Chastney et al. use multiplexed proximity-dependent labeling to define the topology of integrin adhesion complexes. The data support current models of adhesome architecture and provide a resource to examine proximal relationships within the adhesome and to identify novel interactors.
Abstract
Integrin adhesion complexes (IACs) bridge the extracellular matrix to the actin cytoskeleton and transduce signals in response to both chemical and mechanical cues. The composition, interactions, stoichiometry, and topological organization of proteins within IACs are not fully understood. To address this gap, we used multiplexed proximity biotinylation (BioID) to generate an in situ, proximity-dependent adhesome in mouse pancreatic fibroblasts. Integration of the interactomes of 16 IAC-associated baits revealed a network of 147 proteins with 361 proximity interactions. Candidates with underappreciated roles in adhesion were identified, in addition to established IAC components. Bioinformatic analysis revealed five clusters of IAC baits that link to common groups of prey, and which therefore may represent functional modules. The five clusters, and their spatial associations, are consistent with current models of IAC interaction networks and stratification. This study provides a resource to examine proximal relationships within IACs at a global level.
Introduction
The ability of cells to adhere to the ECM and respond to its chemical and mechanical properties is essential for multicellular life. This adhesion is primarily mediated by integrin receptors, which bridge the ECM to the contractile actomyosin cytoskeleton via a range of integrin adhesion complexes (IACs; Campbell and Humphries, 2011; Barczyk et al., 2010). In addition to providing a mechanical interface, IACs form a signaling hub from which many biochemical and biomechanical signaling pathways are transduced to guide cellular fate (Green and Brown, 2019; Humphries et al., 2019).
The complement of adaptors, enzymes, and cytoskeletal components that associate at IACs has been termed the adhesome (Zaidel-Bar et al., 2007). Extensive literature mining led to the construction of an in silico network of >200 IAC-associated proteins, forming a “literature-curated adhesome” (Winograd-Katz et al., 2014; Zaidel-Bar et al., 2007). The scale of the reported complexity of IACs is consistent with their substantial functional diversity. Although empirical analysis of IACs has been hampered by their lability and vicinity to the plasma membrane, advances in mass spectrometry (MS) coupled to the development of protocols to isolate IACs and adjacent material have greatly facilitated the identification of IAC-associated proteins (Kuo et al., 2012; Jones et al., 2015; Schiller et al., 2011, 2013; Atkinson et al., 2018; Horton et al., 2015; Humphries et al., 2009). The large-scale examination of isolated IACs enabled assembly of adhesome datasets, and the bioinformatic integration of seven such analyses (of fibronectin substrate-induced IACs) defined a “meta-adhesome” of >2,400 proteins (Horton et al., 2015). This dataset was further refined to a “consensus adhesome” of 60 commonly identified components postulated to represent the core adhesion machinery. These 60 components were organized into an interaction network containing four interconnected, hypothetical signaling modules. How closely this theoretical interaction network represents IAC organization and protein–protein interactions (PPIs) in situ has yet to be experimentally defined.
Dynamic PPIs underpin the transmission of biochemical and biomechanical information across IACs and are therefore dependent on the organization of adhesome components. Microscopic analyses have enabled the close examination of the ultrastructure of IACs, revealing a high degree of lateral and vertical organization (Kanchanawong et al., 2010; Case et al., 2015; Spiess et al., 2018; Xu et al., 2018). For example, superresolution light microscopy revealed the vertical stratification of a number of components within IACs at a 10–20-nm resolution, revealing three distinct, but overlapping, functional layers, albeit with some differences in the localization of specific components (Kanchanawong et al., 2010; Case et al., 2015; Liu et al., 2015; Stubb et al., 2019). However, the organization of adhesome components on a larger scale has yet to be experimentally defined, and an empirical view of protein interactions within IACs is lacking.
Recently, proximity-dependent biotinylation techniques, such as BioID, have offered attractive alternatives to affinity purification approaches to examine protein proximal associations. BioID uses a mutated biotin ligase, BirA*, fused to a protein of interest (the bait), to promiscuously biotinylate proximal proteins (the prey) over the course of several hours, with an estimated labeling range of 10–15 nm (Kim et al., 2014). As labeling occurs in situ, and purification takes advantage of the high-affinity bond between biotin and avidin, proximity biotinylation circumvents the need to retain PPIs throughout processing. BioID has been used to probe the structure of labile and membrane-associated complexes that are difficult to study using more traditional techniques, including nuclear pore complexes, the centrosome, and cell–cell contacts (Van Itallie et al., 2013; Gupta et al., 2015; Kim et al., 2014). BioID has also been used to examine the proximity interactomes of individual IAC-associated proteins and has revealed a number of potential new adhesome candidates (Dong et al., 2016; Mekhdjian et al., 2017; Rahikainen et al., 2019). For example, KANK2 was identified as a paxillin- and kindlin-2–proximal protein in U2OS cells and was shown to localize to IACs (Dong et al., 2016). To date, however, a large-scale analysis of protein proximal networks in IACs has not been performed.
In this study, we multiplexed BioID data from a set of 16 IAC component baits to generate a proximity-dependent adhesome, representing both the core adhesome machinery and proximal interactors more peripheral to IACs. The resulting resource enables the interrogation of the proximal relationships between adhesome components, in addition to providing insights into the architecture of IACs. Bioinformatic analysis of the data revealed five clusters of bait proteins that linked to common groups of proteins with diverse, but overlapping, functional roles, which may represent functional modules. The grouping of these proteins was consistent with current literature-based models of IAC interaction networks (Horton et al., 2015; Green and Brown, 2019). Interrogation of the topological organization of the proximal interaction network identified a bait-prey organization that is consistent with the reported stratified arrangement of components within IACs. A number of well-characterized adhesome components were identified among a group of 11 proteins with multiple links to a range of bait proteins, which may be part of the core adhesome machinery. This group also contained several proteins that may have underappreciated roles in adhesion regulation. This empirically defined adhesome network provides a valuable tool to interrogate the proximity interaction networks within IACs and to drive further hypothesis generation.
Results
Generation of a proximity-dependent adhesome
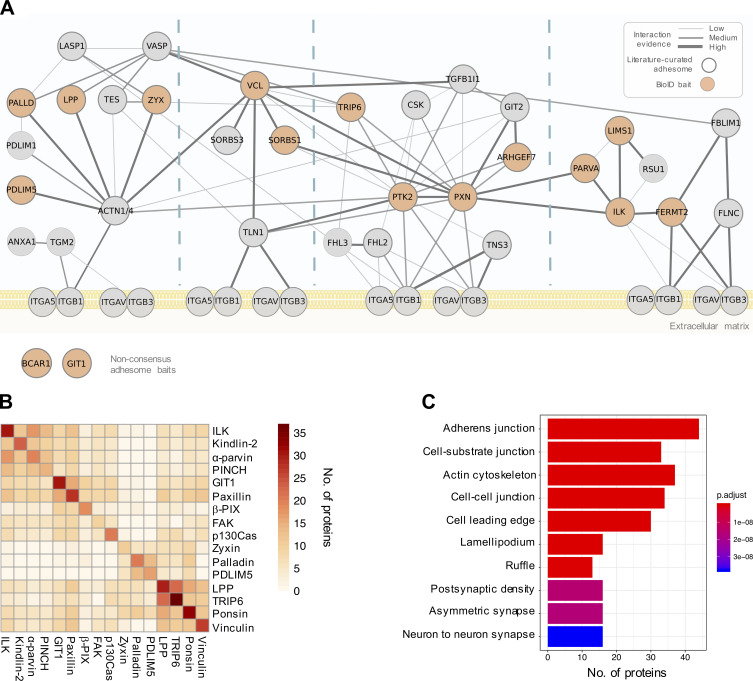
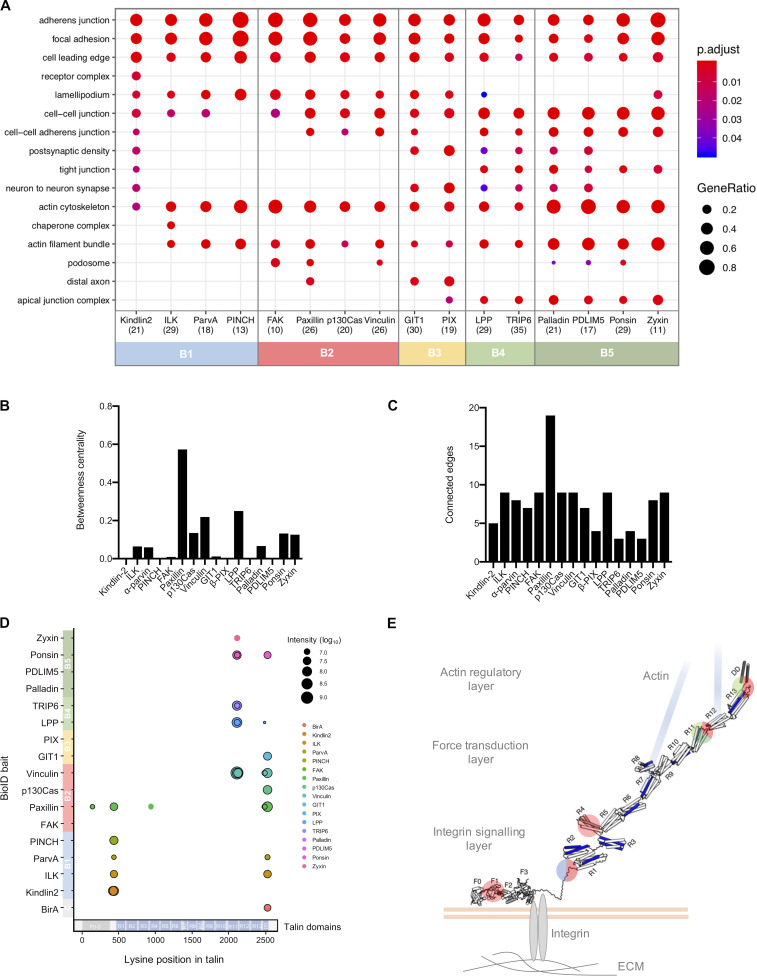
To maximize the capture of proximity interactions within IACs, 16 commonly identified adhesome components were selected as BioID baits. The 16 baits represent a broad range of functions within the adhesome and span all four putative signaling axes of the consensus adhesome (α-actinin-zyxin-VASP, talin-vinculin, FAK-paxillin, and kindlin-ILK-PINCH-parvin; Fig. 1 A). There is evidence for all four of these axes contributing to integrin-dependent signaling and mechanotransduction, but as yet, no systems-level understanding of their integration has been generated. The extent to which these axes assemble independently or, conversely, whether they are all interconnected, is also unclear. The selected proteins were cloned into the pCDH lentiviral vector containing myc-BirA* with a self-cleaving BFP and stably expressed in an immortalized mouse pancreatic fibroblast cell line (imPSC). Immunofluorescence microscopy confirmed colocalization of BirA*-tagged adhesome baits and biotinylated proteins with paxillin-positive structures (Fig. S1 and Fig. S2), confirming that subcellular targeting to IACs was not inhibited by the myc-BirA* tag. All baits strongly colocalized with paxillin, but in addition BirA*-PDLIM5, -palladin, -ponsin, and -zyxin also stained IAC-proximal actin filaments, and BirA*–β-Pix, -GIT1, and -p130Cas staining was slightly more diffuse than other baits. Cells expressing the BirA*-only control showed no specific subcellular localization of bait or biotinylated proteins.
Figure 1.
