Moon et al., demonstrate a new role for ribosome-associated quality control (RQC) factors in mediating mRNA release from ribosomes during stress to promote their localization to stress granules.
Abstract
Stress granules are dynamic assemblies of proteins and nontranslating RNAs that form when translation is inhibited in response to diverse stresses. Defects in ubiquitin–proteasome system factors including valosin-containing protein (VCP) and the proteasome impact the kinetics of stress granule induction and dissolution as well as being implicated in neuropathogenesis. However, the impacts of dysregulated proteostasis on mRNA regulation and stress granules are not well understood. Using single mRNA imaging, we discovered ribosomes stall on some mRNAs during arsenite stress, and the release of transcripts from stalled ribosomes for their partitioning into stress granules requires the activities of VCP, components of the ribosome-associated quality control (RQC) complex, and the proteasome. This is an unexpected contribution of the RQC system in releasing mRNAs from translation under stress, thus identifying a new type of stress-activated RQC (saRQC) distinct from canonical RQC pathways in mRNA substrates, cellular context, and mRNA fate.
Introduction
Stress granules (SGs) are evolutionarily conserved assemblies of nontranslating RNAs, translation factors, and RNA-binding proteins that form during diverse stress conditions that cause translation inhibition (Anderson and Kedersha, 2008; Ivanov et al., 2019; Protter and Parker, 2016). These stress-induced RNA–protein granules are cytoplasmic membrane-less condensates similar to neuronal transport granules, processing bodies, and germ cell granules (Anderson and Kedersha, 2006; Buchan, 2014). SGs are enriched in long, inefficiently translated RNAs, but other key determinants of mRNA recruitment to, and retention within, SGs remain to be established (Khong et al., 2017; Moon et al., 2019; Namkoong et al., 2018). While the function of SGs is not well understood, they are thought to contribute to cell survival in part by sequestration of signaling proteins to either enhance or limit signaling activity (Kedersha et al., 2013). The dysregulation of SGs is implicated in an array of neurodegenerative and neuromuscular disorders including frontotemporal dementia and amyotrophic lateral sclerosis (Buchan and Parker, 2009; Kedersha et al., 2013; Li et al., 2013; Ramaswami et al., 2013; Taylor et al., 2016). Thus, understanding the fundamental mechanisms that dictate SG composition and regulation could provide insight into how cells respond to stress and how this process goes awry in disease contexts.
The ubiquitin–proteasome system (UPS) is a key mechanism in the cell that mediates protein homeostasis through targeted ubiquitin-mediated protein degradation by the 26S proteasome (Nandi et al., 2006). Certain ubiquitinated protein substrates must be processed by the ubiquitin segregase valosin-containing protein (VCP; also known as p97 or Cdc48) to enable proteasome-mediated degradation (Bard et al., 2018; Ye et al., 2017). The UPS limits the accumulation of cytotoxic, misfolded, aggregation-prone proteins in the cell and serves important cotranslational quality control functions such as degrading nascent proteins that fail to fold properly (Goldberg, 2003). The ribosome-associated quality control (RQC) pathway is one such mechanism that targets nascent proteins and mRNAs for degradation when ribosomes stall on aberrant mRNAs (such as mRNAs lacking stop codons, mRNAs that are prematurely polyadenylated, or mRNAs have truncated ORFs, strong secondary structures, rare codons, or are oxidized; Brandman and Hegde, 2016; Brandman et al., 2012; Doma and Parker, 2006; Ito-Harashima et al., 2007; Meaux and Van Hoof, 2006; Shoemaker and Green, 2012; Simms et al., 2014; Wilson and Beckmann, 2011). Importantly, defects in the UPS are associated with aging and a wide range of neurodegenerative disorders (de Bot et al., 2012; Chu et al., 2009; Gonzalez et al., 2014; Johnson et al., 2010; Watts et al., 2004).
Altered UPS activity interferes with SG induction upon stress and their disassembly when stress is resolved. Proteasome inhibition causes SGs to assemble after several hours (Mazroui et al., 2007). However, inhibition of VCP or proteasome activity perturbs SG formation in response to acute stress (Seguin et al., 2014). In addition, VCP inhibition can cause constitutive SGs and limits SG disassembly (Buchan et al., 2013; Rodriguez-Ortiz et al., 2016; Turakhiya et al., 2018; Wang et al., 2019). Thus, SG disassembly requires the activities of VCP and the proteasome, potentially acting either by removing misfolded or aberrant proteins from SGs for proteasomal decay (Turakhiya et al., 2018) or by limiting granulophagy, the degradation of SGs via autophagy (Buchan et al., 2013).
In this study, we applied single mRNA imaging approaches (Khong et al., 2017; Moon et al., 2019; Morisaki et al., 2016) to evaluate the role of VCP and the proteasome in regulating mRNA–SG interactions. Our goal was to directly assess how the UPS impacts the release of mRNAs from translation and their partitioning into SGs. Surprisingly, we uncovered a new role for VCP, the proteasome, and RQC factors listerin (LTN1) and nuclear export mediator factor (NEMF) for resolving stalled ribosomes on mRNAs and facilitating mRNA partitioning into SGs. Disease-associated alleles of VCP (Watts et al., 2004; Johnson et al., 2010) with enhanced ubiquitin segregase activity (Zhang et al., 2015, 2017) increased mRNA partitioning into SGs. This work suggests a new role for a stress-activated RQC (saRQC) pathway in mediating mRNA recruitment to SGs.
Results
Inhibition of VCP or proteasome function increases mRNAs associated with nascent protein chains during arsenite stress
To evaluate whether and how VCP or proteasome perturbation impacts mRNA regulation during stress, we simultaneously examined the translation status and interaction of individual mRNAs with SGs in living cells using nascent chain tracking (Morisaki et al., 2016). This assay allows visualization of three distinct aspects of individual mRNA molecules being targeted to SGs: their release from polysomes, the duration of transient docking interactions with SGs, and the kinetics of stable mRNA association with SGs (Moon et al., 2019). We examined how N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ), a VCP inhibitor (Chou et al., 2011), and MG132, a proteasome inhibitor (Figueiredo-Pereira et al., 1994), affected mRNA–nascent chain interactions and mRNA interactions with SGs in U-2 OS cells stably expressing GFP-G3BP1 (Kedersha et al., 2008) during acute arsenite stress, which inhibits translation initiation and leads to ribosome run-off (McEwen et al., 2005).
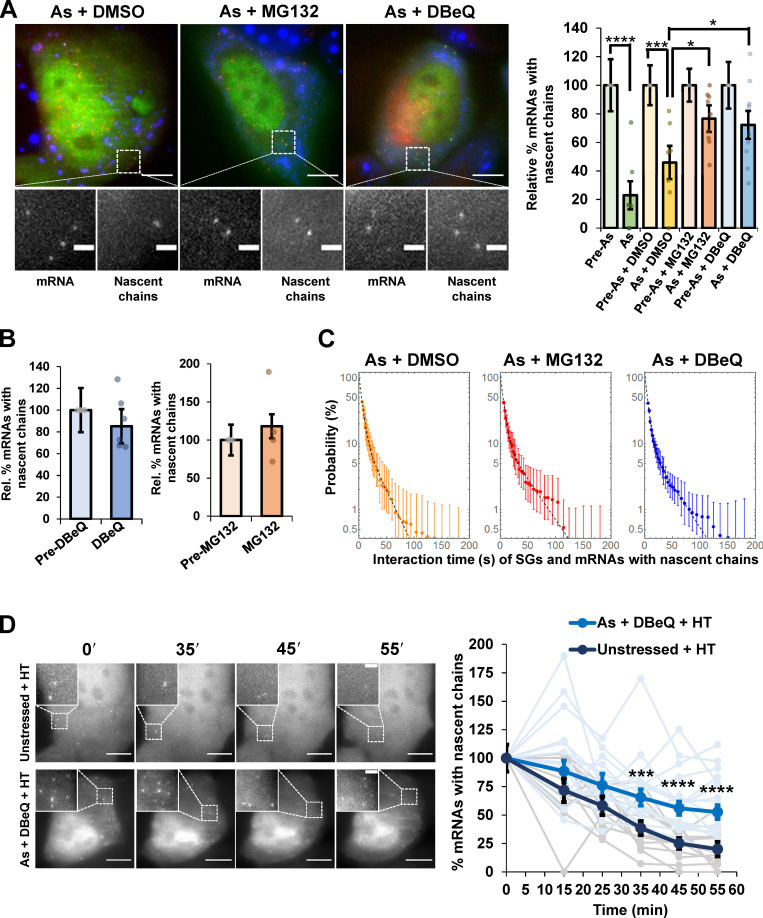
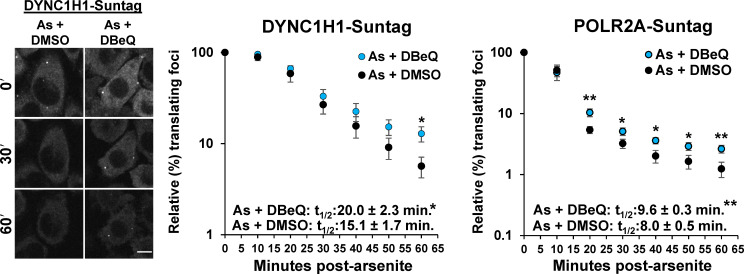
An important observation was that cells treated with DBeQ or MG132 during arsenite stress exhibited an increased number of mRNAs associated with nascent protein chains despite the presence of SGs (Fig. 1 A, Video 1, Video 2, and Video 3). Cells were cotreated simultaneously with arsenite and either DMSO, DBeQ, or MG132, and all images were taken pre-cotreatment and after SGs formed (15–75 min after stress) as described previously (Moon et al., 2019). Treatment with DBeQ or MG132 alone for a similar duration (10–67 min) did not alter the relative number of mRNAs associated with nascent protein chains (Fig. 1 B). The mRNAs associated with nascent chains, in both stressed cells treated with carrier (DMSO) and in the presence of DBeQ or MG132, only interacted with SGs in a transient manner and could not enter into a stable interaction state with them (Fig. 1 C). The transient interaction of SGs with mRNAs associated with nascent protein chains and ribosomes is consistent with previous observations showing the mRNAs associated with 80S subunits can only interact with SGs for short periods (Moon et al., 2019). A complementary approach showed a similar increase in mRNAs associated with nascent chains for the DYNC1H1 and POLR2A mRNAs visualized with the SunTag system upon DBeQ treatment during arsenite stress (Fig. S1; Pichon et al., 2016). These observations suggest that on some mRNA molecules, VCP and proteasome function is required for efficient ribosome release during arsenite stress.
Figure 1.
Translating ribosomes stall on reporter mRNAs during arsenite stress and require VCP and proteasome activity to be resolved. (A) Left: Representative images of SM-KDM5B reporter mRNAs tagged with MS2 coat protein (MCP)–Halo JF646 (red) with nascent chains visualized with Cy3–anti-FLAG antibody fragments (green) in cells expressing the SG marker GFP-G3BP1 (blue) cotreated with arsenite (As) and DMSO, MG132, or DBeQ. Scale bars, 10 µm. Magnified panels show individual mRNAs and nascent chains. Scale bars, 2 µm. Right: Average relative percent SM-KDM5B reporter mRNAs with nascent chains ± SEM with individual points representing a single cell relative to prestress levels for As (0.5 mM, n = 7 cells), As + DMSO (0.1%, n = 7 cells), As + MG132 (10 µM, n = 9 cells), and As + DBeQ (10 µM, n = 9 cells). Student’s t test was done to assess significance, with *, P ≤ 0.05; ***, P ≤ 0.005; and ****, P ≤ 0.001. (B) The percentage of SM-KDM5B reporter mRNAs visualized as in A in cells before or after treatment with DBeQ (left; 10 µM, n = 6 cells) or MG132 (right; 10 µM, n = 4 cells) alone. (C) Average binding-time survival probability (±SEM) of SM-KDM5B mRNAs associated with nascent chains and SGs in cells cotreated as in A with As + DMSO (n = 6 cells), As + DBeQ (n = 14 cells), and As + MG132 (n = 10 cells). (D) Cells harboring SM-KDM5B reporter mRNAs, FLAG antibody fragments, and MCP–Halo JF646 were either unstressed and treated with HT or stressed with As and cotreated with DBeQ and HT. Images were acquired prestress and then in 10-min intervals starting 15 min later. HT was added 15 min after As + DBeQ (or no treatment). Left: Representative time series of unstressed cells (top) and As + DBeQ cells (bottom) treated with HT. Scale bars, 10 µm (in whole cell) and 2 µm (in insets). Right: The average ± SEM of the percent SM-KDM5B mRNAs with nascent chains at each time point relative to prestress conditions is shown in unstressed cells (black) and As-stressed cells treated with DBeQ (blue), with individual cells shown in gray (unstressed + HT, n = 13 cells) and light blue (As stress + DBeQ + HT, n = 17 cells). Student’s t test was done to assess significance between untreated + HT and As + DBeQ + HT conditions at each time point, with ***, P ≤ 0.005 and ****, P ≤ 0.001. Exact P values and source data are listed in Table S1.
Video 1.
Representative video showing nascent chain tracking of SM-KDM5B mRNAs in U-2 OS cells treated with arsenite and DMSO, related to Fig. 1, A and C. Nascent protein chains are shown in green, mRNAs (Halo-MS2 coat protein labeled with JF646) are shown in red, and SGs (GFP-G3BP1) are shown in blue. Images were acquired every ∼2 s for 10 min. Scale bar, 10 µm.
Video 2.
Representative video showing nascent chain tracking of SM-KDM5B mRNAs in U-2 OS cells treated with arsenite and DBeQ, related to Fig. 1, A and C. Nascent protein chains are shown in green, mRNAs (Halo-MS2 coat protein labeled with JF646) are shown in red, and SGs (GFP-G3BP1) are shown in blue. Images were acquired every ∼2 s for 10 min. Scale bar, 10 µm.
Video 3.
Representative video showing nascent chain tracking of SM-KDM5B mRNAs in U-2 OS cells treated with arsenite and MG132, related to Fig. 1, A and C. Nascent protein chains are shown in green, mRNAs (Halo-MS2 coat protein labeled with JF646) are shown in red, and SGs (GFP-G3BP1) are shown in blue. Images were acquired every ∼2 s for 10 min. Scale bar, 10 µm.
