Zhang and colleagues summarize phase separation and transition in the assembly of autophagosome formation sites and triage of protein condensates for degradation.
Abstract
Liquid–liquid phase separation (LLPS) compartmentalizes and concentrates biomacromolecules into distinct condensates. Liquid-like condensates can transition into gel and solid states, which are essential for fulfilling their different functions. LLPS plays important roles in multiple steps of autophagy, mediating the assembly of autophagosome formation sites, acting as an unconventional modulator of TORC1-mediated autophagy regulation, and triaging protein cargos for degradation. Gel-like, but not solid, protein condensates can trigger formation of surrounding autophagosomal membranes. Stress and pathological conditions cause aberrant phase separation and transition of condensates, which can evade surveillance by the autophagy machinery. Understanding the mechanisms underlying phase separation and transition will provide potential therapeutic targets for protein aggregation diseases.
Introduction
Emerging evidence has revealed that numerous membraneless structures, including signaling clusters, processing bodies (P-bodies), nuclear speckles, stress granules (SGs), and germline granules, are assembled via liquid–liquid phase separation (LLPS; Banani et al., 2017; Bergeron-Sandoval et al., 2016; Shin and Brangwynne, 2017). LLPS refers to a process in which key molecules, when they reach a threshold concentration, become concentrated together with other proteins or RNAs into confined liquid-like compartments. The constituents exhibit high mobility and undergo dynamic exchange with the surrounding liquid milieu (Banani et al., 2017; Shin and Brangwynne, 2017; Wang and Zhang, 2019). LLPS-assembled structures are collectively called condensates in this review. These condensates ensure that a variety of cellular activities take place in a spatially and temporally controlled manner. LLPS is mediated by multivalent weak interactions conferred by intrinsically disordered regions (IDRs) and/or modular interacting domains of the constituent proteins (Fig. 1 A; Banani et al., 2017; Li et al., 2012; Shin and Brangwynne, 2017; Zeng et al., 2016). IDRs lack a stable tertiary structure and often have a biased amino acid composition that is enriched in a limited subset of residues, especially glycine, polar residues (e.g., glutamine and serine), and aromatic residues (e.g., tyrosine and phenylalanine; Boeynaems et al., 2018; Wang and Zhang, 2019). These domains are also referred to as low-complexity domains (LCDs). Protein–RNA or RNA–RNA interactions also contribute to LLPS-mediated formation of RNP granules such as SGs and P-bodies (Lin et al., 2015; Molliex et al., 2015). Liquid-like condensates can transition over time into more stable states, such as the gel-like state, in which the constituents show reduced mobility, and the inert solid state (amorphous or fibrillar; Fig. 1 B; Bergeron-Sandoval et al., 2016; Kaganovich, 2017). During phase transition, weak and transient intermolecular interactions in liquid droplets become strong and stable interactions in gel and solid structures (Bergeron-Sandoval et al., 2016; Kaganovich, 2017). The material properties of condensates are essential for their distinct cellular functions (Kaganovich, 2017).
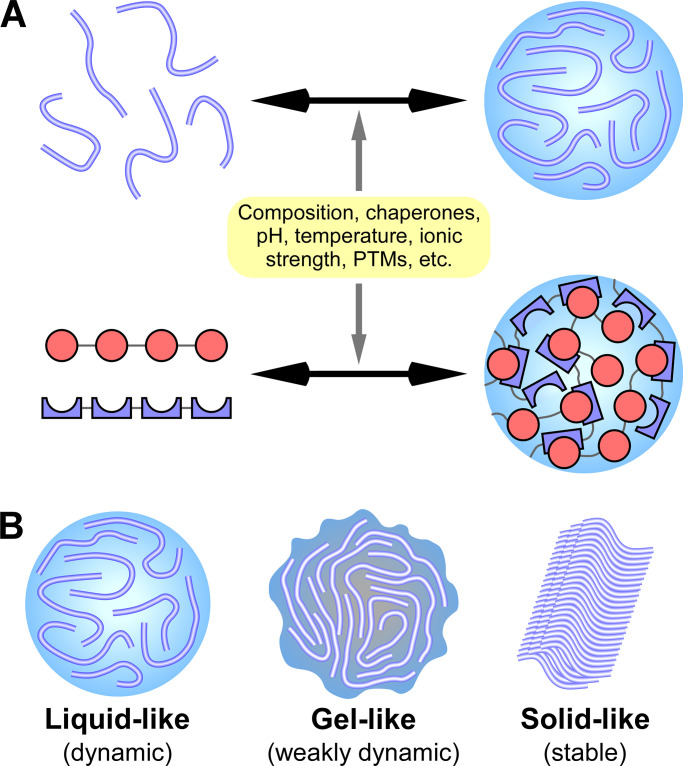
Figure 1.
Phase separation and transition mediate the assembly of protein condensates with distinct material properties. (A) Multivalent interactions, mediated by proteins containing IDRs (top) or tandem modular interacting domains (bottom, represented by the red circles and blue crescents in two different proteins), drive the formation of phase-separated liquid condensates. Factors affecting interactions, including composition, chaperones, pH, temperature, and various PTMs, modulate LLPS. (B) Liquid-like phase-separated condensates (with highly dynamic constituents) can transition into more stable states, such as the gel-like state (with less mobile constituents) and the inert solid state.
Protein LLPS and phase transition are modulated by numerous factors that affect multivalent interactions, including the concentration of key components, composition, mutations in key molecules, chaperones, and environmental factors such as pH and salt concentration (Banani et al., 2017; Shin and Brangwynne, 2017; Snead and Gladfelter, 2019). Various protein posttranslational modifications (PTMs), including phosphorylation, methylation, ubiquitination, sumoylation, and oxidation, affect the valency and strength of the interaction (Snead and Gladfelter, 2019). PTMs thus serve as a general mechanism to integrate various signals and stresses into phase separation and transition under physiological and pathological conditions.
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved lysosome-mediated degradation process involving the formation of a double-membrane autophagosome that engulfs a portion of the cytosolic constituents and delivers them to lysosomes (Fig. 2 A; Feng et al., 2014; Mizushima et al., 2011; Nakatogawa et al., 2009). By degrading and recycling the sequestrated materials, autophagy provides energy and building blocks for cell survival in response to stresses (Mizushima, 2018; Zhang and Baehrecke 2015). Autophagy also selectively removes protein aggregates formed by misfolded proteins, disease-associated proteins, or unwanted proteins to maintain cellular homeostasis, a process referred to aggrephagy (Stolz et al., 2014). Aberrant accumulation of protein aggregates as a result of impaired autophagic degradation has been linked with aging and pathogenesis of various diseases (Levine et al., 2011; Menzies et al., 2017; Zhao and Zhang, 2019a). In this review, we will discuss the involvement of LLPS in the autophagy pathway. LLPS mediates the assembly of autophagosome formation sites and also serves as a new mechanism for regulating the activity of TORC1 kinase, which functions as a key nutrient and energy sensor to control autophagy activity. LLPS triages protein substrates into condensates for selective autophagy (Wang and Zhang, 2019). Stress or pathological insults can modulate phase separation and transition so that protein condensates can evade surveillance by the autophagy machinery. This can have positive consequences, by helping cells adapt to stress, or negative consequences, by facilitating the pathological accumulation of aggregates (Guzikowski et al., 2019; Riback et al., 2017; Zhang et al., 2018). We mainly focus on LLPS-mediated assembly of two autophagy substrates, namely PGL granules and SGs. Autophagic degradation of PGL granules during Caenorhabditis elegans embryogenesis provides a framework for us to understand how phase separation and transition are precisely controlled to ensure that specific condensates are efficiently removed under normal growth conditions but allowed to accumulate under heat stress (Zhang et al., 2018). The assembly of SGs serves as an example to unravel how genetic mutations and cellular stresses lead to aberrant phase transition in the pathogenesis of a variety of diseases (Alberti and Hyman, 2016; Wang and Zhang, 2019).
