Autophagosomes engulf a variety of targets, from a portion of cytosol to large organelles, by regulating the size of the autophagosomal membrane. Yamamoto et al. identify ERdj8, a novel ER membrane protein that affects the size of autophagosomes.
Abstract
In macroautophagy, membrane structures called autophagosomes engulf substrates and deliver them for lysosomal degradation. Autophagosomes enwrap a variety of targets with diverse sizes, from portions of cytosol to larger organelles. However, the mechanism by which autophagosome size is controlled remains elusive. We characterized a novel ER membrane protein, ERdj8, in mammalian cells. ERdj8 localizes to a meshwork-like ER subdomain along with phosphatidylinositol synthase (PIS) and autophagy-related (Atg) proteins. ERdj8 overexpression extended the size of the autophagosome through its DnaJ and TRX domains. ERdj8 ablation resulted in a defect in engulfing larger targets. C. elegans, in which the ERdj8 orthologue dnj-8 was knocked down, could perform autophagy on smaller mitochondria derived from the paternal lineage but not the somatic mitochondria. Thus, ERdj8 may play a critical role in autophagosome formation by providing the capacity to target substrates of diverse sizes for degradation.
Introduction
In macroautophagy, an intracellular degradation pathway, a double-membrane organelle called the autophagosome engulfs a variety of targets and delivers them to the lysosome/vacuole for degradation (Yang and Klionsky, 2010; Galluzzi et al., 2017). In the case of nonselective macroautophagy, autophagosomes are relatively homogenous in size (Mizushima, 2004). In selective autophagy, autophagosome size is rather heterogeneous. In the Cvt pathway in yeast, which enwraps the super complex of aminopeptidase I in an autophagy-related structure, the diameter is smaller (∼150 nm; Baba et al., 1997). By contrast, in the case of bacteria-targeted autophagy, larger autophagosomes (5–10 µm) can form (Singh et al., 2006; Yamaguchi et al., 2009). These observations raise two important questions: How is the size of selective autophagosomes determined, and why are nonselective autophagosomes mostly uniform in size? One study proposed that autophagosome size is determined by the total expression level of Atg8 (Xie et al., 2008). Another group proposed that actin assembly inside the autophagosome determines its shape (Mi et al., 2015). Despite these advances, we still do not fully understand how autophagosome size is determined.
The ER plays an important role as a platform for autophagosome formation (Axe et al., 2008; Ylä-Anttila et al., 2009; Hayashi-Nishino et al., 2009). ERdj proteins, a family of ER-localized DnaJ-like proteins, contain a J-domain that binds binding immunoglobulin protein (BiP/GRP-78) and promotes its ATPase activity (Kampinga and Craig, 2010; Otero et al., 2010). We have shown that some of the ERdjs are involved in protein folding, ER-associated protein degradation, or Ca2+ homeostasis (Yamamoto et al., 2010; Ushioda et al., 2016). In this study, we characterized ERdj8, the novel eighth member of the ERdj family that affects the size of the autophagosome membrane.
Results
ERdj8 is concentrated in a subdomain of the ER with the autophagic machineries
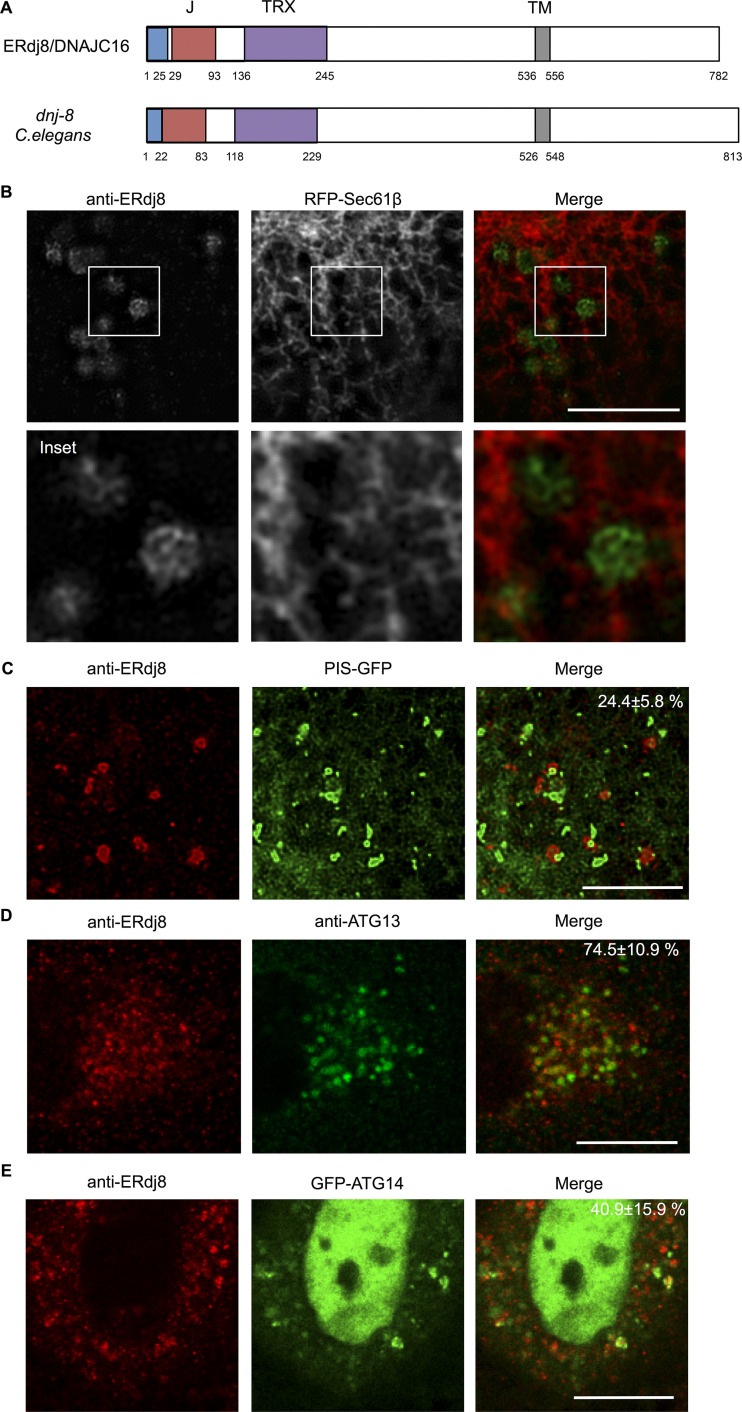
ERdj8/DNAJC16 is a type 1 membrane protein that contains a single transmembrane stretch along with DnaJ and thioredoxin-like domains (Fig. 1 A and data not shown). It was distributed within the ER network along with the ER luminal marker DsRed-KDEL and formed prominent puncta (Fig. S1 A, white arrows). Observation using a specific antibody against ERdj8 and structured illumination microscopy revealed that these punctate ERdj8-positive structures exhibited a dense meshwork morphology (Fig. 1 B). Importantly, these structures slightly excluded, but were still positive for, the typical ER marker protein GFP-Sec61β, indicating they were connected to the ER network and constituted a part of it (Fig. 1 B). These data suggest the existence of a specialized subdomain of the ER where ERdj8 primarily resides.
Figure 1.
ERdj8 is concentrated in ER subdomains. (A) Schematic diagram of ERdj8/DNAJC16 (Human) and dnj-8 (C. elegans). Blue, signal peptide; red, DnaJ domain (J); purple, thioredoxin-like domain (TRX); and gray, transmembrane region (TM). (B) COS-7 cells transfected with RFP-Sec61β were stained with anti-ERdj8 and imaged on SpinSR10. Insets, enlargements of framed regions. Scale bar, 10 µm. (C) COS-7 cells stably expressing PIS-GFP were starved for 2 h, stained with anti-ERdj8, and imaged on SpinSR10. Scale bar, 5 µm. The number is the percentage and SD of PIS-GFP–positive structures among ERdj8 structures per cell (n = 7). (D) HeLa cells were starved for 1 h, immunolabeled with ERdj8 and ATG13, and imaged on an SP-8. The number is the percentage and SD of ATG13-positive puncta among ERdj8 structures per cell (n = 10). Scale bar, 10 µm. (E) HeLa cells transfected with GFP-ATG14 were starved for 1 h, immunolabeled with ERdj8, and imaged on SP-8. The number is the percentage and SD of GFP-ATG14–positive structures among ERdj8 structures per cell (n = 10). Scale bar, 10 µm.
Figure S1.