Overview of the proximity-dependent adhesome. (A) 16 adhesome proteins were selected as BioID baits, 14 of which were present in the consensus adhesome and span the four putative signaling axes of the PPI network (Horton et al., 2015). p130Cas (Bcar1) and GIT1 are present in the literature-curated adhesome (and the GIT1 homologue GIT2 is in the consensus adhesome; Zaidel-Bar et al., 2007). Baits are shown in orange, and edges represent evidence of PPIs. Thick gray borders indicate literature-curated adhesome proteins. Gene names are shown. Consensus adhesome components unconnected to the main network are not shown. (B) Pairwise comparisons of proximal proteins (BFDR ≤ 0.05) identified by each BioID bait are displayed as a heatmap. Protein names not matching the gene names in A are FAK, PTK; kindlin-2, FERMT2; palladin, PALLD; α-parvin, PARVA; paxillin, PXN; β-Pix, ARHGEF7; PINCH, LIMS1; ponsin, SORBS1; vinculin, VCL; and zyxin, ZYX. (C) GO enrichment analysis of the 147 proteins in the proximity-dependent adhesome. The top 10 overrepresented terms under the cellular component category are shown.
Figure S1.
Subcellular localization of BirA* and BirA*-tagged adhesome components and biotinylation of proximal proteins. imPSC cells stably expressing BirA* and BirA*-tagged adhesome constructs or untransfected control cells were incubated with biotin for 24 h before being fixed and stained for myc and paxillin or vinculin and biotinylated proteins (using fluorophore-conjugated streptavidin). Scale bar: 30 µm.
Figure S2.
Subcellular localization of BirA*-tagged adhesome components and biotinylation of proximal proteins. imPSC cells stably expressing BirA*-tagged adhesome constructs were incubated with biotin for 24 h before being fixed and stained for myc and paxillin or vinculin and biotinylated proteins (using fluorophore-conjugated streptavidin). Scale bar: 30 µm.
To determine proximal interactors of each BioID adhesome bait, label-free quantitative MS was performed on affinity-purified biotinylated proteins from three independent experiments, and raw data were analyzed by MaxQuant using ion intensity-based quantification. SAINTexpress was used to identify high-confidence bait–prey proximity interactions, with BirA* as a negative control for nonspecific interactions (Table S1). The number of proteins predicted to be within each proximity interactome varied by bait, with 10–37 proteins predicted to be “true” proximity interactors at a Bayesian false discovery rate (BFDR) of ≤0.05. Pairwise comparisons were performed to visualize the number of proximal proteins common to each bait (Fig. 1 B). Whereas some BioID baits, such as BirA*-LPP and -TRIP6, shared a large number of proteins, others showed little similarity to the majority of baits, notably BirA*-tagged zyxin, palladin, and PDLIM5.
The 16 individual BioID datasets were integrated into a single network to generate a proximity-dependent adhesome. A total of 147 proximal proteins were found across all datasets, which is likely to represent a combination of core IAC proteins, IAC-associated proteins, and proteins with a proximal association with the BioID baits in more distal subcellular localizations. 361 proximity interactions were identified (excluding bait–bait interactions), the majority of which are absent from published PPI databases (292; 81%; see Materials and methods for details). These associations may represent unknown direct interactions, indirect proximity interactions, or nonspecific background interactions. Excluding BirA*-tagged bait proteins, more than half of the prey identified (77) were unique to a specific bait, whereas eight proteins were identified by at least half of the 16 BioID baits (and may therefore represent core adhesome components).
Gene ontology (GO) analysis of the network revealed an overrepresentation of multiple terms related to cell–ECM adhesion, including “cell-substrate junction,” “actin cytoskeleton,” and “cell leading edge” (Fig. 1 C). Terms relating to cell–cell adhesion were also identified (“adherens junction” and “cell–cell junction”), which may reflect the shared components between cell–cell and cell–ECM adhesion and/or the subcellular targeting of multiple baits to cell–cell contacts (e.g., as reported for vinculin, LPP, and zyxin; Zaidel-Bar, 2013). Furthermore, a relatively large number of the proteins identified by proximity biotinylation were identified in published adhesomes, with 24.5% (36) and 19.7% (23) of the 147 proteins in the proximity-dependent adhesome identified in the literature-curated (232 components) and consensus (60 components) adhesomes, respectively (Table S1; Horton et al., 2015; Winograd-Katz et al., 2014). The majority of prey proteins (96; 65.3%) were also identified in at least one of the seven datasets comprising the meta-adhesome (Table S1). There was also substantial overlap between the prey identified for BirA*-paxillin and –kindlin-2, with those reported by Dong et al. (2016), with 75% (18) of BirA*-paxillin proximal interactors and 75% (21) of BirA*–kindlin-2 interactors also identified in U2OS cells (Table S1). In the study by Dong et al. (2016), 14 prey were identified that associated with both BirA*-tagged kindlin-2 and paxillin. In our study, this was true for six of these proteins (talin-1, KANK2, PEAK1, tensin-1, tensin-3, and lamellipodin/Raph1). ILK, α-parvin, EphA2, and RN-Tre (Usp6nl) associated with one of the baits, whereas RASnGAP (Rasal2), liprin-α1 (Ppfia1), PINCH, and Bcar3 were not found (although some did associate with other baits). It is conceivable that the absence of some prey may be due to cell type–specific differences. This high degree of similarity, despite differences in experimental methodology, is indicative of common proximal interactors across multiple cell lines. Together, these findings indicate that a large proportion of the proteins identified are likely to be highly relevant to IACs.
Functional modules within IACs
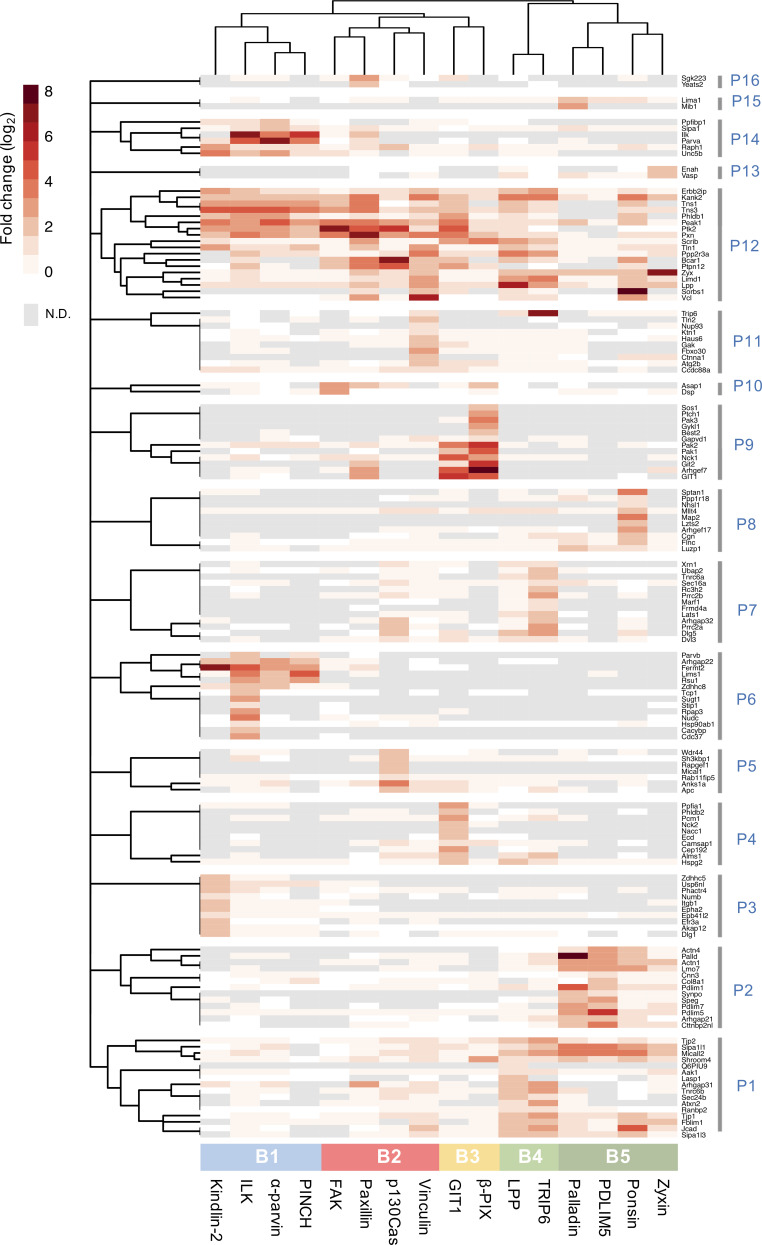
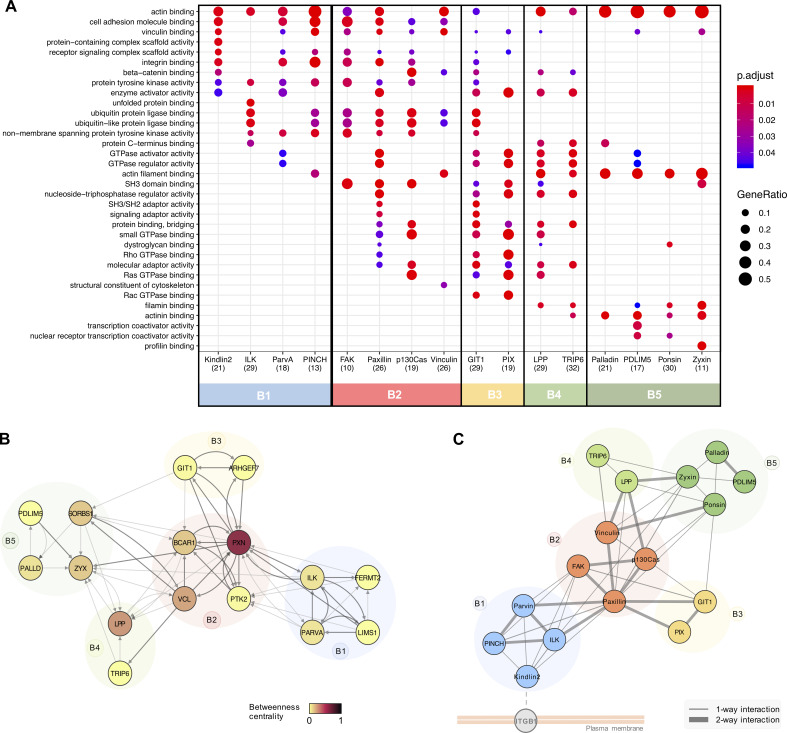
2D hierarchical clustering was performed to provide an unbiased interrogation of the relationships between baits and the prey identified. This analysis revealed five clusters of bait proteins (B1–B5) and 16 clusters of prey (P1–P16) that are likely to represent groups of spatially linked protein subcomplexes (Fig. 2 and Table S2). B1 contained BirA*-tagged kindlin-2 and the members of the IPP complex (ILK, α-parvin, and PINCH); B2 comprised BirA*-FAK, -paxillin, -p130Cas, and -vinculin; and B3 contained BirA*-GIT1 and β-Pix. The remaining two clusters contained actin-associated/regulatory baits (BirA*-LPP and -TRIP6 in B4; BirA*-palladin, -PDLIM5, -ponsin, and -zyxin in B5). These five bait clusters broadly correlated with theoretical interaction networks in the literature (Horton et al., 2015; Green and Brown, 2019) and with preassembled ternary complexes of ILK-PINCH-parvin and FAK-p130Cas-paxillin previously identified by fluorescence cross-correlation spectroscopy and fluorescence recovery (Hoffmann et al., 2014; Zamir et al., 2008). These findings provide evidence not only that published theoretical IAC networks are largely reflective of protein interactions in situ, but also that BioID captures relevant interactions within IACs.
Figure 2.
