutes to Bacteroidetes ratio was not significantly different between the two groups. (C) Alpha diversity analyzed using the Chao1 index, Shannon index, and phylogenetic diversity index. No significant differences were observed between before and after the PBS and IRT5 treatments. (D) Scatterplot showing the first principal coordinate (PC) versus the second PC using Bray-Curtis and UniFrac analysis. Percentages shown are the percentages of variation explained by the components.
LEfSe analysis and Kruskal-Wallis tests were performed to compare the differences in gut microbiota pre- and posttreatment and between the IRT5 and PBS groups. In the IRT5-treated group, the taxonomic relative abundance of 33 different OTUs, including a Lactobacillus helveticus group, Lactobacillus hamsteri, a L. reuteri group, a L. casei group, a Lactobacillus brantae group, a Lactobacillus amylovorous group, Akkermansiamuciniphila, an Aerococcusviridans group, B. bifidum, and a Streptococcus salivarius group, was significantly higher than in the PBS-treated group. In contrast, the relative abundance of 53 OTUs, including an Escherichia coli group, was significantly lower in the IRT5-treated group than in the PBS-treated group (Supplementary Table S4).
IRT5 Probiotics Treatment without Antibiotic Pretreatment Also Improves Autoimmune Dry Eye and Modifies Gut Microbiome Composition
To rule out the effect of antibiotics, we conducted an additional experiment administering IRT5 probiotics or PBS without pretreatment with antibiotics. After IRT5 treatment without the antibiotic pretreatment, the ocular staining scores significantly decreased compared with the pretreatment group (P = 0.0042, Wilcoxon matched-pairs signed rank test) and the PBS treatment group (P = 0.0011, Mann-Whitney U test). However, tear secretion was not significantly different in the IRT5-treated group when compared with the PBS-treated group (P = 0.6334, Mann-Whitney U test) or the pretreatment group (P = 0.5566, Wilcoxon matched-pairs signed rank test) (Fig. 5A).
Figure 5.
Effect of the IRT5 treatment without antibiotics pretreatment in an autoimmune dry eye mouse model. (A) Ocular staining score was significantly lower in the IRT5-treated group (n = 5, 10 eyes) than in the PBS-treated group (n = 4, 8 eyes). However, the phenol red thread test results were not significantly changed after IRT5 treatment. (B) In the conjunctiva and cornea of the IRT5-treated group, IL-1β was significantly lower (P = 0.0434) and IL-10 was significantly higher (P = 0.0343) than in the PBS-treated group. (C) Average taxonomic composition at the family level before and after the PBS (n = 4) and IRT5 (n = 5) treatment is shown. (D) The Firmicutes to Bacteroidetes ratio and alpha diversity analyzed using the Chao1 index were not significantly different between the two groups. (E) Compared to the PBS-treated group, Lactobacillus intestinalis was lower, and the Lactobacillus helveticus group, Lactobacillus hamsteri, and the Staphylococcus saprophyticus group were higher in the IRT5-treated group.
The IL-10 was significantly higher (P = 0.0343, Mann-Whitney U test) and the IL-1β significantly lower (P = 0.0434, Mann-Whitney U test) in the conjunctiva and cornea of the IRT5-treated group (Fig. 5B).
The averaged taxonomic composition changed in both groups (Fig. 5C). Neither the Firmicutes to Bacteroidetes ratio nor alpha diversity showed any significant differences between the IRT5-treated group and either the pretreatment group or the PBS-treated group (Fig. 5D). Including the L. helveticus group, L. hamsteri, and the Staphylococcus saprophyticus group, the proportion of 12 OTUs was increased in the IRT5-treated group compared with the PBS treated group. In contrast, 15 OTUs, including Lactobacillus intestinalis, were decreased in the IRT5-treated group compared with the PBS-treated group (Fig. 5E, Table 3).
Table 3.