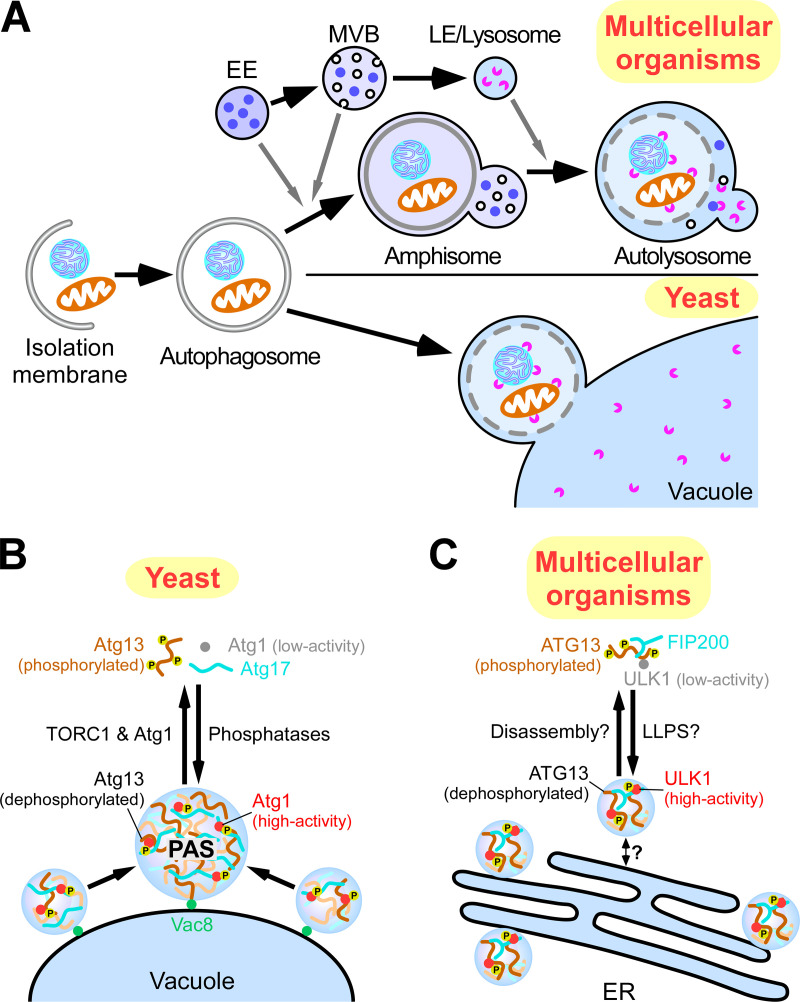
Figure 2.
Formation of autophagosomes in yeast and multicellular organisms. (A) Schematic illustration of the autophagy pathway in yeast and multicellular organisms. In multicellular organisms, closed autophagosomes undergo maturation by fusing with early endosomes (EEs) and/or multivesicular bodies (MVBs) to form amphisomes and with late endosomes/lysosomes (LE/lysosomes) to form autolysosomes. In yeast, autophagosomes directly fuse with the vacuole. (B) Organization of autophagosome formation sites by LLPS. In yeast, upon autophagy induction, Atg13 is dephosphorylated, and the ATG1 complex is formed, which undergoes LLPS to organize PAS precursors on the vacuolar membrane. PAS precursors coalescence into one PAS. (C) In mammals, upon autophagy induction, the ULK complex moves to the ER via an unknown mechanism and induces multiple autophagosome formation sites. This process might be caused by LLPS of the ULK complex.
Autophagosome formation requires coordinated actions of multiple autophagy protein complexes
The core step of autophagy is the formation of the double-membrane autophagosome. It starts with nucleation of a cup-shaped isolation membrane (IM; also known as the phagophore), which further expands and seals (Fig. 2 A; Feng et al., 2014; Ktistakis and Tooze, 2016; Lamb et al., 2013; Melia et al., 2020). A set of autophagy-related (ATG) proteins form different complexes that act at different steps of autophagosome formation. The ATG1 Ser/Thr kinase complex and the ATG14-containing VPS34 phosphatidylinositol-3-phosphate kinase complex are required for initiation and nucleation of IMs. Vesicles carrying the membrane-spanning protein ATG9 have been suggested to serve as the initial source of membrane for IMs (Ktistakis and Tooze, 2016; Lamb et al., 2013; Zhao and Zhang, 2018). The ATG2–ATG18 complex is involved in IM expansion, possibly by supplying phospholipids via the lipid transfer activity of the complex (Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019; Zhang, 2020). Two ubiquitin-like conjugation systems, ATG7(E1 enzyme)/ATG3(E2 enzyme)/ATG8(ubiquitin-like protein) and ATG7(E1)/ATG10(E2)/ATG12(ubiquitin-like protein), participate in multiple steps of autophagosome formation, including IM expansion, shaping, and closure (Mizushima et al., 2011; Nakatogawa et al., 2009).
Despite the involvement of conserved ATG proteins, the spatial and temporal organization of autophagosome formation sites is different in yeast and multicellular organisms. In yeast, the ATG proteins are targeted to a single site at the vacuolar membrane, called the preautophagosomal structure (PAS). Upon closure, autophagosomes directly fuse with the vacuole (Feng et al., 2014; Nakatogawa et al., 2009). In multicellular organisms, upon autophagy induction, the ULK1 complex (counterpart of the ATG1 complex) is targeted to multiple sites on the ER, where it further recruits the VPS34 complex to generate phosphatidylinositol-3-phosphate–enriched subdomains of the ER, known as omegasomes (Axe et al., 2008; Itakura and Mizushima, 2010). The omegasomes act as platforms for recruiting ATG proteins for the initiation and expansion of the IMs (Ktistakis and Tooze, 2016; Lamb et al., 2013; Zhao and Zhang, 2018). Nascent autophagosomes undergo maturation by fusing with vesicles originating from the endolysosomal compartments before forming degradative autolysosomes (Fig. 2 A; Zhao and Zhang, 2019b). The unique steps of autophagy in multicellular organisms require the actions of metazoan-specific autophagy proteins, such as EPG-3, EPG-4, and EPG-5, which were identified from worm genetic screens (Tian et al., 2010; Wang et al., 2016; Zhao et al., 2017).
Phase separation in the assembly of autophagosome formation sites
Organization of the PAS by LLPS
Upon autophagy induction in yeast, the ATG1 complex acts in the most upstream step to organize the autophagosome formation site (Noda and Fujioka, 2015). The yeast ATG1 complex is composed of a Ser/Thr kinase, Atg1; an intrinsically disordered protein, Atg13; a scaffold protein, Atg17; and the accessory proteins Atg29 and Atg31 (Noda and Fujioka, 2015). PAS organization is regulated by TORC1. Under nutrient-rich conditions, Atg13 is hyperphosphorylated by TORC1, which impairs PAS organization by inhibiting the formation of the ATG1 complex. When TORC1 is inhibited by starvation or rapamycin treatment, Atg13 is rapidly dephosphorylated by PP2C phosphatases, leading to the formation of the ATG1 complex (Fujioka et al., 2014; Kamada et al., 2000; Memisoglu et al., 2019). The multivalent interaction of Atg13 with Atg17 dimers triggers phase separation of the ATG1 complex (Fig. 2 B; Fujioka et al., 2020; Yamamoto et al., 2016). The resultant condensates are tethered to the vacuolar membrane through the specific interaction of Atg13 with Vac8, a vacuolar membrane protein important for autophagy (Fujioka et al., 2020; Hollenstein et al., 2019). These condensates move rapidly on the vacuolar membrane and coalesce to form one large condensate termed the early PAS (Fig. 2 B). Downstream ATG proteins are then recruited to the early PAS in a hierarchical order, thereby building up the PAS for initiation of autophagosome formation (Suzuki et al., 2007). The distinguishing feature of the PAS is its high liquidity, which enables dynamic exchange of the components with those in the cytoplasm to facilitate autophagosome generation (Fujioka et al., 2020; Yamamoto et al., 2016).
In multicellular organisms, the ULK1 complex contains ATG13, which possesses abundant IDRs and is phosphoregulated by mTORC1, similar to yeast Atg13, although the ULK1 complex forms constitutively (Noda and Fujioka, 2015; Noda and Mizushima, 2016). ATG proteins are recruited to the autophagosome formation sites in a hierarchical manner, which is in principle similar to the PAS recruitment of ATG proteins in yeast (Fig. 2 C; Itakura and Mizushima, 2010). Further studies are required to determine whether autophagosome formation sites in multicellular organisms are also initiated by LLPS of the ULK1 complex. The restricted movement of autophagosome formation sites on the ER membrane might inhibit their coalescence, resulting in the simultaneous presence of multiple sites in multicellular organisms. mTORC1 also regulates the organization of autophagosome formation sites, but the mechanism has yet to be identified.