Hierarchical clustering of BioID data. Hierarchical clustering was performed on the 147 proteins identified in the proximity-dependent adhesome and the results are displayed as a heatmap. BioID baits were clustered using the Jaccard distance of the presence (BFDR ≤ 0.05) or absence (BFDR > 0.05) of prey. Prey proteins were clustered using the Euclidean distance of log2 fold-change enrichment over BirA* control. Log2 fold-changes of prey proteins are displayed as a heatmap. Dendrograms were split to identify clusters of baits (B1–B5, color coded) and prey (P1–P16). N.D., not detected.
GO analysis of the prey identified by each bait revealed a number of overrepresented terms relating to IACs and their associated structures. Many of these terms under the cellular component category were common to all baits, including “focal adhesion,” cell leading edge, and actin cytoskeleton, confirming that each of the baits identified proteins relevant to cell–ECM adhesion (Fig. S3 A). Some GO terms were unique to a single bait, such as “receptor complex” with BirA*–kindlin-2 and “chaperone complex” with BirA*-ILK, suggestive of specific roles for these proteins. A broad range of GO terms was also identified across all baits under the molecular function domain, many of which were again shared (Fig. 3 A). For example, “actin binding” was overrepresented across baits in all clusters. However, other actin-related GO terms such as “actin filament binding” and actinin binding were predominantly restricted to B4 and B5, in accordance with their roles in actin regulation. Similarly, GO terms relating to GTPase binding/regulation were found by multiple baits (e.g., “GTPase activator activity,” “GTPase regulator activity,” and “small GTPase binding”) but were predominantly identified by B2, B3, and B4, supporting their reported roles in signaling within IACs. The similarity of GO terms identified within bait clusters indicated that these groups of proteins have similar functions, and that the clusters may have a functional relevance.
Figure S3.
Ontological analysis of proximity interactomes and biotinylation of talin. (A) Functional enrichment analysis of proteins identified by each of the 16 BirA*-tagged adhesome proteins (BFDR ≤ 0.05). The top three overrepresented GO terms under the “cellular component” category for each bait are listed and displayed for all baits (if identified) with an adjusted P value ≤ 0.05. The number of proteins recognized per interactome is shown in brackets. Baits are ordered and color coded according to hierarchical clustering as in Fig. 2. p.adjust, adjusted P value. GeneRatio, proportion of total proteins identified in each GO term. (B and C) Betweenness centrality (B) and number of connected edges (C) of each bait from network analysis of bait–prey interactions between BirA*-tagged baits in Fig. 3 B. (C) Biotinylated lysines from talin identified by each adhesome bait were mapped onto the talin sequence. Black borders indicate biotinylated peptides present in a minimum of two of three repeats. Baits are colored according to the hierarchical clustering, and the domain structure of talin is indicated at the bottom (Gough and Goult, 2018). Note that the most C-terminal biotinylated peptide in the DD domain is also found in BirA* alone and therefore likely to be nonspecific. (D) Schematic of biotinylated lysines mapped onto the tertiary structure of talin in its extended active form in IACs. Vinculin binding sites are shown in blue. The position of biotinylated lysines are highlighted and color coded according to the BioID bait clusters in Fig. 2. Blue, B1; red, B2; light green, B4; dark green, B5. Talin structure adapted from Yao et al. (2016). F0–F3, FERM domains 0–3; R1–R13, rod domains 1–13; DD, dimerization domain.
Figure 3.
Functional roles and subcomplex organization of functional bait modules. (A) Functional overrepresentation analysis of proximal proteins identified by each of the 16 BirA*-tagged adhesome proteins (BFDR ≤ 0.05). The top 3 GO terms in the “molecular function” category for each bait are listed and displayed for all baits if identified with an adjusted P value ≤ 0.05. The number of proteins recognized is shown in brackets. Baits are color coded according to hierarchical clustering shown in Fig. 2. p.adjust, adjusted P value; GeneRatio, proportion of total proteins identified in each GO term. (B) Network analysis of bait–prey interactions between BirA*-tagged baits. Nodes represent BirA*-tagged baits, which are color coded according to betweenness centrality and grouped into bait clusters B1–B5, according to the hierarchical clustering (Fig. 2). Edges indicate bait–prey proximity interactions, with arrows indicating direction of interaction (bait to prey). Dark gray edges indicate PPIs also present in a published PPI database (see Materials and methods). (C) Schematic showing potential organization of adhesome bait modules within IACs in relation to the membrane. Positioning of modules was guided by the functional roles of prey identified by each cluster (i.e., transmembrane protein or actin-regulatory; see Table S1 for details).
Close examination of the interconnectivity between baits revealed highly specific associations of bait clusters. Although a large number of bidirectional proximal associations were observed within individual bait clusters, particularly for B1, B2, and B3, very few interactions were observed between clusters (e.g., no proximity interactions were observed between B1 and B3 or between B1 and B4/5; Fig. 3 B). The exception to this was B2, which was highly connected with components from the other clusters. Even then, connections between B2 and B1 were mediated only by paxillin and FAK, connections between B2 and B3 were via paxillin and p130Cas, and connections between B2 and B4 were via p130Cas, paxillin, and vinculin. These findings may be indicative of a spatial distinction between the clusters and suggest that B2 may form a central link between other clusters, either in space (i.e., all modules form a single structure, with B2 in the center) or time (i.e., B2 interacts with other bait clusters separately in different structures, such as nascent adhesions and fibrillar adhesions). Network analysis revealed that paxillin had a particularly large number of proximal interactions with BioID baits (19) and a high betweenness centrality (0.57; Fig. 3 B and Fig. S3, B and C), which may be indicative of a central role of paxillin as a key adaptor, mediating connections with other proteins and coordinating interactions between different bait clusters. This is consistent with the model proposed by Green and Brown (2019), in which paxillin is described as an “über-linker,” interacting with all functional modules. Some baits, such as PDLIM5 and β-Pix, had few connected nodes and a low betweenness centrality and are therefore likely to be more peripheral in the adhesome network. How the bait clusters identified here link to spatially defined functions requires further investigation.
Various studies have provided evidence for functional modules within IACs, which represent groups of proteins that perform a similar function, such as signaling or mechanotransduction (Horton et al., 2015; Green and Brown, 2019; Stutchbury et al., 2017). The five bait clusters identified in this study are unlikely to represent distinct, separate structures that operate individually and are more likely to represent dynamic, interconnected groups of proteins that interact to form IACs. Additionally, it is possible that cytosolic interactions contribute toward the network of proximity interactions described here. For example, various adhesome components have been shown to form preassembled dimeric or trimeric complexes in the cytosol, which are thought to facilitate the assembly of IACs in a modular manner (Hoffmann et al., 2014). Indeed, many of the multimolecular interactions previously described were also identified in this study, including the trimeric complexes of ILK, PINCH, and α-parvin and FAK, p130Cas, and paxillin. Each of these trimers was shown to have bidirectional bait–bait proximity interactions and was present within the same bait cluster identified by hierarchical clustering.
The five bait clusters bear a striking resemblance to current models and provide further evidence of functional modules within IACs (Green and Brown, 2019; Horton et al., 2015). The members of the IPP complex and kindlin-2, which form B1, are commonly grouped together in literature-based models, together with Rsu1 (a prey identified by BirA*-tagged ILK, PINCH, and α-parvin in this study). Although the exact role of this functional module is not fully understood, it has been proposed to play a role in the recruitment of regulatory components to IACs to regulate Rac1 GTPase activity and other signals at the cell cortex (Green and Brown, 2019; Böttcher et al., 2017). B2 comprises well-studied IAC regulators and adaptors (FAK, paxillin, vinculin, and p130Cas). Each of the baits within this highly connected central cluster has been shown to play a role in mechanotransduction, and future experiments examining the effects of force on the proximity-dependent adhesome will be instructive (Pasapera et al., 2010; Sawada et al., 2006; Hirata et al., 2014). The components of B3 (GIT1 and β-Pix) have been shown to form a stable complex and regulate various signaling pathways and cytoskeletal dynamics via effectors such as Rho family GTPases and PAK family kinases (prey identified by BirA*-GIT1 and –β-Pix in this study; Zhou et al., 2016). Although they are often treated as a single entity, GIT and Pix have different interacting partners, and there is evidence that they function independently of one another (Zhou et al., 2016). This may account for the differential proximal interactors identified by each component in this study. The final two bait clusters represent two actin-regulatory modules and contain a number of components responsible for mediating the connection between actin filaments and IACs through the recruitment of actin regulatory and bundling proteins, such as α-actinin. The inclusion of additional BioID baits, such as integrins, talin, and tensins, may provide useful information about the interconnectivity between these modules through the detection of connecting proteins.
Substructure and stratification of the proximity-dependent adhesome
Although superresolution microscopy has revealed the organization of a small number of components within IACs, little is known about the localization of the majority of IAC components, and a more comprehensive view could provide insights into IAC regulation and signaling outputs. A key advantage of BioID is that it provides a means to interrogate spatial relationships between groups of proteins. Examination of the prey identified by each bait cluster may therefore provide insights into the spatial organization of IACs and build on previous evidence for substructure (Dong et al., 2016; Kanchanawong et al., 2010). As multiple membrane-associated proteins and transmembrane receptors, such as integrin β1 (Itgb1) and netrin receptor (Unc5b), were predominantly identified by BirA*–kindlin-2 and other members of B1 (Table S1), it is likely that these baits lie within close proximity to the plasma membrane (Fig. 3 C). Conversely, the large proportion of actin-regulatory proteins detected by the actin-associated baits in B4 and B5 indicates that these proteins lie more distal from the membrane, in the proximity of actomyosin filaments. As members of B2 have multiple links to B1 and B4/B5, these proteins may lie within or between these two regions. This organization broadly correlates with the stratified architecture of IACs determined by superresolution microscopy (Kanchanawong et al., 2010), with zyxin (B5) and prey identified by B4 and B5 (VASP and α-actinin) localized to a membrane-distal actin-regulatory layer, and members of B2 (FAK, paxillin, and vinculin) distributed across the force-transduction layer and integrin-signaling layer. Although the stratified organization of members of B1 has not yet been examined in mammalian cells in culture, recent work in Drosophila melanogaster has localized them to the membrane-proximal integrin signaling layer (Green et al., 2018).
The stratified organization of components determined by BioID is further supported by the detection of biotinylation within specific domains of talin by each of the BioID baits (Table S3). Talin has a polarized orientation within IACs and spans the three layers of the stratified model of IAC architecture, with its N-terminal FERM domain being located proximal to the plasma membrane, and the end of its C-terminal rod domain mediating attachment to actin filaments (Kanchanawong et al., 2010; Liu et al., 2015). The peptides within talin that were biotinylated by its associating baits were mapped to its primary and tertiary structure (Fig. S3, D and E) and found to correlate with the organization of bait clusters relative to the plasma membrane outlined in Fig. 3 C. The actin-associated baits BirA*-LPP, -TRIP6, -zyxin, and -ponsin, and BirA*-vinculin, biotinylated peptides in the C-terminal R11 and R13/DD domains of talin, which lie proximal to the ABS3 actin-binding site (Gough and Goult, 2018). Despite the many reported vinculin binding sites in talin, and localization to multiple layers of IACs (Case et al., 2015), vinculin biotinylated only peptides at the C-terminus of talin (Gough and Goult, 2018). No biotinylated peptides from B4 or B5 baits were found in the ABS1 or ABS2 domains of talin, despite their reported roles in actin binding (Atherton et al., 2015; Kumar et al., 2016; Lee et al., 2004). By contrast, B1 baits biotinylated peptides located in the membrane-proximal linker domain of talin, close to the IBS1 integrin-binding site and consistent with direct kindlin-integrin binding (Harburger et al., 2009). BirA*-paxillin biotinylated peptides from various domains across the length of talin, which may indicate that paxillin is localized across multiple layers of IACs. Alternatively, these may represent interactions with talin in its autoinhibited, inactive conformation (Atherton et al., 2020). Paxillin has been reported to interact with the talin R7/R8 domain via its leucine-aspartic acid (LD) domains (Zacharchenko et al., 2016), although it is likely that there are additional binding sites (Goult, B., personal communication).