Differences in Abundance of Bacterial Communities as Assessed by LEfSe
| Taxon Name | Post PBS | Post IRT | LDA Effect Size | q Value |
|---|---|---|---|---|
| Increased | ||||
| PAC001071_s | 1.40115 | 2.54230 | 3.75640 | 0.01452 |
| PAC001112_s | 1.14062 | 2.00330 | 3.63493 | 0.01453 |
| PAC001118_s | 0.00739 | 0.69573 | 3.53691 | 0.02864 |
| Lactobacillus helveticus group | 0.00000 | 0.21799 | 3.03857 | 0.01054 |
| PAC002441_s | 0.25844 | 0.38583 | 2.80774 | 0.01466 |
| PAC001244_s | 0.00341 | 0.12922 | 2.80012 | 0.01401 |
| Staphylococcus saprophyticus group | 0.00213 | 0.06748 | 2.51554 | 0.02861 |
| EU511112_s | 0.00487 | 0.05132 | 2.37711 | 0.02846 |
| PAC001550_s | 0.01650 | 0.05921 | 2.34357 | 0.01461 |
| PAC001370_s | 0.00000 | 0.00775 | 2.12212 | 0.01056 |
| Lactobacillus hamsteri | 0.00604 | 0.02909 | 2.10369 | 0.02840 |
| PAC001549_s | 0.01097 | 0.02841 | 2.01551 | 0.01459 |
| Decreased | ||||
| Lactobacillus intestinalis | 10.72607 | 4.84257 | 4.46862 | 0.02843 |
| PAC001128_s | 0.58433 | 0.33275 | 3.10076 | 0.02849 |
| PAC001121_s group | 0.25399 | 0.01503 | 3.07787 | 0.01400 |
| PAC001554_s | 0.22915 | 0.08702 | 2.85228 | 0.01462 |
| PAC001560_s | 0.14342 | 0.01524 | 2.80823 | 0.01464 |
| PAC002039_s | 0.13548 | 0.00990 | 2.79886 | 0.01404 |
| PAC002419_s | 0.12160 | 0.05311 | 2.53967 | 0.02855 |
| PAC001586_s | 0.05931 | 0.00502 | 2.43771 | 0.02562 |
| PAC001235_s | 0.08000 | 0.02974 | 2.41812 | 0.01456 |
| PAC002314_s | 0.04626 | 0.00175 | 2.35018 | 0.02568 |
| PAC001588_s | 0.05335 | 0.01130 | 2.33173 | 0.02565 |
| PAC001402_s | 0.05175 | 0.01300 | 2.29307 | 0.01458 |
| PAC001543_s | 0.03263 | 0.00800 | 2.12408 | 0.05199 |
| PAC001536_s | 0.02481 | 0.00000 | 2.10187 | 0.03048 |
| PAC001772_s | 0.02204 | 0.00468 | 2.07747 | 0.01403 |
IRT5 Treatment Downregulates Antigen Presentation by the Immune Cells
Notably, the IRT5 and PBS treatment showed more differences in the CD11c+ and CD11b+ cells of the spleen than in the CD3+ cells of the lymph node, and the splenic CD11c+ cells showed the largest difference between the PBS treatment group and the IRT5 treatment group (Supplementary Fig. S1). A total of 574 DEPs were identified in the spleen CD11c+ cells. Proteins associated with antigen presentation were significantly decreased in the spleen CD11c+ cells of the IRT5-treated group (Table 4).
Table 4.
Differentially Expressed Proteins Associated with Antigen Presentation in the Spleen CD11c+ Cells
| Protein Name | Gene Name | Fold Changes in CD11c+ Cells (IRT5/PBS) | Fold Changes in CD11b+ Cells (IRT5/PBS) | P Value | P Value |
|---|---|---|---|---|---|
| Class II histocompatibility antigen, M beta 1 chain (H2-M beta 1 chain) | H2-DMb1 | 0.468 | 0.000 | ||
| H-2 class I histocompatibility antigen, D-37 alpha chain | H2-T23 | 0.576 | 0.026 | ||
| Immunoglobulin J chain | Jchain | 0.488 | 0.281 | 0.001 | 0.006 |
| Ig alpha chain C region | IGHA1 | 0.507 | 0.003 | ||
| Ig gamma-2B chain C region | Igh-3 | 0.469 | 0.000 |
Discussion
The present study indicates that the application of IRT5 probiotics changes the protein expression associated with immunomodulation in the extraorbital lacrimal gland, resulting in the improvement of dry eye signs. This is interesting as it is the first experimental study to report that the proteome of the lacrimal gland can be altered by modulating the gut microbiome.