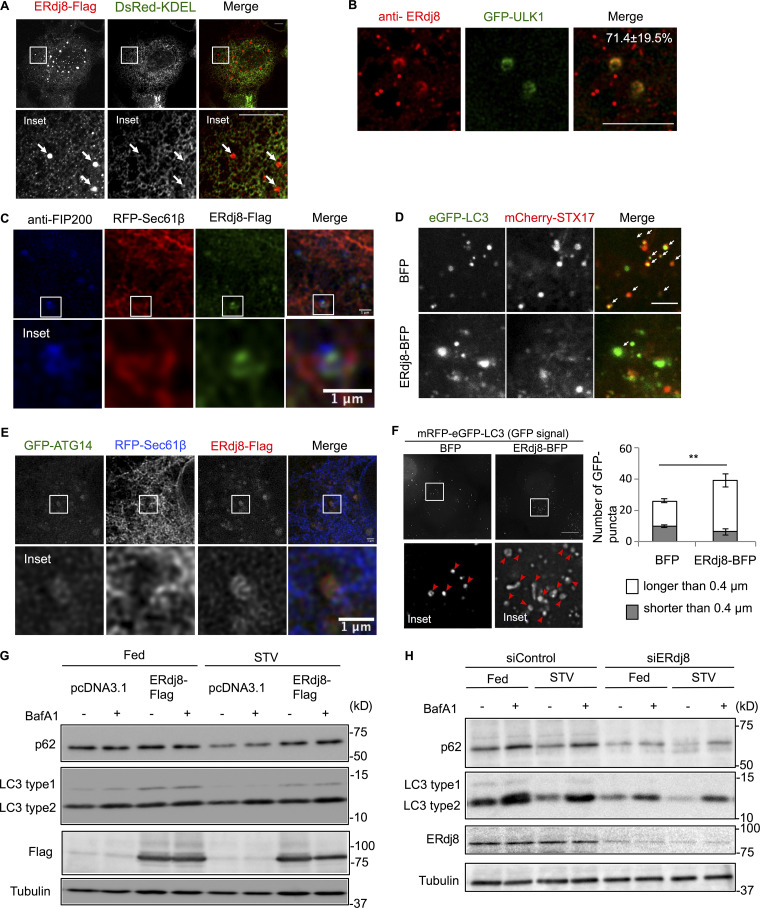
ERdj8 localization and ERdj8 overexpression or knockdown on autophagy flux. (A) COS-7 cells were cotransfected with ERdj8-Flag and DsRed-KDEL, immunostained with anti-FLAG, and observed on the LSM700. Insets, enlargements of framed regions. White arrows show ERdj8-Flag puncta. Scale bar, 10 µm. (B) HeLa cells stably expressing GFP-ULK1 were starved for 1 h, immunolabeled for ERdj8, and imaged on an SP-8. The number is the percentage and SD of GFP-ULK1–positive structures among ERdj8 structures per cell (n = 14). Scale bar, 10 µm. (C) HeLa cells transfected with RFP-Sec61β and ERdj8-Flag were stained with anti-FIP200 and imaged on an SP-8. Insets, enlargements of framed regions. Scale bar, 1 µm. (D) HeLa cells stably expressing eGFP-LC3 were transfected with mCherry-STX17 and ERdj8-BFP or BFP only, starved for 2 h, and imaged on an SP-8. White arrows show co-localization of eGFP-LC3 and mCherry-STX17. Scale bar, 5 µm. (E) HeLa cells transfected with RFP-Sec61β, GFP-ATG14, and ERdj8-Flag were imaged on an SP-8. Insets, enlargements of framed regions. Scale bar, 1 µm. (F) HeLa cells stably expressing mRFP-eGFP-LC3 were transfected with ERdj8-BFP or BFP alone and then cultured under starvation conditions for 4 h. Images were acquired on a DeltaVision system and subjected to deconvolution. For deconvolution, five 0.2-µm–slice images were processed using SoftWoRx software (Applied Precision). Numbers of larger (red arrowheads, white bars) or smaller (gray bars) GFP-positive LC3 puncta are shown. Insets, enlargements of framed regions. Scale bar, 10 µm. Length is the distance to the most distal point in each of the eGFP-positive puncta. Graph shows numbers of eGFP-positive puncta longer or shorter than 0.4 µm in all cells. **, P < 0.005 by t test. Results are reported as the means of 33 cells ± SEM. (G) LC3 turnover monitored by Western blot in HeLa cells overexpressing ERdj8-Flag, either fed or starved for amino acids (STV), with (+) or without (−) BafilomycinA1 (BafA1). (H) LC3 turnover was monitored by Western blot in HeLa cells following knockdown of ERdj8 using siRNA (siERdj8no1 and siERdj8no2). Cells were either fed or starved for amino acids (STV), with (+) or without (−) BafilomycinA1 (BafA1).
An ER subdomain enriched in the glycerophospholipid biosynthesis enzyme phosphatidylinositol synthase (PIS) exists (Kim et al., 2011; English and Voeltz, 2013). ERdj8-positive puncta were overlapped with PIS-GFP or closely adjacent (∼24%; Fig. 1 C). As this PIS-enriched domain is associated with autophagic machinery (Nishimura et al., 2017), we investigated their relationship. ATG13 forms a complex with ULK1, FIP200, and ATG101 that acts as a scaffold for ATG proteins (Mizushima et al., 2011; Karanasios et al., 2013), and 74% of the ATG13-positive structures were associated with endogenous ERdj8 (Fig. 1 D). The autophagy-specific class III PI3-kinase complex, which contains ATG14, is recruited to the ER, where it generates PI3P (Matsunaga et al., 2010; Itakura and Mizushima, 2010). 41% of ATG14 signals were associated with ERdj8 upon induction of autophagy (Fig. 1 E). Moreover, a part of ERdj8 puncta colocalized with FIP200 or ATG14 on the ER (Fig. S1, C and E). Together, these results led us to hypothesize that ERdj8 is associated with autophagosome formation.
Overexpression of ERdj8 leads to enlargement of autophagosomes
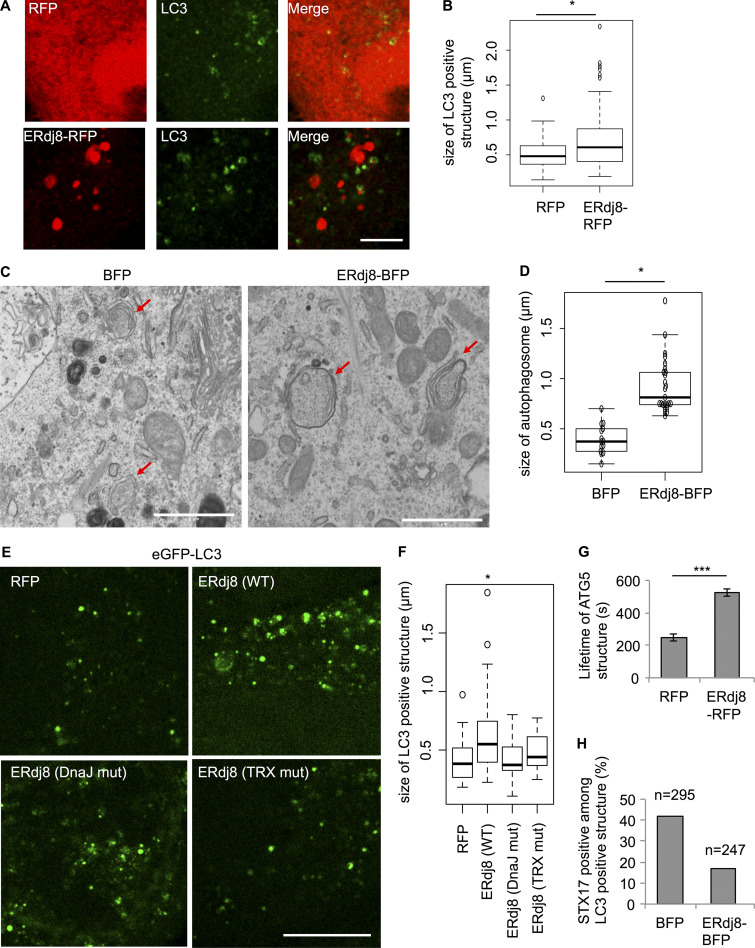
We then examined the effect of ERdj8 overexpression by observing LC3, a marker of autophagosomes (Fig. 2 A, bottom panels; Kabeya et al., 2000). The size of LC3-positive structures was significantly increased by ERdj8 overexpression (Fig. 2, A and B) but not other members of the ERdj family (not shown). We noticed that some of the GFP-LC3–positive structures became elongated when ERdj8 was overexpressed (Fig. S1 F, bottom panels). Hence, we measured the distance to the distal tips of GFP-LC3–positive structures after refining the images through a deconvolution process. This analysis revealed that the population of larger GFP-LC3–positive structures (>0.4 µm) was significantly increased by ERdj8 overexpression (Fig. S1 F, red arrowheads). Electron microscopic analysis also revealed that overexpression of ERdj8 increased the size of autophagosomes (Fig. 2, C and D). Unlike wild-type ERdj8, point mutants in the ERdj8 DnaJ domain (H57Q) or TRX domain (C174A, C177A) did not increase the size of GFP-LC3–positive structures when overexpressed (Fig. 2, E and F).