Evidently, inferring IAC substructure from proximity-dependent labeling relies on making a number of assumptions (i.e., shared proximal interactions occur in the same time and place), and further experiments are required to confirm such speculations. Nevertheless, despite capturing interactions from a heterogeneous population of IACs, the organization of adhesome proteins inferred from proximity biotinylation correlates with models of IAC architecture, such as the stratified organization of IACs in mammalian cells and myotendinous junctions in Drosophila (Green et al., 2018; Kanchanawong et al., 2010). The specific biotinylation of talin domains by BioID baits provides further evidence for a high degree of organization. Although the stratified arrangement of BioID baits needs to be further validated, these data could be used to infer the localization of prey proteins based on the baits with which it was proximally associated.
Topological organization of the proximity-dependent adhesome
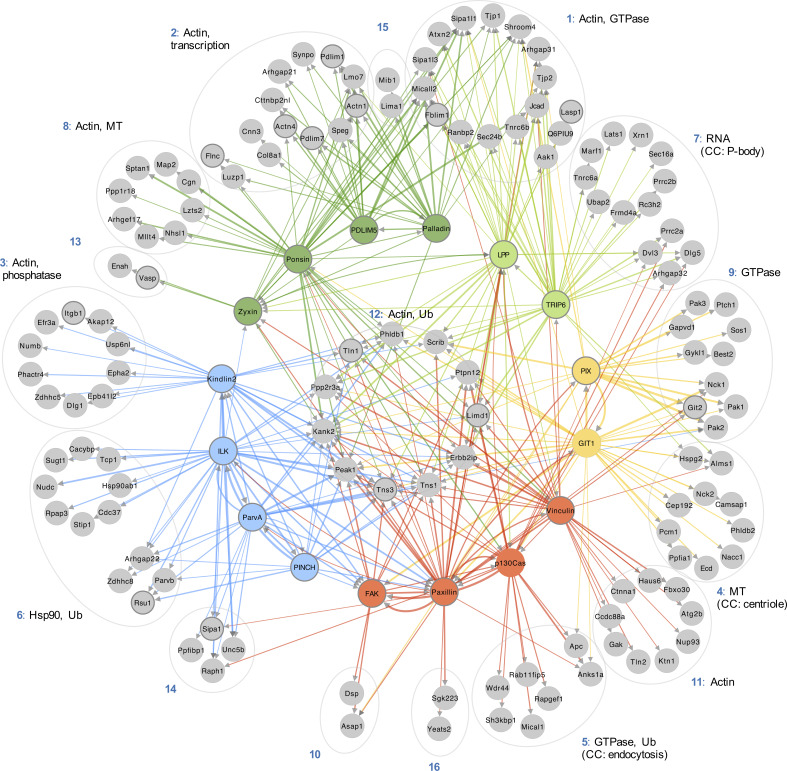
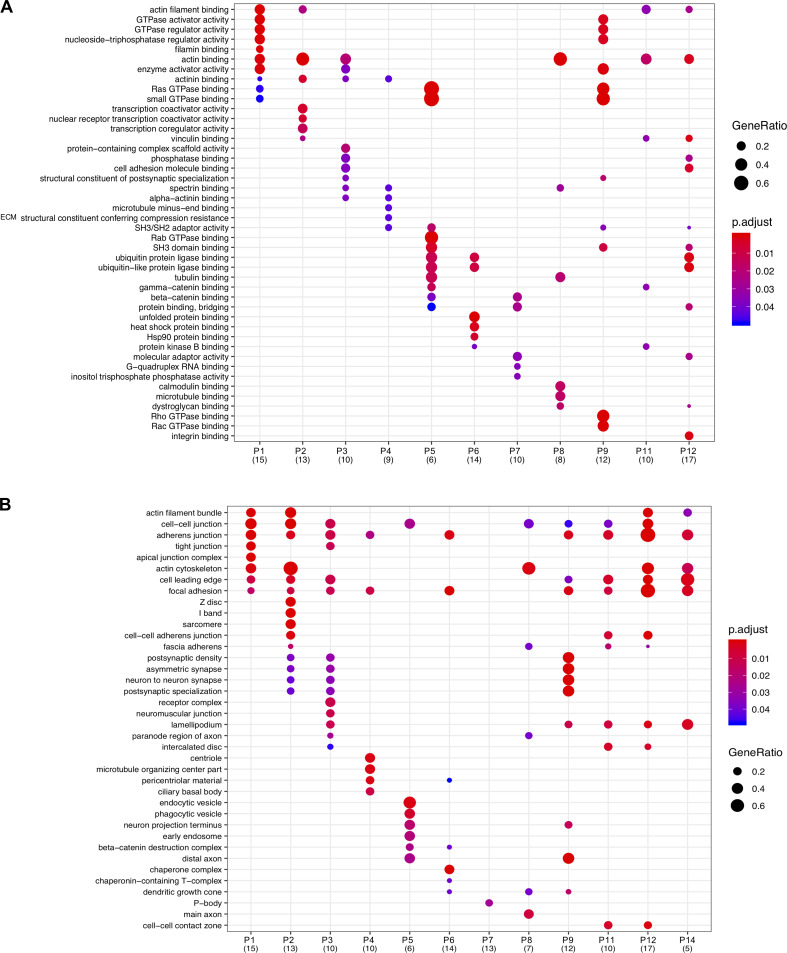
The hierarchical cluster analysis of bait proteins and prey was then used to interrogate the topology of the proximity-dependent adhesome network, and GO analysis was performed to determine the functional relevance of prey clusters (Fig. 4 and Fig. S4, A and B). Organization of the network was driven by hierarchical cluster analysis, with baits excluded from the prey clusters. Whereas some proteins had shared interactions with multiple baits and bait clusters, others were uniquely identified by a single bait, indicating underappreciated links to more distal roles. For example, a subgroup of eight proteins in prey cluster P6, exclusively identified by BirA*-ILK, contains a large number of Hsp90-binding chaperone/cochaperone proteins. The association of ILK with Hsp90, and their colocalization at IACs, has been previously reported (Radovanac et al., 2013), but this link may be more significant for IACs than previously thought, involving a number of additional chaperones. Similarly, BirA*-GIT1 uniquely identified a number of microtubule-associated proteins, with overrepresented GO terms such as “microtubule binding” in the molecular function category and those relating to the centriole in the cellular component category. These associations are in line with the reported role of GIT1 in microtubule nucleation at the centrosome (Zhao et al., 2005; Černohorská et al., 2016). Furthermore, BirA*-TRIP6 has a number of unique links to proteins involved in RNA binding and regulation, and the overrepresented GO terms from P7 under the cellular component category include “P-body.” TRIP6 has been reported to localize to the nucleus, and other zyxin family members LIMD1, ajuba, and WTIP were shown to associate with processing-bodies in U2OS cells (James et al., 2010). Although TRIP6 itself was shown to have poor colocalization with processing bodies, it is possible that it plays a role in RNA regulation (James et al., 2010).
Figure 4.
Topological organization of the proximity-dependent adhesome. Network of proximity interactions within the proximity-dependent adhesome. Network organization was driven by hierarchical clustering of BirA*-tagged adhesome baits and proximal prey proteins (BFDR ≤ 0.05; Fig. 2). Gray nodes represent prey proteins, and nodes indicating BirA*-tagged adhesome bait proteins are color coded according to the hierarchical clustering in Fig. 2. Consensus adhesome components are indicated with thick gray outlines (Horton et al., 2015). Edges indicate bait–prey proximity interactions, with color representing source node and width representing fold-change over BirA* control. The top GO terms under the molecular function category for each prey cluster are indicated. Gene names are shown. CC, cellular component; MT, microtubule; Ub, ubiquitination.
Figure S4.
Functional enrichment analysis of prey clusters. (A and B) GO analysis of the prey clusters identified from hierarchical clustering of the proximity-dependent adhesome (Fig. 2) with a minimum of five proteins. The top five overrepresented GO terms under the molecular function (A) or cellular component (B) category for each prey cluster are listed and displayed for each cluster if identified with an adjusted P value ≤ 0.05. The number of proteins per prey cluster is shown in brackets. p.adjust, adjusted P value. GeneRatio, proportion of total proteins identified in each GO term.
Other prey clusters were associated with multiple bait proteins. The actin-associated baits in B4 and B5 were highly connected to prey in P1 and P2. GO terms relating to actin regulation were well represented within these clusters and may be indicative of an actin filament regulatory module. Furthermore, a central group of 11 highly connected proteins (P12) had multiple links to all five bait clusters and may represent core IAC components. Multiple well-established adhesome proteins were identified within this central group, including talin-1 and tensin-1 and -3, in addition to the more recently identified IAC component, KANK2, and the cortical microtubule stabilizing complex (CMSC) component LL5-α (Phldb1; Noordstra and Akhmanova, 2017; Bouchet et al., 2016).
KANK2 was robustly identified by almost all the BioID baits in this study, and as a proximity interactor of both paxillin and kindlin-2 by proximity biotinylation in U2OS cells (Dong et al., 2016). The role of KANK proteins in cell–ECM adhesion has become apparent in recent years, as they have been shown to be involved in the turnover of IACs through the recruitment of CMSCs to IACs and the uncoupling of mechanical transduction between integrins and the actomyosin network, resulting in sliding focal adhesions (Bouchet et al., 2016; Sun et al., 2016). Although KANK is known to bind talin, a direct interaction with any of the baits used in this study has yet to be described (Bouchet et al., 2016). As KANK2 was identified in a number of proximity interactomes, it is possible that additional direct interactions exist (Stubb et al., 2019). In addition to KANK2 and LL5-α (Phldb1), three other CMSC components were identified in the proximity-dependent adhesome: LL5-β (Phldb2), liprin-α1 (Ppfia1), and liprin-β1 (Ppfibp1). This provides further evidence of the association of CMSCs and microtubule-associated structures with IACs (Bouchet et al., 2016; Lansbergen et al., 2006; Stehbens et al., 2014). Some of these components exhibited a restricted set of binding partners. For example, liprin-α1 and LL5-β were uniquely linked to BirA*-GIT1, and liprin-β1 was uniquely detected by BirA*-α-parvin, which may indicate specific roles for GIT1 and α-parvin in CMSC regulation and microtubule-targeting to IACs.
PEAK1, PTP-PEST (Ptpn12), and LIMD1 have previously been identified as adhesome components, though their precise roles in IAC regulation are less well studied (Winograd-Katz et al., 2014; Horton et al., 2015; Wang et al., 2010; Huggins and Andrulis, 2008; Garton and Tonks, 1999). PEAK1 (also known as SgK269) is a pseudokinase that functions as a scaffolding protein to recruit various signaling molecules, and its overexpression has been linked to progression of various cancers, including pancreatic ductal adenocarcinoma (Kelber et al., 2012; Croucher et al., 2013; O’Rourke and Daly, 2018). PEAK1 localizes to IACs and the actin cytoskeleton following growth factor receptor stimulation and cellular attachment, where it regulates cell motility, spreading, and IAC turnover (Wang et al., 2010; Bristow et al., 2013). The signaling activity of PEAK1 is mediated by phosphorylation by Src family kinases, such as Src and Lyn (Kelber et al., 2012; Croucher et al., 2013). In turn, PEAK1 regulates phosphorylation of adhesome components such as paxillin and p130Cas, although the mechanisms are unknown (Wang et al., 2010; Bristow et al., 2013). Indeed, both BirA*-paxillin and -p130Cas detected PEAK1 as a proximal interactor. PEAK1 is also identified by many other baits and may act as an adaptor to recruit other adhesome components.