Figure S1.
Nascent peptides visualized with SunTags are elevated in arsenite-stressed cells upon VCP inhibition. HeLa cells with endogenously tagged SunTagX32-DYNC1H1 (DYNC1H1-Suntag) and SunTagX56-POLR2A (POLR2A-Suntag) genes and stably expressing scFv-sfGFP (Pichon et al., 2016) were imaged every 10 min in a humidified, 37ºC chamber on a spinning disc microscope at 40× for 1 h following treatment with 0.5 mM arsenite (As), cotreated with 0.1% DMSO or 10 µM DBeQ. A representative time series (0, 30, and 60 min after stress) is shown at left for the SunTagX32-DYNC1H1 cell line. Scale bar, 10 µm. Shown at right is the average ± SEM of the relative percent SunTag foci in each frame from n = seven independent experiments for each cell line. Student’s t test was done to assess significance between the relative number of translating foci in DMSO- versus DBeQ-treated cells at each time point, with *, P < 0.05 and **, P < 0.01. Insets: Data were fit with exponential trendlines to calculate the time when 50% of translation foci (t1/2) were present compared with prestress images for each replicate (DYNC1H1-Suntag: As + DBeQ R2 = 0.9527, As + DMSO R2 = 0.9616; POLR2A-Suntag: As + DBeQ R2 = 0.8405, As + DMSO R2 = 0.8334). Student’s t test was used to assess significance, with *, P < 0.05 and **, P < 0.01. Exact P values and source data are provided in Table S1.
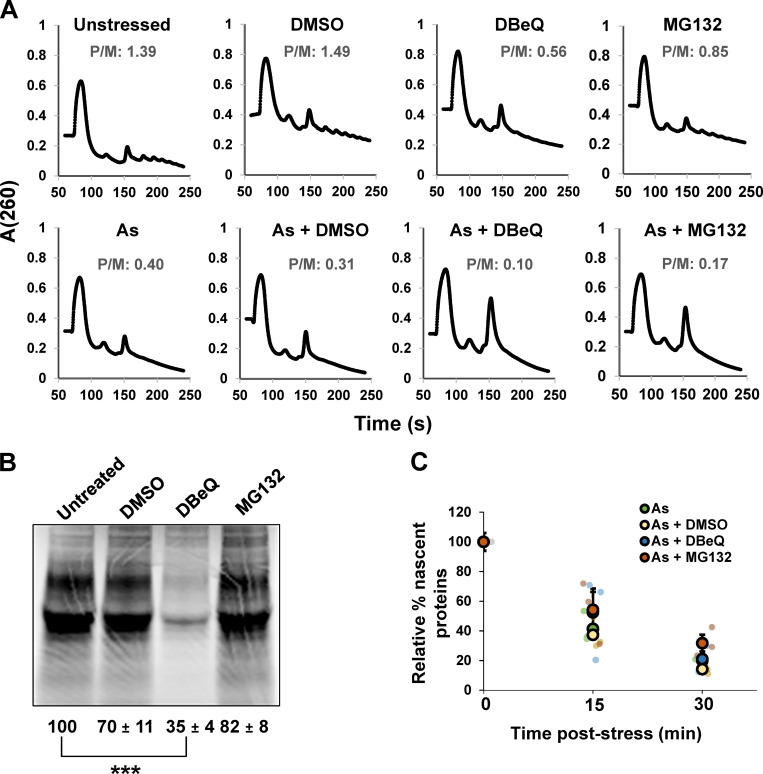
One possibility is that DBeQ and MG132 block translation repression during stress, resulting in the observed increase in nascent polypeptides associated with mRNAs. However, polysome profiling revealed polysomes collapse, and incorporation of 35S-labeled amino acids into nascent proteins showed translation is similarly suppressed during arsenite stress in the presence or absence of DBeQ or MG132 (Fig. S2). Thus, VCP and proteasome inhibition do not block global translation shutoff during arsenite stress. Moreover, since polysomes collapse, we infer that only a subfraction of mRNA molecules requires VCP and proteasome function for efficient ribosome release.
Figure S2.
Translation activity is globally suppressed during arsenite stress when VCP or the proteasome are inhibited. (A) U-2 OS cells were unstressed or stressed with 0.5 mM arsenite (As) and/or DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM) for 30 min. Cells were placed on ice and immediately pelleted and frozen at −80ºC and then lysed and polysome profiles were obtained following ultracentrifugation on sucrose gradients. Shown are representative profiles and fraction RNA in polysome:monosome (P/M) from n = two independent experiments. (B) Metabolic labeling of U-2 OS cells with 35S-Met and 35S-Cys for 30 min was done in the presence or absence of 0.1% DMSO, 10 µM DBeQ, or 10 µM MG132. Cells were lysed, and proteins were run on 4–12% gradient NuPAGE SDS-PAGE gels and exposed to phosphor screens. Total lane intensity was quantified with ImageJ. The average relative percent translation activity from n = two independent replicates is shown ± SEM. Student’s t test was done to assess significance between untreated and treated cells, with ***, P < 0.005. (C) U-2 OS cells were metabolically labeled with 35S-Met and 35S-Cys for 15 min immediately before collection following 0, 15, or 30 min under unstressed or stressed conditions (0.5 mM As) in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM). Translation activity was quantified relative to untreated, unstressed cells as described in B from n = three independent experiments. The average ± SEM is shown for each condition with individual replicates shown in green (As), yellow (As + DMSO), blue (As + DBeQ), and red (As + MG132). Exact P values and source data are provided in Table S1.
A second possibility is that those mRNAs associated with nascent chains in stressed cells treated with DBeQ or MG132 are stalled in translation elongation during arsenite stress, and ribosome run-off is inhibited. Ribosome run-off can be measured by blocking translation initiation with harringtonine (HT) and measuring the time it takes for nascent peptide chains to dissociate from the mRNA (Fresno et al., 1977; Ingolia et al., 2011). Cotreatment of cells with the translation initiation inhibitor HT revealed a significant increase in the number of mRNAs associated with nascent chains in stressed, DBeQ-treated cells compared with cells treated with HT alone (Fig. 1 D and Video 4). However, the number of translating mRNAs decreased over time in cells cotreated with arsenite, DBeQ, and HT (Fig. 1 D and Video 5), suggesting ribosomes may be slowed rather than stopped during elongation in this context, or there are additional mechanisms for stalled ribosome removal. These results argue that ribosomes pause, or are substantially slowed, on a subset of mRNA molecules during arsenite stress and require VCP and the proteasome for mRNA release.
Video 4.
Representative video showing nascent SM-KDM5B proteins (gray) in unstressed U-2 OS cells treated with HT, related to Fig. 1 D. Frame 1 was acquired pretreatment, and then 15 min later images were obtained every 10 min up to 55 min. HT was added immediately after capturing the 15-min image. Scale bar, 10 µm.
Video 5.
Representative video showing nascent SM-KDM5B proteins (gray) in arsenite-stressed U-2 OS cells treated with HT, related to Fig. 1 D. Frame 1 was acquired prestress and then arsenite was added to stress cells. 15 min later, images were acquired every 10 min for up to 55 min after arsenite addition. HT was added immediately after acquiring the image at 15 min after stress. Scale bar, 10 μm.
Inhibition of VCP or proteasome function affects mRNA partitioning into SGs
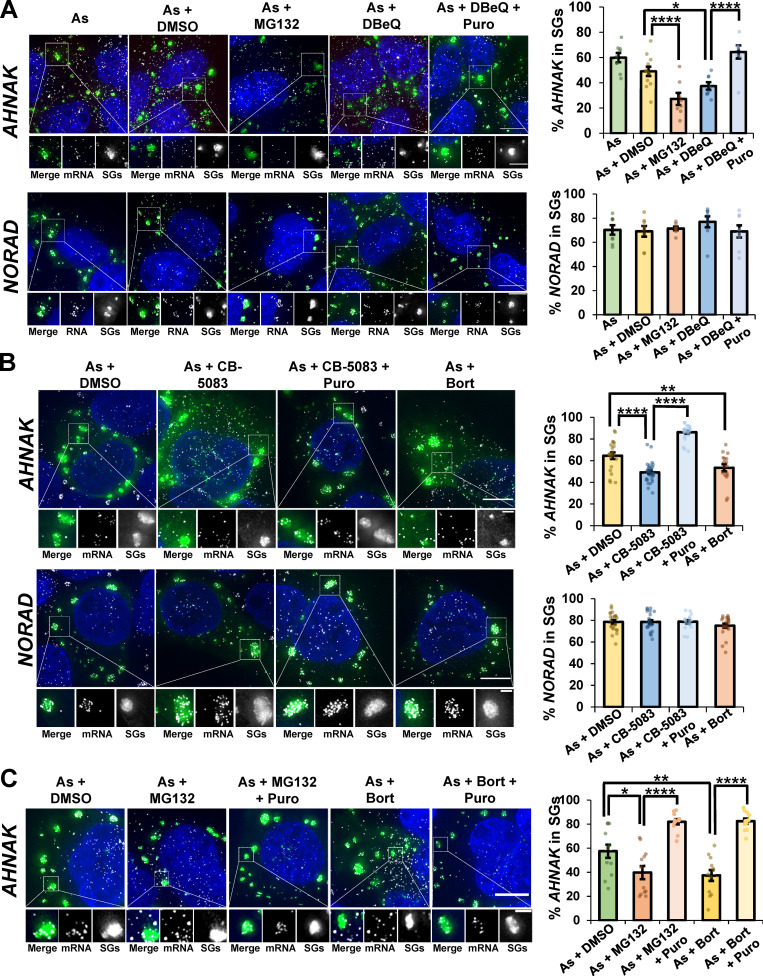
If mRNAs are trapped in stalled translation complexes during stress upon VCP or proteasome inhibition, recruitment of at least some endogenous mRNAs to SGs should be reduced by VCP or proteasome inhibition. Previous studies demonstrated that treatment of stressed cells with cycloheximide or emetine, which inhibits translation elongation and traps mRNAs in polysomes, reduces mRNA recruitment to SGs (Khong and Parker, 2018; Namkoong et al., 2018; Zurla et al., 2011). Therefore, we examined the recruitment of AHNAK mRNAs to SGs using single molecule FISH (smFISH; Khong et al., 2017) in stressed cells treated with VCP or proteasome inhibitors. Strikingly, we observed that the recruitment of AHNAK mRNAs to SGs was significantly reduced in the presence of MG132 or DBeQ during arsenite stress (Fig. 2 A).
Figure 2.
VCP and the proteasome are required for efficient release of endogenous mRNAs from ribosomes for partitioning into SGs. (A) U-2 OS cells expressing the SG marker GFP-G3BP1 were stressed with 0.5 mM sodium arsenite (As; AHNAK n = 9 cells, NORAD n = 8 cells) for 45 min with or without 0.1% DMSO (As + DMSO, AHNAK n = 12 cells, NORAD n = 8 cells), 10 µM MG132 (As + MG132, AHNAK n = 8 cells, NORAD n = 8 cells), 10 µM DBeQ (As + DBeQ, AHNAK n = 8 cells, NORAD n = 8 cells), or 10 µM DBeQ + 10 µg/ml puromycin (As + DBeQ + Puro, AHNAK n = 8 cells, NORAD n = 8 cells). (B) Cells were stressed with As and cotreated as in A except in the presence or absence of CB-5083 or bortezomib (Bort). For As + DMSO AHNAK n = 21 cells, and for NORAD n = 23 cells; for As + CB-5083 AHNAK n = 31 cells, and for NORAD n = 22 cells; for As + CB-5083 + puromycin (Puro) AHNAK n = 25 cells and for NORAD n = 17 cells; for As + Bort AHNAK n = 17 cells, and for NORAD n = 21 cells. (C) Cells were stressed with As and cotreated as in A with DMSO, MG132, or Bort in the presence or absence of puromycin (n = 12 cells for all conditions). Cells were fixed, smFISH was performed to detect the mRNA AHNAK or lncRNA NORAD, and maximum intensity projections assembled from z-stacks were imaged at 100× on a DeltaVision microscope. SGs are shown in green, RNAs in white, and nuclei in blue (DAPI). Representative photomicrographs with the average percent AHNAK or NORAD RNAs colocalizing with SGs ± SEM are shown (right). Two independent experiments were performed with individual points representing a single cell, and Student’s t test was done to assess significance, with *, P ≤ 0.05; **, P ≤ 0.01; and ****, P ≤ 0.001. Scale bars, 10 µm (whole cell), 5 µm (magnified panels in A and C), or 2 µm (magnified panels in B). Exact P values and source data are provided in Table S1.
To determine whether the reduced accumulation of mRNAs in SGs was directly due to inhibition of VCP or the proteasome, we tested additional inhibitors. Importantly, we observed a similar depletion of AHNAK mRNAs associated with SGs in arsenite-stressed cells treated with the VCP inhibitor CB-5083 or the proteasome inhibitor bortezomib (Fig. 2 B). Since multiple inhibitors of these complexes gave a similar phenotype, we interpret the reduction in AHNAK mRNAs localized to SGs as being due to loss of VCP or proteasome function.