Figure 2.
Overexpression of ERdj8 increases autophagosome size. (A and B) HeLa cells were transfected with RFP or ERdj8-RFP, incubated under starved condition for 1 h, immunolabeled for LC3, and imaged on an SP-8. Scale bar, 10 µm. (B) Length of the most distal point in each of the LC3-positive structures. *, P < 0.05 by t test. *, P < 0.05 by ANOVA, Tukey–Kramer test. Mean of five cells ± SD. (C and D) HeLa cells stably expressing eGFP-LC3 were transfected with ERdj8-BFP or BFP and starved for 1 h. CLEM analysis was conducted. Scale bar, 1 µm. Red arrows show autophagosomes. (D) Length of the most distal point in each of the autophagosomes.Diameters of GFP-LC3–positive autophagosomes were measured in BFP (14 autophagosomes) and ERdj8-BFP (25 autophagosomes). *, P < 0.05 by t test. Mean of GFP-LC3–positive autophagosomes ± SD. (E and F) HeLa cells stably expressing eGFP-LC3 were transfected with ERdj8(WT)-RFP, ERdj8 (DnaJ muta)-RFP, ERdj8 (TRX mut)-RFP, or RFP only and imaged on an SP-8. Scale bar, 10 µm. (F) Length of the most distal point in each of the eGFP-positive puncta. *, P < 0.05 by ANOVA, Tukey Kramer test. Mean of five cells ± SD. (G) COS-7 cells stably expressing YFP-ATG5 were transfected with ERdj8-RFP or RFP and starved for 2 h. Live images of YFP-ATG5–positive structures were acquired on a DeltaVision system at intervals of 10 s. The average of 12 lifetimes of each YFP-ATG5 structure is shown. Mean of YFP-ATG5–positive puncta ± SEM. ***, P < 0.001. (H) HeLa cells stably expressing eGFP-LC3 were transfected with mCherry-STX17 and ERdj8-BFP or BFP only (as a control), starved for 2 h, and imaged on an SP-8. Percentage of mCherry-STX17–positive among GFP-LC3–positive structures. Total numbers of eGFP-LC3–positive structures counted are shown as n.
We then examined the effect of ERdj8 overexpression on the isolation membrane, the precursor of the autophagosome. ATG5 associates with the isolation membrane, but upon completion of autophagosome formation, it detaches from the membrane (Mizushima et al., 2001). In COS-7 cells stably expressing YFP-ATG5, overexpression of ERdj8 doubled the lifetime of YFP-ATG5–puncta signal relative to control cells (Fig. 2 G). Syntaxin17 (STX17) is mostly associated with completed autophagosomes, and some GFP-LC3 puncta were colocalized with mCherry-STX17 (Fig. S1 D, upper panels, white arrows; Tsuboyama et al., 2016; Kumar et al., 2018; Kumar et al., 2019). ERdj8-BFP overexpression diminished the colocalization, implying that autophagosome formation was less efficient (Fig. 2 H). Therefore, the overall effect on autophagy flux by overexpression of ERdj8 was marginal (Fig. S1 G). Collectively, these observations indicate that ERdj8 overexpression delayed the transition from isolation membranes/phagophores to autophagosomes.
ERdj8 knockdown yields a less-expanded autophagosome
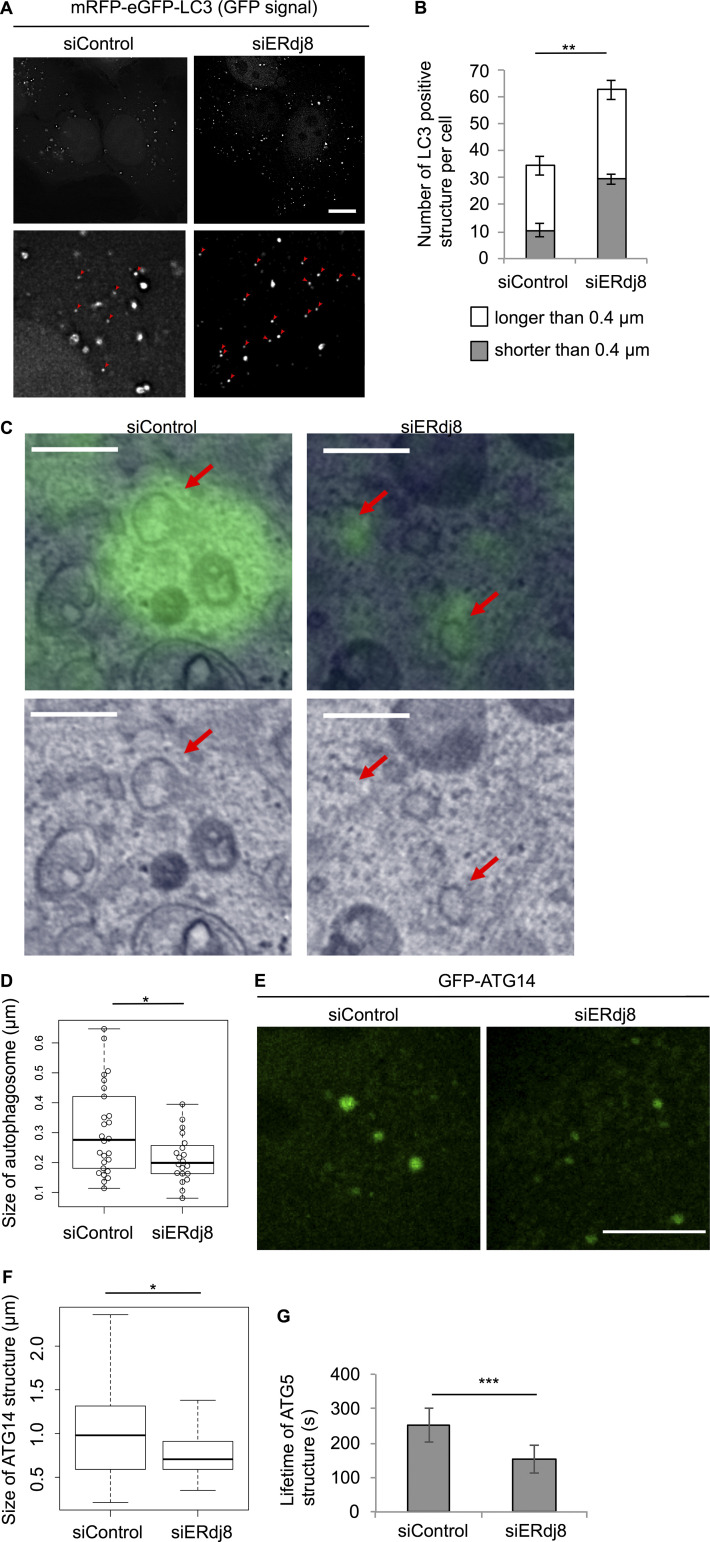
On the other hand, ERdj8 knockdown led to a marked increase in smaller LC3-positive structures (<0.4 µm in diameter) under starvation (Fig. 3 A, red arrowheads; and Fig. 3 B). Indeed, autophagosomes, which are labeled with GFP-LC3 by correlative light electron microscopy (CLEM), were significantly decreased in size by ERdj8 knockdown (Fig. 3, C and D). GFP-ATG14–positive puncta were also smaller (Fig. 3, E and F), and the lifetime of ATG5-positive puncta was reduced (Fig. 3 G), although autophagy flux was scarcely affected (Fig. S1 H). These data indicate that ERdj8 ablation led to formation of less-expanded autophagosomes.
Figure 3.
