Abstract
Severe acute respiratory syndrome virus 2 (SARS-CoV-2) belongs to the single-stranded positive-sense RNA family. The virus contains a large genome that encodes four structural proteins, small envelope (E), matrix (M), nucleocapsid phosphoprotein (N), spike (S), and 16 nonstructural proteins (nsp1-16) that together, ensure replication of the virus in the host cell. Among these proteins, the interactions of N and Nsp3 are essential that links the viral genome for processing. The N proteins reside at CoV RNA synthesis sites known as the replication–transcription complexes (RTCs). The N-terminal of N has RNA-binding domain (N-NTD), capturing the RNA genome while the C-terminal domain (N-CTD) anchors the viral Nsp3, a component of RTCs. Although the structural information has been recently released, the residues involved in contacts between N-CTD with Nsp3 are still unknown. To find the residues involved in interactions between two proteins, three-dimensional structures of both proteins were retrieved and docked using HADDOCK. Residues at N-CTD were detected in interaction with L499, R500, K501, V502, P503, T504, D505, N506, Y507, I508, T509, K529, K530K532, S533 of Nsp3 and N-NTD to synthesize SARS-CoV-2 RNA. The interaction between Nsp3 and CTD of N protein may be a potential drug target. The current study provides information for better understanding the interaction between N protein and Nsp3 that could be a possible target for future inhibitors.
Graphic abstract
Keywords: SARS-CoV-2, N-CTD, Nsp3, interactions
Introduction
SARS-CoV-2 belongs to the single-stranded positive-sense RNA family (Anand et al. 2005; Chang et al. 2014). This virus family has a large genome that encodes four structural proteins, small envelope (E), matrix (M), nucleocapsid phosphoprotein (N), spike (S), and 16 nonstructural proteins (nsp1-16) that together, ensure replication of the virus in the host cell. The nonstructural proteins, mostly associated with RNA replication, carry out the enzymatic function required for viral replication (Anand et al. 2005; Zhang et al. 2005; Ahmed et al. 2020). The genome of SARS-CoV-2 also encodes for nsp7, nsp8, and nsp12 that together form a complex called RNA-dependent RNA-polymerase, nsp10, nsp13, nsp14 and 16 complexes called RNA capping machinery, and nsp3, PLpro, and nsp5 (3CLpro) known as proteases that impede innate immunity and also essential for cleaving viral polyproteins (Snijder et al. 2016).
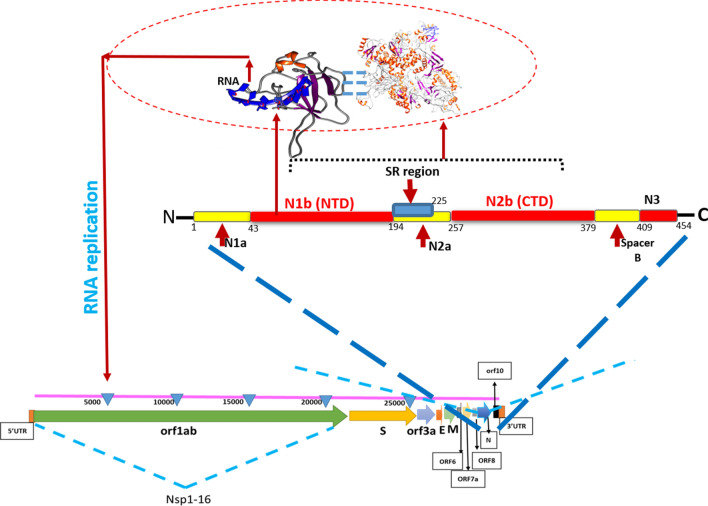
Among these, phosphoprotein (N) is an essential component that links the viral genome with a membrane called nucleocapsid. The N-terminal RNA-binding domain (N-NTD) captures the RNA genome while the C-terminal domain anchors the ribonucleoprotein complex to the viral membrane via its interaction with the M protein. The four structural proteins together with the viral + RNA genome and the envelope constitute the complete virion. The nucleocapsid phosphoprotein consists of an N-terminal (NTD) and a C-terminal (CTD) domain. Both of these domains have the RNA binding affinity, while the CTD binds the M protein, establishing the physical linkage between the envelope and + RNA. The SARS N proteins also play regulatory roles in the viral life cycle through the host intracellular machinery (Chang et al. 2014). A more recent study shows the structure of N protein, right hand-like fold, composed of a β-sheet core with an extended central loop. The core region adopts a five-stranded U-shaped right-handed antiparallel β-sheet platform with the topology β4–β2–β3–β1–β5, flanked by two short α-helices. A prominent feature of the structure is a large extending loop between β2–β3 that forms a long basic β-hairpin (β2′ and β3′) (Dinesh et al. 2020).