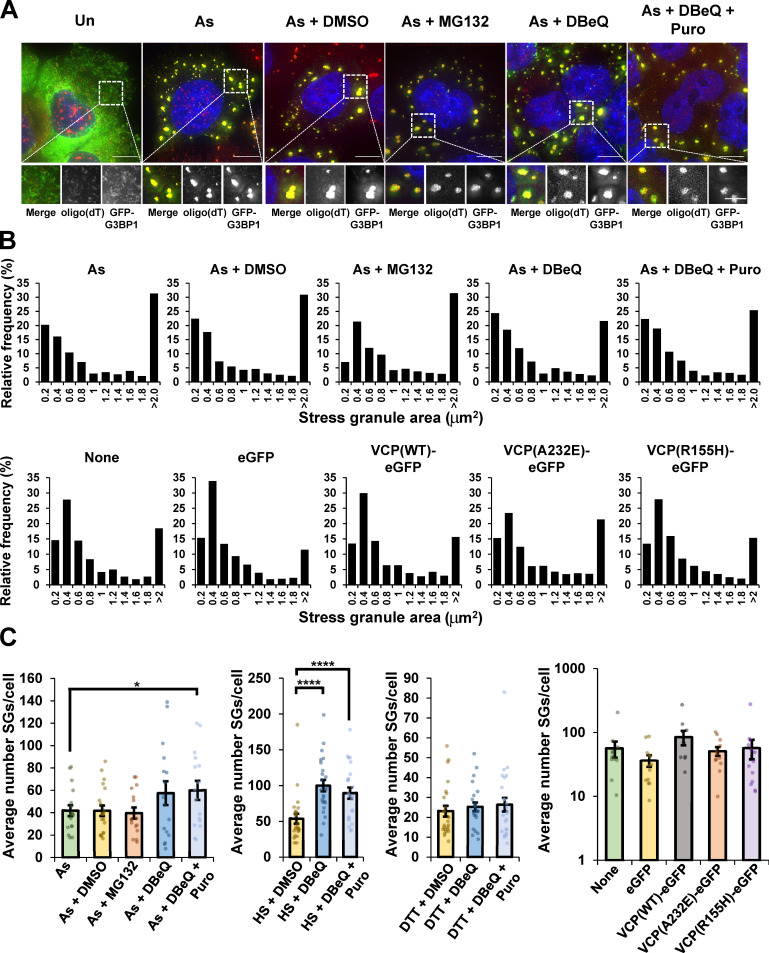
Two observations suggest that AHNAK mRNAs are limited in their recruitment to SGs in the presence of VCP or proteasome inhibitors due to ribosome association. First, treatment with puromycin, which releases ribosomes from mRNAs and increases their recruitment to SGs (Khong and Parker, 2018), restored targeting of AHNAK mRNAs to SGs in stressed cells treated with DBeQ (Fig. 2 A), CB-5083 (Fig. 2 B), MG132, or bortezomib (Fig. 2 C). Second, the accumulation of the nontranslating long noncoding RNA (lncRNA) NORAD into SGs was unaffected by VCP inhibition (Fig. 2, A and B). In addition, analyzed cells had similar size distributions of SGs regardless of treatment conditions (Fig. S3), and while there was a small increase in SG number upon DBeQ treatment in the presence or absence of puromycin during arsenite stress, no change in SG number was observed with MG132 and arsenite cotreatment (Fig. S3). Therefore, changes in SG properties due to VCP or proteasome inhibition are unlikely to underlie the observed differences in AHNAK recruitment to SGs.
Figure S3.
SGs contain polyadenylated mRNAs, and SG area and number are not substantially altered when VCP or the proteasome are inhibited, or when pathogenic VCP alleles are expressed during arsenite stress. (A) U-2 OS cells stably expressing the SG marker protein GFP-G3BP1 (green) were stressed for 45 min with 0.5 mM sodium arsenite (As) in the presence or absence of DMSO (0.1%), MG132 (10 µM), DBeQ (10 µM), or DBeQ (10 µM) and puromycin (Puro; 10 µg/ml), and then FISH was performed with oligo(dT)-Cy3 probes to detect polyadenylated transcripts (red). Nuclei are stained with DAPI (blue); these cells were also assessed for AHNAK mRNAs by smFISH (Fig. 2 A). Shown are representative maximum intensity projections of 25 z-stacks acquired at 100× on a DeltaVision microscope. Scale bars, 10 µm (whole cell) or 5 µm (magnified panels). Results represent n = two independent experiments. (B and C) Cells were treated as described in A, and the relative distribution of SG areas (B) and the number of SGs per cell (C) were determined using ImageJ/Fiji in the images used to quantify the fraction of AHNAK mRNAs (B) and both AHNAK and NORAD RNAs (C) colocalized with SGs (Fig. 2 and Fig. 3). For cells expressing VCP alleles (Fig. 7), results represent n = three independent experiments with average number of SGs per cell ± SEM shown in C. Student’s t test was done to assess significance, with *, P < 0.05 and ****, P < 0.001. Exact P values and source data are provided in Table S1.
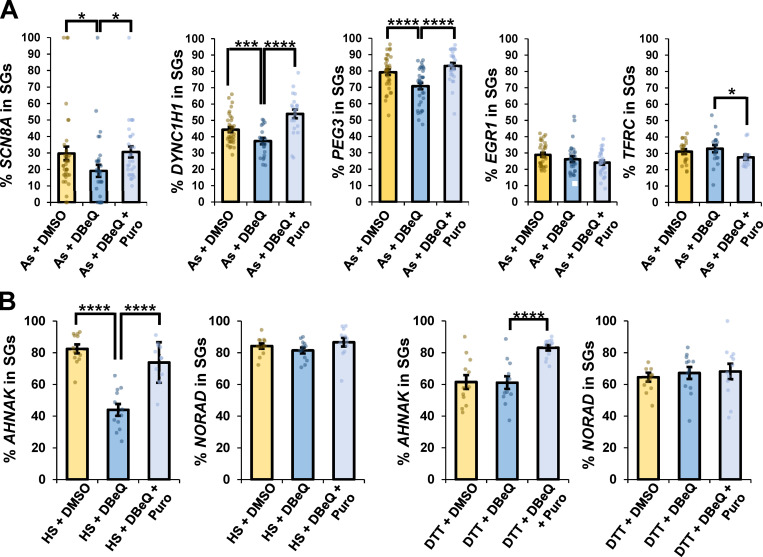
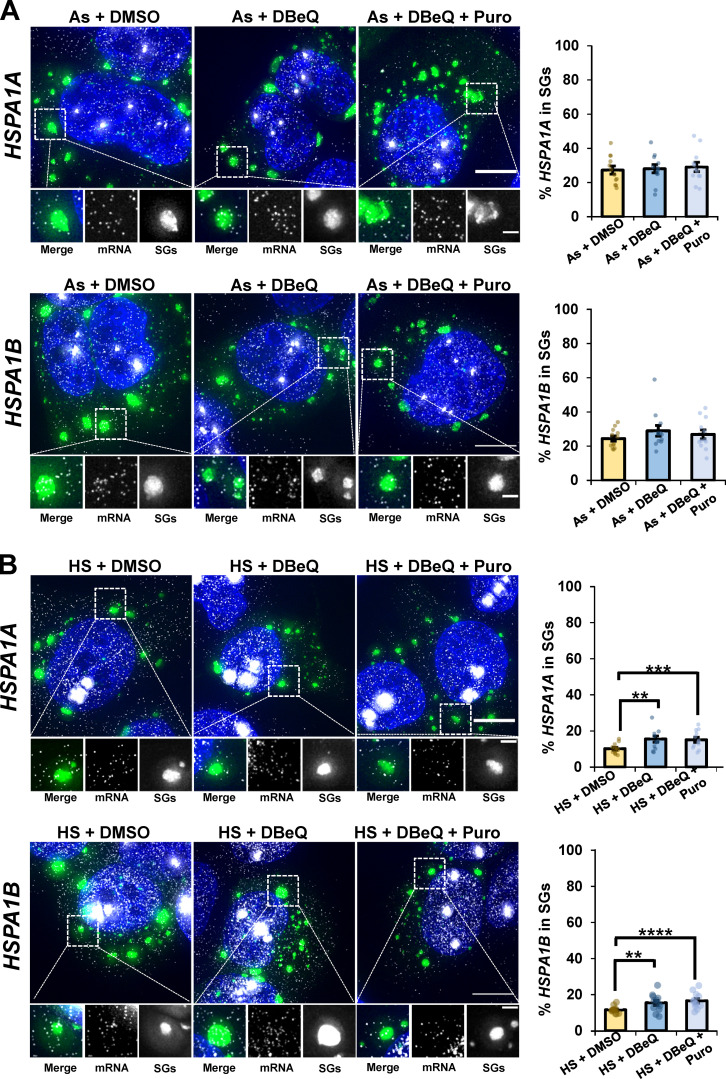
Three other mRNAs, SCN8A, DYNC1H1, and PEG3, were significantly depleted from SGs in the presence of DBeQ, by ∼35, ∼16, and ∼11%, respectively, and their localization to SGs was restored with puromycin cotreatment (Fig. 3 A). However, not all mRNAs were depleted from SGs upon VCP inhibition, as we observed EGR1 and TFRC were not depleted from SGs upon VCP inhibition (Fig. 3 A). Furthermore, the mRNAs of stress-induced genes HSPA1A and HSPA1B were not depleted from SGs upon VCP inhibition (Fig. S4). These observations argue ribosomes stall in translation during arsenite stress on some, but not all, mRNAs, and those mRNAs require VCP and proteasome activity for efficient ribosome release and targeting to SGs. It should be noted that since some mRNA molecules of affected transcripts still accumulate in SGs, we anticipate that not every mRNA molecule from a given gene undergoes ribosome pausing, which might be affected by stochastic events on a given mRNA.
Figure 3.
VCP inhibition affects the targeting of specific transcripts to SGs during arsenite stress, and VCP is required for complete AHNAK localization to SGs upon heat stress, but not ER stress. (A) U-2 OS cells were stressed with arsenite (As; 0.5 mM) in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or DBeQ (10 µM) plus puromycin (Puro; 10 µg/ml) for 45 min. Cells were fixed, smFISH was performed, and cells were imaged at 100× on a DeltaVision microscope. The average percent SCN8A (As + DMSO n = 36 cells; As + DBeQ n = 32 cells; As + DBeQ + Puro n = 30 cells), DYNC1H1 (As + DMSO n = 34 cells; As + DBeQ n = 23 cells; As + DBeQ + Puro n = 22 cells), PEG3 (As + DMSO n = 34 cells; As + DBeQ n = 34 cells; As + DBeQ + Puro n = 28 cells), EGR1 (As + DMSO n = 33 cells; As + DBeQ n = 28 cells; As + DBeQ + Puro n = 32 cells), and TFRC (As + DMSO n = 22 cells; As + DBeQ n = 19 cells; As + DBeQ + Puro n = 15 cells) mRNAs in SGs ± SEM are shown, with *, P ≤ 0.05; ***, P ≤ 0.005; and ****, P ≤ 0.001. (B) U-2 OS cells were subjected to heat stress (HS; 42°C for 45 min) or DTT (2 mM for 45 min) and processed as in A. For all DTT and HS conditions, AHNAK and NORAD n = 12 cells. The average percent AHNAK or NORAD RNAs in SGs ± SEM from maximum intensity projections of 25 z-stacks is shown, with individual points representing a single cell from two independent replicates. Student’s t test was done to assess significance, with ****, P ≤ 0.001. Exact P values and source data are provided in Table S1.
Figure S4.
Stress-induced HSPA1A and HSPA1B mRNAs are not dependent on VCP to localize to SGs. (A and B) U-2 OS cells stably expressing the SG marker G3BP1-GFP were stressed for 45 min with arsenite (As; 0.5 mM; A) or heat stress (HS; 42°C; B) in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or DBeQ (10 µM) plus puromycin (Puro; 10 µg/ml). Cells were fixed, and smFISH was performed to detect HSPA1A (top panels) and HSPA1B (bottom panels), and samples were imaged at 100× on a DeltaVision microscope. Representative maximum intensity projections of 25 z-stacks are shown at left with G3BP1-GFP in green, mRNAs in white, and nuclei in blue. Scale bars, 10 µm (whole cell) and 2 µm (magnified panels). The average ± SEM from two independent experiments is shown at right with individual data points shown for single cells (n = 12 cells for every condition). Student’s t test was performed to assess significance, with **, P < 0.01; ***, P < 0.005; and ****, P < 0.001. Exact P values and the source data are provided in Table S1.
To address whether VCP impacts mRNA recruitment to SGs during other stresses, we assessed the localization of AHNAK and NORAD RNAs to SGs upon heat stress and ER stress. The recruitment of AHNAK to SGs was reduced by ∼47% in cells exposed to heat stress in the presence of DBeQ and restored by puromycin cotreatment, while NORAD was unaffected (Fig. 3 B). However, AHNAK recruitment to SGs is not dependent on VCP under all stresses. For example, under a stress response induced by DTT, which triggers ER stress, DBeQ treatment did not result in a significant reduction in AHNAK recruitment to SGs (Fig. 3 B). Therefore, the VCP-dependent release of mRNAs from ribosomes for partitioning into SGs occurs in specific stress conditions, including arsenite and heat stress, but not ER stress caused by DTT.