Activation of Atg1 kinase at autophagosome formation sites
The kinase activity of Atg1 is low under nutrient-rich conditions but is activated upon autophagy induction (Kamada et al., 2000; Torggler et al., 2016). Activated Atg1 phosphorylates various Atg proteins, such as Atg2 and Atg9, that are important for autophagy initiation (Noda and Fujioka, 2015). As is the case with other protein kinases, activation of Atg1 requires intermolecular autophosphorylation at the activation loop, especially at Thr226 (Yeh et al., 2010). Under starvation conditions, LLPS of the ATG1 complex promotes autophosphorylation of Atg1 through concentration at the PAS (Fujioka et al., 2020). Activated Atg1 has the ability to dissociate the PAS by phosphorylating Atg13. Loss of the kinase activity of Atg1 leads to an enlarged PAS, with aberrant accumulation of some ATG proteins, which cannot mediate autophagosome formation (Cheong et al., 2008). The balance of Atg13 phosphorylation by Atg1 and dephosphorylation by phosphatases may be important for maintenance of the liquid-like PAS, which dynamically recruits ATG proteins as autophagy progresses.
Under nutrient-rich conditions, the vacuolar enzyme aminopeptidase I (Ape1) is delivered to the vacuole through selective autophagy, in a process known as cytoplasm-to-vacuole targeting (Cvt; Lynch-Day and Klionsky, 2010). As described below, Ape1 undergoes LLPS to form a gel-like condensate, which is the substrate for the Cvt pathway (Yamasaki et al., 2020). Atg19 is the receptor protein for Ape1 and links Ape1 condensates to Atg8 and Atg11, a scaffold protein essential for selective autophagy (Shintani et al., 2002). A small population of Atg1 is recruited to Ape1 condensates via Atg19 and Atg11 and is locally activated by autophosphorylation, which is important for the progression of the Cvt pathway (Kamber et al., 2015). Thus, phase-separated condensates (the PAS during starvation-induced autophagy and Ape1 condensates in the Cvt pathway) play a critical role in Atg1 activation.
LLPS modulates TORC1 activity in autophagy regulation
TORC1 integrates the availability of energy and nutrients to control autophagy activity at multiple steps. In mammalian cells, TORC1, via direct phosphorylation, inhibits the activities of the ULK1 complex and the VPS34 complex to block autophagosome initiation and also maintains the cytoplasmic localization of TFEB/TFE3, the master transcription factors of the autophagy–lysosome pathway (Puertollano et al., 2018; Russell et al., 2014). Multiple factors such as the Ragulator complex and the Rag complex have been identified that couple the availability of amino acids, glucose, and other nutrients with targeting of mTORC1 to lysosomes, where it is activated (Liu and Sabatini, 2020). TORC1 activity is also modulated by phase-separated structures triggered by diverse stresses. Under stress conditions (e.g., heat, osmotic, or oxidative stress), TORC1 is partitioned into SGs with concurrent dissociation from the vacuolar/lysosomal membrane to blunt TORC1 signaling (Takahara and Maeda, 2012; Wippich et al., 2013). TORC1 is reactivated upon disassembly of SGs in the stress recovery phase (Takahara and Maeda, 2012). In mammalian cells, the dual-specificity kinase DYRK3, whose LCD mediates dynamic partitioning between SGs and the cytosol, regulates SG disassembly and thus mTORC1 release (Wippich et al., 2013). Sequestration of TORC1 into SGs acts as an adaptive response (Takahara and Maeda, 2012).
Phase separation of yeast Pbp1 (polyA-binding protein–binding protein 1) integrates the cellular redox state into TORC1 regulation. Under conditions that demand intense mitochondrial respiration, such as growth on a nonfermentable carbon source, the methionine-rich LCD of Pbp1 drives phase separation, forming assemblies near the mitochondria (Yang et al., 2019). Pbp1 condensates inhibit TORC1 activity via a yet-to-be-determined mechanism, possibly by affecting the conformation or oligomeric status of TORC1, to promote autophagy activity (Yang et al., 2019). Phase separation of Pbp1 in vitro and in living cells is inhibited by hydrogen peroxide (H2O2)–mediated oxidation of the methionine residues in the LCD (Kato et al., 2019). Therefore, stress-induced assembly of condensates via LLPS is involved in spatial and temporal control of TORC1 activity for autophagy regulation.
Phase separation mediates the assembly of Ape1 condensates for trafficking via the double-membrane Cvt vesicle
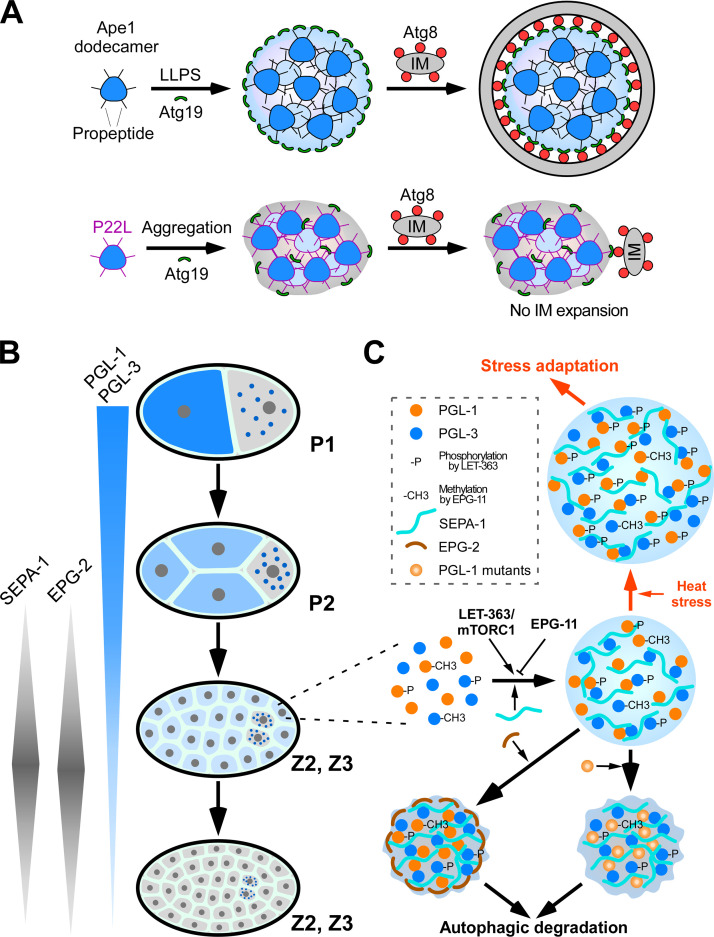
In budding yeast, some vacuole-resident hydrolases are constitutively delivered to the vacuole through the Cvt pathway. Although the Cvt pathway is biosynthetic, it is mechanistically similar to selective autophagy (Lynch-Day and Klionsky, 2010). The aminopeptidase Ape1 is translated as a precursor with a 45–amino acid propeptide and forms dodecamers (Kim et al., 1997). The propeptide mediates Ape1 interaction with the receptor protein Atg19 and is also required for clustering of Ape1 dodecamers. The propeptide is intrinsically disordered but assumes a helical conformation when self-assembling or interacting with Atg19 (Yamasaki et al., 2016). Weak multivalent interactions between propeptides trigger LLPS of Ape1 to form Ape1 condensates, which are historically known as the Ape1 complex (Fig. 3 A; Yamasaki et al., 2020). Ape1 condensates possess gel-like properties. They are spherical in shape, coalesce slowly, are resistant to high salt concentrations, and have low internal mobility (Yamasaki et al., 2020). Atg19 binding is restricted to Ape1 located in the outer layer of the condensates, so that Atg19 coats the Ape1 condensates (Fig. 3 A; Yamasaki et al., 2016; 2020). In experiments with Atg8-conjugated giant unilamellar vesicles (Atg8-GUVs) as a proxy for IMs, clustering of Atg19 at the surface of Ape1 condensates tethers the condensates with Atg8-GUVs via Atg19–Atg8 interaction and also elicits shape changes of the membrane, thereby enabling selective sequestration in the Cvt pathway (Yamasaki et al., 2020).
Figure 3.
LLPS mediates assembly of protein condensates for selective autophagy. (A) Weak multivalent interactions between Ape1 dodecamers induce LLPS to form gel-like Ape1 condensates. Atg19 is localized at the condensate surface. Autophagosome formation proceeds along the surface of the condensates using the Atg8–Atg19 interaction. In the case of the Ape1 P22L mutant, strong multivalent interactions cause aggregation of Ape1 with little liquidity, and autophagosome formation does not proceed. (B) Schematic illustration showing the degradation of oocyte-loaded PGL-1/PGL-3 by autophagy in somatic cells. SEPA-1 and EPG-2 are zygotically synthesized, and their levels exhibit temporal variation. P1 and P2 are germline blastomeres. Z2 and Z3 are germline precursor cells derived from the germline blastomere P4. Z2 and Z3 remain quiescent during embryogenesis. (C) Phase separation and transition control autophagic degradation or accumulation of PGL granules under different growth conditions. The concerted actions of SEPA-1 and EPG-2, together with EPG-11– and LET-363–mediated modifications of PGL-1 and PGL-3, ensure that the assembly rate, size, and material state of PGL granules are coordinated with autophagic flux during embryogenesis. Under heat stress conditions, this combination of enhanced PGL granule assembly and normal EPG-2 degradation result in insufficient EPG-2 to render PGL granules amenable to degradation.