IL-1β was significantly lower and IL-10 significantly higher in the conjunctiva and cornea of the IRT5-treated group than in those of the PBS-treated group. Similarly, there was a tendency toward an increase of IL-10, an anti-inflammatory cytokine, and decreases of IL-6 and IL-1β in the extraorbital lacrimal glands of the IRT5-treated group, despite the fact that the trend was not significant due to the small sample size (Supplementary Fig. S2). These findings are in line with our previous report that the inflammation foci score of the extraorbital lacrimal gland was significantly lower in the IRT5 group than in the PBS group.25 Changes in the lacrimal glands can affect the ocular surface, possibly through tears containing anti- and proinflammatory cytokines, growth factors, and other proteins. Although we did not evaluate the tear directly, our results suggest that changes in the lacrimal gland may affect the ocular surface.39,40
In particular, Psmb9, PTPRC, and TAP2, which are known to be upregulated in Sjögren syndrome,41–46 were downregulated in IRT5-treated mice. Psmb8 and Psmb9 activate NF-κ B in B cells and process the numerous MHC class I restricted T-cell epitopes.47 PTPRC modulates signaling mediated by the T-cell receptor and activates B-cell antigen receptor signaling, which modulates susceptibility to autoimmune diseases.48 TAP2 is involved in the transportation of antigens from the cytoplasm to the endoplasmic reticulum for association with MHC class I epitopes, and TAP2 polymorphism has been associated with ankylosing spondylitis.49 Psmb8 and PTPRC also showed strong centralities within immune system process and could be regarded as key molecules in the anti-inflammatory role of IRT5 treatment. This finding corresponds well with the results of a previous meta-analysis showing these proteins to be key contributors to Sjögren syndrome in humans. Therefore, we reasoned that IRT5 treatment ameliorates dry eye signs by reducing the level of these proteins and their related biological processes. Although its association with Sjögren syndrome is not clear, the expression of intracellular adhesion molecule 1 was also attenuated. HMGB2, as a probable factor causing or aggravating Sjögren syndrome,50–54 showed moderate centrality between leukocyte-cell adhesion and the phagocytosis network, and its expression decreased with IRT5 treatment. Cox7c, which was downregulated, reduces electron transfer and decreases the mitochondrial H+ gradient. Therefore, Cox7c downregulation may reduce adenosine triphosphate (ATP) production and decrease activation of P2 purinergic receptors, which may contribute to inflammatory processes.55
Among the upregulated DEPs was Chmp4c, one of the components of transport complex III. Chmp4c sorts endosomal cargo proteins into multivesicular bodies (MVBs) and is involved in MVB formation. Chmp4c showed a higher rate of expression when treated with IRT5, which may result in the formation of MVBs that may, in turn, affect degradation processes. Pex6, a member of the AAA (ATPases associated with diverse cellular activities) family of ATPases, also helps with protein import into peroxisomes. Peroxisomes have two functions, diverse reactions in the lipid metabolism and defense systems for scavenging peroxides and reactive oxygen species.56 Upregulation of Pex6 may reduce oxidative stress by promoting the import of peroxisomal protein.
In addition, Ube2d3, encoding a member of the E2 ligase family, was downregulated, although genes encoding the other DEPs associated with proteolysis and degradation were upregulated after IRT5 treatment. The connection between proteolysis and dry eye is not clear. Given that the ubiquitin proteasome pathway is responsible for generating the precise C termini of MHC-presented peptides, proteolytic changes may be associated with immune response.57
Gut dysbiosis plays an important role in autoimmune disease. Sjögren syndrome also shows reduced diversity in the gut microbiota.11 A recent clinical study reported that administration of Bifidobacterium may attenuate dry eye syndrome.21 In agreement with this observation, our previous25 and current preclinical studies both showed that changes in the gut microbiome after IRT5 treatment were associated with improvements in the clinical signs of autoimmune dry eye disease.
Interestingly, the L. helveticus group and L. hamsteri were higher in the IRT5 group than in the PBS group, regardless of antibiotic pretreatment. The R0052 strain of L. helveticus is a component of Lacidofil (Rosell Institute, Montreal, Canada) and has been used as a probiotic since 1995.58 It has been reported that it inhibits the adhesion of bacteria and modulates immune function by downregulating proinflammatory cytokines.59–61 Additionally, the NS8 strain of L. helveticus has also been reported to exhibit immunomodulatory properties by inducing higher levels of IL-10.62,63 Although no clinical studies have reported that Lactobacillus intake increases IL-10 expression on the ocular surface, an increase of serum IL-10 with an immunosuppressive effect was shown after Lactobacillus intake in atopic dermatitis.64 Consistent with previous reports, IL-10 on the ocular surface was increased in the IRT5-treated group, suggesting that the immunomodulatory effect might be related to L. helveticus. Therefore, we believe that probiotics may alter the gut microbiome to show systemic anti-inflammatory effects that could reach the ocular mucosal surface as well as other targets of autoimmune disease.