Members of the RQC pathway also affect mRNA partitioning into SGs
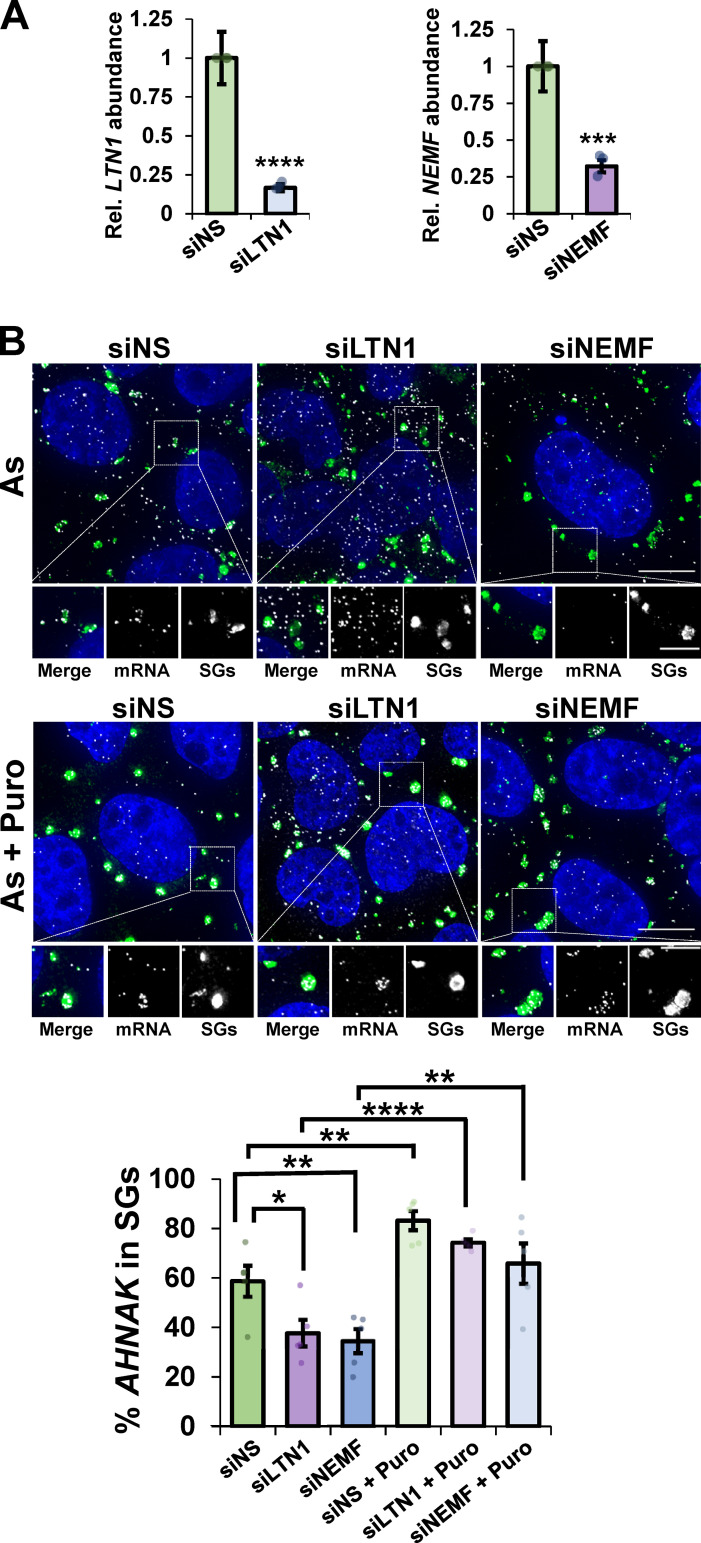
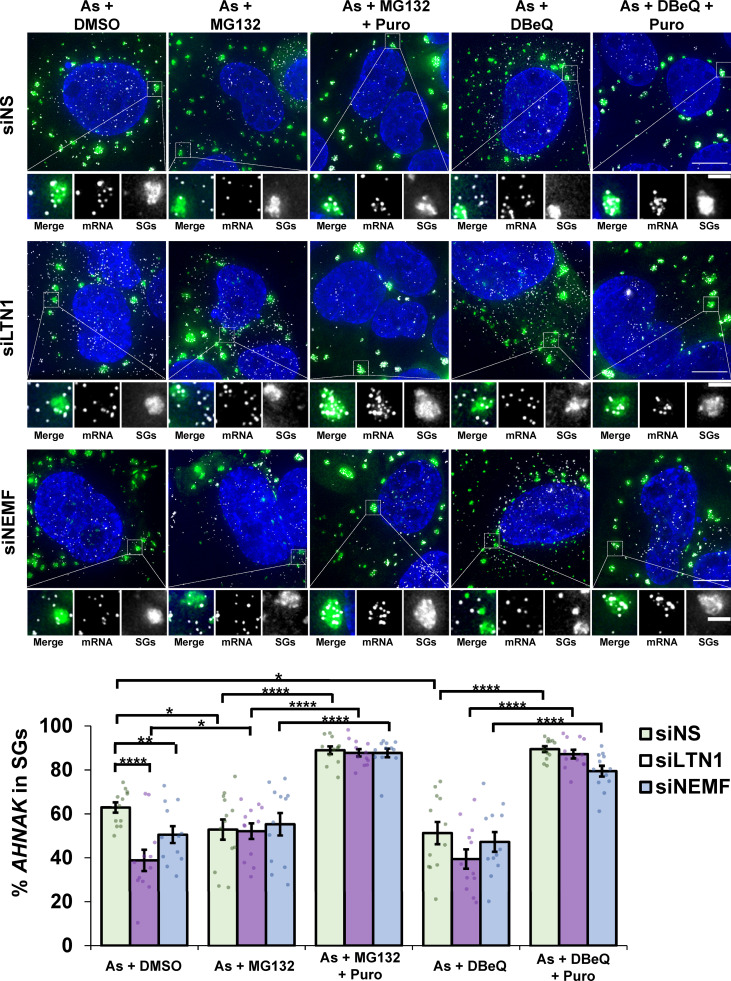
VCP is known to function in the RQC pathway where it is thought to extract ubiquitinated nascent chains from free 60S subunits after ribosome splitting (Brandman et al., 2012; Shao et al., 2015). To determine whether ribosome release and targeting of mRNAs to SGs involved other components of the RQC pathway, we examined whether LTN1, which ubiquitinates aberrant nascent chains, and its cofactor NEMF (Brandman et al., 2012; Shao et al., 2015; Verma et al., 2013) affected mRNA targeting to SGs. We observed siRNA depletion of LTN1 or NEMF (Fig. 4 A) reduced AHNAK recruitment to SGs, which was restored by puromycin cotreatment (Fig. 4 B). Taken together, the observations that LTN1 and NEMF depletion reduces AHNAK partitioning into SGs and puromycin cotreatment restores AHNAK in SGs demonstrate that components of the RQC pathway also influence the targeting of mRNAs to SGs.
Figure 4.
Canonical RQC factors LTN1 and NEMF are required for efficient mRNA partitioning into SGs. U-2 OS cells were transfected with nonspecific siRNAs (siNS), siRNAs to LTN1 (siLTN1), or siRNAs to NEMF (siNEMF). (A) The levels of LTN1 and NEMF relative to GAPDH were detected by RT-qPCR (n = three independent experiments) with average ± SEM shown. Student’s t test was done to assess significance, with ***, P ≤ 0.005 and ****, P ≤ 0.001. (B) Cells were stressed for 45 min with arsenite (As; 0.5 mM) in the presence (As + puromycin [Puro], bottom panels) or absence (As, top panels) of Puro (10 µg/ml) with n = five frames counted in all conditions (42–57 total cells per condition). IMF staining to detect G3BP (green) and smFISH to detect AHNAK mRNA (white) was performed, and nuclei were visualized with DAPI (blue). Top: Representative photomicrographs. Scale bars, 10 µm (whole cell) or 5 µm (magnified panels). Bottom: The average percent AHNAK mRNA in SGs ± SEM is shown with individual points representing a single frame. Student’s t test was done to evaluate significance between siNS and siLTN1 or siNEMF conditions and between siRNA treatments or siRNA + Puro treatments, with *, P ≤ 0.05; **, P ≤ 0.01; and ****, P ≤ 0.001. Exact P values and source data are provided in Table S1.
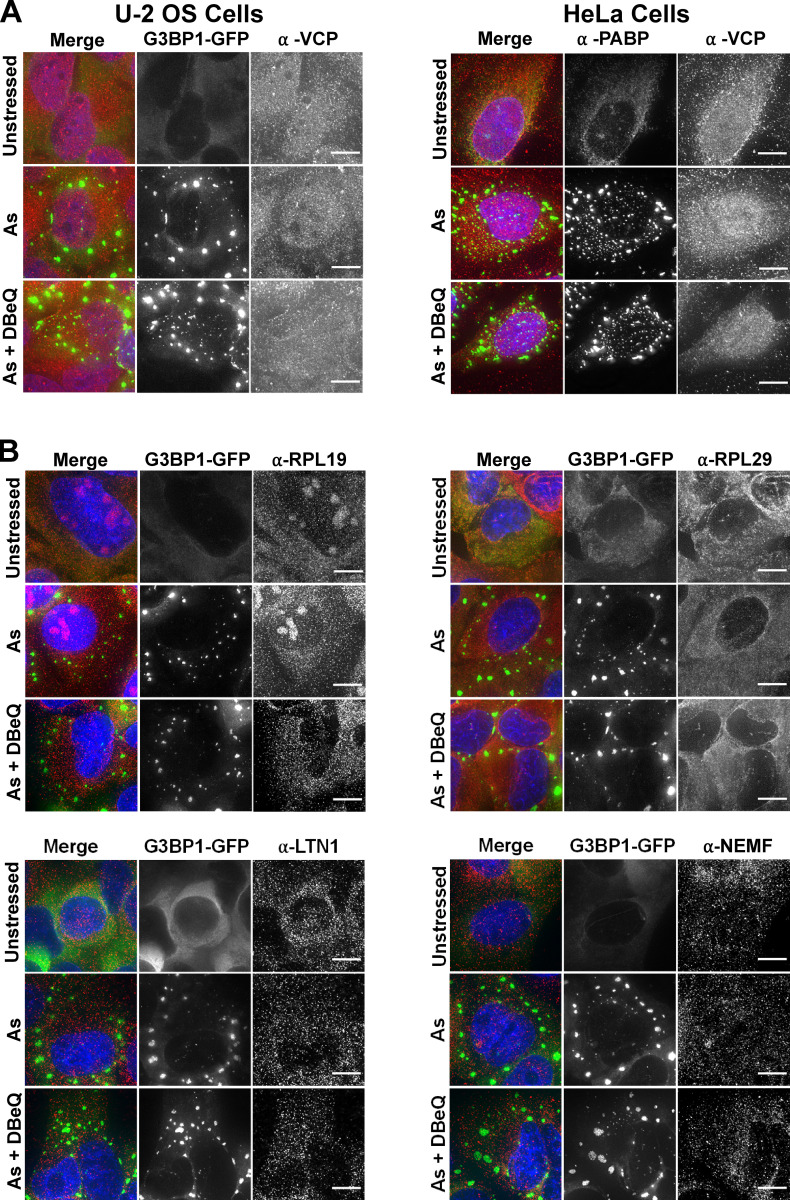
VCP, LTN1, and NEMF were not enriched in SGs and were diffusely localized in the cytoplasm with two ribosomal proteins (RPL19 and RPL29) 45 min following arsenite stress in the presence or absence of DBeQ (Fig. S5). This argues for a function of these RQC factors outside of SGs. The observation that VCP did not partition into SGs was in contrast to prior studies (Buchan et al., 2013; Turakhiya et al., 2018); however, it is possible that VCP is only recruited to SGs during late-stage stress or recovery from stress. In sum, these results point to a new function of RQC factors in releasing mRNAs stalled in translation during acute stress before their localization to SGs.
Figure S5.
VCP, LTN1, NEMF, and ribosomal proteins Rpl19 and Rpl29 are not enriched in SGs 45 min after arsenite stress. (A) U-2 OS cells stably transfected with the SG marker GFP-G3BP1 (green; left) or HeLa cells (right) were unstressed or stressed for 45 min with 0.5 mM sodium arsenite (As) in the presence or absence of DBeQ (10 µM). Cells were fixed, and IMF staining was performed to detect endogenous VCP (red); SGs in HeLa cells were visualized with anti–poly(A)-binding protein (PABP; green). Results represent n = three independent experiments. (B) U-2 OS cells expressing GFP-G3BP1 (green) were treated as described in A, and IMF staining was done to detect Rpl29, Rpl19, LTN1, or NEMF (red). Nuclei are shown in blue (DAPI). Cells were imaged at 100× with a DeltaVision elite microscope, and representative maximum intensity projections of 25 z-stacks are shown with 10-µm scale bars. Results represent n = two independent experiments.
The effect of VCP on mRNA partitioning into SGs is through an RQC pathway
In principle, the effect of VCP on mRNA partitioning into SGs could be through the RQC pathway, where VCP is known to play a role (Brandman et al., 2012; Ye et al., 2017), or through a parallel pathway. If VCP and RQC factors affect independent pathways, we would expect to observe an additive reduction in AHNAK partitioning to SGs when inhibiting or depleting two factors at once. Conversely, if VCP and RQC factors work in the same pathway, we would expect simultaneous inhibition of both RQC factors and VCP should give a similar phenotype as single depletions or inhibitions. Given this, we examined the recruitment of AHNAK mRNA to SGs using a combination of siRNA depletion and chemical inhibition treatments.
We observed that when LTN1 or NEMF is knocked down, treatment with DBeQ or MG132 to inhibit VCP or the proteasome did not lead to a further reduction in AHNAK recruitment to SGs during arsenite stress (Fig. 5). Puromycin rescued AHNAK localization to SGs in all cases, again supporting the interpretation that LTN1, NEMF, VCP, and the proteasome aid mRNA release from ribosomes during arsenite stress. These results argue that LTN1, NEMF, VCP, and the proteasome are all functioning as part of the RQC pathway to mediate mRNA recruitment to SGs.
Figure 5.
Treatment of cells depleted of LTN1 or NEMF with VCP or proteasome inhibitors does not further deplete AHNAK mRNAs from SGs during arsenite stress. U-2 OS cells expressing the SG marker G3BP1-GFP were depleted of LTN1 (siLTN1) or NEMF (siNEMF) using siRNAs or transfected with nonspecific control siRNAs (siNS). Cells were stressed with 0.5 mM arsenite (As) for 45 min with 0.1% DMSO (As + DMSO), 10 µM MG132 (As + MG132), 10 µM MG132 with 10 µg/ml puromycin (As + MG132 + Puro), 10 µM DBeQ (As + DBeQ), or 10 µM DBeQ with 10 µg/ml Puro (As + DBeQ + Puro), fixed, and smFISH for AHNAK mRNA was performed. Samples were imaged at 100× on a DeltaVision microscope. Representative image maximum intensity projections of 25 z-stacks are presented (top), with scale bars representing 10 µm (whole cells) or 2 µm (magnified panels), with G3BP1-GFP in green, AHNAK mRNAs in white, and nuclei in blue. The percent AHNAK mRNAs that colocalized with SGs was determined in individual cells, and the average ± SEM is shown (bottom) from two independent experiments. For siNS, As + DMSO n = 12 cells, As + MG132 n = 13 cells, As + MG132 + Puro n = 12 cells, As + DBeQ n = 12 cells, As + DBeQ + Puro n = 12 cells; for siLTN1, n = 12 cells for all conditions; for siNEMF, n = 12 cells for all conditions. Student’s t tests were done to assess significance, with *, P ≤ 0.05; **, P ≤ 0.01; and ****, P ≤ 0.001. Exact P values and source data are provided in Table S1.