A Pro-to-Leu mutation at residue 22 (P22L) in the Ape1 propeptide has been shown to impair the Cvt pathway. This mutation neither impairs the formation of Ape1 condensates nor severely reduces the interaction with Atg19 (Oda et al., 1996). The P22L mutation enhances the self-association of propeptides, resulting in the formation of amorphous condensates with almost no mobility (Fig. 3 A; Yamasaki et al., 2020). In the presence of Atg19, the Ape1 P22L condensates fail to deform Atg8-GUV membranes (Yamasaki et al., 2020), which suggests that the condensates must be in a gel-like state for selective sequestration of Ape1 condensates by IMs. The selective transport of Ape1 condensates to the vacuole is not severely impaired by the P22L mutation under starvation conditions, which activate autophagy. This difference is due to the different cargo dependency of the Cvt pathway and bulk autophagy; the former, but not the latter, absolutely requires Ape1 condensates for autophagosome formation (Shintani and Klionsky, 2004). These observations suggest that the gel-like property of the cargo condensate is important for its selective engulfment by the autophagosome-like double-membrane Cvt vesicles.
Phase separation and transition specify PGL granules for autophagic degradation
During C. elegans embryogenesis, specialized RNPs in the oocyte, known as P granules, are partitioned into both germline and somatic blastomeres during early asymmetric cell divisions (Strome, 2005). P granule proteins in somatic blastomeres are quickly disassembled and removed, resulting in exclusive localization of P granules in the germline blastomeres and eventually in the germline precursor cells Z2 and Z3 (Fig. 3 B; Strome, 2005; Zhang and Baehrecke, 2015). The RGG box–containing P granule proteins PGL-1 and PGL-3 are degraded by autophagy in somatic cells. In autophagy mutant embryos, PGL-1 and PGL-3 accumulate into a large number of aggregates in somatic cells, called PGL granules (Zhang et al., 2009). In wild-type C. elegans embryos, autophagy activity occurs at a basal level, and presumably, the autophagosomes are uniformly sized. Complete removal of the diffuse oocyte-loaded PGL-1/PGL-3 proteins, therefore, requires precisely controlled mechanisms to ensure that the assembly rate and size of PGL granules are coordinated with autophagic flux during embryogenesis (Wang and Zhang, 2019).
The receptor protein SEPA-1 facilitates LLPS-mediated assembly of PGL granules
The zygotically synthesized SEPA-1 acts as the receptor for formation and degradation of PGL granules (Zhang et al., 2009). In sepa-1 mutant embryos, PGL-1 and PGL-3 fail to be removed and are diffusely localized in the cytoplasm of somatic cells. SEPA-1 directly interacts with PGL-3 and also with the C. elegans Atg8 homologue LGG-1, bridging the PGL granule with autophagic structures (Zhang et al., 2009).
PGL-1 and PGL-3 proteins undergo LLPS in vitro. Mixing of PGL-1 and PGL-3 reduces the critical concentration of each protein for LLPS. LLPS of PGL-1/PGL-3 is further promoted by SEPA-1 in a concentration-dependent manner (Zhang et al., 2018). Co-addition of SEPA-1 greatly reduces the threshold concentration of PGL-1/PGL-3 for LLPS. SEPA-1 is homogenously distributed in the condensates (Zhang et al., 2018). PGL-1/PGL-3 droplets or PGL-1/PGL-3/SEPA-1 droplets possess liquid-like properties; they are spherical in shape, exhibit high internal mobility, fuse with each other, and become deformed when encountering a physical barrier (Zhang et al., 2018). During embryogenesis, SEPA-1 levels display a temporal pattern, becoming evident at the ∼24-cell stage, peaking at the ∼100-cell stage, and disappearing at the comma stage (Li et al., 2013; Zhang et al., 2009). PGL-1/PGL-3 levels are gradually reduced by degradation as development proceeds in early stage embryos (Zhang et al., 2009). Increased levels of SEPA-1 can promote condensation of low levels of diffuse PGL-1 and PGL-3 into aggregates for degradation.
EPG-11/PRMT1-mediated arginine methylation and LET-363/mTORC1-mediated phosphorylation modulate LLPS of PGL-1/-3
Degradation of PGL granules in embryos is modulated by PTMs. Loss of function of the C. elegans PRMT1 homologue EPG-11 causes a defect in autophagic degradation of PGL granules (Li et al., 2013). Loss of epg-11 activity also renders the formation of PGL granules independent of SEPA-1 (Li et al., 2013). EPG-11–mediated arginine methylation of PGL-1 and PGL-3 increases the threshold concentration of PGL-1/PGL-3 proteins for LLPS and reduces the droplet size (Zhang et al., 2018). PGL-1 and PGL-3 are also phosphorylated by the mTORC1 kinase LET-363 (Zhang et al., 2018). Phosphorylation of PGL-1 and PGL-3 reduces the critical concentrations required for LLPS. In let-363/mTORC1 mutant embryos, low levels of diffuse PGL-1 and PGL-3 proteins persist in the cytoplasm of somatic cells (Zhang et al., 2018). The tightly controlled levels of arginine methylation and phosphorylation ensure assembly of PGL granules in a strictly SEPA-1–dependent manner, which is essential for subsequent autophagic degradation and also for condensation and efficient removal of low levels of PGL-1 and PGL-3.
The scaffold protein EPG-2 mediates gelation of PGL granules
Degradation of PGL granules requires the scaffold protein EPG-2 (Tian et al., 2010). EPG-2 is zygotically synthesized, and its temporal expression pattern overlaps with SEPA-1, with which it directly interacts (Tian et al., 2010; Li et al., 2013). Co-addition of EPG-2 leads to transition of liquid PGL-1/PGL-3/SEPA-1 droplets to a gel-like state. The EPG-2–containing PGL-1/PGL-3/SEPA-1 droplets are spherical, have low mobility, and remain small in size over time due to very slow coalescence. Interestingly, EPG-2 does not mix into the droplets but rather coats the droplet surface (Zhang et al., 2018). In autophagy mutant embryos, EPG-2 encircles PGL granules and reduces the mobility of PGL proteins (Zhang et al., 2018). PGL granules do not colocalize with LGG-1–labeled autophagic structures in epg-2 mutants (Tian et al., 2010). This indicates that the biophysical properties of PGL granules specified by EPG-2 are essential for tethering the granules to autophagic structures.
The gel-like state of PGL granules is essential for their autophagic degradation
Gelation of PGL granules can be induced by mutations in the PGL-1 protein, including P55S, L82P, and E360K. Droplets composed of PGL-3, SEPA-1, and mutant PGL-1(P55S, L82P, or E360K) have gel-like properties, resembling EPG-2-containing droplets. PGL-1(P55S) and PGL-1(L82P) are incorporated into the droplets, while PGL-1(E360K) concentrates into a few small dots that localize on the surface and inside the droplets (Zhang et al., 2018). In embryos expressing mutant PGL-1(P55S, L82P, or E360K), autophagic degradation of PGL granules becomes largely independent of EPG-2 (Zhang et al., 2018). This indicates that the gelation status of PGL granules, which can be modulated by wild-type EPG-2 or mutant PGL-1, is essential for autophagic degradation of the granules. Therefore, the concerted actions of temporally expressed SEPA-1 and EPG-2, together with the effects of PTMs, modulate phase separation and transition of PGL-1 and PGL-3 for their efficient autophagic degradation (Fig. 3 C). Germline P granules exhibit liquid-like properties, which may be because they lack SEPA-1 and EPG-2 but contain multiple other RNA-binding proteins (RBPs) not present in PGL granules (Brangwynne et al., 2009; Strome, 2005).
LLPS mediates the assembly of aggregates containing p62 and polyubiquitinated proteins
Misfolded proteins are constitutively produced in cells. Their levels are elevated by genetic mutations, impairment of the folding machinery, and alterations of the intracellular environment (Alberti and Hyman, 2016; Mateju et al., 2017). Molecular chaperones assist refolding of proteins in aberrant conformations. When the capacity of the folding machinery is overwhelmed, misfolded proteins are removed by the ubiquitin-proteasome system (UPS) and the autophagy pathway to maintain cellular homeostasis. For degradation by autophagy, misfolded proteins are ubiquitinated and form aggregates, a process that is mediated by a family of receptor proteins such as p62/sequestosome 1 (Komatsu et al., 2007). p62 contains a self-polymerization PB1 domain, a ubiquitin-associating (UBA) domain, and an LC3/Atg8-interacting region (Bjørkøy et al., 2005; Komatsu et al., 2007; Pankiv et al., 2007).
