TAN1 is a microtubule-binding protein required for the spatial control of plant division plane orientation. TAN1 mediates both lateral and end-on microtubule interactions in vitro. These activities may promote proper division plane orientation in vivo.
Abstract
The microtubule cytoskeleton serves as a dynamic structural framework for mitosis in eukaryotic cells. TANGLED1 (TAN1) is a microtubule-binding protein that localizes to the division site and mitotic microtubules and plays a critical role in division plane orientation in plants. Here, in vitro experiments demonstrate that TAN1 directly binds microtubules, mediating microtubule zippering or end-on microtubule interactions, depending on their contact angle. Maize tan1 mutant cells improperly position the preprophase band (PPB), which predicts the future division site. However, cell shape–based modeling indicates that PPB positioning defects are likely a consequence of abnormal cell shapes and not due to TAN1 absence. In telophase, colocalization of growing microtubules ends from the phragmoplast with TAN1 at the division site suggests that TAN1 interacts with microtubule tips end-on. Together, our results suggest that TAN1 contributes to microtubule organization to ensure proper division plane orientation.
Introduction
The proper organization of microtubule networks during interphase and mitosis is important to promote growth and development at both the cell and organismal levels (Wasteneys and Ambrose, 2009; Elliott and Shaw, 2018; Ehrhardt and Shaw, 2006; Baskin et al., 2004). Mechanisms for achieving and modulating microtubule organization are driven by microtubule–microtubule or microtubule–protein interactions, including zippering at low contact angles (Ho et al., 2012; Tulin et al., 2012; Smertenko et al., 2004; Shaw et al., 2003), contact-mediated catastrophe (Dixit and Cyr, 2004), severing (Lindeboom et al., 2013; Zhang et al., 2013; Panteris et al., 2018; Komis et al., 2017), and stabilization at cell edges (Ambrose et al., 2011). These processes alter microtubule dynamics and organization. Mitotic microtubule structures are formed and modified by these activities to perform a distinct role in DNA segregation and separation of daughter cells. In plants, the key mitotic structures are the preprophase band (PPB), metaphase spindle, and phragmoplast. Proteins that regulate the formation and function of these structures are localized along these different structures as well as the cortical plant division site.
During the G2 phase of the cell cycle, the PPB is formed as a ring-shaped arrangement of microtubules, actin, and associated proteins that localize just under the plasma membrane to form the cortical division zone (Smertenko et al., 2017; Van Damme et al., 2007). The PPB is an early marker of the future division site in land plants; it indicates the location where the developing new cell wall will fuse with the mother cell (Rasmussen and Bellinger, 2018; Facette et al., 2019; Pickett-Heaps and Northcote, 1966). Several microtubule-associated proteins play an important role in division plane orientation by promoting PPB formation. A large family of proteins with microtubule-binding motifs recruit a protein phosphatase type 2A complex to form the PPB (Spinner et al., 2010, 2013; Wright et al., 2009; Traas et al., 1995; Drevensek et al., 2012; Schaefer et al., 2017). The proper formation and positioning of the PPB may orient the metaphase spindle to promote rapid mitotic progression (Chan et al., 2005; Ambrose and Cyr, 2008; Schaefer et al., 2017). As cells enter metaphase, the PPB is completely disassembled; however, a handful of proteins that colocalize with the PPB continue to label the division site until the end of cytokinesis (Walker et al., 2007; Xu et al., 2008; Lipka et al., 2014; Martinez et al., 2017; Li et al., 2017; Buschmann et al., 2015).
During telophase, the phragmoplast is assembled from microtubules, actin, and associated proteins to aid in the formation of the cell plate via vesicle delivery (Smertenko et al., 2017; Smertenko, 2018; Lee and Liu, 2013; Jürgens, 2005b). The phragmoplast expands outwards to the cell cortex through the polymerization of new microtubules from existing leading-edge microtubules and depolymerization at the lagging edge as the cell plate is assembled (Murata et al., 2013). The direction of phragmoplast expansion is thought to be guided by proteins that continuously label the division site (Rasmussen and Bellinger, 2018; Livanos and Müller, 2019). Once the phragmoplast reaches the cortex, it is disassembled and the cell plate fuses with the plasma membrane, completing cytokinesis (Jürgens, 2005a; Worden et al., 2012). Mutants with defects in maintaining division plane orientation place new cell walls outside the location originally specified by the PPB. In maize, tangled1 (tan1) mutants have division plane defects in both symmetric and asymmetric divisions (Smith et al., 1996) caused by a failure of the phragmoplast to return to the division site originally indicated by the PPB (Martinez et al., 2017). TAN1-YFP localizes to the cortical division site throughout mitosis in Arabidopsis thaliana and maize (Martinez et al., 2017; Walker et al., 2007). TAN1 also colocalizes with mitotic microtubule arrays in vivo when fused to YFP (Martinez et al., 2017) and using a nonspecific TAN1 antibody (Smith et al., 2001). TAN1 is a highly basic protein without any obvious known domains (Smith et al., 2001). Structure–function analysis identified two highly conserved regions of TAN1 that separately promoted its localization to the division site either during late G2 or telophase (Rasmussen et al., 2011). The protein region promoting TAN1 localization during telophase was subsequently shown to be critical for its function in vivo (Mir et al., 2018).
Double mutants for two kinesin 12 paralogs in A. thaliana, phragmoplast orienting kinesin 1 (pok1) and pok2, display a severe division plane defect (Müller et al., 2006). POK1 interacts directly with TAN1 and localizes to the division site (Walker et al., 2007; Lipka et al., 2014; Rasmussen et al., 2011). Both POK1 and POK2 are required for TAN1 localization to the division site after metaphase (Walker et al., 2007; Lipka et al., 2014). POK2 acts as a weak microtubule plus-end–directed motor in vitro (Chugh et al., 2018). Interestingly, in addition to its division site localization, POK2 also accumulates in the phragmoplast midline, where it may interact with Microtubule-associated protein 65-3 (MAP65-3) or other MAP65 proteins (Herrmann et al., 2018; Ho et al., 2011). Another closely related MAP65, MAP65-4, is localized to the PPB, spindle, phragmoplast, and the division site (Li et al., 2017). The map65-3 map65-4 double mutant in A. thaliana has a cytokinesis defect, but it is not yet clear whether it has a division plane defect (Li et al., 2017). MAP65-4 regulates microtubule stability by increasing microtubule elongation phases during bundling (Fache et al., 2010), while another related MAP65, MAP65-1, increases microtubule stability by protecting against severing and promoting microtubule flexibility during bundling (Portran et al., 2013; Stoppin-Mellet et al., 2013; Burkart and Dixit, 2019). Microtubule binding and bundling proteins therefore may contribute to the assembly of the mitotic microtubule structures, but they also serve as important effectors for the establishment, timely progression, and execution of properly oriented plant cell divisions.
In addition to division plane defects, the tan1 mutant has mitotic progression delays and reduced plant stature (Martinez et al., 2017; Smith et al., 1996). Mitotic progression delays and phragmoplast guidance defects were mostly uncoupled using a partially rescued tan1 mutant expressing TAN1-YFP fused to the CYCLIN B–destruction box motif (Martinez et al., 2017). In this partially rescued line, mitotic delays are observed, but division plane defects are rare, coinciding with TAN1-YFP signal at the division site but lack of detectable TAN1-YFP signal in the spindle and phragmoplast. We hypothesize that TAN1 is a multifunctional protein that aids in timely mitotic progression when it localizes to mitotic microtubule structures and maintains division plane orientation via phragmoplast guidance when it is localized to the division site. Here, we report an in vitro function for TAN1 in mediating microtubule interactions and an in vivo function in spindle organization and phragmoplast microtubule interactions at the division site.