However, not many studies on L. hamsteri have been published. Therefore, the immunomodulating function of L. hamsteri needs to be further evaluated. Interestingly, three (the L.s reuteri group, the L. casei group, and B. bifidum) of the five strains constituting IRT5 were increased in the IRT5-treated group compared with the PBS-treated group only when antibiotic pretreatment was provided. Correspondingly, tear secretion increased in the IRT5-treated group only when antibiotic pretreatment was performed. These results suggest that pretreatment with antibiotics promotes IRT5 establishment and maximizes the therapeutic effects without severely diminishing microbial diversity. Suez et al.65 also reported that antibiotic treatment partially alleviated resistance to probiotic species and mildly enhanced probiotic colonization.
However, there is controversy about pretreatment with antibiotics before taking probiotics. Manichanh et al.66 reported a twofold decrease in microbial load after administration of imipenem and vancomycin for 3 days. They also reported a significant decrease of Bacteroidetes and an increase of Firmicutes. Although not statistically significant, the Firmicutes to Bacteroidetes ratio in our study increased more than 10-fold in both the PBS- and IRT5-treated groups after antibiotic pretreatment, while the ratio remained the same without the antibiotic pretreatment. In addition, antibiotic intake changed the taxonomic composition but did not reduce microbial diversity. Previous studies have reported a wide range of losses of alpha diversity after antibiotic exposure, ranging from 10% to 80%.67–69 This might be due to differences in the type of antibiotics used or the vulnerability of the host. Since it is known that changes in the gut microbiome caused by antibiotics increase the risk of diabetes and allergies, further research on the immune system changes caused by antibiotics should be conducted.70
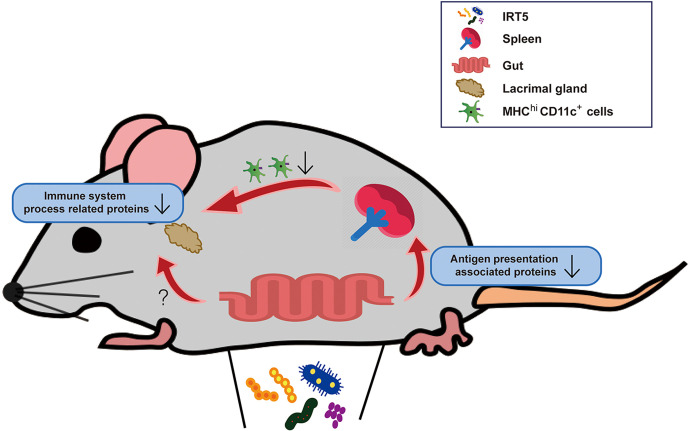
It is well known that the gut microbiome is involved in the host immune system, but the mechanism is unclear.71 We could not directly correlate proteomic changes in the lacrimal gland with compositional changes in the gut microbiome. Given the previous report that probiotics cause a large change in intestinal transcription without significantly changing the composition of the microbiome,72 it can be inferred that the changes in the microbiome after IRT5 administration influenced the lacrimal protein changes in this study. Using an additional mechanistic study, we revealed that the CD11c+ cells of the spleen showed larger proteomic changes than the CD3+ cells of the lymph nodes. We found that the proteins related to the antigen presentation pathway were significantly decreased in the IRT5-treated group. This is consistent with previous reports that the gut microbiome affects intestinal dendritic cell functioning and impacts immune homeostasis.73 To support these findings, we retrospectively reanalyzed flow cytometry data from the previous experiments.25 The proportion of MHCIIhi cells in the cervical lymph node was also lower in the IRT5-treated group (n = 10, 24.01 ± 3.69) than in the PBS-treated group (n = 10, 28.58 ± 2.22) (P = 0.0052, Mann-Whitney U test) (Supplementary Fig. S3). From these results, it can be deduced that the downregulation of antigen presentation is one of the mechanisms of the immunomodulation caused by the administration of IRT5 (Fig. 6). Another speculation is that short-chain fatty acid or tryptophan derivatives, which are known to affect neuroinflammation in the central nervous system, may also contribute to the immune modulation of the eye.74 Further studies are warranted to identify the metabolites that affect the microbiome-gut-eye axis.