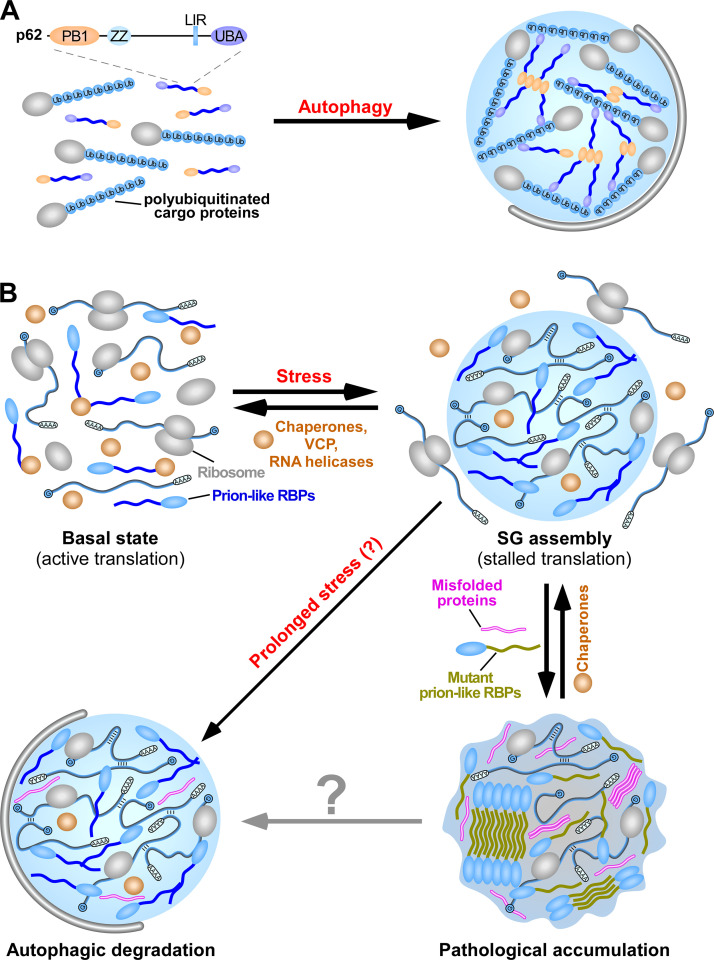
Phase separation occurs when p62 is mixed in vitro with substrates carrying two or more ubiquitin chains (each chain containing three or more ubiquitin moieties) or with long ubiquitin chains that are not attached to proteins (Fig. 4 A; Sun et al., 2018; Zaffagnini et al., 2018). The phase separation requires PB1 domain–mediated polymerization and the p62–ubiquitin interaction and is modulated by the valency of the polyubiquitin chains and the p62–polyubiquitin chain binding affinity (Sun et al., 2018; Zaffagnini et al., 2018). p62 condensate formation is facilitated by increasing the binding affinity of p62, such as through phosphorylation of Ser 403 in the UBA domain, and it is attenuated by reducing the binding affinity of p62, such as through the M404T and G411S mutations in the UBA domain, which are found in Paget’s disease of bone (Danieli and Martens, 2018; Sun et al., 2018; Zaffagnini et al., 2018). In condensates formed in vitro, p62 forms filaments that are cross-linked by polyubiquitin chains. p62 is relatively static, while polyubiquitin chains exhibit high mobility (Sun et al., 2018; Zaffagnini et al., 2018). In living cells, p62 shows a higher mobility in condensates (Sun et al., 2018; Zaffagnini et al., 2018), which suggests that other cellular factors such as chaperones and/or modifications modulate the fluidity of condensates.
Figure 4.
Phase separation of p62-polyubiquitinated protein aggregates and SGs. (A) LLPS mediates the assembly of condensates containing p62 and polyubiquitinated proteins. p62 polymerizes and interacts with ubiquitin to drive LLPS. In the condensates, p62 forms filaments via PB1 polymerization, while the polyubiquitinated proteins cross-link the p62 filaments via ubiquitin–UBA domain interaction. p62 aggregates are selectively enclosed by autophagosomal membranes. LIR, LC3-interacting region; PB1, self-polymerization Phox and Bem1 (PB1) domain; UBA, UBA domain; ZZ, ZZ-type zinc-finger domain. (B) SGs are assembled via LLPS. SGs contain translationally stalled mRNAs, associated preinitiation factors, and various prion-like RBPs. Once the SG-inducing stress is mitigated, SGs rapidly disassemble with the assistance of chaperone proteins, VCP, and RNA helicases. A subset of SGs that fail to disassemble are subjected to autophagic degradation. Incorporation of misfolded proteins or mutations in SG-resident prion-like RBPs causes aberrant transition of SGs into solid fibrillar structures, resulting in their accumulation.
Degradation of p62 condensates is promoted by Alfy, the BEACH-containing protein WDR81, and the PB1 and UBA domain–containing protein NBR1, all of which associate with p62 and LC3 (Clausen et al., 2010; Filimonenko et al., 2010; Kirkin et al., 2009; Liu et al., 2017). These factors may facilitate phase separation and/or their interactions with IMs. The p62 condensates show gel-like behaviors, namely slow fusion and low mobility (Sun et al., 2018; Zaffagnini et al., 2018), which suggests that the gel-like state may ensure efficient autophagic degradation of these condensates. Consistent with the notion that higher liquidity is not favorable for degradation, overexpression of NBR1, which promotes p62-mediated phase separation and also dramatically increases the mobility of p62 in the condensates, impairs autophagic degradation of p62 condensates (Sánchez-Martín et al., 2020; Zaffagnini et al., 2018). Aberrant p62-positive aggregates, such as Mallory–Denk bodies, α1 antitrypsin aggregates, Lewy bodies, and huntingtin aggregates, accumulate in a diversity of diseases (Yamamoto and Simonsen, 2011). These aggregates are enriched in disease-specific constituents as well as p62. It remains largely unknown how the different constituents specify the formation and material properties of p62 condensates that can escape from autophagic degradation.
Phase separation mediates the assembly of SGs
When cells experience adverse conditions such as heat, osmotic stress, oxidative stress, and proteasome inhibition, a stress-adaptive response occurs in which translationally stalled mRNAs, associated preinitiation factors, and other proteins assemble into SGs (Guzikowski et al., 2019; Kedersha et al., 2013; Panas et al., 2016; Riback et al., 2017). SGs are heterogeneous in composition. The scaffold factor that nucleates SG assembly varies in different cell types and under different stresses (Guzikowski et al., 2019). For example, the RBP G3BP1 is essential for SG assembly in response to arsenite treatment, but not to heat shock or osmotic stress (Yang et al., 2020). Prion-like RBPs, including hnRNPA1, hnRNPA2, TIA1, TDP-43, and FUS, undergo stress-triggered nucleus-to-cytoplasm transport and accumulate in SGs (Guzikowski et al., 2019). Overexpression of SG-resident prion-like RBPs can also lead to SG formation. Upon stress cessation, the translationally stalled mRNAs can resume translation.
SGs are assembled via LLPS that is driven by protein–protein, RNA–protein, and RNA–RNA intermolecular interactions (Fig. 4 B). The LCD-harboring prion-like RBPs phase separate into liquid droplets that further transition into hydrogels and solid fibrillar structures in vitro (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015; Riback et al., 2017). RNAs play dual roles in regulating SG assembly. Low RNA/protein ratios facilitate, while high RNA/protein ratios inhibit, LLPS of prion-like RBPs (e.g., TDP-43, FUS, and hnRNPA1; Lin et al., 2015; Maharana et al., 2018; Molliex et al., 2015).