Results and discussion
TAN1 binds to microtubules in vitro
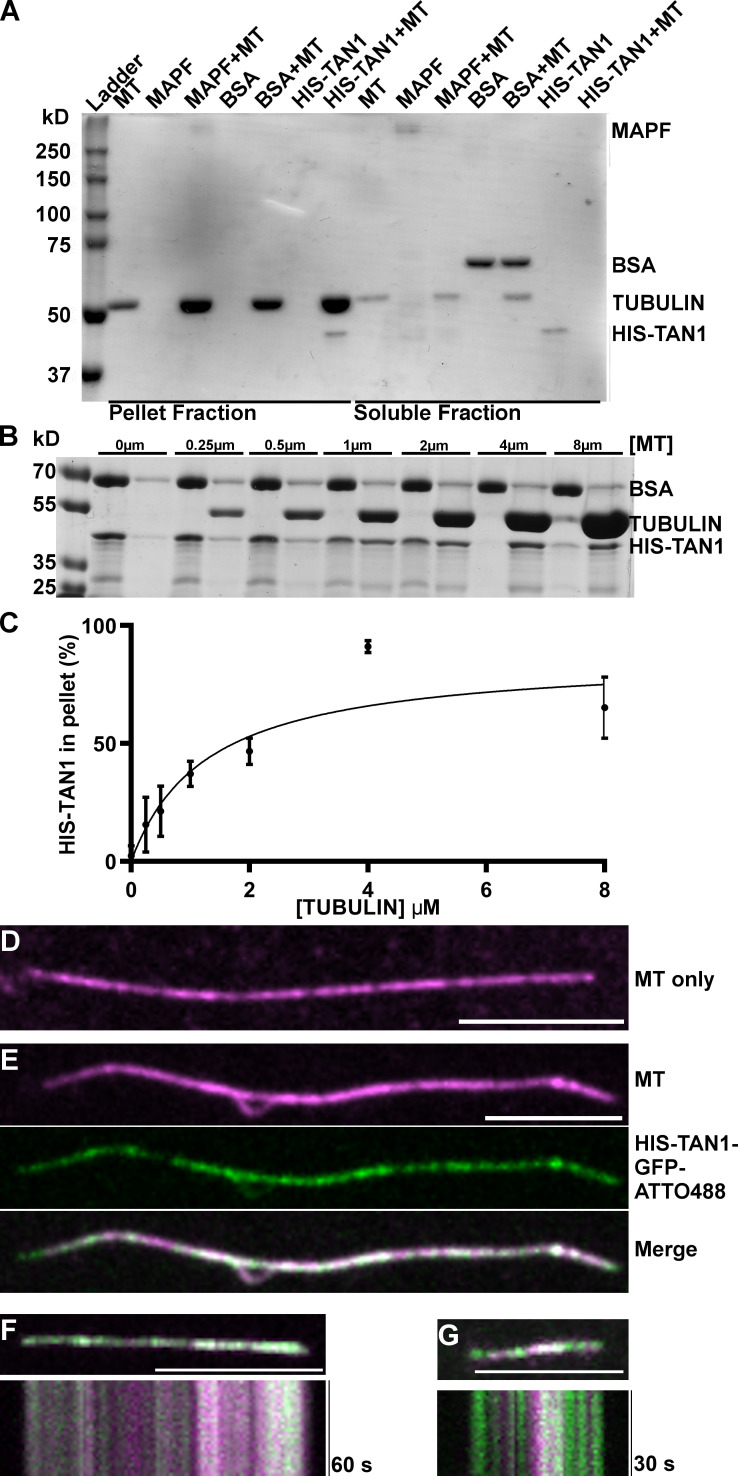
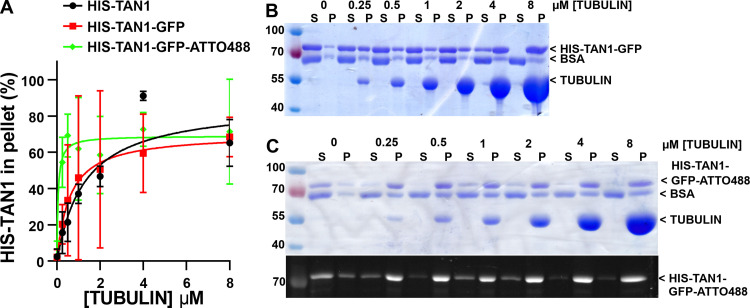
TAN1 protein has been shown to bind to Taxol-stabilized microtubules in a blot overlay assay (Smith et al., 2001). To quantitatively assess the binding of TAN1 to microtubules, we recombinantly expressed 6xHIS-tagged ZmTAN1 (HIS-TAN1) protein and tested its ability to bind to microtubules. HIS-TAN1 protein bound to Taxol-stabilized microtubules in cosedimentation experiments (Fig. 1 A). Titration of microtubules against a fixed concentration of HIS-TAN1 resulted in saturable TAN1–microtubule binding. Fitting the binding data hyperbolically as in similar studies (Tulin et al., 2012; Wong and Hashimoto, 2017) yielded a K0.5 value of 1.08 µM (95% confidence interval [CI], 0.722 to 1.43 µM) and suggested that at least 70% of the HIS-TAN1 was active in binding microtubules. This calculated affinity is similar to that of other microtubule-binding proteins (Tulin et al., 2012; Portran et al., 2013; Wong and Hashimoto, 2017). Significantly less than 100% TAN1 saturation was observed at the maximal available microtubule concentration. This could be explained in terms of an inactive protein fraction, but alternatively by a model such as multi-site binding with negative cooperativity (Table S1). To directly visualize the binding of TAN1 to microtubules in vitro, we purified recombinant HIS-TAN1-GFP. Unfortunately, this fusion protein was not fluorescent, potentially because GFP did not fold correctly during renaturation of recombinant protein from bacterial inclusion bodies. Since HIS-TAN1-GFP still bound to microtubules with similar affinity as HIS-TAN1 (Fig. S1 A), we labeled it with the organic fluorophore Atto488 to visualize it using fluorescence microscopy. When coincubated with Taxol-stabilized, rhodamine-labeled microtubules, Atto488-tagged HIS-TAN1-GFP (100 nM) localized along the microtubule lattice (Fig. 1, D and E). Kymographs of Atto488-tagged HIS-TAN1-GFP showed that it did not move on biotinylated guanosine-5′-(α,β-methylene)triphosphate (GMPCPP) rhodamine-labeled microtubules over ∼2 min of imaging (Fig. 1, F and G). Atto488-tagged HIS-TAN1-GFP unfortunately aggregated over the course of microtubule cosedimentation assays (Fig. S1 B), and therefore, we did not use it in further experiments.
Figure 1.
Recombinantly expressed TAN1 binds to microtubules (MTs). (A) Coomassie-stained SDS PAGE results from microtubule cosedimentation with HIS-TAN1, positive control (microtubule-associated protein fraction [MAPF], 70% MAP2) and negative control (BSA) controls separated into corresponding pellet and soluble fractions. (B) Coomassie-stained SDS-PAGE results from microtubule cosedimentation assay using 2 µM HIS-TAN1 and from 0 to 8 µM tubulin, alternating soluble and pellet fractions. (C) Hyperbolic fit of microtubule cosedimentation data for HIS-TAN1 at varying concentrations of microtubules determines an apparent K0.5 value of 1.08 µM (95% CI, 0.722-1.43 µM), removing the outlier at 4 µM tubulin; error bars are 95% confidence intervals. (D and E) Rhodamine-labeled, Taxol-stabilized microtubule in buffer only control (magenta; D) or incubated with HIS-TAN1-GFP-Atto488 (green). (F and G) Time lapse and kymograph of rhodamine-labeled GMPCPP-stabilized microtubules (magenta) and HIS-TAN1-GFP-Atto488 (green). Scale bars, 10 µm.
Figure S1.
HIS-TAN1-GFP and HIS-TAN1-GFP-Atto488 binding affinity to Taxol-stabilized microtubules. (A) HIS-TAN1-GFP cosedimentation binding data with fits to hyperbolic binding isotherms for HIS-TAN1 (replotted from Fig. 1 C), HIS-TAN1-GFP, and HIS-TAN1-GFP-Atto488. Apparent affinity for HIS-TAN1-GFP is 0.595 µM ± 0.389 to 0.800 µM, while HIS-TAN1-GFP-Atto488 is 0.05 µM ± 0.0009 to 0.129 µM corrected for the average pelleting in samples without microtubules added (average ± 95% CI). (B) Coomassie-stained SDS-PAGE experiment from spindown of HIS-TAN1-GFP in the presence of varying concentrations of tubulin (0–8 µM), alternating soluble (S) and pellet (P) fractions . (C) Coomassie-stained SDS-PAGE experiment from spindown of HIS-TAN1-GFP-Atto488 in the presence of varying concentrations of tubulin (0–8 µM). Below the Coomassie-stained SDS-PAGE experiment, HIS-TAN1-GFP-Atto488 was excited using a UV light source to confirm Atto488 maleimide conjugation with HIS-TAN1-GFP used in the spindown assays.
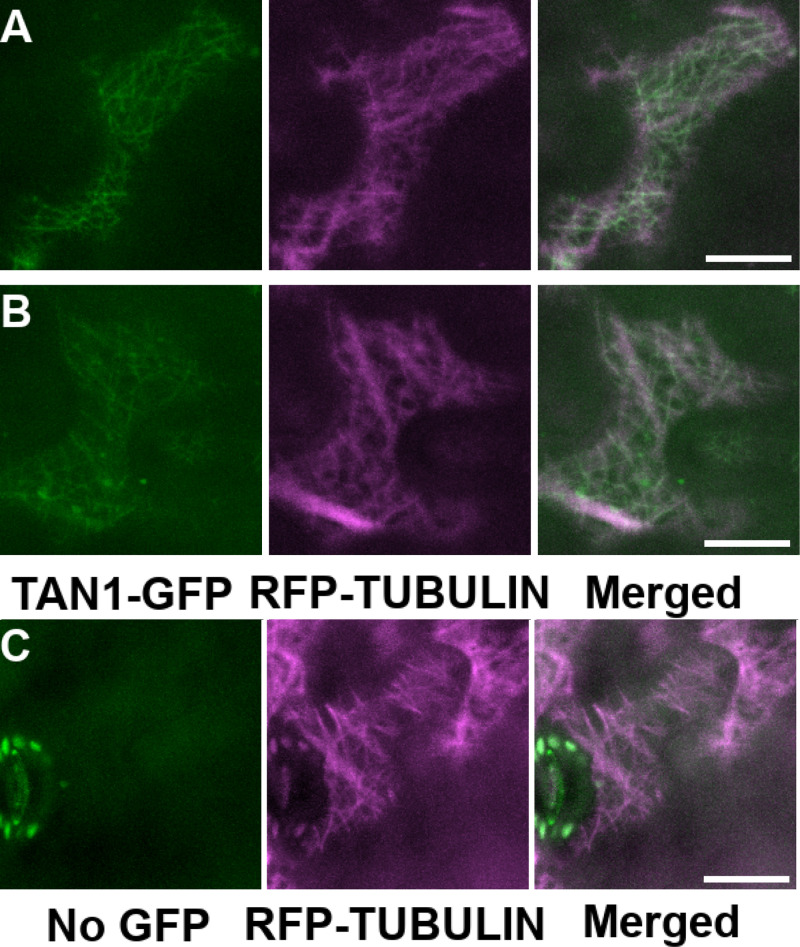
Previous results showed that TAN1-YFP colocalizes with microtubules in the PPB, spindle, and phragmoplast (Martinez et al., 2017). However, direct TAN1–microtubule binding data suggested that TAN1 will interact with microtubules regardless of cell cycle stage. To examine TAN1–microtubule interaction in interphase, we transiently expressed both TAN1-GFP and RFP-TUBULIN in nondividing Nicotiana benthamiana epidermal cells using the constitutive 35S promoter. After 3 d of incubation, we imaged TAN1-GFP and RFP-TUBULIN using confocal microscopy. TAN1-GFP colocalized with RFP-TUBULIN (Fig. S2, A and B), indicating that mitosis-specific proteins are not necessary for TAN1 interaction with microtubules, consistent with our in vitro cosedimentation assays. No obvious differences in microtubule arrays in interphase epidermal cells were observed between those infiltrated with RFP-TUBULIN and TAN1-GFP (Fig. S2, A and B) or RFP-TUBULIN only (Fig. S2 C). This lack of obvious changes in microtubule organization contrasts with overexpression of other MAPs such as MAP65-1 (Ho et al., 2012) and CLASP (Kirik et al., 2007). However, the TAN1-GFP fluorescent signal was also low, consistent with the hypothesis that TAN1 levels may be posttranslationally regulated by degradation (Rasmussen et al., 2011).
Figure S2.
Colocalization of TAN1 and cortical microtubules in N. benthamiana (tobacco). (A and B) TAN1-GFP (green) and RFP-TUBULIN–labeled microtubules (magenta) colocalize (merged image) when expressed transiently in tobacco epidermal cells. (C) RFP-TUBULIN was only expressed transiently in tobacco cells. Scale bars, 20 µm. Bright oval signals in guard cells in C are chloroplast autofluorescence.