ERdj8 knockdown decreases autophagosome size. (A) HeLa cells stably expressing tandem fluorescent-tagged LC3 (tfLC3) were treated with small interfering RNA (siRNA; siERdj8no1 or control), incubated for 96 h, and imaged on a DeltaVision system. Insets are enlargements of framed regions. Smaller GFP-positive puncta are indicated by red arrowheads. Scale bar, 10 µm. (B) Length of the most distal point in each of the eGFP-positive puncta. Numbers of eGFP-positive puncta longer or shorter than 0.4 µm in all cells are shown. **, P < 0.005 by t test. Results are shown as means of a total of 10 cells ± SEM. (C and D) HeLa cells stably expressing eGFP-LC3 were treated with siRNA (siERdj8no1 or control), starved for 1 h, and subjected to CLEM analysis. Scale bar, 0.5 µm. Red arrows show autophagosomes. Diameters of GFP-LC3–positive autophagosomes were measured in control (26 autophagosomes) and siERdj8 (20 autophagosomes). *, P < 0.05 by t test. Mean of GFP-ATG14–positive puncta ± SD. Median: lines, upper and lower quartiles: boxes; 1.5-interquartile range: whiskers. Significance of differences was evaluated by unpaired two-tailed t test. (E) HeLa cells stably expressing GFP-ATG14 were treated with siRNA (siERdj8 no1 or control), incubated for 96 h, and imaged on an SP-8. Scale bar, 10 µm. (F) Length of the most distal point in each of the eGFP-positive puncta. *, P < 0.05 by ANOVA, Tukey–Kramer test. Mean of five cells ± SD. Median: lines; upper and lower quartiles: boxes; 1.5-interquartile range: whiskers. (G) HeLa cells stably expressing GFP-ATG5 were treated with either siRNA (siERdj8no1 or control), incubated for 72 h, and imaged on a DeltaVision system. Live images were acquired at intervals of 10 s. Lifetime of each GFP-ATG5–positive structure. An average of 18 signals is shown. ***, P < 0.001. Error bars indicate SEM.
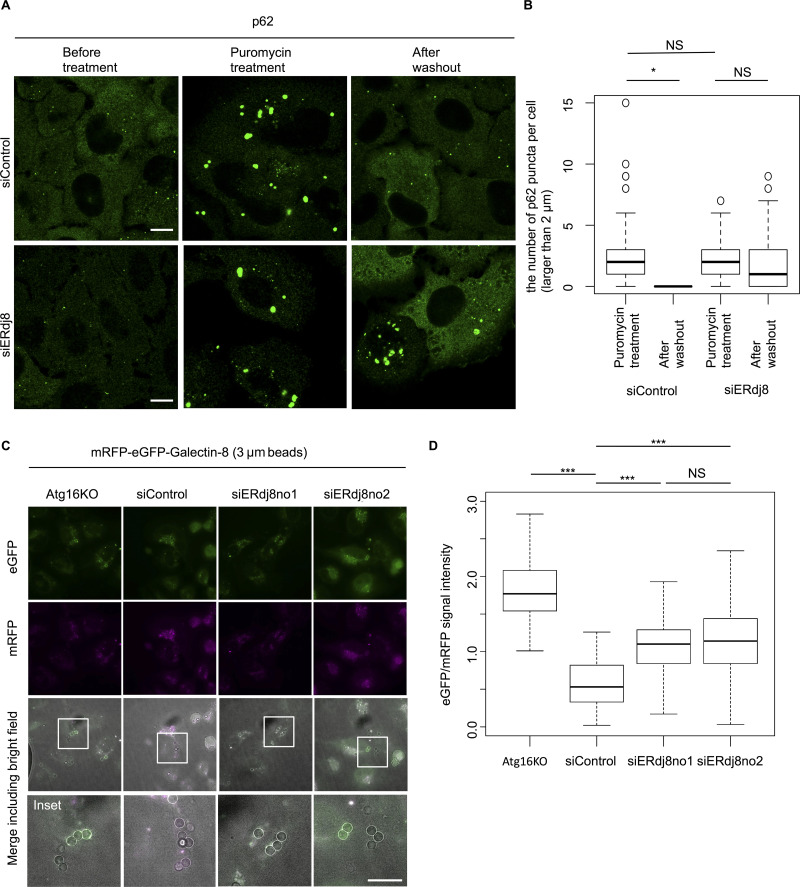
To confirm this observation, we investigated whether ERdj8 knockdown affected the size capacity of autophagic targets by monitoring clearance of large protein aggregates (>2 µm in diameter) induced by mild puromycin treatment (Kirkin et al., 2009). As shown in Fig. 4, A and B, ERdj8 knockdown decreased the efficiency of clearance of large p62/SQSTM–positive protein aggregates relative to control.
Figure 4.
ERdj8 knockdown allows enwrapping of only smaller target. (A) HeLa cells were treated with siRNA (siERdj8no1 or control), incubated for 96 h, and then treated with 5 µg/ml puromycin for 6 h. After washout, cells were cultured for 17 h, stained with anti-p62, and imaged on an SP-8. Scale bar, 10 µm. (B) Percentage of numbers of p62-positive puncta longer than 2 µm in a cell after washout versus before washout. *, P < 0.05 by ANOVA, Tukey Kramer test. NS means P > 0.05. Median: lines; upper and lower quartiles: boxes; 1.5-interquartile range: whiskers. (C) HeLa subjected to the indicated siRNA or ATG16L-knockout stably expressing mRFP-eGFP-Galectin8 was incubated with 3-µm beads for 24 h and imaged on a DeltaVision system. Scale bar, 10 µm. Insets, enlargements of framed regions. (D) GFP and RFP signal intensities associated with 3-µm beads were measured; the GFP/RFP ratio of 61 beads is shown. ***, P < 0.001. NS means P > 0.05. Median: line; upper and lower quartiles: boxes; 1.5-interquartile range: whiskers.
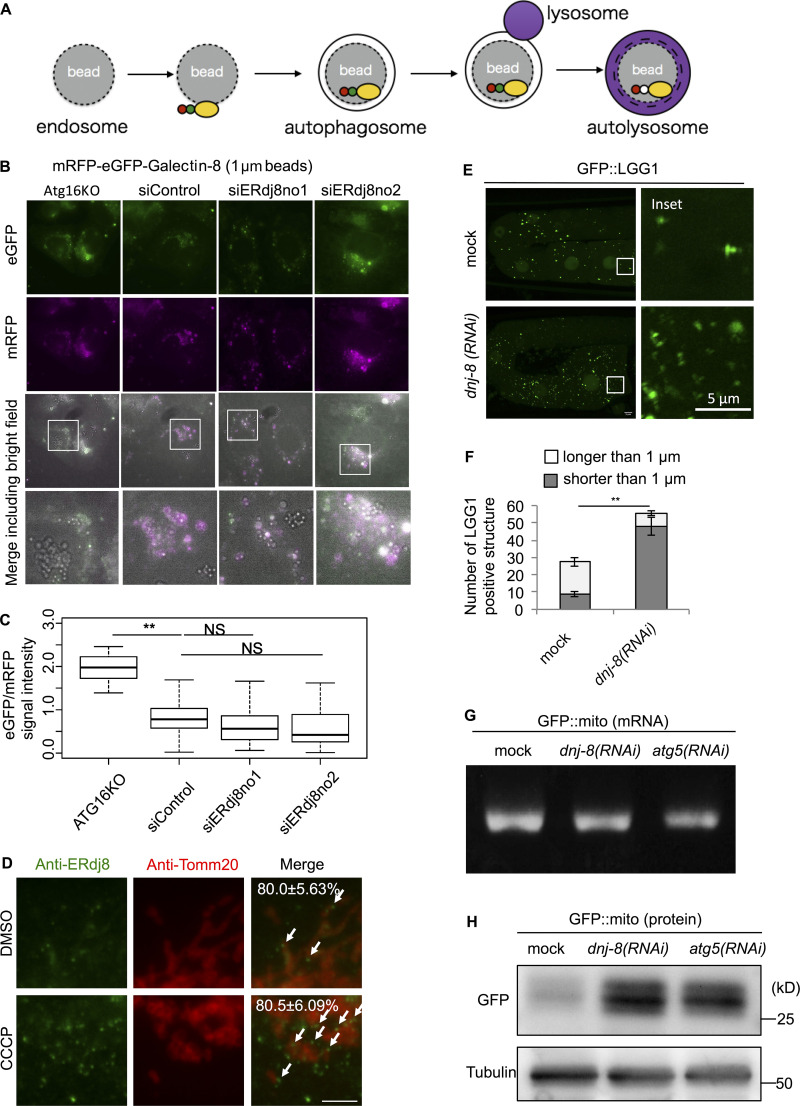
To further support this notion, we employed latex beads coated with transfection reagents, which are introduced into cells via the endocytic pathway and are ultimately engulfed by autophagosomes; this process mimics the xenophagy pathway, which targets invading bacteria (Kobayashi et al., 2010). We used the mRFP-enhanced GFP (eGFP)-Galectin8 system to monitor engulfment of latex beads by autophagosomes (Fig. S2 A; Fujita et al., 2013; Maejima et al., 2013). Galectin8, a kind of lectin, is expressed in the cytoplasm and targeted to exposed carbohydrate chains within the damaged endosomal luminal side, which contains latex beads, and is eventually engulfed by the autophagosome, leading to fusion with the lysosome (Thurston et al., 2012). In the low-pH environment of the lysosome, the eGFP signal is attenuated due to its high pKa (acidity constant) of the protein (∼6.0; Kneen et al., 1998). Therefore, if mRFP-eGFP-Galectin8 surrounding the beads exhibits a weakened eGFP signal, it would indicate that the autophagic process had successfully completed in the lysosome. Two kinds of latex beads with different sizes (1 and 3 µm in diameter) were used for this assay. First, the 1-µm beads were introduced into HeLa. In Atg16L1-knockout cells, the GFP signal surrounding mRFP-positive beads was significantly stronger than in wild-type HeLa (control siRNA), indicating that bead autophagy was defective (Fig. S2, B and C). In contrast, eGFP signals in ERdj8-knockdown cells were attenuated relative to control, indicating successful autophagic engulfment (Fig. S2, B and C). However, when 3-µm beads were introduced, attenuation of eGFP-positive signals was suppressed in ERdj8-knockdown cells (Fig. 4, C and D). These data indicate that ERdj8 knockdown resulted in a defect in enwrapping of larger targets, but it preserved the cell’s ability to enwrap smaller ones.