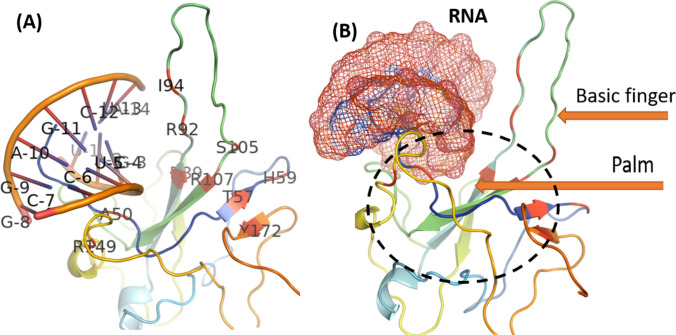
We docked the crystal structure of RNA (Sheng et al. 2014) and nucleocapsid protein using HADDOCK (Dominguez et al. 2003). The RNA duplex was found to be bound between the basic finger and the palm of the N-NTD with highly positive arginine residues (R92, R107, R149) that directly contact the RNA. The model predicts several hydrophobic interactions with side-chains of residues I94 and L104, contributing the RNA binding. Residues A50, T57, H59, R92, I94, S105, R107, R149, Y172 in the surrounding may form interactions with RNA. However, no experimental or long-term molecular dynamic simulation (MD) was performed to confirm what kind of interaction existed.
The nucleocapsid N-terminal residues (N-NTD) interact with genomic RNA of SARS-CoV-2 and incorporate it into progeny particles (Tang et al. 2005; Cong et al. 2020; Kang et al. 2020). The replication–transcription complexes (RTCs) sites are the place where CoV RNA is synthesized. The N proteins are also present at RTCs sites and help viral replication, interacting with nonstructural protein 3 (Nsp3), a component of the RTCs. These two aspects of the CoV life cycle, however, have not been linked. The N protein was found to bind exclusively to nsp3 and to no other RTCs components. The questions arise that what kind of interactions and how these interactions will be affected in the case of mutations. To address these questions, we docked N proteins with Nsp3 using HADDOCK servers to unveil the residues involved in interactions between Nsp3 and N proteins complex formation for future inhibitor designing. This study will provide a better understanding of rapid drug designing to control the global epidemic of SARS-CoV-2.
Methods
Protein structure retrieval
PDB is a database of biomolecule structure, providing crystal structure for proteomics and in silico studies. Only structure relevant to the current study was chosen. The crystal structure of nucleocapsid N-terminal domain (N-NTD) was retrieved from Protein Data Bank (PDB ID: 6yi3). However, for a better understanding of interactions, the full structure of N protein (QHD43423) was download from I-TASSER (Zheng et al. 2019) and that of Nsp3 (6w6y) were downloaded from Protein Data Bank (Berman 2008).
Protein preparation
The structures were subjected to Protein Preparation Wizard in the molecular operating environment (MOE) (Vilar et al. 2008) and also in the Chimera (Pettersen et al. 2004). The missing hydrogens were added, and partial charges were assigned. The protein structure was energy minimized using default value in Amber 10 forcefield in MOE. Selenomethionines were changed into methionine. The prepared was saved in PDB format for docking.
The N protein and Nsp3 docking
To gain insight into the interactions, N proteins and Nsp3 were docked using HADDOCK server (de Vries et al. 2010; van Zundert et al. 2016). The server supports nucleic acids, small molecules, and can perform tasks with experimental data. HADDOCK is widely used in protein–protein, protein–DNA/RNA complexes. The server allows the users for conformational modification of the side chain and also of the backbone of the biomolecules during complex formation. Besides, the server directly supports the docking of PDB and NMR data that contain multiple structural models. Default parameters were used for docking except correlation was set to shape and electrostatics. Intermolecular bonds were analyzed using UCSF Chimera (Pettersen et al. 2004), MOE, and PyMOL (Lill and Danielson 2011).