TAN1 does not markedly alter microtubule dynamics in vitro
To determine whether TAN1 regulates microtubule polymerization dynamics, we conducted in vitro microtubule polymerization experiments. Microtubules were nucleated from GMPCPP-stabilized microtubule seeds, and their polymerization and depolymerization were promoted by adding 17.5 µM tubulin. Microtubule dynamics were visualized using rhodamine-labeled tubulin and total internal reflection fluorescence (TIRF) microscopy (Materials and methods). At lower concentrations of HIS-TAN1 (<1 µM), no significant effect on microtubule dynamics was observed (Table 1). At a concentration of 2 µM HIS-TAN1, which is close to the apparent K0.5 of TAN1 for Taxol-stabilized microtubules, we observed small decreases in both microtubule plus-end growth and plus-end shrinkage rates (compared with 0 µM HIS-TAN1, using the Mann–Whitney test; Table 1). HIS-TAN1 addition did not alter the amount of time microtubules spent growing or the frequency of catastrophes. However, small but significant differences in time spent shrinking were observed (compared with 0 µM HIS-TAN1; Table 1). Under the experimental conditions used, rescue events were rare and the minus ends were not dynamic; therefore, these parameters were not quantified. Together, these results suggest that regulation of microtubule polymerization dynamics is unlikely to be the primary function of TAN1.
Table 1. Summary of microtubule dynamics and microtubule interactions at different concentrations of HIS-TAN1.
| Plus-end dynamics | 0 µM HIS-TAN1 | 0.1 µM HIS-TAN1 | 0.5 µM HIS-TAN1 | 1 µM HIS-TAN1 | 2 µM HIS-TAN1 |
|---|---|---|---|---|---|
| Growth events (n) | 156 | 180 | 166 | 214 | 196 |
| Growth rate (μm/s, mean ± SD) | 1.8 ± 0.4 | 1.8 ± 0.3 | 1.8 ± 0.3 | 1.7 ± 0.5* | 1.5 ± 0.3*** |
| Shrinkage events (n) | 109 | 127 | 113 | 153 | 149 |
| Shrinkage rate (μm/s, mean ± SD) | 31.5 ± 15.6 | 27.7 ± 10.8 | 26.2 ± 8.8* | 27.8 ± 9.7 | 24.2 ± 10.0*** |
| Time growing (%) | 94.9 | 93.8 | 94.5 | 94.6 | 95.2 |
| Time shrinking (%) | 5.1 | 6.2** | 5.5* | 5.4 | 4.8 |
| Catastrophe frequency (events/min) | 0.3 | 0.4 | 0.4 | 0.4 | 0.5 |
| Crossovers (n) | 445 | 346 | 334 | 334 | 506 |
| Bundling events (n) | 2 | 0 | 0 | 3 | 139 |
| Bundling frequency (%) | 0.5 | 0 | 0 | 0.9 | 27.5 |
Bundling includes both zippering and pulling. Significance was calculated by comparing values to 0 µM HIS-TAN1 determined by Mann–Whitney test (*, P > 0.05; **, P > 0.01; ***, P > 0.001). Three trials were performed for each concentration of HIS-TAN1.
HIS-TAN1 mediates lateral and end-on microtubule interactions in vitro
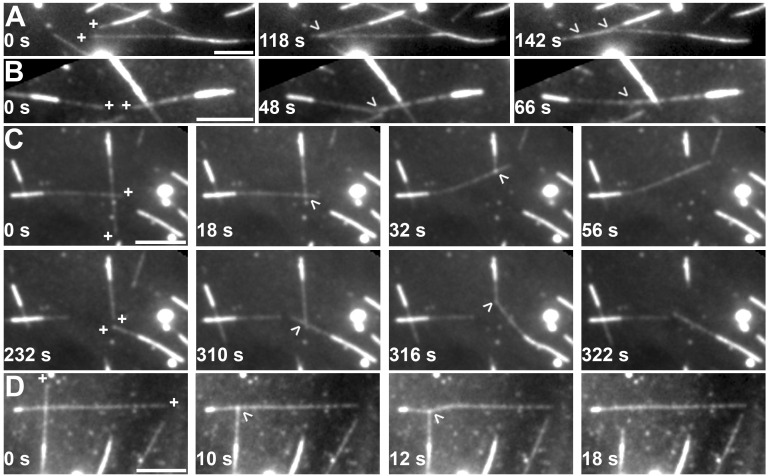
During the course of our in vitro microtubule dynamics experiments, we observed that at high concentrations of HIS-TAN1 (2 µM), microtubules that contacted each other transiently interacted. To promote microtubule interactions, we conducted experiments with a higher concentration of GMPCPP-stabilized seeds and free tubulin dimers (22.5 µM concentration) to generate more microtubules that grew longer and hence encountered each other more frequently. We used 2 µM HIS-TAN1 because it resulted in microtubule interactions (139 interaction events resulting from 506 crossovers) in dynamic microtubule assays, whereas no interactions were observed at lower concentrations of HIS-TAN1 (Table 1). We observed two kinds of microtubule bundling interactions depending on the microtubule contact angle. At small or shallow contact angles (angle = 19.6° ± 7.6° [average ± SD]), the microtubules progressively zippered together to produce bundles (n = 47 bundling events out of a total of 139 interactions observed [34% of bundling events]; Fig. 2, A and B). Zippering of microtubules in parallel and antiparallel configurations occurred with similar frequencies (n = 13/27 and 14/27 where orientation was unambiguous, respectively). Therefore, TAN1 does not preferentially bundle microtubules in specific orientations. In contrast, MAP65 microtubule bundling proteins preferentially bundle antiparallel microtubules (Gaillard et al., 2008; Tulin et al., 2012). At high contact angles (angle = 60° ± 20° [average ± SD]), transient “end-on” microtubule interactions were observed during microtubule depolymerization (Fig. 2, C and D; Video 1). As one microtubule depolymerized past a previous crossover site, TAN1 mediated an interaction at the crossover point. The depolymerizing end stayed bound to the sidewall of the second microtubule, resulting in a pulling force on the stable microtubule (n = 92 end-on interactions out of a total of 139 interactions observed [66% of interaction events]). Interestingly, highly basic peptides linked together to form an artificial polypeptide capable of multivalent electrostatic interactions with microtubules displayed similar microtubule pulling and bundling activities as TAN1 (Drechsler et al., 2019). The intrinsically disordered microtubule-associated protein tau also results in similar microtubule interactions that are thought to depend on tau’s multivalent microtubule binding (Kellogg et al., 2018). Based on the similarities in the types of microtubule interactions mediated by the artificial polypeptide tau and TAN1 and their shared biochemical characteristics of net positive charge and intrinsically disordered regions, we hypothesize that TAN1 likely contains multiple microtubule-binding sites that enable interaction between microtubules. This property would also allow TAN1 to bundle microtubules without requiring dimerization or multimerization in contrast to the bundling protein MAP65-1 (Ho et al., 2011). Based on these data, we conclude that the outcomes of TAN1–microtubule interactions depend on the initial contact or crossover angle between the microtubules and that at high contact angles, TAN1–microtubule interactions lead to transient pulling or catching.
Figure 2.
HIS-TAN1 contact angle–independent dynamic microtubule interactions. (A–D) Dynamic rhodamine-labeled microtubules nucleated from GMPCPP-stabilized seeds with plus ends indicated by a (+) and a crossover indicated by arrowheads. Microtubule seeds are identified by their brighter signal compared with the growing microtubule ends. 2 µM HIS-TAN1 is present in the assay. (A) Two microtubule plus ends are indicated with their plus ends polymerizing in the same direction. These microtubules encounter each other in a parallel orientation and are zippered together. (B) Two microtubule plus ends are indicated with their plus ends growing toward each other. These microtubules are zippered together in an antiparallel orientation. (C) Two microtubule plus ends are indicated at the start (0 s). These microtubules crossover, and at 18 s, one of them depolymerizes. The depolymerizing end of this microtubule appears to pull on the other microtubule over the course of depolymerization. At 232 s (new plus-end growth indicated), a new crossover is formed, followed by a depolymerization event, which again pulls at the crossover with the nondepolymerizing microtubule (316 s). Time lapse is shown in Video 1. (D) Two microtubule plus ends are indicated at the start (0 s), which cross over at a high angle (∼90°). Depolymerization of one microtubule leads to transient deformation of the other microtubule at the crossover point. Scale bars, 10 µm.
Video 1.
In vitro microtubule interactions mediated by HIS-TAN1. Video of microtubule–microtubule interaction (pulling) with 2 µM HIS-TAN1; 20 frames per second for 3 s.
Microtubule zippering is a well-characterized form of microtubule bundling in plants, animals, and fungi (Dixit and Cyr, 2004; Tulin et al., 2012; Janson et al., 2007; Subramanian et al., 2010; Gaillard et al., 2008). Microtubule end-on interactions have been studied extensively in animals and fungi and typically involve forces generated by motor proteins (Laan et al., 2012a,b). For example, end-on microtubule capture by motor proteins is important for spindle positioning in animals (Kiyomitsu, 2019) and yeast (Gupta et al., 2006). Non–motor-dependent mechanisms, such as harnessing the energy of a depolymerizing microtubule, also generate pulling forces (Dogterom et al., 2005; Grishchuk et al., 2005). TAN1, because it lacks canonical motor domains, is unlikely to be a motor protein. However, similar to the microtubule binding protein tau, it is both highly basic and is predicted to contain intrinsically disordered regions when analyzed by the prediction software DisEMBL (Linding et al., 2003).