RQC activity during stress is different from canonical RQC pathways
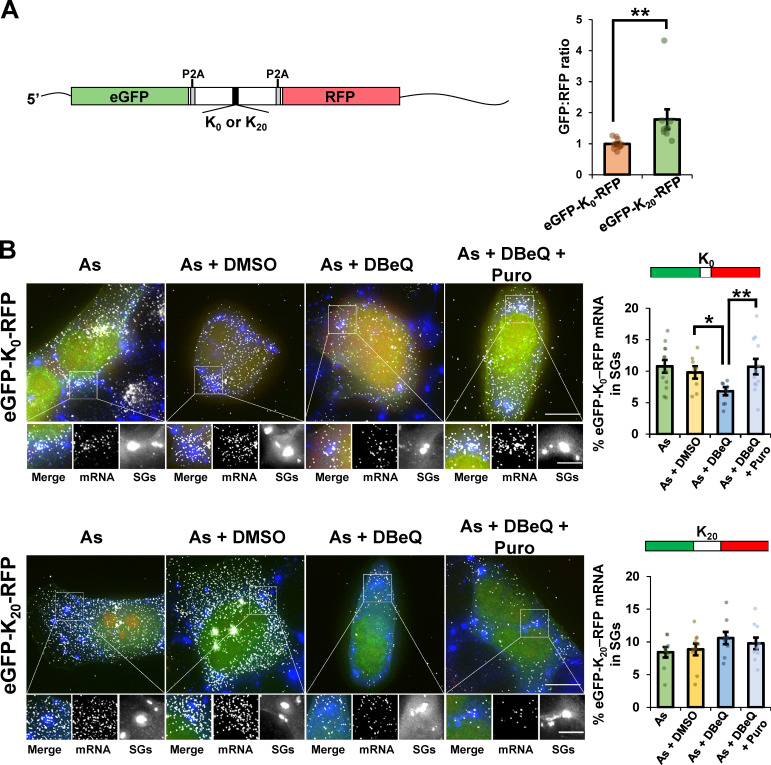
The observation that RQC factors would be required for the release of nascent chains from the mRNA and targeting the mRNA to SGs was surprising since RQC proteins are thought to only interact with the 60S subunit after ribosome dissociation and release from mRNAs with strong ribosomes stall sites (Brandman and Hegde, 2016; Shao et al., 2015). Given this difference, we examined whether the effect of VCP on mRNA targeting to SGs was influenced by the same translation perturbations that trigger a canonical RQC response. For this experiment, we assessed the localization of a dual-fluorescence stalling reporter mRNA with EGFP encoded upstream of RFP, with or without a stalling poly(A) tract, which triggers canonical RQC, inserted between them (Fig. 6 A, left; Juszkiewicz and Hegde, 2017). The EGFP:RFP signal intensity was higher (1.8 ± 0.3) in cells expressing the stalling reporter than in the control reporter construct (1.0 ± 0.04; Fig. 6 A, right), confirming lower ribosome readthrough past the poly(A) sequence.
Figure 6.
A canonical RQC substrate reporter mRNA does not depend on VCP activity for localization to SGs. Reporter mRNAs encoding eGFP and RFP separated by a linker region that encodes 20 lysine residues in a 60-nt poly(A) tract (K20), or no lysines (K0; from Juszkiewicz and Hegde, 2017) were expressed in U-2 OS cells. Cells were stressed for 45 min with 0.5 mM arsenite (As) in the absence or presence of 0.1% DMSO (As + DMSO), 10 µM DBeQ (As + DBeQ), or 10 µM DBeQ and 10 µg/ml puromycin (As + DBeQ + Puro). Cells were fixed and G3BP detected by IMF staining (blue) to mark SGs. The reporter mRNA was detected by smFISH using probes against the eGFP ORF (white); eGFP is shown in green and RFP in red. Cells expressing eGFP were imaged at 100× using a DeltaVision microscope, and maximum intensity projections of 25 z-stacks are shown. (A) Left: Diagram of reporter mRNAs. Right: Quantification of the relative eGFP and RFP intensities in individual cells (shown as dots) was performed from cells treated with As, with the average ± SEM reported from two independent experiments (K0 n = 12 cells; K20 n = 9 cells). (B) Representative images are shown at left with average ± SEM of the percent eGFP mRNA that colocalized with SGs shown at right from two independent experiments. Scale bars, 10 µm (whole cell) or 5 µm (magnified panels). Individual points represent a single frame with one to five cells counted per frame. For K0, As n = 12 frames; As + DMSO n = 8 frames; As + DBeQ n = 8 frames; As + DBeQ + Puro n = 12 frames; for K20, As n = 9 frames; As + DMSO n = 10 frames; As + DBeQ n = 9 frames; As + DBeQ + Puro n = 10 frames. Student’s t test was done to assess significance, with *, P ≤ 0.05 and **, P ≤ 0.01. Exact P values and source data are provided in Table S1.
A key observation was that while a control reporter transcript was depleted from SGs upon VCP inhibition, the partitioning of the stalling reporter mRNA into SGs was unaffected by VCP inhibition (Fig. 6 B). Therefore, the fate of typical mRNAs stalled in translation during arsenite stress is different from that of canonical RQC substrate mRNAs. This highlights a difference between saRQC complex and RQC pathways (see Discussion).
mRNA partitioning into SGs is increased by pathogenic VCP alleles
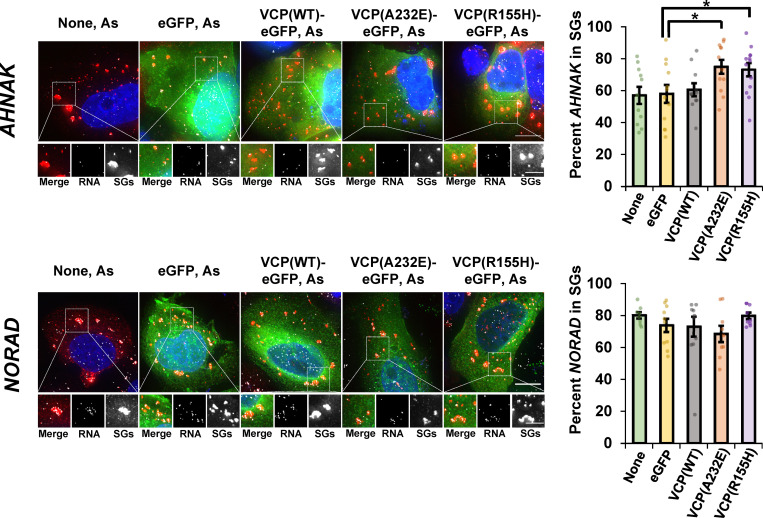
The results mentioned above show that inhibition of VCP function leads to a decrease in some mRNAs accumulating in SGs. Interestingly, there are a series of pathogenic VCP alleles that cause neurodegeneration and inclusion body myopathies (Johnson et al., 2010; Watts et al., 2004). These are thought to be hypermorphic alleles, at least in some cases, as the VCP-R155H and VCP-A232E alleles have increased ubiquitin segregase activity (Zhang et al., 2015, 2017), and to act in a dominant manner in patients (Johnson et al., 2010; Watts et al., 2004). Given that we had identified a new function of VCP in affecting mRNA partitioning into SG, we tested how this process was affected by pathogenic VCP alleles by examining AHNAK localization in cells expressing WT or EGFP-tagged pathogenic VCP variants (Zhang et al., 2015; Tresse et al., 2010; Halawani et al., 2009; Manno et al., 2010; Niwa et al., 2012). The common VCP-R155H variant affects the N-terminal domain that mediates ubiquitin and cofactor binding, and the severe A232E variant alters the N-terminal AAA-ATPase domain D1 that mediates homo-hexamerization of VCP and catalysis (Watts et al., 2004; Wang et al., 2003).
We observed that both pathogenic VCP variants, but not the WT protein, caused increased recruitment of AHNAK mRNAs to SGs (Fig. 7). On average, the VCP-R155H allele caused a 26% increase and the VCP-A232E allele caused a 29% increase in AHNAK localization to SGs (Fig. 7). Partitioning of NORAD into SGs (Fig. 7) and the size and number of SGs per cell were not altered (Fig. S3), suggesting global changes in SG properties did not account for the increased AHNAK localization to SGs. It was previously shown that exogenous expression of pathogenic VCP alleles caused constitutive SGs in a subset of unstressed cells (Buchan et al., 2013) and delayed SG disassembly following heat stress (Wang et al., 2019). Therefore, pathogenic VCP variants can increase the targeting of some mRNAs to SGs, which would alter the SG transcriptome, and may contribute to the formation of aberrant or constitutive SGs in disease contexts.
Figure 7.
Pathogenic VCP alleles increase recruitment of AHNAK mRNAs, but not NORAD lncRNAs, to SGs. U-2 OS cells stably expressing the SG marker mRuby2-G3BP1 were either untransfected (None) or transiently transfected with empty vector peGFP-N1 (eGFP), WT VCP fused to eGFP (VCP(WT)-eGFP), or VCP alleles (VCP(A232E)-eGFP and VCP(R155H)-eGFP). Cells were stressed for 45 min with 0.5 mM arsenite (As) and then fixed, and smFISH was performed to detect AHNAK mRNA (top) or NORAD lncRNA (bottom). Representative photomicrographs for are shown at left, with RNAs shown in white, SGs in red, and eGFP in green. Images were acquired on a DeltaVision microscope at 100×, and maximum intensity projections of 25 z-stacks are shown. Scale bars, 10 µm (whole cell) or 5 µm (magnified panels). At right, average percent AHNAK mRNA or NORAD lncRNA ± SEM that colocalize with SGs. Three independent experiments were done with individual dots representing one frame, and only cells expressing eGFP were counted for all transfected conditions. For AHNAK, None n = 11 frames, GFP n = 12 frames, VCP(WT) n = 11 frames, VCP(A232E) n = 12 frames, VCP(R155H) n = 13 frames; for NORAD, None n = 8 frames, GFP n = 10 frames, VCP(WT) n = 10 frames, VCP(A232E) n = 9 frames, VCP(R155H) n = 9 frames. Student’s t test was done to assess significance between GFP and VCP(WT), VCP(A232E), or VCP(R155H) for AHNAK and NORAD, with *, P ≤ 0.05. Exact P values and source data are provided in Table S1.
Discussion
In this work, we present evidence that the machinery involved in the RQC pathway, including the VCP complex and the proteasome, affects the partitioning of mRNAs into SGs by mediating ribosome release from mRNAs. This is based on the observation that inhibiting VCP, the proteasome, LTN1, or NEMF leads to a reduced level of accumulation of some mRNAs in SGs. This affect is related to ribosome release since mRNAs are restored in SGs following puromycin treatment, which releases ribosomes from mRNAs, and the lncRNA NORAD, which also accumulates in SGs, is not affected by inhibition of VCP or the proteasome. Moreover, we observe the persistence of nascent peptide chains associated with mRNAs, directly demonstrating the persistence of elongating ribosomes associated with mRNAs when VCP or the proteasome is inhibited. This study demonstrates that ribosomes stall on mRNAs during translation in at least some acute stress conditions and are released, at least in part, by a noncanonical saRQC for partitioning into SGs.
The saRQC differs from canonical RQC in two manners. First, when VCP or the proteasome is inhibited during arsenite treatment, we observed the persistence of nascent peptide chains associated with mRNAs, suggesting the continued presence of an 80S subunit on the mRNA. This is distinct from the current model of RQC wherein 80S ribosomes are split before nascent peptide release and decay from the 60S subunit, with the mRNA undergoing endonucleolytic cleavage (Brandman and Hegde, 2016; Joazeiro, 2017; Shao et al., 2015). However, prior studies demonstrated that depletion of ltn1 or cdc48 causes nascent polypeptide chains encoded by mRNAs lacking stop codons to accumulate in 80S ribosome fractions, which suggests in certain contexts LTN1 and VCP can interact with intact ribosomes (Bengtson and Joazeiro, 2010; Verma et al., 2013). Second, saRQC and RQC act on different mRNA substrates since a traditional RQC substrate with a poly(A) tract in the coding region was not altered in its SG partitioning by VCP inhibition, while a control reporter mRNA without a ribosome stall site was affected. This observation is consistent with the interpretation that ribosome splitting and endonucleolytic cleavage of mRNAs with strong stall sites occur before VCP activity on the nascent polypeptide chain and 60S ribosome (Shao et al., 2013, 2015; Verma et al., 2013). In sum, these experiments demonstrate the saRQC shares components of the canonical RQC pathway but diverges in molecular mechanism, mRNA substrates, and mRNA fate.
An interesting question is the nature of the perturbation that leads to delays in translation elongation during arsenite or heat stress in mammalian cells. One possibility, based on ribosomal profiling during heat stress or proteotoxic stress, is that defects in the folding of the nascent protein trigger a feedback inhibition of translation elongation (Liu et al., 2013; Shalgi et al., 2013). Ribosomes could stall due to aberrant cotranslational protein processing, such as that observed in cases of ZNF598 (Hel2)-dependent ribosome stalling where aberrant nascent proteins limit translation elongation by blocking incoming aminoacyl-tRNAs (Chandrasekaran et al., 2019).
Importantly, the saRQC would likely be distinct from ribosome stalling at internal poly(A) tracts, which triggers RQC in a ZNF598 (Hel2)-dependent mechanism (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017; Matsuo et al., 2017) for two reasons. First, we observe the saRQC under conditions where translation initiation is globally suppressed, which would limit the likelihood of ribosome collisions, and limits ZNF598-mediated RQC (Juszkiewicz et al., 2018). Second, mRNAs targeted by the ZNF598-mediated RQC pathway undergo endonucleolytic cleavage and are rapidly degraded in the no-go decay pathway (Doma and Parker, 2006; D’Orazio et al., 2019; Simms et al., 2017). Because mRNAs released from translation are partitioned into SGs during acute stress, the saRQC pathway appears to be distinct from the ZNF598-mediated RQC pathway.
A second possibility, since chemical damage to mRNAs such as oxidation can cause elongation pauses (Simms et al., 2014; Yan et al., 2019), is that damage to the mRNA causes elongation arrest and triggers a requirement for the RQC complex to allow ribosome release. Interestingly, work in yeast has shown that translation elongation can be inhibited during oxidative stress induced by hydrogen peroxide (Gerashchenko et al., 2012; Wu et al., 2019). We suggest the possibility that translation elongation may be altered in numerous stress conditions but would generally not be observed in WT or untreated cells due to the action of the RQC machinery.