Other proteins within the central cluster have very few reported associations with IACs, including scribble (Scrib), erbin (Erbb2ip), and the protein phosphatase 2A (PP2A) regulatory subunit, Ppp2r3a. These proteins may represent underappreciated IAC components and regulators. Scribble is known as an adaptor protein that regulates cell polarity, but it has also been reported to interact with a number of adhesome components, including LPP, TRIP6, and β-Pix, and to coimmunoprecipitate with others, such as GIT1, PAK, and integrin α5 (Nola et al., 2008; Michaelis et al., 2013; Lim et al., 2017; Petit et al., 2005; Bonello and Peifer, 2019). Consistent with these reports, scribble was identified as a proximal interactor by BirA*-tagged LPP, TRIP6, β-Pix, and GIT1 (together with kindlin-2 and paxillin). Although scribble is typically localized to cell–cell junctions, a number of studies have reported its recruitment to the leading edge of migrating nonfibroblastic cells, where it colocalized with β-Pix and Cdc42 and regulated directional cell migration (Dow et al., 2007; Osmani et al., 2006; Michaelis et al., 2013; Nola et al., 2008). It is therefore conceivable that scribble also plays a role in IACs and directional cell migration in fibroblasts. Scribble was also detected in the meta-adhesome (two of seven datasets) and was identified in a phosphoproteomic analysis of IACs (Horton et al., 2015; Robertson et al., 2015). Similar to scribble, erbin also localizes to the basolateral membrane at cell–cell junctions in epithelial cells and is also found at synapses (Jaulin-Bastard et al., 2002; Kravic et al., 2016). Other than evidence for an interaction of erbin with integrin β4 in hemidesmosomes, it has few published links to adhesome components, and a potential role in IAC regulation has yet to be explored (Favre et al., 2001). Like scribble, erbin has been localized to the leading edge of cells and was identified in the meta-adhesome (one of seven datasets; Jin et al., 2019; Dan et al., 2010; Horton et al., 2015). However, considering that erbin is identified as a proximal interactor by a different subset of adhesome baits, and is not detected by BirA*–β-Pix and -GIT1, it is likely that the two proteins have different binding partners and perform different roles.
Two phosphatases, PTP-PEST and a regulatory subunit of PP2A (Ppp2r3a), were also identified and may represent underappreciated core regulators of IAC dynamics and signaling outputs. PTP-PEST, a tyrosine phosphatase, is reported to have a key role in IAC turnover and cell motility through the dephosphorylation of a number of core adhesome components, including p130Cas, FAK, and paxillin (Angers-Loustau et al., 1999; Shen et al., 2000; Zheng et al., 2009). In line with this, PTP-PEST was identified as a proximal interactor by BirA*-tagged paxillin, FAK, and p130Cas. PTP-PEST was also identified as a proximal interactor by BirA*-vinculin, -ILK, and -GIT1, although these proteins have not been identified as substrates for PTP-PEST and may represent indirect interactors. Ppp2r3a, a regulatory subunit of the serine threonine phosphatase heterotrimer PP2A, is part of the PR72/PR130 subgroup of PP2A isoforms (Ruvolo, 2016). Recently, ppp2r3a was shown to regulate cell migration via interaction with LPP LIM domains (Janssens et al., 2016). Although it colocalized with LPP at the cell periphery in spreading cells, ppp2r3a was excluded from mature IACs. It is thought that LPP may bind ppp2r3a to target PP2A to early IACs, bringing it within close proximity to enable dephosphorylation of substrates to regulate dynamic IAC turnover and enable effective cell migration. However, although it is well established that PP2A can regulate IACs via dephosphorylation of paxillin (Ito et al., 2000), the potential PR72/PR130 family-specific PP2A substrates have yet to be identified. In this study, ppp2r3a was identified as a proximal interactor by BirA*-tagged LPP, TRIP6, vinculin, p130Cas, GIT1, and ILK. Whether these represent PP2A substrates or adaptor proteins that recruit PP2A to IACs via ppp2r3a is unknown. Recently, ppp2r3a and LPP were identified in a proteome-wide screen to identify novel LD motifs (Alam et al., 2020). The LD motifs in paxillin interact with various adhesome proteins containing LD binding domains, including ILK, vinculin, GIT, and talin, among others, and it is feasible that the LD motifs in ppp2r3a and LPP also facilitate interactions with such proteins (Nikolopoulos and Turner, 2001; Brown et al., 1996; Schmalzigaug et al., 2007; Zacharchenko et al., 2016).
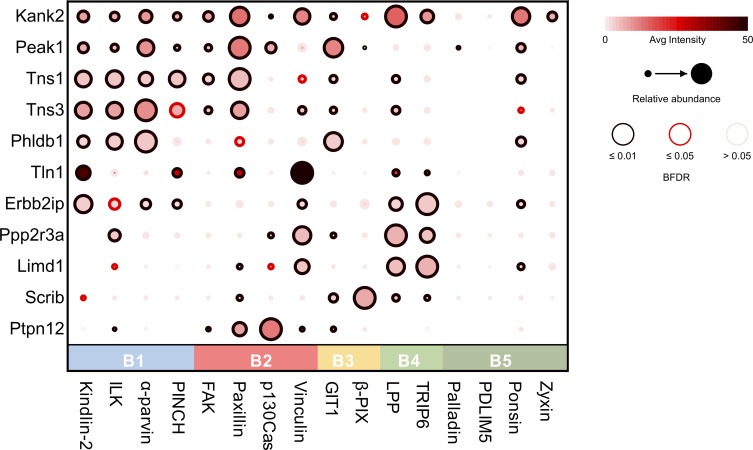
A closer examination of the relative abundances of the central components revealed differential detection. Whereas some proteins were detected at relatively similar levels across baits in a range of clusters (e.g., KANK2, PEAK1, and talin), others were detected with high relative abundance by one or two baits within a single cluster (Fig. 5). For example, PTP-PEST was detected with high relative abundance by BirA*-p130Cas and -paxillin, which may suggest a more specific association. Both paxillin (Shen et al., 1998) and p130Cas (Garton et al., 1996, 1997) have been reported to be substrates for PTP-PEST. Some prey proteins showed similar patterns of bait ID and relative abundance (e.g., Ppp2r3a/LIMD1 and KANK2/PEAK1/Tns1/Tns3), which may suggest that these proteins have similar roles. Future experiments examining the role of these central components in IAC function and regulation will be informative.
Figure 5.
Proximity interactions of the central prey cluster. Dot plot of proteins within the central prey cluster (P12) of the proximity-dependent adhesome. Prey also used as baits found in the central cluster were excluded (Ptk2, Pxn, Vcl, Bcar1, Zyx, Lpp, and Sorbs1). Baits were organized into clusters defined by hierarchical clustering in Fig. 2. Average intensity generated from SAINTexpress. Dot plot was generated by ProHits-Viz (Knight et al., 2017).
Discussion
In this study, multiplexed proximity-dependent biotinylation was used to generate an empirically defined network of proximal associations within IACs. Unbiased bioinformatic analysis of the network revealed five groups of baits that link to common groups of prey and may represent functional modules within IACs. The interconnectivity between these modules and their inferred stratified organization are consistent with current models of adhesome PPI networks and IAC architecture. A large number of IAC-associated proteins were identified within the proximity-dependent adhesome, in addition to a range of prey that may represent novel IAC components or underappreciated links to other cellular organelles. Because of the multifunctionality of many of the BioID baits, it is not possible to conclude that proximal interactors associate with IACs directly, or localize to more distal structures, without further experiments. For example, vinculin, LPP, TRIP6, and zyxin also localize to cell–cell contacts, and it is possible that a proportion of the proximity interactions identified lie at cell–cell junctions. Nonetheless, a central group of 11 highly connected prey was identified that may represent core adhesome components, some of which have few reported associations with IACs. The detection of these proteins by a number of adhesome baits suggests that they may play a more central role in IACs than currently appreciated, and future studies should focus on their role.
Although proximity-dependent labeling methods, such as BioID, have become increasingly popular to examine individual protein interactomes and in large-scale initiatives to map protein interaction networks, there are limitations that must be kept in mind when interpreting data. For example, although highly stringent analyses were performed, it is possible that a number of nonspecific contaminants were identified. For example, despite being extracellular proteins, perlecan (Hspg2) and collagen α-1 (VIII) chain (Col8a1) were identified in the proximity-dependent adhesome and are likely to represent false positives. Similarly, some proximal interactors may be missed due to the restricted labeling radius of BirA* (10–15 nm) and the dependence of labeling on accessible lysines. Indeed, some reported adhesome components were not identified in this study, such as Csk and Hic-5. Finally, due to the differential availability of lysine residues, protein turnover rates, and MS detection of individual peptides and proteins, proximity-dependent biotinylation is unable to differentiate between the degree of proximity, protein abundance, or frequency/permanence of interactions.
Nevertheless, this study has generated insights into the topological organization of the adhesome and has highlighted some underappreciated components that may play a central role in IAC function and regulation. The study therefore provides a useful resource to drive further hypothesis generation and demonstrates that proximity-dependent labeling is a valuable addition to the tools currently available to examine IAC composition and protein–protein relationships. Future studies that focus on how this network is altered when the composition of the extracellular matrix varies and under disease-relevant conditions (e.g., under different force conditions [Horton et al., 2016] or throughout the cell cycle [Jones et al., 2019, 2018]) may further our understanding of the role of IACs in governing cellular behavior in health and disease.
Materials and methods
Reagents
All reagents were acquired from Sigma-Aldrich unless otherwise specified. Primary antibodies used for immunofluorescence microscopy were mouse anti-vinculin (hVin-1; 1:200), rabbit anti-paxillin (GeneTex; 1:200), and mouse anti-c-myc (9B11, Cell Signaling Technologies; 1:200). Alexa Fluor 680–conjugated streptavidin was from Life Technologies, and secondary antibodies (anti-mouse IgG Alexa Fluor 488 and anti-rabbit IgG Alexa Fluor 488) were from Invitrogen.
Cell culture
Mouse pancreatic fibroblasts (im-PSC; Tape et al., 2016) and HEK 293 cells were cultured in D5796 DMEM supplemented with 10% (vol/vol) FBS (Life Technologies) and 2 mM L-glutamine. Cells were maintained at 37°C in a humidified atmosphere with 5% (vol/vol) CO2.