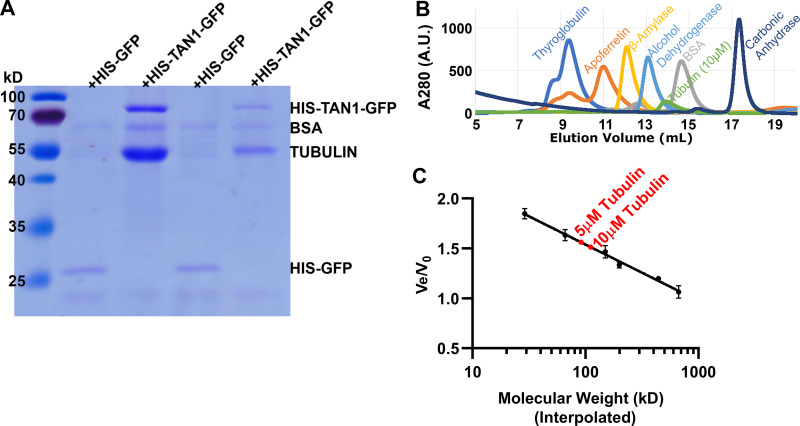
We were surprised that significant numbers of microtubule interactions were detectable in vitro only with relatively high concentrations of TAN1 (2 µM), when TAN1–MT interactions were detected using GMPCPP-stabilized microtubule seeds at low TAN1 concentrations (100 nM). One potential reason for this apparent discrepancy in binding or interaction could be due to TAN1 binding tubulin dimers in addition to microtubules. Tubulin dimer binding in addition to microtubule binding occurs with proteins such as tau (Fauquant et al., 2011) or Clasp (Al-Bassam et al., 2010). Therefore, we tested whether TAN1 binds soluble tubulin using in vitro affinity chromatography. Tubulin was incubated with HIS-TAN1-GFP and anti-GFP agarose beads. HIS-TAN1-GFP pulled down tubulin while HIS-GFP did not, indicating that TAN1 interacts with tubulin in addition to microtubule polymers (Fig. S3 A). By densitometry analysis, we estimate that one HIS-TAN1-GFP molecule binds to approximately two tubulin dimers (n = 3 replicates), indicating that TAN1 contains at least two distinct tubulin-binding regions. We used size exclusion chromatography to assess whether tubulin was dimeric in the affinity chromatography buffer (BRB80) and temperature conditions (∼4°C). Tubulin eluted with an apparent size of ∼110 kD, consistent with tubulin dimerization using both the same concentration of tubulin used for affinity chromatography (5 µM, 91.45 ± 12.32 kD [average ± SD]) and twice as much (10 µM, 111.13 ± 14.18 kD [average ± SD]; Fig. S3, B and C). Overall, this suggests that TAN1 binds tubulin in two distinct regions. TAN1–tubulin binding may potentially sequester TAN1 both in dynamic microtubule assays and in vivo. Alternatively, TAN1–tubulin binding may promote microtubule rescue, similar to Clasp (Al-Bassam et al., 2010). Further experiments would need to be performed to determine whether TAN1 dimerizes or multimerizes, whether tubulin binding occurs in vivo, and whether tubulin and microtubule binding sites overlap, as well as their relative affinities.
Figure S3.
HIS-TAN1-GFP binds to soluble tubulin dimers. (A) Agarose beads fused with an anti-GFP antibody were used to pull down HIS-TAN1-GFP or HIS-GFP in the presence of tubulin dimers. A tubulin band is detected after pull-downs in the presence of HIS-TAN1-GFP (1 µM) and not detected in the pull-down with HIS-GFP (1 µM). Coomassie-stained SDS-PAGE results from two independent in vitro pull-down are shown. Band intensity differences reflect differences in the relative amount of immunoprecipitated HIS-TAN1-GFP between experiments. (B) Size exclusion chromatography of one representative replicate of molecular weight standards and tubulin using fast protein liquid chromatography. The A280 (a.u.) for each standard and tubulin was plotted against elution volume (Ve) for the same running conditions to determine the Ve for each protein. (C) Interpolated molecular weights for tubulin were plotted on a semilog Ve/V0 curve determined from molecular weight standards (void volume determined by running blue dextran through column). Two replicates for 5 µM tubulin (91.45 ± 12.32 kD [average ± SD]) and 10 µM tubulin (111.13 ± 14.18 kD) and protein standards were run, indicating that tubulin elutes as a dimer.
Abnormal cell shape is likely responsible for spatial positioning defects of the PPB in the tan1 mutant
Defects in division plane orientation can occur early in the cell cycle, before the formation of the PPB, or later, after the PPB has already formed. We showed using live-cell imaging that tan1 mutant phragmoplasts did not return to the division site previously marked by the PPB, indicating a later defect in division plane orientation (Martinez et al., 2017). In contrast, previous work indicated that the orientation of the PPB is more variable in tan1 mutant compared with WT cells, indicative of a potential PPB placement defect (Cleary and Smith, 1998; Mir et al., 2018). However, whether TAN1 contributes to proper PPB placement is unclear, because TAN1 protein does not accumulate at the division site until late G2, after the PPB has already formed (Martinez et al., 2017).
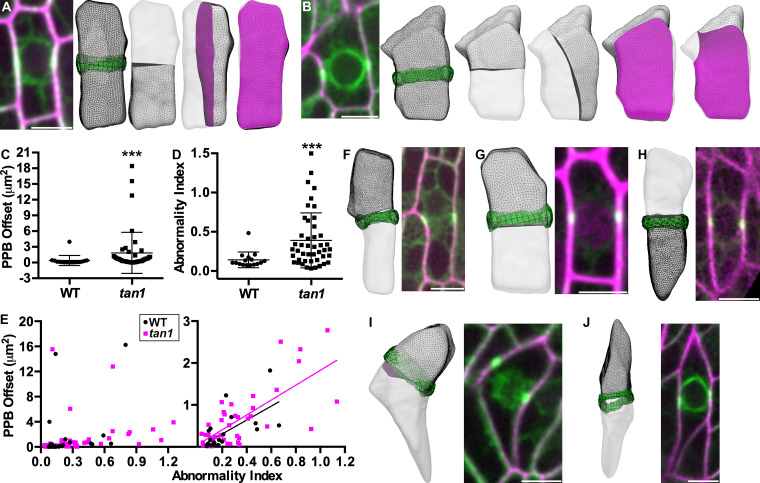
Previous measurements of PPB placement were obtained from 2D micrographs, which might not accurately reflect the position of the PPB in 3D, particularly in cells with irregular shapes. To overcome this shortcoming, we used our recently developed mathematical modeling approach to accurately predict 3D division planes (Martinez et al., 2018). This model generates soap-film minima from real, 3D cell shapes and allows us to compare purely geometric predictions to in vivo cell division sites (Martinez et al., 2018). The majority of predicted divisions closely match in vivo animal and plant cell divisions (Martinez et al., 2018). We collected confocal Z-stacks and used the image processing software MorphoGraphX (Barbier de Reuille et al., 2015) to extract WT (Fig. 3 A) and tan1 mutant 3D cell shapes (Fig. 3 B). We then used Surface Evolver to generate 3D reconstructions of the cells. Then, the gradient descent function in Surface Evolver was used to generate soap-film minima that divided the volume into two equal halves. These soap-film minima are division planes predictions (Martinez et al., 2018; Brakke, 1992). The predicted division planes were then compared with the in vivo PPB location (Fig. 3, A and B). To measure the offset between the predicted division and the location of the PPB, we compared the location of the midplane of the PPB to the outer edge of the predicted division. When the value of the PPB offset is low, the prediction matches the in vivo division plane. For WT cells, the average PPB offset from the predicted divisions was 0.40 µm2 ± 0.96 (average ± SD, n = 16), while PPB offset was higher in tan1 mutants (PPB offset = 1.85 µm2 ± 3.93, average ± SD, n = 45; P = 0.0012, Mann–Whitney; Fig. 3 C).
Figure 3.
Abnormally shaped cells have higher PPB offset in WT and tan1 cells. (A) On the left is a micrograph of WT maize leaf epidermal cell expressing YFP-TUBULIN (green) stained with propidium iodide (magenta) next to the 3D cell shape reconstruction generated with Surface Evolver (gray mesh). The green band is the 3D reconstruction of the PPB. Predicted soap-film minimum divisions for the cell are shown with magenta meshes, from left to right transverse, longitudinal, periclinal division planes. (B) Micrograph of tan1 maize leaf epidermal cell next to cell shape reconstruction with PPB overlaid and (from left to right) transverse, longitudinal, periclinal, and other division planes. (C) PPB offset in tan1 mutants is significantly higher than WT (WT n = 16 PPB offset = 0.40 µm2 ± 0.96, tan1 n = 45 1.85 µm2 ± 3.93 [average ± SD]; ***, P = 0.0012, Mann–Whitney). (D) Abnormality index in tan1 mutants is significantly higher compared with WT (WT: n = 16, abnormality index = 0.14 ± 0.1 [average ± SD]; tan1: n = 45, abnormality index = 0.37 ± 0.32; ***, P < 0.0008, Mann–Whitney). (E) Abnormality index versus PPB offset suggests abnormal cell shapes generally show higher offsets (WT: r = 0.57, P = 0.007; tan1: r = 0.59; P < 0.0001, Spearman correlation coefficient). A subset of data (values of PPB offset < 3) is displayed adjacent with a linear fit of WT (r = 0.7, P = 0.0003, n = 22) and tan1 (r = 0.6, P < 0.0001, n = 40; Spearman correlation coefficient). (F–J) Best-fit predicted divisions overlaid with in vivo PPB location next to corresponding micrograph of maize epidermal cells expressing YFP-TUBULIN (green) and either expressing membrane marker PIP2-CFP (H, magenta) or stained with propidium iodide (F, G, I, and J, magenta) to outline the cell shape. (F) Example of a WT cell with an abnormality index of 0.59 and a PPB offset of 1.82 µm2. (G) Example of a WT cell with an abnormality index of 0.09 and a PPB offset of 0.39 µm2. (H) Example of a tan1 mutant cell with an abnormality index of 0.32 and a PPB offset of 0.26 µm2. (I) Example of a tan1 mutant cell with an abnormality index of 1.25 and a PPB offset of 3.92 µm2. (J) Example of a tan1 mutant cell with an abnormality index of 1.14 and a PPB offset of 1.08 µm2. Scale bars, 10 µm.
To determine whether the increased PPB offset in tan1 mutants is due to improper PPB placement or an indirect consequence of abnormal cell shapes in the tan1 mutant, we developed a quantitative method to compare cell shapes called the “abnormality index” by measuring the distance between the surface area center and volume center (see Materials and methods). WT cells had an approximately threefold lower and more consistent abnormality index compared with tan1 mutant cells (Fig. 3 D; WT cells: n = 16, abnormality index [average ± SD] = 0.14 ± 0.1; tan1: n = 45, abnormality index = 0.39 ± 0.35; P < 0.0008, Mann–Whitney). These data confirm that WT plants tend to have normally shaped cells, while tan1 mutants have cells with both normal and abnormal shapes, consistent with our imaging data.
If TAN1 plays a direct role in PPB placement, we would expect abnormal PPB placement in tan1 mutants regardless of variations in cell shape abnormality index. In contrast, we found a significant positive correlation between abnormality index and PPB offset in tan1 mutant cells (Spearman correlation coefficient = 0.59, P < 0.0001, n = 45 cells), suggesting that PPB placement deviated from predicted divisions more in highly abnormally shaped cells. To address whether this trend was similar in WT cells, we specifically looked for and modeled additional WT cells which displayed aberrant cell shapes with high abnormality indices (Spearman correlation coefficient = 0.57, P value = 0.003 n = 25 cells). Both WT and tan1 mutant cells with higher abnormality indices typically had higher PPB offsets for the whole dataset (Fig. 3 E, left panel) as well as the dataset removing outliers (Fig. 3 E, right panel), with examples of cells with high abnormality indices shown in (Fig. 3, F–J). Due to the correlation between PPB placement defects and aberrant cell shapes in tan1 mutants, we hypothesize that defects in PPB placement are a consequence of cell shape abnormalities and not directly related to TAN1 function during G2.