The dynamics and material state of SGs are influenced by molecular composition, chaperone-mediated surveillance system, RNA, and PTMs (Fig. 4 B; Kroschwald et al., 2015). Misfolded proteins, such as defective ribosomal products resulting from prematurely terminated polypeptides, are gradually recruited into SGs under severe or prolonged stress conditions. Incorporation of misfolded proteins reduces the dynamics and disassembly kinetics of SGs (Ganassi et al., 2016; Kroschwald et al., 2015; Mateju et al., 2017). The activity of chaperones such as HSP70 and HSP27 counteracts the accumulation of misfolded proteins in SGs and facilitates SG disassembly when the stress is mitigated (Ganassi et al., 2016; Kroschwald et al., 2015; Mateju et al., 2017; Panas et al., 2016). The SG-resident AAA-ATPase VCP promotes SG disassembly, probably by facilitating extraction of ubiquitinated proteins from SGs (Buchan et al., 2013; Mateju et al., 2017). The autophagy proteins ULK1 and ULK2 also accumulate in SGs, in which they phosphorylate and activate VCP to promote SG dynamics (Wang et al., 2019). The nuclear transport receptors Karyopherin-β2 (Kapβ2; also known as transportin-1) for proteins harboring the C-terminal Pro-Tyr NLS, and the Importin-α/Kapβ1 complex for proteins harboring a classic NLS also act as chaperones to prevent and/or reverse phase separation and fibrillization of Pro-Tyr NLS–containing RBPs (e.g., FUS, TAF15, EWSR1, hnRNPA1, and hnRNPA2), and classic NLS-containing protein such as TDP-43, respectively (Guo et al., 2018; Hofweber et al., 2018; Qamar et al., 2018; Yoshizawa et al., 2018). In living cells, Kapβ2 inhibits FUS, hnRNPA1, and hnRNPA2 accumulation in SGs (Guo et al., 2018; Hofweber et al., 2018). By modulating intermolecular RNA–RNA interactions, ATP-dependent DEAD-box RNA helicases such as eIF4A act as RNA chaperones to counteract the recruitment of RNAs to SGs and reduce SG formation (Tauber et al., 2020). The SG-localized protein Ubiquillin-2 (UBQLN2) promotes the dynamics of protein–RNA interaction and thus negatively regulates SG formation (Alexander et al., 2018). PTMs of SG components also modulate phase separation and transition of SGs. Multivalent poly(ADP-ribose) promotes LLPS of poly(ADP-ribose)-binding prion-like RBPs such as FUS and TDP-43 and their accumulation in SGs (Leung et al., 2011; McGurk et al., 2018; Patel et al., 2015). DNA-dependent protein kinase–mediated phosphorylation of the FUS LCD inhibits phase separation (Monahan et al., 2017; Murray et al., 2017). Arginine methylation of RG/RGG repeats, which disrupts arginine-mediated cation-π intermolecular interactions, also impairs phase separation and solidification of FUS and hnRNPA2 and reduces their SG association (Hofweber et al., 2018; Qamar et al., 2018; Ryan et al., 2018). Thus, multiple mechanisms are employed to modulate the formation and dynamics of SGs.
SGs are selectively degraded by autophagy
Once the adverse stress is alleviated, the majority of SGs rapidly disassemble with the assistance of chaperone proteins such as the HSPB8–BAG3–HSP70 complex (Ganassi et al., 2016). A subset of SGs, which may undergo slow or no disassembly, are targeted for autophagic degradation (Fig. 4 B; Buchan et al., 2013; Ganassi et al., 2016; Chitiprolu et al., 2018; Mateju et al., 2017). p62, which is dispensable for SG formation, associates with C9ORF72 to mediate autophagic degradation of SGs (Chitiprolu et al., 2018). Ubiquitinated proteins are not enriched in SGs. Instead, the p62–C9ORF72 complex, together with the Tudor protein SMN, recognizes symmetrically methylated arginines in SG-resident proteins such as FUS (Chitiprolu et al., 2018). VCP also facilitates autophagic degradation of SGs via an unknown mechanism (Buchan et al., 2013; Mateju et al., 2017). Dysfunctional autophagy, or mutations in genes encoding proteins involved in SG degradation, such as p62 and VCP, causes persistence of SGs (Buchan et al., 2013). The mechanism that makes a specific subset of SGs amenable to autophagic degradation is unknown. The material state may be a key determining factor.
Accumulation of SGs has been associated with pathogenesis of a group of degenerative diseases, including inclusion body myopathy, amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and multisystem proteinopathy (Aguzzi and Altmeyer, 2016; Alberti and Hyman, 2016). Mutations in SG-resident prion-like RBPs, including TDP-43, FUS, hnRNPA1, hnRNPA2B1, hnRNPDL, and TIA-1, accelerate the aberrant transition of SGs into solid fibrillar structures, resulting in their accumulation (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Niaki et al., 2020; Patel et al., 2015; Riback et al., 2017; Ryan et al., 2018). For FUS, different ALS/FTD-linked mutations promote phase transition via distinct mechanisms such as diminishing dynamic protein–RNA binding (e.g., R244C missense mutation in FUS), accelerating aberrant phase transition (e.g., G156E missense mutation in FUS), or increasing its cytoplasmic concentration and rendering it less sensitive to the chaperone activity of Kapβ2 (e.g., mutations in the Pro-Tyr NLS of FUS; Hofweber et al., 2018; Niaki et al., 2020; Patel et al., 2015). ALS-linked mutations in UBQLN2 impair the ability of UBQLN2 to regulate FUS–RNA complex dynamics (Alexander et al., 2018). The ALS-associated repeat expansions in an intron of C9ORF72 result in transcription and translation of repetitive RNAs and dipeptide repeats. The arginine-rich dipeptide repeats promote liquid-to-solid transition of FUS droplets and reduce the dynamics of SGs (Boeynaems et al., 2017; Lee et al., 2016). SGs in ALS and FTD also contain high levels of RBPs with altered PTMs that facilitate phase transition, such as hyperphosphorylation of TDP-43 and loss of arginine methylation of FUS (Hofweber et al., 2018). Aberrant phase transition renders SGs less susceptible to autophagic degradation and more susceptible to subsequent pathological accumulation.
Assembly of other autophagy substrates via LLPS
Amyloids formed by the microtubule-associated protein Tau accumulate in several neurodegenerative diseases such as Alzheimer’s disease. Multivalent homotypic interactions of the lysine-rich microtubule-binding repeats (three or four repeats resulting from alternative splicing) of Tau drive LLPS. Phosphorylation of Ser residues in the repeat by microtubule-associated protein/microtubule affinity–regulating kinase or coacervation with negatively charged molecules such as heparin or RNA facilitates LLPS of Tau repeats and also enhances their transition into amyloid-like structures (Ambadipudi et al., 2017). A polyglutamine-expanded exon 1 fragment of the huntingtin protein (HTTex1) forms aggregates in Huntington’s disease. LLPS of HTTex1 is driven by weak hydrophobic interactions conferred by the polyQ tract and a Pro-rich region (Peskett et al., 2018). When the polyQ tract in HTTex1 reaches disease-associated lengths, the condensates transition into solid-like fibrillar assemblies in living cells. Autophagy also mediates degradation of Tau and HTTex1 both in cell lines and animal models (Iwata et al., 2005; Krüger et al., 2012; Qin et al., 2003; Schaeffer et al., 2012). Aberrant phase transition of Tau repeats and polyQ tracts may impair their autophagic degradation.
Phase separation and transition control autophagic degradation or accumulation of protein condensates under different developmental or stress conditions
Phosphorylation-mediated disassembly of amyloid RIM4 fibrils for degradation
The dynamic assembly and clearance of amyloid-like aggregates of the translational repressor Rim4 in yeast is essential for progression through meiotic divisions (Berchowitz et al., 2015). During meiosis I, monomeric Rim4 converts into amyloid-like aggregates, a process that is mediated by its IDRs. These aggregates repress the translation of mRNAs critical for meiosis II progression such as the B-type cyclin CLB3 and the meiotic ubiquitin ligase activator AMA1 (Berchowitz et al., 2015). At the onset of meiosis II, amyloid-like Rim4 aggregates are disassembled, a process triggered by multisite phosphorylation in its IDRs by Ime2 (Carpenter et al., 2018). Rim4 is then degraded by autophagy and/or the UPS to enable translation of target mRNAs (Carpenter et al., 2018; Wang et al., 2020). The number of phosphorylated sites in the IDRs may cause the disassembly of Rim4 fibers into monomers or oligomers that are degraded by the UPS and autophagy, respectively (Carpenter et al., 2018). The coordinated actions of the UPS and autophagy ensure the rapid removal of Rim4 during the short interval between meiosis I and II.
Modulation of phase separation of PGL granules for stress adaptation
Protein condensates that are normally removed by autophagy can escape from degradation under harsh growth conditions. In C. elegans embryos laid by animals grown under heat stress, PGL-1/PGL-3 proteins fail to be degraded and instead accumulate into a large number of granules (Fig. 3 C). LET-363/mTORC1 signaling is required for PGL granules to evade degradation under heat stress (Zhang et al., 2018). Phosphorylation of PGL-1 and PGL-3 by LET-363, whose levels are greatly elevated under heat stress, promotes assembly of PGL granules; the droplet size is increased and the critical concentration of PGL-1/PGL-3 undergoing LLPS is decreased (Zhang et al., 2018). EPG-2, as in embryos at ambient temperatures, undergoes autophagic degradation during heat stress. This combination of enhanced PGL granule formation and normal EPG-2 degradation means that there is insufficient EPG-2 to render PGL granules amenable to degradation. Accumulation of PGL granules in embryos under heat stress is suppressed by inactivation of mTORC1 or overexpression of EPG-2. Heat-stress–triggered accumulation of PGL granules promotes embryonic survival (Zhang et al., 2018). PGL granules may participate in RNA biogenesis or sequestrate cell death factors. Thus, levels of mTORC1-mediated phosphorylation of PGL-1/PGL-3 are tightly controlled to ensure their degradation or accumulation to provide a fitness advantage under heat stress.