The N-NTD and RNA docking
Crystal structure of N-NTD (PDB ID 6M3M) and dsRNA (PDB structure 2LK2), a 21-base pair-long dsRNA(Liu et al. 2008) was extracted from PDB and docked with the fully optimized 6yi3 using the HEX docking server (Macindoe et al. 2010). The HEX docking method considers the electrostatic potential, and is based on the rigid body docking algorithm (Macindoe et al. 2010). The default parameters were used for docking except correlation was set to shape and electrostatics. Intermolecular bonds were analyzed using UCSF Chimera (Pettersen et al. 2004) and PyMOL (Lill and Danielson 2011).
Results and discussion
Docking results
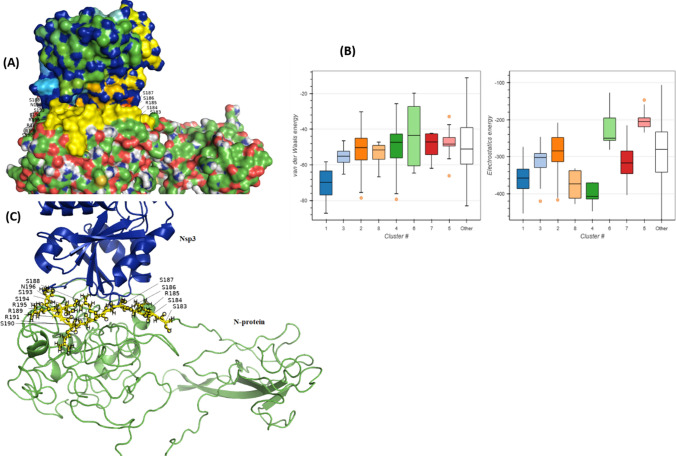
The top clusters based on HADDOCK score are shown in Table 1. Among the seven clusters, cluster 1 was selected, containing 39 structures. However, the tope one based on HADDOCK score, van der Waals, and electrostatic energies has been selected for the current investigation. In the majority of protein–protein dockings, the van der Waals energy, and electrostatic energies combine finds accurate results with promising levels as the scoring function (Mandell et al. 2001). Van der Waals (dispersion forces) contribute to protein interactions with surfaces or other biomolecules.
Table 1.
HADDOCK Nsp3-N protein docking results
| Cluster 1 | Cluster 3 | Cluster 2 | Cluster 8 | Cluster 6 | Cluster 7 | Cluster 5 | |
|---|---|---|---|---|---|---|---|
| HADDOCK score | 90.9 ± 2.5 | 113.7 ± 11.9 | 124.4 ± 6.5 | 129.7 ± 25.7 | 147.6 ± 19.6 | 153.9 ± 13.4 | 175.9 ± 14.9 |
| Cluster size | 39 | 18 | 29 | 4 | 6 | 4 | 7 |
| RMSD of lowest energy structure | 12.3 ± 0.0 | 12.7 ± 0.3 | 10.8 ± 0.0 | 7.7 ± 0.1 | 10.9 ± 0.3 | 12.5 ± 0.1 | 9.6 ± 0.3 |
| Van der Waals energy | −76.9 ± 9.6 | −59.2 ± 3.8 | −61.9 ± 2.3 | −54.3 ± 7.5 | −53.8 ± 11.7 | −49.6 ± 8.0 | −51.8 ± 8.8 |
| Electrostatic energy | −357.2 ± 41.0 | −318.9 ± 28.3 | −342.2 ± 56.4 | −376.4 ± 41.6 | −229.6 ± 60.1 | −313.0 ± 67.0 | −219.5 ± 14.2 |
| Desolvation energy | 19.8 ± 4.3 | 19.7 ± 1.1 | 12.8 ± 3.5 | 26.1 ± 2.8 | 20.8 ± 1.1 | 26.7 ± 5.7 | 20.0 ± 2.0 |
| Restraints violation energy | 2194.4 ± 76.4 | 2170.6 ± 116.2 | 2419.4 ± 34.5 | 2331.3 ± 172.3 | 2265.3 ± 136.4 | 2394.4 ± 81.6 | 2516.0 ± 145.8 |
| Buried surface area | 2532.2 ± 96.3 | 2423.9 ± 134.1 | 2449.7 ± 118.1 | 2310.0 ± 152.6 | 1985.7 ± 115.9 | 2356.0 ± 135.5 | 2006.6 ± 115.0 |
| Z-score | −1.7 | −0.8 | −0.4 | −0.1 | 0.6 | 0.8 | 1.8 |
A total 116 structures have been clustered in HADDOCK server which was further clustered into 8 groups, representing 57% of the water-refined models. The statistics of the topmost reliable clusters are given in Table 1. Its Z-score (the more negative the better) specifies the number of standard deviations from the average this cluster is positioned in the relation of the score. However, van der Waals and electrostatic interactions are important to observe the protein–protein docking where a more negative van der Waals (vdW) and electrostatic interactions show well-docked molecules. The strength of binding of the two proteins relies on the number of residues present at the interface between the two proteins and its area corresponding to interface. Large interfaces depict high binding energy (sum of vdW, H-bonds, electrostatics) (Roth et al. 1996; Nilofer et al. 2017). Docking results were examined with the PyMOL, MOE, and Chimera (DeLano 2002; Pettersen et al. 2004; Vilar et al. 2008), for interactions between the N protein and Nsp3 the closest interacting residues were labeled for better understanding.