Cloning
The BioID vectors pCDH-TagBFP-T2A-myc-BirA* (Pedley et al., 2019) and pCDNA3.1-BirA*-paxillin were gifts from A. Gilmore (University of Manchester) and E. Manser (IMCB, A*STAR, Singapore). The plasmids containing ILK, ponsin, kindlin-2, vinculin (Humphries et al., 2007), and α-parvin were a gift from C. Ballestrem (University of Manchester), and the plasmid containing zyxin was a gift from A. Sharrocks (University of Manchester). The pcDNA3.1-myc-BirA*-LPP and pcDNA3.1-myc-BirA*-TRIP6 plasmids from which LPP and TRIP6 were amplified, respectively, were generated by J. Askari and J. Zha (University of Manchester) from plasmids containing LPP and TRIP6 that were a gift from A. Sharrocks (University of Manchester). Flag-ECFP-betaPixa (plasmid 15235), mEmerald-PINCH-C-14 (plasmid 54229), mCherry-Palladin-C-7 (plasmid 55113), pEGFP-GIT1 (plasmid 15226), and pGFP-Cas (plasmid 50729) were purchased from Addgene. Full-length ORFs of target adhesome proteins were amplified by PCR and cloned into the pCDH-TagBFP-T2A-myc-BirA* vector using Gibson assembly (vinculin, ponsin, and p130Cas), HiFi DNA assembly (FAK, kindlin-2, β-Pix, palladin, α-parvin, PINCH, PDLIM5, and zyxin), or restriction enzymes (BirA*-paxillin, BirA*-LPP, and BirA*-TRIP6, BspEI and SalI-HF; BirA*-ILK, XhoI and SalI-HF; see Table S4 for primer pairs and annealing temperatures). During PCR amplification, two different annealing temperatures were used to promote efficient primer annealing, first to the plasmid template (10 cycles) then to the PCR product template (25 cycles). All constructs included a 5-aa linker (LERPL) between BirA* and the protein of interest. Primers for Gibson assembly and HiFi assembly were designed using SnapGene (GSL Biotech), and primers were manufactured by Integrated DNA Technologies. ORF sequences were confirmed by sequencing.
Generation of stable cell lines
Lentiviruses containing BirA* constructs were produced by transient cotransfection of HEK 293 cells with pCDH-TagBFP-T2A-myc-BirA* plasmids and packaging vectors (psPAX2 and pM2G) using polyethylenimine (PEI)-mediated transfection. 250 µl DNA mix containing 6 µg pCDH-TagBFP-T2A-myc-BirA* plasmid, 4.5 µg psPAX2, and 3 µg pM2G in Opti-MEM reduced serum medium (Thermo Fisher Scientific) was added to 250 µl PEI mix (44.4 µM PEI and 1.5 mM NaCl in Opti-MEM) and incubated at RT for 20 min. HEK 293 cells (T75 flask, ∼60% confluence) were incubated with 5 ml Opti-MEM and PEI/DNA mix for 6 h before the medium was replaced with fresh medium. 3 d after transfection, filter-sterilized viral medium was added to im-PSC cells for 24 h before being replaced with fresh medium, and cells were passaged 24–48 h later. Cells expressing BFP were selected using FACS and sorted into high-, medium-, and low-expressing populations. Western blotting and immunofluorescence microscopy were used to confirm expression of full-length constructs and select appropriate cell populations with clear subcellular targeting of bait proteins (and biotinylated proteins) to IACs with minimal background localization and biotinylation for use in subsequent experiments.
Proximity biotinylation and affinity purification of biotinylated proteins
To induce proximity biotinylation, cells expressing BirA* constructs were seeded onto plastic tissue culture plates for 8 h to allow for robust IAC formation, and then incubated in medium with 50 µM biotin for 24 h. Biotinylated proteins were affinity purified following a protocol adapted from Roux et al. (Roux et al., 2012, 2018). Three 10-cm plates of cells were washed three times in PBS, and cells were lysed with 400 µl lysis buffer (50 mM Tris HCl, pH 7.4, 250 mM NaCl, 0.1% [wt/vol] SDS, 0.5 mM DTT, and 1× cOmplete Protease Inhibitor Cocktail) at RT. 120 µl 20% (vol/vol) Triton X-100 was added, and cell lysates were maintained at 4°C. DNA was sheared by passing cell lysates through a 19-G needle four times and through a 21-G needle four times before 360 µl chilled 50 mM Tris-HCl, pH 7.4, was added, and then passing through a 27-G needle four times. Cell lysates were centrifuged at full speed for 10 min at 4°C, and supernatant was rotated with 45 µl MagReSyn streptavidin beads (ReSyn Biosciences) at 4°C overnight. Beads were washed twice with 500 µl wash buffer 1 (10% [wt/vol] SDS), once with 500 µl wash buffer 2 (0.1% [wt/vol] deoxycholic acid, 1% [wt/vol] Triton X-100, 1 mM EDTA, 500 mM NaCl, and 50 mM Hepes), and once with 500 µl wash buffer 3 (0.5% [wt/vol] deoxycholic acid, 0.5% [wt/vol] NP-40, 1 mM EDTA, and 10 mM Tris/HCl, pH 7.4). Proteins were eluted in 100 µl of 2× reducing sample buffer with 100 µM biotin for 10 min at 70°C. The presence of biotinylated proteins was confirmed using Western blotting, and samples were analyzed using liquid chromatography–tandem MS (LC-MS/MS).
MS sample preparation
Eluted proteins were briefly subjected to SDS-PAGE (3 min at 200 V, 4–12% Bis-Tris gel, Life Technologies) and stained with InstantBlue Coomassie protein stain before being washed with ddH2O overnight at 4°C. Bands were excised and transferred to wells in a perforated 96-well plate, and in-gel tryptic digestion was performed as previously described (Humphries et al., 2009). Peptides were desalted using 1 mg POROS Oligo R3 beads (Thermo Fisher Scientific). Beads were washed with 50 µl 0.1% (vol/vol) formic acid (FA) before the peptide solution was added. Beads were washed twice with 100 µl 0.1% (vol/vol) FA, and peptides were eluted twice with 50 µl 50% (vol/vol) acetonitrile (ACN) and 0.1% (vol/vol) FA. Peptides were dried using a vacuum centrifuge and resuspended in 11 µl 5% (vol/vol) ACN and 0.1% (vol/vol) FA before analysis by LC-MS/MS.
MS data acquisition
Peptides were analyzed using LC-MS/MS on a 3000 Rapid Separation LC (Dionex Corp.) Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Mobile phase A was 0.1% (vol/vol) FA in water, mobile phase B was 0.1% (vol/vol) FA in ACN, and a 75 mm × 250 µm internal diameter 1.7-mM CSH C18 analytical column (Waters) was used. 3 µl of sample was transferred to a 5-µl loop and loaded onto the column at a flow rate of 300 nl/min for 13 min at 5% (vol/vol) mobile phase B. The loop was taken out of line and the flow was reduced to 200 nl/min in 30 s. Peptides were separated using a gradient of 5% to 18% B in 34.5 min, from 18% to 27% B in 8 min, and from 27% to 60% B in 1 min. The column was washed in 60% B for 3 min before reequilibration to 5% B in 1 min. Flow was increased at 55 min to 300 nl/min until the end of the run at 60 min. MS data were acquired in a data-directed manner for 60 min in positive mode. Peptides were automatically selected for fragmentation by data-dependent analysis on the basis of the top 12 peptides with m/z 300–1750 Th and a charge state of 2, 3, or 4 with a dynamic exclusion set at 15 s. The MS resolution was set at 120,000 with an AGC target of 3e6 and a maximum fill time set at 20 ms. The MS2 resolution was set to 30,000 with an AGC target of 2e5, a maximum fill time of 45 ms, isolation window of 1.3 Th, and collision energy of 28.
Raw data were processed using MaxQuant (v1.6.2.10, available from Max Planck Institute of Biochemistry; Tyanova et al., 2016). All experiments using mouse BioID baits were searched against the mouse proteome obtained from Uniprot (August 2018; The UniProt Consortium, 2017). Experiments involving nonmouse BioID baits were run individually against the same mouse proteome with the relevant nonmouse BioID bait protein sequence appended. Default parameters were used in MaxQuant, with the addition of biotinylation of lysine as a variable modification, match between runs turned on, LFQ quantification selected, and unique peptides only for protein quantification. All mass proteomic data are available via ProteomeXchange with identifier PXD017241.
Bioinformatic analyses
MaxQuant protein LFQ intensities were used to assess the confidence of bait–prey interactions by MS1 intensity–based SAINTexpress (Teo et al., 2014; v3.6.3). Default parameters were used, and a BFDR of ≤ 0.05 was used as a stringency threshold to identify high-confidence bait–prey proximity interactions.
Pairwise comparisons, hierarchical cluster analyses, and visualization of talin biotinylated peptides were performed in R. Hierarchical clustering of baits was performed using the Jaccard distance of proximal prey (BFDR ≤ 0.05, present; BFDR > 0.05, absent), and prey were clustered using the Euclidean distance of fold-change enrichment over control. Results were displayed as a hierarchically clustered heatmap (log2 fold-change values visualized).
Network visualization and analyses were performed using Cytoscape (v3.7.1; Su et al., 2014). Proteins were mapped onto an interaction network compiled from mouse, rat, and human interaction databases from the Biological General Repository for Interaction Datasets (BioGRID; 3.5.166, November 2018), the MatrixDB (April 2012), and the literature-curated adhesome (Chautard et al., 2011; Naba et al., 2012; Winograd-Katz et al., 2014). Network analysis was performed using the NetworkAnalyzer plugin in Cytoscape (Assenov et al., 2008). GO analyses were performed and visualized using the clusterProfiler package in R (Yu et al., 2012). Biotinylated peptides were searched against the mouse talin-1 sequence from UniProt (P26039) to identify biotinylated lysine sequence positions. Highly confident biotinylated lysine positions were selected for mouse talin-1 with MaxQuant localization probability >0.75. The dot plot in Fig. 5 was generated using ProHits-Viz (Knight et al., 2017), using the average intensity generated by SAINTexpress as a measure of abundance.
Immunofluorescence microscopy
Cells expressing BirA* constructs were plated onto glass coverslips for 24 h and incubated with 50 µM biotin for 24 h to initiate biotinylation of proximal proteins. Cells were fixed with 4% (wt/vol) PFA for 15 min at RT and permeabilized with 300 µl of 0.2% (wt/vol) Triton X-100 for 20 min at RT. Coverslips were incubated with primary antibodies directed against proteins indicated in 2% (wt/vol) BSA in PBS for 1 h at RT. Cells were then incubated with fluorophore-conjugated secondary antibodies at RT for 20 min and stained with 1 µg/ml DAPI for 1 min before washing and mounting onto glass slides. Images were acquired using an Olympus BX51 upright microscope with a 60×/0.65-1.25 UPlanFLN or 10×/0.30 UPlanFLN objective and captured using a Coolsnap EZ camera (Photometrics) through MetaVue software (Molecular Devices).
Data deposition
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017241 (Perez-Riverol et al., 2019).
Online supplemental material
Fig. S1 and Fig. S2 show the subcellular targeting of BirA*-tagged adhesome components to IACs and the localization of biotinylated proximal proteins. Fig. S3 shows functional enrichment analysis of proteins identified by each BioID bait, alongside network analysis of bait–bait interactions and the mapping of biotinylated lysines onto the secondary and tertiary structure of talin. Fig. S4 shows functional enrichment analyses of the 16 prey clusters identified by hierarchical cluster analysis of the prey identified by the 16 BioID baits. Table S1 lists all bait–prey interactions identified, with prey annotated according to their functional role and presence in published adhesomes (consensus, meta, and literature-curated) and proximity-interactomes (kindlin-2 and paxillin; Dong et al., 2016). Table S2 contains the results from the hierarchical cluster analysis of BioID baits and prey. Table S3 lists the biotinylated lysines present in talin-1 identified by the BioID baits and their domain localization. Table S4 lists the primer sequences and primer pairs used to generate the 16 BioID constructs, alongside the specific cloning and PCR conditions.
Supplementary Material
describes the proximity-dependent adhesome.
shows the hierarchical clustering of BioID baits and prey.
describes the biotinylation of lysines in talin-1.
lists primers and primer pairs used to generate BioID constructs.
Acknowledgments
The support of the Bio-MS mass spectrometry and Bioimaging core facilities in the Faculty of Biology, Medicine and Health at the University of Manchester is gratefully acknowledged.