Figure 6.
Changes in the gut microbiome after IRT5 administration is suggested to contribute to the immune modulation of the eye via the downregulation of the antigen presentation process in the spleen. Changes in metabolites caused by the gut microbiome and transmitted to the eye through the bloodstream can also be hypothesized as a plausible mechanism.
Our study has some limitations. First, the analysis method, which targeted the V3 to V4 regions of the 16S rRNA gene, could not identify the exact strain of the microbiome. As it is known that actions can vary depending on the type of strain even within one species, whole-genome sequencing is needed. Similarly, the current study is based on OTUs with a cutoff value of 97%. Even if the organisms share more than 97% of the entire 16S rRNA gene sequence, they may or may not represent the same species.75,76 Second, the number of animals used in the experiment was small due to difficulties in breeding them. Third, even if mice are bred under the same food and environmental conditions, there are already microbiome variations between the mice before the experiment. Only male NOD.B10.H2b mice were used in this experiment because females tend to develop sialadenitis instead of dacryoadenitis.77 This sex restriction limited co-housing before the experiments because males fight to the death when co-housed. Additional pretreatment, such as fecal microbiota transplantation, should be considered in further experiments. Fourth, we could not invasively obtain samples directly from the gut but instead collected noninvasive fecal samples. Given that the microbiome composition is different in each part of the intestinal tract, the taxonomic composition measured in the feces can be different.78,79 Therefore, using the fecal microbiome alone is limited in determining the effects of IRT5 on the whole microbiome of the host.
In this study, we investigated changes in the gut microbial community and the proteome of the lacrimal gland after IRT5 treatment in an animal model of Sjögren syndrome. Our findings can be summarized as follows. First, ocular inflammation and tear secretion improved after IRT5 treatment. Second, proteins in defense response and immune system process were downregulated in the extraorbital lacrimal gland after IRT5 treatment. Third, IRT5 treatment induced changes in the composition of several OTUs, including the L. helveticus group and L. hamsteri. Our observations may be beneficial in understanding the pathophysiology of the gut-microbiota-eye axis in dry eye disease.
Supplementary Material
Acknowledgments
The authors thank Editage (www.editage.co.kr) for English language editing. IRT-5 probiotics were kindly provided by Young-Tae Ahn (Korea Yakult Co., Giheung, Korea).
Supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No.2017R1A2B2007209).
Disclosure: S.H. Choi, None; J.W. Oh, None; J.S. Ryu, None; H.M. Kim, None; S.-H. Im, None; K.P. Kim, None; M.K. Kim, None
References
- 1. Gill SR, Pop M, Deboy RT, et al.. Metagenomic analysis of the human distal gut microbiome. Science. 2006; 312: 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Group NHW, Peterson J, Garges S, et al.. The NIH Human Microbiome Project. Genome Res. 2009; 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kahrstrom CT, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature. 2016; 535: 47. [DOI] [PubMed] [Google Scholar]
- 4. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016; 535: 75–84. [DOI] [PubMed] [Google Scholar]
- 5. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016; 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 6. Reyes JL, Vannan DT, Eksteen B, et al.. Innate and adaptive cell populations driving inflammation in dry eye disease. Mediators Inflamm. 2018; 2018: 2532314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Meulen TA, Harmsen H, Bootsma H, Spijkervet F, Kroese F, Vissink A. The microbiome-systemic diseases connection. Oral Dis. 2016; 22: 719–734. [DOI] [PubMed] [Google Scholar]