Conclusion and perspectives
Recent studies demonstrate that phase separation and transition act at different steps of autophagy. LLPS of Atg proteins mediates the organization of autophagosome formation sites in yeast. In multicellular organisms, it has yet to be determined whether the autophagy initiation complexes are also assembled via LLPS and, if so, how the autophagy induction signals trigger LLPS of the ULK1 complex and how distinct ATG protein complexes are dynamically recruited. LLPS also triages protein cargoes for selective autophagic degradation, which involves the tight association of condensates with the surrounding IMs. Many questions about LLPS-mediated assembly of autophagy substrates remain to be addressed. Different types of protein condensates have distinct compositions. For example, in C. elegans, PGL granules are distinct from SQST-1/p62 aggregates (Tian et al., 2010). How is the composition of distinct phase-separated condensates specified in living cells? Phase-separated condensates occur in a range of states, from liquid-like to solid structures, which are modulated by composition, PTMs, and mutations in key components. Delivery of Ape1 condensates and PGL condensates to the vacuole/lysosomes indicates that the gel-like state is essential for building up surrounding membranes. The gel-like state may be a common theme in selective cargo clearance by autophagy. Liquid-like and solid condensates may fail to elicit membrane shape changes during IM expansion. The rapid coalescence of liquid-like condensates may also lead to excessively large structures that exceed the relatively fixed autophagosome size. How do animal cells sense and integrate stresses into specification of the material properties of protein condensates? Phase separation and transition determine the autophagic degradation or accumulation of PGL granules under normal and stress conditions. Is this mechanism generally employed in other systems under certain growth conditions to induce accumulation of proteins that are normally degraded?
Studies of selective autophagy of protein condensates in yeast and C. elegans model systems also provide insights into the regulation and function of LLPS in diseases. Aberrant solidification of protein condensates, resulting from mutations in a key constituent protein or impaired chaperone surveillance, inhibits their efficient removal by autophagy. Accumulation of solid protein condensates is an early and critical process in the pathogenesis of protein aggregation diseases. Investigating the specification and regulation of the biophysical properties of protein condensates will help the design of therapeutic interventions for mitigation of aberrant protein aggregate accumulation.
Acknowledgments
We are grateful to Dr. Isabel Hanson for editing work.
Work in H. Zhang’s laboratory was supported by the Beijing Municipal Science and Technology Commission (grant Z181100001318003), the National Natural Science Foundation of China (grant 31871426 to Z. Wang and grants 31421002, 31561143001, 31630048, and 31790403 to H. Zhang), the Chinese Ministry of Science and Technology (grant 2017YFA0503401), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB19000000), and the Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (grant QYZDY-SSW-SMC006). Work in N.N. Noda’s laboratory was supported by the Japan Society for the Promotion of Science KAKENHI (grants 18H03989, 18H02099, and 19H05707) and Japan Science and Technology Agency CREST (grant JPMJCR13M7).
The authors declare no competing financial interests.
Author contributions: N.N. Noda, Z. Wang, and H. Zhang wrote and edited the manuscript.
References
- Aguzzi A., and Altmeyer M.. 2016. Phase Separation: Linking Cellular Compartmentalization to Disease. Trends Cell Biol. 26:547–558. [DOI] [PubMed] [Google Scholar]
- Alberti S., and Hyman A.A.. 2016. Are aberrant phase transitions a driver of cellular aging? BioEssays. 38:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander E.J., Ghanbari Niaki A., Zhang T., Sarkar J., Liu Y., Nirujogi R.S., Pandey A., Myong S., and Wang J.. 2018. Ubiquilin 2 modulates ALS/FTD-linked FUS-RNA complex dynamics and stress granule formation. Proc. Natl. Acad. Sci. USA. 115:E11485–E11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M.. 2017. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., and Rosen M.K.. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L.E., Kabachinski G., Walker M.R., Carlile T.M., Gilbert W.V., Schwartz T.U., and Amon A.. 2015. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 163:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron-Sandoval L.P., Safaee N., and Michnick S.W.. 2016. Mechanisms and Consequences of Macromolecular Phase Separation. Cell. 165:1067–1079. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T.. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V.H., et al. 2017. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell. 65:1044–1055.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., et al. 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.M., Taylor J.P., and Parker R.. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 153:1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K., Bell R.B., Yunus J., Amon A., and Berchowitz L.E.. 2018. Phosphorylation-Mediated Clearance of Amyloid-like Assemblies in Meiosis. Dev. Cell. 45:392–405.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., and Klionsky D.J.. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 19:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitiprolu M., Jagow C., Tremblay V., Bondy-Chorney E., Paris G., Savard A., Palidwor G., Barry F.A., Zinman L., Keith J., et al. 2018. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 9:2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.H., Lamark T., Isakson P., Finley K., Larsen K.B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., et al. 2010. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 6:330–344. [DOI] [PubMed] [Google Scholar]
- Danieli A., and Martens S.. 2018. p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J. Cell Sci. 131 jcs214304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M., Isakson P., Finley K.D., Anderson M., Jeong H., Melia T.J., Bartlett B.J., Myers K.M., Birkeland H.C., Lamark T., et al. 2010. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 38:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Suzuki S.W., Yamamoto H., Kondo-Kakuta C., Kimura Y., Hirano H., Akada R., Inagaki F., Ohsumi Y., and Noda N.N.. 2014. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 21:513–521. [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Alam J.M., Noshiro D., Mouri K., Ando T., Okada Y., May A.I., Knorr R.L., Suzuki K., Ohsumi Y., et al. 2020. Phase separation organizes the site of autophagosome formation. Nature. 578:301–305. [DOI] [PubMed] [Google Scholar]
- Ganassi M., Mateju D., Bigi I., Mediani L., Poser I., Lee H.O., Seguin S.J., Morelli F.F., Vinet J., Leo G., et al. 2016. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol. Cell. 63:796–810. [DOI] [PubMed] [Google Scholar]
- Guo L., Kim H.J., Wang H., Monaghan J., Freyermuth F., Sung J.C., O’Donovan K., Fare C.M., Diaz Z., Singh N., et al. 2018. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell. 173:677–692.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzikowski A.R., Chen Y.S., and Zid B.M.. 2019. Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA. 10 e1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.D., Simons M., Niessing D., Madl T., et al. 2018. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell. 173:706–719.e13. [DOI] [PubMed] [Google Scholar]
- Hollenstein D.M., Gómez-Sánchez R., Ciftci A., Kriegenburg F., Mari M., Torggler R., Licheva M., Reggiori F., and Kraft C.. 2019. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J. Cell Sci. 132 jcs235002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., and Mizushima N.. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A., Riley B.E., Johnston J.A., and Kopito R.R.. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280:40282–40292. [DOI] [PubMed] [Google Scholar]
- Kaganovich D. 2017. There Is an Inclusion for That: Material Properties of Protein Granules Provide a Platform for Building Diverse Cellular Functions. Trends Biochem. Sci. 42:765–776. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., and Ohsumi Y.. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber R.A., Shoemaker C.J., and Denic V.. 2015. Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol. Cell. 59:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Yang Y.S., Sutter B.M., Wang Y., McKnight S.L., and Tu B.P.. 2019. Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell. 177:711–721.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Scott S.V., Oda M.N., and Klionsky D.J.. 1997. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163. [DOI] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S.. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 4 e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger U., Wang Y., Kumar S., and Mandelkow E.M.. 2012. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging. 33:2291–2305. [DOI] [PubMed] [Google Scholar]