We suggest two possible, potentially overlapping, mechanisms that could explain how VCP, the proteasome, and RQC factors might affect 80S ribosome release and/or mRNA recruitment to SGs. In the first model, these factors would have previously unrecognized roles in releasing a stalled 80S subunit from mRNAs. For example, VCP, perhaps in a complex with the proteasome (Isakov and Stanhill, 2011), might be involved in extracting ubiquinated eS10 from stalled ribosomes (Juszkiewicz and Hegde, 2017), thereby promoting ribosome release from mRNAs. In the simplest sense, this model would require LTN1 and NEMF to have new roles since they are currently thought to only interact with the 60S subunit after 80S splitting and release from the mRNA (Brandman and Hegde, 2016; Joazeiro, 2017; Shao et al., 2015). While this requires new roles, we note that often proteins have multiple related functions. For example, the Ski2–Ski3–Ski8 complex was recently shown to be able to extract and degrade mRNAs associated with stalled 80S subunits without disruption of the 80S complex (Zinoviev et al., 2020; Schmidt et al., 2016).
An alternative model is that the observed effects on mRNA release from 80S ribosomes are due to a titration of RQC components during a stress response, perhaps because of a large number of stalled 80S subunits. For example, components required for 80S release, such as Hbs1 and Dom34/Pelota (Pisareva et al., 2011; Guydosh and Green, 2014; Shoemaker et al., 2010), might become limiting due to their sequestration in intermediates of the RQC pathway, thus creating a negative feedback loop that limits 80S subunit release from mRNA. However, since these ribosome rescue factors are thought to be released from the ribosome during ribosome dissociation, it is unclear how such a mechanism would occur. This feedback mechanism would also fail to explain why the saRQC mechanism appears to affect “normal” mRNAs and not an RQC substrate with an internal poly(A) tract. Defining the molecular mechanism of the saRQC pathway will be an important goal of future work.
Another important insight of this work is the demonstration that RQC components and VCP can affect mRNA accumulation in SGs. This would be in addition to other roles of VCP affecting SG clearance and/or disassembly (Buchan et al., 2013; Seguin et al., 2014; Rodriguez-Ortiz et al., 2016; Turakhiya et al., 2018; Wang et al., 2019). One implication of these results is that SGs could serve as storage depots for mRNAs that are modified in response to stress and undergo processing by the saRQC. A second implication is that defects in saRQC factors might contribute to degenerative disease states through perturbed release of mRNAs from stalled ribosomes to enable a rapid, reversible response to cellular stress. We propose that in addition to VCP playing a potential role in mediating SG disassembly via granulophagy (Buchan et al., 2013) and/or proteasome-mediated degradation of SG components (Turakhiya et al., 2018), altered RNA recruitment to SGs upon VCP dysfunction could also contribute to SG dysregulation.
An additional implication of this work is that defects in the removal of mRNAs from stalled elongation complexes could contribute to degenerative disease. Several recent studies support the notion that aberrant translation elongation or termination can contribute to pathogenesis (reviewed in Kapur et al., 2017). Furthermore, aberrant cellular responses to stress, such as a chronic stress response, are implicated in a wide range of neurological disorders (Kapur et al., 2017; Moon et al., 2018; Tahmasebi et al., 2018). The results of this study suggest that one detrimental consequence of pathogenic alleles of VCP would be increased mRNA targeting to SGs, which could alter SG properties and the cellular response to stress. Pathogenic VCP alleles cause inclusion body myopathies, Paget’s disease of the bone, Charcot–Marie–Tooth type 2 disease, amyotrophic lateral sclerosis, and frontotemporal dementia (de Bot et al., 2012; Gonzalez et al., 2014; Haubenberger et al., 2005; Johnson et al., 2010; Watts et al., 2004), and genetic perturbation of LTN1 is associated with an amyotrophic lateral sclerosis–like phenotype in mice (Chu et al., 2009). Because these disorders are associated with a wide degree of phenotypic variability, it is possible that a defect in rapid mRNA recruitment to SGs promoted by the saRQC, and later disassembly of SG, could contribute to unique yet related disease states partially dependent on environmental factors or contexts. Future studies will aim to determine the consequences of aberrant VCP function in the saRQC pathway on SG properties and function.
Materials and methods
Cell culture and treatment conditions
U-2 OS and HeLa cells were grown in DMEM with 10% FBS and 1% streptomycin/penicillin at 37°C under 5% CO2. U-2 OS cells stably expressing EGFP-G3BP1 and mRFP-DCP1a were a kind gift of Dr. N. Kedersha (Division of Rheumatology, Inflammation, and Immunity, Brigham and Women's Hospital and Harvard Medical School, Boston, MA; Kedersha et al., 2008) and were selected by flow cytometry in the BioFrontiers Institute Flow Cytometry Core facility. U-2 OS cells stably expressing mRuby2-G3BP1 (described below) were created by G418 selection and clonal expansion after plasmid transfection. HeLa cells with endogenously tagged SunTagX56-POLR2A and SunTagX32-DYNC1H1 genes stably expressing scFv-sfGFP were a generous gift of Drs. X. Pichon and E. Bertrand (Institut de Génétique Moléculaire de Montpellier and Equipe labélisée Ligue Nationale Contre le Cancer, University of Montpellier, CNRS, Montpellier, France; Pichon et al., 2016). Cells were authenticated by morphological assessment and/or short tandem repeat profiling (ATCC) and periodically confirmed negative for mycoplasma by DAPI staining. Arsenite stress was done using 0.5 mM sodium arsenite, and samples were collected at 45 min after stress unless otherwise indicated. For stress experiments, cells were treated with arsenite alone, DTT (2 mM), or heat stress (42°C in L-15 Leibovitz medium in a CO2-free environment) and cotreated with DMSO (0.1%; Sigma-Aldrich), MG132 (10 µM in DMSO; Sigma-Aldrich), DBeQ (10 µM in DMSO; Thermo Fisher Scientific), CB-5083 (10 µM in DMSO; Cayman Chemical), or bortezomib (10 µM in DMSO; VWR) with or without puromycin (10 µg/ml; Sigma Aldrich).
Generation of the mRuby2-G3BP1 plasmid
The mRuby2-G3BP1 plasmid was generated by insertion of codon-optimized mRuby2 lacking a stop codon into pEGFP-C1-G3BP1 plasmid (a gift of N. Kedersha) to replace the EGFP coding sequence. A gBlock (shown below; Integrated DNA Technologies) was designed containing the mRuby2 sequence with 40-nt annealing arms to the pEGFP-C1 G3BP1 plasmid cut with BglII and AgeI to remove the EGFP coding sequence. mRuby2 was inserted using the HiFi DNA Assembly reagent (New England Biolabs).
mRuby2 gBlock sequence
5′-GCAGAGCTGGTTTAGTGAACCGTCAGATCCGCTAGCGCTACCGGTCGCCACCATGGTAAGCAAAGGCGAAGAACTGATTAAGGAAAACATGCGAATGAAGGTCGTAATGGAGGGCTCAGTCAATGGTCACCAGTTTAAGTGCACAGGTGAAGGTGAAGGTAGACCATACGAGGGGACACAGACCATGAGGATCAAGGTCATTGAGGGTGGCCCACTCCCATTCGCATTTGATATATTGGCGACCAGCTTTATGTACGGCTCCAGAACGTTTATTAAATATCCCGCCGATATTCCCGATTTTTTTAAGCAATCATTCCCAGAGGGATTTACTTGGGAAAGAGTCACAAGATATGAAGATGGCGGTGTCGTCACGGTTACCCAGGACACAAGTCTCGAAGATGGCGAGTTGGTTTACAACGTTAAAGTGCGGGGCGTAAATTTTCCTTCAAATGGTCCAGTGATGCAAAAGAAGACTAAAGGCTGGGAACCGAATACCGAGATGATGTACCCGGCAGACGGGGGACTGCGAGGTTACACCGATATTGCGTTGAAGGTAGATGGTGGTGGACATTTGCATTGCAACTTTGTGACCACGTATCGCAGCAAAAAGACGGTCGGAAATATAAAAATGCCTGGCGTCCACGCAGTGGACCACCGCCTCGAGCGAATTGAGGAGTCTGACAATGAAACGTATGTGGTGCAGCGCGAAGTAGCAGTGGCTAAATATTCCAATCTCGGGGGGGGAATGGACGAGTTGTATAAATCCGGACTCAGATCTATGGTGATGGAGAAGCCTAGTCCCCTGCTGGTCGG-3′.
Nascent chain tracking
Nascent chain tracking was performed as previously described (Moon et al., 2019) using a reporter mRNA encoding the human KDM5B ORF with 10× FLAG tags at the N terminus and 24× MS2 stem loops inserted in the 3′ untranslated region to create the spaghetti monster (SM) KDM5B reporter (Morisaki et al., 2016). Purified recombinant MS2 coat protein fused with Halo and anti-FLAG antibody fragments labeled with Cy3 or Alexa Fluor 488 were bead loaded with the reporter plasmid into U-2 OS cells stably expressing GFP-G3BP1 or mRuby2-G3BP1 to mark SGs. Two hours later, cells were incubated with the far-red Halo ligand JF646 for 30 min and washed thrice before imaging. Cells were imaged in complete maintenance medium lacking phenol red in a humidified chamber at 37°C under 5% CO2 using a custom-built semi-TIRF microscope with two electron multiplying charge-coupled device cameras (iXon Ultra 888, Andor) with a 60× (NA 1.49) Olympus oil immersion objective (Morisaki et al., 2016). Cells were imaged first in the red channel, and then GFP and far-red channels were imaged simultaneously. The entire cell volume was imaged with 13 z-stacks with a step size of 500 nm on a piezoelectric stage. Images were captured before stress and then were imaged every 2 s for 10 min starting ∼15–75 min after arsenite addition (to 0.5 mM) when SGs were visible as described in Moon et al. (2019). For all drug treatment experiments, drugs resuspended in DMSO (10 µM DBeQ, 10 µM MG132), 0.1% DMSO, or puromycin were added at the same time as arsenite, except for those cells treated with DBeQ or MG132 alone.
The percentage of mRNAs associated with nascent peptides was quantified in a single frame of maximum intensity projections from time-lapse images obtained before and after stress using the Cell Counter plugin in ImageJ. The percent mRNAs associated with nascent peptides before stress in each cell was set to 100%, and the relative percent mRNAs associated with nascent peptides after stress is presented in Fig. 1, with the number of cells and mRNAs analyzed and percent mRNAs associated with nascent peptides in each cell reported in Table S1. One-tailed t tests were done to assess significance between the percent mRNAs associated with nascent peptides in cells before and after stress and between cells treated with arsenite + DMSO and arsenite + DBeQ or arsenite + MG132, with exact P values provided in Table S1. The times after stress when cells were imaged are reported in Table S1.
The duration of mRNA–SG interactions was quantified as previously described using previously published (Moon et al., 2019) custom Mathematica (version 11.2.0.0) code (accessible on GitHub at https://raw.githubusercontent.com/TatsuyaMorisaki/Translation-Stress/master/Translation-Stress.nb). The mean ± SEM binding-time survival probability of individual mRNAs associated with nascent peptides with SGs is shown in Fig. 1.
For HT chase experiments, U-2 OS cells harboring GFP-G3BP1 or mRuby2-G3BP1 were bead loaded as described above and stressed with arsenite in the presence or absence of DBeQ (10 µM); 15 min later, HT (3 µg/ml; Cayman Chemical) was added. Cells were imaged as described above every 10 min beginning immediately before HT addition for up to 55 min after stress. The number of mRNAs associated with nascent peptides in each frame from maximum intensity projections was quantified in ImageJ using the Cell Counter plugin from two independent experiments with n = 13 cells (unstressed + HT) and n = 17 cells (arsenite + DBeQ + HT) analyzed. Student’s t test (one tailed) was done to assess significance between the percent mRNAs associated with nascent peptides in unstressed cells treated with HT versus cells stressed with arsenite + DBeQ + HT at each time point. Exact P values are reported in Table S1. The average ± SEM of the percent mRNAs translating at each time point is shown in bold colors (blue and black), with data from individual cells shown in light blue and gray in Fig. 1.
Live cell imaging of SunTag reporter transcripts was performed using HeLa cells harboring endogenously tagged SunTagX32-DYNC1H1 (DYNC1H1-SunTag) and SunTagX56-POLR2A, (POLR2A-SunTag) genes that were stably expressing scFv-sfGFP (Pichon et al., 2016). Cells were imaged every 10 min for 1 h in a humidified, 37°C Oko Labs chamber on a Nikon Ti-E spinning disc confocal microscope with a 40×/0.95-NA air objective (488 nm, excitation laser; emission filter, 525/50 nm) and an Andor 888 Ultra electron multiplying charge-coupled device camera and Nikon Elements software following treatment with 0.5 mM arsenite either cotreated with 0.1% DMSO or 10 µM DBeQ. The number of SunTag foci in each frame was determined using the ImageJ Cell Counter plugin, and Student’s t test was done to assess significance between arsenite + DMSO and arsenite + DBeQ conditions. Source data are provided in Table S1.
Immunofluorescence (IMF) microscopy and smFISH
To determine protein localization (RPL19, RPL29, VCP, LTN1, NEMF, and G3BP) in U-2 OS cells, and VCP and poly(A)-binding protein localization in HeLa cells, cells were stressed and treated as indicated, rinsed with PBS, and then fixed with 4% PFA for 10 min. Cells were washed with PBS and then permeabilized with Triton X-100 (0.5%) for 5 min. Cells were washed with PBS for 5 min and then blocked in 3% BSA at 4°C overnight or at room temperature for 1 h. Cells were washed thrice with PBS and incubated with primary antibody (Table S2) in 3% BSA in PBS at 4°C overnight or at room temperature for 1 h. Cells were washed thrice with PBS and then incubated with secondary antibodies (Table S2) in 3% BSA in PBS for 1 h at room temperature and then washed thrice with PBS. Coverslips were mounted using VECTASHIELD antifade mounting medium with DAPI to visualize nuclei.
smFISH was performed as described in Khong et al. (2017) using protocols adapted from Stellaris, Khong and Parker (2018), Gaspar et al. (2017), and Dunagin et al. (2015). Following stresses and treatments, U-2 OS cells plated on coverslips were rinsed with PBS and then fixed with 10% formaldehyde or 4% PFA in PBS for 10 min, washed twice with PBS, and then incubated under 70% ethanol for ≥1 h at 4°C. Cells were incubated in Wash Buffer A (10% formamide in nuclease-free 2× SSC) for 5 min and hybridized with probes to AHNAK, PEG3, TFRC, or NORAD generated by Biosearch Technologies labeled in far-red (Quasar 670) or red (Quasar 570) or to HSPA1A, HSPA1B, SCN8A, or EGR1 (oligonucleotide sequences listed in Table S3) generated as in Gaspar et al. (2017), or the 5′ end of DYNC1H1 from Khong and Parker (2018) in hybridization buffer (10% dextran sulfate [wt/vol] in 10% formamide in 2× SSC; Dunagin et al., 2015) for 16 h at 37°C in humidified chambers. Cells were washed with Wash Buffer A for 30 min at 37°C twice and then with nuclease-free 2× SSC for 5 min. Coverslips were mounted onto slides with VECTASHIELD antifade mounting medium with or without DAPI, stored at 4°C overnight, and imaged as described below.