- 8. Zarate-Blades CR, Horai R, Caspi RR. Regulation of autoimmunity by the microbiome. DNA Cell Biol. 2016; 35: 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horai R, Sen HN, Caspi RR. Commensal microbiota as a potential trigger of autoimmune uveitis. Expert Rev Clin Immunol. 2017; 13: 291–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T cell epitope mimicry between Sjogren's syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin Immunol. 2014; 152: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Paiva CS, Jones DB, Stern ME, et al.. Altered mucosal microbiome diversity and disease severity in Sjogren syndrome. Sci Rep. 2016; 6: 23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalyana Chakravarthy S, Jayasudha R, Sai Prashanthi G, et al.. Dysbiosis in the gut bacterial microbiome of patients with uveitis, an inflammatory disease of the eye. Indian J Microbiol. 2018; 58: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin P. The role of the intestinal microbiome in ocular inflammatory disease. Curr Opin Ophthalmol. 2018; 29: 261–266. [DOI] [PubMed] [Google Scholar]
- 14. Kang HJ, Im SH. Probiotics as an immune modulator. J Nutr Sci Vitaminol (Tokyo). 2015; 61(suppl): S103–S105. [DOI] [PubMed] [Google Scholar]
- 15. Alok A, Singh ID, Singh S, Kishore M, Jha PC, Iqubal MA. Probiotics: a new era of biotherapy. Adv Biomed Res. 2017; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He J, Zhang F, Han Y. Effect of probiotics on lipid profiles and blood pressure in patients with type 2 diabetes: a meta-analysis of RCTs. Medicine (Baltimore). 2017; 96: e9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Athalye-Jape G, Rao S, Patole S. Effects of probiotics on experimental necrotizing enterocolitis: a systematic review and meta-analysis. Pediatr Res. 2018; 83: 16–22. [DOI] [PubMed] [Google Scholar]
- 18. Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther. 2017; 46: 389–400. [DOI] [PubMed] [Google Scholar]
- 19. Mansfield JA, Bergin SW, Cooper JR, Olsen CH. Comparative probiotic strain efficacy in the prevention of eczema in infants and children: a systematic review and meta-analysis. Mil Med. 2014; 179: 580–592. [DOI] [PubMed] [Google Scholar]
- 20. Aqaeinezhad Rudbane SM, Rahmdel S, Abdollahzadeh SM, Zare M, Bazrafshan A, Mazloomi SM. The efficacy of probiotic supplementation in rheumatoid arthritis: a meta-analysis of randomized, controlled trials. Inflammopharmacology. 2018; 26: 67–76. [DOI] [PubMed] [Google Scholar]
- 21. Chisari G, Chisari EM, Francaviglia A, Chisari CG. The mixture of bifidobacterium associated with fructo-oligosaccharides reduces the damage of the ocular surface. Clin Ter. 2017; 168: e181–e185. [DOI] [PubMed] [Google Scholar]
- 22. Kwon HK, Kim GC, Kim Y, et al.. Amelioration of experimental autoimmune encephalomyelitis by probiotic mixture is mediated by a shift in T helper cell immune response. Clin Immunol. 2013; 146: 217–227. [DOI] [PubMed] [Google Scholar]
- 23. Chae CS, Kwon HK, Hwang JS, Kim JE, Im SH. Prophylactic effect of probiotics on the development of experimental autoimmune myasthenia gravis. PLoS One. 2012; 7: e52119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeong JJ, Woo JY, Ahn YT, et al.. The probiotic mixture IRT5 ameliorates age-dependent colitis in rats. Int Immunopharmacol. 2015; 26: 416–422. [DOI] [PubMed] [Google Scholar]
- 25. Kim J, Choi SH, Kim YJ, et al.. Clinical effect of IRT-5 probiotics on immune modulation of autoimmunity or alloimmunity in the eye. Nutrients. 2017; 9: E1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verma R, Lee C, Jeun EJ, et al.. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018; 3: eaat6975. [DOI] [PubMed] [Google Scholar]
- 27. Speciale I, Verma R, Di Lorenzo F, Molinaro A, Im SH, De Castro C. Bifidobacterium bifidum presents on the cell surface a complex mixture of glucans and galactans with different immunological properties. Carbohydr Polym. 2019; 218: 269–278. [DOI] [PubMed] [Google Scholar]
- 28. Jung JH, Ji YW, Hwang HS, et al.. Proteomic analysis of human lacrimal and tear fluid in dry eye disease. Sci Rep. 2017; 7: 13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horwath-Winter J, Schneider MR, Wackernagel W, et al.. Influence of single-fraction Gamma-Knife radiosurgery on ocular surface and tear function in choroidal melanoma patients. Br J Ophthalmol. 2013; 97: 466–470. [DOI] [PubMed] [Google Scholar]
- 30. Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009; 6: 359–362. [DOI] [PubMed] [Google Scholar]