This work was supported by a Cancer Research UK Program Grant (C13329/A21671 to M.J. Humphries), Institute Award (A19258 to C. Jorgensen), and Experimental Medicine Program Award (A25236 to C. Jorgensen), the Rosetrees Trust (M286 to C. Jorgensen), and a European Research Council Consolidator Award (ERC-2017-COG 772577 to C. Jorgensen). M.R. Chastney was supported by a PhD studentship from Biotechnology and Biological Sciences Research Council. The work was conducted within the Wellcome Centre for Cell-Matrix Research (core award 203128/Z/16/Z).
The authors declare no competing financial interests.
Author contributions: M.R. Chastney, C. Jorgensen, and M.J. Humphries conceived the project. M.R. Chastney, J.D. Humphries, M.C. Jones, and M.J. Humphries designed the experimental strategy. S. Warwood and D. Knight performed MS analyses. M.R. Chastney performed all other experiments. C. Lawless and M.R. Chastney performed bioinformatic analyses. M.R. Chastney and M.J. Humphries wrote the manuscript, with critical comments and feedback from all authors.
References
- Alam T., Alazmi M., Naser R., Huser F., Momin A.A., Astro V., Hong S., Walkiewicz K.W., Canlas C.G., Huser R., et al. 2020. Proteome-level assessment of origin, prevalence and function of leucine-aspartic acid (LD) motifs. Bioinformatics. 36:1121–1128. 10.1093/bioinformatics/btz703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers-Loustau A., Côté J.F., Charest A., Dowbenko D., Spencer S., Lasky L.A., and Tremblay M.L.. 1999. Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J. Cell Biol. 144:1019–1031. 10.1083/jcb.144.5.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y., Ramírez F., Schelhorn S.-E., Lengauer T., and Albrecht M.. 2008. Computing topological parameters of biological networks. Bioinformatics. 24:282–284. 10.1093/bioinformatics/btm554 [DOI] [PubMed] [Google Scholar]
- Atherton P., Lausecker F., Carisey A., Gilmore A., Critchley D., Barsukov I., and Ballestrem C.. 2020. Relief of talin autoinhibition triggers a force-independent association with vinculin. J. Cell Biol. 219 e201903134 10.1083/jcb.201903134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton P., Stutchbury B., Wang D.-Y., Jethwa D., Tsang R., Meiler-Rodriguez E., Wang P., Bate N., Zent R., Barsukov I.L., et al. 2015. Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 6:10038 10.1038/ncomms10038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S.J., Gontarczyk A.M., Alghamdi A.A., Ellison T.S., Johnson R.T., Fowler W.J., Kirkup B.M., Silva B.C., Harry B.E., Schneider J.G., et al. 2018. The β3-integrin endothelial adhesome regulates microtubule-dependent cell migration. EMBO Rep. 19 e44578 10.15252/embr.201744578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M., Carracedo S., and Gullberg D.. 2010. Integrins. Cell Tissue Res. 339:269–280. 10.1007/s00441-009-0834-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello T.T., and Peifer M.. 2019. Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell Biol. 218:742–756. 10.1083/jcb.201810103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher R.T., Veelders M., Rombaut P., Faix J., Theodosiou M., Stradal T.E., Rottner K., Zent R., Herzog F., and Fässler R.. 2017. Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216:3785–3798. 10.1083/jcb.201701176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet B.P., Gough R.E., Ammon Y.C., van de Willige D., Post H., Jacquemet G., Altelaar A.M., Heck A.J.R., Goult B.T., and Akhmanova A.. 2016. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife. 5 e18124 10.7554/eLife.18124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow J.M., Reno T.A., Jo M., Gonias S.L., and Klemke R.L.. 2013. Dynamic phosphorylation of tyrosine 665 in pseudopodium-enriched atypical kinase 1 (PEAK1) is essential for the regulation of cell migration and focal adhesion turnover. J. Biol. Chem. 288:123–131. 10.1074/jbc.M112.410910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.C., Perrotta J.A., and Turner C.E.. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135:1109–1123. 10.1083/jcb.135.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.D., and Humphries M.J.. 2011. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3 a004994 10.1101/cshperspect.a004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., Baird M.A., Shtengel G., Campbell S.L., Hess H.F., Davidson M.W., and Waterman C.M.. 2015. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17:880–892. 10.1038/ncb3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černohorská M., Sulimenko V., Hájková Z., Sulimenko T., Sládková V., Vinopal S., Dráberová E., and Dráber P.. 2016. GIT1/βPIX signaling proteins and PAK1 kinase regulate microtubule nucleation. Biochim. Biophys. Acta. 1863(6, 6 Pt A):1282–1297. 10.1016/j.bbamcr.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Chautard E., Fatoux-Ardore M., Ballut L., Thierry-Mieg N., and Ricard-Blum S.. 2011. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 39(Database issue):D235–D240. 10.1093/nar/gkq830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher D.R., Hochgräfe F., Zhang L., Liu L., Lyons R.J., Rickwood D., Tactacan C.M., Browne B.C., Ali N., Chan H., et al. 2013. Involvement of Lyn and the atypical kinase SgK269/PEAK1 in a basal breast cancer signaling pathway. Cancer Res. 73:1969–1980. 10.1158/0008-5472.CAN-12-1472 [DOI] [PubMed] [Google Scholar]
- Dan L., Shi M., Duan H., Han C., and Guo N.. 2010. Erbin, a negative regulator in diverse signal pathways. Curr. Protein Pept. Sci. 11:759–764. 10.2174/138920310794557673 [DOI] [PubMed] [Google Scholar]
- Dong J.-M., Tay F.P.-L., Swa H.L.-F., Gunaratne J., Leung T., Burke B., and Manser E.. 2016. Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal. 9:rs4 10.1126/scisignal.aaf3572 [DOI] [PubMed] [Google Scholar]
- Dow L.E., Kauffman J.S., Caddy J., Zarbalis K., Peterson A.S., Jane S.M., Russell S.M., and Humbert P.O.. 2007. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 26:2272–2282. 10.1038/sj.onc.1210016 [DOI] [PubMed] [Google Scholar]
- Favre B., Fontao L., Koster J., Shafaatian R., Jaunin F., Saurat J.-H., Sonnenberg A., and Borradori L.. 2001. The hemidesmosomal protein bullous pemphigoid antigen 1 and the integrin β 4 subunit bind to ERBIN. Molecular cloning of multiple alternative splice variants of ERBIN and analysis of their tissue expression. J. Biol. Chem. 276:32427–32436. 10.1074/jbc.M011005200 [DOI] [PubMed] [Google Scholar]
- Garton A.J., and Tonks N.K.. 1999. Regulation of fibroblast motility by the protein tyrosine phosphatase PTP-PEST. J. Biol. Chem. 274:3811–3818. 10.1074/jbc.274.6.3811 [DOI] [PubMed] [Google Scholar]
- Garton A.J., Flint A.J., and Tonks N.K.. 1996. Identification of p130(cas) as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol. Cell. Biol. 16:6408–6418. 10.1128/MCB.16.11.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton A.J., Burnham M.R., Bouton A.H., and Tonks N.K.. 1997. Association of PTP-PEST with the SH3 domain of p130cas; a novel mechanism of protein tyrosine phosphatase substrate recognition. Oncogene. 15:877–885. 10.1038/sj.onc.1201279 [DOI] [PubMed] [Google Scholar]
- Gough R.E., and Goult B.T.. 2018. The tale of two talins - two isoforms to fine-tune integrin signalling. FEBS Lett. 592:2108–2125. 10.1002/1873-3468.13081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H.J., and Brown N.H.. 2019. Integrin intracellular machinery in action. Exp. Cell Res. 378:226–231. 10.1016/j.yexcr.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Green H.J., Griffiths A.G.M., Ylänne J., and Brown N.H.. 2018. Novel functions for integrin-associated proteins revealed by analysis of myofibril attachment in Drosophila. eLife. 7 e35783 10.7554/eLife.35783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T., et al. 2015. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 163:1484–1499. 10.1016/j.cell.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger D.S., Bouaouina M., and Calderwood D.A.. 2009. Kindlin-1 and -2 directly bind the C-terminal region of β integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284:11485–11497. 10.1074/jbc.M809233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Tatsumi H., Lim C.T., and Sokabe M.. 2014. Force-dependent vinculin binding to talin in live cells: a crucial step in anchoring the actin cytoskeleton to focal adhesions. Am. J. Physiol. Cell Physiol. 306:C607–C620. 10.1152/ajpcell.00122.2013 [DOI] [PubMed] [Google Scholar]
- Hoffmann J.-E., Fermin Y., Stricker R.L., Ickstadt K., and Zamir E.. 2014. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. eLife. 3 e02257 10.7554/eLife.02257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton E.R., Astudillo P., Humphries M.J., and Humphries J.D.. 2016. Mechanosensitivity of integrin adhesion complexes: role of the consensus adhesome. Exp. Cell Res. 343:7–13. 10.1016/j.yexcr.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Horton E.R., Byron A., Askari J.A., Ng D.H.J., Millon-Frémillon A., Robertson J., Koper E.J., Paul N.R., Warwood S., Knight D., et al. 2015. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 17:1577–1587. 10.1038/ncb3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins C.J., and Andrulis I.L.. 2008. Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Lett. 267:55–66. 10.1016/j.canlet.2008.03.015 [DOI] [PubMed] [Google Scholar]
- Humphries J.D., Byron A., Bass M.D., Craig S.E., Pinney J.W., Knight D., and Humphries M.J.. 2009. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2:ra51 10.1126/scisignal.2000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.D., Chastney M.R., Askari J.A., and Humphries M.J.. 2019. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 56:14–21. 10.1016/j.ceb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Humphries J.D., Wang P., Streuli C., Geiger B., Humphries M.J., and Ballestrem C.. 2007. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179:1043–1057. 10.1083/jcb.200703036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A., Kataoka T.R., Watanabe M., Nishiyama K., Mazaki Y., Sabe H., Kitamura Y., and Nojima H.. 2000. A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J. 19:562–571. 10.1093/emboj/19.4.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James V., Zhang Y., Foxler D.E., de Moor C.H., Kong Y.W., Webb T.M., Self T.J., Feng Y., Lagos D., Chu C.-Y., et al. 2010. LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA. 107:12499–12504. 10.1073/pnas.0914987107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Zwaenepoel K., Rossé C., Petit M.M.R., Goris J., and Parker P.J.. 2016. PP2A binds to the LIM domains of lipoma-preferred partner through its PR130/B2 subunit to regulate cell adhesion and migration. J. Cell Sci. 129:1605–1618. 10.1242/jcs.175778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin-Bastard F., Arsanto J.-P., Le Bivic A., Navarro C., Vély F., Saito H., Marchetto S., Hatzfeld M., Santoni M.-J., Birnbaum D., et al. 2002. Interaction between Erbin and a Catenin-related protein in epithelial cells. J. Biol. Chem. 277:2869–2875. 10.1074/jbc.M109652200 [DOI] [PubMed] [Google Scholar]
- Jin X., Li B., Zhao Y., Liu X., Li Y., Song L., Cui L., Xie D., Li T., Zhang X., et al. 2019. Erbin plays a critical role in human umbilical vein endothelial cell migration and tubular structure formation via the Smad1/5 pathway. J. Cell. Biochem. 120:4654–4664. 10.1002/jcb.27754 [DOI] [PubMed] [Google Scholar]
- Jones M.C., Askari J.A., Humphries J.D., and Humphries M.J.. 2018. Cell adhesion is regulated by CDK1 during the cell cycle. J. Cell Biol. 217:3203–3218. 10.1083/jcb.201802088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.C., Humphries J.D., Byron A., Millon-Frémillon A., Robertson J., Paul N.R., Ng D.H.J., Askari J.A., and Humphries M.J.. 2015. Isolation of integrin-based adhesion complexes. Curr. Protoc. Cell Biol. 66:9.8.1–9.8.15. [DOI] [PMC free article] [PubMed]