Nsp3 and N protein interactions
Here in the current investigations, we detected that residues at N-CTD might be involved in interacting with Nsp3 (Fig. 1a), a member of RTCs for SARS-CoVs RNA processing. The interacting residues of Nsp3, V207, N208, S209, F210, S211, G212, Y213, L214, K215, L216, T217, and D218 might be an important target of inhibitors to prevent viral RNA synthesis, mediated by N protein and Nsp3 interactions (Fig. 1a–c). The surface representation of Nsp3-N complex revealed that the contacts are stable to serve in tethering the genome at a very early stage of infection.
Fig. 1.
Residues interactions between N proteins and Nsp3. (a) surface representation of N proteins and Nsp3 (PDB ID: 6w6y, residues 207–379). Yellow represents N protein residues involved in interactions. (b) The number of clusters predicted by HADDOCK server and their van der Waals and electrostatic energies. Ribbon representation of loop residues of N. The N-NTD is involved with viral RNA binding during replication
Protein–protein interactions are important in every life activity, leading to the execution of numerous elementary roles inside the cell (Koegl and Uetz 2007; Wang et al. 2019). The CoVs N protein plays a vital role in virions through interactions with the positive-strand RNA, the M protein, and other nsp. Previous studies report that the N-terminal end of Nsp3 (1 to amino acid 233) may be central for interactions with the N protein (Hurst et al. 2010, 2013) which induces RTCs-facilitated CoV RNA replication. However, specific residues have not been highlighted, interacting with RTCs for viral RNA synthesis.
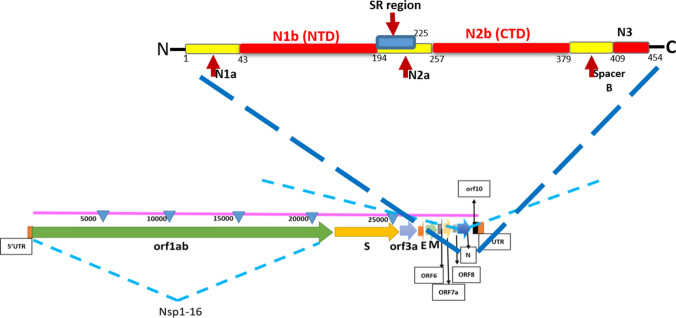
Residues, S188, S190, R191, N192, R195, S197, T198, P199, G200, S201, K237, G238, Q239, Q241, G243, Q244, T245, V246, T247, K248, F314, P309, S310, A311, S312, and A313 of Nsp3 have been detected, interacting with N protein (S183, S184, R185, S186, S187, S188, R189, S190, R191, S193, S194, R195, and N196). Previous studies also confirmed that the N-terminal part of nsp3, from residues 1 to 233 is essential for interaction with N protein (Hurst et al. 2010, 2013). In a more recent study (Cong et al. 2020), it has been shown that interactions with Nsp3 are mediated by N1a–N1b (amino acids 1 to 194) and to a minor extent N2a (amino acids 195 to 257) (Fig. 2). Although there are some other important targets (Khan et al. 2020b), designing potential inhibitor against these interacting sites of both, N and Nsp3 might be useful to block the SARS-CoV-2 replication in host cells.
Fig. 2.