Modeling approaches based on microtubule organization suggest that interphase cortical microtubule arrangements may be an important modulator in PPB positioning (Chakrabortty et al., 2018; Mirabet et al., 2018). The orientation of the PPB typically follows the orientation of the prior interphase microtubule array (Flanders et al., 1989; Gunning and Sammut, 1990). Our result suggests that intrinsically abnormally shaped cells may lead, in the next round of cell division, toward less geometrically accurately placed PPBs. This effect may explain why other division plane mutants have offset or oblique PPBs (Müller et al., 2006; Pietra et al., 2013). Additionally, mutants with cell expansion defects that cause aberrant cell shapes may also lead first to misoriented PPBs and then apparent division plane defects.
Spindle organization is disrupted in the tan1 mutant
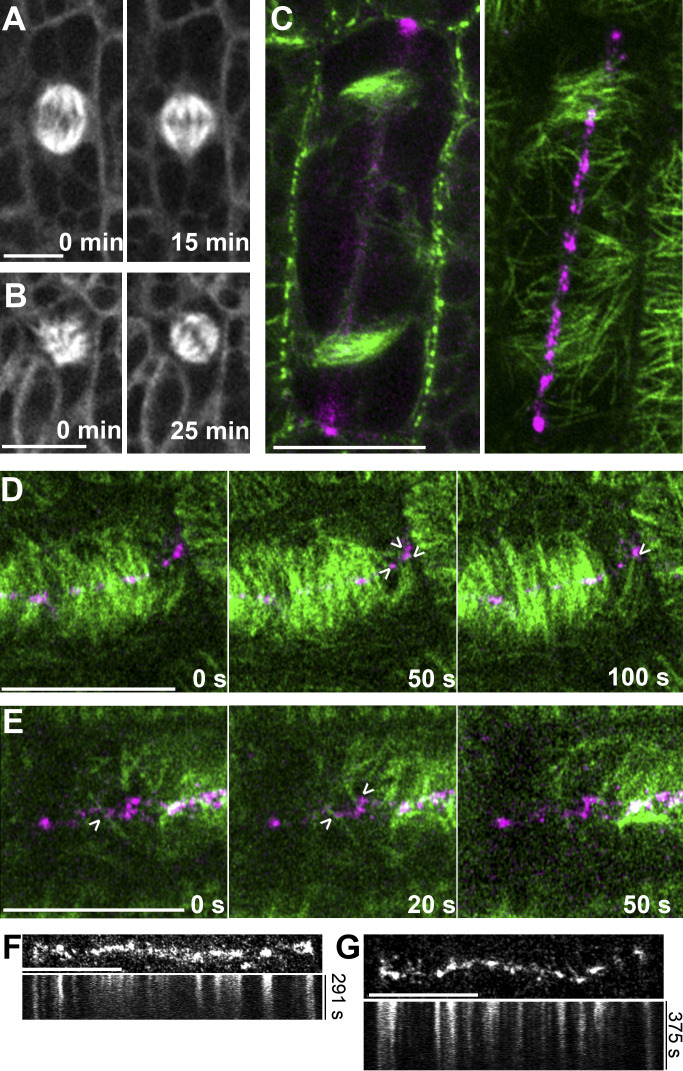
Previously, we showed that tan1 mutant cells had mitotic progression delays during metaphase and telophase, but we did not propose a specific hypothesis to explain why delays occurred (Martinez et al., 2017). If TAN1 plays a significant role in cross-linking spindle microtubules, then metaphase delays may reflect defective spindle organization. Using time-lapse imaging, we assessed overall spindle morphology in maize leaf cells expressing YFP-TUBULIN. In WT cells, we always observed bipolar spindles (n = 38; Fig. 4 A). In tan1 mutant cells, spindles occasionally displayed delayed bipolar organization (13.5%; n = 5/35), but recovered after ∼20 ± 8 min (average ± SD) following nuclear envelope breakdown into typical bipolar spindles (Fig. 4 B and Video 2). Metaphase delays previously described in tan1 mutants occurred frequently, leading to an average 1.5× time delay compared with WT (Martinez et al., 2017), whereas delayed bipolar spindle organization defects were more rare. This suggests that defects in microtubule organization only occasionally lead to detectable defects in spindle organization in the tan1 mutant, consistent with redundant mechanisms for spindle assembly. Metaphase spindle microtubules cross-linking or bundling is important for proper and timely spindle assembly (Masoud et al., 2013; Mullen and Wignall, 2017; Ambrose and Cyr, 2007; Winters et al., 2019). Based on in vitro microtubule zippering by TAN1, it is possible that TAN1 mediates bundling of spindle microtubules as they encounter each other at shallow angles. Thus, TAN1 localization to the spindle might be important for correct spindle assembly and mitotic progression through metaphase.
Figure 4.
TAN1-mediated microtubule interactions may organize spindles and promote proper phragmoplast guidance. (A) WT maize epidermal cell expressing YFP-TUBULIN displays normal bipolar spindle morphology over time. (B) tan1 maize epidermal cell expressing YFP-TUBULIN has a disorganized spindle that recovers to canonical bipolar organization. Time lapse of spindles is shown in Video 2. (C and D) WT maize epidermal cells expressing CFP-TUBULIN (green) and TAN1-YFP (magenta). (C) Micrographs display both at the midplane and cortex of cell undergoing a longitudinal division. Phragmoplast and leading-edge microtubules at the cortex are positioned at the division site and are partially colocalized with TAN1. (D and E) Time lapse showing potential phragmoplast leading-edge microtubule contact and interaction with TAN1 at the division site (arrowheads). Fig. 4 D is shown in Video 3. (F and G) Time lapse and kymograph of TAN1-YFP at the division site. Scale bars, 10 µm.
Video 2.
Spindle morphology during WT and tan1 mutant epidermal cells. Video of spindles is 10 frames per seccond for 5 min.
Microtubules and TAN1 colocalize at the division site during telophase
To understand how TAN1 might mediate phragmoplast guidance during telophase (Martinez et al., 2017; Mir et al., 2018), we imaged TAN1 and microtubules at the division site. CFP-TUBULIN–labeled microtubules and TAN1-YFP were imaged together in cells undergoing longitudinal divisions, where phragmoplast guidance is more readily visualized. Colocalization of CFP-TUBULIN and TAN1-YFP at the division site was assessed at the cell cortex after initial phragmoplast contact. A small number of phragmoplast microtubules colocalize with TAN1 puncta (Pearson’s correlation coefficient = 0.23 ± 0.078 [average ± SD], n = 21), but approximately half of the TAN1 puncta were associated with microtubules (Manders overlay coefficient [C] = 0.41 ± 0.1 [average ± SD]; Fig. 4 C). Together, these results suggest that a small subpopulation of microtubules from the leading edge of the phragmoplast interact with cortical TAN1 puncta as the phragmoplast expands across the division site (Fig. 4, D and E; and Video 3). These TAN1 puncta at the division site do not appear to be mobile over imaging of ∼5 min (n = 8 cells; Fig. 4, F and G).
Video 3.
Phragmoplast leading-edge microtubule interactions with TAN1-YFP at the division site. TAN1-YFP is magenta and microtubules are green; two frames per second for 3 s.
Models for phragmoplast guidance previously proposed that leading-edge phragmoplast microtubules interact with proteins at the cortical division site either through specific protein–protein interactions or microtubule–protein interactions (Herrmann et al., 2018; Lipka et al., 2014; Li et al., 2017). POK2, which is localized to the division site, was shown to be a plus-end–directed kinesin (Chugh et al., 2018). POK2 may effectively push against the plus ends of microtubules that encounter the division site (Chugh et al., 2018). POK2 also directly interacts with MAP65-3, which is localized to bundled microtubules both at the phragmoplast midzone and leading edge, serving as another potential type of interaction between the phragmoplast and the division site. The localization of TAN1 at the division site is important for its function in phragmoplast guidance (Mir et al., 2018; Martinez et al., 2017). Based on the results from this study, we propose that end-on interactions between the plus ends of phragmoplast leading-edge microtubules and TAN1-YFP puncta at the division site may exert pulling forces on these microtubules to guide phragmoplast trajectory.
While TAN1 has long been characterized as a microtubule-binding protein, the functional significance of this finding remained elusive. Our in vitro analysis of TAN1–microtubule activities combined with live-imaging observations of TAN1 localization on spindle microtubules and at phragmoplast leading-edge microtubule tips suggests that TAN1–microtubule interactions may depend on the geometry of microtubule encounters. This provides a plausible explanation for how TAN1 contributes to spindle organization and phragmoplast guidance.
Materials and methods
HIS-TAN1 and HIS-TAN1-GFP purification and labeling
A codon-optimized cDNA encoding the maize HIS-TAN1 and HIS-TAN1-GFP was synthesized in vitro, followed by protein expression and purification, all performed by Genscript. Escherichia coli strain SHuffle was transformed with recombinant plasmid encoding HIS-TAN1. After cell pellets were sonicated and centrifuged, the precipitate was dissolved using urea, followed by affinity purification. E. coli strain BL21 Star (DE3) was transformed with recombinant plasmid encoding HIS-TAN1-GFP. After cell pellets were sonicated and centrifuged, the precipitate was dissolved using urea, followed by affinity purification (Genscript). Proteins were refolded and sterilized by filtering. HIS-TAN1 and HIS-TAN1-GFP concentrations were checked with a BCA protein assay (Genscript). After refolding, HIS-TAN1-GFP was no longer fluorescent. HIS-TAN1-GFP therefore was tagged with an Atto488 dye. HIS-TAN1-GFP was conjugated with Atto488-maleimide (Sigma; 28562). 4 µM HIS-TAN1-GFP in 80 mM Pipes, 1 mM MgCl2, and 1 kmM EGTA buffer was reduced with 12.5 µM Tris(2-carboxyethyl)phosphine hydrochloride for 10 min followed by a 4-h incubation with 250 µM Atto488 dissolved in DMSO (10 mM) at room temperature. Unreacted excess dye was removed by running the sample through a 10DG desalting column (Bio-Rad; 732–2010) and concentrating with a 30-kD molecular weight cut-off polyethersulfone membrane (30K MWCO PES) concentrator (Thermo; 88521). HIS-TAN1-GFP and HIS-TAN1-GFP-Atto488 (∼80% degree of labeling) activity was confirmed by microtubule cosedimentation assay. Conjugation of Atto488 dye was determined by imaging the results of the microtubule cosedimentation assay on a SDS-PAGE experiment using a UV light source showing fluorescent bands corresponding to a Atto488-tagged HIS-TAN1-GFP.