For consecutive IMF-smFISH, cells were treated and stressed as indicated and then fixed with 4% PFA for 10 min. Cells were rinsed with PBS twice and then permeabilized with Triton X-100 (0.1%) and Ribolock RNase inhibitor for 5 min. Cells were rinsed with PBS and incubated with mouse anti-G3BP1 (1:200, ab56574; Abcam) in PBS with Ribolock RNase inhibitor for 1 h at room temperature in a humidified chamber. Cells were washed thrice with PBS and then incubated with goat anti-mouse Alexa Fluor 405 (1:200, A-31553; Thermo Fisher Scientific) and Ribolock RNase inhibitor for SG detection for 1 h at room temperature in a humidified chamber. Cells were washed thrice with PBS and then fixed with PFA for 10 min. Cells were rinsed with PBS, and then the smFISH protocol described above was performed for mRNA detection. In general, SGs were detected via expression of fusion proteins GFP-G3BP1 or mRuby2-G3BP1, or via IMF detection of G3BP as indicated.
Image acquisition and analysis of smFISH and IMF samples
Image acquisition was performed using a DeltaVision Elite widefield microscope with a 100×, 1.40-NA Olympus UPlanSApo oil immersion objective and PCO Edge sCMOS camera at room temperature. At least 25 z-stacks were acquired (200-nm step sizes), and two to three frames were captured per sample in each experiment. Images were acquired and deconvolved with softWoRx 6.5.2, and maximum intensity projections were created in ImageJ/Fiji (Schindelin et al., 2012). For smFISH experiments, the total number of individual smFISH foci in the cytoplasm per cell and the number of smFISH foci that colocalized with SGs were quantified using the ImageJ/Fiji (Schindelin et al., 2012) Cell Counter plugin. The percent RNA in SGs was calculated for each cell or frame from two to three frames per replicate, and the average ± SEM from two to three independent experimental replicates is presented. Student’s t test (one tailed) was done to assess significance, with *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.005, and ****, P ≤ 0.001 for all experiments. Exact P values are reported in Table S1 for all experiments. SG areas were quantified in maximum intensity projections using ImageJ/Fiji (Schindelin et al., 2012) from two independent experimental replicates (two frames per condition) in cells stressed with arsenite and probed for AHNAK mRNA and oligo(dT), and SGs were defined as having areas of 0.2 to >2 µm2 and reported (45 min after arsenite addition). The number of SGs per cell was quantified using the Cell Counter plugin in ImageJ/Fiji from cells stained for AHNAK and NORAD RNAs during arsenite stress (Fig. 2 A), heat stress, or DTT stress (Fig. 3 B) and AHNAK for transfected cells (Fig. 7). All source data are provided in Table S1.
siRNA-mediated depletion of LTN1 or NEMF
To deplete LTN1 or NEMF, 10 nM siRNA SMARTpools (Dharmacon) siGENOME Human LTN1 (26046, 5′-GCAGUGGUGUGAAGAAUUA-3′, 5′-GAGAGUACCUUCCUUACAUU-3′, 5′-GCACUUACCUCACCCAUCA-3′, 5′-CAAACUUCCUGCAAGAUUA-3′), siGENOME Human NEMF (9147, 5′-GGGAAGAGACAUUAAAUUA-3′, 5′-GAAGAAGACCGUGAACUUA-3′, 5′-GGACGAACCUGUGAAGAAA-3′, 5′-GCACAAUGGCACUUUGCUA-3′), or nonspecific siGENOME nontargeting siRNA (pool #2, Dharmacon, 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-AUGAACGUGAAUUGCUCAA-3′, 5′-UGGUUUACAUGUCGACUAA-3′) was transfected into U-2 OS cells with INTERFERin Polyplus Reagent (VWR) according to the manufacturer’s protocol. 3 d later, cells were either collected for RT-quantitative PCR (qPCR; described below) or were unstressed or stressed (0.5 mM arsenite), in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM), in the presence or absence of puromycin (10 µg/µl) for 45 min and fixed for smFISH-IMF. SGs were detected by G3BP staining, and AHNAK mRNA localization was assessed as described above. Source data are provided in Table S1.
Transient VCP-EGFP transfections and treatments
U-2 OS cells stably expressing mRuby2-G3BP1 were plated on coverslips and untransfected or transiently transfected using Lipofectamine LTX with Plus reagent (Invitrogen) with peGFP-N1 (empty vector), VCP(WT)-EGFP, VCP(A232E)-EGFP, or VCP(R155H)-EGFP (gifts from Nico Dantuma, Addgene nos. 23971, 23973, and 23972; Tresse et al., 2010). Cells were treated with 0.5 mM sodium arsenite in normal growth medium for 45 min at 24 h after transfection. Cells were fixed and smFISH performed to detect AHNAK or NORAD RNAs as described above. Results are from three independent experiments. Student’s t test (one tailed) was done to assess significance between the percent AHNAK or NORAD RNAs in SGs in cells expressing EGFP alone versus VCP(WT)-EGFP, VCP(A232E)-EGFP, or VCP(R155H)-EGFP, with exact P values reported in Table S1. The frequency of observing SGs with an area of 0.2 to >2 µm2 in the transfected cells analyzed for AHNAK localization described above was obtained using ImageJ/Fiji using maximum intensity projections (histograms represent the relative frequency of each indicated SG size), and the average number of SGs per cell was derived using the Cell Counter plugin as described above. Source data are provided in Table S1.
Metabolic labeling of nascent proteins
Nascent proteins in U-2 OS cells expressing GFP-G3BP1 were labeled with 35S-Met and 35S-Cys (EXPRE35S35S Protein Labeling Mix; Perkin Elmer) for 30 min in the presence of DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM) following a 30-min incubation in labeling medium with 10% dialyzed FBS and 1% streptomycin/penicillin. Cells were lysed in NP-40 buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40 substitute, and 5 mM EDTA) with protease inhibitor cocktail (Sigma-Aldrich), and equal volumes were run on 4–12% NuPAGE protein gels (Thermo Fisher Scientific). Gels were exposed to phosphor screens and imaged on a Typhoon FLA 9500 phosphorimager. ImageJ/Fiji (Schindelin et al., 2012) was used to quantify the signal intensity in each lane. A representative image and the average relative nascent protein abundance ±SEM are shown from two independent experiments. Student’s t test (one tailed) was done to assess significance between untreated and treated samples, with ***, P ≤ 0.005, and exact P values are reported in Table S1. Stressed cells (0.5 mM sodium arsenite) in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM) were pulse labeled for 15 min starting at 0 and 15 min after stress. The nascent protein abundance in each sample relative to an unstressed, untreated control was determined, and individual measurements and the average ± SEM from n = three independent experiments are reported, with exact values reported in Table S1.
Polysome profiles
U-2 OS cells stably expressing GFP-G3BP1 were stressed with 0.5 mM sodium arsenite or unstressed in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or MG132 (10 µM) for 30 min. Translation inhibitors (e.g., cycloheximide) were not used. Cells were scraped into cold PBS with protease inhibitors and RiboLock RNase inhibitors, pelleted, and frozen at −80°C. Cells were resuspended in lysis buffer (500 µM Tris, pH 7.5, 250 µM MgCl2, 150 µM KCl, 1 mM DTT, cOmplete ULTRA mini EDTA-free protease inhibitor cocktail [Sigma-Aldrich], RiboLock RNase inhibitor, and diethylpyrocarbonate-treated water) and vortexed. Triton X-100 (to 0.5%) and sodium deoxycholate (to 0.5%) were added, and lysates were passed through a 25G needle 10 times. Lysates were clarified by brief centrifugation, and total RNA was measured by A260 to load equal RNA amounts for each sample onto 15–50% sucrose gradients (15–50% sucrose in 20 mM Hepes, pH 7.6, 100 mM KCl, 5 mM MgCl2, and 1 mM DTT). Samples were ultracentrifuged at 36,000 rpm for 2 h. Gradients were fractionated and A260 detected using a Teledyne ISCO system. The ratio of RNA in polysome and monosome fractions was determined by measuring the area under the polysome and monosome curves using ImageJ/Fiji. Two independent experiments were performed and results from one representative experiment are presented.
RQC substrate analysis
Two constructs encoding EGFP upstream of RFP separated by a linker region encoding two P2A sites and either no lysines (K0) or 20 lysines (K20) encoded by poly(A) tracts described in Juszkiewicz and Hegde (2017), called pmGFP-P2A-K0-P2A-RFP and pmGFP-P2A-KAAA20-P2A-RFP, were a gift from Ramanujan Hegde (Addgene plasmids no. 105686 and no. 105688; http://n2t.net/addgene:105686 and http://n2t.net/addgene:105688; RRID:Addgene_105686 and RRID:Addgene_105688). Plasmids were electroporated (BTX Harvard Apparatus Gemini System) into U-2 OS cells and 18–24 h later, cells were unstressed or stressed (0.5 mM arsenite) in the presence or absence of DMSO (0.1%), DBeQ (10 µM), or DBeQ (10 µM) plus puromycin (10 µg/µl = l) for 45 min. Cells were fixed, and IMF staining was performed to detect SGs using mouse anti–G3BP (Table S2) and goat anti-mouse 405 (Table S2). SmFISH was performed to detect reporter mRNAs using custom-made DNA probes (Integrated DNA Technologies) against EGFP designed using the LGC Biosearch Technologies Stellaris Designer (Table S3). Probes were labeled via Terminal Deoxynucleotidyl Transferase (Thermo Fisher Scientific) with ddUTP-ATTO-633 (far-red) and then purified by phenol–chloroform isoamyl alcohol extraction and ethanol precipitation and resuspended to ∼12.5 µM for hybridizations (1:100). Those cells expressing EGFP were imaged, and the percent EGFP mRNA in SGs was calculated as described above from two independent replicates. The number of cells and mRNAs analyzed is reported in Table S1. Student’s t test (one tailed) was done to determine significance (*, P ≤ 0.05 and **, P ≤ 0.01), and exact P values are reported in Table S1.
RT-qPCR
To determine the percent knockdown of LTN1 and NEMF, cells were transfected with siRNAs as described above, and total RNA was extracted using TRIzol Reagent according to the manufacturer’s instructions at 72 h after transfection. cDNA was generated using 1 µg of total RNA and random hexamers with SuperScript III reverse transcription, and qPCR was performed with iQ Supermix (Bio-Rad) using a CFX96 Real-Time Thermocycler (Bio-Rad) with GAPDH as a reference gene. GAPDH primers were previously described (Moon et al., 2012; GAPDH forward [Fw]: 5′-TCTTTTGCGTCGCCAGCCGA-3′; GAPDH reverse [Rv]: 5′-ACCAGGCGCCCAATACGACC-3′), LTN1 primers (efficiency of 91.6%) that recognized both isoforms were used (LTN1 Fw: 5′-AGCAACCTGAAGCCATAGCA-3′; LTN1 Rv: 5′-CTCTGGAACAGTTTGCGGGT-3′), and NEMF primers (efficiency of 87.7%) that recognized both isoforms were used (NEMF Fw: 5′-GGTGGACGAGATCAGCAACA-3′; NEMF Rv: 5′-CAGCTTCAGTCAAGGTCCGT-3′). ΔΔCq values were calculated with the Bio-Rad CFX Manager software, and the average abundance of LTN1 or NEMF relative to samples transfected with the nonspecific control siRNAs ± SEM are reported from n = three independent experiments.
Code availability
Custom Mathematica (version 11.2.0.0) code previously described in Moon et al. (2019) was deposited on GitHub and is accessible at https://raw.githubusercontent.com/TatsuyaMorisaki/Translation-Stress/master/Translation-Stress.nb.
Data availability
Data supporting the findings of this study are provided in the supplemental tables and are also available from the corresponding authors upon request.
Online supplemental material
Fig. S1 shows the abundance of nascent DYNC1H1 or POLR2A proteins tagged with the SunTag system during arsenite stress and VCP inhibition. Fig. S2 shows global translation activity by 35S-amino acid labeling and polysome profiling in cells treated with VCP or proteasome inhibitors during arsenite stress. Fig. S3 shows oligo(dT) staining to detect polyadenylated mRNAs, SG area, and number upon VCP or proteasome inhibition, or with exogenous expression of pathogenic VCP alleles during stress. Fig. S4 shows localization of stress-induced gene mRNAs HSPA1A and HSPA1B and SGs during arsenite stress or heat stress in the presence or absence of VCP inhibitors. Fig. S5 shows VCP, LTN1, NEMF, and ribosomal protein subcellular localization and SGs by IMF staining during arsenite stress. Videos 1, 2, 3, 4, and 5 are representative time series related to Fig. 1. Table S1 contains all source data, statistics, and the number of experimental and technical replicates for all figures. Table S2 lists antibodies used for nascent chain tracking and IMF microscopy. Table S3 lists oligonucleotides used to make smFISH probes.
Supplementary Material
shows source data for all figures and supplemental figures.
lists antibodies used for nascent chain tracking and IMF microscopy.
shows oligonucleotides for smFISH probes used to detect EGFP, SCN8A, EGR1, HSPA1A, and HSPA1B transcripts.