Figure S2.
Scheme of mRFP-eGFP-Galectin8 engulfment assay, additional effect of ERdj8 knockdown on mRFP-eGFP-Galectin8 1-µm bead assay, and data on dnj-8 knockdown on C. elegans. (A) Scheme of mRFP-eGFP-Galectin8 engulfment assay. (B and C) HeLa subjected to the indicated siRNA treatment or ATG16L-knockout stably expressing mRFP-eGFP-Galectin8 was treated with 1-µm beads for 24 h and imaged on a DeltaVision system. Insets, enlargements of framed regions. Scale bar, 10 µm. (C) GFP and RFP signal intensities associated with beads were measured; the GFP/RFP ratio of 92 beads is shown. **, P < 0.005. Median: line; upper and lower quartiles: boxes; 1.5-interquartile range: whiskers. (D) The cells were treated with CCCP or DMSO, immunostained with ERdj8 and Tomm20 antibodies, and imaged on a DeltaVision system. Scale bar, 10 µm. The number is the percentage and SD of ERdj8–positive structures associating with Tomm20 per cell (n = 10). White arrows show ERdj8–positive structures associating with Tomm20. Scale bar, 10 µm. (E and F) C. elegans expressing GFP::lgg-1 with or without dnj-8 RNAi treatment was grown to the adult stage, and GFP::LGG-1 in oocytes was imaged on an FV1000. Insets, enlargements of framed regions. (F) Length of the most distal point in each of the eGFP-positive puncta. The average number of eGFP-positive puncta longer or shorter than 1 µm in a total of 29 cells is shown. Error bars indicate SEM. **, P < 0.005. (G and H) GFP::mito mRNA levels (G) and protein levels (H) in mock, dnj-8–knockdown, or atg-5–knockdown. C. elegans embryos from worms expressing GFP::mito around the 1.5-fold stage were measured by RT-PCR or by Western blotting.
ERdj8 depletion allows enwrapping of small paternal mitochondria, but not normal somatic mitochondria
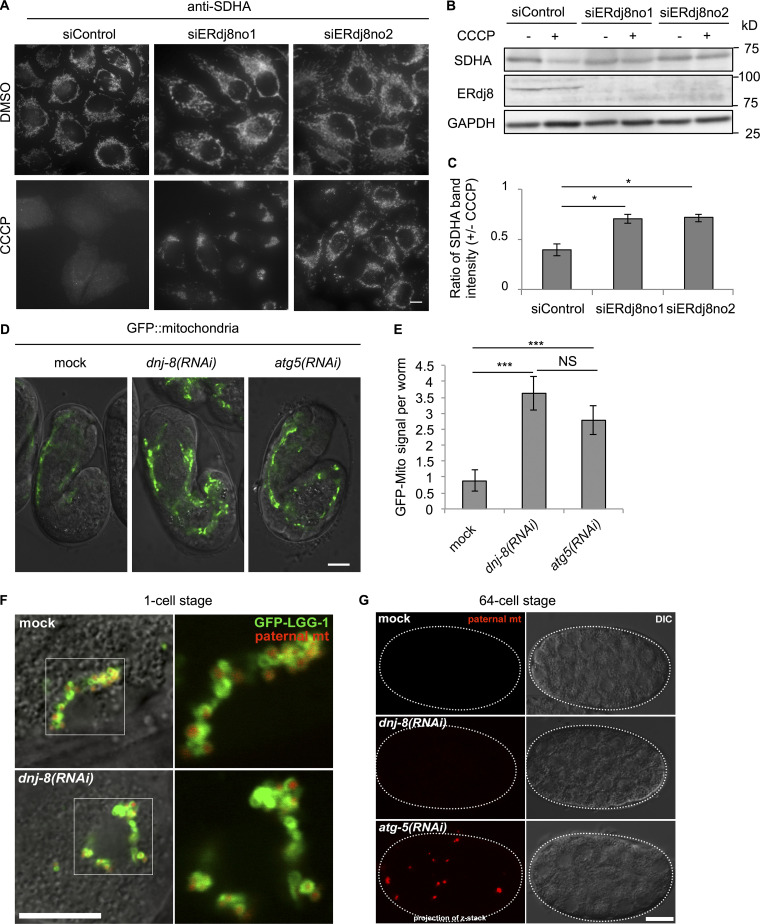
Next, we explored the role of ERdj8 in the engulfment of physiologically relevant autophagy targets: damaged mitochondria (Youle and Narendra, 2011). Mitochondrial autophagy (mitophagy) occurs in response to loss of mitochondrial membrane potential following treatment with the uncoupler Carbonyl cyanide m-chlorophenyl hydrazone (Ding and Yin, 2012). After CCCP treatment, ∼80% of endogenous ERdj8-positive puncta were associated with Tomm20, a mitochondrial protein (Fig. S2 D, white arrows). CCCP treatment attenuated the signal from the mitochondria inner membrane protein succinate dehydrogenase A (SDHA), but this attenuation was suppressed in ERdj8-knockdown cells (Fig. 5, A–C). The survival rate was reduced after CCCP treatment of ERdj8-knockdown cells (data not shown). Thus, mitophagy does not proceed normally in ERdj8 knockdown cells.
Figure 5.
ERdj8 depletion causes a defect in the enwrapping of larger mitochondria. (A–C) HeLa cells expressing mCherry-Parkin were treated with siRNA (siERdj8no1, siERdj8no2, and control). The cells were treated with CCCP or DMSO for 20 h, immunostained with SDHA antibody, and imaged on a DeltaVision system. Scale bar, 10 µm. (B) The lysates were subjected to immunoblotting. (C) The average and SEM of three independent experiments of the ratio of SDHA to GAPDH band intensity. *, P < 0.05. (D and E) C. elegans expressing myo-3::GFP(mito) were treated with or without dnj-8 and/or atg-5 RNAi. Embryos around the 1.5-fold stage were observed on an LSM700. Scale bar, 10 µm. (E) Total GFP signal intensity per worm. The average of 129 worms is shown. ***, P < 0.005. Results are reported as means ± SEM. (F and G) GFP::LGG-1 (green) and paternal mitochondria (mt; HSP-6::mCherry; red) in mock or dnj-8 (RNAi)-treated 1 cell–stage C. elegans embryos at the pronuclear expansion stage (F) or at the 64-cell stage (G) were observed on an FV1000. Fluorescence images merged with DIC images are shown. Insets, enlargements of framed regions. Scale bar, 10 µm.
Finally, we explored the role of ERdj8 at the organismal level in Caenorhabditis elegans. As with mammalian ERdj8 knockdown, knockdown of dnj-8 increased the abundance of smaller GFP::LGG-1 puncta (<1 µm in diameter); LGG-1 is an Atg8/LC3 paralog (Fig. S2, E and F; Meléndez and Levine, 2009). Next, we examined the effect of dnj-8 knockdown on mitophagy in C. elegans. The signal intensity and protein level of mitochondrially targeted GFP (GFP::Mito) in the body wall muscle was increased by dnj-8 knockdown as in atg-5 knockdown, while the mRNA level was unchanged (Fig. 5, D and E; and Fig. S2, G and H). In the fertilized egg of C. elegans, sperm-derived paternal mitochondria are selectively degraded by mitophagy during early embryogenesis (Sato and Sato, 2011; Al Rawi et al., 2011). The paternal mitochondria are spherical and have an average diameter of 400–500 nm, significantly smaller than the materially derived somatic mitochondria (Zhou et al., 2016). Hence, we monitored the fate of the paternal GFP mitochondria. At the one-cell stage, GFP::LGG-1 signals surrounded sperm-derived paternal mitochondria in both mock and dnj-8 (RNAi) embryos (Fig. 5 F). At the 32- to 64-cell stages, sperm-derived paternal mitochondria disappeared in both mock and dnj-8 (RNAi) embryos but accumulated in atg-5 (RNAi) embryos (Fig. 5 G). Together, these data indicate that ERdj8 is responsible for enwrapping larger mitochondria but is dispensable for enwrapping smaller mitochondria, even at the organismal level.