Microtubule binding and cosedimentation
A microtubule-binding assay kit was used to assess HIS-TAN1 microtubule binding in relation to positive and negative controls, according to manufacturer conditions (Cytoskeleton; MK029). For determining affinity of HIS-TAN1 to microtubules, microtubules were polymerized from a 50-µM starting concentration of tubulin in the presence of 1 mM GTP for 2 h at 37°C followed by the addition of 10 µM Taxol. HIS-TAN1 and microtubules were incubated for 25 min and spun down at 39,000 g at 25°C. HIS-TAN1-GFP and HIS-TAN1-GFP-Atto488 protein was incubated with microtubules at room temperature for 25 min and spun down at 21,000 g at 25°C. Equal volumes of soluble and pellet samples were loaded into an SDS-PAGE (10% gel) and stained with Coomassie. The percentage of TAN1 cosedimentation was determined by measuring the ratio between TAN1 protein found in the pellet over the total TAN1 protein found in both the pellet and soluble as determined by densitometry analysis using ImageJ Gel Analysis tool. Correction by subtracting TAN1 from the pellet fraction samples without microtubules was applied to spindowns due to some TAN1 precipitation during the assay in samples without microtubules. Spindowns were performed at least three times for each concentration tested. Curve-fitting and statistical analysis was performed using MATLAB and GraphPad Prism. Figure construction was performed using GraphPad Prism. To assess microtubule binding by microscopy, rhodamine-labeled microtubules (1:25 rhodamine tubulin/unlabeled tubulin) were polymerized from 50 µM starting concentration of tubulin in the presence of 1 mM GTP for 2 h at 37°C followed by the addition of 10 µM Taxol. 100 nM rhodamine-labeled microtubules were incubated with 50 nM HIS-TAN1-Atto488 for 5 min and then pipetted onto a coverslip and imaged. Time-lapse analysis was performed using 1 µM rhodamine-labeled GMPCPP-stabilized microtubules (1:25 rhodamine tubulin/unlabeled tubulin) incubated with 1 nM HIS-TAN1-Atto488 for 10 min and then imaged at 1- or 2-s intervals.
Transient expression in N. benthamiana
5-wk-old N. benthamiana plants grown under standard 16-h light, 8-h dark conditions were used for transient colocalization experiments. Plasmids for constitutively expressing the viral protein p19, RFP-TUBULIN6 (Ambrose et al., 2011), and TAN1-GFP (Walker et al., 2007) were transformed into Agrobacterium tumefaciens strain GV3101. Agrobacteria were grown to stationary phase, spun down at 1,000 rpm, and then resuspended for 1 h at room temperature in infiltration buffer containing 10 mM MES (pH 5.7), 10 mM MgCl2, 0.5% D-glucose (wt/vol), and 200 µM acetosyringone. Equal amounts of agrobacteria (with and without the TAN1-GFP) were mixed together, and a 1-ml syringe without a needle was used to infiltrate the abaxial side of N. benthamiana leaves. After 3 d of incubation, the leaves were removed, and the abaxial epidermal cells were imaged using the spinning confocal disk microscope with the 60× objective described above. Maximum intensity projections and automatic background subtraction in FIJI were used in Fig. S2.
Reconstitution of in vitro microtubule dynamics
In vitro microtubule dynamics were conducted according to previous protocols (Dixit and Ross, 2010). Flow chambers were assembled using silanized coverslips and double-sided sticky tape with a chamber volume of ∼20 µl. A 20% monoclonal anti-biotin antibody (Sigma; clone BN-34) was used to coat the surface followed by blocking with 5% pluronic F-127 (Sigma; #P2443) for 5 min each step. Rhodamine and GMPCPP microtubule seeds were then flowed into the cell. Microtubule growth was initiated using 17.5-µM 1:25 rhodamine-labeled bovine tubulin in 80 mM Pipes, 1 mM MgCl2, 1 mM EGTA with 0.15% methylcellulose (wt/vol), 100 mM DTT, oxygen scavengers (250 µg/ml glucose oxidase, 25 µg/ml catalase), 5 mg/ml glucose, and 2 mM GTP along with the specified amount of HIS-TAN1 protein. To assess microtubule bundling, a higher concentration of tubulin (22.5 µM, 1:25 rhodamine tubulin/unlabeled tubulin) was used in the reaction to promote microtubule growth and crossovers. At least two slides were prepared for each concentration and experimental condition. The samples were excited with a 561-nm (at 4 mW output) diode-pumped solid-state laser (Melles Griot) and visualized through a 100× objective (NA 1.45) and back-illuminated electron-multiplying charge-coupled device camera with a 582–636-nm emission filter set using TIRF (Hammamatsu; ImageEM). Images were collected every 2 s. Kymographs were used to analyze data in FIJI (Schindelin et al., 2012).
In vitro pull-down of HIS-TAN1-GFP and tubulin
10 µl agarose beads bound to anti-GFP (MBL D153-8) was incubated with 500 nM HIS-TAN1-GFP (for calculation of stoichiometry) or 1 µM HIS-TAN1-GFP (for determining ability to pull down tubulin) or 1 µM HIS-GFP (ABM; 00033P) and 5 µM of TUBULIN (Cytoskeleton; T240) in BRB80 buffer with 50 mM NaCl, 0.2 mM PMSF, 10 mM DTT, and 0.05% Tween-20. Samples were incubated for 3 h at 4°C and subsequently washed three times using BRB80 supplemented with 50 mM NaCl and 0.05% Tween-20. Beads were then transferred to a new tube and washed four more times with BRB-80 supplemented with 50 mM NaCl and 0.05% Tween-20. Coomassie-stained SDS-PAGE was used for protein visualization and analyzed by densitometry using ImageJ to determine protein amounts that were pulled down.
Size exclusion chromatography
Molecular weight standards were prepared according to manufacturer specifications, except they were resuspended in BRB80 buffer (Sigma; MWGF1000). Equal sample volume of standard was run twice on a Superdex 200 Increase 10/300 GL column (GE Lifesciences) using an NGC Chromatography System (Bio-Rad; 2-ml injection volume and 0.25-ml/min flow rate) with absorbance at 280 nm recorded during the experiment (Fig. S3 B). Blue Dextran (Sigma; MWGF1000) was used to determine void volume of the column. A semilog plot of elution volume over void volume for each standard versus molecular weight was constructed to calculate a standard curve to determine tubulin molecular weight (GraphPad Prism 8.4; Fig. S3 C). An equal volume of 1 mg/ml (10 µm) and 0.5 mg/ml (5 µm) tubulin (Cytoskeleton; T240) in BRB80 buffer was run on column to determine likely oligomerization state of tubulin used for experiments.
Predicting division planes from WT and tan1 cell shapes using Surface Evolver
Samples from WT and tan1 mutant maize plants expressing YFP-TUBULIN (α-tubulin fused to the citrine variant of YFP; Mohanty et al., 2009) were dissected to the symmetrically dividing leaf zones to identify PPB location. To identify the cell outlines for 3D reconstruction, samples were either stained with 0.1 mM propidium iodide or expressed Plasma membrane intrinsic protein 2-1 fused to CFP to outline the plasma membranes (Mohanty et al., 2009). 3D cell shape reconstructions were generated using MorphoGraphX, while 3D PPB reconstructions were generated using Trainable Weka Segmentation (Barbier de Reuille et al., 2015; Arganda-Carreras et al., 2017). Cells were collected from more than three individual plants for each genotype. A previous protocol was followed for modeling symmetric divisions by soap-film minimization using Surface Evolver (Brakke, 1992; Martinez et al., 2018). This model generates soap-film minima from real, 3D cell shapes to explicitly test the hypothesis that plant cell divisions mimic mathematically predicted soap-film minima (Errera, 1888). As we previously demonstrated for both plant and animal cells, the majority of predicted divisions closely match in vivo divisions (Martinez et al., 2018). This model does not take into account cell–cell interactions, mechanical or developmental cues. Briefly, cell outlines were smoothed using 30th-degree spherical harmonics followed by surface area minimization from 241 starting planes with normals uniformly distributed over a sphere. For PPB offset measurements, the distance between the midplane of the PPB and the surface of the predicted division was measured in microns squared. The abnormality index was defined by the distance between the area surface center and the volume center for the cell. The Surface Evolver pipeline can be downloaded from GitHub (https://github.com/jdhayes/predictive_division/).
Colocalization analysis
Maize plants were dissected to reveal the symmetrically dividing leaf zones to image TAN1-YFP and CFP-TUBULIN at the cortex of maize epidermal cells during telophase using a Zeiss 880 LSM. Airyscan super resolution mode was used and the images were processed using default settings. Three separate plants were imaged for the collection of cells. Micrographs were imported into FIJI and cropped to the cell of interest, where colocalization was assessed. Just Another Colocalization Plugin was used in order to determine the Pearson correlation coefficient and Manders overlap coefficient for each cell (Bolte and Cordelières, 2006). Data generated were analyzed using GraphPad (Prism).
Microscopy for in vitro and in vivo imaging
Taxol-stabilized, rhodamine-labeled microtubules and HIS-TAN1-GFP-Atto488 were visualized on an inverted Nikon Ti stand with a W1 spinning disk (Yokogawa) and a motorized stage (ASI Piezo) run with Micromanager software (micromanager.org) and built by Solamere Technology. Time lapse of rhodamine-labeled GMPCPP stabilized microtubules and HIS-TAN1-Atto488 was also imaged on this microscope. Solid-state lasers (Obis) and emission filters (Chroma Technology) used had excitation 561 nm, emission 620/60 nm for rhodamine-tubulin and excitation 488 nm, emission 520/50 nm for HIS-TAN1-GFP-Atto488. A 100× oil lens (1.45 NA) and Immersion Oil Type FF (Cargille; immersion oil, 16212) was used. Maize epidermal cells used for modeling were visualized using a 60× water-immersion objectives with 1.2 NA. An excitation of 561 nm and an emission of 620/60 nm were used for propidium iodide, and an excitation of 514 nm and an emission of 540/30 nm were used for YFP-TUBULIN. Perfluorocarbon immersion liquid (Cargille; RIAAA-678) was used on the objective.