- 31. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012; 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011; 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee B, Moon T, Yoon S, Weissman T. DUDE-Seq: fast, flexible, and robust denoising for targeted amplicon sequencing. PLoS One. 2017; 12: e0181463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 36. Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988; 4: 11–17. [DOI] [PubMed] [Google Scholar]
- 37. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011; 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lv Z, Wang Y, Yang T, et al.. Vitamin A deficiency impacts the structural segregation of gut microbiota in children with persistent diarrhea. J Clin Biochem Nutr. 2016; 59: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ji YW, Mittal SK, Hwang HS, et al.. Lacrimal gland-derived IL-22 regulates IL-17-mediated ocular mucosal inflammation. Mucosal Immunol. 2017; 10: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conrady CD, Joos ZP, Patel BC. Review: the lacrimal gland and its role in dry eye. J Ophthalmol. 2016; 2016: 7542929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khuder SA, Al-Hashimi I, Mutgi AB, Altorok N. Identification of potential genomic biomarkers for Sjogren's syndrome using data pooling of gene expression microarrays. Rheumatol Int. 2015; 35: 829–836. [DOI] [PubMed] [Google Scholar]
- 42. Altorok N, Coit P, Hughes T, et al.. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjogren's syndrome. Arthritis Rheumatol. 2014; 66: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fang K, Zhang K, Wang J. Network-assisted analysis of primary Sjogren's syndrome GWAS data in Han Chinese. Sci Rep. 2015; 5: 18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen CQ, Sharma A, She JX, McIndoe RA, Peck AB. Differential gene expressions in the lacrimal gland during development and onset of keratoconjunctivitis sicca in Sjogren's syndrome (SJS)-like disease of the C57BL/6.NOD-Aec1Aec2 mouse. Exp Eye Res. 2009; 88: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anaya JM, Correa PA, Mantilla RD, Arcos-Burgos M. TAP, HLA-DQB1, and HLA-DRB1 polymorphism in Colombian patients with primary Sjogren's syndrome. Semin Arthritis Rheum. 2002; 31: 396–405. [DOI] [PubMed] [Google Scholar]
- 46. Fox RI, Tornwall J, Michelson P. Current issues in the diagnosis and treatment of Sjogren's syndrome. Curr Opin Rheumatol. 1999; 11: 364–371. [DOI] [PubMed] [Google Scholar]
- 47. Basler M, Kirk CJ, Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Curr Opin Immunol. 2013; 25: 74–80. [DOI] [PubMed] [Google Scholar]
- 48. Trop S, Charron J, Arguin C, Lesage S, Hugo P. Thymic selection generates T cells expressing self-reactive TCRs in the absence of CD45. J Immunol. 2000; 165: 3073–3079. [DOI] [PubMed] [Google Scholar]
- 49. Qian Y, Wang G, Xue F, et al.. Genetic association between TAP1 and TAP2 polymorphisms and ankylosing spondylitis: a systematic review and meta-analysis. Inflamm Res. 2017; 66: 653–661. [DOI] [PubMed] [Google Scholar]
- 50. Kim KH, Kim DH, Jeong HJ, et al.. Effects of subconjunctival administration of anti-high mobility group box 1 on dry eye in a mouse model of Sjogren's syndrome. PLoS One. 2017; 12: e0183678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu B, Zhu Q, Li N, Wu T, Liu S, Liu S. Association of serum/plasma high mobility group box 1 with autoimmune diseases: a systematic review and meta-analysis. Medicine (Baltimore). 2018; 97: e11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kanne AM, Julich M, Mahmutovic A, et al.. Association of high mobility group box chromosomal protein 1 and receptor for advanced glycation end products serum concentrations with extraglandular involvement and disease activity in Sjogren's syndrome. Arthritis Care Res (Hoboken). 2018; 70: 944–948. [DOI] [PubMed] [Google Scholar]
- 53. Ek M, Popovic K, Harris HE, Naucler CS, Wahren-Herlenius M. Increased extracellular levels of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in minor salivary glands of patients with Sjogren's syndrome. Arthritis Rheum. 2006; 54: 2289–2294. [DOI] [PubMed] [Google Scholar]
- 54. Dupire G, Nicaise C, Gangji V, Soyfoo MS. Increased serum levels of high-mobility group box 1 (HMGB1) in primary Sjogren's syndrome. Scand J Rheumatol. 2012; 41: 120–123. [DOI] [PubMed] [Google Scholar]