Discussion
In this study, we showed that a novel membrane protein ERdj8 is localized mainly in an ER subdomain involved in autophagosome formation. Overexpression resulted in enlargement of the autophagosome. In contrast, knockdown limited the target size of autophagic engulfment. However, the overall effect on autophagy flux by overexpression and/or knockdown of ERdj8 was marginal. On the basis of these findings, we propose that ERdj8 may play a critical role in fine-tuning the autophagosome formation process.
ERdj8 is concentrated in a subdomain of the ER that we named the ERdj8 domain. Because ERdj8 is a membrane-spanning protein, it must be laterally translocated from the conventional ER tubular network, and we can reasonably conclude that the ERdj8 domain is connected to the ER network. Sec61β, an abundant ER-resident membrane protein, is scarce in the ERdj8 domain, although not totally absent (Fig. 1 B). PIS partly overlapped with ERdj8, but mainly seemed to surround the periphery of the ERdj8 domain (Fig. 1 C). In light of a recent report that autophagosome formation is required for de novo phospholipid synthesis (Schütter et al., 2020; Andrejeva et al., 2019) and takes place adjacent to a PIS-enriched domain (Nishimura et al., 2017), colocalization with PIS provided us with an important clue about the function of ERdj8. Indeed, ATG13, ULK1, FIP200, and ATG14L were associated with the ERdj8 domain (Fig. 1, D and E; and Fig. S1, B, C, and E). ATG2, which bridges the isolation membrane and the ER, acts as a lipid transfer protein, possibly by transferring lipids to the isolation membrane (Gómez-Sánchez et al., 2018; Kotani et al., 2018; Osawa et al., 2019; Valverde et al., 2019). Because the ERdj8 domain associates with the phosphoglycerolipid synthesis enzyme PIS and possibly CEPT1, abundant phosphoglycerolipid should be generated in its vicinity. Furthermore, overexpression of ERdj8 enlarges the PIS1-containing ER subdomain (data not shown), which may facilitate lipid supply and autophagosome enlargement. It is tempting to speculate that ERdj8 regulates some of these processes. The DnaJ and TRX domains of ERdj8 on the ER luminal side were important for the function of ERdj8 in the regulation of autophagy (Fig. 2, E and F), and unidentified ERdj8 effector proteins and/or BiP may play important roles in this regulation. We are currently seeking to identify these proteins, with the goal of understanding the underlying mechanism.
In summary, our study revealed that ERdj8 plays some critical role in regulation of autophagosome formation, shedding light on the mechanism by which autophagosome size is determined, and these observations provide novel insight into the determination of autophagosome size.
Experimental model and subject details
Cell culture and transfection
All cell lines were cultured in DMEM high glucose (GIBCO) supplemented with 10% FBS (Invitrogen). For starvation treatment, cells were cultured in Earle’s balanced salt solution (EBSS; Sigma) for the indicated periods. Lipofectamine 2000 (Invitrogen, P/N 52887) was used for transfection.
The following cell lines were described previously: COS-7; HeLa stably expressing eGFP-LC3; HeLa stably expressing mRFP-eGFP-LC3 (Kageyama et al., 2011); HeLa stably expressing GFP-ATG5 (Fujita et al., 2008); HeLa stably expressing ULK1-GFP (Kageyama et al., 2011); HeLa stably expressing mCherry-Parkin (Narendra et al., 2008); HeLa stably expressing eGFP-DFCP1; and COS-7 stably expressing YFP-ATG5 (Hamasaki et al., 2013).
HeLa stably expressing mRFP-eGFP-Galectin8, Atg16-knockout HeLa cells stably expressing mRFP-eGFP-Galectin8, and COS-7 cells stably expressing PIS-GFP were constructed using the pMRX retroviral vector (Saitoh et al., 2003). For retrovirus preparation, plasmids mRFP-eGFP-Galectin8 and PIS-GFP were cloned and transiently transfected into Plat-E cells using Lipofectamine 2000. Transfection medium was removed 4 h after transfection, and fresh medium was added to the plate. Cell supernatants were collected at 36 h and filtered through a 0.45-µm filter. Cells to be transduced were seeded 24 h before infection and then transduced with virus-containing supernatant supplemented with Polybrene. Cells were left to recover for 24 h in growth media before puromycin selection (1 µg/ml; Wako, 160–23151).
Atg16L1 KO HeLa cells were generated as follows. CRISPR gRNA sequences designed against the ATG16L1 gene were cloned into px330 (Cong et al., 2013). The target sequence was 5′-GCGCCGCTGACTTCCCCCGC-3′. HeLa cells were transfected with px330 encoding the gRNA. After 24 h, cells were diluted and seeded in 96-well plates for isolation of single clones. Clones with mutations in both alleles were identified by immunoblotting and confirmed by sequencing of genomic DNA.
Immunoblotting
Cells were lysed in 1% Triton-X100 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 0.1% [vol/vol] PIC-2 [ISTI, A-0014-20]) for 30 min on ice. The suspension was sedimented by centrifugation at 20,400 g for 20 min at 4°C. Lysates were mixed with SDS loading buffer (200 mM Tris-HCl, pH 6.8, 5% β-mercaptoethanol, 20% SDS, and 40% glycerol), incubated at 65°C for 20 min, and subjected to SDS-PAGE and Western blotting according to standard protocols.
Antibodies
The following primary antibodies were obtained from the indicated suppliers: rabbit anti-DNAJC16 (ERdj8; Proteintech, 17599–1-AP; Western blotting [WB], 1/500); mouse anti-ATG13 (Merck, MABC46; immunocytochemistry [IC], 1/100), mouse anti-SDHA (Novex, 459200; WB, IC, 1/1,000); mouse anti-GAPDH (HyTest, 5G4; WB, 1/1,000); mouse anti-Tomm20 (Abcam, ab56783; WB, 1/1,000); mouse anti-LC3 (MBL, M186-3; IC, 1/500; WB, 1/1,000); mouse anti-tubulin α (MBL, PM054; WB, 1/1,000); mouse anti-Tomm20 (Abcam, ab56783; IC, 1/1,000); rabbit anti-p62 (MBL, PM066; IC, 1/500); rabbit anti-ATG16L1 (MBL, PM040Y; WB, 1/1,000); rabbit anti-DDDDK (MBL, PM020; WB, IC, 1/1,000); rabbit anti-FIP200 (Proteintech; 17250–1-AP, IC, 1/100); mouse anti-Calnexin, C-terminal (Enzo, ADI-SPA-860; IC, 1/1,000); rabbit anti-Calnexin, N-terminal (MBL, PM060; IC, 1/1,000), and rabbit anti-DNAJC16_744 (IBL; IC, 1/100). Rabbit anti-DNAJC16_744 was raised against the ERdj8 peptide CGLGSRPIKGKLSKLSL (Immuno-Biological Laboratories Co., Ltd.; IC, 1/100).
Secondary antibodies were as follows: HRP-conjugated goat anti-rabbit IgG (H+L; Invitrogen, 656120; WB, 1/2,000); HRP-conjugated goat anti-mouse IgG (H+L; Invitrogen, 626520; WB, 1/2,000); Alexa Fluor 488 goat anti-mouse IgG (H+L; Invitrogen, A11029; IC, 1/1,000); Alexa Fluor 647 goat anti-mouse IgG (H+L; Invitrogen, M21235; IC, 1/1,000); and Alexa Fluor 488 goat anti-mouse IgG (H+L; Invitrogen, A11034; IC, 1/1,000).
siRNAs and plasmids
siRNA duplexes targeting ERdj8 (no. 1: sense, 5′-CCUGCAAAUUCUGUCUGCGUUGGAU-3′, antisense, 5′-AUCCAACGCAGACAGAAUUUGCAGG-3′; no. 2: sense, 5′-GAUCCUGGAGCAGAAGACAAGUUCA-3′, antisense, 5′-UGAACUUGUCUUCUGCUCCAGGAUC-3′) and siRNA negative control (Stealth RNAi) were purchased from Invitrogen. The Stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAi MAX (Invitrogen). After 2 d, the cells were again transfected with the same siRNA and cultured for an additional 2 d before analysis.