Dynamic rhodamine-labeled microtubules were excited with a 561-nm (at 4 mW output) diode-pumped solid-state laser (Melles Griot) using a 100× (NA 1.45) objective and TIRF microscopy, described above. Images were acquired with a back-illuminated electron-multiplying charge-coupled device camera (Hamamatsu ImageEM) and rhodamine filter sets (582–636-nm emission).
Colocalization data on TAN1-YFP and CFP-TUBULIN in Fig. 4 were collected using a Zeiss LSM 880 Elyra, Axio Observer and a 100×/1.46 NA oil lens (Cargille; immersion oil, 16212). TAN1-YFP was excited using 514 nm, while CFP-TUBULIN was excited using 458 nm and imaged using superresolution Airyscan mode with a main beam splitter 458/514 and 420–480 band pass + long pass 605 filter set. Airyscan images were processed using default settings using Zen Black software (Zeiss).
Online supplemental material
Fig. S1 shows HIS-TAN1-GFP and HIS-TAN1-GFP-Atto488 microtubule binding and affinity using a quantitative microtubule cosedimentation assay. Fig. S2 shows colocalization of TAN1 and cortical microtubules in N. benthamiana (tobacco). Fig. S3 shows HIS-TAN1-GFP and tubulin pull-down and confirmation of tubulin dimerization by size exclusion chromatography. Video 1 shows HIS-TAN1–mediated microtubule cross-linking events observed during in vitro dynamic microtubule reconstitution assays imaged using TIRF microscopy. Video 2 displays examples of spindle organization of WT (left) and two tan1 mutant cells as they progress through mitosis. Video 3 shows potential microtubule interactions between the phragmoplast leading edge and TAN1-YFP protein localized at the cortical division site in maize epidermal leaf cells. Table S1 displays Model fitting for TAN1–microtubule binding data.
Supplementary Material
shows model fitting for TAN1–microtubule binding data.
Acknowledgments
Thanks to Ms. Jocelyne Aranda, Mr. Christopher Hoyt, and Ms. Sukhmani Sidhu for collecting some images used for the modeling work presented. Thanks to Dr. Hong Liang for help with transient tobacco assays. We thank Dr. David Carter (University of California, Riverside, Riverside, CA) for Zeiss LSM 880 training. We thank Prof. Laurie Smith (University of California, San Diego, La Jolla, CA) for the TAN1-GFP plasmid and Prof. Chris Ambrose (University of Saskatchewan, Saskatoon, Canada) for the RFP-TUBULIN plasmid. We also thank the University of California, Riverside Agricultural Operations for greenhouse and field space and David Wetovick (University of California, Riverside Agricultural Operations, Riverside, CA) for helpful field management.
This work was funded by the National Science Foundation (grant MCB1716972 to C.G. Rasmussen and grant MCB1453726 to R. Dixit) and the U.S. Department of Agriculture National Institute of Food and Agriculture (C.G. Rasmussen). Funding from a Ford Foundation dissertation fellowship to P. Martinez is gratefully acknowledged.
The authors declare no competing financial interests.
Author contributions: C.G. Rasmussen, R. Dixit, A. Zhang, S.E. O’Leary, and K.A. Brakke provided equipment, reagents, and experimental guidance. P. Martinez and R.S. Balkunde performed in vitro experiments, with R.S. Balkunde offering assistance on microtubule cosedimentation. P. Martinez captured images used for modeling and performed live-cell time lapse. P. Martinez and A. Zhang performed size exclusion chromatography. C.G. Rasmussen, S.E. O’Leary, and R. Dixit supervised experiments. P. Martinez and C.G. Rasmussen analyzed data and made figures, P. Martinez and C.G. Rasmussen wrote the manuscript, with comments and edits from coauthors. C.G. Rasmussen, P. Martinez, and R. Dixit acquired funding.
References
- Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., and Chang F.. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 19:245–258. 10.1016/j.devcel.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., and Cyr R.. 2007. The kinesin ATK5 functions in early spindle assembly in Arabidopsis. Plant Cell. 19:226–236. 10.1105/tpc.106.047613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., and Cyr R.. 2008. Mitotic spindle organization by the preprophase band. Mol. Plant. 1:950–960. 10.1093/mp/ssn054 [DOI] [PubMed] [Google Scholar]
- Ambrose C., Allard J.F., Cytrynbaum E.N., and Wasteneys G.O.. 2011. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2:430 10.1038/ncomms1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., and Seung S.H.. 2017. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 33:2424–2426. 10.1093/bioinformatics/btx180 [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P., Routier-Kierzkowska A.-L., Kierzkowski D., Bassel G.W., Schüpbach T., Tauriello G., Bajpai N., Strauss S., Weber A., Kiss A., et al. 2015. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 4 e05864 10.7554/eLife.05864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I., Beemster G.T.S., Judy-March J.E., and Marga F.. 2004. Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135:2279–2290. 10.1104/pp.104.040493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Brakke K.A. 1992. The Surface Evolver. Exp. Math. 1:141–165. 10.1080/10586458.1992.10504253 [DOI] [Google Scholar]
- Burkart G.M., and Dixit R.. 2019. Microtubule bundling by MAP65-1 protects against severing by inhibiting the binding of katanin. Mol. Biol. Cell 30:1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Dols J., Kopischke S., Peña E.J., Andrade-Navarro M.A., Heinlein M., Szymanski D.B., Zachgo S., Doonan J.H., and Lloyd C.W.. 2015. Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain. J. Cell Sci. 128:2033–2046. 10.1242/jcs.156570 [DOI] [PubMed] [Google Scholar]
- Chakrabortty B., Blilou I., Scheres B., and Mulder B.M.. 2018. A computational framework for cortical microtubule dynamics in realistically shaped plant cells. PLOS Comput. Biol. 14 e1005959 10.1371/journal.pcbi.1005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., and Lloyd C.. 2005. Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell. 17:1737–1748. 10.1105/tpc.105.032615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh M., Reißner M., Bugiel M., Lipka E., Herrmann A., Roy B., Müller S., and Schäffer E.. 2018. Phragmoplast Orienting Kinesin 2 Is a Weak Motor Switching between Processive and Diffusive Modes. Biophys. J. 115:375–385. 10.1016/j.bpj.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary A.L., and Smith L.G.. 1998. The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell. 10:1875–1888. 10.1105/tpc.10.11.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., and Ross J.L.. 2010. Studying plus-end tracking at single molecule resolution using TIRF microscopy. Methods Cell Biol. 95:543–554. 10.1016/S0091-679X(10)95027-9 [DOI] [PubMed] [Google Scholar]
- Dixit R., and Cyr R.. 2004. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell. 16:3274–3284. 10.1105/tpc.104.026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., Kerssemakers J.W.J., Romet-Lemonne G., and Janson M.E.. 2005. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 17:67–74. 10.1016/j.ceb.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Drechsler H., Xu Y., Geyer V.F., Zhang Y., and Diez S.. 2019. Multivalent electrostatic microtubule interactions of synthetic peptides are sufficient to mimic advanced MAP-like behavior. Mol. Biol. Cell. 30:2953–2968. 10.1091/mbc.E19-05-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevensek S., Goussot M., Duroc Y., Christodoulidou A., Steyaert S., Schaefer E., Duvernois E., Grandjean O., Vantard M., Bouchez D., et al. 2012. The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell. 24:178–191. 10.1105/tpc.111.089748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D.W., and Shaw S.L.. 2006. Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57:859–875. 10.1146/annurev.arplant.57.032905.105329 [DOI] [PubMed] [Google Scholar]
- Elliott A., and Shaw S.L.. 2018. Update: Plant Cortical Microtubule Arrays. Plant Physiol. 176:94–105. 10.1104/pp.17.01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errera L. 1888. Über Zellformen und Siefenblasen. Bot. Centralblatt. 34:395–399. [Google Scholar]
- Facette M.R., Rasmussen C.G., and Van Norman J.M.. 2019. A plane choice: coordinating timing and orientation of cell division during plant development. Curr. Opin. Plant Biol. 47:47–55. 10.1016/j.pbi.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Fache V., Gaillard J., Van Damme D., Geelen D., Neumann E., Stoppin-Mellet V., and Vantard M.. 2010. Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell. 22:3804–3815. 10.1105/tpc.110.080606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquant C., Redeker V., Landrieu I., Wieruszeski J.-M., Verdegem D., Laprévote O., Lippens G., Gigant B., and Knossow M.. 2011. Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J. Biol. Chem. 286:33358–33368. 10.1074/jbc.M111.223545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanders D.J., Rawlins D.J., Shaw P.J., and Lloyd C.W.. 1989. Computer-aided 3-D reconstruction of interphase epidermal cells of Datura stramonium reveals assembly. Development. 106:531–541. [Google Scholar]
- Gaillard J., Neumann E., Van Damme D., Stoppin-Mellet V., Ebel C., Barbier E., Geelen D., and Vantard M.. 2008. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell. 19:4534–4544. 10.1091/mbc.e08-04-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., and McIntosh J.R.. 2005. Force production by disassembling microtubules. Nature. 438:384–388. 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Gunning B., and Sammut M.. 1990. Rearrangements of Microtubules Involved in Establishing Cell Division Planes Start Immediately after DNA Synthesis and Are Completed just before Mitosis. Plant Cell. 2:1273–1282. 10.2307/3869345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L. Jr., Carvalho P., Roof D.M., and Pellman D.. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923. 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Herrmann A., Livanos P., Lipka E., Gadeyne A., Hauser M.T., Van Damme D., and Müller S.. 2018. Dual localized kinesin-12 POK2 plays multiple roles during cell division and interacts with MAP65-3. EMBO Rep. 19:46085 10.15252/embr.201846085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-M.K., Hotta T., Guo F., Roberson R.W., Lee Y.-R.J., and Liu B.. 2011. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell. 23:2909–2923. 10.1105/tpc.110.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-M.K., Lee Y.-R.J., Kiyama L.D., Dinesh-Kumar S.P., and Liu B.. 2012. Arabidopsis microtubule-associated protein MAP65-3 cross-links antiparallel microtubules toward their plus ends in the phragmoplast via its distinct C-terminal microtubule binding domain. Plant Cell. 24:2071–2085. 10.1105/tpc.111.092569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson M.E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F.J., and Tran P.T.. 2007. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 128:357–368. 10.1016/j.cell.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Jürgens G. 2005a Cytokinesis in higher plants. Annu. Rev. Plant Biol. 56:281–299. 10.1146/annurev.arplant.55.031903.141636 [DOI] [PubMed] [Google Scholar]