- Ktistakis N.T., and Tooze S.A.. 2016. Digesting the Expanding Mechanisms of Autophagy. Trends Cell Biol. 26:624–635. [DOI] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A., et al. 2016. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 167:774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., and Chang P.. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 42:489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., and Virgin H.W.. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature. 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yang P., Tian E., and Zhang H.. 2013. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol. Cell. 52:421–433. [DOI] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., and Parker R.. 2015. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.Y., and Sabatini D.M.. 2020. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21:183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li Y., Wang X., Xing R., Liu K., Gan Q., Tang C., Gao Z., Jian Y., Luo S., et al. 2017. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J. Cell Biol. 216:1301–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M.A., and Klionsky D.J.. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584:1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Otomo C., and Otomo T.. 2019. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife. 8 e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M., et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 360:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D., Franzmann T.M., Patel A., Kopach A., Boczek E.E., Maharana S., Lee H.O., Carra S., Hyman A.A., and Alberti S.. 2017. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36:1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R.G., Shorter J., and Bonini N.M.. 2018. Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol. Cell. 71:703–717.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T.J., Lystad A.H., and Simonsen A.. 2020. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 219 e202002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu G., Eapen V.V., Yang Y., Klionsky D.J., and Haber J.E.. 2019. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc. Natl. Acad. Sci. USA. 116:1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies F.M., Fleming A., Caricasole A., Bento C.F., Andrews S.P., Ashkenazi A., Füllgrabe J., Jackson A., Jimenez Sanchez M., Karabiyik C., et al. 2017. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron. 93:1015–1034. [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2018. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20:521–527. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. [DOI] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H., O’Meally R., Dignon G.L., Conicella A.E., Zheng W., et al. 2017. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36:2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T., Michel C.H., et al. 2015. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 88:678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D.T., Kato M., Lin Y., Thurber K.R., Hung I., McKnight S.L., and Tycko R.. 2017. Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell. 171:615–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y.. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467. [DOI] [PubMed] [Google Scholar]
- Niaki A.G., Sarkar J., Cai X., Rhine K., Vidaurre V., Guy B., Hurst M., Lee J.C., Koh H.R., Guo L., et al. 2020. Loss of Dynamic RNA Interaction and Aberrant Phase Separation Induced by Two Distinct Types of ALS/FTD-Linked FUS Mutations. Mol. Cell. 77:82–94.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., and Fujioka Y.. 2015. Atg1 family kinases in autophagy initiation. Cell. Mol. Life Sci. 72:3083–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., and Mizushima N.. 2016. Atg101: Not Just an Accessory Subunit in the Autophagy-initiation Complex. Cell Struct. Funct. 41:13–20. [DOI] [PubMed] [Google Scholar]
- Oda M.N., Scott S.V., Hefner-Gravink A., Caffarelli A.D., and Klionsky D.J.. 1996. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 132:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., and Noda N.N.. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26:281–288. [DOI] [PubMed] [Google Scholar]
- Panas M.D., Ivanov P., and Anderson P.. 2016. Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to PTM/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. [DOI] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- Peskett T.R., Rau F., O’Driscoll J., Patani R., Lowe A.R., and Saibil H.R.. 2018. A Liquid to Solid Phase Transition Underlying Pathological Huntingtin Exon1 Aggregation. Mol. Cell. 70:588–601.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Ferguson S.M., Brugarolas J., and Ballabio A.. 2018. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Ströhl F., et al. 2018. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell. 173:720–734.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z.H., Wang Y., Kegel K.B., Kazantsev A., Apostol B.L., Thompson L.M., Yoder J., Aronin N., and DiFiglia M.. 2003. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Genet. 12:3231–3244. [DOI] [PubMed] [Google Scholar]
- Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., and Drummond D.A.. 2017. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 168:1028–1040.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R.C., Yuan H.X., and Guan K.L.. 2014. Autophagy regulation by nutrient signaling. Cell Res. 24:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan V.H., Dignon G.L., Zerze G.H., Chabata C.V., Silva R., Conicella A.E., Amaya J., Burke K.A., Mittal J., and Fawzi N.L.. 2018. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell. 69:465–479.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Martín P., Sou Y.S., Kageyama S., Koike M., Waguri S., and Komatsu M.. 2020. NBR1-mediated p62-liquid droplets enhance the Keap1-Nrf2 system. EMBO Rep. 21 e48902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer V., Lavenir I., Ozcelik S., Tolnay M., Winkler D.T., and Goedert M.. 2012. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 135:2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., and Brangwynne C.P.. 2017. Liquid phase condensation in cell physiology and disease. Science. 357 eaaf4382. [DOI] [PubMed] [Google Scholar]
- Shintani T., and Klionsky D.J.. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.P., Stromhaug P.E., and Klionsky D.J.. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead W.T., and Gladfelter A.S.. 2019. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Mol. Cell. 76:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. [DOI] [PubMed] [Google Scholar]
- Strome S. 2005. Specification of the germ line. WormBook. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wu R., Zheng J., Li P., and Yu L.. 2018. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., and Ohsumi Y.. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218. [DOI] [PubMed] [Google Scholar]
- Takahara T., and Maeda T.. 2012. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell. 47:242–252. [DOI] [PubMed] [Google Scholar]
- Tauber D., Tauber G., Khong A., Van Treeck B., Pelletier J., and Parker R.. 2020. Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell. 180:411–426.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055. [DOI] [PubMed] [Google Scholar]
- Torggler R., Papinski D., Brach T., Bas L., Schuschnig M., Pfaffenwimmer T., Rohringer S., Matzhold T., Schweida D., Brezovich A., et al. 2016. Two Independent Pathways within Selective Autophagy Converge to Activate Atg1 Kinase at the Vacuole. Mol. Cell. 64:221–235. [DOI] [PubMed] [Google Scholar]
- Valverde D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., and Melia T.J.. 2019. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218:1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., and Zhang H.. 2019. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends Cell Biol. 29:417–427. [DOI] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., et al. 2016. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell. 63:781–795. [DOI] [PubMed] [Google Scholar]
- Wang B., Maxwell B.A., Joo J.H., Gwon Y., Messing J., Mishra A., Shaw T.I., Ward A.L., Quan H., Sakurada S.M., et al. 2019. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell. 74:742–757.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhang R., Feng W., Tsuchiya D., Ballew O., Li J., Denic V., and Lacefield S.. 2020. Autophagy of an Amyloid-like Translational Repressor Regulates Meiotic Exit. Dev. Cell. 52:141–151.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., and Pelkmans L.. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 152:791–805. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., and Simonsen A.. 2011. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol. Dis. 43:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Fujioka Y., Suzuki S.W., Noshiro D., Suzuki H., Kondo-Kakuta C., Kimura Y., Hirano H., Ando T., Noda N.N., et al. 2016. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell. 38:86–99. [DOI] [PubMed] [Google Scholar]
- Yamasaki A., Watanabe Y., Adachi W., Suzuki K., Matoba K., Kirisako H., Kumeta H., Nakatogawa H., Ohsumi Y., Inagaki F., et al. 2016. Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates. Cell Rep. 16:19–27. [DOI] [PubMed] [Google Scholar]
- Yamasaki A., Alam J.M., Noshiro D., Hirata E., Fujioka Y., Suzuki K., Ohsumi Y., and Noda N.N.. 2020. Liquidity Is a Critical Determinant for Selective Autophagy of Protein Condensates. Mol. Cell. 77:1163–1175.e9. [DOI] [PubMed] [Google Scholar]
- Yang Y.S., Kato M., Wu X., Litsios A., Sutter B.M., Wang Y., Hsu C.H., Wood N.E., Lemoff A., Mirzaei H., et al. 2019. Yeast Ataxin-2 Forms an Intracellular Condensate Required for the Inhibition of TORC1 Signaling during Respiratory Growth. Cell. 177:697–710.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Mathieu C., Kolaitis R.M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q., et al. 2020. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell. 181:325–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.Y., Wrasman K., and Herman P.K.. 2010. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 185:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T., Ali R., Jiou J., Fung H.Y.J., Burke K.A., Kim S.J., Lin Y., Peeples W.B., Saltzberg D., Soniat M., et al. 2018. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell. 173:693–705.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., Peterbauer T., Sztacho M., Trapannone R., Tarafder A.K., et al. 2018. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37 e98308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M., Shang Y., Araki Y., Guo T., Huganir R.L., and Zhang M.. 2016. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell. 166:1163–1175.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. 2020. Lipid transfer at ER-isolation membrane contacts. Nat. Rev. Mol. Cell Biol. 21:121. [DOI] [PubMed] [Google Scholar]
- Zhang H., and Baehrecke E.H.. 2015. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 25:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321. [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang Z., Du Z., and Zhang H.. 2018. mTOR Regulates Phase Separation of PGL Granules to Modulate Their Autophagic Degradation. Cell. 174:1492–1506.e22. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., and Zhang H.. 2018. Formation and maturation of autophagosomes in higher eukaryotes: a social network. Curr. Opin. Cell Biol. 53:29–36. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., and Zhang H.. 2019a Core autophagy genes and human diseases. Curr. Opin. Cell Biol. 61:117–125. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., and Zhang H.. 2019b Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 218:757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell. 67:974–989.e6. [DOI] [PubMed] [Google Scholar]