- 55. Guzman-Aranguez A, Gasull X, Diebold Y, Pintor J. Purinergic receptors in ocular inflammation. Mediators Inflamm. 2014; 2014: 320906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Peroxisomes. Molecular Biology of the Cell. New York, NY: Garland Science; 2002. [Google Scholar]
- 57. Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006; 3: 255–261. [PubMed] [Google Scholar]
- 58. Foster LM, Tompkins TA, Dahl WJ. A comprehensive post-market review of studies on a probiotic product containing Lactobacillus helveticus R0052 and Lactobacillus rhamnosus R0011. Benef Microbes. 2011; 2: 319–334. [DOI] [PubMed] [Google Scholar]
- 59. Atassi F, Brassart D, Grob P, Graf F, Servin AL. In vitro antibacterial activity of Lactobacillus helveticus strain KS300 against diarrhoeagenic, uropathogenic and vaginosis-associated bacteria. J Appl Microbiol. 2006; 101: 647–654. [DOI] [PubMed] [Google Scholar]
- 60. Easo JG, Measham JD, Munroe J, Green-Johnson JM. Immunostimulatory actions of lactobacilli: mitogenic induction of antibody production and spleen cell proliferation by Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus acidophilus. Food Agricult Immunol. 2002; 14: 73–83. [Google Scholar]
- 61. Wallace TD, Bradley S, Buckley ND, Green-Johnson JM. Interactions of lactic acid bacteria with human intestinal epithelial cells: effects on cytokine production. J Food Protection. 2003; 66: 466–472. [DOI] [PubMed] [Google Scholar]
- 62. Rong J, Zheng H, Liu M, et al.. Probiotic and anti-inflammatory attributes of an isolate Lactobacillus helveticus NS8 from Mongolian fermented koumiss. BMC Microbiol. 2015; 15: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rong J, Liu S, Hu C, Liu C. Single probiotic supplement suppresses colitis-associated colorectal tumorigenesis by modulating inflammatory development and microbial homeostasis. J Gastroenterol Hepatol. 2019; 34: 1182–1192. [DOI] [PubMed] [Google Scholar]
- 64. Pessi T, Sutas Y, Hurme M, Isolauri E. Interleukin-10 generation in atopic children following oral Lactobacillus rhamnosus GG. Clin Exp Allergy. 2000; 30: 1804–1808. [DOI] [PubMed] [Google Scholar]
- 65. Suez J, Zmora N, Zilberman-Schapira G, et al.. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018; 174: 1406–1423.e1416. [DOI] [PubMed] [Google Scholar]
- 66. Manichanh C, Reeder J, Gibert P, et al.. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010; 20: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fouhy F, Guinane CM, Hussey S, et al.. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012; 56: 5811–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes. 2010; 1: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008; 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015; 17: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Spasova DS, Surh CD. Blowing on embers: commensal microbiota and our immune system. Front Immunol. 2014; 5: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zmora N, Zilberman-Schapira G, Suez J, et al.. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018; 174: 1388–1405.e1321. [DOI] [PubMed] [Google Scholar]
- 73. Owen JL, Mohamadzadeh M. Microbial activation of gut dendritic cells and the control of mucosal immunity. J Interferon Cytokine Res. 2013; 33: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Haase S, Haghikia A, Wilck N, Muller DN, Linker RA. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018; 154: 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fox GE, Wisotzkey JD, Jurtshuk P Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992; 42: 166–170. [DOI] [PubMed] [Google Scholar]
- 76. Mysara M, Vandamme P, Props R, et al.. Reconciliation between operational taxonomic units and species boundaries. FEMS Microbiol Ecol. 2017; 93:4: fix029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Doyle ME, Boggs L, Attia R, et al.. Autoimmune dacryoadenitis of NOD/LtJ mice and its subsequent effects on tear protein composition. Am J Pathol. 2007; 171: 1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Falony G, Vieira-Silva S, Raes J. Richness and ecosystem development across faecal snapshots of the gut microbiota. Nat Microbiol. 2018; 3: 526–528. [DOI] [PubMed] [Google Scholar]
- 79. Ericsson AC, Gagliardi J, Bouhan D, Spollen WG, Givan SA, Franklin CL. The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci Rep. 2018; 8: 4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.