Schematic structural organization of SARS-CoV-2 genome and N proteins. The region between NTD and CTD (light blue) is SR-rich region (amino acid 194–225) located inside N2a. N1b and N2a domains mediate the binding to Nsp3. The N1b is required for RNA binding during replication in which N2b interacts with M proteins
The N plays a key role to link the viral + RNA to the membrane. There are two domains, N-terminal RNA-binding domain (N-NTD) that binds the RNA while the C-terminal domain (CTD) after interaction with the M protein, is involved in anchoring the ribonucleoprotein to the viral membrane. A more recent study also reported that amino acid residues A50, T57, H59, R92, I94, S105, R107, R149, Y172 are important in the establishment of interactions with SARS-CoV-2 RNA (Dinesh et al. 2020) (Fig. 3). For a better result of inhibitors, conserved residues may be identified for better management of CoVs infections. Residues S105 and R107 are conserved among all SARS-CoV N-NTD (SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-OC43) (Kang et al. 2020). Inhibitors may be designed to block the ssRNA binding N-NTD site. Binding of drugs at protein interfaces is mostly controlled by some specific residues contributing disproportionately to the Gibbs free energy of binding and dynamics of proteins (Massova and Kollman 1999; Weiss et al. 2000; Arkin and Wells 2004; Zhao and Chmielewski 2005; Moreira et al. 2007; Wells and McClendon 2007; Boukharta et al. 2014; Ibarra et al. 2019; Khan et al. 2020a).
Fig. 3.
Crystal structure of nucleocapsid (PDB ID: 6YI3) and suggested interaction site residues with RNA. The interaction site was predicted in a more recent study. (a) Residues predicted, interacting with RNA. Positively charged cleft between the basic finger and the palm creating a putative RNA-binding site in the hinge/junction region between the palm and basic finger. (b) RNA-binding site
A previous study reports the importance of N-nsp3 interaction with N proteins for SARS-CoV-2 replication. A wide range of supporting evidence confirmed the interaction between N proteins and Nsp3 replicase–transcriptase complex (Hurst et al. 2013). These findings suggest that some peptide inhibitor may be designed to prevent the interaction of Nsp3 and N proteins, required of viral replication and propagation.
In conclusion, the N protein seems the most potent drug target in SARS-CoV-2. Its N-NTD interacts with the viral RNA while N-CTD interacts with the Nsp3 of RTCs during RNA synthesis. Domains, N1b, N2a, and N2b, all are a potential target. Residues involved in interaction may be the potential target of drug and peptides inhibitors. Drug development and screening against interacting residues may be useful for better management of SARS-CoV-2 infections.
Acknowledgements
The current study was technically supported by Director PTRL KP, Peshawar, Dr. Sajid Ali, Molecular biologist and Dr Taj Ali, Associate professor, Kohat University of Science and Technology, Pakistan.
Abbreviations
- E
Envelope
- M
Matrix
- Nsp1-16
Sixteen nonstructural proteins
- N
Nucleocapsid phosphoprotein
- N-CTD
Nucleocapsid C-terminal domain
- Nsp
Nonstructural proteins
- PDB
Protein Data Bank
- RMSD
Root means square fluctuation (RMSF)
- Rg
Radius of gyration
- RMSF
Root means square fluctuation
- SARS-CoV-2
S: Spike, Severe acute respiratory syndrome coronavirus-2
Authors’ contributions
Conceptualization: DQW, MTK, ZC, MI. Data curation: MTK, AA, AA, MTZ, MA. Experimental work: MTK, SIM, AA, AK. Formal analysis: AA, MTK, HA, ASK, SA. Funding acquisition: DQW. Approval: DQW, GS.
Funding
Dong-Qing Wei is supported by the Natural Science Foundation of Henan Province (162300410060), the Ministry of Science and Technology of China (2016YFA0501703), the Science and Technology Commission of Shanghai Municipality (19430750600), the National Natural Science Foundation of China (61503244,61832019), and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Muhammad Tahir Khan, Email: pi133001@cust.pk.
Muhammad Tariq Zeb, Email: drtariqzeb@gmail.com.
Hina Ahsan, Email: hina.ahsan@riphah.edu.pk.
Abrar Ahmed, Email: Abrar22@sjtu.edu.cn.
Arif Ali, Email: arifali@sjtu.edu.cn.
Khalid Akhtar, Email: drkhalidakhtar@smme.edu.pk.
Shaukat Iqbal Malik, Email: drshaukat@cust.edu.pk.
Zhilei Cui, Email: zhileicui@126.com.