The plasmids encoding mCherry-STX17, RFP-Sec61β, GFP-ATG14 (Hamasaki et al., 2013), and DsRed-KDEL (Ito and Nagata, 2016) were described previously. ERdj8-mRFP and ERdj8-BFP were purchased (OriGene Technologies, Inc.). To construct ERdj8-Flag, KIAA0962 (ERdj8/DNAJC16) cDNA was purchased (Kazusa DNA Research Institute). The primers used were forward, 5′-GCCGCAGGTACCGGAAGAGAAATGGAAGTGAGAAAGTTG-3′ and reverse, 5′-TCCGCCGGGCCCTCACTTATCGTCGTCATCCTTGTAATCGTCTAGTTCAGGGGA-3′. Amplification with these primers fused a FLAG tag to the C-terminus of ERdj8. This insert was subcloned into the KpnI–ApaI site of pcDNA3.1(+) (Invitrogen). The H57Q for DnaJ domain mutant (CAT to CAG) and the TRX domain mutant C174A (TGC to GCC), C177A (TGC to GCC) were created using the PrimeSTAR Mutagenesis Basal Kit (TaKaRa).
Fluorescence microscopy
Fluorescence microscopy was performed using an FV1000 confocal microscope system equipped with a 60×, 1.35 NA UPlanSApo or a 100×, 1.40 NA UPlanSApo oil objective lens (Olympus Corp.), a TCS SP-8 conformal laser-scanning fluorescence microscope (Leica) equipped with a 63× objective (HC PL APO 63×/1.40 OIL CS2; Leica) with Leica HyVolution Deconvolution Imaging (Huygens), a DeltaVision Elite fluorescence microscope (GE Healthcare Life Science) equipped with a 60× PlanAPO oil immersion objective lens (Olympus; NA 1.42), and a scientific complementary metal oxide semiconductor (CMOS) camera, a SpinSR10 Super Resolution Imaging System (Olympus) equipped with a 60× PlanAPO oil immersion objective lens (Olympus; NA 1.42), or a LSM700 (Zeiss) confocal laser scanning microscope equipped with an α PlanAPO 100×/1.46Oil differential interference contrast (DIC) microscope M27 (Zeiss).
For immunostaining, cells were fixed with 4% PFA for 10 min, quenched with PBS, permeabilized with 20 µg/ml digitonin, blocked with 5% normal goat serum, and mounted in mounting reagent (Mowiol 4–88, nonfluorescent glycerol, and 0.2 M Tris-Cl, pH 8.5). For observation of proteins fused to fluorescent tags, the permeabilization step was omitted.
For live cell imaging, cells were placed on a glass-bottom dish (Matsunami glass) and maintained at 37°C with 5% CO2.
mRFP-eGFP-Galectin8 bead assay
After 2 d of knockdown, 1.5 × 105 cells were seeded on a coverslip (for immunofluorescence) or directly (for immunoblotting) in a six-well plate. After 48 h, beads were prepared by mixing 1-µm or 3-µm beads (PolySciences, Inc.) with Effectene transfection reagent (QIAGEN; Kobayashi et al., 2010). The bead mixture (100 µl) was further mixed with 1 ml of growth medium and then added to cells by replacing the medium. After incubation with the bead mixture for 1 h, the cells were washed twice with fresh medium to remove unattached beads, incubated for an additional 24 h, and imaged. The area corresponding to each bead was selected as the region of interest (ROI), and the signal intensity of GFP and RFP within the ROI was quantified using ImageJ.
p62 body degradation assay
After 2 d of knockdown, 1.0 × 105 HeLa cells were seeded in six-well plates on a coverslip. After 48 h, the cells were treated with 5 µM puromycin (Wako, 160–23151) for 6 h to induce the formation of large p62 bodies. Puromycin was washed out and cultured for 17 h. Cells were fixed with 4% PFA, stained with p62 antibody, and subjected to microscopy.
Mitophagy assay
After 2 d of knockdown, 1.5 × 105 mCherry-Parkin stable HeLa cells were seeded in six-well plates on a coverslip (for immunofluorescence) or directly (for immunoblotting). After 48 h, the cells were treated with 5 µM CCCP (Sigma, C2759-250MG) for 20 h to induce mitophagy. Cells were fixed with 4% PFA, stained with Cox-II antibody, and subjected to microscopy or subjected to Western blotting.
CLEM
After 2 d of knockdown of ERdj8 or overexpression of ERdj8-BFP or BFP, HeLa cells stably expressing eGFP-LC3 were cultured on glass-bottom dishes with a grid pattern (MatTek, P35G-2–14-C-GRID) and starved for 1 or 2 h. The cells were fixed with 4% (wt/vol) formaldehyde in 30 mM Hepes buffer (pH 7.4) containing 100 mM NaCl, 2 mM CaCl2, and 1 µg/ml Hoechst 33342 for 30 min at room temperature; washed with 30 mM Hepes buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2; and examined using a confocal laser scanning microscope (Leica, SP-8). The same specimens were further incubated with 2% (wt/vol) formaldehyde and 2.5% (wt/vol) glutaraldehyde in 30 mM Hepes buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2 at 4°C for overnight. After three washes in 30 mM Hepes buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2, the samples were postfixed with 1% (wt/vol) osmium tetroxide in 30 mM Hepes buffer (pH 7.4) containing 0.5% (wt/vol) potassium ferrocyanide, 100 mM NaCl, and 2 mM CaCl2 for 1 h, washed three times in distilled water, dehydrated in ethanol, and embedded in Epon812 (TAAB Laboratories Equipment). Ultrathin sections (70-nm thick) were stained with saturated uranyl acetate and Reynolds lead citrate solution. The electron micrographs were taken with a JEOL JEM-1011 transmission electron microscope.
Worm experiments
The transgenic lines used in this study were dkIs398 (Ppie-1::GFP::lgg-1, unc-119(+)) and dkIs698 (Pspe-11::hsp-6::mCherry, unc-119(+); Sato and Sato, 2011) and SJ4103 (myo-3::GFP(mit); Benedetti et al., 2006). RNAi experiments were conducted using the feeding method (Kamath and Ahringer, 2003). L4 larvae were treated with RNAi, and embryos dissected from the F1 adults or oocytes of the F1 adults were scored. For RNAi experiments, L4440 containing a cDNA fragment of dnj-8 or atg-5 was used. For imaging, adult worms, eggs, and dissected embryos were mounted on agarose pads with M9 buffer containing 10 mM levamisole and observed using a laser scanning confocal microscope LSM700 or FV1000. For immunoblotting, eggs were lysed in SDS loading buffer (200 mM Tris-HCl, pH 6.8, 20% SDS, and 40% glycerol) at −80°C and then at 100°C for 15 min. The suspension was sedimented by centrifugation at 20,400 g for 10 min at 4°C. Lysates were mixed with 5% β-mercaptoethanol, incubated at 100°C for 5 min, and subjected to SDS-PAGE and Western blotting according to standard protocols. RT-PCR was performed as described previously for worm experiments (Williams et al., 1992). The sequences of the oligonucleotides for GFP::mito were as follows: forward, 5′-GGAGAAGAACTTTTCACTGG-3′; reverse, 5′-CCATGCCATGTGTAATCC-3′. Each reaction was repeated at least five times to assess reproducibility.
Online supplemental material
Fig. S1 shows additional ERdj8 localization and ERdj8 overexpression or knockdown on autophagy flux data. Fig. S2 shows a scheme of mRFP-eGFP-Galectin8 engulfment assay, additional effect of ERdj8 knockdown on mRFP-eGFP-Galectin8 1-µm bead assay, and data on dnj-8 knockdown on C. elegans. Video 1 shows intracellular localization of ERdj8-RFP and YFP-ATG5.
Video 1.
Timelapse imaging of ERdj8 and ATG5. COS-7 cells stably expressing YFP-ATG5 were seeded in a glass-bottom dish. After 24 h, the cells were transfected with ERdj8-RFP or RFP for 24 h. The cells were starved for 2 h. Live images of YFP-ATG5–positive structures were acquired on a DeltaVision system at intervals of 10 s. Frame rate is 10 s per frame.
Acknowledgments
We thank Dr. S. Yamaoka (Tokyo Medical and Dental University, Tokyo, Japan) for pMRX-IRES-puro vector; T. Kitamura (The University of Tokyo, Tokyo, Japan) for Plat-E cells; and R. Yamashita (Kyoto Sangyo University, Kyoto, Japan) for worm experiments. SpinSR10 at the Advanced Cell imaging Core, Osaka University, was utilized in this study.
This work is supported by the Japanese Society for the Promotion of Science KAKENHI (16K07347 to T. Noda and 18H04002 to K. Nagata); a grant from the Takeda Science Foundation (to K. Nagata); the Japan Science and Technology Agency CREST (JPMJCR13M6 to K. Nagata); and the joint research program of the Institute for Molecular and Cellular Regulation, Gunma University (16026 to Y.-H. Yamamoto).