Acknowledgments
We thank N. Kedersha for U-2 OS cell lines, X. Pichon and E. Bertrand for SunTag HeLa cell lines, and E. Braselmann and T. Nahreini for purifying the U-2 OS GFO-G3BP1 cell line in the BioFrontiers Institute Flow Cytometry Core facility. We thank Denise Muhlrad and Ken Lyon for technical assistance; Anne Webb and the T.J. Stasevich and R. Parker laboratories, B. Dodd, and A. McCorkindale for helpful suggestions; and Jeffrey Wilusz’s laboratory for peGFP-N1. Spinning disc confocal microscopy was performed on a Nikon Ti-E microscope supported by the BioFrontiers Institute and the Howard Hughes Medical Institute at the BioFrontiers Institute Advanced Light Microscopy Core.
S.L. Moon was funded by the Anna and John J. Sie Foundation and Howard Hughes Medical Institute, T.J. Stasevich was funded by the National Institutes of Health (R35GM119728) and the Boettcher Foundation’s Webb-Waring Biomedical Research Program, and R. Parker was funded by Howard Hughes Medical Institute.
R. Parker is a consultant for Third Rock Ventures. The authors declare no competing financial interests.
Author contributions: Conceptualization: R. Parker and S.L. Moon; Methodology: S.L. Moon, T. Morisaki, R. Parker, and T.J. Stasevich; Software: T. Morisaki and T.J. Stasevich; Validation: S.L. Moon; Formal analysis: S.L. Moon, T. Morisaki, and T.J. Stasevich; Investigation: S.L. Moon and T. Morisaki; Resources: S.L. Moon, R. Parker, T. Morisaki, and T.J. Stasevich; Data curation: S.L. Moon and T. Morisaki; Writing - original draft preparation: S.L. Moon; Writing - review and editing: S.L. Moon, R. Parker, T. Morisaki, and T.J. Stasevich; Visualization: S.L. Moon and T. Morisaki; Supervision: R. Parker and T.J. Stasevich; Project administration: S.L. Moon; Funding acquisition: R. Parker, T.J. Stasevich, and S.L. Moon.
References
- Anderson P., and Kedersha N.. 2006. RNA granules. J. Cell Biol. 172:803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., and Martin A.. 2018. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 87:697–724. 10.1146/annurev-biochem-062917-011931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson M.H., and Joazeiro C.A.P.. 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 467:470–473. 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., and Hegde R.S.. 2016. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23:7–15. 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.-W., Zhou S., King D., Shen P.S., Weibezahn J., et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11:1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.-M., Taylor J.P., and Parker R.. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 153:1461–1474. 10.1016/j.cell.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V., Juszkiewicz S., Choi J., Puglisi J.D., Brown A., Shao S., Ramakrishnan V., and Hegde R.S.. 2019. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol. 26:1132–1140. 10.1038/s41594-019-0331-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.-F., Brown S.J., Minond D., Nordin B.E., Li K., Jones A.C., Chase P., Porubsky P.R., Stoltz B.M., Schoenen F.J., et al. 2011. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. USA. 108:4834–4839. 10.1073/pnas.1015312108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C., Glynne R.J., Wu H., Masliah E., Joazeiro C.A.P., et al. 2009. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. USA. 106:2097–2103. 10.1073/pnas.0812819106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bot S.T., Schelhaas H.J., Kamsteeg E.-J., and van de Warrenburg B.P.C.. 2012. Hereditary spastic paraplegia caused by a mutation in the VCP gene. Brain. 135:e223–e224. 10.1093/brain/aws201 [DOI] [PubMed] [Google Scholar]
- Doma M.K., and Parker R.. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 440:561–564. 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio K.N., Wu C.C.-C., Sinha N., Loll-Krippleber R., Brown G.W., and Green R.. 2019. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. eLife. 8 e49117 10.7554/eLife.49117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunagin M., Cabili M.N., Rinn J., and Raj A.. 2015. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. Methods Mol. Biol. 1262:3–19. 10.1007/978-1-4939-2253-6_1 [DOI] [PubMed] [Google Scholar]
- Figueiredo-Pereira M.E., Berg K.A., and Wilk S.. 1994. A new inhibitor of the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) induces accumulation of ubiquitin-protein conjugates in a neuronal cell. J. Neurochem. 63:1578–1581. 10.1046/j.1471-4159.1994.63041578.x [DOI] [PubMed] [Google Scholar]
- Fresno M., Jiménez A., and Vázquez D.. 1977. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72:323–330. 10.1111/j.1432-1033.1977.tb11256.x [DOI] [PubMed] [Google Scholar]
- Gaspar I., Wippich F., and Ephrussi A.. 2017. Enzymatic production of single-molecule FISH and RNA capture probes. RNA. 23:1582–1591. 10.1261/rna.061184.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko M.V., Lobanov A.V., and Gladyshev V.N.. 2012. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA. 109:17394–17399. 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Gonzalez M.A., Feely S.M., Speziani F., Strickland A.V., Danzi M., Bacon C., Lee Y., Chou T.-F., Blanton S.H., Weihl C.C., et al. 2014. A novel mutation in VCP causes Charcot-Marie-Tooth Type 2 disease. Brain. 137:2897–2902. 10.1093/brain/awu224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh N.R., and Green R.. 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 156:950–962. 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D., LeBlanc A.C., Rouiller I., Michnick S.W., Servant M.J., and Latterich M.. 2009. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell. Biol. 29:4484–4494. 10.1128/MCB.00252-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubenberger D., Bittner R.E., Rauch-Shorny S., Zimprich F., Mannhalter C., Wagner L., Mineva I., Vass K., Auff E., and Zimprich A.. 2005. Inclusion body myopathy and Paget disease is linked to a novel mutation in the VCP gene. Neurology. 65:1304–1305. 10.1212/01.wnl.0000180407.15369.92 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., and Weissman J.S.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakov E., and Stanhill A.. 2011. Stalled proteasomes are directly relieved by P97 recruitment. J. Biol. Chem. 286:30274–30283. 10.1074/jbc.M111.240309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S., Kuroha K., Tatematsu T., and Inada T.. 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21:519–524. 10.1101/gad.1490207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Kedersha N., and Anderson P.. 2019. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 11 a032813 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A.P. 2017. Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu. Rev. Cell Dev. Biol. 33:343–368. 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. ; ITALSGEN Consortium . 2010. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 68:857–864. 10.1016/j.neuron.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S., and Hegde R.S.. 2017. Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol. Cell. 65:743–750. 10.1016/j.molcel.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S., Chandrasekaran V., Lin Z., Kraatz S., Ramakrishnan V., and Hegde R.S.. 2018. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell. 72:469–481. 10.1016/j.molcel.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M., Monaghan C.E., and Ackerman S.L.. 2017. Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron. 96:616–637. 10.1016/j.neuron.2017.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Tisdale S., Hickman T., and Anderson P.. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552. 10.1016/S0076-6879(08)02626-8 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38:494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., and Parker R.. 2018. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 217:4124–4140. 10.1083/jcb.201806183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., and Parker R.. 2017. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell. 68:808–820. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., King O.D., Shorter J., and Gitler A.D.. 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201:361–372. 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Han Y., and Qian S.-B.. 2013. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 49:453–463. 10.1016/j.molcel.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno A., Noguchi M., Fukushi J., Motohashi Y., and Kakizuka A.. 2010. Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Genes Cells. 15:911–922. 10.1111/j.1365-2443.2010.01428.x [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K., et al. 2017. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8:159 10.1038/s41467-017-00188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Di Marco S., Kaufman R.J., and Gallouzi I.-E.. 2007. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell. 18:2603–2618. 10.1091/mbc.e06-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.-J., Anderson P., and Kaufman R.J.. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- Meaux S., and Van Hoof A.. 2006. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 12:1323–1337. 10.1261/rna.46306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.L., Anderson J.R., Kumagai Y., Wilusz C.J., Akira S., Khromykh A.A., and Wilusz J.. 2012. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 18:2029–2040. 10.1261/rna.034330.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.L., Sonenberg N., and Parker R.. 2018. Neuronal Regulation of eIF2α Function in Health and Neurological Disorders. Trends Mol. Med. 24:575–589. 10.1016/j.molmed.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Moon S.L., Morisaki T., Khong A., Lyon K., Parker R., and Stasevich T.J.. 2019. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 21:162–168. 10.1038/s41556-018-0263-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T., Lyon K., DeLuca K.F., DeLuca J.G., English B.P., Zhang Z., Lavis L.D., Grimm J.B., Viswanathan S., Looger L.L., et al. 2016. Real-time quantification of single RNA translation dynamics in living cells. Science. 352:1425–1429. 10.1126/science.aaf0899 [DOI] [PubMed] [Google Scholar]
- Namkoong S., Ho A., Woo Y.M., Kwak H., and Lee J.H.. 2018. Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell. 70:175–187. 10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi D., Tahiliani P., Kumar A., and Chandu D.. 2006. The ubiquitin-proteasome system. J. Biosci. 31:137–155. 10.1007/BF02705243 [DOI] [PubMed] [Google Scholar]
- Niwa H., Ewens C.A., Tsang C., Yeung H.O., Zhang X., and Freemont P.S.. 2012. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J. Biol. Chem. 287:8561–8570. 10.1074/jbc.M111.302778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X., Bastide A., Safieddine A., Chouaib R., Samacoits A., Basyuk E., Peter M., Mueller F., and Bertrand E.. 2016. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 214(6):769–781. 10.1083/jcb.201605024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva V.P., Skabkin M.A., Hellen C.U.T., Pestova T.V., and Pisarev A.V.. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30:1804–1817. 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S.W., and Parker R.. 2016. Principles and Properties of Stress Granules. Trends Cell Biol. 26:668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M., Taylor J.P., and Parker R.. 2013. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 154:727–736. 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ortiz C.J., Flores J.C., Valenzuela J.A., Rodriguez G.J., Zumkehr J., Tran D.N., Kimonis V.E., and Kitazawa M.. 2016. The Myoblast C2C12 Transfected with Mutant Valosin-Containing Protein Exhibits Delayed Stress Granule Resolution on Oxidative Stress. Am. J. Pathol. 186:1623–1634. 10.1016/j.ajpath.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Kowalinski E., Shanmuganathan V., Defenouillère Q., Braunger K., Heuer A., Pech M., Namane A., Berninghausen O., Fromont-Racine M., et al. 2016. The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science. 354:1431–1433. 10.1126/science.aaf7520 [DOI] [PubMed] [Google Scholar]
- Seguin S.J., Morelli F.F., Vinet J., Amore D., De Biasi S., Poletti A., Rubinsztein D.C., and Carra S.. 2014. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 21:1838–1851. 10.1038/cdd.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., and Burge C.B.. 2013. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 49:439–452. 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., von der Malsburg K., and Hegde R.S.. 2013. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 50:637–648. 10.1016/j.molcel.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Brown A., Santhanam B., and Hegde R.S.. 2015. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell. 57:433–444. 10.1016/j.molcel.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., and Green R.. 2012. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19:594–601. 10.1038/nsmb.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., Eyler D.E., and Green R.. 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 330:369–372. 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., and Zaher H.S.. 2014. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 9:1256–1264. 10.1016/j.celrep.2014.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C.L., Yan L.L., and Zaher H.S.. 2017. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell. 68:361–373.e5. 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., and Bennett E.J.. 2017. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell. 65:751–760. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S., Khoutorsky A., Mathews M.B., and Sonenberg N.. 2018. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 19:791–807. 10.1038/s41580-018-0034-x [DOI] [PubMed] [Google Scholar]
- Taylor J.P., Brown R.H. Jr., and Cleveland D.W.. 2016. Decoding ALS: from genes to mechanism. Nature. 539:197–206. 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.-P., Dantuma N.P., and Taylor J.P.. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 6:217–227. 10.4161/auto.6.2.11014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhiya A., Meyer S.R., Marincola G., Böhm S., Vanselow J.T., Schlosser A., Hofmann K., and Buchberger A.. 2018. ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol. Cell. 70:906–919.e7. 10.1016/j.molcel.2018.04.021 [DOI] [PubMed] [Google Scholar]
- Verma R., Oania R.S., Kolawa N.J., and Deshaies R.J.. 2013. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2 e00308 10.7554/eLife.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Song C., and Li C.-C.H.. 2003. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 300:253–260. 10.1016/S0006-291X(02)02840-1 [DOI] [PubMed] [Google Scholar]
- Wang B., Maxwell B.A., Joo J.H., Gwon Y., Messing J., Mishra A., Shaw T.I., Ward A.L., Quan H., Sakurada S.M., et al. 2019. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell. 74:742–757. 10.1016/j.molcel.2019.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts G.D.J., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., and Kimonis V.E.. 2004. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36:377–381. 10.1038/ng1332 [DOI] [PubMed] [Google Scholar]
- Wilson D.N., and Beckmann R.. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 21:274–282. 10.1016/j.sbi.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Wu C.C.-C., Zinshteyn B., Wehner K.A., and Green R.. 2019. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell. 73:959–970. 10.1016/j.molcel.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.L., Simms C.L., McLoughlin F., Vierstra R.D., and Zaher H.S.. 2019. Oxidation and alkylation stresses activate ribosome-quality control. Nat. Commun. 10:5611 10.1038/s41467-019-13579-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Tang W.K., Zhang T., and Xia D.. 2017. A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front. Mol. Biosci. 4:39 10.3389/fmolb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gui L., Zhang X., Bulfer S.L., Sanghez V., Wong D.E., Lee Y., Lehmann L., Lee J.S., Shih P.-Y., et al. 2015. Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. USA. 112:E1705–E1714. 10.1073/pnas.1418820112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Mishra P., Hay B.A., Chan D., and Guo M.. 2017. Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. eLife. 6 e17834 10.7554/eLife.17834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A., Ayupov R.K., Abaeva I.S., Hellen C.U.T., and Pestova T.V.. 2020. Extraction of mRNA from Stalled Ribosomes by the Ski Complex. Mol. Cell. 77:1340–1349. 10.1016/j.molcel.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurla C., Lifland A.W., and Santangelo P.J.. 2011. Characterizing mRNA interactions with RNA granules during translation initiation inhibition. PLoS One. 6 e19727 10.1371/journal.pone.0019727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows source data for all figures and supplemental figures.
lists antibodies used for nascent chain tracking and IMF microscopy.
shows oligonucleotides for smFISH probes used to detect EGFP, SCN8A, EGR1, HSPA1A, and HSPA1B transcripts.
Data Availability Statement
Data supporting the findings of this study are provided in the supplemental tables and are also available from the corresponding authors upon request.