Sajid Ali, Email: sajidjan@live.com.
Anwar Sheed Khan, Email: anwar786kp@hotmail.com.
Manzoor Ahmad, Email: manzoor@mail.sysu.edu.cn.
Dong-Qing Wei, Email: dqwei@sjtu.edu.cn.
Muhammad Irfan, Email: irfanmuhammad@ufl.edu.
References
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K, Yang H, Bartlam M, et al. Coronavirus main proteinase: target for antiviral drug therapy. Coronaviruses with Special Emphasis on First Insights Concerning SARS. 2005 doi: 10.1007/3-7643-7339-3_9. [DOI] [Google Scholar]
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Berman HM. The protein data bank: a historical perspective. Acta Crystallogr A. 2008;64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- Boukharta L, Gutiérrez-de-Terán H, Åqvist J. Computational prediction of alanine scanning and ligand binding energetics in G-protein coupled receptors. PLOS Comput Biol. 2014;10:e1003585. doi: 10.1371/journal.pcbi.1003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Hou M-H, Chang C-F, et al. The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Dinesh DC, Chalupska D, Silhan J, et al. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. bioRxiv. 2020 doi: 10.1101/2020.04.02.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst KR, Ye R, Goebel SJ, et al. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra AA, Bartlett GJ, Hegedüs Z, et al. Predicting and experimentally validating hot-spot residues at protein-protein interfaces. ACS Chem Biol. 2019;14:2252–2263. doi: 10.1021/acschembio.9b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Yang M, Hong Z, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. bioRxiv. 2020 doi: 10.1101/2020.03.06.977876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Khan M, Saleem S, et al. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscip Sci Comput Life Sci. 2020 doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Ali A, Wang Q, et al. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genomic Proteomic. 2007;6:302–312. doi: 10.1093/bfgp/elm035. [DOI] [PubMed] [Google Scholar]
- Lill MA, Danielson ML. Computer-aided drug design platform using PyMOL. J Comput Aided Mol Des. 2011;25:13–19. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newslett on protein crystallogr. 2002;40(1):82–92. [Google Scholar]
- Liu L, Botos I, Wang Y, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macindoe G, Mavridis L, Venkatraman V, et al. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Roberts VA, Pique ME, et al. Protein docking using continuum electrostatics and geometric fit. Protein Eng Des Sel. 2001;14:105–113. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- Massova I, Kollman PA. Computational alanine scanning to probe protein−protein interactions: a novel approach to evaluate binding free energies. J Am Chem Soc. 1999;121:8133–8143. doi: 10.1021/ja990935j. [DOI] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Computational alanine scanning mutagenesis—an improved methodological approach. J Comput Chem. 2007;28:644–654. doi: 10.1002/jcc.20566. [DOI] [PubMed] [Google Scholar]
- Nilofer C, Sukhwal A, Mohanapriya A, Kangueane P. Protein-protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity. Bioinformation. 2017;13:164–173. doi: 10.6026/97320630013164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Roth CM, Neal BL, Lenhoff AM. Van der Waals interactions involving proteins. Biophys J. 1996;70:977–987. doi: 10.1016/S0006-3495(96)79641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Larsen A, Heuberger BD, et al. Crystal structure studies of RNA duplexes containing s(2)U: A and s(2)U: U base pairs. J Am Chem Soc. 2014;136:13916–13924. doi: 10.1021/ja508015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus rna synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T-K, Wu MP-J, Chen S-T, et al. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert GCP, Rodrigues JPGLM, Trellet M, et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- Wang Y, You Z-H, Yang S, et al. A high efficient biological language model for predicting protein-protein interactions. Cells. 2019;8:122. doi: 10.3390/cells8020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, et al. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zheng N, Hao P, et al. A molecular docking model of SARS-CoV S1 protein in complex with its receptor, human ACE2. Comput Biol Chem. 2005;29:254–257. doi: 10.1016/j.compbiolchem.2005.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chmielewski J. Inhibiting protein-protein interactions using designed molecules. Curr Opin Struct Biol. 2005;15:31–34. doi: 10.1016/j.sbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Zheng W, Li Y, Zhang C, et al. Deep-learning contact-map guided protein structure prediction in CASP13. Proteins. 2019;87:1149–1164. doi: 10.1002/prot.25792. [DOI] [PMC free article] [PubMed] [Google Scholar]