- Jürgens G. 2005b Plant cytokinesis: fission by fusion. Trends Cell Biol. 15:277–283. 10.1016/j.tcb.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Kellogg E.H., Hejab N.M.A., Poepsel S., Downing K.H., DiMaio F., and Nogales E.. 2018. Near-atomic model of microtubule-tau interactions. Science. 360:1242–1246. 10.1126/science.aat1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J.C., Ehrhardt D.W., and Hülskamp M.. 2007. CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell. Sci. 120:4416–4425. 10.1242/jcs.024950 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T. 2019. The cortical force-generating machinery: how cortical spindle-pulling forces are generated. Curr. Opin. Cell Biol. 60:1–8. 10.1016/j.ceb.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Komis G., Luptovčiak I., Ovečka M., Samakovli D., Šamajová O., and Šamaj J.. 2017. Katanin Effects on Dynamics of Cortical Microtubules and Mitotic Arrays in Arabidopsis thaliana Revealed by Advanced Live-Cell Imaging. Front Plant Sci. 8:866 10.3389/fpls.2017.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., and Dogterom M.. 2012a Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Roth S., and Dogterom M.. 2012b End-on microtubule-dynein interactions and pulling-based positioning of microtubule organizing centers. Cell Cycle. 11:3750–3757. 10.4161/cc.21753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-R.J., and Liu B.. 2013. The rise and fall of the phragmoplast microtubule array. Curr. Opin. Plant Biol. 16:757–763. 10.1016/j.pbi.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Li H., Sun B., Sasabe M., Deng X., Machida Y., Lin H., Julie Lee Y.R., and Liu B.. 2017. Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol. 215:187–201. 10.1111/nph.14532 [DOI] [PubMed] [Google Scholar]
- Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., Emons A.M.C., Mulder B.M., Kirik V., and Ehrhardt D.W.. 2013. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science. 342 1245533 10.1126/science.1245533 [DOI] [PubMed] [Google Scholar]
- Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., and Russell R.B.. 2003. Protein disorder prediction: implications for structural proteomics. Structure. 11:1453–1459. 10.1016/j.str.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Lipka E., Gadeyne A., Stöckle D., Zimmermann S., De Jaeger G., Ehrhardt D.W., Kirik V., Van Damme D., and Müller S.. 2014. The Phragmoplast-Orienting Kinesin-12 Class Proteins Translate the Positional Information of the Preprophase Band to Establish the Cortical Division Zone in Arabidopsis thaliana. Plant Cell. 26:2617–2632. 10.1105/tpc.114.124933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos P., and Müller S.. 2019. Division Plane Establishment and Cytokinesis. Annu. Rev. Plant Biol. 70:239–267. 10.1146/annurev-arplant-050718-100444 [DOI] [PubMed] [Google Scholar]
- Martinez P., Luo A., Sylvester A., and Rasmussen C.G.. 2017. Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc. Natl. Acad. Sci. USA. 114:2759–2764. 10.1073/pnas.1619252114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P., Allsman L.A., Brakke K.A., Hoyt C., Hayes J., Liang H., Neher W., Rui Y., Roberts A.M., Moradifam A., et al. 2018. Predicting division planes of three-dimensional cells by soap-film minimization. Plant Cell. 30:2255–2266. 10.1105/tpc.18.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud K., Herzog E., Chabouté M.-E., and Schmit A.-C.. 2013. Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J. 75:245–257. 10.1111/tpj.12179 [DOI] [PubMed] [Google Scholar]
- Mir R., Morris V.H., Buschmann H., and Rasmussen C.G.. 2018. Division Plane Orientation Defects Revealed by a Synthetic Double Mutant Phenotype. Plant Physiol. 176:418–431. 10.1104/pp.17.01075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabet V., Krupinski P., Hamant O., Meyerowitz E.M., Jönsson H., and Boudaoud A.. 2018. The self-organization of plant microtubules inside the cell volume yields their cortical localization, stable alignment, and sensitivity to external cues. PLOS Comput. Biol. 14 e1006011 10.1371/journal.pcbi.1006011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A., Luo A., DeBlasio ST., Ling X., Yang Y., Tuthill D.E., Williams K.E., Hill D., Zadrozny T., Chan A., et al. 2009. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 149:601–605. 10.1104/pp.108.130146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T.J., and Wignall S.M.. 2017. Interplay between microtubule bundling and sorting factors ensures acentriolar spindle stability during C. elegans oocyte meiosis. PLoS Genet. 13 e1006986 10.1371/journal.pgen.1006986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Han S., and Smith L.G.. 2006. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16:888–894. 10.1016/j.cub.2006.03.034 [DOI] [PubMed] [Google Scholar]
- Murata T., Sano T., Sasabe M., Nonaka S., Higashiyama T., Hasezawa S., Machida Y., and Hasebe M.. 2013. Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 4:1967 10.1038/ncomms2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteris E., Diannelidis B.-E., and Adamakis I.S.. 2018. Cortical microtubule orientation in Arabidopsis thaliana root meristematic zone depends on cell division and requires severing by katanin. J. Biol. Res. (Thessalon.). 25:12 10.1186/s40709-018-0082-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., and Northcote D.H.. 1966. Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J. Cell Sci. 1:109–120. [DOI] [PubMed] [Google Scholar]
- Pietra S., Gustavsson A., Kiefer C., Kalmbach L., Hörstedt P., Ikeda Y., Stepanova A.N., Alonso J.M., and Grebe M.. 2013. Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 4:2779 10.1038/ncomms3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D., Zoccoler M., Gaillard J., Stoppin-Mellet V., Neumann E., Arnal I., Martiel J.L., and Vantard M.. 2013. MAP65/Ase1 promote microtubule flexibility. Mol. Biol. Cell. 24:1964–1973. 10.1091/mbc.e13-03-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C.G., and Bellinger M.. 2018. An overview of plant division-plane orientation. New Phytol. 219:505–512. 10.1111/nph.15183 [DOI] [PubMed] [Google Scholar]
- Rasmussen C.G., Sun B., and Smith L.G.. 2011. Tangled localization at the cortical division site of plant cells occurs by several mechanisms. J. Cell Sci. 124:270–279. 10.1242/jcs.073676 [DOI] [PubMed] [Google Scholar]
- Schaefer E., Belcram K., Uyttewaal M., Duroc Y., Goussot M., Legland D., Laruelle E., de Tauzia-Moreau M.-L., Pastuglia M., and Bouchez D.. 2017. The preprophase band of microtubules controls the robustness of division orientation in plants. Science. 356:186–189. 10.1126/science.aal3016 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., and Ehrhardt D.W.. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 300:1715–1718. 10.1126/science.1083529 [DOI] [PubMed] [Google Scholar]
- Smertenko A. 2018. Phragmoplast expansion: the four-stroke engine that powers plant cytokinesis. Curr. Opin. Plant Biol. 46:130–137. 10.1016/j.pbi.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Chang H.-Y., Wagner V., Kaloriti D., Fenyk S., Sonobe S., Lloyd C., Hauser M.-T., and Hussey P.J.. 2004. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell. 16:2035–2047. 10.1105/tpc.104.023937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A., Assaad F., Baluška F., Bezanilla M., Buschmann H., Drakakaki G., Hauser M.-T., Janson M., Mineyuki Y., Moore I., et al. 2017. Plant Cytokinesis: Terminology for Structures and Processes. Trends Cell Biol. 27:885–894. 10.1016/j.tcb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Hake S., and Sylvester A.W.. 1996. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 122:481–489. [DOI] [PubMed] [Google Scholar]
- Smith L.G., Gerttula S.M., Han S., and Levy J.. 2001. Tangled1: a microtubule binding protein required for the spatial control of cytokinesis in maize. J. Cell Biol. 152:231–236. 10.1083/jcb.152.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner L., Pastuglia M., Belcram K., Pegoraro M., Goussot M., Bouchez D., and Schaefer D.G.. 2010. The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development. 137:2733–2742. 10.1242/dev.043810 [DOI] [PubMed] [Google Scholar]
- Spinner L., Gadeyne A., Belcram K., Goussot M., Moison M., Duroc Y., Eeckhout D., De Winne N., Schaefer E., Van De Slijke E., et al. 2013. A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 4:1863 10.1038/ncomms2831 [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Fache V., Portran D., Martiel J.-L., and Vantard M.. 2013. MAP65 coordinate microtubule growth during bundle formation. PLoS One. 8 e56808 10.1371/journal.pone.0056808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., and Kapoor T.M.. 2010. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 142:433–443. 10.1016/j.cell.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J., Bellini C., Nacry P., Kronenberger J., Bouchez D., and Caboche M.. 1995. Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature. 375:676–677. 10.1038/375676a0 [DOI] [Google Scholar]
- Tulin A., McClerklin S., Huang Y., and Dixit R.. 2012. Single-molecule analysis of the microtubule cross-linking protein MAP65-1 reveals a molecular mechanism for contact-angle-dependent microtubule bundling. Biophys. J. 102:802–809. 10.1016/j.bpj.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D., Vanstraelen M., and Geelen D.. 2007. Cortical division zone establishment in plant cells. Trends Plant Sci. 12:458–464. 10.1016/j.tplants.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Walker K.L., Müller S., Moss D., Ehrhardt D.W., and Smith L.G.. 2007. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17:1827–1836. 10.1016/j.cub.2007.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G.O., and Ambrose J.C.. 2009. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19:62–71. 10.1016/j.tcb.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Winters L., Ban I., Prelogović M., Kalinina I., Pavin N., and Tolić I.M.. 2019. Pivoting of microtubules driven by minus-end-directed motors leads to spindle assembly. BMC Biol. 17:42 10.1186/s12915-019-0656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.H., and Hashimoto T.. 2017. Novel Arabidopsis microtubule-associated proteins track growing microtubule plus ends. BMC Plant Biol. 17:33 10.1186/s12870-017-0987-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden N., Park E., and Drakakaki G.. 2012. Trans-Golgi Network: An Intersection of Trafficking Cell Wall Components. J. Integr. Plant. Biol. 54:875–876. [DOI] [PubMed] [Google Scholar]
- Wright A.J., Gallagher K., and Smith L.G.. 2009. discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell. 21:234–247. 10.1105/tpc.108.062810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Zhao Q., Rodrigo-Peiris T., Brkljacic J., He C.S., Müller S., and Meier I.. 2008. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. USA. 105:18637–18642. 10.1073/pnas.0806157105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fishel E., Bertroche T., and Dixit R.. 2013. Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23:2191–2195. 10.1016/j.cub.2013.09.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows model fitting for TAN1–microtubule binding data.