The authors declare no competing financial interests.
Author contributions: Conceptualization, Y.-H. Yamamoto, T. Noda., and K. Nagata; Writing – Original Draft, Y.-H. Yamamoto, S. Bar-Nun, K. Nagata, and T. Noda. Writing – Review and Editing, T. Noda. Investigation, Y.-H. Yamamoto (most aspects of the study); S. Bar-Nun (topology analysis); A. Kasai (mitophagy analysis). T. Takino, M. Sugihara, and M. Sato. (worm experiments); T. Umemoto (bead assays); T. Hatta (mass spec); H. Omori, R. Arai., S. Waguri, (EM analysis); resources, R.I. Morimoto, M. Hamasaki, T. Yoshimori, K. Sato, and T. Natsume.
References
- Al Rawi, S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., Hajjar C., Boyd L., Legouis R., and Galy V.. 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 334:1144–1147. 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- Andrejeva, G., Gowan S., Lin G., Wong Te Fong A.L., Shamsaei E., Parkes H.G., Mui J., Raynaud F.I., Asad Y., Vizcay-Barrena G., et al. 2019. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy. 13:1–17. 10.1080/15548627.2019.1659608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe, E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Osumi M., Scott S.V., Klionsky D.J., and Ohsumi Y.. 1997. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139:1687–1695. 10.1083/jcb.139.7.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, C., Haynes C.M., Yang Y., Harding H.P., and Ron D.. 2006. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 174:229–239. 10.1534/genetics.106.061580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, W.-X., and Yin X.-M.. 2012. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 393:547–564. 10.1515/hsz-2012-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A.R., and Voeltz G.K.. 2013. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15:169–178. 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., Itoh T., Omori H., Fukuda M., Noda T., and Yoshimori T.. 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 19:2092–2100. 10.1091/mbc.e07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., Morita E., Itoh T., Tanaka A., Nakaoka M., Osada Y., Umemoto T., Saitoh T., Nakatogawa H., Kobayashi S., et al. 2013. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 203(1):115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi, L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., et al. 2017. Molecular definitions of autophagy and related processes. EMBO J. 36:1811–1836. 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez, R., Rose J., Guimarães R., Mari M., Papinski D., Rieter E., Geerts W.J., Hardenberg R., Kraft C., Ungermann C., et al. 2018. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217:2743–2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki, M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino, M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., and Yamamoto A.. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Itakura, E., and Mizushima N.. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776. 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, S., and Nagata K.. 2016. Mutants of collagen-specific molecular chaperone Hsp47 causing osteogenesis imperfecta are structurally unstable with weak binding affinity to collagen. Biochem. Biophys. Res. Commun. 469:437–442. 10.1016/j.bbrc.2015.12.028 [DOI] [PubMed] [Google Scholar]
- Kabeya, Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, S., Omori H., Saitoh T., Sone T., Guan J.-L., Akira S., Imamoto F., Noda T., and Yoshimori T.. 2011. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell. 22:2290–2300. 10.1091/mbc.e10-11-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kampinga, H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios, E., Stapleton E., Manifava M., Kaizuka T., Mizushima N., Walker S.A., and Ktistakis N.T.. 2013. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. Journal of Cell Science. 126(22):5224–5238. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J., Guzman-Hernandez M.L., and Balla T.. 2011. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 21:813–824. 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin, V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kneen, M., Farinas J., Li Y., and Verkman A.S.. 1998. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 74:1591–1599. 10.1016/S0006-3495(98)77870-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S., Kojidani T., Osakada H., Yamamoto A., Yoshimori T., Hiraoka Y., and Haraguchi T.. 2010. Artificial induction of autophagy around polystyrene beads in nonphagocytic cells. Autophagy. 6:36–45. 10.4161/auto.6.1.10324 [DOI] [PubMed] [Google Scholar]
- Kotani, T., Kirisako H., Koizumi M., Ohsumi Y., and Nakatogawa H.. 2018. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci USA. 115(41):10363–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Jain A., Farzam F., Jia J., Gu Y., Choi S.W., Mudd M.H., Claude-Taupin A., Wester M.J., Lidke K.A., et al. 2018. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 217:997–1013. 10.1083/jcb.201708039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Gu Y., Abudu Y.P., Bruun J.A., Jain A., Farzam F., Mudd M., Anonsen J.H., Rusten T.E., Kasof G., et al. 2019. Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev. Cell. 49:130–144.e6. 10.1016/j.devcel.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima, I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., et al. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, K., Morita E., Saitoh T., Akira S., Ktistakis N.T., Izumi T., Noda T., and Yoshimori T.. 2010. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190:511–521. 10.1083/jcb.200911141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez, A., and Levine B.. 2009. Autophagy in C. elegans. WormBook. 1–26. 10.1895/wormbook.1.147.1 [DOI] [PubMed] [Google Scholar]
- Mi, N., Chen Y., Wang S., Chen M., Zhao M., Yang G., Ma M., Su Q., Luo S., Shi J., et al. 2015. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat. Cell Biol. 17:1112–1123. 10.1038/ncb3215 [DOI] [PubMed] [Google Scholar]
- Mizushima, N.. 2004. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36:2491–2502. 10.1016/j.biocel.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., and Yoshimori T.. 2001. Dissection of Autophagosome Formation Using Apg5-Deficient Mouse Embryonic Stem Cells. Journal of Cell Biology. 152(4):657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Narendra, D., Tanaka A., Suen D.-F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Journal of Cell Biology. 183(5):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Tamura N., Kono N., Shimanaka Y., Arai H., Yamamoto H., and Mizushima N.. 2017. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36:1719–1735. 10.15252/embj.201695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., and Noda N.N.. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26:281–288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Otero, J.H., Lizák B., and Hendershot L.M.. 2010. Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21:472–478. 10.1016/j.semcdb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, T., Nakayama M., Nakano H., Yagita H., Yamamoto N., and Yamaoka S.. 2003. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J. Biol. Chem. 278:36005–36012. 10.1074/jbc.M304266200 [DOI] [PubMed] [Google Scholar]
- Sato, M., and Sato K.. 2011. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 334:1141–1144. 10.1126/science.1210333 [DOI] [PubMed] [Google Scholar]
- Schütter, M., Giavalisco P., Brodesser S., and Graef M.. 2020. Local Fatty Acid Channeling into Phospholipid Synthesis Drives Phagophore Expansion during Autophagy. Cell. 180:135–149.e14. 10.1016/j.cell.2019.12.005 [DOI] [PubMed] [Google Scholar]
- Singh, S.B., Davis A.S., Taylor G.A., and Deretic V.. 2006. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 313:1438–1441. 10.1126/science.1129577 [DOI] [PubMed] [Google Scholar]
- Thurston, T.L.M., Wandel M.P., von Muhlinen N., Foeglein A., and Randow F.. 2012. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 482:414–418. 10.1038/nature10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama, K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., and Mizushima N.. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 354:1036–1041. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- Ushioda, R., Miyamoto A., Inoue M., Watanabe S., Okumura M., Maegawa K.I., Uegaki K., Fujii S., Fukuda Y., Umitsu M., et al. 2016. Redox-assisted regulation of Ca2+ homeostasis in the endoplasmic reticulum by disulfide reductase ERdj5. Proc. Natl. Acad. Sci. USA. 113:E6055–E6063. 10.1073/pnas.1605818113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., and Melia T.J.. 2019. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218:1787–1798. 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.D., Schrank B., Huynh C., Shownkeen R., and Waterston R.H.. 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 131:609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Nair U., and Klionsky D.J.. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 19:3290–3298. 10.1091/mbc.e07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, H., Nakagawa I., Yamamoto A., Amano A., Noda T., and Yoshimori T.. 2009. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 5. e1000670. 10.1371/journal.ppat.1000670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y.H., Kimura T., Momohara S., Takeuchi M., Tani T., Kimata Y., Kadokura H., and Kohno K.. 2010. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct. Funct. 35:107–116. 10.1247/csf.10023 [DOI] [PubMed] [Google Scholar]
- Yang, Z., and Klionsky D.J.. 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12:814–822. 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila, P., Vihinen H., Jokitalo E., and Eskelinen E.-L.. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185. 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Youle, R.J., and Narendra D.P.. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Li H., Li H., Nakagawa A., Lin J.L., Lee E.S., Harry B.L., Skeen-Gaar R.R., Suehiro Y., William D., et al. 2016. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 353:394–399. 10.1126/science.aaf4777 [DOI] [PMC free article] [PubMed] [Google Scholar]