- Jones M.C., Zha J., and Humphries M.J.. 2019. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374 20180227 10.1098/rstb.2018.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelber J.A., Reno T., Kaushal S., Metildi C., Wright T., Stoletov K., Weems J.M., Park F.D., Mose E., Wang Y., et al. 2012. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res. 72:2554–2564. 10.1158/0008-5472.CAN-11-3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V., and Roux K.J.. 2014. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA. 111:E2453–E2461. 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J.D.R., Choi H., Gupta G.D., Pelletier L., Raught B., Nesvizhskii A.I., and Gingras A.-C.. 2017. ProHits-viz: a suite of web tools for visualizing interaction proteomics data. Nat. Methods. 14:645–646. 10.1038/nmeth.4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravic B., Huraskin D., Frick A.D., Jung J., Redai V., Palmisano R., Marchetto S., Borg J.-P., Mei L., and Hashemolhosseini S.. 2016. LAP proteins are localized at the post-synaptic membrane of neuromuscular junctions and appear to modulate synaptic morphology and transmission. J. Neurochem. 139:381–395. 10.1111/jnc.13710 [DOI] [PubMed] [Google Scholar]
- Kumar A., Ouyang M., Van den Dries K., McGhee E.J., Tanaka K., Anderson M.D., Groisman A., Goult B.T., Anderson K.I., and Schwartz M.A.. 2016. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213:371–383. 10.1083/jcb.201510012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.-C., Han X., Yates J.R. III, and Waterman C.M.. 2012. Isolation of focal adhesion proteins for biochemical and proteomic analysis. Methods Mol. Biol. 757:297–323. 10.1007/978-1-61779-166-6_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A.B., Grosveld F., et al. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell. 11:21–32. 10.1016/j.devcel.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Lee H.-S., Bellin R.M., Walker D.L., Patel B., Powers P., Liu H., Garcia-Alvarez B., de Pereda J.M., Liddington R.C., Volkmann N., et al. 2004. Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 343:771–784. 10.1016/j.jmb.2004.08.069 [DOI] [PubMed] [Google Scholar]
- Lim K.Y.B., Gödde N.J., Humbert P.O., and Kvansakul M.. 2017. Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor β-PIX. J. Biol. Chem. 292:20425–20436. 10.1074/jbc.M117.799452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang Y., Goh W.I., Goh H., Baird M.A., Ruehland S., Teo S., Bate N., Critchley D.R., Davidson M.W., et al. 2015. Talin determines the nanoscale architecture of focal adhesions. Proc. Natl. Acad. Sci. USA. 112:E4864–E4873. 10.1073/pnas.1512025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhdjian A.H., Kai F., Rubashkin M.G., Prahl L.S., Przybyla L.M., McGregor A.L., Bell E.S., Barnes J.M., DuFort C.C., Ou G., et al. 2017. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell. 28:1467–1488. 10.1091/mbc.e16-09-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis U.R., Chavakis E., Kruse C., Jungblut B., Kaluza D., Wandzioch K., Manavski Y., Heide H., Santoni M.J., Potente M., et al. 2013. The polarity protein Scrib is essential for directed endothelial cell migration. Circ. Res. 112:924–934. 10.1161/CIRCRESAHA.112.300592 [DOI] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., and Hynes R.O.. 2012. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics. 11 M111.014647 10.1074/mcp.M111.014647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulos S.N., and Turner C.E.. 2001. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 276:23499–23505. 10.1074/jbc.M102163200 [DOI] [PubMed] [Google Scholar]
- Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., et al. 2008. Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 17:3552–3565. 10.1093/hmg/ddn248 [DOI] [PubMed] [Google Scholar]
- Noordstra I., and Akhmanova A.. 2017. Linking cortical microtubule attachment and exocytosis. F1000 Res. 6:469 10.12688/f1000research.10729.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke R.L., and Daly R.J.. 2018. The pseudokinases SgK269 and SgK223: A novel oncogenic alliance in human cancer. Cell Adhes. Migr. 12:524–528. 10.1080/19336918.2017.1394570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N., Vitale N., Borg J.-P., and Etienne-Manneville S.. 2006. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr. Biol. 16:2395–2405. 10.1016/j.cub.2006.10.026 [DOI] [PubMed] [Google Scholar]
- Pasapera A.M., Schneider I.C., Rericha E., Schlaepfer D.D., and Waterman C.M.. 2010. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188:877–890. 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley R., King L., Mallikarjun V., Wang P., Swift J., Brennan K., and Gilmore A.P.. 2019. BioID based proteomic analysis of the Bid interactome identifies novel proteins involved in cell cycle dependent apoptotic priming. bioRxiv. (Preprint posted February 7, 2020.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1):D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.M.R., Meulemans S.M.P., Alen P., Ayoubi T.A.Y., Jansen E., and Van de Ven W.J.M.. 2005. The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 6:1 10.1186/1471-2121-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovanac K., Morgner J., Schulz J.-N., Blumbach K., Patterson C., Geiger T., Mann M., Krieg T., Eckes B., Fässler R., et al. 2013. Stabilization of integrin-linked kinase by the Hsp90-CHIP axis impacts cellular force generation, migration and the fibrotic response. EMBO J. 32:1409–1424. 10.1038/emboj.2013.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahikainen R., Öhman T., Turkki P., Varjosalo M., and Hytönen V.P.. 2019. Talin-mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J. Cell Sci. 132 jcs226514 10.1242/jcs.226514 [DOI] [PubMed] [Google Scholar]
- Robertson J., Jacquemet G., Byron A., Jones M.C., Warwood S., Selley J.N., Knight D., Humphries J.D., and Humphries M.J.. 2015. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat. Commun. 6:6265 10.1038/ncomms7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Burke B., and May D.G.. 2018. BioID: A Screen for Protein-Protein Interactions. Curr. Protoc. Protein Sci. 91:19.23.1-19.23.15. 10.1002/cpps.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo P.P. 2016. The broken “Off” switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 6:87–99. 10.1016/j.bbacli.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B.J., Cherniavskaya O., Sakai R., Tanaka S., and Sheetz M.P.. 2006. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 127:1015–1026. 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Friedel C.C., Boulegue C., and Fässler R.. 2011. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12:259–266. 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Hermann M.-R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.-E., Théry M., et al. 2013. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15:625–636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Schmalzigaug R., Garron M.-L., Roseman J.T., Xing Y., Davidson C.E., Arold S.T., and Premont R.T.. 2007. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell. Signal. 19:1733–1744. 10.1016/j.cellsig.2007.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Lyons P., Cooley M., Davidson D., Veillette A., Salgia R., Griffin J.D., and Schaller M.D.. 2000. The noncatalytic domain of protein-tyrosine phosphatase-PEST targets paxillin for dephosphorylation in vivo. J. Biol. Chem. 275:1405–1413. 10.1074/jbc.275.2.1405 [DOI] [PubMed] [Google Scholar]
- Shen Y., Schneider G., Cloutier J.F., Veillette A., and Schaller M.D.. 1998. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J. Biol. Chem. 273:6474–6481. 10.1074/jbc.273.11.6474 [DOI] [PubMed] [Google Scholar]
- Spiess M., Hernandez-Varas P., Oddone A., Olofsson H., Blom H., Waithe D., Lock J.G., Lakadamyali M., and Strömblad S.. 2018. Active and inactive β1 integrins segregate into distinct nanoclusters in focal adhesions. J. Cell Biol. 217:1929–1940. 10.1083/jcb.201707075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:561–573. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubb A., Guzmán C., Närvä E., Aaron J., Chew T.-L., Saari M., Miihkinen M., Jacquemet G., and Ivaska J.. 2019. Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. Nat. Commun. 10:4756 10.1038/s41467-019-12611-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutchbury B., Atherton P., Tsang R., Wang D.-Y., and Ballestrem C.. 2017. Distinct focal adhesion protein modules control different aspects of mechanotransduction. J. Cell Sci. 130:1612–1624. 10.1242/jcs.195362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Morris J.H., Demchak B., and Bader G.D.. 2014. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 47:1–24: 24. 10.1002/0471250953.bi0813s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Tseng H.Y., Tan S., Senger F., Kurzawa L., Dedden D., Mizuno N., Wasik A.A., Thery M., Dunn A.R., et al. 2016. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 18:941–953. 10.1038/ncb3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tape C.J., Ling S., Dimitriadi M., McMahon K.M., Worboys J.D., Leong H.S., Norrie I.C., Miller C.J., Poulogiannis G., Lauffenburger D.A., et al. 2016. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 165:910–920. 10.1016/j.cell.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.-C., and Choi H.. 2014. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 100:37–43. 10.1016/j.jprot.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45(D1):D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., and Cox J.. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11:2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Van Itallie C.M., Aponte A., Tietgens A.J., Gucek M., Fredriksson K., and Anderson J.M.. 2013. The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J. Biol. Chem. 288:13775–13788. 10.1074/jbc.M113.466193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kelber J.A., Tran Cao H.S., Cantin G.T., Lin R., Wang W., Kaushal S., Bristow J.M., Edgington T.S., Hoffman R.M., et al. 2010. Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression [corrected]. Proc. Natl. Acad. Sci. USA. 107:10920–10925. 10.1073/pnas.0914776107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd-Katz S.E., Fässler R., Geiger B., and Legate K.R.. 2014. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15:273–288. 10.1038/nrm3769 [DOI] [PubMed] [Google Scholar]
- Xu L., Braun L.J., Rönnlund D., Widengren J., Aspenström P., and Gad A.K.B.. 2018. Nanoscale localization of proteins within focal adhesions indicates discrete functional assemblies with selective force-dependence. FEBS J. 285:1635–1652. 10.1111/febs.14433 [DOI] [PubMed] [Google Scholar]
- Yao M., Goult B.T., Klapholz B., Hu X., Toseland C.P., Guo Y., Cong P., Sheetz M.P., and Yan J.. 2016. The mechanical response of talin. Nat. Commun. 7:11966 10.1038/ncomms11966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharchenko T., Qian X., Goult B.T., Jethwa D., Almeida T.B., Ballestrem C., Critchley D.R., Lowy D.R., and Barsukov I.L.. 2016. LD Motif Recognition by Talin: Structure of the Talin-DLC1 Complex. Structure. 24:1130–1141. 10.1016/j.str.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R. 2013. Cadherin adhesome at a glance. J. Cell Sci. 126:373–378. 10.1242/jcs.111559 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma’ayan A., Iyengar R., and Geiger B.. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867. 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir E., Geiger B., and Kam Z.. 2008. Quantitative multicolor compositional imaging resolves molecular domains in cell-matrix adhesions. PLoS One. 3 e1901 10.1371/journal.pone.0001901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.S., Lim J.P., Ng Y.-W., Lim L., and Manser E.. 2005. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell. 20:237–249. 10.1016/j.molcel.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Xia Y., Hawke D., Halle M., Tremblay M.L., Gao X., Zhou X.Z., Aldape K., Cobb M.H., Xie K., et al. 2009. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol. Cell. 35:11–25. 10.1016/j.molcel.2009.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Li X., and Premont R.T.. 2016. Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 129:1963–1974. 10.1242/jcs.179465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
describes the proximity-dependent adhesome.
shows the hierarchical clustering of BioID baits and prey.
describes the biotinylation of lysines in talin-1.
lists primers and primer pairs used to generate BioID constructs.